Abstract
Melanocortin-4 receptor (MC4R)-induced anorexigenic signaling in the hypothalamus controls body weight and energy homeostasis. So far, MC4R-induced signaling has been exclusively attributed to its coupling to Gs proteins. In line with this monogamous G protein coupling profile, most MC4R mutants isolated from obese individuals showed a reduced ability to activate Gs. However, some mutants displayed enhanced Gs coupling, suggesting that signaling pathways independent of Gs may be involved in MC4R-mediated anorexigenic signaling. Here we report that the Gs signaling-deficient MC4R-D90N mutant activates G proteins in a pertussis toxin-sensitive manner, indicating that this mutant is able to selectively interact with Gi/o proteins. Analyzing a hypothalamic cell line (GT1-7 cells), we observed activation of pertussis toxin-sensitive G proteins by the wild-type MC4R as well, reflecting multiple coupling of the MC4R to Gs and Gi/o proteins in an endogenous cell system. Surprisingly, the agouti-related protein, which has been classified as a MC4R antagonist, selectively activates Gi/o signaling in GT1-7 cells. Thus, the agouti-related protein antagonizes melanocortin-dependent Gs activation not only by competitive antagonism but additionally by initiating Gi/o protein-induced signaling as a biased agonist.
The melanocortin system has been shown to play a pivotal role in food intake and energy homeostasis. Therefore, dysfunction of the melanocortin system inevitably leads to an obese phenotype in mammals. Accordingly, targeted disruption of the melanocortin-4 receptor (MC4R)2 gene in mice causes an obesity-diabetes syndrome characterized by hyperphagia, hyperinsulinemia, and hyperglycemia (1). The importance of MC4R signaling in the regulation of human metabolism has been highlighted by the finding that mutations in the MC4R gene are the most frequent monogenic cause of severe obesity (2–7).
Signaling pathways involved in MC4R-mediated regulation of energy homeostasis have been attributed to its coupling to Gs proteins and the resulting activation of the protein kinase A pathway (8, 9). Agouti and agouti-related protein (AGRP) are the only known endogenously occurring neuropeptides that block GPCR activity and are, therefore, classified as MCR antagonists. AGRP has been shown to block melanocortin signaling at MC3R and MC4R subtypes (10). In addition, it has been proposed that AGRP decreases basal as well as forskolin-promoted adenylyl cyclase activity, thus also acting as an inverse agonist on basal MCR activity (11). However, recent studies revealed that the effects of AGRP on appetite control are independent of melanocortin signaling (12, 13). For example, in mice deficient of the melanocortin precursor proopiomelanocortin starvation after AGRP neuron ablation is independent of melanocortin signaling (13). Thus, the orexigenic effects induced by AGRP appear to be mediated in a melanocortin-independent manner by a so far unknown mechanism.
Interestingly, the aforementioned MC4R mutants isolated from obese patients exerted inconsistent effects on Gs signaling. For example, the MC4R-G181D or -S94R mutants showed a loss-of-function phenotype, whereas the MC4R-P78L or -R165W variants exhibited reduced function, whereas other mutants (MC4R-G253S, -I317T, -I251L) showed no functional alterations. Even more surprisingly, some mutants (MC4R-S127L, -P230L) constitutively increased Gs-dependent adenylyl cyclase activity (5). Therefore, no clear correlation could be drawn between the cellular phenotype resulting from these mutations and obesity observed in vivo.
Melanocortin-independent actions of AGRP and non-uniform effects of obesity-associated mutations on Gs signaling suggest that the MC4R receptor may interact with G proteins other than Gs. The D90N mutation of the MC4R has also been associated with severe early onset obesity (14). This MC4R variant binds melanocortins with unchanged high affinity, but agonist binding does not initiate Gs signaling (14). Thus, the D90N variant represents an excellent tool to analyze putative Gs-independent signaling pathways of the MC4R.
Directly measuring incorporation of GTPγ35S, we show herein that the wild-type MC4R and the MC4R-D90N mutant activate pertussis toxin (PTX)-sensitive Gi/o proteins. Multiple coupling of the MC4R to Gs and Gi/o proteins in HEK293 cells is reflected by cAMP accumulation, as treatment of cells with PTX significantly increased melanocortin-induced cAMP accumulation, indicative of simultaneous activation of adenylyl cyclase stimulating and inhibiting pathways. Using a hypothalamic cell line (GT1-7 cells) endogenously expressing MC4R, we demonstrate PTX-sensitive melanocortin-induced GTPγ35S incorporation and an increase in MC4R-mediated cAMP accumulation in response to toxin treatment. In addition, we show that AGRP, assumed to be an antagonist of the MC4R, promotes PTX-sensitive signaling in GT1-7 cells and, thus, exhibits biased agonistic actions on MC4R in hypothalamic cells. These data may explain melanocortin-independent AGRP signaling and define the MC4R as an interface integrating anorexigenic and orexigenic signaling depending on the stimulus received.
EXPERIMENTAL PROCEDURES
Materials
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum, penicillin/streptomycin, and l-glutamine were purchased from PAA Inc. (Pasching, Austria). Metafectene was obtained from Biontex (München, Germany). Anti-mouse horseradish peroxidase-conjugated secondary antibodies from sheep were purchased from Amersham Biosciences. [3H]Adenine and GTPγ35S were from PerkinElmer Life Sciences. α-MSH and HSO-14 ([acetyl-Cys11,β-(2-naphthyl)-d-Ala14,Cys18]-β-melanocyte hormone fragment 11–22) were purchased from Sigma. The human AGRP fragment (amino acids 86–132) was obtained from Peptides International (Louisville, KY). Gallein was obtained from Calbiochem/Merck.
Plasmid Construction
For enzyme-linked immunosorbent assay experiments we fused the coding sequence of the Xpress peptide (Asp-Leu-Tyr-Asp-Asp-Asp-Asp-Lys) to the 5′-end of both receptor variants. To this end, PCR fragments containing the entire coding sequences of the mouse MC4R or the MC4R-D90N mutant were subcloned into the pcDNA4 vector (from Invitrogen) in a way that fused the Xpress peptide to the N terminus of the MCR. The pAD-CRE-Fluc plasmid was kindly provided by Dr. Himmler, Bender GmbH (Vienna, Austria).
Cell Culture and Transfection
HEK293 and GT1-7 cells (kindly provided by Dr. Weiner, University of California, San Francisco) were cultured in DMEM supplemented with 10% fetal bovine serum and 2 mm l-glutamine. For transient expression of recombinant proteins, cells were seeded at a density of 2 × 106 cells in 10-cm dishes, cultured for 24 h, and then transfected with the appropriate amount of plasmid DNA using the metafectene reagent as described by the manufacturer's protocol. Twenty-four hours post-transfection, cells were detached and seeded in new dishes as required for the following experiment. For stable protein expression, transfected cells were selected for 2–3 weeks with 400 μg/ml zeocin and then tested for protein expression as described under “Results.”
cAMP Accumulation
To determine agonist-promoted cAMP accumulation, ∼200,000 HEK293 or ∼300,000 GT1-7 cells were seeded in 12-well dishes coated with 0.1% poly-l-lysine 24 h before the experiment and labeled in serum-free DMEM containing 2 μCi/ml [3H]adenine for 2–4 h. Cells were stimulated for 45 min at 37 °C in DMEM containing 2.5 μm 3-isobutyl-1-methylxanthine along with various concentrations of ligand. To block βγ-subunits, cells were preincubated with 3-isobutyl-1-methylxanthine and 10 μm gallein (2-(3,4,5-trihydroxy-6-oxoxanthen-9-yl)-benzoic acid dehydrate) for 15 min at 37 °C. The reaction was terminated by removing the medium and adding ice-cold 5% trichloroacetic acid to the cells. [3H]ATP and [3H]cAMP were then purified by sequential chromatography (Dowex resin/aluminum oxide columns), and the accumulation of [3H]cAMP was expressed as the ratio of [3H]cAMP/([3H]cAMP + [3H]ATP).
