Abstract
Inflammasomes have been extensively characterized in monocytes and macrophages, but not in epithelial cells, which are the preferred host cells for many pathogens. Here we show that cervical epithelial cells express a functional inflammasome. Infection of the cells by Chlamydia trachomatis leads to activation of caspase-1, through a process requiring the NOD-like receptor family member NLRP3 and the inflammasome adaptor protein ASC. Secretion of newly synthesized virulence proteins from the chlamydial vacuole through a type III secretion apparatus results in efflux of K+ through glibenclamide-sensitive K+ channels, which in turn stimulates production of reactive oxygen species. Elevated levels of reactive oxygen species are responsible for NLRP3-dependent caspase-1 activation in the infected cells. In monocytes and macrophages, caspase-1 is involved in processing and secretion of pro-inflammatory cytokines such as interleukin-1β. However, in epithelial cells, which are not known to secrete large quantities of interleukin-1β, caspase-1 has been shown previously to enhance lipid metabolism. Here we show that, in cervical epithelial cells, caspase-1 activation is required for optimal growth of the intracellular chlamydiae.
Chlamydia trachomatis is the most common cause of bacterial sexually transmitted disease in the United States, and it is the leading cause of preventable blindness in the world (1–5). Untreated, C. trachomatis infection in women can cause pelvic inflammatory disease, which can lead to infertility and ectopic pregnancy because of scarring of the ovaries and the Fallopian tubes (6). Infection by the lymphogranuloma venereum (LGV)2 strain of C. trachomatis, which has become more common in North America and Europe (7, 8), is characterized by swelling and inflammation of the lymph nodes in the groin (9).
Chlamydiae are intracellular pathogens that preferentially infect epithelial mucosa and have a biphasic infection cycle (10). A metabolically inactive form, the elementary body, infects the epithelial host cells through entry vesicles that avoid fusion with host cell lysosomes and develop into a membrane-bound inclusion (11–13). Despite their intravacuolar localization, chlamydiae are still able to acquire nutrients from the host cell and interact with host-cell signaling pathways (13–23). Within a few hours, the elementary bodies differentiate into larger, metabolically active reticulate bodies, which proliferate but are noninfectious. Depending on the strain of C. trachomatis, the reticulate bodies transform back into elementary bodies after 1–3 days and are released into the extracellular medium to infect other cells (11, 24, 25). Chlamydial species possess a type III secretion (T3S) system that secretes bacterial virulence factors into host cell cytosol and may control interactions between the inclusion and host-cell compartments (26).
Long before the adaptive immune response is activated, infected epithelial cells produce proinflammatory cytokines and chemokines, including interleukin (IL)-6, IL-8, and granulocyte-macrophage colony-stimulating factor (27), which recruit neutrophils to the site of infection and activate other immune effector cells. However, in many cases the immune system fails to clear the infection, and the chronic release of cytokines becomes a major contributor to the scarring and damage associated with the infection (28–30).
The innate immune response during C. trachomatis infection is initiated by chlamydial pathogen-associated molecular patterns, including lipopolysaccharides, which bind to pattern recognition receptors such as Toll-like receptors and cytosolic NOD-like receptors (NLRs), ultimately promoting pro-inflammatory cytokine gene expression and secretion of the cytokine proteins (31–37). However, secretion of the key pro-inflammatory cytokine IL-1β is tightly regulated (38). First, pro-IL-1β is produced following activation of pattern recognition receptor, and the precursor is then cleaved into the mature form by the pro-inflammatory cysteine protease, caspase-1 (also known as interleukin-1 converting enzyme or ICE). The mechanism by which caspase-1 is activated in response to infection or tissue damage was found to be modulated by a macromolecular protein complex termed the “inflammasome,” which consists of an NLR family member, an adaptor protein (apoptosis-associated speck-like protein containing a caspase activation recruitment domain or ASC), and an inactive caspase-1 precursor (pro-caspase-1) (39, 40). Previous studies demonstrated that IL-1β is produced in response to chlamydial infection in dendritic cells, macrophages, and monocytes (41–44). Moreover, C. trachomatis or Chlamydia caviae infection activates caspase-1 in epithelial cells or monocytes (43, 45, 46). However, whether caspase-1 activation during chlamydial infection requires the formation of an inflammasome remains unclear.
Previous studies have shown that different pathogens can cause inflammasome-mediated caspase-1 activation in macrophages and monocytes (47). However, epithelial cells lining mucosal surfaces are not only the preferred target for chlamydial infection and other intracellular pathogens but also play an important role in early host immune response to infection by secreting proinflammatory cytokines and chemokines (27). Although epithelial cells are not known to secrete large amounts of IL-1β, inflammasome-dependent caspase-1 activation in epithelial cells is known to contribute to lipid metabolism and membrane regeneration in epithelial cells damaged by the membrane-disrupting toxin, aerolysin (48). As lipids are sorted from the Golgi apparatus to the chlamydial inclusion (13, 15, 49), we therefore investigated whether C. trachomatis induces caspase-1 activation in epithelial cells via the assembly of an inflammasome. We demonstrated that C. trachomatis-induced caspase-1 activation is mediated by an inflammasome containing the NLR member, NLRP3. Several studies have demonstrated the involvement of T3S apparatus in inflammasome-mediated caspase-1 activation by different pathogens in macrophages and monocytes (50–56). Therefore, we further investigated the mechanism by which C. trachomatis triggers the formation of the NLRP3 inflammasome. Our results showed that metabolically active chlamydiae, relying on their T3S apparatus, cause K+ efflux, which in turn leads to formation of reactive oxygen species (ROS) and ultimately NLRP3-dependent caspase-1 activation. Epithelial cells do not typically secrete large amounts of IL-1β; instead, caspase-1 activation in cervical epithelial cells contributes to development of the chlamydial inclusion.
