Abstract
Collagen, known for its structural role in tissues and also for its participation in the regulation of homeostatic and pathological processes in mammals, is assembled from triple helices that can be either homotrimers or heterotrimers. High resolution structural information for natural collagens has been difficult to obtain because of their size and the heterogeneity of their native environment. For this reason, peptides that self-assemble into collagen-like triple helices are used to gain insight into the structure, stability, and biochemistry of this important protein family. Although many of the most common collagens in humans are heterotrimers, almost all studies of collagen helices have been on homotrimers. Here we report the first structure of a collagen heterotrimer. Our structure, obtained by solution NMR, highlights the role of electrostatic interactions as stabilizing factors within the triple helical folding motif. This addresses an issue that has been actively researched because of the predominance of charged residues in the collagen family. We also find that it is possible to selectively form a collagen heterotrimer with a well defined composition and register of the peptide chains within the helix, based on information encoded solely in the collagenous domain. Globular domains are implicated in determining the composition of several collagen types, but it is unclear what their role in controlling register may be. We show that is possible to design peptides that not only selectively choose a composition but also a specific register without the assistance of other protein constructs. This mechanism may be used in nature as well.
Collagens constitute an important structural protein family. They are found in the extracellular matrix and undergo a hierarchical self-assembly into large supramolecular structures with specific morphologies carefully crafted by nature to fulfill diverse structural and functional roles in a wide variety of tissues. In total, there are 28 known isoforms of collagen in humans arranged in a variety of structures and in a wide range of tissues. The feature defining this protein family is the presence of domains with uninterrupted Xaa-Yaa-Gly sequence repeats. These domains adopt a left-handed polyproline type II conformation because of the predominance of proline in the X position and hydroxyproline (Hyp = O), a post-translationally modified amino acid with a hydroxyl group on the γ-carbon of the proline side chain, in the Y position. Three such domains associate to form tightly packed right-handed triple helices in a folding motif commonly known as the collagen triple helix.
Collagens are also implicated in pivotal homeostatic events in mammals such as the production of new vascular tissue and pathological conditions such as cancer metastasis (1). These processes are notoriously governed by interactions at the molecular level between cell surface proteins and the collagen triple helix and not by the morphology of the collagen aggregates (2, 3). Thus, an understanding of the collagen molecule and its interactions with other proteins at the atomic level has been actively pursued. However, because of its complex hierarchical self-assembly, and the scale of the resulting supramolecular structures, it is difficult to obtain information at atomic resolution for collagenous proteins (4). An approach developed to overcome this limitation is the use of short model peptides that adopt a triple helical fold (5). Such peptides have been used to study the structure (6, 7), folding (8), and dynamics (9) of the triple helix. These peptides have been shown to retain the biochemical properties of the higher assemblies found in their natural counterparts, binding to cell surface proteins such as integrins (10).
Most of the studies performed on collagen mimetic peptides utilize triple helices with three identical chains called homotrimers (8, 11–13). Such systems are good models for some types of collagen, like type II. However, many of the most abundant types of collagen such as type I, IV, and IX are heterotrimeric species containing two (AAB) or three (ABC) different chains. Recently we introduced a new method to prepare heterotrimeric collagen like triple helices via noncovalent interactions (14–16). These systems have been primarily characterized through CD spectroscopy, which is a good indicator for the fold and stability of the peptides but lacks the ability to give detailed structural information. There are few studies available in the literature that utilize NMR to examine collagen heterotrimers; however, none of them use the technique to examine the structures of the assemblies in detail (17–20), and none result in a complete structural determination.
A system of particular interest is composed of three peptides, (Pro-Lys-Gly)10, (Asp-Hyp-Gly)10, and (Pro-Hyp-Gly)10, which we abbreviate K, D, and O respectively. Upon mixing and annealing of the peptides, CD studies indicate that an ABC triple helix with a surprisingly high thermal stability is formed (16). We hypothesized that the high thermal stability of these systems comes from the formation of charge pairs between lysine and aspartic acid. Homotrimeric model peptides that contain the sequence KGD, which occurs both in mammalian collagen (21) and bacterial collagen-like proteins (22), apparently also use this charge pairing. However, no structural information has been available to confirm the nature of the interactions.
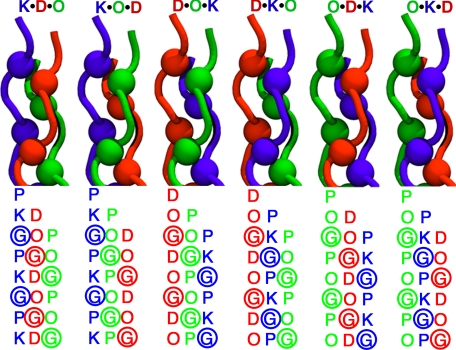
Here we study the K·D·O system using two- and three-dimensional NMR techniques to determine the composition and fold of the components of an annealed mixture of these three peptides. Also, for the first time, we are able to study the register or relative stagger of the peptide chains in the triple helix. In a collagen triple helix, the chains assemble staggered by one amino acid, so that there is always a glycine residue in every cross-section of the helix taken perpendicular to the helical axis. This allows the peptide chains to pack tightly while avoiding steric clashes in the center of the assembly. Depending on the nature of the leading, middle, and trailing chain, a total of six different assemblies, or registers, are possible for an ABC system (Fig. 1). Given the high thermal stability of the system, which allows for the recording of high quality NMR spectra, we found that our peptides preferentially populate one register, and using samples with strategically placed isotopically labeled amino acids, we are able to determine which one. Furthermore, we are able to obtain the first structure of a collagen triple helix in solution and give direct evidence of ionic hydrogen bonds as a stabilizing factor within the triple helical folding motif.
FIGURE 1.
Schematic N-terminal representation of the six possible registers for the heterotrimeric triple helix. The difference in the sequence is highlighted below each representation, where the position of glycine residues is marked by colored spheres.
