Abstract
Production of major diterpenoid phytoalexins, momilactones and phytocassanes, is induced in rice upon recognition of pathogenic invasion as plant defense-related compounds. We recently showed that biosynthetic genes for momilactones are clustered on rice chromosome 4 and co-expressed after elicitation, mimicking pathogen attack. Because genes for most metabolic pathways in plants are not organized in gene clusters, examination of the mechanism(s) regulating the expression of such clustered genes is needed. Here, we report a chitin oligosaccharide elicitor-inducible basic leucine zipper transcription factor, OsTGAP1, which is essential for momilactone biosynthesis and regulates the expression of the five genes in the cluster. The knock-out mutant for OsTGAP1 had almost no expression of the five clustered genes (OsCPS4, OsKSL4, CYP99A2, CYP99A3, and OsMAS) or production of momilactones upon elicitor treatment. Inductive expression of OsKSL7 for phytocassane biosynthesis was also largely affected in the ostgap1 mutant, although phytocassane accumulation still occurred. Conversely, OsTGAP1-overexpressing lines exhibited enhanced expression of the clustered genes and hyperaccumulation of momilactones in response to the elicitor. Furthermore, enhanced expression of OsKSL7 and hyperaccumulation of phytocassanes was also observed. We also found that OsTGAP1 overexpression can influence transcriptional up-regulation of OsDXS3 in the methylerythritol phosphate pathway, eventually leading to inductive production of diterpenoid phytoalexins. These results indicate that OsTGAP1 functions as a key regulator of the coordinated transcription of genes involved in inductive diterpenoid phytoalexin production in rice and mainly exerts an essential role on expression of the clustered genes for momilactone biosynthesis.
Plants attacked by pathogenic microorganisms respond with a variety of defensive reactions, including the production of antimicrobial secondary metabolites known as phytoalexins (1). These compounds are transiently generated in response to signal molecules called elicitors, which are usually derived from pathogens. In rice, 14 diterpenoid phytoalexins have been identified to date in suspension-cultured cells treated with biotic elicitors, such as chitin oligosaccharide, and in leaves infected with the blast fungus Magnaporthe grisea or irradiated with UV light. Phytoalexins can be classified into four groups, based on the structure of their hydrocarbon precursors: phytocassanes A–E, oryzalexins A–F, momilactones A and B, and oryzalexin S (2–10).
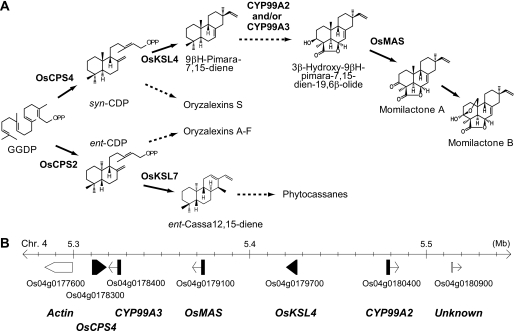
Geranylgeranyl diphosphate, a common precursor of diterpenoid phytoalexins, is first cyclized by OsCPS2/OsCyc2 and OsCPS4/OsCyc1 to yield ent-copalyl diphosphate and syn-copalyl diphosphate (11, 12). Next, OsKSL7/OsDTC1, OsKSL10, OsKSL4, and OsKSL8/OsDTC2 catalyze the second cyclization of ent-copalyl diphosphate or syn-copalyl diphosphate to four distinct diterpene hydrocarbons: ent-cassa-12,15-diene, ent-sandaracopimaradiene, 9βH-pimara-7,15-diene, and stemar-13-ene, respectively (13–15). For momilactone biosynthesis, two cytochrome P450 monooxygenase (P450) genes (CYP99A2 and CYP99A3) and a dehydrogenase gene (OsMAS) are also involved in the downstream oxidation steps of the diterpene hydrocarbons. Intriguingly, five genes for momilactone biosynthesis are localized in a narrow region of chromosome 4, creating a functional gene cluster (Fig. 1) (16).
FIGURE 1.
A, proposed biosynthetic pathway for the diterpenoid phytoalexins in rice. The enzymes involved in momilactone and phytocassane biosyntheses are indicated. The dashed arrows show steps required for multiple reactions. B, map of the momilactone biosynthetic gene cluster on rice chromosome 4. The black boxes indicate the five momilactone biosynthetic genes.
Genes for most metabolic pathways in plants are not organized in gene clusters. However, there are some examples of gene clusters for the biosynthesis of isoprenoids and plant defense compounds other than momilactones in rice. For example, the genes responsible for the biosynthesis of thalianol in Arabidopsis, benzoxazinoids in maize, and avenacins in oat are all organized in clusters (17–19). The spatial expression of the avenacin biosynthetic genes is tightly regulated and occurs only in the root epidermis, the site of accumulation of the end product (20). In contrast, the rice momilactone biosynthetic gene cluster exhibits a temporally coordinated expression pattern of mRNAs, peaking at 6–12 h after elicitor treatment of rice cell suspension cultures (11, 21). Such coordinated, stress-inducible clustered gene expression, responsible for the biosynthesis of one particular compound, has not previously been reported in plants. Thus, it is interesting to examine the mechanism(s) regulating the coordinated expression of the momilactone biosynthetic gene cluster. Here, we report the identification of an elicitor-inducible basic leucine zipper (bZIP)3 transcription factor, OsTGAP1, which is essential for the biosynthesis of momilactone in rice and which coordinately regulates the expression of all five genes in the cluster. We also show that OsTGAP1 can influence the expression of a phytocassane biosynthetic gene and the upstream methylerythritol phosphate (MEP) pathway gene, leading to production of phytocassanes. These results indicate that OsTGAP1 functions as a key regulator of the coordinated transcription of genes involved in inductive diterpenoid phytoalexins production in rice.
