Abstract
Nucleostemin (NS) is a nucleolar protein involved in the regulation of cell proliferation. Both overexpression and knockdown of NS increase the activity of the tumor suppressor protein p53, resulting in cell cycle arrest. In addition, NS regulates processing of pre-rRNA and consequently the level of total protein synthesis. Here, we describe a previously uncharacterized function of NS in the maintenance of the tripartite nucleolar structure as well as the integrity of small nucleolar ribonucleoproteins (snoRNPs). NS is also necessary to maintain the telomerase complex which shares common protein subunits with the H/ACA box snoRNPs. First, immunofluorescence microscopy and electron microscopy demonstrated that knockdown of NS disorganized the nucleolar architecture, in particular, the dense fibrillar component where snoRNPs are localized. Second, gel filtration chromatography and immunoprecipitation indicated that NS depletion leads to dissociation of the components of snoRNPs and the telomerase complex. Third, NS depletion reduced both telomerase activity and the cellular level of pseudouridine, an H/ACA snoRNP-mediated modification of rRNA and other RNAs that are important for their folding and stability. These morphological, biochemical and functional studies demonstrate that NS plays an important role to maintain nucleolar structure and function on a more fundamental level than previously thought.
Nucleostemin (NS)2 is a GTP-binding nucleolar protein highly expressed in many types of proliferating cells and downregulated upon their exit from the cell cycle (1–3). Cells in NS-null mouse embryos fail to enter the S phase, resulting in embryonic death at the blastocyst stage (4, 5). NS is generally interpreted to function as a cell cycle regulator located upstream of the p53-mediated signaling pathway (1, 6–8). However, this does not appear to be the only function of NS because NS-null mouse embryos will die even when p53 is not expressed (5). This observation indicates that the role of NS is not limited to p53-mediated cell cycle regulation. Indeed, we have recently found that NS is important for ribosome biogenesis, in particular, the processing of 32S pre-rRNA into 28S rRNA (9).
In the current study we focused on an additional novel role of NS in the maintenance of the integrity of nucleolar structure and nucleolar RNA-protein complexes, such as snoRNPs and the telomerase complex. snoRNPs can be divided into two major groups, C/D snoRNPs and H/ACA snoRNPs, based on the conserved motifs of the RNA component in each group (10–12). Each C/D snoRNP contains one of more than 100 unique C/D RNAs in addition to the four conserved core proteins, fibrillarin, Nop56, Nop58, and 15.5K, as well as other less characterized proteins. C/D RNAs are small RNAs (60–150 nucleotides in length) containing the conserved C box and the D box sequences, which serve as binding signals for the protein subunits of snoRNPs. Each C/D RNA also contains a short unique sequence that hybridizes to a specific target RNA sequence of 15–20 bases, serving as a guide for recruiting C/D snoRNPs to their target RNAs. Once recruited, fibrillarin methylates a 2′-hydroxyl group of a ribose in the nearby ribonucleotide (2′-O-methylation), which is important for folding, stability, and processing of target RNAs, such as pre-rRNA and spliceosomal small nuclear RNAs. The binding of 15.5 K to the C/D boxes is necessary for the assembly of C/D snoRNPs (13). While Nop58 is required for the stability of snoRNAs (14), Nop56 is not essential for this purpose (15). There is also a small group of C/D snoRNPs that induces proper folding and cleavage of target RNAs without 2′-O-methylation.
Similar to C/D snoRNPs, each H/ACA snoRNP contains a unique H/ACA RNA and four conserved proteins, dyskerin, NHP2, GAR1, and Nop10, in addition to other less defined proteins. H/ACA RNAs contain the conserved H and ACA boxes and a unique sequence which guides each H/ACA RNP to a specific sequence on its target RNA. Pre-rRNA is again one of the best studied target RNAs. Dyskerin has the catalytic activity of converting uridine to pseudouridine (pseudouridylation), which is important for folding and stability of pre-rRNA. Human pre-rRNA is modified by 2′-O-methylations or pseudouridylations at ∼100 sites each before processing (16). As for the protein subunits, while GAR1 and Nop10 bind to the catalytic domain of dyskerin, NHP2 directly interacts with snoRNAs (10). snoRNPs are assembled as inactive pre-snoRNPs on nascent snoRNAs in the nucleoplasm and then transported to Cajal bodies in the nucleus for maturation to functional complexes. snoRNPs are eventually transported to the nucleolus and function within this structure (10).
The human telomerase complex shares several structural features with H/ACA snoRNPs. The telomerase complex contains two essential components, the catalytic subunit TERT (telomerase reverse transcriptase) and the RNA component TR (telomerase RNA component) (17, 18). TERT uses the TTAGGG sequence of TR as a template to synthesize the telomeric DNA repeat. TR contains the H and the ACA boxes, which are important for the assembly and stability of the telomerase complex. The telomerase complex also contains dyskerin, NHP2, GAR1, and Nop10 (19, 20). These proteins interact with TR and stabilize the level of TR. Although GNL3L, a nucleolar protein structurally related to NS, is a component of the telomerase complex, NS is not directly incorporated into the telomerase complex (19). In human cells TR remains in Cajal bodies throughout most of the cell cycle, while TERT is localized in distinct nuclear foci, which are different from Cajal bodies and telomeres (10, 21). TR and TERT move to telomeres during S phase and are assembled into functional telomerase complexes. TR and TERT are temporarily localized in nucleoli during these processes but the details are not known.
In this study we first investigated the morphological alteration of the nucleolus after knockdown of NS in HeLa cells. We then studied the effects of NS knockdown on the integrity and function of snoRNPs and the telomerase complex. The components of these complexes are localized in a nucleolar subdomain that was morphologically altered by NS knockdown. This study revealed that loss of NS causes unexpectedly significant disruptions in nucleolar structure and functions.
EXPERIMENTAL PROCEDURES
Cell Culture and NS Knockdown
HeLa cells were cultured in Minimum Essential Medium with 10% fetal bovine serum (FBS) supplemented with non-essential amino acids and sodium pyruvate. The following short-interfering RNAs (siRNAs) were used to knock down NS and B23. NS siRNA-1: 5′-GAACUAAAACAGCAGCAGAdTdT-3′ (9). NS siRNA-2: 5′-AGCUGGUACUUAUAUUAAAdTdT-3′. B23 siRNA: 5′-AGUGGAAGCCAAAUUCAUCdTdT-3′ (22). We also used the scrambled sequence 5′-AGUACUGCUUACGAUACGGdTdT-3′ (Ambion) and called it control siRNA. HeLa cells were transfected with 60 nm siRNA and Lipofectamine 2000 (Invitrogen) on day 1 and day 4 and harvested on day 6 for all the analyses in the current work unless specified otherwise. In a separate experiment, HeLa cells were treated with 0.4 μg/ml cycloheximide for 7 days without siRNA, and total protein amount was measured with a Quant-iT Protein Assay kit (Invitrogen).
