Abstract
Detailed structural and biochemical studies with the human cytomegalovirus (HCMV UL54) DNA polymerase are hampered by difficulties to obtain this enzyme in large quantities. The crystal structure of the related RB69 DNA polymerase (gp43) is often used as a model system to explain mechanisms of inhibition of DNA synthesis and drug resistance. However, here we demonstrate that gp43 is ∼400-fold less sensitive to the pyrophosphate analog foscarnet, when compared with UL54. The RB69 enzyme is also able to discriminate against the nucleotide analog inhibitor acyclovir. In contrast, the HCMV polymerase is able to incorporate this compound with similar efficiency as observed with its natural counterpart. In an attempt to identify major determinants for drug activity, we replaced critical regions of the nucleotide-binding site of gp43 with equivalent regions of the HCMV enzyme. We show that chimeric gp43-UL54 enzymes that contain residues of helix N and helix P of UL54 are resensitized against foscarnet and acyclovir. Changing a region of three amino acids of helix N showed the strongest effects, and changes of two segments of three amino acids in helix P further contributed to the reversal of the phenotype. The engineered chimeric enzyme can be produced in large quantities and may therefore be a valuable surrogate system in drug development efforts. This system may likewise be used for detailed structural and biochemical studies on mechanisms associated with drug action and resistance.
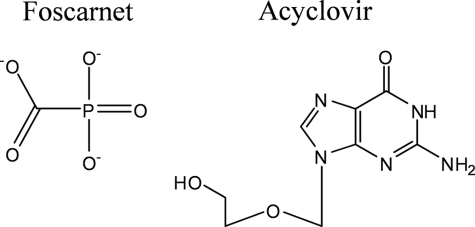
Infection with the human cytomegalovirus (HCMV),2 which belongs to the Herpesviridae, remains an important health problem in immunocompromised persons (1–7). Several drugs that target the viral DNA polymerase (UL54) have been developed to treat the infection (8–12). Cidofovir (CDV), ganciclovir (GCV), or its prodrug valganciclovir are nucleotide or nucleoside analog inhibitors, respectively, that are intracellularly phosphorylated to their triphosphate form and compete with natural nucleotide pools for incorporation (13–20). These compounds are characterized by an acyclic sugar moiety with the equivalent of a 3′-hydroxyl group that is required for the next nucleotide incorporation event (21). Thus, once incorporated, these compounds interfere with DNA synthesis at various positions downstream (18, 22, 23). In contrast, compounds that lack the 3′-hydroxyl group, such as the antiherpetic drug acyclovir (ACV) (Fig. 1), act as chain terminators (24, 25). Although active against HCMV, ACV is not approved for treatment of HCMV infection, and its efficacy is inferior to GCV or CDV (8, 26, 27). The pyrophosphate analog foscarnet (phosphonoformic acid, PFA) is the third approved anti-HCMV drug that inhibits UL54 (Fig. 1) (28, 29). However, toxicity, problems with oral bioavailability, and the rapid development of resistance can limit the clinical utility of each of the approved drugs.
FIGURE 1.
Structures of foscarnet and acyclovir.
PFA is a broad spectrum antiviral agent that was shown to inhibit various polymerases, including enzymes encoded by herpes simplex virus (HSV), human herpesvirus, HCMV, and the reverse transcriptase (RT) of the human immunodeficiency virus type 1 (HIV-1) (28, 29). Progress has been made in elucidating the mechanism of inhibition of HIV-1 RT (30, 31). Site-specific footprinting experiments revealed that the enzyme can oscillate between two conformations, referred to as pre- and post-translocation (32). The 3′ end of the primer still occupies the nucleotide-binding site in the pre-translocated complex (33, 34). Binding of the next nucleotide requires translocation of the enzyme relative to its nucleic acid substrate (35). The dNTP substrate can bind to and is incorporated in the post-translocated complex. In contrast, PFA traps the pre-translocational complex, which provides a plausible mechanism for inhibition (30, 32). The mechanism of action might be similar with the HCMV enzyme; however, the limited solubility of UL54 makes it difficult to produce the purified enzyme in sufficient amounts required for detailed biochemical and structural studies (36, 37). Combined in vitro transcription/translation systems and the baculovirus expression system have proven successful for the expression of UL54 and the related HSV polymerase (UL30) (38–43). The UL30 apoenzyme has been crystallized (44); however, crystallographic data for UL54 are not available (45).
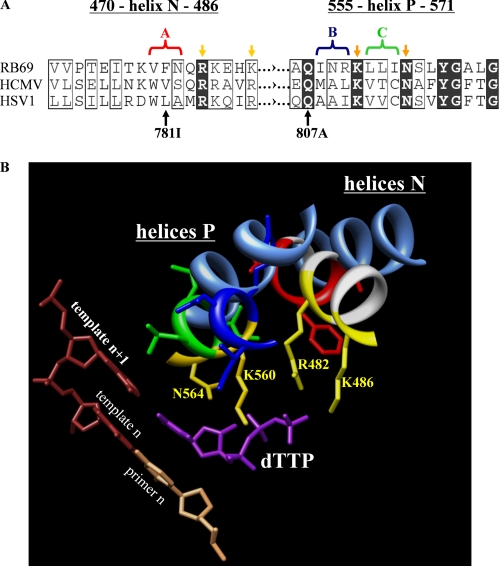
Like the related phage RB69 DNA polymerase (gp43), UL54 and UL30 belong to the polymerase α family (46). The RB69 polymerase can be expressed in its soluble form in Escherichia coli, which facilitates protein production at high yields (47, 48). This enzyme has also been crystallized in various forms, with and without the bound primer-template and the nucleotide substrate (48–50). The ternary complex serves as a model system that is often used to explain mechanisms of drug action and resistance associated with UL54 (45). The structure of the ternary complex shows that helix N and helix P provide important contacts that help to trap the incoming nucleotide (49). Enzyme kinetic data confirmed the roles played by conserved amino acids in nucleotide binding and catalysis (51, 52). Phenotypic drug susceptibility assays suggest that the equivalent regions in UL54 can affect susceptibility to GCV, CDV, and also to PFA (7, 53). Our recent biochemical data have shown that changes at residues of helix P implicated in PFA resistance or hypersusceptibility can either increase or decrease the inhibitory effects of this compound (42). A comparison with the crystal structure of gp43 suggests that the binding sites for PFA and the γ-phosphate of the bound nucleotide might overlap. However, although the existence of conserved residues within helix P points to a similar functional role in both enzymes, several residues in this region are different in nature, which may in turn affect sensitivity to PFA (Fig. 2A). Moreover, the contribution of helix N in PFA binding remains to be determined.
