Abstract
The protozoan parasite Toxoplasma gondii relies on post-translational modification, including proteolysis, of proteins required for recognition and invasion of host cells. We have characterized the T. gondii cysteine protease cathepsin L (TgCPL), one of five cathepsins found in the T. gondii genome. We show that TgCPL is the primary target of the compound morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl (LHVS), which was previously shown to inhibit parasite invasion by blocking the release of invasion proteins from microneme secretory organelles. As shown by fluorescently labeled LHVS and TgCPL-specific antibodies, TgCPL is associated with a discrete vesicular structure in the apical region of extracellular parasites but is found in multiple puncta throughout the cytoplasm of intracellular replicating parasites. LHVS fails to label cells lacking TgCPL due to targeted disruption of the TgCPL gene in two different parasite strains. We present a structural model for the inhibition of TgCPL by LHVS based on a 2.0 Å resolution crystal structure of TgCPL in complex with its propeptide. We discuss possible roles for TgCPL as a protease involved in the degradation or limited proteolysis of parasite proteins involved in invasion.
The recent completion of many genome-sequencing projects has allowed an unprecedented view of the complete set of proteases in biologically or medically important organisms (1). Of the five mechanistically distinct catalytic types (serine, cysteine, aspartyl, metallo, and threonine), cysteine proteases are the second largest group. In particular, cysteine proteases of the C1 papain family of “lysosomal” cathepsins have garnered intense scrutiny because of their key roles in cancer, embryogenesis, heart disease, osteoporosis, immunity, and infectious diseases. Microbial cathepsins, particularly those expressed by parasites, have also attracted attention recently because of their potential as targets for treatment of helminthic and protozoal infections (2, 3).
The protozoan parasite Toxoplasma gondii infects virtually all warm-blooded animals and approximately one-third of the human population worldwide. Although most Toxoplasma infections are benign, severe opportunistic disease is seen in immunodeficient or immunosuppressed individuals or congenitally infected babies. T. gondii is an obligate intracellular organism that uses an actin-myosin-based motility system to actively invade nucleated host cells (4, 5). The parasite secretes a variety of proteins during and after cell invasion that contribute to recognition of the host cell, formation of an adhesive “moving” junction, modulation of host signaling pathways and gene expression, and remodeling of the parasitophorous vacuole in preparation for parasite growth (6, 7). Although it has been known for some time that many Toxoplasma secretory proteins are post-translationally modified by proteolysis before and/or after secretion, in most cases, the consequences of proteolysis or the specific protease involved are unclear.
Analysis of the T. gondii genome indicates the existence of five genes encoding cathepsin proteases of the papain family, including three cathepsin C proteases (TgCPC1, TgCPC2, and TgCPC3), one cathepsin B (Toxopain-1 or TgCPB), and one cathepsin L (TgCPL). TgCPC1 and TgCPC2 are secreted into the parasitophorous vacuole after parasite invasion and are proposed to function in nutrient acquisition (8). TgCPC3 is not expressed in tachyzoites, a rapidly dividing form of the parasite that is most commonly studied in the laboratory. TgCPB is localized in club-shaped invasion organelles called rhoptries, where it may act as a maturase for rhoptry proteins involved in modulation of the host cell (9). TgCPL is predicted to be a type II membrane protein, and a recent report by Reed and co-workers (10) showed that it has enzymatic activity with a low pH optimum and that it occupies a membrane-bound structure in the apical region of extracellular parasites. This same study revealed that T. gondii expresses two endogenous inhibitors of cysteine proteases (TgICP1 and TgICP2), but their role in regulating parasite or host cysteine proteases remains to be determined. Similar inhibitors are expressed by other parasites, including Trypanosoma cruzi, that act on host proteases, and the crystal structure of an inhibitor (chagasin)-enzyme (human cathepsin L) complex was recently reported (11).
In a recent study, we screened a small library of cathepsin and proteasome inhibitors and identified two compounds that substantially impair Toxoplasma cell invasion (12). The most effective of these compounds, morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl (LHVS),2 inhibited invasion with a 50% inhibitory concentration (IC50) of ∼10 μm. Further analysis revealed that LHVS blocks parasite attachment and gliding motility by impairing the release of proteins from a distinct set of apical secretory organelles called micronemes. Here we definitively show, using a variety of biochemical, genetic, and structural approaches, that TgCPL is the primary target of LHVS in the parasite.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, Purification, Refolding, and Autoactivation
A 0.95-kb fragment coding for 94 carboxyl terminal amino acids of the prodomain and the complete 224-amino acid mature domain of TgCPL (100TgCPL) was amplified by PCR from a T. gondii RH cDNA library3 with primers TgCatL.313(BamH1)f and TgCatL.1269(HindIII)r (supplemental Table S1) using ExpandTM High Fidelity enzyme mix containing Taq DNA polymerase and Tgo DNA polymerase with proofreading activity (Roche Applied Science). The PCR product was gel-purified using a Qiagen gel extraction kit, initially ligated into pGEM-T Easy Vector (pGEM-T/100TgCPL), and transformed into the DH5α Escherichia coli strain. Clones were sequenced in both directions and digested with the restriction enzymes BamHI and HindIII. The complete cDNA encoding TgCPL was also amplified, cloned, sequenced, and deposited in GenBankTM (accession number DQ407191).
For recombinant protein expression, the 100TgCPL cDNA was ligated into BamHI- and HindIII-digested pQE30 vector, which provided an in-frame His6 tag at the N terminus (Qiagen), thereby generating pQE30/100TgCPL. Competent E. coli M15[pRep4] cells (Qiagen) were transformed with pQE30/100TgCPL and grown at 37 °C in Terrific Broth containing 100 μg/ml ampicillin and 25 μg/ml kanamycin to A600 ∼0.6 when 1 mm isopropyl-β-d-thiogalactopyranoside was added before additional culturing for 5 h. Inclusion bodies were purified from the harvested cells and resuspended in denaturing buffer (20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10 mm imidazole, and 8 m urea). Solubilized inclusion bodies were then incubated with Ni2+-nitrilotriacetic acid resin (Qiagen), equilibrated with the same buffer, overnight at room temperature. The mixture was loaded into a column and washed with 20–30 column volumes of denaturing buffer, and denatured proform TgCPL was eluted with elution buffer (100 mm NaH2PO4, 10 mm Tris, 8 m urea, pH 4.5). Elution fractions were pooled and concentrated ∼5-fold using a spin concentrator (Millipore) and dialyzed overnight back into denaturing buffer. The denatured protein was then reduced with 10 mm dithiothreitol for 45 min at 37 °C and diluted to 20 μg/ml with ice-cold refolding buffer (100 mm Tris-HCl, pH 8.0, 1 mm EDTA, 20% glycerol, 250 mm l-arginine, 2 mm reduce glutathione, 1 mm oxidized glutathione). After incubation at 4 °C for 48 h with moderate stirring, proform recombinant TgCPL (rTgCPL) was concentrated ∼10-fold and dialyzed overnight into proform buffer (50 mm Tris-HCl, pH 6.8, 900 mm NaCl, 2 mm EDTA). To activate the protease, proform rTgCPL was exchanged into activation buffer (100 mm sodium acetate, pH 5.5, 900 mm NaCl, 2 mm EDTA), followed by an increase of the dithiothreitol concentration to 5 mm and incubation at 37 °C for 4–5 h. The efficiency of maturation was monitored using SDS-PAGE. Autoactivated rTgCPL was then concentrated to 4.7 mg/ml, supplemented with 0.825 mm LHVS, and immediately flash frozen in liquid nitrogen prior to storage at −80 °C to await crystallization.