EXPERIMENTAL PROCEDURES
Peptide Synthesis and Sample Preparation
All peptides were synthesized with an Advanced Chemtech Apex 396 solid phase peptide synthesizer using standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and a Rink 4-methylbenzhydrylamine-amide resin and were N-terminally acetylated and C-terminally amidated. The uniformly labeled amino acids were purchased form Cambridge Isotope Laboratories. Purification was performed on a Varian PrepStar220 high pressure liquid chromatograph using a preparative reverse phase C-18 column with a linear gradient of water and acetonitrile each containing 0.5% trifluoroacetic acid and analyzed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry on a Bruker Autoflex II (supplemental Fig. S1). Further details on the synthesis protocol used are available in the supplemental material.
NMR samples were prepared in a 9:1 ratio of H2O to D2O and a 10 mm phosphate buffer to maintain a neutral pH. The concentration used for samples containing only one peptide strand was 1.2 mm, determined by mass. For experiments including all three chains, the peptides were mixed in a 1:1:1 ratio, with a total peptide concentration of 3.6 mm, unless otherwise noted. Heterotrimer samples were annealed at 85 °C for 15 min and then incubated for at least 72 h at room temperature before beginning the NMR measurements. Samples of the following composition were prepared: K, D, O, K/D/O, and K*/D*/O*. A fourth sample with composition K/D/O* was prepared, in which the peptides were mixed in a 1.5:1:1 ratio. The asterisks indicate peptides that containing amino acids uniformly labeled with 15N and 13C, for details on their sequence refer to Table 1.
TABLE 1.
Peptide abbreviations and chemical sequences
| Abbreviation | Sequencea |
|---|---|
| K | (PKG)10 |
| D | (DOG)10 |
| O | (POG)10 |
| K* | (PKG)4PKGPKG(PKG)4 |
| D* | (DOG)4DOGDOG(DOG)4 |
| O* | (POG)4POGPOG(POG)4 |
a The underlined amino acids are uniformly 15N,13C-labeled.
NMR Spectroscopy
All NMR experiments were recorded in an 800-MHz Varian spectrometer equipped with a cryogenic probe. The spectra were processed using the NMRpipe software (23) and analyzed using Sparky (24) and ccpnmr (25). Square cosine bell windows were used as apodization functions, and the data were zero-filled to the next power of 2 in both dimensions. Linear base-line corrections and forward-backward linear prediction were applied when necessary.
Monomeric samples (K and D) were analyzed exclusively through two-dimensional total correlated spectroscopy (TOCSY)2 at 25 °C. In contrast, TOCSY and nuclear Overhauser effect spectroscopy (NOESY) experiments at 15 and 25 °C were recorded for the triple helical samples (O and K/D/O). 1H,15N- and 1H,13C-heteronuclear single quantum coherence experiments (HSQC) were recorded for the labeled samples (K*/D*/O* and K/D/O*) at 25 °C.
To determine the register of the triple helix, a two-dimensional version of a four-dimensional 1H,13C-HMQC-NOESY-1H,15N-HSQC experiment was recorded at 25 °C (26); to ease further discussion of this experiment, it will be referred to as a two-dimensional 13C,15N-edited NOESY. Details on this experiment are available in the supplemental material.
A three-dimensional HNHA experiment was also recorded at 25 °C to compute 3JHNHα coupling constants (27). Details about the computation of the coupling constants from the spectrum are available in the supplemental material. A three-dimensional HNHB experiment was also recorded at 25 °C to estimate the 3JNHβ coupling constants (28). A qualitative approach to the estimation of the coupling constants and side chain rotamers was taken (29). The detailed processing algorithms and parameters for all experiments are available in the supplemental material online.
Molecular Modeling
Homology models were built starting from the crystal structure of a triple helical peptide (Protein Data Bank code 1k6f) (30). The necessary sequence changes were made using PyMOL (31) to generate a preliminary structure for each of the six possible registers. Each structure was then minimized using the AMBER99 (32) force field with implicit water (generalized Born approximation). Additional force field parameters to account for the stereo electronic effects of the hydroxyl group on the proline side chain conformation were included (33). Short constant temperature Langevin dynamics runs at 300, 200, and 100 K were used within the minimization algorithm to equilibrate the structures and obtain low energy conformers.
Conformational Restraints and Structure Calculation
Distance restraints were generated from the two-dimensional NOESY experiments. The peaks were mapped onto the shortest stretch of the chemical sequence that could unambiguously accommodate all inter- and intra-strand resonances (PKGPKG for K, OGDO for D, and POG for O). A qualitative approach was taken, and the peaks were divided into four categories (very strong, strong, medium, and weak) according to their intensity. The restraints were propagated along the sequence from triplet 2 to triplet 9, assuming that all those amino acids have an identical conformation contributing equally to the observed peaks and leaving the N- and C-terminal triplets unconstrained because those amino acids have been shown to populate a less ordered conformation in homotrimeric triple helices (11).
Three types of dihedral restraints were used in the calculations. The χ1 angle of K and D were loosely constrained based on the results of the HNHB experiment, and the χ1 and ϕ dihedrals of proline and hydroxyproline were constrained according to the ring puckering of the side chain, as determined by the intensity ratio of the β- and δ-protons (11). The third type of dihedral restraints corresponds to the ϕ backbone dihedral angles for K, D, and G, constrained using values derived from the coupling constants obtained from the HNHA experiment. Because the Karplus equation generates up to four possible dihedral values for each coupling constant, a complementary strategy is needed to obtain a single value to use in the refinement procedure. In the case of glycine residues, this is straightforward as each of the methylene protons affords a different coupling constant, one being shifted by a phase factor of 120°. Solving the equation using the coupling constant measured for each proton and comparing the solutions yields only one pair of angles that satisfies this condition. To obtain a value for aspartic acid and lysine, we used a preliminary simulated annealing round starting from unfolded chains using distance restraints supplemented by dihedrals for all residues type except K and D, with coupling constants restraints for the charged residues (all possible solutions to the Karplus equation). We looked at the low energy structures of the calculation and picked the solution of the Karplus equation that best agreed with the observed ϕ distribution for K and D residues (supplemental Fig. S5). As with the distance constraints, all dihedral constraints were applied for the residues in triplets 2–9. Further details on the restraints are available in the supplemental material. This resulted in the unambiguous selection of one set of angles that were used for further refinement.
