Abstract
The telosome/shelterin, a six-protein complex formed by TRF1, TRF2, RAP1, TIN2, POT1, and TPP1, functions as the core of the telomere interactome, acting as the molecular platform for the assembly of higher order complexes and coordinating cross-talks between various protein subcomplexes. Within the telosome, there are two oligonucleotide- or oligosaccharide-binding (OB) fold-containing proteins, TPP1 and POT1. They can form heterodimers that bind to the telomeric single-stranded DNA, an activity that is central for telomere end capping and telomerase recruitment. Through proteomic analyses, we found that in addition to POT1, TPP1 can associate with another OB fold-containing protein, OBFC1/AAF44. The yeast homolog of OBFC1 is Stn1, which plays a critical role in telomere regulation. We show here that OBFC1/AAF44 can localize to telomeres in human cells and bind to telomeric single-stranded DNA in vitro. Furthermore, overexpression of an OBFC1 mutant resulted in elongated telomeres in human cells, implicating OBFC1/AAF4 in telomere length regulation. Taken together, our studies suggest that OBFC1/AAF44 represents a new player in the telomere interactome for telomere maintenance.
Telomeres are specialized linear chromosome end structures, which are regulated and protected by networks of protein complexes (1–4). Telomere length, structure, and integrity are critical for the cells and the organism as a whole. Telomere dysregulation can lead to DNA damage response, cell cycle checkpoint, genome instability, and predisposition to cancer (5–9). Mammalian telomeres are composed of double-stranded (TTAGGG)n repeats followed by 3′-single-stranded overhangs (10). In addition to the telomerase that directly mediates the addition of telomere repeats to the end of chromosomes (11, 12), a multitude of telomere-specific proteins have been identified that form the telosome/shelterin complex and participate in telomere maintenance (9, 13). The telosome in turn acts as the platform onto which higher order telomere regulatory complexes may be assembled into the telomere interactome (14). The telomere interactome has been proposed to integrate the complex and labyrinthine network of protein signaling pathways involved in DNA damage response, cell cycle checkpoint, and chromosomal end maintenance and protection for telomere homeostasis and genome stability.
Of the six telomeric proteins (TRF1, TRF2, RAP1, TIN2, POT1, and TPP1) that make up the telosome, TRF1 and TRF2 have been shown to bind telomeric double-stranded DNA (15, 16), whereas the OB3 fold-containing protein POT1 exhibits high affinities for telomeric ssDNA in vitro (17, 18). Although the OB fold of TPP1 does not show appreciable ssDNA binding activity, heterodimerization of TPP1 and POT1 enhances the POT1 ssDNA binding (17, 18). More importantly, POT1 depends on TPP1 for telomere recruitment, and the POT1-TPP1 heterodimer functions in telomere end protection and telomerase recruitment. Notably, the OB fold of TPP1 is critical for the recruitment of the telomerase (18). Disruption of POT1-TPP1 interaction by dominant negative inhibition, RNA interference, or gene targeting could lead to dysregulation of telomere length as well DNA damage responses at the telomeres (18–21).
In budding yeast, the homolog of mammalian POT1, Cdc13, has been shown to interact with two other OB fold-containing proteins, Stn1 and Ten1, to form a Cdc13-Stn1-Ten1 (CST) complex (22, 23). The CST complex participates in both telomere length control and telomere end capping (22, 23). The presence of multiple OB fold-containing proteins from yeast to human suggests a common theme for telomere ssDNA protection (4). Indeed, it has been proposed that the CST complex is structurally analogous to the replication factor A complex and may in fact function as a telomere-specific replication factor A complex (23). Notably, homologs of the CST complex have been found in other species such as Arabidopsis (24), further supporting the notion that multiple OB fold proteins may be involved in evolutionarily conserved mechanisms for telomere end protection and length regulation. It remains to be determined whether the CST complex exists in mammals.
