Abstract
Cyclic AMP is a fundamentally important second messenger for numerous peptide hormones and neurotransmitters that control gene expression, cell proliferation, and metabolic homeostasis. Here we show that cAMP works with the POU homeodomain protein Oct-1 to regulate gene expression in pancreatic and intestinal endocrine cells. This ubiquitously expressed transcription factor is known as a stress sensor. We found that it also functions as a repressor of Cdx-2, a proglucagon gene activator. Through a mechanism that involves the activation of exchange protein activated by cyclic AMP, elevation of cAMP leads to enhanced phosphorylation and nuclear exclusion of Oct-1 and reduced interactions between Oct-1 or nuclear co-repressors and the Cdx-2 gene promoter, detected by chromatin immunoprecipitation. In rat primary pancreatic islet cells, cAMP elevation also reduces nuclear Oct-1 content, which causes increased proglucagon and proinsulin mRNA expression. Our study therefore identifies a novel mechanism by which cAMP regulates hormone-gene expression and suggests that ubiquitously expressed Oct-1 may play a role in metabolic homeostasis by functioning as a sensor for cAMP.
Many peptide hormones and neurotransmitters use the second messengers, such as cyclic AMP (cAMP), to exert their biological functions, including regulation of gene expression and metabolic homeostasis (1–7). Extensive studies have shown that in addition to the activation of protein kinase A (PKA),5 cAMP is able to trigger intracellular signaling events via other mechanisms, including Epac (the activation of exchangeprotein directly activated by cAMP) (8–13). As non-kinase effectors of cAMP, Epac molecules are evidently involved in regulating gene expression, cell adhesion, and pancreatic peptide hormone secretion (4, 10, 12, 14).
In pancreatic islet and intestinal endocrine L cells, cAMP elevation is associated with increased expression of proglucagon (gcg) or proinsulin genes (15, 16). Expression of these two hormone-encoding genes is also controlled by transcriptional activators, including certain homeodomain proteins such as the caudal homeodomain protein Cdx-2 (17–22). We have demonstrated previously that in both PKA-active and PKA-deficient pancreatic and intestinal proglucagon-producing endocrine cell lines, cAMP elevation leads to increased Cdx-2 expression (23). Furthermore, Cdx-2 expression in a PKA-deficient pancreatic islet InR1-G9 cell line can be activated by an Epac pathway-specific cAMP analogue 8-pMeOPT-2′-O-Me-cAMP (23). More recently, we have observed expression of Epac2 in pancreatic and intestinal proglucagon-producing cells and demonstrated that Epac signaling serves as the mediator of cAMP in regulating the expression of gcg (14).
In this study, we further explored mechanistically how cAMP-Epac signaling activates Cdx-2 expression in pancreatic and intestinal endocrine cells. Our observations suggest the existence of a novel mechanism by which cAMP regulates pancreatic and intestinal hormone-gene expression. It is likely that this regulation involves nuclear-cytoplasmic shuttling of the POU homeodomain protein Oct-1.
Oct-1 is a ubiquitously expressed transcriptional regulator with a POU-type DNA binding domain (24, 25). It exerts multiple biological functions via up- or down-regulating the expression of many target genes in different cell lineages, including endocrine and neuroendocrine cells (13, 25–33). In this study, we found that in proglucagon-expressing endocrine cells, activation of Epac signaling in response to cAMP elevation reduced nuclear Oct-1 content, an event associated with enhanced Cdx-2 and gcg expression. In rat primary pancreatic islet cells, reduction in nuclear levels of Oct-1 in response to cAMP elevation was shown to be associated with enhanced gcg and proinsulin I mRNA expression.
EXPERIMENTAL PROCEDURES
Reagents, Plasmids, Cell Cultures, and DNA/siRNA Transfection
Forskolin and 3-isobutyl-1-methylxanthine (IBMX) were purchased from Sigma. The MEK inhibitor PD98059 was purchased from Calbiochem. The PKA inhibitor H89 was the product of Calbiochem, and the Epac pathway-specific cAMP analogue 8-pMeOPT-2′-O-Me-cAMP was provided by BIOLOG Life Sciences Institute (Bremen, Germany). The plasmid construct pcDNA-Oct-1-Myc (Oct-1-Myc) was generated by inserting a copy of the human Oct-1 coding sequence (34) into pcDNA3.1/Myc/HisA (Invitrogen). Hamster pancreatic α cell line InR1-G9, mouse large intestinal L cell line GLUTag, mouse small intestinal STC-1, as well as human colon cancer Caco-2 cell lines were maintained as described previously (18, 35–37). We have demonstrated previously that InR1-G9 cell line is deficient in the activity of PKA (23).
Real Time RT-PCR
cDNAs were generated using a Superscript First-strand RT-PCR kit (Invitrogen). Real time RT-PCR was conducted using the QuantiTect SYBR green PCR kit from Qiagen (Mississauga, Ontario, Canada). DNA sequences of the primers utilized for quantitatively assessing mRNA expression by real time RT-PCR are as follows: hamster Cdx-2, forward, 5′-CCTAGACAAGGACGTGAGCA-3′, and reverse, 5′-CCTAGACAAGGACGTGAGCA-3′; hamster and rat gcg, forward, 5′-AGAAGAAGTCGCCATTGCTG-3′, and reverse, 5′-CGCAGAGATGTTGTCAAGA-3′; and rat proinsulin I, forward, 5-GGACCCACAAGTGGAACAAC-3′, and reverse, 5′-GGTGGGCCTTAGTTCCAGTA-3′.
Nuclear and Cytosolic Protein Extraction
Nuclear and cytosolic proteins were extracted based on the method by Schreiber et al. (38). Briefly, ∼1 × 106 cells collected were washed with phosphate-buffered saline and pelleted by centrifugation (1500 × g for 5 min). The pellet was then resuspended in 500 μl of cold buffer A (10 mm HEPES (pH 8.0), 10 mm KCl, 0.1 mm EDTA, 0.1 mm EGTA, 1 mm dithiothreitol, and 0.5 mm phenylmethylsulfonyl fluoride) and incubated on ice for 15 min. After addition of 25 μl of 10% Nonidet P-40, the cells were vigorously votexed for 10 s. Following centrifugation for 30 s, the supernatant was collected and treated as the cytoplasmic fraction. The nuclear pellet was then washed three times with buffer A and resuspended in 60 μl of ice-cold buffer C (20 mm HEPES (pH 8.0), 0.4 m NaCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, and 1 mm phenylmethylsulfonyl fluoride), and the tube was vigorously rocked at 4 °C for 15 min. Nuclear proteins were then collected by a 5-min centrifugation at 4 °C.
Western Blotting, Immunoprecipitation, and in Vivo Metabolic Labeling
The polyclonal Cdx-2 antibody was generated as described previously (39). Antibodies against actin, ERK (sc-94), phosphorylated ERK (sc-7383), cAMP-response element-binding protein (CREB, sc-240), phosphorylated CREB (sc-101662), proliferating cell nuclear antigen, and horseradish peroxidase-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). The Oct-1 antibody sc-8024 was also a product of Santa Cruz Biotechnology, and three other Oct-1 antibodies were described in supplemental Fig. 1. The anti-phosphoserine antibody (ab9332) was purchased from Abcam Inc. (Cambridge, MA), and the anti-phosphothreonine antibody (9381) was from Cell Signaling Technology, Inc. (Danvers, MA). Preparation of whole cell lysates, Western blotting, and immunoprecipitation were carried out as described previously (23, 36). In vivo metabolic labeling was performed to examine the effect of cAMP-Epac activation on Oct-1 phosphorylation. Briefly, InR1-G9 cells were incubated with 8-orthophosphate (H332PO4, 1 mCi per 10-cm plate) in the presence or absence of the indicated reagents for 2 h. Cell lysates were collected for immunoprecipitation against Oct-1. The amount of 32P-labeled Oct-1 in the immunoprecipitation eluate was then assessed by SDS-PAGE, followed by x-ray film exposure (40).
