Abstract
Protein degradation via the ubiquitin proteasome system has been shown to regulate changes in synaptic strength that underlie multiple forms of synaptic plasticity. It is plausible, therefore, that the ubiquitin proteasome system is itself regulated by synaptic activity. By utilizing live-cell imaging strategies we report the rapid and dynamic regulation of the proteasome in hippocampal neurons by synaptic activity. We find that the blockade of action potentials (APs) with tetrodotoxin inhibited the activity of the proteasome, whereas the up-regulation of APs with bicuculline dramatically increased the activity of the proteasome. In addition, the regulation of the proteasome is dependent upon external calcium entry in part through N-methyl-d-aspartate receptors and L-type voltage-gated calcium channels and requires the activity of calcium/calmodulin-dependent protein kinase II (CaMKII). Using in vitro and in vivo assays we find that CaMKII stimulates proteasome activity and directly phosphorylates Rpt6, a subunit of the 19 S (PA700) subcomplex of the 26 S proteasome. Our data provide a novel mechanism whereby CaMKII may regulate the proteasome in neurons to facilitate remodeling of synaptic connections through protein degradation.
Synaptic plasticity is a complex process that requires the selective remodeling of synaptic connections. A majority of this remodeling occurs post-synaptically, where neurotransmitter receptors, scaffold proteins, and signaling molecules are modified in response to neuronal activity (1). The incorporation of newly synthesized proteins is required for long-lasting changes in synaptic efficacy (2, 3). Protein degradation, on the other hand, provides an additional mechanism to modify the stoichiometry of synaptic proteins to promote, limit, or restrict plasticity.
The ubiquitin proteasome system (UPS)4 is a major pathway for protein turnover in eukaryotic cells. The selective degradation of proteins via the UPS involves three steps; they are recognition of the target protein via specific signals, marking of the target protein with ubiquitin chains by ubiquitin ligases, and delivery of the target protein to the 26 S proteasome for degradation (4). The 26 S proteasome is a large energy-dependent protease consisting of two multiprotein subcomplexes; that is, the 20 S proteolytic core and 19 S cap (PA700). The 20 S consists of 28 subunits (α and β), 6 of which are catalytic, whereas the 19 S consists of Rpt (ATPase) and Rpn (non-ATPase) regulatory subunits (5).
The UPS plays a crucial role in the development, maintenance, and remodeling of synaptic connections (6–9). Additionally, pharmacological inhibition of the proteasome inhibits various forms of synaptic plasticity (10–17). Several studies have identified key synaptic proteins such as PSD-95, Shank, GKAP, AKAP, SPAR, RIM-1, and GRIP1 that are regulated by UPS-dependent protein turnover (10, 18–21). In addition, it appears that cohorts of synaptic proteins are degraded in response to chronic activity blockade or up-regulation (18, 22).
Although ubiquitination has been widely studied, much less is known about regulation of the proteasome itself. There is increasing evidence that the proteasome may be regulated by interacting proteins or posttranslational modifications such as glycosylation or phosphorylation (23–26). Here, we set out to determine how the proteasome is regulated in neurons. We utilized live-cell imaging strategies to uncover the dynamics of proteasome activity in cultured hippocampal neurons. We found that both the blockade of action potentials (APs) with tetrodotoxin (TTX) and the up-regulation of APs with bicuculline (BIC) had rapid and opposing effects on proteasome activity. In addition, increased proteasome activity was found to be dependent upon external calcium entry in part through N-methyl-d-aspartate (NMDA) receptors and L-type voltage-gated calcium channels (VGCCs) and required the activity of calcium/calmodulin-dependent protein kinase II (CaMKII). To investigate the involvement of CaMKII in proteasome regulation, we overexpressed the constitutively active CaMKII (T286D) in HEK293T cells. CaMKII robustly stimulated the activity of the proteasome. Additionally, CaMKIIα directly phosphorylated Rpt6, a 19 S proteasome subunit in vitro. Together, our data indicate proteasome function can be rapidly controlled by neuronal activity, with CaMKII as a key regulator.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
α2 proteasome (MCP21 monoclonal antibody (mAb)), αβ core proteasome (polyclonal antibody), and Rpt6 (mAb) antibodies were purchased from Biomol. GFP antibody (full-length, polyclonal antibody) was from Santa Cruz Biotechnology. The following pharmacological reagents were used. 2-Amino-5-phosphonopentanoic acid (APV), BIC, nimodipine, TTX, and 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) were from Tocris Bioscience. MG132 was from Peptide Institute. Cycloheximide was from MP Biomedicals. Adamantane-acetyl-(6-aminohexanoyl)(3)-(leucinyl)(3)-vinyl-(methyl)-sulfone (vinyl sulfone), epoxomicin, (±)-α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), KN-93, and myristoylated autocamtide-2-related inhibitory peptide (AIP) were from Biomol. Calpeptin was from EMD Biosciences. Suc-LLVY-AMC fluorogenic substrate was from Biomol. Purified calmodulin (human, recombinant) and CaMKIIα (full-length human recombinant) were from Biomol.
DNA and Sindbis Constructs
RSV-CaMKII T286D was a kind gift from Anirvan Ghosh (University of California San Diego, La Jolla, CA). CaMKII T286D was amplified by PCR with primers containing XbaI and StuI with 5′ and 3′ overhangs, respectively. The PCR products were ligated into the XbaI and StuI sites of SinRep5 (Invitrogen). GFPu (in EGFP-C1 plasmid backbone; Clontech, Palo Alto, CA), a fusion of the CL1 degron (degradation signal) on the carboxyl terminus of GFP, was kindly provided by Ron Kopito (Stanford University, Palo Alto, CA). GFPu is ubiquitinated and specifically degraded by the proteasome (27–29). GFP-odc (in EGFP-C1 plasmid backbone; Clontech), a fusion of the ornithine decarboxylase (odc) degron on the carboxyl terminus of GFP, was also kindly provided by Ron Kopito. GFP-odc is targeted directly to the proteasome for degradation in an ubiquitin-independent manner (30, 31). The AgeI-BsrGI fragment from photoactivatable (pa) GFP (a kind gift provided by Jennifer Lipponcott-Schwartz, National Institutes of Health, Bethesda, MD) was subcloned into the AgeI-BsrGI sites of the GFP, GFPu, and GFP-odc plasmids. paGFPu, paGFP-odc, and the paGFP (no degron control) open reading frames were amplified by PCR with primers containing NheI and BamHI 5′ and 3′ overhangs, respectively. The PCR products were ligated into the NheI-BamHI sites of pCMV-IRES-mCherry. pCMV-IRES-mCherry was created by subcloning mCherry (pRSETB-mCherry; a kind gift from Roger Tsien, University of California San Diego, La Jolla, CA) into IRES2-EGFP (Clontech). This was done by ligating the BamHI (blunted)-BsrGI mCherry-containing fragment into BstXI (blunted)-BsrGI sites. For Sindbis virus construction, the NheI-XbaI reporter-IRES-mCherry containing fragment from pCMV-paGFP, paGFPu, and paGFP-odc plasmids were subcloned into the XbaI site of SinRep5. All constructs were confirmed by DNA sequencing. For production of recombinant Sindbis virions, RNA was transcribed using the SP6 mMessage mMachine kit (Ambion, Austin, TX) and electroporated into baby hamster kidney cells using a BTX ECM 600 electroporator (BTX, Holliston, MA) at 220 V, 129 ohms, and 1050 microfarads. Virion were collected after 24–32 h and stored at −80 °C until use.
Neuronal Cultures
Rat dissociated hippocampal neurons from postnatal day 1 or 2 were plated at a density of 45,000 cells/cm2 onto poly-d-lysine-coated coverslips or glass-bottom 35-mm dishes (Mattek, Ashland, MA) and maintained in B27-supplemented Neurobasal media (Invitrogen) for 17 days or more as previously described (32). High density rat cortical or hippocampal neurons from postnatal day 1 were plated onto poly-d-lysine-coated 6-well dishes (∼500,000 cells/well) and maintained in B27 supplemented Neurobasal media.
