Abstract
Pancreatic β-cells couple the oxidation of glucose to the secretion of insulin. Apart from the canonical KATP-dependent glucose-stimulated insulin secretion (GSIS), there are important KATP-independent mechanisms involving both anaplerosis and mitochondrial GTP (mtGTP). How mtGTP that is trapped within the mitochondrial matrix regulates the cytosolic calcium increases that drive GSIS remains a mystery. Here we have investigated whether the mitochondrial isoform of phosphoenolpyruvate carboxykinase (PEPCK-M) is the GTPase linking hydrolysis of mtGTP made by succinyl-CoA synthetase (SCS-GTP) to an anaplerotic pathway producing phosphoenolpyruvate (PEP). Although cytosolic PEPCK (PEPCK-C) is absent, PEPCK-M message and protein were detected in INS-1 832/13 cells, rat islets, and mouse islets. PEPCK enzymatic activity is half that of primary hepatocytes and is localized exclusively to the mitochondria. Novel 13C-labeling strategies in INS-1 832/13 cells and islets measured substantial contribution of PEPCK-M to the synthesis of PEP. As high as 30% of PEP in INS-1 832/13 cells and 41% of PEP in rat islets came from PEPCK-M. The contribution of PEPCK-M to overall PEP synthesis more than tripled with glucose stimulation. Silencing the PEPCK-M gene completely inhibited GSIS underscoring its central role in mitochondrial metabolism-mediated insulin secretion. Given that mtGTP synthesized by SCS-GTP is an indicator of TCA flux that is crucial for GSIS, PEPCK-M is a strong candidate to link mtGTP synthesis with insulin release through anaplerotic PEP cycling.
β-Cells in pancreatic islets of Langerhans make and release insulin in response to changes in blood glucose levels. The mechanisms by which high concentrations of glucose stimulate insulin release from islets remain unclear. The canonical explanation for GSIS2 is that glucose metabolism increases mitochondrial ATP production, thereby raising the cytosolic ATP:ADP ratio that triggers the closure of ATP-sensitive K+ channels. This, in turn, depolarizes the membrane and stimulates the opening of voltage-dependent Ca2+ channels with increased Ca2+ influx promoting the exocytosis of insulin. Although KATP channels certainly have an important role in β-cells, KATP-independent signals are implicated to play a fundamental role in GSIS. In particular, β-cells are known to have notably elevated rates of anaplerotic flux of the carbon from glucose into the mitochondria and back out to pyruvate (pyruvate cycling) that is tightly correlated with insulin secretion (1–4).
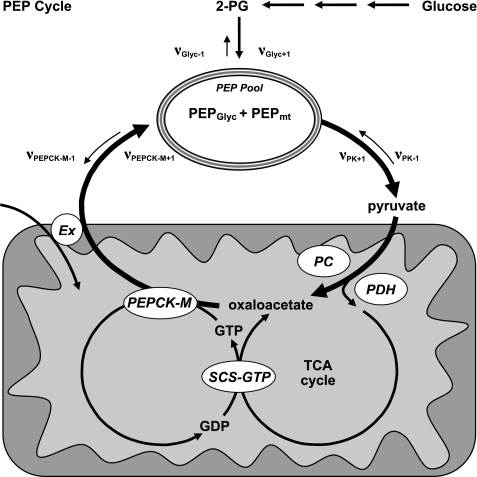
Recently, mtGTP synthesis was identified as a novel KATP-independent mitochondrial signal for insulin secretion (5). mtGTP is synthesized as a product of glucose metabolism by the GTP-specific isoform of the matrix enzyme SCS. mtGTP synthetic rates are determined by the rate of TCA cycle flux as well as by the ratio of activities of the ATP-specific and GTP-specific isoforms of SCS. The mtGTP signal is trapped within the matrix of the mitochondria, suggesting that another GTPase in the matrix transmits the mtGTP signal to the cytosol. Because both mtGTP synthesis and anaplerotic flux correlate with insulin secretion, we investigated whether the GTP-dependent mitochondrial isoform of PEPCK, an enzyme that lies at the intersection of anaplerosis and mtGTP metabolism (see Fig. 1A), is important for GSIS.
FIGURE 1.
PEP cycle. PEP is produced during glycolysis and is further metabolized to pyruvate by PK. Pyruvate that enters the TCA cycle by pyruvate dehydrogenase will generate GTP via direct synthesis by SCS-GTP. Anaplerotic pyruvate entry by PC will generate oxaloacetate. PEPCK-M will then consume oxaloacetate and GTP to produce PEP, GDP, and CO2. PEP is then transported out of the mitochondrial matrix by an anion transporter (Ex) in exchange for another metabolite depending on the transporter. Mitochondrial PEP, thus, contributes to the PEP pool that is determined by the rate of appearance (νPEPCK-M+1+ νGlyc+1) of PEP minus the rate of disappearance (νPK+1). One turn of the PEP cycle will result in the net exchange of one ion into the mitochondrial matrix. Unidirectional fluxes are indicated by ν followed by the enzyme with the forward direction being +1 and the reverse −1. GDP in turn can be reused by SCS-GTP. PDH, pyruvate dehydrogenase.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
INS-1 832/13 cells were cultured and siRNA-transfected, GSIS assays were performed, and islets were isolated as previously described (5). Primary hepatocytes were isolated by standard procedures from Sprague-Dawley rats (300g) from the Yale University Liver Center. As a control, a non-silencing siRNA 5′-AATTCTCCGAACGTGTCACGT-3′ (Qiagen) was used. Mitochondrial PEPCK (PEPCK-M) was targeted by two different siRNAs (Qiagen) with the following DNA templates: #1, 5′-AACGTGAACAATTTGACATTA-3′; #2, 5′-TCCCATTGGGCTCGTACCAAA-3′. Quantitation of mRNA by reverse transcription and real-time PCR was performed using the following primers: cytosolic PEPCK (PEPCK-C) (5′-ATGACACCCTCCTCCTGCAT-3′, 5′-CAGGAAGTGAGGAAGTTTGTGG-3′), PEPCK-M (5′-TTATGCACGATCCCTTTGCCATGC-3′, 5′-TCCTTCCTTTGGTACGAGCCCAAT-3′), and GAPDH (5′-GTTACCAGGGCTGCCTTCTC-3′, 5′-GGGTTTCCCGTTGATGACC-3′) as well as β-actin and SCS-GTP as previously described (5).
Western Blotting
Proteins were detected by Western blot analysis after separating 30–50 μg of total protein lysate on a 4–12% Tris-glycine gel (Invitrogen) and transferring to a polyvinylidene difluoride membrane (Immobilon-P 0.45 μm, Millipore). The sheep polyclonal PEPCK-C antibody was a kind gift from Daryl Granner (Vanderbilt University Medical Center). Goat anti-PEPCK-M (Abcam) and rabbit anti-GAPDH (Abcam) were used to compare PEPCK-M versus PEPCK-C in cultured cells, islets, and liver. A rabbit PEPCK-M antibody (Santa Cruz) was used in knockdown experiments.
Enzyme Activities
Sample Preparation
Cells or islets were homogenized in 1 ml of ice-cold isolation buffer (10 mm Hepes, pH 7.4, 250 mm sucrose, 1 mm EDTA, and 1 mm dithiothreitol) using a Potter-Elvehjem Teflon pestle by 50 vertical passes on ice. The supernatant from a 5-min 2000 rcf spin of the lysates was used for whole cell assays. Mitochondria were isolated from this supernatant by centrifugation at 10,000 rcf for 10 min. The pellet was solubilized in 0.4% deoxycholate on ice for 20 min before the insoluble material was pelleted by a second spin at 16,100 rcf for 10 min. To determine the percent mitochondrial activity, mitochondria were isolated from 1 ml of the post-nuclear supernatant as described above and dissolved in 100 μl of 0.4% deoxycholate on ice for 20 min, and 900 μl of the isolation buffer was then added for a final volume of 1 ml. 4% deoxycholate was added to the cytosol-containing supernatant from the mitochondrial spin to a final concentration of 0.04%. Cytosolic and mitochondrial fractions were assayed simultaneously.
PEPCK Activities
PEPCK activities were measured in the direction of oxaloacetate (OAA) formation as previously described with some modifications (6). The reaction was coupled to malate dehydrogenase for detection of NADH oxidation by fluorescence. Deoxy-GDP (dGDP) was used as a reactant in place of GDP or IDP because it discriminates against pyruvate kinase (PK), which is known to be high in insulin-secreting tissues (7). The reactions were performed in quadruplicate in 96-well plates with a 200-μl final volume containing 110 mm imidazole-Cl, pH 6.8, 3 mm MgSO4, 3 mm MnCl2, 13 mm NaF, 10 mm phenylalanine, 1 μm rotenone, 30 mm NaHCO3, 0.15 mm NADH, 6 units/ml malate dehydrogenase, 2 mm PEP, 0.5 mm dGDP, and cell homogenate containing ∼5–50 μg of protein. Magnesium was left out when comparing cytosolic to mitochondrial activities because it favors PEPCK-C. Control samples were run simultaneously in the absence of HCO3−/CO2, and this background slope was subtracted from the slope of the complete reaction. Before the experiment, the reaction mixture was freshly gassed with 100% CO2 for 10 min. The reaction was initiated with 0.5 mm dGDP, and the drop in NADH signal was assayed at 37 °C for 10–20 min at 10-s intervals by fluorescence using 335 nm for excitation and 460 nm for emission (Flex Station 3, Molecular Devices).