FIGURE 2.
Structure-based correlation between the nucleotide-binding sites of related RB69, HSV-1, and HCMV polymerases. A, sequence alignment of critical regions of helix N and helix P of DNA polymerases from HSV-1, HCMV, and RB69. Conserved residues are highlighted in black, and similar residues are boxed (61). Yellow arrows point to the amino acid residues of RB69 gp43 that interact with the triphosphate. Light and dark yellow color code distinguishes between residues of helix N and P, respectively. The amino acid substitutions V781I and Q807A illustrate previously reported mutations associated with resistance to foscarnet in HCMV (42, 62). The color-coded brackets and the corresponding captions “A, B, and C” illustrate the groups of three amino acids of the HCMV polymerase that were replaced in this study. The color code used in A is the same as in B. B, crystal structure of RB69 DNA polymerase in complex with a nucleotide substrate and primer-template (PDB code 1IG9). The amino acid residues of RB69 gp43 that interact with the triphosphate are highlighted in yellow. The cornflower blue ribbons represent the helices N and P of the HSV-1 polymerase. The crystal structure of HSV-1 polymerase in its apo form (PDB code 1GV9) was aligned with the crystal structure of RB69 polymerase (PDB code 1IG9).
In light of the limitations with respect to the expression of UL54, and the general shortcomings of surrogate systems such as the RB69 or UL30 enzymes, we engineered a chimeric RB69 DNA polymerase in which critical components of helix N and helix P were replaced by equivalent regions of the related HCMV enzyme (Table 1 and Fig. 2B). The aim of this study was to characterize the roles of the two helices in drug susceptibility to PFA and ACV and, in turn, to improve model systems for more detailed biochemical and structural analyses of UL54 and its interaction with antiviral drugs. The chimeric enzyme retained properties that facilitate expression and purification, and at the same time, the enzyme facilitates the study of mechanisms involved in drug action and drug resistance associated with the clinically relevant HCMV system. Most importantly, we show that gp43 is resistant to PFA and ACV, whereas the chimeric gp43-UL54 enzyme is almost as sensitive to both drugs as seen with UL54.
TABLE 1.
RB69-HCMV DNA polymerase chimera
| Nomenclature for chimera (block) | Region | RB69 residues | HCMV residues | Comments |
|---|---|---|---|---|
| A | Helix N | Val-478 | Trp-780 | None |
| Phe-479 | Val-781 | |||
| Asn-480 | Ser-782 | |||
| A/V781I | Helix | Chimera A V479I | Chimera A V781I | Foscarnet resistance conferring mutationa |
| B | Helix P | Ile-557 | Met-808 | None |
| Asn-558 | Ala-809 | |||
| Arg-559 | Leu-810 | |||
| C | Helix P | Leu-601 | Val-812 | None |
| Leu-602 | Thr-813 | |||
| Ile-603 | Cys-814 | |||
| ABC/R784A | Helix N | Chimera A | Chimera A | |
| R482A | R784A | Conserved residue | ||
| Helix P | Chimera BC | Chimera BC | ||
| ABC/Q807A | Helix N | Chimera A | Chimera A | |
| Helix P | Chimera BC | Chimera BC | ||
| Q556A | Q807A | Conserved residue conferring resistance to foscarnetb |
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Wild-type HCMV polymerase (UL54) was derived from recombinant viruses generated by overlapping cosmids as described previously (40). The UL54 coding sequence was kindly provided by Dr. Guy Boivin (Laval University). The UL54 coding sequence was cloned into pCITE4b (Novagen) by use of the EcoRI and HindIII sites to generate pCITE4b/UL54. We also generated a 3′–5′-exonuclease negative construct that contains the D542A substitution. The RB69 DNA polymerase (gp43) coding sequence was kindly provided by Dr. Sylvie Doublié (University of Vermont) and Dr. Jim Karam (Tulane University). The gp43 coding sequence was cloned into pPR-IBA1 (IBA) using the BsaI site to generate pPR-IBA1/gp43. This construct facilitates protein purification through Strep-tag affinity chromatography (IBA). D222A and D327A substitutions were introduced to remove the 3′–5′-exonuclease activity. Constructs for the production of mutant enzymes were generated by site-directed mutagenesis. The amino acid substitutions were introduced with PfuUltra DNA polymerase (Stratagene) according to the manufacturer's recommendations.
Protein Expression
The HCMV polymerase UL54 was expressed in rabbit reticulocyte lysate with a coupled in vitro transcription-translation system (Promega). Reactions were conducted essentially as described previously (41, 42). The RB69 DNA polymerase and chimeric RB69/HCMV enzymes were expressed as described previously (47). All enzymes were purified using Strep-tag affinity chromatography (IBA) according to the manufacturer's recommendations. Heterodimeric reverse transcriptase p66/p51 was expressed and purified as described (54).
Nucleic Acids and Chemicals
Oligodeoxynucleotides used in this study were chemically synthesized and purchased from Invitrogen. The following sequences were used as templates: T1, 5′GTAACTAGAGATCCCTCAGACCCTTTTAGTCAGAAT, and T2, 5′CCAATATTCACCATCAAGGCTTGATGAAACTTCACTCCACTATACCACTC. The underlined nucleotides are the portion of the templates annealed to the primer. The following primers were used in this study: P1, 5′TTCTGACTAAAAGGGTCTGAGGGAT, and P2, 5′GAGTGGTATAGTGGAGTGAA. Deoxynucleotides were purchased from Fermentas Life Sciences, and PFA was purchased from Sigma.