Structure calculations were done using cycles of simulated annealing (SA) followed by a refinement in implicit solvent. In the SA stage, 300 trial structures were calculated using a combination of torsional and Cartesian dynamics with the standard protocol available in the Crystallography and NMR System (CNS) software (34). The refinement stage was done in AMBER99, performing a minimization in implicit solvent subjected to the same constraints utilized in the SA stage on the 150 conformers that showed the lowest CNS target function. In the initial cycle, structure calculations were started from extended polypeptide chains, and only backbone dihedral constrains were used. The minimum energy conformer was then used to start a new cycle, in which only Cartesian dynamics were used in the SA stage, but all the constraints available were included. The 15 conformers with the lowest energy, as calculated by AMBER, were then selected for the final ensemble.
RESULTS
Spin System Identification
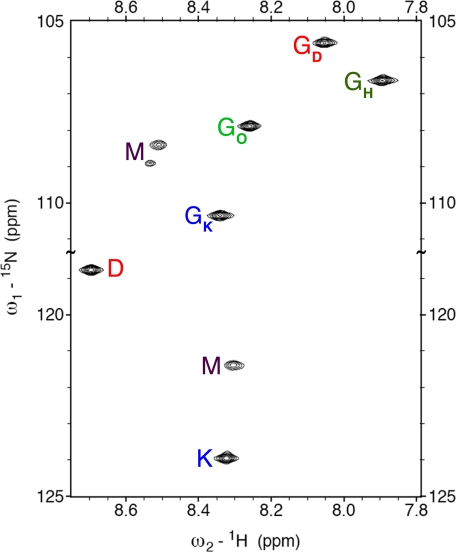
The number of species present in the sample was determined from a nitrogen 1H,15N-HSQC experiment using the peptides K*, D*, and O* with uniformly 15N,13C-labeled amino acids (chemical sequences shown in Table 1 and spectrum in Fig. 2). Some of the peaks can easily be identified as the monomeric forms of the highly charged D and K peptides using the information from TOCSY spectra of samples composed of each peptide separately. These are labeled M in the spectrum. The O peptide readily forms O·O·O homotrimers in solution, and the presence of this species in the mixture was identified using homonuclear spectra containing exclusively this peptide (11). That peak is labeled as GH in the spectrum. The remaining five resonances correspond to the novel assembly formed when the three peptides come together and represent our subset of interest.
FIGURE 2.
Two-dimensional 1H,15N-HSQC spectrum of K*·D*·O*. Triple helical resonances are labeled using single letter codes. For glycine, the chain to which it belongs is specified as a subscript (H stands for homotrimer). Monomeric resonances are labeled M.
To simplify further discussion, the following notation will be used when referring to a particular atom: ZN(B), where Z is the amino acid single letter code; N is the peptide chain (either D, O, K, or H for the O·O·O homotrimer), and B specifies which particular atom in the amino acid is being discussed. In cases where the peptide chain is unambiguous or amino acids from multiple chains are being discussed, the index specifying the register is omitted. For example P(CαH) refers to all proline α-protons.
The spin systems belonging to each chain were determined by homonuclear sequential assignment using TOCSY and NOESY spectra at 15 and 25 °C. Intra-residue connectivity can be readily identified in the TOCSY spectra, and all possible inter-residue NOEs from the NH of residue i to the CαH of residues i–l are present. Even though they lack amide protons, all P(CαH) and O(CαH) resonance frequencies were identified through the NOEs with the NH of the next amino acid, except for the proline on the O chain. In that case the sequential following of two imino acids makes it impossible to determine the PO(CαH) chemical shift this way. Thus this residue was assigned based solely on the resonances present in the aliphatic region of the spectra. This approach was feasible because the cross-peaks necessary for the sequential assignment can be uniquely identified as intra-chain peaks, because their inter-chain analogs are not present.
Most methylene groups presented unique chemical shifts for both their diastereotopic protons with the exception of the γ-protons of proline and the δ- and ϵ-protons of lysine. Stereospecific assignments for the methylene groups with nondegenerate chemical shifts for the proline and hydroxyproline residues were carried out using the NOE intensities of the cross-peaks between the β-, δ-, and α-protons and the β-, δ-, and γ-protons, respectively. Because of conformational restrictions placed on the methylene groups by the proline rings, these assignments are straightforward. In the case of the α-protons of the glycine residues, a combination of NOE data and the cross-peak intensity in the HNHA spectrum was used. A similar approach was taken for the β-protons of lysine and aspartic residues but using the information from the HNHB spectrum instead. The γ-protons of the lysine residues were assigned exclusively based on NOE cross-peak intensity.
