Abstract
Signal-regulatory protein α (SIRPα) is a myeloid membrane receptor that interacts with the membrane protein CD47, a marker of self. We have solved the structure of the complete extracellular portion of SIRPα, comprising three immunoglobulin superfamily domains, by x-ray crystallography to 2.5 Å resolution. These data, together with previous data on the N-terminal domain and its ligand CD47 (possessing a single immunoglobulin superfamily domain), show that the CD47-SIRPα interaction will span a distance of around 14 nm between interacting cells, comparable with that of an immunological synapse. The N-terminal (V-set) domain mediates binding to CD47, and the two others are found to be constant (C1-set) domains. C1-set domains are restricted to proteins involved in vertebrate antigen recognition: T cell antigen receptors, immunoglobulins, major histocompatibility complex antigens, tapasin, and β2-microglobulin. The domains of SIRPα (domains 2 and 3) are structurally more similar to C1-set domains than any cell surface protein not involved in antigen recognition. This strengthens the suggestion from sequence analysis that SIRP is evolutionarily closely related to antigen recognition proteins.
Signal-regulatory protein α (SIRPα)4 is a membrane receptor present on myeloid cells and neurons that interacts with the widely distributed cell surface protein CD47 (reviewed in Refs. 1 and 2). Absence of CD47 leads to uptake of cells via macrophages, indicating that CD47 acts as a marker of self (3). SIRPα gives inhibitory signals through immunoreceptor tyrosine-based inhibition motifs in the cytoplasmic region that interact with phosphatases SHP-1 and SHP-2 (4). Binding of the N-terminal immunoglobulin superfamily (IgSF) V-set domain of SIRPα (SIRPα d1) to the single IgSF domain of CD47 is mediated by the loops of the SIRPα IgSF domain, analogous to the interactions mediated by antigen receptors, albeit involving only a single domain (5, 6). This type of binding distinguishes the CD47-SIRPα interaction from that of many interactions at the cell surface involving IgSF domains such as CD2-CD58, where the face of the IgSF domain is involved (7). SIRPα domains 2 and 3 (d2 and d3) show amino acid sequence similarity to IgSF C1-set domains (8). Since IgSF C1-set domains have only been confirmed in vertebrate antigen receptors and associated proteins (Ig light and heavy chains, T cell receptor chains, MHC class I and II and related proteins, β2-microglobulin, and very recently tapasin (9)) of the vertebrate adaptive immune system, it was suggested that SIRPα might have evolved from a precursor of the antigen receptors (8).
We describe here the crystal structure of the full three-domain extracellular region of SIRPα, revealing that the topology of the CD47-SIRPα interaction is compatible with productive engagement occurring when cells come together in synapse-like contacts. We show that the two membrane-proximal IgSF domains are particularly close in structure to C1-set IgSF domains. This, together with the presence of an IgSF V-set domain mediating ligand recognition, suggests that SIRPα is related to a key precursor in the evolution of vertebrate antigen receptors.
EXPERIMENTAL PROCEDURES
Recombinant extracellular SIRPα comprising the 30-residue N-terminal leader sequence and all three extracellular domains (residues 1–319 of the mature protein; accession number CAA71403) followed by the sequence TRHHHHHH was produced, deglycosylated, and crystallized as for SIRPα d1 (6). Crystallization experiments were performed in 96-well nanoliter-scale sitting drops (100 nl of 18.6 mg/ml SIRPα d1–d3 plus 100 nl of precipitant) equilibrated at either 5 or 20.5 °C against 95-μl reservoirs of precipitant and were monitored via an automated storage and imaging system (10). Diffraction quality crystals grew at 20.5 °C against a reservoir of 1.0 m trisodium citrate, 0.1 m sodium cacodylate, pH 6.5, within 2 weeks. Crystals were cryoprotected by a quick sweep through perfluoropolyether PFO-X125/03 (Lancaster Synthesis) before being flash-cryocooled by transfer directly into a cold stream of nitrogen gas (100 K). Diffraction data were recorded from a single frozen (100 K) crystal of SIRPα d1–d3 at European Synchrotron Radiation Facility beamline ID14-2 (λ = 0.933 Å) on an ADSC Quantum4R CCD detector. Diffraction data were indexed, integrated, and scaled using XDS (11) and SCALA (12) via the xia2 automated data processing pipeline5 (Table 1).
TABLE 1.