EXPERIMENTAL PROCEDURES
Plant Material and Chemical Treatment
Purified chitooctaose (Yaizu Suisankagaku Industry Co., Ltd., Tokyo, Japan) was re-N-acetylated to give N-acetylchitooctaose, as described (22), and used as a chitin oligosaccharide elicitor throughout this study. Oryza sativa L. cv. Nipponbare was used as the wild type. Calli of O. sativa L. cv. Nipponbare were cultured as described previously (13). Six days after transfer to fresh culture medium, the cultured rice cells were used for assays with a chitin oligosaccharide elicitor treatment (N-acetylchitooctaose, 1 ppm) throughout this study. Tos17-inserted mutants of H0155 (for AK073715) and NC0005 (for AK102690) were obtained from the National Institute of Agrobiological Sciences of Japan (Tos17 mutant panel project; available on the World Wide Web). The insertion site of Tos17 is indicated in supplemental Fig. S2A. The Tos17 homozygous mutants were selected by PCR genotyping, and generated cultured cells of the mutant were used for the experiments.
Cloning and Transformation
The OsKSL4 promoter fragments were amplified by PCR, using genomic DNA prepared from suspension-cultured rice cells (cv. Nipponbare) as template. The mutated constructs were generated via a two-step PCR process (primer overlapping mutagenesis), and the core TGAC sequence was converted to CCTA. The OsKSL4 promoter deletions and mutated constructs were cloned into the KpnI and XhoI sites of the pGL3 basic vector (Promega, Madison, WI) containing the firefly luciferase gene (LUC).
For transactivation assays, the OsTGAP1 coding sequence was amplified using OsTGAP1 cDNA obtained from the Rice Genome Resource Center. The OsTGAP1 cDNA was fused to the DNA-binding domain (1–147 amino acids) of the yeast transcription factor GAL4 (GAL4-DBD) to prepare the effector construct. LUC, which is under the control of a promoter containing six repeats of the GAL4 binding site (6× UAS), was used as a reporter (23). For the production of recombinant GST-OsTGAP1 protein and the generation of T7-OsTGAP1-overexpressing plants, the OsTGAP1 coding sequence was cloned into pENTR/D/Topo (Invitrogen). For protein purification, the entry clone was confirmed by sequencing, recombined (with Invitrogen LR GATEWAY recombinase) into pDEST15 (Invitrogen), and introduced into Escherichia coli strain Rosetta (DE3). The entry clone was also recombined into pGWB27 (24) and used for generating transgenic rice cells by Agrobacterium-mediated transformation, as described by Kaku et al. (25). Overexpression of T7-OsTGAP1 was confirmed by qRT-PCR and immunoblot analysis with an anti-T7 antibody (Chemicon, Temecula, CA).
Expression Analysis
RT-PCR and qRT-PCR were used to confirm the levels of expression. For the RT reaction, a SuperScript reverse transcriptase kit (Invitrogen) and Quantitect kit (Qiagen K.K., Tokyo, Japan) were used. qRT-PCR was performed using SYBR Green technology on an ABI PRISM 7300 real time PCR system (Applied Biosystems, Foster City, CA). Raw data from qRT-PCR were analyzed using the ΔCT (difference in threshold cycles) method, and the results were expressed as relative mRNA values normalized to the expression level of UBQ, as described previously (21).
Luciferase Activity Assay
Particle bombardment was carried out with the Biolistic PDS-1000/He particle delivery system (Bio-Rad) according to the manufacturer's protocol. Suspension-cultured rice cells, 6 days after transfer to fresh culture medium, were used for bombardment. The OsKSL4 promoter-LUC reporter construct was mixed in a 2:1 molar ratio with the internal control and precipitated onto gold particles (1.6-μm diameter). Approximately 1 μg of DNA was delivered into the rice cells per bombardment. Following bombardment, the samples were incubated in N6 medium with or without N-acetylchitooctaose (1 mg/liter) for 15 h at 25 °C in darkness, collected, and then homogenized in cell lysis buffer (Promega). The firefly LUC and Renilla LUC luminescence was monitored using a Centro LB960 plate reader (Berthold Japan, Tokyo, Japan) according to the manufacturer's instructions. To normalize the values after each assay, the ratio of LUC activity (firefly LUC/Renilla LUC) was calculated.
Electrophoretic Mobility Shift Assays
Double-stranded oligonucleotides spanning the TGACG motif upstream of OsKSL4 were synthesized, annealed, and digoxigenin-labeled. The OsTGAP1 protein was overexpressed in E. coli as a glutathione S-transferase (GST) fusion protein (GST-OsTGAP1). The E. coli Rosetta-gami (DE3), harboring a plasmid for the expression of GST-OsTGAP1 (pDEST17-OsTGAP1), was cultured in Overnight Express Instant TB medium (30 °C, 24 h) (Merck). The fusion protein was affinity-purified on glutathione-Sepharose 4B (Amersham Biosciences) and used for electrophoretic mobility shift assays. Probe fragments were amplified by PCR using the −1928 construct (for normal probes) or the m3 construct (for mutated probes) as template. Electrophoretic mobility shift assays were carried out using a digoxigenin gel shift kit, second generation (Roche Applied Science) according to the manufacturer's instructions.
Metabolite Analysis
Diterpene hydrocarbons were extracted from elicitor-induced suspension-cultured rice cells using ethyl acetate and detected by gas chromatography-mass spectrometry (GC-MS) as described previously (13). Phytoalexins were extracted from suspension-cultured rice cells after elicitation and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described previously (26).
Oligonucleotide Primers
The oligonucleotides used for genotyping, RT-PCR, qRT-PCR, and cloning are listed in supplemental Table S1.