Although the circuitry of interactions among telosome components has been well documented and studied, how core telosome subunits such as TPP1 help to coordinate the cross-talks between telomere-specific signaling pathways and other cellular networks remains unclear. To this end, we carried out large scale immunoprecipitations and mass spectrometry analysis of the TPP1 protein complexes in mammalian cells. Through these studies, we identified OB fold-containing protein 1 (OBFC1) as a new TPP1-associated protein. OBFC1 is also known as α-accessory factor AAF44 (36). Sequence alignment analysis indicates that OBFC1 is a homolog of the yeast Stn1 protein (25). Further biochemical and cellular studies demonstrate the association of OBFC1 with TPP1 in live cells. Moreover, we showed that OBFC1 bound to telomeric ssDNA and localized to telomeres in mammalian cells. Dominant expression of an OBFC1 mutant led to telomere length dysregulation, indicating that OBFC1 is a novel telomere-associated OB fold protein functioning in telomere length regulation.
MATERIALS AND METHODS
Preparation of Nuclear Extracts
AB2.2 mouse embryonic stem (ES) cells (kindly provided by the Darwin Core facility, Baylor College of Medicine, Houston, TX) were grown in ES medium with 15% fetal bovine serum and leukemia inhibitory factor (26). ES cells (1–2 × 1010) were washed in ice-cold phosphate-buffered saline and resuspended in hypotonic buffer (10 mm Tris (pH = 7.3), 10 mm KCl, 1.5 mm MgCl2, 0.2 mm phenylmethylsulfonyl fluoride, and 10 mm 2-mercaptoethanol). The cells were then homogenized until cell lysis reached ∼80%. The lysates were centrifuged at 25,000 × g for 10 min at 4 °C, and the pellets were resuspended in low salt buffer (one-half volume) (20 mm KCl, 20 mm Tris (pH = 7.3), 25% glycerol, 1.5 mm MgCl2, and 0.2 mm EDTA). An equal volume of high salt buffer (1.2 m KCl, 20 mm Tris (pH = 7.3), 25% glycerol, 1.5 mm MgCl2, and 0.2 mm EDTA) was added subsequently and mixed for 30 min at 4 °C. The mixture was centrifuged at 100,000 × g for 30 min, and the supernatant was dialyzed in BC100 buffer (20 mm Tris (pH = 7.3), 20% glycerol, 0.1 m KCl, 0.2 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, and 10 mm 2-mercaptoethanol) for 2 h followed by centrifugation at 100,000 × g for 30 min. Aliquots of cleared supernatant were then stored at −80 °C.
Affinity Purification and Mass Spectrometry
Nuclear extracts were incubated with anti-mTPP1 antibodies (20 μg) (Bethyl Laboratories) at 4 °C for 2 h. Protein A beads (20 μl) (Santa Cruz Biotechnology) were then added, and the mixtures were incubated at 4 °C for 1 h. The immunoprecipitates were washed four times with ice-cold NETN buffer (20 mm Tris (pH = 8.0), 100 mm NaCl, 1 mm EDTA, and 0.5% Nonidet P-40). The bound proteins were eluted in 2× Laemmli buffer, resolved by SDS-PAGE, and stained with Coomassie Blue. The lanes were sectioned and excised for sequencing on a liquid chromatography tandem mass spectrometer.
Telomere Restriction Fragment Assay (TRF Assay)
Sequences encoding full-length and mutant human OBFC1 (hOBFC1) were cloned into pBabe-based retroviral vectors that contained FLAG tag sequences. HTC75 cells stably expressing these constructs were generated by retroviral infection followed by puromycin selection. The selected cells were allowed to recover (designated as “P0”) and passaged for collection at various time points.
The TRF assay was performed as described previously (28). Briefly, genomic DNA was purified from the cells using the DNeasy kit (Qiagen), digested with HinF1 and Rsa1, resolved by agarose gel electrophoresis, and transferred to nylon membranes. Southern blotting was then performed using the radiolabeled telomeric probe (TTAGGG)3. The data were then analyzed using the Telorun analysis tool (29).
GST Protein Purification and Electrophoretic Mobility Shift Assay
Bacterially expressed GST-OBFC1 fusion proteins were purified using glutathione agarose beads and eluted in 10 mm reduced glutathione (Sigma) and 50 mm Tris (pH = 8.0). For electrophoretic mobility shift assays, different concentrations of purified GST-OBFC1 were incubated with radiolabeled (32P) probes (TelG31 and TelC31) at room temperature for 1 h followed by electrophoresis and autoradiography (18). The sequences used were: TelG31, 5′-GTTAGGGTTAGGGTTAGGGTTAGGGTTAGGG-3′, and TelC31, 5′-CCCTAACCCTAACCCTAACCCTAACCCTAAC-3′.