Firefly Luciferase Reporter Gene Assay
A pAD-CRE-Fluc plasmid containing the coding sequence of the firefly (Photinus pyralis) luciferase (Fluc) under the control of the CRE promoter was transfected into HEK293 cells using the metafectene reagent according to the manufacturer's protocol. Twenty-four hours after transfection cells were seeded in 12-well dishes coated with 0.1% poly-l-lysine. After 12 h, cells were serum-starved for 12 h and stimulated in serum-free DMEM with increasing concentrations of α-MSH. After 12 h of ligand stimulation, cells were lysed, and Fluc activity was determined using a luciferase reporter system (Promega, Mannheim, Germany) according to the manufacturer's protocol in a PolarSTAR plate reader from BMG (Offenburg, Germany).
Enzyme-linked Immunosorbent Assay to Detect Cell Surface Receptors
HEK293 cells stably expressing MCR were seeded directly on 12-well dishes. Dishes were then placed on ice, and cells were washed twice with ice-cold PBS containing 1% BSA. After blocking unspecific binding sites for 5 min on ice with the same buffer, Xpress epitope-MCR fusion proteins on the cell surface were detected by incubating the cells with 400 ng/ml anti-Xpress antibody (Invitrogen) in PBS, 1% BSA for 1 h at 4 °C. After washing the cells twice with PBS containing 1% BSA, cells were fixed for 15 min with 3–10% paraformaldehyde at room temperature. Then cells were washed twice with PBS and incubated for 45–60 min with anti-mouse horseradish peroxidase-conjugated secondary antibodies from sheep (1:2000) in PBS with 1% BSA at room temperature. Thereafter, cells were washed twice for 20 min with PBS containing 1% BSA and once with pure PBS. The substrate o-phenylenediamine dihydrochloride (Sigma) was added according to the manufacturer's instructions. After 5–10 min the reaction was stopped with 3 n HCl, and extinction was measured at 492 nm. To be able to compare MC4R expression in HEK239-Ex-MC4R and in GT1-7 cells, cell surface enzyme-linked immunosorbent assay experiments were performed with the following modifications; anti-MC4R-peptide antibody (ab24233: 400 ng/ml) from Abcam plc (Cambridge, UK) was used, and the concentrations of the secondary antibody (1:1000) and of the horseradish peroxidase substrate were increased (5-fold). The exposure time of the substrate to horseradish peroxidase was also increased (5-fold). Background values due to unspecific binding of the secondary antibody to GT1-7 cells were determined in the absence of the anti-MC4R-peptide antibody and subtracted from the signal obtained in the presence of the anti-MC4R-peptide antibody.
Radioligand Binding Assay
For ligand binding studies, ∼100,000 cells were seeded in 48-well dishes coated with 0.1% poly-d-lysine. After 24 h binding of [α-125I]MSH (0.1–5 nm) was determined in the presence or absence of 1 μm α-MSH. These radioligand binding assays were carried out on attached cells for 2 h on ice in DMEM with 100 mm HEPES, pH 7.4, and 0.1% of BSA. The amount of cell-bound radioactivity was measured after washing the cells twice with 0.5 ml of ice-cold binding buffer and lysing them with 0.1% SDS and 0.1 n NaOH.
Real-time quantitative PCR (RTQ-PCR)
To compare expression levels from MC3R and MC4R subtypes in GT1-7 cells, total RNA was isolated using the TriFast Reagent (PeqLab, Erlangen, Germany). First-strand synthesis was carried out with random hexamers as primers (pdN6, Amersham Biosciences) using REVERTAID reverse transcriptase (MBI-Fermentas, Sankt Leon-Roth, Germany). Products were amplified using mouse MC3R (forward, 5′-tgggcaccctatatatccaca-3′; reverse, 5′-cccttcatgcaggagtgc-3′)-, mouse MC4R (forward, 5′-ttccctccacctctggaas-3′; reverse, 5′-gggggaaacaaaaagttg-3′)-, or as a control, mouse β-actin (forward, 5′-ccaaccgtgaaaagatgacc-3′; reverse, 5′-gtggtacgaccagaggcatac-3′)-specific primer pairs. RTQ-PCR was done using the Quantitect SYBR Green PCR kit (Qiagen, Hilden, Germany) containing a HotStar Taq polymerase, the corresponding buffer, nucleotides, MgCl2 (final concentration, 2.5 mm), and SYBR Green. 10 pmol of each primer pair and 0.2 μl from the first-strand synthesis were added to the reaction mixture. RTQ-PCR reactions were carried out using the following conditions: initial denaturation for 3 min at 94 °C, 45 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min followed by a final extension at 72 °C for 7 min using a LightCycler®2.0 from Roche Applied Science. To compare expression levels of various adenylyl cyclase (AC) subtypes in GT1-7 cells, RTQ-PCR has been performed as described above using an aliquot of the same freshly prepared cDNA for each sample and the following primer pairs: AC1 (forward, 5′-agatgggacttgacatgatcg-3′; reverse, 5′-cgcatgttcaggtctacttcag-3′); AC2 (forward, 5′-ctgctcgccgtcttcttc-3′; reverse, 5′-tgggaacggttattaaaaatgc-3′); AC3 (forward, 5′-ctcaatggcactgacagc-3′; reverse, 5′-ctcagcatcatgacgaacatc-3′); AC4 (forward, 5′-cgggaggctcttagctctct-3′; reverse, 5′-gcaggaaggatagacaagagga-3′); AC5 (forward, 5′-gggagaaccagcaacagg-3′; reverse, 5′-catctccatggcaacatgac-3′); AC6 (forward, 5′-catctccatggcaacatgac-3′; reverse, 5′-aggtgctaccgatggtcttg-3′); AC7 (forward, 5′-aagctggatgggatcaacag-3′; reverse, 5′-acaggcccgtggtttatg-3′). Fluorescence intensities were recorded after the extension step at 80 °C of each cycle to exclude fluorescence of primer dimers melting at temperatures lower than 80 °C. Crossing points were determined by the software provided by the manufacturer. The relative gene expression was quantified using the formula (2(crossing point of β-actin − crossing point of gene of interest)) × 100 = % of reference gene expression (β-actin).
GTPγ35S Assay
To directly monitor G protein activation, the incorporation of non-hydrolysable GTPγ35S into total membrane fractions was detected. MC4R stably expressing HEK293 or GT1-7 cells were detached on ice with 2 ml of ice-cold PBS containing 2 mm EDTA, and the total membrane fraction was prepared as follows; cells were homogenized in ice-cold membrane buffer (5 mm Tris/HCl, pH 7.4, 2 mm EDTA, 5 mg/ml leupeptin, 10 mg/ml benzamidine, and 5 mg/ml soybean trypsin inhibitor) using a Polytron (ultra-turrax T24, IKA) for 10–15 s at maximum speed. Lysates were centrifuged at 500 × g for 10 min at 4 °C. The resulting supernatant containing the total membrane fraction was again centrifuged (20,000 × g for 20 min), and the resulting pellet was collected in 20 ml of membrane buffer. After repetition of this centrifugation step and resuspension of the pellet in 0.5–1.0 ml of assay buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 10 mm MgCl2, and 1 mm CaCl2), the amount of total proteins was determined according to the protocol of Bradford. 20 μg (HEK293 cells) or 40 μg (GT1-7 cells) of total membranes were used in a total volume of 1 ml per sample. 10 min before agonist stimulation, 5 μm GDP was added to the membranes to allow sufficient pre-coupling of the receptor to G proteins. Finally, the reaction was started by adding 0.1 nm GTPγ35S and a given amount of the ligand. After 30 min at 30 °C, the reaction was stopped by separating bound from free GTPγ35S by filtration of the samples through glass fiber filters in a cell harvester. After washing the filters twice with PBS, the amount of GTPγ35S bound was detected by scintillation counting in a β-counter.
Data Analysis
Data obtained in reporter gene and cAMP accumulation assays were analyzed using Prism3.0. Statistical significance of differences in all assays was assessed by the two-tail Student's t test.
