Abstract
We carried out a test sample study to try to identify errors leading to irreproducibility, including incompleteness of peptide sampling, in LC-MS-based proteomics. We distributed a test sample consisting of an equimolar mix of 20 highly purified recombinant human proteins, to 27 laboratories for identification. Each protein contained one or more unique tryptic peptides of 1250 Da to also test for ion selection and sampling in the mass spectrometer. Of the 27 labs, initially only 7 labs reported all 20 proteins correctly, and only 1 lab reported all the tryptic peptides of 1250 Da. Nevertheless, a subsequent centralized analysis of the raw data revealed that all 20 proteins and most of the 1250 Da peptides had in fact been detected by all 27 labs. The centralized analysis allowed us to determine sources of problems encountered in the study, which include missed identifications (false negatives), environmental contamination, database matching, and curation of protein identifications. Improved search engines and databases are likely to increase the fidelity of mass spectrometry-based proteomics.
Introduction
Liquid chromatography mass spectrometry (LC-MS) has become the most popular technique for proteomics analysis. In this strategy, proteins of a sample are typically separated by PAGE and then digested with trypsin. Following extraction from the gel, peptides are separated by LC, and upon elution are ionized via electrospray into the mass spectrometer for characterization by mass analysis. The mass spectrometer subsequently selects peptides for fragmentation to yield mass values that are then used to identify the peptide and the corresponding protein by searching sequence databases. This technique, termed tandem MS, is repeated to continuously select ionized peptides from the LC column. Depending on protein abundance and complexity, the mass spectrometer type and its set-up, up to about 15,000 peptides, and up to about 4,000 proteins can be identified in a single experiment1.
Despite the high-mass accuracy of modern mass spectrometers, the general perception of the reliability of MS-based proteomics is that it is low. Previous test sample studies have demonstrated that there is both a lack of reproducibility between different laboratories as well as a general inability to identify purified proteins in samples of low complexity2 (http://www.abrf.org/ResearchGroups/ProteomicsStandardsResearchGroup/EPosters/ABRFs PRGStudy2006poster.pdf). This is in part due to the stochastic nature of peptide sampling by the mass spectrometer and the inherent bias towards peptides of higher concentrations, which further confounds the statistical challenges and pitfalls associated with MS-based analyses, particularly when samples are rich in protein complexity. Protein solubilization, protein separation, protease digestion, peptide separation and peptide selection, all involve steps and protocols that vary greatly among laboratories, and different commercially available tandem mass spectrometers have different mass accuracies and different rates of peptide selection for fragmentation. The use of different search engines to decode tandem mass spectra and match them to databases of theoretical tryptic peptides is also a source of variability3, due to differences in the search engines themselves as well as different levels of false discovery rates4,5. Furthermore, the matching of high quality tandem mass spectra to different databases may lead to irreproducibility since protein databases vary greatly in terms of their curation, completeness, and comprehensiveness6-8. Despite variability in instruments, search engines, and databases, the high mass accuracy of modern mass spectrometers9 should assure a 100% success rate of protein identification for those tryptic peptides that readily ionize and for which high quality tandem mass spectra can be obtained.
Prior work in analytical chemistry and genomics10-14 has demonstrated the benefits of standardized test sample efforts for testing the reproducibility of technology platforms. To address the question of reproducibility in LC-MS-based proteomics, 15 the Human Proteome Organization (HUPO) created a test samples working group to carry out a controlled study involving 27 different labs. We produced a test sample made up of 20 human proteins of high purity and at equimolar ratios. To test for any potential stochastic bottleneck as a consequence of current data-dependent acquisition methods16, all 20 proteins were selected to contain at least one unique tryptic peptide of 1250 ± 5 Da each with a different amino acid sequence. The primary task given to the 27 labs was to identify all 20 human proteins and all unique peptides (22) of mass 1250 ± 5 Da, and to report these to the lead investigator, AWB. We encouraged the labs to use whatever optimized procedures and instrumentation they routinely employed, without constraints, which would allow us to assess any trends in those procedures or instruments which were the most effective. We had the labs utilize the same version of the NCBI nr human protein database (Nov 27, 2006) so as to minimize variability in data matching and reporting.