Antibody Production
A primary injection of rabbits with 200 μg of proform rTgCPL in Freund's complete adjuvant (Sigma) was followed by four boosts of 200 μg each in Freund's incomplete adjuvant (Sigma) at 2-week intervals. A polyclonal mouse antiserum also was raised against proform rTgCPL by immunizing mice with a single injection of 25 μg of recombinant protein mixed with TiterMax Gold adjuvant (Cytex). Serum was collected 4 weeks postimmunization.
Activity-based Profiling
RH strain tachyzoites, grown in human foreskin fibroblasts and Dulbecco's minimal essential medium, containing 10% fetal bovine serum, 2 mm glutamine, were filter-purified, washed with Dulbecco's minimal essential medium-glutamine-HEPES medium (Dulbecco's minimal essential medium, 2 mm glutamine, 10 mm HEPES), and resuspended at 5 × 108 ml−1. Fifty μl of parasite suspension was added to each well of a round bottom microwell plate containing 0.5 μl of DMSO or 20 μm BODIPY-LHVS (BO-LHVS) in DMSO. After labeling for 30 min in a humidified 37 °C, 5% CO2 incubator, the plate was cooled on ice and centrifuged 5 min at 4 °C. Forty-five μl of supernatant was removed, and parasites were lysed in 50 μl of SDS-PAGE sample buffer containing 2% 2-mercaptoethanol and placed in a 100 °C water bath for 5 min. Samples were vortexed vigorously, microcentrifuged to remove insoluble material, and resolved by 12.5% SDS-PAGE. Gels were imaged on a Typhoon Trio PhosphorImager (GE Healthcare) and quantified using ImageQuant software.
For activity-based protein profiling of rTgCPL, BO-LHVS (4 μm, 400 nm, 40 nm, 4 nm, or 400 pm) was incubated with 400 nm active enzyme or heat-inactivated (90 °C, 5 min) enzyme at 37 °C for 30 min. Additionally, rTgCPL (4 μm, 400 nm, 40 nm, 4 nm, or 400 pm) was incubated with 400 nm BO-LHVS at 37 °C for 30 min. Samples were resolved and imaged as described above.
Targeted Disruption of TgCPL
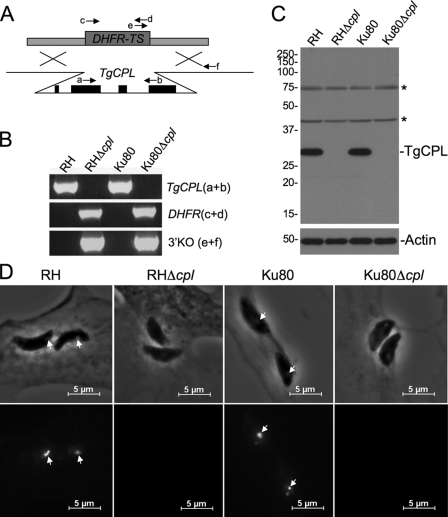
Genomic flanking sequences of the TgCPL gene were obtained from the Toxoplasma gondii data base (ToxoDB (39) accession number 645.m 00037). Primers 3′dhfr-ts.PacI.5′TgCat.L.F, 3′TgCatL.1749411.F, 5′HXG.NsiI.3′TgCatL.R, and 5′TgCatL.1756165.R (supplemental Table S1) were used to amplify ∼1.5 kb of the 5′- and 3′-sequence flanking the gene. The flanks were amplified to have overlap on one end with the dihydrofolate reductase-thymidylate synthase (DHFR-TS) selectable marker cassette, and similarly, the DHFR-TS marker cassette was amplified to contain TgCPL sequences on the ends. RH genomic DNA was used as template for amplifying the 5′- and 3′-flanks, and pDHFR-TSc plasmid (13) was used as template for amplifying the DHFR-TS selectable marker. A fusion PCR product was then amplified using primers, and the three individual 5′, 3′, and DHFR-TS fragments as templates. The TgCPL knock-out construct (13 μg) was electroporated into RH and Ku80 tachyzoites using conditions described previously (14). Parasites were grown in 24-well plates for 2 days in the absence of drug selection before transferring them to new wells containing 1 μm pyrimethamine (Sigma). After 2 weeks of selection, parasite clones were derived by limited dilution in 96-well plates. Clones were screened for the absence of TgCPL expression by immunoblotting and fluorescence microscopy. Clones were also assessed for correct integration of the construct at the TgCPL locus by PCR using the following primers depicted in Fig. 2A: TgCpl.213.F (a), TgCpl.853.R (b), dhfrHXGPRTdhfr.1.F (c), CplDHFRko.847.R (d), 3′TgCatL.1749205.F (e), and dhfr5′end.105.R (f) (supplemental Table S1).
FIGURE 2.
Targeted deletion of TgCPL. A, schematic illustration of the TgCPL knock-out strategy. A knock-out construct consisting of ∼3 kb of 5′- and 3′-flanking sequence from the TgCPL gene appended to either side of a DHFR-TS-selectable marker cassette was transfected into RH and Ku80 parasites for double crossover gene replacement of TgCPL. The arrows indicate PCR primers used in B. B, agarose gel electorphoresis of PCR products derived from parental (RH and Ku80) and knock-out (RHΔcpl and Ku80Δcpl) strains by amplification with the indicated primers. C, immunoblot analysis of parental and knock-out strains probed with RαTgCPL. Note the absence of the TgCPL reactivity. Asterisks denote nonspecific bands. A parallel blot was probed with anti-actin as a loading control. D, indirect immunofluorescence assay of newly invaded intracellular tachyzoites showing MαTgCPL reactivity with RH and Ku80 (arrows) but lack of reactivity with RHΔcpl or Ku80Δcpl.
Immunoblotting, Immunoprecipitation, and Fluorescence Microscopy
Immunoblotting, immunoprecipitation, and indirect immunofluorescence assay were performed essentially as described previously (15). Antibodies were diluted 1:5,000 for immunoblotting and 1:250 for indirect immunofluorescence assay. Tachyzoites were labeled with 200 nm BO-LHVS for 30 min in a humidified 37 °C, 5% CO2 incubator before fixation with 4% paraformaldehyde and antibody staining. Images were acquired on a Nikon E800 upright fluorescence microscope with a SpotRT slider camera or a Zeiss Axiovert Observer Z1 inverted fluorescence microscope and an AxioCAM MRm camera and processed using Compix software or Zeiss Axiovision 4.3 software, respectively.
