Abstract
Despite extensive characterization of the μ-opioid receptor (MOR), the biochemical properties of the isolated receptor remain unclear. In light of recent reports, we proposed that the monomeric form of MOR can activate G proteins and be subject to allosteric regulation. A μ-opioid receptor fused to yellow fluorescent protein (YMOR) was constructed and expressed in insect cells. YMOR binds ligands with high affinity, displays agonist-stimulated [35S]guanosine 5′-(γ-thio)triphosphate binding to Gαi, and is allosterically regulated by coupled Gi protein heterotrimer both in insect cell membranes and as purified protein reconstituted into a phospholipid bilayer in the form of high density lipoprotein particles. Single-particle imaging of fluorescently labeled receptor indicates that the reconstituted YMOR is monomeric. Moreover, single-molecule imaging of a Cy3-labeled agonist, [Lys7, Cys8]dermorphin, illustrates a novel method for studying G protein-coupled receptor-ligand binding and suggests that one molecule of agonist binds per monomeric YMOR. Together these data support the notion that oligomerization of the μ-opioid receptor is not required for agonist and antagonist binding and that the monomeric receptor is the minimal functional unit in regard to G protein activation and strong allosteric regulation of agonist binding by G proteins.
Opioid receptors are members of the G protein-coupled receptor (GPCR)2 superfamily and are clinical mainstays for inducing analgesia. Three isoforms of opioid receptors, μ, δ, and κ, have been cloned and are known to couple to Gi/o proteins to regulate adenylyl cyclase and K+/Ca+ ion channels (1–3). An ever growing amount of data suggests that many GPCRs oligomerize (4, 5), and several studies have suggested that μ-opioid receptors (MORs) and δ-opioid receptors heterodimerize to form unique ligand binding and G protein-activating units (6–10). Although intriguing, these studies utilize cellular overexpression systems where it is difficult to know the exact nature of protein complexes formed between the receptors.
To study the function of isolated GPCRs, our laboratory and others have utilized a novel phospholipid bilayer reconstitution method (11–16). In this approach purified GPCRs are reconstituted into the phospholipid bilayer of a high density lipoprotein (HDL) particle. The reconstituted HDL (rHDL) particles are monodispersed, uniform in size, and preferentially incorporate a GPCR monomer (14, 15). Previous work in our lab has shown that rhodopsin, a class A GPCR previously proposed to function as a dimer (17–19), is fully capable of activating its G protein when reconstituted as a monomer in the rHDL lipid bilayer (15). Moreover, we have demonstrated that agonist binding to a monomeric β2-adrenergic receptor, another class A GPCR, can be allosterically regulated by G proteins (14). This led us to determine whether a monomer of MOR, a class A GPCR that endogenously binds peptide ligands, is the minimal functional unit required to activate coupled G proteins. We additionally investigated whether agonist binding to monomeric MOR is allosterically regulated by inhibitory G protein heterotrimer.
To study the function of monomeric MOR we have purified a modified version of the receptor to near homogeneity. A yellow fluorescent protein was fused to the N terminus of MOR, and this construct (YMOR) was expressed in insect cells for purification. After reconstitution of purified YMOR into rHDL particles, single-molecule imaging of Cy3-labeled and Cy5-labeled YMOR determined that the rHDL particles contained one receptor. This monomeric YMOR sample binds ligands with affinities nearly equivalent to those observed in plasma membrane preparations. Monomeric YMOR efficiently stimulates GTPγS binding to Gi2 heterotrimeric G protein. Gi2 allosteric regulation of agonist binding to rHDL·YMOR was also observed. Single-particle imaging of binding of [Lys7, Cys8]dermorphin-Cy3, a fluorophore-labeled agonist, to rHDL·YMOR supports the notion that the rHDL particles contain a single YMOR. Taken together, these results suggest that a monomeric MOR is the minimal functional unit for ligand binding and G protein activation and illustrate a novel method for imaging ligand binding to opioid receptors.
EXPERIMENTAL PROCEDURES
Materials
G protein baculoviruses encoding rat Gαi2, His6-Gβ1, and Gγ2 were provided by Dr. Alfred G. Gilman (University of Texas Southwestern, Dallas, TX). DNA encoding human μ-opioid receptor (NM 000914.2) was provided by Dr. John R. Traynor (University of Michigan, Ann Arbor, MI). Expired serum was generously provided by Dr. Bert La Du (University of Michigan, Ann Arbor, MI). Spodoptera frugiperda (Sf9) and Trichoplusia ni (HighFiveTM) cells, pFastBacTM baculovirus expression vectors and Sf900TM serum-free medium were from Invitrogen. InsectExpressTM medium was purchased from Lonza (Allendale, NJ). N-Dodecyl-β-d-maltoside was from Dojindo (Rockville, MD). All of the lipids were from Avanti Polar Lipids (Alabaster, AL). [3H]diprenorphine (DPN, 54.9 Ci/mmol) and [35S]GTPγS (1250 Ci/mmol) were obtained from PerkinElmer Life Sciences. EZ-LinkTM NHS-Biotin reagent was from Pierce. Ovomucoid trypsin inhibitor was purchased from United States Biological (Swampscott, MA). GF/B and BA85 filters, Cy3 and Cy5 NHS-ester mono-reactive dyes, Cy3-maleimide dye, and Source 15Q and Superdex 200 chromatography resins were from GE Healthcare. TalonTM resin was from Clontech. Bio-BeadsTM SM-2 absorbant resin was from Bio-Rad. Chromatography columns were run using a BioLogic Duo-Flow protein purification system from Bio-Rad. Amicon Ultra centrifugation filters were from Millipore (Billerica, MA). Amino acids for ligand synthesis were obtained from Advanced ChemTech (Louisville, KY) or Sigma-Aldrich. All other chemicals and ligands were from either Sigma-Aldrich or Fisher Scientific.
YFP-μ-Opioid Receptor Fusion Protein Expression and Purification
Baculoviruses were created using transfer vectors (pFastBacTM) that encoded a fusion protein of an N-terminal cleavable hemagglutinin signal sequence (MKTIIALSYIFCLVF), a FLAG epitope (DYKDDDD), a decahistidine tag, the monomeric and enhanced yellow fluorescent protein (Clontech), and the human MOR. High titer viruses (107–108 plaque-forming units/ml) were used to infect Sf9 or HighFiveTM suspension cultures at a multiplicity of infection of 0.25 to 1. FLAG-His10-mEYFP-MOR (YMOR) was expressed for 48–52 h in the presence of 1 μm naltrexone (NTX). The cells were resuspended in Buffer A (50 mm Tris·HCl, pH 8.0, 50 mm NaCl, 100 nm NTX, and protease inhibitors (3.2 μg/ml leupeptin, 3.2 μg/ml ovomucoid trypsin inhibitor, 17.5 μg/ml phenylmethanesulfonyl fluoride, 16 μg/ml tosyl-l-lysine-chloromethyl ketone (TLCK), 16 μg/ml tosyl-l-phenylalanine chloromethyl ketone (TPCK)) and lysed by nitrogen cavitation. Supernatants from a 500 × g spin were then subjected to 125,000 × g spin for 35 min, and pellets containing membrane fractions were resuspended in Buffer A and either stored at −80 °C for later binding assays or further processed to purify receptor.
