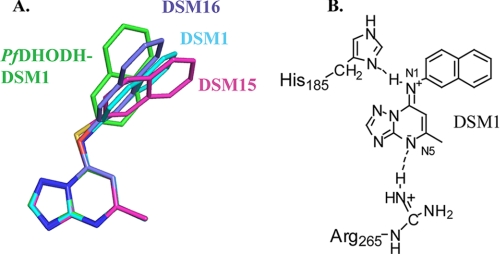
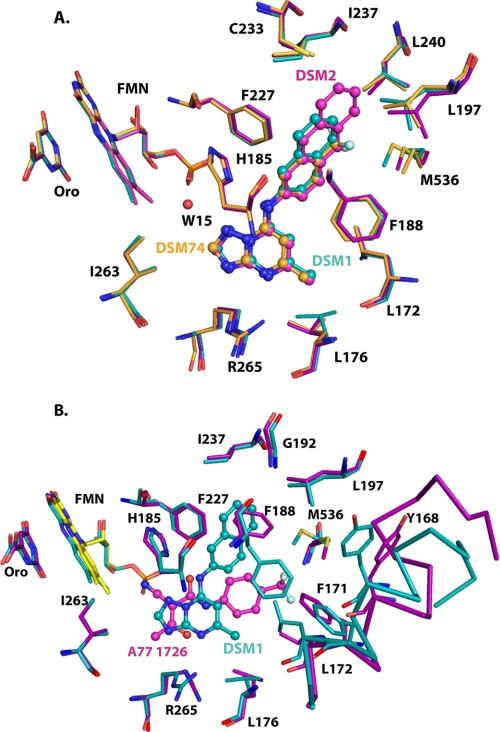
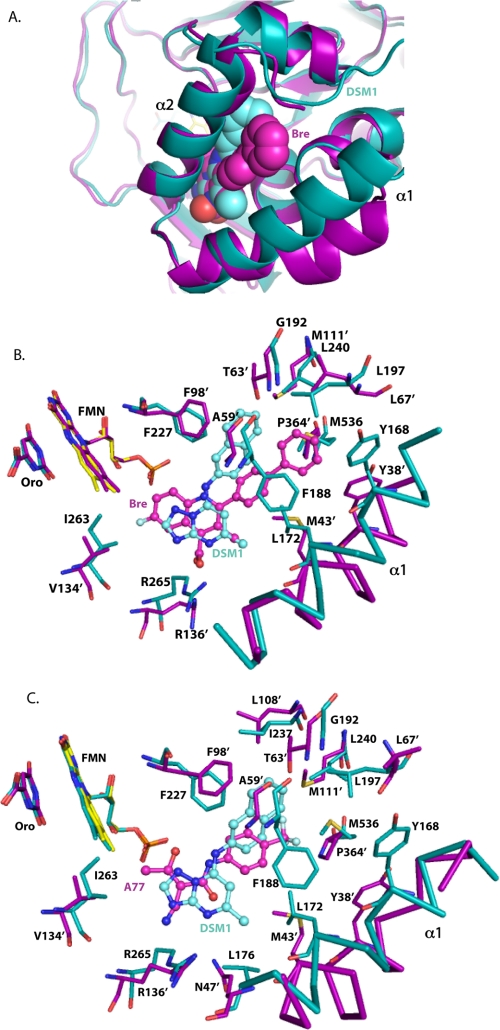
Abstract
Malaria remains a major global health burden and current drug therapies are compromised by resistance. Plasmodium falciparum dihydroorotate dehydrogenase (PfDHODH) was validated as a new drug target through the identification of potent and selective triazolopyrimidine-based DHODH inhibitors with anti-malarial activity in vivo. Here we report x-ray structure determination of PfDHODH bound to three inhibitors from this series, representing the first of the enzyme bound to malaria specific inhibitors. We demonstrate that conformational flexibility results in an unexpected binding mode identifying a new hydrophobic pocket on the enzyme. Importantly this plasticity allows PfDHODH to bind inhibitors from different chemical classes and to accommodate inhibitor modifications during lead optimization, increasing the value of PfDHODH as a drug target. A second discovery, based on small molecule crystallography, is that the triazolopyrimidines populate a resonance form that promotes charge separation. These intrinsic dipoles allow formation of energetically favorable H-bond interactions with the enzyme. The importance of delocalization to binding affinity was supported by site-directed mutagenesis and the demonstration that triazolopyrimidine analogs that lack this intrinsic dipole are inactive. Finally, the PfDHODH-triazolopyrimidine bound structures provide considerable new insight into species-selective inhibitor binding in this enzyme family. Together, these studies will directly impact efforts to exploit PfDHODH for the development of anti-malarial chemotherapy.
The human malaria parasite is endemic in 87 countries putting 2.5 billion people in the poorest nations of the tropics at risk for the disease (1, 2). Despite intensive efforts to control malaria through combination drug therapy and insect control programs, malaria remains one of the largest global health problems. The most severe form of the disease is caused by Plasmodium falciparum, which kills 1–2 million people yearly, primarily children and pregnant woman. Effective vaccines have not been developed, and chemotherapy remains the mainstay of both treatment and prevention of the disease. Unfortunately widespread drug resistance to almost every known anti-malarial agent has compromised the effectiveness of malaria control programs (3). The introduction of artemisinin combination chemotherapy has provided new treatment options to combat drug-resistant parasites (4). However, recent reports by the World Health Organization suggest that resistance to artemisinin is developing along the Thai-Cambodian border, underscoring the need for a continual pipeline of new drug development to combat this disease.
The malaria parasite relies exclusively on de novo pyrimidine biosynthesis to supply precursors for DNA and RNA biosynthesis (5, 6). In contrast, the human host cells contain the enzymatic machinery for both de novo pyrimidine biosynthesis and for salvage of preformed pyrimidine bases and nucleosides. The lack of a redundant mechanism to acquire pyrimidines in malaria has raised interest in this pathway as a potential source for new therapeutic targets. Dihydroorotate dehydrogenase (DHODH)4 is a flavin mononucleotide (FMN)-dependent mitochondrial enzyme that catalyzes the oxidation of dihydroorotate (DHO) to produce orotate, the fourth step in de novo pyrimidine biosynthesis (7, 8). Coenzyme Q (CoQ) is required to catalyze the reoxidation of the flavin cofactor, and recent genetic studies suggest that the main function of mitochondrial electron transport in the parasite is to supply CoQ for this reaction (9). These studies provide genetic evidence that PfDHODH is an essential enzyme to the malaria parasite. An inhibitor of human DHODH (hDHODH) (A77 1726 the active metabolite of leflunomide) is marketed for the treatment of rheumatoid arthritis, illustrating that DHODH is a druggable target (10, 11). Finally, biochemical (12, 13) and structural studies (14, 15) suggested that the identification of species-selective inhibitors against this target was feasible.
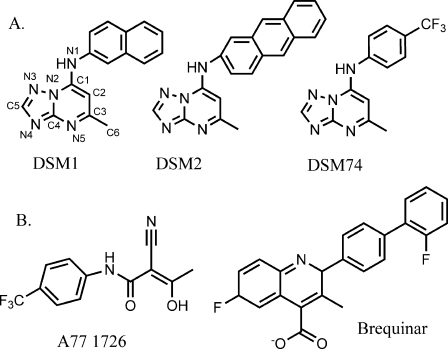
Our recent studies have since directly led to the validation of PfDHODH as a new target for the discovery of anti-malarials. We utilized a high throughput screen to identify several classes of potent and species selective inhibitors of PfDHODH (12, 16–18). These compounds are competitive with CoQ and inhibit the CoQ-dependent oxidation of FMN while not affecting the FMN-dependent oxidation of DHO. Site-directed mutagenesis data supported a model whereby the CoQ-binding site does not overlap with the inhibitor site, but instead inhibitors either block electron transfer between FMN and CoQ or stabilize a conformation that excludes CoQ binding (18, 19). Of the identified inhibitors, one promising series has emerged based on a triazolopyrimidine core structure (see Fig. 1 and Table 1). PfDHODH inhibitors in this class show potent nanomolar activity against P. falciparum in vitro, with excellent correlation observed between inhibition of PfDHODH and activity against the parasite (17, 18). We identified a metabolically stable derivative of this series (DSM74) that is able to suppress Plasmodium berghei infections in the malaria mouse model, providing the first proof that PfDHODH inhibitors can have anti-malarial activity in vivo (17).