EXPERIMENTAL PROCEDURES
Cells, Bacteria, and Chemical Reagents
Cervical epithelial cells (HeLa 229 cells) and the LGV/L2 strain of C. trachomatis were obtained from American Type Culture Collection (ATCC). HeLa cells were cultured in a humidified incubator at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (Dulbecco's modified Eagle's medium/F-12) (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), and 10 μg/ml gentamycin (Omega Scientific, Tarzana, CA). The number of bacterial inclusion forming units was determined as described previously (18). N-Acetylcysteine (NAC), glibenclamide, penicillin G, and cycloheximide were from Sigma. KCl was from Fisher; chloramphenicol was from Calbiochem, and Z-YVAD-fmk was from Biovision (Mountain View, CA). Z-WEHD-fmk was purchased from R & D Systems (Minneapolis, MN). INPs (0341 and 0406) were kind gifts from Dr. Pia Keyser (Innate Pharmaceuticals, Umeå, Sweden).
Cell Culture, Infection, and Treatments
HeLa cells growing at 50% confluency on tissue culture flasks (Costar, Corning, NY) were infected with the LGV/L2 strain of C. trachomatis at a multiplicity of infection (m.o.i.) of 3.0, unless specified otherwise, and incubated for the indicated times in an incubator at 37 °C with 5% CO2. Treatment with inhibitors or other reagents was performed at the indicated times and concentrations.
Generation of HeLa Cells Expressing shRNA
HeLa cells stably expressing shRNA against NLRP3 and ASC were obtained by transducing HeLa cells with lentiviral particles. The sequences 5′-CCGGGCGTTAGAAACACTTCAAGAACTCGAGTTCTTGAAGTGTTTCTAACGCTTTTTG-3′ for human NLRP3 (Sigma; catalog number NM_004895) and 5′-CCGGCGGAAGCTCTTCAGTTTCACACTCGAGTGTGAAACTGAAGAGCTTCCGTTTTTG-3′ for human ASC (Sigma; catalog number NM_013258) were used separately to silence gene expression following the manufacturer's instructions. Nontarget shRNA control cells were also generated using an irrelevant sequence (Sigma; catalog number SHC002V).
FLICA Staining
During the last hour of incubation, cells were labeled with FAM-YVAD-fmk caspase-1 FLICATM kit (Immunochemistry, Bloomington, IN), which binds activated caspase-1. Flow cytometric analysis was performed according to manufacturer's manual. In brief, cells were detached using TrypLETM Express (Invitrogen) and then incubated with 1× FLICA for 1 h followed by two washes and analyzed with a Guava EasyCyte (Guava Technologies, Hayward, CA).
Western Blotting
Samples were lysed using RIPA Lysis Buffer (Millipore) and loaded onto a 15% SDS-polyacrylamide gel and then transferred to a polyvinylidene difluoride membrane (Millipore). Blots were blocked for 1 h with 5% (w/v) nonfat dried milk in TBST. The membrane was incubated overnight at 4 °C with rabbit anti-human caspase-1 antibody (Millipore) and then incubated again with conjugated anti-rabbit IgG horseradish peroxidase (Millipore). For confirmation of NLRP3 depletion by RNA interference, a 9% gel was used, and the blot was incubated with rabbit anti-human NLRP3 antibody (Sigma; catalog number HPA012878). Immunoreactive proteins were detected with ECL Plus Western blotting detection reagents (Amersham Biosciences) using a gel doc system (Bio-Rad).
Measurement of Production of ROS
Cells were labeled with the cell-permeant ROS indicator dihydrocalcein-AM (Molecular Probes, Eugene, OR), following the manufacturer's instructions. Briefly, cells were plated in phenol red-free Dulbecco's modified Eagle's medium (Invitrogen) and then infected and/or treated for the indicated times. Cells were loaded with 10 μm dihydrocalcein-AM in phosphate-buffered saline for 45 min at 37 °C, then recovered in growth media for 20 min, and finally analyzed by flow cytometry with a Guava EasyCyte.
RNA Isolation and Real Time PCR
mRNA was isolated from HeLa cells using the Qiagen RNeasy kit (Qiagen, Valencia, CA) following the manufacturer's instructions, and total RNA was converted into cDNA by standard reverse transcription with Taqman® reverse transcriptase kit (Applied Biosystems, Foster City, CA). Quantitative PCR was performed with 1:50 of the cDNA preparation in the Mx3000P (Stratagene, La Jolla, CA) in a 25-μl final volume with Brilliant QPCR master mix (Stratagene). The primers for human GAPDH were 5′-CTTCTCTGATGAGGCCCAAG-3′ forward and 5′-GCAGCAAACTGGAAAGGAAG-3′ reverse. Primers for human NLRP3 were 5′-CTTCCTTTCCAGTTTGCTGC-3′ forward and 5′-TCTCGCAGTCCACTTCCTTT-3′ reverse. Primers for human ASC were 5′-AGTTTCACACCAGCCTGGAA-3′ forward and 5′-TTTTCAAGCTGGCTTTTCGT-3′ reverse. Real time PCR included initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min, and one cycle of 95 °C for 1 min, 55 °C for 30 s, 95 °C for 30 s.
Chlamydia Quantitation and Fluorescence Microscopy
Following 24 h of infection, cells were harvested using a cell scraper, frozen at −80 °C, thawed, and thoroughly vortexed before titrating on 50% confluent HeLa cells. Cells were then stained by Hoechst stain (Sigma) and anti-chlamydial antibody (Argene, North Massapequa, NY), mounted on slides, and quantified by immunofluorescence on a wide field fluorescence microscope (Leica, Deerfield, IL).
Statistical and Flow Cytometric Analyses
The statistical analysis was performed using GraphPad Instat software (GraphPad Software Inc, La Jolla, CA) by Student's t test and was considered significant at p < 0.05. Flow cytometry data were analyzed using FlowJo® software (Tree Star Inc, Ashland, OR).