Antibodies
Sources of antibodies are as follows: NS, Nop58, dyskerin, NHP2, GAR1, upstream binding factor (UBF) and B23 (Santa Cruz Biotechnology), fibrillarin and TERT (Abcam), NS and histone H2B (Millipore), Nop56 and Nop58 (gift from Dr. Reinhard Lührmann) (13, 23), coilin (Abcam), horseradish peroxidase-conjugated secondary antibodies, and Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories) and Alexa Fluor 488-conjugated secondary antibodies (Invitrogen).
Western Blotting
Protein in an SDS-PAGE gel was transferred onto an Immobilon P membrane (Millipore). After incubation with primary antibodies and then horseradish peroxidase-conjugated secondary antibodies for 1 h each, chemiluminescence signal was detected with the SuperSignal West Dura substrate (ThermoFisher Scientific).
Immunofluorescence Staining
HeLa cells cultured on coverslips were fixed with 4% formaldehyde in phosphate-buffered saline (PBS). The plasma membrane was permeabilized with 0.5% Triton X-100 in PBS for 5 min, and the cells were washed with washing solution (10% fetal bovine serum and 0.2% Tween 20 in PBS). The cells were incubated with primary and secondary antibodies diluted in Washing Solution for 1 h each and counterstained with Topro 3 (Invitrogen) for 15 min. Mounting solution was composed of 50 mg/ml 1,4-diazabicyclo[2.2.2]octane (DABCO, Sigma) and 50% glycerol in PBS. Fluorescence images were captured with a 63x oil Plan Apochromat objective (numerical aperture 1.4) attached to an Axioskop 2 Plus microscope and the Radiance 2000 confocal system (both Bio-Rad). A 488-nm argon laser and a 543-nm green helium-neon laser were used to detect fluorescence signal. Photoshop 7.0 (Adobe Systems) was used for image processing.
Electron Microscopy
Cells were grown on 12-mm round coverslips with no. 1.5 thickness. Cells were washed in PBS and fixed in 1% glutaraldehyde and 3% formaldehyde in 0.1 m phosphate buffer, pH 7.4. The cells were washed in 0.1 m phosphate buffer pH 7.4 and post-fixed in 1% osmium tetroxide for 1 h at 25 °C. The cells were rinsed in distilled water and enbloc stained in 2% uranyl acetate followed by a wash with distilled water. The cells were dehydrated in an ascending ethanol series up to 100% and embedded in EmBed812 epoxy resin (Electron Microscopy Sciences) according to the manufacturer's protocol. Ultrathin sections (90 nm) were mounted on carbon-coated formvar copper grids and stained for 5 min with uranyl acetate and 5 min with lead citrate. The sections were observed with a CM12 transmission electron microscope (Philips). 12-bit images were captured with an LC3 digital camera (Scientific Instruments and Applications) and processed with Photoshop 7.0.
Gel Filtration Chromatography of HeLa Cell Extracts
HeLa cells were treated with NS siRNA-1 or control siRNA, and whole cell extract was prepared from these cells. 100-μl cytoplasmic buffer (10 mm HEPES pH 7.4, 10 mm KCl, 1.5 mm MgCl2, 0.5 mm dithiothreitol, 0.03% Nonidet P-40, 0.2 mm phenylmethylsulfonyl fluoride, 2 mm leupeptin, and 1.5 mm pepstatin A) was added to 1 × 107 cells, and the cells were incubated on ice for 20 min. The cell suspension was centrifuged at 6,000 × g for 10 min at 4 °C, and the supernatant was recovered as the cytoplasmic extract. The pellet was resuspended in 50-μl nuclear buffer (10 mm HEPES pH 7.4, 420 mm NaCl, 1.5 mm MgCl2, 0.5 mm dithiothreitol, 0.2 mm EDTA, 25% glycerol, 0.2 mm phenylmethylsulfonyl fluoride, 2 mm leupeptin, and 1.5 mm pepstatin A) and incubated on ice for 20 min. After centrifugation at 15,000 × g for 15 min at 4 °C, supernatant was collected as the nuclear extract. The cytoplasmic and nuclear extracts were combined and resolved with a Superdex 200 gel filtration column (GE Healthcare). The column was run with 10 mm HEPES pH 7.8, 150 mm NaCl, 4 mm MgCl2, 1 mm dithiothreitol, 10% glycerol, 0.003% Triton X-100, 0.2 mm phenylmethylsulfonyl fluoride, 2 mm leupeptin, and 1.5 mm pepstatin A. The elution patterns of snoRNP proteins, and TERT were analyzed by Western blotting.
Immunoprecipitation
HeLa cells were treated with 2 mm dithiobis[succinimidylpropionate] (DSP, Fisher Scientific) for 2 h at 4 °C to cross-link proteins and RNAs (24). DSP was quenched by addition of 20 mm Tris-HCl pH 8.0 at 25 °C. Whole cell extract was prepared from these cells by incubating 1 × 107 cells with 50 mm Tris-HCl pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 2 mm MgCl2, 0.2 mm phenylmethylsulfonyl fluoride, 2 mm leupeptin, 1.5 mm pepstatin A, and 1 unit/μl RNaseOUT (Invitrogen). The extract was precleared with 2 μg of rabbit control IgG and 30 μl of GammaBind G-Sepharose beads (GE Healthcare) for 1 h at 4 °C with rotation. The extract was then incubated with 3 μg of antibody against the target protein or rabbit control IgG for 1 h at 4 °C and subsequently with 20 μl of the GammaBind G-Sepharose for additional 1 h at 4 °C. After extensive washing, the beads were separated into half. The first half was incubated with 2× SDS-PAGE sample buffer (25) at 98 °C for 5 min for the analysis of co-precipitated protein by Western blotting. The second half was used to prepare RNA for quantitative RT-PCR (qRT-PCR) as described below.