Glutamate Dehydrogenase Activities
Samples were assayed for glutamate dehydrogenase activity in the direction of glutamate formation from α-ketoglutarate (α-KG) in the presence of NH3 at 37 °C and pH 7.4 in a 250-μl total reaction mixture containing 70 mm triethanolamine, 3 mm EDTA, 125 mm ammonium acetate, 1.25 mm ADP, 0.25 mm NADH, 1.3 units/ml lactate dehydrogenase, and cell homogenate. The reactions were initiated with addition of 10 μl of 233 mm α-KG or water as a control and the decrease in absorption at 340 nm was monitored. Activities were obtained by subtracting the background slope of the samples without α-KG from those with α-KG.
LC/MS/MS Isotopologue Analysis
Mass isotopologue analysis of PEPCK activity was performed in INS-1 832/13 cells or rat islets preincubated in DMEM base without glucose for 1 h to minimize the amount of unlabeled glycolytic carbon in the cells. For the INS-1 832/13 cells, the medium was then aspirated and subsequently replaced with G0, G3, G7, or G15 mm unlabeled glucose containing either 5 mm unlabeled or [3-13C]pyruvate (Cambridge Isotope Laboratories. As such, [3-13C]pyruvate was used as a tracer entering distal to the irreversible PK step to label the mitochondrial OAA precursor pool of PEPCK-M and kept at a constant concentration with varying concentrations of glucose. To ensure that the [3-13C]pyruvate could contribute label to the PEP precursor pool in the face of increasing glucose concentrations, an extracellular concentration of 5 mm was applied. β-Cells in intact islets do not take up pyruvate but do take up glutamine, and even low glucose concentrations sufficiently increase mtGTP synthesis to prevent islet glutamine metabolism (via allosteric inhibition of glutamate dehydrogenase) (8). Thus, after preincubation, the islets were incubated in either unlabeled or 1,2-13C-labeled glutamine in the absence of glucose. Parallel studies using unlabeled substrate were performed to account for natural abundance contribution to the signal.
After substrate incubations, INS 832/13 cells that were cultured in 6-well plates were quenched by rapid wash with ice-cold water and then collected in 200 μl of ice-cold buffer A (5% acetonitrile, 2 mm ammonium acetate, 10 μm EDTA) with 10 μm d4-taurine (CDN Isotopes) as a load control (n = 6). For experiments with islets, after the incubation 100 islets per replicate were transferred to microcentrifuge tubes and centrifuged at 2000 rcf for 5 min to pellet the islets which were immediately frozen and lyophilized before resuspension in buffer A. The collected cells or islets were centrifuged to pellet bulk insoluble materials before filtration (MultiScreen BV, Millipore). The lysates were separated on a C16 5-μm 120 Å 4.6 × 250-mm column (Acclaim® Polar Advantage, Dionex) before ionization for multiple reaction monitoring (MRM) analysis by LC/MS/MS (Applied Biosystems MDS SCIEX, 4000 Q-TRAP).
Each analyte was eluted isocratically in buffer A at a flow rate of 400 μl/min with a single gaussian-shaped peak whose retention time was confirmed with known standards. Individual MRM transition pairs (Q1/Q3) were designated for the natural abundance (M) and mass + 1 (M +1) isotopologues of each analyte (see Table 1). In addition to malate and PEP, aspartate and combined 2-phosphoglycerate/3-phosphoglycerate (2-PG/3-PG) enrichments were measured for comparison. Endogenous taurine and d4-taurine were used as internal and external controls. 13C incorporation into taurine did not occur under the experimental conditions nor did the absolute concentration of taurine change during the incubations. 2-PG and 3-PG were unable to be separated either chromatographically or by MRM analysis. So, for comparison with the atomic percent excess (APE) of PEP, they were treated as the same analyte for calculation of APEs because of equilibrium of both phosphoglycerate species across phosphoglycerate mutase. Phosphate was the daughter anion for both PEP and 2-PG/3-PG and sulfate for taurine as well as d4-taurine and did not vary with 13C incorporation. For the remaining analytes, because the daughter fragment contained two or more carbons, the M and M+1 of the daughter anions were summed. Under the conditions of these experiments, M + 2 and greater labeling were minimal. When multiple Q3 daughter anions were measurable for a single parent, the one with the highest signal to noise ratio and minimal spectral overlap were chosen and coincidence of both MRM pairs was confirmed in the chromatogram. The KEGG data base of metabolites was screened for potential significant M and M+1 mono- and di-anions that might give similar MRM pairs. Each analyte was determined to have either a unique MRM pair and/or retention time. Concentrations of each metabolite are presented relative to taurine calibrated by standard curves and referenced to an internal taurine concentration of 12.0 nmol/mg of protein (5).
TABLE 1.
LC/MS/MS masses, retention times, and natural abundance measurements
| Compound | Mass/fragment | Natural abundance | Retention time |
|---|---|---|---|
| % M+1 | min | ||
| Malate | |||
| M0 | 133/71 | 4.45 ± 0.027 | 3.89 ± 0.002 |
| M+1 | 134/71, 134/72 | ||
| Aspartate | |||
| M0 | 132/88 | 5.55 ± 0.246 | 3.70 ± 0.093 |
| M+1 | 133/88, 133/89 | ||
| PEP | |||
| M0 | 166.6/79 | 3.62 ± 0.08 | 3.85 ± 0.002 |
| M+1 | 167.6/79 | ||
| 2-PG/3-PG | |||
| M0 | 185/79 | 3.52 ± 0.089 | 3.85 ± 0.003 |
| M+1 | 186/79 | ||
| Taurine | 123.6/80 | 5.52 ± 0.002 | |
| d4-Taurine | 127.6/80 | 5.54 ± 0.002 |
The signal from natural abundance 13C in standards or in samples to which 13C-enriched substrates were not added (Table 1) was as predicted from 1.11% background enrichment (3.33% for PEP and 4.44% for malate) and did not change with increasing glucose concentration (supplemental Fig. S1). As such, the similarities of the natural abundance enrichments of the unlabeled samples with increasing glucose concentrations to the predicted values further supports that the peaks analyzed are the mono-anions of PEP and malate. At low concentrations, PEP had higher than predicted natural abundance from the decreased signal to noise ratio for M+1. Calculation of APEs was as traditionally defined, APE = 100 × (rsa − rbk)/((rsa − rbk) + 1), where r is the ratio of (M+1)/(M) for the sample, rsa, and background, rbk.
Calculations of the contribution of PEPCK-M to relative glycolytic flux use the following definitions and assumptions (Fig. 1). 1) Glycolytic flux is defined as the contribution of unlabeled carbon into PEP in the glycolytic direction from enolase (νGlyc+1). 2) PEPCK-M is the only significant source of PEPCK activity in islets and INS-1 832/13 cells (νPEPCK-M+1 ≫ νPEPCK-C+1). 3) The only reactions contributing significant carbon to PEP synthesis are enolase (νGlyc+1) and PEPCK-M (νPEPCK-M+1), and there is negligible reverse flux from pyruvate kinase into PEP (νPK-1). 4) The only reaction contributing to significant disappearance of PEP is PK (i.e. there is minimal net reverse enolase (νGlyc-1)or PEPCK (νPEPCK-C-1 + νPEPCK-M-1) flux). 5) The size of the mitochondrial anion pools (i.e. of malate and PEP) are small relative to and in rapid communication with the cytosolic pool (9). 6) PEP derived from PEPCK-M (PEPmt) and from enolase (PEPGlyc) is metabolically indistinguishable by PK. 7) Malate is primarily of mitochondrial origin and is in mass isotopologue equilibrium with OAA in both pools (10, 11).
Thus, PEP is treated as a single pool whose composition is determined by the rate of appearance of PEP (νGlyc+1 + νPEPCK-M+1) minus the rate of disappearance of PEP (νPK+1). Because νPK+1 for PEPmt and PEPGlyc is proportional to their population of this pool at any given time, the fractional contribution of PEPCK-M flux relative to glycolytic flux (ΦPEPCK-M) during the course of the incubations can be estimated to be ΦPEPCK-M = 100 × νPEPCK-M+1/(νPEPCK-M+1 + νGlyc+1). The 13C enrichment of the product of an enzymatic reaction is determined by the 13C enrichment of the precursor pool. Because the 13C enrichment of PEP (PEPAPE) and its precursor pool (malateAPE) can be measured, then the ΦPEPCK-M is determined simply by 100 × PEPAPE/MalateAPE as long as there is a 1:1 relationship between the product and precursor labeling.
Statistics and Data Analysis
All data are reported as the mean ± S.E. Unpaired two-tailed Student t tests and one-way ANOVA were performed using the prism software package version 5 (GraphPad). Differences were considered to be significant at p < 0.05.
RESULTS
PEPCK mRNA and Protein
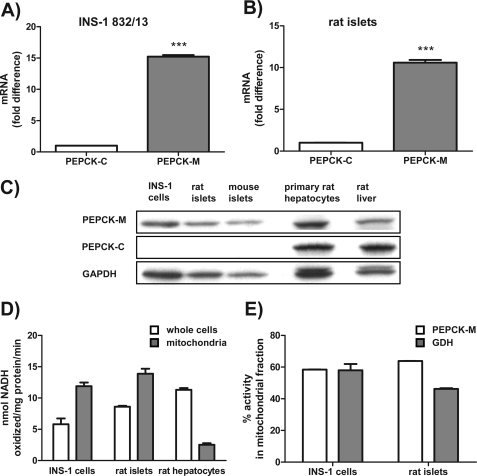
Previous reports have confirmed the absence of any significant PEPCK-C activity in islets, but little is known about PEPCK-M (12, 13). PEPCK-M mRNA was 15-fold higher relative to PEPCK-C in INS-1 832/13 cells (Fig. 2A) and 11-fold higher level in rat islets (Fig. 2B). The relative abundance of PEPCK-M message was 97.1 ± 0.3% of SCS-GTP and 20 ± 0.4% of β-actin in INS-1 832/13 cells. Western blotting demonstrated no significant PEPCK-C in INS-1 832/13 cells, mouse, or rat islets and confirmed its presence in primary rat hepatocytes and whole rat liver (Fig. 2E). In contrast, robust PEPCK-M expression was detected in INS-1 832/13 cells, rat, and mouse islets at a level comparable with that observed in the hepatocytes and liver.
