Abstract
Free fatty acids (FFA) induce hepatocyte lipoapoptosis by a c-Jun N-terminal kinase (JNK)-dependent mechanism. However, the cellular processes by which JNK engages the core apoptotic machinery during lipotoxicity, especially activation of BH3-only proteins, remain incompletely understood. Thus, our aim was to determine whether JNK mediates induction of BH3-only proteins during hepatocyte lipoapoptosis. The saturated FFA palmitate, but not the monounsaturated FFA oleate, induces an increase in PUMA mRNA and protein levels. Palmitate induction of PUMA was JNK1-dependent in primary murine hepatocytes. Palmitate-mediated PUMA expression was inhibited by a dominant negative c-Jun, and direct binding of a phosphorylated c-Jun containing the activator protein 1 complex to the PUMA promoter was identified by electrophoretic mobility shift assay and a chromatin immunoprecipitation assay. Short hairpin RNA-targeted knockdown of PUMA attenuated Bax activation, caspase 3/7 activity, and cell death. Similarly, the genetic deficiency of Puma rendered murine hepatocytes resistant to lipoapoptosis. PUMA expression was also increased in liver biopsy specimens from patients with non-alcoholic steatohepatitis as compared with patients with simple steatosis or controls. Collectively, the data implicate JNK1-dependent PUMA expression as a mechanism contributing to hepatocyte lipoapoptosis.
Non-alcoholic fatty liver disease is commonly associated with the metabolic syndrome and affects up to one-third of the American population (1). 5–10% of non-alcoholic fatty liver disease patients develop hepatic inflammation, a syndrome referred to as non-alcoholic steatohepatitis (NASH)2 (2), which can progress to cirrhosis and hepatocellular carcinoma (3, 4). Insulin resistance, a hallmark of the metabolic syndrome, is a major risk factor for NASH and is characterized by an increase in circulating free fatty acids (FFA) (5). These circulating FFA are transported into hepatocytes by the fatty acid transporter protein 5 and CD36 (6–8); within the hepatocyte, these FFA can be esterified to form neutral triglycerides resulting in hepatic steatosis. Esterification of FFA appears to be a detoxification process (9, 10), as non-esterified FFA are inherently toxic to hepatocytes and induce apoptosis, a phenomenon termed lipoapoptosis (11). Saturated and unsaturated FFA differ in regard to their potential for lipoapoptosis; saturated long-chain FFA are significantly more toxic than unsaturated FFA (12, 13). Therefore, it is not surprising that NASH is characterized by both an elevation of serum FFA levels and hepatocyte apoptosis, and the magnitude of circulating FFA correlates with disease severity (14, 15).
Activation of the c-Jun N-terminal kinase (JNK) signaling pathway has been implicated as a central mediator of FFA-induced hepatocyte lipoapoptosis in both rodent and human steatohepatitis (16–18). Of the three mammalian JNK genes, only JNK1 and JNK2 are expressed in the liver (19). These two isozymes are alternatively spliced to yield α and β isoforms of both a p54 and p46 protein (20). JNK activation is mediated through sequential kinase cascade that includes MAPK kinase (MAPKKs) and MAPK kinase kinase (MAPKKKs). Among the MAPKKKs, the mixed lineage kinase 3 (MLK3) when activated phosphorylates and activates MAPKK4 and -7, which in turn phosphorylates and activates JNK. JNK activation can be further self-amplified via a feed forward phosphorylation and activation of MLK3 by JNK (21, 22).
Both JNK1 and -2 have been implicated in liver injury, although JNK1 is more strongly associated with steatohepatitis (17, 18). JNK can cause cell death signals by both transcriptional and post-transcriptional mechanisms (23). JNK1, but not JNK2, phosphorylates c-Jun, a critical member of the activator protein 1 (AP-1) transcription factor complex (24, 25). This transcription factor can induce expression of death mediators (23). Alternatively, JNK can post-transcriptionally activate the pro-apoptotic members of the Bcl-2 family Bim, Bad, and Bax (26–28) or inactivate the anti-apoptotic members of this family Bcl-2 and Bcl-XL (29). Translocation of JNK to mitochondria with associated mitochondrial dysfunction can also cause cell death (30). Although JNK plays a pivotal role in many models of cell death by a variety of mechanisms, its contribution to lipotoxicity remains undefined.
Apoptosis is regulated by BH3-only proteins which constitute a subset of pro-apoptotic members of the Bcl-2 protein family. This group of proteins includes Bad, Bid, Bik, Bim, Bmf, Hrk, NOXA, and PUMA, which display sequence conservation exclusively in the short (9–16 amino acids) BH3 (Bcl-2 homology 3) region, which is necessary for their ability to induce apoptosis. BH3-only proteins such as Bid, Bim, and PUMA can directly activate the multidomain pro-apoptotic members of the Bcl-2 family Bax and Bak (31–34). The oligomerization of Bax and Bak in the outer mitochondrial membrane results in mitochondrial dysfunction, downstream activation of the effector caspases 3, 6, and 7, and ultimately cell death by apoptosis (35). Among the BH3-only proteins, Bim has been identified as contributing to lipoapoptosis (12). Indeed, saturated FFA up-regulates Bim expression, and small interfering RNA targeted knockdown of Bim partially attenuates FFA-mediated apoptosis (12). Because the reduction in apoptosis was incomplete, it appears that in addition to Bim, other BH3-only proteins with complementary functions contribute to lipoapoptosis.
PUMA (p53 up-regulated modulator of apoptosis) is a potent pro-apoptotic protein whose cellular levels are transcriptionally regulated by both p53-dependent and -independent mechanisms (36–42). PUMA is an attractive candidate BH3-only protein to participate in apoptotic cascades with Bim as cooperation between PUMA and Bim during apoptosis has been reported (43). Two BH3-domain-containing isoforms of PUMA have been identified in humans, an α isoform (23 kDa) and a β isoform (18 kDa), both capable of efficiently inducing apoptosis (41). Although PUMA has been extensively studied in several organs including colon cancer cells, thymocytes, and neurons (32, 40, 44, 45), its contribution to liver injury remains unexplored.
The findings of this study suggest that PUMA contributes to FFA-induced lipoapoptosis in liver cells. The saturated FFA palmitate induces PUMA expression by a JNK1/AP-1 signaling cascade. PUMA up-regulation is also demonstrated in human liver samples from patients with NASH, strengthening the in vitro observations. These data provide further mechanistic insights linking JNK activation to the core apoptotic machinery during FFA-mediated lipotoxicity.
EXPERIMENTAL PROCEDURES
Cells
Huh-7 cells, a human hepatoma cell line, were cultured in Dulbecco's modified Eagle's medium containing glucose (25 mm), 100,000 units/liter penicillin, 100 mg/liter streptomycin, and 10% fetal bovine serum. Mouse hepatocytes were isolated from C57BL/6 wild type, p53−/−, Jnk1−/−, Jnk2−/− (The Jackson Laboratory, Bar Harbor, ME), and Puma−/− mice by collagenase perfusion, purified by Percoll gradient centrifugation, and plated as primary cultures. Development and characterization of the Puma−/− mice have been previously described in detail (39). Human hepatocytes were prepared from liver resection specimens of two consenting adult patients undergoing partial hepatectomy for clinical indications. Tumor-free margins of the liver were selected, and hepatocytes were isolated by a two-step collagenase perfusion method as previously described in detail (46).
Fatty Acid Treatment
Palmitic and oleic acids were dissolved in isopropyl alcohol at a stock concentration of 80 mm. FFAs were added to Dulbecco's modified Eagle's medium containing 1% bovine serum albumin to assure a physiologic ratio between bound and unbound FFA in the media, approximating the molar ratio present in plasma (47). The concentration of FFA used in the experiments (800 μm) was similar to the fasting FFA plasma concentrations observed in human non-alcoholic steatohepatitis, ranging from 700 to 1000 μm (15, 48, 49). The concentration of the vehicle, isopropyl alcohol, was 1% in final incubations.
Immunoblot Analysis
Whole cell lysates were prepared as previously described (12), and equal amounts of protein (50 μg) were resolved by SDS-PAGE on a 12.5 or 15% acrylamide gel. Antibodies used were obtained from the following sources: anti-PUMA (Ab 54288; Abcam Inc., Cambridge, MA); anti-NOXA (ProSci Inc., Poway, CA); anti-phospho-SAPK (stress-activated protein kinase)/JNK (Thr183/Tyr185) and anti-SAPK/JNK (Cell Signaling Technology, Danvers, MA); anti-phospho-c-Jun (Ser63), anti-c-Jun, and anti-β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA); anti-S-peptide (a gift from Dr. Scott Kaufmann, Mayo Clinic, Rochester, MN).
Quantitative Real Time Polymerase Chain Reaction
Total RNA was extracted from the cells using the Trizol reagent (Invitrogen) and was reverse-transcribed into complementary DNA with Moloney leukemia virus reverse transcriptase and random primers (both from Invitrogen). Quantification of the complementary DNA template was performed by real-time PCR using SYBR green fluorescence on a LightCycler 480 instrument (Roche Applied Science) as previously described by us in detail (12). Primers used are displayed in Table 1.
TABLE 1.
