Abstract
Laminaripentaose-producing β-1,3-glucanase (LPHase), a member of glycoside hydrolase family 64, cleaves a long-chain polysaccharide β-1,3-glucan into specific pentasaccharide oligomers. The crystal structure of LPHase from Streptomyces matensis DIC-108 was solved to 1.62 Å resolution using multiple-wavelength anomalous dispersion methods. The LPHase structure reveals a novel crescent-like fold; it consists of a barrel domain and a mixed (α/β) domain, forming a wide-open groove between the two domains. The liganded crystal structure was also solved to 1.80 Å, showing limited conformational changes. Within the wide groove, a laminaritetraose molecule is found to sit in an electronegatively charged central region and is proximal to several conserved residues including two carboxylates (Glu154 and Asp170) and four other sugar-binding residues (Thr156, Asn158, Trp163, and Thr167). Molecular modeling using a laminarihexaose as a substrate suggests roles for Glu154 and Asp170 as acid and base catalysts, respectively, whereas the side chains of Thr156, Asn158, and Trp163 demarcate subsite +5. Site-directed mutagenesis of Glu154 and Asp170 confirms that both carboxylates are essential for catalysis. Together, our results suggest that LPHase uses a direct displacement mechanism involving Glu154 and Asp170 to cleave a β-1,3-glucan into specific α-pentasaccharide oligomers.
Glycoside hydrolases (GHs,3 EC 3.2.1.x) hydrolyze the glycosidic bond between two or more carbohydrates or between a carbohydrate and non-carbohydrate moiety (1). These enzymes play diverse roles in nature; they breakdown cellulose into smaller carbohydrates (i.e. during biomass degradation by cellulases), they function during pathogenesis such as the activity of influenza virus neuraminidase (2), and they are engaged in normal cellular metabolic processes that involve the formation and breakage of glycosidic bonds along with glycosyl transferase (3). GHs can be classified as exo- or endo-type of glycoside hydrolases that catalyze the hydrolysis of the glycosidic bond from the end or at the middle, respectively, of a polysaccharide chain. GHs can also be classified as the inverting or the retaining enzymes with respect to their distinct stereochemical mechanisms during catalysis (3). Sequence-based classification of GHs into various families has been proposed by Henrissat et al. (4, 5). Additionally, numerous structures of GHs have revealed details of their catalytic mechanisms as well as the basis for their diverse substrate specificity (6). Based on sequence comparisons and structural analyses, the carbohydrate-active enzymes data base (CAZy) provides continuously updated information on the GH families (7).
Two key residues among GHs, generally found as carboxylates, are involved in the hydrolysis of the glycosidic bond: a proton donor and a nucleophile/base (3). In either the retaining or the inverting enzymes, the position of the proton donor is found within hydrogen-bonding distance of the glycosidic oxygen. Active sites that consist of the key residues have been classified into three topologies by Davies and Henrissat (1): (i) a pocket or a crater that preferentially recognizes a saccharide molecule with a non-reducing end, presenting the exo-type hydrolysis, (ii) a cleft or groove that accommodates a large substrate for endo-type cleavage, and (iii) a tunnel that enables a polysaccharide chain to be threaded through for efficient endo-hydrolase processivity.
Among the current 114 families of GHs, β-1,3-glucanases, namely exo-β-1,3-glucanases, (E.C. 3.2.1.58) and endo-β-1,3-glucanases (E.C 3.2.1.39) that degrade β-1,3-glucans into smaller biological response modifiers (8) are found in seven families. GH-3 and GH-5 are found to be exo-type; GH-16 and GH-17 are in the endo-type category. Both endo- and exo-type enzymes are found in GH-55. However, GH-64 and GH-81 remain unclear (7). Three-dimensional structures of members from GH-3, GH-5, GH-16, GH-17, and GH-55 have been solved, providing detailed structure-activity information (9–13). Members of the GH-5 and GH-17 families contain a (β/α)8 architecture, whereas GH-16 family members fold as a β-jelly roll. Barley β-d-glucan exohydrolase, a member of the GH-3 family has an N-terminal TIM-barrel domain and a C-terminal 6-stranded β-sandwich. Notably, glucanases from these families are retaining enzymes. The newly resolved structure of Lam55A, an inverting enzyme in GH-55 family (13), has two β-helical domains, separated by a long linker region, that form a ribcage-like structure. To date, no structure has been reported for GH-64 and GH-81, and thus detailed information on the mode of catalysis is lacking.
Laminaripentaose-producing β-1,3-glucanase (LPHase) cleaves a long-chain polysaccharide, β-1,3-glucan, including laminarin, into a specific pentasaccharide oligomer “laminaripentaose” (14, 15). Of interest, β-1,3-glucans such as laminarin, which constitute the cell walls of plants and fungi have interesting biological roles in immune modulation (8, 16, 17). Biochemical characterization of LPHase from Streptomyces matensis DIC-108 showed that the enzyme belongs to the GH-64 family and is an inverting enzyme (14, 15). This enzyme is unique that it releases mainly laminaripentaose as the end product, as compared with that exo-type enzymes produce much smaller sugars (monosaccharides or disaccharides) (18–20) while endo-type enzymes yield heterogeneous forms of oligosaccharides. This atypical product specificity, to our knowledge, has not been reported for other glucanases. Here we report the three-dimensional structures for the apo and complex LPHase of S. matensis. Structural analysis, modeling, and mutagenesis results revealed a novel crescent-fold structure containing Glu154 and Asp170 involved in the cleavage of a long-chain oligosaccharide from the reducing end.