For the first time in a proteomics test sample study, each of the participating laboratories is publicly identified here, though all data have been rendered anonymous to prevent tracking to any individual lab. This test sample experiment goes beyond previous efforts as after the 27 labs initially reported their findings to us, we communicated back to them the potential sources of misidentification such that most errors could be corrected. Furthermore, we requested that each lab deposit all raw data, methodology, peak lists, peptide statistics, and protein identifications into Tranche17 for subsequent submission to PRIDE18. The availability of the raw data enabled us to perform a centralized analysis of all data. Such subsequent analysis showed that even though most participating labs initially failed to report all 20 proteins and the 22 1250-Da peptides correctly, their raw data clearly indicates that most participants should have been able to identify all 20 proteins as well as most of the 22 1250-Da peptides.
Results
Test sample proteins
To create the test sample, we selected 20 proteins in the MW range 32-110kDa from the ORF19 and MGC20 collections (Supplementary Methods online). The criteria (Supplementary Fig. 1a online) for selection included a purity of ca. 95%, unique tryptic peptide sequences, and the presence of at least one tryptic peptide of 1250 ± 5 Da (Supplementary Fig. 1b and c online). We expressed the candidate proteins in E. coli and purified them following a production strategy by employing ion exchange and reverse phase chromatography or by preparative electrophoresis purification from inclusion bodies (Supplementary Methods). 1D-SDS PAGE revealed the purity of the 20 purified proteins (Supplementary Fig. 1d online) at 95% or greater (Supplementary Table 1 online) as evaluated by densitometry (Supplementary Fig. 2 and Supplementary Table 2 online). MS analysis of the 20 purified proteins revealed a vector derived N-terminal extension of 7 amino acids present on each of the proteins (Supplementary Fig. 3 online). MS analysis of the test sample confirmed quality (Supplementary Fig. 4 and Supplementary Tables 2 and 3 online) and stability (Supplementary Fig. 5 and Supplementary Table 4 online) prior to distribution to the 27 labs.
Protein identification
We selected the NCBInr human protein database of November 27, 2006 with exact matches for all 20 test sample proteins (see Supplementary Fig. 6 and Supplementary Table 5 online) for protein identification. We instructed the 27 selected labs to use this database. The individual results from the labs are reported in Supplementary Table 6 online and are summarized in Table 1. Analysis of the reports revealed clear differences in the number of tandem MS assigned based on the instrument employed (Supplementary Fig. 7 online) however, incorrect reporting of false positive and contaminating proteins were not specifically linked to any mass spectrometry platform or search engine.
Table 1. Initial Results of Test Sample reporting of 24 academic laboratories (1-24) and 3 vendors (A-C).
Groups I-IV identify those labs who scored 100% (group I), those with naming (N) errors (group II), and those with naming errors as well as false positive, contaminant and redundant identifications (group III). Group IV includes labs with these errors as well as errors attributed to acrylamide alkylation (AC), database searching (DB), excessive stringency (ST), under-sampling (A) or trypsinization (TR) related errors.
| Laboratory / Vendor | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | B | 9 | 1 0 |
11 | 1 2 |
1 3 |
1 4 |
1 5 |
1 6 |
1 7 |
1 8 |
1 9 |
2 0 |
2 1 |
2 2 |
C | 2 3 |
2 4 |
||
| Gene Symbol |
MW (kDa) |
Group I | Group II | Group III | Group IV | |||||||||||||||||||||||
| KHK | 33 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | D B |
+ | + | + | + | + |
| ATPAF2 | 33 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | S T |
N | T R |
+ | T R |
| SETD3 | 34 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | D B |
+ | + | + | + | + |
| SPRY2 | 35 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | T R |
| GLB1L3 | 35 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | N | + | N | + | + | T R |
+ |