Primers used for Quantitative real-time PCR
F, forward; R, reverse.
| Gene name | Accession number | Primer sequence (5′ → 3′) | Product size |
|---|---|---|---|
| bp | |||
| 18 S | X03205 | F: cgttcttagttggtggagcg | 212 |
| R: cgctgagccagtcagtgtag | |||
| NOXA | NM_021127 | F: GCAGAGCTGGAAGTCGAGTG | 101 |
| R: GAGCAGAAGAGTTTGGATATCAG | |||
| PUMA | NM_001127240 | F: GACGACCTCAACGCACAGTA | 101 |
| R: AGGAGTCCCATGATGAGATTGT | |||
| Bad | NM_004322 | F: CGGAGGATGAGTGACGAGTT | 123 |
| R: ccaccaggactggaagactc | |||
| Bid | NM_197966 | F: CTACGATGAGCTGCAGACTG | 145 |
| R: GATGCTACGGTCCATGCTGTC | |||
| Bik | NM_001197 | F: GACCATGGAGGTTCTTGGCA | 141 |
| R: AGGCTCACGTCCATCTCGTC | |||
| Bmf | NM_001003940 | F: ccaccagccaggaagacaaag | 172 |
| R: tgctccccaatgggcaagact | |||
| Hrk | NM_003806 | F: TGCTCGGCAGGCGGAACTTGTAG | 100 |
| R: CTTTCTCCAAGGACACAGGG |
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear cell extracts were prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce) according to the manufacturer's instructions. For the EMSA, 20 μg of nuclear proteins were incubated at room temperature for 20 min in binding buffer (25 mm HEPES, pH 7.5, 0.1 m NaCl, 2 mm EDTA, 6% glycerol, 0.1% Triton X-100, 0.1 mm phenylmethylsulfonyl fluoride, 0.4 mm dithiothreitol, 0.5 μg/μl poly(dI-dC), 0.5 μg/μl salmon sperm) with 0.04 pmol of CY 5.5-labeled double-stranded DNA oligonucleotide containing either the wild-type or the mutated putative AP-1 binding sequence within the promoter region or the intronic region of human PUMA gene (Table 2). Protein-DNA complexes were separated from the unbound DNA probe by electrophoresis through 5% native polyacrylamide gels containing 0.5× Tris borate-EDTA. Fluorescence was visualized directly on the gel using an Odyssey fluorescent imager (Licor Biosciences, Lincoln, NE). For competition assays, a 200-fold molar excess of unlabeled double-stranded oligonucleotide was added to the reaction mixture 20 min before the addition of the fluorescent probe. In the supershift assays, nuclear cell extract was first incubated at room temperature for 25 min with 2 μg of anti-phospho-c-Jun or anti-pan-Fos (Santa Cruz Biotechnology). The entire antibody/protein mixture was then incubated with CY 5.5-labeled probe and processed for the gel shift as described above.
TABLE 2.
Sequence of CY 5.5-labeled oligonucleotides used for EMSA (sense sequence)
CY 5.5-labeled forward sense oligonucleotides were annealed with the respective antisense oligonucleotides. Bold sequences indicate the putative AP-1 binding site within the promoter region or the intronic region of human PUMA gene; the underlined sequences indicate the altered nucleotides.
| Oligonucleotide name | Oligonucleotide sequence (5′ → 3′) |
|---|---|
| Promoter AP-1 | CACTGGAAGGTGAGTCACTCTGGTGAGGCG |
| Intronic AP-1 | aagctgagaaattgactcactagcgatggg |
| Promoter AP-1 mutant | CACTGGAAGGAACTGCACTCTGGTGAGGCG |
| Intronic AP-1 mutant | aagctgagaaattgcagttctagcgatggg |
Luciferase Reporter Assay
pAP1-Luc vector (containing multiple copies of the cis-acting enhancer AP-1 binding site) was purchased from Panomics (Fremont, CA). The luciferase reporter construct was transfected into cells, and transfection efficiency was corrected by a Renilla luciferase vector (pRL-TK, Promega, Madison WI). Luciferase vectors were transfected using FuGENE HD transfection reagent (Roche Applied Science), and treatments were carried out 24 h later. Both firefly and Renilla luciferase activities were quantitated using the dual luciferase reporter assay system (Promega) according to the manufacturer's instructions. Luciferase was quantified using a luminometer (TD-20/20, Turner Designs, Sunnyvale, CA).
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assay was performed using a commercially available assay kit (ChIP-IT enzymatic kit, Active Motif, Carlsbad, CA) according to the manufacturer's protocol. Chromatin immunoprecipitation was performed with 3 μg of anti-c-Jun polyclonal antibody (39309, Active Motif) incubated with protein G-coated magnetic beads at 4 °C overnight with rotation. A mock immunoprecipitation without antibody was also performed. DNA fragments co-immunoprecipitated with the target protein c-Jun were subjected to quantitative real-time PCR analysis using various primer sets (Table 3) spanning either the putative AP-1 sequence in the human PUMA promoter, a nonspecific sequence in the human PUMA promoter (used as a negative control), and the AP-1-like sequence in the human JUN promoter (used as a positive control) (50). The amount of promoter sequence present in chromatin immunoprecipitates was expressed relative to the amount of input DNA according to the comparative threshold cycle (Ct) method.
TABLE 3.
Primer sets used for amplification of chromatin immunoprecipitation DNA
TSS, transcription start site; F, forward; R, reverse.
| Promoter region | Location from TSS | Primer sequence (5′ → 3′) | Product size |
|---|---|---|---|
| bp | |||
| PUMA promoter #1 AP-1 | −121 | F: AGATTACCTGCATCTCTTGG | 229 |
| +108 | R: CCCTGCTCTGGTTTGGTGAGT | ||
| PUMA promoter #2, nonspecific | −8372 | F: GCTCACCAAGTCTGGAGCT | 125 |
| −8247 | R: CGAGAGACCGTAACGTGCAT | ||
| JUN promoter | −140 | F: tgtaggagggcagcggagcattacc | 197 |
| AP-1 | +57 | R: gcccgagctcaacacttatctgctaccag |
Plasmid and Transfection
Huh-7 cells were transfected with 1 μg/ml DNA plasmid (PUMA, MISSION short hairpin RNA lentiviral plasmid; Sigma Aldrich) using Lipofectamine (Invitrogen). Stably transfected clones were selected in medium containing 1200 mg/liter G418 and screened by immunoblot analysis. The cDNA encoding the full-length human c-Jun open reading frame (J04111) was generated by PCR using the primers 5′-ACGGGATCCAACTGCAAAGATGGAAACGAC-3′ (forward) and 5′-ACGGAATTCTCAAAATGTTTGCAACTGCTGCG-3′ (reverse). A BamHI and EcoRI restriction site were added, respectively, at the 5′ end of the forward and reverse primer (underlined sequence), and RSV-c-Jun plasmid (kindly provided by Dr. R. Janknecht, Mayo Clinic, Rochester, MN) was used as a template in PCR amplification. PCR amplified full-length human c-Jun was then cloned into BamHI/EcoRI sites of pcDNA3 empty vector which encodes an S-tag peptide at the N terminus (c-Jun plasmid). A dominant-negative transactivation deletion mutant of c-Jun (DN-c-Jun plasmid) was generated using the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) based on the technique described by Hansson et al. (51). Briefly, 50 ng of c-Jun plasmid and 125 ng of each primer were mixed with reaction buffer, which contained dNTPs and Pfu DNA polymerase. Reaction was performed on a thermal cycler (18 cycles) using an annealing temperature of 55 °C. Parental DNA was removed by digestion with DpnI restriction enzyme. Positive clones were selected in the presence of ampicillin and confirmed by sequencing. The following primers pairs were used to delete the first 122 amino acids of the human c-Jun open reading frame (DN-c-Jun): forward primer, 5′-ACCGGCGCGCCGGATCCAAGCCAGAACACGCTGCCC-3′, and reverse primer, 5′-GGGCAGCGTGTTCTGGCTTGGATCCGGCGCGCCGGT-3′. S-tagged c-Jun and DN-c-Jun plasmids were transfected using FuGENE HD transfection reagent (Roche Applied Science), and treatments were carried out 24 h later.
Immunocytochemistry
Cells were cultured on collagen-coated coverslips. After FFA treatment, mitochondria were labeled by the addition of Mito Tracker Red CMXRos to the cultures (100 nm final concentration; Molecular Probes, Eugene, OR) for 30 min at 37 °C in the incubator. The medium was aspirated, and the cells were washed 3 times with phosphate-buffered saline (PBS) and then fixed with freshly prepared 4% paraformaldehyde in PBS containing 0.1 m PIPES, 1 mm EGTA, and 3 mm MgSO4 for 15 min at 37 °C. After a second washing step with PBS, cells were permeabilized using 0.0125% (w/v) CHAPS in PBS at 37 °C for 10 min. Next, the cells were incubated in PBS containing 5% goat serum and 0.1 N NaN3 at room temperature for 30 min. After incubation with anti-Bax antisera (clone 6A7; Exalpha Biologicals, Watertown, MA) overnight at 4 °C, cells were washed 3 times with PBS and incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Molecular Probes) for 1 h at 37 °C. All antibodies were diluted in PBS plus 5% fetal bovine serum. Cells were then washed 3 times in PBS and 3 times in H2O, mounted onto slides using a ProLong Antifade kit (Molecular Probes), and imaged by confocal microscopy with excitation and emission wavelengths of 488 and 507 nm. Fluorescence was quantified using Zeiss KS400 image analysis software (Carl Zeiss, Inc., Oberkochen, Germany). 6A7-immunoreactive cells were quantified and expressed as a percentage of total cells counted.
