Abstract
Neuroblastoma cells having stem cell-like qualities are widely employed models for the study of neural stem/progenitor cell proliferation and differentiation. We find that human BE(2)C neuroblastoma cells possess a signaling cascade initiated by Ca2+ influx via voltage-dependent calcium channels and the N-methyl-d-aspartate (NMDA) receptor and culminating in nuclear calmodulin-dependent protein kinase IV (CaMKIV)-mediated phosphorylation and activation of the transcription factors Ca2+/cyclic AMP-response element-binding protein (CREB) and ATF1 (activating transcription factor-1). This pathway functions to maintain BE(2)C cells in an undifferentiated, proliferative state. Parallel to this Ca2+-dependent pathway is a hormone-responsive program by which retinoic acid (RA) initiates the differentiation of BE(2)C cells toward a neuronal lineage. This is evidenced by RA-dependent induction of the cell cycle inhibitor p21/Cip1 (Cdk-interacting protein 1) and cell cycle arrest, induction of the neuroblastic marker doublecortin and of the neuron-specific intermediate filament protein, peripherin, and by RA-stimulated extension of neuritic processes. During neuronal differentiation there is a complex antagonistic interplay between these two major signaling pathways. RA down-regulates expression of CaMKIV and one of its upstream activators, CaMKK1 (calmodulin-dependent protein kinase kinase 1). This is accompanied by RA-induced suppression of activating phosphorylation of CREB with a time course paralleling that of CaMKIV down-regulation. RA-induced repression of the Ca2+/calmodulin-dependent protein kinase kinase/CaMKIV/CREB pathway appears to be involved in regulating the timing of neuronal differentiation, as shown by the effect of RNA interference of CaMKIV to markedly accelerate RA-dependent up-regulation of p21/Cip1 and doublecortin expression and RA-promoted neurite outgrowth. RA-induced repression of the CaMKIV signaling pathway may represent an early event in retinoid-dependent neuronal differentiation.
Intracellular Ca2+ fulfills a pleiotropic role in the physiology of neural cells through its regulation of events such as neurotransmitter release, synaptic plasticity, dendrite and axon morphogenesis, and neuronal migration (recently reviewed in Refs. 1–3). Although the functions of intracellular Ca2+ have been most intensively studied in postmitotic neurons, recent evidence has raised the possibility of developmental roles of intracellular Ca2+ prior to or during neuronal differentiation via regulation of the proliferation and/or differentiation of neural stem cells (NSCs)2 and restricted neural progenitors (4–10).
Many of the biochemical events underlying intracellular Ca2+ action in developing and mature neural tissues are mediated by Ca2+ complexed with calmodulin (CaM) and activation of members of the CaM kinase subfamily comprising CaMKI, -II, and -IV (reviewed in Refs. 11 and 12). CaMKIV is characterized by its mainly nuclear localization (13), its requirement of phosphorylation by CaMKKs 1(α) and/or 2(β) (14–17) for optimal activity, and by its acquisition of Ca2+/CaM-independent (autonomous) activity as a consequence of CaMKK-dependent phosphorylation and subsequent autophosphorylation (18–20). The downstream targets of CaMKIV are thought to be primarily nuclear due to its sequestration in the cytoplasm at resting intracellular Ca2+ levels by binding to Ser/Thr protein phosphatase type 2A, which maintains CaMKIV in a dephosphorylated and inactive state (21). Elevation of intracellular Ca2+ leads to the accumulation of CaMKIV in a phosphorylated, activated, and Ca2+-autonomous state in the nucleus (18, 19, 22, 23).
In the nucleus, CaMKIV regulates the transactivating abilities of a range of sequence-specific transcription factors and transcriptional co-regulators, including CREB (24–26), the related CREB family member, ATF1 (27), and CREB-binding protein (28, 29). In multiple cell types, including T-cells and neurons, intracellular Ca2+ elevation provokes CREB phosphorylation at its transcriptionally activating site, Ser-133, by CaMKIV (30, 31). In addition to CaMKIV, protein kinase A and downstream kinases in the mitogen-activated protein kinase cascade (ribosomal S6 kinase and mitogen- and stress-activated protein kinase) can also effect CREB phosphorylation as a result of intracellular Ca2+-elevating stimuli. In depolarized neurons, CaMKIV is reported to mediate more rapid and transient phosphorylation of CREB than does activation of the mitogen-activated protein kinase cascade (32). However, for the most part, the roles and kinetics of the various CREB kinases in intracellular Ca2+-dependent CREB phosphorylation and transcriptional activation are undefined (recently reviewed in Refs. 12 and 33).
In the adult, CaMKIV is expressed most abundantly in brain, testis, ovary, and T-lymphocytes (11). CaMKIV expression appears to be regulated during embryonic development. In some cell types, its expression correlates with distinct periods of cellular proliferation, differentiation, or survival (34). Consonant with potential developmental roles, CaMKIV is found in different types of multipotent stem and progenitor cells, including immature neutrophils, embryonic stem cells, hematopoietic stem cells, and osteoclast progenitors (35–38). Potential developmental roles of CaMKIV are largely unexplored, although several recent studies have documented participation of CaMKIV in spermiogenesis and hematopoietic stem cell maintenance (37, 39, 40), and it has been observed that CaMKIV null mice exhibit reduced numbers of Purkinje neurons following cerebellar development (41).
The research described herein was conducted to explore the role of the CaMKIV signaling pathway in the proliferation, differentiation, and survival of multipotent neural cells. As a cellular model, we utilized human BE(2)C neuroblastoma cells. BE(2)C cells phenotypically resemble malignantly transformed NSCs, having the capacity for both self-renewal and multipotency. BE(2)C cells differentiate to a substrate-adherent/glial phenotype with long term bromodeoxyuridine (BrdUrd) treatment or to a neuroblastic phenotype when cultured with the vitamin A derivative, all-trans-retinoic acid (RA) (42–44). We report here that RA represses the CaMKIV cascade during its induction of neuronal differentiation, an event that may be important for temporal coordination of the transition from neural stem/progenitor cell proliferation to a neuronally differentiated state.
EXPERIMENTAL PROCEDURES
Materials
The following reagents were obtained from Sigma: STO-609 (7H-benzimidazo[2,1-a]benz[de]isoquinoline-7-one-3-carboxylic acid), RA, NMDA, 4′,6-diamidino-2-phenylindole (DAPI), tri-iodothyronine (thyroid hormone), BrdUrd, 1,25-dihydroxyvitamin D3, and forskolin. Insulin-like growth factor 1 was obtained from BIOSOURCE. All other reagents were obtained from standard sources and were of the highest grade available.
Cell Culture
Human BE(2)C neuroblastoma cells were obtained from ATCC. Cells were routinely cultured in Dulbecco's modified Eagle's medium/F-12 (1:1 ratio) and containing 10% fetal bovine serum (Foundation, Gemini Bio-products), 1.0 mm non-essential amino acids (Invitrogen), and, unless otherwise indicated, 1× penicillin/streptomycin (Invitrogen) and maintained in a 5% CO2, 95% air humidified incubator at 37 °C. For routine culturing, cells were passaged once every 7 days, and complete medium was replaced 4 days later.
Plasmids, siRNAs, and Transfection
The following plasmids were used: pcDNA 3.1-EGFP (a gift of M. K. Stachowiak), Ca2+/cAMP-response element (CRE)-firefly luciferase reporter (CRE-Luc; Stratagene), SV40-Renilla luciferase (pRL; Promega), and constitutively active CaMKIV (CA-CaMKIV; a gift of G. A. Wayman and T. R. Soderling). The following siRNA duplexes (Ambion) were used: CaMKIV siRNA targeting exon 11 (CaMKIVsi-11), CaMKIV siRNA targeting exon 1 (CaMKIVsi-1), and negative control siRNA 1 (NSsi). BE(2)C cells were plated at an appropriate density in antibiotic-free complete medium 24 h prior to transfection. Plasmids or siRNA duplexes were transfected using Lipofectamine 2000 as the transfection agent according to the manufacturer's instructions (Invitrogen). After a 5–6-h incubation period in serum-free OPTIMEM I, transfection components were removed and replaced with complete medium lacking antibiotics.
Gene Reporter Assays
CRE-Luc, pRL, and, where indicated, CA-CaMKIV were transfected into BE(2)C cells plated on 100-mm plates at concentrations of 150, 3.48, and 300 ng/ml, respectively. After 48 h, the dual luciferase assay was performed according to the manufacturer's instructions (Promega).
Subcellular Fractionation
Subcellular fractionation of BE(2)C neuroblastoma cells growing in monolayer culture was performed with the ProteoextractTM subcellular proteome extraction kit according to the manufacturer's instructions (BD Biosciences). Nuclear and cytosolic compartments were subjected to immunoblotting for their respective markers, histone H3 and lactate dehydrogenase, to assess the effectiveness of the fractionation procedure.
Immunoblotting
BE(2)C cells were lysed in harvest buffer (50 mm Tris, pH 8.0, 2% SDS, 1× protease inhibitor mixture (Sigma), 25 mm NaF, 100 μm Na3VO4, 100 μm okadaic acid, 5 mm EGTA, and 5 mm EDTA) and chilled on ice for 10 min, and lysate and cellular debris were removed with a rubber policeman. Cell lysate was sonicated on ice to shear DNA to reduce viscosity. Cellular debris was then pelleted by centrifugation at 14,000 rpm at 4 °C for 10 min, and supernatant was transferred to a fresh prechilled tube. Equal protein amounts were subjected to standard electrophoresis conditions on 7.5, 10, or 12.5% SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Membranes were rinsed in TBST (Tris-buffered saline, 0.1% Tween 20) twice, for 5 min each time, at room temperature and incubated in blocking buffer (5% (w/v) nonfat milk or 5% bovine serum albumin in TBST) for 18 h at 4 °C or 1 h at room temperature. Membranes were rinsed three times for 5 min each time in TBST and then incubated for 18 h at 4 °C or for 1 h at room temperature in blocking buffer containing the following primary antibodies at their respective dilutions: p21/Cip1 (1:500; Cell Signaling), pan-CREB (1:5,000; Cell Signaling), phosphoserine 133 CREB (1:1,000; Cell Signaling), CaMKIV (1:3,000 (Cell Signaling) or 1:1,000 (Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), GAPDH (1:10,000–20,000; Santa Cruz Biotechnology), Bcl-2 (1:500–1,000; Cell Signaling), poly(ADP)-ribose polymerase (PARP) (1:500–1:5,000; Cell Signaling), pro-caspase-3 (1:1,000; Cell Signaling), DCX (1:500–1,000; Cell Signaling), lactate dehydrogenase (1:500; U.S. Biologicals), histone H3 (1:10,000; AbCam), or vimentin (1:1,000; BD Biosciences). Following three 5-min washes in TBST, blots were incubated with appropriate secondary antibodies in blocking buffer for 1 h and then subjected to four 15-min washes in TBST. Immunoreactive bands were visualized by ECL.