qRT-PCR
qRT-PCR was used to quantify the amount of specific RNA within HeLa cells and the amount of immunoprecipitated RNA. Total RNA was purified from whole HeLa cells and immunoprecipitated materials with a PureLink Micro to Midi Total RNA Purification System (Invitrogen) and treated with DNase I as indicated by the manufacturer. qRT-PCR was performed with a Power SYBR Green RNA-to-CT 1-step kit (Applied Biosystems) on a Mastercycler ep gradient S (Eppendorf). The primer sequences are listed in Table 1. The relative total amount of each RNA in Figs. 4, E and G and 6E was calculated as follows. ΔCT (threshold cycle) of each RNA = average CT value of each RNA obtained from three PCRs − average CT value of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA obtained from three PCRs. ΔΔCT of each RNA = ΔCT of NS siRNA sample − ΔCT of control siRNA sample. Relative total value (%) = 2−ΔΔCT × 100. Relative co-precipitated amount of each RNA in Figs. 4, F and H and 6F was calculated as follows. In the case of anti-fibrillarin antibody, ΔCT of each RNA = average CT with anti-fibrillarin antibody obtained from three PCRs − average CT with control IgG obtained from three PCRs. ΔΔCT of each RNA = ΔCT with NS siRNA − ΔCT with control siRNA. Relative co-precipitated amount (%) = 2−ΔΔCT × 100. For all these analyses mean and standard deviation (S.D.) were calculated from the values obtained from three independent experiments.
TABLE 1.
Primer sequences used for qRT-PCR
| Gene namea | Primer sequence (5′–3′) |
|---|---|
| GAPDH F | TGCACCACCAACTGCTTAG |
| GAPDH R | GATGCAGGGATGATGTTC |
| TR F | TCTAACCCTAACTGAGAAGGGCGT |
| TR R | TGCTCTAGAATGAACGGTGGAAGG |
| U3 F | TTCTCTGAACGTGTAGAGCACCGA |
| U3 R | GATCATCAATGGCTGACGGCAGTT |
| U8 F | CGTCAGGTGGGATAATCCTT |
| U8 R | GGGTGTTGCAAGTCCTGATT |
| U13 F | CTTTTGTAGTTCATGAGCGTGAT |
| U13 R | GGTCAGACGGGTAATGTGC |
| U15A F | ATGTTCTCTTTGCCCAGGTG |
| U15A R | CCT*TCTCAGACAAATGCCTCT |
| U16 F | GCAATGATGTCGTAATTTGCGTCT |
| U16 R | CGTCAACCTTCTGTACCAGCTTAC |
| U19 F | ATGTGGTGCCTGTGATGGTGTTAC |
| U19 R | ACACTGCCCAAAGGTACTCAGCTA |
| U23 F | GTCTTCTCATTGAGCTCCTTTCTGTC |
| U23 R | GCTAGTTCTGTGAATTCTCTCTTCTTCTCG |
| U67 F | TCAGTGCCTCTCAGGAAAGTAGCA |
| U67 R | GGCAGAGGAAATAAGCATCCCAGT |
| U70 F | AAGCCGACTGAGTTCCTTTCCTCA |
| U70 R | ACAACCAACAGGCTGCGTACACTA |
| U71A F | GCCTGTGCCTAGGTCATTGATAGT |
| U71A R | AAAGCTTCAGGGTTCGGATGGGAT |
a F and R indicate forward and reverse primers, respectively.
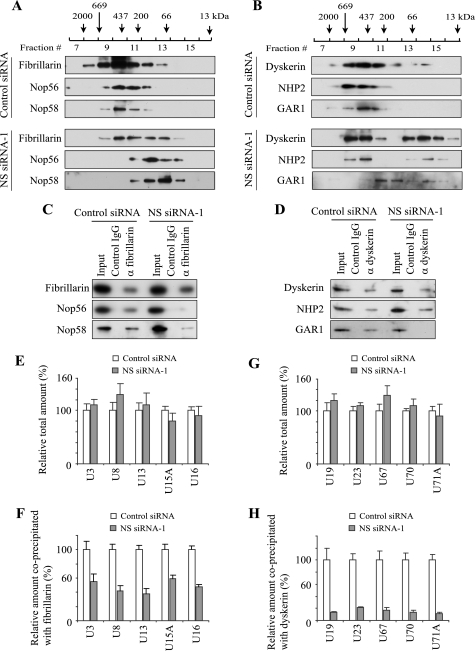
FIGURE 4.
Disruption of snoRNPs by NS knockdown. A and B, Western blotting of fractions obtained from Superdex 200 gel filtration. Protein components of C/D snoRNPs were detected in A and those of H/ACA snoRNPs in B. Whole cell extract prepared from HeLa cells transfected with NS siRNA-1 or control siRNA was applied. Elution peaks of the standard marker proteins are indicated at the top. C and D, Western blotting of co-immunoprecipitated proteins with anti-fibrillarin antibody (C) and those with anti-dyskerin antibody (D) from HeLa cell extract after siRNA transfection. Input wells contain 10% of the extract used for immunoprecipitation. E and G, relative total amounts of snoRNAs in HeLa cells transfected with NS siRNA-1 in comparison to the amount in HeLa cells transfected with control siRNA. The amounts in the control cells were defined as 100%. E shows C/D snoRNAs, and G shows H/ACA snoRNAs. Mean + S.D. obtained from three independent experiments is shown. F and H, relative amounts of snoRNAs co-immunoprecipitated with anti-fibrillarin antibody (F) and those with anti-dyskerin antibody (H) from HeLa cells after transfection with NS siRNA-1.
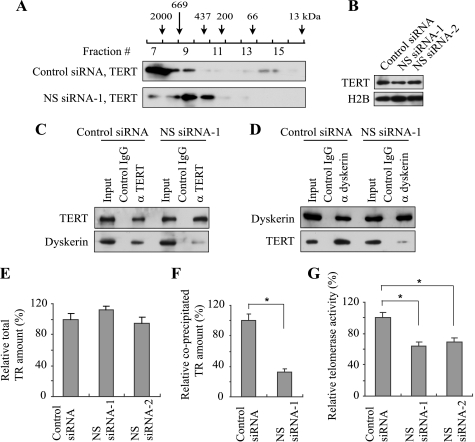
FIGURE 6.
Disruption of the telomerase complex by NS knockdown. A, Western blotting of fractions obtained from Superdex 200 gel filtration. Anti-TERT antibody was used. B, Western blotting of TERT in HeLa cells transfected with siRNAs. C and D, Western blotting of co-immunoprecipitated proteins with anti-TERT antibody (C) and those with anti-dyskerin antibody (D) from HeLa cell extracts after siRNA transfection. E, relative total amount of TR in HeLa cells transfected with NS siRNAs. The amount in the cells transfected with control siRNA was defined as 100%. F, relative amount of TR co-immunoprecipitated with anti-TERT antibody from HeLa cells transfected with NS siRNA-1. The value obtained with control siRNA was defined as 100%. The differences of the mean values between control and NS siRNAs are statistically significant in F and G (asterisk, p < 0.05). G, relative telomerase activity, measured as TPG values, in HeLa cells transfected with NS siRNAs in comparison to that in control cells.