| FYTTD1 | 36 | + | + | + | + | + | + | + | + | + | + | N | N | N | N | + | + | N | N | N | N | + | + | + | + | T R |
+ | T R |
| IHPK1 | 50 | + | + | + | + | + | + | + | + | + | N | + | + | + | N | + | + | + | + | N | N | + | + | + | + | + | S T |
T R |
| IFRD1 | 50 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | N | + | + | N | + | + | N |
| GCNT3 | 51 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | T R |
+ |
| EIF2S3 | 51 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | A | + | + | + |
| F2 | 70 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | N | + |

|
+ | + | A | T R |
S T |
T R |
| FARP2 | 73 | + | + | + | + | + | + | + | + | + | + | + | + | + | N | + | + | + | + | + | N | + | + | + | + | + | + | + |
| ENOX1 | 73 | + | + | + | + | + | + | + | N | + | N | + | N | N | + | + | + | N | + | N | N | + | N | + | + | + | + | N |
| KLHL13 | 74 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | N | + | + | + | N | + | S T |
+ |
| NIBP | 101 | + | + | + | + | + | + | + | + | N | + | N | + | + | + | + | N | + | N | + | + | + | + | + | + | + | + | + |
| MARS | 101 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | T R |
+ |
| NUP210 | 106 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| THBS4 | 106 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | S T |
A | T R |
+ | + |
| KIAA0746 | 112 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | N | N | S T |
+ | T R |
T R |
+ |
| HIRA | 112 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | N |
| Reported Correct False Positives Contaminants Redundant |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
20 | 2 0 |
2 4 |
2 2 |
2 1 |
2 2 |
3 0 |
2 5 |
2 2 |
2 2 |
2 1 |
2 1 |
2 0 |
1 8 |
1 8 |
| 2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
2 0 |
1 9 |
1 9 |
1 8 |
1 8 |
1 8 |
18 | 1 7 |
2 0 |
1 9 |
1 8 |
1 8 |
1 6 |
1 5 |
1 6 |
1 6 |
1 6 |
1 4 |
1 5 |
1 3 |
1 2 |
|
| 1 | 1 | 1 | 1 | ||||||||||||||||||||||||
| 3 | 2 | 1 | 2 | 1 | 1 | 1 | 3 | 1 | 3 | 5 | 5 | 3 | |||||||||||||||
| 1 | 8 | 3 | 2 | 1 | 2 | ||||||||||||||||||||||
| Score (%) | 100 | 9 0 |
9 0 |
8 1 |
8 1 |
8 1 |
81 | 7 2 |
8 3 |
8 2 |
7 7 |
7 4 |
4 2 |
4 5 |
5 8 |
5 8 |
6 1 |
4 7 |
5 6 |
4 7 |
4 0 |
||||||
| SDS-PAGE Mass Spect Peaklist Sofware Search Engine |
+ | + | + | + | + | + | + | + | + | ||||||||||||||||||
| Q T |
I T |
I T |
Q T |
I T |
H | T T |
Q T |
Q T |
Q T |
I T |
I T |
H | Q T |
H | H | Q T |
H | I T |
H | IT | IT | I T |
T T |
Q T |
H | I T |
|
| P | D | E | M | X | E | E x |
D i |
Di | P P |
E | B | DT A |
P P |
X c |
E x |
D i |
X c |
B | E | S p |
B | X c |
E x |
S p |
S p |
S p |
|
| O | M | S | S p |
M | M | M | M | M | O | M | S | M | O | S | M | M | O | S | S | S p |
S | O | M | S p |
S p |
S | |
| Turn-around | 1 1 |
1 6 |
6 1 |
7 0 |
9 0 |
1 1 4 |
5 1 |
4 1 |
1 0 3 |
2 2 |
5 3 |
4 2 |
70 | 4 4 |
8 9 |
9 3 |
8 5 |
9 2 |
6 3 |
1 0 8 |
4 6 |
4 8 |
6 9 |
5 7 |
1 4 |
4 8 |
6 3 |
Initially, only 7 labs (classified as Group I) correctly identified all 20 proteins (Table 1). The labs classified as Group II encountered naming errors. Labs classified as Group III encountered naming errors, false positive and redundant identifications (Supplementary Fig. 8 and Supplementary Table 7 online). No redundant identifications were reported by any lab that used the Mascot (Matrix Science) search engine (n=11) whereas labs using Sequest and SpectrumMill did report redundant identifications. Labs classified as Group IV encountered a number of problems. We distributed fresh samples to labs which had indicated trypsinization problems (labs C, 23, 24; Supplementary Table 8 online). Lab 22, which had a problem with undersampling, (Supplementary Table 9 online) performed a further analysis with their remaining sample. Other errors encountered by Group IV included incomplete matching of tandem MS due to acrylamide alkylation (Supplementary Fig. 9 online), database search errors (Supplementary Table 10 online), and overly stringent identification criteria (Supplementary Table 11 online), all of which resulted in missed identifications. We devised a scoring system to take incorrect reporting into account. After we discussed the problems with each laboratory (Supplementary Table 12 online) and in some cases had them perform repeat analyses, all labs identified all 20 proteins, achieving a uniform score of 100% (not shown).