Synthesis of ACV-TP
ACV (1.5 mmol) was dissolved in 200 μl of dry 1,3-dimethyl-2-oxohexahydropyrimidine, N,N′-dimethylpropylene urea with 12–15 molecular sieves under nitrogen and stirred for 24 h. The mixture was chilled with an ice-water bath and stirred for 1 h, followed by slow addition of phosphorus oxychloride (3 eq) and stirred for an additional 25 min. A solution of tributylammonium pyrophosphate (4 eq) in 200 μl of N,N′-dimethylpropylene and tributyl amine (15 eq) was simultaneously added to the reaction. After 45 min the reaction was quenched with ice-cold water. The reaction was washed with chloroform, and the aqueous layer was collected and co-evaporated with deionized water three times. The residue was resuspended in 100 μl of deionized water and purified on ion-exchange column by high performance liquid chromatography λmax = 253 nm. The final product was co-evaporated with water five times, giving a total yield of ACV-TP (NH3)4 of 18% with purity ≥95%. The molecular weight of the ACV-TP was confirmed by liquid chromatography-mass spectrometry/tandem mass spectrometry m/z (M + 1) 466 → 152 (55).
Enzyme Kinetics
100 nm DNA/DNA primer-template hybrid T1/P1 (100 nm) was preincubated for 5–10 min at 37 °C with a given DNA polymerase in a buffer containing 25 mm Tris-HCl (pH 8), 50 mm NaCl, 0.5 mm dithiothreitol, 0.2 mg/ml bovine serum albumin, and 5% glycerol. To compare different enzymes in single nucleotide incorporation assays, we adjusted the enzyme concentration and the time point of the reaction such that ∼40% of the primer was used at the saturating concentration of dNTP (Fig. 3A). For Km and Ki measurements, the range of the dNTP substrate and/or inhibitor was chosen such that the values were in the middle of the chosen concentration range. Nucleotide incorporation was initiated by the addition of MgCl2 to a final concentration of 10 mm, and the reactions were allowed to proceed for 5 min. The reactions were stopped by the addition of three reaction volumes of formamide containing traces of bromphenol blue and xylene cyanol. Samples were then subjected to 15% denaturing PAGE followed by phosphorimaging. The incorporation of single nucleotides was quantified as the fraction of the DNA substrate (primer n) converted to product (primer n + 1).
FIGURE 3.
PFA inhibition of DNA synthesis by viral polymerases under steady-state conditions for single nucleotide incorporation events. A, single nucleotide incorporation events in the presence of constant concentrations of dCTP and increasing concentrations of PFA. The migration pattern of the 5′ end radioactively labeled primer (s) and product (p) is illustrated by corresponding arrows. B, graph of data shown below A. IC50 values for PFA were calculated by fitting the data points to a sigmoidal dose response. Dotted lines illustrate the respective IC50 values for PFA.
Km and Ki Experiments
The rate of the reaction was plotted versus the concentration of nucleotide substrate. The data points of a 42-data point Km/Ki experiment were fit to the general mixed model of inhibition using GraphPad Prism (version 5.0) to calculate kcat, Km, and Ki values. kcat is defined as the enzyme turnover number that is calculated by normalizing the maximum rate of the single nucleotide incorporation reaction to the enzyme concentration. Km is defined as the dNTP substrate concentration at half-maximum rate of the reaction. Ki is the inhibitor dissociation constant (56, 57). Significant figures for the fitted data of all experiments are as reported by the software. Standard deviations for all experiments were determined on the basis of at least three independent replicates.
IC50 Experiments
The incorporation of dCTP was determined by plotting the percentage of incorporation against the concentration of PFA (Fig. 3). IC50 values were calculated by fitting at least 10 data points to a sigmoidal dose-response (variable slope) equation using GraphPad Prism (version 5.0). Significant figures for the fitted data of all experiments are as reported by the software. Standard deviations for all experiments were determined on the basis of at least three independent replicates.
Bioinformatics
The secondary structure prediction-based alignment between UL54 and UL30 (PDB code 2GV9) was generated through ESyPred3D Web Server 1.0 (58). The result was uploaded into the crystal structure-based alignment between UL30 (PDB code 2GV9) and gp43 (PDB code 1IG9 (49)) using the chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (59) The sequence alignment output was graphically prepared with ESPRIPT software (60). Conserved residues are highlighted in black, and similar residues are boxed (61).
RESULTS
Experimental Design
The goal of this study was to engineer and to characterize a chimeric RB69/UL54 enzyme that facilitates the study of PFA-mediated inhibition of DNA synthesis and drug resistance. We focused on segments of helix N and helix P that are located in close proximity to the phosphates of the bound dNTP substrate in the ternary complex with gp43 (Table 1 and Fig. 2) (49). Several amino acids in these regions are likewise implicated in binding of PFA by UL54. Phenotypic susceptibility assays, and our previous biochemical data suggest that the region between 807 and 815 in helix P (556–564 in gp43) plays an important role in this regard (7, 42). Some of the amino acids of this segment can interact with the bound nucleotide, whereas others appear to be involved in interhelical interaction with residues 779–784 (477–482 in gp43) of helix N (49, 50) (Fig. 2B). In an attempt to closely mimic the structure of UL54, we replaced amino acids that differ at equivalent positions in gp43 according to the sequence alignment shown in Fig. 2A. These changes involve the block of three amino acids between the conserved residues Lys-477 and Gln-481/Lys-482 of helix N, referred to as block A, and two blocks of three amino acids between conserved residues Gln-556 and Asn-564 of helix P, referred to as block B and block C, respectively. Of note, the alignment also points to several amino acid changes in equivalent blocks A, B, and C of UL54 and UL30, respectively. All enzymes designed and characterized in this study are listed in Table 1.