RESULTS
Chlamydia-mediated Caspase-1 Activation in Epithelial Cells Requires the NLRP3 Inflammasome
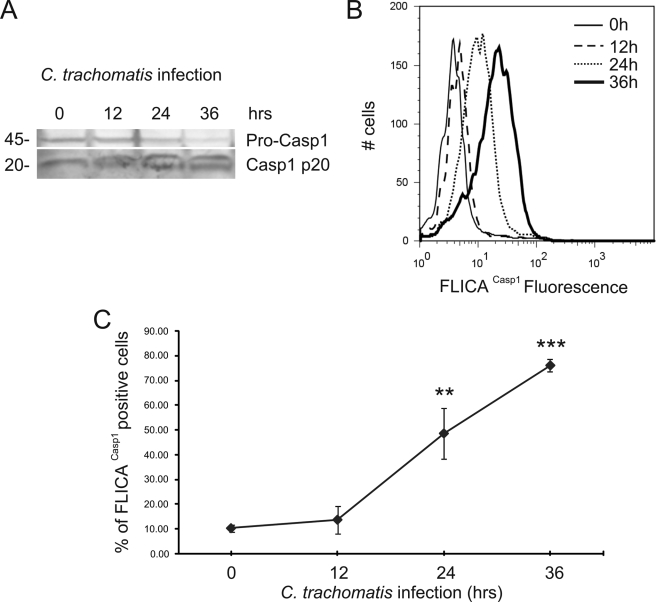
To characterize caspase-1 activation during C. trachomatis infection, we infected human cervical epithelial (HeLa) cells with C. trachomatis (L2) at a multiplicity of infection (m.o.i.) of 3 for 12, 24, and 36 h. The 45-kDa procaspase-1 was cleaved into the active 20-kDa caspase-1 at 24 h post infection, and processing was further increased at 36 h post infection, as shown by the disappearance of the procaspase-1 zymogen and the concomitant appearance of the active caspase-1 (Fig. 1A). Similar results were obtained when we quantified caspase-1 activation in HeLa cells using a cell-permeable fluorescent reagent, FLICACasp1, which can specifically bind to the active form of caspase-1 (Fig. 1, B and C). Caspase-1 activation was barely noticeable at 12 h post infection but became significant after a day of infection (Fig. 1C), in agreement with recent studies (45).
FIGURE 1.
C. trachomatis induces caspase-1 activation in HeLa cells. HeLa cells were infected with C. trachomatis at an m.o.i. = 3 for 12, 24, and 36 h. A, Western blot analysis of HeLa cell lysates was performed to monitor caspase-1 (Casp1) activation using an antibody that detects pro-caspase-1 (p45, upper band) and active caspase-1 (p20, lower band). B, caspase-1 activation was quantified using fluorescent FLICACasp1 reagent and analyzed by flow cytometry. Nonfluorescent cells were gated in the first log-decade, and the fluorescence intensity was proportional to the level of caspase-1 activation. C, column chart of FLICACasp1 flow cytometry data, showing the % of cells with activated caspase-1 as a function of infection time. Error bars represent standard deviation (n = 3). ** indicate p < 0.01; *** indicate p < 0.001, compared with uninfected cells.
Next we sought to determine whether an inflammasome may be required for caspase-1 activation. The adaptor protein ASC can be coupled to different NLR family members and therefore seemed likely to play a role in inflammasome activation during C. trachomatis infection. As IPAF (also known as NLRC4 (57)) was shown in previous studies to recognize mainly bacterial flagellin (58, 59), which is not expressed in the chlamydial genome (60), we focused our initial attention on NLRP3 (also known as cryopyrin and Nalp3 (57)).
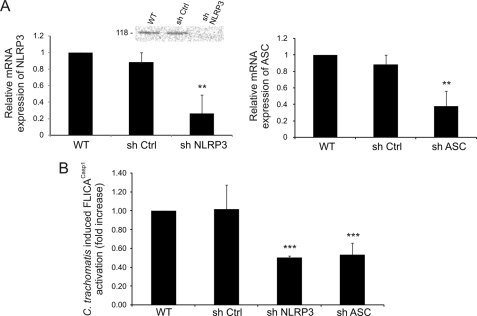
A role for either ASC or NLRP3 during chlamydial infection was determined by silencing NLRP3 or ASC by shRNA in HeLa cells. mRNA expression of either inflammasome component was significantly reduced in comparison with nontarget shRNA, as measured by real time PCR (Fig. 2A), although higher levels of NLRP3 protein depletion were observed by Western blotting (Fig. 2A). Consistent with a role for the NLRP3 inflammasome in caspase-1 activation, individual knockdown of NLRP3 or ASC caused a reduction of ∼50% in the activation of caspase-1 after 24 h of C. trachomatis infection, when compared with wild type HeLa cells or cells that were transfected with nontarget shRNA (Fig. 2B). The reduction of caspase-1 activation was not complete (Fig. 2B) and was similar to the level of mRNA depletion by shRNA but lower than the level of NLRP3 protein depletion (Fig. 2A). These results imply that C. trachomatis infection induces caspase-1 activation through a process that requires, at least partially, the assembly of the NLRP3 inflammasome.
FIGURE 2.
C. trachomatis-induced caspase-1 activation requires NLRP3 inflammasome. A, HeLa cells were stably transfected with shRNAs that target NLRP3 or ASC, and mRNA expressions of NLRP3 (left panel) and ASC (right panel) were quantified by real time PCR and compared with wild type (WT) and nontarget control (sh Ctrl). Inset, Western blot analysis of wild type HeLa cells, cells treated with nontarget control, and cells treated with shNLRP3, confirming decreased expression of the NLRP3 protein after mRNA knockdown. Western blot was performed with an anti-NLRP3 antibody, which detects the 118-kDa protein. B, NLRP3, ASC, or nontarget control knockdown HeLa cells were infected with L2 at an m.o.i. = 3 for 24 h, and C. trachomatis-induced caspase-1 activation was measured by FLICACasp1. The fold increase in caspase-1 activation in infected nontarget controls, shNLRP3-treated cells, and shASC-treated cells with respect to uninfected cells was compared with the increase in 24 h-infected wild type cells. Error bars represent standard deviation of an experiment performed in triplicate on three separate occasions. ** indicates p < 0.01; *** indicates p < 0.001.
