Abstract
JAK2 (Janus kinase 2) is essential for cytokine receptor signaling, and several lines of evidence support a causal role of an activating JAK2 mutation in myeloproliferative disorders. JAK2 activity is autoinhibited by its pseudokinase domain in the basal state, and the inhibition is released by cytokine stimulation; how engagement of the cognate receptor triggers this release is unknown. From a functional screen for gain-of-function JAK2 mutations, we discovered 13 missense mutations, nine in the pseudokinase domain and four in the Src homology 2 (SH2)-pseudokinase domain linker. These mutations identified determinants for autoinhibition and inducible activation in JAK2. Two of the mutants, K539I and N622I, resulted in erythrocytosis in mice. Scanning mutagenesis of the SH2-pseudokinase domain linker indicated that its N-terminal part was essential for interaction of JAK2 with the Epo receptor, whereas certain mutations in the C-terminal region conferred constitutive activation. We further showed that substitutions for Glu543-Asp544 in this linker or Leu611, Arg683, or Phe694 in the hinge proximal region of the pseudokinase domain resulted in activated JAK2 mutants that could not be further stimulated by Epo. These results suggest that the SH2-pseudokinase domain linker acts as a switch that relays cytokine engagement to JAK2 activation by flexing the pseudokinase domain hinge.
The Janus family of tyrosine kinases (JAKs)2 are key regulators of cytokine receptor signaling in hematopoiesis and immune responses (1). Of the four mammalian JAK kinases, JAK2 transmits signals for a variety of cytokine receptors, including the erythropoietin receptor (EpoR) that is essential for red blood cell production (2). Upon Epo stimulation, JAK2 activates downstream signaling, such as STAT5, Ras/mitogen-activated protein kinase, and phosphatidylinositol 3-kinase/AKT pathways (2). Mice deficient in Epo, EpoR, or JAK2 die embryonically due to the absence of definitive erythropoiesis (3–5).
In addition to regulation by phosphatases and suppressors of cytokine signaling (6, 7), JAK2 kinase activity is critically controlled by an autoinhibitory mechanism. Like other JAK members, JAK2 contains an N-terminal segment followed by a pseudokinase domain and a C-terminal tyrosine kinase domain. The N-terminal segment, consisting of a FERM (protein 4.1, ezrin, moezin, radixin homologous) domain and an atypical SH2 domain (1), mediates association with the membrane-proximal region of the cytokine receptors (8). Binding of JAK2 through its N-terminal segment to the EpoR is essential for EpoR surface expression (9). The pseudokinase domain is predicted to adopt a kinase fold but lacks residues essential for catalysis (10). Deletion of the pseudokinase domain leads to a marked increase in JAK2 kinase activity and loss of response to cytokine stimulation (11–13). Therefore, this domain is essential for JAK2 autoinhibition and is essential for JAK2 activation upon cytokine stimulation. Consistent with this notion, a point mutation in the JAK2 pseudokinase domain was identified in the majority of myeloproliferative disorder patients, including 90% of polycythemia vera (PV) patients (14–18). This mutation, V617F, in the presence of a dimerized receptor scaffold, such as the EpoR, resulted in the constitutive activation of JAK2 and downstream signaling effectors (19, 20) and caused erythrocytosis in a murine bone marrow transplant model (14, 21–23). Recently, mutations immediately adjacent to the JAK2 pseudokinase domain in the SH2-pseudokinase domain linker were identified in PV patients and shown to cause constitutive activation of JAK2 and a PV-like phenotype in mice (24–26). The molecular mechanisms underlying the control of JAK2 activity (i.e. the swift augmentation of its activity upon receptor activation) are poorly understood. The residues involved in the autoinhibition in JAK2 are unknown.
In this work, we sought to characterize the regulatory mechanisms controlling JAK2 kinase activity. Using a functional screen for activating JAK2 mutations that signal constitutively, we discovered 13 mutations in the pseudokinase domain and in the SH2-pseudokinase domain linker. These mutations identified specific residues that are important for the inhibition of basal JAK2 kinase activity and for cytokine-induced JAK2 activation. In addition, we showed that the SH2-pseudokinase domain linker is essential for interaction with the EpoR, autoinhibitory regulation, and Epo-inducible JAK2 activation and may act as a switch in relaying cytokine receptor engagement to JAK2 activation by flexing the pseudokinase domain hinge.
EXPERIMENTAL PROCEDURES
cDNA Constructs and Reagents
For the JAK2 mutant library, murine JAK2 cDNA was subcloned into retroviral vector pEYK3.1 to generate pEYK3.1-JAK2. cDNAs of wild-type or HA-tagged murine JAK2, JAK2 mutants, or JAK2 domains were subcloned into pMX-IRES-CD4, pMX-IRES-GFP, or the pEBG vector that expresses GST upstream (9). HA-tagged or V5-tagged domains of JAK2 were subcloned into the pcDNA3.1 vector. All mutants were generated using the QuikChange site-directed mutagenesis kit (Stratagene) and verified by sequencing. Antibodies were from the following sources: HA (Covance); phosphotyrosine 4G10, JAK2, phospho-JAK2, and GST (Millipore); phospho-STAT5, phospho-ERK, AKT, and phospho-AKT (Cell Signaling Technology); V5 (Invitrogen); STAT5 and ERK (Santa Cruz Biotechnology, Inc., Santa Cruz, CA). HA affinity resin was from Roche Applied Science. Glutathione Sepharose 4B, horseradish peroxidase-coupled secondary antibodies, and the ECL chemiluminescence system were from Amersham Biosciences. Interleukin (IL)-3, IL-6, and murine stem cell factor were from PeproTech.
Construction of JAK2 Mutant Library and Screening
pEYK3.1-JAK2 was used as a template for library construction. Two methods were used to generate the JAK2 mutant library. In one method, we used XL-1 Red (Stratagene), a bacterial strain deficient in three major pathways for DNA repair. pEYK3.1-JAK2 was transformed into XL-1 Red, and colonies were collected from bacterial plates and propagated for 14 h at 30 °C in LB broth supplemented with antibiotics. Randomly mutated plasmids were isolated and used to generate retroviruses to infect Ba/F3 cells (9). In the other method, random mutagenesis of the SH2 and pseudokinase domains of JAK2 was performed by error-prone PCR using the Gene Morph II Kit (Stratagene). The manufacturer's conditions for generation of one or two mutations per molecule were used. Two PCRs were used, since shorter products have higher ligation efficiency into pEYK3.1 than do longer ones. The region encompassing the SH2 and pseudokinase domains (residues 393–840) was mutagenized. An internal XhoI site exists at residue 393 of JAK2. An NcoI site was engineered at residue 717, and a BamHI site was engineered at residue 840. One PCR product spanned residues 393–717, whereas the other spanned residues 717–840. PCR products were digested with XhoI/NcoI or NcoI/BamHI and subcloned into pEYK3.1-JAK2 to replace the corresponding region. Ligated products were transformed into Escherichia coli strain ElectroMAX Stbl4 (Invitrogen). Transformants (1.2 × 105) were selected on Zeocin agar plates. Bacterial colonies were collected, and plasmid DNA was isolated. Retroviruses were produced in HEK293T cells and used to infect Ba/F3 cells. The infected Ba/F3 cells were incubated for 16 h and then washed with RPMI media containing 1% fetal bovine serum. Cells (2.5 × 105/well) were plated in 6-well plates in RPMI media with 20% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.24% agar. After 10 days, isolated colonies were picked and amplified in liquid culture as described (27). Genomic DNA was isolated, and integrated provirus was recovered (28). Retroviruses were generated from rescued genomic DNAs and used in the second round of screening. Plasmids from the second round of screening were sequenced. Clones harboring multiple mutations were further dissected by examining each mutation individually.