Confocal Microscopy
For all imaging purposes, we used a Leica (Wetzlar, Germany) DMI6000 inverted microscope outfitted with a Yokogawa (Tokyo, Japan) spinning disk confocal head, an Orca ER high Resolution B&W Cooled CCD camera (6.45 μm/pixel at 1×) (Hamamatsu, Sewickley, PA), Plan Apochromat 40×/1.25 NA and 63x/1.4 NA objective, and a Melles Griot (Carlsbad, CA) argon/krypton 100-milliwatt air-cooled laser for 488/568/647-nm excitations. Confocal z-stacks were acquired in all experiments. Photoactivation of reporter constructs was achieved with 100-watt Hg2+ lamp through a D405/40× excitation filter and 440 DCLP dichroic mirror (Chroma, Rockingham, VT) (10–15-s exposure times for photoactivation).
Live Imaging
For Sindbis virus infection of mature hippocampal neurons (>21 days in vitro) on glass-bottom 35-mm dishes, paGFPu, paGFP-odc, or paGFP reporter virion were added directly to culture media. Expression was allowed to continue for only 12–14 h to prevent cytotoxicity. The medium was then replaced with HEPES-based saline solution containing 119 mm NaCl, 5 mm KCl, 2 mm CaCl2, 2 mm MgCl2, 30 mm glucose, 10 mm HEPES with the appropriate pharmacological treatments. To determine basal rates of reporter degradation, cells were preincubated for 20 min in HEPES-based saline with either vehicle (DMSO) or proteasome inhibitors (MG132 25 μm, vinyl sulfone 1 μm, or epoxomicin 1 μm) at ∼35 °C using a ceramic heat lamp (ZooMed, San Luis Obispo, CA). The bath temperature was continually monitored by a digital probe thermometer. Infected neurons (identified by mCherry expression) were then photoactivated for 10–15 s with a 100-watt Hg2+ lamp and a D405/40× with 440 DCLP dichroic filter set (Chroma). For live imaging of neurons, the primary dendrites from pyramidal-like neurons were selected. Confocal Z-stack images (with 0.5-μm sections) were acquired with a 63× objective every 2, 3, or 4 min (as indicated) for the duration of the time lapse experiment. In some experiments (as indicated) both GFP (excitation 488 nm) and mCherry (568 nm) images were acquired. In others, GFP was acquired every time point, whereas pre- and post-mCherry time lapse images were acquired. For treatment of neurons with APV (50 μm), CNQX (40 μm), calpeptin (10 μm), KN-93 (10 μm), AIP (5 μm), or Ca2+-free containing HEPES-based saline, cells were similarly preincubated, selected by mCherry expression, photoactivated, and imaged by time-lapse microscopy. All pretreatments were for 15 min except for AIP (30 min). For treatment of neurons with TTX or BIC alone or with various combinations of other pharmacological treatments in HEPES-based saline, TTX or BIC was added to the bath after an initial 10-minute imaging period (12 min for paGFP-odc). Time lapse imaging followed for an additional 20-min period (48 min for paGFP-odc). Imaging of control and BIC-treated cells was interspersed between different treatments to verify that the degradation rates of our reporters under control and BIC-stimulated conditions in individual dendrites performed throughout the study did not vary significantly from the grouped data.
Florescence Intensity Quantitation
All imaging was acquired in the dynamic range of 8 or 12 bit acquisition (0–255 and 0–1024 pixel intensity units, respectively) with Simple PCI (Hamamatsu) imaging software. For quantitation of reporter degradation, straightened dendrites from maximum projected confocal z-stacks were analyzed with NIH ImageJ. Images were thresholded above background. Total integrated fluorescence intensity was measured from entire (proximal and distal regions) dendrites at each time interval and expressed as the percent change from time 0. Grouped analysis of dendritic fluorescence decay over time from each treatment group is plotted as line graphs (mean ± S.E.). The degradation rate of reporter fluorescence decay is obtained by taking the difference of total fluorescence loss (arbitrary units (AU)) over time (Fi − Fn/timen) from individual experiments. The mean degradation rate ± S.E. per treated group is then divided by the control rate to obtain the “relative to control” value. In some cases the degradation rate was extracted from smaller defined time windows after the addition of activity-altering pharmacological drugs to highlight and compare dynamic changes in reporter degradation. For statistical analysis, degradation rates were compared using Student's t tests. p values are listed in Table 1 and supplemental Table S1.
TABLE 1.
paGFPu rates of degradation
p values were determined by two-tailed unpaired Student's t test.
| Conditions | Rate of degradation | p value | No. imaged dendrites |
|---|---|---|---|
| % Fluorescence decay/2 min | |||
| Controla | 2.43 ± 0.50 | 21 | |
| MG132 | 0.58 ± 0.39 | 0.009 | 16 |
| Vinyl sulfone | 0.55 ± 0.29 | 0.025 | 9 |
| Epoxomicin | 0.87 ± 0.18 | 0.035 | 11 |
| Calcium-free | 0.67 ± 0.19 | 0.012 | 13 |
| APV | 1.34 ± 0.27 | 0.115 | 13 |
| Nimodipine | 1.70 ± 0.05 | 0.418 | 7 |
| CNQX | 2.14 ± 0.28 | 0.704 | 10 |
| BIC, t 10 min | 5.67 ± 0.51 | <0.001 | 27 |
| BIC, t 20 min | 4.06 ± 0.37 | 0.01 | 27 |
| BIC, t 60 min | 1.65 ± 0.07 | 0.47 | 5 |
| AMPA | 5.90 ± 0.34 | <0.001 | 11 |
| TTX | 1.13 ± 0.10 | 0.02 | 19 |
| KN-93 | 1.39 ± 0.12 | 0.076 | 16 |
| paGFPb | 0.25 ± 0.25 | 0.003 | 13 |
| BICa | 4.06 ± 0.37 | 27 | |
| MG132 + BIC | 1.54 ± 0.31 | 0.001 | 8 |
| Calpeptin + BIC | 3.92 ± 0.40 | 0.814 | 15 |
| paGFPb + BIC | 1.27 ± 0.12 | <0.001 | 12 |
| APV + BIC | 1.90 ± 0.26 | <0.001 | 14 |
| Nimodipine + BIC | 1.67 ± 0.30 | 0.001 | 10 |
| CNQX + BIC | 1.25 ± 0.16 | 0.003 | 5 |
| KN-93 + BIC | 1.58 ± 0.11 | <0.001 | 17 |
a Indicates reference category.
b paGFP, no-degron control reporter.
26 S Proteasome Fluorogenic Peptidase Assays
In vitro assay of 26 S proteasome chymotryptic activity was performed as previously described (33) with slight modifications. HEK293T cells were transfected with Rous sarcoma virus CaMKII T286D for 24–36 h. Neuronal cultures were transduced with CaMKII T286D by Sindbis virus. Cells were lysed in proteasome activity assay buffer (50 mm Tris-HCl (pH 7.5), 250 mm sucrose, 5 mm MgCl2, 0.5 mm EDTA, 2 mm ATP, 1 mm dithiothreitol, and 0.025% digitonin). 100 μm fluorogenic substrate Suc-LLVY-AMC was then added to lysates in a 96-well microtiter plate. Fluorescence (380-nm excitation, 460-nm emission) was monitored on a microplate fluorometer (HTS 7000 Plus, PerkinElmer Life Sciences) every 4 min for 2 h at room temperature.