FIGURE 2.
PEPCK-M but not PEPCK-C is present in INS-1 832/13 cells, rat islets, and mouse islets. A and B, PEPCK-M mRNA expression in comparison to PEPCK-C mRNA expression by quantitative PCR in INS-1 832/13 cells (A) and rat islets (B). C, Western blots using an antibody specific for PEPCK-C and a peptide antibody specific for PEPCK-M in INS-1 832/13 cells, rat, and mouse islets. D, PEPCK activity in whole cells as well as isolated mitochondria of INS-1 832/13 cells, rat islets, and hepatocytes using a traditional method measuring NADH oxidation. E, PEPCK activity in the mitochondrial fraction of INS-1 832/13 cells and rat islets compared with glutamate dehydrogenase (GDH) activity. Error bars are S.E. and significance by t test; ***, p < 0.001.
PEPCK Enzyme Activities
There is conflicting evidence among PEPCK enzymatic activity determinations for the significant presence versus the absence of PEPCK in rodent islets (12–15). Where no PEPCK activity was detected, the assay was a single end point measurement of the forward reaction in the direction of PEP formation using Mg-ITP as reactant in the absence of any inhibitors (13). A particular concern of the forward reaction is that the product can be rapidly depleted by either contaminating enolase or PK activity leading to gross underestimations of enzyme activity. This is especially important because the IDP formed by PEPCK can lead to consumption of PEP by PK whose activity is extremely high in insulin-secreting cells (13, 16–18). For all our measurements, the reverse reaction (in the direction of OAA formation) was used whereby OAA production is coupled to malate dehydrogenase where NADH oxidation to NAD+ is observed in real time. Specificity of these reactions is conferred by comparison of the activities in the presence and absence of concentrated CO2. dGDP was used because of its preferential use by PEPCK versus PK. Inhibitors were used to avoid nonspecific NADH oxidation by Complex 1 (rotenone) and lactate dehydrogenase (by blocking pyruvate production via inhibition of PK with phenylalanine (Phe)) (6, 7). The enolase inhibitor NaF was used to prevent loss of PEP because of enolase activity.
Using this strategy, substantial PEPCK activity was measured in INS-1 832/13 (51 ± 8%) cells and rat islets (76 ± 1%) compared with primary rat hepatocytes (Fig. 2D). In contrast to whole cell activities, mitochondria PEPCK activities were higher in insulin-secreting cells than hepatocytes; that is, INS-1 832/13 cells having PEPCK activity 470 ± 20% of mitochondria from hepatocytes, although islets were 550 ± 30% higher.
PEPCK-M and PEPCK-C have many similarities in enzyme activities but display notable differences in νmax depending on the pH and the counter ion(s) utilized as well as the Km of CO2 as might be expected for their unique subcellular environments (19–24). Of note, Mg2+ reduces PEPCK-M activity, whereas Mn2+ is the preferred divalent. Mn2+ was used for comparisons of subcellular localization of PEPCK to prevent underestimation of PEPCK-M activity. Direct measurements of PEPCK activity were made in the cytosolic and mitochondrial fractions with glutamate dehydrogenase as a marker for contamination of mitochondrial matrix components. Virtually all of the PEPCK activity from cells and islets was localized to the mitochondrial fraction when adjusted for leakage of mitochondrial contents into the cytosol fraction by parallel measurements of glutamate dehydrogenase activity from the same samples (Fig. 2E).
Intracellular PEPCK-M Flux in INS-1 832/13 Cells
Simply the presence of enzyme activity does not necessarily connote an important physiologic role; therefore, the intracellular function of PEPCK-M was queried. Because PEPCK-M is compartmentalized in the mitochondrial matrix where the concentrations of substrate and products may be locally high and different from the cytosol, direct comparison of these activities ex vivo in lysates may be misleading. Thus, a labeling strategy was developed to measure PEPCK-M flux in intact cells to assess the functional contribution of PEPCK-M to metabolism.
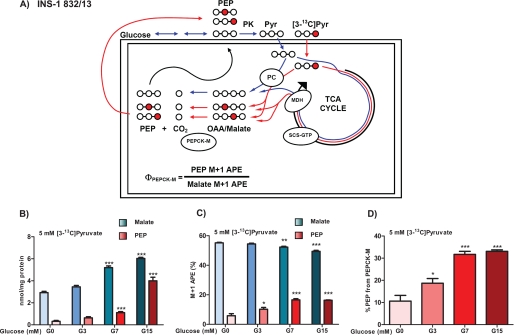
The two primary sources of PEP synthesis are the glycolytic enolase reaction of 2-PG and the cataplerotic PEPCK reaction (Figs. 1 and 3A). Because insulin-secreting cells lack PEPCK-C (Fig. 2 and Refs. 12 and 13)), any PEPCK activity measured in β-cells will arise from PEPCK-M. INS-1 832/13 cells, unlike islets, have high monocarboxylate transporter activity, so 13C-labeled pyruvate can be used to assess PEPCK-M flux in living cells. After entry into the mitochondria, pyruvate carboxylase (PC) can metabolize [3-13C]pyruvate into OAA that becomes racemized across carbons 2 and 3 as a consequence of rapid equilibration across fumarase (Fig. 3A). Pyruvate dehydrogenase metabolism of [3-13C]pyruvate with continued TCA cycle flux will lead to a similar labeling pattern of OAA. Any labeled pyruvate that spontaneously decarboxylated to form [2-13C]acetate would similarly label OAA if incorporated into [2-13C]acetyl-CoA. Therefore, regardless of the direction of mitochondrial metabolism (PC versus pyruvate dehydrogenase), the OAA pool will be enriched equally in the 2 and 3 position during the first cycle. Because OAA and malate are in isotopologue equilibrium with each other, the 13C enrichment of the more abundant and less labile malate reflects the enrichment of OAA. Thus, [3-13C]pyruvate can specifically mass-label the PEPCK precursor pool as determined by measuring the M+1 APE of malate by LC/MS/MS. As the OAA is metabolized to PEP by PEPCK, 100% of the 13C label will be transferred to the respective 2 or 3 position with the loss of unlabeled CO2. The newly synthesized PEP will have the same mass isotopologue enrichment as its precursor OAA. Thus, by knowing the precursor pool enrichment of OAA (as measured by the APE of its surrogate, malate) it is possible to calculate the fractional contribution of PEPCK-M to the cellular PEP pool (ΦPEPCK-M).
FIGURE 3.
Mitochondrial PEPCK makes PEP in INS-1 832/13 cells. A, schematic of PEP labeling strategy. Glucose is metabolized via glycolysis to PEP and then to pyruvate (Pyr) by PK before mitochondrial uptake where it is metabolized by the TCA cycle producing OAA via malate dehydrogenase (MDH) or by PC directly to OAA. Unlabeled glucose or pyruvate will produce malate and PEP that has only natural abundance carbon in all positions (white circles and blue lines). Metabolism of [3-13C]pyruvate (red circles and lines) will label OAA equally in the 2 or 3 position regardless of pyruvate dehydrogenase (PDH) or PC metabolism. B–D, PEPCK-M flux in INS-1 832/13 cells is enhanced by glucose. INS-1 832/13 cells were preincubated in DMEM base without glucose for 45 min. Then 5 mm [3-13C]pyruvate with 0, 3, 7, or 15 mm glucose was added for 45 min before cells were quenched. B, the LC/MS/MS measured concentrations of total malate and PEP (labeled and unlabeled). C, the LC/MS/MS calculated APE of label in malate and PEP M+1 isotopologues. D, the contribution of PEPCK-M to PEP synthesis relative to glycolysis. Error bars are S.E. and significance was by ANOVA; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
INS-1 832/13 cells were incubated in 5 mm [3-13C]pyruvate with G0, G3, G7, or G15 mm unlabeled glucose. As the glucose concentration increases, both glycolytic and mitochondrial metabolite pools expand. As expected the concentration (combined labeled and unlabeled) of PEP and malate increased with increasing concentrations of glucose. Compared with 0 mm glucose, malate concentrations doubled, whereas PEP increased more than 20-fold at 15 mm glucose (Fig. 3B). Similarly, the -fold increase from G0 in 2-PG/3-PG that is in exchange with PEP was 2.0 ± 0.3-fold at G3, 3.9 ± 0.4-fold at G7, and 14.4 ± 1.0-fold at G15. Unlike malate, the aspartate pool that is not in direct chemical exchange with malate shrunk as expected to 0.76 ± 0.02 at G3, 0.47 ± 0.02-fold at G7, and to 0.26 ± 0.2-fold at G15 compared with G0. In contrast to the large changes in concentrations noted above, in the absence of 13C label, the enrichment of the M+1 isotopologues of malate, aspartate, PEP, and 2-PG/3-PG approximated natural abundance predictions (supplemental Fig. S1) with the exception of PEP at G0, where the base-line contribution to M+1 was higher because of the low signal-to-noise ratio.