All of the purification steps were performed sequentially and at 4 °C or on ice. Membrane preparations were diluted to 5 mg/ml in Buffer A plus 1% DDM (w/v) and 0.01% cholesteryl hemisuccinate (CHS) (w/v) and gently stirred for 1 h to solubilize YMOR out of the membrane. Detergent extracted YMOR was enriched via TalonTM metal affinity chromatography resin (Clontech) in Buffer A plus 0.1% DDM and 0.01% CHS and eluted with Buffer A plus 0.1% DDM, 0.01% CHS, and 150 mm imidazole. Fractions containing YMOR (based on Coomassie staining after SDS-PAGE separation) were pooled, diluted 5-fold in Buffer B (20 mm Hepes, pH 8.0, 5 mm MgCl2, 0.1% DDM, 0.01% CHS, 100 nm NTX, and phenylmethanesulfonyl fluoride-TLCK-TPCK protease inhibitors), and eluted from a 1-ml Source 15Q strong anion exchange column with a 50–300 mm NaCl linear gradient. Peak fractions were identified by radioligand binding assays using [3H]DPN (2–4 nm). Peak fractions were then pooled and concentrated on Amicon Ultra centrifugation filters (10-kDa molecular mass cut-off). As a final purification step this YMOR sample was resolved based on size using a Superdex 200 gel filtration column (GE Healthcare) in Buffer C (20 mm Hepes, pH 8.0, 100 mm NaCl, 0.1% DDM, 0.01% CHS, 100 nm NTX, and protease inhibitors). Coomassie staining after SDS-PAGE was used to identify fractions containing YMOR, which were pooled and concentrated to ∼1–2 μm. Glycerol was added to a final concentration of 10% (v/v), and the samples were flash-frozen in liquid nitrogen and then stored at −80 °C until further use.
ApolipoproteinA-1 Purification
Wild type human apoA-1 was purified from expired serum as previously described (14). A recombinant apoA-1 with an N-terminal 43-amino acid deletion and a histidine tag (Δ(1–43)-His6-ApoA-1) was expressed using a pET15b vector to transform competent Escherichia coli cells (BL21). The cells were resuspended and lysed by gentle vortexing in 10 mm Tris·HCl, pH 8.0, 100 mm NaH2PO4, 6 m guanidine hydrochloride, 1% Triton X-100. Lysate was fractionated by centrifugation at 10,000 × g, and the supernatant was loaded onto a nickel-nitrilotriacetic acid column by gravity flow. The column was washed with 10 mm Tris·HCl, pH 8.0, 100 mm NaH2PO4, 6 m guanidine hydrochloride, 1% Triton X-100 and then with 50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 1% Triton X-100. Bound Δ(1–43)-His6-apoA-1 was eluted with 50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 250 mm imidazole, 1% Triton X-100. Peak fractions were further purified on a Superdex 75 gel filtration chromatography column in 20 mm Hepes, pH 8.0, 100 mm NaCl, 1 mm EDTA, 20 mm sodium cholate. Pooled apoA-1 was then dialyzed against 20 mm Hepes, pH 8.0, 100 mm NaCl, 1 mm EDTA, 5 mm sodium cholate. Purified apoA-1 was concentrated to ∼10 mg/ml and stored at −80 °C until use.
In Vitro HDL Reconstitution
HDL particles were reconstituted according to previously reported protocols (14). Briefly, 21 mm sodium cholate, 7 mm lipids (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (POPG) at a molar ratio of 3:2), purified YMOR (DDM-solubilized), and 100 μm purified apoA-1 were solubilized in 20 mm Hepes, pH 8.0, 100 mm NaCl, 1 mm EDTA, 50 mm sodium cholate. The final concentration of YMOR varied from 0.2 to 0.4 μm, but YMOR always comprised 20% of the total reconstitution volume. In some reconstitutions the lipid component was modified such that porcine polar brain lipid extract (Avanti Polar Lipids) was used in addition to POPC and POPG for a final concentration of 7 mm lipids at a molar ratio of 1.07:1.5:1 brain lipid:POPC:POPG. Following incubation on ice (1.5–2 h), the samples were added to Bio-BeadsTM (Bio-Rad, 0.05 mg/ml of reconstitution volume) to remove detergent and to form HDL particles. Particles containing YMOR were purified via M1 anti-FLAG immunoaffinity chromatography resin (Sigma) and eluted with 1 mm EDTA plus 200 μg/ml FLAG peptide. To assess the efficiency of HDL reconstitution, the total protein concentration of rHDL·YMOR was compared with the concentration of active reconstituted YMOR. FLAG affinity column-purified rHDL·YMOR was resolved from BSA and FLAG peptide on a Superdex 200 gel filtration column, and the peak fraction was analyzed for protein content with Amido Black staining (20), whereas [3H]DPN saturation assays were used to measure active YMOR content. YMOR reconstituted into HDL was stored on ice until further use.
G Protein Addition to rHDL·YMOR Particles
Gi2 heterotrimer (Gαi2-His6Gβ1-Gγ2) was expressed in Sf9 cells and purified as previously described (21). The concentration of active G protein was determined by filter binding of 20 μm [35S]GTPγS (isotopically diluted) in 30 mm NaHepes, pH 8.0, 100 mm NaCl, 50 mm MgCl2, 1 mm EDTA, 0.05% C12E10, and 1 mm dithiothreitol after 1 h of incubation at room temperature. Purified G protein heterotrimer was added to preformed, FLAG-purified rHDL·YMOR particles at a molar ratio of 1:10 receptor to G protein. Gi2 was purified in 0.7% CHAPS, and the final volume of G protein added to rHDL·YMOR was such that the final CHAPS concentration was well below its critical micelle concentration. Reconstituted receptor and G protein samples were then incubated in Bio-BeadsTM for 30–45 min at 4 °C to remove residual detergent.
[3H]DPN Saturation and Agonist Competition Binding Assays
Binding reactions were performed in 100-μl volumes. Membrane fractions prepared from Sf9 or HighFiveTM cells expressing YMOR (0.5–5 μg total protein, prepared as above) were incubated with [3H]DPN (0.25 to 4 nm; 54.9 Ci/mmol) for 1 h at room temperature in 25 mm Tris·HCl, pH 7.7, 136 mm NaCl, 2.7 mm KCl (Tris-buffered saline) buffer. Nonspecific binding was determined in the presence of 20 μm NTX. Bound [3H]DPN was separated from free by rapid filtration through GF/B filters and three 200-μl washes of ice-cold Tris-buffered saline. [3H]DPN saturation binding reactions on YMOR incorporated into rHDL particles were prepared in Tris-buffered saline, pH 7.7, plus 0.1% BSA, and Sephadex G-50 Fine (GE Healthcare) gravity flow columns were used to separate bound from free [3H]DPN. Agonist competition assays in insect membranes and rHDL particles were performed in 25 mm Tris·HCl, pH 7.7, 6–7 mm NaCl. Competition assays in rHDL particles also included 0.1% BSA. [3H]DPN binding assays on detergent-solubilized YMOR were performed in 50 mm Tris·HCl, pH 7.7, 136 mm NaCl, 0.1% DDM, 0.01% CHS and separated on Sephadex G-50 gravity flow columns. For agonist competition assays, receptor samples were incubated with 0.5–1 nm [3H]DPN and increasing concentrations of agonist (1 pm to 1 mm) in the absence or presence of 10 μm GTPγS. The samples were measured for radioactivity on a liquid scintillation counter, and the data were fit with one-site saturation, one-site competition, or two-site competition binding models using Prism 5.0 (GraphPad, San Diego, CA).
[35S]GTPγS Binding Assay
100-μl volume reactions were prepared containing 1 μg of total membrane protein from YMOR expressing HighFiveTM cells or ∼50–60 fmol of YMOR incorporated into rHDL particles in 30 mm Tris·HCl, pH 7.4, 100 mm NaCl, 5 mm MgCl2, 0.1 mm dithiothreitol, 10 or 1 μm GDP (membranes or HDL particles), and 0.1% BSA. Membrane assays and rHDL assays were incubated with 10 nm isotopically diluted [35S]GTPγS (12.5 Ci/mmol). YMOR samples were incubated with increasing concentrations of agonists (1 pm to 1 mm) for 1 h at room temperature, then rapidly filtered through GF/B (membrane samples) or BA85 filters (HDL samples), and washed three times with 2 ml of ice-cold 30 mm Tris·HCl, pH 7.4, 100 mm NaCl, 5 mm MgCl2. The samples were measured for radioactivity on a liquid scintillation counter, and the data were fit with a log dose-response model using Prism 5.0.