Thin Layer Chromatography (TLC)
HeLa cells were transfected with NS siRNA or control siRNA as described above. On day 6, the culture medium was replaced with phosphate-free Dulbecco's modified Eagle medium supplemented with 10% dialyzed FBS, and the cells were cultured for 2 h. [32P]orthophosphate was added to a final concentration of 25 μCi/ml, and the cells were incubated for an additional 6 h before harvest. Total RNA was prepared from the cells as described above and digested with nuclease P1 (US Biological) in 30 mm ammonium acetate pH 5.3 for 24 h at 37 °C. Digested RNA (30,000 cpm) was resolved by two-dimensional TLC using polygram polyester sheet (CEL300, Macherey-Nagel). Isobutyric acid/5% ammonium hydroxide/water (66:1:33 (v/v/v)) was used for the first dimension and 100 mm sodium phosphate buffer pH 6.8/ammonium sulfate/n-propanol (100:60:2 (v/w/v)) was used for the second dimension (26). The TLC sheets were dried and analyzed by autoradiography. In addition, spots corresponding to uridine and pseudouridine were cut out from the TLC sheets and applied for Cerenkov counting to obtain the pseudouridine/uridine ratio of radioactivity.
Telomeric Repeat Amplification Protocol (TRAP) Assay
Telomerase activity of HeLa cells transfected with NS siRNA or control siRNA was measured with TRAP assay using a TRAPEZE XL Telomerase Detection Kit (Millipore) following the manufacturer's instruction. The values of Total Products Generated (TPG), which represents telomerase activity, were obtained from three independent experiments and used to calculate the mean value and S.D.
RESULTS
Dispersal of snoRNP Clusters by NS Knockdown
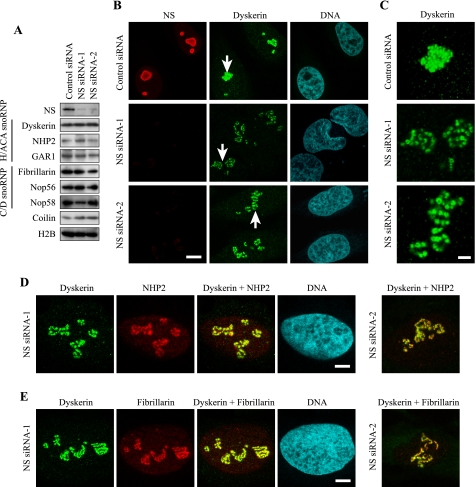
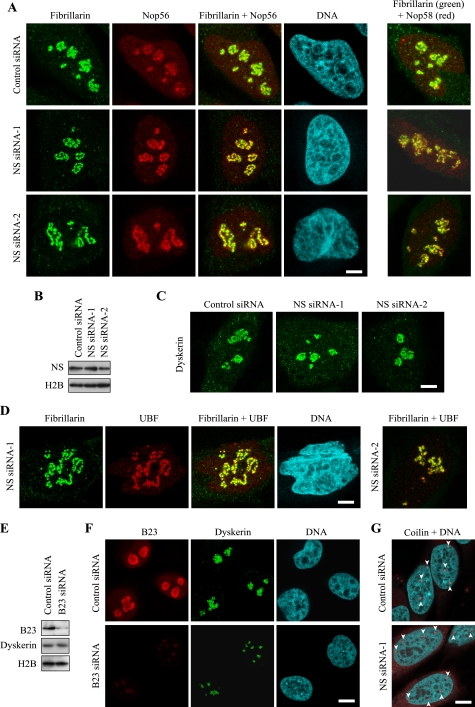
Our previous finding on the role of NS for the processing of pre-rRNA led us to investigate if nucleolar localization of snoRNPs, which are involved in the processing, was also affected by NS depletion. HeLa cells were used as target cells in the current study, because the expression level of p53 is negligible in the cells (27, 28), which allowed us to observe the effects of NS knockdown without immediate cell cycle arrest (9). We used two independent siRNA sequences to knock down NS. The first sequence we used had been previously shown to deplete NS in HeLa cells (NS siRNA-1) (9). The second siRNA sequence (NS siRNA-2) also significantly depleted NS from HeLa cells (Fig. 1A). These two siRNAs did not substantially affect the protein levels of the components of H/ACA snoRNPs (dyskerin, NHP2, and GAR1) or C/D snoRNPs (fibrillarin, Nop56, and Nop58) (Fig. 1A). The amount of coilin (a main component of Cajal bodies) was not significantly affected by NS knockdown, either. In previous studies neither we nor others could detect a major alteration in the distribution of several nucleolar proteins, such as B23 and nucleolin, by NS knockdown, which was interpreted that NS depletion leaves the nucleolar structure largely intact (6, 7, 9). However, we found that snoRNP proteins became dispersed within the nucleoli upon NS knockdown. Dyskerin-containing round structures (called beads hereafter) were tightly packed in HeLa cells transfected with control siRNA (Fig. 1B). In contrast, following transfection with NS siRNA-1 and NS siRNA-2, the beads were clearly dispersed, and the space between each bead became apparent in more than 50% of the cells (Fig. 1, B and C). Dispersed dyskerin was co-localized with NHP2, another component of H/ACA snoRNPs, and fibrillarin, a component of C/D snoRNPs, within each bead (Fig. 1, D and E). As expected, dispersed fibrillarin was co-localized with two other C/D RNP proteins, Nop56 and Nop58 (Fig. 2A). These findings suggest that NS knockdown loosens the connections between the snoRNP clusters (beads) without significantly diminishing the amounts of snoRNP proteins within the clusters. The loosening of the clusters was reversible. Five days after the second transfection of NS siRNAs, the protein levels of NS (Fig. 2B) and the distribution pattern of dyskerin (Fig. 2C) once again became indistinguishable from the results with control siRNA. Because casein kinase 2 inhibitors are known to reversibly disperse RNA polymerase I machinery, such as UBF, in a similar way (29), we immunostained UBF and found that this protein was also redistributed with fibrillarin 2 days after the second transfection (Fig. 2D). The protein level of casein kinase 2 was not substantially affected by NS knockdown (not shown), indicating that simple decrease of casein kinase 2 is not the mechanism for the relocalization of snoRNP proteins. A previous study could not detect such redistribution of fibrillarin after NS knockdown (6). The discrepancy between this report and our observation may be due to differences in the cell types tested or the degree of NS depletion.