EXPERIMENTAL PROCEDURES
Expression and Purification of LPHase
The artificial LPHase gene without the sequence of signal peptide (the first 35 residues) was reconstructed by PCR using a set of primers (sequences not shown). The amplified DNA fragment was inserted into pRSET A (designated pRSET_lph) and expressed in Escherichia coli BL21 (DE3). The resulting protein had an extra methionine residue at the N terminus. After overnight induction by isopropyl-1-thio-β-d-galactopyranoside, cells (from 1 liter of culture) were harvested and resuspended in 20 ml of acetate buffer (20 mm, pH 5.8). Cells were then disrupted by sonication for 10 min with 30 s for each interval. After centrifugation to remove the cell debris, the supernatant (20 ml) was loaded onto a cation exchange HiTrap SP column (2.6 × 40 cm, Amersham Biosciences) that was pre-equilibrated with the acetate buffer, pH 5.8. Column was eluted by a linear gradient of 0–1 m NaCl with 10 mm/min at a flow rate of 3 ml/min. The active fractions eluted in the range of 100–250 mm NaCl were collected and concentrated 20-fold before loading onto a anion exchange HiTrap Q column (1.6 × 20 cm, Amersham Biosciences), which was equilibrated with phosphate buffer (20 mm, pH 6.8). LPHase was eluted by a linear gradient of 0–1 m NaCl with 5 mm/min at a flow rate of 1.5 ml/min. The active fractions eluted in the range of 200–350 mm NaCl were collected and concentrated 10-fold. The finally step of purification was to load 5 ml of crude protein (derived from the previous step) onto high performance CM ion exchange column (1.6 × 20 cm, Amersham Biosciences) that was equilibrated with sodium acetate buffer (20 mm, pH 5.0). LPHase (in 20 mm acetate buffer, pH 5.0) was eluted by a linear pH gradient with phosphate buffer (0.1 m, pH 7.2) at a flow rate of 1.5 ml/min. The active fractions were eluted at the range of pH 5.5–6.5. The eluted protein was concentrated for 25-fold and kept in 4 °C for further study. The purity of LPHase was verified by SDS-PAGE analysis. Expression of selenomethionine-labeled LPHase (Se-Met-LPHase) was done essentially described by Yang et al. (29) and Van Duyne et al. (30) and purified with the same protocol described above. The protein sample was concentrated to 20 mg/ml in a solution containing 40 mm sodium acetate (pH 6.5) by centrifugal filter device, Amicon Ultra (10000 MWCO) (Millipore, Carrigtwohill, Co. Cork, Ireland).
Crystallization
Crystallization was performed by the hanging-drop vapor-diffusion method at 20 °C by mixing equal volumes of protein sample and reservoir solution. Initial crystallization conditions were screened using Crystal Screen I and II kits (Hampton Research) and Clear Strategy Screen I and II kits (Molecular Dimension). Crystals were initially observed from two conditions: (1) using 0.2 m ammonium sulfate, 30% polyethylene glycol (PEG) 8000, 0.1 m sodium cacodylate (pH 6.5), and (2) 0.2 m ammonium sulfate, 30% PEG monomethyl ether (MME) 5000, 0.1 m MES (pH 6.5). Optimized crystals grew in a modified solution containing 0.15 m ammonium sulfate, 30% PEG MME 5000, and 0.1 m MES (pH 6.5). The native crystal diffracted to 1.62 Å and belonged to space group P212121 with the unit cell dimensions a = 46.16, b = 60.68, c = 149.40 Å. There was one molecule per asymmetric unit.
Crystals of the Se-Met-derivatized protein were obtained in a solution containing 0.1 m lithium sulfate, 25% PEG MME 2000, and 0.1 m Tris (pH 7.0) using Se-Met-LPHase (20 mg/ml) at 4 °C. The SeMet-LPHase crystal also belonged to space group P212121 with unit cell dimensions a = 46.27, b = 60.36, c = 150.13, and one molecule per asymmetric unit.
The liganded LPHase crystals were obtained by the co-crystallization method in a solution containing 0.05 m ammonium sulfate, 20% PEG 8000, 0.1 m sodium cacodylate (pH 6.5), 20 mg/ml LPHase, and 20 mm laminaripentaose. The complex crystals were characterized as space group P212121, with unit cell dimensions a = 45.98, b = 60.35, c = 149.12, and one molecule per asymmetric unit.