[32P]Orthophosphate Live Labeling and Immunoprecipitation
HEK293T cells were transfected with CaMKII (T286D) for 24–36 h. Cultured cortical neurons or HEK293T cells were live-labeled for 2 h in a low phosphate HEPES-buffered solution (34) (5.6 mm KCl, 0.2 mm KH2P04, 137.6 mm NaCl, 2.4 mm NaHCO3, 5.6 mm glucose, 0.4 mm MgSO4, 0.5 mm MgCl2, 1.26 mm CaCl2, 20 mm HEPES (pH 7.4)) with 0.40 mCi/ml [32P]orthophosphate added to each well (of a 6-well culture dish). Cells were lysed in proteasome immunoprecipitation buffer (25 mm Tris (pH 7.5), 15% glycerol, 2.5 mm MgCl2, 5 mm ATP, 5 mm sodium pyrophosphate, 50 mm sodium fluoride, 1 mm sodium vanadate, 1 mm dithiothreitol, 0.5% Nonidet P-40). Lysates were immunoprecipitated with the indicated antibodies and resolved by SDS-PAGE. 32P labeling was detected with a Typhoon Scanner (GE Healthcare) and analyzed using ImageQuant TL (Amersham Biosciences).
In Vitro Phosphorylation of PA700, the 19 S Regulator of the 26 S Proteasome
19 S proteasome was purified from bovine red blood cells as described previously (35). PS-1 and PS-2 subcomplexes of the 19 S were purified as described previously (36). Phosphorylation reactions were conducted with autoactivated CaMKIIα. CaMKIIα was activated in reactions containing 100 μm ATP, 1.2 μm calmodulin, 10 mm MgCl2, and 2 mm CaCl2. After incubation for 10 min at 30 °C, activated kinase was incubated with 19 S. Assays contained 5 μm CaMKIIα, 45 μm 19 S, 100 μm [γ-32P]ATP (500 mCi/m mol), 50 mm Tris-HCl (pH 7.6), 2 mm dithiothreitol, and 10% glycerol in a final volume of 50 μl. Control reactions contained either no kinase or no 19 S. After various times of incubation at 30 °C samples were treated with SDS sample buffer and subjected to SDS-PAGE and autoradiography.
RESULTS
Action Potential Blockade and Up-regulation Produce Rapid and Opposite Effects on Proteasome Activity in Hippocampal Dendrites
To determine the effects of neuronal activity on proteasome function, we utilized a GFP-based reporter imaging strategy to monitor the rate of proteasome activity in the dendrites of cultured hippocampal neurons. GFPu is a fusion of the CL1 degron, a 16-amino acid hydrophobic sequence first identified in yeast (29), with the carboxyl terminus of GFP (28). It has a very short half-life (<30 min) in mammalian cells and is constitutively ubiquitinated and degraded by the proteasome (27–29). We modified GFPu by replacing GFP with a pa variant (37) (Fig. 1A) to monitor the fluorescence decay of an isolated population of reporter proteins after photoactivation. paGFPu was expressed in mature hippocampal neurons (>21 days in vitro) for 12–14 h by Sindbis viral delivery. mCherry, a monomeric variant of red fluorescent protein (38), was simultaneously expressed by an internal ribosomal entry site (IRES) sequence to identify infected neurons before photoactivation of paGFPu (Fig. 1, A and B). After photoactivation, paGFPu fluorescence decay was monitored by confocal time-lapse imaging. paGFP (control reporter that lacks the CL1 degron) was utilized to account for the effects of diffusion and photobleaching.
FIGURE 1.
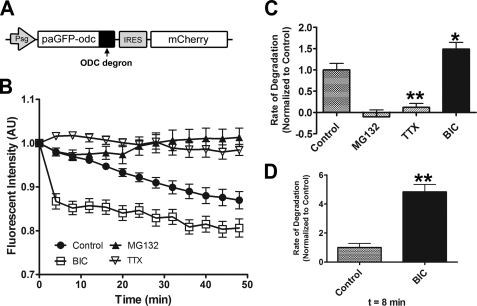
Action potential blockade and up-regulation produce opposite effects on proteasome activity in hippocampal neurons. A, schematic of the pa UPS reporter Sindbis constructs. The CL1 degradation signal (degron) is fused to the carboxyl terminus of paGFP (non CL1 containing control reporter), converting it into an ubiquitin-dependent proteasomal degradation reporter (paGFPu). mCherry is co-expressed using an IRES signal. B, representative image of a hippocampal neuron infected with paGFPu before (PRE) and after (POST) photoactivation. mCherry is expressed to locate infected cells before photoactivation. C, CL1 degron promotes degradation of paGFPu. Straightened dendrites from representative time-lapse experiments of cultured hippocampal neurons expressing paGFPu alone, paGFPu plus MG132 (25 μm), or paGFP. paGFPu fluorescence decay is observed in dendrites and is blocked by the addition of the proteasome inhibitor MG132. No significant fluorescence loss is observed in paGFP (no degron control)-expressing dendrites. D, representative straightened dendrites from time-lapse experiments in which cultured hippocampal neurons expressing paGFPu were treated with TTX (2 μm), BIC (40 μm), or BIC plus MG132 (25 μm), and imaged every 2 min for 20 min. Time of treatment is indicated by black arrows. E, grouped analysis (plotted as line graphs, mean ± S.E.) of dendritic fluorescent intensity normalized to time 0 for control (paGFPu alone), MG132, TTX, BIC, BIC plus MG132-treated neurons, or paGFP (no degron). F, bar graph depicting the mean degradation rate ± S.E. (AU) per treated group normalized to the control (paGFPu) rate of fluorescence decay. BIC dramatically increased paGFPu rate of degradation, most significantly in the first 10 min, whereas TTX blocks paGFPu degradation (*, p < 0.05; **, p < 0.001, relative to paGFPu (control)). See Table 1 for rates of degradation and p and n values. The scale bar for all images is 10 μm.
We found that paGFPu was rapidly degraded (as monitored by its fluorescence decay) in hippocampal dendrites (Fig. 1C), which is consistent with reported findings of GFPu degradation in mammalian cells (28). When paGFPu-expressing cells were treated with three different proteasome inhibitors (25 μm MG132, 1 μm vinyl sulfone, and 1μm epoxomicin), the degradation rate of paGFPu was significantly blocked to levels of paGFP, which lacked the CL1 degron (mean rates of degradation ± S.E. relative to control: MG132, 23.9 ± 16.1%; vinyl sulfone, 22.6 ± 11.9%; epoxomicin, 35.8 ± 7.4%; paGFP, 10.3 ± 10.3%; Fig. 1, C, E, and F and Table 1).
We next asked what the effect of blocking APs with TTX or up-regulating APs with BIC would be on proteasome activity. After monitoring paGFPu degradation in hippocampal dendrites for a 10-min control period, BIC (40 μm final) or TTX (2 μm final) was added to the bath solution. paGFPu-expressing dendrites were then imaged for an additional 20 min (2-min intervals). Relative to control-treated neurons, BIC significantly increased the paGFPu rate of degradation (167 ± 15.2% relative to control; Fig. 1, D–F, and Table 1). The rate of paGFPu degradation in BIC-treated neurons was most pronounced within the first 10 min after the addition of BIC (233 ± 15.2% relative to control; Fig. 1F, Table 1). This effect was specific to proteasome-mediated turnover of paGFPu as the BIC-induced increase in paGFPu degradation was blocked in cells that were pretreated with the proteasome inhibitor MG132 (25 μm), but not with a specific calpain inhibitor, calpeptin (10 μm) (relative to BIC: MG132 + BIC, 37.9 ± 7.6%; calpeptin + BIC, 96.6 ± 9.9%; Fig. 1, D–F, Table 1). Furthermore, minimal effects of BIC were observed on the fluorescence decay of the paGFP control reporter (31.3 ± 3.0% relative to BIC; Table 1). Interestingly, we also observed in the majority of BIC-treated neurons that the highest rates of paGFPu degradation initiated and persisted in spines for more than 10 min (supplemental Movie S1).
In contrast, we observed opposite effects on paGFPu degradation by AP blockade. We found the rate of paGFPu degradation to be significantly attenuated in cells that were treated with TTX (46.5 ± 4.1% relative to control; Fig. 2, D–F, Table 1). Furthermore, application of TTX rapidly increased ubiquitin conjugate levels in neurons in a manner similar to cells treated with proteasome inhibitors (supplemental Fig. S1). These results indicate that the activity of the proteasome is rapidly tuned to increased or decreased neuronal activity.