Co-immunoprecipitation, Western Blotting, and Immunostaining
Nuclear extracts from 108 mouse ES cells were used for co-immunoprecipitation and Western blotting analysis as described previously (28). U2OS cells were plated on poly-d-lysine-coated glass coverslips, fixed with 3% paraformaldehyde, and permeabilized in 0.5% Triton X-100 (in phosphate-buffered saline) buffer. Immunostaining analysis was then carried out as described in Ref. 28. Fluorescence microscopy was performed on a Nikon TE 200 microscope equipped with a Coolsnap camera.
Antibodies used are listed below. Rabbit polyclonal anti-mTPP1, anti-RAP1, and anti-OBFC1 antibodies were from Bethyl Laboratories. Rabbit monoclonal anti-GFP (green fluorescent protein) antibody was from Epitomics, and mouse monoclonal anti-GST antibodies were from Qiagen. Mouse monoclonal anti-FLAG antibody was from Sigma. Mouse monoclonal anti-OBFC1 antibody was from Abnova.
Bimolecular Fluorescence Complementation Assay (BiFC)
BiFC was performed essentially as described previously (26, 27). Briefly, the N-terminal domain (residues 1–155) of Venus YFP (YFPn) was fused to full-length TPP1 and OBFC1 and cloned into pBabe-based retroviral vectors. The C-terminal domain (residues 156–239) of YFP (YFPc) was fused to telosome subunits (TRF1, TRF2, TIN2, TPP1, POT1, and RAP1), full-length OBFC1, and the various deletion mutants of TPP1 and OBFC1 and also cloned into retroviral vectors. HTC75 or U2OS cells that stably co-express the various pairs of the constructs were subsequently generated by retroviral infection. The cells were allowed to recover and then analyzed by flow cytometry using a Guava PCA system (Guava Technologies) and microscopy.
RESULTS
Identification of a Novel Telomere-associated Protein, OBFC1
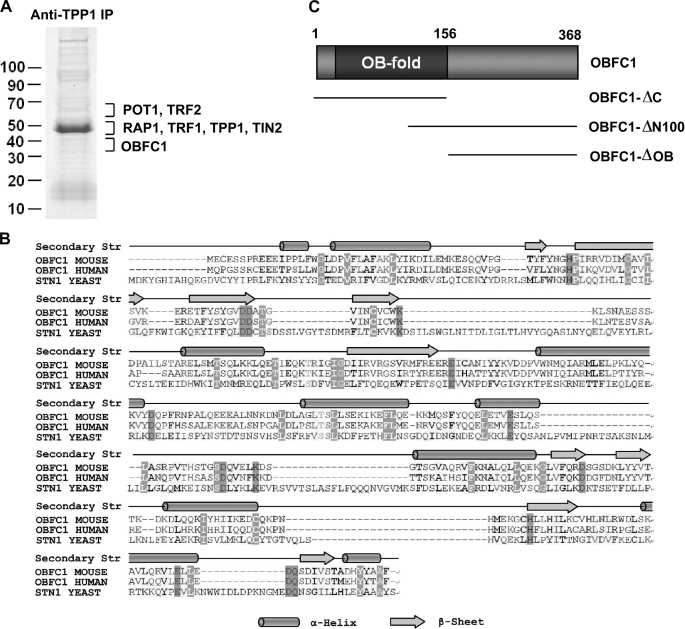
Protein complexes of telosome subunits have been examined in a number of mammalian somatic cell lines, the majority of which are tumor cell lines such as HeLa cells (13, 28, 30). We decided to examine the telomere protein complexes in mouse ES cells by large scale co-immunoprecipitation (IP) and mass spectrometry sequencing. Anti-TPP1 antibodies were used to affinity purify TPP1 and its associated proteins using nuclear extracts prepared from mouse ES cells. As shown in Fig. 1A, all the known core telomere complex proteins including TRF1, TRF2, TIN2, RAP1, TPP1, and POT1 were found in the TPP1 complex, indicating that anti-TPP1 immunoprecipitation successfully isolated the TPP1 complexes.
FIGURE 1.