FIGURE 1.
Inhibitors of dihydroorotate dehydrogenase. A, selective inhibitors of PfDHODH. DSM1, DSM2, and DSM74. B, selective inhibitors of human DHODH, A77 1726, and brequinar.
TABLE 1.
Inhibition kinetics of wild-type and mutant PfDHODH
The error represents the standard error of the mean for n = 4–5. The fold change is shown in parentheses. The IC50 values for A77 1726 against PfDHODH and hDHODH are 180 μm (19) and 0.3–1 μm (54–56), respectively. Brequinar binds hDHODH with an IC50 of 0.006–0.01 μm (54–56).
| Inhibitor enzyme | IC50 |
||
|---|---|---|---|
| DSM1 | DSM2 | DSM74 | |
| μm | |||
| PfDHODH | 0.047a | 0.056a | 0.28b |
| hDHODH | >100a | >100a | >100b |
| PfDHODH-H185A | 1.3 ± 0.17 (28) | 1.3 ± 0.17 (23) | 25 ± 9.0 (90) |
| PfDHODH-R265A | 2.1 ± 0.70 (45) | 2.4 ± 1.1 (43) | 23 ± 6.7 (82) |
| PfDHODH-F227A | 1.3 ± 0.086 (28) | 4.4 ± 1.1 (79) | 6.3 ± 1.1 (23) |
| PfDHODH-F188A | 2.1 ± 1.4 (45) | 0.21 ± 0.044 (4) | 1.4 ± 0.37 (5) |
Interestingly, despite a vast number of ongoing attempts, PfDHODH represents one of only a few truly new targets for the development of anti-malarial agents since the discovery that atovoquone targets the cytochrome bc1 complex in the mitochondria (20). This has led to a substantial effort to target PfDHODH for drug discovery programs and to the identification of diverse scaffolds showing species-selective inhibition of the enzyme (17, 18, 21–24). The prior structure of PfDHODH complexed to A77 1726 (14), a hDHODH-specific inhibitor with poor affinity for PfDHODH (19), neither explains the ability of PfDHODH to bind the array of identified inhibitors nor provides an understanding of the developing SAR for the triazolopyrimidine-based inhibitor series.
Here we report the x-ray structures of PfDHODH bound to three triazolopyrimidine-based inhibitors with different sized substituents bound to the triazolopyrimidine core. In addition, we examined the small molecule x-ray structures of these inhibitors and compared them with the protein-bound ligand structures. Finally, the inhibitor-bound PfDHODH structures were compared with the structures of hDHODH bound to A77 1726 and to brequinar, a potent 4-quinolinecarboxylic acid inhibitor of the human enzyme (25, 26). Together these studies explain both the high affinity binding and species selectivity of this important class of PfDHODH inhibitors, thus laying the foundation for future lead optimization programs for the present anti-malarial agents. More broadly, this study provides new, unexpected insight into why PfDHODH is a highly attractive drug target for a large set of diverse chemical entities with potential for sustainable, robust lead optimization programs.
EXPERIMENTAL PROCEDURES
Gene Cloning of PfDHODH
N-terminally truncated PfDHODH (amino acids 159–569) was PCR-amplified with primers 1 (5′-AAGGATCCGTTTGAATCTTATAACCCG-3′) and 4 (5′-GGGTCGACTTCCATGGTACCAGCTGCAG-3′) from plasmid pRSET-pfDHODH encoding the codon optimized gene (provided by Jon Clardy) (14) and ligated into the pET28b (Novagen) expression vector at the BamH1/SalI sites. PfDHODHΔ384–413 was generated by PCR amplification of two fragments with compatible restriction sites that excluded the loop sequence. Fragment 1 (amino acids 159–383) containing 5′-BamHI and 3′-EcoRI was generated using primers 1 and 2 (5′-GGGAATTCGTCATTCATAATGTTATTTT-3′) and fragment 2 (amino acids 414–569) containing 5′-EcoRI and 3′-SalI was PCR-amplified with primers 3 (5′-GGGAATTCCTGTGGTTTAATACCACGAA-3′) and 4. Both PCR products were subcloned separately into pCR-Blunt II-TOPO vector (Invitrogen). The resulting clones were used to generate the final expression clone containing loop-truncated PfDHODHΔ384–413 in pET28b in frame with an N-terminal His6 tag.
Protein Expression and Purification
Escherichia coli BL21 phage-resistant cells (Novagen) were used for the expression of pET28b wild-type PfDHODH and PfDHODHΔ384–413 constructs using a modification of methods described previously (19). Proteins were expressed in Terrific Broth medium containing kanamycin (50 μg/ml). The cells were grown to 0.8 A600 at 37 °C, 0.2 mm isopropyl-β-d-thiogalactoside was added to induce protein expression, and the cells were grown overnight at 16 °C. The cells were pelleted by centrifugation (4000 × g) and resuspended in lysis buffer (100 mm HEPES, pH 8.0, 150 mm NaCl, 10% glycerol, and 0.05% THESIT detergent (Fluka)), containing protease-inhibitor mixture for His tag protein (Sigma). The cells were lysed by three passes through an EmulsiFlex-C5 high pressure homogenizer (Avestin Inc.), the lysate was clarified by centrifugation (20,000 × g), and the resulting supernatant was applied to a HisTrap HP column (GE Healthcare) precharged with Ni+2. The column was sequentially washed with lysis buffer and lysis buffer containing 20 mm imidazole. PfDHODHs were eluted from the column using a linear gradient from 20 to 400 mm imidazole. Fractions containing PfDHODH were pooled, concentrated with Amicon Ultra concentrator (Millipore), and then purified by gel filtration column chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare) equilibrated with crystallization buffer (10 mm HEPES, pH 7.8, 100 mm NaCl, 1 mm N,N-dimethyldodecylamine N-oxide (Fluka), 5% glycerol, 10 mm dithiothreitol). Fractions containing PfDHODH were pooled and concentrated to 20 mg/ml. The construction and purification of the H185A, F188A, F227A, and R265A mutant PfDHODH enzymes were described previously (12, 19).
Enzyme Kinetic Analysis
Steady-state kinetic analysis was performed as described previously (12, 19). To determine the kcat and Km of PfDHODHΔ384–413 in comparison with the wild-type enzyme the direct assay that follows the oxidation of DHO at 296 nm (ϵ = 4.3 mm−1 cm−1) was used (Supplemental Table S1). The reactions were performed in assay buffer (100 mm HEPES, pH 8.0, 150 mm NaCl, 10% glycerol, 0.1% Triton) at 20 °C for a range of CoQD (0.025–0.15 mm) and DHO concentrations (0.01–0.5 mm) with PfDHODH (2 nm). Inhibitor kinetics for both wild-type (10 nm) and mutant (10 nm) PfDHODH were followed using the indirect assay that couples the oxidation of DHO (0.2 mm) in the presence of CoQD (20 μm) to the reduction 2,6-dichloroindophenol (60 μm) at 600 nm (ϵ = 18.8 mm−1 cm−1). The data were fitted to the Michaelis-Menten equation to determine the steady-state kinetic parameters or to the following equation to determine the IC50 values using Graph Pad Prism (Graph Pad), vi = Bottom + (Top − Bottom)/(1 + 10(Log[I]−LogIC50)), where, vi is the initial velocity (s−1), and Top and Bottom represent the plateaus in velocity units.
Crystallization and Data Collection of PfDHODHΔ384–413 Bound to Inhibitors
Random crystallization screen Cryo suite (Nextal) and detergent screen kits (Hampton Research) were utilized to determine preliminary crystallization conditions. Subsequent refinement of pH, precipitant, detergent, and protein concentrations was then done to find optimal conditions. N,N-Dimethyldodecylamine N-oxide (1 mm) was found to improve crystallization, and it was subsequently added during the final gel filtration purification step. The crystals of PfDHODHΔ384–413 were grown by vapor diffusion in hanging drop at 20 °C. Reservoir solution (0.16 m ammonium sulfate, 0.1 m sodium acetate, pH 4.6, 18–20% polyethylene glycol 4000, 25% glycerol, and 10 mm dithiothreitol) was mixed with an equal volume of PfDHODHΔ384–413 (15 mg/ml) containing inhibitor (DMS1 (0.6 mm); DSM2 (0.6 mm), or DSM74 (1 mm)) and DHO (2 mm). The crystals typically grew in 4–7 days.