FIGURE 2.
Action potential blockade and up-regulation produce opposite effects on proteasome activity in hippocampal neurons as monitored by our ubiquitin-independent proteasome reporter (paGFP-ODC). A, schematic of the paGFP-odc reporter Sindbis construct. The degradation sequence (degron) of ODC is fused to the carboxyl terminus of paGFP, converting it into an ubiquitin-independent reporter of proteasome activity. mCherry is co-expressed using an IRES signal. B, grouped analysis (plotted as line graphs ± S.E.) of dendritic paGFP-odc fluorescence intensity normalized to time 0. Cultured hippocampal neurons expressing paGFP-odc were treated with DMSO (Control), MG132 (25 μm), BIC (40 μm), or TTX (2 μm) and imaged every 4 min for 48 min. C, bar graph depicting the mean degradation rate ± S.E. (AU) per treated group relative to the control (paGFP-odc) rate of fluorescence intensity. TTX significantly blocks paGFP-odc degradation, whereas BIC significantly increases the rate of degradation. As expected, the rate of paGFP-odc degradation is blocked by MG132 (*, p < 0.05; **, p < 0.001, relative to control). D, the BIC induced degradation was most dramatic in the first 8 min (**, p < 0.001, relative to control for 0–8-min time point). See supplemental Table 1 for rates of degradation and p and n values.
To confirm these results we utilized an additional GFP-based degradation reporter. Ornithine decarboxylase (ODC) is a well characterized cellular protein subject to ubiquitin-independent proteasomal degradation (31, 39–40). Fusion of amino acids 422–461 of the degradation domain of mouse ornithine decarboxylase to the carboxyl-terminal end of GFP promotes its degradation by the proteasome (30). As with paGFPu, we modified the GFP-odc proteasome reporter with the pa variant of GFP (37) (Fig. 2A). Degradation of paGFP-odc is independent of the ubiquitination cascade, allowing us to monitor the activity of the proteasome exclusively. We found paGFP-odc to be degraded in hippocampal dendrites in a proteasome-dependent manner (Fig. 2, B and C, supplemental Table S1). We then examined the effect of blocking or up-regulating APs on proteasome activity by monitoring the degradation of paGFP-odc. As observed with paGFPu, we found that BIC increased the rate of paGFP-odc degradation, whereas TTX significantly attenuated its degradation (BIC, 149 ± 15.6% relative to control; TTX, 11.9 ± 9.2% relative to control; Fig. 2, B–D, supplemental Table S1). The largest increase in the rate of paGFP-odc degradation occurred immediately after the addition of BIC (679 ± 70.6% relative to control; Fig. 2D, supplemental Table S1). Taken together, these results suggest that the activity of the proteasome can be rapidly and dynamically regulated in hippocampal dendrites by changes in neuronal activity.
Regulation of Proteasome Activity by External Calcium
Because of the rapid and dynamic nature of proteasome regulation by neuronal activity, we hypothesized that calcium (Ca2+) might be a regulator of proteasome function in neurons. During action potentials, extracellular Ca2+ enters postsynaptic dendritic and spine compartments largely through NMDA receptors and L-type VGCC (41, 42). Furthermore, the influx of Ca2+ into postsynaptic compartments regulates many biochemical signaling pathways that are involved in controlling synaptic strength (43). To determine whether proteasome activity required the influx of external Ca2+, we monitored the degradation rate of our reporters in neurons incubated with Ca2+-free media for 15 min. The rates of paGFPu and paGFP-odc degradation were attenuated to levels comparable with neurons treated with proteasome inhibitors (relative to control: paGFPu, 27.6 ± 7.8%; paGFP-odc, 20.2 ± 21.1%; Table 1 and supplemental Table S1). This suggests that external Ca2+ entry into dendrites is required for proteasome activity.
To determine the mode of Ca2+ entry that was involved in regulation of the proteasome, we monitored paGFPu degradation in neurons treated with NMDA receptor antagonist APV or L-type VGCC antagonist nimodipine. Application of APV or nimodipine slightly attenuated the rate of paGFPu degradation when compared with the control paGFPu degradation rate (relative to control: APV, 55.1 ± 11.1%; nimodipine, 70 ± 2.1%; Table 1). However, both APV and nimodipine significantly attenuated BIC-stimulated degradation of paGFPu (relative to BIC: APV, 46.8 ± 6.4%; nimodipine, 41.1 ± 7.4%; Table 1). This suggests that NMDA receptors and L-type VGCCs are required for the BIC-induced increase in proteasome activity. Furthermore, we also found that blocking synaptic activity with the competitive AMPA receptor antagonist, CNQX, also significantly attenuated BIC-stimulated paGFPu degradation (30.1 ± 3.9% relative to BIC; Table 1). Indeed it has been previously reported that post-synaptic activity-dependent Ca2+ influx is highly sensitive to AMPA receptor blockade (44, 45). Taken together, these results suggest that external calcium influx through postsynaptic NMDA receptors and L-type VGCCs is important for activity-dependent proteasome function in neurons.
Regulation of Proteasome Activity by CaMKII
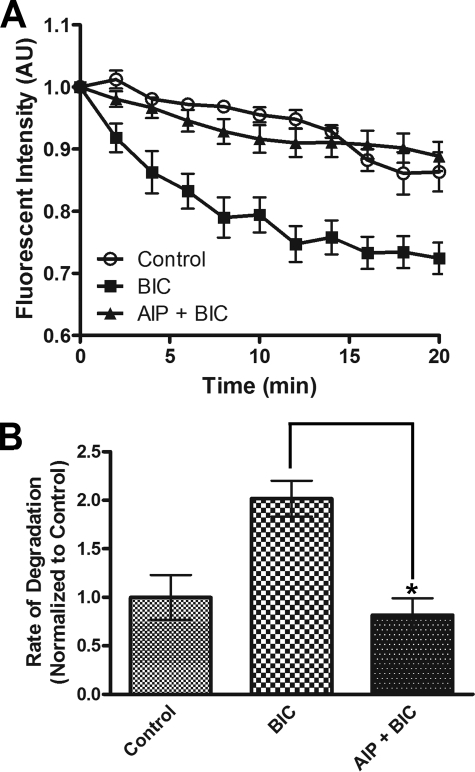
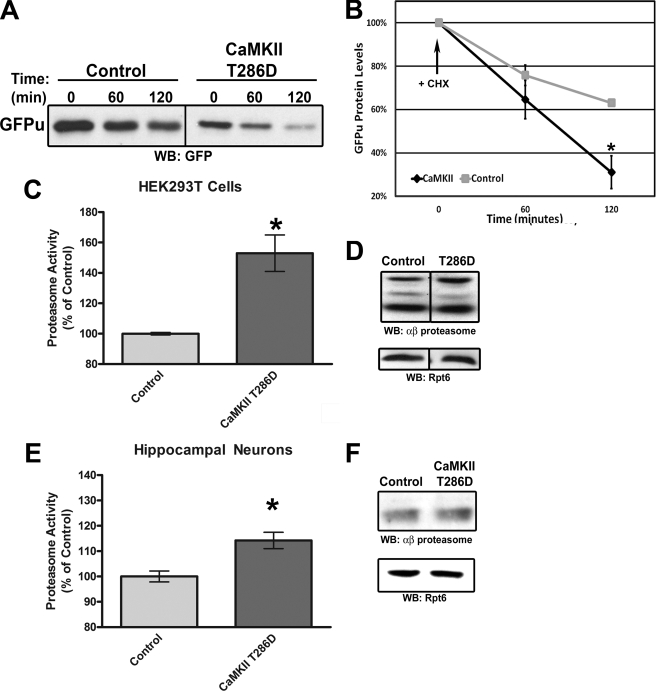
Ca2+ influx through postsynaptic compartments leads to a subsequent activation of Ca2+-dependent kinases, such as CaMKII, to modulate changes in synaptic strength (46). Phosphorylation of substrates can regulate targeting of these proteins for ubiquitination and proteasomal degradation (47). Additionally, the proteasome itself has been shown to be phosphorylated (24). We wondered if CaMKII, a key plasticity kinase, regulates proteasome function in neurons. We found that the BIC-stimulated degradation of paGFPu was significantly attenuated in hippocampal neurons when pretreated for 10 min with a CaMKII inhibitor, KN-93 (10 μm) (38.9 ± 2.7% relative to BIC; Table 1). Additionally, KN-93 blocked BIC-stimulated degradation of paGFP-odc, especially 8 min after stimulation (19.7 ± 4.6% relative to BIC at 8 min; supplemental Table S1). Treatment with KN-93 alone only slightly attenuated the degradation of paGFPu or paGFP-odc (relative to control: paGFPu, 57.2 ± 4.9%; paGFP-odc, 80.7 ± 3.7%; Table 1 and supplemental Table S1). One potential caveat to these results is that KN-93 has also been shown to inhibit other calcium/calmodulin kinase family members (48). To obtain specificity we treated neurons with myristoylated-AIP, a cell-permeable, highly specific, and potent inhibitor of CaMKII (49). BIC-stimulated degradation of paGFPu was significantly blocked