Single-molecule Imaging of Reconstituted Cy3- and Cy5-YMOR
Purified YMOR (∼200 pmol) was incubated with NHS-ester Cy3 or Cy5 mono-reactive dye (GE Healthcare) in 20 mm Hepes, pH 8.0, 100 mm NaCl, 5 mm MgCl2, 6 mm EDTA, 0.1% DDM, 0.01% CHS, 100 nm NTX for 30 min at 25 °C and then 1 h at 4 °C. Conjugation reactions were quenched by the addition of Tris·HCl, pH 7.7 buffer (10 mm final). Cy-labeled YMOR was then separated from free dye using a 12-cm Sephadex G-50 column. The final dye to protein molar ratio was 3.8:1 for Cy3-YMOR and 3.7:1 for Cy5-YMOR, determined by absorbance at 280 nm for total protein, 550 nm for Cy3 dye, and 650 nm for Cy5 dye as measured on a NanoDrop ND-1000 spectrophotometer. A separate aliquot of YMOR was co-labeled with both Cy3 and Cy5 for final molar ratios of 2:1 Cy3:YMOR and 1.8:1 Cy5:YMOR. HDL reconstitutions of Cy3-YMOR alone, Cy5-YMOR alone, a mixture of Cy3- and Cy5-YMOR, and Cy3-Cy5-YMOR were then performed. Reconstitutions were performed as previously described, using apoA-1 that had been biotinylated at a 3:1 molar ratio using EZ-Link NHS-Biotin according to the manufacturer's protocol (Pierce). Reconstituted samples were diluted 5000-fold in 25 mm Tris·HCl, pH 7.7, and injected into a microfluidic channel on a quartz slide that was previously coated with biotinylated polyethylene glycol and treated with 0.2 mg/ml streptavidin to generate a surface density of ∼0.05 molecules/μm2. An oxygen scavenging system of 10 mm Trolox (Sigma-Aldrich), 100 mm protocatechuic acid, and 1 μm protocatechuate-3,4-dioxygenase was included in the sample dilution (22). Following a 10-min incubation to allow binding of the biotin-HDL·Cy-YMOR complex to the streptavidin-coated slide, the channel was washed with ice-cold 25 mm Tris·HCl, pH 7.7, containing the oxygen scavenging system. An Olympus IX71 inverted microscope configured for prism-based total internal reflection fluorescence (TIRF) and coupled to an intensified CCD camera was used to image Cy3 and Cy5 fluorophores (CrystaLaser, 532 nm; Coherent CUBE laser, 638 nm; Chroma band pass filters HQ580/60 and HQ710/130 nm). Fluorophore intensity time traces were collected for 30–100 s at 10 frames/s. Time traces were analyzed for photobleaching with in-house software (MatLab 7.0).
Synthesis of [Lys7, Cys8]Dermorphin and Labeling with Cy3 Dye
[Lys7, Cys8]dermorphin (Tyr-d-Ala-Phe-Gly-Tyr-Pro-Lys-Cys-NH2) was synthesized on Rink resin (solid support) using an Applied Biosystems 431A peptide synthesizer and standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. The samples were characterized on a Waters reverse phase HPLC using a Vydac C18 (Protein and Peptide) 10-micron column. The samples were run on a linear gradient of 0–45% acetonitrile in an aqueous phase containing 0.1% trifluoroacetic acid at 35 °C and monitored at 254 and 230 nm (supplemental Fig. S3). Peptides were similarly purified on a Waters semi-preparative reverse phase HPLC using a Vydac C18 10-micron column at room temperature. The purified peptide was labeled with Cy3-maleimide (GE Healthcare) according to the manufacturer's instructions using a ratio of 1.5:1 peptide to fluorophore and repurified by HPLC as before. The labeled peptide was further purified via semi-preparative HPLC using a 5-micron Vydac C18 column as described above. The potency and efficacy of [Lys7, Cys8]dermorphin-Cy3 at MOR were confirmed in radiolabeled [3H]DPN competition and [35S]GTPγS binding assays.
Prism-based Single-molecule TIRF and Step Photobleaching Analysis of [Lys7, Cys8]Dermorphin-Cy3 Binding to rHDL·YMOR + Gi2
Purified recombinant apoA-1 was biotinylated at a 3:1 molar ratio of biotin:apoA-1. Purified YMOR was then reconstituted with biotin-apoA-1, POPC, POPG, and brain lipid extract as above. Purified Gi2 heterotrimer was added to reconstituted receptor as above, and coupling was confirmed by observing high affinity competition of [3H]DPN binding by the agonist DAMGO. Five nanomolar biotin-rHDL·YMOR+Gi2 was then incubated with 5 μm [Lys7, Cys8]dermorphin-Cy3 for 45 min at 25 °C in 25 mm Tris·HCl, pH 7.7. The samples were diluted 100-fold in 25 mm Tris·HCl, pH 7.7, and imaged as described above with a surface density of ∼0.25 molecules/μm2. Fluorophore intensity time traces were collected for 30–100 s at 10 frames/s and analyzed for photobleaching with in-house software (MatLab 7.0).
RESULTS
Expression of a Functional μ-Opioid Receptor Fusion Protein in Insect Cells
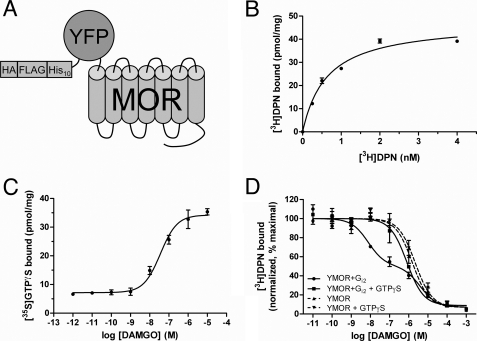
Human YMOR (Fig. 1A) expressed in Sf9 and HighFiveTM insect cells bound the nonspecific opioid antagonist [3H]diprenorphine ([3H]DPN) with high affinity in a saturable manner (Kd = 0.6 ± 0.1 nm; Fig. 1B). Maximal receptor levels of 12–40 pmol/mg were routinely observed.
FIGURE 1.
Functional expression of a modified MOR in insect cells. A, schematic representation of the modified μ-opioid receptor expressed in insect cells. N-terminal modifications included a hemagglutinin signal sequence (HA), FLAG epitope, His10, and a YFP. This construct, termed YMOR, was expressed in Sf9 and HighFiveTM insect cells using a recombinant Baculovirus system. B, YMOR expressed in insect cells exhibits high affinity ligand binding. Plasma membrane preparations (5 μg) of HighFiveTM cells expressing YMOR were used in [3H]diprenorphine saturation binding assays. YMOR bound [3H]DPN with a Bmax of ∼40 pmol/mg and a Kd of 0.6 ± 0.1 nm. C, YMOR couples to G protein in insect cell membranes. Membrane preparations (2 μg) of HighFiveTM cells co-expressing YMOR and Gi2 heterotrimer were incubated with increasing concentrations of the μ-opioid agonist DAMGO in the presence of 10 nm [35S]GTPγS (isotopically diluted). DAMGO stimulated [35S]GTPγS exchange on Gαi2 with an EC50 of 36 ± 0.1 nm. D, agonist binding to YMOR expressed in insect cells is allosterically regulated by G proteins. YMOR/Gi2 membrane preparations (2–3 μg) were incubated with 0.8 nm [3H]DPN and increasing concentrations of DAMGO in the absence or presence of 10 μm GTPγS. A high affinity binding site for DAMGO (●, Ki hi = ∼2.9 nm) was disrupted in the absence of G protein coupling (with GTPγS, ■, Ki = ∼300 nM). DAMGO also displayed low affinity binding to membranes not expressing Gi2 (YMOR, ▴, Ki = ∼580 nm; and YMOR + GTPγS, ▾, Ki = ∼800 nm). Binding data were normalized to the curve fit maximum. For all panels the data are representative of at least two experiments performed in duplicate, and the error bars represent the standard error of the mean.