Diffraction data were collected at 100 K on Beamline 19ID at Advanced Photon Source using an ADSC Q315 detector. PfDHODH-DSM1 diffracted to 2.0 Å and has a space group of P64 with the cell dimension of a = b = 85.9, c = 138.4; PfDHODH-DSM2 diffracted to 2.4 Å with space group P64 and cell dimension of a = b = 85.9, c = 138.7; and PfDHODH-DSM74 diffracted to 2.7 Å with space group P64 and cell dimension of a = b = 85.4, c = 138.6 (supplemental Table S2). All three structures have one molecule of PfDHODH in the asymmetric unit. Diffraction data were integrated, and intensities were scaled with the HKL2000 package (27).
Structure Determination and Refinement of PfDHODH Bound to Inhibitors
Crystallographic phases for PfDHODH-DSM1 were solved by molecular replacement with Phaser (28) using the previously reported structure of PfDHODH bound to A77 1726 (Protein Data Bank code 1tv5) (14) as a search model. PfDHODH-DSM1 was used as the search model to find solutions for PfDHODH-DSM2 and PfDHODH-DSM74. Structures were rebuilt with COOT (29) and refined with REFMAC (30), and phases were improved with DM (31) (supplemental Table S2). All of the residues were within the allowed section of the Ramachandran plot (supplemental Table S2). Water molecules were added if the density was stronger than 3.4 σ and removed if the density was weaker than 1 σ in the density map with ARP/warp (32). For PfDHODH-DSM1 the final structure contains residues Ser160–Ser567, with the exception that density was not observed for a loop formed by residues 348–355, 124 water molecules, and a bound LDHO detergent molecule. The final DSM2- and DSM74-bound PfDHODH structures contain all residues between Glu159–His566 (PfDHODH-DSM2) and Phe161–His566 (PfDHODH-DSM74), plus 63 and 38 water molecules, respectively. No density was observed for the detergent molecule in these structures. In the DSM1-, DSM2-, and DSM74-bound structures the shortened surface loop at position 384 is observed in its entirety.
Molecular Modeling
The structures were displayed using the graphics program PyMol (26). The PfDHODH-DSM1 structure was superimposed with the PfDHODH-DSM2, PfDHODH-DSM74, PfDHODH-A77 (Protein Data Bank code 1tv5), hDHODH-A77 (Protein Data Bank code 1D3H) (15), and hDHODH-bre (Protein Data Bank code 1D3G) (15) structures by aligning only backbone atoms of the β/α domain in LSQKAB (33). For the alignment between pfDHODH-DSM1 and hDHODH, the following sequences were superimposed: Pf 217–232 to h88–103; Pf271–278 to h142–149; Pf337–343 to h207–213; Pf436–460 to h262–286; Pf473–494 to h301–322; Pf501–529 to h329–357; and Pf541–552 to h369–380. The same PfDHODH residues were used to superimpose the various PfDHODH structures. RMSD values were calculated for the superimposed structures based on the Cα positions using LSQMAN (32, 34). Moleman2 (35) was used to manipulate the Protein Data Bank files before the analysis (Software was obtained from the Uppsala Software Factory.).
Small Molecule X-ray Structure Determination
Crystallization of DSM1 from CH2Cl2/CH3OH was described previously (18). DSM15, DSM16, and DSM74 were crystallized similarly to CH2Cl2. Single crystals were mounted on a glass capillary with oil. The data were collected at −143 °C with a Nonius Kappa CCD FR590 single crystal x-ray diffractometer. The crystal-to-detector distance was 30 mm. Other data collection parameters are summarized in supplemental Table S3. The data were integrated and scaled using hkl-SCALEPACK (27). Solution by direct methods (SIR97) (36) produced a complete heavy atom phasing model (supplemental Table S4). All of the hydrogen atoms were located using a riding model with the exception of H1. All of the non-hydrogen atoms were refined anisotropically by full matrix least squares utilizing SHELXL97 (37). Scattering factors are from Waasmaier and Kirfel (38), and the absolute structure assignment for DSM1 was established via anomalous scattering. The Flack enantiopole parameter (39) is −0.06 ± 0.11. ORTEP drawings (40) are shown in supplemental Fig. S4.
The crystals of DSM15 were twinned such that two lattices of very similar intensities were apparent in the diffraction pattern. One lattice was indexed. In a first refinement cycle, intensities of reflections of type {0 k l} (10% of total data) appeared systematically larger than those calculated. Excluding {0 k l} reflections from the refinement allowed a useful structure to be determined. The ratio between the calculated and measured intensities of the excluded data was close to 2, consistent with a total overlap of two data sets. Such twining can arise from 2-fold twinning along [1 0 0] or a mirror on the [0 k l] plane. No additional twin law was found for the reduced data set, indicating that the twinning did not create any detectable intensity overlaps for other peaks. Thus the excluded intensity data were corrected for the factor 2 and merged with the other data for the final structural refinement.
For DSM74 some of the intermolecular fluoride-fluoride distances were too short. This problem arises from disorder of the CF3 group. CF3 groups are very often found to rotate or be arranged in two alternating geometries. This is the case in this structure where two conformations of the CF3 (atoms F1–3 and F4–6) were refined at half-site occupancies including the carbon. However the disorder, which is represented by to a 2-fold rotation around the axis C7-N1, extended to the phenyl group, which is also represented in the refinement by two rings of half-site occupancies. When F1 (F4) is present, the symmetry related F2 (F5) of the same disordered moiety is absent, which, if there were no defects or additional voids in the crystal, would require a structure with the disordered CF3 alternating, which in turn would cause a doubled c axis length. There is evidence in the scattering for this to be partially the case because there are very faint reflections in the diffraction images pointing toward a doubled c axis; however, the order is not persistent throughout the whole crystal. The disorder required a special refinement where the phenyl ring was constrained in its angles, but the average distance between the atoms was freely refined. The distances of the disordered rings to the neighboring atoms are as follows: N-1–C-7, 1.42 ± 0.02; N-1–C-7′, 1.44 ± 0.02; C-10–C-13, 1.50 ± 0.02; and C-10′–C-13′, 1.51 ± 0.02.
Synthetic Methods: General Chemistry and Analysis
The reagents and solvents were obtained from commercial suppliers and were used without further purification. The reaction progress was monitored by TLC using silica gel 60 F-254 (0.25 mm) plates and detection with UV light. Flash chromatography was carried out with silica gel (32–63 μm). 1H NMR spectra were recorded in CDCl3 at 300 MHz. Chemical shifts are reported in parts per million (δ) downfield from tetramethylsilane. Coupling constants (J) are reported in Hz. Spin multiplicities are described as s (singlet), brs (broad singlet), d (doublet), t (triplet), q (quartet), and m (multiplet). Electrospray ionization mass spectra were acquired on a Bruker Esquire liquid chromatograph ion trap mass spectrometer. Melting points (Pyrex capillary) were determined on a Mel-Temp apparatus and are uncorrected.
DSM1, DSM2, and DSM74 were prepared as described previously (17, 18). DSM15 and DSM16 were prepared as shown in supplemental Fig. S3. Condensation of 3-amino-[1,2,4]triazole with ethyl acetoacetate in acetic acid yielded the 7-hydroxy-[1,2,4]-triazolo[1,5-a]pyrimidine. Chlorination with phosphorous oxychloride gave the corresponding 7-chloro-[1,2,4]triazolo[1,5-a]pyrimidine as described previously (18), which upon treatment with 2-thionaphthol/2-naphthol (1.2 eq) in 5 ml of DMF, K2CO3 (1.2 eq) stirred under N2 atmosphere at room temperature for a 20-h yielded product. The crude products were purified by column chromatography using ethyl acetate/hexane.
Physical Properties
The physical properties are as follow: 5-methyl-7-(naphthalene-2-ylthio)-[1,2,4]triazolo[1,5-a]pyrimidine (DSM15), mp 189 °C; 1H NMR (300 MHz, CDCl3): δ 8.50 (s, 1H), 8.30 (s, 1H), 8.10–7.94 (m, 3H), 7.74–7.62 (m, 3H), 6.16 (s, 1H), 2.48 (s, 3H); MS m/z 293.1 [M + H]+; and 5-methyl-7-(naphthalene-2-yloxy)-[1,2,4]triazolo[1,5-a]pyrimidine (DSM16); mp 170 °C; 1H NMR (300 MHz, CDCl3): δ 8.51 (s, 1H), 8.05 (d, J = 9 Hz, 1H), 8.01–7.88 (m, 2H), 7.78 (m, 1H), 7.68–7.58 (m, 2H), 7.45–7.38 (m, 1H), 6.08 (s, 1H), 2.58 (s, 3H). MS m/z 277.2 [M + H]+.