RESULTS
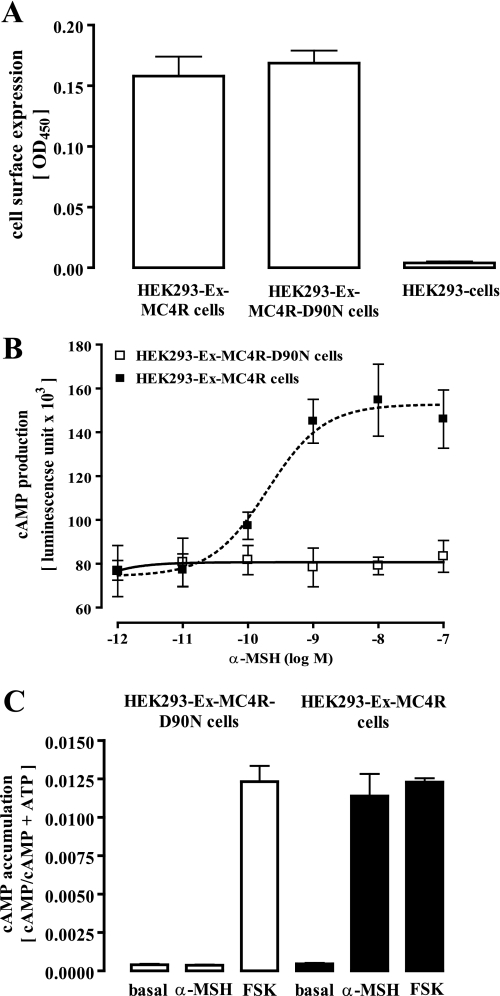
To analyze the G protein coupling profile of the MC4R, we stably expressed the mouse wild-type MC4R or the obesity-associated D90N mutant fused with the sequence of the Xpress (Ex) epitope to their 5′-ends. Both cell pools showed a comparable amount of Ex epitopes at the cell surface (Fig. 1A), indicating similar cell membrane expression levels of both receptor variants. The MC4R-D90N mutant has been shown to bind α-MSH with unchanged high affinity, but no activation of Gs proteins could be detected (14). In line with this previous finding, we did not observe any α-MSH-promoted signaling in MC4R-D90N-expressing cells when employing reporter gene or cAMP accumulation assays, although MC4R-expressing cells strongly reacted under the these conditions (Fig. 1, B and C).
FIGURE 1.
Stable expression of the mouse MC4R and the MC4R-D90N mutant in HEK293 cells. A, cell surface expression in HEK293 cells stably expressing the MC4R wild type or the D90N mutant fused on the N terminus with the Xpress epitope was monitored by the enzyme-linked immunosorbent assay technique with intact cells. B, HEK293-Ex-MC4R or HEK293-Ex-MC4R-D90N cells were transiently transfected with a reporter gene construct harboring the firefly luciferase gene under the control of the CRE promoter. Thirty-six hours after transfection (including 12 h of cell starvation), cells were stimulated with increasing concentrations of α-MSH. After lysis of the cells, firefly luciferase activity was determined. Dose-response curves were drawn from data obtained in three independent experiments carried out in triplicate. C, cAMP accumulation in HEK293-Ex-MC4R or HEK293-Ex-MC4R-D90N cells was assessed after labeling of the cells with [3H]adenine followed by the purification of [3H]cAMP and [3H]ATP by sequential chromatography and expressed as the ratio of [3H]cAMP/([3H]cAMP + [3H]ATP). Cells were stimulated or not with 1 μm α-MSH for 30 min at 37 °C. As a control, cells were also stimulated with the MC4R-independent direct adenylyl cyclase activator forskolin (FSK, 20 μm). Results are expressed as the mean ± S.E. of five independent experiments performed in triplicate.
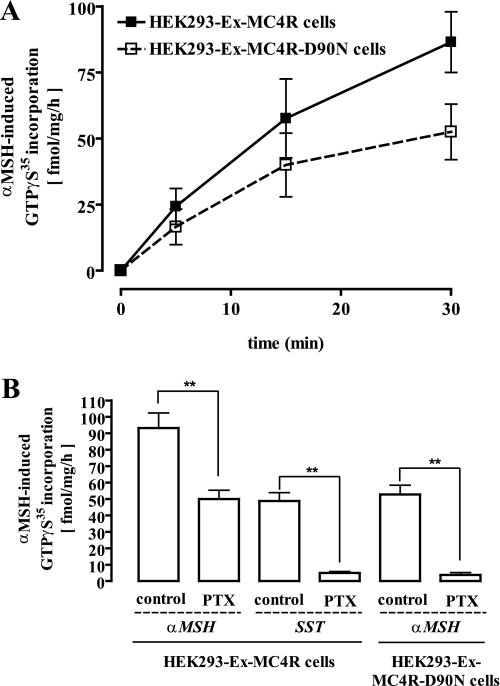
Next, we measured GTPγ35S incorporation into total membrane fractions derived from HEK293 cells. As shown in Fig. 2A, a saturating concentration of α-MSH (1 μm) induced GTPγ35S incorporation of 92 ± 9 fmol/mg/h (at 30 min) in membranes of HEK293-Ex-MC4R cells. However, despite the absence of Gs signaling, α-MSH promoted GTPγ35S incorporation in membranes of HEK293-Ex-MC4R-D90N cells. Agonist-promoted GTPγ35S incorporation amounted to 53 ± 6 fmol/mg/h (Fig. 2A, 30 min), indicating that the Asp to Asn substitution at position 90 of the MC4R did not preclude G-protein coupling of the receptor. To investigate the effects of PTX (50 ng/ml for 16 h), we took advantage of the endogenously expressed somatostatin receptor-2 subtype (15, 16). As expected, PTX treatment completely blocked somatostatin-promoted GTPγ35S incorporation (Fig. 2B). The same membranes showed a partial block (∼50%) of α-MSH-induced (1 μm) GTPγ35S incorporation (Fig. 2B). In contrast, the D90N variant behaved like the SST-2 receptor, as α-MSH-induced (1 μm) GTPγ35S incorporation was completely abolished after toxin treatment (Fig. 2B). Hence, these data are indicative of the dual coupling of the MC4R to Gs and Gi/o proteins and demonstrate that a single amino acid exchange in an obesity-associated MC4R mutant is sufficient to support a distinct active receptor conformation that redirects coupling of the receptor specifically to PTX-sensitive G proteins.
FIGURE 2.
α-MSH-induced activation of pertussis toxin-sensitive G proteins in HEK293-Ex-MC4R or HEK293-Ex-MC4R-D90N cells. A, 20 μg of total membrane fractions derived from HEK293-Ex-MC4R or HEK293-Ex-MC4R-D90N cells were incubated with 0. 1 nm non-hydrolysable GTPγ35S for 5, 15, or 30 min at 30 °C. Signals from non-stimulated membranes are subtracted from the signals of stimulated (1 μm α-MSH) cells. Results are expressed as the mean ± S.E. of two independent experiments performed in quadruplicate. B, incorporation of GTPγ35S (30 min) was monitored as described under A. Additionally, membranes derived from control HEK293-Ex-MC4R cells or from cells treated with the Gi/o-specific inhibitor PTX (50 ng/ml) for 16–24 h at 37 °C were stimulated with 1 μm somatostatin (SST) to activate endogenously expressed somatostatin-2 receptor subtypes as a control for a Gi/o-coupled receptor. Results are expressed as the mean ± S.E. of four independent experiments carried out in quadruplicate. Asterisks indicate a significant (**, p < 0.01) difference between PTX-treated and non-treated cells.