Modeling of JAK2 Pseudokinase Domain
To build a structure model of the JAK2 pseudokinase domain, JAK2 sequence homologs were collected using PSI-BLAST (29) (default values, gi|2499668 as a query). Sequences that included all domains found in Jak2 (FERM, SH2, pseudokinase, and kinase) fell into four orthologous groups in vertebrates that included JAK2, JAK1, JAK3, and TYK2. These sequences were aligned using MAFFT (30) to define residue conservations and hydrophobicity profiles for use in refining alignments to structure templates. Appropriate structure templates were identified using 3D-Jury consensus of fold prediction metaserver results (31) for the mouse Jak2 query sequence (gi|2499668). Initial alignments produced by fold prediction servers to two top-scoring templates (Protein Data Bank codes 1opl and 1k9a) were adjusted manually based on 3D-Jury consensus alignments, secondary structure assignments, hydrophobicity profiles, and residue conservations. Based on the resulting alignments, models for the JAK2 sequence (gi|2499668, range 525–815) were built using the alignment mode (1opl_A, 1opl_B, and 1k9a_A) of the SWISS-MODEL workspace (32, 33) combined with the Swiss-PdbViewer (34). Alignment of the JAK2 sequence insert regions were further refined using the most favorable reported final energies of the models (e.g. 1oplA, −10967 kJ/mol). The final model depicting the JAK2 pseudokinase domain was based on the autoinhibited c-Abl tyrosine kinase structure, which also includes an N-terminal SH2 domain not modeled here.
Generation of Ba/F3 Cell Lines
Ba/F3 or Ba/F3-EpoR cells stably expressing JAK2 constructs at a predetermined level were isolated by fluorescence-activated cell sorting based on CD4 (9). To examine factor-independent growth, cells were washed extensively in RPMI medium with 1% bovine serum albumin and then grown in RPMI medium with 10% fetal bovine serum without IL-3.
Immunoprecipitation and Immunoblotting
HA-tagged or untagged JAK2 and JAK2 mutants were expressed in the pRK5 or pcDNA3.1 vectors. γ2A cells transiently transfected with wild-type or JAK2 mutants were lysed in 1% Nonidet P-40 lysis buffer with phosphatase and protease inhibitors. The lysates were immunoblotted with antibodies to phospho-JAK2 or JAK2. Bound antibodies were detected by the ECL chemiluminescence system after incubation with horseradish peroxidase-coupled secondary antibodies. For immunoprecipitations, HA-tagged JAK2 or JAK2 mutants were transiently expressed in HEK293T cells. The lysates were immunoprecipitated with HA affinity resin. The precipitates were eluted with SDS sample buffer, separated on SDS-PAGE, transferred to nitrocellulose membranes, and probed with anti-phospho-JAK2 or monoclonal antibodies to HA. To determine stimulation by Epo, wild-type or JAK2 mutants were transiently expressed in γ2A cells with the EpoR. After 48 h, cells were starved for 2 h and then stimulated with 40 units/ml Epo for 7 min. Lysates were immunoblotted with anti-phospho-JAK2 antibodies. To examine the ability of a fragment of JAK2 containing the SH2 and pseudokinase domains to inhibit JAK2 kinase domain in trans, the fragment was expressed as HA-tagged protein in pcDNA3.1. Mutations were generated in this background. Two concentrations of wild-type or mutant HA-JAK2(SH2+pseudo) were transiently co-expressed with V5-tagged JAK2 kinase domain, JAK2(K)-V5, in HEK293T cells. Lysates from these cells were immunoblotted with antibodies to phospho-JAK2, V5, or HA. To examine signaling, Ba/F3 cells stably expressing EpoR together with wild-type or mutant JAK2s were starved in RPMI containing 1% bovine serum albumin for 4 h (35). Cells expressing wild-type JAK2 were stimulated with Epo (100 units/ml) for 7 min as a positive control. The cells were lysed in 1% Nonidet P-40 lysis buffer with phosphatase and protease inhibitors. The lysates were immunoblotted with antibodies to phospho-STAT5, phospho-ERK, phospho-AKT, STAT5, ERK, or AKT.
In Vitro JAK2 Kinase Assay
HA-tagged JAK2 proteins were immunoprecipitated using HA affinity resin. Immunoprecipitants were washed three times with lysis buffer and twice with kinase buffer (10 mm HEPES, pH 7.4, 50 mm NaCl, 5 mm MnCl2, 5 mm MgCl2, 1 mm dithiothreitol) and resuspended in kinase buffer supplemented with 100 μm ATP, 2 μCi of [γ-32P]ATP (PerkinElmer Life Sciences), and phosphatase and protease inhibitors. A peptide derived from STAT5 (AKAADGY694VKPQIKQVV) containing the JAK2 phosphorylation site was used as a substrate at 1 mm. Kinase reactions were stopped at 30 min, a time within the linear reaction range, by the addition of SDS sample buffer. Samples were then boiled at 98 °C for 5 min, separated on SDS-PAGE, and analyzed by Typhoon Trio scanning (Amersham Biosciences).
JAK2-dependent EpoR Surface Expression
HA-tagged EpoR surface expression was measured in the presence of wild-type or mutant full-length JAK2 or GST-JAK2(N) constructs in half a million γ2A cells, as described previously (36).
Retroviral Transduction of Murine Bone Marrow Cells
Murine JAK2 or JAK2 mutants were expressed in MSCV-IRES-GFP. Retroviral supernatants were generated from HEK293T cells as described (9). To transduce murine bone marrow cells, 5-fluorouracil (150 mg/kg) was injected intravenously into 6-week-old BALB/c mice (Charles River Laboratories). After 5 days, marrow cells were harvested by flushing the femurs and tibias, and cells were cultured overnight in Iscove's modified Dulbecco's medium containing 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 6 ng/ml IL-3, 10 ng/ml IL-6, and 10 ng/ml murine stem cell factor in a 37 °C, 5% CO2 incubator. Cells were subsequently transduced by spin infection (1800 × g, 90 min, 33 °C) with retroviral particles in the presence of Polybrene (10 μg/ml) 24 h before and again on the day of transplantation on RetroNectin-coated plates.
Bone Marrow Transplantation and Hematopoietic Analysis
Transduced bone marrow cells were washed twice and resuspended in phosphate-buffered saline. Cells (0.5 × 106) were injected into the lateral tail vein of lethally irradiated (2 × 450 centigrays) BALB/c recipient mice (7–8 weeks old; Charles River Laboratories). Blood counts were determined by analysis of retro-orbital blood samples anti-coagulated with potassium EDTA (Antech Diagnostics). An unpaired Student's t test was used to compare peripheral blood counts among recipients of bone marrow cells expressing wild-type or mutant JAK2. All animal studies were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center at Dallas.