Data collection and refinement statistics
| Parameters | Values |
|---|---|
| Data collection | |
| Resolution limits (Å)a | 38.1-2.5 (2.55-2.49) |
| Space group | P212121 |
| Unit cell dimensions (Å) | 47.4, 84.4, 98.9 |
| Unique reflectionsa | 14,246 (1,037) |
| Redundancya | 4.8 (4.8) |
| Completeness (%)a | 98.5 (98.2) |
| I/σ(I)a | 14.2 (2.0) |
| Rmerge (%)a,b | 6.9 (85.4) |
| Rmeas (%)a,c | 7.7 (95.7) |
| Refinement | |
| Resolution limits (Å)a | 27.0-2.5 (2.64-2.49) |
| No. of reflections in working seta | 14,207 (2,133) |
| No. of reflections in test seta | 692 (106) |
| Rxpct (%)a,d | 21.8 (28.1) |
| Rfree (%)a,e | 27.1 (32.8) |
| No. of atoms (protein/carbohydrate/water) | 2,350/14/49 |
| Residues in Ramachandran favored region (%) | 98.7 |
| Ramachandran outliers (%) | 0.0 |
| r.m.s. bond lengths (Å) | 0.004 |
| r.m.s. bond angles (degrees) | 0.875 |
| Average B factors (Å2) (protein/carbohydrate/water) | 62.7/85.1/54.9 |
a Numbers in parentheses refer to the highest resolution shell.
b Rmerge = 100 × (ΣhklΣi|I(hkl;i) − 〈I(hkl)〉|/ΣhklΣiI(hkl;i)), where I(hkl;i) is the intensity of an individual measurement of a reflection, and 〈I(hkl)〉 is the average intensity of that reflection.
c Rmeas is the redundancy-independent merging R factor (40). Rmeas = 100 × (Σhkl((n(hkl)/(n(hkl) − 1))½ Σi|I(hkl;i) − 〈I(hkl)〉|)/ΣhklΣiI(hkl;i)), where n is the number of times a given reflection has been observed.
d Rxpct = 100 × (Σhkl(‖Fo(hkl)| − |Fxpct(hkl)‖)/Σhkl|Fo(hkl)|), where |Fo(hkl)| and |Fxpct(hkl)| are the observed structure facture amplitude and the expectation of the model structure factor amplitude, respectively (16).
e Rfree equals Rxpct of test set (5% of the data not used during refinement).
The structure of SIRPα d1–d3 was solved by molecular replacement using MOLREP (13), the structures of SIRPα d1 (Protein Data Bank code 2uv3), the light chain constant domain of monoclonal antibody YTS 105.18 (Protein Data Bank code 2ajr), and the α3 domain of rat MHC class I (Protein Data Bank code 1ed3) being used as search models for domains 1, 2, and 3, respectively. Manual model building was performed in COOT (14), and the model was refined using a combination of phenix.refine (15) for coordinate refinement and BUSTER/TNT (16) for refinement of B values (17). At all stages, building and refinement were informed by the validation tools present in COOT and by the MolProbity Web server (18).
Searches for similar protein structures were performed using the protein structure comparison service SSM at the European Bioinformatics Institute (19) against the IgSF domain subsets present in version 1.73 of the SCOP data base (20), using default parameters. SIRPα d1, d2, and d3 (residues 1–114, 115–220, and 221–317, respectively) were used as query structures. Additional query structures were CD80 d2 (SCOP domain d1dr9a2), VCAM-1 d2 (SCOP domain d1vscc1), constant domains (d2) of MHC class II β chain (SCOP domain d1s9ve1) and of Ig κ (Protein Data Bank code 1q9l chain L residues 109–221), lutheran d2 (Protein Data Bank code 2pet chain A residues 115- 231), and tapasin d2 (Protein Data Bank code 3f8u chain B residues 270–381). Representative V-, C1-, C2-, and I-set IgSF domains (non-redundant at a level of 70% sequence identity, 271 structures in total) were extracted from the SCOP data base with the assistance of the ASTRAL compendium (21). Pairwise superposition of the 271 representative IgSF domains plus SIRPα d1–d3; d1 from SIRPβ, -β(2), and -γ; CD80 d2; VCAM-1 d2; MHC class II β chain d2; Ig κ d2; lutheran d2; and tapasin d2 was performed using SSM. A metric for defining the relatedness of domains was obtained using Equation 1,
where R represents the relatedness, D is the Cα r.m.s. deviation of the two domains, 〈n〉 is the mean number of residues in the two domains, and M is the number of structurally equivalent Cα atoms. A two-dimensional graph depicting the relatedness of IgSF domains, where edges (lines) connecting highly related structures are short and edges connecting unrelated structures are long, was generated from the matrix of 283 × 282 relatedness values using the Fruchterman-Reingold graph layout algorithm (22) as implemented by CLANS (23) with default parameters. Results depicted are representative of multiple minimizations with random initial starting conditions. For illustrations, multiple structures were superposed using Theseus (24), and molecular graphics were prepared using PyMOL (DeLano Scientific).