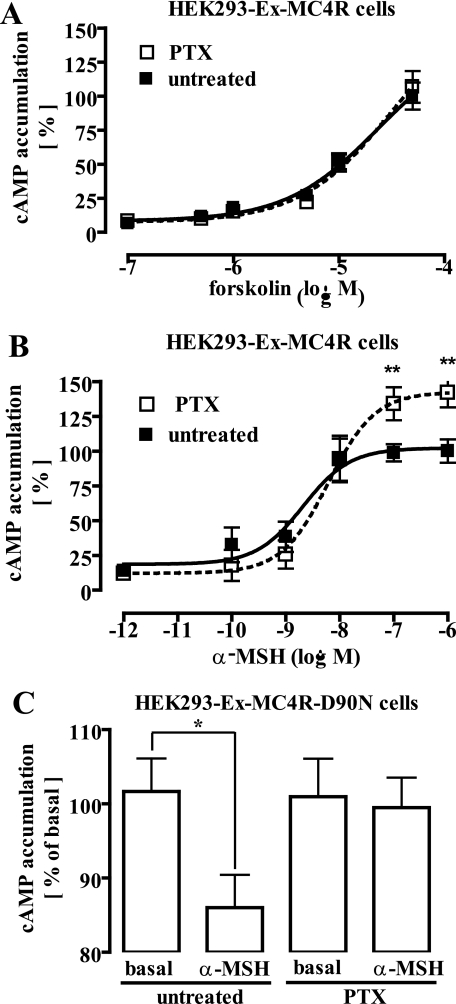
Given the opposing roles of Gs and Gi/o proteins in the regulation of the intracellular cAMP concentration, we next analyzed the effect of PTX (50 ng/ml for 16 h) on α-MSH-induced cAMP accumulation. As shown in Fig. 3A, PTX did not impact cAMP accumulation induced by direct forskolin-mediated activation of AC, indicating that PTX had no overall detrimental effects on the ability of HEK293 cells to accumulate cAMP. In contrast, toxin treatment increased α-MSH-induced cAMP accumulation as evidenced by a 30% increase in the Emax value of α-MSH concentration-response curves (Fig. 3B). Thus, it appears that dual coupling of the MC4R to PTX-insensitive (Gs) and -sensitive (Gi/o) proteins leads to counteracting effects on AC. Given the selective Gi/o coupling of the D90N mutant, we predicted that α-MSH would decrease basal cAMP concentrations in HEK293-Ex-D90N cells. Accordingly, stimulation of MC4R-D90N-expressing cells with α-MSH (1 μm) significantly decreased basal cAMP concentrations in a PTX-sensitive manner (Fig. 3C).
FIGURE 3.
α-MSH-induced regulation of adenylyl cyclase activity is sensitive to pertussis toxin in HEK293-Ex-MC4R or HEK293-Ex-MC4R-D90N cells. A, cAMP accumulation in HEK293-Ex-MC4R cells was assessed after labeling of the cells with [3H]adenine followed by the purification of [3H]cAMP by sequential chromatography over Dowex resin and aluminum oxide columns. Cells were treated or not with 50 ng/ml PTX for 16–24 h at 37 °C and then stimulated with increasing concentrations of forskolin for 30 min at 37 °C. Results are expressed as the mean ± S.E. of three independent experiments carried out in triplicate. B, ligand-promoted cAMP accumulation in HEK293-Ex-MC4R cells was assessed as described in A, but here cells were stimulated with increasing concentrations of α-MSH. Results are expressed as the mean ± S.E. of three independent experiments carried out in triplicate. Maximal cAMP accumulation measured in non-PTX treated cells was set to 100%. Asterisks indicate a significant (**, p < 0.01) difference between PTX-treated and untreated cells. C, basal cAMP accumulation in HEK293-Ex-MC4R-D90N cells was assessed as described in A. PTX-treated or untreated cells were stimulated with 1 μm α-MSH for 1 h at 37 °C. Data are given as percentage of untreated and non-stimulated cells. Results are expressed as the mean ± S.E. of five independent experiments carried out in triplicate. Asterisks indicate a significant (*, p < 0.05) difference between PTX-treated and non-treated cells.
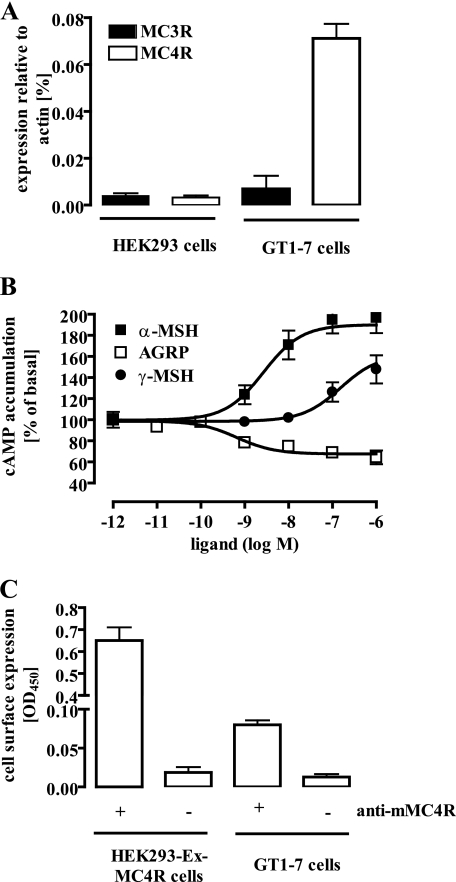
To exclude that multiple G protein coupling of the MC4R is caused by overexpression of the recombinant protein, we took advantage of the recently described GT1-7 cell line derived from cultured murine hypothalamic neurons. Because endogenous expression of MC3R and MC4R in GT1-7 cells has been suggested (17–19), we first identified the MCR subtype present in these cells. Using RTQ-PCR, we demonstrated that GT1-7 cells almost exclusively express the MC4R subtype (Fig. 4A). Measuring cAMP accumulation in these cells, we obtained a concentration-response curve for α-MSH with a potency of 2.6 ± 1.5 nm and an efficacy of 90 ± 8% over basal values (Fig. 4B). α- and γ-MSH have been shown to bind the MC3R with a similar affinity, whereas the MC4R subtype binds α-MSH with a higher affinity than γ-MSH. Thus, we determined concentration-response curves with γ-MSH and GT1-7 cells. These experiments revealed that γ-MSH is less potent (153 ± 11.2 nm) in GT1-7 cells than α-MSH, confirming that functional MC3R were not detectable in these cells. Finally, to compare the MC4R cell surface expression levels in GT1-7 and in HEK293-Ex-MC4R cells, we employed an anti-MC4R-peptide antibody raised against the N terminus of the mouse MC4R. This antibody revealed an ∼10-fold higher receptor density in HEK293-Ex-MC4R cells when compared with GT1-7 cells (Fig. 4C). Performing saturation binding experiments with [α-125I]MSH and HEK293-Ex-MC4R cells (supplemental Fig. 1), we obtained a Bmax value of 688 ± 89 fmol/mg. Thus, we estimate the Bmax value of GT1-7 cells to be ∼68 fmol/mg. This value is in good agreement with the previously reported Bmax value of 43 fmol/mg obtained in saturation binding experiments with [α-125I]NDP-MSH and GT1-7 cells (17).
FIGURE 4.
Hypothalamic GT1-7 cells exclusively express the MC4R subtype. A, the presence of MC3R or MC4R transcripts in GT1-7 cells was detected by RTQ-PCR and is given relative to the expression of β-actin. As a control, cDNA from HEK-293 cells was used. B, cAMP accumulation in GT1-7 cells was assessed after labeling of the cells with [3H]adenine followed by the purification of [3H]cAMP and [3H]ATP by sequential chromatography and expressed in percentage of basal cAMP production. Cells were stimulated with increasing concentrations of α-MSH, γ-MSH, or AGRP for 30 min at 37 °C. Results are expressed as the mean ± S.E. of three independent experiments carried out in triplicate. C, cell surface expression of the MC4R in HEK293 or GT1-7 cells was monitored by performing an enzyme-linked immunosorbent assay with intact cells using an anti-MC4R-peptide antibody raised against an N-terminal peptide of the murine MC4R.