RESULTS
X-ray Structure Determination of PfDHODH Bound to Triazolopyrimidine Analogs
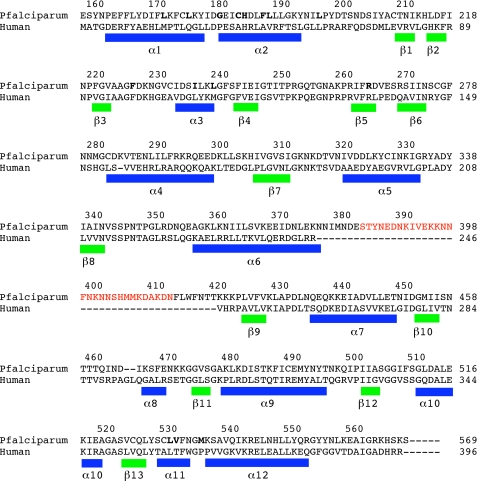
Three triazolopyrimidine analogs containing naphthyl (DSM1), anthracenyl (DSM2), and phenyl-trifluoromethyl (DSM74) substituents, which span a range of inhibitor potency (0.05–0.3 μm), were chosen for crystallographic analysis (Fig. 1 and Table 1). Proteolysis of a P. falciparum-specific surface loop, which is not present in the enzymes from human or other Plasmodium species (residues 384–413; Fig. 2), led to difficulties obtaining diffraction quality crystals with these inhibitors. To improve crystallization we generated a PfDHODH construct lacking amino acid residues 384–413 (PfDHODHΔ384–413). This region of the structure was missing or disordered in the PfDHODH structure described previously (14). Steady-state kinetic analysis of PfDHODHΔ384–413 demonstrated that the catalytic efficiency and inhibitor binding properties of the loop-minus protein were similar to the wild-type enzyme (supplemental Table S1). PfDHODHΔ384–413 was co-crystallized with DSM1, DSM2, or DSM74 in the presence of substrate DHO, and 2.0, 2.4, and 2.7 Å resolution data sets were collected, respectively. The three structures were solved by molecular replacement and refined to Rfac of 23.1, 22.5, and 23.6 and Rfree of 24.8, 26.7, and 27.6, respectively (supplemental Table S2).
FIGURE 2.
Sequence alignment of P. falciparum and human DHODH. Secondary structure elements are defined based on the PfDHODH-DSM1 structure: α-helices are indicated by blue bars, and β-strands are indicated by green bars. Residues within 4 Å of DSM1 are displayed in bold type. The truncated surface loop in the PfDHODHΔ384–413 construct used for structure determination is shown in red.
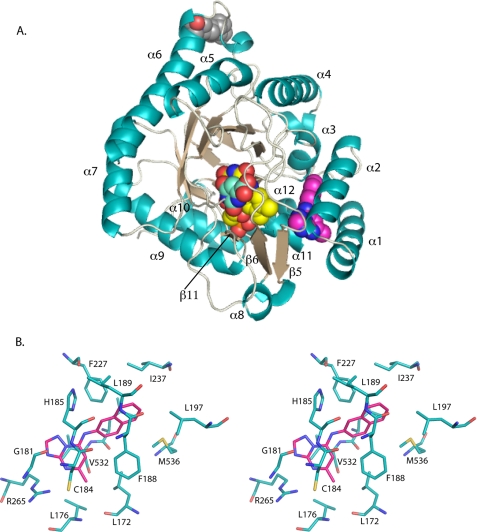
The three-dimensional fold of PfDHODH in all three inhibitor bound complexes is similar to the previously reported structures of PfDHODH bound to A77 1726 (14) and of human DHODH (hDHODH) (15). The enzyme consists of a short N-terminal helical domain (residues 163–194) followed by a classic β/α barrel domain that begins with β-strand 3 (Fig. 2 and 3A). The electron density maps of the active sites of all three structures showed strong, interpretable density for the bound inhibitors (DSM1, DSM2, or DSM74) (supplemental Fig. S1).
FIGURE 3.
A, ribbon diagram of DSM1 bound to PfDHODH. α-Helices are displayed in teal, β-strands are displayed in sand, ligands are displayed as space filling balls with FMN in yellow, DSM1 is in pink, orotate is in turquoise, and the bound detergent molecule is in gray. The position of the detergent corresponds to the position of the shortened surface loop truncated in the PfDHODHΔ384–413 construct. B, stereo diagram of the DSM1-binding site. Residues within 4 Å of the bound inhibitor are displayed. DSM1 is displayed in pink, and residues are displayed in teal. Nitrogen is displayed in blue, oxygen is in red, and sulfur is in yellow.
Triazolopyrimidine-binding Site
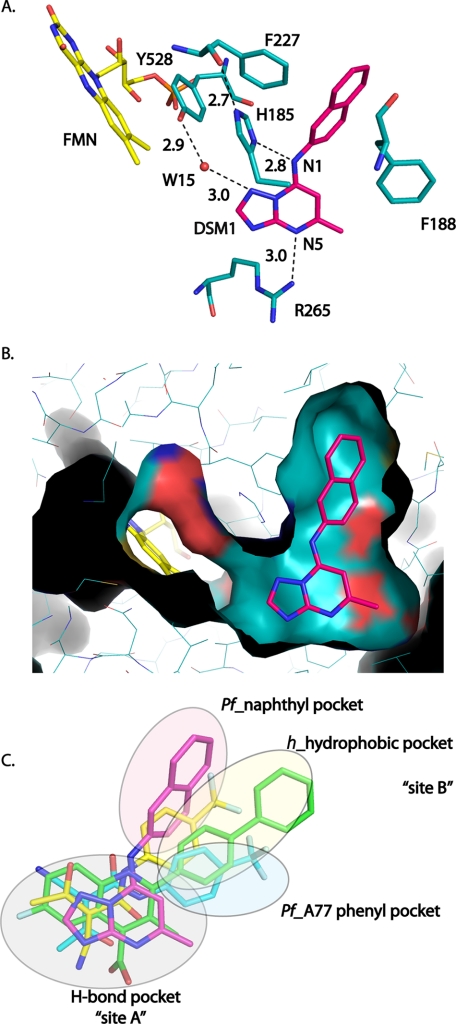
The inhibitor-binding site is formed adjacent to the FMN site between the two N-terminal helices (α1 and α2), the top of helix α3 in the β/α barrel, the 310-helix α11, and strand β5 (Fig. 3A). The naphthyl of DSM1 interacts with α3, whereas the triazolopyrimidine group lies against α11. DSM1, DSM2, and DSM74 bind in the same site. The inhibitors are oriented such that the C-5 position of the triazolopyrimidine ring is the closest atom to FMN at a distance of 6 Å (Fig. 3). The inhibitors are bound in an extended conformation with the naphthyl (DSM1), anthracenyl (DSM2), or phenyl-trifluoromethyl (DSM74) groups oriented away from the triazolopyrimidine core and from FMN. The triazolopyrimidine ring in all three structures binds to a largely hydrophobic pocket formed by Val532, Leu172, Leu176, Cys184, and Gly181 (Fig. 3B and supplemental Fig. S2) that also contains two residues that form the only nonhydrophobic contacts in this pocket. These nonhydrophobic contacts include ion pair H-bonds between His185 and the bridging nitrogen N-1 and between Arg265 and the pyridine nitrogen N-5 (Fig. 4, A and B). We have defined this pocket as the H-bond pocket (Fig. 4C). The naphthyl (DSM1), anthracenyl (DSM2), or phenyl-trifluoromethyl (DSM74) groups bind in a completely hydrophobic pocket formed by residues Ile237, Leu189, Leu197, Met536, Phe227, and Phe188 (Fig. 3B and supplemental Fig. S2), which we will define as the Pfnaphthyl pocket (Fig. 4C). An edge-to-face stacking interaction between Phe227, the aromatic group bound in the Pfnaphthyl pocket, and Phe188, is present (Fig. 4A), suggesting that these interactions likely contribute to the potent binding of the inhibitor series. These interactions involve the inhibitor in an extended aromatic stacking network that projects from FMN through Tyr528 to Phe227 on one face of the inhibitor and from Phe188 to Phe171 and beyond on the other side.