BrdUrd Labeling and Immunocytochemistry
BE(2)C cells were plated on coverslips in wells of a 12-well plate at a density of ∼104 cells/coverslip, 24 h prior to drug treatments or transfections and then labeled with 10 μm BrdUrd for 1–3 h, as indicated in the figure legends (Fig. 3, Table 1). Following treatment, cells were fixed with 1 ml of fixation solution (8% paraformaldehyde in PBS, pH 7.2, and containing 300 mm sucrose) at 37 °C, which was replaced with fixation solution but containing 4% paraformaldehyde. Cells were rinsed three times in PBS, pH 7.2, and then permeabilized with 0.5% Triton X-100 in PBS for 5 min. They were then rinsed three times with PBS and incubated in 2 n HCl at 37 °C for 15 min in PBS to denature chromatin. Cells were rinsed five times with PBS, pH 10.0, followed by three rinses with PBS, pH 7.2, and then incubated in PBS containing 2% goat serum for 18 h at 4 °C to block nonspecific binding. Following three 5-min washes, cells were incubated with PBS, 2% goat serum containing BrdUrd monoclonal antibody (Sigma) at a 1:1000 dilution for 18 h at 4 °C. Goat anti-mouse Alexa 587 (Molecular Probes) in PBS, 2% goat serum was used at a 1:2,000 dilution to detect BrdUrd labeling.
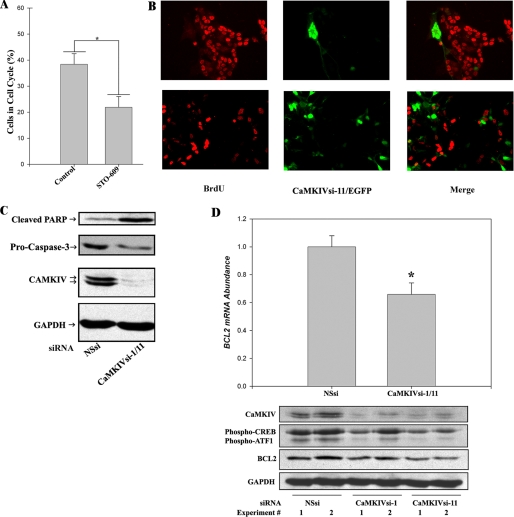
FIGURE 3.
The CaMKIV signaling cascade promotes the proliferation and survival of BE(2)C neuroblastoma cells. A, CaMKK inhibition by STO-609 reduces the percentage of cells in the cell cycle. BE(2)C cells were cultured for 48 h in the absence (vehicle control) or presence of 10 μm STO-609. Cells were labeled by incubation with BrdUrd during the final 3 h of incubation and fixed with 4% paraformaldehyde. Cells were identified by nuclear staining with DAPI, and cycling cells were visualized by BrdUrd immunocytochemistry. The percentage of cycling cells is calculated as the number of cells incorporating BrdUrd divided by the number of DAPI-stained (total) cells. *, p < 0.001. B, RNAi of CaMKIV induces cell cycle arrest. BE(2)C cells were co-transfected with 100 nm CaMKIVsi-11 and pcDNA 3.1-EGFP for 48 h and labeled with BrdUrd (BrdU) for 1 h. Cells incorporating CaMKIVsi-11 are identified by EGFP fluorescence (green) in the center and right panels. Cycling cells are identified by BrdUrd immunocytochemistry (red) in the left and right panels. The upper and lower rows are two representative fields of cells. C, RNAi of CaMKIV induces markers of apoptosis. BE(2)C cells were transfected with 100 nm NSsi or 100 nm (total) CaMKIVsi-1/11 and analyzed by immunoblotting after 48 h for the apoptotic markers cleaved PARP and (loss of) intact pro-Caspase-3. Blots were also probed for CaMKIV and GAPDH to confirm silencing of CaMKIV and equivalent protein loading, respectively. D, Bcl-2 mRNA and protein expression are down-regulated by RNAi of CaMKIV. Top, BE(2)C cells were transfected with 100 nm NSsi or 100 nm (total) CaMKIVsi-1/11 for 48 h, following which lysates were prepared, and Bcl-2 mRNA abundance was determined by qRT-PCR. *, p < 0.01. Bottom, BE(2)C cells were transfected with either 100 nm NSsi, CaMKIVsi-1, or CaMKIVsi-11 for 48 h and analyzed by immunoblotting for expression of Bcl-2 and CREB/ATF-1 phosphorylation. Blots were also probed for CaMKIV and GAPDH to confirm silencing of CaMKIV and equivalent protein loading, respectively.
TABLE 1.
RNAi of CaMKIV induces cell cycle arrest
BE(2)C cells were transfected with 100 nm CaMKIVsi-11 and p cDNA 3.1-EGFP for 48 h and labeled with BrdUrd for 1 h. Cells incorporating CaMKIVsi-11 or NSsi were identified by EGFP fluorescence. EGFP-positive cycling (BrdUrd-positive) and arrested (BrdUrd-negative) cells were identified by BrdUrd immunocytochemistry, as described under “Experimental Procedures.” Images were analyzed using Image J with cell counting by an observer blind as to conditions. χ2 = 25.407, 1 degree of freedom (p < 0.001).
| BrdUrd+/GFP+ | BrdUrd−/GFP+ | |
|---|---|---|
| CaMKIVsi-11 | 24 | 538 |
| NSsi | 102 | 725 |
For detection of neuronal microtubules, coverslips were incubated with βIII tubulin antibody (Sigma) at a 1:1000 dilution for 18 h at 4 °C. Goat anti-rabbit fluorescein isothiocyanate (Molecular Probes) was then used at a 1:1,000 dilution in PBS, 2% goat serum. After 18 h of incubation, coverslips were incubated with 300 nm DAPI for 5 min (where indicated), rinsed three times in PBS and then H2O, and mounted in ProLong Gold antifade reagent (Molecular Probes). Images were captured with a Zeiss Axioimager Z1 Axiophot wide field fluorescence microscope and analyzed using the program Image J.
RNA Isolation and cDNA Synthesis
BE(2)C cells were lysed, and total RNA was isolated using Qiashredder columns and RNeasy kits, respectively, according to the manufacturer's instructions (Qiagen). Genomic DNA was removed by on column DNase digestion during RNA isolation with an RNase-free DNase set (Qiagen). RNA was quantified, and purity was assessed spectrophotometrically. cDNA was synthesized using Superscript III reverse transcriptase and other reagents as recommended by the manufacturer (Invitrogen). Briefly, RNA (2.12 μg) was combined with 30 ng of random hexamers and 0.4 mm dNTPs, heated at 65 °C for 5 min, and rapidly chilled on ice for 5 min. First strand buffer, 0.1 m dithiothreitol, RNase out, and Superscript III reverse transcriptase were added. Samples were incubated at 25 °C for 10 min, 50 °C for 60 min, and 70 °C for 15 min, at which point the reaction was stopped by holding at 4 °C. RNase H was added to each tube, and remaining RNA was digested at 37 °C for 20 min.
End Point PCR
End point (non-quantitative) PCR was performed using Platinum Taq polymerase under standard reaction conditions as recommended by the manufacturer (Invitrogen) with BE(2)C cell cDNA (2 μg) and 0.2 μm forward and 0.2 μm reverse primers. PCR was performed using a PerkinElmer Life Sciences or Bio-Rad thermocycler. Thermocycling parameters were as follows: 10 min at 95 °C, 30–35 cycles of 95 °C for 30 s, 58–60 °C for 60 s, and 72 °C for 60 s, followed by 10 min at 72 °C.
Quantitative Reverse Transcription PCR (qRT-PCR)
mRNA was quantified by real time qRT-PCR of BE(2)C cDNA with SYBR Green detection (Bio-Rad) using a Bio-Rad iCycler. Thermocycling parameters were as follows: 2 min at 95 °C and 45 cycles at 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s. The amplification specificity was verified by melt curve analysis. The average starting quantity for each condition was calculated by the standard curve method using GAPDH as normalizer.
Primers
Primers were designed using the program PRIME (Genetics Computer Group). Specificity of the primers was established by a preliminary end point PCR (in the presence and absence of reverse transcriptase) and melt curve analysis following qRT-PCR. In most cases, primers spanned at least one exon/intron junction. Primers are listed by target cDNA, forward primer (5′ to 3′), and reverse primer (5′ to 3′), respectively: Bcl-2, GCCGTTGGCCCCCGTTGCTTTTC and CGCCCACATCTCCCGCATCCCAC; CaMKIV, TGCAACTCCAGCCCCAGATGCAC and ACAGACCACATGTCCACCTCAGGTCC; CaMKK1, GCAACCAGTTTGAGGGGAACGACGC and CAACGTGACGCCAGTGGCCCATAC; CaMKK2, CCAGACCAGCCCGACATAGCTGAGGA and CGTGACCCAGGGGTGCAGCTTGATT; DCX, CGTTTCTACCGCAATGGGGACCGC and ACGCACTCCCTGAGGCAGGTTGATG; GAPDH, TGGCATTGCCCTCAACGACCACTTTG and TCCTCTTGTGCTCTTGCTGGGGCT; p21/Cip1, CGAAAACGGCGGCAGACCAGCATGA and TGAGGCCCTCGCGCTTCCAGGAC; Peripherin, GGAGGAGCTGCGACAGCTAAAAGAGG and AGGAGGCAAAAGAATGGACGGGCAC.
RESULTS
In neurons, CaMKIV is localized primarily to the nucleus (13). In experiments using T-lymphocytes and embryonic kidney 293 cells, a proportion of the enzyme was shown to be cytoplasmic due to interaction with Ser/Thr protein phosphatase 2A (21). Moreover, the CaMKIV-Ser/Thr protein phosphatase 2A interaction is disrupted by Ca2+/CaM, suggesting that the nuclear/cytoplasmic ratio of CaMKIV is responsive to variations over time in the level of intracellular Ca2+ (45). We therefore assessed the subcellular distribution of CaMKIV in BE(2)C neuroblastoma cells without the addition of an intracellular Ca2+-elevating agonist using a selective extraction/differential centrifugation method. As shown in Fig. 1A, CaMKIV was almost exclusively recovered in the nuclear fraction. This suggests that either the localization of CaMKIV to the nuclear compartment in BE(2)C cells does not require elevation of intracellular Ca2+ or that intracellular Ca2+ is elevated to a sufficient extent under basal conditions to maintain nuclear retention of CaMKIV.