RESULTS
Identification of an Elicitor-inducible cis-Acting Element in the OsKSL4 Gene Promoter
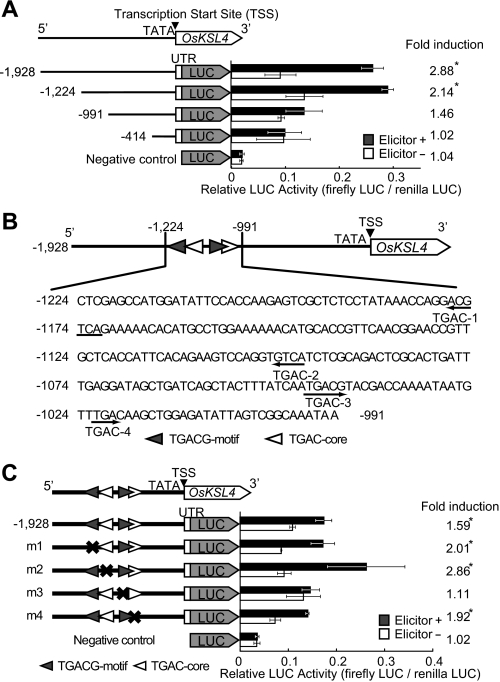
To identify regulating factors for momilactone biosynthesis, we first examined a chitin oligosaccharide elicitor-inducible promoter activity within a 2-kb region upstream of the OsKSL4 gene, responsible for the first committed step specific to momilactone biosynthesis, using a LUC reporter assay. Fig. 2A shows that deletion from −1224 to −991 bp and further deletion to −414 bp rendered the promoter incapable of responding to the elicitor. A search for known consensus recognition sites for transcription factors in the 233-bp region between −1224 and −992 bp revealed the presence of two TGACG motifs (as-1-like elements, TGACG(T/G)) that are known to be bound by bZIP transcription factors, including TGA factors (27), and two TGAC core-containing W-box elements, which are putative recognition sites for WRKY transcription factors (28). These elements were tentatively designated TGAC-1 to TGAC-4 (Fig. 2B). Subsequent mutation analyses of the promoter identified a TGACG motif in the region from −1045 to −1040 bp, upstream of the translation start site of OsKSL4 (TGAC-3), as a cis-acting element required for the elicitor-inducible expression of OsKSL4 (Fig. 2C). This suggested that a bZIP transcription factor was probably involved in the regulatory process.
FIGURE 2.
A, deletion analysis of the OsKSL4 promoter between −1928 and −414 bp. The promoter fragment located −1928 bp upstream of the transcription start site (TSS) of OsKSL4 and the 5′-untranslated region (UTR; 59 bp) were successively deleted from −1928 to −414, and the resulting fragments were fused to the firefly LUC gene. A putative TATA box is located at −39 to −31 bp. B, nucleotide sequence of the region from −1224 to −991 upstream of the transcription start site of OsKSL4 and known cis-acting elements existing in this region. C, mutation analysis of candidate cis-acting elements in the promoter region from −1224 to −991. Positions of mutated TGAC sequences, in which TGAC was altered to CCTA, are indicated by X. LUC activity was normalized against that of Renilla LUC. The black and open bars indicate the relative LUC activities of the deletion derivatives after 15 h of incubation of the rice cells with or without the elicitor, respectively. The data are means ± S.D. of three replicates. *, significant differences (p < 0.01) between the relative LUC activities in the elicited and non-elicited cells.
Elicitor-inducible bZIP Transcription Factors Are Candidates for Regulating OsKSL4
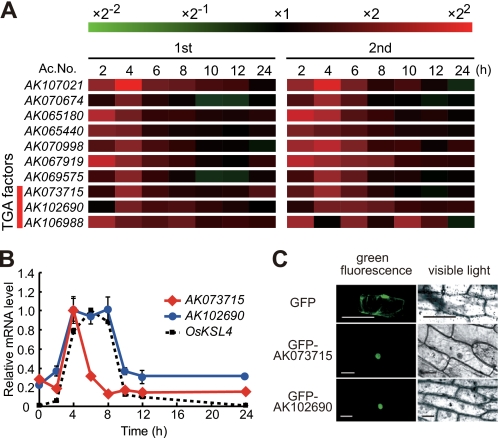
We focused on elicitor-inducible bZIP transcription factors with expression profiles similar to that of OsKSL4 or those expressed prior to the OsKSL4 gene by examining time course microarray data following the elicitor treatment (21). From more than 100 transcription factors having features of bZIP transcription factors, 10 candidate genes were selected, based on their expression profiles after elicitor treatment (Fig. 3A). Among them, the AK073715, AK102690, and AK106988 proteins were TGA-type bZIP transcription factors, having the typical conserved motifs of TGA factors (supplemental Fig. S1). Arabidopsis TGA factors can bind to the TGACG motif and are involved in defense responses (29). Thus, we further analyzed these three TGA-type bZIP transcription factors as promising candidates to regulate OsKSL4 gene expression. Expression analyses of these genes by qRT-PCR revealed that the AK073715 gene was maximally induced 4 h after elicitor treatment (slightly earlier than OsKSL4), expression returned to basal levels by 8 h, and the AK102690 gene was induced for 4–8 h after treatment, similar to OsKSL4 (Fig. 3B). We could not clearly detect inducible expression of the AK106988 gene and thus discarded it as a candidate. Phylogenetic analysis revealed that the AK073715 product was relatively close to the node containing soybean sTGA1, tobacco TGA1a, and Arabidopsis TGA1 and TGA4 and that the AK102690 product was classified in the node containing AtbZIP21 (supplemental Fig. S1). Tobacco TGA1a is shown to bind the as-1 element involved in salicylic acid-induced activation of transcription (30), and TGA4 is involved in defense responses (31), but the function of AtbZIP21 is still unknown. Nuclear localization of the AK073715 and AK102690 products was confirmed using green fluorescent protein as a reporter (Fig. 3C). Thus, we focused on the AK073715 and AK102690 genes to determine whether they were responsible for the inducible expression of OsKSL4.
FIGURE 3.