Protein Crystallization
Purified protein was screened at the high throughput facility at the Hauptman Woodward Institute to identify initial crystallization conditions (16). Twenty-four crystallization leads were identified at the 4-week time point. One of these leads was further optimized in house using sitting drop vapor diffusion to produce crystals suitable for x-ray diffraction data collection. Upon thawing, 1 μl of autoactivated protein solution (4.7 mg/ml) was mixed with an equivalent amount of reservoir solution (40% polyethylene glycol 8000, 0.1 m ammonium bromide, and 0.1 m sodium citrate at pH 4.0). The resultant drop was equilibrated over a 100-μl reservoir at 25 °C. American football-shaped crystals measuring ∼30 μm from point to point appeared within 2 weeks. The crystals were cryoprotected by adding 2.5 μl of reservoir solution followed by 0.5 μl of ethylene glycol directly to the drop. Crystals were then mounted in cryoloops and frozen in liquid nitrogen in preparation for x-ray diffraction experiments.
Data Collection and Structure Determination
Crystals of TgCPL were screened at the Stanford Synchrotron Radiation Lightsource on beamline 11-1 using the Stanford Synchrotron Radiation Lightsource automated mounting system (17). Data from a single crystal maintained at 100 K were collected at a wavelength of 0.98 Å using the Blu-Ice software package (18) and processed using HKL2000 (19) to a resolution of 2.0 Å. The crystals belong to space group P43212 with unit cell dimensions of 65.6 × 65.6 × 149.8 Å. One complex of the active protease with its propeptide is present in the asymmetric unit, giving a solvent content of ∼43% and a Matthews coefficient of ∼2.2 Å3/Da. However, the asymmetric unit was initially believed to contain one or, remotely possible, two copies of only the active protease in complex with LHVS, which would correspond to Matthews coefficients of ∼3.3 or 1.65 Å3/Da, respectively.
The structure of TgCPL was solved by molecular replacement with the program Phaser (20), using the full resolution range of the data and a search model derived from human procathepsin L (Protein Data Bank code 1cs8). The sequence identity between the active protease domains of TgCPL and 1cs8 is about 50%, whereas the identity between the full-length constructs of the two proteins is about 42%. Because it was expected that the crystal would contain the protease domain in complex with the inhibitor LHVS, the search model was modified prior to molecular replacement to be consistent with this expectation (21). First, the propeptide domain of 1cs8 was manually removed, and then, using a pairwise sequence alignment created with ClustalW (22), the non-conserved amino acids were truncated or removed using the program CHAINSAW (23). The resulting Phaser-placed model was refined as a rigid body using Refmac5 (24) and manually edited using Coot (25). Refinement continued by iteration of manual editing in Coot, followed by restrained refinement in Refmac5. For all steps from data preparation through refinement, the CCP4 suite of programs (26, 27) was used.
After several building/refinement cycles of nearly full-length protease, large polypeptide-like blobs of positive difference density remained, and R factors were stalled near 40%. This suggested that the asymmetric unit contained more than the single copy of the protease that was placed by molecular replacement, but there did not appear to be enough room to place a second copy. LHVS was present in the protein solution, and although it is somewhat peptide-like in appearance, the difference density extended well beyond the active site and was much too large to be accounted for solely by inhibitor. It was then realized that the propeptide domain was not purified away from the active proteases. The model of the protease domain was submitted to ARP/wARP (28) to use for initial phase calculations in the automated rebuilding of the full-length proform of the TgCPL construct. This step also served to reduce bias toward the search model. Indeed, ARP/wARP was able to build 64 amino acids belonging to the propeptide in addition to rebuilding most of the existing model, which resulted in a drop of the R factor by ∼15%. The iteration of building and refinement continued as before with this more complete model. In the final cycles of refinement, the propeptide and the protease were described by four translation/libration/screw groups each, with group boundaries suggested by the TLSMD server (29, 30). Translation/libration/screw parameters were refined for each group prior to restrained refinement in Refmac5. Model quality was monitored and validated using Coot, MolProbity (31), and Rfree. Data collection and model refinement statistics are presented in Tables 1 and 2, respectively.
TABLE 1.
Data collection statistics
| Data set | Tgon077550AAB |
|---|---|
| Beamline | SSRL 11-1 |
| Space group | P43212 |
| Unit cell parameters (Å) (α = β = γ = 90°) | a = 65.6, b = 65.6, c = 149.8 |
| Wavelength (Å) | 0.979 |
| Resolution (Å) | 34.0-1.99 (2.06-1.99)a |
| Unique reflections | 23,266 (2,242) |
| Completeness (%) | 99.9 (99.2) |
| Rmerge | 0.122 (0.744) |
| 〈I/σ(I)〉 | 18.1 (2.8) |
| Redundancy | 8.7 (8.5) |
| Wilson B factor (Å2) | 25.0 |
a Values in parenthesis are for the highest resolution shell.
TABLE 2.
Model refinement statistics
| Data set | Tgon077550AAB |
|---|---|
| Resolution (Å) | 32.8–1.99 |
| No. of reflections | 23,178 |
| No. of reflections in test set | 1,186 |
| Rwork | 0.165 |
| Rfree | 0.206 |
| RMSD bonds (Å) | 0.01 |
| RMSD angles (degrees) | 1.16 |
| Protein atoms | 2,370 |
| Non-protein atoms | 174 |
| Ramachandran statistics (%)a | |
| Residues in favored regions | 96.6 |
| Residues in allowed regions | 100 |
| Unmodeled residuesb | −11-0, 105p–107p, 183p–198p, 1 |
| TLS groups (residues)c | A: 2–15, 16–118, 119–135, 136–224; P: 108–123, 124–136, 137–168, 169–182 |
| Mean Biso + BTLS protein atoms (Å2) | 30.9 |
| Mean Biso non-protein atoms (Å2) | 26.7 |
| Protein Data Bank entry | 3f75 |
a Determined using the MolProbity Server (50).
b Residues −11 to 0 are an N-terminal His tag and linker that replaced the first 104 propeptide residues of the full-length protein sequence (ToxoDB (39) accession number 645.m00037) during cloning.
c Determined using the TLSMD server (30).
One hundred sixty-five water molecules, one ethylene glycol molecule from the cryoprotectant, and two halide atoms originating from either the protein storage buffer (chloride) or the mother liquor (bromide) were placed in the final model. The last two corresponded to strong, round difference density peaks that are surrounded primarily by nitrogen atoms but also a few oxygen atoms and do not appear to be water molecules or metal ions because the distances to the closest potential hydrogen bond donors/acceptors or coordinating ligands are too great. The surrounding atoms are contributed by symmetry-related protein molecules, so the modeled halides are involved in crystal contacts.