In the presence of 13C-labeled pyruvate, the enrichment of malate was 55%, with a small but significant decrease in enrichment as the glucose concentration was increased to 15 mm, presumably as a consequence of dilution of the 13C-labeled pyruvate by unlabeled carbon from glucose (Fig. 3C). The 13C enrichment of the aspartate pool, which also is in chemical exchange with the OAA pool, was labeled to a lesser extent (∼43%) and did not change as glucose increased (supplemental Fig. S2), suggesting it is in a different metabolic pool than malate. In sharp contrast, the enrichment of 13C label into PEP steadily grew with increasing glucose concentration, reaching nearly 20% enrichment despite decreasing enrichment of the malate precursor pool. As was seen with the concentrations of PEP and 2-PG/3-PG, the M+1 isotopologue APEs were indistinguishable between these two metabolites (supplemental Fig. S2B), suggesting that νGlyc-1 is not negligible and the two metabolic pools are in rapid exchange with each other.
ΦPEPCK-M was calculated from the relative M+1 enrichment of PEP with respect to its precursor pool reflected in malate 13C enrichment. In the absence of glucose, ΦPEPCK-M was 10%, suggesting most of the carbon in this small PEP pool was unlabeled (Fig. 3D). Remarkably, as the glucose and PEP pool size increase, ΦPEPCK-M climbed to more than 30%. Thus, despite increasing glycolytic precursors, there was a more than 3-fold increase in ΦPEPCK-M, indicating that PEPCK-M activity was stimulated by glucose despite dilution of the precursor pool.
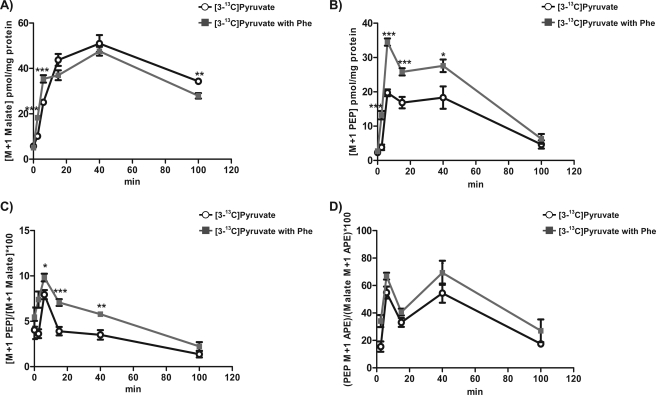
Even though reverse PK flux (νPK-1) is believed to be negligible because of the large free energy, and the rate should decrease with increasing glucose, a cell-permeable PK inhibitor, phenylalanine, was used to rule out any significant contribution of label from the added pyruvate via PK into PEP (25). If there were significant νPK-1, as opposed to νPEPCK-M+1, leading to PEP labeling, Phe would decrease νPK-1 and, thus, decrease the concentration of M+1 PEP in the cells. On the other hand, if PEP labeling comes predominantly from νPEPCK-M+1, then Phe would decrease νPK+1 and lead to an accumulation of M+1 PEP. INS-1 832/13 cells were preincubated in DMEM base without glucose for 60 min to minimize glycolytic metabolites (and, thus, maximize the chance of reverse flux) and then changed to DMEM base with 5 mm [3-13C]pyruvate with or without 5 mm Phe and quenched at the indicated times. PK inhibition initially led to an increase in M+1 malate concentration that peaked at 40 min but did not affect the overall rate of label incorporation of 13C from pyruvate into malate (Fig. 4A). Rather, PK inhibition significantly increased M+1 PEP (as well as total PEP concentrations, not shown) that peaked at 6 min and declined during the remainder of the incubation (Fig. 4B), consistent with synthesis of labeled PEP via νPEPCK-M+1 rather than νPK-1. Similarly, the ratio of the M+1 concentration of PEP to malate was higher in the Phe-treated cells, reflecting accumulation of more PEP (Fig. 4C). Because ΦPEPCK-M is determined by the 13C enrichments of both malate and PEP, the lack of a difference with Phe treatment further supports νPEPCK-M+1 as the primary metabolic pathway of M+1 PEP from [13C]pyruvate (Fig. 4D).
FIGURE 4.
Inhibition of pyruvate kinase by phenylalanine in INS-1 832/13 cells. A–D, INS-1 832/13 cells were preincubated in DMEM base without glucose for 60 min. Then the medium was changed to DMEM base with 5 mm [3-13C]pyruvate with (gray squares) or without (black circles) 5 mm phenylalanine and quenched at the indicated times. The concentration of 13C-labeled M+1 isotopologue of malate (A) and PEP (B) with PK inhibition. C, the ratio of [PEP M+1] concentration to [Malate M+1] concentration. D, ratio of PEP M+1 APE to malate M+1 APE (ΦPEPCK-M). Error bars are S.E. and significance was by ANOVA; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Intracellular PEPCK-M Activity in Rat Islets
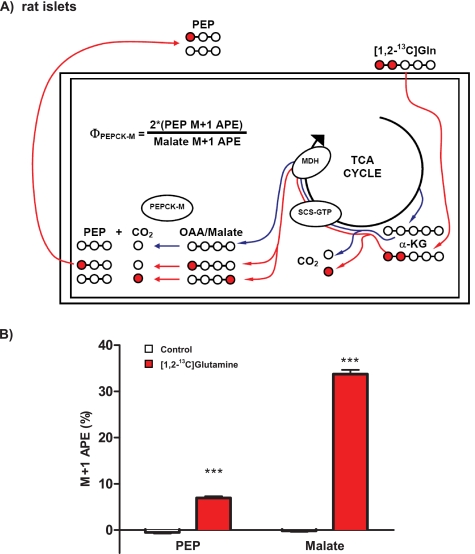
Because islets lack substantial monocarboxylate transporter activity and, therefore, do not take up pyruvate (8), a different labeling strategy using [1,2-13C]glutamine was implemented (Fig. 5A). Glutamine requires mitochondrial metabolism to generate PEP; hence, any contribution of label to PEP from reverse synthesis from pyruvate by PK is eliminated. Glutamine is transported into the islet and deaminated to α-KG then metabolized to succinyl-CoA, losing the labeled carbon in position 1 as 13CO2. The 13C label is evenly split between the 1 and the 4 carbon of OAA because of racemization by SCS and while generating GTP in the process (10, 11). Thus, when CO2 is lost to make PEP, only half of the labeled carbon remains so that the ΦPEPCK-M, when [1,2-13C]glutamine is substrate, is calculated as 100 × [2(PEP M+1 APE)/(Malate M+1 APE)].
FIGURE 5.
Mitochondrial PEPCK makes PEP in intact rat islets. A, schematic of PEP labeling strategy for islets. [1,2-13C]Glutamine (red circles and lines) is transported into the islet and deaminated to α-KG and then metabolized to succinyl-CoA loosing the label in carbon 1 to CO2. Racemization across SCS and fumarase will evenly split the 13C label between carbons 1 and 4 of OAA. Thus, only half of the PEP produced by PEPCK-M will retain its label, whereas half is lost to CO2. Unlabeled glutamine will produce malate and PEP that has only natural abundance carbon in all positions (white circles and blue lines). B, PEPCK-M activity in intact rat islets using [1,2-13C]glutamine. Rat islets were preincubated in DMEM base without glucose for 45 min and then with or without 5 mm [1,2-13C]glutamine for 45 min. The 13C enrichment of the M+1 isotopologue of malate M+1 and PEP are shown. Error bars are S.E. and significance by t test and ANOVA; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Islets were preincubated in DMEM base in the absence of glucose for 45 min before incubation with 5 mm [1,2-13C]glutamine or unlabeled glutamine for 45 min. There were no differences in the concentration of malate or PEP (combined labeled and unlabeled) in the islets after the incubation. Islets incubated in unlabeled glutamine had no enrichment in the M+1 isotopologues of either malate or PEP. In contrast, labeled glutamine enriched the M+1 isotopologue of malate to ∼33%, whereas that of the PEP M+1 isotopologue reached nearly 7% (Fig. 5B). Thus, the PEPCK-M contribution to islet PEP metabolism was calculated as 41 ± 3%. Importantly, as in INS-1 832/13 cells, flux through PEPCK-M in islets is also quite substantial compared with glycolysis.
PEPCK-M in Insulin Secretion
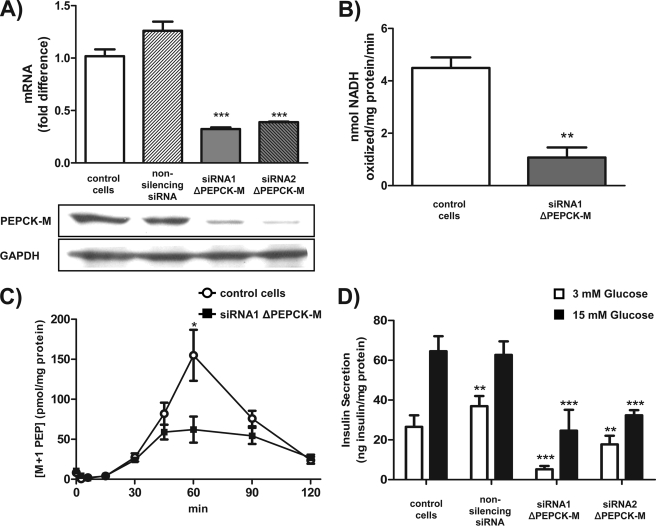
To assess whether the observed high PEPCK-M metabolic flux is related to insulin secretion, PEPCK-M was silenced in INS-1 832/13 cells with two separate siRNAs. After transfection, there were 68 and 61% reductions, respectively, in PEPCK-M message for siRNA1 and siRNA2 in INS-1 832/13 cells (Fig. 6A, top). Silencing was confirmed at the protein level by Western blotting (Fig. 6A, bottom) as well as by a 75% reduction in the PEPCK activity in whole cells (Fig. 6B).