A, heat map of elicitor-inducible bZIP transcription factor genes isolated from microarray data after elicitor treatment. The results of two independent experiments are shown. The ratio of the level of gene expression at various time points in untreated cells compared with elicitor-treated cells was used to compile the heat map, which represents increases (red) and decreases (green) in expression relative to the untreated levels. The color bar represents the range of ratios. Three TGA-type transcription factor genes are indicated. B, transcription profiling of AK073715, AK102690, and OsKSL4 after elicitation, using qRT-PCR. Values indicate relative mRNA levels of AK073715 (red diamond), AK102690 (blue circles), and OsKSL4 (black box), normalized to the expression level of the UBQ gene. The maximal value in each experiment using different primers was arbitrarily set at 1.0. The results are the average of at least three independent experiments; bars indicate the means ± S.D. C, nuclear localization of the green fluorescent protein (GFP)-fused AK073715 or AK102690 protein in onion epidermal cells. Top, localization of the green fluorescent protein control. Middle, localization of AK073715 tagged with green fluorescent protein at its N terminus. Bottom, localization of AK102690 tagged with green fluorescent protein at its N terminus. The pictures represent green fluorescence (left) or visible light (right). Bars, 100 μm.
OsTGAP1 Is Essential for Elicitor-inducible OsKSL4 Gene Expression and Production of Momilactones
We used the rice Tos17 insertion mutants H0155 (for AK073715) and NC0005 (for AK102690) to examine their physiological functions. Two mutants obtained from the Rice Genome Research Program contained the Tos17 insertion in the first intron of AK073715 and the fifth intron of AK102690 (supplemental Fig. S2A) (32–34). They were both shown to be null mutants, lacking expression in response to elicitation (supplemental Fig. S2B).
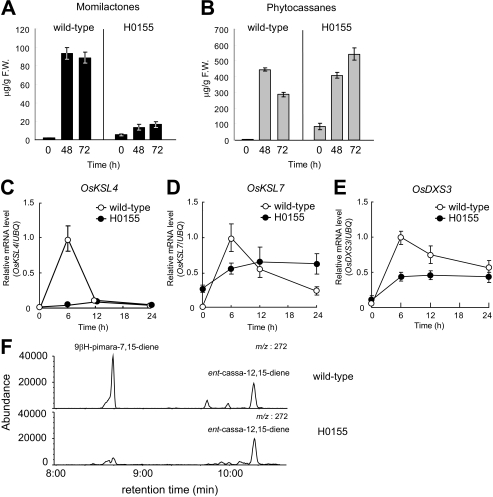
LC-MS/MS of phytoalexins accumulated in the culture medium 0, 48, and 72 h after elicitor treatment revealed that the level of momilactones in H0155 mutant cells severely decreased to less than 20% of that in wild-type cells (Fig. 4A), whereas the phytocassanes levels were almost the same between the mutant and wild-type cells at 48 h; thereafter, the phytocassanes levels were somewhat higher than that in wild-type cells at 72 h in the H0155 mutant (Fig. 4B). The inductive expression of OsKSL4 after elicitation was severely suppressed in the H0155 mutant compared with wild-type cells (Fig. 4C). The inductive expression of OsKSL7 after the elicitation seen in wild-type cells was also weakened in the H0155 mutant, but the expression levels at 0 h (basal level) and 24 h after the elicitation were rather higher than that of wild-type cells (Fig. 4D). In addition to expression analysis of specific genes for phytoalexin biosynthesis, the upstream gene expression responsible for plastidial geranylgeranyl diphosphate substrate production was also analyzed in the H0155 cells. The elicitor-induced expression of OsDXS3 in rice, which encodes deoxyxylulose phosphate synthase involved in the first committed step in the MEP pathway in plastids, was significantly suppressed in the H0155 mutant (Fig. 4E). GC-MS of diterpene hydrocarbons accumulated after elicitation revealed that significant accumulation of 9βH-pimara-7,15-diene, a precursor for momilactones, was observed only in the wild-type cells, whereas accumulation of ent-cassa-12,15-diene, a precursor for phytocassanes, was detected to the same extent in both the H0155 mutant and wild-type cells (Fig. 4F). These results suggest that AK073715 is essential for the elicitor-inductive momilactone biosynthesis through the up-regulation of OsKSL4 gene expression and has a role in regulation of OsKSL7 and OsDXS3 expression induced by elicitor treatment, which normally leads to transient production of phytocassanes after the elicitor recognition. Hence, the AK073715 gene was designated OsTGAP1 (Oryza sativa TGA factor for phytoalexin production 1). In the NC0005 mutant, both diterpenoid phytoalexin production and the biosynthetic gene expression (OsKSL4) were comparable with that in wild-type cells (supplemental Fig. S3), suggesting that AK102690 is unlikely to be involved in regulating phytoalexin biosynthesis.
FIGURE 4.
Physiological function of AK073715 in momilactone biosynthesis. A, accumulation of momilactones in wild-type and H0155 mutant cells after elicitor treatment. B, accumulation of phytocassanes in wild-type and H0155 mutant cells after elicitor treatment. Phytoalexin levels in the culture medium collected 0, 48, and 72 h after elicitor treatment were determined by LC-MS/MS. The results are the average of at least three independent experiments. Bars, mean ± S.D. C, expression analysis of OsKSL4 in elicitor-treated cells, by qRT-PCR. Total RNA was prepared 0, 6, 12, and 24 h after elicitor treatment. The results are the average of at least three independent experiments, and values for OsKSL4 mRNA expression were normalized to the expression of the UBQ gene. Bars, means ± S.D. D, expression analysis of OsKSL7 in elicitor-treated cells after qRT-PCR as in C. E, expression analysis of OsDXS3 in elicitor-treated cells after qRT-PCR as in C. F, GC-MS analysis of accumulated diterpene hydrocarbons in the mutant and wild-type cells after elicitation. Selected ion chromatograms at a mass/charge ratio of 272 showing 9βH-pimara-7,15-diene and ent-cassa-12,15-diene are indicated.