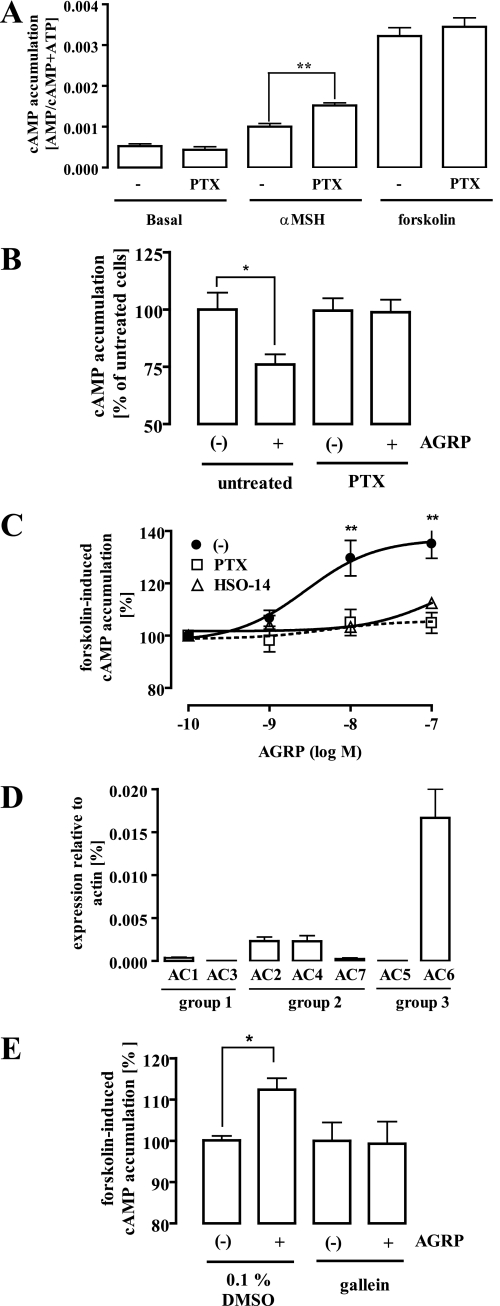
As mentioned before, MC4R activity is not only regulated by the agonistic actions of melanocortins but also by the inverse agonistic effects of AGRP (11). However, so far no data are available, demonstrating inverse agonistic effects of AGRP on endogenously expressed MC4R. Thus, we determined AGRP-dependent changes in cAMP levels in GT1-7 cells. As expected, AGRP decreased basal cAMP production in GT1-7 cells in a concentration-dependent manner (Fig. 4B). Next, we compared α-MSH- (1 μm) and forskolin-induced (5 μm) cAMP accumulation in PTX-treated and untreated cells. The efficacy of α-MSH to induce cAMP production increased due to the toxin treatment by 53 ± 6% over basal conditions (no PTX treatment), whereas the ability of forskolin to activate AC was not affected by PTX (Fig. 5A). These data suggest that the MC4R couples to Gi/o and Gs proteins in hypothalamic GT1-7 cells. Therefore, we next analyzed whether blocking Gi/o signaling interferes with the ability of AGRP to reduce basal cAMP levels. As shown in Fig. 5C, treatment of GT1-7 cells with PTX (50 ng/ml for 16 h) inhibited AGRP-promoted (100 nm) reduction of basal cAMP levels. However, given the low expression level of the MC4R in an endogenous expression system, weak effects of PTX on AGRP-independent cAMP accumulation (PTX decreased basal cAMP accumulation about 25%) may have also contributed to this observation. Because a previous report demonstrated strong effects of AGRP on forskolin-induced cAMP production after overexpression of the MC4R in B16/G4F cells (11), we tested the effects of PTX on putative AGRP-promoted inhibition of forskolin-promoted cAMP accumulation in GT1-7 cells. Surprisingly, in contrast to the recombinant overexpression system, AGRP did not decrease but increased forskolin-induced cAMP production in GT1-7 cells (Fig. 5C). At first glance, these results appear to be paradoxical. However, it should be taken into account that AC isoforms can be grouped into 3 subfamilies: group 1 (AC1, -3, and -8), group 2 (AC2, -4, and -7), and group 3 (AC5 and -6) (20). Members of group 2 have been shown to be less sensitive to Gi/o-mediated inhibition but are sensitized by βγ-subunits released from Gi/o (21–24). Thus, expression of AC2, -4, or -7 in GT1-7 cells could account for the stimulatory effects of AGRP on forskolin-induced cAMP production. As shown in Fig. 5D, the widespread AC6 isoform is the predominant AC isoform expressed in GT1-7 cells. However, transcripts of AC2 and AC4 are also detectable in GT1-7 cells. Transcripts of group 2 isoforms roughly amount to around one-third of all AC subtypes analyzed. Thus, in GT1-7 cells AGRP-mediated activation of Gi/o-signaling might increase forskolin-induced cAMP production by sensitization of group 2 AC isoforms. To test this hypothesis, we first analyzed the effect of PTX on AGRP-mediated sensitization of forskolin-promoted AC activation. Pretreatment of GT1-7 cells with PTX blocked AGRP effects on forskolin-induced cAMP accumulation (Fig. 5C), indicating that in GT1-7 cells PTX-sensitive G proteins are involved in AGRP-mediated regulation of cAMP production. Second, we examined the effect of the recently described cell-permeable β1γ2-subunit inhibitor gallein (25) on AGRP-induced enhancement of forskolin-promoted cAMP accumulation (Fig. 5E). Pretreatment of GT1-7 cells with 10 μm gallein for 15 min completely blocked the ability of AGRP to sensitize ACs for forskolin by releasing βγ-subunits from Gi/o proteins. Finally, to identify the receptor involved in AGRP-mediated signaling, we took advantage of the MC4R-specific antagonist HSO-14 (26, 27). HSO-14 (500 nm) abrogated the ability of AGRP to block forskolin-induced cAMP production (Fig. 5C), defining the MC4R subtype as the GPCR-mediating PTX-sensitive AGRP-induced signaling in GT1-7 cells.
FIGURE 5.
Dual coupling of the MC4R to Gs and Gi/o proteins in hypothalamic GT1-7 cells. A, cAMP accumulation in GT1-7 cells was assessed after labeling of the cells with [3H]adenine followed by the purification of [3H]cAMP and [3H]ATP by sequential chromatography and expressed as the ratio of [3H]cAMP/([3H]cAMP + [3H]ATP). Cells were treated or not with 50 ng/ml PTX for 16 -24 h at 37 °C and then stimulated with 1 μm α-MSH or 5 μm forskolin for 30 min at 37 °C. Results are expressed as the mean ± S.E. of five independent experiments carried out in triplicate. Asterisks indicate a significant (**, p < 0.01) difference between PTX-treated and non-treated cells. B, cAMP accumulation in GT1-7 cells was assessed as described above. Cells were treated or not with 50 ng/ml PTX for 16–24 h at 37 °C and then stimulated with 100 nm AGRP for 30 min at 37 °C. Results are expressed in percentage of untreated and non-stimulated cells. Basal cAMP accumulation of each condition was set to 100%. Asterisks indicate a significant (*, p < 0.05) difference between PTX-treated and non-treated cells. C, cAMP accumulation in GT1-7 cells was assessed as described above and presented as percentage of forskolin-promoted cAMP accumulation. Cells were treated or not with 50 ng/ml PTX for 16–24 h at 37 °C and then stimulated with increasing concentrations of AGRP alone or together with 500 nm HSO-14 in the presence of 5 μm forskolin for 30 min at 37 °C. Results are expressed as a percentage of forskolin-induced cAMP accumulation in the absence of AGRP and represent the mean ± S.E. of five independent experiments carried out in triplicate. Asterisks indicate a significant (*, p < 0.05) difference between PTX-treated and non-treated cells. D, expression analysis of adenylyl cyclase isoforms (AC1–7) in GT1-7 cells was performed by RTQ-PCR and is given relative to the expression of β-actin. Results are expressed as the mean ± S.D. of two independent experiments carried out in duplicate. E, cAMP accumulation in GT1-7 cells was assessed as described above. Cells were pretreated or not with 10 μm gallein for 15 min at 37 °C and then stimulated with 5 μm forskolin alone or along with 100 nm AGRP for 30 min at 37 °C. Results of four experiments are compiled and expressed in the percentage of untreated and non-stimulated cells. cAMP accumulation of with forskolin (alone)-treated cells was set to 100%. Asterisks indicate a significant (*, p < 0.05) difference between AGRP-treated and non-treated cells.