RESULTS
A Functional Mutagenesis Screen for Activating JAK2 Mutation
Substituting Val617 with several residues other than Phe did not constitutively activate JAK2 (supplemental Fig. S1). Therefore, residues in addition to Val617 are critical for maintenance of JAK2 in the inactive state. We chose to identify activating residues using a functional screen. Overexpression of the constitutively activated JAK2V617F at high levels in the IL-3-dependent hematopoietic cell line Ba/F3 leads to factor-independent growth (19). We therefore expressed a library of JAK2 mutants in Ba/F3 cells and isolated those that grew in the absence of IL-3. After two rounds of selection, we identified 14 distinct amino acid substitutions at 12 residues in JAK2 that conferred factor-independent growth of Ba/F3 cells. IL-3-independent growth was verified in Ba/F3 cells stably expressing these mutants at similar expression levels, and cells expressing vector alone or wild-type JAK2 died within 48 h (supplemental Fig. S2). These mutations are located either in the pseudokinase domain like V617F or in the linker region between the SH2 and pseudokinase domains. Specifically, R588M, S591L, K607E, L611S, V617F, V617I, N622I, R683S, F694L, and F694S reside in the pseudokinase domain, whereas N531I, M535I, H538L, and K539I are located in the SH2-pseudokinase linker (Fig. 1). All of these residues are conserved between murine and human JAK2 with one exception; position 531 is Asn in mouse and His in human JAK2. None of the mutations corresponds to any known JAK2 single-nucleotide polymorphisms.
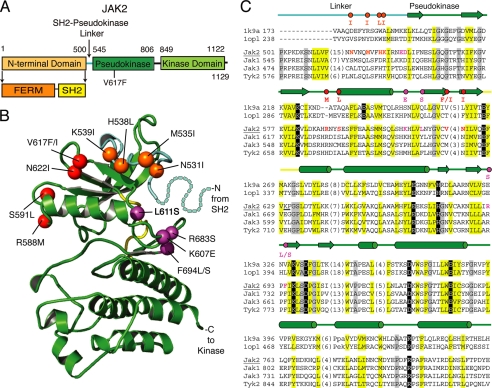
FIGURE 1.
JAK2 activating mutations are located in the pseudokinase domain and in the SH2-pseudokinase linker. A, domain organization of JAK2. B, ribbon diagram representing the JAK2 pseudokinase structure model rendered using PyMOL. The pseudokinase domain is in green, the linker region is in cyan, and the kinase hinge is in yellow. Positions of activating mutations are indicated with spheres; linker region mutations are in orange, V617F surface mutations are in red, and hinge proximal mutations are in magenta. Residues corresponding to a portion of the linker not modeled are depicted with dots. C, vertebrate JAK sequence representatives (JAK2, JAK1, JAK3, and TYK2) aligned with two kinase structure sequences (Protein Data Bank codes 1k9a and 1opl). Residue numbers are indicated to the left and right of the sequence. Common secondary structure elements are noted above the alignment and are colored according to functional domains: cyan, linker; green, pseudokinase domain; yellow, hinge. Residues within the alignment are highlighted according to conserved properties: yellow, generally hydrophobic positions; gray, generally small positions; black, polar conservations that probably contribute to function. Residues in positions of activating mutations are colored as above, with the activating mutations indicated above the alignment. Hinge region residues are underlined. The JAK2 sequence used to build the model structure is underlined.
To date, a three-dimensional structure is available for the JAK2 kinase domain (37) but not the entire protein. We therefore performed homology-based molecular modeling of the JAK2 pseudokinase domain to help us visualize the location of the activating mutations. Our model (Fig. 1B) differs only slightly from a previous model (38). The 12 critical residues identified in our screen map to the SH2-pseudokinase linker (N531I, M535I, H538L, and K539I), the same pseudokinase domain surface as V617F (R588M, S591L, V617F, V617I, and N622I), or the pseudokinase domain hinge-proximal region (K607E, L611S, R683S, F694L, and F694S).
JAK2 Mutants Have Elevated Kinase Activity and Exhibit Constitutive Downstream Signaling
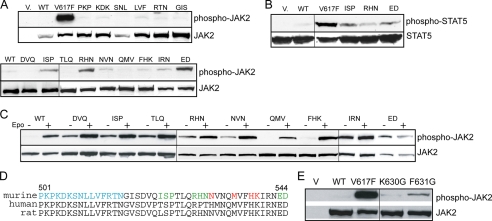
To characterize these JAK2 mutants, we first examined their autophosphorylation activity. HA-tagged wild-type or mutant JAK2 proteins were transiently expressed in HEK293T cells. After immunoprecipitation with HA affinity resin, the precipitants were run on SDS-PAGE and analyzed using antibodies that recognize JAK2 phosphorylated on Tyr1007 and Tyr1008. A kinase-deficient mutant JAK2 in which Lys882 is replaced by Glu, JAK2(KD), was used as a negative control. As shown in Fig. 2A, JAK2 mutants except for R588M and S591L showed a higher level of autophosphorylation than wild-type JAK2. These results were confirmed with untagged versions of these mutants in γ2A cells that do not express endogenous JAK2 (data not shown). To examine the ability of these mutants to phosphorylate exogenous substrates, HA-tagged JAK2 proteins were incubated with a peptide substrate derived from STAT5 in in vitro kinase assays. Compared with wild-type JAK2, JAK2 mutants phosphorylated the STAT5 peptide to a much higher level (Fig. 2B). Although R588M and S591L had little detectable autophosphorylation, they conferred IL-3-independent Ba/F3 growth (supplemental Fig. S2) and had a reproducible albeit modest increase in kinase activity compared with wild-type JAK2 in in vitro kinase assays (Fig. 2B). They may hyperactivate JAK2 to a lesser extent than the other mutations.
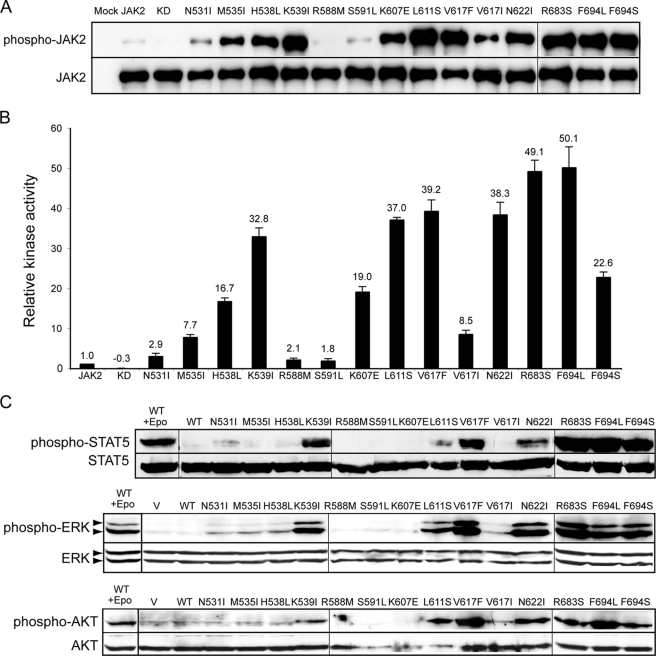
FIGURE 2.