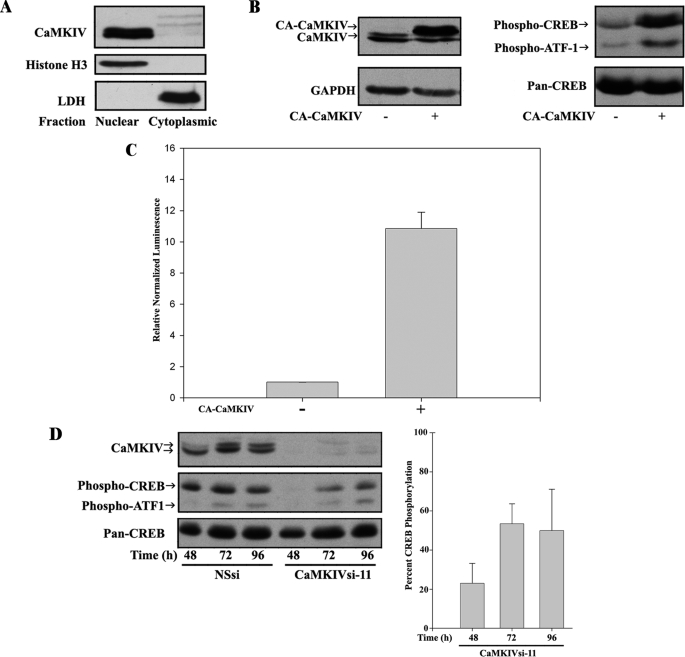
FIGURE 1.
CaMKIV is necessary and sufficient for CREB/ATF1 activation in response to Ca2+ influx in BE(2)C neuroblastoma cells. A, CaMKIV is expressed and predominantly nuclearly localized in human BE(2)C neuroblastoma cells. BE(2)C cells were fractionated into cytoplasmic and nuclear compartments, and the levels of expression of CaMKIV (top) and the subcellular markers histone H3 (Nuclear; middle) and lactate dehydrogenase (LDH) (Cytoplasmic; bottom) were determined by immunoblotting. Note that, depending on electrophoretic conditions, the two isoforms of CaMKIV (α and β) are resolved (e.g. see B and D) or unresolved (A). B, constitutively active CaMKIV (CA-CaMKIV) induces CREB/ATF1 phosphorylation. BE(2)C cells were serum-starved and mock-transfected or transfected with CA-CaMKIV. Right, CREB/ATF-1 phosphorylation and expression of total CREB (Pan-CREB) were determined by immunoblotting. Left, blots were also probed for CaMKIV and GAPDH to confirm expression of CA-CaMKIV and equivalent protein loading, respectively. Note that CA-CaMKIV electrophoreses at a higher molecular size than endogenous CaMKIV due to a plasmid-encoded His tag. C, CA-CaMKIV is sufficient to drive CRE-mediated transcriptional activity. Serum-starved BE(2)C cells were transfected with pRL and pCRE-Luc with or without CA-CaMKIV. Transcriptional activity was calculated by normalizing firefly luciferase to Renilla luciferase activities and then calculating normalized activity in CA-CaMKIV-transfected cells relative to cells transfected with luciferase plasmids alone. D, endogenous CaMKIV is required for CREB/ATF1 phosphorylation in BE(2)C cells. BE(2)C cells were transfected with 50 nm NSsi or CaMKIVsi-11, and levels of CaMKIV expression and CREB/ATF-1 phosphorylation were determined by immunoblotting after the indicated times. Blots were also probed for total CREB (Pan-CREB). Right, CREB phosphorylation was quantified by scanning densitometry of phospho-CREB immunoreactivity normalized to total CREB immunoreactivity in CaMKIVsi-11-transfected cells and expressed as a percentage of normalized CREB phosphorylation in NSsi-transfected cells.
The nuclear transcription factor CREB is the most thoroughly documented in vivo target of CaMKIV (24–27, 29–31). However, given the observations of other CREB kinases mediating its phosphorylation in response to intracellular Ca2+ influx (32), we examined the role of nuclear CaMKIV in enhancing transcription by phosphorylating and activating CREB in BE(2)C cells. Transfection of CA-CaMKIV led to increased phosphorylation of CREB at its transcriptionally activating phosphorylation site (Ser-133) in approximate proportion to the increase in the intracellular concentration of CaMKIV (Fig. 1B). The Ser(P)-133-specific anti-CREB antibody also recognizes the phosphorylation site known to be essential for CaMKIV-mediated activation of the CREB family member ATF1 (Ser-63) (27). As shown, phosphorylation of ATF1 was also increased by CA-CaMKIV. To verify that CaMKIV is capable of promoting transcriptional activation of endogenous CREB, we examined the ability of CA-CaMKIV to enhance transcription of a luciferase reporter driven by a CRE. CA-CaMKIV produced a 10.85 ± 3.15-fold (p < 0.001) increase in CRE-dependent transcription (Fig. 1C). Thus, active CaMKIV is sufficient to induce CREB and ATF1 phosphorylation and CRE-mediated transcription.
We then used RNAi to examine a potential requirement of endogenous CaMKIV for CREB and ATF1 activation in BE(2)C cells. In preliminary experiments, the concentration dependences and targeting specificities of siRNAs targeting exon 11 or exon 1 of CaMKIV (CaMKIVsi-11 or CaMKIVsi-1, respectively) were established (supplemental Fig. S1). Of the two, CaMKIVsi-11 was somewhat more efficacious, with the ability to reduce CaMKIV protein and mRNA levels by ∼80–90%. CaMKIVsi-11 was therefore preferentially used in this study, either alone or in equimolar combination with CaMKIVsi-1 (CaMKIVsi-1/11). Neither siRNA was capable of silencing protein expression of the related CaMKs, CaMKII β (supplemental Fig. S1A) or CaMKIα (data not shown). In addition, CaMKIVsi-11 was ineffective in reducing mRNA levels of the upstream activators of CaMKIV, CaMKK1, and CaMKK2 (data not shown).
We first investigated the role of CaMKIV in cells cultured in the presence of serum but in the absence of added intracellular Ca2+-elevating agonists. Compared with a nonspecific siRNA (NSsi), CaMKIVsi-11 was highly effective in reducing CaMKIV protein expression for up to 96 h with a maximal reduction at 48 h of incubation (Fig. 1D). Concomitantly, CREB and ATF1 activation site phosphorylation was readily detected during incubation with NSsi, whereas their phosphorylation was greatly reduced by incubation with CaMKIVsi-11, also with maximal reduction at 48 h. CREB phosphorylation, determined by scanning densitometry and normalized to total (pan-)CREB expression was decreased by 77.1 ± 17.8, 46.6 ± 17.7, and 50.1 ± 30.0% at 48, 72, and 96 h, respectively (Fig. 1D, right). Thus, the time point of maximal reduction and partial rebound in CREB phosphorylation paralleled the kinetics of CaMKIV silencing. Since the levels of CaMKIV available for CREB phosphorylation in the nucleus are not precisely known, increase in the latter may be the result of the gradual return in CaMKIV expression between the 48 and 96 h time points or alternatively due to the participation in CREB phosphorylation of other kinases. Regardless, it may be concluded based on these results that CaMKIV is required for optimal activation site CREB/ATF1 phosphorylation in BE(2)C neuroblastoma cells.
We next probed the role of Ca2+ influx in CaMKIV-mediated CREB/ATF1 phosphorylation under both unstimulated and stimulated conditions. For the experiment illustrated in Fig. 2A, cells were serum-starved to reduce the influence of serum components on Ca2+ influx and then incubated in one of four types of media: Ca2+ (∼1 mm)-containing medium, Ca2+-depleted (EGTA added) medium, medium with a 10 μm concentration of the dihydropyridine voltage-dependent Ca2+ channel (VDCC) blocker nimodipine, or medium containing both EGTA and nimodipine. After 15 min, media were replaced as indicated, and cells were incubated for an additional 15 min to determine whether any observed effects were reversible. CREB and ATF1 phosphorylation were then assessed by immunoblotting. Continuous exposure throughout both incubation periods to Ca2+-containing medium induced robust CREB and ATF1 phosphorylation (leftmost lane 1). However, removal of extracellular Ca2+ or VDCC blockade or the combination of the two treatments greatly attenuated both CREB and ATF1 phosphorylation (lanes 2, 4, and 5). These treatments did not induce irreversible, toxic effects, since CREB and ATF1 phosphorylation was fully restored by shifting the cells back to Ca2+-containing medium for an additional 15 min of incubation (lanes 3 and 6, respectively). Thus, CREB and ATF1 phosphorylation is rapidly and reversibly responsive to Ca2+ influx without the need for the addition of an intracellular Ca2+-elevating agonist. This is probably due to a basal influx mediated by either L-type or T-type VDCCs, given the relatively depolarized resting potential of BE(2)C cells of ∼−55 mV (46). Continuous Ca2+ influx is consistent with only a slight increase in CREB and ATF1 phosphorylation after exposure of BE(2)C cells to depolarizing concentrations of KCl (data not shown) and with the nuclear accumulation of CaMKIV under resting conditions (Fig. 1A).
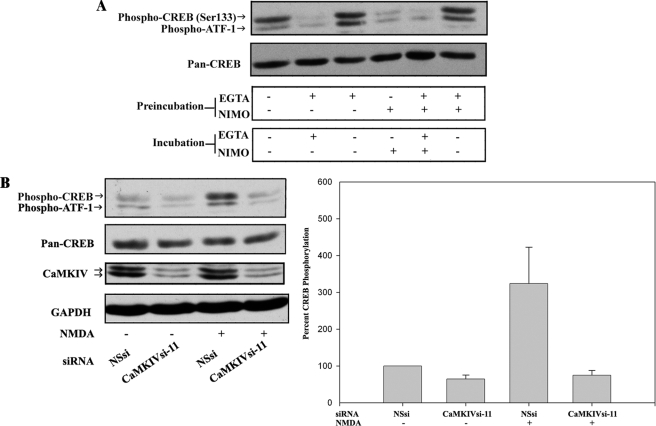
FIGURE 2.