Chlamydia-induced Caspase-1 Activation Is Caused by ROS Production as a Result of K+ Efflux
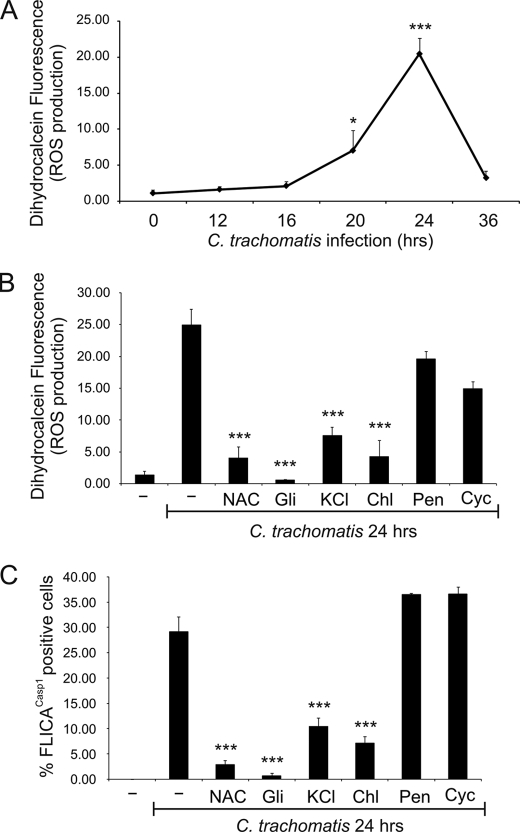
We investigated the mechanism by which C. trachomatis induces caspase-1 activation in HeLa cells. Previous studies have shown that ATP-dependent ROS production in macrophages can activate caspase-1 (61), and that asbestos and silica can activate the NLRP3 inflammasome by increasing ROS production (62, 63). We therefore evaluated whether C. trachomatis increases ROS production in HeLa cells, using a commercially available fluorescent reagent, dihydrocalcein, to measure intracellular ROS production. Similarly to the time course for caspase-1 activation, there was essentially no ROS production during the first 12 h of chlamydial infection, but ROS production increased at 20 h post infection and continued to increase at 24 h post infection. However, unlike the case for caspase-1 activation, ROS levels returned to basal levels at 36 h post infection (Fig. 3A).
FIGURE 3.
Caspase-1 activation following C. trachomatis infection is caused by K+ efflux and ROS production. A, HeLa cells were infected with C. trachomatis at an m.o.i. = 3 for 0, 12, 16, 20, and 24 h, and intracellular ROS levels were measured with the fluorescent dihydrocalcein reagent and analyzed by flow cytometry. Data are plotted as a line chart. B and C, HeLa cells were infected or not with C. trachomatis at an m.o.i. = 3 for 24 h, and treated or not with 10 mm NAC, 50 μm glibenclamide (Gli), 70 mm KCl during the last 15 h of infection, or 60 μg/ml chloramphenicol (Chl), 100 μg/ml penicillin (Pen), 10 μg/ml cycloheximide (Cyc) during the last 6 h of infection. ROS production was quantified with dihydrocalcein (B), or caspase-1 activation was measured with FLICACasp1 (C). Error bars represent standard deviation from at least three separate experiments. * indicates p < 0.05; *** indicates p < 0.001, compared with uninfected cells (A) or untreated infected cells (B and C).
To confirm whether chlamydia-induced ROS production may be involved in activation of caspase-1, we used the anti-oxidant NAC, which was previously shown to inhibit caspase-1 activation and NLRP3 assembly (61, 63). Interestingly, the antioxidant NAC significantly diminished both ROS production and caspase-1 activation induced by 24 h of chlamydial infection (Fig. 3, B and C), suggesting that ROS production is upstream from caspase-1 activation.
Besides ATP binding to the purinergic receptor, P2X7, the NLRP3 inflammasome can be activated by ligands as varied as asbestos, alum, monosodium urate, bacterial toxins, and K+ ionophores (64, 65). What all of these disparate ligands have in common is their ability to induce K+ efflux from cells (63, 66). Given that an older study had shown that chlamydial infection causes loss of intracellular potassium (67), we examined whether K+ efflux results in ROS production and/or caspase-1 activation during chlamydial infection. Indeed, specifically blocking potassium channels with glibenclamide or limiting K+ release by addition of extracellular potassium (KCl) was able not only to significantly reduce caspase-1 activation but also to diminish ROS production induced by 24 h of infection with C. trachomatis (Fig. 3, B and C). Thus, our results demonstrate that C. trachomatis infection leads to loss of intracellular potassium, which in turn causes production of ROS and subsequently caspase-1 activation.
Chlamydial Protein Synthesis and the T3S System Are Required for ROS Production and Caspase-1 Activation
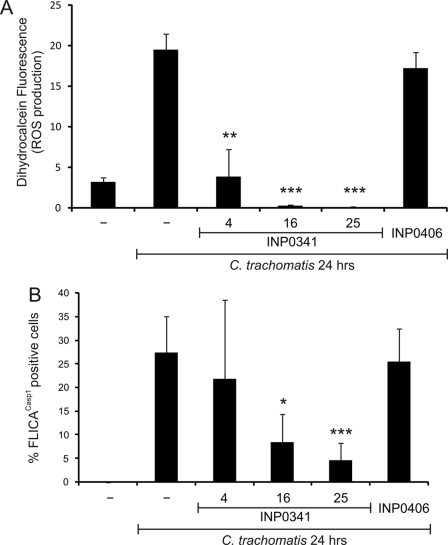
To further investigate the mechanism by which Chlamydia activates caspase-1, we explored the possibility that chlamydiae may secrete proteins into the host cells via T3S, leading to ROS production and activation of caspase-1. We first checked whether chlamydial protein synthesis is important for ROS production and caspase-1 activation during infection. Indeed, chloramphenicol (chlamydial protein synthesis inhibitor), but neither penicillin (inhibitor of chlamydial division) nor cycloheximide (host cell protein synthesis inhibitor), was able to significantly decrease ROS production and caspase-1 activation following 24 h of infection with C. trachomatis (Fig. 3, B and C). To determine whether the newly synthesized bacterial proteins must be secreted, we used a chemically synthesized compound that can inhibit chlamydial T3S (INP0341) and a closely related control compound INP (INP0406) (68). INP0341, but not INP0406, was able to abrogate Chlamydia-induced ROS production in a dose-dependent manner (Fig. 4A). Moreover, INP0341, unlike INP0406, significantly diminished caspase-1 activation after 24 h of C. trachomatis infection (Fig. 4B). These results highlight the need for chlamydial T3S in causing ROS production and the subsequent activation of caspase-1.