RESULTS
Crystallization and Structure Determination
The extracellular region of SIRPα was expressed in a mutant Chinese hamster ovary cell line with defective glycosylation machinery that renders glycoproteins sensitive to endoglycosidase H treatment and more homogenous, thereby promoting crystallization (6, 25, 26). Crystals of deglycosylated SIRPα were obtained by sitting drop vapor diffusion, and diffraction data were recorded to 2.5 Å (Table 1). The structure was solved by molecular replacement using the N-terminal V-set domain of SIRPα (Protein Data Bank code 2uv3) and the C1-set domains of an Fab light chain (Protein Data Bank code 2ajr) and an MHC class I α3 domain (Protein Data Bank code 1ed3) as starting models for domains 1–3, respectively. The structure has been refined to 2.5 Å resolution with residuals R = 21.8% and Rfree = 27.1% (Table 1).
The Structure of the Extracellular Region of SIRPα
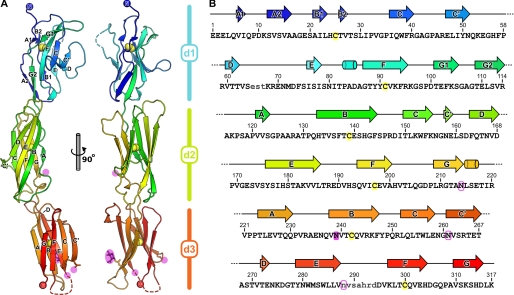
The extracellular region of SIRPα is shown in Fig. 1. The final refined structure comprises residues 1 (Glu) to 317 (Lys), the loops between residues 65–67 (d1) and 288–294 (d3) being excluded from the model due to the absence of well resolved electron density, presumably arising from disorder. An N-acetylglucosamine adduct was observed attached to the side chain of residue Asn239. Residual electron density consistent with the presence of N-linked carbohydrate was also evident at residue Asn214, but the electron density was not sufficiently well resolved to allow modeling of the carbohydrate moiety. As in previous determinations of the SIRPα d1 structure, no electron density was observed for N-acetylglucosamine at Asn80. The side chains of the other two potential glycosylation sites, Asn261 and Asn288, were not sufficiently well resolved to determine their glycosylation states. The N-terminal domain (d1) of the three-domain SIRPα structure can be superposed on the higher resolution (1.85 Å) structure of SIRPα in complex with CD47 with 1.1 Å r.m.s. deviation over 111 Cα atoms (residues 1–114, excluding residues 65–67 not observed in electron density). This allows the topology of the interaction to be determined (see below). In addition, there are now seven independent structures for SIRPα d1, which, when overlaid, illustrate the previously noted flexibility in the CC′, C′D, DE, and FG loops (supplemental Fig. S1) (5, 6). Excluding these loops and the first residue, all SIRPα d1 structures overlay extremely well (0.4 Å r.m.s. deviation over 101 Cα atoms).
FIGURE 1.
Structure of the extracellular region of SIRPα. A, the extracellular region of SIRPα is shown in ribbon representation colored from blue (N terminus) to red (C terminus). Disulfide bonds are shown as yellow spheres, the observed N-acetylglucosamine moiety is shown as magenta sticks, and other potential sites of N-linked glycosylation are indicated with magenta spheres. B, secondary structure is shown above the sequence of the three IgSF domains of SIRPα, arrows and cylinders representing β sheets and α helices, respectively. Residues not modeled due to poorly defined electron density are in lowercase type, the cysteine residues that form disulfide bond between β sheets are highlighted in yellow, the residue where N-linked glycosylation was observed in electron density is colored magenta, and other potential sites of N-linked glycosylation are outlined in magenta.