Immunofluorescence Microscopy
InR1-G9 cells were grown in coverslips and transfected with pcDNA-Oct-1-Myc using Lipofectamine 2000 according to the manufacturer's instructions. The transfected cells were serum-starved for 12 h and treated with forskolin/3-isobutyl-1-methylxanthine (IBMX, 10 μm each), or 8-bromo-cAMP (100 μm), or the Epac pathway-specific cAMP analogue 8pMeOPT-2′-O-ME-cAMP (20 μm) for 1 h. After fixation of the cells with 3.7% paraformaldehyde for 20 min, nonspecific binding sites were blocked with 5% bovine serum albumin in phosphate-buffered saline. The proteins were stained using primary monoclonal antibody against the Myc tag (Cell Signaling Technology, clone 9B11, 1:500 dilution) and secondary Cy3-conjugated anti-mouse IgG; the nuclei were stained by DAPI and visualized by a Nikon fluorescence microscope (Eclipse TE 200).
Chromatin Immunoprecipitation (ChIP)
The method for ChIP has been described previously (37, 41, 42). Briefly, formaldehyde was added to the InR1-G9 cells (with or without a given treatment) at the final concentration of 1% to cross-link chromatin and nuclear proteins. After a sonication procedure, a designated antibody was added to precipitate the sheared chromatin. Following a reverse cross-link procedure, one-tenth of the final precipitated DNA (2 μl) was used in each PCR. The primers used in the ChIP assays are as follows: forward, 5′- GGAAGGAGGAAACTCTTTAAC-3′, and reverse, 5′-CCTTACGTGATTAACGAGTG-3′. This pair of primers amplifies a 214-bp DNA fragment that contains hamster Cdx-2pOCT (Fig. 1B). The control primers are as follows: forward, 5′-GGCTCACCGGCCGCCGCTATG-3′, and reverse, 5′-CGGCTGCGCGGGCTTCCGCAT-3′. This pair of primers amplifies a 231-bp fragment representing exon I of hamster Cdx-2, located >1.2 kb downstream of the OCT-binding site.
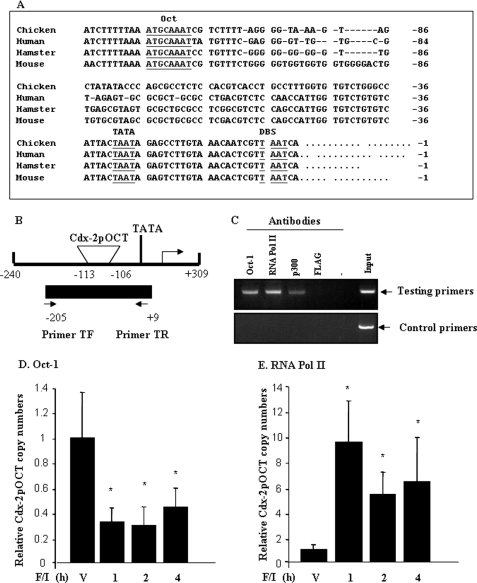
FIGURE 1.
Oct-1 binds to Cdx-2 promoter in vivo. A, comparison of the nucleotide sequences of the mouse Cdx-2 gene proximal promoter region with that of its homologues in hamsters (Cdx-3), humans (CDX2), and chickens (Cdx-C). TATA, TATA box; DBS, downstream binding site (36). B, schematic representation of the hamster Cdx-2 gene 5′-flanking region (GenBankTM accession number EF530070), positions of Cdx-2pOCT, and two primers utilized in the ChIP assay. C, Oct-1, RNA polymerase II (RNA pol II), and p300 antibody-precipitated Cdx-2 promoter containing genomic DNA (but not the control DNA) in InR1-G9 cells detected by ChIP. D, forskolin/IBMX treatment (F/I, 10 μm each) reduced in in vivo binding of Oct-1 to hamster Cdx-2 promoter, detected by quantitative ChIP. V, vehicle for forskolin/IBMX. E, forskolin/IBMX treatment (F/I, 10 μm each) enhanced binding of RNA pol II to Cdx-2 promoter, detected by quantitative ChIP.
RESULTS
Oct-1 Serves as a Transcriptional Repressor of Cdx-2
A typical OCT consensus sequence has been located within the proximal Cdx-2 gene promoter region in the rodent species and in humans (17, 43, 44). This OCT-binding site, designated as Cdx-2pOCT (35), and its flanking sequences are also well conserved in the Cdx-2 homologue, Cdx-C, in chickens (45) (Fig. 1A). We have demonstrated previously by electrophoretic mobility shift assay that Oct-1 binds to this site (35). To investigate whether the binding occurs in vivo (in intact cells) and whether the binding plays a role in regulating Cdx-2 and therefore gcg expression in the PKA-deficient hamster pancreatic α cell line InR1-G9 (23), we performed ChIP analyses.
For this purpose, we determined a piece of 5′-flanking sequence of the hamster Cdx-2 gene (GenBankTM accession number EF530070) (Fig. 1B) and conducted ChIP for the InR1-G9 cells treated with and without the cAMP promoting agents, forskolin and IBMX. The supplemental Fig. 1A shows that two of the Oct-1 antibodies were able to precipitate the genomic DNA fragment that contains the Cdx-2 gene promoter in our ChIP assay. We then chose one of them (sc-8024) for our remaining experiments. Fig. 1C shows that we were able to detect interactions between the Cdx-2 promoter and Oct-1 and between the Cdx-2 promoter and RNA polymerase II (RNA pol II). The interaction between the Cdx-2 promoter and p300 was also detected, although the sensitivity appeared relatively low. The control anti-FLAG antibody, however, did not precipitate the Cdx-2 promoter containing genomic DNA, and the control primers did not generate the PCR product from Oct-1, RNA pol II, or p300 antibody precipitated genomic DNA (Fig. 1C).
To determine the effect of cAMP elevation on the interactions, InR1-G9 cells were then treated with forskolin/IBMX for 1, 2, and 4 h, followed by ChIP analyses. We observed repeatedly a reduction in the Cdx-2 promoter-derived PCR product in cells treated with forskolin/IBMX, indicating that cAMP elevation attenuated Oct-1 binding (supplemental Fig. 1B). Furthermore, to quantify the effect of forskolin/IBMX treatment on the association of Oct-1 and RNA pol II with the Cdx-2 promoter, chromatin DNA obtained by ChIP was assessed by real time PCR (quantitative ChIP). As shown in Fig. 1, D and E, forskolin/IBMX-dependent reduction in Oct-1 binding varied from 48 to 67% over a 4-h test period, whereas the reciprocal increase in RNA pol II binding ranged from 5- to 8-fold. These results indicate that Oct-1 indeed binds to the octamer-containing Cdx-2 promoter in live proglucagon-producing InR1-G9 cells. Notably, the binding can be attenuated by cAMP elevation, with a concomitant enhancement of RNA pol II association.