Quantitation of Apoptosis
Cells were stained with 5 μg/ml 4′,6-diamidine-2′-phenylindole dihydrochloride for 30 min at room temperature and visualized under fluorescence microscopy (Nikon Eclipse TE200, Nikon Corp., Japan). Apoptotic cells were quantified by counting 300 random cells per study. Cells with the characteristic nuclear changes of chromatin condensation and nuclear fragmentation were considered apoptotic. Apoptosis was expressed as a percentage of total cells counted. For caspase 3/7 activity, cells were plated in 96-well plates. The assay was performed using the commercially available Apo-ONE homogeneous caspase 3/7 assay (Promega) according to the manufacturer's instructions as previously described by us (12, 13).
Patient Population
The study was approved by the Mayo Institutional Review Board, and all patients gave written informed consent for participation in medical research. Our cohort consisted of 48 patients who underwent liver biopsy at the time of bariatric surgery for medically complicated obesity at Mayo Clinic, Rochester, MN. The diagnosis of non-alcoholic fatty liver disease was based on liver biopsy features as assessed by an experienced hepatopathologist. Patients whose body mass index of >30 kg/m2, were subdivided into three histologic groups: obese normal (normal liver biopsies, n = 16), simple steatosis (n = 17), and NASH (n = 16) (52). Patients with non-alcoholic fatty liver disease who had secondary causes of steatohepatitis (drugs, prior gastric surgery for obesity) and patients with other etiologies of chronic liver disease (excessive alcohol consumption, viral hepatitis (B, C), cholestatic liver disease, hemochromatosis, Wilson disease, drug-induced liver disease, and α1-antitrypsin deficiency) were excluded from this study. For molecular analysis, a 10–15-mg piece of liver biopsy sample was placed in a microcentrifuge tube that contained 300 μl of HEPES protein extraction buffer (50 mm HEPES, pH 7.4, 0.05% SDS) and 20 μl of protease inhibitor mixture III (Calbiochem® EMD Biosciences, Inc., La Jolla, CA). The tissue sample was then emulsified using a PRO250 Homogenizer (Pro Scientific, Oxford, CT). This tissue lysate was centrifuged at 750 × g for 6 min. The supernatant was removed and centrifuged a second time at 10,000 × g for 5 min. Messenger RNA was extracted using RNeasy Mini kit from Qiagen (Valencia, CA).
Reagents
Palmitic acid (P5585) and oleic acid (O1008) were from Sigma-Aldrich. The JNK inhibitor SP600125 (420119) and the mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK) inhibitor PD98059 (513000) were from Calbiochem. The pan-caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (FK-009) was from MP Biomedicals (Solon, OH).
Statistical Analysis
All data represent at least three independent experiments and are expressed as the means ± S.E. of the mean. Differences between groups were compared using Student's t tests and one-way analysis of variance with post hoc Dunnett test, and significance was accepted at p < 0.05.
RESULTS
Palmitate Induces PUMA Expression
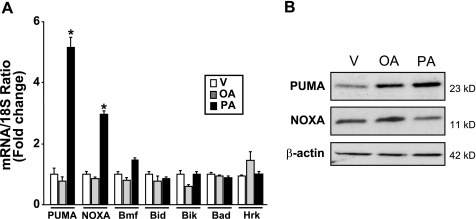
Our prior studies have identified the BH3-only protein Bim as contributing to lipotoxicity (12, 13). Given the fact that BH3-only proteins cooperate in cell death (31), the other members of this family were profiled in Huh-7 cells by real-time PCR and immunoblot analysis. Palmitate induced a 5- and 3-fold increase in PUMA and NOXA mRNA expression, respectively, but less than a 2-fold increase in the remainder BH3-only proteins Bmf, Bid, Bik, Bad, and Hrk (Fig. 1A). Under identical conditions, the non-toxic monounsaturated FFA, oleate, did not induce PUMA or NOXA mRNA expression (Fig. 1A). Consistent with the real-time PCR data, an increase in cellular protein levels of the PUMA α isoform (the β isoform was weakly and inconsistently identified) was observed by immunoblot analysis as early as 12 h after treatment with palmitate (supplemental Fig. S1A). This increase in PUMA protein levels was prolonged and persisted at 16 h (Fig. 1B), whereas no or minimal increase in PUMA protein levels was detected after treatment with oleate (supplemental Fig. S1B and Fig. 1B). In contrast, cellular NOXA protein levels were not consistently increased by either palmitate or oleate treatments (Fig. 1B and supplemental Fig. S1, A and B). These results demonstrate that the saturated FFA palmitate increases PUMA expression both at the message and protein levels in Huh-7 cells.
FIGURE 1.
Palmitate increases PUMA mRNA and protein levels. Total RNA and whole cell lysates were prepared from Huh-7 cells treated with palmitic acid (PA), oleic acid (OA) at 800 μm, and vehicle (V) was used as control. A, BH3-only protein mRNA expression was profiled by real-time PCR 8 h after FFA treatment. -Fold induction was determined after normalization to 18 S. Data represent the mean and S.E. of four independent experiments; *, p < 0.01, palmitate-treated cells versus vehicle- or oleate-treated cells. B, immunoblot analysis was performed for PUMA and NOXA protein expression 16 h after treatment with FFA. β-Actin was used as control for protein loading.
Palmitate-induced PUMA Expression Is JNK-dependent
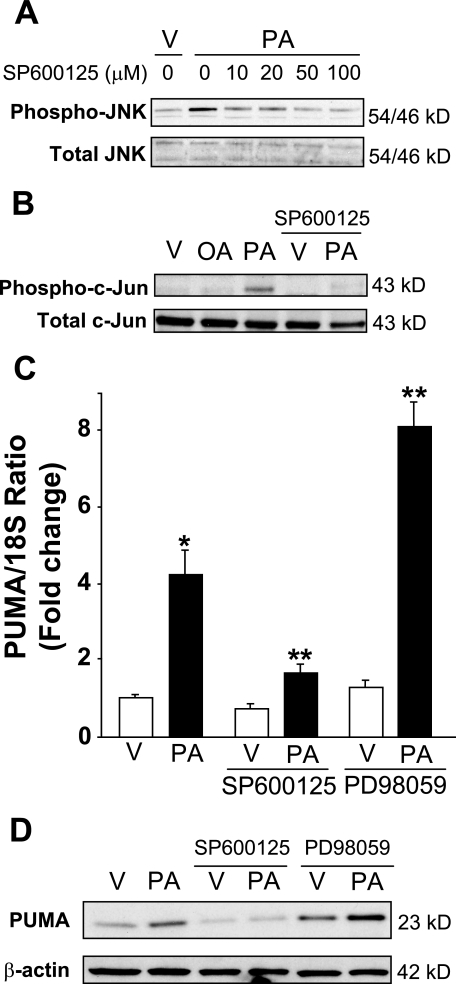
Because JNK activation has been implicated as a key mediator of lipoapoptosis, we next assessed if PUMA expression is also JNK-dependent. Palmitate induced JNK phosphorylation in Huh-7 cells (Fig. 2A), whereas oleate treatment led to a minimal increase in phospho-JNK activity (13, 53). Additionally, palmitate induced c-Jun phosphorylation (Fig. 2B), a known target of JNK1 (18), and the pharmacologic JNK inhibitor, SP600125, prevented palmitate-induced c-Jun activation in Huh-7 cells (Fig. 2B). Although SP600125 is a known potent inhibitor of JNK signaling activity, it also decreases palmitate-induced JNK phosphorylation (Fig. 2A), an observation consistent with a prior report by Bennett et al. (54); this effect is likely mediated by the inhibition of JNK self-amplifying phosphorylation and activation via MLK3 (21, 22). SP600125 treatment also reduced by 70% the palmitate-induced increase in PUMA mRNA and protein levels in Huh-7 cells (Fig. 2, C and D). Pharmacological inhibition of another MAPK, extracellular receptor-stimulated kinase (ERK), by PD98059 did not inhibit, but rather, increased palmitate-mediated PUMA induction at the mRNA and protein levels (Fig. 2, C and D).
FIGURE 2.