FIGURE 4.
Chlamydial T3S is responsible for ROS production and caspase-1 activation. HeLa cells were infected or not with C. trachomatis at an m.o.i. = 3 for 24 h and treated or not with 4, 16, or 25 μm T3S inhibitor (INP0341) or 25 μm control INP (INP0406) for 9 h. ROS production was quantified by staining cells with dihydrocalcein (A), or caspase-1 activation was measured by staining cells with FLICACasp1 (B). Error bars represent standard deviation of at least three separate experiments. * indicates p < 0.05; ** indicates p < 0.01; *** indicates p < 0.001, compared with infected untreated cells.
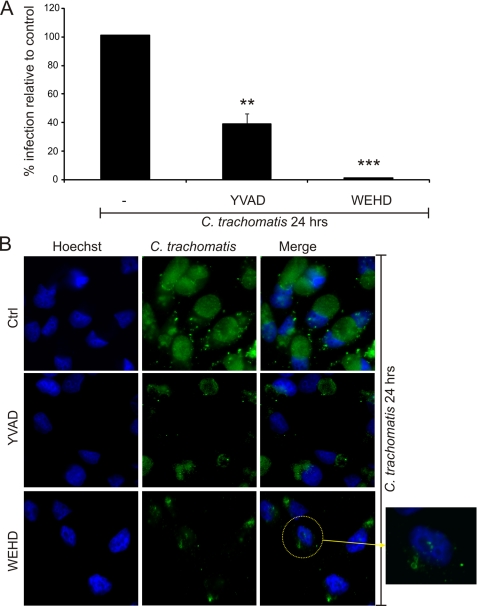
Caspase-1 Activation Is Required for Efficient Development of the Chlamydial Inclusion
NLRP3-dependent caspase-1 activation during plasma membrane damage because of treatment with the bacterial toxin, aerolysin, has been shown to play an indispensable role in promoting lipid synthesis and membrane repair (48). As this process also involved K+ efflux from the toxin-treated cells, we investigated the possibility that caspase-1 activation may be required for efficient chlamydial infection. For this purpose, epithelial cells were infected with C. trachomatis at an m.o.i. of 3 for 24 h and treated with the irreversible caspase-1 inhibitor Z-YVAD-fmk or the caspase-1/caspase-5 inhibitor Z-WEHD-fmk for the last 15 h of infection (the inhibitors were added after 9 h post infection). When the chlamydiae were retitrated on a fresh monolayer of epithelial cells, a dramatic decrease in the infectious activity of the chlamydiae was observed, especially in cell samples that were treated with the caspase-1/caspase-5 inhibitor (Fig. 5A). Furthermore, addition of either inhibitor had a remarkable effect on the chlamydial inclusions; smaller, fragmented inclusions were observed in the epithelial cells treated with either inhibitor (Fig. 5B). These results suggest that caspase-1 activation is required for efficient chlamydial infection.
FIGURE 5.
Caspase-1 activation is required for efficient C. trachomatis infection. HeLa cells were infected with C. trachomatis at an m.o.i. = 3 for 24 h and treated or not with two different caspase inhibitors as follows: 100 μm caspase-1 inhibitor (Z-YVAD-fmk) or 100 μm caspase-1/caspase-5 inhibitor (Z-WEHD-fmk) at 9 h post infection. A, cells were harvested and retitrated on a fresh monolayer of HeLa cells for 24 h and stained with Hoechst stain for DNA and anti-chlamydial antibody, and infected cells were counted. B, cells were stained with Hoechst stain (blue) and anti-chlamydial antibody (green) and visualized on a fluorescence microscope. Ctrl, control. Error bars represent standard deviation of three separate experiments. ** indicates p < 0.01; *** indicates p < 0.001, compared with infected untreated cells.
DISCUSSION
Processing and secretion of IL-1β and IL-18 require the activity of caspase-1, which in turn is activated following assembly of an inflammasome (37). Infection of monocytes and macrophages by Chlamydia in vitro leads to IL-1β secretion, and a requirement for caspase-1 was shown (43, 45, 46, 69). This suggests that an inflammasome is assembled in monocytes and macrophages during chlamydial infection.
However, the preferred host cells for most chlamydial species and strains are epithelial cells. More specifically, sexually transmitted strains of C. trachomatis infect mainly epithelial cells of the urogenital tract. Both C. trachomatis and the murine strain, Chlamydia muridarum, were recently shown to activate caspase-1 in human epithelial cells (45, 69), with a time course similar to that observed by us. Neither the mechanism for caspase-1 activation nor the consequences of caspase-1 activation for development of chlamydial infection were investigated. Clearance of infection of vaginally infected mice was not affected by caspase-1 deficiency in vivo (45), but as caspase-1 could both inhibit infection through its role in secretion of IL-1β by monocytes/macrophages and enhance infection in infected epithelial cells, the two effects could have cancelled each other. A direct role for IL-1β in resolution of chlamydial infection therefore needs to be evaluated using IL-1β-deficient mice. Nonetheless, caspase-1 deficiency did affect upper urogenital tract pathology in Chlamydia-infected mice (45).