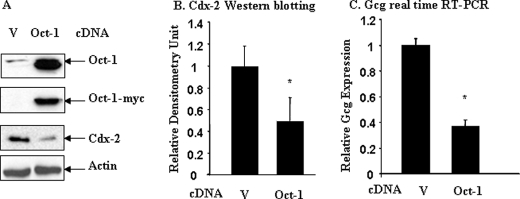
It is well known that cAMP elevation leads to increased Cdx-2 and gcg expression (23, 46, 47). The repressive effect of cAMP elevation on in vivo binding of Oct-1 to the Cdx-2 gene promoter therefore would suggest that Oct-1 may function as a negative regulator of Cdx-2 expression. To examine this hypothesis, we assessed the effect of Oct-1 overexpression on endogenous Cdx-2 protein and gcg mRNA expression in InR1-G9 cells transfected with pcDNA3.1-(Oct-1-Myc). The transfection efficiency was determined to be higher than 60%, based on the expression of the green fluorescent proteins in the InR1-G9 cells transfected with CMV-EGFP using the same transfection strategy (data not shown). A representative immunoblot in Fig. 2A shows the levels of total and exogenous Oct-1 and endogenous Cdx-2 in cells transfected with the empty pcDNA3.1 vector versus cells transfected with Oct-1-Myc. Endogenous Cdx-2 expression was substantially reduced in response to Oct-1-Myc overexpression (Fig. 2A). A quantitative densitometric analysis of four independent experiments indicates that Oct-1 overexpression resulted in a 52% inhibition of endogenous Cdx-2 protein expression (Fig. 2B). Consistent with the reduction of Cdx-2 expression, real time RT-PCR results showed that gcg mRNA expression in Oct-1 overexpressing InR1-G9 cells was reduced by 62% (Fig. 2C). It is notable that in our Western blotting (Fig. 2A) transfected Oct-1-Myc migrated faster than endogenous Oct-1. This is due to the fact that the Oct-1 expression plasmid utilized in this study lacking the first 34 amino acids (34). Our observations therefore suggest that this N-terminal region is not essential for its repressive effect on Cdx-2 expression.
FIGURE 2.
Oct-1 overexpression inhibits Cdx-2 and gcg expression. A, InR1-G9 cells were transfected with the pCDNA3.1 (V) or pcDNA-Oct-1-Myc (Oct-1) for 24 h. Whole cell lysates were immunoblotted with the indicated antibodies. B, results of densitometric analysis of four independent experiments described in A. C, InR1-G9 cells were transfected with pCDNA3.1 or pcDNA-Oct-1-Myc. The effect of Oct-1 overexpression on gcg expression was assessed by real time RT-PCR. Values are mean ± S.E. (n = 3). *, p < 0.05.
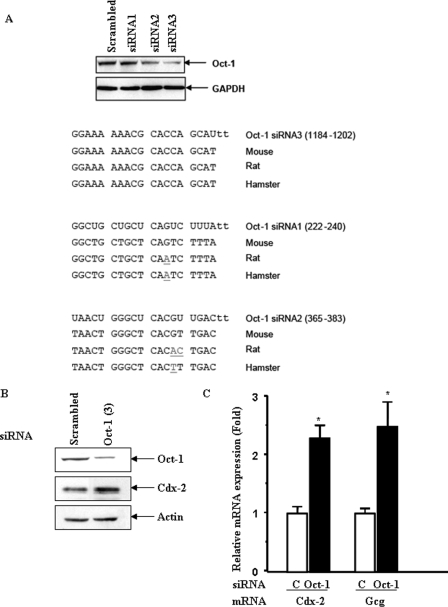
To further verify the trans-repressive effect of Oct-1 on Cdx-2 expression, we employed siRNA to “knock down” Oct-1 expression in the hamster pancreatic α cell line InR1-G9. Among the three siRNAs we have tested, one of them, Oct-1 siRNA3, shows 100% sequence homology with the hamster Oct-1 cDNA, whereas the other two (Oct-1 siRNA1 and Oct-1 siRNA2) carry 1 or 2 bp mismatches (Fig. 3A). Oct-1 siRNA3 was shown to significantly repress Oct-1 expression in the InR1-G9 cell line (Fig. 3A), and this specific siRNA was then demonstrated to enhance the expression of endogenous Cdx-2 protein expression, as detected by Western blotting (Fig. 3B). Quantitative real time RT-PCR showed that this Oct-1-specific siRNA increased endogenous Cdx-2 and gcg mRNA expression by 2.2- and 2.6-fold, respectively (Fig. 3C). These data therefore further suggest that Oct-1 functions as a repressor of Cdx-2 in the proglucagon-expressing InR1-G9 cell line.
FIGURE 3.
Oct-1 knockdown stimulates Cdx-2 and gcg expression. A, Oct-1 siRNA3 significantly repressed Oct-1 expression. InR1-G9 cells were transfected with 50 nm of control (scrambled) or an indicated Oct-1 siRNA for 36 h. Cells were harvested, and whole cell lysates were assessed for Oct-1 expression by Western blotting. B, reduced Oct-1 expression in InR1-G9 cells was accompanied by increased Cdx-2 protein expression, detected by Western blotting. C, Oct-1 knockdown led to increased Cdx-2 and gcg mRNA expression. InR1-G9 cells were transfected with 50 nm control or Oct-1 siRNA3. Total RNA was isolated and assessed for Cdx-2 and gcg mRNA expression by real time RT-PCR. Values are mean ± S.E. (n > or = 3). *, p < 0.05.
Oct-1 Recruits Nuclear Co-repressors to Cdx-2 Promoter
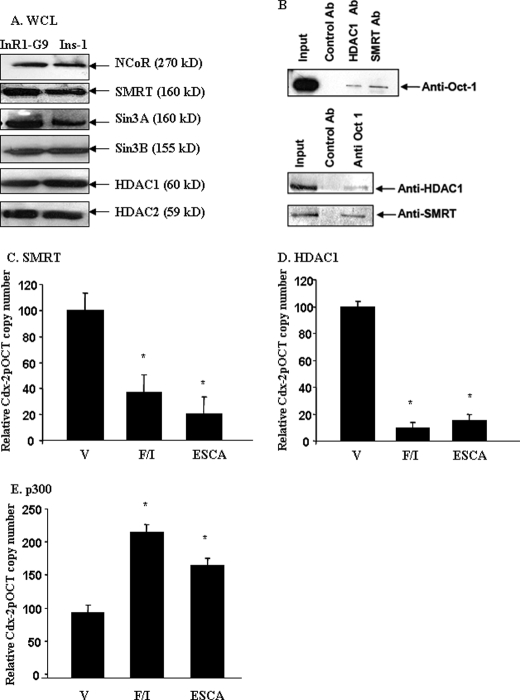
To investigate mechanisms underlying the repressive effect of Oct-1 on Cdx-2 expression, we asked whether Oct-1 recruits nuclear co-repressors to the Cdx-2 gene promoter. We have first assessed the interactions between Oct-1 and the nuclear co-repressors by co-immunoprecipitation. The detection of the expression of a panel of nuclear co-repressors in InR1-G9 cells and the pancreatic β cell line Ins-1 by Western blotting is shown in Fig. 4A. Fig. 4B shows the detection of, by co-immunoprecipitation, the interactions between Oct-1 and silencing mediator of retinoid and thyroid hormone receptors (SMRT), and between Oct-1 and histone deacetylase 1 (HDAC1). However, we could not detect interactions between Oct-1 and HDAC2, NCoR, Sin3A, or Sin3B (data not shown). Our ChIP assays showed that anti-SMRT and anti-HDAC1 antibodies precipitated Cdx-2pOCT containing chromatin DNA in InR1-G9 cells, indicating that interactions between Oct-1 and these two nuclear co-repressors could occur in intact live cells (data not shown). Furthermore, in the InR1-G9 cells, cAMP-Epac activation profoundly inhibited these interactions. Forskolin/IBMX and 8-pMeOPT-2′-O-Me-cAMP inhibited the interaction between Cdx-2pOCT and SMRT by 63 and 78%, respectively (Fig. 4C), and inhibited the interaction between Cdx-2-pOCT and HDAC1 by 91 and 86%, respectively (Fig. 4D). In contrast, cAMP-Epac activation increased the association between the nuclear co-activator p300 and the Cdx-2 promoter (Fig. 4E). These observations, along with the results presented in Figs. 1–3, suggest that Oct-1 may inhibit Cdx-2 expression by recruiting certain nuclear co-repressors to the Cdx-2 gene promoter.
FIGURE 4.