Construction of Mutants
Mutagenesis was performed by the QuikChange method (Stratagene Co.), with the pRSET_lph vector as template and the following oligonucleotides as primers: E154Q, 5′-CTTCAACTGGTCCCAGTACACGCTCAACG-3′ and 5′-CGTTGAGCGTGTACTGGGACCAGTTGAAG-3′; E154D, 5′-CTTCAACTGGTCCGACTACACGCTCAACG-3′ and 5′-CGTTGAGCGTGTAGTCGGACCAGTTGAAG-3′; D170N, 5′-CAGTACGCAGGTCAACATGTTCTCAGCTC-3′ and 5′-GAGCTGAGAACATGTTGACCTGCGTACTG-3′; D170E, 5′-CAGTACGCAGGTCGAGATGTTCTCAGCTC-3′ and 5′-GAGCTGAGAACATCTCGACCTGCGTACTG-3′ (underlining shows the location of the mutation). All mutations were confirmed by DNA sequencing. For those mutations creating significant losses in enzymatic activity, the entire mutated genes were sequenced to confirm that only the intended mutations had occurred.
Activity Assays
For preparation of gel-form curdlan, 5 g of curdlan was dissolved in 150 ml phosphate buffer (100 mm, pH 10.5) and stirred at room temperature for 30 min. The solution was neutralized by 0.1 n HCl. The precipitant, collected by centrifugation, was then resuspended in 100 ml of water and heated at 70 °C for 2 h. The resulting solution was subjected to centrifugation to obtain the precipitant, which was then resuspended in 200 ml of phosphate buffer (50 mm, pH 7.5) to form 2% (w/v) gel-form curdlan solution for subsequent assay. The activity of LPHase was analyzed by determining the amount of reducing ends of sugars using 3,5-dinitrosalicylic acid (DNS) method (31). In brief, an appropriate amount of enzyme (0.2 ml) was incubated with curdlan (0.3 ml, 2%, pH 7.5) at 37 °C for 2 h, followed by adding 0.5 ml of 3,5-dinitrosalicylic acid reagent to stop the reaction. The mixture was boiled for 15 min, chilled, and centrifuged to separate the precipitates. The resulting adducts were measured spectrophotometrically at 540 nm. The absorption coefficient of the resulting adducts was determined to be 780 m−1·cm−1 when d-glucose was used as reducing sugar.
X-ray Data Collection and Processing
Prior to data collection, crystals were dipped into Fomblin® cryoprotectant oil for several seconds and then flash-frozen in a liquid nitrogen stream. All protein crystals were initially screened and characterized using an RU-300 rotating-anode x-ray generator (Rigaku/MSC Inc.) at the Macromolecular x-ray Crystallographic Laboratory of National Tsing Hua University, Taiwan. The 1.62 Å native dataset and the 1.80 Å liganded dataset were collected on the SPXF beamline BL13C1 and BL13B1, respectively, at the National Synchrotron Radiation Research Center in Taiwan using an ADSC Quantum 4R CCD detector. The multi-wavelength anomalous dispersion (MAD) data set was collected from a single Se-Met-LPHase crystal on the BL12B2 Taiwan beamline at SPring-8 in Japan using an ADSC Quantum 4R CCD detector. Data were collected at three wavelengths: peak (0.9794 Å), inflection of selenium K-edge (0.9796 Å), and remote (0.9642 Å). All data sets were indexed, integrated, and scaled using HKL-2000 (32). Data collection statistics are shown in Table 1.
TABLE 1.
X-ray data and refinement statistics
| Data collection and phasing | |||||
| Data set | Native LPHase | SeMet-LPHase (MAD) |
LPHase Complex | ||
| Peak | Edge | High remote | |||
| Beamline | NSRRC_BL13C1 | JASRI_SPring-8_BL12B2 |
NSRRC_BL13B1 | ||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | |||||
| a (Å) | 46.16 | 46.27 | 46.27 | 46.26 | 45.98 |
| b (Å) | 60.68 | 60.36 | 60.36 | 60.36 | 60.35 |
| c (Å) | 149.40 | 150.13 | 150.13 | 150.11 | 149.12 |
| Wavelength (Å) | 0.9762 | 0.9794 | 0.9796 | 0.9642 | 0.9795 |
| Resolution (Å) | 30.00-1.62 | 30.00-2.29 | 30.00-2.30 | 30.00-2.26 | 30.00-1.80 |
| Highest resolution shell (Å) | 1.68-1.62 | 2.37-2.29 | 2.38-2.30 | 2.34-2.26 | 1.86-1.80 |
| Completeness (%)a | 100.0 (100.0) | 98.5 (95.4) | 98.7 (98.1) | 98.4 (96.2) | 97.0 (89.2) |
| Average I/σ (I)a | 43.9 (8.4) | 36.4 (34.0) | 38.1 (35.1) | 41.0 (34.1) | 41.7 (19.5) |
| No. of unique reflections | 54,410 | 19,889 | 19,770 | 20,788 | 39,457 |
| Redundancya | 14.0 (12.3) | 10.3 (9.2) | 10.3 (9.3) | 10.1 (9.0) | 8.3 (8.2) |
| Rmerge (%)a,b | 6.0 (33.6) | 5.1 (6.7) | 4.9 (6.2) | 4.8 (6.1) | 4.0 (11.7) |
| Overall figure of meritc | 0.87 | ||||
| Solvent content (%) | 42.2 | 42.1 | |||
| Refinement | |||||
| Resolution range (Å) | 30.00–1.62 | 30.00–1.80 | |||
| Number of atoms | |||||
| Protein | 2757 | 2757 | |||
| Solvent | 730 | 652 | |||
| Average B-factor (Å2) | 17.1 | 18.7 | |||
| R factord | 0.179 | 0.166 | |||
| Rfreee | 0.218 | 0.217 | |||
| R.m.s.d. bond lengths (Å)f | 0.010 | 0.014 | |||
| R.m.s.d. bond angles (°)f | 1.323 | 1.185 | |||
| Estimated coordinate error (Å) | 0.053 | 0.071 | |||
| Ramachandran analysis (%)g | |||||
| Favored/Allowed/Generous/Disallowed | 89.2/10.4/0.0/0.3 | 89.6/9.7/0.3/0.3 | |||
a Values in parentheses refer to statistics in the highest-resolution shell.