We generated chimeric gp43-based enzymes in which block A of helix N and blocks B/C of helix P have been replaced by equivalent regions of UL54. Several conserved amino acids within these regions interact with the phosphates of the nucleotide (Fig. 2B). Arg-482 of helix N as well as Gln-556 of helix P appear to contact the γ-phosphate of the bound nucleotide. The same region is also implicated in PFA binding. The Q807A mutation in helix P in UL54 increases the IC50 value for PFA (42). However, a potential role of helix N in PFA resistance remains to be defined. Of note, Phe-479 of helix N in gp43 is equivalent to Val-781 in UL54, and the V781I substitution in the HCMV enzyme shows decreased phenotypic susceptibility to PFA (62). This residue is likewise located in the vicinity of the γ-phosphate, and this region appears to be important for PFA binding. Thus, to test how close the chimeric enzyme may mimic the natural HCMV polymerase, we introduced V781I and alanine substitutions at conserved residues Arg-784 (helix N) and Gln-807 (helix P), respectively, against the background of the chimera.
Efficiency of Nucleotide Incorporation
We initially determined steady-state kinetic parameters for single nucleotide incorporation events, and we compared several chimeric enzymes with WT gp43 (Table 2). Throughout this study, we used 3′–5′-exonuclease negative mutants to prevent potentially confounding effects through the editing activity. Replacing block A (helix N) with the equivalent UL54 region caused ∼2–3-fold reductions in the efficiency of single nucleotide incorporation events. Introducing the V781I mutation against this background caused similar reductions in kinetic parameters. Chimeric enzymes containing block A (helix N) as well as block B/C (helix P) of UL54 showed 10-fold reductions in the same assay. Alanine mutations at conserved positions Arg-784 and Gln-807 that were introduced against this mutational background did not cause further significant reductions in the efficiency of nucleotide incorporation. Overall, these findings show that replacing critical elements of helix N and helix P with their UL54 counterparts can cause reductions in enzymatic activity. Because of the lack of a corresponding expression/purification protocol for UL54, it is at this point difficult to assess whether the activity of the HCMV enzyme is intrinsically reduced or whether the chimeric nature of the enzyme may cause such deficits. However, the enzymatic activities are sufficiently high to measure potential changes in drug sensitivity, provided that the concentrations of each of the enzymes to be compared are appropriately adjusted as outlined under “Experimental Procedures”.
TABLE 2.
Kinetic constants for dCTP incorporation
Values were calculated by fitting the data points to Michaelis-Menten equation using GraphPad Prism (version 5.0).
| Enzyme | kcata | Kmb | kcat/Km |
|---|---|---|---|
| s−1 | μm | ||
| gp43c | 0.61d | 0.49 | 1.25 |
| A | 0.39 | 0.92 | 0.42 |
| A/V781I | 0.51 | 0.92 | 0.55 |
| ABC | 0.46 | 2.7 | 0.17 |
| ABC/R784A | 0.39 | 3.3 | 0.12 |
| ABC/Q807A | 0.50 | 3.6 | 0.14 |
a kcat is the enzyme turnover number that is calculated by normalizing the maximum velocity of the reaction to the enzyme concentration.
b Km is the substrate concentration at half-maximum velocity of the reaction.
c The previously reported gp43 values for dCTP kcat and Km are 1.0 s−1 and 0.57 μm, respectively (51).
d Standard deviations were determined on the basis of at least three independent experiments and represent a maximum of 12% of the reported value.
Sensitivity to PFA
The ability to inhibit single nucleotide incorporation events with PFA was expressed in both IC50 and Ki values (Table 3). Fig. 3 shows an example of the assay used to evaluate the extent of inhibition. We compared gp43 with UL54 and HIV-1 RT as another control. UL54 and HIV-1 RT are sensitive to inhibition with PFA and have IC50 values between 0.6 and 0.9 μm (Table 3). In contrast, gp43 has an IC50 value greater than 300 μm, which renders the enzyme ∼400-fold less susceptible to PFA when compared with UL54. Thus, despite the evidence for structural and functional links between gp43 and UL54, these data point to important differences between the two enzymes.
TABLE 3.
Kinetic constants for inhibition of nucleotide incorporation by PFA
| Enzyme | IC50a | Foldb change | Kic | Fold change |
|---|---|---|---|---|
| μm | μm | |||
| UL54d | 0.87e | Referencef | 0.076 | Reference |
| RT | 0.60 | 1.4Sg | 1.1 | 14 |
| gp43 | 333 | 380R | NAh | NA |
| gp43 | 333 | Reference | NA | NA |
| A | 7.5 | 44S | NA | NA |
| ABC | 3.3 | 100S | NA | NA |
| A | 7.5 | Reference | 0.99 | Reference |
| A/V781I | 19.7 | 2.6R | 1.8 | 1.8R |
| ABC | 3.3 | Reference | 0.55 | Reference |
| ABC/R784A | 37 | 11R | 5.9 | 11R |
| ABC/Q807A | 27 | 8.2R | 6.1 | 11R |
a IC50 is the inhibitory concentration of PFA that reduced the nucleotide incorporation activity of enzyme by 50%. Values were calculated by fitting at least 10 data points to a sigmoidal dose-response (variable slope) equation using GraphPad Prism (version 5.0).
b Fold change is calculated as the ratio of the IC50 or Ki values of the reference and the query enzyme in the subgroup such that the ratio is more than 1.
c Ki is the inhibitor dissociation constant. Values were calculated by globally fitting the 42-data point Ki experiment to the general mixed model of inhibition using GraphPad Prism (version 5.0).
d The HCMV polymerase UL54 was expressed in the in vitro transcription/translation system.
e Standard deviations were determined on the basis of at least three independent experiments and represent a maximum of 25% of the reported value.
f Reference defines the enzyme whose inhibitory constants were used as a reference value in the subgroup. Subgroups of enzymes are separated by an extra row.
g Superscript R and S indicate resistance and sensitivity, respectively, and illustrate fold changes in resistance or sensitivity to PFA with respect to the reference enzyme in the subgroup.
h NA means not available.