in neurons pretreated with myristoylated AIP (5 μm) for 30 min (40.1 ± 8.6% relative to BIC; Fig. 3). These results suggest that CaMKII regulates proteasome activity in response to increased APs. Because inhibition of CaMKII blocked increased neuronal activity-dependent proteasome function, we next asked if the activation of CaMKII could increase proteasome activity. To do this we overexpressed a constitutively active form of CaMKII (T286D mutation (50, 51)) in HEK293T cells and performed in vitro and in vivo proteasome activity assays. To test in vivo proteasome activity we co-transfected our proteasome reporter GFPu along with either CaMKII T286D or a control vector in HEK293T cells. GFPu protein levels were significantly decreased in cells expressing CaMKII T286D after 2 h of protein synthesis inhibition (GFPu protein levels at time 120 min: control, 63.1 ± 1.6%; CaMKII T286D, 31.1 ± 7.5; Fig. 4, A and B). Steady state levels of GFPu were also dramatically lower in CaMKII T286D-transfected cells (GFPu fluorescence relative to control: 45.6 ± 2.5%; supplemental Fig. S2, A and B). This is likely because of increased proteasome activity and not an impairment of protein synthesis, as the addition of proteasome inhibitor MG132 increases the levels of GFPu in both control and CaMKII-expressing cells (supplemental Fig. S2, A and C). To determine whether this was a direct effect on proteasome function by CaMKII, we assayed lysates for in vitro proteasome activity with the fluorogenic proteasome substrate Suc-LLVY-AMC. Lysates from HEK293T cells transfected with CaMKII T286D had significantly increased proteasome activity as measured by AMC hydrolysis (153 ± 12% relative to control; Fig. 4C). This was not merely because of an increase in total proteasome levels, as Western blot analysis showed similar amounts of core and cap proteasome subunit levels (Fig. 4D). Furthermore, hippocampal neurons infected with CaMKII T286D Sindbis virion also had a slight but significant increase in proteasome activity as measured by Suc-LLVY-AMC cleavage (114 ± 3.2% relative to control; Fig. 4E) despite less than 50% viral infection. Together these results suggest CaMKII is a regulator of proteasome function.
FIGURE 3.
CaMKII is required for activity-dependent proteasome function in hippocampal neurons. A, the increased degradation of paGFPu induced by BIC is significantly blocked by CaMKII inhibitor myristoylated AIPII (AIP, 10 μm). A, line graphs display grouped analysis (±S.E.) of dendritic proteasome reporter fluorescence intensity normalized to time zero. B, bar graphs depict the mean degradation rate ± S.E. (AU) per treated group relative to the control rate of fluorescence intensity (n = 8, 19, and 20 imaged dendrites for control, BIC, and AIP + BIC respectively; *, p < 0.01).
FIGURE 4.
CaMKII regulates proteasome function. A, HEK293T cells were cotransfected with GFPu and either CaMKII T286D or control vector (β-galactosidase). Cells were treated with protein synthesis inhibitor cycloheximide (CHX) for the indicated time lengths, and equal amounts of protein were resolved by SDS-PAGE and probed for total GFPu protein levels. WB, Western blot. B, quantification of total GFPu protein levels in A. CaMKII stimulates the rapid degradation of the GFPu proteasome reporter. *, p < 0.05, t test; n = 3. C, HEK293T cells were transfected with either CaMKII T286D or control vector, and lysates were assayed for 26 S proteasome activity with fluorogenic substrate Suc-LLVY-AMC. A bar graph depicting final fluorescent values (mean ± S.E.) relative to control is shown. CaMKII significantly stimulates 26 S proteasome activity in vitro (*, p < 0.05, t test; n = 10). D, Western blotting shows similar amount of total core (αβ proteasome) and cap (Rpt6) proteasome subunits for HEK293T cells transfected with either CaMKII T286D or control vector. E, cultured rat hippocampal neurons were infected with control (GFP only) or CaMKII T286D (coexpressing GFP) Sindbis virion for 20 h (∼50% infection rate). Proteasome activity was measured by Suc-LLVY-AMC hydrolysis. AMC fluorescence increased by 14.2 ± 3.2% in neurons overexpressing CaMKII T286D (*, p < 0.01, t test; n = 15). F, Western blotting similar equal amount of total core (αβ proteasome) and cap (Rpt6) proteasome subunits for neurons infected with either CaMKII T286D or control virus.
Phosphorylation of Proteasome by CaMKII
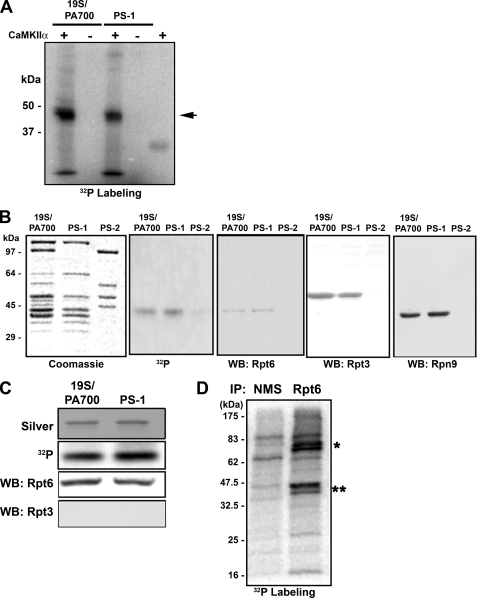
Because inhibition of CaMKII blocked activity-dependent proteasome activity, and overexpression of a catalytically active CaMKII increased proteasome activity, we hypothesized that phosphorylation of the proteasome may regulate its function. Phosphorylation of proteasomes has been reported in non-neuronal cell types by both casein kinase 2 and protein kinase A (26, 52–55). This phosphorylation is believed to regulate proteasome function by affecting assembly, activity, or interactions (24, 26, 53). To test if proteasomes are directly phosphorylated by CaMKII, we performed in vitro kinase assays using highly purified 19 S (PA700) and 19 S subcomplexes (PS-1 and PS-2) as substrates (36). CaMKII catalyzed the incorporation of [32P]phosphate from ATP into a single band of both 19 S and PS-1, a subcomplex of 19 S that contains only 2 of the 6 Rpt subunits found in intact 19 S. This band had an approximate molecular mass of about 45 kDa and was indistinguishable between 19 S and PS-1 (Fig. 5A). After long incubations a second, minor band appeared in some experiments. The sizes of both the major and minor labeled proteins were similar to those of subunits Rpt3, Rpn9, or Rpt6. Kinase reactions with PS-2 and PS-3, two additional 19 S subcomplexes that do not contain Rpt3, Rpn9, or Rpt6 subunits, produced no labeled proteins (Fig. 5B and data not shown). Interestingly, Rpt3 and Rpt6 previously have been reported to be phosphorylated (53, 56). To determine the identity of the labeled protein we performed Western blotting with antibodies against Rpt3, Rpn9, and Rpt6 as well as other 19 S subunits with this approximate size. The 32P-labeled protein co-migrated with Rpt6 (Fig. 5B). To further demonstrate the identity of the labeled protein in 19 S, we excised the band from the SDS gel, solubilized the protein, and subjected it to a second round of SDS-PAGE. The recovered labeled protein was detected only by antibodies against Rpt6 (Fig. 5C). Moreover, mass spectrometric analysis of tryptic peptides identified the protein as Rpt6 (supplemental Table S2). Together, these data strongly indicate that Rpt6 is the proteasome subunit phosphorylated by CaMKII.