Modeling of the LHVS Inhibitor into the TgCPL Active Site
A search of the Protein Data Bank (32) for “vinyl sulfone” yielded 14 hits, all of which were inhibitors bound to papain-like proteases or to the ATP-dependent protease, HslV. These vinyl sulfone inhibitor structures were then manually examined for a scaffold that approximated that of LHVS. Through this search, we identified the inhibitor N-[1S-(2-phenylethyl)-3-phenylsulfonylallyl]-4-methyl-2R-piperazinyl carbonylaminovaleramide (APC3328) bound to human cathepsin K (Protein Data Bank code 1mem) (33), which differs from LHVS only in that it contains a piperazine ring in place of the morpholine ring of LHVS; a single-atom difference of nitrogen versus oxygen. Structure factors were deposited in the Protein Data Bank with the 1mem coordinates, so it was possible to inspect the electron density of the APC3328 inhibitor. Difference electron density shows that the rotamer selected for the inhibitor's leucine side chain is not ideal, so the side chain was altered from the deposited 1mem coordinates to a rotamer that more favorably fits the density.
APC3328 was initially placed in the active site of TgCPL by performing a least squares superposition of all the catalytic triad atoms of TgCPL and 1mem using Lsqkab (34), after first removing the propeptide residues from TgCPL. The root mean square deviation (RMSD) between the catalytic triads is 0.28 Å, with differences between the side chain atoms of the histidine/asparagine residues contributing the most to this value. Nitrogen N4 of the piperazine ring in the superimposed inhibitor was substituted with oxygen to change APC3328 into LHVS, and the side chains of the catalytic triad residues of TgCPL were substituted with those of the superimposed 1mem structure. The side chain amide of Gln69 was flipped 180° so that Nϵ could hydrogen-bond with O4 of the LHVS morpholine ring; Oϵ of Gln69 is 3.2 Å from Nζ of Lys181p in the propeptide-bound structure. To achieve a more meaningful model of the binding mode, energy minimization of this superimposed, modified ligand was performed. To this end, water molecules were removed, and hydrogen atoms were added to protein and ligand polar groups. The active site was defined as residues within 7.0 Å of the starting position of the superimposed ligand. Energy minimization calculations were carried out with QXP/FLO (35). Protein atoms were fixed during the calculations, with the exception of the side chain of Asn69.
RESULTS
LHVS Reacts with Recombinant and Native TgCPL
LHVS is a dipeptide vinyl sulfone that was originally designed as a selective inhibitor of human cathepsins S and V. However, extensive studies of its specificity have revealed that, although most potent against cathepsins S and V, it is also a moderate inhibitor of human cathepsins K, L, and O2 and a weak inhibitor of human cathepsin B (36, 37). Since LHVS is an order of magnitude more potent toward human cathepsin L than human cathepsin B, we expected that T. gondii cathepsin L should likewise be much more sensitive to inhibition by this compound than the cathepsin B homolog. To test TgCPL for reactivity with LHVS, we used a fluorescently labeled derivative of LHVS termed BO-LHVS (Fig. 1A). rTgCPL was expressed in E. coli, extracted, purified, refolded, and activated, as described previously (38) (supplemental Fig. S1). BO-LHVS incubation with rTgCPL and analysis by fluorescence-scanned SDS-polyacrylamide gels (activity-based protein profiling) showed saturated covalent labeling at molar ratios of >1:1 (Fig. 1B). BO-LHVS failed to react with heat-inactivated rTgCPL, demonstrating that labeling is dependent on enzymatic activity. To determine if LHVS also reacts with native TgCPL, we performed activity-based protein profiling of live purified tachyzoites incubated with solvent (DMSO) or 200 nm BO-LHVS (Fig. 1C). BO-LHVS strongly labeled a 30-kDa protein and weakly labeled proteins of 29 and 24 kDa. Immunoprecipitation with anti-rTgCPL confirmed that the 30-kDa species is TgCPL. The 29-kDa species may have also been immunoprecipitated by anti-rTgCPL but was only very faintly detected, whereas the 24-kDa species was not observed in the immunoprecipitate and therefore is probably unrelated to TgCPL.
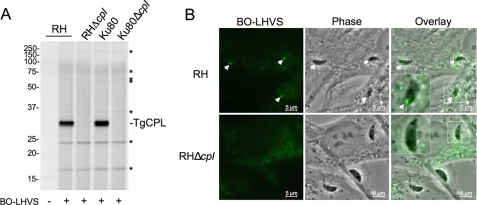
FIGURE 1.
BO-LHVS reacts with recombinant and native TgCPL. A, chemical structure of LHVS and BO-LHVS. Major chemical groups of LHVS from left to right are morpholinurea leucine, homophenylalanine, vinyl sulfone, phenyl. For BO-LHVS, BODIPY 493/503 is substituted for morpholinurea. B, activity-based protein profiling of rTgCPL with BO-LHVS in vitro showing dose-dependent labeling of the 30-kDa active enzyme but not labeling of the heat-inactivated (HI) enzyme. rTgCPL was incubated with BO-LHVS at the indicated ratios, resolved by 12.5% SDS-PAGE, and imaged by laser-scanning fluorometry. Molecular mass markers are indicated (kDa). C, activity-based protein profiling of native TgCPL in live cells. Tachyzoites were incubated with solvent (DMSO) or BO-LHVS and lysed. Samples were either analyzed directly (left two lanes) or immunoprecipitated (IP) with MαrTgCPL or normal mouse serum (NMS) (right two lanes) before electrophoresis and imaging as in B. The arrows denote the TgCPL bands, and an asterisk indicates the 24-kDa minor labeled product. D, competition assay with LHVS. Tachyzoites were preincubated with LHVS before exposure to 200 nm BO-LHVS and analysis as above. Asterisks denote bands that are not blocked by pretreatment with LHVS. E, subcellular distribution of active TgCPL. Extracellular or replicating intracellular tachyzoites were incubated with BO-LHVS before fixation and staining with MαrTgCPL and an Alexa 592 (red)-conjugated secondary antibody. Most of the labeled structures show dual fluorescence of the active (BO-LHVS) and total (MαrTgCPL) enzyme, with the exceptions indicated by arrows.
Since BO-LHVS differs from LHVS by the presence of the BODIPY fluorophore and the absence of the morpholinurea group (Fig. 1A), either of which could affect specificity, it is important to confirm that LHVS is also capable of inhibiting TgCPL in live parasites using a competition assay (Fig. 1D). Tachyzoites were preincubated with increasing concentrations of LHVS before exposure to BO-LHVS and gel analysis. LHVS effectively blocked subsequent TgCPL labeling by BO-LHVS, exhibiting an IC50 of ∼20 nm for the 30-kDa species. Interestingly, the 29-kDa species appeared to be slightly more refractory to inhibition by LHVS, suggesting that it is either a distinct protein or a modified form of TgCPL with lower affinity for LHVS.
To examine the subcellular distribution of active TgCPL within the parasite, we labeled live extracellular and replicating intracellular tachyzoites with BO-LHVS before fixing and staining with anti-rTgCPL (Fig. 1E). Although BO-LHVS labeling was much weaker than that of the antibody, the two staining patterns were nearly identical, with extracellular parasites showing one or two discrete structures usually in the apical region but occasionally posterior to the nucleus. A few of these structures were labeled with anti-rTgCPL but not BO-LHVS, possibly indicating the presence of a pool of proform or otherwise inactive enzyme in some parasites. Intracellular replicating tachyzoites tended to display a greater number of TgCPL-associated structures distributed throughout the parasite. Parasites preincubated with LHVS showed a dose-dependent reduction in labeling of the TgCPL-associated structures (data not shown), confirming that the labeling is principally specific. Properties of the TgCPL-associated organelle will be described in greater detail in a separate study.4 Together, these findings establish that TgCPL is a major reactive target of BO-LHVS in vitro and in live parasites.