Peptide Sampling
We also assessed the completeness of peptide sampling and selection in the mass spectrometer by assessing the ability of the 27 labs to detect the 22 designed tryptic peptides of mass 1250 ± 5 Da (Supplementary Table 13 online), 6 of which contained cysteine residues whose mass increases as a consequence of reduction and alkylation as routinely employed prior to protein trypsinization. Initially, only one lab reported detection of all 22 peptides (Table 2) and only a further 3 reported detecting any peptides that contained cysteines. Peptides of mass 1250 ± 5 Da derived from contaminating proteins were incorrectly reported by several groups. Several groups also reported peptides in the 1250 ± 5 Da mass range as a result of a single missed trypsin cleavage (denoted as a true positive). We requested that these labs perform a reassessment as described above for protein reporting.
Table 2. Designed Peptide Mass Complexity Reporting of 24 academic laboratories and 3 vendors.
Initial (grey shading) and updated (black shading) reporting of peptides of mass 1250 ± 5 Da. Analysis scoring was calculated from the fraction of correct peptide identifications and the accuracy of reporting peptides of mass 1250 Da, whereas the Report Scoring was based on the fraction of correct peptide identifications reported ÷ the number identified by the centralized analysis. Results not reported (NR); no raw data (NRD); submitted and data reprocessing difference (DRD). DRDs are indicated by fewer peptides identified by the centralized analysis as compared to the number reported.
| Laboratory / Vendor | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol |
A | 1 | 2 | 3 | 4 | 5 | 6 b |
7 | 8 | B | 9 | 1 0 |
1 1 |
1 2 |
1 3 |
1 4 |
1 5 |
1 6 |
1 7 |
1 8 |
1 9 |
2 0 |
2 1 |
2 2 |
C | 2 3 |
2 4 |
2 2 R |
C R |
2 3 R |
2 4 R |
|
| KHK | + | + | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | +c d |
+ | |||||||||||||||
| ATPAFa | +c | + c |
+c | + c |
+ c |
+ c |
+ c |
+ c |
+ c |
+c | ||||||||||||||||||||||
| SETD3 | + | + e |
+ | + | + e |
+ | + | + | + | |||||||||||||||||||||||
| SPRY2 | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||||||
| GLB1L3 | + | + | + e |
+ e |
+ e |
+ | + | + | + | + | ||||||||||||||||||||||
| FYTTD1 | +e | + | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||||
| IHPK1a | +c | +c | + c |
+ c |
+ c |
+ e |
+ cd |
+ e |
+c d |
+c d |
||||||||||||||||||||||
| IFRD1a | +c | + c |
+ c |
+ e |
+ e |
+ c |
+ c |
+c | +c | |||||||||||||||||||||||
| GCNT3 | + | + | + | + | + | + | + e |
+ e |
+ | + | + | + | + | +f | + | |||||||||||||||||
| EIF2S3 | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||
| F2 | + | + | + | + | + | + | + | + | + | + | + | + | + | +c d |
+ | |||||||||||||||||
| FARP2 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| ENOX1 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| KLHL13 | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| NIBPa | +c | + e |
+ e |
+ cd |
+c d |
|||||||||||||||||||||||||||
| MARS | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||
| NUP210 | + | + | + | + | + | + | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | |||||||
| NUP210a | +c d |
+c | + e |
+ c |
+ c |
+ c |
||||||||||||||||||||||||||
| THBS4 | + | + | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | + | + | |||||||||||||||
| KIAA0746 | +e | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||||||
| HIRA | + | + | + | + e |
+ | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||||||||||
| HIRAa | +c | + c |
+ c |
+ c |
+ c |
+c | ||||||||||||||||||||||||||
| Total (initial) |
1 5 |
9 | 3 | 1 6 |