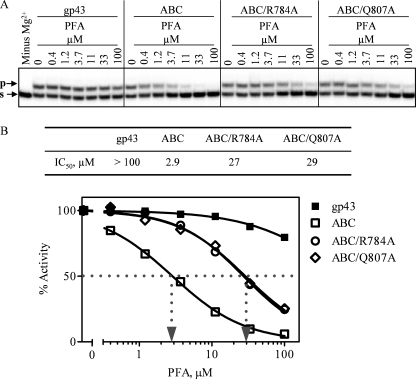
IC50 and Ki measurements show that helix N is a critical structural determinant that mediates PFA sensitivity (Table 3). The chimeric enzyme containing block A (helix N) from UL54 shows ≈50-fold reductions in IC50 values when compared with WT gp43. This value is 3-fold increased when the V781I mutation is introduced against this mutational background. A similar trend is observed for the Ki values. The enzyme with blocks A (helix N) and B/C (helix P) of UL54 showed further subtle increases in sensitivity to PFA. Overall, we measured a 100-fold increase as compared with WT gp43. Moreover, alanine changes at conserved residues Gln-807 and Arg-784 confer 8–11-fold resistance in the context of our biochemical measurements. Together, these findings identify helix N as an important determinant for PFA sensitivity, whereas helix P, in relation to helix N, appears to be a minor contributor. However, the resistance data unambiguously indicated that changes in either one of the two helices can affect PFA susceptibility.
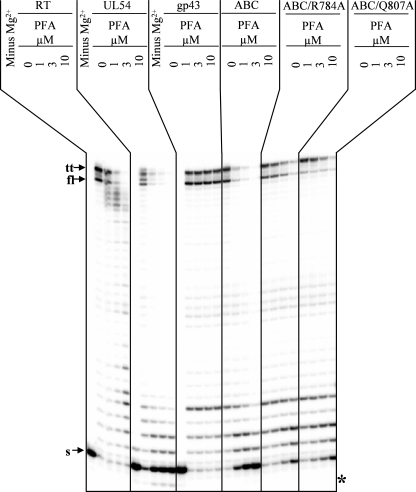
To confirm these phenotypes, we devised a gel-based assay that allowed us to monitor the effects of PFA over multiple template positions (Fig. 4). We focused on the chimeric enzyme with blocks A and B/C of UL54, referred to as gp43-UL54ABC, that showed the lowest IC50 values. In agreement with the aforementioned data, we demonstrated that both HIV-1 RT and UL54 were sensitive to PFA. Concentrations as low as 1 μm had significant levels of inhibition as judged by full-length product formation, although gp43 is not inhibited at concentrations of 10 μm. The chimeric enzyme gp43-UL54ABC shows a similar inhibition profile as observed with HIV-1 RT and UL54. Amino acid substitutions Q807A and R784A, introduced against this mutational background, confer significant levels of resistance to PFA. Each of the aforementioned changes at positions Val-781, Gln-807, and Arg-784 that are located in close proximity to the γ-phosphate of an incoming nucleotide can in turn affect binding of PFA (Fig. 2B). Thus, the data obtained under conditions that allow either single or multiple nucleotide incorporation events are in good agreement and demonstrate that critical elements of helix N and helix P in UL54 can sensitize the RB69 enzyme to PFA.
FIGURE 4.
PFA inhibition of DNA synthesis for multiple nucleotide incorporation events. Reactions were monitored in the presence of constant concentrations of dNTPs and increasing concentrations of PFA. The reactions conditions were chosen such that the maximum of the available primer-template substrate was used in the absence of PFA. PFA-mediated inhibition of DNA synthesis is illustrated by the reduction of the intensity of the signal corresponding to the migration pattern of a full-length product (fl) that is indicated by the corresponding arrow. The migration pattern of the 5′ end radioactively labeled primer (s) and the product of the terminal transferase activity (tt) is illustrated by corresponding arrows. The asterisk points to the exogenous background exonuclease activity present in samples with HCMV polymerase UL54 expressed in vitro transcription/translation system.
Efficiency of Incorporation of ACV-MP
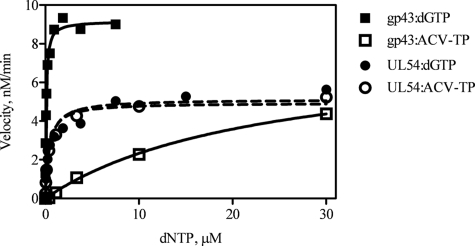
The large difference in PFA sensitivity between WT gp43 and the gp43-UL54ABC mutant enzyme suggests that the critical helical elements of UL54 may also affect the precise positioning of the triphosphate moiety of an incoming nucleotide. Such effects may be exacerbated with acyclic nucleotide analog inhibitors that rely even more on specific contacts through their phosphates. To test this hypothesis, we asked whether gp43, UL54, and gp43-UL54ABC showed differences in efficiency of incorporation of ACV-TP. To assess the ability of viral enzymes to use ACV-TP as a substrate for DNA synthesis, we determined the steady-state parameters for incorporation of ACV-MP and its natural counterpart dGMP (Fig. 5 and Table 4). The ratio of Vmax/Km is indicative of the efficiency of nucleotide incorporation, and the ratio of Vmax/Km(dGTP) and Vmax/Km(ACV-TP) defines the selectivity for the inhibitor. Among the three enzymes, the RB69 enzyme demonstrated the highest efficiency for incorporation of dGMP but the lowest for ACV-MP. The gp43-UL54ABC showed significantly reduced rates of incorporation of dGMP as compared with UL54. The selective advantage of incorporation of the natural nucleotide over the inhibitor was >300. Of significance, the gp43-UL54ABC mutant enzyme does not show such a selective advantage. The mutant enzyme behaved almost exactly like UL54. The selectivity values for UL54 and gp43-UL54ABC were close to 1, suggesting that these enzymes accept ACV-TP and dGTP as substrates for DNA synthesis with similar efficiencies.
FIGURE 5.
Efficiency of nucleotide incorporation for ACV-TP and dGTP. Graphical representation of single nucleotide incorporation events monitored in the presence of increasing concentrations of dGTP or ACV-TP.
TABLE 4.