Assessment of the Triple Helical Topology
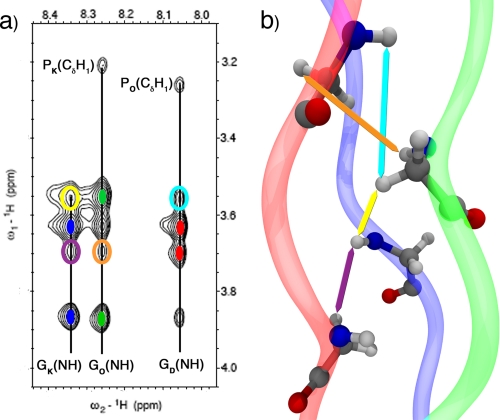
The amide region of the NOESY spectrum (supplemental Fig. S2) shows a set of resonances at the chemical shifts corresponding to the position of the GK(NH), GD(NH), GO(NH), GH(NH), D(NH), and K(NH) peaks in the 1H dimension of the 1H,15N-HSQC spectrum, indicating that most amino acids in each peptide show an ordered structure that is very similar to that of the central triplets, thus having identical chemical shifts. This phenomenon is characteristic of triple helical peptides, where the majority of the triplets show an identical chemical environment because of the symmetry of the helix. However, to ensure that are our peptides are indeed folded in a triple helical conformation, we need to compare the NOEs observed to those expected from a triple helix. To this effect, we compared all unique inter-chain NOEs expected for the O·O·O homotrimer (11) and our heterotrimer (supplemental Table S1). We were able to find analogous peaks, and although not all of them can be resolved unequivocally, because of the overlap of the PO(CγH), OO(CβH1) and OD(CβH1) resonances, this comparison gives us confidence that the observed NOEs are indeed consistent with a triple helical fold of the peptides.
Because of the symmetry breaking induced by the heterogeneity of the chemical composition in our assembly, some of the resonances that are degenerate in the case of a homotrimer can be easily resolved in our system. Such resonances are of interest because they are not only characteristic of a collagen triple helix but also demonstrate that the three chains are in close proximity, as expected for an ABC heterotrimer. An illustrative set of cross-peaks of these two facts is the one arising from the G(NH) to the G(CαH) protons. Besides the intra-chain NOEs identified in the TOCSY spectrum, a set of inter-strand NOEs is present (Fig. 3A). Although all possible G(NH)-G(CαH) inter-chain correlations are present, some of them overlap, and only the ones that can be unambiguously assigned are highlighted. These peaks confirm the spatial proximity of the α-protons and the amide protons on all the chains in the core of the helix, as expected from the collagen model (Fig. 3B). The same set of resonances cannot be observed in the homotrimer spectrum as the G(NH) and G(CαH) chemical shifts are indistinguishable between the different chains. Another way to probe the conformation of peptides in solution is to measure the 3JHNHα coupling constant because it can be directly linked to the ϕ backbone dihedral angle via the Karplus relation. We measured the coupling constant for our heterotrimeric helix using the HNHA experiment (see “Experimental Procedures” for details) and for the residual O·O·O homotrimer in our system. The values obtained range from 4 to 7 Hz and agree with previously measured values for homotrimeric triple helices (12). Table 2 shows a comparison between the ϕ dihedral angles computed from these values and those of a high resolution O·O·O crystal structure (35) and two model helices, one with 7/2 symmetry (6) and one with 10/3 symmetry (36). The angles obtained for aspartic acid and lysine agree with those expected from amino acids in the X and Y position of a collagen triple helix. The values obtained for the glycines of all three chains also agree with those determined for the homotrimer using x-ray crystallography and our NMR measurements.
FIGURE 3.
Two-dimensional NOESY spectrum and homology model. a, NOESY spectrum shows the resonances from the glycine amide hydrogen to the α-protons. The vertical lines mark the NH chemical shifts; the solid colored ellipses indicate the position of the intra-strand cross-peaks (red stands for D, green for O, and blue for K), and the open ellipses indicate the unambiguous inter-strand interactions. The P(CδH1)-G(NH) cross-peaks are also shown. b, homology model highlighting the atoms that that give rise to the inter-stand cross-peaks in the spectrum using colored arrows.
TABLE 2.
Dihedral angles calculated from the 3JHNHα coupling constants
The ratio between the homotrimeric and heterotrimeric species was determined using the peak intensity observed for each triple helix in the 1H,15N-HSQC experiment. For the K*/D*/O* sample, which contains a 1:1:1 mixture of the peptides, the ratio of heterotrimer to homotrimer is ∼3 to 1. Changing the relative amount of one of the peptide strands in the mixture can shift the equilibrium toward the formation of the heterotrimer. This was observed in the K/D/O* sample, which contains an excess of the K peptide (1.5:1:1), and has a ratio of the heterotrimer to homotrimer of ∼11 to 1.
Register of the Chains in the Triple Helix
The glycine amide nitrogen and hydrogen chemical shifts of amino acids in triple helical conformation are known to be very sensitive to subtle changes in their hydrogen bonding network (19, 20). Because we observe only one set of resonances for each amino acid type in each peptide chain of the heterotrimeric triple helix (Fig. 2), we believe that that the peptides preferentially assemble in only one of the six possible registers. We would expect to see a more heterogeneous spectrum, for example as observed by Slatter et al. (20), had there been more species present in solution. We are able to determine the presence of several peptide assemblies (including the (POG)10 homotrimer, monomeric (DOG)10, and monomeric (PKG)10), but only one assembly corresponds to a heterotrimeric triple helix. We do not rule out the possibility of other registers being present; however, we are sure that if present they are below the level of detection by NMR.
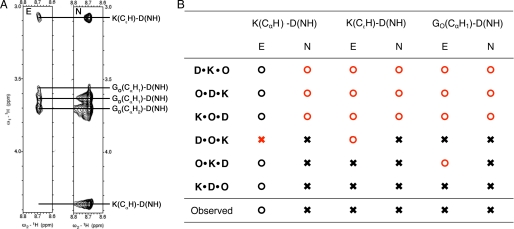
To determine which register is predominantly populated, we built a homology model for each possible assembly (see under “Experimental Procedure” for details) and compared the NOEs observed with those expected from each model. A set of resonances that is very useful when analyzing this problem arises between the K(CαH1)-D(NH), K(CϵH)-D(NH), and GO(CαH1)-D(NH) protons. These are depicted in the strip corresponding to the NOESY spectrum shown in Fig. 4A (labeled N). Fig. 4B summarizes the expected results of this experiment for each register. When an inter-proton distance less than 5 Å is observed for any of the aforementioned pairs in the model, an × is placed in the column corresponding to the NOESY spectrum (N) of that register; otherwise an ○ is placed in that spot. The result of the actual NMR experiment is summarized in the last row of the table in Fig. 4B, where an × has been placed for each of the observed resonances. Any inconsistencies between the spectra and the models are highlighted in red. Using this comparison, the three registers D·K·O, O·D·K, and K·O·D can be discarded. However, because of the periodic nature of our sequences, unambiguously determining the register, using only two-dimensional NOESY experiments is not possible.