Targeted Disruption of TgCPL
To validate the specificity of LHVS for TgCPL, we generated parasites deficient in TgCPL expression by gene ablation. The TgCPL gene is composed of four exons, is present in a single copy on chromosome 1b, and is transcribed at a moderate level in a variety of Toxoplasma strains as annotated in ToxoDB (39). We generated TgCPL-deficient strains by double homologous gene replacement with a mutant allele of DHFR-TS conferring resistance to pyrimethamine (13) (Fig. 2A). TgCPL was deleted in RH strain parasites and in a RH-derived strain called Ku80, which is more amenable to targeted genetic manipulation (40). Both knock-out strains, RHΔcpl and Ku80Δcpl, showed the expected pattern of PCR products consistent with targeted deletion of TgCPL (Fig. 2B). The absence of TgCPL expression in the knock-out strains was confirmed by immunoblotting (Fig. 2C), showing the loss of the 30-kDa major immunoreactive species. An immunofluorescence assay (Fig. 2D) also showed the lack of TgCPL staining in newly invaded intracellular RHΔcpl or Ku80Δcpl tachyzoite. Similar results were seen in extracellular and replicating intracellular knock-out tachyzoites (data not shown). Collectively, these results confirm the abrogation of TgCPL expression in the RHΔcpl and Ku80Δcpl strains.
BO-LHVS Labeling of TgCPL Knock-out Strains
To determine if TgCPL is the primary LHVS reactive species in tachyzoites, we incubated wild-type and knock-out parasites with BO-LHVS for activity-based protein profiling. Neither knock-out strain showed reactivity in the 30-kDa region above background, strongly suggesting that TgCPL is both the 30- and 29-kDa reactive species (Fig. 3A). Although several additional minor reactive bands remained visible in RHΔcpl and Ku80Δcpl lysates, including 24 and 16 kDa bands, integration of the fluorescence scans showed that collectively these labeled species constitute only 14.8% of the TgCPL reactivity in the wild-type strains. Moreover, as described above (Fig. 1D), BO-LHVS labeling of these species is not blocked by pretreatment with unlabeled LHVS, indicating their nonspecific reactivity with BO-LHVS. Whereas BO-LHVS labeling of newly invaded intracellular RH parasites showed staining of discrete puncta, these structures were not labeled in RHΔcpl parasites. RHΔcpl showed faint residual staining with BO-LHVS that seems to be associated with the internal periphery of the parasite. The identity of this structure is unknown but could be the parasite's tubular mitochondrion or a subdomain of the endoplasmic reticulum. Similar results were seen with Ku80Δcpl (data not shown). Staining of numerous puncta within the cytoplasm of both infected and non-infected host cells is consistent with BO-LHVS labeling of host cathepsins within lysosomes, which conveniently serve as an internal control for reactivity. Collectively, these findings definitively establish that TgCPL is the primary target of BO-LHVS.
FIGURE 3.
BO-LHVS labeling of TgCPL knock-out strains. A, activity-based protein profiling gel analysis of parental and knock-out strains with BO-LHVS. Parasites were incubated without (−) or with (+) 200 nm BO-LHVS, lysed, and analyzed by SDS-PAGE and laser-scanning fluorometry. Note the lack of BO-LHVS reactivity in the 30 kDa region of RHΔcpl and Ku80Δcpl. Asterisks indicate minor bands reactive with BO-LHVS in all strains. B, APB microscopy analysis of parental and knock-out strains with BO-LHVS. Newly invaded parasites were incubated with 200 nm BO-LHVS, fixed with paraformaldehyde, and viewed by fluorescence microscopy. Note the labeling of structures in RH parasites (arrows) and lack of reactivity in RHΔcpl parasites. The inset shows an enlargement of a representative tachyzoite.
X-ray Crystal Structure of TgCPL and Its Propeptide Reveals the Canonical Catalytic Triad and Active Site Cleft
To determine whether the active site architecture of TgCPL is consistent with susceptibility to LHVS, we determined the x-ray crystal structure of rTgCPL as a complex with much of its propeptide at a resolution of 2.0 Å (Fig. 4). The enzyme was autoproteolytically activated prior to crystallization, as evidenced by SDS-PAGE. An attempt was made to separate the propeptide from the activated protease by chromatography, but it was unsuccessful due to low recovery of material. Thus, by necessity, the crystals were grown from a protein solution containing the active protease and the cleaved propeptide. The final model of TgCPL consists of residues 108p–182p of the propeptide and residues 2–224 of the protease. The propeptide is missing the N-terminal His tag and linker through the first three cloned residues (Ile105p–Glu107p) and the last 16 residues (Ser183p–Leu198p), presumably due to disorder in these regions of the protein. The protease model is essentially complete, with only the first residue (Asn1) not visible in the model. The refined structural model for TgCPL is available in the Protein Data Bank with accession code 3f75.
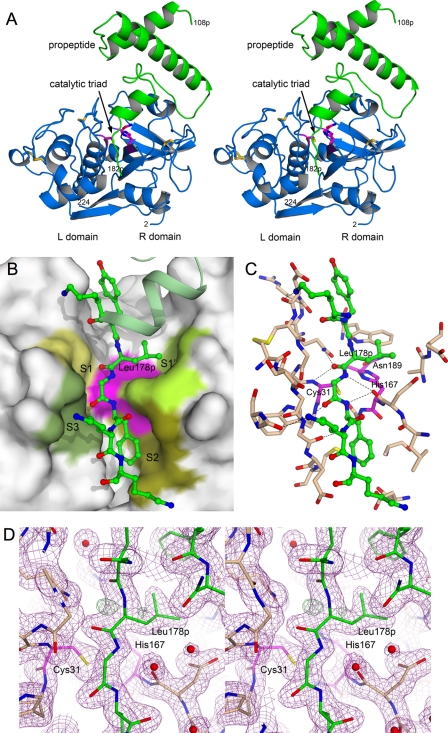
FIGURE 4.
Structure of rTgCPL in complex with its propeptide. A, stereoview looking into the active site cleft with the left (L) domain on the left, the right (R) domain on the right, and the propetide on the top. The protease is colored blue, and the propeptide is colored green. N- and C-terminal residues of each polypeptide are labeled with the corresponding amino acid number. The catalytic triad (Cys31, His167, and Asn189) is colored magenta with side chains shown as sticks. The side chains of the cysteines comprising the five disulfide bonds are also shown as sticks. B and C, detail of the propeptide residues (Lys176p–Lys182p) that occupy the active site cleft. Propeptide amino acids in the cleft are shown as balls and sticks. The orientation matches that in A. B, surface representation of the occupied cleft. Substrate-binding subsites are labeled. The catalytic triad is colored magenta. C, the surface is removed, and TgCPL residues within 5 Å of propeptide residues 176p–182p are shown as sticks. D, stereoview of the electron density around the TgCPL active site. Electron density maps are calculated using the final refined model. Purple mesh is the σA-weighted 2Fo − Fc map contoured at 1.2 σ. The green mesh is the σA-weighted Fo − Fc difference map contoured at 3.2 σ, whereas the red mesh is the Fo − Fc difference map contoured at −3.2 σ. The positive difference density peak near the backbone of propeptide residue Leu178p described under “Results” is shown. (Figs. 4 and 5 were created with PyMOL (51).)