JNK pharmacological inhibition prevents palmitate-induced PUMA expression. A, whole cell lysates were prepared from Huh-7 cells treated with palmitic acid (PA) at 800 μm in the presence of serial dose of SP600125 (10–100 μm) for 2 h. Vehicle (V) was used as control. Immunoblot analysis was performed for phosphorylated JNK protein expression and total JNK, a control for protein loading. B, whole cell lysates were prepared from Huh-7 cells treated with vehicle (V), palmitic acid (PA), or oleic acid (OA) at 800 μm and from Huh-7 cells treated with vehicle or palmitic acid in the presence of SP600125 (50 μm). Immunoblot analysis was performed for phosphorylated c-Jun protein expression 16 h after treatment with FFA. Total c-Jun was used as a control for protein loading. C, total RNA were prepared from Huh-7 cells treated with vehicle or palmitic acid (800 μm) in the presence of SP600125 (50 μm) or PD98059 (50 μm) for 8 h. PUMA mRNA expression was profiled by real time PCR. -Fold induction was determined after normalization to 18 S. Data represent the mean and S.E. of three independent experiments. *, p < 0.01, palmitate-treated cells versus vehicle-treated cells. **, p < 0.01, palmitate-treated cells in the presence of inhibitors versus palmitate-treated cells. D, whole cell lysates were prepared from Huh-7 cells treated with vehicle or palmitic acid (800 μm) in the presence of SP600125 (50 μm) or PD98059 (50 μm) for 16 h. Immunoblot analysis was performed for PUMA protein expression, and β-actin was used as control for protein loading.
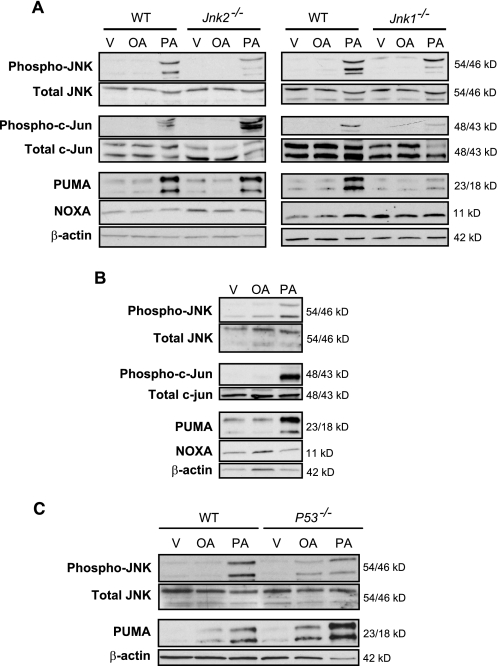
Palmitate also increased JNK phosphorylation and PUMA expression in isolated primary mouse and human hepatocytes (Fig. 3, A and B), demonstrating conservation of this response to palmitate in both primary and transformed hepatocytes and across species. Both 23- and 18-kDa isoforms of PUMA were identified in the primary mouse hepatocytes as previously recognized by Jeffers et al. (39). We next examined the contribution of each JNK isoform to PUMA up-regulation in primary murine hepatocytes obtained from Jnk1−/− or Jnk2−/− mice. The genetic deletion of Jnk1 attenuated palmitate-induced increases of PUMA protein (Fig. 3A), whereas the genetic deletion of Jnk2 did not alter palmitate-mediated increases of PUMA protein. Further evidence for JNK1 in this process was obtained by demonstrating that palmitate induces phosphorylation of the JNK1 substrate, c-Jun, in Huh-7 cells (Fig. 2A) and primary human and mouse hepatocytes (Fig. 3, A and B). Palmitate failed to induce c-Jun phosphorylation in Jnk1−/− primary murine hepatocytes, although activating phosphorylation of c-Jun was observed in Jnk2−/− hepatocytes (Fig. 3A), an observation consistent with prior studies indicating that JNK1, but not JNK2, phosphorylates c-Jun (18). Taken together, these results suggest that palmitate-induced PUMA expression is likely predominantly JNK1-dependent.
FIGURE 3.
JNK1-mediated c-Jun phosphorylation induces PUMA expression in primary hepatocytes during lipoapoptosis. Whole cell lysates were obtained from isolated primary mouse and human hepatocytes treated with palmitic acid (PA) or oleic acid (OA) at 800 μm for 8 h. Vehicle (V) was used as control. Immunoblot analysis was performed for phosphorylated and total JNK, phosphorylated and total c-Jun, PUMA, NOXA, and β-actin. A, primary murine hepatocytes isolated from WT, Jnk1−/−, and Jnk2−/− mice. B, primary human hepatocytes. C, primary murine hepatocytes isolated from WT and p53−/− mice.
The Transcription Factor AP-1 Complex Contributes to PUMA mRNA Up-regulation
Although PUMA was originally identified as a p53 transcriptional target (40–42), p53 is mutated and functionally inactive in Huh-7 cells, making it unlikely that p53 mediates PUMA expression by palmitate. Consistent with this concept, palmitate increased PUMA protein levels in primary murine hepatocytes from p53−/− animals (Fig. 3C).
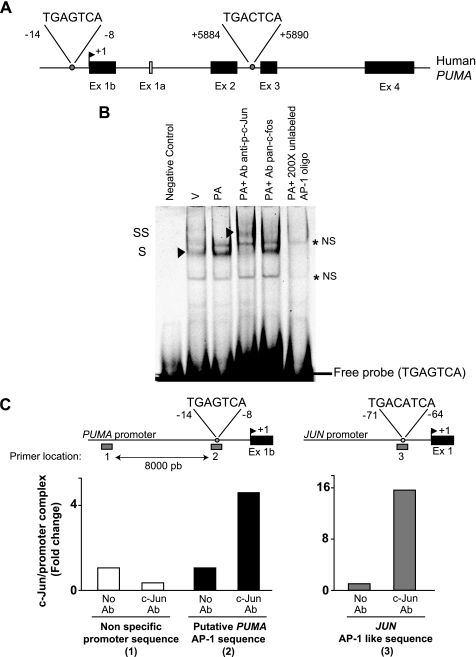
Next, we examined the potential role of the JNK-activated c-Jun containing AP-1 complex in PUMA induction. To ascertain if AP-1 complexes may directly up-regulate PUMA transcription, the human PUMA gene sequence was examined to identify potential consensus AP-1 binding sites (TGA(G/C)TCA). Two such binding sites were identified by computational analysis; one in the promoter region and another in the intronic region between the exons 2 and 3 (Fig. 4A). To test if AP-1 complexes bind to these sites within the human PUMA gene, an EMSA was performed. No binding to the putative intronic AP-1 sequence was identified (data not shown). In contrast, binding to the putative promoter AP-1 sequence was observed in nuclear extracts from palmitate-treated Huh-7 cells, which was supershifted with anti-phospho-c-Jun antisera (Fig. 4B). No supershift was observed in the presence of anti-pan-Fos antisera (Fig. 4B), indicating that this AP-1 complex is likely composed of c-Jun/Jun family protein dimers. The mobility shift of the CY 5.5-labeled probe could be prevented by adding a 200-fold molar excess of the unlabeled probe (Fig. 4B). Binding was also abolished when using a mutated probe in which the critical AP-1 binding residues had been altered (data not shown).
FIGURE 4.
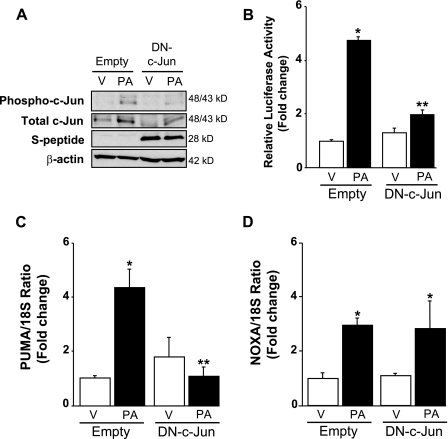
Palmitate-induced AP-1 complex binds to the PUMA promoter. A, the genomic structure of PUMA showing the exon-intron organization and 2 potential AP-1 binding sites. B, nuclear protein extracts isolated from Huh-7 cells treated with vehicle (V) or palmitic acid (PA) at 800 μm for 12 h. EMSA was performed using CY 5.5-labeled double-stranded oligonucleotide containing the putative AP-1 binding sequence within the human PUMA promoter (located at −14/−8 nucleotides from the transcriptional start codon), supershift experiments using antibody against phospho-c-Jun or pan-c-Fos, and competition experiments with 200-fold molar excess cold oligonucleotide as described under “Experimental Procedures.” Retarded complexes are indicated by arrows (S, shift; SS, supershift; NS, nonspecific band). C, Huh-7 cells were treated with palmitic acid (800 μm) for 2 h. Next, the cells were fixed and lysed, and DNA fragments co-immunoprecipitated with the target protein c-Jun were subjected to quantitative real-time PCR analysis using various primer sets as indicated by the gray bars. The primer set 1 (P1) and 2 (P2) were used to amplify, respectively, an upstream nonspecific sequence or the putative AP-1 sequence within the human PUMA promoter; the primer set 3 (P3) was used to amplify the AP-1 like sequence in the human JUN promoter. As a control, a mock immunoprecipitation without antibody (Ab) was also performed. Representative results of three independent experiments are shown.