The μ-selective agonist DAMGO ([d-Ala2, N-MePhe4, Gly5-ol]enkephalin) stimulated [35S]GTPγS binding to membranes co-expressing YMOR and Gαi2-His6Gβ1-Gγ2 G protein heterotrimer (Gi2) in a concentration-dependent manner (Fig. 1C). DAMGO elicited strong activation of Gi2, stimulating [35S]GTPγS binding nearly 4-fold over basal levels with an EC50 of 36 ± 0.1 nm. The potency and efficacy of DAMGO at insect cell expressed YMOR is well in line with its observed pharmacological characteristics in mammalian cell expression systems (23–25).
Allosteric regulation of agonist binding to opioid receptors by G proteins has been well established in plasma membrane preparations of brain homogenates and overexpression systems (26–30) and was also observed for YMOR expressed in insect cells. In the absence of Gi2, DAMGO competed [3H]DPN (0.5 nm) binding in a concentration-dependent manner with a Ki of ∼580 nm (Fig. 1D). In membranes expressing both YMOR and Gi2, DAMGO exhibited a biphasic mode of inhibition of [3H]DPN binding with a Ki hi of ∼2.9 nm and a Ki lo of ∼1.5 μm (fraction of Ki hi ∼0.53). The addition of 10 μm GTPγS to the YMOR + Gi2 membranes eliminated the high affinity DAMGO-binding site (Ki = ∼300 nm), illustrating that YMOR is allosterically regulated by G proteins. Therefore, YMOR expressed in HighFiveTM cells proved fully functional in regards to G protein coupling.
YMOR Purification
The capacity of a variety of detergents (zwitterionic, polar, and nonionic detergents such as Triton X-100, digitonin, Nonidet P-40, CHAPS, C12E10, and n-octyl-β-glucoside) to solubilize YMOR from insect cell membranes was assessed by anti-FLAG Western blot analysis. Extraction of YMOR with DDM proved to be most efficient (data not shown). The addition of CHS (0.01% w/v) improved the stability of the solubilized receptor as measured by [3H]DPN binding, consistent with the stabilizing effects of cholesterol moieties observed for solubilized β2AR (31) (supplemental Fig. S1). The expression levels of YMOR and the efficiency of DDM solubilization, assessed by [3H]DPN binding and anti-FLAG Western blots, were enhanced in the presence of naltrexone. Therefore this antagonist was present (100 nm) during all subsequent purification steps.
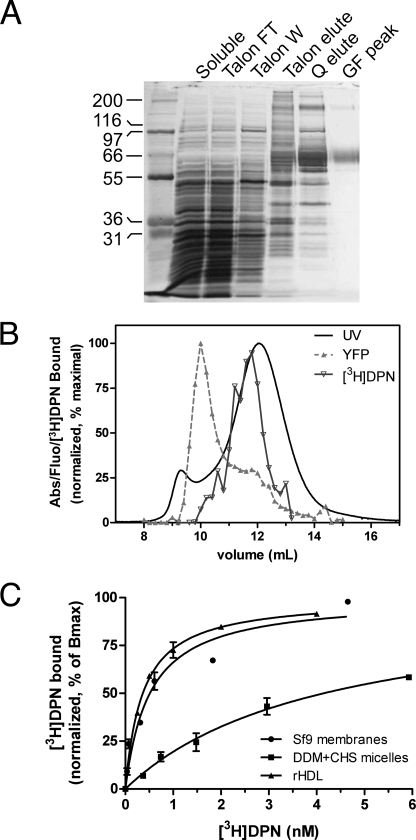
DDM-extracted YMOR was purified through a series of chromatographic steps including metal chelate (TalonTM), anion exchange (Source 15Q), and size exclusion (Superdex 200) chromatography columns. Peak fractions from each column were determined by Coomassie staining, anti-FLAG antibody Western blotting, and [3H]DPN binding. Fractions displaying the highest [3H]DPN binding capacity and appropriate molecular weight were pooled and subjected to the next chromatographic step. Superdex 200 peak fractions were pooled, concentrated, flash-frozen in liquid N2, and then stored at −80 °C until later use. A representative silver-stained SDS-PAGE separation of each enrichment step is shown in Fig. 2A. Yields of YMOR were typically ∼50 μg of >95% pure receptor/liter of insect culture. YMOR, deemed pure based on silver staining, bound [3H]DPN at ∼20% of the predicted binding sites calculated from the molecular mass. This loss of activity is likely due to the detrimental freeze/thaw process.
FIGURE 2.
Purification of YMOR from HighFiveTM insect cells and reconstitution into HDL particles. A, YMOR was extracted from membranes in the presence of 100 nm naltrexone with 1% n-dodecyl-β-d-maltoside and 0.01% cholesteryl hemisuccinate and enriched on a TalonTM metal affinity column. The TalonTM pool containing YMOR (predicted 73-kDa molecular mass) was applied to a Source 15Q anion exchange column followed by a size exclusion gel filtration column (Superdex 200). Samples of the purification steps were resolved by SDS-PAGE and silver-stained. YMOR was enriched to ∼95% purity (GF peak). B, purified YMOR was reconstituted into HDL particles (see “Experimental Procedures”) and resolved by size exclusion chromatography (Superdex 200). Fractions were analyzed for total protein content (UV absorbance), active YMOR ([3H]DPN binding), and YFP fluorescence. UV absorbance showed a major peak corresponding to rHDL particles (stokes diameter of ∼10.5 nm). The elution volume of active YMOR corresponded with the rising slope of the rHDL peak. YFP fluorescence eluted as two peaks, corresponding to the active YMOR and a larger YMOR aggregate that was not incorporated into rHDL particles. This aggregated YMOR was not active based on [3H]DPN binding. The data were normalized such that the maximal value for each parameter was set to 100%. [3H]DPN binding measurements were performed in duplicate, and the error bars are omitted for clarity. C, high affinity [3H]DPN binding to YMOR is disrupted in detergent micelles but restored following reconstitution into rHDL particles. [3H]DPN binding to YMOR in insect cell membranes (●, Kd = ∼0.5 nm), purified in DDM micelles (■, Kd = ∼4.1 nm), and in rHDL particles (▴, Kd = ∼0.4 nm). The data are representative of at least three experiments performed in duplicate, and the error bars represent the standard error of the mean. The binding data were normalized to the maximal binding level (Bmax) as calculated by a one-site saturation curve fit (Prism 5.0, GraphPad).
Reconstitution of YMOR into High Density Lipoprotein Particles
The reconstitution of YMOR into high density lipoprotein (rHDL) particles was performed with a 500-fold molar excess of apoA-1 to YMOR (250-fold excess of rHDL) to favor reconstitution of a single YMOR molecule/rHDL particle. Resolution of rHDL particles containing YMOR (rHDL·YMOR) with size exclusion chromatography (SEC) suggested an apparent Stokes diameter of 10.5 nm (Fig. 2B). [3H]DPN binding to the SEC fractions confirmed the presence of functional YMOR in particles that eluted slightly earlier than the main UV absorbance peak (Stokes diameter of ∼10.3 nm), indicating the slightly larger size of rHDL·YMOR compared with rHDL. The YFP fluorescence of the SEC fractions indicated an additional peak that elutes near in the Superdex 200 void volume, suggestive of aggregated YMOR. Because these fractions did not bind [3H]DPN, we surmised that this receptor population was inactive. This aggregated and inactive receptor is consistent with the previous observation that ∼80% of the purified YMOR is inactive post freeze/thaw and prior to reconstitution into HDL.