1 2 |
1 5 |
9 | 1 4 |
3 | 1 1 |
1 2 |
1 0 |
1 5 |
4 | 1 0 |
2 2 |
1 4 |
8 | 1 1 |
1 5 |
1 | 8 | 8 | 4 | 0 | 7 | 1 2 |
0 | 1 3 |
|||
| Total (final) | 1 8 |
1 0 |
2 2 |
1 6 |
1 9 |
1 1 |
1 8 |
1 1 |
2 2 |
1 1 |
2 | 1 1 |
1 6 |
|||||||||||||||||||
| True Positived |
3 | 1 | 3 | 1 | 3 | |||||||||||||||||||||||||||
| False Positives |
||||||||||||||||||||||||||||||||
| Contamina nts |
1 | 1 | 6 | 1 | 3 | 1 | 3 | |||||||||||||||||||||||||
| Analysis Scoring |
||||||||||||||||||||||||||||||||
| Initial score (%) |
6 8 |
4 1 |
1 4 |
6 8 |
5 5 |
6 4 |
4 1 |
5 9 |
1 4 |
5 0 |
5 5 |
4 5 |
4 9 |
1 8 |
4 1 |
1 0 0 |
6 4 |
3 6 |
5 0 |
5 7 |
2 | 2 6 |
3 6 |
1 8 |
0 | N R |
3 2 |
5 5 |
0 | 5 9 |
||
| Final score (%) |
8 2 |
4 5 |
1 4 |
1 0 0 |
7 3 |
6 4 |
4 1 |
8 6 |
1 4 |
5 0 |
5 5 |
4 5 |
8 2 |
1 8 |
5 0 |
1 0 0 |
6 4 |
3 6 |
5 0 |
5 7 |
2 | 2 6 |
3 6 |
1 8 |
9 | N R |
5 0 |
7 3 |
0 | 5 9 |
||
| Report Scoring |
||||||||||||||||||||||||||||||||
| Centralized (count) |
1 0 |
1 5 |
2 0 |
1 9 |
1 8 |
2 1 |
1 0 |
1 9 |
5 | 2 1 |
1 9 |
2 2 |
2 2 |
6 | 1 7 |
2 2 |
1 8 |
1 3 |
1 4 |
2 2 |
6 | 1 6 |
1 6 |
3 | N R D |
2 | 1 4 |
1 5 |
1 1 |
1 8 |
||
| Score (%) | D R D |
6 7 |
1 5 |
D R D |
8 9 |
7 1 |
9 0 |
1 0 0 |
6 0 |
5 2 |
6 3 |
4 5 |
8 2 |
6 7 |
6 5 |
1 0 0 |
7 8 |
6 2 |
8 5 |
6 8 |
1 7 |
5 0 |
5 0 |
D R D |
N R D |
N R |
7 9 |
D R D |
0 | 7 2 |
||
Cysteine containing peptides.
iTRAQ labeling employed.
Alkylated peptide reported.
Reported peptide contains 1 missed trypsin cleavage site.
Peptides assigned at <95% confidence reported.
Methionine residue oxidized.
We used our scoring system to assess both the analysis and the reporting of the 1250 ± 5 Da tryptic peptides. Initially, only lab 14 achieved 100%. After guidance, lab 3 achieved 100% success by correcting for cysteine containing peptides and excluding peptides derived from contaminants. All other labs reported insufficient data. To distinguish between incomplete reporting and incomplete sampling, we compared the 1250-Da peptides that were reported to those that were identified by the centralized analysis (see below). Labs 10, 11, 14, and 18 (but not lab 3) were found to have data for all 22 1250-Da peptides. However, labs 10, 11, and 18 were unable to report the peptides and our centralized analysis failed to identify the 22 peptides in the data from lab 3 (Table 2). Besides lab 14, only lab 7 achieved 100% reporting of all 1250-Da peptides in their data set (a total of 19 peptides, as assessed by our centralized analysis of the data) (Table 2, Supplementary Table 13).
Data deposition to Tranche and PRIDE
We asked the 27 labs to transfer their raw MS data, the methodologies used, peak lists, peptide statistics, and protein identifications to Tranche, a repository for raw data. Initial problems related to the transfer of data to Tranche were all overcome. Tranche hash and passphrase codes are available in Supplementary Table 14 online. A copy of all data was transferred from Tranche to PRIDE, a centralized public data repository for the standardized reporting of proteomics results, by PRIDE personnel. As evaluated by PRIDE personnel, the initially deposited data had several problems including incomplete files, proprietary software formats and screenshots of data displays in software rather than actual data files. The wide variety of data formats encountered faithfully represents the heterogeneity in the field concerning proteomics bioinformatics. It also appears that the implementation of community standards for data reporting and exchange is not yet at a level that accommodated the minimal requirements for these 20 test proteins.