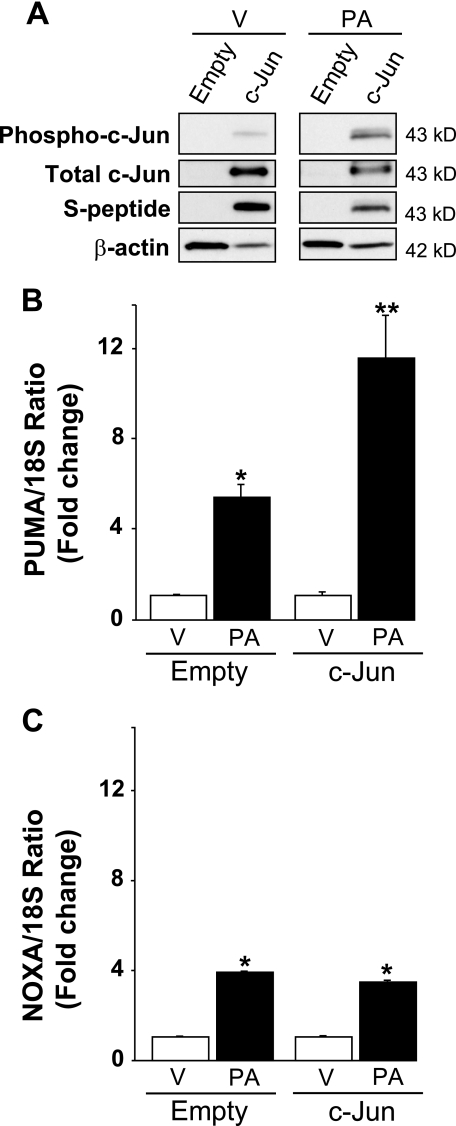
To determine whether a phosphorylated c-Jun containing AP-1 complex binds to the PUMA promoter, a chromatin immunoprecipitation assay was performed. c-Jun immunoprecipitation captured the putative AP-1 sequence of the PUMA promoter and to a greater extent the AP-1-like sequence of the JUN promoter (employed as a positive control) in nuclear extracts obtained from palmitate-treated Huh-7 cells (Fig. 4C). No binding of c-Jun to an unrelated upstream region of the PUMA promoter was observed (Fig. 4C). These data indicate that c-Jun directly binds in vivo to the AP-1 sequence in the PUMA promoter located at −14/−8 nucleotides from the transcriptional start codon in the exon 1b but not the upstream control region. To exclude the involvement of other JNK-dependent mechanisms and provide functional evidence that a phosphorylated c-Jun containing AP-1 complex controls PUMA transcription, Huh-7 cells were transiently transfected with an S-peptide-tagged dominant negative c-Jun (DN-c-Jun), which lacks the transactivation domain (1–122 amino acids) but contains the dimerization and DNA binding domains. Enforced expression of this dominant negative c-Jun decreased palmitate-induced activation (phosphorylation) of endogenous c-Jun (Fig. 5A). The loss of AP-1 function by the dominant-negative c-Jun was further examined using an AP-1 luciferase reporter, pAP1-Luc, which contains six AP-1 sites upstream of the luciferase promoter. Twenty-four hours of treatment with palmitate resulted in a 4.5-fold increase in reporter activity in Huh-7 cells as compared with cells treated with vehicle (Fig. 5B), and palmitate-induced AP-1 transcriptional activity was markedly suppressed by expression of DN-c-Jun (Fig. 5B). Along with an inhibition of AP-1 transactivation, enforced expression of DN-c-Jun prevented palmitate induction of PUMA mRNA (Fig. 5C) but did not modify palmitate induction of NOXA mRNA (Fig. 5D). Conversely, transient transfection of Huh-7 cells with a plasmid containing the full-length human c-Jun open reading frame increased c-Jun expression and phosphorylation (Fig. 6A) and enhanced PUMA mRNA up-regulation by palmitate (Fig. 6B). Enforced expression of c-Jun activation did not, however, alter palmitate-induced NOXA mRNA expression (Fig. 6C). Thus, a JNK1-activated AP-1 complex induces PUMA transcription likely by binding to a consensus AP-1 binding site within its promoter region.
FIGURE 5.
Enforced expression of dominant negative c-Jun decreased phosphorylation of endogenous c-Jun and AP-1 transcriptional activity and prevents induction of PUMA mRNA level in response to palmitate. Huh-7 cells were transiently transfected for 24 h with an S-peptide-tagged dominant negative c-Jun encoding plasmid (DN-c-Jun) or the control empty plasmid. Next, cells were treated with palmitic acid (PA) at 800 μm. Vehicle (V) was used as control. A, whole cell lysates were prepared 8 h after FFA treatment, and effective expression of dominant negative c-Jun was identified by immunoblot analysis using an anti-S-peptide antibody (which reveals a band at approximately 28 kDa). Immunoblot analysis was also performed for phosphorylated and total c-Jun and β-actin. B, using pAP1-Luc vector, relative luciferase activity was assessed 24 h after FFA treatment in the presence of the pan-caspase inhibitor benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone (50 μm) to mitigate cell death, as described under “Experimental Procedures.” Data represent the mean and S.E. of three independent experiments; *, p < 0.01, palmitate-treated empty vector versus vehicle-treated empty vector; **, p < 0.01, palmitate-treated DN-c-Jun versus palmitate-treated empty vector. Total RNA was extracted 8 h after FFA treatment, and PUMA (C) and NOXA (D) mRNA expression was profiled by real-time PCR. -Fold induction was determined after normalization to 18 S. Data represent the mean and S.E. of three experiments; *, p < 0.01, palmitate-treated cells versus vehicle-treated cells; **, p < 0.01, palmitate-treated DN-c-Jun versus palmitate-treated empty vector.
FIGURE 6.
Enhanced expression of c-Jun exacerbates palmitate induction of PUMA mRNA levels. Huh-7 cells were transiently transfected for 24 h with a plasmid containing full-length of human c-Jun open reading frame tagged with S-peptide (c-Jun) or the empty vector. Total RNA and whole cell lysates were prepared 8 h after treatment with palmitic acid (PA) at 800 μm, and vehicle (V) was used as the control. A, effective overexpression of c-Jun protein level was verified by immunoblot analysis using an anti-S-peptide antibody and an anti-total c-Jun antibody. Immunoblot analysis was also performed for phosphorylated c-Jun and β-actin. PUMA (B) and NOXA (C) mRNA expression was profiled by real-time PCR. -Fold induction was determined after normalization to 18 S. Data represent the mean and S.E. of three experiments. *, p < 0.05, palmitate-treated cells versus vehicle-treated cells; **, p < 0.01, palmitate-treated c-Jun versus palmitate-treated empty vector.
PUMA Contributes to Bax Activation and Apoptosis by Palmitate
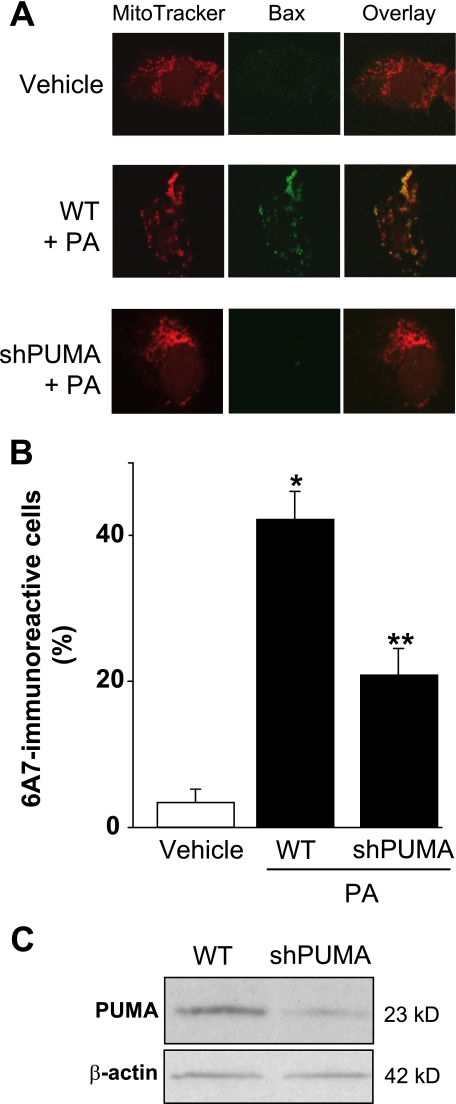
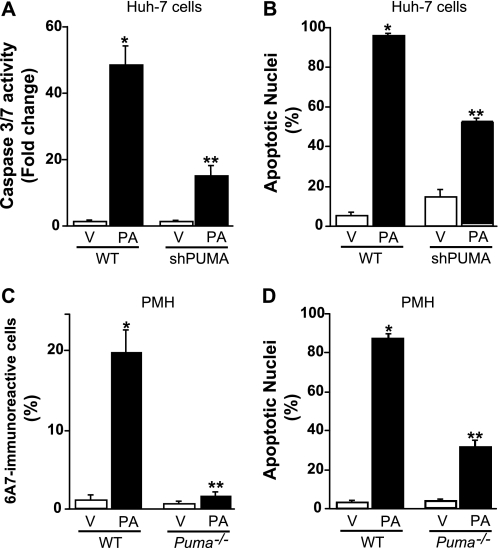
Prior studies by us have implicated Bax activation as a pivotal step in lipoapoptosis (13). Although Bax activation was JNK-dependent (13), the mechanism whereby JNK resulted in Bax activation was unclear. Because PUMA can promote Bax activation (32, 34, 38), we next examined the potential relationship between PUMA induction and Bax activation during palmitate-induced lipoapoptosis. Upon activation, Bax undergoes a conformational change, which is specifically recognized by a monoclonal antibody directed against amino acids 12–24 of Bax (6A7). Under basal conditions no Bax activation was observed in Huh-7 cells, whereas palmitate treatment for 12 h induced readily apparent Bax activation (Fig. 7, A and B). To ascertain whether PUMA is directly involved in palmitate-induced Bax activation, Huh-7 cells were stably transfected with a lentivirus construct containing a PUMA targeted short hairpin RNA. An efficient knockdown of PUMA expression was verified by immunoblot analysis (Fig. 7C); this knockdown of PUMA reduced by 50% the number of 6A7-immunoreactive cells after treatment with palmitate (Fig. 7, A and B). PUMA knockdown also conferred cytoprotection against palmitate-induced apoptosis as assessed by caspase 3/7 activity and nuclear morphology (Fig. 8, A and B). Similarly, hepatocytes isolated from mice genetically deficient in Puma (Puma−/−) were more resistant to palmitate-induced Bax activation (Fig. 8C) and subsequent apoptosis (Fig. 8D) than hepatocytes obtained from wild-type mice. Collectively, these results suggest that PUMA contributes to Bax activation and lipoapoptosis by palmitate.