The effectiveness of YMOR reconstitution into HDL is clear when comparing [3H]DPN binding between detergent-solubilized receptor and rHDL·YMOR particles. Prior to reconstitution into HDL, DDM-soluble YMOR bound [3H]DPN with a Kd of ∼4.1 nm, whereas reconstituted YMOR bound [3H]DPN with a Kd of ∼0.4 nm (Fig. 2C). This significant disruption of high affinity ligand binding for the soluble receptor is not surprising, because detergents are known to decrease the ligand affinity of opioid receptors (32) and disrupt GPCRs in general (33). Replacement of detergents with phospholipids reverses the deleterious effects of the detergent and restores YMOR conformation to one that binds [3H]DPN with native, membrane-bound affinity.
YMOR Is Monomeric When Incorporated into rHDL
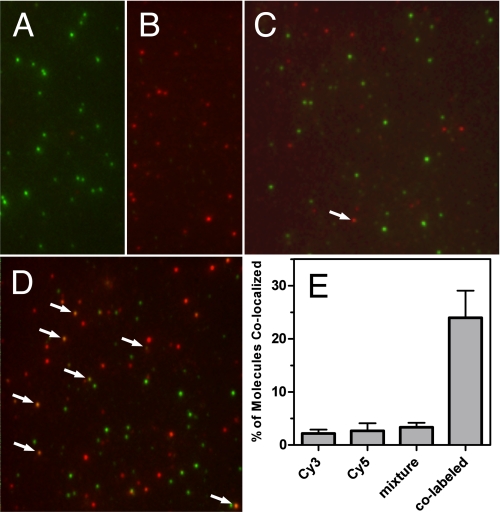
To determine the number of YMOR molecules present in each rHDL particle, we assessed the degree of receptor co-localization between Cy3- and Cy5-labeled YMOR using single-particle imaging. Purified YMOR was labeled with Cy3- or Cy5-reactive fluorescent dyes and reconstituted into HDL using biotinylated Δ(1–43)-His6-apoA-1. This biotin-rHDL·Cy-YMOR complex was then incubated on a streptavidin-coated microfluidic slide and excited with 532- and 638-nm lasers. Fluorescence emission was detected using prism-based SM-TIRF. Reconstituted Cy3-YMOR and Cy5-YMOR were visualized as mono-disperse fluorescent foci (Fig. 3, A and B). When Cy3-YMOR and Cy5-YMOR were mixed prior to reconstitution (Cy3-YMOR+Cy5-YMOR), a low level of co-localization of the two fluorophores (3.4%) was observed (Fig. 3, C and E). A false-positive co-localization signal of 2.8 and 2.2% was observed for the Cy3-YMOR and Cy5-YMOR samples, respectively (Fig. 3E). As a positive control, co-localization for YMOR labeled with both Cy3 and Cy5 was also measured (Fig. 3D). Considering that our method does not allow for the differentiation between YMOR labeled with multiple Cy probes of the same fluorophore versus co-localization of multiple YMORs labeled with the same Cy probe, we may underestimate co-localization by as much as a factor of 2. Thus we estimate an upper limit for co-localization, i.e. incorporation of two or more YMORs into a single rHDL particle, of ∼1.8% ((3.4% × 2) − (2.8% + 2.2%)). These data suggest that HDL reconstitution resulted in a sample containing mostly monomeric YMOR. Therefore the ligand binding and G protein coupling characteristics of the HDL-reconstituted YMOR presented here are indicative of the properties of a monomeric receptor.
FIGURE 3.
YMOR is monomeric when reconstituted into HDL particles. Purified YMOR was labeled with Cy3 or Cy5 fluorescent dyes and reconstituted separately or together into biotin-labeled rHDL. The particles were imaged using single-molecule total internal reflection fluorescence microscopy on quartz slides coated with streptavidin. Representative overlay images of reconstituted rHDL·Cy3-YMOR (A), rHDL·Cy5-YMOR (B), rHDL·(Cy3-YMOR + Cy5-YMOR) (C), and rHDL·Cy3-Cy5-YMOR (D) are shown. Quantification of Cy3 and Cy5 co-localization (E) showed that when both Cy3-YMOR and Cy5-YMOR were mixed together prior to reconstitution (mixture, 4c), only ∼3.4% of rHDL particles contained two labeled receptors, compared with a false-positive co-localization signal of 2.8 and 2.2% observed for the rHDL·Cy3-YMOR and rHDL·Cy5-YMOR samples. YMOR co-labeled with both Cy3 and Cy5 was also imaged as a positive control for co-localization. Approximately 24% of rHDL·Cy3-Cy5-YMOR particles (co-labeled, 4d) exhibited co-localization, indicating that not all YMOR received a secondary fluorescent dye under the labeling conditions.
To assess the amount of active versus inactive YMOR incorporated into rHDL, we compared the total protein concentration of purified rHDL·YMOR particles to the maximal concentration of YMOR based on [3H]DPN saturation binding assays. Reconstituted YMOR was purified using FLAG affinity resin and resolved by SEC into a sample containing only apoA-1 and YMOR (based on SDS-PAGE and Coomassie staining). Amido Black staining of these peak fractions indicated a protein concentration of ∼5.2 μg/ml. Considering that the protein sample consists of two apoA-1 molecules (molecular mass, ∼25,500 Da) and one YMOR (molecular mass, ∼73,500 Da), the rHDL·YMOR sample has a total molecular mass of 124,500 Da and therefore a molar concentration of ∼42 nm. Saturation binding assays on the rHDL·YMOR SEC peak indicated a molar concentration of ∼31 nm for YMOR which bound ligand. Given the inherent limitations of protein detection assays at the low concentration of our samples, these data suggest that at least 75% of the YMOR in rHDL is active based on [3H]DPN binding.
Monomeric YMOR Binds Antagonists with High Affinity
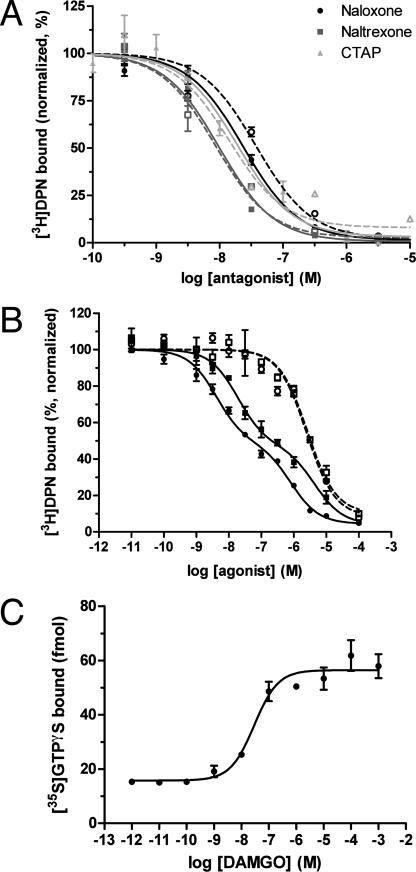
Reconstituted and monomeric YMOR binds antagonists with affinities similar to those observed in the plasma membrane. The antagonists naloxone and naltrexone and the μ-specific peptide CTAP (d-Phe-Cys-Tyr-d-Trp-Arg-Thr-Pen-Thr-NH2) compete [3H]DPN binding in similar fashion when in HighFiveTM cell membrane preparations or in reconstituted HDL particles (Fig. 4A). Therefore reconstitution into HDL allows YMOR to adopt a conformation that appears identical to receptor in plasma membranes in terms of high affinity antagonist binding.