FIGURE 5.
CaMKIIα phosphorylates Rpt6, a specific subunit of 19 S/PA700. A, equimolar amounts of 19 S or PS-1 (19 S subcomplex) were subjected to phosphorylation reactions with CaMKIIα and [32P]ATP under identical conditions and subjected to SDS-PAGE and autoradiography. CaMKII phosphorylates a 19 S subunit of ∼45 kDa (arrowhead). B, phosphorylated 19 S, PS-1, or PS-2 (19 S subcomplex that does not contain Rpt6) were subjected to SDS-PAGE, and samples were analyzed by protein staining, autoradiography, and Western blotting (WB) for the indicated 19 S subunits. Gels are aligned for direct comparison. C, the 32P-labeled proteins were excised from the gel, solubilized, and re-subjected to SDS-PAGE. The gel was analyzed for labeled protein and subjected to Western blotting for the indicated proteins. D, representative autoradiogram of [32P]orthophosphate labeling for immunoprecipitations (IP) of Rpt6 from cortical neuron lysates. Normal mouse serum (NMS) served as a control immunoprecipitate. The two highly phosphorylated bands at 45–47 kDa (**) present in the RPT6 IP are likely Rpt6 and another 19 S Rpt subunit. Two highly phosphorylated bands at 70–80 kDa (*) do not correspond to any proteasome subunits.
We next asked whether this in vitro phosphorylation had a direct effect on proteasome function. Purified proteasomes were mixed with purified CaMKIIα and then tested for activity by fluorogenic peptidase assay. Although CaMKII phosphorylated both the control myelin basic protein substrate and proteasomes in vitro, no significant changes in proteasome activity were observed (supplemental Fig. S3, A and B). Additionally we measured 26 S proteasome assembly from isolated 20 S proteasome and 19 S subcomplexes and again observed no effect after CaMKII phosphorylation (supplemental Fig. S3C). This suggests that CaMKII phosphorylation of proteasomes likely affects proteasome activity via regulation of interactions of additional factors not present in our highly purified in vitro system.
We next asked whether CaMKII phosphorylates Rpt6 in vivo. HEK293T cells transfected with CaMKII T286D were labeled with [32P]orthophosphate. Proteasomes were immunoprecipitated with an antibody to the α2 subunit previously shown to effectively pull down human proteasomes (57). We observed a CaMKII-dependent phosphorylation of a protein with the approximate molecular mass of 45 kDa, similar to that of Rpt6 (supplemental Fig. S4, A and B). Indeed, we also observed highly phosphorylated proteins that migrated at ∼45 kDa in Rpt6 immunoprecipitates from 32P live labeled rat cortical neurons (Fig. 5D). Together with our in vitro phosphorylation experiments, these data indicate that Rpt6 is phosphorylated by CaMKII.
DISCUSSION
In vertebrates changes in the composition of the postsynaptic density are regulated by synaptic activity. These changes are bidirectional and require the activity of the UPS (18, 22). It is quite plausible that neuronal activity activates regulates UPS function to control the stoichiometry of proteins at synapses. Regulated proteolysis by the UPS has been typically studied at the level of the ubiquitination. However, there are hundreds of E3 ubiquitin ligases identified in the mammalian genome, each which may be dynamically regulated by various cellular signals. Therefore, mapping E3-substrate relationships remains a difficult task in all cell types. This is even more evident in neurons, as the distribution of UPS components in functionally distinct compartments is likely regulatory for their function. For instance, subcellular regulation of an SCF-type E3 complex in flies contributes to precise synaptic connectivity through selective synapse elimination (58).
However, the proteasome itself is also dynamic in nature. For instance, subunit composition, core association with regulatory cap complexes, and interactions with accessory proteins have all been shown to control the specificity and activity of the proteasome (23, 59–61). Furthermore, there is a growing body of literature that indicates proteasomes to be modified by post-translational events such as phosphorylation (53, 62). Recently, it was shown that proteasomes rapidly redistribute from dendritic shaft to dendritic spine compartments in response to synaptic stimulation. This redistribution was found to be dependent upon NMDA receptor activation (63). A subsequent report showed that NAC1, a cocaine-regulated transcriptional protein that interacts with the proteasome, regulates the trafficking of the proteasome in an activity-dependent manner (64). These data suggest that synaptic activity can promote the recruitment and sequestration of proteasomes to locally remodel the protein composition of synapses. However, a clear understanding of the synaptic molecular mechanisms involved to regulate proteasome trafficking and activity is far from understood.
In our current study we report the rapid and dynamic regulation of proteasome function by neuronal activity. Action potential blockade or up-regulation produced rapid and opposite effects on the activity of the proteasome as monitored by our fluorescent proteasome reporters. The activation of proteasome by increased action potentials required external calcium influx through NMDA receptors and L-type VGCCs. UPS components may be regulated directly by Ca2+ (65–68), and it has been reported that the release of Ca2+ from intracellular stores transiently activates the 26 S proteasome (69). Our findings are in line with these studies, and they provide evidence for Ca2+ signaling in the regulation of the proteasome activity in neurons.
To further explore the role of calcium signaling in regulation of the proteasome, we investigated the involvement of calcium activated kinase cascades. We identified CaMKII as a novel regulator of proteasome activity in neurons. CaMKII is a calcium-dependent protein kinase that plays a key role in neuronal behavior, development, and plasticity (48). We found that BIC-stimulated activation of the proteasome to require the activity of CaMKII. Additionally, overexpression of a constitutively active form of CaMKII robustly stimulated the proteolytic activity of the proteasome. We have identified Rpt6, a 19 S regulatory subunit of the proteasome, to be phosphorylated by CaMKII. This is the first time to our knowledge that CaMKII has been shown to phosphorylate the proteasome. Interestingly, Rpt6 has also been shown to be phosphorylated by protein kinase A in non-excitable cells (53). We, therefore, cannot rule out a role for protein kinase A in regulation of proteasome in neurons. CaMKII and protein kinase A both phosphorylate targets at similar substrate recognition motifs (70). Furthermore, CaMKII has been shown to be activated in neurons stimulated with the protein kinase A agonist forskolin (71, 72). However, both Zhang et al. (53) and our findings identify the phosphorylation of Rpt6 in the regulation of proteasome function. This is significant because components of the 19 S complex have been shown to be important regulators of 26 S proteasome assembly and activation (73). Because CaMKII-dependent phosphorylation of Rpt6 did not directly regulate proteasome activity in vitro, we hypothesize that other regulatory factors, not present in our reconstituted and highly purified in vitro assay, may be required. Associated proteins have previously been found to regulate various aspects of proteasome function (23, 64, 74). Interestingly, both proteasome and CaMKII translocate from dendritic shaft to dendritic spine compartments in response to neuronal activity in a similar timeframe (63, 75, 76). This potentially indicates that the phosphorylation of the proteasome by CaMKII may regulate its trafficking in neurons.