FIGURE 1.
Dispersal of snoRNP protein clusters upon NS knockdown in HeLa cells. A, Western blotting of proteins related to snoRNPs, comparing HeLa cells transfected with NS siRNAs and control siRNA. Histone H2B was examined as a loading control. B, double immunofluorescence staining of HeLa cells transfected with NS siRNAs or control siRNA. Antibodies against NS and dyskerin were used. DNA was counterstained with Topro 3. Bar, 10 μm. C, areas indicated by arrows in A were magnified 3-fold. Bar, 3 μm. D and E, double immunofluorescence staining of HeLa cells with antibodies against dyskerin and NHP2 (D) and with antibodies against dyskerin and fibrillarin (E) after transfection with NS siRNAs. Bar, 5 μm in D and E.
FIGURE 2.
Distribution patterns of snoRNP proteins and other nuclear proteins in NS knockdown cells. A, double immunofluorescence staining of HeLa cells transfected with NS siRNAs or control siRNA. Antibodies against fibrillarin and Nop56 were used in the left set of the panels. Different cells were double-stained with antibodies against fibrillarin and Nop58 in the right set. Bar, 5 μm in A, C, and D. B, Western blotting of HeLa whole cell extracts 5 days after the second transfection of siRNAs. C, immunostaining of HeLa cells with anti-dyskerin antibody 5 days after the second transfection of siRNAs. D, HeLa cells transfected with NS siRNAs were double immunofluorescence stained with antibodies against fibrillarin and UBF. E, Western blotting of proteins prepared from HeLa cells transfected with B23 siRNA or control siRNA. F, HeLa cells transfected with B23 siRNA or control siRNA were double immunofluorescence stained with antibodies against B23 and dyskerin. Bar, 10 μm in F and G. G, HeLa cells transfected with NS siRNA-1 or control siRNA were stained with the anti-coilin antibody and Topro 3. Arrowheads indicate Cajal bodies.
NS is primarily localized in the granular component (GC) of the nucleolus (30), where late steps of pre-ribosome assembly take place (31). All human H/ACA snoRNP proteins are localized in the dense fibrillar component (DFC) of the nucleolus (32), which is involved in rRNA transcription along the border of the fibrillar center (FC), as well as in the early steps of pre-ribosome assembly. Because the DFC is surrounded by the GC (Fig. 3B, left panel), depletion of any abundant proteins in the GC might nonspecifically disperse the DFC due to a loss of structural support. To test this possibility, we knocked down B23, another major protein in the GC, which has multiple functions, including acting as a molecular chaperon and a ribonuclease against pre-rRNA (33). B23 also interacts with NS although the functional implication of this remains to be studied (34). B23 was depleted from HeLa cells with siRNA as described before (22) (Fig. 2E). This knockdown did not alter the protein level of dyskerin but the sizes of the areas containing dyskerin became substantially smaller than those in control cells (Fig. 2F). This observation indicates that the redistribution pattern of snoRNP proteins is dependent on which protein is depleted in the GC.
FIGURE 3.
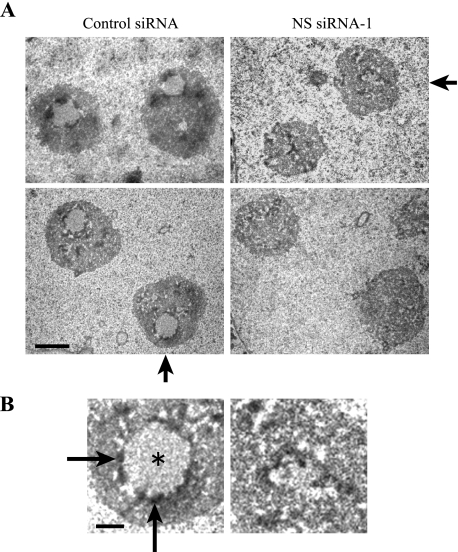
Disruption of the DFC and the FC of the nucleolus in HeLa cells by NS knockdown. A, electron micrographs of nucleoli in HeLa cells transfected with NS siRNA-1 or control siRNA. Bar, 2 μm. B, nucleoli indicated by arrows in A are magnified 3-fold. Asterisk (round light gray area) and long arrows (dark rim surrounding the light gray area) indicate the FC and the DFC, respectively. The rest of the area corresponds to the GC. Bar, 0.5 μm.
NS depletion decreased total protein amount by 12.1 ± 2.2% as measured by a Quant-iT Protein Assay kit. Therefore, disruption of the nucleolar structure and snoRNPs might be a nonspecific effect of the decreased levels of unidentified nucleolar proteins that are necessary to maintain the integrity of nucleolar structure and snoRNPs. This possibility was examined by treating HeLa cells for 7 days with cycloheximide, an inhibitor of translational elongation; however, this treatment did not disperse snoRNP clusters (not shown) despite the decrease of the total protein level by 21.7 ± 4.2%. Thus, simple down-regulation of protein synthesis does not appear to be the reason for the nucleolar disorganization by NS knockdown.
We also examined if Cajal bodies are affected by NS depletion because snoRNPs are temporarily localized in this subnuclear structure during their maturation process as described earlier. We evaluated Cajal bodies using antibody against coilin. Immunofluorescence staining did not demonstrate any changes in the sizes or numbers of Cajal bodies after depletion of NS (Fig. 2G), suggesting that Cajal bodies remain grossly intact in the NS knockdown cells. Consistent with this, NS was not localized in Cajal bodies in untreated HeLa cells (not shown).
Electron Microscopy of NS-depleted Cells
We applied electron microscopy to further examine structural disorganization of the nucleolus induced by the depletion of NS. The three nucleolar subdomains (the FC, the DFC, and the GC) were clearly distinguishable in control cells (Fig. 3, A and B). The FC (Fig. 3B, asterisk), where RNA polymerase I machinery is stored (31), is usually surrounded by a thin electron dense area of the DFC (Fig. 3B, arrows). The GC occupies the remaining volume which is more than 70% of the nucleolus (31). Among more than 100 control nuclei we observed under an electron microscope, around 30% of the nuclei clearly displayed the three domains. However, the area of the FC normally surrounded by the DFC was not obvious in any of 100 nuclei in NS knockdown cells. Because of the fact that the H/ACA snoRNPs are localized in the DFC, disorganization of this area by NS knockdown is consistent with the loosely distributed snoRNP proteins described above.