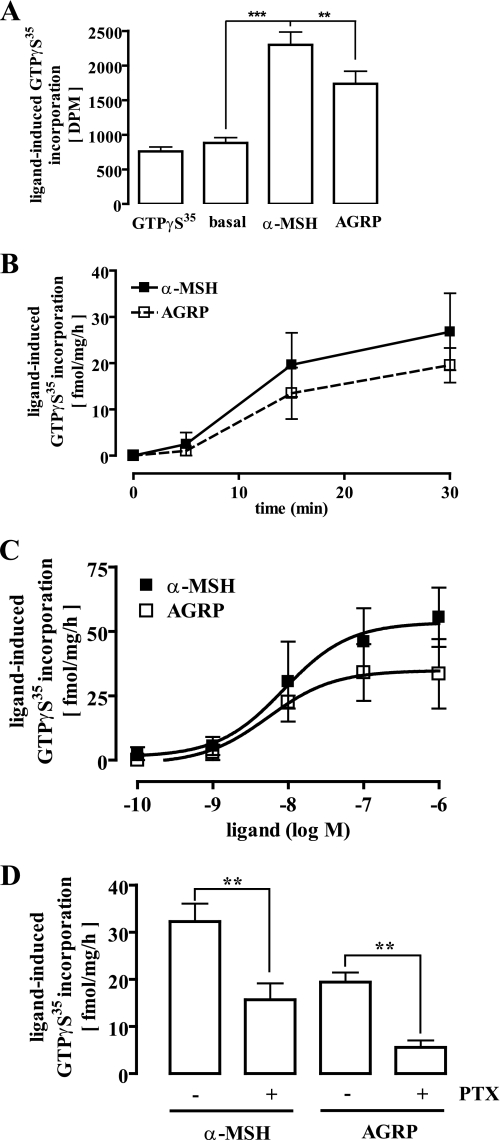
To evaluate melanocortin- and AGRP-promoted activation of Gi/o proteins, we directly assessed G protein activation by measuring the amount of GTPγ35S incorporated in freshly prepared membrane fractions of GT1-7 cells. As shown in Fig. 6m A and B, despite the low expression level of MC4R in GT1-7 cells, the GTPγ35S assay is suitable to detect ligand-mediated G protein activation. Performing dose-response curves with α-MSH, we obtained a Bmax value of 26 ± 6 fmol/mg/h and an EC50 value of 5.4 ± 2.6 nm (Fig. 6C). Although AGRP has been assumed to be an inverse agonist, it promoted the activation of G proteins in GT1-7 cells, characterized by a Bmax value of 18 ± 3 fmol/mg/h and an EC50 value of 8.7 ± 3.1 nm (Fig. 6C). PTX treatment decreased α-MSH-induced GTPγ35S incorporation to 13 ± 4 fmol/mg/h, clearly indicating that in line with our results obtained in the cAMP accumulation assay, the MC4R activates members of the Gs and Gi/o subfamily in a hypothalamic cell line (Fig. 6D). Finally, AGRP almost completely lost its propensity to activate G proteins in membranes derived from PTX-treated cells (Fig. 6D). Thus, in addition to its inverse agonistic actions on MC4R-promoted Gs signaling, AGRP is able to activate PTX-sensitive G proteins in GT1-7 cells and, thus, can be classified as one of the first naturally occurring biased agonists identified so far.
FIGURE 6.
Direct activation of Gi/o proteins by AGRP in hypothalamic GT1-7 cells. 40 μg of total membrane fractions derived from GT1-7 cells were incubated with 0. 1 nm non-hydrolysable GTPγ35S for 30 min at 30 °C. A, membranes were incubated with either 10 μm GTPγS, 1 μm α-MSH, or 100 nm AGRP. Incorporated GTPγ35S is given as total dpm. Data from one representative experiment performed in triplicate are shown. Asterisks indicate a significant (**, p < 0.01; ***, p < 0.005) difference between ligand-treated and non-treated cells. B, membranes were incubated with either 1 μm α-MSH or 100 nm AGRP for 5, 15, or 30 min. Results are expressed as the mean ± S.D. of two independent experiments carried out in duplicate. C, membranes were incubated with increasing concentrations of either α-MSH or AGRP for 30 min. Results are expressed as the mean ± S.D. of two independent experiments carried out in quadruplicate. D, before the membrane preparation cells were treated or not with 50 ng/ml PTX for 16–24 h at 37 °C. Incorporated GTPγ35S of membranes stimulated with 1 μm α-MSH or 100 nm AGRP is given as fmol/mg/h. Results are expressed as the mean ± S.E. of five independent experiments performed in quadruplicate. Asterisks indicate a significant (**, p < 0.01) difference between PTX-treated and non-treated cells.
DISCUSSION
The goal of the present study was to systematically dissect MC4R-dependent signaling pathways that impinge on the regulation of energy homeostasis. Monitoring direct G protein activation and cAMP production, we provide conclusive evidence that in addition to coupling to Gs proteins the MC4R also interacts with PTX-sensitive G proteins. Furthermore, we show that an obesity-associated MC4R variant and the orexigenic neuropeptide AGRP selectively engage MC4R-mediated Gi/o signaling.
Although the G protein coupling profile of a given GPCR may vary according to the cellular context, most GPCRs have been classified based on their predominant coupling to one given G protein subfamily. Interestingly, based on data obtained from A Guide to Receptors and Channels (from Stephen P. H. Alexander), we estimate that 17 members of 47 GPCR families (thus, ∼36%) interact with more than one G protein. Hence, coupling of a GPCR to multiple G proteins appears to be a common phenomenon. At first sight, dual coupling of a receptor to opposing G proteins such as Gs and Gi/o appears to be illogical and counterproductive. However, this kind of coupling pattern has been reported for glycoprotein hormone receptors (lutropin/chorionic gonadotropin receptor and thyroid-stimulating hormone receptor) (28), the lysophosphatidic acid receptor-3 (29), relaxin family peptide receptors (30), sphingosine 1-phosphate receptors (31), and for the β2-adrenergic receptor (32). For the β2-adrenergic receptor, in particular, sequential dual coupling has been proposed, as Gs-dependent phosphorylation of the receptor protein by protein kinase A increases the affinity to Gi/o proteins, leading to dual G protein coupling of the β2-adrenergic receptor (33). In the case of the MC4R, inhibition of protein kinase A activity with a selective inhibitor did not affect G protein coupling selectivity (supplemental Fig. 2), suggesting that dual coupling of the MC4R is not dependent on its signaling and occurs in parallel.
Directly measuring G protein activation in GT1-7 cells, we observed that even more than 50% of the activated G protein population is sensitive to PTX. However, the GTPγ35S assay is much more suitable for members of the Gi/o subfamily than for Gs proteins, as Gi/o proteins are more abundant than other heterotrimeric GTPases and have a higher nucleotide exchange rate (34, 35). Thus, a direct correlation between the proportion of GTPγ35S incorporated into different G protein subpopulations and the proportion of actually activated G protein subunits cannot be drawn. At the cAMP level, α-MSH strongly increased the production of this second messenger; thus, Gs coupling appears to dominate the Gi/o coupling of the receptor. However, treatment of GT1-7 cells with PTX increased α-MSH-mediated cAMP accumulation, indicating that a significant amount of Gi/o proteins is activated by α-MSH.