Kinetic constants for dGTP and ACV-TP incorporation
Values were calculated by fitting the data points to Michaelis-Menten function using GraphPad Prism (version 5.0).
| Enzyme | dGTP |
ACV-TP |
SELc | ||||
|---|---|---|---|---|---|---|---|
| Vmaxa | Kmb | Vmax/Km | Vmax | Km | Vmax/Km | ||
| nm/min | μm | nm/min | μm | ||||
| UL54d | 2.2 ± 0.086e | 0.041 ± 0.0067 | 54 | 1.2 ± 0.13 | 0.039 ± 0.023 | 31 | 1.7 |
| gp43f | 9.2 ± 0.12 | 0.079 ± 0.0053 | 117 | 8.6 ± 0.45 | 24 ± 2.8 | 0.36 | 325 |
| ABC | 5.1 ± 0.23 | 0.47 ± 0.10 | 11 | 5.1 ± 0.14 | 0.41 ± 0.059 | 12.4 | 0.87 |
a Vmax is the maximum velocity of the reaction.
b Km is the substrate concentration at half-maximum velocity of the reaction.
c SEL means selectivity, which is calculated as a ratio of kpol/Kd for dGTP over kpol/Kd for ACV-TP.
d UL54, the HCMV polymerase, was expressed in the in vitro transcription/translation system. The previously reported UL54 value for dGTP Km is 0.11 μm (42).
e Standard deviations were determined on the basis of at least two independent experiments.
f The previously reported gp43 value for dGTP Km is 0.17 μm (51).
DISCUSSION
In light of the problems associated with the development of an expression system for the HCMV DNA polymerase (UL54) (37), we engineered and characterized chimeric enzymes derived from the related RB69 polymerase (gp43) with critical elements of its UL54 counterpart. We replaced regions of helix N and helix P that are located in close proximity to the triphosphate moiety of a bound nucleotide, according to the available crystallographic data obtained with gp43. Our biochemical data point to profound differences in sensitivity and selectivity to antiviral drugs when UL54 was compared with gp43. The phage enzyme appears to be resistant to PFA and ACV, respectively, whereas the HCMV enzyme was sensitive to inhibition with either one of the two drugs. These findings reveal limitations of the ternary gp43 complex as a model system to study mechanisms associated with drug action and drug resistance (45).
In general, mutations that affect susceptibility to antiviral drugs often involve subtle structural changes that do not include conserved amino acids, which makes it difficult to model such effects if only the structures of related enzymes are available. The chimeric enzymes designed in this study may help to tackle this problem.
Replacing the critical regions of helix N of gp43 with the equivalent region of UL54 caused marked increases in sensitivity to PFA (Table 3). The introduction of naturally occurring, resistance-conferring mutations against this mutational background translated this phenotype in biochemical terms. Thus, a limited number of amino acids can be changed to produce a chimeric gp43-UL54 enzyme that shows a similar behavior to an antiviral drug as observed with WT UL54. The substitution of the three amino acids of helix N showed the strongest PFA resensitization effects, although the contribution of changes in helix P are minor. However, helix P contains various amino acids that are involved in HCMV resistance to PFA, CDV, and GCV (7). Changes at these amino acids may directly affect binding of these inhibitors or indirectly through intermolecular interactions between helix N and helix P. Thus, future studies aimed at elucidating mechanisms of resistance to the approved anti-HCMV enzymes may be performed with the gp43-UL54ABC enzyme in which important regions of both helices are replaced.
A structural model of T7 DNA polymerase bound to a primer-template and nucleotide substrate provided a better understanding of the biochemical consequences of specific ACV resistance conferring mutations in the HSV-1 polymerase (UL30) (63); however, the detailed role played by helix N and helix P in drug susceptibility remained elusive. Modeling studies based on the crystal structure of UL30 in its apo form suggest an equivalent role for the two helices gp43 (44), although a comparison of the structures of the apo form of gp43 and the ternary complex revealed significant conformational changes upon nucleotide binding (52). We have superimposed the structures of UL30 (apo form) and of gp43 (ternary complex) and arrive to the same conclusion (Fig. 2B). Moreover, there are also several amino acid changes in the relevant regions of the two helices of UL30 and UL54 that can affect susceptibility to antiviral drugs. The resistance profile of the two enzymes is not identical in this region (64). Thus, the approach to engineer gp43-based chimeric enzymes may likewise provide a valuable tool to study differences in drug susceptibility among the various polymerases that belong to the Herpesviridae.
We have further demonstrated that changes of residues that are located in close proximity to the γ-phosphate of a bound nucleotide, either as part of helix N or as part of helix P, diminish the inhibitory effects of PFA. These findings are consistent with our previous mutational studies on UL54, and point to this region as a possible binding site for PFA (42). Binding of PFA to the pre-translocated complex of HIV-1 RT depends on the presence of Mg2+ ions, which suggests that the inhibitor binds in close proximity to the location that occupies the β- and γ-phosphates of the bound nucleotide in the post-translocated complex (30, 32, 35). Like the nucleotide, the presence of PFA stabilizes the complex, presumably as a result of a conformational change that traps the ligand. The structures of binary and ternary complexes of the RB69 enzyme show a similar conformational change that involves helix N and helix P (49, 50). Thus, the differences between gp43 and UL54 in sensitivity to PFA may also be linked to potential differences in the ability of the two enzymes to trap the inhibitor.
The incorporation of nucleotides and nucleotide analogs follow the sequence of binding, conformational change, and catalysis (65–68). Sensitivity to a certain nucleotide analog can likewise be affected by these parameters (21, 34, 69–72). Our results show that gp43 is literally resistant to ACV, whereas UL54 and gp43-UL54ABC are both, and to the same degree, sensitive to the inhibitor.
Taken together, these findings/results indicate that the chimeric gp43-UL54 enzyme can provide a valuable tool to study the mechanisms of action and resistance to PFA and to nucleotide analog inhibitors. Although various other regions of the enzyme, including its 3′–5′-exonuclease activity can also contribute to differences in drug susceptibility, our findings reveal that helix N and helix P play an important role in this context (7, 40). gp43-UL54ABC and the various resistant mutant enzymes can be expressed and purified at high yields, which facilitates detailed structural and biochemical studies.
Acknowledgments
We thank Suzanne McCormick for excellent technical assistance. The Center for AIDS Research, Department of Pediatrics, Emory University School of Medicine and Veterans Affairs Medical Research, Atlanta, GA, was the recipient of National Institutes of Health Grant 5P30-AI-50409.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R37-AI-041980 and 4R37-AI-025899 (to R. F. S.). This work was also supported by the Canadian Institutes for Health Research (to M. G.) and by the United States Department of Veterans Affairs (to R. F. S.).