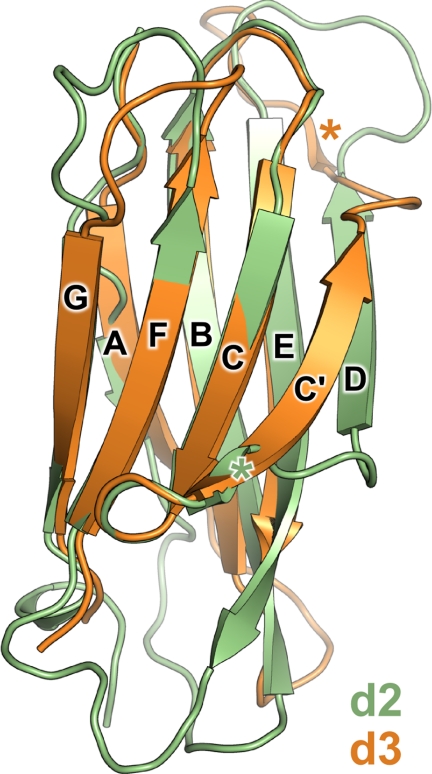
Domains 2 and 3 (d2 and d3) of SIRPα adopt typical IgSF folds, with eight β strands in two sheets linked by the canonical disulfide bond (Fig. 1). Both domains are more closely related in structure to each other than to SIRPα d1, superposing with 2.1 Å r.m.s. deviation over 77 Cα atoms versus 2.9 and 2.8 Å r.m.s. deviation over 77 and 69 Cα atoms when superposing d1 onto d2 and d3, respectively. Strands ABE and GFC of the two β sheets overlay extremely well (Fig. 2), although a single residue “insertion” and the presence of a proline residue causes a slight “bulge” immediately preceding the shorter strand A in d2. Strands C′ and D are markedly different in length between the two domains, d3 having an unusually long C′ strand and a short D strand. The conformations of the loops between β strands AB, C′D, DE, and FG differ significantly between the two domains, and the electron density of the EF loop in d3 is too poorly resolved to be modeled. Interestingly, the conformations of the loops linking strands B and C′ to strand C are very well conserved between d2 and d3. Sequence and structural conservation of the BC loop is a hallmark of IgSF C1-set domains, and it appears that SIRPα d2 and d3 are indeed members of this subset of IgSF domains (discussed below).
FIGURE 2.
Superposition of SIRPα domains 2 and 3. The superposed structures of SIRPα d2 (residues 115–220; green) and d3 (residues 221–317; orange) are displayed as ribbons. The short C′ strand of d2 and short D strand of d3 are marked with green and orange asterisks, respectively.
Topology of the CD47-SIRPα Interaction at the Cell Surface
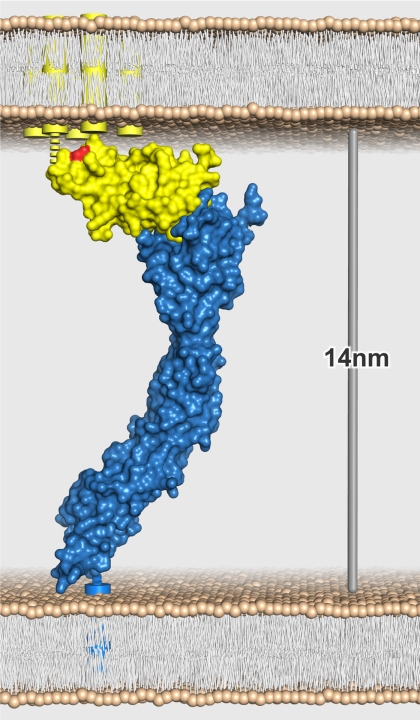
The new structure for the full extracellular region of SIRPα together with the previous structure of the complex between SIRPα d1 and CD47 allows the topology of the full CD47-SIRPα interaction to be visualized (Fig. 3). The overall dimensions of the modeled complex are 13.7 × 6.5 × 3.7 nm. Although the distance between the last residues of SIRPα and CD47 is only 12.0 nm, our structures do not include the links between the final residues of the extracellular domains and the predicted transmembrane helices, comprising 8 residues in CD47 and 23 residues in SIRPα. Further, the orientation of the complex is constrained by the presence in vivo of a disulfide bond between residues Cys15 and Cys245 of CD47, the latter being predicted to lie in a 9-residue extracellular loop between the fourth and fifth transmembrane helices of CD47 (27). It is therefore likely that the distance between cells engaged in a productive CD47-SIRPα interaction will be ∼14 nm, similar to the cell-cell distances found in other interactions involved in immune recognition, such as MHC-T cell receptor, CD2-CD58, and CD28-CD80 (7, 28, 29).
FIGURE 3.
Model for the CD47-SIRPα complex formed between interacting cells. The entire extracellular CD47-SIRPα complex, modeled by superposing the structure of SIRPα domains 1–3 onto the structure of CD47 in complex with SIRPα domain 1 (5), is shown as a blue (SIRPα) and yellow (CD47) molecular surface. Schematic representations of the five transmembrane helices of CD47 (yellow cylinders) and single transmembrane helix of SIRPα (blue cylinder) are shown. The regions between transmembrane helices and resolved structures for CD47 and SIRPα are illustrated as dotted lines. The location of CD47 residue Cys15, which forms a disulfide bond with Cys245 (proposed to reside in the extracellular loop between transmembrane helices 4 and 5), is highlighted in red.