JAK2 mutants have enhanced tyrosine kinase activity. A, HA-tagged wild-type or mutant JAK2 proteins were expressed in HEK293T cells and isolated by HA affinity resin. The precipitants were run on SDS-PAGE and probed with antibodies that recognize JAK2 phosphorylated on Tyr1007 and Tyr1008 (anti-phospho-JAK2) or anti-HA antibodies. B, HA-tagged proteins were incubated with STAT5 peptide, and in vitro kinase activities were normalized to wild-type JAK2. The number above each bar represents the average relative activity from four experiments. C, signaling of activating JAK2 mutants was evaluated in Ba/F3-EpoR cells stably expressing HA-tagged wild-type or JAK2 mutant proteins after starvation for 4 h. Ba/F3-EpoR cells expressing wild-type JAK2 were stimulated with Epo as a positive control. Activated STAT5, ERK, or AKT was detected using phospho-specific antibodies in cell lysates. WT, wild type; KD, kinase-deficient. Vertical lines have been inserted to indicate repositioned gel lanes.
We next assessed whether these JAK2 mutants could constitutively activate downstream signaling in hematopoietic cells. Co-expression of a cytokine receptor scaffold, such as the EpoR, is necessary for JAK2V617F to recruit downstream signaling molecules (19, 20). We thus examined signaling in Ba/F3 cells that stably co-expressed the EpoR and JAK2 mutants. The STAT5, Ras/mitogen-activated protein kinase, and phosphatidylinositol 3-kinase/AKT pathways were examined. In contrast to cells expressing wild-type JAK2, cells expressing the majority of JAK2 mutants showed constitutive activation of STAT5, ERK, and AKT after IL-3 deprivation for 4 h (Fig. 2C). Therefore, JAK2 mutants we identified were constitutively active and could constitutively activate downstream signaling pathways.
For the majority of JAK2 mutants, autophosphorylation correlated with in vitro kinase activity and downstream signaling; however, there was some discordance between the results of different methods. L611S was heavily autophosphorylated but caused poor STAT5 phosphorylation in Ba/F3-EpoR cells. K607E and F694S had similar kinase activity values but different phosphorylation levels of downstream signaling proteins in Ba/F3-EpoR cells. One reason for these discrepancies may be that for autophosphorylation and in vitro kinase assays, wild-type or mutant JAK2 was expressed and examined in the absence of cytokine receptors, whereas downstream signaling of these JAK2 proteins was determined in Ba/F3-EpoR cells where the EpoR was overexpressed. Signaling measured in Ba/F3-EpoR cells most likely originated from a EpoR-JAK2 complex. L611S and K607E may affect the conformation of JAK2 such that they are less efficient in phosphorylating substrates (e.g. EpoR cytoplasmic tyrosines and/or STAT5) in the context of an EpoR-JAK2 complex, resulting in lower phosphorylation of downstream signaling proteins. In addition, differences in assay sensitivity may play a role in these discrepancies.
JAK2 Fragments Containing Gain-of-function Mutations Did Not Efficiently Inhibit JAK2 Kinase Activity
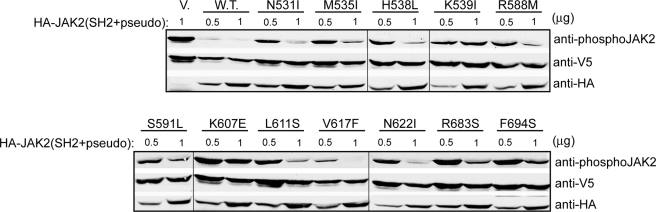
The pseudokinase domain inhibits activity of the JAK2 kinase domain in trans (12). To examine the mechanism underlying our activating mutations, we first tested an HA-tagged JAK2 fragment encompassing the SH2 domain, the SH2-pseudokinase linker, and the pseudokinase domain for its ability to inhibit JAK2 kinase domain in trans. Since some of the activating mutations were found in the SH2-pseudokinase linker, we included the SH2 domain in hopes of preserving the conformation of the linker region. Consistent with previous findings, co-expression of HA-JAK2(SH2+pseudo) with V5-tagged JAK2 kinase domain drastically inhibited JAK2 kinase activity in HEK293T cells. In contrast, HA-JAK2(SH2+pseudo) constructs harboring activating mutations expressed at a similar level (0.5 μg of DNA/transfection) were defective in their inhibitory ability (Fig. 3). At a higher expression level (1 μg of DNA/transfection), levels of inhibition differed among the mutant constructs. K607E seemed to have the most effect on releasing the inhibition posed by HA-JAK2(SH2+pseudo) toward the kinase domain, consistent with its high autophosphorylation and in vitro kinase activities. However, as discussed above, this mutation may be less efficient in phosphorylating substrates in the context of a EpoR-JAK2 complex, resulting in low downstream signaling in Ba/F3-EpoR cells. The lack of effective inhibition of JAK2 kinase activity by the activating mutations suggests that there was a looser interaction between HA-JAK2(SH2+pseudo) mutants and the kinase domain than between the two wild-type domains. Alternatively, these mutations may lock the pseudokinase domain in a lower inhibitory conformation.
FIGURE 3.
Activating mutations decreased the ability of a JAK2 fragment encompassing the SH2 and pseudokinase domains to inhibit JAK2 kinase domain. HA-tagged wild-type or mutant fragments were expressed at two amounts (0.5 and 1 μg of DNA) with V5-tagged JAK2 kinase domain in HEK293T cells. JAK2 kinase activity was determined using anti-phospho-JAK2 antibodies. V, vector.
Hinge Region JAK2 Mutants Were Not Further Stimulated by Epo
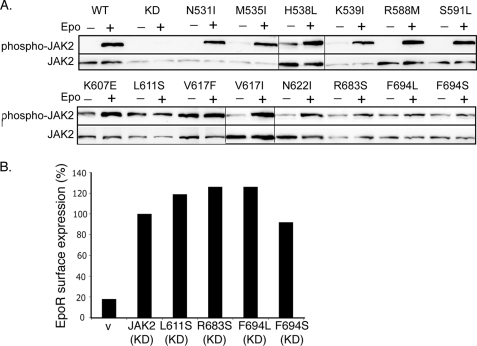
The pseudokinase domain is essential for cytokine-induced JAK2 activation (11, 13), indicating its importance in communication between JAK2 and the cognate receptor. We examined whether our JAK2 mutants were further stimulated by Epo. Using JAK2-deficient γ2A cells to avoid effects from endogenous JAK2, most of the JAK2 mutants, although partially active, showed increased autophosphorylation upon Epo stimulation (Fig. 4A). Interestingly, L611S, R683S, F694L, and F694S mutants showed little or no response to Epo. To rule out the possibility that their lack of Epo inducibility was due to lack of interaction with the EpoR, we examined their ability to promote EpoR surface expression. Consistent with previous reports (14, 39), we found that total EpoR protein levels were greatly diminished in cells expressing activating mutations (data not shown), possibly because EpoR degradation is linked to JAK2 activation (40). Since kinase activity is dispensable for JAK2 interaction with the EpoR (9), we tested these activating mutations in the context of a kinase-dead allele, JAK2K882E. As shown in Fig. 4B and supplemental Fig. S3, L611S, R683S, F694L, and F694S did not interfere with the ability of JAK2K882E to promote EpoR surface expression, indicating that these mutants did not disrupt the interaction with the EpoR. Therefore, these residues are not essential for EpoR interaction but might instead be part of the machinery that activates JAK2 in response to Epo engagement of the EpoR. In our model (Fig. 1B), all four residues are near the pseudokinase domain hinge, a region that connects the two pseudokinase domain lobes and mediates alternative conformations of protein kinases.
FIGURE 4.