FIGURE 4.
Two-dimensional edited NOESY (E) and NOESY (N) spectra of K·D·O. A, strips from both experiments (A) corresponding to the D(NH) chemical shift. B, table showing the expected outcome of both spectra for the six possible registers of the assembly according to our models. The × indicates that a peak should be observable in the spectrum, and ○ indicates that no peak is expected. The last row summarizes the results of the strips on the right. Inconsistencies between the models and the spectra are highlighted in red.
To distinguish between the three remaining registers, knowledge about which triplet along the sequence of the amino acids gives rise to these resonances is required, i.e. we need to know if the ϵ-protons of lysine in triplet n are close to the amide proton of aspartic acid in triplet n, n + 1, or n + 2. To obtain this information we used a two-dimensional 13C,15N-edited NOESY spectrum (refer to supplemental material), where the observed resonances occur only between labeled amino acids (Lys-14, Asp-16, and GO15). A strip of the spectrum corresponding to the D(NH) chemical shift is shown in Fig. 4A (labeled E). The main difference between the edited and regular spectrum is the absence of the K(CαH1)-D(NH) peak in the edited spectrum. This means that the α-proton of Lys-14 (fifth triplet, K chain) is not close to amide proton of Asp-16 (sixth triplet, D chain). Meanwhile, the presence of the K(CϵH)-D(NH) resonance indicates that the ϵ-protons of Lys-14 are near the amide proton of Asp-16 and the GO(CαH1)-D(NH) peak that Asp-16 is within 5 Å of GO15 (fifth triplet, O chain). Using the same convention as before, the results of the experiment expected for each register are summarized in Fig. 4B under the column corresponding to the edited NOESY spectrum (E). Because of the arrangement of the chains, the K(CαH1)-D(NH) resonance would be expected instead of K(CϵH)-D(NH) for the D·O·K register, and the GO(CαH1)-D(NH) peak should be absent for O·K·D according to the model (supplemental Fig. S5). This comparison yields, in agreement with our hypothesis based on the number of peaks seen in the spectra, only one register, K·D·O.
Solution Structure
With knowledge about the register, the NOEs observed can be unambiguously assigned to proton pairs (or groups in the case of overlapping methylene resonances) along the chemical sequence of the peptides and, together with the constraints obtained from the HNHA and HNHB experiments, used to calculate an a ensemble of structures that are representative of the solution conformation of the triple helix. A summary of the constraints and structural statistics is provided in Table 3, and details about the protocol used for structural determination are given under “Experimental Procedures.”
TABLE 3.
NMR and refinement statistics
| NMR constraintsa | |
| Distance constraints | 771 |
| Intra-residue | 253 |
| Sequential (|i − j| = 1) | 180 |
| Interchain | 338 |
| Dihedral angle restraints | 120 |
| ϕ | 72 |
| χ1 | 48 |
| Structure statisticsb | |
| Violations (mean ± S.D.) | |
| Distance constraints | 0.07 ± 0.05 Å |
| Dihedral angle constraints | 1.86 ± 1.65° |
| Maximum dihedral angle violation | 4.97° |
| Maximum distance constraint violation | 0.244 Å |
| Deviations from idealized geometry | |
| Bond lengths | 0.0097 ± 0.0001 Å |
| Bond angles | 2.34 ± 0.04° |
| Average pairwise root mean square deviation, 15 structures | |
| Heavy | 0.68 Å |
| Backbone | 0.53 Å |
a Constraints observed were replicated by symmetry to all identical triplets between 2 and 9, see text for details.
b Statistics were calculated over the 72 restrained amino acids in the final structure.
The backbone of our refined NMR structure (Fig. 5A) behaves in a similar way to the homotrimeric system. Most of the points in the Ramachandran plot for the ensemble (supplemental Fig. S3) are grouped in a narrow region corresponding to the poly-proline type II helix, and only the unconstrained residues populate different regions of the (ϕ, ψ) space. The hydrogen-bonding network along the backbone of the peptides, which goes from the carbonyl of the amino acid in position X to the amide proton of glycine in a neighboring chain, is also conserved, although no explicit hydrogen bonding-type restraints were used during the refinement procedure. The helical pitch can very hard to determine by NMR because of its long range character, but the coupling constants measured indicate that our helix is probably closer to a 7/2 helix than to a 10/3 helix, like the O·O·O homotrimer.
FIGURE 5.
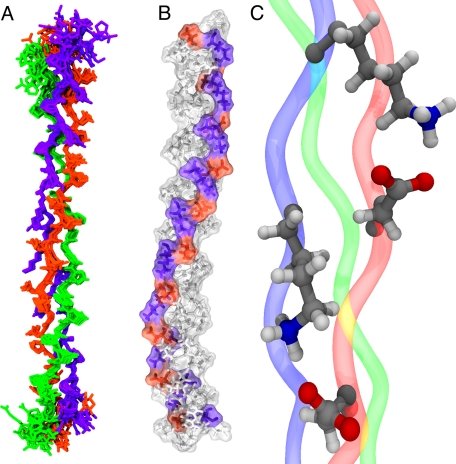
NMR structure of K·D·O. A, superposition of the 15 lowest energy conformers. The K peptide is blue; D is red, and O is green. B, surface and CPK representation of the lowest energy conformer highlighting the position of charged amino acids, lysine in red, and aspartic acid in blue, along the triple helix. C, expanded view of two of the salt bridges observed in B with distinct conformations.