Relationship of SIRPα d2 and d3 to Ig C1-set Domains
Overall Features
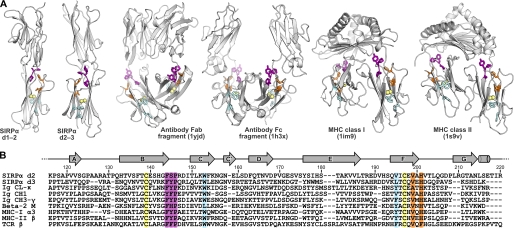
At the primary amino acid sequence level, IgSF domains are recognized by characteristic sequence patterns, although the overall sequence identity is often low (30, 31). Some features are typical of most IgSF domains, such as the two Cys residues that form a disulfide bridge between the β sheets and a Trp in β strand C that forms part of the hydrophobic core of the domain. The IgSF domain comprises 7–9 β strands. V-set domains, which are particularly common in biology and include the domains responsible for antigen recognition, have nine β strands that are identified at the primary sequence level by additional sequence between the two Cys residues. These domains often have other characteristic sequence motifs, such as DX(G/A)XYXC in β strand F. Domain 1 of the SIRPs is an IgSF V-set domain and contains this sequence motif (5, 6). C1-set domains are discussed below, whereas C2-set domains often share the sequence patterns of V-set domains but are considerably smaller, lacking strands C″ and D. Further subdivisions of the IgSF, such as the I-set family of domains, have been made on the basis of structural data (32). SIRPα domains 2 and 3 share the sequence properties that distinguish C1-sets from other IgSF domains. These include the characteristic sequence (F/Y)(F/Y/H)P at the beginning of the BC loop (FSP in d2, FYP in d3) and the sequence XVXH after the Cys residue in the F strand (Fig. 4). It is interesting to note that the two conserved sequence motifs are in close proximity in the three-dimensional structures of C1-set domains and of SIRPα d2 and d3 (Fig. 4) and may be important in maintaining the C1-set fold. Both d2 and d3 have exactly 13 residues between the Cys in strand B and the Trp in strand C. The presence of 13 residues between these Cys and Trp residues is also highly characteristic of C1-set domains (present in 1242 of 1295 C1-set sequences), whereas it varies in length considerably in V-set domains and C2-set domains (the distance of 13 residues was only found in 14% of 350 IgSF domains from non-antigen receptors) (33).
FIGURE 4.
Structural features conserved across SIRPα d2-d3 and the C1-set family of IgSF domains. A, structures of SIRPα, MHC class II, MHC class I (with β2-microglobulin), Fab regions, and Fc regions of IgG are shown as white ribbons (with the Protein Data Bank codes indicated in parentheses). Conserved residues in the BC and FG loops characteristic of C1-set domains are colored purple and orange, respectively, disulfide bonds are shown as yellow spheres, and conserved residues that form part of the β-sandwich hydrophobic core are colored light blue. B, sequence alignments of IgSF C1-set domains illustrated in A plus a T-cell receptor β chain constant domain (Protein Data Bank code 1tcr). The secondary structure of SIRPα d2 is shown above the sequences with arrows and cylinders representing β sheets and α helices, respectively. Conserved features characteristic of the C1-set family of IgSF domains are colored as in A.
Structural Homology
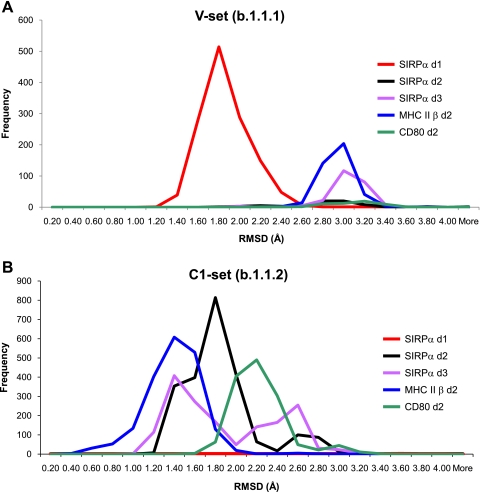
The structural similarity of SIRPα domains 2 and 3 to other IgSF domains was investigated using the protein structure comparison service SSM at the European Bioinformatics Institute (19) to compare the structures for SIRPα d2 and d3 with panels of V- C1-, C2- and I-set IgSF domains, as classified by the SCOP data base (20). Searches performed using SIRPα d2 and d3 gave large numbers of hits with the C1-set domains but very few with C2-, V-, or I-set domains (Table 2 and Fig. 5), comparable with searches performed using bona fide C1-set domains. This provides strong evidence that SIRPα d2 and d3 are more similar to C1-set domains than other subsets of IgSF domains. It has been suggested that CD80 domain 2 and lutheran domain 2 also possess C1-set domains (34, 35). However, these structures do not possess the conserved sequence patterns discussed above and do not share structural similarity with large numbers of C1-set IgSF domains (Table 2). By comparison, an SSM search of the SCOP-classified IgSF domain panels using SIRPα d1 identified 1128 V-set IgSF domains and 44 I-set domains but no C1- or C2-set domains, consistent with the classification of SIRPα d1 as a V-set IgSF domain and illustrating the specific features of the different subsets. The recent x-ray crystallography structure of tapasin confirmed that it had a C1-set domain (9). This domain of tapasin gave scores comparable with that of SIRPα d2 or 3 or bona fide C1-set (Table 2). Tapasin and MHC antigens may be closely related in evolution (9, 36).