FIGURE 4.
Monomeric YMOR binds ligands with high affinity and functionally couples to Gi2 heterotrimer. A, opioid antagonists bind YMOR in rHDL with affinities equivalent to those observed in membrane preparations. Competition binding assays were performed using plasma membranes of HighFiveTM cells expressing YMOR and Gi2 heterotrimer (dotted lines, open symbols) or rHDL·YMOR (solid lines, closed symbols). For CTAP binding assays in rHDL, YMOR was coupled to Gi2 heterotrimer. Increasing concentrations of naloxone, naltrexone, or CTAP competed the binding of 1 nm [3H]DPN in the rHDL system or 0.5 nm [3H]DPN in membrane preparations. The observed binding constants were: naloxone Ki mem = ∼19 nm, Ki rHDL = ∼7.8 nm; naltrexone Ki mem = ∼4.3 nm, Ki rHDL = ∼3.3 nm; CTAP Ki mem = ∼7.1 nm, Ki rHDL = ∼7.6 nm. B, binding of morphine to rHDL·YMOR is allosterically regulated by G proteins. Purified Gi2 heterotrimer was added to rHDL·YMOR at a molar ratio of 10:1, G protein to receptor. DAMGO and morphine bound rHDL·YMOR with high affinity, competing the binding of 0.75 nm [3H]DPN in a biphasic manner (DAMGO (■), Ki hi = ∼7.6 nm, Ki lo = ∼1.8 μm; morphine (●), Ki hi = ∼1.7 nm, Ki lo = ∼320 nm). High affinity agonist binding was lost with the addition of 10 μm GTPγS (DAMGO (□), Ki GTPγS = ∼1 μm; morphine (○), Ki GTPγS = ∼1 μm). In both A and B, data were normalized to the maximal binding level as calculated by a one- or two-site competition curve fit (Prism 5.0, GraphPad). C, HDL-reconstituted YMOR activates Gi2 in response to agonist binding. YMOR (an estimated 50–60 fmol) and associated Gi2 heterotrimer were incubated with 10 nm [35S]GTPγS (isotopically diluted) in the presence of increasing concentrations of DAMGO. DAMGO activated Gi2 with an EC50 of 29 ± 1.4 nm, stimulating ∼40 fmol of [35S]GTPγS binding to Gαi2 over basal levels, suggesting approximately a 1:0.7 coupling between YMOR and G protein. These results correlate with the estimated 53% high state observed in B. The data are representative of three experiments performed in duplicate. The error bars represent the standard error of the mean.
Monomeric YMOR Functionally Couples Inhibitory G Proteins
We next analyzed the capacity of monomeric YMOR to functionally couple to heterotrimeric G protein. Functional measurements included allosteric regulation of agonist binding (Fig. 4B) and agonist-mediated stimulation of [35S]GTPγS binding (Fig. 4C). Anti-FLAG affinity column purification of rHDL·YMOR was performed to remove “empty” rHDL particles, and purified Gi2 heterotrimer was added at a 10:1 Gi2:rHDL·YMOR molar ratio. The concentration of purified G protein was determined by [35S]GTPγS binding assays, and receptor concentration was measured with [3H]DPN saturation assays of rHDL·YMOR samples. Gi2 addition results in high affinity agonist binding (Fig. 4B). Morphine competed [3H]DPN binding with a Ki hi of ∼1.7 nm and a Ki lo of ∼320 nm, whereas DAMGO inhibited with a Ki hi of ∼7.6 nm and a Ki lo of ∼1.8 μm. As in membranes the addition of 10 μm GTPγS to the monomeric rHDL·YMOR+Gi2 uncouples the G protein and results in a single-site, low affinity agonist competition curve. Similarly, when rHDL·YMOR was not coupled to Gi2, DAMGO competed [3H]DPN binding with micromolar affinity (supplemental Fig. S2).
Just as YMOR membrane preparations displayed DAMGO-induced [35S]GTPγS binding to Gi2, monomeric YMOR in rHDL particles showed strong DAMGO stimulation of nucleotide exchange (∼4.7-fold stimulation with an EC50 of 31 ± 2.2 nm; Fig. 4C). A fluorescently labeled agonist, [Lys7, Cys8]dermorphin-Cy3, was also tested and showed concentration-dependent stimulation of [35S]GTPγS binding to Gi2 by monomeric YMOR (EC50 of 32 ± 9.1 nm; supplemental Fig. S4B).
Although Gi2 was added to rHDL·YMOR at a 10:1 molar ratio in these reconstitutions, the observed high and low agonist affinities illustrate that not all YMOR was coupled to Gi2. In fact, the agonist competition assays indicate that ∼54% of the receptor was coupled to Gi2. These data are consistent with previous studies on the β2AR where the addition of purified G protein heterotrimer to rHDL particles in the absence of detergents resulted in 90–95% G protein loss, largely because of aggregation (14). Indeed, SEC analysis of rHDL·YMOR before and after Gi2 addition confirmed that a large amount of the heterotrimer aggregates during its addition (data not shown). As such, the two populations of ∼54% coupled and ∼46% uncoupled YMOR correspond well with the expected final G protein to YMOR ratio of 0.5–1:1.
Single-particle Visualization of Agonist Binding to rHDL·YMOR
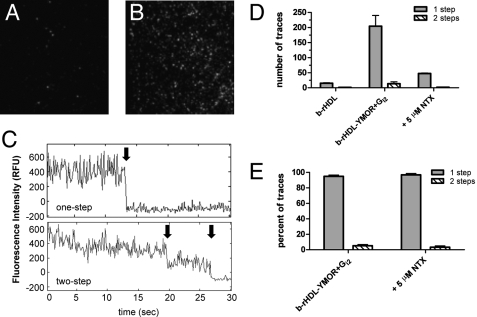
[Lys7, Cys8]dermorphin was used as an agonist probe to visualize binding to monomeric YMOR in rHDL at the single-molecule level with TIRF (Fig. 5). [Lys7, Cys8]dermorphin was labeled with a Cy3 fluorophore (supplemental Fig. S3), and its ability to bind reconstituted YMOR with high affinity and stimulate [35S]GTPγS binding to Gαi2 was confirmed (supplemental Fig. S4). YMOR was reconstituted with biotinylated Δ(1–43)-His6-apoA-1 (b-rHDL·YMOR), enriched on a FLAG affinity column, and added to Gi2 heterotrimer at a 30:1 Gi2:YMOR molar ratio to ensure complete receptor coupling to G protein. Binding of a saturating concentration (500 nm) of [Lys7, Cys8]dermorphin-Cy3 to b-rHDL·YMOR+Gi2 was then imaged with SM-TIRF. Fifty picomolar b-rHDL or b-rHDL·YMOR+Gi2 (Fig. 5, A and B) samples were continuously excited at 532 nm and fluorescence intensity traces (Fig. 5C) were analyzed for step photobleaching. Approximately 95% of the b-rHDL·YMOR+Gi2 fluorescence traces exhibited single-step photobleaching, suggesting that a single [Lys7, Cys8]dermorphin-Cy3 was localized within each fluorescent spot (Fig. 5, D and E). Having shown that rHDL particles contain a single receptor, these photobleaching data indicate that only one dermorphin bound per receptor.
FIGURE 5.