FIGURE 6.
PEPCK-M is required for PEP synthesis and insulin secretion. A, two different siRNAs were used to silence PEPCK-M expression in INS-1 832/13 cells. -Fold change in PEPCK-M mRNA levels after 36 h of transfection with a control siRNA (non-silencing siRNA), siRNA1, or siRNA2 against PEPCK-M normalized to GAPDH. Western blot analysis of PEPCK-M after knockdown using siRNA in INS-1 832/13 cells is shown below. B, PEPCK activity measurement in INS-1 832/13 cells after silencing of PEPCK-M with siRNA1 compared with non-treated cells. C, time course of M+1 labeled PEP by PEPCK-M. INS-1 832/13 cells were preincubated in DMEM base without glucose for 60 min and then 15 mm glucose with 5 mm [3-13C]pyruvate. Shown are the concentrations of the M+1 isotopologue of PEP metabolized by PEPCK-M in non-transfected and siRNA1-transfected cells at the indicated times. D, glucose-stimulated insulin secretion in INS-1 832/13 cells that were non-transfected or transfected with a control siRNA, siRNA1, or siRNA2 against PEPCK-M. Cells were preincubated in KRBH with 3 mm glucose for 90 min and then with either 3 or 15 mm glucose for 90 min. Error bars are S.E. and significance by t test and ANOVA; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To confirm functional loss of PEP synthesis from PEPCK-M silencing in intact cells, labeling studies were performed in control and silenced INS 832/13 cells. Cells were incubated simultaneously with 15 mm glucose and 5 mm [3-13C]pyruvate, and then PEP enrichment was measured at different time points. When PEPCK-M was silenced, both total and 13C-labeled PEP concentrations were significantly reduced, confirming the impact of silencing on metabolism (Fig. 6C).
Because mtGTP had been previously demonstrated to be essential for GSIS, and PEPCK-M is dependent on mtGTP for mitochondrial PEP synthesis, the role of PEPCK-M in insulin secretion was assessed. Compared with control cells, basal insulin secretion in 3 mm glucose was significantly suppressed (Fig. 6D). Furthermore, insulin secretion in response to 15 mm glucose was completely reduced to that of control basal secretion when PEPCK-M was silenced by either siRNA, illustrating the importance of both mtGTP and PEPCK in GSIS.
DISCUSSION
Pancreatic β-cells couple the oxidation of glucose to the secretion of insulin. Although KATP channels are definitively involved in GSIS, this mechanism does not account for the entirety of the metabolic signal. Indeed, in the absence of any measurable KATP channel activity in the islets of SUR−/− mice, glucose-induced calcium oscillations and insulin secretion are observed (26, 27). Investigations of KATP-independent mechanisms of GSIS have identified important roles for both anaplerotic metabolism from high PC flux and SCS-GTP-produced mtGTP (1–5). In addition to previous reports suggesting the presence of PEPCK activity, data presented herein indicate 1) there is significant mRNA expression, immunodetectable protein, and enzyme activity of PEPCK-M but not PEPCK-C, 2) there is a substantial metabolic flux of substrate through PEPCK-M in intact cells and islets, and 3) that PEPCK-M is involved in GSIS. Given that mtGTP synthesized by SCS-GTP is an indicator of TCA flux and crucial for GSIS, PEPCK-M is a strong candidate for connecting this metabolic signal to insulin release by hydrolyzing mtGTP to make PEP (Fig. 1).
There are conflicting reports whether or not significant PEPCK activity exists in pancreatic islets. A similar controversy has arisen in studies of gluconeogenesis in the liver where both PEPCK isoforms are reported to varying extents. Unlike most eutherian mammals where PEPCK-M is significant compared with PEPCK-C, the prevailing opinion is that the rat has little or no detectable hepatic PEPCK-M activity. Compartmentalization of PEPCK to the cytosol and mitochondria of the rat has been compared mostly by measuring enzymatic activity either directly or indirectly in fractions of tissue homogenate with the percentage of PEPCK-M activity ranging from undetectable (6, 28), 5% (29), 6% (30), 10% (31),18% (32), 25% (33, 34), to 50% (35). This approach has inherent weaknesses inasmuch as the assumption is made that enzymatic activities under the assayed condition, are representative of metabolic flux within each of these compartments in a living cell. There is no clear explanation for why there is such a wide range of ratios of mitochondrial versus cytosolic PEPCK activity. Some of the differences might be attributed to the age of the rodent (PEPCK-M is prominent early in life) (31, 32, 35), whether they were fed or fasted (fasting or stress favors PEPCK-C since it is glucagon and glucocorticoid driven), mitochondrial isolation techniques (differences arise from hypotonic lysis, sonication, freeze thaw, detergent, and lyophilization) (28, 30, 36), whether leakage of mitochondrial contents into cytosolic fractions was accounted for, or whether the mitochondrial activity was measured directly or as a difference between total and cytosolic. Moreover, direct comparisons of PEPCK-M and PEPCK-C enzymatic activity favor one or the other isoform based on the type of assay (whether the assay is measured as the forward reaction, reverse reaction or by CO2 fixation), the pH, the counter ion (Mn2+, Mg2+, or both), and different Km of products and reactants for the different directions (19–24). A third confounding factor in these comparisons is that of competing enzyme activities. In particular when inositol nucleotides are used as a reactant, then PK becomes a concern as both islets and rat liver (compared with other species) have particularly high levels of PK, and both PK as well as PEPCK can utilize IDP in reactions with PEP to lead to either false positive or false negative activities depending on the type of assay performed (13, 16–18).
Early reports of a PEPCK enzymatic activity were described in mouse islets with many of the same characteristics as that of PEPCK from liver (including the dissociation constants and a dependence on Mn2+ but lacking “ferroactivator” or hormonal/nutrient stimulation) (14, 15). Later, while investigating why glyceraldehyde but not pyruvate itself stimulated insulin secretion in islets, no significant PEPCK activity in rat islets was detected despite a seemingly exhaustive analysis (13). Subsequently, as further support for the absence of PEPCK activity, the same group found no demonstrable evidence for gluconeogenesis from labeled pyruvate nor mRNA from PEPCK-C in islets (12). Lack of monocarboxylate transporter activity in islet β-cells, unlike other islet cell types, later proved to be the explanation for non-responsiveness to pyruvate (8). Despite later reports that pancreatic islets contain PEPCK activity as well as PEPCK protein by Western blot and immunohistochemistry (37, 38), the prevailing conclusion remains that insulin-secreting cells do not contain substantial PEPCK activity.
Retrospectively, these discrepancies may be reconciled on the basis 1) of the presence of two distinct PEPCK gene products (mitochondrial and cytosolic), 2) that islets contain little or no PEPCK-C, and 3) the different assays variably detected the cytosolic and mitochondrial isoforms. In the two reports of observed PEPCK activity, the total soluble cellular fraction (i.e. containing both cytosolic and mitochondrial fractions) was assayed (14, 15). Activity was measured in the forward direction from islets that were sonicated and dialyzed in a hypotonic buffer using ITP. NaF was added as an inhibitor to block enolase, and the addition of Mn2+ led to a large stimulation of activity. When PEPCK activity was found lacking in rat islets cytosol, a “true” cytosolic fraction devoid of PEPCK-M activity was assayed (13). Because islets lack PEPCK-C, minimal activity was observed in this fraction. When the mitochondrial fraction was studied, intact mitochondria were added to a similar reaction mixture that contained ITP and Mg2+ but with no inhibitors. The lack of activity in the mitochondrial fraction could be because either the mitochondria were still intact and reactants could not get in or contaminating mitochondrial outer membrane-associated enolase activity consumed the PEP. Mn2+ or Fe2+ did not further stimulate PEPCK activity, but because the combination of Mg2+ and Mn2+ can blunt enzyme activity measurements and ferroactivator (later identified to be glutathione peroxidase) does not activate the mitochondrial fraction, this is not surprising (39).
Here significant PEPCK-M enzyme activity is reported in islets and INS-1 832/13 cells, comparable with the total activity found in hepatocytes. The assay to measure PEPCK activity is a modified version of Petrescu et al. (7) where dGDP is used as nucleotide because PK cannot utilize it, rotenone is used to inhibit Complex 1, and background activity is accounted for by parallel reactions lacking HCO3−. In addition, NaF was added to inhibit contaminating enolase and because PK activity is so high in islets an additional inhibitor of PK activity (Phe) was used (25). Reduction in PEPCK activity after siRNA silencing (Fig. 6B) confirms that the activity measured is that of PEPCK-M.
PEPCK-M mRNA has previously been detected in the pancreas by Northern blotting, whereas PEPCK-C was absent (40). Here we demonstrate by quantitative PCR that PEPCK-M and not PEPCK-C are present in INS-832/13 cells and rat islets (Fig. 2A). In addition, silencing PEPCK-M by two different siRNA reduces PEPCK-M message (Fig. 6A), confirming the identity of the message.
Previously, PEPCK has also been detected in islets using a rabbit polyclonal antibody by Western blotting and immunohistochemistry. This antibody was raised against PEPCK protein purified from whole liver by GTP affinity chromatography (37). Because both PEPCK isoforms have high sequence homology, similar affinities for GTP, and the same electrophoretic mobilities, it is likely that this antibody was raised against both PEPCK-M and PEPCK-C as antigens. Furthermore, given the high sequence homology of the two isoforms, cross-reactivity might be expected. Using an isoform-specific antibody from a peptide unique to PEPCK-M, PEPCK-M was clearly identified in INS-1 832/13 cells as well as rat and mouse islets at comparable levels with those found in hepatocytes. Silencing PEPCK-M led to a reduction in the immunodetectable PEPCK-M, confirming the identity of the band on Western blot.