Identification of the telomere-associated protein OBFC1/AAF44. A, anti-TPP1 antibodies were used for IP of the TPP1 protein complex using nuclear extracts from mouse ES cells. The IP products were resolved by SDS-PAGE and stained with Coomassie Blue. The lanes were sectioned for subsequent trypsin digestion, peptide extraction, and mass spectrometry sequencing. B, human and mouse OBFC1/AAF44 share sequence and structural homology with the yeast OB fold protein Stn1. The alignment was generated by ClustalX with manual editing. The secondary structure (Secondary Str) prediction above the alignment was based on the Jpred program. C, schematic diagram of OBFC1/AAF44 and its deletion mutants.
In addition, we identified a 40-kDa protein, OBFC1, that has not been previously linked to telomere regulation in mammalian cells. OBFC1 is also known as AAF44, the 44-kDa subunit of DNA polymerase α-primase (36). Sequence analysis revealed the presence of an OB fold in the N terminus of OBFC1/AAF44 and a helical domain in the C terminus that has no significant homology with other known motifs (Fig. 1, B and C). Sequence alignment indicates that the N-terminal two-thirds of OBFC1/AAF44 including the OB fold is most similar to the yeast telomere-associated OB fold protein Stn1 (Fig. 1C) (25). Stn1 is part of the CST complex that also includes Cdc13 and Ten1 and regulates telomere function (23, 31). The finding of OBFC1/AAF44 in the TPP1 complex and its homology to Stn1 suggest a potential role of OBFC1/AAF44 in telomere regulation.
OBFC1/AAF44 Associates with TPP1
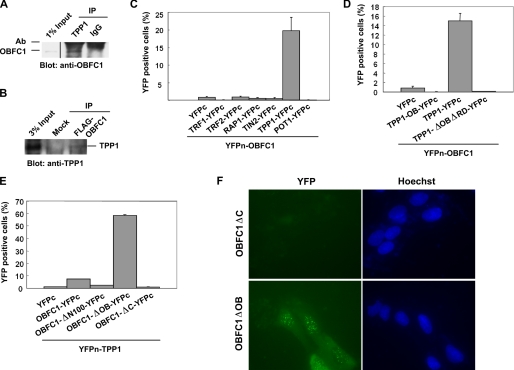
To confirm our large scale IP findings, we first carried out co-IP experiments in mouse ES cells. As shown in Fig. 2A, anti-TPP1 immunoprecipitation brought down endogenous OBFC1/AAF44 in mouse ES cells. Similarly, anti-FLAG immunoprecipitation pulled down endogenous TPP1 in human cells expressing FLAG-tagged OBFC1/AAF4 (Fig. 2B). However, the antibody for OBFC1/AAF44 cannot immunoprecipitate endogenous OBFC1/AAF44, thereby precluding reciprocal IPs. To further probe the interaction between human OBFC1 (hOBFC1) and TPP1 (hTPP1), we utilized the BiFC assay that enables examination of protein-protein interactions in live cells (32). In this assay, interactions between two proteins that are tagged respectively with N- or C-terminal YFP fragments would bring the YFP fragments to close proximity and allow fluorescence complementation to occur. OBFC1/AAF44 and the six telomeric proteins were first tagged respectively with the N-terminal half of Venus YFP (YFPn-OBFC1) and C-terminal domain of YFP (e.g. TPP1-YFPc). These proteins were then stably co-expressed in HTC75 cells for fluorescence complementation analysis. As shown in Fig. 2C, whereas ∼20% of YFPn-OBFC1 and YFPc-TPP1 co-expressing cells appeared YFP-positive as measured by flow cytometry, none of the other five telomeric proteins tested exhibited fluorescence complementation over background. These results suggest that the interaction between OBFC1/AAF44 and TPP1 is specific and that these two proteins are in close proximity to each other in vivo.
FIGURE 2.