- HCMV
- human cytomegalovirus
- UL54
- catalytic subunit of the HCMV DNA polymerase
- CDV
- cidofovir
- GCV
- ganciclovir
- ACV
- acyclovir
- ACV-TP
- acyclovir triphosphate
- PFA
- foscarnet
- HSV
- herpes simplex virus
- RT
- reverse transcriptase
- HIV-1
- human immunodeficiency virus, type 1
- UL30
- catalytic subunit of the HSV DNA polymerase
- gp43
- catalytic subunit of the RB69 DNA polymerase
- WT
- wild type
- PDB
- Protein Data Bank.
REFERENCES
- 1.Deayton J. R., Prof , Sabin C. A., Johnson M. A., Emery V. C., Wilson P., Griffiths P. D. (2004) Lancet 363, 2116–2121 [DOI] [PubMed] [Google Scholar]
- 2.Gandhi M. K., Khanna R. (2004) Lancet Infect. Dis. 4, 725–738 [DOI] [PubMed] [Google Scholar]
- 3.Ljungman P. (2002) J. Infect. Dis. 186, S99–S109 [DOI] [PubMed] [Google Scholar]
- 4.Hassan J., Connell J. (2007) Clin. Exp. Immunol. 149, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boppana S. B., Pass R. F., Britt W. J., Stagno S., Alford C. A. (1992) Pediatr. Infect. Dis. J. 11, 93–99 [DOI] [PubMed] [Google Scholar]
- 6.Griffiths P. D., Walter S. (2005) Curr. Opin. Infect. Dis. 18, 241–245 [DOI] [PubMed] [Google Scholar]
- 7.Gilbert C., Boivin G. (2005) Antimicrob. Agents Chemother. 49, 873–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercorelli B., Sinigalia E., Loregian A., Palù G. (2008) Rev. Med. Virol. 18, 177–210 [DOI] [PubMed] [Google Scholar]
- 9.Biron K. K. (2006) Antiviral Res. 71, 154–163 [DOI] [PubMed] [Google Scholar]
- 10.Andrei G., De Clercq E., Snoeck R. (2008) Curr. Opin. Investig. Drugs 9, 132–145 [PubMed] [Google Scholar]
- 11.De Clercq E. (2003) J. Antimicrob. Chemother. 51, 1079–1083 [DOI] [PubMed] [Google Scholar]
- 12.Coen D. M., Schaffer P. A. (2003) Nat. Rev. Drug Discov. 2, 278–288 [DOI] [PubMed] [Google Scholar]
- 13.De Clercq E., Holý A. (2005) Nat. Rev. Drug Discov. 4, 928–940 [DOI] [PubMed] [Google Scholar]
- 14.Pescovitz M. D., Rabkin J., Merion R. M., Paya C. V., Pirsch J., Freeman R. B., O'Grady J., Robinson C., To Z., Wren K., Banken L., Buhles W., Brown F. (2000) Antimicrob. Agents Chemother. 44, 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Littler E., Stuart A. D., Chee M. S. (1992) Nature 358, 160–162 [DOI] [PubMed] [Google Scholar]
- 16.Sullivan V., Talarico C. L., Stanat S. C., Davis M., Coen D. M., Biron K. K. (1992) Nature 358, 162–164 [DOI] [PubMed] [Google Scholar]
- 17.Biron K. K., Stanat S. C., Sorrell J. B., Fyfe J. A., Keller P. M., Lambe C. U., Nelson D. J. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 2473–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reid R., Mar E. C., Huang E. S., Topal M. D. (1988) J. Biol. Chem. 263, 3898–3904 [PubMed] [Google Scholar]
- 19.Cihlar T., Chen M. S. (1996) Mol. Pharmacol. 50, 1502–1510 [PubMed] [Google Scholar]
- 20.Ho H. T., Woods K. L., Bronson J. J., De Boeck H., Martin J. C., Hitchcock M. J. (1992) Mol. Pharmacol. 41, 197–202 [PubMed] [Google Scholar]
- 21.Deval J. (2009) Drugs 69, 151–166 [DOI] [PubMed] [Google Scholar]
- 22.Xiong X., Smith J. L., Chen M. S. (1997) Antimicrob. Agents Chemother. 41, 594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reardon J. E. (1989) J. Biol. Chem. 264, 19039–19044 [PubMed] [Google Scholar]
- 24.Derse D., Cheng Y. C., Furman P. A., St Clair M. H., Elion G. B. (1981) J. Biol. Chem. 256, 11447–11451 [PubMed] [Google Scholar]
- 25.Furman P. A., St Clair M. H., Spector T. (1984) J. Biol. Chem. 259, 9575–9579 [PubMed] [Google Scholar]
- 26.Talarico C. L., Burnette T. C., Miller W. H., Smith S. L., Davis M. G., Stanat S. C., Ng T. I., He Z., Coen D. M., Roizman B., Biron K. K. (1999) Antimicrob. Agents Chemother. 43, 1941–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann A., Michel D., Paviæ I., Hampl W., Lüske A., Neyts J., De Clercq E., Mertens T. (1997) Antiviral Res. 36, 35–42 [DOI] [PubMed] [Google Scholar]
- 28.Oberg B. (1989) Pharmacol. Ther. 40, 213–285 [DOI] [PubMed] [Google Scholar]
- 29.Wagstaff A. J., Bryson H. M. (1994) Drugs 48, 199–226 [DOI] [PubMed] [Google Scholar]
- 30.Marchand B., Tchesnokov E. P., Götte M. (2007) J. Biol. Chem. 282, 3337–3346 [DOI] [PubMed] [Google Scholar]
- 31.Eriksson B. F., Schinazi R. F. (1989) Antimicrob. Agents Chemother. 33, 663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marchand B., Götte M. (2003) J. Biol. Chem. 278, 35362–35372 [DOI] [PubMed] [Google Scholar]
- 33.Boyer P. L., Sarafianos S. G., Arnold E., Hughes S. H. (2001) J. Virol. 75, 4832–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Götte M. (2004) Expert Rev. Anti Infect. Ther. 2, 707–716 [DOI] [PubMed] [Google Scholar]