CaMKIV-dependent CREB/ATF-1 phosphorylation is downstream of VDCC and NMDA receptor activities. A, removal of extracellular Ca2+ and inhibition of VDCC activity reversibly inhibit CREB/ATF-1 phosphorylation. BE(2)C cells were serum-starved for 3 h and then preincubated for 15 min in medium (containing ∼1 mm calcium) without additions or in the same medium with the addition of either 2 mm EGTA, 10 μm nimodipine (NIMO), or 2 mm EGTA plus 10 μm nimodipine. Media were replaced as indicated, and cells were incubated for an additional 15 min and then analyzed by immunoblotting for CREB/ATF-1 phosphorylation and total CREB (Pan-CREB) expression. B, NMDA-induced CREB/ATF-1 phosphorylation is blocked by RNAi of CaMKIV. BE(2)C cells were transfected with 50 nm NSsi or CaMKIVsi-11 siRNAs for 48 h, serum-starved for 3 h, and then preincubated with HEPES-Tyrode's buffer with glycine substituted for Mg2+ (119 mm NaCl, 2.5 mm CaCl2, 2 mm glycine, 25 mm HEPES, 30 mm glucose) for 15 min. Cultures were then treated with 7.5 μm NMDA or vehicle control (DMSO; −) for 2 min in the same buffer and then analyzed by immunoblotting for CREB/ATF1 phosphorylation and total CREB expression. Blots were also probed for CaMKIV and GAPDH to confirm silencing of CaMKIV and equivalent protein loading, respectively. Right, CREB phosphorylation was quantified by scanning densitometry of phospho-CREB immunoreactivity normalized to total CREB (Pan-CREB) immunoreactivity and expressed as a percentage of normalized CREB phosphorylation in vehicle-treated/NSsi-transfected cells.
In mature neurons, activation of NMDA-type glutamate receptors (NMDARs) results in Ca2+ influx and activation of CaMKIV and CREB phosphorylation (29). To ascertain whether BE(2)C cells are similarly responsive to NMDA, cells were incubated with NMDA at various concentrations for time periods between 2 and 15 min (supplemental Fig. S2). NMDA induced a concentration-dependent increase in CREB phosphorylation, which was maximal at the earliest time point of 2 min. The transient nature of this response suggests that rapid CREB phosphorylation may be counteracted over longer periods of time by NMDAR inactivation and/or phosphatase activation. To determine whether NMDA-induced CREB phosphorylation is mediated by CaMKIV, NSsi- or CaMKIVsi-11-transfected cells were incubated with 7.5 μm NMDA or vehicle control for 2 min. As illustrated and quantified in Fig. 2B, CREB phosphorylation was increased 323% by NMDA as compared with vehicle control in NSsi-treated cells. No increase, however, was observed in CaMKIVsi-11-transfected cells. Together, the results shown in Fig. 2 indicate that phosphorylation of CREB and ATF1 elicited by Ca2+ influx via VDCC or NMDAR activities is CaMKIV-dependent within the examined time frames.
In the following series of experiments, we investigated the role of CaMKIV in the survival, proliferation, and differentiation of BE(2)C cells. Initially we assessed the effects of inhibition of the upstream activators of CaMKIV (i.e. CaMKK1 and/or CaMKK2), using the selective CaMKK inhibitor, STO-609 (47). As determined by BrdUrd labeling, CaMKK inhibition by STO-609 reduced the percentage of cycling cells from 42.3 ± 18.2 to 23.8 ± 8.1% (p < 0.001) (Fig. 3A). In addition, the overall proliferative rate was decreased by STO-609 over a 6-day period in culture (supplemental Fig. S3).
These results are consistent with the involvement of CaMKIV in maintaining continued BE(2)C cell cycling. However, CaMKI and AMPK are also documented targets of CaMKKs (48–51) and could be, at least in part, responsible for these effects. Therefore, we directly tested the role of CaMKIV in cell cycle progression. Cells were co-transfected with either NSsi or CaMKIVsi-11 and EGFP to identify siRNA transfectants. After a brief labeling period of 1 h, cycling cells were identified by BrdUrd immunocytochemistry. As shown in Fig. 3B, a high proportion of cells transfected with CaMKIVsi-11 failed to enter S-phase. Quantification of this effect is shown in Table 1. During this labeling period, the BrdUrd+EGFP+/BrdUrd−EGFP+ ratio for NSsi-transfected cells was 0.141 (102/725). However, for CaMKIVsi-11-transfected cells, this ratio was decreased to 0.045 (24/538), a 3-fold reduction (χ2 = 25.4, 1 degree of freedom, p < 0.001) confirming that silencing of CaMKIV expression promotes cell cycle arrest.
We also investigated the potential for CaMKIV to function in a prosurvival capacity in BE(2)C cells as in other cell types, as has been reported (31, 37, 52, 53). Silencing of CaMKIV expression in BE(2)C cells by RNAi increased the level of processed (cleaved) PARP and reduced the level of intact pro-caspase 3, events associated with the induction of apoptosis (Fig. 3C). Similarly, inhibition of CaMKK activity reduced cell viability (supplemental Fig. S4) and induced markers of apoptosis (data not shown). In addition, RNAi of CaMKIV reduced mRNA and protein levels of the prosurvival oncoprotein Bcl-2 (Fig. 3D), suggesting that CaMKIV-mediated transcriptional control of Bcl-2 expression may be a mechanism for promotion of cell survival. Together, these results are consistent with the CaMKIV cascade acting to maintain the capacity of undifferentiated BE(2)C cells for continued cell cycling.
As noted above, it is known that BE(2)C neuroblastoma cells will differentiate and assume a neuronal phenotype when cultured in the presence of low concentrations of RA (42–44). Since cell cycle exit accompanies terminal neuronal differentiation (54), we determined the effect of RA on BE(2)C cell cycle progression. Treatment of BE(2)C cells with 1 μm RA led to a marked decrease in BrdUrd incorporation during the labeling period (Fig. 4A). RA reduced the percentage of cycling cells from 42.3 ± 18.2 to 24.3 ± 12.01%, a 42.6% reduction (p < 0.005), indicating that RA promotes cell cycle exit. BE(2)C neuroblastoma cells in the absence of a differentiation-inducing stimulus typically grow as clumps of proliferating cells. However, the addition of RA resulted in their extending neuritic processes, which stained positively for the neuron-specific cytoskeletal protein βIII tubulin (Fig. 4B).
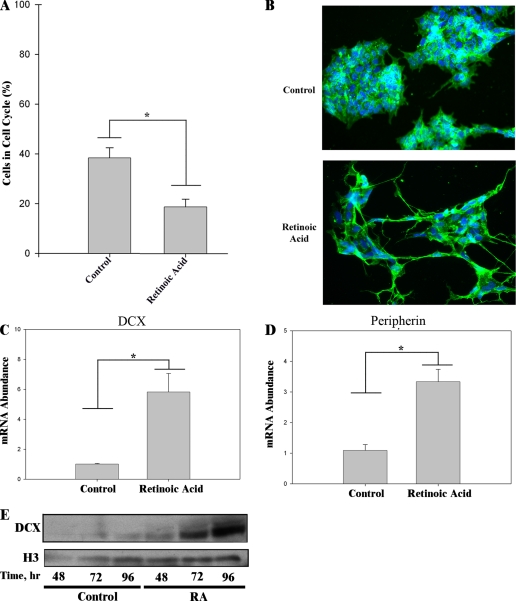
FIGURE 4.
RA induces neuronal differentiation of BE(2)C neuroblastoma cells. A, RA induction of neuronal differentiation is accompanied by cell cycle exit. BE(2)C cells were grown for 48 h in the absence (vehicle control) or presence of 1 μm RA. Percentages of cycling cells were determined as in the legend to Fig. 3A. *, p < 0.005. B, RA induces morphological changes characteristic of neuronal differentiation. BE(2)C cells were incubated the absence (vehicle control) or presence of 1 μm RA for 48 h. Cells were analyzed for βIII tubulin (green) by immunocytochemistry, nuclei were visualized with DAPI (blue), and images were acquired by fluorescence microscopy. C and D, RA induces markers of neuronal differentiation. BE(2)C cells were cultured in the absence (vehicle control) or presence of 1 μm RA for 168 h. Transcript levels of DCX (C) and peripherin (D) were quantified by qRT-PCR. *, p < 0.005. E, time course of induction of DCX protein expression by RA. BE(2)C cells were cultured for various time periods in the absence (vehicle control) or presence of 1 μm RA and analyzed by immunoblotting for expression of DCX. The blot shown was also probed for histone (H3) to confirm equivalent protein loading.
Besides RA-induced changes in cell physiology and morphology, we examined the expression of two neurogenesis-related genes, DCX and peripherin, to monitor the progress of neuronal differentiation. DCX is a neuron-specific microtubule-binding protein that has been employed as a marker of neurogenesis in both fetal and adult mammalian brain (55, 56) (reviewed in Ref. 54). Its expression sharply increases at around the time of cell cycle exit of progenitor cells in cortex and hippocampus. It is therefore used here as an early marker of neuronal differentiation. Induction of DCX mRNA or protein by RA in neuroblastoma cells has not been previously reported. Expression of the neuronal type III intermediate filament protein peripherin is also activated during neuronal differentiation (57). Cui et al. (43) studied the time course of increase of peripherin protein in BE(2)C cells during RA treatment. Increased peripherin protein was seen at 7 days, and a maximum was reached at ≥14 days. Since abundant neurite extension is seen at 48 h of RA treatment (Fig. 4B), peripherin is used here as a late marker of neuronal differentiation. RA induction of peripherin mRNA in neural cells has not been previously reported.
As shown in Fig. 4C, extended (7-day) treatment with RA led to increased expression of the mRNAs encoding both the early (DCX) and late (peripherin) neuronal markers. We also determined the time course of induction of DCX protein after RA addition. DCX protein, although essentially undetectable in the proliferative state, was detected by 72 h of RA treatment (Fig. 4E).
Figs. 1–4 document the presence in BE(2)C human neuroblastoma cells of two major signaling pathways, one Ca2+-dependent and involved in maintaining cells in a proliferative state and the other hormone-dependent with the capacity to terminate proliferation and induce differentiation. The ability of silencing of CaMKIV expression to phenocopy the effect of RA to promote cell cycle exit is consistent with antagonism between these two pathways at the molecular level.