The most interesting feature of the structure is the side chain conformation of the charged aspartic acid and lysine residues, which form a network of ionic hydrogen bonds spiraling along the helical axis (Fig. 5B), following the helicity of a single peptide strand within the triple helix with an axial repeat of ∼60 Å (7). These ionic hydrogen bonds are formed exclusively between lysine in triplet n of one peptide chain and aspartate in triplet n + 1 of the adjacent peptide. The salt bridges formed are highly dynamic, which can be seen in the structure of the lysine side chain resonances. The β- and γ-protons show distinct chemical shifts for each of the diastereotopic hydrogen atoms, in contrast to the δ- and ϵ-methylene groups, which present a single chemical shift for both protons of the methylene group. This indicates that the χ1 and χ2 dihedral angles have well defined values, but the χ3 and χ4 dihedrals sample a wider variety of conformations. Because of the length of the lysine side chain, the different conformers are still able to interact effectively with aspartic acid, which is primarily locked in a single conformation (Fig. 5C).
DISCUSSION
Because of the extended nature of the collagen triple helix and the repetitive primary sequences of the peptides, the set of NOEs that is available as structural constraints is small compared with those measured for globular proteins of similar size. In fact, only intra-residue, sequential, and inter-strand NOEs were observed. This is in agreement with the information available from crystal structures and NMR studies of homotrimeric collagen triple helices. However, this leaves out medium and long range contacts, which provide crucial information in traditional NMR structure calculations of globular proteins. To overcome this challenge, our structure determination protocol relies heavily on information about the conformation of the peptide backbone measured directly, such as those derived from coupling constants, or indirectly, using the NOE intensities of the amino acid side chains to determine their conformation. Furthermore, we take advantage of the periodic nature of the helix, as evidenced by the identical chemical shifts of the majority of the amino acids within the helix, which allows us to effectively multiply all of the constraints obtained by a factor of 8 and assign them to all equivalent triplets in the sequence.
The combination of sequential NOE cross-peaks, dihedral angles, and coupling constants allows us to obtain the expected secondary structure (a right-handed polyproline type II helix) for each peptide chain, whereas the inter-chain NOEs determine the register and pitch of the left-handed superhelix. The structure determination procedure is described in detail under the “Experimental Procedures” and converged after two iterations. The final ensemble presents little variation within the backbone of the eight constrained triplets giving an root mean square of 0.68 Å (Table 3). As expected the N- and C-terminal triplets show greater variation as they were not constrained during the refinement procedure (Fig. 5A).
The structural information obtained from our NMR experiments provides a new level of detail that has not been achieved for collagen heterotrimers before, building from the detailed NMR work available for homotrimers (11–13). We owe a great deal of our success to the inherent characteristics of the system such as its high thermal stability and the specificity of our assembly. Previous studies of triple helical heterotrimers have been hindered by stability issues, generating partially unfolded helices (18), or by the lack of specificity with respect to composition (19) and register (20).
The results obtained are significant because collagens have a high proportion of charged amino acids. In fact, Asp, Glu, Lys, and Arg constitute 15–20% of the sequence of the proteins in this family (37). For this reason, their role in the stabilization of the triple helix (38), the supramolecular assembly of collagen (39), and its interactions with other macromolecules (10) has been actively researched in the past few years. Experiments focused on determining the relationship between sequence and thermal stability of the triple helix found that model homotrimeric peptides containing the motif XKGDY′G or XKGEY′G are particularly stable (21). Changing the order of the charged amino acids or replacing Lys with Arg results in a decrease in the melting temperature of the assembly. However, none of the crystal structures of triple helices available have been able to give a molecular basis for this macroscopic observation. Recently, the structure of a homotrimer composed of a sequence from collagen type III that included the motif NRGERG showed that oppositely charged residues in that arrangement within the triple helix are able to form a network of intra- and inter-chain hydrogen bonds (40). The extent to which those interactions are present in solution, and not exclusively as a result of crystal packing, is unclear as model peptides with the RGE sequence do not exhibit any unusual stabilization (21).
The salt bridges observed in our solution structure are able to explain the surprisingly high thermal stability of homotrimeric triple helices with the KG(D/E) sequence (21), as well as our own heterotrimer (16). The inter-chain charge pairs that can be formed in those systems, homotrimers or heterotrimers, are equivalent and occur from the K in triplet n of one chain and interact with the D in triplet n + 1 of the next chain.
The association of peptides chains into triple helices is well understood for the (POG)n homotrimer. The presence of glycine as every third amino acid results in enthalpic contributions to the stability of the triple helical conformation, allowing for a tight packing in the core of the helix that provides a large surface area for van der Waals contacts and the donor in the backbone hydrogen-bonding network. The imino acids make an entropic contribution, making the single peptide chains prone to adopt the poly-proline conformation found within the helix. Further stabilization can be achieved by adding an electronegative atom with the appropriate stereochemistry at the γ-carbon of the imino acid in the Y position, such as hydroxyproline found in mammalian collagens. The stabilization mechanism of such a substitution is debated to arise from the stereoelectronic effects that direct the puckering of the ring and induce a backbone dihedral angle ideal for the triple helical conformation (41) or through water-mediated hydrogen bonds (5). Although the stabilization mechanism is not completely clear, an increase in the melting temperature of a homotrimer is observed when going from a (PPG)n sequence to a (POG)n sequence and a point mutation of P or O has been shown to result in a decrease in the thermal stability of the collagen mimetic homotrimer (42). In the K·D·O system, the inclusion of 20 substitutions from the ideal POG sequence permits the formation of inter-chain ionic hydrogen bonds leading to a heterotrimeric helix with a thermal stability similar to O·O·O homotrimer. Our interpretation of these observations is that the enthalpic contribution arising from the charge pair hydrogen bonding interactions is sufficient to offset the free energy cost incurred by the point mutations in the different peptide chains through a cooperative effect.