Oct-1 recruits nuclear co-repressors to the Cdx-2 promoter. A, detection of a panel of nuclear co-repressors in pancreatic InR1-G9 and Ins-1 cells by Western blotting. NCoR, nuclear co-repressor; Sin3, histone deacetylase complex subunit 3. Nuclear extracts were utilized for detecting NCoR, whereas whole cell lysates (WCL) were utilized for detecting other transcriptional regulators. Approximately 10 μg of nuclear proteins from nuclear extracts or 50 μg of proteins from whole cell lysates were loaded for each lane. B, detection of the interactions between Oct-1 and SMRT and between Oct-1 and HDAC1 by co-immunoprecipitation. Ab, antibody. C and D, quantitative ChIP shows that forskolin/IBMX (F/I) (10 μm each) or 8-pMeOPT-2′-O-Me-cAMP (ESCA, Epac-specific cAMP analogue, 20 μm) for 2 h inhibited the interactions between SMRT or HDAC and Cdx-2 promoter. V, vehicle. E, quantitative ChIP shows that forskolin/IBMX or 8-pMeOPT-2′-O-Me-cAMP (ESCA) enhanced the interaction between the nuclear co-activator p300 and Cdx-2 promoter. For C–E, values are mean ± S.E. (n = 3). *, p < 0.05.
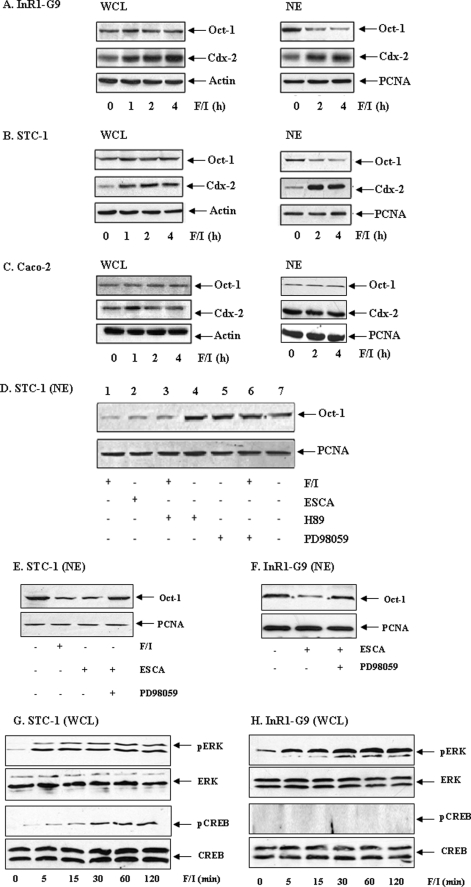
Forskolin/IBMX Treatment Reduces Nuclear Oct-1 Content in Pancreatic and Intestinal Endocrine Cells via Epac-ERK Activation
Elevation of cAMP may affect the binding of Oct-1 to Cdx-2 promoter by reducing its binding affinity or the amount of Oct-1 in nuclei. To investigate the underlying mechanisms, we then examined the effect of forskolin/IBMX treatment on Oct-1 and Cdx-2 expression in the pancreatic islet (InR1-G9), intestinal endocrine (STC-1), and control colon cancer (Caco-2) cell lines by Western blotting. Consistent with our previous observations (23), forskolin/IBMX significantly increased Cdx-2 levels in whole cell lysates of InR1-G9 (Fig. 5A) and STC-1 (Fig. 5B) cells but not of the control Caco-2 cells (Fig. 5C). Fig. 5, A and B, also shows substantial increase of nuclear Cdx-2 contents in response to forskolin/IBMX treatment in the two endocrine cell lines. Interestingly, we found that the amount of nuclear Oct-1 in both InR1-G9 and STC-1 cells was considerably reduced by forskolin/IBMX treatment (Fig. 5, A and B, right panels), although Oct-1 contents in whole cell lysates were not notably affected. In the non-endocrine cell line Caco-2, no substantial effect on Oct-1 contents in either the whole cell lysate or nuclear fraction was observed after forskolin/IBMX treatment. These observations suggest that cAMP elevation stimulates Oct-1 nuclear exclusion in a cell-specific manner.
FIGURE 5.
cAMP-Epac stimulates Oct-1 nuclear exclusion in pancreatic and intestinal endocrine cells. A–C, forskolin/IBMX (F/I, 10 μm each) reduced the nuclear content of Oct-1 in InR1-G9 (A) and STC-1 (B), but not in Caco-2 (C). WCL, whole cell lysate; NE, nuclear extract. Reduced nuclear Oct-1 content was associated with increased Cdx-2 expression (A and B). D–F, PKA-active STC-1 cells (D and E) or PKA-deficient InR1-G9 cells (F) were treated with the indicated chemicals for 2 h. 8-pMeOPT-2′-O-Me-cAMP, defined as ESCA (20 μm); H89 is a PKA inhibitor (10 μm); and PD98059 is an MEK inhibitor (50 μm). Nuclear extracts were assessed by Western blotting. G and H, STC-1 (G) and InR1-G9 (H) cell lines were treated with forskolin/IBMX (10 μm each) for the indicated time before harvesting. Whole cell lysates were utilized in assessing total and phosphorylated ERK and CREB.
InR1-G9 is a PKA-deficient cell line (23), and our previous study has suggested that forskolin/IBMX treatment stimulates Cdx-2 expression in this cell line via MEK-ERK stimulation in response to Epac activation (23). To investigate whether the Epac pathway and ERK activation are involved in cAMP-mediated Oct-1 nuclear exclusion, we examined nuclear Oct-1 contents in PKA-active STC-1 cells that were treated with 8-pMeOPT-2′-O-Me-cAMP or forskolin/IBMX, with or without a PKA or MEK inhibitor. As shown Fig. 5D, forskolin/IBMX or 8-pMeOPT-2′-O-Me-cAMP reduced nuclear Oct-1 content (lanes 1 and 2), whereas the effect of forskolin/IBMX was not blocked by the PKA inhibitor H89 (lane 3). Furthermore, H89 and the MEK inhibitor PD98059 did not affect nuclear Oct-1 content on their own (Fig. 5D, lanes 4 and 5), whereas MEK inhibition substantially attenuated the effect of forskolin/IBMX treatment (lane 6). These observations suggest that the Epac pathway is involved in cAMP elevation-mediated Oct-1 nuclear exclusion. To further verify that ERK mediates the effect of Epac, we conducted additional examinations in both PKA-deficient InR1-G9 and PKA-active STC-1 cell lines. As shown in Fig. 5, E and F, the effect of 8-pMeOPT-2′-O-Me-cAMP on reducing nuclear Oct-1 expression was attenuated in these two cell lines by MEK inhibition. Finally, we compared the effect of forskolin/IBMX treatment on the activation of CREB, an important downstream effector of PKA in regulating gene expression. In both the PKA active STC-1 (Fig. 5G) and the PKA-deficient InR1-G9 (Fig. 5H) cell lines, forskolin/IBMX treatment stimulated ERK phosphorylation. More importantly, the treatment stimulated CREB phosphorylation in STC-1 cells (Fig. 5G) but not in InR1-G9 cells (Fig. 5H). These observations further confirm the PKA-deficient nature of InR1-G9 cells and suggest that the stimulation of Cdx-2 expression and the attenuation of the repressive effect of Oct-1 by cAMP elevation do not require activities of PKA and its downstream effector CREB.