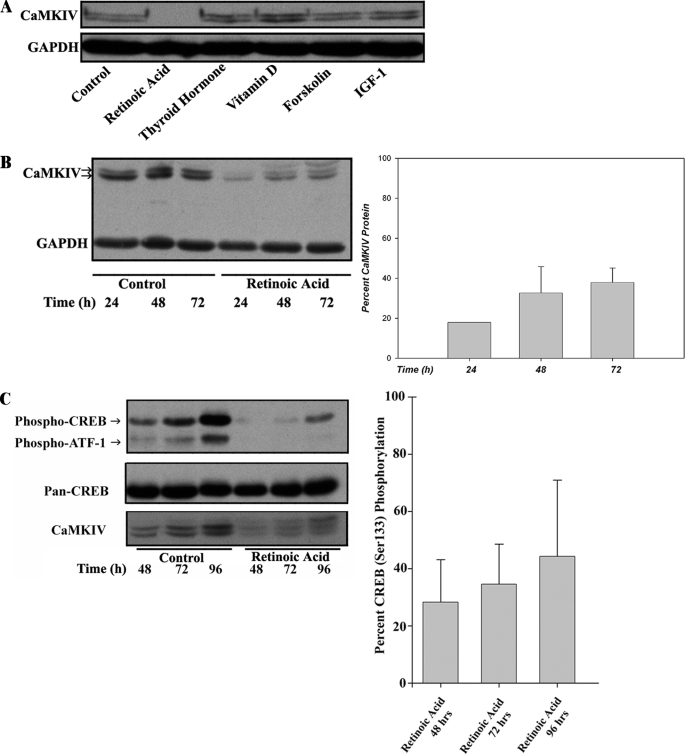
We therefore sought to determine whether such apparent antagonism occurs due to independent effects that are opposite in sign on downstream targets or whether it is due to direct negative interactions (cross-talk) between the two pathways. Although these two potential mechanisms are not mutually exclusive, strong evidence for the latter was obtained by the experiment illustrated in Fig. 5A. Treatment of cells with RA produced virtually complete down-regulation of CaMKIV protein expression, whereas a variety of other hormones and agonists capable of inducing differentiation in neuroblastoma cells (58–63) were without effect. By contrast to the prominent down-regulation of CaMKIV, treatment with RA for up to 96 h produced no detectable change in protein expression of another CaMKK-regulated protein AMPK, thereby confirming that the observed down-regulation of CaMKIV is not due to a transcriptionally nonspecific or otherwise toxic effect (data not shown). These results raise the possibility of a unique relationship between the RA and CaMKIV-mediated signaling pathways, a hypothesis that will be of interest to explore in future investigations.
FIGURE 5.
The CaMKIV signaling cascade is repressed by RA in BE(2)C neuroblastoma cells. A, a screen of neuronal differentiation-inducing agents reveals RA as a specific repressor of CaMKIV protein expression. BE(2)C cells were cultured for 7 days in the absence (vehicle control) or presence, as indicated, of the following: RA (10 μm), thyroid hormone (10 μm), vitamin D (dihydroxyvitamin D3) (10 μm), forskolin (10 μm), or insulin-like growth factor 1 (IGF-1; 100 ng/ml). Treated cells were analyzed by immunoblotting for expression of CaMKIV and GAPDH as a loading control. B, RA rapidly down-regulates CaMKIV protein expression. BE(2)C cells were cultured for various time periods in the absence (vehicle control) or presence of 1 μm RA and analyzed by immunoblotting for expression of CaMKIV and GAPDH as a loading control. Right, CaMKIV protein expression as a function of time of incubation in the presence of RA is quantified by scanning densitometry, normalized to GAPDH expression, and expressed as a percentage of normalized CaMKIV expression in the time-matched vehicle controls. C, RA down-regulates CREB/ATF-1 phosphorylation. BE(2)C cells were cultured for various time periods in the absence (vehicle control) or presence of 1 μm RA and analyzed by immunoblotting for CREB/ATF-1 phosphorylation and for expression of total CREB (Pan-CREB) and CaMKIV. Right, CREB phosphorylation was quantified by scanning densitometry, normalized to total CREB expression in RA-treated cells, and expressed as a percentage of normalized CREB phosphorylation in the respective time-matched vehicle controls.
We then determined the time course of CaMKIV down-regulation by RA. Down-regulation occurred rapidly with a maximal effect (∼80% reduction) achieved by 24 h of incubation with RA (Fig. 5B). Although this reduction in expression abated to some degree over time, CaMKIV protein was still reduced by ∼70% after 72 h of exposure to RA. The rapidity of this effect is to be contrasted with induction of DCX (itself an early marker of RA action), which did not achieve maximally induced levels until ≥96 h of RA treatment (Fig. 4E). This suggests that down-regulation of CaMKIV may represent an “immediate early” action of RA.
We verified that the down-regulation of CaMKIV by RA would lead to corresponding reductions in CREB and ATF1 phosphorylation. As shown in Fig. 5C, treatment with RA yielded reductions in CREB phosphorylation of ∼70% after 48 h or 72 h of incubation, similar to the percentage reduction of CaMKIV at these time points. Thus, RA-induced down-regulation of CaMKIV protein is reflected in lowered nuclear CREB phosphorylation.
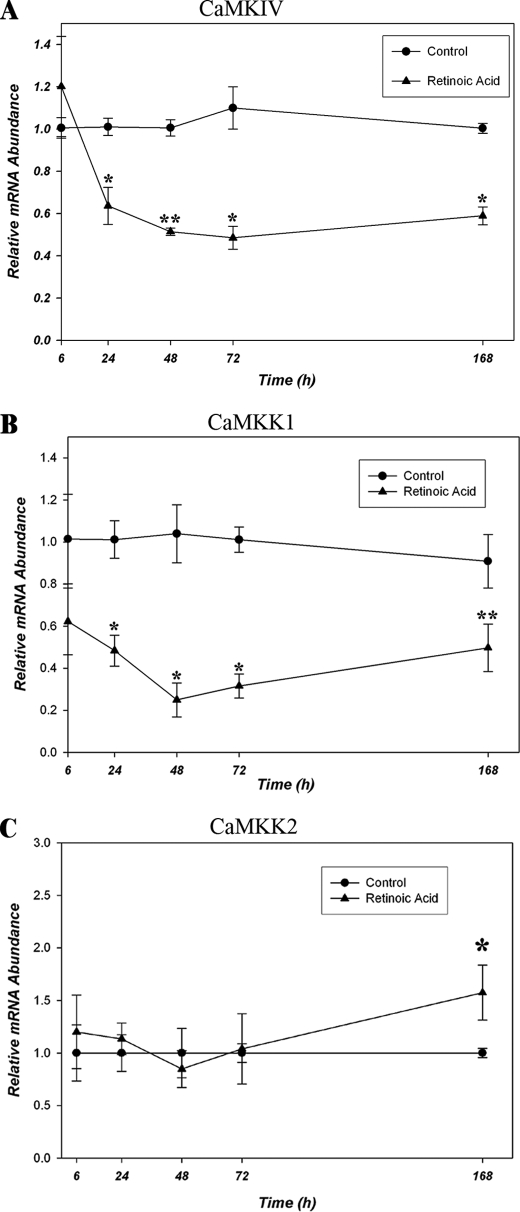
A potential mechanism by which CaMKIV protein might be decreased would be through inhibition of transcription. We therefore quantified CaMKIV mRNA levels in vehicle and RA-treated cells by qRT-PCR. RA had no effect at 6 h but produced significant decreases in CaMKIV mRNA abundance between 24 and 168 h of RA treatment (Fig. 6A).
FIGURE 6.
RA represses the CaMKIV signaling cascade at the mRNA level in BE(2)C neuroblastoma cells. A, RA down-regulates CaMKIV mRNA levels. BE(2)C cells were cultured for various time periods in the absence (vehicle control) or presence of 1 μm RA, following which lysates were prepared, and CaMKIV mRNA was quantified by qRT-PCR. CaMKIV mRNA concentrations are normalized to GAPDH at the respective time points. Relative mRNA abundance is defined as the concentration of CaMKIV mRNA in RA-treated cells divided by the respective time-matched vehicle control values. *, p < 0.001; **, p = 0.002. B, RA down-regulates CaMKK1 mRNA levels. CaMKK1 mRNA abundance in BE(2)C cells cultured in the absence (vehicle control) or presence of 1 μm RA was quantified by qRT-PCR and calculated as described in A. *, p < 0.001; **, p = 0.030. C, RA does not down-regulate CaMKK2 mRNA levels. CaMKK2 mRNA abundance in BE(2)C cells cultured in the absence (vehicle control) or presence of 1 μm RA was quantified by qRT-PCR and calculated as described in A. *, p = 0.010.
CaMKIV in the absence of CaMKK phosphorylation has ≤5% of the activity of its phosphorylated form (18). Therefore, in addition to RA regulation of its expression, it was theoretically possible that CaMKIV could also be regulated by RA-mediated decreases in CaMKK1 and/or CaMKK2 levels of expression. To examine this possibility and whether CaMKK expression over time is correlated with that of CaMKIV, CaMKK1 and CaMKK2 steady state mRNA levels were determined by qRT-PCR in cells incubated in the absence or presence of 1 μm RA over the same time period. A rapid and prominent down-regulation of CaMKK1 but not of CaMKK2 was observed (Fig. 6, B and C). These data indicate that RA down-regulation of CaMKK1 expression represents a potential mechanism for suppression of CaMKIV functional activity.
The stringent down-regulation of the CaMKIV cascade by RA raised the question of whether RA-dependent down-regulation of CaMKIV is in some way mechanistically linked to RA induction of neuronal differentiation. Hypothetically, by maintaining cells in a proliferative state, CaMKIV nuclear activity represents an impediment to RA-induced mitotic exit. In this scheme, RA-mediated suppression of CaMKIV activity, by removing a block to growth arrest, allows differentiation to proceed in an efficient manner. We therefore predicted that silencing of CaMKIV expression by RNAi prior to RA addition would, by mimicking an obligatory early action of RA, hasten RA-induced neuronal differentiation. To test this hypothesis, cells were transfected with NSsi or CaMKIVsi-1/11 for 24 h, after which they were treated with RA for an additional 24–168 h, and a gene expression versus time profile was generated by qRT-PCR (Fig. 7A).
FIGURE 7.