FIGURE 4.
DHODH inhibitor-binding site interactions. A, H-bonding interactions between DSM1 and PfDHODH. DSM1 is displayed in pink, and residues are displayed in teal. Distances are shown for H-bond interactions observed between the inhibitor and residues in the binding site. B, van der Waal's surface representation of DSM1 bound to PfDHODH (cyan) as calculated by PyMol. Ligands were removed from the structure prior to calculation of the surface. The figure shows a narrow channel is present extending from C-5 of DSM1 to FMN. DSM1 is displayed in pink, FMN is displayed in yellow, and amino acid residues and the surface are displayed in teal. C, structural alignment of PfDHODH and hDHODH inhibitor binding modes. The structures of PfDHODH-DSM1 (pink), PfDHODH-A77 (turquoise), hDHODH-bre (green), and hDHODH-A77 (yellow) were superimposed as described in methods. Only the bound inhibitor is displayed.
The van der Waal's surface of the DSM1-binding pocket shows that the inhibitor is buried in the interior of the protein (Fig. 4B). The naphthyl is bound snuggly within a dead end hydrophobic pocket and the triazolopyrimidine ring, and the methyl side chain at C-6 also fit in close contact with the protein. A narrow channel leads from C-5 of the triazolopyrimidine ring toward FMN. An ordered water molecule (Wat15) in this channel is observed in all three structures forming an H-bond with N-3 of the inhibitor and with the hydroxyl of Tyr528, which in turn stacks against the FMN cofactor (Fig. 4A). This channel then extends from the FMN to the protein surface providing the only access to solvent for the bound ligands (FMN, orotate, and inhibitor). However, this channel is too narrow to allow passage of either substrate or inhibitor, and thus significant structural movement must accompany ligand binding.
Mutagenesis of Inhibitor-binding Pocket
The contribution to binding affinity of residues involved in H-bond (His185 and Arg265) or π-stacking interactions (Phe227 and Phe188) (Figs. 3 and 4 and supplemental Fig. S2) were probed by site-directed mutagenesis. We previously published the effects of mutating these four residues to Ala and demonstrated that these mutations had only minimal effects on catalytic activity (kcat and Km remain within 2–4-fold of the wild-type enzyme) (19). In contrast, mutation of each of these residues to Ala increased the IC50 for DSM1 by 30–50-fold, demonstrating that each residue contributes significant binding energy to the enzyme inhibitor interaction (Table 1). The relative contribution of the mutated residues differs for the three inhibitors described in this study. For DSM2, the contribution of His185 and Arg265 is similar to DSM1; however Phe188 appears to play a reduced role in binding of this inhibitor. For DSM74, the H-bonds/ion pairs between His185 and Arg265 and inhibitor contribute more energy to the binding interaction than Phe188. The IC50 is increased by 80–90-fold for mutation of His185 and Arg265, but only by 5-fold upon mutation of Phe188. We previously analyzed only the F227A and R265A mutant enzymes against DSM1 (18); the IC50 reported for R265A was similar to the value reported here in Table 1. However, the previously measured IC50 for F227A was 30-fold higher; solubility problems may have contributed to this elevated value.
Small Molecule Structures of DSM Derivatives
Complementary insight into the importance of N-1 in the bridge position came from small molecule crystal structures of DSM1 and DSM74 in comparison with DSM1 analogs that contained an S (DSM15) or O (DSM16) in place of N-1 (Fig. 5, supplemental Figs. S3 and S4, and supplemental Tables S3–S5). DSM15 and DSM16 were tested against PfDHODH, and they showed no inhibitory activity; thus the substitution of a single atom at the bridging position can eliminate binding. The small molecule x-ray structures of DSM1 and DSM74 show partial double-bond character of the C-1—N-1 bond connecting the bridging nitrogen to the triazolopyrimidine ring with observed C-1—N-1 bond lengths of 1.313 ± 0.006 and 1.344 ± 0.004 Å, respectively (supplemental Table S5) (typical C–N or C N bond lengths are 1.38 and 1.28 Å, respectively (41)). In contrast, for analogs containing a bridging S (DSM15) or O (DSM16) in place of N-1, the C-1–S-1 and C-1—O-1 bond lengths were 1.740 ± 0.004 and 1.338 ± 0.003 Å, respectively, typical of single bonds (e.g. C—S/C—O bond lengths are 1.75/1.34 Å (27), whereas C
N bond lengths are 1.38 and 1.28 Å, respectively (41)). In contrast, for analogs containing a bridging S (DSM15) or O (DSM16) in place of N-1, the C-1–S-1 and C-1—O-1 bond lengths were 1.740 ± 0.004 and 1.338 ± 0.003 Å, respectively, typical of single bonds (e.g. C—S/C—O bond lengths are 1.75/1.34 Å (27), whereas C S/C
S/C O bond lengths are 1.67/1.21 Å (41), respectively). Additionally, partial positive charge character of N-1 is further suggested by hydrogen bonding to a chloride ion in the DSM1 structure. These data suggest that when the bridging atom is N-1, resonance contributes to greater electron delocalization to N-5, intrinsically establishing long range dipoles in the inhibitor. In the protein, electron delocalization of the inhibitor is exploited by the H-bond with His185, which results in sufficient electron density on N-5 to allow formation of an ion pair with Arg265 (Figs. 4A and 5B). Such electron delocalization is more limited, or absent, for molecules containing O or S in the bridging position, providing an explanation for their inactivity as PfDHODH inhibitors. Thus the intrinsic dipole of the triazolopyrimidine ring is a key factor in the potency of this compound class toward PfDHODH.
O bond lengths are 1.67/1.21 Å (41), respectively). Additionally, partial positive charge character of N-1 is further suggested by hydrogen bonding to a chloride ion in the DSM1 structure. These data suggest that when the bridging atom is N-1, resonance contributes to greater electron delocalization to N-5, intrinsically establishing long range dipoles in the inhibitor. In the protein, electron delocalization of the inhibitor is exploited by the H-bond with His185, which results in sufficient electron density on N-5 to allow formation of an ion pair with Arg265 (Figs. 4A and 5B). Such electron delocalization is more limited, or absent, for molecules containing O or S in the bridging position, providing an explanation for their inactivity as PfDHODH inhibitors. Thus the intrinsic dipole of the triazolopyrimidine ring is a key factor in the potency of this compound class toward PfDHODH.
FIGURE 5.
Comparison of protein bound inhibitor conformations with free ligand structures. A, comparison of the conformation of DSM1 bound to PfDHODH (green) with the conformations of DSM1 (cyan), DSM15 (pink), and DSM16 (purple) as observed in the small molecule x-ray structures. B, schematic resonance structure of DSM1 forming H-bonding ion pair interactions with PfDHODH. H-bonds are depicted by the dashed line.
Not all aspects of the small molecule conformation translate to what is observed on the protein. The plane of the naphthyl group in the protein-bound state is twisted by 48° relative to its unbound conformation (Fig. 5A). This conformational difference is necessary to position the naphthyl group of DSM1 to interact productively with the enzyme.
Plasticity to Accommodate Different Sized Triazolopyrimidine Inhibitors
Globally, the DSM1, DSM2, and DSM74-bound PfDHODH structures are very similar with RMSD for superimposition of the Cα atoms of ∼0.4 Å among them (Figs. 6A). The inhibitors occupy very similar positions in all three structures. The different size N-1 substituents are accommodated by conformational flexibility in the binding pocket. In both the PfDHODH-DSM2 and PfDHODH-DSM74 structures the triazolopyrimidine ring tilts slightly toward the bottom of the H-bond pocket relative to PfDHODH-DSM1. Leu176 reorients to accommodate this change. Further for PfDHODH-DSM74, this slight shift (0.2 Å) brings Ile263 into van der Waal's contact with the C-5 position of the inhibitor. For PfDHODH-DSM2, the anthracenyl extends further into the binding site than naphthyl of DSM1. The larger aromatic group is accommodated by small rotational changes in Leu197 and Met536 that enlarge the Pfnaphthyl pocket. New contacts are also made with Leu240 and Cys233. The smaller phenyl-trifluoromethyl group of DSM74 does not completely fill the pocket and makes fewer van der Waal's contacts. In particular Ile237 and Leu189 are no longer within the 4 Å van der Waal's shell of DSM74. However, a new contact is made between the CF3 group and Leu240 in this structure. The reduced surface area of contact likely explains why DSM74 is 10-fold less potent than DSM1 or DSM2.