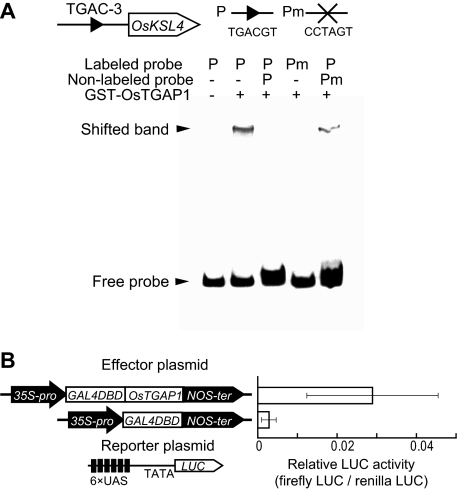
OsTGAP1 Requires the TGACG Motif to Bind to the OsKLS4 Promoter and Has Transactivation Capacity
Gel mobility shift assays revealed that GST-tagged OsTGAP1 could bind the DNA fragment containing the TGAC-3 region (TGACG motif) in the OsKSL4 promoter, in a TGACG motif-dependent manner (Fig. 5A). We also found that OsTGAP1 functioned as a transcriptional activator, using a transient assay system in which the GAL4 binding sequence is fused to LUC as a reporter and the GAL4 DNA-binding domain is fused to OsTGAP1 as an effector (Fig. 5B). On the basis of these data, we concluded that OsTGAP1 regulates momilactone biosynthesis through positive control of OsKSL4 expression, apparently by directly binding to the cis-element in the OsKSL4 promoter.
FIGURE 5.
A, sequence-specific DNA-binding activity of OsTGAP1. Electrophoretic mobility shift assays show the binding of recombinant OsTGAP1 protein to probes (P) containing the TGACG motif (TGACGT), but not to the mutated probe (Pm, with the TGAC sequence mutated to CCTA). For the assay, 240 ng of GST-OsTGAP1 were loaded with the probes. Nonlabeled probe was added in a 250-fold molar excess. B, transactivation activity of OsTGAP1. The scheme represents effector constructs fused to the GAL4 DNA-binding domain (GAL4-DBD) and reporter construct. A non-fused construct was used as a negative control. LUC activity was normalized against that of Renilla LUC. The bar represents relative LUC activity after 15 h of incubation of the rice cells with or without elicitor. The data are means ± S.D. of three replicates.
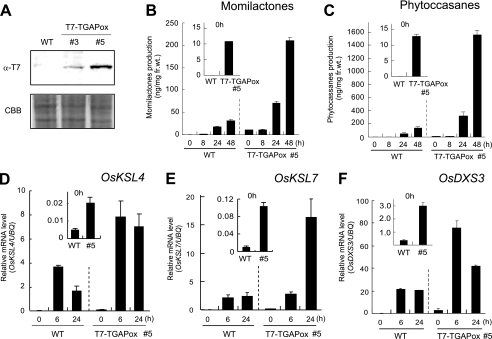
Overexpression of OsTGAP1 Enhances Elicitor-inducible Production of Phytoalexins
To address the manner in which OsTGAP1 regulates expression of phytoalexin biosynthetic genes, transformed rice plants constitutively expressing OsTGAP1 (fused to a T7 epitope tag) were generated (Fig. 6A). In calli overexpressing T7-OsTGAP1, low but significant accumulation of momilactones was observed by LC-MS/MS analysis, without any elicitation (Fig. 6B, inset). Furthermore, OsTGAP1-overexpressing lines had enhanced accumulation of momilactones compared with wild-type cells after treatment with elicitor (Fig. 6B). Similarly, accumulation of phytocassanes in the overexpressing line was detected without elicitation (Fig. 6C, inset). When the overexpressing line was treated with the elicitor, enhanced accumulation of phytocassanes was also evident in the overexpressing lines as in hyperaccumulation of momilactones (Fig. 6C). The level of accumulated phytoalexins correlated well with the expression level of the T7-OsTGAP1 protein (data not shown). OsKSL4, OsKSL7, and OsDXS3 expression in the OsTGAP1-overexpressing lines was assessed by qRT-PCR. Although the T7-OsTGAP1-overexpressing lines exhibited slightly induced expression of these genes without elicitation (Fig. 6, D–F, inset), a striking induction of expression was detected in the overexpressing cells compared with wild-type cells 6 h after elicitation, consistent with the hyperaccumulation of momilactones and phytocassanes in elicitor-treated T7-OsTGAP1-overexpressing lines. High levels of mRNA expression of these genes were maintained for at least 24 h after elicitation in the overexpressing lines (Fig. 6, D–F). These results suggest that OsTGAP1 can influence the biosynthetic pathway for diterpenoid phytoalexins, including the upstream MEP pathway, at a transcriptional level, eventually leading to production of a large amount of the elicitor-inductive diterpenoid phytoalexins.
FIGURE 6.
Phytoalexin accumulation in T7-OsTGAP1-overexpressing lines. A, immunoblot analysis of T7-OsTGAP1-overexpressing lines using an anti-T7 antibody. Two overexpressing lines with different expression levels are indicated. Shown is LC-MS/MS analysis of momilactone (B) and phytocassane (C) production in T7-OsTGAP1-overexpressing lines after elicitation. Phytoalexin accumulation before elicitation is indicated in the inset. qRT-PCR analyses indicate constitutive expression of the OsKSL4 gene (D) and OsKSL7 (E) in the overexpressing lines without elicitation (inset). Hyperinduction of OsKSL4 and OsKSL7 expression is observed in the overexpressing lines after elicitation. F, expression of OsDXS3 involved in production of the geranylgeranyl diphosphate substrate through the MEP pathway, which is located upstream of the momilactone biosynthetic pathway in plastids. The expression profiles at the indicated time points after elicitation in wild-type and T7-OsTGAP1-overexpressing lines were determined by qRT-PCR. Expression levels are shown relative to the levels of the UBQ gene (n > 3).