TABLE 2.
Similarity of SIRPα domains to other IgSF domain families; SSM analysis
The number of domains within the families of IgSF as classified in the SCOP database ((20)) that can be overlaid on the query structure with an r.m.s. deviation of 2 Å or less is shown (frequency histograms of r.m.s. deviation versus SCOP domain for each query are given in Fig. 5). The identification code of each SCOP family is given in parentheses. The regions of the query domains are indicated by residue number (SIRPα d2 and d3), SCOP domain, or Protein Data Bank code with chain and residue numbers.
| SCOP families | SIRPα d2(115–220) | SIRPα d3(221–317) | MHC class II βd2 (d1s9ve1) | Ig LC-κ d2(1yjd; L:109–221) | CD80 d2(d1dr9a2) | Lutheran d2(2pet; A:115–231) | Tapasin d2(3f8u; B:270–381) | VCAM-1 d2(d1vcaa1) | SIRPα d1(2uv3; A:1–114) |
|---|---|---|---|---|---|---|---|---|---|
| V-set (b.1.1.1) | 4 | 3 | 0 | 0 | 0 | 13 | 33 | 0 | 1,128 |
| C1-set (b1.1.2) | 1,988 | 1,015 | 1,911 | 1,515 | 486 | 127 | 1,031 | 514 | 0 |
| C2-set (b1.1.3) | 10 | 22 | 8 | 1 | 6 | 1 | 3 | 10 | 0 |
| I-set (b.1.1.4) | 2 | 4 | 0 | 1 | 1 | 0 | 0 | 1 | 44 |
FIGURE 5.
Similarity of SIRPα domains to IgSF V- and C1-set domains. SSM superposition Cα r.m.s. deviations for query domain structures versus the IgSF SCOP families used to generate Table 2 are shown as frequency histograms in A for the V-set (b.1.1.1) and in B for the C1-set (b.1.1.2). Frequency intervals of 0.2 Å r.m.s. deviation were used.
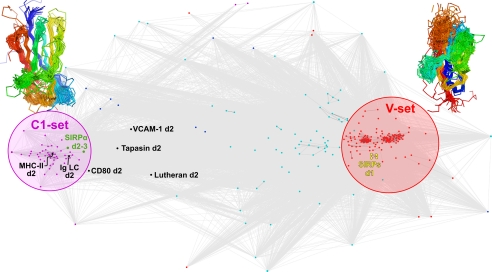
Structure-based clustering analysis (Fig. 6) demonstrates very clearly that SIRPα d2 and d3 are C1-set IgSF domains, whereas domains 1 from SIRPα, -β, -β(2), and -γ are all V-set IgSF domains. Superposition of 46 representative C1-set domains (Fig. 6) shows a striking amount of structural similarity, given the low level of sequence identity shared (Fig. 4). Although the β-sheets that form the core of the domain are well conserved, the loops that connect them are highly divergent. The BC loop is the best conserved of the sheet-connecting loops, consistent with the conserved length and sequence of the BC loop among C1-set domains (see above). Given the high degree of amino acid sequence conservation that SIRPβ and SIRPγ share with SIRPα (about 90% identity) the d2 and d3 of these proteins will almost certainly also be C1-set IgSF domains.
FIGURE 6.
Similarity of SIRP domains to other members of the immunoglobulin superfamily. Cluster analysis of the structural similarities shared by SIRPα domains 1–3; domains 1 of SIRPβ, SIRPβ(2), and SIRPγ; and 277 representative IgSF domains is shown. Each dot represents a single IgSF domain, colored according to its structural classification within the immunoglobulin superfamily (red, V-set; purple, C1-set; blue, C2-set; cyan, I-set; yellow, SIRP domains 1; green, SIRPα domains 2 and 3; black, CD80 d2, VCAM-1 d2, MHC class II β chain d2, Ig κ d2, lutheran d2, and tapasin d2). The distance between domains is proportional to their structural similarity (see “Experimental Procedures”). Superposed C1-set and V-set structures are shown as Cα traces, color-ramped from blue (start of domain) to red (end of domain).