FIGURE 6.
Comparison of PfDHODH inhibitor complexes. A, PfDHODH-DSM1 (teal) superimposed with PfDHODH-DSM2 (purple) and with PfDHODH-DSM74 (orange). B, PfDHODH-DSM1 (teal) superimposed with PfDHODH-A77 (purple). A subset of residues within 4 Å of the bound inhibitor are displayed. FMN (yellow) and orotate (Oro), which are outside this shell, are included to provide orientation. Inhibitors are displayed as ball and stick.
Conformational Flexibility of PfDHODH Accounts for Binding to Diverse Chemical Scaffolds
Comparison of the PfDHODH-DSM1 with PfDHODH-A77 structures shows that PfDHODH can present two alternative binding sites for interactions with different inhibitor classes. The PfDHODH-DSM1 and PfDHODH-A77 structures superimpose with a Cα RMSD of 1.03 Å, much larger than the 0.4 Å RMSD observed between the three triazolopyrimidine class inhibitors. Both inhibitors occupy the H-bond pocket such the triazolopyrimidine ring and the β-hydroxy enamide portion of A77 1726 overlap in this site (Figs. 4C and 6B). In contrast, the aromatic N-1 substituent in the triazolopyrimidines binds in an entirely different position from the phenyl-trifluoromethyl of A77 1726. These different binding modes are created by a large conformational change in the position of Phe188 between the two structures. In PfDHODH-A77, Phe188 occupies the Pfnaphthyl pocket, and in the PfDHODH-DSM1 Phe188 rotates to overlap the position of the phenyl-trifluoromethyl in PfDHODH-A77 (defined as the PfA77 phenyl pocket; Fig. 4C). In addition, Met536, Cys175, Leu176, and residues on helix α1 (Tyr168, Asp169, Phe171, and Leu172) show conformational flexibility between the two structural classes of inhibitors. Further, helix α1 shifts toward the interior of the protein in PfDHODH-DSM1, further restricting the PfA77 phenyl pocket. Similar structural differences are observed between PfDHODH-A77 and PfDHODH-DSM2 and PfDHODH-DSM74.
Host-Parasite Differences
To understand the species-selective binding of the various inhibitor classes, PfDHODH bound to the triazolopyrimidines was compared with hDHODH in complex with brequinar and A77 1726 (Fig. 7). Alignment of the Cα backbone of PfDHODH-DSM1 with hDHODH-bre or with hDHODH-A77 gave an overall RMSD of 1.8 or 1.9 Å, respectively. Brequinar, which is a potent inhibitor of hDHODH, forms some analogous interactions to the triazolopyrimidine core of DSM1 (Fig. 7, A and B). The 4-quinolinecarboxylic acid of brequinar binds the same site (H-bond pocket) as the triazolopyrimidine ring, forming a salt bridge between the carboxylate and hArg136 (equivalent to P. falciparum Arg265 interacting with N-5 of DSM1). This interaction is thought to play a major role in driving binding affinity (15, 42). In contrast, the biphenyl portion of brequinar binds to a distinct and different hydrophobic site (defined as the h_hydrophobic pocket) from the naphthyl-group of DSM1, which binds PfDHODH at the adjacent malaria-specific site (Pfnaphthyl pocket). The h_hydrophobic pocket lies between the DSM1 Pfnaphthyl pocket and the PfA77 phenyl pocket of A77 1726 when bound to PfDHODH (Fig. 4C).
FIGURE 7.
Comparison of PfDHODH-DSM1 to hDHODH inhibitor complexes. A, ribbon diagram of PfDHODH-DSM1 (teal) aligned with hDHODH-bre (purple). Inhibitors are displayed as space filling balls. B, inhibitor-binding site alignment of PfDHODH-DSM1 (teal) with hDHODH-bre (purple). C, inhibitor-binding site alignment of PfDHODH-DSM1 (teal) with hDHODH-A77 (purple). Residue numbers for hDHODH are marked with a prime symbol, whereas PfDHODH numbers are not. Inhibitors are displayed as ball and stick.
These sites represent alternative pockets that are species-specific and are created by the different amino acid composition of the pockets (Fig. 7, A and B). The h_hydrophobic pocket that binds the brequinar biphenyl group is occluded in PfDHODH by replacement of the smaller hAla59 and hPro364 in hDHODH with the larger PfPhe188 and PfMet536 residues in PfDHODH and by the repositioning of PfTyr168 (hTyr38) closer to the inhibitor-binding pocket by the movement of the N-terminal α1 helix. Additionally hLeu68 is positioned further from the biphenyl pocket than the equivalent residue PfLeu197 in PfDHODH. On the flip side, in PfDHODH the naphthyl pocket is opened up relative to hDHODH by the replacement of hThr63 and hMet111 with PfGly192 and PfLeu240. Further the positions of PfPhe227 and PfLeu531 relative to the equivalent residues in hDHODH (hPhe98 and hLeu359) have swung away from the naphthyl position in PfDHODH-DSM1, enlarging the pocket relative to hDHODH. Globally the N-terminal helix (Ser160 to Tyr178) in PfDHODH is packed more closely to the inhibitor-binding site than the analogous helix (residues Met30 to Gly48) in hDHODH (Fig. 7A). This difference contributes to closing of the h_hydrophobic pocket in PfDHODH.
A77 1726 is smaller than brequinar and does not extend as deeply into the h_hydrophobic pocket (Fig. 7C). Furthermore, the phenyl-trifluoromethyl group binds slightly closer to the Pfnaphthyl pocket then brequinar. The β-hydroxy enamide portion of A77 1726 is flipped over in the hDHODH structure relative to when bound to PfDHODH. This changes the angle of projection of the phenyl-trifluoromethyl group into the hydrophobic pocket, allowing it to access the h_hydrophobic pocket instead of the PfA77 phenyl pocket. Similar residues contribute to the differential binding as was observed for brequinar. Again the replacement of hAla59 with PfPhe188 and hPro364 with PfMet536 closes off the phenyl-trifluoromethyl pocket of A77 1726 in PfDHODH relative to the hDHODH structure.
DISCUSSION
Malaria remains one of the most significant global health problems because of wide spread drug resistance compromising current chemotherapies. One of the greatest challenges in developing new anti-malarial agents is the identification of novel targets that will allow the discovery of unexplored chemical scaffolds. Our discovery of the triazolopyrimidine-based inhibitors of PfDHODH has led to the validation of PfDHODH as a target for the development of new anti-malarials and to the identification of a novel chemical class with in vivo anti-malarial activity (17, 18). Here we report the x-ray structures of PfDHODH bound to three triazolopyrimidine-based inhibitors. First, these data show that structural plasticity allows the triazolopyrimidine-based inhibitors to access an unexpected hydrophobic binding pocket that was not observed in previous structures. This structural flexibility allows the target to accommodate a range of inhibitors from different structural classes. A second unexpected insight was gained by comparison of the small molecule x-ray structures of the inhibitors with the protein-bound ligand structures. The intrinsic electronic configuration of the triazolopyrimidine ring favors charge delocalization from N-1 to N-5, providing significant insight into the emerging SAR of the triazolopyrimidine-based series. Finally, the variability in amino acid sequence between the human and malaria DHODH inhibitor-binding sites leads to species-selective inhibitor binding modes that explain the selectivity of the triazolopyrimidine-based inhibitors.