- 35.Götte M. (2006) Curr. Pharm. Des. 12, 1867–1877 [DOI] [PubMed] [Google Scholar]
- 36.Ducancelle A., Gravisse J., Alain S., Fillet A. M., Petit F., Pors M. J., Mazeron M. C. (2005) J. Virol. Methods 125, 145–151 [DOI] [PubMed] [Google Scholar]
- 37.Picard-Jean F., Bougie I., Bisaillon M. (2007) Biochem. J. 407, 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loregian A., Rigatti R., Murphy M., Schievano E., Palu G., Marsden H. S. (2003) J. Virol. 77, 8336–8344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loregian A., Appleton B. A., Hogle J. M., Coen D. M. (2004) J. Virol. 78, 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cihlar T., Fuller M. D., Cherrington J. M. (1998) J. Virol. 72, 5927–5936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cihlar T., Fuller M. D., Mulato A. S., Cherrington J. M. (1998) Virology 248, 382–393 [DOI] [PubMed] [Google Scholar]
- 42.Tchesnokov E. P., Gilbert C., Boivin G., Götte M. (2006) J. Virol. 80, 1440–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaudhuri M., Song L., Parris D. S. (2003) J. Biol. Chem. 278, 8996–9004 [DOI] [PubMed] [Google Scholar]
- 44.Liu S., Knafels J. D., Chang J. S., Waszak G. A., Baldwin E. T., Deibel M. R., Jr., Thomsen D. R., Homa F. L., Wells P. A., Tory M. C., Poorman R. A., Gao H., Qiu X., Seddon A. P. (2006) J. Biol. Chem. 281, 18193–18200 [DOI] [PubMed] [Google Scholar]
- 45.Shi R., Azzi A., Gilbert C., Boivin G., Lin S. X. (2006) Proteins 64, 301–307 [DOI] [PubMed] [Google Scholar]
- 46.Braithwaite D. K., Ito J. (1993) Nucleic Acids Res. 21, 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogg M., Wallace S. S., Doublié S. (2004) EMBO J. 23, 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Sattar A. K., Wang C. C., Karam J. D., Konigsberg W. H., Steitz T. A. (1997) Cell 89, 1087–1099 [DOI] [PubMed] [Google Scholar]
- 49.Franklin M. C., Wang J., Steitz T. A. (2001) Cell 105, 657–667 [DOI] [PubMed] [Google Scholar]
- 50.Shamoo Y., Steitz T. A. (1999) Cell 99, 155–166 [DOI] [PubMed] [Google Scholar]
- 51.Yang G., Lin T., Karam J., Konigsberg W. H. (1999) Biochemistry 38, 8094–8101 [DOI] [PubMed] [Google Scholar]
- 52.Yang G., Franklin M., Li J., Lin T. C., Konigsberg W. (2002) Biochemistry 41, 2526–2534 [DOI] [PubMed] [Google Scholar]
- 53.Baldanti F., Lurain N., Gerna G. (2004) Hum. Immunol. 65, 403–409 [DOI] [PubMed] [Google Scholar]
- 54.Le Grice S. F., Cameron C. E., Benkovic S. J. (1995) Methods Enzymol. 262, 130–144 [DOI] [PubMed] [Google Scholar]
- 55.Burgess K., Cook D. (2000) Chem. Rev. 100, 2047–2060 [DOI] [PubMed] [Google Scholar]
- 56.Copeland R. A. (2005) Evaluation of Enzyme Inhibitors in Drug Discovery: A Primer for Medicinal Chemists and Pharmacologists, pp. 34–37, 48–77, John Wiley & Sons Inc., Hoboken, NJ: [PubMed] [Google Scholar]
- 57.Copeland R. A. (2000) Enzymes, 2nd Ed., pp. 109–145, 266–304, John Wiley & Sons Inc., New York [Google Scholar]
- 58.Lambert C., Léonard N., De Bolle X., Depiereux E. (2002) Bioinformatics 18, 1250–1256 [DOI] [PubMed] [Google Scholar]
- 59.Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 60.Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 62.Baldanti F., Sarasini A., Silini E., Barbi M., Lazzarin A., Biron K. K., Gerna G. (1995) Scand. J. Infect. Dis. Suppl. 99, 103–104 [PubMed] [Google Scholar]
- 63.Huang L., Ishii K. K., Zuccola H., Gehring A. M., Hwang C. B., Hogle J., Coen D. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gilbert C., Bestman-Smith J., Boivin G. (2002) Drug Resist. Updat. 5, 88–114 [DOI] [PubMed] [Google Scholar]
- 65.Kati W. M., Johnson K. A., Jerva L. F., Anderson K. S. (1992) J. Biol. Chem. 267, 25988–25997 [PubMed] [Google Scholar]
- 66.Kuchta R. D., Mizrahi V., Benkovic P. A., Johnson K. A., Benkovic S. J. (1987) Biochemistry 26, 8410–8417 [DOI] [PubMed] [Google Scholar]
- 67.Patel S. S., Wong I., Johnson K. A. (1991) Biochemistry 30, 511–525 [DOI] [PubMed] [Google Scholar]
- 68.Spence R. A., Kati W. M., Anderson K. S., Johnson K. A. (1995) Science 267, 988–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Deval J., Alvarez K., Selmi B., Bermond M., Boretto J., Guerreiro C., Mulard L., Canard B. (2005) J. Biol. Chem. 280, 3838–3846 [DOI] [PubMed] [Google Scholar]
- 70.Sarafianos S. G., Marchand B., Das K., Himmel D. M., Parniak M. A., Hughes S. H., Arnold E. (2009) J. Mol. Biol. 385, 693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Diallo K., Götte M., Wainberg M. A. (2003) Antimicrob. Agents Chemother. 47, 3377–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Götte M., Wainberg M. A. (2000) Drug Resist. Updat. 3, 30–38 [DOI] [PubMed] [Google Scholar]