L611S, R683S, F694L, and F694S were not Epo-inducible. A, γ2A cells transfected with EpoR and wild-type or mutant JAK2s were starved for 2 h and stimulated with Epo. Lysates were immunoblotted with antibodies to phospho-JAK2 or JAK2. Vertical lines have been inserted to indicate repositioned gel lanes. B, kinase-dead JAK2 with mutation L611S, R683S, F694L, or F694S interacted with the EpoR to promote its surface expression. V, vector; KD, kinase-deficient.
The JAK2 SH2-Pseudokinase Linker Is Essential for Interaction with the EpoR, for Autoinhibition, and for Epo-dependent JAK2 Activation
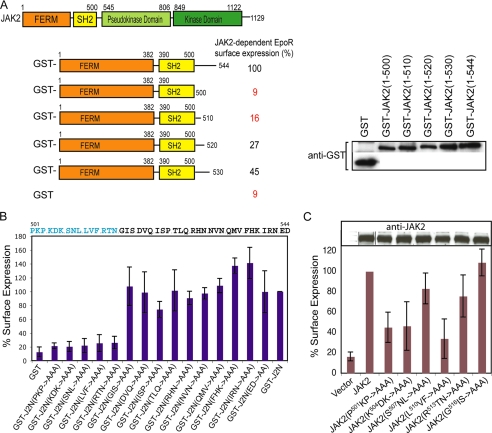
Four activating mutations lie in the SH2-pseudokinase linker, previously demonstrated to be part of the JAK2 N-terminal segment necessary and sufficient for interaction with cytokine receptors to promote their surface expression (9, 36). We hypothesized that this linker may act to coordinate ligand stimulation of the cytokine receptor to JAK2 activation. We first tested if the SH2-pseudokinase linker is required for JAK2 to interact with wild-type EpoR to promote EpoR surface expression. Serial deletions of the SH2-pseudokinase linker were made in the JAK2 N-terminal segment (JAK2(N)) as GST fusions. As shown in Fig. 5A and supplemental Fig. S4, complete deletion of this linker abolished JAK2-dependent EpoR surface expression, and restoring increasing length of this linker resulted in increasing EpoR surface expression. We then performed alanine-scanning mutagenesis between residues 501 and 544 in GST-JAK2(N) and examined the ability of these mutants to promote EpoR surface expression. As shown in Fig. 5B, GST- JAK2(N, P501A/K502A/P503A), GST-JAK(N, K504A/D505A/K506A), GST-JAK2(N, S507A/N508A/L509A), GST-JAK2(N, L510A/V511A/F512A), and GST-JAK2(N, R513A/T514A/N515A) interacted poorly, if at all, with the EpoR, as determined by lack of EpoR surface expression. The remaining mutants promoted EpoR expression on the plasma membrane at levels similar to GST-JAK2(N). We further confirmed that residues P501KP, K504DK, and L510VF were essential in the context of full-length JAK2 for EpoR interaction and surface expression, whereas S507NL and R513TN had lesser effects (Fig. 5C). Therefore, SH2 proximal linker residues P501KPKDKSNLLVF were essential for the interaction between JAK2 and the EpoR.
FIGURE 5.
Residues in the SH2-pseudokinase linker are essential for interaction with EpoR. A, surface expression of HA-EpoR in cells co-expressing GST-JAK2(N) or serial truncations in the SH2-pseudokinase linker was normalized against that obtained with GST-JAK2(N). Immunoblots with anti-GST antibodies showed that the expression levels of these proteins were comparable. B, surface expression of HA-EpoR co-expressed with GST-JAK2(N) or alanine scanning mutations in the SH2-pseudokinase linker was analyzed as described above. C, full-length JAK2s harboring mutations in the SH2-pseudokinase linker were defective in EpoR interaction. Surface expression of HA-EpoR co-expressed with full-length wild-type or mutant JAK2 proteins was analyzed. Immunoblots of wild-type and mutant JAK2 proteins showed comparable expression levels. Vertical lines have been inserted to indicate repositioned gel lanes.
We also examined the kinase activity of the alanine-scanning mutants. As shown in Fig. 6A, JAK2(I522A/S523A/P524A), JAK2(R528A/H529A/N530A), and JAK2(E543A/D544A) exhibited significantly higher JAK2 kinase activity than wild-type JAK2, indicating that these residues participate in autoinhibition. Consistently, they also phosphorylated the substrate STAT5 to a higher level than wild type JAK2 (Fig. 6B). JAK2(N531A/V532A/N533A) and JAK2(I540A/R541A/N542A) also exhibited low levels of constitutive activation (Fig. 6A). We then evaluated alanine mutants that interacted with the EpoR normally for their ability to be further induced by Epo. In the presence of the EpoR as a dimerizing scaffold, weakly activated mutants (D519A/V520A/Q521A, T525A/L526A/Q527A, N531A/V532A/N533A, and I540A/R541A/N542A) had higher basal activity (Fig. 6C). Interestingly, of the mutants tested, JAK2(E543A/D544A) was not further activated upon Epo stimulation (Fig. 6C). These results suggest that the SH2-pseudokinase linker plays a role in both autoinhibition and cytokine-stimulated JAK2 activation, consistent with previous results indicating a regulatory role of the SH2-pseudokinse linker on the pseudokinase domain (Fig. 3). As summarized in Fig. 6D, we identified residues in the N-terminal region of the SH2-pseudokinase domain linker that were essential for EpoR interaction and residues in the C-terminal region that contributed to autoinhibition and inducible activation. This linker appears to function as a switch to transmit conformational changes in the receptor-JAK2 complex upon cytokine stimulation, resulting in JAK2 kinase activation.
FIGURE 6.
Residues in the SH2-pseudokinase linker are important for JAK2 activity and Epo-inducible JAK2 activation. A, full-length JAK2 containing alanine-scanning mutations in the SH2-pseudokinase linker were expressed in γ2A cells, and kinase activities were examined by Western analysis using anti-phospho-JAK2 antibodies. Vertical lines have been inserted to indicate repositioned gel lanes. B, the same mutants were co-expressed with STAT5 in γ2A cells. Phosphorylated STAT5 proteins were detected using anti-phospho-STAT5 antibodies. C, mutation of E543D abolished Epo-inducible JAK2 activation. γ2A cells expressing the EpoR and wild-type (WT) JAK2 or JAK2 mutants were starved for 2 h and stimulated with Epo. D, alignment of the SH2-pseudokinase linker regions of mouse, human, and rat sequences. Residues in the SH2-pseudokinase linker essential for EpoR interaction are colored blue. Residues corresponding to activating mutations that conferred factor-independent growth are colored red. Residues where mutation to alanines enhanced JAK2 activity are colored green. E, full-length JAK2 with mutations at K630G or F631G were expressed in γ2A cells, and phosphorylated JAK2 proteins were detected using anti-phospho-JAK2 antibodies.