Single-molecule imaging of Cy3-labeled agonist binding to rHDL·YMOR + Gi2 confirms that the majority of YMOR is monomeric when reconstituted into HDL. Binding of [Lys7, Cys8]dermorphin-Cy3, a fluorescently labeled MOR-specific agonist, to rHDL·YMOR+Gi2 was observed with prism-based total internal reflection fluorescence microscopy. YMOR was reconstituted into HDL particles using biotinylated apoA-1, followed by Gi2 heterotrimer addition at a 30:1 G protein to receptor molar ratio. Reconstituted HDL (A) or rHDL·YMOR+Gi2 (B) were then incubated with a saturating concentration (500 nm) of [Lys7, Cys8]dermorphin-Cy3, adhered to a streptavidin-coated quartz slide, and washed with ice-cold 25 mm Tris·HCl, pH 7.7 buffer. Bound [Lys7, Cys8]dermorphin-Cy3 was continuously excited at 532 nm to observe photobleaching of the fluorophore. C, representative fluorescence intensity traces for a one- and two-step photobleach event are shown. The arrows indicate photobleach events. D and E, quantification of photobleaching showed that ∼95% of the bound [Lys7, Cys8]dermorphin-Cy3 bleached in a single step, suggesting that 95% of rHDL particles contained monomeric YMOR. [Lys7, Cys8]dermorphin-Cy3 binding was reversible, as shown the addition of 5 μm NTX. A minimum of four slide regions and 200 fluorescent spots were counted for each sample.
DISCUSSION
Intense research over the past decade has focused on the existence and functional consequences of GPCR oligomerization. Recently, our laboratory and others have taken advantage of a unique phospholipid bilayer platform in the form of rHDL particles (13–16). Using this system we previously demonstrated the functional reconstitution of two prototypical GPCRs, rhodopsin and β2AR (14, 15). Having illustrated that these receptors are monomeric following reconstitution, we observed complete functional coupling to their respective G proteins. For the β2AR, agonist binding to monomeric receptor is allosterically regulated by the stimulatory heterotrimeric G protein Gαsβγ. Therefore GPCR dimerization is not required for functional G protein coupling in the prototypical class A GPCRs rhodopsin and β2AR.
In contrast, the accumulation of considerable biochemical and biophysical evidence suggests that the μ-, δ-, and κ-opioid receptors may function in a fashion that is dependent on their oligomerization. Analysis of receptor overexpression in co-transfected cells suggested the existence of δ-δ homodimers (34) and demonstrated unique pharmacology resulting from μ-δ (6, 7, 10) and δ-κ heterodimers (35). Bioluminescence resonance energy transfer studies have shown that homo- and heterodimers can be formed by all three opioid receptor isoforms (8). These findings have promoted the notion that opioid receptor oligomerization is important for function (36, 37). However, with co-transfection systems the final profiles of μ and δ monomers, homodimers, and heterodimers are unknown. In light of these reports, we rationalized that it was vital to determine whether the monomeric form of the MOR behaves in a pharmacologically similar manner as the native, membrane-bound form.
The HDL reconstitution system was used to investigate the function of monomeric MOR. Reconstitution of MOR required the purification of active receptor in large enough quantities for subsequent biochemical manipulation and analysis. Previous reports of MOR purification from endogenous or recombinant sources have yielded either low quantities (38–42) or poor agonist binding affinities (43–45). Several aspects were key to the expression and purification of YFP-MOR (YMOR): a cleavable hemagglutinin signal sequence at the N terminus of the receptor (46), the presence of naltrexone during expression, and the inclusion of cholesteryl hemisuccinate and naltrexone during the entire purification process. These modifications contributed to the stabilization of YMOR, leading to an increase in yields during the chromatography process as well as increasing the specific activity of the detergent solubilized receptor. Reconstitution into HDL facilitated the isolation of a monomeric form of YMOR that couples to and activates heterotrimeric G proteins. Furthermore, we illustrate that the inhibitory G protein Gαiβγ can allosterically regulate agonist binding to monomeric MOR. These results suggest that G protein regulation of monomeric receptors is likely a phenomenon common to all G proteins and class A GPCRs. The capacity of MOR to couple to various isoforms of Gi/o heterotrimers in a differential manner are currently under investigation in our laboratory.
Taken together our results demonstrate the functionality of monomeric MOR, illustrating that the opioid GPCR does not require dimerization to bind ligands and signal to G proteins. However, these data do not refute the existence of opioid receptor oligomerization in cells. It remains possible that homo- and heterodimerization create unique ligand binding entities that may be subject to differential regulation, desensitization, and internalization. One of our future goals is to isolate various oligomeric states of opioid receptors and other GPCRs in rHDL particles and compare their activities toward ligands and signaling partners directly. A major technical obstacle is the formation of “anti-parallel” receptor dimers, where the N termini for the reconstituted GPCRs in the oligomer are on opposite sides of the phospholipid bilayer. Indeed, reconstitution of two GPCRs in a single rHDL particle has been previously demonstrated for rhodopsin (13, 16), but a detailed analysis by nano-gold labeling and single-particle electron microscopy of the reconstituted “dimers” reveals that a significant fraction were anti-parallel (16). It is plausible, and even likely, that an anti-parallel GPCR dimer will result in suboptimal or disrupted G protein coupling. The development of approaches to ensure parallel GPCR dimer incorporation into rHDL particles to confidently study the functional relevance of opioid receptor dimerization is currently a major priority in the laboratory.
HDL reconstitution of YMOR also provided a platform for analysis of ligand binding using single-molecule microscopy. In this study we examined the reversible binding of a μ-opioid receptor specific agonist [Lys7, Cys8]dermorphin (47) with SM-TIRF. To the best of our knowledge these data represent the first reported observation of a peptide agonist binding to an isolated GPCR in a lipid bilayer using single-particle imaging. We are currently refining our methods to resolve the kinetics of ligand binding to YMOR, as well as visualizing ligand binding to opioid receptors in intact cells. Peptide receptors such as the opioid family are particularly amenable to fluorescence spectroscopy because the relatively large ligand size can tolerate the incorporation of fluorophores without drastically impairing binding affinities. This single-molecule visual approach of studying ligand binding to opioid receptors, utilizing labeled agonists and antagonists with unique binding affinities toward receptor homo- and heterodimers, may potentially address opioid receptor oligomerization in physiologically relevant tissue preparations.
Supplementary Material
Acknowledgments
We thank Drs. John Traynor and Richard Neubig for critical discussion of this work.
This work was supported, in whole or in part, by National Institutes of Health Grants GM068603 (to R. K. S.), GM07767 (to A. J. K. and J. P. A.), GM081025 (to S. P. and N. G. W.), and P60-DK020572 (to R. K. S.). This work was also supported by a pilot and feasibility grant from the Michigan Diabetes Research and Training Center.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- GPCR
- G protein-coupled receptor
- MOR
- μ-opioid receptor
- YMOR
- yellow fluorescent protein-tagged MOR
- HDL
- high density lipoprotein
- rHDL
- reconstituted HDL particles
- apoA-1
- apolipoproteinA-1
- DPN
- diprenorphine
- DAMGO
- [D-Ala2, N-MePhe4, Gly5-ol]enkephalin
- GTPγS
- guanosine 5′-(γ-thio)triphosphate
- NTX
- naltrexone
- DDM
- N-dodecyl-β-d-maltoside
- CHS
- cholesteryl hemisuccinate
- POPC
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
- 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]
- BSA
- bovine serum albumin
- SM-TIRF
- single-molecule total internal reflection fluorescence
- YFP
- yellow fluorescent protein
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- HPLC
- high pressure liquid chromatography
- SEC
- size exclusion chromatography
- β2AR
- β2-adrenergic receptor
- CTAP
- d-Phe-Cys-Tyr-d-Trp-Arg-Thr-Pen-Thr-NH2.