Interestingly, recent studies on fear memory in rodents indirectly support our hypothesis that CaMKII regulates activity-dependent proteasome function in neurons. First, Lee et al. (17) showed that infusion of a proteasome inhibitor into the CA1 region of the hippocampus immediately after memory retrieval prevented protein synthesis inhibition-induced memory impairment as well as the extinction of fear memory. This suggests proteasome-dependent protein degradation underlies the destabilization processes after fear memory retrieval. Second, Cao et al. (77) showed that transient CaMKIIα overexpression at the time of recall impairs the retrieval of new and old fear memories. Their analysis suggested that excessive CaMKIIα activity-induced recall deficits are caused by the active erasure of the stored memories rather than disrupting the retrieval access to the stored information (77). Therefore, proteasome inhibition and excessive CaMKIIα activity produce opposite effects on retrieval and extinction of fear memory. In their discussion, Cao et al. (77) postulate the involvement of the UPS. Indeed, our data suggest that CaMKII-dependent regulation of the proteasome may play a key role in active erasure of stored memories.
Proteasome function has been shown to be required for various learning and memory and behavior related paradigms such as long term potentiation (11, 12, 17, 78, 79). Our findings support a role for the direct modulation of proteasome activity in neurons to be important for synaptic plasticity. This is substantiated by our findings that CaMKII regulates proteasome activity which provides a mechanistic link for the regulation of proteasome function by neuronal activity. Phosphorylation of proteasomes by CaMKII may directly affect the interactions of regulatory proteins to modulate the assembly, activity, or localization of the proteasome. This in turn can facilitate protein degradation and alterations in the stoichiometry of synaptic proteins to promote, limit, or restrict synaptic plasticity. We predict neurons to utilize this mechanism to synergize tunable proteasome activity with the dynamic ubiquitination of substrates for degradation. Moreover, because both UPS dysfunction and aberrant Ca2+ signaling have been attributed to several neurodegenerative diseases such as Alzheimer and Huntington diseases, it will be interesting to determine whether misregulation of proteasome phosphorylation is involved in the pathogenesis of these diseases (80, 81).
Supplementary Material
Acknowledgments
We thank M. Scanziani and R. Hampton for advice and critical review of the manuscript, A. Ghosh, M. Sutton, and D. Berg for helpful discussion, A. Hoffmann and S. Werner for help with 32P experiments, D. Immenhausen for hippocampal cultures, and the rest of the Patrick laboratory for support. We also thank R. Kopito (Stanford) for GFPu and GFP-odc constructs, J. Lipponcott-Schwartz (National Institutes of Health) for the paGFP construct, A. Ghosh for the CaMKII construct, and R. Tsien for the mCherry construct. Some of the impetus for these experiments was conceived in the laboratory of Erin Schuman (CalTech).
This work was supported, in whole or in part, by National Institutes of Health Grants DK46181 (to G. N. D.) and NS054732 (to G. N. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4, Tables S1 and S2, and Movie S1.
- UPS
- ubiquitin proteasome system
- AP
- action potential
- TTX
- tetrodotoxin
- BIC
- bicuculline
- NMDA
- N-methyl-d-aspartate
- VGCC
- voltage-gated calcium channels
- CaMKII
- calcium/calmodulin-dependent protein kinase II
- CNQX
- 6-cyano-7-nitroquinoxaline-2,3-dione
- vinyl sulfone
- adamantane-acetyl-(6-aminohexanoyl)(3)-(leucinyl)(3)-vinyl-(methyl)-sulfone
- AMPA
- (±)-α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AIP
- autocamtide-2-related inhibitory peptide
- pa-
- photoactivatable
- odc
- ornithine decarboxylase
- AU
- arbitrary units
- IRES
- internal ribosomal entry site
- E3
- ubiquitin-protein isopeptide ligase
- GFP
- green fluorescent protein
- APV
- 2-amino-5-phosphonopentanoic acid
- suc-LLVY-AMC
- N-succinyl-Leu-Leu-Val-Tyr-AMC (7-amino-4-methylcoumarin).
REFERENCES
- 1.Sheng M., Kim M. J. (2002) Science 298, 776–780 [DOI] [PubMed] [Google Scholar]
- 2.Martin K. C., Zukin R. S. (2006) J. Neurosci. 26, 7131–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sutton M. A., Schuman E. M. (2006) Cell 127, 49–58 [DOI] [PubMed] [Google Scholar]
- 4.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 5.Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 6.DiAntonio A., Hicke L. (2004) Annu. Rev. Neurosci. 27, 223–246 [DOI] [PubMed] [Google Scholar]
- 7.Hegde A. N. (2004) Prog. Neurobiol. 73, 311–357 [DOI] [PubMed] [Google Scholar]
- 8.Patrick G. N. (2006) Curr. Opin. Neurobiol. 16, 90–94 [DOI] [PubMed] [Google Scholar]
- 9.Yi J. J., Ehlers M. D. (2007) Pharmacol. Rev. 59, 14–39 [DOI] [PubMed] [Google Scholar]
- 10.Colledge M., Snyder E. M., Crozier R. A., Soderling J. A., Jin Y., Langeberg L. K., Lu H., Bear M. F., Scott J. D. (2003) Neuron 40, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonseca R., Vabulas R. M., Hartl F. U., Bonhoeffer T., Nägerl U. V. (2006) Neuron 52, 239–245 [DOI] [PubMed] [Google Scholar]
- 12.Karpova A., Mikhaylova M., Thomas U., Knöpfel T., Behnisch T. (2006) J. Neurosci. 26, 4949–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speese S. D., Trotta N., Rodesch C. K., Aravamudan B., Broadie K. (2003) Curr. Biol. 13, 899–910 [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y., Hegde A. N., Martin K. C. (2003) Curr. Biol. 13, 887–898 [DOI] [PubMed] [Google Scholar]
- 15.Willeumier K., Pulst S. M., Schweizer F. E. (2006) J. Neurosci. 26, 11333–11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haas K. F., Miller S. L., Friedman D. B., Broadie K. (2007) Mol. Cell. Neurosci. 35, 64–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S. H., Choi J. H., Lee N., Lee H. R., Kim J. I., Yu N. K., Choi S. L., Lee S. H., Kim H., Kaang B. K. (2008) Science 319, 1253–1256 [DOI] [PubMed] [Google Scholar]
- 18.Ehlers M. D. (2003) Nat. Neurosci. 6, 231–242 [DOI] [PubMed] [Google Scholar]
- 19.Pak D. T., Sheng M. (2003) Science 302, 1368–1373 [DOI] [PubMed] [Google Scholar]
- 20.Yao I., Takagi H., Ageta H., Kahyo T., Sato S., Hatanaka K., Fukuda Y., Chiba T., Morone N., Yuasa S., Inokuchi K., Ohtsuka T., Macgregor G. R., Tanaka K., Setou M. (2007) Cell 130, 943–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo L., Wang Y. (2007) Neuroscience 145, 100–109 [DOI] [PubMed] [Google Scholar]
- 22.Piccoli G., Verpelli C., Tonna N., Romorini S., Alessio M., Nairn A. C., Bachi A., Sala C. (2007) J. Proteome Res. 6, 3203–3215 [DOI] [PubMed] [Google Scholar]
- 23.Schmidt M., Hanna J., Elsasser S., Finley D. (2005) Biol. Chem. 386, 725–737 [DOI] [PubMed] [Google Scholar]