b Rmerge = Σ|Iobs − <I>|/ΣIobs.
c Figure of merit = |Fbest|/|F|.
d r = Σ|Fobs − Fcalc|/ ΣFobs, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
e Rfree was computed using 5% of the data assigned randomly.
f R.m.s.d., root mean square deviation.
g Estimated standard uncertainties based on maximum likelihood.
Structure Determination and Refinement
The apo-LPHase structure was solved by the MAD method. Four selenium atom sites were located with the program PHENIX (33). PHENIX was further used for phasing, density modification, and automated model building. The final model included 98.6% of the residues. Five residues (Met35, Ala36, Asn314, Asp315, and Gln316) were not built into the model because of the negative or broken density map. 2Fo − Fc and Fo − Fc maps were produced and inspected manually using the program “O”, version 9.0.7 (34). Crystallographic refinement was carried out using the maximum likelihood target function embedded in program REFMAC5 (35, 36). The apo-LPHase model (1.62 Å) was obtained using MOLREP (35, 37) and refined using REFMAC5 (35) coupled to ARP/wARP (38), which was used to add water molecules automatically.
The LPHase complex structure was determined by molecular replacement methods using the apo-LPHase structure as a search model by MOLREP and refined by REFMAC5 coupled to ARP/wARP. The apo and complex LPHase models were validated using the program PROCHECK (39). The final models of the apo and liganded LPHase had R values of 17.9% (Rfree = 21.8%) and 16.6% (Rfree = 21.7%), respectively. Refinement statistics of the final models are given in Table 1.
Structural Comparisons and Modeling
The structure of LPHase was compared with all protein structures available in the DALI server. Structural comparisons with the allergenic thaumatin-like protein (PDB code: 1Z3Q; Ref. 23), and the sweet protein, thaumatin (PDB code: 1THW, Ref. 22), were carried out using the program LSQMAN in O (34) to superimpose Cα atoms. Combined sequence and secondary structure alignments and figure preparation were done with the program ESPript (40). Structural figures were prepared with the program PyMol. To build the LPHase-substrate complex, the substrate laminarihexaose was docked using the program GOLD, version 2.1 (CCDC Software Limited, Cambridge, UK) (41). The x-ray structure of the LPHase complex was chosen as the template for docking study. The binding pocket for the docking study was defined as a 10 Å radius sphere centered on the Oϵ2 atom of Glu154. The three glucose moieties from the reducing end of laminarihexaose were manually placed at the site surrounding by Glu154, Thr156, and Thr167 according to the tetraose seen in the LPHase-tetraose structure, and the glycosidic oxygen was positioned between Glu154 and Asp170. The default parameters of GOLD were used. The three best solutions were obtained until a root mean square tolerance of 1.5 Å. This complex model was subjected to energy minimization using the Tripos force field (42) with the SYBYL 8.0 program (The Tripos Associates, St. Louis, MO).
RESULTS AND DISCUSSION
Structure Description
The 1.62 Å resolution electron-density map of apo LPHase reveals one molecule per asymmetric unit. The main-chain and side-chain atoms are well defined except for two segments having negative or weak density: the Met35-Ala36 segment and a loop region (residues 314–316). Only one residue, Arg308, lies in the disallowed region of the Ramachandran plot, despite its low B factor. The unusual geometry of this region is stabilized by three hydrogen bonds: Arg308 O···N Leu310, Arg308 O···N Asp311, and Arg308 O···Oδ1 Asp311.
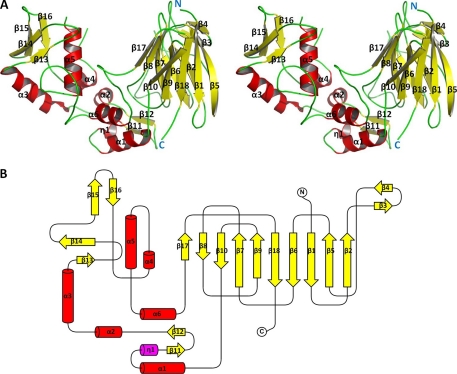
The final model was refined to an R value of 17.9% (Rfree = 21.8%). The mean B-factor of protein atoms is 17.1 Å2 (Table 1). The apo-LPHase crystal structure is composed of two domains, a barrel domain and a mixed (α/β) domain (Fig. 1, A and B). The barrel domain starts from the N terminus and consists of 10 consecutive β-strands (β1-β10). Two additional strands from the C-terminal region (β17-β18) join up, together assembling into a barrel structure. The mixed (α/β) domain contains six helices packed inside. A β-sheet (β13-β16) covers one exterior face of the α/β domain, and a β11 β12 sheet occupies the other side of the α/β domain. Along with three helices (α1, α4, and α6), this region interacts with residues from one end of the barrel domain, forming a crescent-like architecture.