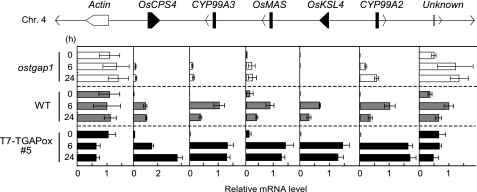
OsTGAP1 Coordinately Regulates the Clustered Genes for Momilactone Biosynthesis
As previously discussed, the momilactone biosynthetic genes are organized in a gene cluster, exhibiting coordinated, inducible expression upon elicitor treatment. OsTGAP1 overexpression led to constitutive production of momilactones even without elicitation, indicating that the five momilactone biosynthetic genes, including OsKSL4, may be coordinately regulated by OsTGAP1. To verify this, we analyzed expression of the genes in the momilactone biosynthetic gene cluster (with neighboring genes) in both the ostgap1 mutant and the T7-OsTGAP1-overexpressing lines. Elicited expression of the five momilactone biosynthetic genes (OsCPS4, OsKSL4, CYP99A2, CYP99A3, and OsMAS) was severely suppressed in ostgap1 cells, whereas expression of the Os04g0177600 gene (encoding actin protein) and the Os04g0180900 gene (unknown function), both of which are located outside the cluster, were virtually unaffected (Fig. 7). The clustered genes, but not the neighboring genes, were hyperinductively expressed in the overexpressing cells stimulated by an elicitor (Fig. 7). The results indicate that clustered genes for momilactone biosynthesis are coordinately regulated by OsTGAP1.
FIGURE 7.
OsTGAP1 regulates expression of the momilactone biosynthetic gene cluster. Expression of the five momilactone biosynthetic genes in the cluster and two neighboring genes located outside of the cluster that is illustrated in Fig. 1B are shown. The expression profiles at the indicated time points after elicitation in the ostgap1 mutant, wild type, and T7-OsTGAP1-overexpressing line (#5) were determined by qRT-PCR. Values for each mRNA expression were normalized to expression of the UBQ gene. Expression levels are shown relative to the levels 6 h after elicitation in wild-type cells (n > 3).
DISCUSSION
In this study, we identified an elicitor-inducible rice bZIP transcription factor, OsTGAP1, which is essential for elicitor-inducible production of momilactones and which coordinately regulates the expression of all five genes in the momilactone biosynthetic gene cluster. OsTGAP1 was also shown to be involved in the transcriptional regulation of OsKSL7 for phytocassane biosynthesis and OsDXS3 in the MEP pathway. Overexpression analysis clearly demonstrated that OsTGAP1 can influence both momilactone and phytocassane production through up-regulation of both the biosynthetic genes and the upstream MEP pathway gene under the elicitor treatment. Since OsTGAP1 expression itself is induced by treatment with a chitin oligosaccharide elicitor, OsTGAP1 would be a crucial master regulator that controls the inducible expression of biosynthetic genes and upstream pathway genes required for diterpenoid phytoalexin production as part of the plant's defensive response, acting through the detection of a chitin oligosaccharide elicitor (25).
In Arabidopsis, genes encoding enzymes that biosynthesize the phytoalexin camalexin are coordinately expressed, and their expression is probably the result of an unidentified key transcription factor(s) (35). Although the transcription factor AtWRKY33, which regulates the expression of the two P450 genes (PAD3 and CYP71A13) involved in camalexin biosynthesis, has been reported (36), whether a key transcription factor exists that is involved in the coordinated up-regulation of multiple camalexin biosynthetic genes is still unknown. Additionally, the PAD3 and CYP71A13 genes are not located in a gene cluster. Thus, such a function of OsTGAP1 in regulating chitin oligosaccharide-inducible expression of the momilactone biosynthetic gene cluster seems to be characteristic of rice plants.
Phylogenetic analysis showed that OsTGAP1 is located close to the node, including Arabidopsis OBF4/TGA4 (sharing 58% amino acid identity), which has been shown to positively regulate basal resistance in Arabidopsis (31). The similarity suggested that OsTGAP1 may function in the defensive response in rice. As a result of a homology search using the RICE cDNA data base and the Knowledge-based Oryza Molecular Biological Encyclopedia (KOME; available on the World Wide Web), at least 13 TGA-type transcription factor genes are expressed in rice. Of these 13, only three genes, including OsTGAP1, were selected as having elicitor responsiveness by our microarray analysis. Thus, our attempt to narrow down the candidate elicitor-inducible TGA factors regulating the OsKSL4 expression using the microarray data was apparently valid in this case.
We used the rice Tos17 insertion mutant H0155 for physiological analysis of OsTGAP1. Although a genetic complementation test of H0155 has not yet been carried out, the phenotype of the mutant is most likely linked to a mutation of the OsTGAP1 locus, because four ostgap1/ostgap1 homozygous plants that we tested, segregated by self-pollination of a heterozygous OsTGAP1/ostgap1 plant, all exhibited the momilactone phenotype. Loss-of-function analyses showed that OsTGAP1 was responsible for elicitor-inducible production of momilactones and the expression of the biosynthetic gene. Because accumulation of momilactones 48 h after the elicitation, but not phytocassanes, was affected by the OsTGAP1 mutation, OsTGAP1 was shown to exert a strong influence on momilactone biosynthetic gene expression; in fact, OsKSL4 gene expression was severely suppressed up to 24 h after elicitation in the ostgap1 mutant, whereas OsKSL7 expression in the mutant was still observed 6 h after the elicitation, which was about 50% suppression compared with the expression level in wild-type cells. Moreover, the basal level of OsKSL7 expression was detected in the mutant without elicitation. Although why the basal level of OsKSL7 expression in the ostgap1 mutant increases is unknown at present, this constitutive OsKSL7 expression with slight up-regulation is consistent with the accumulation of phytocassanes in the ostgap1 mutant (Fig. 4, B and D). With regard to expression of OsDXS3 in the upstream MEP pathway, significant but incomplete suppression of OsDXS3 gene expression was detected in the ostgap1 mutant after elicitation. Thus, the inductive expression of momilactone biosynthetic genes appears to be regulated by OsTGAP1 in a manner different from that of OsKLS7 and OsDXS3 gene expression.