DISCUSSION
The Dimensions of the CD47-SIRPα Interaction Are Compatible with the Immunological Synapse
The structure of SIRPα indicated that the CD47-SIRPα interaction is likely to span ∼14 nm. This is the distance spanned by well characterized components of the immunological synapse. Since SIRPα is present on macrophages and neurons and its ligand CD47 is present on most cell types, it seems likely that productive engagement of SIRPα by CD47 will occur only at regions of close contact, analogous to the immunological synapse, as shown for phagocytosis of red blood cells (37). It is likely that these CD47-SIRPα-containing synapse-like interactions will occur between many cell types, as also suggested previously in the case of the CD200-CD200R interaction (38).
Implications for the Evolution of Antigen Receptors
During vertebrate evolution the adaptive immune system developed into that of mammals today, one that can generate a diverse set of T cell receptors and immunoglobulins to recognize a vast number of different antigens through rearrangement of gene segments and a recognition system involving MHC antigens. Most of these components are present in jawed but not in jawless fish, and there is considerable interest in how these proteins evolved. Characteristic IgSF C1-set domains are present exclusively in antigen receptors and certain proteins closely associated with antigen recognition, such as MHC antigens, tapasin, and β2-microglobulin. Our structure shows that domains 2 and 3 of SIRPα are C1-set domains, strengthening suggestions based on sequence analysis that the SIRPs are closely related to the precursors of antigen receptors (8). C1-set domains are always found in association with other domains, with cis interactions being present between heavy- and light-chain constant domains in antibodies, β2-microglobulin and the C1-set domain in MHC Class I and related proteins, the two domains in MHC class II, and the adjacent chains in the T cell receptors. Within T cell receptor and Ig, C1-set domains are C-terminal to the V-set domain and present as linear arrays in Ig H chains. The SIRPs maintain this linear array and the V-set-C1-set topology.
No SIRP genes have been identified in organisms predating the adaptive immune system (8, 39); thus, one cannot distinguish between SIRPs evolving directly from a precursor of the antigen receptors or from an antigen receptor itself. However, it seems reasonable that the primitive antigen receptor was expressed on myeloid cells and contained both V- and C1-set IgSF domains.
Supplementary Material
Acknowledgments
We are grateful to the staff of beamline ID14-2 at the European Synchrotron Radiation Facility for assistance with data collection and to T. S. Walter for assistance with crystallization.
This work was supported in part by Medical Research Council Grant G0400808 and Wellcome Trust Grant WT084194 (to A. N. B., D. H., K. H., D. I. S., and the Oxford Protein Production Facility). This is a contribution of the European Community project SPINE2-Complexes (LSHG-CT-2006-031220).
The atomic coordinates and structure factors (code 2WNG) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
G. Winter, manuscript in preparation.
- SIRP
- signal-regulatory protein
- IgSF
- immunoglobulin superfamily
- MHC
- major histocompatibility complex
- r.m.s.
- root mean square
- SSM
- secondary structure matching.
REFERENCES
- 1.Barclay A. N., Brown M. H. (2006) Nat. Rev. Immunol. 6, 457–464 [DOI] [PubMed] [Google Scholar]
- 2.Brown E. J., Frazier W. A. (2001) Trends Cell Biol. 11, 130–135 [DOI] [PubMed] [Google Scholar]
- 3.Oldenborg P. A., Zheleznyak A., Fang Y. F., Lagenaur C. F., Gresham H. D., Lindberg F. P. (2000) Science 288, 2051–2054 [DOI] [PubMed] [Google Scholar]
- 4.Fujioka Y., Matozaki T., Noguchi T., Iwamatsu A., Yamao T., Takahashi N., Tsuda M., Takada T., Kasuga M. (1996) Mol. Cell. Biol. 16, 6887–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatherley D., Graham S. C., Turner J., Harlos K., Stuart D. I., Barclay A. N. (2008) Mol. Cell 31, 266–277 [DOI] [PubMed] [Google Scholar]
- 6.Hatherley D., Harlos K., Dunlop D. C., Stuart D. I., Barclay A. N. (2007) J. Biol. Chem. 282, 14567–14575 [DOI] [PubMed] [Google Scholar]