FIGURE 1.
Structure of LPHase. A, stereo view of the LPHase crystal structure. N and C termini are labeled in blue as N or C, and α-helices and β-sheets are labeled with α or β and numbered. B, topology of LPHase. β-sheets (yellow) and α-helices (red) are numbered.
Crystal Structure of LPHase Complex
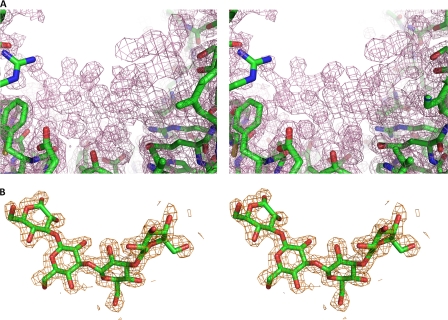
To obtain liganded structures, extensive crystallization trials were attempted in the presence of various concentrations of laminaripentaose. The complex structure was only obtained by the co-crystallization method in a solution containing 20 mm laminaripentaose. The structure was solved by the molecular replacement method. Residues that could not be defined include Met35, Ala36, and a loop region (314–316). As seen in Fig. 2, the electron density map clearly shows a large piece of non-peptide electron density that consists of several rings inside the groove of the crescent shape LPHase. The final complex model was refined to an R value of 16.6% (Rfree = 21.7%) (Table 1). Only four glucoses (laminaritetraose) that are linked by three β-1,3 glycosidic bonds could be built into this density. The absence of the density for the fifth glucose moiety in the pentaose may be due to its flexibility in this region. The other possibility is that laminaripentaose was slowly cleaved by LPHase during the crystallization process. In support of this view, we found that LPHase had slight residual activity toward laminaripentaose, which produced tetraose in an overnight reaction (data not shown).
FIGURE 2.
The electron density map of LPHase-complex structural models. A, stereo view of the initial Fo − Fc electron density map of the groove region, contoured at the 1.0-σ level. B, stereo view of the 2 Fo − Fc electron density map, contoured at the 1.0-σ level, shows the bound laminaritetraose. Carbon, oxygen, and nitrogen atoms are colored green, red, and blue, respectively.
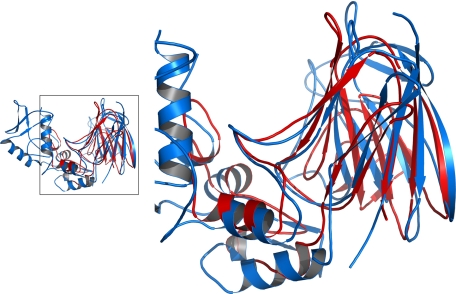
The apo and liganded LPHases share the two-domain crescent architecture. Superposition of the structures reveals a root mean square deviation of 0.24 Å for the Cα atoms, demonstrating an overall identical conformation. Based on analysis with DALI (21), there is no strong structural homology with any GH family member. Only structures of thaumatin (22) or thaumatin-like protein (23) show similarity with a partial region of LPHase, particularly the barrel domain (Fig. 3). These proteins are noted to present the surface with an electronegative cleft, implicating a possible evolutionary relationship.
FIGURE 3.
Structural comparison of LPHase with thaumatin. The structures of LPHase and thaumatin-like protein (PDB code: 1Z3Q) are superimposed and plotted in blue, and red, respectively. The framed region is magnified in the right panel.