Disassembly of snoRNPs by NS Knockdown
To understand how relaxation of the snoRNP protein clusters affects the integrity of snoRNPs, we first compared the size distribution of snoRNPs between control and NS knockdown cells using gel filtration. We prepared cell extract from HeLa cells after siRNA treatment, applied it to a gel filtration column and analyzed the distribution of each snoRNP protein by Western blotting. Although snoRNP proteins are not necessarily exclusively localized within snoRNPs, this approach provided a crude profile of the size distribution of the complexes containing snoRNP proteins. In control extract fibrillarin (34 kDa) was widely eluted from the column in the fractions corresponding to a molecular mass of around 100–2000 kDa, with a peak fraction around 400–500 kDa (Fig. 4A). Nop56 (56 kDa) and Nop58 (60 kDa) were also eluted in these fractions. However, these proteins were invariably eluted in later fractions (corresponding to smaller molecular mass) in NS knockdown cells. Proteins in the H/ACA snoRNPs demonstrated a similar trend. Dyskerin (58 kDa), NHP2 (17 kDa), and GAR1 (28 kDa) were all mainly eluted in the fractions corresponding to molecules of 200–700 kDa with control cell extract (Fig. 4B). With the extract of NS knockdown cells, however, dyskerin and NHP2 were eluted in the fractions of less than 100 kDa as well. The entire elution pattern of GAR1 was shifted to smaller masses and its main elution peak (fractions 11 and 12) was separated from that of dyskerin and NHP2. These elution patterns could be affected not only by molecular mass but also by surface charges and molecular conformations; nonetheless, these findings indicate that a radical change took place in snoRNPs upon NS knockdown.
To investigate the integrity of snoRNPs more specifically, fibrillarin was immunoprecipitated from extract prepared after siRNA transfection and associated components were evaluated by Western blotting and qRT-PCR. While similar amounts of fibrillarin were precipitated from NS knockdown and control cell extracts, much less Nop56 and Nop58 were co-precipitated from the cell extract of NS knockdown than from control cell extract (Fig. 4C). Immunoprecipitation of dyskerin co-precipitated NHP2 from the two types of cell extract but GAR1 was not co-precipitated from the extract of NS knockdown cells (Fig. 4D), in accordance with the gel filtration result. In these experiments NS was not co-precipitated with fibrillarin or dyskerin (not shown), suggesting that NS is not a stably incorporated component of snoRNPs.
We next studied how NS knockdown affected co-precipitation of C/D snoRNAs with fibrillarin. U3, U8, and U13 snoRNA are involved in the processing of pre-rRNA without 2′-O-methylation and U15A and U16 snoRNA are important for 2′-O-methylation of pre-rRNA. The total amounts of these snoRNAs were not significantly decreased by NS knockdown as shown by qRT-PCR (Fig. 4E). In this experiment we used the amount of GAPDH mRNA to normalize the level of each snoRNA because total amount of RNA was decreased by 20–30% by NS knockdown (9). When β-actin mRNA was used to normalize the snoRNA amounts, similar results were obtained (not shown). NS knockdown decreased the levels of these co-precipitated snoRNAs to less than 60% of the control experiment (Fig. 4F). We have previously shown that NS knockdown delays the processing of 32S pre-rRNA to 28S rRNA (9), which may be relevant to the observed disruption of the snoRNPs. In particular, U8 snoRNP is known to be essential for the processing of mammalian 32S pre-rRNA to 28S pre-rRNA (35, 36).
We used the same approach to study co-precipitation of H/ACA snoRNAs with dyskerin. Five H/ACA snoRNAs, U19, U23, U67, U70, and U71A, are all involved in pseudouridylation of rRNA. Total amounts of these snoRNAs were not diminished by NS knockdown (Fig. 4G) but the amounts of these snoRNAs co-precipitated with similar amounts of dyskerin were less than 30% of those in control cells (Fig. 4H). Because co-immunoprecipitation of H/ACA snoRNAs was more severely affected by NS knockdown than co-immunoprecipitation of C/D snoRNAs, we investigated the total level of pseudouridine as a functional consequence of H/ACA snoRNP disruption in the next section.
Decreased Pseudouridylation by NS Knockdown
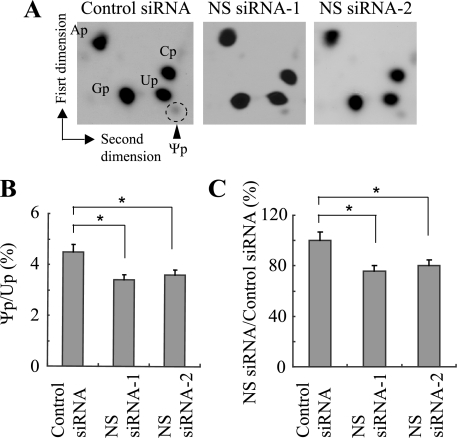
We compared the amount of pseudouridine between NS knockdown cells and control cells. Radiolabeled pseudouridine can be readily detected as a distinct spot by thin layer chromatography (Fig. 5A). While the radioactivity ratio between pseudouridine and uridine was 4.5 ± 0.3% with control siRNA, the ratio was 3.4 ± 0.2% with NS siRNA-1 and 3.6 ± 0.2% with NS siRNA-2 (Fig. 5B). This translates to 75.6 ± 4.4% and 80.0 ± 4.4% for each NS siRNA when the ratio in the control cells is defined as 100% (Fig. 5C). This drop was statistically significant for both NS siRNAs (p < 0.05). This is evidence that disruption of H/ACA snoRNPs indeed resulted in a functional consequence.
FIGURE 5.
Decreased pseudouridine by NS knockdown detected by thin layer chromatography. A, autoradiographs demonstrating the migration patterns of adenosine 5′-monophosphate (Ap), cytosine 5′-monophosphate (Cp), guanosine 5′-monophosphate (Gp), uridine 5′-monophosphate (Up), and pseudouridine 5′-monophosphate (Ψp, marked by a dotted circle). B, radioactivity ratios between Ψp and Up in the panels in A were calculated. Mean + S.D. obtained from three independent experiments is shown. The differences of the mean values between control and NS siRNAs are statistically significant in B and C (asterisk, p < 0.05). C, values obtained in B were re-calculated using the value obtained with control siRNA defined as 100%.