- 7.Wang J. H., Smolyar A., Tan K., Liu J. H., Kim M., Sun Z. Y., Wagner G., Reinherz E. L. (1999) Cell 97, 791–803 [DOI] [PubMed] [Google Scholar]
- 8.van den Berg T. K., Yoder J. A., Litman G. W. (2004) Trends Immunol. 25, 11–16 [DOI] [PubMed] [Google Scholar]
- 9.Dong G., Wearsch P. A., Peaper D. R., Cresswell P., Reinisch K. M. (2009) Immunity 30, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter T. S., Diprose J. M., Mayo C. J., Siebold C., Pickford M. G., Carter L., Sutton G. C., Berrow N. S., Brown J., Berry I. M., Stewart-Jones G. B., Grimes J. M., Stammers D. K., Esnouf R. M., Jones E. Y., Owens R. J., Stuart D. I., Harlos K. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 12.Evans P. R. (2005) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 13.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 14.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 15.Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsletter 42, contribution 8, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 16.Blanc E., Roversi P., Vonrhein C., Flensburg C., Lea S. M., Bricogne G. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 [DOI] [PubMed] [Google Scholar]
- 17.Tronrud D. (1996) J. Appl. Crystallogr. 29, 100–104 [Google Scholar]
- 18.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 19.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 20.Murzin A. G., Brenner S. E., Hubbard T., Chothia C. (1995) J. Mol. Biol. 247, 536–540 [DOI] [PubMed] [Google Scholar]
- 21.Chandonia J. M., Hon G., Walker N. S., Lo Conte L., Koehl P., Levitt M., Brenner S. E. (2004) Nucleic Acids Res. 32, D189–D192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fruchterman T., Reingold E. (1991) Software Practice Experience 21, 1129–1164 [Google Scholar]
- 23.Frickey T., Lupas A. (2004) Bioinformatics 20, 3702–3704 [DOI] [PubMed] [Google Scholar]
- 24.Theobald D. L., Wuttke D. S. (2006) Bioinformatics 22, 2171–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley P. (1989) Mol. Cell. Biol. 9, 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis S. J., Puklavec M. J., Ashford D. A., Harlos K., Jones E. Y., Stuart D. I., Williams A. F. (1993) Protein Eng. 6, 229–232 [DOI] [PubMed] [Google Scholar]
- 27.Rebres R. A., Vaz L. E., Green J. M., Brown E. J. (2001) J. Biol. Chem. 276, 34607–34616 [DOI] [PubMed] [Google Scholar]
- 28.Garboczi D. N., Ghosh P., Utz U., Fan Q. R., Biddison W. E., Wiley D. C. (1996) Nature 384, 134–141 [DOI] [PubMed] [Google Scholar]
- 29.Evans E. J., Esnouf R. M., Manso-Sancho R., Gilbert R. J., James J. R., Yu C., Fennelly J. A., Vowles C., Hanke T., Walse B., Hünig T., Sørensen P., Stuart D. I., Davis S. J. (2005) Nat. Immunol. 6, 271–279 [DOI] [PubMed] [Google Scholar]
- 30.Barclay A. N. (2003) Semin. Immunol. 15, 215–223 [DOI] [PubMed] [Google Scholar]
- 31.Williams A. F., Barclay A. N. (1988) Annu. Rev. Immunol. 6, 381–405 [DOI] [PubMed] [Google Scholar]
- 32.Harpaz Y., Chothia C. (1994) J. Mol. Biol. 238, 528–539 [DOI] [PubMed] [Google Scholar]
- 33.Smith D. K., Xue H. (1997) J. Mol. Biol. 274, 530–545 [DOI] [PubMed] [Google Scholar]
- 34.Mankelow T. J., Burton N., Stefansdottir F. O., Spring F. A., Parsons S. F., Pedersen J. S., Oliveira C. L., Lammie D., Wess T., Mohandas N., Chasis J. A., Brady R. L., Anstee D. J. (2007) Blood 110, 3398–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikemizu S., Gilbert R. J., Fennelly J. A., Collins A. V., Harlos K., Jones E. Y., Stuart D. I., Davis S. J. (2000) Immunity 12, 51–60 [DOI] [PubMed] [Google Scholar]
- 36.Mayer W. E., Klein J. (2001) Immunogenetics 53, 719–723 [DOI] [PubMed] [Google Scholar]
- 37.Tsai R. K., Discher D. E. (2008) J. Cell Biol. 180, 989–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright G. J., Puklavec M. J., Willis A. C., Hoek R. M., Sedgwick J. D., Brown M. H., Barclay A. N. (2000) Immunity 13, 233–242 [DOI] [PubMed] [Google Scholar]
- 39.van Beek E. M., Cochrane F., Barclay A. N., van den Berg T. K. (2005) J. Immunol. 175, 7781–7787 [DOI] [PubMed] [Google Scholar]
- 40.Diederichs K., Karplus P. A. (1997) Nat. Struct. Biol. 4, 269–275 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.