As expected, TgCPL adopts the papain-like fold (reviewed in Refs. 41 and 42) and looks very similar to previously determined papain-like cysteine protease structures, particularly other cathepsin Ls (Fig. 4A). The full TgCPL model, including the propeptide, superimposes by secondary structure matching (43) onto human procathepsin L (Protein Data Bank code 1cs8) with a RMSD of 1.04 Å for 282 aligned Cα atoms of 297 for TgCPL and 316 for human procathepsin L. Considering only the catalytic domain, TgCPL superimposes on mature human cathepsin L (Protein Data Bank code 3bc3) (44) with a RMSD of 0.96 Å over 210 aligned Cα atoms, essentially the entire polypeptide chain.
The protease consists of two domains divided by the deep active site cleft; the left (L) domain is primarily α-helical, whereas the right (R) domain contains a β-barrel-like motif that is decorated by a few short α-helices. The canonical catalytic triad is composed of the catalytic cysteine Cys31, positioned at the N terminus of the long central helix in the L domain, and His167 and Asn189 in the R domain. TgCPL contains the three stabilizing disulfide bonds that are highly conserved among other papain-like cysteine proteases. They link cysteines Cys28 to Cys71 and Cys62 to Cys104 in the L domain and Cys161 to Cys213 in the R domain. Two additional disulfide bonds are also present, namely Cys90-Cys104 and Cys161-Cys213, with the latter possibly being a mixture of free and disulfide-bonded cysteines. These two additional disulfide bonds are not highly conserved among the papain-like cysteine proteases, so they are not likely to be critical for proper folding or activity of TgCPL.
As observed in other cathepsin L structures containing the propeptide (e.g. Protein Data Bank codes 1cjl and 1cs8) (45), the propeptide of TgCPL is composed of an N-terminal globular domain of three α-helices followed by an extended C-terminal tail that occupies the active site cleft (Fig. 4). The propeptide tail lies in the cleft in the opposite orientation to that of a natural polypeptide substrate and, in a true proenzyme, continues around the R domain and links to the N terminus of the protease. Electron density for the final 16 residues of the propeptide is not observed in the structure. These residues could have been removed by further proteolysis, or they might not form an ordered structure and thus are not visible in the electron density map.
Propeptide residues Lys176p–Lys182p occupy the majority of the active site cleft (Fig. 4, B and C). Although the orientation of the propeptide is reversed relative to the peptide backbone of a natural substrate, the environments of the substrate-binding subsites can still be observed in relation to these residues. Lys181p sits roughly in the S3 subsite. Phe180p occupies the S2 subsite, the major specificity-determining subsite, and indeed it has been observed that cathepsin L favors substrates with an aromatic residue at P2 (41).4 Gly179p is adjacent to S1, and the peptide bond between Leu178p and Gly179p lies directly above the catalytic cysteine, approximating the position of the scissile bond of the substrate. Leu178p occupies the S1′ pocket of the protease active site, with its backbone carbonyl oxygen forming a hydrogen bond with Nϵ of Gln25 while being positioned only 3 Å from the Sγ of the catalytic cysteine (Cys31). Interestingly, Leu178p exhibits a somewhat strained backbone conformation (ψ = −110°, ϕ = −116°, ω = 167°). Residual electron density near the carbonyl oxygen (Fig. 4D) suggests that the true conformation of this peptide bond is further distorted from the refined position, which is biased by refinement restraints describing typical peptide geometry. We note that the equivalent propeptide residue in the human procathepsin L (Protein Data Bank code 1cjl) and procathepsin K (Protein Data Bank codes 1by8 and 7pck) structures also displays a strained backbone conformation, although the potential significance is not clear.
Modeling the Binding Mode of LHVS
Although the TgCPL recombinant protein used for structural studies was treated with LHVS, the enzyme remained in a complex with its natural inhibitor, the propeptide, presumably because crystallization conditions were unfavorable for its displacement by LHVS. Despite this, several structures of cathepsin-vinyl sulfone inhibitor complexes have been characterized, thus enabling us to model the TgCPL·LHVS complex to ensure that a favorable binding mode is possible (Fig. 5). The structure of human cathepsin K in complex with APC3328 (Protein Data Bank code 1mem) (33) was used to aid the initial positioning of LHVS in the active site. APC3328 is the closest structural homolog to LHVS available in the Protein Data Bank with only a single atom difference of nitrogen versus oxygen. The conservation of the catalytic triads between the two cathepsins, with a RMSD of only 0.28 Å, allows for an excellent initial placement of the inhibitor. After this initial placement and substitution of the piperazine nitrogen in APC3328 by an oxygen atom to create the morpholine group of LHVS, the binding mode model was improved by energy minimization.
FIGURE 5.
Structural basis of LHVS inhibition. A, surface representation of the TgCPL active site cleft with modeled LHVS shown as balls and sticks. The catalytic triad is colored magenta, and the view is the same as that depicted in Fig. 4, B and C. B, stereoview of the LHVS binding mode. The surface of TgCPL has been removed, and amino acids within 5 Å of modeled LHVS are shown as sticks, with hydrogen bonds between the enzyme and the inhibitor shown as dashed lines. C, two-dimensional representation of the LHVS binding mode. LHVS is shown as black lines, whereas the TgCPL amino acids that surround it are shown as gray lines. Hydrogen bonds are shown as dashed lines. (C was created with ChemDraw 11.)
Overall, the model predicts that LHVS fits quite well in the active site, establishing several hydrogen bonds with residues lining the active site cleft (Fig. 5, B and C). The morpholine group of LHVS mimics position P3 of the natural substrate and occupies the S3 subsite of the protease, with the morpholine oxygen forming a hydrogen bond with Nϵ of Gln69. This favorable interaction is afforded by allowing the side chain amide of Gln69 to rotate ∼180° during energy minimization from its orientation in the propeptide-bound complex, where the side chain carboxyl oxygen is about 3.2 Å from Nζ of propeptide residue Lys181p. The leucyl moiety of LHVS mimics substrate position P2 and occupies the largely hydrophobic S2 pocket of the protease. The amide and carbonyl of the P2 leucine form antiparallel hydrogen bonds with TgCPL residue Gly74 as would be expected for the backbone of the P2 residue in the natural polypeptide substrate. Homophenylalanine mimics the P1 position of the substrate, and its side chain is situated along the fairly shallow S1 protease subsite, whereas its backbone amide forms a hydrogen bond with the carbonyl oxygen of Asp166. The catalytic Cys31 nucleophilically attacks the vinyl group, which mimics the scissile bond, to form a covalent complex with the inhibitor. The phenyl sulfone of LHVS mimics P1′ of the natural substrate and thus occupies the S1′ subsite in the active site cleft. One of the sulfonyl oxygen atoms is oriented toward the catalytic site, where it is stabilized by Nϵ of Gln25 and Nϵ of Trp191 and also by Nδ of His167 in the catalytic triad. The binding mode predicted by this model strongly suggests that a covalent complex between TgCPL and LHVS is favorable and that, similar to other cathepsin L proteases, LHVS will inhibit TgCPL activity.