OBFC1/AAF44 associates with TPP1. A, mouse ES cell nuclear extracts were immunoprecipitated with antibodies (Ab) against TPP1. Western blots were performed using anti-OBFC1 antibodies. Rabbit IgG was used as IP controls. B, nuclear extracts from parental or FLAG-OBFC1-expressing HeLa cells were immunoprecipitated with anti-FLAG antibodies. Western blots were performed using anti-TPP1 antibodies. C–E, bimolecular fluorescence complementation analysis of OBFC1/AAF44 and TPP1 interaction. HTC75 cells expressing the construct pairs as indicated were generated and analyzed by flow cytometry (26). OBFC1 was tagged on the N terminus with the YFPn fragment (YFPn-OBFC1) and co-expressed with YFPc-tagged telosome components (C) or the TPP1 full-length and deletion mutants (D). Similarly, YFPn-tagged TPP1 (YFPn-TPP1) was co-expressed with YFPc-tagged full-length OBFC1 and its truncation mutants (E). Error bars indicate S.E. (n ≥ 3). F, fluorescence microscopy images of live U2OS cells co-expressing YFPn-tagged TPP1 with YFPc-tagged OBFC1 deletion mutants. Nucleus was visualized by Hoechst dye staining.
We then went on to map the sequence requirement for OBFC1-TPP1 interaction and performed BiFC assays using different deletion mutants of OBFC1/AAF44 (Fig. 1C) and TPP1 (18). As shown in Fig. 2D, the OB fold or the C-terminal half of TPP1 alone appeared insufficient to mediate such interactions, indicating that the TPP1 middle domain or the full-length protein may be necessary for the interaction. In comparison, the C-terminal region (residues 101–368) of OBFC1/AAF44 appeared to be important for TPP1 association with OBFC1/AAF44 in this assay (Fig. 2E). Deletion of OBFC1 OB fold (OBFC1ΔOB) increased percentages of YFP-positive cells, raising the possibility that the OB region may inhibit TPP1-OBFC1 interaction. In U2OS cells, BiFC assay revealed punctate YFP signal in the nucleus of cells that co-expressed OBFC1ΔOB and TPP1 (Fig. 2F), a pattern similar to what we observed with TIN2-TPP1 interaction (27). These data suggest that TPP1 interacts with OBFC1/AAF44 through the helical domain of OBFC1/AAF44.
OBFC1/AAF44 Binds to Telomeric ssDNA and Localizes to Telomeres
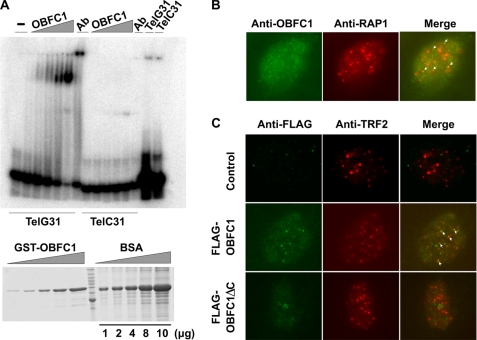
The yeast homolog of OBFC1/AAF44, Stn1, has been shown to bind telomeric ssDNA through its interactions with Cdc13 and Ten1 and localize on telomeres (22, 23). In mammalian cells, the OB folds of both TPP1 and POT1 are critical for their function. POT1 contains at least three OB folds. The two N-terminal OB folds mediate its interaction with ssDNA (33), whereas the C-terminal OB fold is involved in its interaction with TPP1 (28, 30). The OB fold of TPP1 does not appear to interact with ssDNA, but it is important for its interaction with the telomerase and telomere regulation (18). To understand OBFC1/AAF44 function and mode of action, we tested its ability to directly bind single-stranded telomeric sequences, using recombinant GST-tagged human OBFC1/AAF44 proteins purified from bacteria in mobility shift assays. Interestingly, we found that hOBFC1/AAF44 bound to the G-strand telomeric ssDNA in a dose-dependent manner (Fig. 3A). OBFC1/AAF44 also weakly bound the C-strand telomeric ssDNA. To further confirm that OBFC1/AAF44 proteins form a complex with telomeric ssDNA, we included anti-GST antibodies in the reaction mixtures containing GST-OBFC1 and ssDNA probes. As shown in Fig. 3A, the addition of anti-GST antibodies led to slower migration of the protein-DNA complex, indicating that OBFC1/AAF44 can bind to telomeric DNA.
FIGURE 3.