FIGURE 7.
PUMA contributes to Bax activation. A and B, Huh-7 (WT) and Huh-7 cells stably expressing short hairpin RNA complementary to PUMA (shPUMA) were incubated for 12 h with palmitic acid (PA) at 800 μm; vehicle (V)-treated cells were used as control. Cells were stained with Mito Tracker Red to visualize mitochondria. Next, cells were fixed, and Bax activation was assessed using conformation specific antisera (6A7), which only recognizes active Bax and using immunofluorescence microscopy. A, representative images of three independent experiments are depicted. B, 6A7-immunoreactive cells were quantified in 10 random 40× objective fields for each condition at the indicated time point with automated software. Data represent the mean and S.E. of three experiments; *, p < 0.01, palmitate-treated cells versus vehicle-treated cells; **, p < 0.01, palmitate-treated shPUMA versus palmitate-treated WT. C, effective down-regulation of PUMA protein levels in shPUMA Huh-7 cells compared with WT Huh-7 cells was verified by immunoblot analysis on whole cell lysates.
FIGURE 8.
PUMA contributes to palmitate-mediated apoptosis. A and B, Huh-7 (WT) and Huh-7 cells stably expressing short hairpin RNA complementary to PUMA (shPUMA) were incubated for 24 h with palmitic acid (PA) at 800 μm; Vehicle (V)-treated cells were used as control. A, caspases 3 and 7 catalytic activity was assessed as described under “Experimental Procedures.” B, apoptotic nuclei were counted according to morphological criteria after 4′,6-diamidino-2-phenylindole dihydrochloride staining. Data represent the mean and S.E. of three experiments; *, p < 0.05, palmitate-treated cells versus vehicle-treated cells; **, p < 0.01, palmitate-treated shPUMA versus palmitate-treated WT. C, primary murine hepatocytes (PMH) isolated from WT or Puma−/− mice were treated for 8 h with vehicle or palmitic acid (400 μm). Bax activation was assessed as described under “Experimental Procedures,” and 6A7-immunoreactive cells were quantified in 12 random 40× objective fields for each condition with automated software. Data represent the mean and S.E. of three experiments; *, p < 0.01, palmitate-treated cells versus vehicle-treated cells; **, p < 0.01, palmitate-treated Puma−/− versus palmitate-treated WT. D, primary murine hepatocytes isolated from WT or Puma−/− mice were treated for 24 h with vehicle or palmitic acid (200 μm). Apoptotic nuclei were counted according to morphological criteria after 4′,6-diamidino-2-phenylindole dihydrochloride staining. Data represent mean and S.E. of three experiments; *, p < 0.01, palmitate-treated cells versus vehicle-treated cells; **, p < 0.01, palmitate-treated Puma−/− versus palmitate-treated WT.
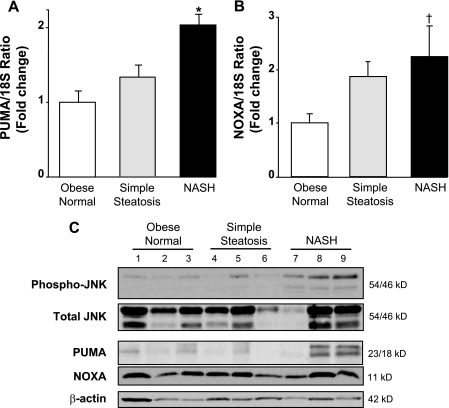
PUMA Is Up-regulated in Human NASH
Clinically, circulating FFA are elevated in the metabolic syndrome and in patients with NASH (15). PUMA expression was assessed in human liver specimens, comparing obese normal control liver specimens to those with simple steatosis and steatosis plus inflammation (NASH). PUMA expression was increased both at the message and protein levels in liver tissue from patients with NASH compared with liver tissue from patients with simple steatosis or obese normal controls (p < 0.01) (Fig. 9, A and C). This increase in PUMA induction was observed in the majority of NASH specimens but not in all, consistent with the heterogeneity of human disease. Along with PUMA up-regulation, an increase of phospho-JNK was observed in NASH patients but not in control or simple steatosis patients (Fig. 9C). An increase in NOXA mRNA was also identified in patients with NASH and to a lesser extent in patients with simple steatosis (Fig. 9B). NASH, however, was not associated with changes in NOXA at the protein level (Fig. 9C). These data suggest that PUMA up-regulation also occurs during chronic hepatic lipotoxicity.
FIGURE 9.
PUMA is up-regulated in human NASH. Homogenates and total RNA were prepared from human liver biopsies of obese normal (n = 16), simple steatosis (n = 17), or NASH patients (n = 16). A and B, PUMA and NOXA mRNA expression was profiled by real-time PCR. -Fold induction was determined after normalization to 18 S. Data represent the mean and S.E.; *, p < 0.01, NASH patients versus obese normal and simple steatosis patients; †, p < 0.05, NASH patient versus obese normal patients. C, immunoblot analysis was performed for phosphorylated and total JNK, phosphorylated and total c-Jun, PUMA, NOXA, and β-actin.
DISCUSSION
The principal findings of this study relate to the mechanisms of saturated FFA-mediated lipoapoptosis. Our results in Huh-7 cells and primary hepatocytes indicate that (i) the toxic saturated FFA palmitate, but not the monounsaturated FFA oleate, induces expression of PUMA mRNA and protein, (ii) saturated FFA-induced JNK1 activates and phosphorylates c-Jun, which directly binds the PUMA promoter and induces PUMA transcription, and (iii) genetic deletion or short hairpin RNA targeted knockdown of PUMA expression attenuates lipoapoptosis by palmitate. Our data also demonstrate that JNK is activated, and PUMA is up-regulated in the liver of patients with NASH. These observations suggest that PUMA up-regulation contributes to JNK1-potentiated cytotoxicity during lipogenic hepatocyte injury.
Pro-apoptotic BH3-only protein family members are critical regulators of lipoapoptosis as small interfering RNA targeted knockdown of Bim partially protects liver cells against lipoapoptosis (12, 13). Although these findings indicate an important role for Bim in FFA-induced apoptosis, the partial protection also suggests that other BH3-only proteins with complementary functions contribute to lipotoxicity. The current study indicates that the saturated FFA palmitate increases PUMA expression in liver cells. In contrast to palmitate, the unsaturated FFA oleate minimally increases cellular PUMA protein levels, below the threshold sufficient to induce cell death (13, 53). These findings are consistent with prior observations that saturated FFA are inherently more toxic than unsaturated FFA (12, 13, 55).
Saturated FFA treatment also induced an increase in NOXA mRNA without affecting the expression of the other BH3-only proteins Bad, Bid, Bik, Bmf, and Hrk. However, the increase in NOXA mRNA did not cause an increase in NOXA protein expression. Whether this discordance is because of post-transcriptional modifications of NOXA by microRNAs, ribosomal occupation, or RNA sequestration remains to be explored. Also, as described for Bim (56), possible post-translational modifications (e.g. by phosphorylation, ubiquitination, etc.) of NOXA and further degradation by the proteasome pathway could shorten the half-life of the protein and mask a potential increase in NOXA protein translation. Given the fact that PUMA is a potent BH3-only protein (33, 40, 57) which cooperates with Bim in cell death processes (43), we focused our study on the contribution and the regulation of PUMA expression during lipoapoptosis.
PUMA has been implicated in mediating cell death by a wide variety of toxic stimuli, including genotoxic stresses, serum withdrawal, or endoplasmic reticulum stress-inducing agents (39, 58). PUMA has been extensively studied in colon cancer cells, thymocytes, and neurons (32, 40, 44, 45, 59), but a potential contribution to liver injury has not been described. Our observations implicate PUMA in hepatic lipotoxicity. For example, short hairpin RNA-targeted knockdown of PUMA in Huh-7 cells or genetic deficiency of Puma in mouse primary hepatocytes renders liver cells partially resistant to FFA cytotoxicity. Saturated FFA induce apoptosis through the mitochondrial death pathway by activating Bax (13), and PUMA promotes apoptosis in a Bax-dependent manner (32, 34, 60, 61). In agreement with these observations, our results suggest that PUMA contributes to Bax activation by saturated FFA, resulting in mitochondrial dysfunction, caspase 3/7 activation, and subsequent cell death.