Here we show that infection of cervical epithelial cells with C. trachomatis leads to caspase-1 activation through a process that requires both inflammasome components, NLRP3 and ASC. In other examples of NLRP3 inflammasome activation, both K+ efflux and ROS production were shown to play a nonredundant role (62, 63, 66). The link between K+ efflux and ROS production (63) could reflect plasma membrane depolarization, which is known to precede ROS production (70). We show that chlamydial infection leads to K+ efflux through K+-specific channels, in agreement with an older study showing that chlamydial infection causes K+ loss from infected cells (67). K+ efflux in turn results in ROS production, because blocking K+ efflux with a K+-channel blocker prevents ROS production. Elevated ROS levels on their own were known to induce NLRP3 inflammasome or caspase-1 activation (61–63), and we find that treating infected cells with antioxidants blocks caspase-1 activation.
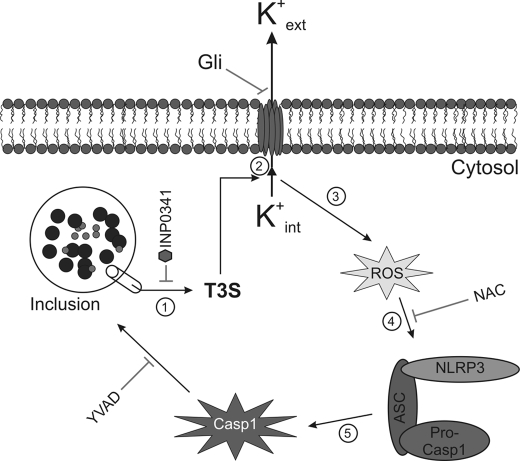
Several studies have demonstrated that T3S is required for different pathogens to cause inflammasome activation (50–56), and C. trachomatis was known to activate caspase-1 through a T3S-dependent mechanism (69). We find that both ROS production and caspase-1 activation require newly synthesized chlamydial proteins, but not host-cell protein synthesis, and both can be blocked with a T3S inhibitor. Taken together, our results suggest that T3S-dependent bacterial protein secretion triggers K+ efflux, which leads to ROS production and subsequently NLRP3-mediated caspase-1 activation in Chlamydia-infected cells (Fig. 6).
FIGURE 6.
C. trachomatis triggers inflammasome-mediated caspase-1 activation through T3S-induced K+ efflux and ROS production. Following infection, chlamydiae inject virulence factors via the T3S apparatus into the host cell cytosol, causing loss of intracellular potassium and resulting in the production of ROS. Elevated ROS levels trigger the assembly of the NLRP3 inflammasome, which subsequently activates caspase-1. Activated caspase-1 plays a role in lipid metabolism and glycolysis and enhances development of the chlamydiae.
Although caspase-1 has been studied mainly in the context of inflammation, pyroptosis, and its role in processing IL-1β and IL-18, a much wider range of caspase-1 substrates has recently been identified through a gel proteomic approach (71). Furthermore, toxins that damage the plasma membrane, causing K+ efflux, also promote caspase-1-dependent activation of the primary regulators of membrane biogenesis, the sterol regulator element-binding proteins (48). As the chlamydial inclusion diverts lipids from the host-cell Golgi apparatus (13, 15, 49), we investigated the possibility that caspase-1 may also modulate chlamydial development. We find in fact that a caspase-1 inhibitor can inhibit chlamydial infection by ∼60%, when included during the last 15 h of a 24-h infection (Fig. 6).
We found a larger effect when using an inhibitor that blocks both caspase-1 and caspase-5, consistent with the presence of both of these inflammatory caspases in an inflammasome in monocytes (39) and the expression of both caspases in cervical epithelial cells (72). Although Z-WEHD-fmk also affects caspase-4, it may also be a more effective caspase-1 inhibitor than Z-YVAD-fmk (73). During the course of this study, another laboratory reported that Z-WEHD-fmk can decrease chlamydial infection in epithelial cells (74). Thus, growth of the inclusion requires fragmentation of the Golgi apparatus, which is because of proteolytic cleavage of a Golgi matrix protein, Golgin-84. Blocking Golgin-84 cleavage with Z-WEHD-fmk prevents Golgi fragmentation and inhibits lipid acquisition and maturation of the chlamydial inclusion (74).
In uninfected cells, caspase-1 can directly cleave 41 proteins, including chaperones, cytoskeletal proteins, glycolytic enzymes, and translation machinery proteins (71). Characterization of NLRP3-dependent caspase-1 activation during chlamydial infection is therefore bound to uncover other effects of infection on host-cell functions, which may include energy metabolism and cytoskeletal integrity.
Acknowledgments
We are grateful to Drs. Pia Keyser (Innate Pharmaceuticals, Umeå, Sweden) and Ellena Peterson (University of California, Irvine) for graciously sharing inhibitors of type III secretion.
This work was supported by the University of California (to D. M. O.) and the Else Kröner-Fresenius-Stiftung (to G. H.).
- LGV
- lymphogranuloma venereum
- ROS
- reactive oxygen species
- IL
- interleukin
- m.o.i.
- multiplicity of infection
- Z
- benzyloxycarbonyl
- fmk
- fluoromethyl ketone
- NAC
- N-acetylcysteine
- shRNA
- short hairpin RNA
- T3S
- type III secretion
- NLR
- NOD-like receptor.