Although identification of PEPCK-M at the message, protein, and enzyme activity level in tissue extracts is important, demonstration of significant functional activity of the enzyme in intact cells is essential to ascribe an important cellular role of the enzyme. Novel 13C labeling strategies were developed for LC/MS/MS measurement of PEPCK-M activity in intact cells and islets. This strategy allows for measurement of ΦPEPCK-M, the fractional contribution of PEPCK-M to PEP synthesis relative to glycolysis. Concordant with the assumptions made for these calculations, PEPCK-M was the only significant source of PEPCK activity (νPEPCK-M+1 ≫ νPEPCK-C+1) (Fig. 2). The theoretical contribution of reverse flux from PK (νPK-1) was confirmed to be negligible (Fig. 4). Because there is not an intramitochondrial pool of 2-PG or 3-PG, the identical 13C enrichment of these analytes as PEP demonstrates that the mitochondrial anion pool is small relative to and in rapid communication with the cytosolic pool of PEP (supplemental Fig. S2B). In addition, the similarity of the enrichment pattern supports PEPmt and PEPGlyc being metabolically indistinguishable by PK. Furthermore, the identical enrichments also suggest that during glucose stimulation there is significant flux in the gluconeogenic direction beyond enolase. Such activity could lead to an underestimation of ΦPEPCK-M. It is of interest that although fructose-1,6-bisphosphosphatase expression is normally low in insulin-secreting cells, it is increased during pathological states of the islet and that overexpression of the enzyme reduces insulin secretion (41). It remains unclear how far in the gluconeogenic direction mitochondrial metabolites can traverse under physiological conditions.
The addition of 13C-labeled substrate to islets resulted in ΦPEPCK-M as high as 41% (Fig. 5), a surprisingly similar percentage compared with maximal PEPCK-M flux in pigeon liver during gluconeogenesis (36). Furthermore, not only is PEPCK-M flux robust compared with glycolysis, but also it is increased further by glucose-stimulated mitochondrial metabolism (Fig. 3D). Thus, multiple lines of compelling data support a crucial physiologic role for PEPCK-M in β-cell metabolism.
There are no known allosteric regulators of either PEPCK isoform, which is unusual for an enzyme at the intersection of important metabolic pathways (42). In contrast to PEPCK-M, much of the control of PEPCK-C appears to be hormonally driven at a transcriptional and translational level. Given the lack of known regulators of PEPCK-M expression, flux through this enzyme is consequently much more susceptible to vary based on product and substrate concentrations. Although, it has been proposed that mitochondrial membrane potential uptake of Mn2+ might be a potential activator of PEPCK-M given 1) the 3000-fold discriminatory activity of Mn2+ over Mg2+ and 2) Mn2+ deficiency in rats leads to decreased insulin secretion, the association remains to be demonstrated (43, 44). The Km of PEPCK-M for HCO3− is an order of magnitude higher than for PEPCK-C (25 versus 2 mm), suggesting that under physiologic conditions, compared with PEPCK-C, metabolism in the direction of PEP may be even more favored for PEPCK-M (45). Catalysis requires two Mn2+ ions, although Mg2+ can substitute, and GTP is the obligate nucleotide that binds before OAA and has a slow off-rate (46, 47). Once GTP is bound, it has a high likelihood of progressing to product (PEP, CO2, and GDP) (45). Whether mitochondrial OAA or mtGTP are rate-determining remains an open question, but the tight association between mtGTP synthesis and PEP production would suggest mtGTP is rate-limiting (23).
In the mitochondrial matrix PEPCK-M is uniquely positioned to integrate the SCS-derived mtGTP TCA flux signal with PC flux. Functional coordination of GTP synthesis by SCS-GTP with PEPCK-M synthesis of PEP has been shown such that at high flux rates there was nearly a 1:1 relationship between mtGTP synthesis and PEP production in isolated mitochondria (23). Moreover, PEP synthesis by PEPCK-M was proposed to be involved in calcium-dependent increases in gluconeogenesis whereby increases in cytosolic calcium activated the 2-OG dehydrogenase to increase net GTP synthesis by SCS-GTP (48). The metabolic role of PEPCK-M in liver is unclear, although it may contribute to hepatic gluconeogenesis in a manner similar to PEPCK-C but may be more predisposed to using substrates preferentially favoring mitochondrial metabolism. Interestingly, although mice lacking PEPCK-C in the liver have some abnormalities in gluconeogenesis, they nevertheless maintain fasting euglycemia, and although extra-hepatic glucose synthesis may compensate, these results may also suggest an undiscovered role for PEPCK-M in gluconeogenesis (49, 50).
Previously it has been demonstrated that PEPCK-M synthesis of PEP in the mitochondria rapidly leads to export of PEP from such mitochondria and has a maximum matrix concentration of 11 nmol/mg of mitochondria (9). PEP itself first emerged as a candidate second messenger when it stimulated insulin release in a reconstituted Cod islet insulin secretion assay (51). The concentration of intra-islet PEP correlated with insulin secretion in response to several fuels (52). PEP was later demonstrated to have the same effect on mitochondrial calcium extrusion as had been seen in other tissues, and it was proposed that PEP stimulated insulin secretion by mobilizing Ca2+ from the mitochondria in an adenine nucleotide transporter-dependent manner (53, 54). However, the emphasis on PEP as a second messenger waned upon the reports of KATP-dependent insulin secretion and that PEPCK (at least PEPCK-C) was not present in islets (13, 55). Furthermore, although phenylalanine nearly doubled PEP levels, it did not increase GSIS, suggesting cytosolic PEP per se was not sufficient to augment insulin release (56). In agreement, overexpression of Escherichia coli PEPCK in the cytosol of islets actually reduced glucose-stimulated increases in insulin synthesis (secretion was not measured) (57). Thus, because total PEP concentrations may not be important for insulin release, the signal from mitochondria-generated PEP is more likely to be either via intramitochondrial allosteric effects or via the transport of PEP out of the matrix in exchange or in lieu of another metabolite. In the event where the cataplerotically synthesized PEP (via PEPCK-M from anaplerotically generated OAA and mtGTP) is transported out of the mitochondria to join the cytosolic PEP pool, a PEP cycle will, thus, be formed, coupling TCA cycle flux with PC flux (Fig. 1).
A PEP cycle is entirely consistent with the compelling observations now from multiple laboratories that anaplerotic pyruvate cycling is an important component of the mechanism regulating GSIS (2–4, 58–63). Because of its dependence upon mtGTP, maximal PEP cycle flux is limited to TCA cycle flux through SCS-GTP, and that may explain why the best correlation of pyruvate cycling with insulin secretion included both PC and TCA flux (4). Just how such cycling is connected to insulin secretion remains a puzzle. Several cycles have been proposed to account for the observed cycling of carbons to pyruvate including 1) the pyruvate-malate cycle, 2) the pyruvate-citrate cycle, and 3) the pyruvate-isocitrate cycle, each of which will be considered below (for review, see Ref. 64). It is particularly important to note that at the time NMR isotopomer modeling techniques were developed to measure pyruvate cycling, malic enzyme was assumed to be the sole path back to pyruvate (i.e. PEPCK-M flux of carbons to PEP and then to pyruvate were assumed to be absent) (2). Thus, any estimate of pyruvate cycling by these techniques includes an indistinguishable component of PEP cycling. It will be, therefore, important in future studies to identify the individual malic enzyme and PEPCK-M contributions when considering the relationship between pyruvate cycling and insulin secretion.
The pyruvate-malate cycle involves the anaplerotic production of malate that is then converted directly to pyruvate via the cytosolic or mitochondrial isoform of malic enzyme. In these reactions mitochondrial malic enzyme reduces NAD+ in the mitochondrial matrix, whereas the cytosolic form reduces NADP+ in the cytosol. There remains considerable controversy about the role of malic enzyme in insulin secretion as overexpression and silencing studies in clonal INS-1 cell lines support a role for malic enzyme in insulin secretion, whereas data supporting such a role in islets is lacking (3, 58–60, 63). Although primary tissue is more physiologically relevant than cell lines, given the discrepancy between the observations in INS cells and islets, interpretation of the negative data from rat islets where the cytosolic isoform of malic enzyme was silenced by adenovirus must be considered with caution (60). This is especially important because only a single titer of one virus was reported versus a scrambled control short hairpin RNA with the absence of supporting functional data such as reductions in enzyme activity or diminished pyruvate cycling and NADPH levels (as had been seen in the cell lines). Given that parallel malic enzyme reactions occur in the cytosol and mitochondria as well as parallel cytosolic NADPH-generating reactions (e.g. via cytosolic isocitrate dehydrogenase), if the malic enzyme reaction is important, these parallel pathways could be compensatory. Similar to the pyruvate-malate cycle, the pyruvate-citrate cycle also requires malic enzyme. In this cycle, anaplerotic citrate is exported to the cytosol, and as such OAA and acetyl-CoA are delivered to the cytosol by the action of citrate lyase. For cycling to occur, the OAA that is metabolized to malate is then decarboxylated to pyruvate by cytosolic malic enzyme, leaving the acetyl-CoA for lipid synthesis pathways. Complementary results were observed in both INS-1 cells and islets where citrate lyase activity was inhibited or silenced with no diminishment of insulin secretion, suggesting a nonessential role for this cycle in GSIS (65). Both the pyruvate-malate and the pyruvate-citrate cycle would be in competition with the PEP cycle for carbons from PC flux, and the maximal rate of PEP cycling would be dependent upon mtGTP production.