OBFC1/AAF44 binds to telomeric ssDNA in vitro and is localized to the telomeres in cells. A, different concentrations of bacterially expressed GST-OBFC1 fusion proteins (0.125, 0.25, 0.5, 1, and 2 μm) were incubated with radiolabeled telomeric G-strand (TelG31) or C-strand (TelC31) ssDNA probes. The reaction mixtures were resolved by non-denaturing PAGE and analyzed by autoradiography. For supershift experiments, anti-GST antibodies were added to the binding mixtures containing the highest concentration of GST-OBFC1 and ssDNA. The lower panel showed the relative amount of GST-OBFC1 used in the assay. The TelC31 probe was used as a negative control. BSA, bovine serum albumin. B, U2OS cells were permeabilized and co-stained with rabbit anti-RAP1 and mouse monoclonal anti-OBFC1 antibodies. C, the localization of FLAG-tagged OBFC1 and OBFC1ΔC was analyzed by immunostaining using anti-FLAG and anti-TRF2 antibodies.
The ability of OBFC1/AAF44 to associate with TPP1 and telomeric DNA entails that OBFC1/AAF44 may localize to the telomeres in mammalian cells. To determine the subcellular localization of OBFC1/AAF44, we carried out indirect immunofluorescence analysis in human cells. We first expressed YFP-tagged hOBFC1 in U2OS cells and examined its co-localization with RAP1. YFP-OBFC1 expression exhibited a punctate pattern that co-localizes with endogenous RAP1 staining (data not shown). Next, we examined the localization of endogenous OBFC1/AAF44 using an antibody generated against human OBFC1/AAF44 in U2OS cells. Again, we were able to consistently observe co-staining of endogenous OBFC1/AAF44 with RAP1 (Fig. 3B). These results collectively indicate that OBFC1/AAF44 can be targeted to the telomeres in human cells. Interestingly, deletion of the C-terminal helical domain of OBFC1 prevented telomere localization of OBFC1 (Fig. 3C), indicating that the OB fold of OBFC1 may not be sufficient for stable telomere localization of OBFC1.
Mutation of OBFC1/AAF44 Impacts Telomere Length Regulation
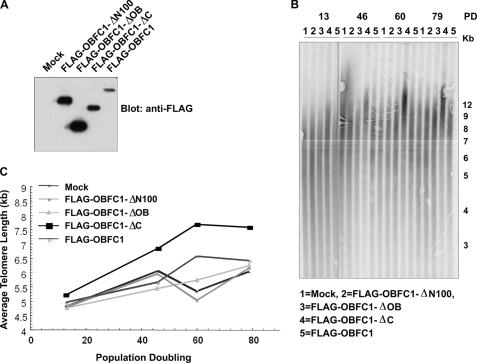
The telosome complex is important for telomere length regulation. Disruption of the interactions among telosome components can lead to dysregulated telomere length (4). Both TPP1 and the OBFC1/AAF44 yeast homolog Stn1 have been shown to be involved in telomere length regulation (25, 28). We reasoned that OBFC1/AAF44 might also play a role in telomere length maintenance. To test this hypothesis, we stably expressed full-length OBFC1 as well as OBFC1 deletion mutants in HTC75 cells (Fig. 4A) and measured the average telomere length over generations by TRF assays. As shown in Fig. 4, B and C, cells expressing the OBFC1 OB fold-only mutant (OBFC1ΔC) exhibited elongated telomeres over time when compared with control cells or cells expressing full-length OBFC1. This result suggests that OBFC1/AAF44 may play a role in telomere length regulation and that OBFC1/AAF44 is a novel telomere regulator.
FIGURE 4.
Expression of the OBFC1/AAF44 OB fold leads to elongated telomeres. A, expression of FLAG-tagged OBFC1 and its deletion mutants in HTC75 cells was examined by Western blotting. B and C, telomere restriction fragment length analysis of mock and FLAG-tagged full-length and mutant OBFC1-expressing cells. Kb, kilobases; PD, population doublings.
DISCUSSION
In this study, we identified the OB fold-containing protein OBFC1/AAF44 in the TPP1 protein complex. OBFC1/AAF44 is a homolog of yeast telomere regulator Stn1 (22, 25). Similar to its yeast homolog, we found that OBFC1/AAF44 bound to telomeric ssDNA in vitro and localized at telomeres in human cells, suggesting evolutionary conservation of telomere-associated OB fold proteins. TPP1 has been shown to interact with the telomerase and the telomere ssDNA-binding protein POT1 to control telomere homeostasis and integrity (17, 18). The association of OBFC1/AAF44 with TPP1 suggests its involvement in telomere regulation as well. Indeed, we found that ectopic expression of the OB fold of OBFC1/AAF44 in HTC75 cells led to telomere length increase over time.