REFERENCES
- 1.Blume A. J., Lichtshtein D., Boone G. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 5626–5630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.North R. A., Williams J. T., Surprenant A., Christie M. J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5487–5491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldhoer M., Bartlett S. E., Whistler J. L. (2004) Annu. Rev. Biochem. 73, 953–990 [DOI] [PubMed] [Google Scholar]
- 4.Bai M. (2004) Cell Signal. 16, 175–186 [DOI] [PubMed] [Google Scholar]
- 5.Milligan G., Canals M., Pediani J. D., Ellis J., Lopez-Gimenez J. F. (2006) Ernst Schering Found. Symp. Proc. 2, 145–161 [DOI] [PubMed] [Google Scholar]
- 6.George S. R., Fan T., Xie Z., Tse R., Tam V., Varghese G., O'Dowd B. F. (2000) J. Biol. Chem. 275, 26128–26135 [DOI] [PubMed] [Google Scholar]
- 7.Martin N. A., Prather P. L. (2001) Mol. Pharmacol 59, 774–783 [DOI] [PubMed] [Google Scholar]
- 8.Wang D., Sun X., Bohn L. M., Sadée W. (2005) Mol. Pharmacol. 67, 2173–2184 [DOI] [PubMed] [Google Scholar]
- 9.Gomes I., Filipovska J., Jordan B. A., Devi L. A. (2002) Methods 27, 358–365 [DOI] [PubMed] [Google Scholar]
- 10.Gomes I., Jordan B. A., Gupta A., Trapaidze N., Nagy V., Devi L. A. (2000) J. Neurosci. 20, RC110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leitz A. J., Bayburt T. H., Barnakov A. N., Springer B. A., Sligar S. G. (2006) BioTechniques 40, 601–606 [DOI] [PubMed] [Google Scholar]
- 12.Nath A., Atkins W. M., Sligar S. G. (2007) Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 13.Bayburt T. H., Leitz A. J., Xie G., Oprian D. D., Sligar S. G. (2007) J. Biol. Chem. 282, 14875–14881 [DOI] [PubMed] [Google Scholar]
- 14.Whorton M. R., Bokoch M. P., Rasmussen S. G., Huang B., Zare R. N., Kobilka B., Sunahara R. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7682–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whorton M. R., Jastrzebska B., Park P. S., Fotiadis D., Engel A., Palczewski K., Sunahara R. K. (2008) J. Biol. Chem. 283, 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banerjee S., Huber T., Sakmar T. P. (2008) J. Mol. Biol. 377, 1067–1081 [DOI] [PubMed] [Google Scholar]
- 17.Liang Y., Fotiadis D., Filipek S., Saperstein D. A., Palczewski K., Engel A. (2003) J. Biol. Chem. 278, 21655–21662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fotiadis D., Liang Y., Filipek S., Saperstein D. A., Engel A., Palczewski K. (2003) Nature 421, 127–128 [DOI] [PubMed] [Google Scholar]
- 19.Jastrzebska B., Fotiadis D., Jang G. F., Stenkamp R. E., Engel A., Palczewski K. (2006) J. Biol. Chem. 281, 11917–11922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffner W., Weissmann C. (1973) Anal. Biochem. 56, 502–514 [DOI] [PubMed] [Google Scholar]
- 21.Kozasa T. (1999) in G Proteins: Techniques of Analysis ( Manning D. R. ed) pp. 23–28, CRC Press, Boca Raton, FL [Google Scholar]
- 22.Patil P. V., Ballou D. P. (2000) Anal. Biochem. 286, 187–192 [DOI] [PubMed] [Google Scholar]
- 23.Emmerson P. J., Clark M. J., Mansour A., Akil H., Woods J. H., Medzihradsky F. (1996) J. Pharmacol. Exp. Ther 278, 1121–1127 [PubMed] [Google Scholar]
- 24.Alt A., Mansour A., Akil H., Medzihradsky F., Traynor J. R., Woods J. H. (1998) J. Pharmacol. Exp. Ther. 286, 282–288 [PubMed] [Google Scholar]
- 25.Clark M. J., Furman C. A., Gilson T. D., Traynor J. R. (2006) J. Pharmacol. Exp. Ther. 317, 858–864 [DOI] [PubMed] [Google Scholar]
- 26.Frances B., Moisand C., Meunier J. C. (1985) Eur. J. Pharmacol. 117, 223–232 [DOI] [PubMed] [Google Scholar]
- 27.Werling L. L., Puttfarcken P. S., Cox B. M. (1988) Mol. Pharmacol. 33, 423–431 [PubMed] [Google Scholar]
- 28.Ott S., Costa T. (1989) Biochem. Pharmacol. 38, 1931–1939 [DOI] [PubMed] [Google Scholar]
- 29.Richardson A., Demoliou-Mason C., Barnard E. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10198–10202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenakin T. (2004) Trends Pharmacol. Sci. 25, 186–192 [DOI] [PubMed] [Google Scholar]
- 31.Rosenbaum D. M., Cherezov V., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Yao X. J., Weis W. I., Stevens R. C., Kobilka B. K. (2007) Science 318, 1266–1273 [DOI] [PubMed] [Google Scholar]
- 32.Cho T. M., Ge B. L., Loh H. H. (1985) Life Sci. 36, 1075–1085 [DOI] [PubMed] [Google Scholar]
- 33.Ramon E., Marron J., del Valle L., Bosch L., Andrés A., Manyosa J., Garriga P. (2003) Vision Res. 43, 3055–3061 [DOI] [PubMed] [Google Scholar]
- 34.Cvejic S., Devi L. A. (1997) J. Biol. Chem. 272, 26959–26964 [DOI] [PubMed] [Google Scholar]
- 35.Jordan B. A., Devi L. A. (1999) Nature 399, 697–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levac B. A., O'Dowd B. F., George S. R. (2002) Curr. Opin Pharmacol. 2, 76–81 [DOI] [PubMed] [Google Scholar]
- 37.Milligan G. (2004) Mol. Pharmacol. 66, 1–7 [DOI] [PubMed] [Google Scholar]
- 38.Gioannini T. L., Howard A. D., Hiller J. M., Simon E. J. (1985) J. Biol. Chem. 260, 15117–15121 [PubMed] [Google Scholar]
- 39.Li L. Y., Zhang Z. M., Su Y. F., Watkins W. D., Chang K. J. (1992) Life Sci. 51, 1177–1185 [DOI] [PubMed] [Google Scholar]
- 40.Talmont F., Sidobre S., Demange P., Milon A., Emorine L. J. (1996) FEBS Lett. 394, 268–272 [DOI] [PubMed] [Google Scholar]
- 41.Obermeier H., Wehmeyer A., Schulz R. (1996) Eur. J. Pharmacol. 318, 161–166 [DOI] [PubMed] [Google Scholar]
- 42.Massotte D., Baroche L., Simonin F., Yu L., Kieffer B., Pattus F. (1997) J. Biol. Chem. 272, 19987–19992 [DOI] [PubMed] [Google Scholar]
- 43.Sarramegna V., Demange P., Milon A., Talmont F. (2002) Protein Expr. Purif. 24, 212–220 [DOI] [PubMed] [Google Scholar]
- 44.Sarramegna V., Talmont F., Seree de Roch M., Milon A., Demange P. (2002) J. Biotechnol. 99, 23–39 [DOI] [PubMed] [Google Scholar]
- 45.Sarramegna V., Muller I., Mousseau G., Froment C., Monsarrat B., Milon A., Talmont F. (2005) Protein Expr. Purif. 43, 85–93 [DOI] [PubMed] [Google Scholar]
- 46.Guan X. M., Kobilka T. S., Kobilka B. K. (1992) J. Biol. Chem. 267, 21995–21998 [PubMed] [Google Scholar]
- 47.Attila M., Salvadori S., Balboni G., Bryant S. D., Lazarus L. H. (1993) Int. J. Pept. Protein Res. 42, 550–559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.