Mechanism of JAK2 Activation
Similar to JAK2(E543A/D544A) in the SH2-pseudokinase linker, JAK2 mutants near the pseudokinase hinge were not further activated by Epo (Fig. 4A). In addition, the SH2-pseudokinase linker and the pseudokinase domain hinge are close in proximity in our model (Fig. 1B). In protein kinases, the hinge region accounts for a variety of conformational modes. Although the JAK2 pseudokinase domain lacks residues essential for phosphorylation activity, it retains the global structure of a kinase and preserves conservations within the hinge region. We reasoned that upon cytokine binding, conformational changes in the receptor-JAK2 complex may propagate through the SH2-pseudokinase linker to the pseudokinase domain hinge. This propagation may flex the hinge to affect the relative position of the two lobes of the pseudokinase domain, in particular the small lobe that includes the V617F surface, to activate kinase activity. To test this hypothesis, we mutated two other hinge residues in the pseudokinase domain, namely Lys630 and Phe631. Substitutions with a Gly were chosen to increase potential flexibility between the two lobes. Both JAK2(K630G) and JAK2(F631G) were constitutively active (Fig. 6E). Therefore, the relative location of the two lobes may be important for regulation of JAK2 kinase activity, and activation upon cytokine stimulation may involve the pseudokinase domain hinge.
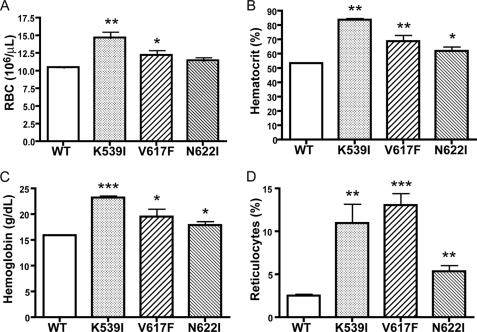
Expression of Mutant JAK2 Proteins in Bone Marrow Cells Resulted in Erythrocytosis in Transplanted Mice
To determine the physiological consequences of JAK2 mutations in vivo, lethally irradiated BALB/c recipient mice were transplanted with bone marrow cells infected with retroviruses expressing wild-type JAK2 or mutant K539I, V617F, or N622I. We chose K539I to represent mutants in the SH2-pseudokinase domain linker, because it is similar to the mutation (K539L) already identified in patients with PV (24). N622I was chosen to represent mutants in the pseudokinase domain, because it had the highest mitogenic activities in Ba/F3 cells (supplemental Fig. S2). Eight weeks after bone marrow transplantation, blood was collected from these mice and analyzed. As shown in Fig. 7, animals that received V617F-transduced bone marrow cells exhibited erythrocytosis, consistent with previous observations (14, 22, 23, 26). Recipients of K539I-transduced cells also had elevated hematocrit and higher red blood cell and reticulocyte count than mice transplanted with wild-type JAK2. Mice transplanted with N622I also showed higher hematocrit, hemoglobin, and reticulocyte count than mice transplanted with wild-type JAK2. Red blood cell counts were also elevated but not to a statistically significant level. This experiment was repeated three times. Therefore, similar to the V617F mutation identified from myeloproliferative disorder patients, our gain-of-function JAK2 mutations can result in hematological abnormalities.
FIGURE 7.
Expression of JAK2 mutants in hematopoietic cells resulted in aberrant hematopoiesis in vivo. BALB/c mice were lethally irradiated and transplanted with syngeneic bone marrow cells transduced with retroviruses expressing wild-type (WT) JAK2 (n = 7), K539I (n = 7), V617F (n = 7), or N622I (n = 8). Two months after transplantation, red blood cell counts (A), hematocrit (B), blood hemoglobin levels (C), and reticulocyte counts (D) were measured. *, p < 0.05 (unpaired t test) between mutant and wild type mice; **, p < 0.005 between mutant and wild type mice; ***, p < 0.0005 between mutant and wild-type mice.
DISCUSSION
The V617F mutation in the JAK2 pseudokinase domain is the major molecular lesion in patients with BCR/ABL-negative myeloproliferative disorder, underlining the importance of regulatory mechanisms controlling this kinase. In this work, we discovered 13 JAK2 mutations that also resulted in its constitutive activation and activation of downstream signaling pathways. Two of the mutants, K539I and N622I, resulted in hyperproliferation of red blood cells in vivo. These mutants identify specific residues important for both the inhibition of basal JAK2 kinase activity and for cytokine-induced JAK2 activation and enable us to propose a model for JAK2 activation upon cytokine stimulation. In this model, the SH2-pseudokinase domain linker plays a critical role in communicating between the cognate receptor and JAK2 kinase activity by changing the pseudokinase domain conformation through its hinge.
Our activating mutations lie exclusively in the pseudokinase domain and in the SH2-pseudokinase domain linker. Based on their location in a molecular model (Fig. 1B), mutations were categorized into three groups. Those in the first group (N531I, M535I, H538L, and K539I) lie in the SH2-pseudokinase linker. Mutations in the second group (R588M, S591L, V617F, V617I, and N622I) surround Val617 and are on the two previously postulated interacting surfaces between the pseudokinase and kinase domains: Arg588 and Ser591 on interface 1 and Val617 and Asn622 on interface 2 (41). The third group (K607E, L611S, R683S, F694L, and F694S) is near the “hinge” region of the pseudokinase domain that controls the relative orientation between the two lobes, on the opposite surface to Val617. These results are in line with previous reports showing that mutations other than V617F in the pseudokinase domain enhanced JAK2 activity (42–48).
A JAK2 fragment encompassing the SH2 domain, the SH2-pseudokinase linker, and the pseudokinase domain inhibits activity of the kinase domain in trans. All activating mutations in the context of this JAK2 fragment resulted in decreased ability to inhibit JAK2 kinase activity relative to the wild-type fragment. Since the pseudokinase domain alone is sufficient to inhibit the kinase domain (12),3 mutations in either the pseudokinase domain or in the SH2-pseudokinase linker may disrupt critical interactions between the pseudokinase and kinase domains. It was proposed that Val617 directly interacts with the kinase activation loop to keep it in an inactive conformation (41). The activating mutations may disrupt this critical interaction and allow the activation loop to break free for trans-phosphorylation/activation. Interestingly, all mutations in the V617F group and in the nearby linker region group involve substitutions with bulky hydrophobic amino acids. Replacing Met535, His538, or Lys539 with Ala did not activate JAK2 (Fig. 6, A and C), and only Phe, Ile, Met, and Trp substitutions at Val617 activate JAK2 (49). Therefore, a bulky hydrophobic surface around this region may be important for the disruption of the critical interaction between Val617 and the activation loop.
Our findings assign three important functions to the SH2-pseudokinase domain linker: interaction with the EpoR, autoinhibitory regulation of JAK2 activity, and cytokine-inducible JAK2 activation. We found that the N-terminal SH2-proximal region of this linker, residues P501KPKDKSNLLVF, was essential for interaction of JAK2 with the EpoR. These residues may be involved in direct binding to the membrane-proximal Box 1 region of cognate receptors or may hold the JAK N-terminal segment in the correct conformation to interact with cognate receptors (9, 36, 50). Four gain-of-function mutations in this linker transformed Ba/F3 cells, and three alanine-scanning mutants resulted in increased JAK2 activity. These residues, like activating mutations identified from PV patients (24–26), are localized to the C-terminal region of the linker. The close proximity of footprints for receptor interaction and for autoinhibition prompts us to propose that the SH2-pseudokinase linker functions as a switch to communicate conformational changes in the cognate cytokine receptors upon stimulation to JAK2 activation. This proposal is consistent with the fact that the footprint of the JAK2 interaction site and a conserved hydrophobic motif essential for JAK2 activation upon stimulation also are adjacent on the EpoR (35). Moreover, a partially active mutant in the C-terminal region of this linker, JAK2(E543A/D544A), was not further stimulated by Epo.