More recently the pyruvate-isocitrate cycle has reached some prominence as a potentially important cycle to regulate insulin secretion. In this cycle, the citrate/isocitrate flux is diverted to the cytosol via the citrate carrier (CIC), and then eventually NADPH and α-KG are made by the action of the cytosolic isoform of isocitrate dehydrogenase (cIDH). The α-KG then is either further metabolized in the cytosol or returns to the TCA cycle. Without a return to the mitochondria to enter the PEP cycle or the pyruvate-malate cycle, the carbons in this pathway cannot cycle back to pyruvate. Thus, unlike the two previously described cycles, this cycle does not involve anaplerotic pyruvate cycling but, rather, is an extra-mitochondrial loop for generation of cytosolic NADPH and/or α-KG that is reminiscent of the well established isocitrate-oxoglutarate shuttle for transporting mitochondrial NADPH equivalents to the cytosol (66, 67). It is, therefore, not surprising that eliminating this extra-mitochondrial loop by silencing cIDH leads to only a ∼20% reduction in total pyruvate cycling compared with controls (62). In support of the importance of this loop, silencing cIDH was accompanied by dramatically diminished GSIS in INS-1 cells and rat islets compared with a scrambled control, although GSIS did not completely correlate with the degree of mRNA suppression. Although there was a significant ∼15% decrease in NADPH/NADP+ ratios when cIDH was silenced, this ratio was the same as that found during maximal GSIS, suggesting that the impaired GSIS under these conditions was not primarily because of the NADPH/NADP+ ratio. Cytosolic aconitase has a dual role as an enzyme catalyzing the interconversion of citrate and isocitrate as well as a transcription factor (regulating among other things iron homeostasis) that translocates to the nucleus upon losing its Fe-S complex (69). Furthermore, both citrate and isocitrate are physiologic chelators of divalent cations such as calcium and manganese. It is, therefore, possible that altering citrate and/or isocitrate fluxes and/or concentrations in the cytosol influence GSIS by a mechanism independent of the proposed cIDH mechanism. Both silencing and overexpressing the CIC transporter also provided further evidence supporting the importance of the cIDH pathway (61). Given that this exchange carrier can transport citrate, isocitrate, malate, and PEP, it is unclear whether the effects on GSIS are because of a pathway involving cIDH or an anaplerotic cycling pathway. Regardless, unlike the two aforementioned pyruvate cycles, the cIDH loop is independent of PEP cycling except in the case where PEP transport via the CIC is interrelated or the carbons from α-KG do not return to the TCA cycle.
Silencing PEPCK-M both reduces νPEPCK-M+1 as well as blocks GSIS (Fig. 6). Unlike other anaplerotic pyruvate cycles that may be involved in insulin secretion, the PEP cycle presented here is unique in that it results in no net reduction of NADP+ to NADPH in the cytosol. However, the PEP cycle could incorporate a cIDH loop to generate cytosolic NADPH, mtGTP, and mitochondrial PEP within a single turn of the cycle. Although intramitochondrial PEP metabolism is a possibility, given the lack of PK in the matrix, transport out of the mitochondria becomes particularly relevant to its signaling role. Mitochondrial PEP transport is carrier-dependent and occurs primarily by the CIC and the adenine nucleotide transporter (in exchange for ADP) (70) (Fig. 1). There may also be a PEP/pyruvate transporter (71). PEP regulation of both mitochondrial calcium transport and ATP/ADP exchange is well documented. The addition of PEP to mitochondria will drive calcium out of the mitochondria in a process that 1) requires an energized membrane potential, 2) is independent of ATP synthesis, 3) is a coupled exchange process that requires transport of phosphate and adenine nucleotides out of the mitochondria, and 4) occurs through the adenine nucleotide transporter (and not the CIC) (38, 48, 68, 72–76). The observation that mtGTP-dependent insulin secretion alters ATP synthesis rates as well as mitochondrial and cytosolic calcium suggests a link between mtGTP and PEPCK-M as well as insulin secretion. An advantage of this interaction would be coupling PC flux with TCA cycle flux to generate PEP in a PEP cycle (Fig. 1) that may then act as a second messenger itself, either as an allosteric modulator (e.g. ATP synthesis or calcium transport) within the mitochondrial matrix or by driving the exchange of another ion into the matrix via mitochondrial exchange transporters (e.g. the CIC or the adenine nucleotide transporter). Although it remains to be determined how mitochondrial PEP synthesis is coupled to insulin secretion, linked co-transport of other ions such as calcium or phosphate may be strong candidates.
In summary, mtGTP and anaplerosis via PC flux are essential components of GSIS that hitherto lacked a clear mechanism linking them together (1–4). Given that PC flux itself involves ATP hydrolysis without increasing electron transport, it is a poor candidate to generate an ATP signal. Similarly, mtGTP synthesis by SCS-GTP, an indicator of TCA flux, is also a potent signal for insulin secretion that leads to reduced ATP levels and ATP synthesis rates (5). Substrate level synthesis of mtGTP by SCS-GTP can provide energy for PEP synthesis from OAA in the mitochondrial matrix by PEPCK-M and thereby link PC flux with TCA cycle flux to stimulate insulin release. Hypoglycemia would occur if the entire insulin content of islets were inappropriately released. As such, requiring simultaneous flux through both PC and the TCA cycle via a transmitochondrial PEP cycle may act as a fail-safe to prevent inappropriate insulin secretion yet allow responsiveness to glucose as the preferred stimulus.
Supplementary Material
Acknowledgments
We thank X. Zhao and L. Thomas for expert technical and administrative assistance.
This work was supported by United States Public Health Service Grants R01 DK-40936, R01 DK-71071, K08 DK-80142, P30 DK-45735, and P30 DK-34989 and by American Diabetes Association Grant 1-09-RA-86.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- GSIS
- glucose-stimulated insulin secretion
- OAA
- oxaloacetic acid
- PEPCK
- phosphoenolpyruvate (PEP) carboxykinase
- PEPCK-M
- mitochondrial PEPCK
- PEPCK-C
- cytosolic
- PK
- pyruvate kinase
- PC
- pyruvate carboxylase
- α-KG
- α-ketoglutarate
- SCS
- succinyl-CoA synthetase
- mtGTP
- mitochondrial GTP
- 2-PG
- 2-phosphoglycerate
- 3-PG
- 3-phosphoglycerate
- MRM
- multiple reaction monitoring
- CIC
- citrate carrier
- cIDH
- cytosolic isocitrate dehydrogenase
- TCA
- tricarboxylic acid
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DMEM
- Dulbecco's modified Eagle's medium
- MRM
- multiple reaction monitoring
- LC/MS/MS
- liquid chromatography-tandem mass spectroscopy
- APE
- atomic percent excess
- ANOVA
- analysis of variance
- rcf
- relative centrifugal force.
REFERENCES
- 1.Newgard C. B., Lu D., Jensen M. V., Schissler J., Boucher A., Burgess S., Sherry A. D. (2002) Diabetes 51, S389–393 [DOI] [PubMed] [Google Scholar]
- 2.Lu D., Mulder H., Zhao P., Burgess S. C., Jensen M. V., Kamzolova S., Newgard C. B., Sherry A. D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2708–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pongratz R. L., Kibbey R. G., Shulman G. I., Cline G. W. (2007) J. Biol. Chem. 282, 200–207 [DOI] [PubMed] [Google Scholar]
- 4.Cline G. W., Lepine R. L., Papas K. K., Kibbey R. G., Shulman G. I. (2004) J. Biol. Chem. 279, 44370–44375 [DOI] [PubMed] [Google Scholar]
- 5.Kibbey R. G., Pongratz R. L., Romanelli A. J., Wollheim C. B., Cline G. W., Shulman G. I. (2007) Cell Metab. 5, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiese T. J., Lambeth D. O., Ray P. D. (1991) Comp. Biochem. Physiol. B 100, 297–302 [DOI] [PubMed] [Google Scholar]
- 7.Petrescu I., Bojan O., Saied M., Bârzu O., Schmidt F., Kühnle H. F. (1979) Anal. Biochem. 96, 279–281 [DOI] [PubMed] [Google Scholar]
- 8.Ishihara H., Wang H., Drewes L. R., Wollheim C. B. (1999) J. Clin. Invest. 104, 1621–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garber A. J., Ballard F. J. (1969) J. Biol. Chem. 244, 4696–4703 [PubMed] [Google Scholar]