The formation of ionic hydrogen bonds, like the ones depicted in this work, can function as a local stabilization mechanism in the fold of triple helical molecules. It has been reported that local fluctuations in the triple helical stability may play a role in the interactions of collagens and other proteins, such as matrix metalloproteases (43). So far, the imino acid content of a particular sequence stretch has been the main factor used in such considerations, but it is known that collagenase labile stretches contain a very low content of charged residues (44, 45). The results presented here suggest that the lack of salt bridges in those regions may provide a complementary destabilization strategy. A statistical analysis of the sequences of human collagen type I–III, V, and type X showed that both the KGD and KGE motifs occur with a higher frequency than expected based on the number of occurrences of the Lys, Asp, and Glu residues in those proteins (21). Specifically the presence of such a motif in homotrimeric collagens, like type II and X, guarantees the ideal spatial arrangement of K/D or K/E pairs to form charge pairs. In the case of the heterotrimeric collagens like type I (AAB) or type V (AAB or ABC), the KG(D/E) motif does not guarantee the formation of inter-chain salt bridges, even for AAB types, because the peptides still need to adopt the proper register for the amino acid side chains to come in close contact. In fact, as demonstrated here, heterotrimeric systems require only the XKG and DYG motifs to be present in consecutive triplets of adjacent chains. The identification of such instances in heterotrimeric collagens is, however, not straightforward because explicit knowledge about the register of the peptide chains in the triple helix is required. Because that information is, to the best of our knowledge, not available, the number of charge pairs that can form in heterotrimeric helices cannot be determined at this point.
Ionic hydrogen bonds also occur in triple helical proteins synthesized by species that lack prolyl hydroxylase and thus require an alternative stabilization mechanism. For example, the Scl1 and Scl2 proteins found in the surface of the bacterium Streptococcus pyogenes contain domains of about 80 YGX repeats, where Y is predominantly Lys and X is Asp or Glu. Those domains fold into homotrimeric triple helices and are thought to meditate the adhesion of bacterium to human cells (22). Salt bridges, similar to those observed in the K·D·O system, serve to stabilize the triple helical conformation in this pathogen.
Besides their role in the stabilization of the fold, the electrostatic interactions present in our structure help determine the composition and register of the triple helix, directed through the information encoded exclusively in the triple helical domain. In natural collagens, it is unclear how the triple helix register is controlled. Globular terminal domains are implicated in determining the composition of several collagen types (46), but it is unclear what their role, if any, in controlling register may be. For fibril-forming collagens, such as type I, those domains are proteolytically cleaved once the helical domain is properly folded, and it has been shown that the collagenous domains of this type preferentially form homotrimers when allowed to fold without direction from the globular domains (46). Thus, it can be concluded that the terminal domains aid in determining the composition of the helices, but their role in determining the register is not well understood. Here we show that is possible to design peptides that not only selectively choose a composition but also a specific register when forming a collagen-like triple helix without the assistance of other protein constructs. A similar mechanism may be used by nature in the selection and registration process of collagens, especially in types that only have small noncollagenous domains, like the fibril-associated collagen type IX.
In a similar way that the O·O·O homotrimer has served as a molecular scaffold to induce the formation of triple helices and study the properties of homotrimeric collagens, the K·D·O system can be use to direct the formation of heterotrimeric triple helices with control over the composition and register of the resulting assembly. This opens up the possibility to design synthetic peptides to probe the conformation, dynamics, and biochemistry of a whole new domain of this protein family, by including sequences occurring in one, two, or three of the triple helical peptide chains (15, 47). Furthermore, the degree of control over the stagger of the peptides may aid in discerning the dependence of the biological activity of collagens on the polypeptide register.
Supplementary Material
Acknowledgments
We thank Dr. Sean Moran and Prof. Kevin MacKenzie for assistance with NMR.
This work was supported in part by Robert A. Welch Foundation Research Grant C1557, National Science Foundation Career Award DMR-0645474, and the Camille Dreyfus Teacher Scholar Awards Program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, Figs. S1–S5, Table S1, and additional references.
- TOCSY
- total correlated spectroscopy
- HSQC
- heteronuclear single quantum coherence
- NOESY
- nuclear Overhauser effect spectroscopy
- SA
- simulated annealing
- NOE
- nuclear Overhauser effect; for abbreviations of peptide chains please refer to Table 1.