How does OBFC1/AAF44 control telomere length? In budding yeast, Stn1 is known to interact with Cdc13 and Ten1 to form the CST complex for telomere maintenance (22, 23). Stn1 and Ten1 can also function independent of CDC13 in telomere capping (34). In line with the function of OBFC1/AAF44 in human cells, it has been shown that cells expressing the stn1 mutant (stn1–13) exhibit deregulation in telomere length control and abnormal increase of telomere length (25). One possibility is that endogenous OBFC1/AAF44 may act in negatively regulating telomerase recruitment as the OB fold-only OBFC1 mutant dominantly interferes with normal OBFC1/AAF44 function. Alternatively, OBFC1/AAF44 may complex with other unknown proteins and form the functionally equivalent of yeast CST complex at mammalian telomeres.
In yeast, the CST complex can bind DNA polymerases via Stn1 to mediate C-strand synthesis of the telomere DNA (35). Consistent with this notion, recently, OBFC1/AAF44 has been reported to be a component of the AAF capable of stimulating the activity of DNA polymerase α-primase (36), indicating the existence of evolutionarily conserved mechanisms for C-strand synthesis and suggesting that OBFC1/AAF44 may regulate telomere length through DNA polymerases in a fashion similar to yeast Stn1. Notably, OBFC1/AAF44 heterodimerizes with AAF132 (36). The connection between AAF132-AAF44 and the CST complex warrants further investigation.
Our data indicate that OBFC1/AAF44 associates with and is in close proximity to TPP1. Whether such interaction is direct remains to be determined. Bacterially expressed TPP1 and OBFC1/AAF44 proteins failed to interact with each other (data not shown), supporting the notion that the interaction between TPP1 and OBFC1/AAF44 may be indirect. However, in vitro binding assays may not recapitulate in vivo conditions. Furthermore, TPP1 and OBFC1/AAF44 interaction may be regulated by post-translational modifications. Regardless of whether the interaction is direct, TPP1 has emerged as an important node in the protein-protein interaction network at telomeres that links the OBFC1/Stn1 complex to the telosome and the telomerase. Although OBFC1/AAF44 may directly bind the 3′-telomere overhang, the affinity of such interactions appears low (Kd ∼1 μm, Fig. 3A). In addition, the OB fold alone failed to localize OBFC1/AAF44 to the telomeres (Fig. 3C). In this regard, TPP1 may help to recruit OBFC1/AAF44 to the telomeres through its association with OBFC1/AAF44. The telomere localization of OBFC1/AAF44 may further control C-strand synthesis at the telomeres. It was shown previously (36) that OBFC1/AAF44 was enriched on replication foci in HeLa cells. This suggests that the association of OBFC1/AAF44 may be controlled by DNA replication as well. It will be interesting to explore in detail the function of OBFC1/AAF44 at telomeres and gain more insight into the mechanisms of telomere capping by multiple OB fold proteins and the telomere interactome.
Acknowledgments
We thank Dr. Quanyuan He for sequence alignment analysis and Dr. Sung Yung Jung for mass spectrometry sequencing.
This work was supported, in whole or in part, by National Institutes of Health Grant. This work was also supported by grants from the American Heart Association, the American Cancer Society, and the Welch Foundation.
- OB fold
- oligonucleotide and oligosaccharide-binding fold
- OBFC1
- OB fold-containing protein 1
- hOBFC1
- human OBFC1
- AAF
- α-accessory factor
- ssDNA
- single-stranded DNA
- IP
- immunoprecipitation
- ES
- embryonic stem
- CST
- Cdc13-Stn1-Ten1
- BiFC
- bimolecular fluorescence complementation assay
- TRF
- telomere restriction fragment
- GST
- glutathione S-transferase
- YFP
- yellow fluorescent protein.