The majority of the activating mutants, like V617F, can be further activated by Epo. This is consistent with the notion that cytokines trigger at least two different effects in JAK2 activation: release of pseudokinase domain autoinhibition and alignment of associated JAK2s into an optimal conformation for maximal activation (19, 46). However, mutating hinge-proximal region residues Leu611, Arg683, and Phe694 in the pseudokinase domain, similar to JAK2(E543A/D544A) in the SH2-pseudokinase linker, resulted in JAK2 mutants that are partially activated but could not be further stimulated by Epo. In line with our results, the pseudokinase domains of JAK2 and JAK3, where Leu611, Arg683, and Phe694 are conserved, can be swapped to generate functional proteins that retain cytokine-inducible signaling (11). Although we cannot rule out the possibility that these mutations caused artificial effects in JAK2 that resulted in their inability to be further stimulated, we favor a model in which these residues are involved in a machinery that senses receptor engagement. We hypothesize that upon cytokine binding, a conformational change in the SH2-pseudokinase linker is transmitted through the pseudokinase domain hinge to release its inhibitory interaction with the kinase domain, causing JAK2 activation. This mechanism probably involves release of the clamp imposed by the SH2-pseudokinase linker on the pseudokinase domain, thus changing the relative conformation of the two lobes and releasing the activation loop of the kinase domain. This hypothesis is supported by the fact that multiple alanine scanning mutants in the SH2-pseudokinase linker activated JAK2 (Fig. 6) and that mutations in this linker, including those with internal deletions or duplications, were isolated from myeloproliferative disorder patients (51). A similar kind of regulation was beautifully demonstrated for ZAP70 kinase. In ZAP70, a tandem SH2 unit inhibits catalytic activity by interacting with the hinge region of the kinase domain, suppressing its flexibility. Phosphorylation of the linker between the SH2 domain and the kinase domain releases the inhibition and activates ZAP70 (52). To test if a similar mechanism is used by JAK2, we generated two mutations in the pseudokinase domain hinge, designed to increase its flexibility. Both JAK2(K630G) and JAK2(F631G) activated JAK2. Further verification of this model will require additional studies and a detailed structure of the full-length JAK2 protein.
To examine the physiological consequences of these mutations, we transplanted bone marrow cells transduced with retroviruses expressing wild-type JAK2 or JAK2 with mutation V617F, K539I, or N622I into lethally irradiated BALB/c recipient mice. All three mutations resulted in erythrocytosis, consistent with previous results (14, 22–24, 26). Thus, the activating mutants we identified elicit hematological effects in vivo. It will be interesting to examine other mutations in this model and to determine whether activating JAK2 mutants can support hematopoiesis in JAK2-null animals.
Like V617F and K539I, some mutations we identified have been implicated in hematological malignancies. For example, L611S was identified in a child with acute lymphoblastic leukemia (53). K607N (K607E was selected in our screen) was identified in patients with acute myelogenous leukemia (54). R683S was identified in Down syndrome patients with acute lymphoblastic leukemia (47, 48). Although it is not known whether the mutation in JAK2 contributed to these diseases, these mutants can be used as tools to generate both biochemical and animal models to test the link between JAK2 activation and myeloproliferative disorders and/or leukemia.
Supplementary Material
Acknowledgments
We thank Drs. Michael White, Rita Sulahian, Huan-you Wang, and Yu-min Shen for helpful discussions. We thank Dr. George Daley for pEYK3.1, Dr. James Ihle for pRK5-JAK2, and Dr. Romano Kroemer for the coordinates of the JAK2 model.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL089966 (to L. J. H.). This work was also supported by the American Cancer Society (to L. J. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
L. Zhao and L. J. Huang, unpublished results.
- JAK
- Janus kinase
- EpoR
- Epo receptor
- PV
- polycythemia vera
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- ERK
- extracellular signal-regulated kinase
- IL
- interleukin
- SH2
- Src homology 2.
REFERENCES
- 1.Yamaoka K., Saharinen P., Pesu M., Holt V. E., 3rd, Silvennoinen O., O'Shea J. J. (2004) Genome Biol. 5, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Constantinescu S. N., Ghaffari S., Lodish H. F. (1999) Trends Endocrinol. Metab. 10, 18–23 [DOI] [PubMed] [Google Scholar]
- 3.Wu H., Liu X., Jaenisch R., Lodish H. F. (1995) Cell 83, 59–67 [DOI] [PubMed] [Google Scholar]
- 4.Neubauer H., Cumano A., Müller M., Wu H., Huffstadt U., Pfeffer K. (1998) Cell 93, 397–409 [DOI] [PubMed] [Google Scholar]
- 5.Parganas E., Wang D., Stravopodis D., Topham D. J., Marine J. C., Teglund S., Vanin E. F., Bodner S., Colamonici O. R., van Deursen J. M., Grosveld G., Ihle J. N. (1998) Cell 93, 385–395 [DOI] [PubMed] [Google Scholar]
- 6.Wormald S., Hilton D. J. (2004) J. Biol. Chem. 279, 821–824 [DOI] [PubMed] [Google Scholar]
- 7.Irie-Sasaki J., Sasaki T., Matsumoto W., Opavsky A., Cheng M., Welstead G., Griffiths E., Krawczyk C., Richardson C. D., Aitken K., Iscove N., Koretzky G., Johnson P., Liu P., Rothstein D. M., Penninger J. M. (2001) Nature 409, 349–354 [DOI] [PubMed] [Google Scholar]
- 8.Haan C., Kreis S., Margue C., Behrmann I. (2006) Biochem. Pharmacol. 72, 1538–1546 [DOI] [PubMed] [Google Scholar]
- 9.Huang L. J., Constantinescu S. N., Lodish H. F. (2001) Mol. Cell 8, 1327–1338 [DOI] [PubMed] [Google Scholar]
- 10.Boudeau J., Miranda-Saavedra D., Barton G. J., Alessi D. R. (2006) Trends Cell Biol. 16, 443–452 [DOI] [PubMed] [Google Scholar]
- 11.Saharinen P., Silvennoinen O. (2002) J. Biol. Chem. 277, 47954–47963 [DOI] [PubMed] [Google Scholar]
- 12.Saharinen P., Takaluoma K., Silvennoinen O. (2000) Mol. Cell. Biol. 20, 3387–3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saharinen P., Vihinen M., Silvennoinen O. (2003) Mol. Biol. Cell 14, 1448–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James C., Ugo V., Le Couédic J. P., Staerk J., Delhommeau F., Lacout C., Garçon L., Raslova H., Berger R., Bennaceur-Griscelli A., Villeval J. L., Constantinescu S. N., Casadevall N., Vainchenker W. (2005) Nature 434, 1144–1148 [DOI] [PubMed] [Google Scholar]