REFERENCES
- 1.Kalluri R. (2003) Nat. Rev. Cancer 3, 422–433 [DOI] [PubMed] [Google Scholar]
- 2.Tuckwell D. S., Ayad S., Grant M. E., Takigawa M., Humphries M. J. (1994) J. Cell Sci. 107, 993–1005 [DOI] [PubMed] [Google Scholar]
- 3.Vandenberg P., Kern A., Ries A., Luckenbill-Edds L., Mann K., Kühn K. (1991) J. Cell Biol. 113, 1475–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orgel J. P., Irving T. C., Miller A., Wess T. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9001–9005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brodsky B., Thiagarajan G., Madhan B., Kar K. (2008) Biopolymers 89, 345–353 [DOI] [PubMed] [Google Scholar]
- 6.Okuyama K., Okuyama K., Arnott S., Takayanagi M., Kakudo M. (1981) J. Mol. Biol. 152, 427–443 [DOI] [PubMed] [Google Scholar]
- 7.Okuyama K., Wu G., Jiravanichanun N., Hongo C., Noguchi K. (2006) Biopolymers 84, 421–432 [DOI] [PubMed] [Google Scholar]
- 8.Baum J., Brodsky B. (1997) Fold. Des. 2, R53–R60 [DOI] [PubMed] [Google Scholar]
- 9.Melacini G., Bonvin A. M., Goodman M., Boelens R., Kaptein R. (2000) J. Mol. Biol. 300, 1041–1049 [DOI] [PubMed] [Google Scholar]
- 10.Emsley J., Knight C. G., Farndale R. W., Barnes M. J., Liddington R. C. (2000) Cell 101, 47–56 [DOI] [PubMed] [Google Scholar]
- 11.Li M. H., Fan P., Brodsky B., Baum J. (1993) Biochemistry 32, 7377–7387 [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Brodsky B., Baum J. (2007) J. Biol. Chem. 282, 22699–22706 [DOI] [PubMed] [Google Scholar]
- 13.Mohs A., Popiel M., Li Y., Baum J., Brodsky B. (2006) J. Biol. Chem. 281, 17197–17202 [DOI] [PubMed] [Google Scholar]
- 14.Gauba V., Hartgerink J. D. (2007) J. Am. Chem. Soc. 129, 2683–2690 [DOI] [PubMed] [Google Scholar]
- 15.Gauba V., Hartgerink J. D. (2008) J. Am. Chem. Soc. 130, 7509–7515 [DOI] [PubMed] [Google Scholar]
- 16.Gauba V., Hartgerink J. D. (2007) J. Am. Chem. Soc. 129, 15304–15341 [DOI] [PubMed] [Google Scholar]
- 17.Madhan B., Xiao J., Thiagarajan G., Baum J., Brodsky B. (2008) J. Am. Chem. Soc. 130, 13520–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiori S., Saccà B., Moroder L. (2002) J. Mol. Biol. 319, 1235–1242 [DOI] [PubMed] [Google Scholar]
- 19.Slatter D. A., Miles C. A., Bailey A. J. (2003) J. Mol. Biol. 329, 175–183 [DOI] [PubMed] [Google Scholar]
- 20.Slatter D. A., Foley L. A., Peachey A. R., Nietlispach D., Farndale R. W. (2006) J. Mol. Biol. 359, 289–298 [DOI] [PubMed] [Google Scholar]
- 21.Persikov A. V., Ramshaw J. A., Kirkpatrick A., Brodsky B. (2005) Biochemistry 44, 1414–1422 [DOI] [PubMed] [Google Scholar]
- 22.Mohs A., Silva T., Yoshida T., Amin R., Lukomski S., Inouye M., Brodsky B. (2007) J. Biol. Chem. 282, 29757–29765 [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 24.Goddard T. D., Kneller D. G. (2008) SPARKY 3, Version 3.115 University of California, San Francisco, CA [Google Scholar]
- 25.Vranken W. F., Boucher W., Stevens T. J., Fogh R. H., Pajon A., Llinas M., Ulrich E. L., Markley J. L., Ionides J., Laue E. D. (2005) Proteins 59, 687–696 [DOI] [PubMed] [Google Scholar]
- 26.Muhandiram D. R., Guang Y. X., Kay L. E. (1993) J. Biomol. NMR, 13, 463–470 [Google Scholar]
- 27.Vuister G. W., Bax A. (1993) J. Am. Chem. Soc. 115, 7772–7777 [Google Scholar]
- 28.Archer S. J., Ikura M., Torchia D. A., Bax A. (1991) J. Magn. Reson. 95, 636–641 [Google Scholar]
- 29.Powers R., Garrett D. S., March C. J., Frieden E. A., Gronenborn A. M., Clore G. M. (1993) Biochemistry 32, 6744–6762 [DOI] [PubMed] [Google Scholar]
- 30.Berisio R., Vitagliano L., Mazzarella L., Zagari A. (2002) Protein Sci. 11, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delano W. L. (2002) The PyMOL Molecular Graphics System, Delano Scientific, San Carlos, CA [Google Scholar]
- 32.Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park S., Radmer R. J., Klein T. E., Pande V. S. (2005) J. Comput. Chem. 26, 1612–1616 [DOI] [PubMed] [Google Scholar]
- 34.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 35.Nagarajan V., Kamitori S., Okuyama K. (1999) J. Biochem. 125, 310–318 [DOI] [PubMed] [Google Scholar]
- 36.Fraser R. D., MacRae T. P., Suzuki E. (1979) J. Mol. Biol. 129, 463–481 [DOI] [PubMed] [Google Scholar]
- 37.Emsley J., Knight C. G., Farndale R. W., Barnes M. J. (2004) J. Mol. Biol. 335, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 38.Kramer R. Z., Bella J., Brodsky B., Berman H. M. (2001) J. Mol. Biol. 311, 131–147 [DOI] [PubMed] [Google Scholar]
- 39.Kramer R. Z., Venugopal M. G., Bella J., Mayville P., Brodsky B., Berman H. M. (2000) J. Mol. Biol. 301, 1191–1205 [DOI] [PubMed] [Google Scholar]
- 40.Boudko S. P., Engel J., Okuyama K., Mizuno K., Bächinger H. P., Schumacher M. A. (2008) J. Biol. Chem. 283, 32580–32589 [DOI] [PubMed] [Google Scholar]
- 41.DeRider M. L., Wilkens S. J., Waddell M. J., Bretscher L. E., Weinhold F., Raines R. T., Markley J. L. (2002) J. Am. Chem. Soc. 124, 2497–2505 [DOI] [PubMed] [Google Scholar]
- 42.Persikov A. V., Ramshaw J. A., Brodsky B. (2005) J. Biol. Chem. 280, 19343–19349 [DOI] [PubMed] [Google Scholar]
- 43.Stultz C. M. (2002) J. Mol. Biol. 319, 997–1003 [DOI] [PubMed] [Google Scholar]
- 44.Fields G. B. (1991) J. Theor. Biol. 153, 585–602 [DOI] [PubMed] [Google Scholar]
- 45.Lauer-Fields J. L., Juska D., Fields G. B. (2002) Biopolymers 66, 19–32 [DOI] [PubMed] [Google Scholar]
- 46.Khoshnoodi J., Cartailler J. P., Alvares K., Veis A., Hudson B. G. (2006) J. Biol. Chem. 281, 38117–38121 [DOI] [PubMed] [Google Scholar]
- 47.Brodsky B., Baum J. (2008) Nature 453, 998–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.