REFERENCES
- 1.Gerbase A. C., Rowley J. T., Mertens T. E. (1998) Lancet 351, 2–4 [DOI] [PubMed] [Google Scholar]
- 2.Miller W. C., Ford C. A., Morris M., Handcock M. S., Schmitz J. L., Hobbs M. M., Cohen M. S., Harris K. M., Udry J. R. (2004) JAMA 291, 2229–2236 [DOI] [PubMed] [Google Scholar]
- 3.Schachter J. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity ( Stephens R. S. ed) pp. 139–170, American Society for Microbiology, Washington, DC [Google Scholar]
- 4.Thylefors B., Négrel A. D., Pararajasegaram R., Dadzie K. Y. (1995) Bull. World Health Organ. 73, 115–121 [PMC free article] [PubMed] [Google Scholar]
- 5.Belland R., Ojcius D. M., Byrne G. I. (2004) Nat. Rev. Microbiol. 2, 530–531 [DOI] [PubMed] [Google Scholar]
- 6.Cohen C. R., Brunham R. C. (1999) Sex. Trans. Infect. 75, 21–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bébéar C., de Barbeyrac B. (2009) Clin. Microbiol. Infect. 15, 4–10 [DOI] [PubMed] [Google Scholar]
- 8.Tinmouth J., Gilmour M. W., Kovacs C., Kropp R., Mitterni L., Rachlis A., Richards S., Salit I., Sikri R., Valencia G. R., Wesson T., Wong T., Wood H. (2008) Int. J. STD AIDS 19, 805–809 [DOI] [PubMed] [Google Scholar]
- 9.Mabey D., Peeling R. W. (2002) Sex Transm. Infect. 78, 90–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moulder J. W. (1991) Microbiol. Rev. 55, 143–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyrick P. B. (2000) Cell. Microbiol. 2, 275–282 [DOI] [PubMed] [Google Scholar]
- 12.Dautry-Varsat A., Subtil A., Hackstadt T. (2005) Cell. Microbiol. 7, 1714–1722 [DOI] [PubMed] [Google Scholar]
- 13.Hackstadt T., Fischer E. R., Scidmore M. A., Rockey D. D., Heinzen R. A. (1997) Trends Microbiol. 5, 288–293 [DOI] [PubMed] [Google Scholar]
- 14.McClarty G. (1994) Trends Microbiol. 2, 157–164 [DOI] [PubMed] [Google Scholar]
- 15.Carabeo R. A., Mead D. J., Hackstadt T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rzomp K. A., Scholtes L. D., Briggs B. J., Whittaker G. R., Scidmore M. A. (2003) Infect. Immun. 71, 5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatch G. M., McClarty G. (1998) Biochem. Biophys. Res. Commun. 243, 356–360 [DOI] [PubMed] [Google Scholar]
- 18.Ojcius D. M., Degani H., Mispelter J., Dautry-Varsat A. (1998) J. Biol. Chem. 273, 7052–7058 [DOI] [PubMed] [Google Scholar]
- 19.Bea F., Puolakkainen M. H., McMillen T., Hudson F. N., Mackman N., Kuo C. C., Campbell L. A., Rosenfeld M. E. (2003) Circ. Res. 92, 394–401 [DOI] [PubMed] [Google Scholar]
- 20.Birkelund S., Johnsen H., Christiansen G. (1994) Infect. Immun. 62, 4900–4908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coombes B. K., Mahony J. B. (2002) Cell. Microbiol. 4, 447–460 [DOI] [PubMed] [Google Scholar]
- 22.Hess S., Peters J., Bartling G., Rheinheimer C., Hegde P., Magid-Slav M., Tal-Singer R., Klos A. (2003) Cell. Microbiol. 5, 785–795 [DOI] [PubMed] [Google Scholar]
- 23.Xia M., Bumgarner R. E., Lampe M. F., Stamm W. E. (2003) J. Infect. Dis. 187, 424–434 [DOI] [PubMed] [Google Scholar]
- 24.Ito J. I., Jr., Lyons J. M., Airo-Brown L. P. (1990) Infect. Immun. 58, 2021–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyairi I., Mahdi O. S., Ouellette S. P., Belland R. J., Byrne G. I. (2006) J. Infect. Dis. 194, 350–357 [DOI] [PubMed] [Google Scholar]
- 26.Peters J., Wilson D. P., Myers G., Timms P., Bavoil P. M. (2007) Trends Microbiol. 15, 241–251 [DOI] [PubMed] [Google Scholar]
- 27.Rasmussen S. J., Eckmann L., Quayle A. J., Shen L., Zhang Y. X., Anderson D. J., Fierer J., Stephens R. S., Kagnoff M. F. (1997) J. Clin. Invest. 99, 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darville T., Andrews C. W., Laffoon K. K., Shymasani W., Kishen L. R., Rank R. G. (1997) Infect. Immun. 65, 3065–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darville T., Andrews C. W., Jr., Sikes J. D., Fraley P. L., Rank R. G. (2001) Infect. Immun. 69, 3556–3561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward M. E. (1999) in Chlamydia: Intracellular Biology, Pathogenesis, and Immunity ( Stephens R. S. ed) pp. 171–210, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 31.Darville T., O'Neill J. M., Andrews C. W., Jr., Nagarajan U. M., Stahl L., Ojcius D. M. (2003) J. Immunol. 171, 6187–6197 [DOI] [PubMed] [Google Scholar]
- 32.O'Connell C. M., Ionova I. A., Quayle A. J., Visintin A., Ingalls R. R. (2006) J. Biol. Chem. 281, 1652–1659 [DOI] [PubMed] [Google Scholar]
- 33.Prebeck S., Brade H., Kirschning C. J., da Costa C. P., Dürr S., Wagner H., Miethke T. (2003) Microbes Infect. 5, 463–470 [DOI] [PubMed] [Google Scholar]
- 34.Welter-Stahl L., Ojcius D. M., Viala J., Girardin S., Liu W., Delarbre C., Philpott D., Kelly K. A., Darville T. (2006) Cell. Microbiol. 8, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 35.Chamaillard M., Girardin S. E., Viala J., Philpott D. J. (2003) Cell. Microbiol. 5, 581–592 [DOI] [PubMed] [Google Scholar]
- 36.Inohara , Chamaillard , McDonald C., Nuñez G. (2005) Annu. Rev. Biochem. 74, 355–383 [DOI] [PubMed] [Google Scholar]
- 37.Martinon F., Tschopp J. (2005) Trends Immunol. 26, 447–454 [DOI] [PubMed] [Google Scholar]