REFERENCES
- 1.Blackburn E. H. (2001) Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 2.Wei C., Price M. (2003) Cell Mol. Life Sci. 60, 2283–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey S. M., Goodwin E. H. (2004) Cytogenet. Genome Res. 104, 109–115 [DOI] [PubMed] [Google Scholar]
- 4.Xin H., Liu D., Songyang Z. (2008) Genome Biol. 9, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granger M. P., Wright W. E., Shay J. W. (2002) Crit. Rev. Oncol. Hematol. 41, 29–40 [DOI] [PubMed] [Google Scholar]
- 6.Harrington L., Robinson M. O. (2002) Oncogene 21, 592–597 [DOI] [PubMed] [Google Scholar]
- 7.Blasco M. A. (2007) Nat. Rev. Genet. 8, 299–309 [DOI] [PubMed] [Google Scholar]
- 8.Verdun R. E., Karlseder J. (2007) Nature 447, 924–931 [DOI] [PubMed] [Google Scholar]
- 9.de Lange T. (2005) Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 10.Blackburn E. H. (1991) Trends Biochem. Sci. 16, 378–381 [DOI] [PubMed] [Google Scholar]
- 11.Hackett J. A., Greider C. W. (2002) Oncogene 21, 619–626 [DOI] [PubMed] [Google Scholar]
- 12.Collins K. (2006) Nat. Rev. Mol. Cell Biol. 7, 484–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu D., O'Connor M. S., Qin J., Songyang Z. (2004) J. Biol. Chem. 279, 51338–51342 [DOI] [PubMed] [Google Scholar]
- 14.Songyang Z., Liu D. (2006) Crit. Rev. Eukaryot. Gene Expr. 16, 103–118 [DOI] [PubMed] [Google Scholar]
- 15.Court R., Chapman L., Fairall L., Rhodes D. (2005) EMBO Rep. 6, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanaoka S., Nagadoi A., Nishimura Y. (2005) Protein Sci. 14, 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 18.Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M. S., Songyang Z. (2007) Nature 445, 559–562 [DOI] [PubMed] [Google Scholar]
- 19.Veldman T., Etheridge K. T., Counter C. M. (2004) Curr. Biol. 14, 2264–2270 [DOI] [PubMed] [Google Scholar]
- 20.Hockemeyer D., Daniels J. P., Takai H., de Lange T. (2006) Cell 126, 63–77 [DOI] [PubMed] [Google Scholar]
- 21.Wu L., Multani A. S., He H., Cosme-Blanco W., Deng Y., Deng J. M., Bachilo O., Pathak S., Tahara H., Bailey S. M., Deng Y., Behringer R. R., Chang S. (2006) Cell 126, 49–62 [DOI] [PubMed] [Google Scholar]
- 22.Martín V., Du L. L., Rozenzhak S., Russell P. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 14038–14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V. (2007) Nat. Struct. Mol. Biol. 14, 208–214 [DOI] [PubMed] [Google Scholar]
- 24.Song X., Leehy K., Warrington R. T., Lamb J. C., Surovtseva Y. V., Shippen D. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19815–19820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandin N., Reed S. I., Charbonneau M. (1997) Genes Dev. 11, 512–527 [DOI] [PubMed] [Google Scholar]
- 26.Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 27.Chen L. Y., Liu D., Songyang Z. (2007) Mol. Cell Biol. 27, 5898–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004) Nat. Cell Biol. 6, 673–680 [DOI] [PubMed] [Google Scholar]
- 29.Ouellette M. M., Liao M., Herbert B. S., Johnson M., Holt S. E., Liss H. S., Shay J. W., Wright W. E. (2000) J. Biol. Chem. 275, 10072–10076 [DOI] [PubMed] [Google Scholar]
- 30.Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. (2004) Genes Dev. 18, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grandin N., Damon C., Charbonneau M. (2001) EMBO J. 20, 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu C. D., Kerppola T. K. (2003) Nat. Biotechnol. 21, 539–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei M., Podell E. R., Cech T. R. (2004) Nat. Struct. Mol. Biol. 11, 1223–1229 [DOI] [PubMed] [Google Scholar]
- 34.Petreaca R. C., Chiu H. C., Eckelhoefer H. A., Chuang C., Xu L., Nugent C. I. (2006) Nat. Cell Biol. 8, 748–755 [DOI] [PubMed] [Google Scholar]
- 35.Grossi S., Puglisi A., Dmitriev P. V., Lopes M., Shore D. (2004) Genes Dev. 18, 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casteel D. E., Zhuang S., Zeng Y., Perrino F. W., Boss G. R., Goulian M., Pilz R. B. (2009) J. Biol. Chem. 284, 5807–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]