The malaria enzyme has the flexibility to form two different inhibitor-binding pockets (Pfnaphthyl pocket and the PfA77 phenyl pocket). This flexibility is likely to underlie the finding that multiple inhibitor series have been reported for PfDHODH that sample diverse chemical space (17, 18, 21–24). Although the triazolopyrimidine derivatives interact with the Pfnaphthyl pocket, other identified inhibitors are likely to bind to the PfA77 phenyl pocket, explaining how the enzyme can accommodate these wide ranging scaffolds. Additionally local flexibility within the Pfnaphthyl pocket contributes to the ability of this pocket to bind ligands of variable size. Flexibility in inhibitor binding modes has also been observed in hDHODH associated with conformational changes in the position of hArg136 and hGln47 (42), the N-terminal helix (α1) (11) and a loop (hLeu68–hArg72) postulated to perform a gate keeper function (43). Thus structural plasticity appears to be an inherent property of the DHODH enzyme family, enhancing the suitability of this enzyme as a drug target by creating many design options and reducing the risks or liabilities that would be associated with only a single chemical class. Additionally the ability of the enzyme to adapt to different analogs within a structural class provides sites for modification during lead optimization to improve pharmacological properties. Protein flexibility has been observed in a wide array of known drug targets (44–46), and protein dynamics is increasingly being incorporated into drug design strategies (47–51). Thus structural plasticity leading to binding of diverse inhibitor classes can be appreciated as an important property of high value drug targets guiding future target selection for drug discovery projects.
Analysis of drug interactions that are involved in conformational changes in the target protein suggests that hydrophobic inhibitors can induce refolding of local regions of the binding pocket leading to high affinity interactions (46). This suggests that the triazolopyrimidine-based inhibitors induce a refolding of the aromatic network by reorienting Phe188 and allowing the naphthyl-functionality to insert into an extended edge-to-face stacking network that runs through the protein. Alternatively the inhibitors may selectively bind and stabilize a pre-existing conformation of the aromatic network that is part of the native state ensemble but that has not previously been observed. Aromatic edge-to-face stacking interactions provide additional binding energy beyond a typical van der Waal's contact (52) as has been observed previously for potent binding of carboxypeptidase inhibitors (53). Our data support a role for these interactions in inhibitor binding to PfDHODH because mutation of residues in this network (Phe227 and Phe188) resulted in a significant reduction in binding affinity.
The intrinsic electronic properties of the triazolopyrimidine ring allow it to populate a resonance form that promotes charge separation, which is key to the formation of H-bond interactions with the enzyme. The importance of these ion pair interactions to inhibitor potency is supported both by the site-directed mutagenesis data and by the chemical modification of the inhibitor. When N-1 is replaced with atoms that are unable to delocalize electrons into the triazolopyrimidine ring (S or O), the IC50 increases by >4.4 kcal/mol. Interestingly mutation of His185 or Arg265 each reduced the binding affinity by 2–3 kcal/mol, suggesting that if the contributions of these two residues are additive, the loss in binding energy by replacing the bridging nitrogen with S or O is equivalent to losing both interactions.
The x-ray structures of PfDHODH bound to the triazolopyrimidine-based inhibitors represent the first structural studies of the malarial enzyme bound to malaria-specific inhibitors, and as such they provide considerable new insight into species selective binding in this enzyme family. The inhibitor-binding pocket can be divided into site A (the H-bond pocket), which forms H-bonding interactions with the bound inhibitors, and site B, which is entirely hydrophobic (Fig. 4C). Both malaria and human-specific inhibitors interact with site A. Species selectivity of inhibitor binding arises from differences in the orientation of site B relative to site A within the protein structure. These differences have their basis in the species-specific amino acid composition of the inhibitor-binding pockets. For the triazolopyrimidine analogs, specificity for PfDHODH arises from the projection of an aromatic side chain from the N-1 position into the Pfnaphthyl pocket, which is blocked in the human structure. Whereas for brequinar the presence of the biphenyl group on the equivalent position to C-2 of DSM1 allows good interaction with the h_hydrophobic pocket, this orientation cannot be accommodated by either the site B-binding pocket on PfDHODH. The structural basis for differential binding of A77 1726 is similar. The fact that residues that form H-bonds with the triazolopyrimidine ring (PfHis185 and PfArg265) are conserved in the human enzyme suggests inhibitors of hDHODH could also be developed based on this core scaffold.
The x-ray structures of PfDHODH bound to the triazolopyrimdine inhibitors provide significant insight into the observed SAR for this series. In addition to explaining the loss of activity upon replacing the bridging nitrogen N-1 with S or O, several other aspects of the SAR (17, 18) can now be readily understood. First we have observed that when the aromatic group is a phenyl, para substitutions yield good binding potency, whereas ortho substitutions are completely inactive. The PfDHODH-DSM74 structure shows that the ortho carbon atoms are 4.1 Å from the carbonyl oxygen of His185 on one side and 4.0Å from the CD1 of Leu172 on the other. Thus based on the current structures there is not space in the pocket to accommodate ortho substituents, nor apparently is this region of the binding pocket capable of the conformational flexibility that would be needed to accommodate this substitution. Second, the completely hydrophobic nature of the Pfnaphthyl pocket explains why aromatic rings containing heteroatoms show reduced potency. Third, the close interaction between His185 and the bridging nitrogen N-1 explains why a secondary nitrogen is required at this position and why the addition of a third substituent on the nitrogen reduces potency. Finally, the potential for stacking interactions between Phe227 and Phe188, in addition to the hydrophobic nature of the pocket, explains why large aromatic groups are favored.
In summary, x-ray structure determination of PfDHODH in complex to three triazolopyrimidine-based inhibitors has provided insight into the structural basis for potent and species-selective binding of this series of inhibitors. It has advanced our understanding of the emerging SAR for this promising lead series. Ongoing efforts to improve the potency and in vivo properties of the triazolopyrimidine-lead series will be greatly aided by these studies. More broadly, the results have provided insight into the structural requirements for designing species-selective inhibitors of either malarial or human DHODH that can be applied to scaffolds beyond the triazolopyrimidine analogs. Finally, the advantages of attacking plastic active sites in lead optimization offer general lessons for prioritizing structure-based protein-ligand interactions for pharmaceutical applications.
Supplementary Material
Acknowledgments
We thank Diana Tomchick and Mischa Machius of the University of Texas Southwestern crystallography core for help with data collection and David Matthews and Jeremy Burrows for insightful and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants U01AI075594 and AI053680 (to M. A. P. and P. K. R.). This work was also supported by a grant from Malaria Medicines Venture (to M. A. P. and P. K. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1–S5 and Figs. S1–S4.
The atomic coordinates and structure factors (codes 3I65, 3I68, and 3I6R) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- DHODH
- dihydroorotate dehydrogenase
- Pf
- P. falciparum
- h
- human
- CoQ
- coenzyme Q
- FMN
- flavin mononucleotide
- DHO
- dihydroorotate
- A77 1726
- 2-cyano-3-hydroxy-N-[4-(trifluoromethyl)phenyl]-2-butenamide
- brequinar
- 6-fluoro-2-(2′-fluoro-1,1′-biphenyl-4-yl)-3-methyl-4-quinolinecarb oxylic acid
- DSM1
- 5-methyl-[1,2,4]triazolo[1, 5-a]pyrimidin-7-yl)-naphthalen-2-yl-amine
- DSM2
- 5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-anthracen-2-yl-amine
- DSM74
- (5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7-yl)-(4-trifluoromethyl-phenyl)-amine
- DSM15
- 5-methyl-7-(naphthalene-2-ylthio)-[1,2,4]triazolo[1,5-a]pyrimidine
- DSM16
- 5-methyl-7-(naphthalene-2-yloxy)-[1,2,4]triazolo[1,5-a]pyrimidine
- PfDHODH-DSM1
- the x-ray structure of PfDHODHΔ384–413 bound to DSM1
- PfDHODH-DSM2
- the x-ray structure of PfDHODHΔ384–413 bound to DSM2
- PfDHODH-DSM74
- the x-ray structure of PfDHODHΔ384–413 bound to DSM74
- PfDHODH-A77
- the x-ray structure of PfDHODH bound to A77 17267
- hDHODH-bre
- the x-ray structure of hDHODH bound to brequinar
- hDHODH-A77
- the x-ray structure of hDHODH bound to A77 1726
- RMSD
- root mean square deviation
- SAR
- structure activity relationship.