- 24.Glickman M. H., Raveh D. (2005) FEBS Lett. 579, 3214–3223 [DOI] [PubMed] [Google Scholar]
- 25.Zhang F., Paterson A. J., Huang P., Wang K., Kudlow J. E. (2007) Physiology 22, 373–379 [DOI] [PubMed] [Google Scholar]
- 26.Lu H., Zong C., Wang Y., Young G. W., Deng N., Souda P., Li X., Whitelegge J., Drews O., Yang P. Y., Ping P. (2008) Mol. Cell. Proteomics 7, 2073–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bence N. F., Bennett E. J., Kopito R. R. (2005) Methods Enzymol. 399, 481–490 [DOI] [PubMed] [Google Scholar]
- 28.Bence N. F., Sampat R. M., Kopito R. R. (2001) Science 292, 1552–1555 [DOI] [PubMed] [Google Scholar]
- 29.Gilon T., Chomsky O., Kulka R. G. (1998) EMBO J. 17, 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C. C., Kain S. R. (1998) J. Biol. Chem. 273, 34970–34975 [DOI] [PubMed] [Google Scholar]
- 31.Murakami Y., Matsufuji S., Kameji T., Hayashi S., Igarashi K., Tamura T., Tanaka K., Ichihara A. (1992) Nature 360, 597–599 [DOI] [PubMed] [Google Scholar]
- 32.Patrick G. N., Bingol B., Weld H. A., Schuman E. M. (2003) Curr. Biol. 13, 2073–2081 [DOI] [PubMed] [Google Scholar]
- 33.Kisselev A. F., Goldberg A. L. (2005) Methods Enzymol. 398, 364–378 [DOI] [PubMed] [Google Scholar]
- 34.Scholz W. K., Palfrey H. C. (1991) J. Neurosci. 11, 2422–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. (1994) J. Biol. Chem. 269, 3539–3547 [PubMed] [Google Scholar]
- 36.Thompson D., Hakala K., DeMartino G. N. (2009) J. Biol. Chem. 284, 24891–24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson G. H., Lippincott-Schwartz J. (2002) Science 297, 1873–1877 [DOI] [PubMed] [Google Scholar]
- 38.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 39.Hoyt M. A., Zhang M., Coffino P. (2003) J. Biol. Chem. 278, 12135–12143 [DOI] [PubMed] [Google Scholar]
- 40.Hoyt M. A., Zhang M., Coffino P. (2005) Methods Enzymol. 398, 399–413 [DOI] [PubMed] [Google Scholar]
- 41.Blackstone C., Sheng M. (2002) Front. Biosci. 7, 872–885 [DOI] [PubMed] [Google Scholar]
- 42.Cavazzini M., Bliss T., Emptage N. (2005) Cell Calcium. 38, 355–367 [DOI] [PubMed] [Google Scholar]
- 43.Kennedy M. B., Beale H. C., Carlisle H. J., Washburn L. R. (2005) Nat. Rev. Neurosci. 6, 423–434 [DOI] [PubMed] [Google Scholar]
- 44.Emptage N., Bliss T. V., Fine A. (1999) Neuron 22, 115–124 [DOI] [PubMed] [Google Scholar]
- 45.Yuste R., Majewska A., Cash S. S., Denk W. (1999) J. Neurosci. 19, 1976–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayford M. (2007) Curr. Opin. Neurobiol. 17, 313–317 [DOI] [PubMed] [Google Scholar]
- 47.Hunter T. (2007) Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 48.Wayman G. A., Lee Y. S., Tokumitsu H., Silva A., Soderling T. R. (2008) Neuron 59, 914–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishida A., Kameshita I., Okuno S., Kitani T., Fujisawa H. (1995) Biochem. Biophys. Res. Commun. 212, 806–812 [DOI] [PubMed] [Google Scholar]
- 50.Fong Y. L., Taylor W. L., Means A. R., Soderling T. R. (1989) J. Biol. Chem. 264, 16759–16763 [PubMed] [Google Scholar]
- 51.Waldmann R., Hanson P. I., Schulman H. (1990) Biochemistry 29, 1679–1684 [DOI] [PubMed] [Google Scholar]
- 52.Fernández Murray P., Pardo P. S., Zelada A. M., Passeron S. (2002) Arch. Biochem. Biophys. 404, 116–125 [DOI] [PubMed] [Google Scholar]
- 53.Zhang F., Hu Y., Huang P., Toleman C. A., Paterson A. J., Kudlow J. E. (2007) J. Biol. Chem. 282, 22460–22471 [DOI] [PubMed] [Google Scholar]
- 54.Bose S., Stratford F. L., Broadfoot K. I., Mason G. G., Rivett A. J. (2004) Biochem. J. 378, 177–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ludemann R., Lerea K. M., Etlinger J. D. (1993) J. Biol. Chem. 268, 17413–17417 [PubMed] [Google Scholar]
- 56.Mason G. G., Murray R. Z., Pappin D., Rivett A. J. (1998) FEBS Lett. 430, 269–274 [DOI] [PubMed] [Google Scholar]
- 57.Hendil K. B., Kristensen P., Uerkvitz W. (1995) Biochem. J. 305, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding M., Chao D., Wang G., Shen K. (2007) Science 317, 947–951 [DOI] [PubMed] [Google Scholar]
- 59.Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X., Chen C. F., Baker P. R., Chen P. L., Kaiser P., Huang L. (2007) Biochemistry 46, 3553–3565 [DOI] [PubMed] [Google Scholar]
- 61.Wang X., Huang L. (2008) Mol. Cell. Proteomics 7, 46–57 [DOI] [PubMed] [Google Scholar]
- 62.Rivett A. J., Bose S., Brooks P., Broadfoot K. I. (2001) Biochimie 83, 363–366 [DOI] [PubMed] [Google Scholar]
- 63.Bingol B., Schuman E. M. (2006) Nature 441, 1144–1148 [DOI] [PubMed] [Google Scholar]
- 64.Shen H., Korutla L., Champtiaux N., Toda S., LaLumiere R., Vallone J., Klugmann M., Blendy J. A., Mackler S. A., Kalivas P. W. (2007) J. Neurosci. 27, 8903–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen H., Polo S., Di Fiore P. P., De Camilli P. V. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14908–14913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Realini C., Rechsteiner M. (1995) J. Biol. Chem. 270, 29664–29667 [DOI] [PubMed] [Google Scholar]
- 67.Santella L., Ercolano E., Nusco G. A. (2005) Cell. Mol. Life Sci. 62, 2405–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santella L., Kyozuka K., De Riso L., Carafoli E. (1998) Cell Calcium. 23, 123–130 [DOI] [PubMed] [Google Scholar]
- 69.Aizawa H., Kawahara H., Tanaka K., Yokosawa H. (1996) Biochem. Biophys. Res. Commun. 218, 224–228 [DOI] [PubMed] [Google Scholar]
- 70.Kemp B. E., Pearson R. B. (1990) Trends Biochem. Sci. 15, 342–346 [DOI] [PubMed] [Google Scholar]
- 71.Makhinson M., Chotiner J. K., Watson J. B., O'Dell T. J. (1999) J. Neurosci. 19, 2500–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Valverde R. H., Tortelote G. G., Lemos T., Mintz E., Vieyra A. (2005) J. Biol. Chem. 280, 30611–30618 [DOI] [PubMed] [Google Scholar]
- 73.Gillette T. G., Kumar B., Thompson D., Slaughter C. A., DeMartino G. N. (2008) J. Biol. Chem. 283, 31813–31822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferrell K., Wilkinson C. R., Dubiel W., Gordon C. (2000) Trends Biochem. Sci 25, 83–88 [DOI] [PubMed] [Google Scholar]
- 75.Rose J., Jin S. X., Craig A. M. (2009) Neuron 61, 351–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee S. J., Escobedo-Lozoya Y., Szatmari E. M., Yasuda R. (2009) Nature 458, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cao X., Wang H., Mei B., An S., Yin L., Wang L. P., Tsien J. Z. (2008) Neuron 60, 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dong C., Upadhya S. C., Ding L., Smith T. K., Hegde A. N. (2008) Learn. Mem. 15, 335–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Artinian J., McGauran A. M., De Jaeger X., Mouledous L., Frances B., Roullet P. (2008) Eur. J. Neurosci. 27, 3009–3019 [DOI] [PubMed] [Google Scholar]
- 80.Ciechanover A., Brundin P. (2003) Neuron 40, 427–446 [DOI] [PubMed] [Google Scholar]
- 81.Bezprozvanny I., Mattson M. P. (2008) Trends Neurosci. 31, 454–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.