Whether PUMA directly or indirectly activates Bax is controversial. Recent studies have demonstrated a direct interaction between PUMA and the first α helix of Bax, which promotes Bax translocation to the mitochondria and triggers apoptosis (34). Also, PUMA may indirectly promote Bax activation by binding to pro-survival Bcl-2 proteins such as Mcl-1 and/or Bcl-XL, which are both expressed in hepatocytes (60, 61). By binding these anti-apoptotic proteins, PUMA may displace or prevent their sequestration of other BH3-only proteins such as Bim. The liberated Bim would then be able to directly activate Bax (62). Alternatively, binding of PUMA to an anti-apoptotic protein such as Bcl-XL can result in the dissociation of Bax from Bcl-XL, thereby indirectly promoting Bax mitochondrial activation (32, 61). Detailed analysis of the mechanisms by which PUMA activates Bax in this model will require further studies.
Although PUMA was originally identified as a p53 transcriptional target (40–42), our results refute any involvement of this transcription factor in our model. Other transcription factors independent of p53 and p73 have been also described to directly regulate PUMA expression. For example, PUMA is a target of the transcription factor FoxO3a (36, 63), and enhanced activity of FoxO3a contributes to lipoapoptosis (12). Whether PUMA up-regulation by saturated FFA depends in part on a FoxO3a-dependent mechanism was not investigated, as this process appears to be strongly linked to JNK activity (see below).
Activation of the mitogen-activated protein kinase JNK has been identified as a pivotal step in FFA-induced Bax activation and hepatocyte apoptosis (13). Prior observations indicate that JNK can be activated by saturated FFA-stimulated MAPK kinase kinase MLK3 (64) and/or secondary to saturated FFA-induced ER stress (55, 65). Although JNK-dependent Bax activation has been reported in lipotoxicity (13), the mechanisms remained obscure. The results herein link JNK to Bax activation by identifying JNK-mediated PUMA up-regulation with subsequent PUMA-dependent Bax activation. Indeed, PUMA was identified as a transcriptional target of the JNK/activated AP-1 transcription factor complex. Both JNK1 and JNK2 have been implicated in liver injury. However, depending on the stimulus, these two JNK isoforms differentially contribute to hepatocyte apoptosis. Whereas JNK2 appears to promote apoptosis in a tumor necrosis factor-induced liver injury model (66), enhanced JNK1 signaling contributes to hepatocyte apoptosis in a murine model of steatohepatitis (17). JNK1 predominantly contributes to the phosphorylation of the transcription factor c-Jun (17, 18, 25), a critical member of the AP-1 complex. Consistent with these observations, our results indicate that saturated FFA-induced JNK1/c-Jun phosphorylation results in the formation of an active AP-1 complex which directly regulates PUMA transcription. In support of this model, we identified AP-1 binding to a consensus site within the human PUMA promoter region; an AP-1 consensus sequence (TGAGTCA) was also identified in the mouse Puma promoter located at −369/−363 nucleotides from the transcriptional start codon. Furthermore, genetic deficiency of Jnk1 in murine hepatocytes, pharmacologic inhibition of JNK, or interference of AP-1 function by a dominant-negative c-Jun attenuated PUMA up-regulation by saturated FFA. Therefore, our study defines the transcriptional regulation of PUMA as an intermediary process linking JNK1 activation and FFA-mediated apoptosis.
Elevated serum FFA levels and hepatocyte apoptosis are hallmarks of NASH, and levels of circulating FFA correlates with liver disease severity (14, 15). Prior studies have identified structural and functional mitochondrial abnormalities (48, 67, 68) as well as an endoplasmic reticulum stress-induced JNK activation in liver cells from NASH patients (16). Consistent with these latter observations, JNK activation was observed in liver biopsies from patient with NASH. Along with JNK activation, PUMA up-regulation was also observed in a subset of NASH patients, suggesting a possible contribution of PUMA to the development of chronic hepatic lipotoxicity.
In summary, the current studies provide mechanistic insights regarding the link between JNK1 activation, Bax activation, and FFA-mediated lipotoxicity. The results support that a pathway comprising JNK1/c-Jun-induced PUMA transcriptional up-regulation with subsequent Bax activation as integral steps promoting saturated FFA-mediated apoptosis. Modulation of PUMA expression and/or function may represent a potential therapeutic strategy to ameliorate lipoapoptosis.
Supplementary Material
Acknowledgments
We thank Dr. Anuradha Krishnan and Kimberly B. Viker for isolating the human primary hepatocytes and Erin Nystuen-Bungum for excellent secretarial assistance. The optical microscopy core was supported by National Institutes of Health Grant DK84567.
This work was supported, in whole or in part, by National Institutes of Health Grants DK41876 (to G. J. G.), DK069757-05 (to M. R. C.), and DK079875 (to J. L. M.). This work was also supported by the Mayo Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NASH
- non-alcoholic steatohepatitis
- FFA
- free fatty acid(s)
- AP-1
- activator protein-1
- BH3
- Bcl-2 homology domain
- EMSA
- electrophoretic mobility shift assay
- JNK
- c-Jun-N-terminal kinase
- MAPK
- mitogen-activated protein kinase
- MAPKK
- MAPK kinase
- MAPKKK
- MAPK kinase kinase
- PUMA
- p53-up-regulated mediator of apoptosis
- WT
- wild type
- MLK3
- mixed lineage kinase 3
- DN
- dominant negative
- PBS
- phosphate-buffered saline
- PIPES
- 1,4-piperazinediethanesulfonic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Browning J. D., Szczepaniak L. S., Dobbins R., Nuremberg P., Horton J. D., Cohen J. C., Grundy S. M., Hobbs H. H. (2004) Hepatology 40, 1387–1395 [DOI] [PubMed] [Google Scholar]
- 2.Adams L. A., Lymp J. F., St Sauver J., Sanderson S. O., Lindor K. D., Feldstein A., Angulo P. (2005) Gastroenterology 129, 113–121 [DOI] [PubMed] [Google Scholar]
- 3.Ekstedt M., Franzén L. E., Mathiesen U. L., Thorelius L., Holmqvist M., Bodemar G., Kechagias S. (2006) Hepatology 44, 865–873 [DOI] [PubMed] [Google Scholar]
- 4.Ratziu V., Poynard T. (2006) Hepatology 44, 802–805 [DOI] [PubMed] [Google Scholar]
- 5.Bookman I. D., Pham J., Guindi M., Heathcote E. J. (2006) Liver Int. 26, 566–571 [DOI] [PubMed] [Google Scholar]
- 6.Donnelly K. L., Smith C. I., Schwarzenberg S. J., Jessurun J., Boldt M. D., Parks E. J. (2005) J. Clin. Invest. 115, 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doege H., Grimm D., Falcon A., Tsang B., Storm T. A., Xu H., Ortegon A. M., Kazantzis M., Kay M. A., Stahl A. (2008) J. Biol. Chem. 283, 22186–22192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J., Febbraio M., Wada T., Zhai Y., Kuruba R., He J., Lee J. H., Khadem S., Ren S., Li S., Silverstein R. L., Xie W. (2008) Gastroenterology 134, 556–567 [DOI] [PubMed] [Google Scholar]
- 9.Listenberger L. L., Han X., Lewis S. E., Cases S., Farese R. V., Jr., Ory D. S., Schaffer J. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi K., Yang L., McCall S., Huang J., Yu X. X., Pandey S. K., Bhanot S., Monia B. P., Li Y. X., Diehl A. M. (2007) Hepatology 45, 1366–1374 [DOI] [PubMed] [Google Scholar]
- 11.Unger R. H., Orci L. (2002) Biochim. Biophys. Acta 1585, 202–212 [DOI] [PubMed] [Google Scholar]
- 12.Barreyro F. J., Kobayashi S., Bronk S. F., Werneburg N. W., Malhi H., Gores G. J. (2007) J. Biol. Chem. 282, 27141–27154 [DOI] [PubMed] [Google Scholar]
- 13.Malhi H., Bronk S. F., Werneburg N. W., Gores G. J. (2006) J. Biol. Chem. 281, 12093–12101 [DOI] [PubMed] [Google Scholar]
- 14.Feldstein A. E., Canbay A., Angulo P., Taniai M., Burgart L. J., Lindor K. D., Gores G. J. (2003) Gastroenterology 125, 437–443 [DOI] [PubMed] [Google Scholar]
- 15.Nehra V., Angulo P., Buchman A. L., Lindor K. D. (2001) Dig. Dis. Sci. 46, 2347–2352 [DOI] [PubMed] [Google Scholar]