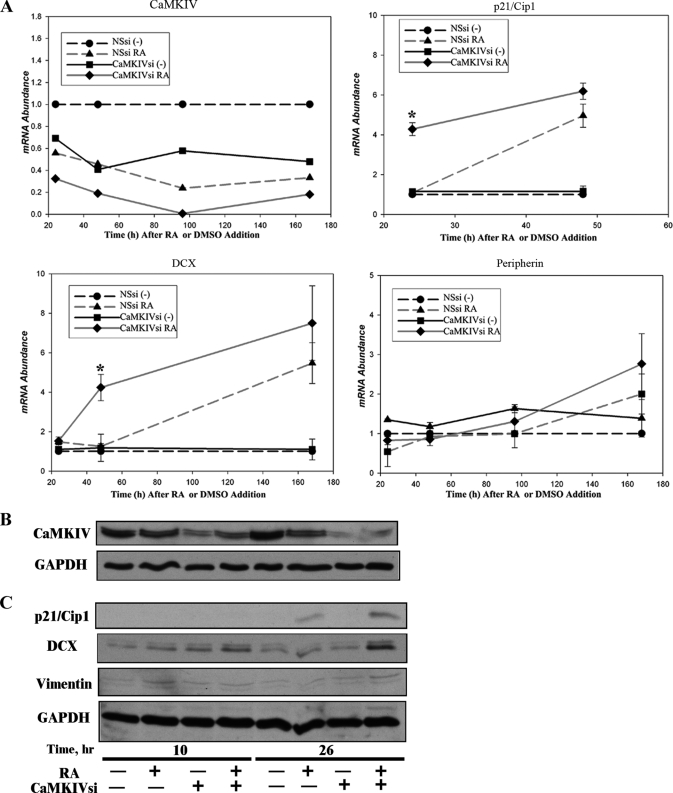
RNAi of CaMKIV accelerates RA induction of markers of neuronal differentiation in BE(2)C neuroblastoma cells. A, variation with time in mRNA levels of CaMKIV and differentiation markers by RNAi of CaMKIV, followed by RA treatment. BE(2)C cells were transfected for 24 h with 100 nm NSsi or 100 nm (total) CaMKIVsi-1/11, following which they were incubated in the absence (−; DMSO) or presence of 10 μm RA. At the indicated time points, mRNA abundances of CaMKIV, p21/Cip1, DCX, and peripherin were quantified by qRT-PCR. *, p < 0.05. B, variation with time in protein levels of CaMKIV by RNAi of CaMKIV followed by RA treatment. BE(2)C cells were transfected for 24 h with 100 nm NSsi or 100 nm (total) of CaMKIVsi-1/11, following which they were incubated in the absence (−; vehicle, DMSO), or presence (+), of 10 μm RA. At the indicated time points, protein levels of CaMKIV were analyzed by immunoblotting. The blot shown was also probed for GAPDH to confirm equivalent protein loading. C, variation with time in protein levels of differentiation markers by RNAi of CaMKIV followed by RA treatment. BE(2)C cells were treated as in B. At the indicated time points, protein levels of p21/Cip1, DCX, and vimentin were analyzed by immunoblotting. The blot shown was also probed for GAPDH to confirm equivalent protein loading.
First, we tested the effect of this regimen on CaMKIV mRNA levels. As shown by the plot in the upper left quadrant of Fig. 7A, RNAi of CaMKIV and RA treatment produced roughly equivalent reductions in CaMKIV mRNA at the 24 and 48 h time points. The treatments also showed additivity of effects, producing a more rapid drop in CaMKIV expression (to ∼20% of control levels) when combined than either treatment alone. We then proceeded to test the effects of this experimentally induced rapid drop in CaMKIV levels on the mRNA abundances of three genes associated with neuronal differentiation, p21/Cip1, DCX, and peripherin (Fig. 7A).
The G1 CDK inhibitor, p21/Cip1, a key regulator of neurogenesis, is reported to mediate RA-dependent cell cycle arrest in neuroprogenitor cells and therefore is expected to be among the first genes induced by RA (64). In control, CaMKIV-expressing cells, induction of p21/Cip1 mRNA required greater than 24 h of exposure to RA but eventually achieved a 4.96 ± 0.82-fold level of induction after 48 h. By contrast, in CaMKIV-silenced cells, the level of p21/Cip1 mRNA was elevated 4.28 ± 0.46-fold at the 24 h point and did not differ significantly from RA-treated, CaMKIV-expressing cells at the 48 h time point. In the absence of RA treatment, RNAi of CaMKIV had no effect on p21/Cip1 mRNA abundance at either time point. In similar fashion, RNAi of CaMKIV had no detectable effect on DCX mRNA levels in the absence of RA but significantly accelerated induction of DCX in its presence. As with p21/Cip1, the level of induction achieved by RA after an extended time of incubation did not differ significantly between CaMKIV-expressing or silenced cells.
As noted above, the intermediate filament protein peripherin is normally induced by RA in BE(2)C cells with a relatively slow time course, with increases detected by day 7 and a maximal response around day 14 (43). Peripherin mRNA showed a trend to be up-regulated after 7 days of RA treatment, although this did not achieve statistical significance at p < 0.05, a result consistent with a time course of RA induction of its expression being significantly delayed relative to those of p21/Cip1 and DCX (Fig. 7A). Similarly to the changes in mRNA levels, the protein levels of the early neuronal markers p21/Cip1 and DCX were increased at an early time point of 26 h of RA treatment when preceded by CaMKIV silencing compared with RA alone (Fig. 7, B and C).
Since multipotent BE(2)C cells are capable of non-neuronal differentiation, we also checked whether vimentin, a marker of neuroblastoma differentiation to a Schwann cell/melanocyte phenotype (42), was up-regulated by RA preceded by CaMKIV silencing. In contrast to the neuronal markers, RA did not up-regulate vimentin with or without silencing of CaMKIV expression (Fig. 7C). Thus, silencing of CaMKIV expression, although accelerating RA-initiated differentiation, does not alter its specification of a neuronal fate.
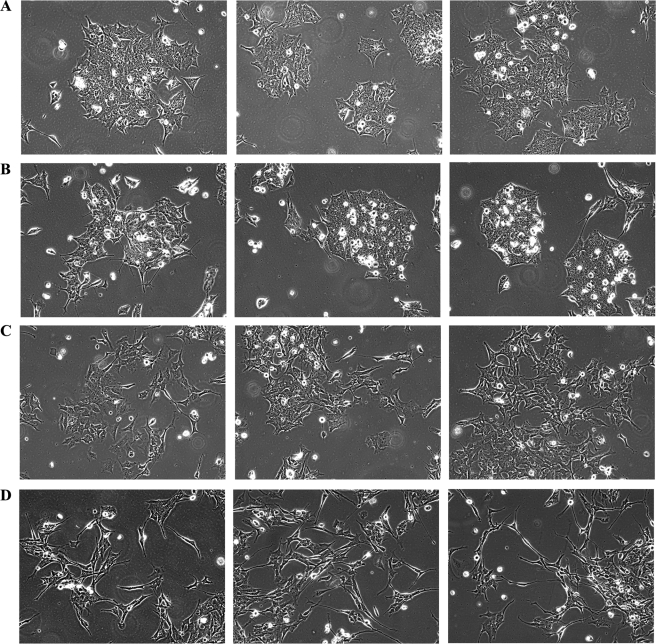
Finally, this paradigm was used to examine the effects of CaMKIV silencing on the morphology of BE(2)C cells during RA-initiated differentiation (Fig. 8). As predicted, silencing of CaMKIV expression did not, by itself, result in morphological differentiation. In the absence of RA, the cells continued to grow in clumps with little evidence of neuritic processes (Fig. 8, A and B). However, when the cells were treated with RA for 26 h, they appeared to lose cellular adhesiveness and began to extend neurites, and these effects were more prominent when RA addition was preceded by siRNA-induced silencing of CaMKIV expression (Fig. 8, C and D).
FIGURE 8.
RNAi of CaMKIV accelerates RA induction of a neuronal morphology of BE(2)C neuroblastoma cells. BE(2)C cells were transfected for 24 h with 100 nm NSsi or 100 nm (total) of CaMKIVsi-1/11. Cells were then incubated in the absence (vehicle; DMSO) or presence of 10 μm RA for an additional 26 h. Phase-contrast images were captured from cells treated with NSsi/vehicle (A), CaMKIVsi-1/11/vehicle (B), NSsi/RA (C), or CaMKIVsi-1/11/RA (D). Shown are three representative fields of cells per condition selected by an observer blind as to experimental conditions.
Taken, together, these results indicate that reduction in activity of the CaMKIV cascade is not by itself sufficient to trigger neuronal differentiation but rather creates an environment permissive for efficient RA-induced cell cycle exit and the initial steps of neuronal differentiation.
DISCUSSION
In this report, we describe the involvement of two signaling pathways in the decisions of stem cell-like BE(2)C cells to either continue to proliferate or to exit the cell cycle and differentiate. In one pathway, a signaling cascade initiated by Ca2+ influx through VDCCs or NMDARs culminates in nuclear CaMKIV-mediated phosphorylation and activation of CREB and ATF1 and is ultimately involved in maintenance of a proliferative state. Whereas in some respects, this pathway in BE(2)C neuroblastoma cells recapitulates elements of CaMKIV signaling in mature neurons or lymphocytes, in other respects, it appears to exhibit novel elements.
In mature hippocampal neurons, CaMKIV phosphorylation of CREB is transient (duration of ∼10–60 min) in response to Ca2+ influx evoked by K+ depolarization or high frequency stimulation (32, 65). Similarly, the response of BE(2)C neuroblastoma cells to NMDA treatment is also transient, being maximal at ∼2 min of treatment (supplemental Fig. S2). Surprisingly, however, silencing of CaMKIV expression in BE(2)C cells produced an 80% decrement of basal CREB phosphorylation over a 2-day period (Fig. 1D). Since, under these conditions, CREB phosphorylation is dependent upon the presence of extracellular Ca2+ and VDCC activity (Fig. 2), we hypothesize that in contrast to differentiated neurons, the maintenance of undifferentiated BE(2)C cells may rely on continuous Ca2+ influx and, at least in part, CaMKIV signaling. We speculate further that this mechanism could have been selected for during formation of the original tumor from which this cell line was ultimately derived.
Previous investigations in both neural and non-neural cell types have documented CaMKIV to be a mediator of cell survival. Apoptosis of cerebellar or spiral ganglia neurons due to withdrawal of trophic support may be counteracted by depolarization-induced Ca2+ influx. However, in the absence of either trophic support or depolarizing medium, neuronal viability can still be maintained by transfection of CA-CaMKIV (52, 53, 66, 67). In transgenic mice expressing catalytically inactive CaMKIV, there is a reduction in the number of thymic T lymphocytes and in T cell survival (31). In CaMKIV null mice, there is a significant loss of hematopoietic stem cells and cerebellar Purkinje neurons (37, 41). In Xenopus, decreased survival of erythroid precursors is observed upon inactivation of CaMKIV (39).
The present data reinforce the association of CaMKIV with cell survival. RNAi of CaMKIV activated pro-caspase-3 processing and PARP degradation and decreased levels of Bcl-2 mRNA and protein (Fig. 3, C and D). This association is also supported by the dose-dependent decrease in cell viability and increased pro-caspase-3 processing and PARP degradation by the action of the selective CaMKK inhibitor STO-609 (supplemental Fig. S4) (data not shown).
However, beyond a role in survival, possible association of CaMKIV with events underlying cell cycle exit and differentiation has received relatively little experimental attention. Kitsos et al. (37) reported that CaMKIV-null hematopoietic progenitor cells proliferate at an aberrantly higher rate than wild type cells but then ultimately suffer exhaustion. Also, mitochondrial dysfunction-induced hypoproliferation in a mouse fibroblast cell line was associated with CaMKIV phosphorylation of CREB (68).