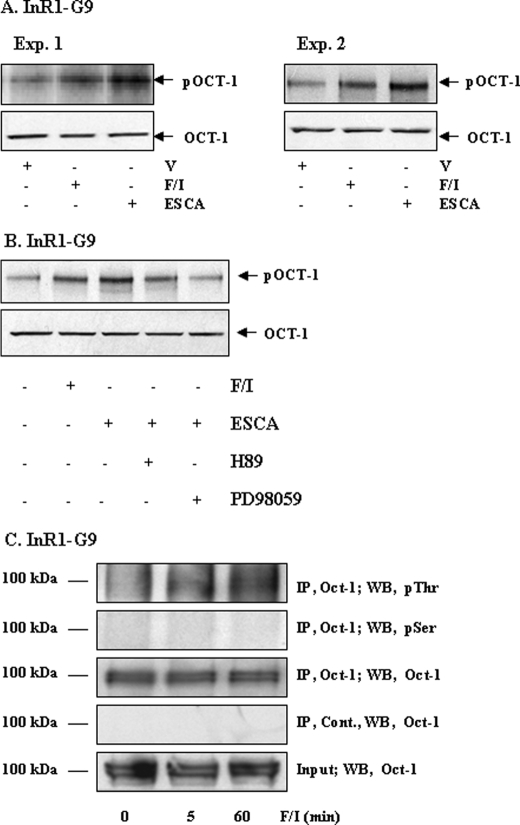
Although a few previous studies have suggested that Oct-1 may be phosphorylated in different cell signaling pathways (40, 48–52), there is no antibody that recognizes phosphorylated Oct-1. To examine whether reduction of nuclear Oct-1 content is mediated by a phosphorylation or dephosphorylation event, we conducted in vivo metabolic labeling in the PKA-deficient InR1-G9 cell line treated with either forskolin/IBMX or 8-pMeOPT-2′-O-Me-cAMP. As shown in Fig. 6A, a 2-h treatment with either forskolin/IBMX or 8-pMeOPT-2′-O-Me-cAMP enhanced the incorporation of orthophosphate into the Oct-1 molecule, suggesting the role of cAMP-Epac signaling in stimulating Oct-1 phosphorylation. Fig. 6B shows that the stimulatory effect of 8-pMeOPT-2′-O-Me-cAMP can be blocked by MEK inhibition but not by PKA inhibition. Furthermore, we examined whether forskolin/IBMX treatment affects Oct-1 phosphorylation at serine or threonine sites. We immunoprecipitated Oct-1 and probed the immunoprecipitated proteins by Western blotting using anti-phosphoserine and anti-phosphothreonine antibodies. Our results suggest that forskolin/IBMX treatment may increase Oct-1 phosphorylation at threonine but not serine residues (Fig. 6C).
FIGURE 6.
Examination of Oct-1 phosphorylation in response to forskolin/IBMX treatment and Epac activation. A, in vivo metabolic labeling shows that a 2-h treatment with forskolin/IBMX (F/I) (10 μm each) or 8-pMeOPT-2′-O-Me-cAMP (ESCA, 20 μm) enhanced the incorporation of orthophosphate into the Oct-1 molecules in InR1-G9 cells (representative blots, n = 3). V, vehicle. B, in vivo metabolic labeling shows that the MEK inhibitor (PD98059) but not the PKA inhibitor (H89) attenuated the stimulatory effect of 8-pMeOPT-2′-O-Me-cAMP on the incorporation of orthophosphate into the Oct-1 molecules in InR1-G9 cells. C, Oct-1 in forskolin/IBMX-treated and untreated InR1-G9 cells was precipitated with the Oct-1 antibody (sc-8024) and detected by Western blotting (WB) using the anti-phosphoserine or anti-phosphothreonine antibodies. Cont., normal rabbit control serum; IP, immunoprecipitated.
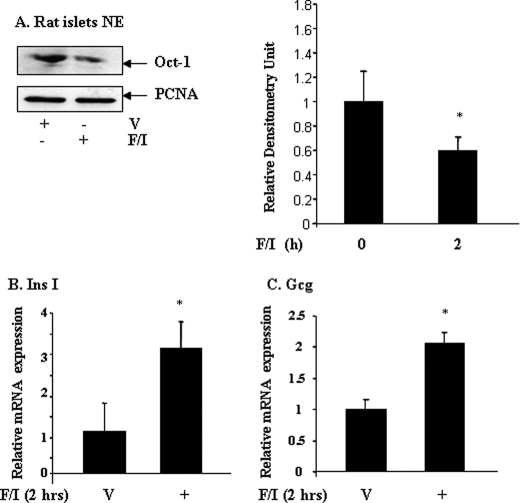
Forskolin/IBMX Reduces Nuclear Oct-1 Content, Associated with Increased gcg and Proinsulin I mRNA Expression in Primary Rat Pancreatic Islet Cells
The above observations demonstrated the existence of a cAMP-Epac-Oct-1 signaling cascade as a putative novel pathway by which cAMP regulates pancreatic and intestinal hormone-gene expression. We next tested whether this signaling cascade functions in primary pancreatic islet cells as well. For this purpose, we have conducted our examinations in primary rat pancreatic islet cells. Approximately 300 pancreatic islets were isolated from each of three adult Wistar rats. The islets were then divided equally into two parts, treated with forskolin/IBMX or with the vehicle for 2 h, followed by assessing nuclear Oct-1 content by Western blotting. Consistent with our pancreatic and intestinal endocrine cell line data, we observed that forskolin/IBMX treatment significantly reduced nuclear Oct-1 content in the primary pancreatic islet cells (Fig. 7A). Next, we isolated ∼300 pancreatic islets from each of four Wistar rats, and we assessed the effect of forskolin/IBMX treatment on proinsulin I and gcg mRNA expression by real time RT-PCR. As shown in Fig. 7, B and C, proinsulin I and gcg mRNA expression were up-regulated by forskolin/IBMX treatment by ∼3- and 2-fold, respectively. These results led us to propose that Oct-1 nuclear exclusion functions as a novel mechanism not only in up-regulating proglucagon production in pancreatic α and intestinal endocrine L cells, but also in up-regulating proinsulin expression in pancreatic β cells.
FIGURE 7.
Forskolin/IBMX treatment reduces nuclear Oct-1 content in primary pancreatic islet cells. A, Wistar rat pancreatic islets were treated with or without forskolin/IBMX (F/I) (10 μm each) for 2 h. V, vehicle. Nuclear proteins were extracted, and Oct-1 contents were assessed by Western blotting (left panel) and quantified by densitometric analysis (right panel) (n = 3). B and C, real time RT-PCR shows that forskolin/IBMX treatment enhanced proinsulin I (Ins I, B) and proglucagon (Gcg, C) mRNA expression. Values are mean ± S.E. (n = 4). *, p < 0.05.
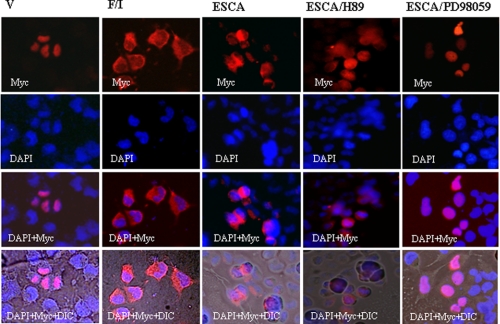
We have tried to detect the effect of forskolin/IBMX and the Epac pathway-specific cAMP analogue on Oct-1 cytosol-nuclear shuttling in primary pancreatic islet cells by immunofluorescence staining without success. All four Oct-1 antibodies we have tested detected a nonspecific signal in the cytosol of the primary pancreatic islet cells by immunostaining (data not shown). We have therefore examined the effect of forskolin/IBMX and 8-pMeOPT-2′-O-Me-cAMP treatment on cellular distribution of Myc-tagged Oct-1 in the pancreatic InR1-G9 cells by immunofluorescence staining. We found that 24 h after transfection of Oct-1-Myc into the InR1-G9 cells, the expression of Myc-tagged Oct-1 could be detected by the anti-Myc tag antibody. As shown in Fig. 8, for transfected cells treated with vehicle, Oct-1-Myc was mainly located within nuclei. When transfected cells were treated with forskolin/IBMX or 8-pMeOPT-2′-O-Me-cAMP for 1 h, Oct-1-Myc was mainly located in cytosol. Furthermore, the effect of 8-pMeOPT-2′-O-Me-cAMP can be blocked by MEK inhibition but not by PKA inhibition (Fig. 8). We have also generated an expression plasmid expressing a fusion protein of Oct-1 and enhanced green fluorescent protein (EGFP), and we observed that forskolin/IBMX treatment stimulated nuclear exclusion of Oct-1/EGFP (data not shown).