Multiple coupling of the MC4R to Gs and Gi/o proteins in hypothalamic GT1-7 cells endogenously expressing the MC4R raises the important question about the physiological relevance of this new signaling pathway. AGRP exclusively activates PTX-sensitive G proteins in GT1-7 cells, suggesting that AGRP antagonizes melanocortin-induced MC4R-mediated Gs activation not only by competitive antagonism but also by promoting G protein-induced signaling on its own. Interestingly, AGRP-mediated activation of PTX-sensitive G proteins sensitizes forskolin-induced cAMP production in GT1-7 cells, revealing the first AGRP-promoted signaling pathway so far.
Distinct AC subtypes are differentially regulated by Gi/o. Some AC subtypes (AC2, AC4, and AC7) have been shown to be less sensitive to Gi/o-mediated inhibition but are sensitized by βγ-subunits released from Gαi/o (21–24). Indeed, RTQ-PCR experiments with cDNA derived from GT1-7 cells suggest that Gβγ-sensitive AC subtypes are expressed in these cells, and the recently described cell-permeable β1γ2-subunit inhibitor gallein (25) blocked AGRP-mediated sensitization of forskolin-induced cAMP accumulation.
These observations clearly contrast with data obtained in an overexpression system (11), where AGRP dramatically decreased forskolin-induced cAMP production. Thus, overexpression of the MC4R allows for the activation of alternative signaling pathways by AGRP. In the HEK293-Ex-MC4R cells used in the present study, AGRP induced GTPγ35S incorporation in a PTX-sensitive manner (supplemental Fig. 3A) but inhibited forskolin-induced cAMP production in a PTX-insensitive way (supplemental Fig. 3B), suggesting that overexpression of the MC4R fundamentally affects the pharmacological properties of AGRP.
Given the opposing physiological role of melanocortins and AGRP on appetite control, we do not assume that sensitizing AC is the biological role of PTX-sensitive AGRP signaling. Although Gi/o proteins have been established as regulators of AC activity, other downstream effectors regulated by Gi/o proteins have been described. In GT1–1 cells, which are closely related to the GT1-7 cell line used in the present study, α-MSH-induced activation of extracellular-regulated kinases (ERK) has been shown (36). This observation is of particular interest, first because a specific inhibitor of ERK-blunted α-MSH-induced reduction of food intake in rats (37), and second, because Gi/o proteins activate the ERK pathway (38–40). However, α-MSH-induced ERK activation in GT1–1 cells is dependent on protein kinase C and is not blocked by PTX (36). Interestingly, in the same study α-MSH-induced ERK activation has been shown to be sensitive to PTX in HEK293 cells, again indicating that the MC4R exhibits different G protein coupling profiles in recombinant and endogenous expression systems.
Other well established downstream effectors of Gi/o signaling are ion channels and phospholipase Cβ. Thus, we analyzed α-MSH- or AGRP-promoted increases in intracellular calcium concentrations due to phospholipase Cβ activation, but activation of this signaling pathway by the MC4R in GT1-7 cells was not detectable (supplemental Fig. 4). The regulation of ion channel activities in MC4R-expressing neurons could impact the release of appetite-regulating peptides by these neurons. Recently, it has been described that melanocortins modulate the excitability of hypothalamic neurons by alterations of resting potassium conductance (41). Interestingly, inward-rectifier potassium channels (KIR3.x) are regulated by βγ-subunits released from Gαi/o (42, 43). Furthermore, AGRP inhibits hypothalamic neurons in a PTX-sensitive manner (44). Based on the exclusive Gs coupling of the MC4R postulated so far, it has been contended by the authors that PTX-sensitive signaling of AGRP cannot be mediated by the MC4R and probably is mediated by another, unidentified GPCR. Based on the data presented in our paper, it will be enlightening to clarify whether AGRP-mediated PTX-sensitive inhibition of hypothalamic neurons is blocked by MC4R-selective antagonists, indicating that this effect is mediated by MC4R-dependent activation of Gi/o proteins as discovered here.
In line with the hypothesis that AGRP induces its orexigenic effects by directly activating PTX-sensitive G proteins, we observed that the D90N mutant, which has been shown to be associated with severe obesity, selectively activates Gi/o. This finding raises the intriguing possibility that the D90N mutant does not only fail to induce anorexigenic Gs-mediated effects but also activates putative Gi/o-induced orexigenic signaling pathways. Thus, signaling pathways other than Gs activation have to be considered when assessing the functional consequences of obesity-associated MC4R mutations.
Besides the importance of our data for the understanding of the physiological effects of melanocortins, they provide new insights into the molecular mechanism of MC4R-mediated G protein activation. It has been proposed that receptors exist in an inactive, G protein-uncoupled (R) and in an active, G protein-coupled (R*) conformation (45). Over the last decade it has been debated whether GPCRs form only one or more R* conformations (46, 47). This is of particular interest in the case of a receptor subtype coupling to different G proteins. Our data obtained with the D90N mutant indicate that a single-point mutation in the coding sequence of the MC4R allows the specific activation of Gi/o but not of Gs. Hence, we postulate that the MC4R exists in R* conformation specific for Gs or Gi/o proteins, indicating that MC4R-propagated signaling is best described in a model consistent with multiple R* conformations. A multi-state model of receptor activation raises the intriguing possibility that distinct ligands might be able to differentially stabilize distinct R* conformations and, therefore, selectively activate a particular signaling pathway. Such so-called “biased agonists” have recently been identified for members of other GPCR families (48–51).
Here, we suggest a new model in which the AGRP peptide acts as a naturally occurring biased agonist that selectively activates potential Gi/o-mediated orexigenic and simultaneously blocks Gs-induced anorexigenic signaling pathways. The discovery of synthetic biased agonists that selectively induce different signaling pathways via the MC4R may offer an interesting strategy for the development of new drugs regulating appetite and energy homeostasis in either direction.
Supplementary Material
Acknowledgment
We are grateful to Dr. Tim Plant from the Institute of Pharmacology and Toxicology of the Philipps-University Marburg (Germany) for critically reading the manuscript.
This work was supported by the “Bundesministerium für Bildung und Forschung” as a part of the NGFN-2 (Nationales Genomforschungsnetzwerk N2NV-S30T09) Network.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- MC4R
- melanocortin-4 receptor
- AC
- adenylyl cyclase
- AGRP
- agouti-related protein
- BSA
- bovine serum albumin
- CRE
- cAMP-response element
- DMEM
- Dulbecco's modified Eagle's medium
- GPCR
- G protein-coupled receptor
- GTPγ35S
- guanosine 5′-(γ-thio)triphosphate
- HEK
- human embryonic kidney
- MCR
- melanocortin receptor
- MSH
- melanocyte stimulating hormone
- PBS
- phosphate-buffered saline
- RTQ-PCR
- real-time quantitative PCR
- PTX
- pertussis toxin
- ERK
- extracellular-regulated kinase
- Ex
- Xpress.