- 38.Dinarello C. A. (2006) Am. J. Clin. Nutr. 83, 447S–455S [DOI] [PubMed] [Google Scholar]
- 39.Martinon F., Burns K., Tschopp J. (2002) Mol. Cell 10, 417–426 [DOI] [PubMed] [Google Scholar]
- 40.Martinon F., Tschopp J. (2004) Cell 117, 561–574 [DOI] [PubMed] [Google Scholar]
- 41.Entrican G., Wilkie R., McWaters P., Scheerlinck J., Wood P. R., Brown J. (1999) Clin. Exp. Immunol. 117, 309–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gervassi A., Alderson M. R., Suchland R., Maisonneuve J. F., Grabstein K. H., Probst P. (2004) Infect. Immun. 72, 7231–7239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ojcius D. M., Souque P., Perfettini J. L., Dautry-Varsat A. (1998) J. Immunol. 161, 4220–4226 [PubMed] [Google Scholar]
- 44.Rothermel C. D., Schachter J., Lavrich P., Lipsitz E. C., Francus T. (1989) Infect. Immun. 57, 2705–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng W., Shivshankar P., Li Z., Chen L., Yeh I. T., Zhong G. (2008) Infect. Immun. 76, 515–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu H., Shen C., Brunham R. C. (2000) J. Immunol. 165, 1463–1469 [DOI] [PubMed] [Google Scholar]
- 47.Yu H. B., Finlay B. B. (2008) Cell Host Microbe 4, 198–208 [DOI] [PubMed] [Google Scholar]
- 48.Gurcel L., Abrami L., Girardin S., Tschopp J., van der Goot F. G. (2006) Cell 126, 1135–1145 [DOI] [PubMed] [Google Scholar]
- 49.Hackstadt T., Scidmore M. A., Rockey D. D. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franchi L., Stoolman J., Kanneganti T. D., Verma A., Ramphal R., Núñez G. (2007) Eur. J. Immunol. 37, 3030–3039 [DOI] [PubMed] [Google Scholar]
- 51.Galle M., Schotte P., Haegman M., Wullaert A., Yang H. J., Jin S., Beyaert R. (2008) J. Cell. Mol. Med. 12, 1767–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao E. A., Alpuche-Aranda C. M., Dors M., Clark A. E., Bader M. W., Miller S. I., Aderem A. (2006) Nat. Immunol. 7, 569–575 [DOI] [PubMed] [Google Scholar]
- 53.Miao E. A., Ernst R. K., Dors M., Mao D. P., Aderem A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2562–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shin H., Cornelis G. R. (2007) Cell. Microbiol. 9, 2893–2902 [DOI] [PubMed] [Google Scholar]
- 55.Sun G. W., Lu J., Pervaiz S., Cao W. P., Gan Y. H. (2005) Cell. Microbiol. 7, 1447–1458 [DOI] [PubMed] [Google Scholar]
- 56.Suzuki T., Franchi L., Toma C., Ashida H., Ogawa M., Yoshikawa Y., Mimuro H., Inohara N., Sasakawa C., Nuñez G. (2007) PLoS Pathog. 3, e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ting J. P., Lovering R. C., Alnemri E. S., Bertin J., Boss J. M., Davis B. K., Flavell R. A., Girardin S. E., Godzik A., Harton J. A., Hoffman H. M., Hugot J. P., Inohara N., Mackenzie A., Maltais L. J., Nunez G., Ogura Y., Otten L. A., Philpott D., Reed J. C., Reith W., Schreiber S., Steimle V., Ward P. A. (2008) Immunity 28, 285–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamkanfi M., Kanneganti T. D., Franchi L., Núñez G. (2007) J. Leukocyte Biol. 82, 220–225 [DOI] [PubMed] [Google Scholar]
- 59.Sutterwala F. S., Flavell R. A. (2009) Clin. Immunol. 130, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephens R. S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R. L., Zhao Q., Koonin E. V., Davis R. W. (1998) Science 23, 754–759 [DOI] [PubMed] [Google Scholar]
- 61.Cruz C. M., Rinna A., Forman H. J., Ventura A. L., Persechini P. M., Ojcius D. M. (2007) J. Biol. Chem. 282, 2871–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B. T., Tschopp J. (2008) Science 320, 674–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., Tschopp J. (2007) Cell Death Differ. 14, 1583–1589 [DOI] [PubMed] [Google Scholar]
- 64.Martinon F., Mayor A., Tschopp J. (2009) Annu. Rev. Immunol. 27, 229–265 [DOI] [PubMed] [Google Scholar]
- 65.Benko S., Philpott D. J., Girardin S. E. (2008) Cytokine 43, 368–373 [DOI] [PubMed] [Google Scholar]
- 66.Franchi L., Kanneganti T. D., Dubyak G. R., Núñez G. (2007) J. Biol. Chem. 282, 18810–18818 [DOI] [PubMed] [Google Scholar]
- 67.Chang G. T., Moulder J. W. (1978) Infect. Immun. 19, 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slepenkin A., Enquist P. A., Hägglund U., de la Maza L. M., Elofsson M., Peterson E. M. (2007) Infect. Immun. 75, 3478–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wolf K., Betts H. J., Chellas-Géry B., Hower S., Linton C. N., Fields K. A. (2006) Mol. Microbiol. 61, 1543–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cameron A. R., Nelson J., Forman H. J. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 3726–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shao W., Yeretssian G., Doiron K., Hussain S. N., Saleh M. (2007) J. Biol. Chem. 282, 36321–36329 [DOI] [PubMed] [Google Scholar]
- 72.Lin X. Y., Choi M. S., Porter A. G. (2000) J. Biol. Chem. 275, 39920–39926 [DOI] [PubMed] [Google Scholar]
- 73.Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 74.Heuer D., Lipinski A. R., Machuy N., Karlas A., Wehrens A., Siedler F., Brinkmann V., Meyer T. F. (2009) Nature 457, 731–735 [DOI] [PubMed] [Google Scholar]