FIGURE 8.
Forskolin/IBMX treatment or Epac activation stimulates nuclear exclusion of Myc-tagged Oct-1. InR1-G9 cells transfected with Oct-1-Myc were treated with vehicle (V), or forskolin/IBMX (10 μm each, F/I), or 8-pMeOPT-2′-O-Me-cAMP (ESCA, 20 μm), or ESCA plus H89, or ESCA plus PD98059 for 1 h. The intracellular distribution of Oct-1-Myc was examined by immunostaining. 4′,6-Diamidino-2-phenylindole (DAPI), nuclear staining; DIC, differential interference contrast.
DISCUSSION
Oct-1 as a Transcriptional Repressor of Cdx-2
Proglucagon gene expression can be activated by cAMP via multiple and complex mechanisms (14, 46, 47, 53–57). The expression of Cdx-2, a transactivator of the gcg gene, can also be stimulated by cAMP (23). This stimulation, however, was shown to involve the activation of the Epac-MEK signaling pathway (23). We have identified the evolutionarily conserved cis element, Cdx-2pOCT, within the proximal promoter region of the mouse Cdx-2 gene (35). This OCT-binding site is highly conserved among divergent species (17, 43–45) (Fig. 1A). Because co-transfection of Oct-1 cDNA was shown to stimulate the expression of a luciferase reporter driven by a Cdx-2 promoter construct in transient transfection assays, we initially surmised that Oct-1 serves as a transactivator of Cdx-2 (35). However, Almeida et al. (58) reported that Oct-1 does not activate Cdx-2 expression in the non-endocrine gastric mucosa cells. In addition, Oct-1 expression levels are usually high in colon cancer cell lines, whereas these cell lines often show a reduction or absence of Cdx-2 expression (59). Finally, the expression of Cdx-2 and the embryonic transcription factor Oct-4 are mutually exclusive during early embryonic development stages (60), raising the possibility that Cdx-2pOCT functions as a silencer element. Following the notification, by ChIP analyses, that forskolin/IBMX treatment attenuated in vivo interaction between Oct-1 and Cdx-2 promoter, we investigated and observed that Oct-1 overexpression inhibited both Cdx-2 and gcg expression, whereas “knocking down” Oct-1 expression generated opposite effects. These findings, along with our observations that cAMP elevation stimulates Cdx-2 expression and reduces nuclear Oct-1 content, and that the binding of Cdx-2pOCT by Oct-1 in the living cells can be attenuated by forskolin/IBMX, collectively suggest that Oct-1 does function as a transcriptional repressor of Cdx- 2 in the proglucagon-expressing endocrine cells. These observations further indicate that reporter gene analyses performed in chromatin-free conditions may not always be reliable. Indeed, Oct-1 has been shown to exert complex regulatory effects on gene expression in different systems depending on its binding sites, the presence of co-factors and nuclear co-regulators, and extracellular stimulators (25–31, 33, 61).
Epac Activation and Oct-1 Nuclear Exclusion
Epac molecules can be directly bound by cAMP and then transfer the signal by interacting with certain small GTPases, such as Rap-1 (8, 9, 11). After the discovery of the Epac molecules (8, 9), studies on Epac expression and function in different cell lineages have revealed its versatile physiological roles, including the regulation of cell adhesion and hormone secretion (4, 5, 7, 10). In kidney cells, calcitonin was shown to stimulate H,K-ATPase through a cAMP/Epac/Rap/ERK signaling cascade (61). Ster et al. (62) found that in cerebellar neurons, Epac mediates cAMP activation of p38 MAPK. Enserink et al. (63), however, demonstrated that in a number of cell lineages, 8-pCPT-2′-O-Me-cAMP fully activated Rap-1 but not ERK and suggested that cAMP-induced activation of ERK and activation of Rap-1 are independent processes. Wang et al. (64) also found that 8-pCPT-2′-O-Me-cAMP did not activate ERK in the PC12 cell line. However, 8-pCPT-2′-O-Me-cAMP was shown to stimulate ERK after the expression of an engineered Epac1 that incorporated a membrane-targeting motif (64). We have reported that Epac2 is expressed in pancreatic and intestinal proglucagon-producing cells, and 8-pCPT-2′-O-Me-cAMP stimulates ERK phosphorylation as well as Cdx-2 and proglucagon expression (14, 23). We suggest that ERK could be activated by Epac in certain cell lineages, although detailed mechanisms underlying this activation need further investigation.
In this study, we found that Epac activation in response to cAMP elevation led to reduced nuclear Oct-1 content. We propose that pERK (either pERK itself or a downstream kinase of pERK) is involved in Oct-1 phosphorylation and subsequent nuclear exclusion, based on our observation that ERK inhibition blocked the stimulatory effect of cAMP on Cdx-2 and gcg expression (23). Indeed, the effect of cAMP-Epac on Oct-1 nuclear content was also repressed by MEK inhibition but not by PKA inhibition (Fig. 5, D–F). Furthermore, because the effects of cAMP elevation on both Cdx-2 expression and nuclear Oct-1 content were observed in the PKA-deficient InR1-G9 cell line, we suggest that these events do not require PKA and its downstream effector CREB (Fig. 5, G and H). To further explore mechanistically how Epac mediates Oct-1 nuclear exclusion and therefore attenuates its repressive effect on Cdx-2 and other target gene expression, it is essential to identify amino acid residues in Oct-1 that are phosphorylated in response to cAMP-Epac activation.
Cyclic AMP-mediated Oct-1 Nuclear Exclusion in Pancreatic Islets
In the primary pancreatic islet cells, we found reduced nuclear Oct-1 content in response to forskolin/IBMX treatment. This reduction is associated with stimulated proinsulin I and gcg mRNA expression. Hence, this novel signaling cascade not only functions in pancreatic α cells but also in pancreatic β cells. Cdx-2 has been shown to activate both proinsulin promoter and endogenous proinsulin mRNA expression in cultured cell lines (17, 19). Proinsulin expression can also be up-regulated by certain other homeodomain transcription factors, including Pdx-1 (65). Whether Pdx-1 expression could also be repressed by Oct-1 and, if so, whether the repression could be attenuated by cAMP/Epac-mediated nuclear exclusion of Oct-1, deserve further investigation. Furthermore, because Epac2 has been shown to function as the mediator of the incretin hormone glucagon-like peptide-1 (GLP-1) in stimulating insulin secretion (66), it will be interesting to know whether Epac2 also mediates the effect of GLP-1 in stimulating proinsulin production via enhancing Oct-1 phosphorylation and nuclear exclusion in pancreatic β cells.
Mechanistic Similarity of Oct-1 and FOXO Proteins
Our observations suggest that the regulation and function of Oct-1 share mechanistic similarities with those of FOXO proteins, a subfamily of forkhead transcription factors (67, 68). In the absence of insulin or growth factors, FOXOs are located mainly in nuclei, regulating a set of target genes that promote cell cycle arrest, stress resistance, and apoptosis (68–70). In the presence of insulin/growth factors, FOXOs are exported into the cytoplasm where they undergo rapid degradation. Such a nuclear exclusion event is mainly mediated through phosphorylation of FOXOs by protein kinase B and serum- and glucocorticoid-regulated protein kinase (67, 68). In this way, FOXOs function to control cellular growth, development, metabolism and possibly life expectancy in response to insulin and growth factors. We suggest that the ubiquitously expressed Oct-1 functions to control hormone-gene expression in response to cAMP elevation via a similar nuclear-cytoplasmic shuttling system, which confines Oct-1 to either the nucleus or the cytoplasm.