The ostgap1 mutant also showed that all five genes for momilactone biosynthesis in the cluster were regulated by OsTGAP1 upon elicitor treatment, whereas expression of genes located outside of the cluster (Os04g0177600 and Os04g0180900) was virtually unaffected. Coordinated regulation of the clustered genes for momilactone biosynthesis by OsTGAP1 was also shown using T7-OsTGAP1-overexpressing lines. These results imply that the OsTGAP1 has a profound effect on regulating the gene cluster for momilactone biosynthesis.
We found that OsTGAP1 overexpression can also influence transcriptional up-regulation of the phytocassane biosynthetic gene (OsKSL7) and the MEP pathway gene (OsDXS3) under elicitation, eventually leading to diterpene phytoalexin production. Since OsTGAP1 was shown to function as a transcriptional activator (Fig. 5B), OsTGAP1 may be able to influence expression of these genes by binding to their regulatory elements. In fact, several TGACG motifs can be found in the possible promoter regions of OsKSL7 and OsDXS3 (supplemental Fig. S4). Although the physical binding of OsTGAP1 to the regulatory elements remains to be demonstrated, our data support the idea that OsTGAP1 could be the master regulator for the production of diterpenoid phytoalexins and transcriptionally controls both phytoalexin biosynthetic genes and the upstream MEP pathway gene responsible for supplying geranylgeranyl diphosphate, the precursor of diterpene phytoalexins, after elicitor recognition.
Determining how OsTGAP1 synchronously controls expression of the clustered genes for momilactone biosynthesis will also be important. A survey of TGACG motifs existing in the broad region encompassing the clustered genes revealed more than 100 possible TGACG motifs that could be occupied by OsTGAP1. However, we also found that a large number of TGACG motifs exist in other regions extending from the momilactone cluster in both directions, indicating that the possible promoter regions (∼3 kb) of not only the five momilactone biosynthetic genes but also non-elicitor-inducible genes contain TGACG-motifs (supplemental Fig. S4). Therefore, OsTGAP1 may bind to the putative promoter regions of all clustered genes to regulate their expression, but it is also possible that OsTGAP1 regulates expression of the clustered genes for momilactone biosynthesis by unknown mechanisms other than binding to all TGACG motifs existing in promoter regions of the clustered genes. Confirmation of OsTGAP-binding sites on the momilactone biosynthetic gene cluster would be the next step leading to understanding of the regulation mechanism.
Overexpression of T7-OsTGAP1 also revealed that OsTGAP1 itself was not sufficient to fully activate the expression of genes for diterpenoid phytoalexin biosynthesis. This may suggest that a posttranslational modification of OsTGAP1 and/or the involvement of an unknown protein factor that synergistically functions with OsTGAP1 upon elicitor treatment can explain the elevated activation of biosynthetic gene expression (Fig. 6, D–F). Redox regulation of Arabidopsis TGA1 has been shown to modify the activity of this protein (37) and might also represent a way to modify OsTGAP1 activity. However, cysteine residues that are involved in TGA1 redox control are not conserved in OsTGAP1 (supplemental Fig. S1), suggesting that this is not the case for OsTGAP1 modification. Identification of unknown factors, which are required to modulate gene expression for diterpenoid phytoalexin biosynthesis with OsTGAP1, would provide potential clues for understanding the mechanisms of OsTGAP1-based regulation. Although the way in which OsTGAP1 coordinately transactivates all genes involved in diterpenoid phytoalexin production, including the momilactone biosynthetic gene cluster, is currently unknown, our results provide evidence for the presence of an effective coordinated regulation system by OsTGAP1 to produce defensive compounds in rice.
Supplementary Material
Acknowledgments
We thank M. Takagi and M. Shikata for the 35S:GAL4(430T1.2) plasmids, T. Nakagawa for the pGWB series vectors, and Y. Nagamura and H. Hirochika for rice cDNAs and Tos17 mutant distribution. We also thank A. Osbourn for critically reading the manuscript.
This work was supported by the Program for the Promotion of Basic Research Activities for Innovative Biosciences (PROBRAIN) and Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research 18580102 (to H. Y.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- bZIP
- basic leucine zipper
- MEP
- methylerythritol phosphate
- RT
- reverse transcription
- qRT
- quantitative reverse transcription
- GST
- glutathione S-transferase
- GC
- gas chromatography
- MS
- mass spectrometry
- LC
- liquid chromatography
- LUC
- luciferase
- TGA
- TGACG-sequence-specific binding protein.