REFERENCES
- 1.Greenwood B. M., Fidock D. A., Kyle D. E., Kappe S. H., Alonso P. L., Collins F. H., Duffy P. E. (2008) J. Clin. Invest. 118, 1266–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guerra C. A., Gikandi P. W., Tatem A. J., Noor A. M., Smith D. L., Hay S. I., Snow R. W. (2008) PLoS Med. 5, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White N. J. (2004) J. Clin. Invest. 113, 1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenthal P. J. (2008) N. Engl. J. Med. 358, 1829–1836 [DOI] [PubMed] [Google Scholar]
- 5.Gutteridge W. E., Trigg P. I. (1970) J. Protozool. 17, 89–96 [DOI] [PubMed] [Google Scholar]
- 6.Reyes P., Rathod P. K., Sanchez D. J., Mrema J. E., Rieckmann K. H., Heidrich H. G. (1982) Mol. Biochem. Parasitol. 5, 275–290 [DOI] [PubMed] [Google Scholar]
- 7.Jones M. E. (1980) Annu. Rev. Biochem. 49, 253–279 [DOI] [PubMed] [Google Scholar]
- 8.Nagy M., Lacroute F., Thomas D. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 8966–8970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Painter H. J., Morrisey J. M., Mather M. W., Vaidya A. B. (2007) Nature 446, 88–91 [DOI] [PubMed] [Google Scholar]
- 10.Goldenberg M. M. (1999) Clin. Ther. 21, 1837–1852 [DOI] [PubMed] [Google Scholar]
- 11.Olsen N. J., Stein C. M. (2004) N. Engl. J. Med. 350, 2167–2179 [DOI] [PubMed] [Google Scholar]
- 12.Baldwin J., Farajallah A. M., Malmquist N. A., Rathod P. K., Phillips M. A. (2002) J. Biol. Chem. 277, 41827–41834 [DOI] [PubMed] [Google Scholar]
- 13.Copeland R. A., Marcinkeviciene J., Haque T. S., Kopcho L. M., Jiang W., Wang K., Ecret L. D., Sizemore C., Amsler K. A., Foster L., Tadesse S., Combs A. P., Stern A. M., Trainor G. L., Slee A., Rogers M. J., Hobbs F. (2000) J. Biol. Chem. 275, 33373–33378 [DOI] [PubMed] [Google Scholar]
- 14.Hurt D. E., Widom J., Clardy J. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 312–323 [DOI] [PubMed] [Google Scholar]
- 15.Liu S., Neidhardt E. A., Grossman T. H., Ocain T., Clardy J. (2000) Structure 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 16.Baldwin J., Michnoff C. H., Malmquist N. A., White J., Roth M. G., Rathod P. K., Phillips M. A. (2005) J. Biol. Chem. 280, 21847–21853 [DOI] [PubMed] [Google Scholar]
- 17.Gujjar R., Marwaha A., El Mazouni F., White J., White K. L., Creason S., Shackleford D. M., Baldwin J., Charman W. N., Buckner F. S., Charman S., Rathod P. K., Phillips M. A. (2009) J. Med. Chem. 52, 1864–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phillips M. A., Gujjar R., Malmquist N. A., White J., El Mazouni F., Baldwin J., Rathod P. K. (2008) J. Med. Chem. 51, 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malmquist N. A., Gujjar R., Rathod P. K., Phillips M. A. (2008) Biochemistry 47, 2466–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srivastava I. K., Morrisey J. M., Darrouzet E., Daldal F., Vaidya A. B. (1999) Mol. Microbiol. 33, 704–711 [DOI] [PubMed] [Google Scholar]
- 21.Boa A. N., Canavan S. P., Hirst P. R., Ramsey C., Stead A. M., McConkey G. A. (2005) Bioorg. Med. Chem. 13, 1945–1967 [DOI] [PubMed] [Google Scholar]
- 22.Heikkilä T., Ramsey C., Davies M., Galtier C., Stead A. M., Johnson A. P., Fishwick C. W., Boa A. N., McConkey G. A. (2007) J. Med. Chem. 50, 186–191 [DOI] [PubMed] [Google Scholar]
- 23.Heikkilä T., Thirumalairajan S., Davies M., Parsons M. R., McConkey A. G., Fishwick C. W., Johnson A. P. (2006) Bioorg. Med. Chem. Lett 16, 88–92 [DOI] [PubMed] [Google Scholar]
- 24.Patel V., Booker M., Kramer M., Ross L., Celatka C. A., Kennedy L. M., Dvorin J. D., Duraisingh M. T., Sliz P., Wirth D. F., Clardy J. (2008) J. Biol. Chem. 283, 35078–35085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakaschus G., Löffler M. (1992) Biochem. Pharmacol. 43, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 26.DeLano W. L.The Pymol Molecular Graphics System (2000) DeLano Scientific, San Carlos, CA [Google Scholar]
- 27.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 28.McCoy A. J. (2007) Acta Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P., Cowtan K. (2004) Acta Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 31.Cowtan K. (1998) Acta Crystallogr. 54, 750–756 [DOI] [PubMed] [Google Scholar]
- 32.Kleywegt G. J., Jones T. A. (1994) CCP4/ESF-EACBM Newsletter on Protein Crystallography 31, 9–14 [Google Scholar]
- 33.Kabsch W., Kabsch H., Eisenberg D. (1976) J. Mol. Biol. 100, 283–291 [DOI] [PubMed] [Google Scholar]
- 34.Kleywegt G. J., Jones T. A. (1997) Methods Enzymol. 277, 525–545 [DOI] [PubMed] [Google Scholar]
- 35.Kleywegt G. J. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1878–1884 [DOI] [PubMed] [Google Scholar]
- 36.Altomare A., Burla C., Camalli M., Cascarano L., Giacovazzo C., Guagliardi A., Moliterni A., Polidori G., Spagna R. (1999) J. Appl. Crystallogr. 32, 115–119 [Google Scholar]
- 37.Sheldrick G. (1997) SHELXL97, University of Gottingen, Gottingen, Germany [Google Scholar]
- 38.Waasmaier D., Kirfel A. (1995) Acta Crystallogr. A 51, 416 [Google Scholar]
- 39.Flack H. (1983) Acta Crystallogr. A39, 876–881 [Google Scholar]
- 40.Farrugia L. (1997) Appl. Crystallogr. 30, 565 [Google Scholar]
- 41.March J. (2006) Advanced Organic Chemistry: Reactions, Mechanisms, and Structure, 4th ed., p. 21, Wiley India Pvt. Ltd. [Google Scholar]
- 42.Baumgartner R., Walloschek M., Kralik M., Gotschlich A., Tasler S., Mies J., Leban J. (2006) J. Med. Chem. 49, 1239–1247 [DOI] [PubMed] [Google Scholar]
- 43.Walse B., Dufe V. T., Svensson B., Fritzson I., Dahlberg L., Khairoullina A., Wellmar U., Al-Karadaghi S. (2008) Biochemistry 47, 8929–8936 [DOI] [PubMed] [Google Scholar]
- 44.McCammon J. A. (2005) Biochim. Biophys. Acta 1754, 221–224 [DOI] [PubMed] [Google Scholar]
- 45.Hornak V., Simmerling C. (2007) Drug Discov. Today 12, 132–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teague S. J. (2003) Nat. Rev. Drug Discov. 2, 527–541 [DOI] [PubMed] [Google Scholar]
- 47.Amaro R. E., Baron R., McCammon J. A. (2008) J. Comput. Aided Mol. Des. 22, 693–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.B-Rao C., Subramanian J., Sharma S. D. (2009) Drug Discov. Today 14, 394–400 [DOI] [PubMed] [Google Scholar]
- 49.Bowman A. L., Lerner M. G., Carlson H. A. (2007) J. Am. Chem. Soc. 129, 3634–3640 [DOI] [PubMed] [Google Scholar]
- 50.Chène P. (2008) Drug Discov. Today 13, 522–529 [DOI] [PubMed] [Google Scholar]
- 51.Crespo A., Fernández A. (2008) Mol. Pharm. 5, 430–437 [DOI] [PubMed] [Google Scholar]
- 52.Burley S. K., Petsko G. A. (1985) Science 229, 23–28 [DOI] [PubMed] [Google Scholar]
- 53.Kim H., Lipscomb W. N. (1991) Biochemistry 30, 8171–8180 [DOI] [PubMed] [Google Scholar]
- 54.Knecht W., Löffler M. (1998) Biochem. Pharmacol. 56, 1259–1264 [DOI] [PubMed] [Google Scholar]
- 55.Ullrich A., Knecht W., Fries M., Loffler M. (2001) Eur. J. Biochem./FEBS 268, 1861–1868 [PubMed] [Google Scholar]
- 56.Copeland R. A., Davis J. P., Dowling R. L., Lombardo D., Murphy K. B., Patterson T. A. (1995) Arch. Biochem. Biophys. 323, 79–86 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.