Oct-1 deficiency in mice (Oct-1−/−) is embryonically lethal (71). Using Oct-1−/− fibroblasts, Tantin et al. (72) found that Oct-1 functions as a stress sensor. More recently, Oct-1 was shown to mediate the effect of oxidized LDL (oxLDL) in repressing the expression of vascular cytochrome P450 monooxygenases (33). In human coronary arterial endothelial cells, knockdown of Oct-1 expression prevented oxLDL-mediated silencing of CYP expression. Furthermore, inhibition of oxLDL receptor attenuated oxLDL-mediated endothelial DNA damage, Oct-1 DNA binding, and reversed impaired production of endothelium-derived hyperpolarization factor. Therefore, Oct-1 activation in response to oxidative stress is among the pathological consequences in metabolic dysfunction of coronary arterial endothelium (33). We have also noticed that H2O2 treatment, which induces oxidative stress, leads to increased nuclear Oct-1 content, although total Oct-1 expression was not notably altered.6 We suggest that oxidative and other types of stress may stimulate kinase pathways that differ from the one that is activated by the cAMP/Epac signaling. Phosphorylation of Oct-1 by these kinase pathways may stimulate its nuclear translocation. Indeed Tantin et al. (72) and Schild-Poulter et al. (50) have shown that in radiation-induced stress, Oct-1 could be phosphorylated by DNA-dependent protein kinase. We therefore propose that Oct-1 plays a central role in signaling cascades triggered by oxidative stress and cAMP-generating hormones/neurotransmitter receptor activation. Because Oct-1 is a ubiquitously expressed protein, these findings are of broader application to other endocrine and non-endocrine cell lineages that act via these signaling cascades.
Supplementary Material
Acknowledgments
We thank Dr. Winship Herr for the original human Oct-1 expression plasmid; Drs. Donald Branch, Weiyang Lu, Allen Volchuk, and Burton Yang for their valuable comments; and Yanchun Wang for technical assistance.
This work was supported in part by Canadian Diabetes Association Grant 2341 (to T. J.) and Canadian Institutes of Health Research Grants 62745 (to T. J.), 79534 (to Q. W.), 86544 (to H. Y. G.), and 77750 (to J. H.).
The nucleotide sequence(s) reported in this paper has been submitted to the Gen-BankTM/EBI Data Bank with accession number(s) EF530070.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
P. Wang and T. Jin, unpublished data.
- PKA
- protein kinase A
- Epac
- exchange protein directly activated by cAMP
- OCT
- octamer-binding site
- Oct-1
- octamer transcription factor-1
- siRNA
- small interfering RNA
- RT
- reverse transcription
- IBMX
- 3-isobutyl-1-methylxanthine
- ChIP
- chromatin immunoprecipitation
- ERK
- extracellular signal-regulated kinase
- MEK
- mitogen-activated protein kinase/ERK kinase
- pol
- polymerase
- oxLDL
- oxidized low density lipoprotein
- EGFP
- enhanced green fluorescent protein
- CREB
- cAMP-response element-binding protein
- CYP
- cytochrome P450.
REFERENCES
- 1.Montminy M. (1997) Annu. Rev. Biochem. 66, 807–822 [DOI] [PubMed] [Google Scholar]
- 2.Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J. R., Emerson B., Hogenesch J. B., Unterman T., Young R. A., Montminy M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards J. S. (2001) Mol. Endocrinol. 15, 209–218 [DOI] [PubMed] [Google Scholar]
- 4.Bos J. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 733–738 [DOI] [PubMed] [Google Scholar]
- 5.Bos J. L. (2006) Trends Biochem. Sci. 31, 680–686 [DOI] [PubMed] [Google Scholar]
- 6.Holz G. G. (2004) Diabetes 53, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bos J. L., de Bruyn K., Enserink J., Kuiperij B., Rangarajan S., Rehmann H., Riedl J., de Rooij J., van Mansfeld F., Zwartkruis F. (2003) Biochem. Soc. Trans. 31, 83–86 [DOI] [PubMed] [Google Scholar]
- 8.Kawasaki H., Springett G. M., Mochizuki N., Toki S., Nakaya M., Matsuda M., Housman D. E., Graybiel A. M. (1998) Science 282, 2275–2279 [DOI] [PubMed] [Google Scholar]
- 9.de Rooij J., Zwartkruis F. J., Verheijen M. H., Cool R. H., Nijman S. M., Wittinghofer A., Bos J. L. (1998) Nature 396, 474–477 [DOI] [PubMed] [Google Scholar]
- 10.Holz G. G., Kang G., Harbeck M., Roe M. W., Chepurny O. G. (2006) J. Physiol. 577, 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehmann H., Arias-Palomo E., Hadders M. A., Schwede F., Llorca O., Bos J. L. (2008) Nature 455, 124–127 [DOI] [PubMed] [Google Scholar]
- 12.Holz G. G., Chepurny O. G., Schwede F. (2008) Cell. Signal. 20, 10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyle K. S., Raaijmakers J. H., Bruinsma W., Bos J. L., de Rooij J. (2008) Cell. Signal. 20, 1104–1116 [DOI] [PubMed] [Google Scholar]
- 14.Lotfi S., Li Z., Sun J., Zuo Y., Lam P. P., Kang Y., Rahimi M., Islam D., Wang P., Gaisano H. Y., Jin T. (2006) Endocrinology 147, 3727–3736 [DOI] [PubMed] [Google Scholar]
- 15.Nielsen D. A., Welsh M., Casadaban M. J., Steiner D. F. (1985) J. Biol. Chem. 260, 13585–13589 [PubMed] [Google Scholar]
- 16.Drucker D. J., Philippe J., Mojsov S., Chick W. L., Habener J. F. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 3434–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.German M. S., Wang J., Chadwick R. B., Rutter W. J. (1992) Genes Dev. 6, 2165–2176 [DOI] [PubMed] [Google Scholar]
- 18.Jin T., Drucker D. J. (1996) Mol. Cell. Biol. 16, 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y., Liu T., Zhang N., Yi F., Wang Q., Fantus I. G., Jin T. (2005) J. Endocrinol. 186, 179–192 [DOI] [PubMed] [Google Scholar]
- 20.Laser B., Meda P., Constant I., Philippe J. (1996) J. Biol. Chem. 271, 28984–28994 [DOI] [PubMed] [Google Scholar]
- 21.Ritz-Laser B., Estreicher A., Klages N., Saule S., Philippe J. (1999) J. Biol. Chem. 274, 4124–4132 [DOI] [PubMed] [Google Scholar]
- 22.Hussain M. A., Habener J. F. (1999) J. Biol. Chem. 274, 28950–28957 [DOI] [PubMed] [Google Scholar]
- 23.Chen L., Wang P., Andrade C. F., Zhao I. Y., Dubé P. E., Brubaker P. L., Liu M., Jin T. (2005) FEBS J. 272, 2746–2759 [DOI] [PubMed] [Google Scholar]
- 24.Herr W., Sturm R. A., Clerc R. G., Corcoran L. M., Baltimore D., Sharp P. A., Ingraham H. A., Rosenfeld M. G., Finney M., Ruvkun G., et al. (1988) Genes Dev. 2, 1513–1516 [DOI] [PubMed] [Google Scholar]