- 15.Levine R. L., Wadleigh M., Cools J., Ebert B. L., Wernig G., Huntly B. J., Boggon T. J., Wlodarska I., Clark J. J., Moore S., Adelsperger J., Koo S., Lee J. C., Gabriel S., Mercher T., D'Andrea A., Fröhling S., Döhner K., Marynen P., Vandenberghe P., Mesa R. A., Tefferi A., Griffin J. D., Eck M. J., Sellers W. R., Meyerson M., Golub T. R., Lee S. J., Gilliland D. G. (2005) Cancer Cell 7, 387–397 [DOI] [PubMed] [Google Scholar]
- 16.Baxter E. J., Scott L. M., Campbell P. J., East C., Fourouclas N., Swanton S., Vassiliou G. S., Bench A. J., Boyd E. M., Curtin N., Scott M. A., Erber W. N., Green A. R. (2005) Lancet 365, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 17.Kralovics R., Passamonti F., Buser A. S., Teo S. S., Tiedt R., Passweg J. R., Tichelli A., Cazzola M., Skoda R. C. (2005) N. Engl. J. Med. 352, 1779–1790 [DOI] [PubMed] [Google Scholar]
- 18.Zhao R., Xing S., Li Z., Fu X., Li Q., Krantz S. B., Zhao Z. J. (2005) J. Biol. Chem. 280, 22788–22792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X., Huang L. J., Lodish H. F. (2008) J. Biol. Chem. 283, 5258–5266 [DOI] [PubMed] [Google Scholar]
- 20.Lu X., Levine R., Tong W., Wernig G., Pikman Y., Zarnegar S., Gilliland D. G., Lodish H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18962–18967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wernig G., Mercher T., Okabe R., Levine R. L., Lee B. H., Gilliland D. G. (2006) Blood 107, 4274–4281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacout C., Pisani D. F., Tulliez M., Gachelin F. M., Vainchenker W., Villeval J. L. (2006) Blood 108, 1652–1660 [DOI] [PubMed] [Google Scholar]
- 23.Zaleskas V. M., Krause D. S., Lazarides K., Patel N., Hu Y., Li S., Van Etten R. A. (2006) PLoS ONE 1, e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott L. M., Tong W., Levine R. L., Scott M. A., Beer P. A., Stratton M. R., Futreal P. A., Erber W. N., McMullin M. F., Harrison C. N., Warren A. J., Gilliland D. G., Lodish H. F., Green A. R. (2007) N. Engl. J. Med. 356, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pietra D., Li S., Brisci A., Passamonti F., Rumi E., Theocharides A., Ferrari M., Gisslinger H., Kralovics R., Cremonesi L., Skoda R., Cazzola M. (2008) Blood 111, 1686–1689 [DOI] [PubMed] [Google Scholar]
- 26.Butcher C. M., Hahn U., To L. B., Gecz J., Wilkins E. J., Scott H. S., Bardy P. G., D'Andrea R. J. (2008) Leukemia 22, 870–873 [DOI] [PubMed] [Google Scholar]
- 27.Azam M., Raz T., Nardi V., Opitz S. L., Daley G. Q. (2003) Biol. Proced. Online 5, 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koh E. Y., Chen T., Daley G. Q. (2002) Nucleic Acids Res. 30, e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K., Misawa K., Kuma K., Miyata T. (2002) Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginalski K., Elofsson A., Fischer D., Rychlewski L. (2003) Bioinformatics 19, 1015–1018 [DOI] [PubMed] [Google Scholar]
- 32.Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 33.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guex N., Peitsch M. C. (1997) Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 35.Constantinescu S. N., Huang L. J., Nam H., Lodish H. F. (2001) Mol. Cell 7, 377–385 [DOI] [PubMed] [Google Scholar]
- 36.Tong W., Sulahian R., Gross A. W., Hendon N., Lodish H. F., Huang L. J. (2006) J. Biol. Chem. 281, 38930–38940 [DOI] [PubMed] [Google Scholar]
- 37.Lucet I. S., Fantino E., Styles M., Bamert R., Patel O., Broughton S. E., Walter M., Burns C. J., Treutlein H., Wilks A. F., Rossjohn J. (2006) Blood 107, 176–183 [DOI] [PubMed] [Google Scholar]
- 38.Giordanetto F., Kroemer R. T. (2002) Protein Eng. 15, 727–737 [DOI] [PubMed] [Google Scholar]
- 39.Vainchenker W., Constantinescu S. N. (2005) Hematology Am. Soc. Hematol. Educ. Program, 195–200 [DOI] [PubMed] [Google Scholar]
- 40.Walrafen P., Verdier F., Kadri Z., Chrétien S., Lacombe C., Mayeux P. (2005) Blood 105, 600–608 [DOI] [PubMed] [Google Scholar]
- 41.Lindauer K., Loerting T., Liedl K. R., Kroemer R. T. (2001) Protein Eng. 14, 27–37 [DOI] [PubMed] [Google Scholar]
- 42.Feener E. P., Rosario F., Dunn S. L., Stancheva Z., Myers M. G., Jr. (2004) Mol. Cell. Biol. 24, 4968–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Argetsinger L. S., Kouadio J. L., Steen H., Stensballe A., Jensen O. N., Carter-Su C. (2004) Mol. Cell. Biol. 24, 4955–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazurkiewicz-Munoz A. M., Argetsinger L. S., Kouadio J. L., Stensballe A., Jensen O. N., Cline J. M., Carter-Su C. (2006) Mol. Cell. Biol. 26, 4052–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishida-Takahashi R., Rosario F., Gong Y., Kopp K., Stancheva Z., Chen X., Feener E. P., Myers M. G., Jr. (2006) Mol. Cell. Biol. 26, 4063–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Funakoshi-Tago M., Pelletier S., Moritake H., Parganas E., Ihle J. N. (2008) Mol. Cell. Biol. 28, 1792–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bercovich D., Ganmore I., Scott L. M., Wainreb G., Birger Y., Elimelech A., Shochat C., Cazzaniga G., Biondi A., Basso G., Cario G., Schrappe M., Stanulla M., Strehl S., Haas O. A., Mann G., Binder V., Borkhardt A., Kempski H., Trka J., Bielorei B., Avigad S., Stark B., Smith O., Dastugue N., Bourquin J. P., Tal N. B., Green A. R., Izraeli S. (2008) Lancet 372, 1484–1492 [DOI] [PubMed] [Google Scholar]
- 48.Malinge S., Ben-Abdelali R., Settegrana C., Radford-Weiss I., Debre M., Beldjord K., Macintyre E. A., Villeval J. L., Vainchenker W., Berger R., Bernard O. A., Delabesse E., Penard-Lacronique V. (2007) Blood 109, 2202–2204 [DOI] [PubMed] [Google Scholar]
- 49.Dusa A., Staerk J., Elliott J., Pecquet C., Poirel H. A., Johnston J. A., Constantinescu S. N. (2008) J. Biol. Chem. 283, 12941–12948 [DOI] [PubMed] [Google Scholar]
- 50.Royer Y., Staerk J., Costuleanu M., Courtoy P. J., Constantinescu S. N. (2005) J. Biol. Chem. 280, 27251–27261 [DOI] [PubMed] [Google Scholar]
- 51.Cazzola M. (2007) Haematologica 92, 1585–1589 [DOI] [PubMed] [Google Scholar]
- 52.Deindl S., Kadlecek T. A., Brdicka T., Cao X., Weiss A., Kuriyan J. (2007) Cell 129, 735–746 [DOI] [PubMed] [Google Scholar]
- 53.Kratz C. P., Böll S., Kontny U., Schrappe M., Niemeyer C. M., Stanulla M. (2006) Leukemia 20, 381–383 [DOI] [PubMed] [Google Scholar]
- 54.Lee J. W., Kim Y. G., Soung Y. H., Han K. J., Kim S. Y., Rhim H. S., Min W. S., Nam S. W., Park W. S., Lee J. Y., Yoo N. J., Lee S. H. (2006) Oncogene 25, 1434–1436 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.