REFERENCES
- 1.VanEtten H. D., Mansfield J. W., Bailey J. A., Farmer E. E. (1994) Plant Cell 6, 1191–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koga J., Shimura M., Oshima K., Ogawa N., Yamauchi T., Ogasawara N. (1995) Tetrahedron 51, 7907–7918 [Google Scholar]
- 3.Koga J., Ogawa N., Yamauchi T., Kikuchi M., Ogasawara N., Shimura M. (1997) Phytochemistry 44, 249–253 [Google Scholar]
- 4.Yajima A., Mori K. (2000) Eur. J. Org. Chem. 24, 4079–4091 [Google Scholar]
- 5.Akatsuka T., Kodama O., Sekido H., Kono Y., Takeuchi S. (1985) Agric. Biol. Chem. 49, 1689–1694 [Google Scholar]
- 6.Kato H., Kodama O., Akatsuka T. (1993) Phytochemistry 33, 79–81 [Google Scholar]
- 7.Kato H., Kodama O., Akatsuka T. (1994) Phytochemistry 36, 299–301 [Google Scholar]
- 8.Cartwright D. W., Langcake P., Pryce R. J., Leworthy D. P., Ride J. P. (1981) Phytochemistry 20, 535–537 [Google Scholar]
- 9.Kato T., Kabuto C., Sasaki N., Tsunagawa M., Aizawa H., Fujita K., Kato Y., Kitahara Y., Takahashi N. (1973) Tetrahedron Lett. 14, 3861–3864 [Google Scholar]
- 10.Tamogami S., Mitani M., Kodama O., Akatsuka T. (1993) Tetrahedron 49, 2025–2032 [Google Scholar]
- 11.Otomo K., Kenmoku H., Oikawa H., König W. A., Toshima H., Mitsuhashi W., Yamane H., Sassa T., Toyomasu T. (2004) Plant J. 39, 886–893 [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto T., Miura K., Itoh H., Tatsumi T., Ueguchi-Tanaka M., Ishiyama K., Kobayashi M., Agrawal G. K., Takeda S., Abe K., Miyao A., Hirochika H., Kitano H., Ashikari M., Matsuoka M. (2004) Plant Physiol. 134, 1642–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho E. M., Okada A., Kenmoku H., Otomo K., Toyomasu T., Mitsuhashi W., Sassa T., Yajima A., Yabuta G., Mori K., Oikawa H., Toshima H., Shibuya N., Nojiri H., Omori T., Nishiyama M., Yamane H. (2004) Plant J. 37, 1–8 [DOI] [PubMed] [Google Scholar]
- 14.Nemoto T., Cho E. M., Okada A., Okada K., Otomo K., Kanno Y., Toyomasu T., Mitsuhashi W., Sassa T., Minami E., Shibuya N., Nishiyama M., Nojiri H., Yamane H. (2004) FEBS Lett. 571, 182–186 [DOI] [PubMed] [Google Scholar]
- 15.Peters R. J. (2006) Phytochemistry 67, 2307–2317 [DOI] [PubMed] [Google Scholar]
- 16.Shimura K., Okada A., Okada K., Jikumaru Y., Ko K. W., Toyomasu T., Sassa T., Hasegawa M., Kodama O., Shibuya N., Koga J., Nojiri H., Yamane H. (2007) J. Biol. Chem. 282, 34013–34018 [DOI] [PubMed] [Google Scholar]
- 17.Qi X., Bakht S., Leggett M., Maxwell C., Melton R., Osbourn A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8233–8238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Field B., Osbourn A. E. (2008) Science 320, 543–547 [DOI] [PubMed] [Google Scholar]
- 19.Frey M., Chomet P., Glawischnig E., Stettner C., Grün S., Winklmair A., Eisenreich W., Bacher A., Meeley R. B., Briggs S. P., Simcox K., Gierl A. (1997) Science 277, 696–699 [DOI] [PubMed] [Google Scholar]
- 20.Qi X., Bakht S., Qin B., Leggett M., Hemmings A., Mellon F., Eagles J., Werck-Reichhart D., Schaller H., Lesot A., Melton R., Osbourn A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18848–18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okada A., Shimizu T., Okada K., Kuzuyama T., Koga J., Shibuya N., Nojiri H., Yamane H. (2007) Plant Mol. Biol. 65, 177–187 [DOI] [PubMed] [Google Scholar]
- 22.Ito Y., Kaku H., Shibuya N. (1997) Plant J. 12, 347–356 [DOI] [PubMed] [Google Scholar]
- 23.Shikata M., Takemura M., Yokota A., Kohchi T. (2003) Biosci. Biotechnol. Biochem. 67, 2495–2497 [DOI] [PubMed] [Google Scholar]
- 24.Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007) J. Biosci. Bioeng. 104, 34–41 [DOI] [PubMed] [Google Scholar]
- 25.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu T., Jikumaru Y., Okada A., Okada K., Koga J., Umemura K., Minami E., Shibuya N., Hasegawa M., Kodama O., Nojiri H., Yamane H. (2008) Phytochemistry 69, 973–981 [DOI] [PubMed] [Google Scholar]
- 27.Lam E., Chua N. H. (1989) Plant Cell 1, 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eulgem T., Rushton P. J., Robatzek S., Somssich I. E. (2000) Trends Plant Sci. 5, 199–206 [DOI] [PubMed] [Google Scholar]
- 29.Lebel E., Heifetz P., Thorne L., Uknes S., Ryals J., Ward E. (1998) Plant J. 16, 223–233 [DOI] [PubMed] [Google Scholar]
- 30.Jupin I., Chua N. H. (1996) EMBO J. 15, 5679–5689 [PMC free article] [PubMed] [Google Scholar]
- 31.Kesarwani M., Yoo J., Dong X. (2007) Plant Physiol. 144, 336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirochika H. (1997) Plant Mol. Biol. 35, 231–240 [PubMed] [Google Scholar]
- 33.Hirochika H. (2001) Curr. Opin. Plant Biol. 4, 118–122 [DOI] [PubMed] [Google Scholar]
- 34.Miyao A., Iwasaki Y., Kitano H., Itoh J., Maekawa M., Murata K., Yatou O., Nagato Y., Hirochika H. (2007) Plant Mol. Biol. 63, 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren D., Liu Y., Yang K. Y., Han L., Mao G., Glazebrook J., Zhang S. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5638–5643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiu J. L., Fiil B. K., Petersen K., Nielsen H. B., Botanga C. J., Thorgrimsen S., Palma K., Suarez-Rodriguez M. C., Sandbech-Clausen S., Lichota J., Brodersen P., Grasser K. D., Mattsson O., Glazebrook J., Mundy J., Petersen M. (2008) EMBO J. 27, 2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P. R. (2003) Plant Cell 15, 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.