- 25.Wysocka J., Herr W. (2003) Trends Biochem. Sci. 28, 294–304 [DOI] [PubMed] [Google Scholar]
- 26.Chandran U. R., Attardi B., Friedman R., Zheng Z., Roberts J. L., DeFranco D. B. (1996) J. Biol. Chem. 271, 20412–20420 [DOI] [PubMed] [Google Scholar]
- 27.Chandran U. R., Warren B. S., Baumann C. T., Hager G. L., DeFranco D. B. (1999) J. Biol. Chem. 274, 2372–2378 [DOI] [PubMed] [Google Scholar]
- 28.Tang Q., Mazur M., Mellon P. L. (2005) Mol. Endocrinol. 19, 2769–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belsham D. D., Mellon P. L. (2000) Mol. Endocrinol. 14, 212–228 [DOI] [PubMed] [Google Scholar]
- 30.Vazquez-Martinez R., Leclerc G. M., Wierman M. E., Boockfor F. R. (2002) Mol. Endocrinol. 16, 2093–2100 [DOI] [PubMed] [Google Scholar]
- 31.Cheng C. K., Yeung C. M., Chow B. K., Leung P. C. (2002) Mol. Endocrinol. 16, 1552–1564 [DOI] [PubMed] [Google Scholar]
- 32.Kiyota T., Kato A., Altmann C. R., Kato Y. (2008) Dev. Biol. 315, 579–592 [DOI] [PubMed] [Google Scholar]
- 33.Thum T., Borlak J. (2008) J. Biol. Chem. 283, 19456–19464 [DOI] [PubMed] [Google Scholar]
- 34.Tanaka M., Herr W. (1990) Cell 60, 375–386 [DOI] [PubMed] [Google Scholar]
- 35.Jin T., Li H. (2001) J. Biol. Chem. 276, 14752–14758 [DOI] [PubMed] [Google Scholar]
- 36.Xu F., Li H., Jin T. (1999) J. Biol. Chem. 274, 34310–34316 [DOI] [PubMed] [Google Scholar]
- 37.Sun J., Khalid S., Rozakis-Adcock M., Fantus I. G., Jin T. (2009) Oncogene, in press [DOI] [PubMed] [Google Scholar]
- 38.Schreiber E., Matthias P., Müller M. M., Schaffner W. (1989) Nucleic Acids Res. 17, 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinh K. Y., Jin T., Drucker D. J. (1999) J. Biol. Chem. 274, 6011–6019 [DOI] [PubMed] [Google Scholar]
- 40.Schild-Poulter C., Shih A., Yarymowich N. C., Haché R. J. (2003) Cancer Res. 63, 7197–7205 [PubMed] [Google Scholar]
- 41.Yi F., Brubaker P. L., Jin T. (2005) J. Biol. Chem. 280, 1457–1464 [DOI] [PubMed] [Google Scholar]
- 42.Yi F., Sun J., Lim G. E., Fantus I. G., Brubaker P. L., Jin T. (2008) Endocrinology 149, 2341–2351 [DOI] [PubMed] [Google Scholar]
- 43.German M. S., Wang J., Fernald A. A., Espinosa R., 3rd, Le Beau M. M., Bell G. I. (1994) Genomics 24, 403–404 [DOI] [PubMed] [Google Scholar]
- 44.James R., Erler T., Kazenwadel J. (1994) J. Biol. Chem. 269, 15229–15237 [PubMed] [Google Scholar]
- 45.Marom K., Shapira E., Fainsod A. (1997) Mech. Dev. 64, 41–52 [DOI] [PubMed] [Google Scholar]
- 46.Lü F., Jin T., Drucker D. J. (1996) Endocrinology 137, 3710–3716 [DOI] [PubMed] [Google Scholar]
- 47.Knepel W., Chafitz J., Habener J. F. (1990) Mol. Cell. Biol. 10, 6799–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts S. B., Segil N., Heintz N. (1991) Science 253, 1022–1026 [DOI] [PubMed] [Google Scholar]
- 49.Segil N., Roberts S. B., Heintz N. (1991) Science 254, 1814–1816 [DOI] [PubMed] [Google Scholar]
- 50.Schild-Poulter C., Shih A., Tantin D., Yarymowich N. C., Soubeyrand S., Sharp P. A., Haché R. J. (2007) Oncogene 26, 3980–3988 [DOI] [PubMed] [Google Scholar]
- 51.Segil N., Roberts S. B., Heintz N. (1991) Cold Spring Harbor Symp. Quant. Biol. 56, 285–292 [DOI] [PubMed] [Google Scholar]
- 52.Grenfell S. J., Latchman D. S., Thomas N. S. (1996) Biochem. J. 315, 889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gevrey J. C., Malapel M., Philippe J., Mithieux G., Chayvialle J. A., Abello J., Cordier-Bussat M. (2004) Diabetologia 47, 926–936 [DOI] [PubMed] [Google Scholar]
- 54.Drucker D. J., Jin T., Asa S. L., Young T. A., Brubaker P. L. (1994) Mol. Endocrinol. 8, 1646–1655 [DOI] [PubMed] [Google Scholar]
- 55.Ni Z., Anini Y., Fang X., Mills G., Brubaker P. L., Jin T. (2003) J. Biol. Chem. 278, 1380–1387 [DOI] [PubMed] [Google Scholar]
- 56.Jin T. (2008) J. Endocrinol. 198, 17–28 [DOI] [PubMed] [Google Scholar]
- 57.Wang P., Liu T., Li Z., Ma X., Jin T. (2006) Endocrinology 147, 1950–1958 [DOI] [PubMed] [Google Scholar]
- 58.Almeida R., Almeida J., Shoshkes M., Mendes N., Mesquita P., Silva E., Van Seuningen I., Reis C. A., Santos-Silva F., David L. (2005) J. Pathol. 207, 396–401 [DOI] [PubMed] [Google Scholar]
- 59.Bonhomme C., Duluc I., Martin E., Chawengsaksophak K., Chenard M. P., Kedinger M., Beck F., Freund J. N., Domon-Dell C. (2003) Gut 52, 1465–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niwa H., Toyooka Y., Shimosato D., Strumpf D., Takahashi K., Yagi R., Rossant J. (2005) Cell 123, 917–929 [DOI] [PubMed] [Google Scholar]
- 61.Hitomi T., Matsuzaki Y., Yasuda S., Kawanaka M., Yogosawa S., Koyama M., Tantin D., Sakai T. (2007) FEBS Lett. 581, 1087–1092 [DOI] [PubMed] [Google Scholar]
- 62.Ster J., De Bock F., Guérineau N. C., Janossy A., Barrère-Lemaire S., Bos J. L., Bockaert J., Fagni L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Enserink J. M., Christensen A. E., de Rooij J., van Triest M., Schwede F., Genieser H. G., Døskeland S. O., Blank J. L., Bos J. L. (2002) Nat. Cell Biol. 4, 901–906 [DOI] [PubMed] [Google Scholar]
- 64.Wang Z., Dillon T. J., Pokala V., Mishra S., Labudda K., Hunter B., Stork P. J. (2006) Mol. Cell. Biol. 26, 2130–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jonsson J., Carlsson L., Edlund T., Edlund H. (1994) Nature 371, 606–609 [DOI] [PubMed] [Google Scholar]
- 66.Kang G., Chepurny O. G., Malester B., Rindler M. J., Rehmann H., Bos J. L., Schwede F., Coetzee W. A., Holz G. G. (2006) J. Physiol. 573, 595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Accili D., Arden K. C. (2004) Cell 117, 421–426 [DOI] [PubMed] [Google Scholar]
- 68.Greer E. L., Brunet A. (2005) Oncogene 24, 7410–7425 [DOI] [PubMed] [Google Scholar]
- 69.Essers M. A., de Vries-Smits L. M., Barker N., Polderman P. E., Burgering B. M., Korswagen H. C. (2005) Science 308, 1181–1184 [DOI] [PubMed] [Google Scholar]
- 70.Essers M. A., Weijzen S., de Vries-Smits A. M., Saarloos I., de Ruiter N. D., Bos J. L., Burgering B. M. (2004) EMBO J. 23, 4802–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang V. E., Schmidt T., Chen J., Sharp P. A., Tantin D. (2004) Mol. Cell. Biol. 24, 1022–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tantin D., Schild-Poulter C., Wang V., Haché R. J., Sharp P. A. (2005) Cancer Res. 65, 10750–10758 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.