We find that in human neuroblastoma cells, the CaMKIV cascade functions as a positive regulator of proliferation. This is shown by the cell cycle arrest and hypoproliferation produced by RNAi of CaMKIV, reduction in proliferation by CaMKK inhibition, and the effect of silencing of CaMKIV expression to accelerate RA-dependent up-regulation of the cell cycle inhibitor p21/Cip1. It seems, therefore, that the role of CaMKIV depends on both the tissue of origin and the state of differentiation of the particular cell type under study. In sum, however, the present evidence supports a role for CaMKIV in maintenance of BE(2)C cells through both promotion of survival and cell cycle re-entry, although their proportional contributions to continued optimal proliferation remain to be established.
As a prominent nuclear CaMKIV target, CREB phosphorylation is used here as a sensitive readout of the activity status of nuclear CaMKIV. However, we have not specifically addressed whether CREB mediates CaMKIV modulation of survival and/or proliferation. CREB is well established as playing an important role in neuronal survival, an effect that may be mediated by Bcl-2 among other effectors (69, 70). CREB may also support cell proliferation in other cell types, as was demonstrated in pituitary somatotrophs, Schwann cells, and chondrocytes (71–73). It would be of interest in subsequent studies to explore the role of CREB in survival/proliferation of NSCs and neural progenitors. Such a role is suggested here by the prominent and sustained suppression of CREB phosphorylation during the early phase of RA-induced cell cycle exit and neuronal differentiation (Fig. 5C).
Parallel to this Ca2+-initiated pathway is an RA-mediated pathway for the promotion of neuronal differentiation. This involves RA-dependent up-regulation of the cell cycle inhibitor p21/Cip1, cell cycle exit, induction of the neuroblastic marker DCX and the neuron-specific intermediate filament protein peripherin, and neurite extension. A key finding of this study is that RA employs multiple mechanisms for repressing the CaMKIV pathway, presumably as a means of removing a proliferative pathway that would act as an impediment to efficient induction of neuronal differentiation. The evidence for this is as follows.
First, RA produces pronounced and sustained down-regulation in the levels of CaMKIV mRNA and protein (Figs. 5–7). The specificity of this effect is shown by its absence when BE(2)C cells were incubated with a variety of other hormones reported to be differentiation-inducing in neuroblastoma cells (57–63). Strikingly, this was the case even with dihydroxyvitamin D3 and insulin-like growth factor 1, which reportedly synergize with RA in the induction of differentiation (59, 63). It is also intriguing that there are relatively few targets reported to be repressed by RA, in comparison with the number of targets known to be induced (74).
Second, RA rapidly down-regulates CaMKK1 mRNA with a time course similar to that of CaMKIV down-regulation (Fig. 7). This finding represents a novel mechanism for regulatory control of CaMKIV activity. Since phosphorylation of CaMKIV results in ≥20-fold enhancement of its activity, modulation of CaMKK expression represents, in principle, a point at which regulatory influence could be effectively exerted on its downstream target, CaMKIV (18). The correspondence in the time courses of CaMKIV and CaMKK1 mRNA repression in response to RA without such down-regulation of CaMKK2 mRNA (Fig. 6) also suggests that activating phosphorylation of CaMKIV in BE(2)C neuroblastoma cells may be regulated by CaMKK1 in preference to CaMKK2, although there remains the possibility of functional redundancy of CaMKKs in activation of CaMKIV in these cells.
There have been several previous investigations of the potential for regulation of the CaMKIV cascade at the mRNA level. Liu and Brent (36, 75) demonstrated that in mouse embryonic stem cells differentiated into neurons by thyroid hormone, CaMKIV mRNA was increased ∼5-fold, but there was no change in response to RA. Similarly, Krebs et al. (76) reported that in fetal rat telencephalic neurons, CaMKIV mRNA and protein are induced by thyroid hormone but not by RA. On the other hand, in a non-neural system, maturing neutrophils (MPRO cells), RA reduces CaMKIV mRNA in concordance with the results reported here but simultaneously up-regulates CaMKK1 (35). These divergent results suggest that control of expression of CaMKIV cascade members may subserve different functions in different species, cell types, and developmental stages.
We also examined the directionality of the interaction between the RA and CaMKIV pathways. Contrary to the results presented here, a number of independent studies are consistent with CaMKIV and RA being positive co-effectors in signaling gene expression. These include stimulatory effects of CA-CaMKIV in gene reporter assays on the transcriptional activities of retinoic acid receptor, thyroid hormone receptor, vitamin D receptor, and the orphan receptors RORα2, RORγ, and COUP-TF1 (77–81). We have replicated in BE(2)C cells the effect of CA-CaMKIV to increase RA-response element luciferase activity. However, neither RNAi of endogenous CaMKIV nor inhibition of endogenous CaMKKs by STO-609 inhibited RA-induced RA-response element luciferase activity (data not shown). These results suggest that at least in BE(2)C cells, CaMKIV does not positively influence RA signaling. The basis for effects of CA-CaMKIV to stimulate transcription in reporter gene assays may rest with an ability of nuclear active CaMKIV at perhaps supraphysiological levels to stimulate export from the nucleus of Class II histone deacetylases, leading to corepressor dissociation from a variety of hormone-responsive transcriptional elements (77–80).
In adult neural stem cells, RA acts early in differentiation to specify a neuronal fate in addition to provoking cell cycle exit by up-regulating p21/Cip1 (64). We show here that silencing of CaMKIV expression does not alter lineage commitment, since there was no effect of CaMKIV silencing on vimentin expression (Fig. 7C) (i.e. it did not lead to specification of a Schwann/melanocyte fate). Rather, we infer from our results that the effect of CaMKIV silencing ahead of RA treatment is to mimic and thereby make more efficient a mechanism by which RA normally facilitates differentiation by down-regulating opposing mechanisms driving cell cycle progression. Such a mechanism could have relevance for decisions of other cell types to re-enter the cell cycle or to exit and differentiate.
Supplementary Material
Acknowledgments
We gratefully acknowledge Loukia Karacosta for scoring of BrdUrd labeling, Gary Wayman and Thomas Soderling for providing CA-CaMKIV, M. K. Stachowiak for a gift of pcDNA3.1-EGFP, and James Boyer and Elaine Goldstein for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R21NS050385.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- NSC
- neural stem cell
- BrdUrd
- bromodeoxyuridine
- CaMKIV
- Ca2+/calmodulin-dependent kinase IV
- CaMKK
- CaMK kinase
- CRE
- Ca2+/cAMP-response element
- CREB
- CRE-binding protein
- DCX
- doublecortin
- EGFP
- enhanced green fluorescent protein
- NMDA
- N-methyl-d-aspartate
- NMDAR
- NMDA receptor
- RA
- all-trans-retinoic acid
- VDCC
- voltage-dependent calcium channel
- CaM
- calmodulin
- PARP
- poly(ADP)-ribose polymerase
- PBS
- phosphate-buffered saline
- DAPI
- 4′,6-diamidino-2-phenylindole
- qRT
- quantitative reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- RNAi
- RNA interference
- siRNA
- small interfering RNA.
REFERENCES
- 1.Bloodgood B. L., Sabatini B. L. (2007) Curr. Opin. Neurobiol. 17, 345–351 [DOI] [PubMed] [Google Scholar]
- 2.Zheng J. Q., Poo M. M. (2007) Annu. Rev. Cell Dev. Biol. 23, 375–404 [DOI] [PubMed] [Google Scholar]
- 3.Flavell S. W., Greenberg M. E. (2008) Annu. Rev. Neurosci. 31, 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owens D. F., Kriegstein A. R. (1998) J. Neurosci. 18, 5374–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs E., Gould E. (2000) Eur. J. Neurosci. 12, 2211–2214 [DOI] [PubMed] [Google Scholar]
- 6.Haydar T. F., Wang F., Schwartz M. L., Rakic P. (2000) J. Neurosci. 20, 5764–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owens D. F., Flint A. C., Dammerman R. S., Kriegstein A. R. (2000) Dev. Neurosci. 22, 25–33 [DOI] [PubMed] [Google Scholar]
- 8.Deisseroth K., Singla S., Toda H., Monje M., Palmer T. D., Malenka R. C. (2004) Neuron 42, 535–552 [DOI] [PubMed] [Google Scholar]
- 9.Weissman T. A., Riquelme P. A., Ivic L., Flint A. C., Kriegstein A. R. (2004) Neuron 43, 647–661 [DOI] [PubMed] [Google Scholar]
- 10.Tozuka Y., Fukuda S., Namba T., Seki T., Hisatsune T. (2005) Neuron 47, 803–815 [DOI] [PubMed] [Google Scholar]
- 11.Colomer J., Means A. R. (2007) Subcell. Biochem. 45, 169–214 [DOI] [PubMed] [Google Scholar]
- 12.Wayman G. A., Lee Y. S., Tokumitsu H., Silva A., Soderling T. R. (2008) Neuron 59, 914–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen K. F., Ohmstede C. A., Fisher R. S., Sahyoun N. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2850–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tokumitsu H., Enslen H., Soderling T. R. (1995) J. Biol. Chem. 270, 19320–19324 [DOI] [PubMed] [Google Scholar]