The Binding Site
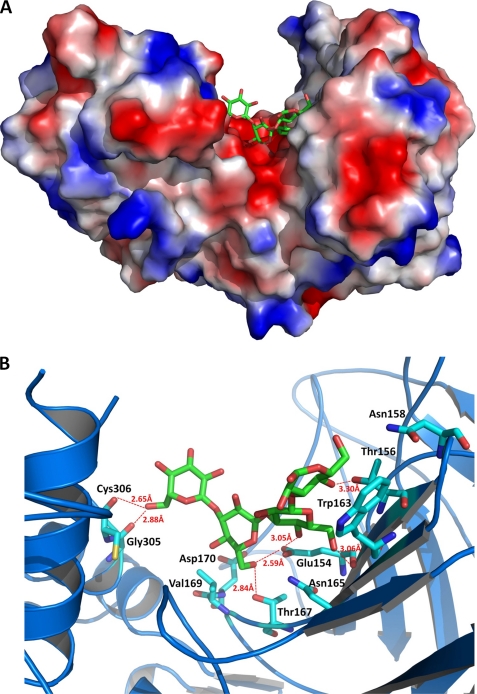
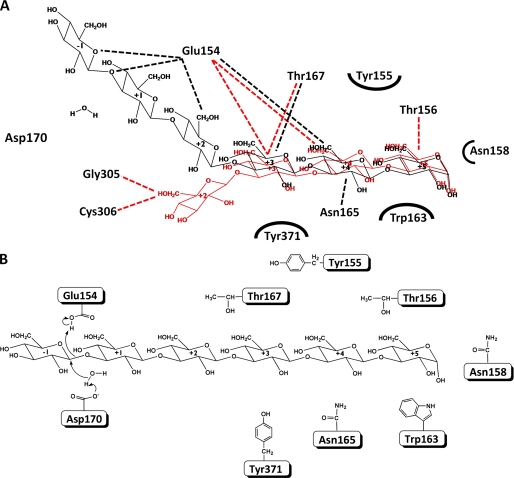
Based on the complex structure, laminaritetraose is situated at a negatively charged cleft of the wide groove between the β-barrel and mixed α/β domains (Fig. 4A). A number of water molecules are observed with or without laminaritetraose within the groove. The first three glucoses (Glc1-Glc2-Glc3) at the reducing end of laminaritetraose interact with a few residues (Glu154, Thr156, Asn158, Trp163, Asn165, Thr167, and Val169) of the barrel domain (Fig. 4B). Glu154 and Thr156 from β7 as well as Thr167 from the β8-β9 loop form hydrogen bonds with O4 or O6 of Glc1, Glc2, and Glc3 (≤3.6 Å) (Glc1 O4···Oγ1 Thr156, Glc2 O4···Oϵ1 Glu154, Glc2 O6···O Glu154, Glc3 O6···Oϵ1 Glu154, and Glc3 O6···Oγ1 Thr167). The outward side chain of Trp163 makes several contacts with Glc1 including three polar interactions (Glc1 O1···Nϵ1 Trp163: 4.30 Å; Glc1 O2···Nϵ1 Trp163: 4.38 Å; Glc1 O5···Nϵ1 Trp163: 4.01 Å). The side chain of Tyr371 from the mixed α/β domain also makes contacts with Glc3 including a polar interaction (Glc3 O5···O Tyr371: 4.23 Å). Glc4, the last moiety, dangles from the barrel domain and faces toward the α4 region of the α/β domain. The farthest O6 atom of Glc4 forms a hydrogen bond with the main-chain O atoms of Gly305 and Cys306, respectively (Glc4 O6···O Gly305 and Glc4 O6···O Cys306).
FIGURE 4.
A, molecular surface of the LPHase·laminaritetraose complex. This figure was prepared with the program PyMol. B, binding pocket of the LPHase·laminaritetraose complex. The bound laminaritetraose is drawn as green sticks, whereas ligand-binding residues of LPHase are shown as cyan sticks. Oxygen, nitrogen, and sulfur atoms are colored red, blue, and yellow, respectively.
Two strictly conserved carboxylates, Glu154 and Asp170 lie at the center of the cleft and a little to the right of the barrel axis and are in the vicinity of laminaritetraose (Fig. 4B); as described above, Glu154 interacts extensively with laminaritetraose including three hydrogen bonds with Glc2 (O4 and O6) and Glc3 (O6), and Asp170, despite being more distant from laminaritetraose (Glc3 O6···Oδ2 Asp170: 7.03 Å; Glc3 O4···Oδ1 Asp170: 6.69 Å), is surrounded by several water molecules. Notably, two waters are situated between laminaritetraose and Asp170 (Asp170 Oδ2···O Wat236: 2.62 Å; Asp170 Oδ1···O Wat456: 2.40 Å). Tyr232 that is nearby Asp170 (Tyr232 Oη···Oδ1 Asp170: 3.77 Å) also interacts with Wat456 (Tyr232 Oη···O Wat456: 2.93 Å). Glu154 and Asp170 adopt very similar conformations in the apo and liganded forms, in which the distance of the side chains is in the range 5.8–8.2 Å, prospectively acting as the catalysts for this LPHase because inverting enzymes require a proton donor and a base, respectively (1).
In parallel with the structural analysis, site-directed mutagenesis studies of Glu154 and Asp170 were carried out. Enzymatic analysis on curdlan showed no detectable activity for E154Q and D170N, suggesting that the presence of the carboxyl moiety is crucial at these sites. We next evaluated the substitution with the other carboxylate for residues 154 and 170, respectively. E154D showed significantly lower relative activity (19%) as compared with the wild-type enzyme, while there was 81% relative activity for D170E. These results suggest the importance of these two carboxylates and that a shorter side chain greatly reduced the catalytic power even with the same carboxyl moiety at residue 154.
The LPHase-Laminarihexaose Model
The topologies of the active sites in the GH superfamily can be classified into three general categories: pocket, tunnel, and cleft (1, 24). The structure of LPHase shows a wide cleft topology, which is an “open” structural feature, allowing binding of polysaccharide substrates and presumably performing an endo-cleavage reaction.