Disrupted Integrity of the Telomerase Complex by NS Knockdown
Finally, we studied if the telomerase complex was also disrupted by NS knockdown because the telomerase complex and H/ACA snoRNPs share structural similarity. The size of the human telomerase complex is estimated to be 550 kDa to 2 MDa depending on the conditions of isolation (37, 38). Consistent with this, our gel filtration of HeLa cell extract indicated that TERT was eluted in the fractions corresponding to proteins from ∼500 kDa to more than 2 MDa (Fig. 6A). After NS knockdown TERT was seen to have an elution pattern consistent with a lower molecular weight, with the elution peak corresponding to around 400–600 kDa. This finding suggests that NS knockdown led to disassembly of the telomerase complex. The total amount of TERT was not altered by NS knockdown (Fig. 6B). Immunoprecipitation of TERT co-precipitated a smaller amount of dyskerin with NS knockdown cell extract than with control cells (Fig. 6C). In reciprocal immunoprecipitation, dyskerin also co-precipitated a diminished amount of TERT from the knockdown extract than from control extract (Fig. 6D). Although the total amount of TR was not decreased by the knockdown (Fig. 6E), the level of TR co-precipitated with TERT became 32.0 ± 4.2% by NS siRNA-1 when the amount co-precipitated after control siRNA transfection was defined as 100% (Fig. 6F). In accordance with these results, telomerase activity was decreased to 64.1 ± 5.2% with NS siRNA-1 and 69.3 ± 4.8% with NS siRNA-2 compared with control siRNA (Fig. 6G). Collectively, these results demonstrate that NS knockdown compromised the integrity of the telomerase complex and decreased its activity. We could not observe a long-term effect of NS depletion on the telomere length because of the transient effect of siRNA. Cells may not be able to repeat enough cell divisions for any potential shortening of telomere length to become apparent due to the decreased protein synthesis by NS knockdown (9). A previous study proposed that NS maintains the telomere length by interacting with and facilitating the degradation of the telomere-binding protein TRF1, which blocks the access of telomerase to telomeres (4). However, we could not detect co-immunoprecipitation of TRF1 with anti-NS antibody from HeLa cell extract (not shown). This could be due to the condition of immunoprecipitation.
DISCUSSION
Our findings can be summarized in two points. First, the immunofluorescence and electron microscopic studies indicated the requirement of NS to maintain the architecture of the FC and the DFC. Second, NS is also required for the integrity of snoRNPs and the telomerase complex whose components are localized in the DFC. This biochemical finding was supported by the functional analyses of the decreased pseudouridylation and telomerase activity in these cells. NS's functions have been studied with an emphasis on its connection with p53; however, our results revealed that NS has a more fundamental role in the maintenance of the nucleolar structure and function than previously appreciated. Since the p53 level is insignificant in HeLa cells due to its degradation by the oncoprotein E6 (27, 28), our findings are unlikely to be linked with p53. If so, a question is raised whether the new roles of NS described in this work can explain already reported p53-independent phenotypes resulting from NS depletion.
The presence of p53-independent functions of NS was suggested by two pieces of evidence. First, loss of p53 cannot rescue embryonic lethality of NS knock-out mice around embryonic day 4 (5). Decreased telomerase activity is unlikely to contribute to the early lethality of the NS knock-out mice; however, disrupted snoRNPs may be mechanistically relevant to the lethality through processing or stability/conformational problems of rRNA and other target RNAs. Testing of this possibility could be initiated by analyzing the level of these target RNAs. Although the immunostaining pattern of fibrillarin appeared normal in the work by Beekman et al., a closer observation of the embryonic cells at a high magnification might have revealed a subtle abnormality of the subnucleolar distribution of fibrillarin and other snoRNP proteins.
The second piece of evidence supporting a p53-independent function of NS was provided when the knockdown of the ribosomal proteins L5 and L11, key activators for p53, only partially rescued the G1 arrest triggered by NS knockdown (7). In this study p53 induction was completely abolished by the knockdown of L5 and L11. It would be informative to investigate how severely the levels of snoRNPs, rRNA, and protein synthesis were affected in these cells.
Whether NS is directly involved in the assembly of snoRNPs and telomerase complex is another question raised by our study. Immunoprecipitation of snoRNPs and TERT did not co-precipitate NS. Subnucleolar localization of NS (the GC) does not overlap with that of snoRNPs (the DFC). Furthermore, NS was not enriched in Cajal body where snoRNPs and components of the telomerase complex reside during or before assembly. Collectively, we could not obtain evidence suggesting direct interaction between NS and snoRNPs or the telomerase complex. It is still possible that NS transiently or weakly interacts with snoRNPs and the telomerase complex; however, the currently available information favors the interpretation that the loss of NS in the GC made the DFC collapse, indirectly preventing the normal assembly process of snoRNPs and the telomerase complex.
The dispersed pattern of the snoRNP clusters we found resembles what is called nucleolar necklace. This pattern is observed upon treatment of cells with the casein kinase 2 inhibitor 5,6-dichloro-1-ribo-furanosylbenzimidazole (DRB) (29). Each bead in the necklace corresponds to a fragment composed of the FC and the DFC. These beads contain rDNA, rRNA, fibrillarin, and UBF. Our beads also contained UBF and several snoRNP proteins, suggesting that the beads dispersed by NS knockdown are quite similar to the nucleolar necklace induced by DRB. Potential regulation of casein kinase 2 activity by NS would be another possibility worth examining to further understand the role of NS in the maintenance of the nucleolar architecture.
NS is highly expressed in rapidly proliferating cells which need robust telomerase activity. NS's involvement in the maintenance of the telomere length has been explained at least partly by the interaction between NS and TRF1, a negative regulator of the telomere length (4). Our discovery that NS is required for integrity of the telomerase complex could provide a second link between NS and telomere length regulation. The essential role of dyskerin for telomerase activity is best demonstrated by the human disease dyskeratosis congenita (DK). DK is a hereditary syndrome characterized by bone marrow failure, abnormal skin pigmentation, nail dystrophy and white patches in the oral cavity called mucosal leukoplakia (39–41). The most common type, X-linked DC, is caused by mutations in DKC1, the gene encoding dyskerin. Mutations in DKC1 destabilize TR and reduce the amount of TR within cells, resulting in shortened telomeres (19, 42). Depletion of NHP2 also reduces the TR level within cells (19). In addition, depletion of one subunit of the telomerase complex diminishes the stability of other subunits (19). Such interdependence among each telomerase subunit underscores the importance of the integrity of the telomerase complex as a whole. Although the TR level was not decreased by NS depletion in our case, and the exact function of each subunit has not been fully elucidated, disassembly of the telomerase complex is highly likely to contribute to diminished telomerase activity.