REFERENCES
- 1.Huszar D., Lynch C. A., Fairchild-Huntress V., Dunmore J. H., Fang Q., Berkemeier L. R., Gu W., Kesterson R. A., Boston B. A., Cone R. D., Smith F. J., Campfield L. A., Burn P., Lee F. (1997) Cell 88, 131–141 [DOI] [PubMed] [Google Scholar]
- 2.Yeo G. S., Lank E. J., Farooqi I. S., Keogh J., Challis B. G., O'Rahilly S. (2003) Hum. Mol. Genet. 12, 561–574 [DOI] [PubMed] [Google Scholar]
- 3.Tao Y. X., Segaloff D. L. (2003) Endocrinology 144, 4544–4551 [DOI] [PubMed] [Google Scholar]
- 4.Hinney A., Bettecken T., Tarnow P., Brumm H., Reichwald K., Lichtner P., Scherag A., Nguyen T. T., Schlumberger P., Rief W., Vollmert C., Illig T., Wichmann H. E., Schäfer H., Platzer M., Biebermann H., Meitinger T., Hebebrand J. (2006) J. Clin. Endocrinol. Metab. 91, 1761–1769 [DOI] [PubMed] [Google Scholar]
- 5.Hinney A., Hohmann S., Geller F., Vogel C., Hess C., Wermter A. K., Brokamp B., Goldschmidt H., Siegfried W., Remschmidt H., Schäfer H., Gudermann T., Hebebrand J. (2003) J. Clin. Endocrinol. Metab. 88, 4258–4267 [DOI] [PubMed] [Google Scholar]
- 6.Govaerts C., Srinivasan S., Shapiro A., Zhang S., Picard F., Clement K., Lubrano-Berthelier C., Vaisse C. (2005) Peptides 26, 1909–1919 [DOI] [PubMed] [Google Scholar]
- 7.Vaisse C., Clement K., Guy-Grand B., Froguel P. (1998) Nat. Genet. 20, 113–114 [DOI] [PubMed] [Google Scholar]
- 8.Gantz I., Miwa H., Konda Y., Shimoto Y., Tashiro T., Watson S. J., DelValle J., Yamada T. (1993) J. Biol. Chem. 268, 15174–15179 [PubMed] [Google Scholar]
- 9.Czyzyk T. A., Sikorski M. A., Yang L., McKnight G. S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ollmann M. M., Wilson B. D., Yang Y. K., Kerns J. A., Chen Y., Gantz I., Barsh G. S. (1997) Science 278, 135–138 [DOI] [PubMed] [Google Scholar]
- 11.Nijenhuis W. A., Oosterom J., Adan R. A. (2001) Mol. Endocrinol. 15, 164–171 [DOI] [PubMed] [Google Scholar]
- 12.Tolle V., Low M. J. (2008) Diabetes 57, 86–94 [DOI] [PubMed] [Google Scholar]
- 13.Wu Q., Howell M. P., Palmiter R. D. (2008) J. Neurosci. 28, 9218–9226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biebermann H., Krude H., Elsner A., Chubanov V., Gudermann T., Grüters A. (2003) Diabetes 52, 2984–2988 [DOI] [PubMed] [Google Scholar]
- 15.Kaupmann K., Bruns C., Hoyer D., Seuwen K., Lübbert H. (1993) FEBS Lett. 331, 53–59 [DOI] [PubMed] [Google Scholar]
- 16.Law S. F., Yasuda K., Bell G. I., Reisine T. (1993) J. Biol. Chem. 268, 10721–10727 [PubMed] [Google Scholar]
- 17.Khong K., Kurtz S. E., Sykes R. L., Cone R. D. (2001) Neuroendocrinology 74, 193–201 [DOI] [PubMed] [Google Scholar]
- 18.Shinyama H., Masuzaki H., Fang H., Flier J. S. (2003) Endocrinology 144, 1301–1314 [DOI] [PubMed] [Google Scholar]
- 19.Dumont L. M., Wu C. S., Aschkenasi C. J., Elmquist J. K., Lowell B. B., Mountjoy K. G. (2001) Mol. Cell. Endocrinol. 184, 173–185 [DOI] [PubMed] [Google Scholar]
- 20.Patel T. B., Du Z., Pierre S., Cartin L., Scholich K. (2001) Gene 269, 13–25 [DOI] [PubMed] [Google Scholar]
- 21.Näsman J., Kukkonen J. P., Holmqvist T., Akerman K. E. (2002) J. Neurochem. 83, 1252–1261 [DOI] [PubMed] [Google Scholar]
- 22.Hanoune J., Defer N. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 145–174 [DOI] [PubMed] [Google Scholar]
- 23.Schallmach E., Steiner D., Vogel Z. (2006) Neuropharmacology 50, 998–1005 [DOI] [PubMed] [Google Scholar]
- 24.Steiner D., Avidor-Reiss T., Schallmach E., Saya D., Vogel Z. (2005) J. Mol. Neurosci. 27, 195–203 [DOI] [PubMed] [Google Scholar]
- 25.Lehmann D. M., Seneviratne A. M., Smrcka A. V. (2008) Mol. Pharmacol. 73, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kask A., Rägo L., Mutulis F., Pähkla R., Wikberg J. E., Schiöth H. B. (1998) Biochem. Biophys. Res. Commun. 245, 90–93 [DOI] [PubMed] [Google Scholar]
- 27.Kask A., Schiöth H. B., Harro J., Wikberg J. E., Rägo L. (2000) Can. J. Physiol. Pharmacol. 78, 143–149 [PubMed] [Google Scholar]
- 28.Laugwitz K. L., Allgeier A., Offermanns S., Spicher K., Van Sande J., Dumont J. E., Schultz G. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C. W., Rivera R., Dubin A. E., Chun J. (2007) J. Biol. Chem. 282, 4310–4317 [DOI] [PubMed] [Google Scholar]
- 30.Halls M. L., Bathgate R. A., Summers R. J. (2007) J. Pharmacol. Exp. Ther. 320, 281–290 [DOI] [PubMed] [Google Scholar]
- 31.Noh S. J., Kim M. J., Shim S., Han J. K. (1998) J. Cell. Physiol. 176, 412–423 [DOI] [PubMed] [Google Scholar]
- 32.Xiao R. P., Ji X., Lakatta E. G. (1995) Mol. Pharmacol. 47, 322–329 [PubMed] [Google Scholar]
- 33.Daaka Y., Luttrell L. M., Lefkowitz R. J. (1997) Nature 390, 88–91 [DOI] [PubMed] [Google Scholar]
- 34.Harrison C., Traynor J. R. (2003) Life Sci. 74, 489–508 [DOI] [PubMed] [Google Scholar]
- 35.Fields T. A., Casey P. J. (1997) Biochem. J. 321, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chai B., Li J. Y., Zhang W., Newman E., Ammori J., Mulholland M. W. (2006) Peptides 27, 2846–2857 [DOI] [PubMed] [Google Scholar]
- 37.Sutton G. M., Duos B., Patterson L. M., Berthoud H. R. (2005) Endocrinology 146, 3739–3747 [DOI] [PubMed] [Google Scholar]
- 38.Pace A. M., Faure M., Bourne H. R. (1995) Mol. Biol. Cell 6, 1685–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferraguti F., Baldani-Guerra B., Corsi M., Nakanishi S., Corti C. (1999) Eur. J. Neurosci. 11, 2073–2082 [DOI] [PubMed] [Google Scholar]
- 40.Shahabi N. A., Daaka Y., McAllen K., Sharp B. M. (1999) J. Neuroimmunol. 94, 48–57 [DOI] [PubMed] [Google Scholar]
- 41.Smith M. A., Hisadome K., Al-Qassab H., Heffron H., Withers D. J., Ashford M. L. (2007) J. Physiol. 578, 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leaney J. L. (2003) Eur. J. Neurosci. 18, 2110–2118 [DOI] [PubMed] [Google Scholar]
- 43.Milovic S., Steinecker-Frohnwieser B., Schreibmayer W., Weigl L. G. (2004) J. Biol. Chem. 279, 34240–34249 [DOI] [PubMed] [Google Scholar]
- 44.Fu L. Y., van den Pol A. N. (2008) J. Neurosci. 28, 5433–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samama P., Cotecchia S., Costa T., Lefkowitz R. J. (1993) J. Biol. Chem. 268, 4625–4636 [PubMed] [Google Scholar]
- 46.Galandrin S., Oligny-Longpré G., Bouvier M. (2007) Trends Pharmacol. Sci. 28, 423–430 [DOI] [PubMed] [Google Scholar]
- 47.Kenakin T. (2007) Mol. Pharmacol. 72, 1393–1401 [DOI] [PubMed] [Google Scholar]
- 48.Galandrin S., Oligny-Longpré G., Bonin H., Ogawa K., Galés C., Bouvier M. (2008) Mol. Pharmacol. 74, 162–172 [DOI] [PubMed] [Google Scholar]
- 49.Schonbrunn A. (2008) Mol. Cell. Endocrinol. 286, 35–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michel M. C., Alewijnse A. E. (2007) Mol. Pharmacol. 72, 1097–1099 [DOI] [PubMed] [Google Scholar]
- 51.Reversi A., Rimoldi V., Marrocco T., Cassoni P., Bussolati G., Parenti M., Chini B. (2005) J. Biol. Chem. 280, 16311–16318 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.