DISCUSSION
Identification of the in vivo target is often the most difficult step in the characterization of small molecule inhibitors. Two properties of LHVS greatly facilitated identification of its main target, TgCPL. First, the vinyl sulfone warhead of LHVS covalently modifies the active site thiol of reactive cathepsin proteases, thus allowing irreversible labeling of its target. Second, the structure of LHVS and its known binding mechanism permitted the synthesis of a functional fluorescent derivative, BO-LHVS, for activity-based protein profiling and target identification. The versatility of this chemical probe is also an asset, since it can label targets in vitro or in live cells, and it allows an assessment of the specificity, abundance, and subcellular distribution of the active enzyme. It should be noted, however, that specificity depends greatly on the concentration of inhibitor used. Incubation of live parasites with 200 nm BO-LHVS principally results in labeling of TgCPL. Several additional minor products are also labeled, but these are not blocked by pretreatment with LHVS, indicating that they are nonspecific targets. LHVS inhibition of parasite microneme secretion, gliding motility, and attachment occurs with an IC50 of 10 μm (12), whereas LHVS inhibits TgCPL activity with an IC50 of ∼20 nm. Therefore, it remains possible that a target in addition to TgCPL contributes to the observed effects on parasite cell entry. Alternatively, the parasite might express a small pool of TgCPL that is only susceptible to high concentrations of LHVS or is maintained in the proform and is thus not reactive with LHVS. Indeed, evidence of this was seen by fluorescence microscopy, where some TgCPL-associated structures failed to label with BO-LHVS (Fig. 1E), although only a small subset of parasites displayed this phenomenon. These antibody-reactive but BO-LHVS-unreactive structures may contain a store of procathepsin L where the propeptide blocks reaction with the catalytic cysteine.
Preliminary phenotypic studies suggest that the RHΔcpl and Ku80Δcpl show different invasion competencies. TgCPB expression is up-regulated to different levels in these strains, which is reminiscent of TgCPC2 up-regulation seen after genetic ablation of TgCPC1 (8). Elevation of TgCPB expression may suppress phenotypes in the TgCPL-deficient strains. Determining the precise relationship between the functions of TgCPL and TgCPB will require more extensive genetic and cell biological studies that are beyond the scope of the current work.
TgCPL is associated with a discrete vesicular structure usually seen in the apical region of extracellular parasites. Intracellular replicating parasites show multiple puncta of TgCPL structures throughout the cytoplasm. Although the exact nature of the TgCPL-associated structure(s) is still being investigated, the available evidence suggests that it is an endocytic organelle possibly related to a lysosome.4 In this case, TgCPL might act in the classical role of a cathepsin (i.e. as a degradative protease involved in protein turnover and nutrient acquisition). Additionally, TgCPL may have a specialized role in the selective proteolysis of substrates, akin to, for example, the function of human cathepsin L in the processing of the MHC II invariant chain in thymic epithelial cells (46) and proenkephalin in neuroendocrine cells (47). In these examples, cathepsin L selectively degrades a regulator of antigen presentation (invariant chain) and performs limited proteolysis of a key neurotransmitter (proenkephalin). Approximately half of the proteins targeted to micronemes and all of those destined for rhoptries undergo limited proteolysis (maturation) en route to these invasion organelles. Indeed, recent studies have revealed that rhoptry and microneme proteins traffic through the parasite endocytic system (15, 48, 49), where they may encounter TgCPL along the way. Many of these proteins are also further processed coincident with their secretion during parasite cell invasion. It is possible that LHVS inhibition of TgCPL (or other reactive targets) interferes with the processing of invasion proteins, thus accounting for the effects of LHVS on parasite entry. TgCPL is expressed in both the tachyzoite stage responsible for acute infection and opportunistic disease and in the bradyzoite cyst stage seen during chronic asymptomatic infection (8).4 Thus, TgCPL could contribute to infection during both stages seen in humans. Identification of TgCPL substrates by comparative analysis of wild type and knock-out parasites or LHVS-treated parasites should provide additional insight into whether it serves as a general degradative protease or has a more selective role in the processing of invasion proteins or both.
The x-ray crystal structure of TgCPL in complex with its propeptide provides a detailed look at the active site of the enzyme and facilitates modeling of the LHVS binding mode. A valid and meaningful model of the LHVS binding in TgCPL is possible from the structure presented here for several reasons. First, the in vitro and in vivo work presented here clearly shows that TgCPL is a primary target of LHVS. Second, the inhibitor is covalently linked to the protein at a defined location and with a known directionality in the active site cleft severely restricting the possible binding mode search space to sample. Also, since comparisons of proform protease structures with structures of their mature enzyme show that the catalytically active form already exists within the proenzyme (41, 42), the presence of the cleaved propeptide in the structure probably has little effect on the active site architecture. The report by Huang et al. (10) that TgCPL favors Leu over other residues in the P2 position is also consistent with the favorable recognition and binding of LHVS. Thus, the model shown in Fig. 5 confirms that a favorable interaction is possible with the TgCPL active site and presents a plausible structural basis for the observed in vitro and in vivo activity of LHVS.
In summary, we identified TgCPL as the primary target of the protease inhibitor LHVS and have shown that the structure of the active site cleft is consistent with recognition of LHVS. Future work will focus on identifying TgCPL substrates and determining its role in parasite microneme secretion and cell invasion.
Supplementary Material
Acknowledgments
We thank Dr. Christophe Verlinde for assistance and advice with LHVS modeling and Tracy Arakaki for assistance with crystal screening and for helpful discussions. We also acknowledge the support and contributions of the entire MSGPP team and thank the Center for High Throughput Structural Biology at the Hauptman-Woodward Institute for continued crystallization support. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. We also gratefully acknowledge Tracey Schultz and Claudia Bordon for technical assistance and Doron Greenbaum, Carolyn Phillips, Viviana Pszenny, Sarah Strickler, Marsha Thomas, and members of the Carruthers laboratory for various contributions and helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants RR020843 (to M. B.), GM64655 (to E. A. M.), AI067921 (to E. A. M.), and AI063263 (to V. B. C.). This work was also supported by Stanley Medical Research Institute Grant 04R-796 (to V. B. C). The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and NIGMS.
The atomic coordinates and structure factors (code 3F75) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
V. B. Carruthers, unpublished results.
F. Parussini, I. Coppens, P. P. Shah, S. L. Diamond, and V. B. Carruthers, manuscript in preparation.
- LHVS
- morpholinurea-leucyl-homophenyl-vinyl sulfone phenyl
- rTgCPL
- recombinant TgCPL
- BO-LHVS
- BODIPY-LHVS
- DHFR
- dihydrofolate reductase
- TS
- thymidylate synthase
- APC3328
- N-[1S-(2-phenylethyl)-3-phenylsulfonylallyl]-4-methyl-2R-piperazinyl carbonylaminovaleramide
- RMSD
- root mean square deviation.