- 10.Wood H. G., Lifson N., Lorber V. (1945) J. Biol. Chem. 159, 475–489 [PubMed] [Google Scholar]
- 11.Shreeve W. W., De Meutter R. C. (1964) J. Biol. Chem. 239, 729–734 [PubMed] [Google Scholar]
- 12.MacDonald M. J., McKenzie D. I., Walker T. M., Kaysen J. H. (1992) Horm. Metab. Res. 24, 158–160 [DOI] [PubMed] [Google Scholar]
- 13.MacDonald M. J., Chang C. M. (1985) Diabetes 34, 246–250 [DOI] [PubMed] [Google Scholar]
- 14.Hedeskov C. J., Capito K., Thams P. (1984) Biochim. Biophys. Acta 791, 37–44 [DOI] [PubMed] [Google Scholar]
- 15.Hedeskov C. J., Capito K. (1980) Horm. Metab. Res. Suppl. 10, 8–13 [PubMed] [Google Scholar]
- 16.Bârzu O., Abrudan I., Proinov I., Kiss L., Ty N. G., Jebeleanu G., Goia I., Kezdi M., Mantsch H. H. (1976) Biochim. Biophys. Acta 452, 406–412 [DOI] [PubMed] [Google Scholar]
- 17.Hohnadel D. C., Cooper C. (1973) FEBS Lett. 30, 18–20 [DOI] [PubMed] [Google Scholar]
- 18.Plowman K. M., Krall A. R. (1965) Biochemistry 4, 2809–2814 [DOI] [PubMed] [Google Scholar]
- 19.Holyoak T., Nowak T. (2004) Biochemistry 43, 7054–7065 [DOI] [PubMed] [Google Scholar]
- 20.Ballard F. J., Hanson R. W., Reshef L. (1970) Biochem. J. 119, 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto Y., Shimizu J., Okazaki T., Shukuya R. (1979) J. Biochem. 86, 71–78 [PubMed] [Google Scholar]
- 22.Holten D. D., Nordlie R. C. (1965) Biochemistry 4, 723–731 [DOI] [PubMed] [Google Scholar]
- 23.Ishihara N., Kikuchi G. (1968) Biochim. Biophys. Acta 153, 733–748 [DOI] [PubMed] [Google Scholar]
- 24.Sato A., Suzuki T., Kochi H. (1986) J. Biochem. 100, 671–678 [DOI] [PubMed] [Google Scholar]
- 25.MacDonald M. J., Chang C. M. (1985) Mol. Cell Biochem. 68, 115–120 [DOI] [PubMed] [Google Scholar]
- 26.Szollosi A., Nenquin M., Aguilar-Bryan L., Bryan J., Henquin J. C. (2007) J. Biol. Chem. 282, 1747–1756 [DOI] [PubMed] [Google Scholar]
- 27.Szollosi A., Nenquin M., Henquin J. C. (2007) J. Biol. Chem. 282, 14768–14776 [DOI] [PubMed] [Google Scholar]
- 28.Söling H. D., Willms B., Kleineke J., Gehlhoff M. (1970) Eur. J. Biochem. 16, 289–302 [DOI] [PubMed] [Google Scholar]
- 29.Cornell N. W., Schramm V. L., Kerich M. J., Emig F. A. (1986) J. Nutr. 116, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 30.Nordlie R. C., Lardy H. A. (1963) J. Biol. Chem. 238, 2259–2263 [PubMed] [Google Scholar]
- 31.Ballard F. J., Hanson R. W. (1967) Biochem. J. 104, 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Usatenko M. S. (1970) Biochem. Med. 3, 298–310 [DOI] [PubMed] [Google Scholar]
- 33.Heitzman R. J., Herriman I. D., Mallinson C. B. (1972) FEBS Lett. 20, 19–21 [DOI] [PubMed] [Google Scholar]
- 34.Saggerson D., Evans C. J. (1975) Biochem. J. 146, 329–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horn D. B., Podolin D. A., Friedman J. E., Scholnick D. A., Mazzeo R. S. (1997) Metab. Clin. Exp. 46, 414–419 [DOI] [PubMed] [Google Scholar]
- 36.Söling H. D., Kleineke J., Willms B., Janson G., Kuhn A. (1973) Eur. J. Biochem. 37, 233–243 [DOI] [PubMed] [Google Scholar]
- 37.Ghanaat-Pour H., Huang Z., Lehtihet M., Sjöholm A. (2007) J. Mol. Endocrinol. 39, 135–150 [DOI] [PubMed] [Google Scholar]
- 38.Boquist L. (1987) Biochem. Int. 14, 531–538 [PubMed] [Google Scholar]
- 39.Punekar N. S., Lardy H. A. (1987) J. Biol. Chem. 262, 6714–6719 [PubMed] [Google Scholar]
- 40.Modaressi S., Brechtel K., Christ B., Jungermann K. (1998) Biochem. J. 333, 359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kebede M., Favaloro J., Gunton J. E., Laybutt D. R., Shaw M., Wong N., Fam B. C., Aston-Mourney K., Rantzau C., Zulli A., Proietto J., Andrikopoulos S. (2008) Diabetes 57, 1887–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suzuki M., Yamasaki T., Shinohata R., Hata M., Nakajima H., Kono N. (2004) Gene 338, 157–162 [DOI] [PubMed] [Google Scholar]
- 43.Baly D. L., Curry D. L., Keen C. L., Hurley L. S. (1984) J. Nutr. 114, 1438–1446 [DOI] [PubMed] [Google Scholar]
- 44.Schramm V. L. (1982) Trends Biochem. Sci. 7, 369–371 [Google Scholar]
- 45.Hanson R. W., Patel Y. M. (1994) Adv. Enzymol. Relat. Areas Mol. Biol. 69, 203–281 [DOI] [PubMed] [Google Scholar]
- 46.Holyoak T., Sullivan S. M., Nowak T. (2006) Biochemistry 45, 8254–8263 [DOI] [PubMed] [Google Scholar]
- 47.Hlavaty J. J., Nowak T. (2000) Biochemistry 39, 1373–1388 [DOI] [PubMed] [Google Scholar]
- 48.Deaciuc I. V., D'Souza N. B., Miller H. I. (1992) Int. J. Biochem. 24, 129–132 [DOI] [PubMed] [Google Scholar]
- 49.She P., Burgess S. C., Shiota M., Flakoll P., Donahue E. P., Malloy C. R., Sherry A. D., Magnuson M. A. (2003) Diabetes 52, 1649–1654 [DOI] [PubMed] [Google Scholar]
- 50.She P., Shiota M., Shelton K. D., Chalkley R., Postic C., Magnuson M. A. (2000) Mol. Cell. Biol. 20, 6508–6517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis B., Lazarus N. R. (1977) J. Physiol. 271, 273–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugden M. C., Ashcroft S. J. (1978) Diabetologia 15, 173–180 [DOI] [PubMed] [Google Scholar]
- 53.Ewart R. B., Yousufzai S. Y., Bradford M. W., Shrago E. (1983) Diabetes 32, 793–797 [DOI] [PubMed] [Google Scholar]
- 54.Sugden M. C., Ashcroft S. J. (1977) Diabetologia 13, 481–486 [DOI] [PubMed] [Google Scholar]
- 55.Cook D. L., Hales C. N. (1984) Nature 311, 271–273 [DOI] [PubMed] [Google Scholar]
- 56.Chatterton T. A., Reynolds C. H., Lazarus N. R., Pogson C. I. (1984) Experientia 40, 1426–1427 [DOI] [PubMed] [Google Scholar]
- 57.German M. S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heart E., Cline G. W., Collis L. P., Pongratz R. L., Gray J. P., Smith P. J. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E1354–E1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guay C., Madiraju S. R., Aumais A., Joly E., Prentki M. (2007) J. Biol. Chem. 282, 35657–35665 [DOI] [PubMed] [Google Scholar]
- 60.Ronnebaum S. M., Jensen M. V., Hohmeier H. E., Burgess S. C., Zhou Y. P., Qian S., MacNeil D., Howard A., Thornberry N., Ilkayeva O., Lu D., Sherry A. D., Newgard C. B. (2008) J. Biol. Chem. 283, 28909–28917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joseph J. W., Jensen M. V., Ilkayeva O., Palmieri F., Alárcon C., Rhodes C. J., Newgard C. B. (2006) J. Biol. Chem. 281, 35624–35632 [DOI] [PubMed] [Google Scholar]
- 62.Ronnebaum S. M., Ilkayeva O., Burgess S. C., Joseph J. W., Lu D., Stevens R. D., Becker T. C., Sherry A. D., Newgard C. B., Jensen M. V. (2006) J. Biol. Chem. 281, 30593–30602 [DOI] [PubMed] [Google Scholar]
- 63.Pongratz R. L., Kibbey R. G., Cline G. W. (2009) Methods Enzymol. 457, 425–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jensen M. V., Joseph J. W., Ronnebaum S. M., Burgess S. C., Sherry A. D., Newgard C. B. (2008) Am. J. Physiol. Endocrinol. Metab. 295, E1287–E1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Joseph J. W., Odegaard M. L., Ronnebaum S. M., Burgess S. C., Muehlbauer J., Sherry A. D., Newgard C. B. (2007) J. Biol. Chem. 282, 31592–31600 [DOI] [PubMed] [Google Scholar]
- 66.Palmieri F. (2004) Pflugers Arch. 447, 689–709 [DOI] [PubMed] [Google Scholar]
- 67.Sies H., Akerboom T. P., Tager J. M. (1977) Eur. J. Biochem. 72, 301–307 [DOI] [PubMed] [Google Scholar]
- 68.Chudapongse P., Haugaard N. (1973) Biochim. Biophys. Acta 307, 599–606 [DOI] [PubMed] [Google Scholar]
- 69.Tong W. H., Rouault T. A. (2007) Biometals 20, 549–564 [DOI] [PubMed] [Google Scholar]
- 70.Passarella S., Atlante A., Valenti D., de Bari L. (2003) Mitochondrion 2, 319–343 [DOI] [PubMed] [Google Scholar]
- 71.Satrústegui J., Pardo B., Del Arco A. (2007) Physiol. Rev. 87, 29–67 [DOI] [PubMed] [Google Scholar]
- 72.Shug A. L., Shrago E. (1973) Biochem. Biophys. Res. Commun. 53, 659–665 [DOI] [PubMed] [Google Scholar]
- 73.Peng C. F., Price D. W., Bhuvaneswaran C., Wadkins C. L. (1974) Biochem. Biophys. Res. Commun. 56, 134–141 [DOI] [PubMed] [Google Scholar]
- 74.Chudapongse P. (1976) Biochim. Biophys. Acta 423, 196–202 [DOI] [PubMed] [Google Scholar]
- 75.Sul H. S., Shrago E., Shug A. L. (1976) Arch. Biochem. Biophys. 172, 230–237 [DOI] [PubMed] [Google Scholar]
- 76.Roos I., Crompton M., Carafoli E. (1978) FEBS Lett. 94, 418–421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.