To understand the catalytic mechanism, we performed a computer simulation to model an enzyme-laminarihexaose structure. The three glucose moieties from the reducing end of laminarihexaose are placed at the site surrounded by Glu154, Thr156, and Thr167 according to the sugar seen in the LPHase-tetraose structure. Assuming that Glu154 and Asp170 mediate catalysis, the glycosidic oxygen will be positioned between these two residues, in which protonation of the glycosidic oxygen by the catalytic acid residue is followed by the nucleophilic attack by water molecule, which is depronoated and activated by the general base. This mechanism would ensure that hydrolysis yields an inverted laminaripentaose structure that can be released from the active site (1, 15). The best models we obtained had the following features (Fig. 5A): (i) only Glu154 is within hydrogen-bonding distance of the glycosidic oxygen (1.94 Å), suggesting its role as the proton donor; (ii) Asp170 is relatively more distant to the anomeric carbon (∼6 Å), allowing the accommodation of a water molecule between Asp170 and the sugar, thus ideally serving as the basic catalyst; (iii) the protruding side chains of Thr156, Asn158, and Trp163 demarcate subsite +5; and (iv) Glu154 and Thr167 form hydrogen bonds with the substrate at subsites +4 and +3, respectively.
FIGURE 5.
A, schematic representation of LPHase·laminarihexaose and LPHase·laminaritetraose interactions. The docked laminarihexaose (black) and laminaritetraose (red) are superimposed and drawn as sticks. The hydrogen-bonding interactions are shown as broken lines. Asp170 that is relatively more distant to the anomeric carbon (∼6 Å) of the docked laminarihexaose interacts with laminarihexaose/laminaritetraose via water molecules. B, scheme of the proposed catalysis of LPHase.
The Catalysis and Substrate Binding Topology
Our structural analyses and modeling suggest a catalytic mechanism for LPHase whereby Glu154 acts as a proton donor to the glycosidic oxygen, and then Asp170 facilitates a base-assisted nucleophilic attack by a water molecule from the opposite side of the sugar ring (Fig. 5B). When a long-chain polysaccharide β-1,3-glucan such as laminarin diffuses into the groove, the glucan is positioned with the reducing end at the +5 subsite, consisting of Thr156, Asn158, and Trp163, and then is bound by the negatively charged region via interactions with several residues including Glu154 and Thr167. Hydrolysis of a β-glycosidic bond from the reducing end of sugar chain by Glu154 and Asp170 thus creates a laminaripentaose product with the α-anomeric configuration. By use of methyl laminarihexaoside and methyl laminariheptaoside as substrate, respectively, methyl laminaripentaoside was indeed detected after enzymatic digestion (data not shown), confirming that the hydrolysis of sugar chain is from the reducing end. A striking feature of LPHase is its unique product specificity that predominantly releases pentaose as product throughout the enzymatic digestion of curdlan, exhibiting an exo-type cleavage manner. Yet, LPHase structure shows a wide groove, which is commonly observed among endo-type GHs. This puzzle remains unresolved even with the LPHase-tetraose complex structure. Early studies on curdlan revealed that the long-chain curdlan forms more complex tertiary structures due to intramolecular and intermolecular hydrogen bonding interactions; the simplest β-1,3-glucan (or curdlan) usually adopts a triple-stranded helical structure in nature (25–27). Treatment of the helical curdlan at high temperature or with alkaline conditions, it rendered single, double, or triple helical structures at its gel form (26, 28). It is thus speculated that the higher-order structure of curdlan may play an important role in binding to LPHase, hence contributing to its product specificity during the catalysis. As shown in the LPHase-tetraose complex structure (Fig. 4) and the simulated laminarihexaose-containing structure (Fig. 5A), the sugar chains are observed to form a distinct kink to fit into the binding site, suggesting that the tertiary structure of curdlan may be a key factor for substrate recognition. The unusually wide groove in LPHase structure also discloses the possibility for a helical curdlan to bind with and consequently results in an exo-type cleavage.
In conclusion, we have determined the structure of LPHase in its apo and liganded forms. The LPHase structure consists of a barrel domain and a mixed (α/β) domain. The most prominent structural feature of the crescent-like LPHase is a wide groove with a predominantly electronegative charge running between the two domains. Analysis of the apo and complex LPHase structures demonstrates a relatively rigid catalytic framework consisting of the β7-β9 region in the barrel domain including two strictly conserved carboxylates (Glu154 and Asp170) and other saccharide-linked residues (Thr156, Thr167, Trp163, Asn165, and Val169). A plausible direct displacement mechanism that involves Glu154 and Asp170 as catalysts is thus proposed to yield a pentaose product with the inverted anomeric configuration.
Acknowledgments
We thank the Macromolecular X-Ray Crystallographic Center of NTHU Instrument Center at Hsinchu, National Tsing Hua University, Taiwan; BL13B1 and BL13C1 beamlines at the National Synchrotron Radiation Research Center (NSRRC), Hsinchu, Taiwan; BL12B2 Taiwan beamline at SPring-8, Japan for access to facilities for data collection. We also thank the staff at NSRRC for their excellent support.
This work was supported, in part, by grants from the National Science Council (NSC 94-2311-B-009-001, NSC96-2313-B-007-001, and NSC97-3112-B-007-005), Taiwan.