In summary, our study revealed novel and profound effects of NS depletion on nucleolar RNA-protein complexes. These effects need to be considered to properly interpret any results obtained by modulation of the protein level of NS.
Acknowledgments
We thank Susan Keirstead and Samuel Rayner for critical reading of the manuscript, Reinhard Lührmann for providing the antibodies, and Robert T. Tranquillo and Stephen Stephens for access to their fluorescence spectrophotometer. Electron microscopy was performed with the support of Mark A. Sanders and Gail Celio at the Imaging Center, College of Biological Sciences, University of Minnesota.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM068027 (to N. K.).
- NS
- nucleostemin
- CT
- threshold cycle
- DFC
- dense fibrillar component
- DK
- dyskeratosis congenita
- DRB
- 5,6-dichloro-1-ribo-furanosylbenzimidazole
- DSP
- dithiobis[succinimidylpropionate]
- FBS
- fetal bovine serum
- FC
- fibrillar center
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GC
- granular component
- PBS
- phosphate-buffered saline
- qRT-PCR
- quantitative RT-PCR
- siRNA
- short interfering RNA
- snoRNP
- small nucleolar ribonucleoprotein
- TERT
- telomerase reverse transcriptase
- TLC
- thin layer chromatography
- TR
- telomerase RNA
- TRAP
- telomeric repeat amplification protocol
- UBF
- upstream binding factor.
REFERENCES
- 1.Tsai R. Y., McKay R. D. (2002) Genes Dev. 16, 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan Y., Liu Z., Zhao S., Lou F., Nilsson S., Ekman P., Xu D., Fang X. (2006) Br. J. Cancer 94, 1658–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma H., Pederson T. (2008) Trends Cell Biol. 18, 575–579 [DOI] [PubMed] [Google Scholar]
- 4.Zhu Q., Yasumoto H., Tsai R. Y. (2006) Mol. Cell. Biol. 26, 9279–9290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J. C. (2006) Mol. Cell. Biol. 26, 9291–9301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma H., Pederson T. (2007) Mol. Biol. Cell 18, 2630–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai M. S., Sun X. X., Lu H. (2008) Mol. Cell. Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng L., Lin T., Tsai R. Y. (2008) J. Cell Sci. 121, 4037–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romanova L., Grand A., Zhang L., Rayner S., Katoku-Kikyo N., Kellner S., Kikyo N. (2009) J. Biol. Chem. 284, 4968–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matera A. G., Terns R. M., Terns M. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 209–220 [DOI] [PubMed] [Google Scholar]
- 11.Meier U. T. (2005) Chromosoma 114, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reichow S. L., Hamma T., Ferré-D'Amaré A. R., Varani G. (2007) Nucleic Acids Res. 35, 1452–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watkins N. J., Dickmanns A., Lührmann R. (2002) Mol. Cell. Biol. 22, 8342–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafontaine D. L., Tollervey D. (1999) RNA 5, 455–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lafontaine D. L., Tollervey D. (2000) Mol. Cell. Biol. 20, 2650–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maden B. E. (1990) Prog. Nucleic Acid Res. Mol. Biol. 39, 241–303 [DOI] [PubMed] [Google Scholar]
- 17.Autexier C., Lue N. F. (2006) Annu. Rev. Biochem. 75, 493–517 [DOI] [PubMed] [Google Scholar]
- 18.Collins K. (2006) Nat. Rev. Mol. Cell Biol. 7, 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu D., Collins K. (2007) Mol. Cell 28, 773–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dragon F., Pogacić V., Filipowicz W. (2000) Mol. Cell. Biol. 20, 3037–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Tomlinson R. L., Lukowiak A. A., Terns R. M., Terns M. P. (2004) Mol. Biol. Cell 15, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonda K., Wudel J., Nelson D., Katoku-Kikyo N., Reed P., Tamada H., Kikyo N. (2006) J. Biol. Chem. 281, 8153–8160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Watkins N. J., Ségault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Lührmann R. (2000) Cell 103, 457–466 [DOI] [PubMed] [Google Scholar]
- 24.Zhang L., Rayner S., Katoku-Kikyo N., Romanova L., Kikyo N. (2007) Biochem. Biophys. Res. Commun. 361, 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26.Grosjean H., Keith G., Droogmans L. (2004) Methods Mol. Biol. 265, 357–391 [DOI] [PubMed] [Google Scholar]
- 27.Scheffner M., Werness B. A., Huibregtse J. M., Levine A. J., Howley P. M. (1990) Cell 63, 1129–1136 [DOI] [PubMed] [Google Scholar]
- 28.Slebos R. J., Lee M. H., Plunkett B. S., Kessis T. D., Williams B. O., Jacks T., Hedrick L., Kastan M. B., Cho K. R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5320–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez-Verdun D. (2006) Histochem. Cell Biol. 126, 135–148 [DOI] [PubMed] [Google Scholar]
- 30.Politz J. C., Polena I., Trask I., Bazett-Jones D. P., Pederson T. (2005) Mol. Biol. Cell 16, 3401–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raska I., Shaw P. J., Cmarko D. (2006) Int. Rev. Cytol. 255, 177–235 [DOI] [PubMed] [Google Scholar]
- 32.Pogacić V., Dragon F., Filipowicz W. (2000) Mol. Cell. Biol. 20, 9028–9040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson M. O., Hingorani K., Szebeni A. (2002) Int. Rev. Cytol. 219, 199–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H., Pederson T. (2008) Mol. Biol. Cell 19, 2870–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peculis B. A., Steitz J. A. (1993) Cell 73, 1233–1245 [DOI] [PubMed] [Google Scholar]
- 36.Gerbi S. A., Borovjagin A. V. (2004) in The Nucleolus ( Olson M. O. J., ed), pp. 170–198, Kluwer Academic, Norwell, MA [Google Scholar]
- 37.Schnapp G., Rodi H. P., Rettig W. J., Schnapp A., Damm K. (1998) Nucleic Acids Res. 26, 3311–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venteicher A. S., Meng Z., Mason P. J., Veenstra T. D., Artandi S. E. (2008) Cell 132, 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vulliamy T. J., Dokal I. (2008) Biochimie 90, 122–130 [DOI] [PubMed] [Google Scholar]
- 40.Kirwan M., Dokal I. (2008) Clin. Genet 73, 103–112 [DOI] [PubMed] [Google Scholar]
- 41.Marrone A., Walne A., Dokal I. (2005) Curr. Opin. Genet. Dev. 15, 249–257 [DOI] [PubMed] [Google Scholar]
- 42.Wong J. M., Collins K. (2006) Genes Dev. 20, 2848–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]