- 16.Puri P., Mirshahi F., Cheung O., Natarajan R., Maher J. W., Kellum J. M., Sanyal A. J. (2008) Gastroenterology 134, 568–576 [DOI] [PubMed] [Google Scholar]
- 17.Singh R., Wang Y., Xiang Y., Tanaka K. E., Gaarde W. A., Czaja M. J. (2009) Hepatology 49, 87–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schattenberg J. M., Singh R., Wang Y., Lefkowitch J. H., Rigoli R. M., Scherer P. E., Czaja M. J. (2006) Hepatology 43, 163–172 [DOI] [PubMed] [Google Scholar]
- 19.Czaja M. J. (2007) Semin. Liver Dis. 27, 378–389 [DOI] [PubMed] [Google Scholar]
- 20.Davis R. J. (2000) Cell 103, 239–252 [DOI] [PubMed] [Google Scholar]
- 21.Xu Z., Kukekov N. V., Greene L. A. (2005) Mol. Cell. Biol. 25, 9949–9959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schachter K. A., Du Y., Lin A., Gallo K. A. (2006) J. Biol. Chem. 281, 19134–19144 [DOI] [PubMed] [Google Scholar]
- 23.Czaja M. J. (2003) Am. J. Physiol. Gastrointest. Liver Physiol. 284, G875–G879 [DOI] [PubMed] [Google Scholar]
- 24.Karin M., Gallagher E. (2005) IUBMB life 57, 283–295 [DOI] [PubMed] [Google Scholar]
- 25.Sabapathy K., Hochedlinger K., Nam S. Y., Bauer A., Karin M., Wagner E. F. (2004) Mol. Cell 15, 713–725 [DOI] [PubMed] [Google Scholar]
- 26.Lei K., Davis R. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2432–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donovan N., Becker E. B., Konishi Y., Bonni A. (2002) J. Biol. Chem. 277, 40944–40949 [DOI] [PubMed] [Google Scholar]
- 28.Kim B. J., Ryu S. W., Song B. J. (2006) J. Biol. Chem. 281, 21256–21265 [DOI] [PubMed] [Google Scholar]
- 29.Yamamoto K., Ichijo H., Korsmeyer S. J. (1999) Mol. Cell. Biol. 19, 8469–8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanawa N., Shinohara M., Saberi B., Gaarde W. A., Han D., Kaplowitz N. (2008) J. Biol. Chem. 283, 13565–13577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 32.Ming L., Wang P., Bank A., Yu J., Zhang L. (2006) J. Biol. Chem. 281, 16034–16042 [DOI] [PubMed] [Google Scholar]
- 33.Neise D., Graupner V., Gillissen B. F., Daniel P. T., Schulze-Osthoff K., Jänicke R. U., Essmann F. (2008) Oncogene 27, 1387–1396 [DOI] [PubMed] [Google Scholar]
- 34.Gallenne T., Gautier F., Oliver L., Hervouet E., Noël B., Hickman J. A., Geneste O., Cartron P. F., Vallette F. M., Manon S., Juin P. (2009) J. Cell Biol. 185, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor R. C., Cullen S. P., Martin S. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 231–241 [DOI] [PubMed] [Google Scholar]
- 36.You H., Pellegrini M., Tsuchihara K., Yamamoto K., Hacker G., Erlacher M., Villunger A., Mak T. W. (2006) J. Exp. Med. 203, 1657–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hershko T., Ginsberg D. (2004) J. Biol. Chem. 279, 8627–8634 [DOI] [PubMed] [Google Scholar]
- 38.Melino G., Bernassola F., Ranalli M., Yee K., Zong W. X., Corazzari M., Knight R. A., Green D. R., Thompson C., Vousden K. H. (2004) J. Biol. Chem. 279, 8076–8083 [DOI] [PubMed] [Google Scholar]
- 39.Jeffers J. R., Parganas E., Lee Y., Yang C., Wang J., Brennan J., MacLean K. H., Han J., Chittenden T., Ihle J. N., McKinnon P. J., Cleveland J. L., Zambetti G. P. (2003) Cancer Cell 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 40.Yu J., Zhang L., Hwang P. M., Kinzler K. W., Vogelstein B. (2001) Mol. Cell 7, 673–682 [DOI] [PubMed] [Google Scholar]
- 41.Nakano K., Vousden K. H. (2001) Mol. Cell 7, 683–694 [DOI] [PubMed] [Google Scholar]
- 42.Han J., Flemington C., Houghton A. B., Gu Z., Zambetti G. P., Lutz R. J., Zhu L., Chittenden T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11318–11323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erlacher M., Labi V., Manzl C., Böck G., Tzankov A., Häcker G., Michalak E., Strasser A., Villunger A. (2006) J. Exp. Med. 203, 2939–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steckley D., Karajgikar M., Dale L. B., Fuerth B., Swan P., Drummond-Main C., Poulter M. O., Ferguson S. S., Strasser A., Cregan S. P. (2007) J. Neurosci. 27, 12989–12999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villunger A., Michalak E. M., Coultas L., Müllauer F., Böck G., Ausserlechner M. J., Adams J. M., Strasser A. (2003) Science 302, 1036–1038 [DOI] [PubMed] [Google Scholar]
- 46.Krishnan A., Viker K., Rietema H., Telgenkamp M., Knudsen B., Charlton M. (2007) Hepatol. Res. 37, 854–862 [DOI] [PubMed] [Google Scholar]
- 47.Richieri G. V., Kleinfeld A. M. (1995) J. Lipid Res. 36, 229–240 [PubMed] [Google Scholar]
- 48.Sanyal A. J., Campbell-Sargent C., Mirshahi F., Rizzo W. B., Contos M. J., Sterling R. K., Luketic V. A., Shiffman M. L., Clore J. N. (2001) Gastroenterology 120, 1183–1192 [DOI] [PubMed] [Google Scholar]
- 49.Belfort R., Harrison S. A., Brown K., Darland C., Finch J., Hardies J., Balas B., Gastaldelli A., Tio F., Pulcini J., Berria R., Ma J. Z., Dwivedi S., Havranek R., Fincke C., DeFronzo R., Bannayan G. A., Schenker S., Cusi K. (2006) N. Engl. J. Med. 355, 2297–2307 [DOI] [PubMed] [Google Scholar]
- 50.Vikhanskaya F., Toh W. H., Dulloo I., Wu Q., Boominathan L., Ng H. H., Vousden K. H., Sabapathy K. (2007) Nat. Cell Biol. 9, 698–705 [DOI] [PubMed] [Google Scholar]
- 51.Hansson M. D., Rzeznicka K., Rosenbäck M., Hansson M., Sirijovski N. (2008) Anal. Biochem. 375, 373–375 [DOI] [PubMed] [Google Scholar]
- 52.Kleiner D. E., Brunt E. M., Van Natta M., Behling C., Contos M. J., Cummings O. W., Ferrell L. D., Liu Y. C., Torbenson M. S., Unalp-Arida A., Yeh M., McCullough A. J., Sanyal A. J. (2005) Hepatology 41, 1313–1321 [DOI] [PubMed] [Google Scholar]
- 53.Malhi H., Barreyro F. J., Isomoto H., Bronk S. F., Gores G. J. (2007) Gut 56, 1124–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bennett B. L., Sasaki D. T., Murray B. W., O'Leary E. C., Sakata S. T., Xu W., Leisten J. C., Motiwala A., Pierce S., Satoh Y., Bhagwat S. S., Manning A. M., Anderson D. W. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13681–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei Y., Wang D., Topczewski F., Pagliassotti M. J. (2006) Am. J. Physiol. Endocrinol. Metab. 291, E275–E281 [DOI] [PubMed] [Google Scholar]
- 56.Ley R., Balmanno K., Hadfield K., Weston C., Cook S. J. (2003) J. Biol. Chem. 278, 18811–18816 [DOI] [PubMed] [Google Scholar]
- 57.Akhtar R. S., Geng Y., Klocke B. J., Latham C. B., Villunger A., Michalak E. M., Strasser A., Carroll S. L., Roth K. A. (2006) J. Neurosci. 26, 7257–7264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo X., He Q., Huang Y., Sheikh M. S. (2005) Cell Death Differ. 12, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 59.Michalak E. M., Villunger A., Adams J. M., Strasser A. (2008) Cell Death Differ. 15, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chipuk J. E., Fisher J. C., Dillon C. P., Kriwacki R. W., Kuwana T., Green D. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 20327–20332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jabbour A. M., Heraud J. E., Daunt C. P., Kaufmann T., Sandow J., O'Reilly L. A., Callus B. A., Lopez A., Strasser A., Vaux D. L., Ekert P. G. (2009) Cell Death Differ. 16, 555–563 [DOI] [PubMed] [Google Scholar]
- 62.Yamaguchi H., Wang H. G. (2002) J. Biol. Chem. 277, 41604–41612 [DOI] [PubMed] [Google Scholar]
- 63.Ekoff M., Kaufmann T., Engström M., Motoyama N., Villunger A., Jönsson J. I., Strasser A., Nilsson G. (2007) Blood 110, 3209–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaeschke A., Davis R. J. (2007) Mol. Cell 27, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 66.Kodama Y., Taura K., Miura K., Schnabl B., Osawa Y., Brenner D. A. (2009) Gastroenterology 136, 1423–1434 [DOI] [PubMed] [Google Scholar]
- 67.Caldwell S. H., Swerdlow R. H., Khan E. M., Iezzoni J. C., Hespenheide E. E., Parks J. K., Parker W. D., Jr. (1999) J. Hepatol. 31, 430–434 [DOI] [PubMed] [Google Scholar]
- 68.Caldwell S. H., Chang C. Y., Nakamoto R. K., Krugner-Higby L. (2004) Clin. Liver Dis. 8, 595–617 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.