REFERENCES
- 1.Puente X. S., Sánchez L. M., Overall C. M., López-Otín C. (2003) Nat. Rev. Genet. 4, 544–558 [DOI] [PubMed] [Google Scholar]
- 2.Rosenthal P. J. (2004) Int. J. Parasitol. 34, 1489–1499 [DOI] [PubMed] [Google Scholar]
- 3.McKerrow J. H., Caffrey C., Kelly B., Loke P., Sajid M. (2006) Annu. Rev. Pathol. 1, 497–536 [DOI] [PubMed] [Google Scholar]
- 4.Keeley A., Soldati D. (2004) Trends Cell Biol. 14, 528–532 [DOI] [PubMed] [Google Scholar]
- 5.Carruthers V., Boothroyd J. C. (2007) Curr. Opin. Microbiol. 10, 83–89 [DOI] [PubMed] [Google Scholar]
- 6.Zhou X. W., Kafsack B. F., Cole R. N., Beckett P., Shen R. F., Carruthers V. B. (2005) J. Biol. Chem. 280, 34233–34244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley P. J., Ward C., Cheng S. J., Alexander D. L., Coller S., Coombs G. H., Dunn J. D., Ferguson D. J., Sanderson S. J., Wastling J. M., Boothroyd J. C. (2005) J. Biol. Chem. 280, 34245–34258 [DOI] [PubMed] [Google Scholar]
- 8.Que X., Engel J. C., Ferguson D., Wunderlich A., Tomavo S., Reed S. L. (2007) J. Biol. Chem. 282, 4994–5003 [DOI] [PubMed] [Google Scholar]
- 9.Que X., Ngo H., Lawton J., Gray M., Liu Q., Engel J., Brinen L., Ghosh P., Joiner K. A., Reed S. L. (2002) J. Biol. Chem. 277, 25791–25797 [DOI] [PubMed] [Google Scholar]
- 10.Huang R., Que X., Hirata K., Brinen L. S., Lee J. H., Hansell E., Engel J., Sajid M., Reed S. (2009) Mol. Biochem. Parasitol. 164, 86–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljunggren A., Redzynia I., Alvarez-Fernandez M., Abrahamson M., Mort J. S., Krupa J. C., Jaskolski M., Bujacz G. (2007) J. Mol. Biol. 371, 137–153 [DOI] [PubMed] [Google Scholar]
- 12.Teo C. F., Zhou X. W., Bogyo M., Carruthers V. B. (2007) Antimicrob. Agents Chemother. 51, 679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donald R. G., Roos D. S. (1993) Proc. Natl. Acad. Sci. U. S. A. 90, 11703–11707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brydges S. D., Harper J. M., Parussini F., Coppens I., Carruthers V. B. (2008) Biol. Cell 100, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper J. M., Huynh M. H., Coppens I., Parussini F., Moreno S., Carruthers V. B. (2006) Mol. Biol. Cell 17, 4551–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luft J. R., Collins R. J., Fehrman N. A., Lauricella A. M., Veatch C. K., DeTitta G. T. (2003) J. Struct. Biol. 142, 170–179 [DOI] [PubMed] [Google Scholar]
- 17.Cohen A. E., Ellis P. J., Miller M. D., Deacon A. M., Phizackerley R. P. (2002) J. Appl. Crystallogr. 35, 720–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) J. Synchrotron Radiat. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 19.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20.McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarzenbacher R. A., Godzik A., Grzechnik S. K., Jaroszewski L.Acta Crystallogr. D Biol. Crystallogr. 60, 1229–1236 [DOI] [PubMed] [Google Scholar]
- 22.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 23.Stein N. (2008) J. Appl. Crystallogr. 41, 641–643 [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 27.Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 28.Perrakis A., Harkiolaki M., Wilson K. S., Lamzin V. S. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 29.Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 30.Painter J., Merritt E. A. (2006) J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 31.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 32.Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath M. E., Klaus J. L., Barnes M. G., Brömme D. (1997) Nat. Struct. Biol. 4, 105–109 [DOI] [PubMed] [Google Scholar]
- 34.Kabsch W. (1976) Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 35.McMartin C., Bohacek R. S. (1997) J. Comput. Aided Mol. Des. 11, 333–344 [DOI] [PubMed] [Google Scholar]
- 36.Brömme D., Klaus J. L., Okamoto K., Rasnick D., Palmer J. T. (1996) Biochem. J. 315, 85–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogyo M., Verhelst S., Bellingard-Dubouchaud V., Toba S., Greenbaum D. (2000) Chem. Biol. 7, 27–38 [DOI] [PubMed] [Google Scholar]
- 38.Bromme D., Nallaseth F. S., Turk B. (2004) Methods 32, 199–206 [DOI] [PubMed] [Google Scholar]
- 39.Gajria B., Bahl A., Brestelli J., Dommer J., Fischer S., Gao X., Heiges M., Iodice J., Kissinger J. C., Mackey A. J., Pinney D. F., Roos D. S., Stoeckert C. J., Jr., Wang H., Brunk B. P. (2008) Nucleic Acids Res. 36, D553–D556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huynh M. H., Carruthers V. B. (2009) Eukaryot. Cell 8, 530–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGrath M. E. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 181–204 [DOI] [PubMed] [Google Scholar]
- 42.Turk D., Guncar G. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 203–213 [DOI] [PubMed] [Google Scholar]
- 43.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 44.Chowdhury S. F., Joseph L., Kumar S., Tulsidas S. R., Bhat S., Ziomek E., Ménard R., Sivaraman J., Purisima E. O. (2008) J. Med. Chem. 51, 1361–1368 [DOI] [PubMed] [Google Scholar]
- 45.Coulombe R., Grochulski P., Sivaraman J., Ménard R., Mort J. S., Cygler M. (1996) EMBO J. 15, 5492–5503 [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa T., Roth W., Wong P., Nelson A., Farr A., Deussing J., Villadangos J. A., Ploegh H., Peters C., Rudensky A. Y. (1998) Science 280, 450–453 [DOI] [PubMed] [Google Scholar]
- 47.Yasothornsrikul S., Greenbaum D., Medzihradszky K. F., Toneff T., Bundey R., Miller R., Schilling B., Petermann I., Dehnert J., Logvinova A., Goldsmith P., Neveu J. M., Lane W. S., Gibson B., Reinheckel T., Peters C., Bogyo M., Hook V. (2003) Proc. Natl. Acad. Sci. U. S. A. 100, 9590–9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoppe H. C., Ngo H. M., Yang M., Joiner K. A. (2000) Nat. Cell Biol. 2, 449–456 [DOI] [PubMed] [Google Scholar]
- 49.Ngo H. M., Yang M., Paprotka K., Pypaert M., Hoppe H., Joiner K. A. (2003) J. Biol. Chem. 278, 5343–5352 [DOI] [PubMed] [Google Scholar]
- 50.Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, LLC, Palo Alto, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.