- 15.Edelman A. M., Mitchelhill K. I., Selbert M. A., Anderson K. A., Hook S. S., Stapleton D., Goldstein E. G., Means A. R., Kemp B. E. (1996) J. Biol. Chem. 271, 10806–10810 [DOI] [PubMed] [Google Scholar]
- 16.Anderson K. A., Means R. L., Huang Q. H., Kemp B. E., Goldstein E. G., Selbert M. A., Edelman A. M., Fremeau R. T., Means A. R. (1998) J. Biol. Chem. 273, 31880–31889 [DOI] [PubMed] [Google Scholar]
- 17.Kitani T., Okuno S., Fujisawa H. (1997) J. Biochem. 122, 243–250 [DOI] [PubMed] [Google Scholar]
- 18.Selbert M. A., Anderson K. A., Huang Q. H., Goldstein E. G., Means A. R., Edelman A. M. (1995) J. Biol. Chem. 270, 17616–17621 [DOI] [PubMed] [Google Scholar]
- 19.Chow F. A., Anderson K. A., Noeldner P. K., Means A. R. (2005) J. Biol. Chem. 280, 20530–20538 [DOI] [PubMed] [Google Scholar]
- 20.Tokumitsu H., Hatano N., Inuzuka H., Yokokura S., Nozaki N., Kobayashi R. (2004) J. Biol. Chem. 279, 40296–40302 [DOI] [PubMed] [Google Scholar]
- 21.Westphal R. S., Anderson K. A., Means A. R., Wadzinski B. E. (1998) Science 280, 1258–1261 [DOI] [PubMed] [Google Scholar]
- 22.Lemrow S. M., Anderson K. A., Joseph J. D., Ribar T. J., Noeldner P. K., Means A. R. (2004) J. Biol. Chem. 279, 11664–11671 [DOI] [PubMed] [Google Scholar]
- 23.Kotera I., Sekimoto T., Miyamoto Y., Saiwaki T., Nagoshi E., Sakagami H., Kondo H., Yoneda Y. (2005) EMBO J. 24, 942–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun P., Enslen H., Myung P. S., Maurer R. A. (1994) Genes Dev. 8, 2527–2539 [DOI] [PubMed] [Google Scholar]
- 25.Enslen H., Sun P., Brickey D., Soderling S. H., Klamo E., Soderling T. R. (1994) J. Biol. Chem. 269, 15520–15527 [PubMed] [Google Scholar]
- 26.Matthews R. P., Guthrie C. R., Wailes L. M., Zhao X., Means A. R., McKnight G. S. (1994) Mol. Cell Biol. 14, 6107–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun P., Lou L., Maurer R. A. (1996) J. Biol. Chem. 271, 3066–3073 [DOI] [PubMed] [Google Scholar]
- 28.Chawla S., Hardingham G. E., Quinn D. R., Bading H. (1998) Science 281, 1505–1509 [DOI] [PubMed] [Google Scholar]
- 29.Impey S., Fong A. L., Wang Y., Cardinaux J. R., Fass D. M., Obrietan K., Wayman G. A., Storm D. R., Soderling T. R., Goodman R. H. (2002) Neuron 34, 235–244 [DOI] [PubMed] [Google Scholar]
- 30.Bito H., Deisseroth K., Tsien R. W. (1996) Cell 87, 1203–1214 [DOI] [PubMed] [Google Scholar]
- 31.Anderson K. A., Ribar T. J., Illario M., Means A. R. (1997) Mol. Endocrinol. 11, 725–737 [DOI] [PubMed] [Google Scholar]
- 32.Wu G. Y., Deisseroth K., Tsien R. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 2808–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen S., Greenberg M. E. (2008) Annu. Rev. Cell Dev. Biol. 24, 183–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S. L., Ribar T. J., Means A. R. (2001) Cell Growth Differ. 12, 351–361 [PubMed] [Google Scholar]
- 35.Lawson N. D., Zain M., Zibello T., Picciotto M. R., Nairn A. C., Berliner N. (1999) Exp. Hematol. 27, 1682–1690 [DOI] [PubMed] [Google Scholar]
- 36.Liu Y. Y., Brent G. A. (2002) Mol. Endocrinol. 16, 2439–2451 [DOI] [PubMed] [Google Scholar]
- 37.Kitsos C. M., Sankar U., Illario M., Colomer-Font J. M., Duncan A. W., Ribar T. J., Reya T., Means A. R. (2005) J. Biol. Chem. 280, 33101–33108 [DOI] [PubMed] [Google Scholar]
- 38.Sato K., Suematsu A., Nakashima T., Takemoto-Kimura S., Aoki K., Morishita Y., Asahara H., Ohya K., Yamaguchi A., Takai T., Kodama T., Chatila T. A., Bito H., Takayanagi H. (2006) Nat. Med. 12, 1410–1416 [DOI] [PubMed] [Google Scholar]
- 39.Wayman G. A., Walters M. J., Kolibaba K., Soderling T. R., Christian J. L. (2000) J. Cell Biol. 151, 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu J. Y., Ribar T. J., Cummings D. E., Burton K. A., McKnight G. S., Means A. R. (2000) Nat. Genet. 25, 448–452 [DOI] [PubMed] [Google Scholar]
- 41.Ribar T. J., Rodriguiz R. M., Khiroug L., Wetsel W. C., Augustine G. J., Means A. R. (2000) J. Neurosci. 20, RC107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross R. A., Spengler B. A., Domènech C., Porubcin M., Rettig W. J., Biedler J. L. (1995) Cell Growth Differ. 6, 449–456 [PubMed] [Google Scholar]
- 43.Cui H., Ma J., Ding J., Li T., Alam G., Ding H.F. (2006) J. Biol. Chem. 281, 34696–34704 [DOI] [PubMed] [Google Scholar]
- 44.Ross R. A., Spengler B. A. (2007) Semin. Cancer Biol. 17, 241–247 [DOI] [PubMed] [Google Scholar]
- 45.Anderson K. A., Noeldner P. K., Reece K., Wadzinski B. E., Means A. R. (2004) J. Biol. Chem. 279, 31708–31716 [DOI] [PubMed] [Google Scholar]
- 46.Yamamori E., Asai M., Yoshida M., Takano K., Itoi K., Oiso Y., Iwasaki Y. (2004) J. Mol. Endocrinol. 33, 639–649 [DOI] [PubMed] [Google Scholar]
- 47.Tokumitsu H., Inuzuka H., Ishikawa Y., Ikeda M., Saji I., Kobayashi R. (2002) J. Biol. Chem. 277, 15813–15818 [DOI] [PubMed] [Google Scholar]
- 48.Aletta J. M., Selbert M. A., Nairn A. C., Edelman A. M. (1996) J. Biol. Chem. 271, 20930–20934 [DOI] [PubMed] [Google Scholar]
- 49.Matsushita M., Nairn A. C. (1999) J. Biol. Chem. 274, 10086–10093 [DOI] [PubMed] [Google Scholar]
- 50.Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 51.Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., Witters L. A. (2005) J. Biol. Chem. 280, 29060–29066 [DOI] [PubMed] [Google Scholar]
- 52.Sée V., Boutillier A. L., Bito H., Loeffler J. P. (2001) FASEB J. 15, 134–144 [DOI] [PubMed] [Google Scholar]
- 53.Hansen M. R., Bok J., Devaiah A. K., Zha X. M., Green S. H. (2003) J. Neurosci. Res. 72, 169–184 [DOI] [PubMed] [Google Scholar]
- 54.Kempermann G., Jessberger S., Steiner B., Kronenberg G. (2004) Trends Neurosci. 27, 447–452 [DOI] [PubMed] [Google Scholar]
- 55.Francis F., Koulakoff A., Boucher D., Chafey P., Schaar B., Vinet M. C., Friocourt G., McDonnell N., Reiner O., Kahn A., McConnell S. K., Berwald-Netter Y., Denoulet P., Chelly J. (1999) Neuron 23, 247–256 [DOI] [PubMed] [Google Scholar]
- 56.Gleeson J. G., Lin P. T., Flanagan L. A., Walsh C. A. (1999) Neuron 23, 257–271 [DOI] [PubMed] [Google Scholar]
- 57.Gorham J. D., Baker H., Kegler D., Ziff E. B. (1990) Dev. Brain Res. 57, 235–248 [DOI] [PubMed] [Google Scholar]
- 58.Ishii K., Adachi Y., Hatanaka M., Sakamoto H., Furuyama J. (1990) J. Cell Physiol. 143, 569–576 [DOI] [PubMed] [Google Scholar]
- 59.Moore T. B., Sidell N., Chow V. J., Medzoyan R. H., Huang J. I., Yamashiro J. M., Wada R. K. (1995) J. Pediatr. Hematol. Oncol. 17, 311–317 [DOI] [PubMed] [Google Scholar]
- 60.Ammer H., Schulz R. (1997) Neurosci. Lett. 230, 143–146 [DOI] [PubMed] [Google Scholar]
- 61.Kim B., Leventhal P. S., Saltiel A. R., Feldman E. L. (1997) J. Biol. Chem. 272, 21268–21273 [DOI] [PubMed] [Google Scholar]
- 62.Perez-Juste G., Aranda A. (1999) J. Biol. Chem. 274, 5026–5031 [DOI] [PubMed] [Google Scholar]
- 63.Perez-Juste G., Aranda A. (1999) Oncogene 18, 5393–53402 [DOI] [PubMed] [Google Scholar]
- 64.Takahashi J., Palmer T. D., Gage F. H. (1999) J. Neurobiol. 38, 65–81 [PubMed] [Google Scholar]
- 65.Kasahara J., Fukunaga K., Miyamoto E. (2001) J. Biol. Chem. 276, 24044–24050 [DOI] [PubMed] [Google Scholar]
- 66.Marshall J., Dolan B. M., Garcia E. P., Sathe S., Tang X., Mao Z., Blair L. A. (2003) Neuron 39, 625–639 [DOI] [PubMed] [Google Scholar]
- 67.Pérez-García M. J., Gou-Fabregas M., de Pablo Y., Llovera M., Comella J. X., Soler R. M. (2008) J. Biol. Chem. 283, 4133–4144 [DOI] [PubMed] [Google Scholar]
- 68.Arnould T., Vankoningsloo S., Renard P., Houbion A., Ninane N., Demazy C., Remacle J., Raes M. (2002) EMBO J. 21, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Riccio A., Ahn S., Davenport C. M., Blendy J. A., Ginty D. D. (1999) Science 286, 2358–2361 [DOI] [PubMed] [Google Scholar]
- 70.Lonze B. E., Ginty D. D. (2002) Neuron 35, 605–623 [DOI] [PubMed] [Google Scholar]
- 71.Struthers R. S., Vale W. W., Arias C., Sawchenko P. E., Montminy M. R. (1991) Nature 350, 622–624 [DOI] [PubMed] [Google Scholar]
- 72.Lee M. M., Badache A., DeVries G. H. (1999) J. Neurosci. Res. 55, 702–712 [DOI] [PubMed] [Google Scholar]
- 73.Long F., Schipani E., Asahara H., Kronenberg H., Montminy M. (2001) Development 128, 541–550 [DOI] [PubMed] [Google Scholar]
- 74.Maden M. (2007) Nat. Rev. Neurosci. 8, 755–765 [DOI] [PubMed] [Google Scholar]
- 75.Liu Y. Y., Brent G. A. (2005) Endocrinology 146, 776–783 [DOI] [PubMed] [Google Scholar]
- 76.Krebs J., Means R. L., Honegger P. (1996) J. Biol. Chem. 271, 11055–11058 [DOI] [PubMed] [Google Scholar]
- 77.Kane C. D., Means A. R. (2000) EMBO J. 19, 691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKinsey T. A., Zhang C. L., Lu J., Olson E. N. (2000) Nature 408, 106–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miska E. A., Langley E., Wolf D., Karlsson C., Pines J., Kouzarides T. (2001) Nucleic Acids Res. 29, 3439–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McKenzie G. J., Stevenson P., Ward G., Papadia S., Bading H., Chawla S., Privalsky M., Hardingham G. E. (2005) J. Neurochem. 93, 171–185 [DOI] [PubMed] [Google Scholar]
- 81.Sucharov C. C., Langer S., Bristow M., Leinwand L. (2006) Am. J. Physiol. Cell Physiol. 291, C1029–C1037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.