The atomic coordinates and structure factors (codes 3GD0 and 3GD9) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GH
- glycoside hydrolase
- LPHase
- laminaripentaose-producing β-1,3-glucanase
- PEG
- polyethyleneglycol
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Davies G., Henrissat B. (1995) Structure 3, 853–859 [DOI] [PubMed] [Google Scholar]
- 2.Colman P. M. (1994) Protein Sci. 3, 1687–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinnott M. L. (1990) Chem. Rev. 90, 1171–1202 [Google Scholar]
- 4.Henrissat B. (1991) Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrissat B., Bairoch A. (1993) Biochem. J. 293, 781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrissat B., Davies G. (1997) Curr. Opin. Struct. Biol. 7, 637–644 [DOI] [PubMed] [Google Scholar]
- 7.Henrissat B., Coutinho P. M. (1999) Carbohydrate-Active Enzymes Server [Google Scholar]
- 8.Bohn J. A., BeMiller J. N. (1995) Carbohydr. Polymer 28, 3–14 [Google Scholar]
- 9.Varghese J. N., Hrmova M., Fincher G. B. (1999) Structure 7, 179–190 [DOI] [PubMed] [Google Scholar]
- 10.Varghese J. N., Garrett T. P., Colman P. M., Chen L., Høj P. B., Fincher G. B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 2785–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fibriansah G., Masuda S., Koizumi N., Nakamura S., Kumasaka T. (2007) Proteins 69, 683–690 [DOI] [PubMed] [Google Scholar]
- 12.Receveur-Bréchot V., Czjzek M., Barre A., Roussel A., Peumans W. J., Van Damme E. J., Rougé P. (2006) Proteins 63, 235–242 [DOI] [PubMed] [Google Scholar]
- 13.Ishida T., Fushinobu S., Kawai R., Kitaoka M., Igarashi K., Samejima M. (2009) J. Biol. Chem. 284, 10100–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakabayashi M., Nishijima T., Ehara G., Nikaidou N., Nishihashi H., Watanabe T. (1998) J. Ferment. Bioeng. 85, 459–464 [Google Scholar]
- 15.Nishimura T., Bignon C., Allouch J., Czjzek M., Darbon H., Watanabe T., Henrissat B. (2001) FEBS Lett. 499, 187–190 [DOI] [PubMed] [Google Scholar]
- 16.Wasser S. P. (2002) Appl. Microbiol. Biotechnol. 60, 258–274 [DOI] [PubMed] [Google Scholar]
- 17.Zeković D. B., Kwiatkowski S., Vrvić M. M., Jakovljević D., Moran C. A. (2005) Crit. Rev. Biotechnol. 25, 205–230 [DOI] [PubMed] [Google Scholar]
- 18.Day A. G., Withers S. G. (1985) Biochem. Biophys. Res. Commun. 133, 628–632 [DOI] [PubMed] [Google Scholar]
- 19.Fukuda K., Hiraga M., Asakuma S., Arai I., Sekikawa M., Urashima T. (2008) Biosci. Biotechnol. Biochem. 72, 3107–3113 [DOI] [PubMed] [Google Scholar]
- 20.Fontaine T., Hartland R. P., Diaquin M., Simenel C., Latgé J. P. (1997) J. Bacteriol. 179, 3154–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bahar I., Atilgan A. R., Erman B. (1997) Fold Des. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 22.Ko T. P., Day J., Greenwood A., McPherson A. (1994) Acta Crystallogr. D Biol Crystallogr. 50, 813–825 [DOI] [PubMed] [Google Scholar]
- 23.Leone P., Menu-Bouaouiche L., Peumans W. J., Payan F., Barre A., Roussel A., Van Damme E. J., Rougé P. (2006) Biochimie 88, 45–52 [DOI] [PubMed] [Google Scholar]
- 24.Henrissat B., Callebaut I., Fabrega S., Lehn P., Mornon J. P., Davies G. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7090–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atkins E. D. T., Parker K. D. (1968) Nature 220, 784–785 [Google Scholar]
- 26.Deslandes Y., Marchessault R. H., Sarko A. (1980) Macromolecules 13, 1466–1471 [Google Scholar]
- 27.Bluhm T. L., Sarko A. (1977) Can. J. Chem. 55, 293–299 [Google Scholar]
- 28.Saitô H., Ohki T., Sasaki T. (1977) Biochemistry 16, 908–914 [DOI] [PubMed] [Google Scholar]
- 29.Yang W., Hendrickson W. A., Kalman E. T., Crouch R. J. (1990) J. Biol. Chem. 265, 13553–13559 [PubMed] [Google Scholar]
- 30.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 31.Miller G. L. (1959) Anal. Chem. 31, 426–429 [Google Scholar]
- 32.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 34.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 35.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 37.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 38.Lamzin V. S., Wilson K. S. (1993) Acta Crystallogr. D Biol. Crystallogr. 49, 129–147 [DOI] [PubMed] [Google Scholar]
- 39.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 40.Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 41.Raghuraman A., Mosier P. D., Desai U. R. (2006) J. Med. Chem. 49, 3553–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark M., Cramer R. D., III, Van Opdenbosch N. (1989) J. Comp. Chem. 10, 982–1012 [Google Scholar]