Centralized Analysis of the Raw Data
To independently assess the individual analyses of the 27 labs, we downloaded all raw data from Tranche. We reanalyzed the collective raw data centrally using a uniform protocol of database searching using X! Tandem21 and post-processing with the Trans Proteomic Pipeline 22 to assign probabilities to all identifications and global false discovery rates as well as to determine the total number of tandem MS assigned, number of distinct peptides and amino acid sequence coverage (Supplementary Tables 13 and 15 online).
We found that the majority of the labs had in fact generated raw data of sufficient quality to identify all 20 proteins and most of the 22 1250-Da peptides. We identified discrepancies between the submitted results (Supplementary Table 12) and the centrally reprocessed results (Supplementary Table 15) for labs 2, 4, 5, 8, 10, 11, 16, 19, 20, 21, 22R, 24 and CR, largely due to the different data analysis strategies that these labs used. The centralized analysis included checks for experimental artifacts including pyroGlu formation, deamidations, and non-tryptic cleavages.
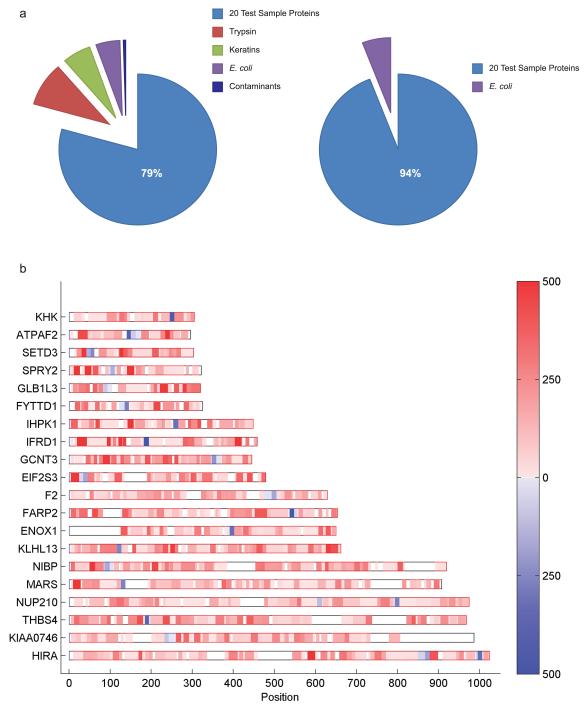
For all 27 labs, the majority of tandem mass spectra (79%) were assigned to the 20 recombinant human proteins, but 21% of the spectra were assigned to contaminants that included E. coli proteins, trypsin, keratins, and other proteins (Fig. 1a left side, Supplementary Table 15). The centralized analysis also revealed that all 22 predicted tryptic peptides of 1250 Da were observed by only 4 labs, three of which used an FTICR instrument (Tables 1 and 2). These instruments reported the highest number of assigned tandem mass spectra, thereby increasing the likelihood of identifying all of the 1250-Da peptides (Supplementary Fig. 7). Tandem mass spectra matching the 1250-Da peptides were variable for each of the 20 proteins (Fig. 1b) and were variably detected in our centralized analysis (Supplementary Fig. 10 online).
Figure 1. Number of tandem mass spectra assigned to tryptic peptides.
(a) Comparison of protein abundances (% total redundant peptides) from the centralized analysis of the raw data collected from the 27 labs (left side) and (right side) after removal of individual lab contaminants including keratins as well as trypsin. (b) Peptide heat map representation for each of the 20 proteins (gene symbol) from the centralized analysis of the raw data from all 27 labs, revealing the frequency of observation of a given peptide as well as its position in the protein sequence. Blue, the 1250 Da peptides; red, all other tryptic peptides.. Raw data from lab 24 was excluded (see Online Methods). Scale bar represents the number of redundant peptides. Scale bar is linear from 1 to 500 peptides.
The centralized analysis also revealed a) that the majority of tandem MS assigned to keratins (human keratins KRT1, KRT2, KRT9 and KRT10 are commonly found in mature epidermal tissue and are also present in laboratory dust and fingerprints, rather than hair or wool derived keratins) were largely attributed to strategies that employed 1D-PAGE (Supplementary Fig. 11, Supplementary Table 15); b) that E. coli proteins were found by all but 2 labs (Supplementary Fig. 11, Supplementary Table 15 online) and most likely were present in the provided sample; c) that other protein contaminants (e.g. albumin, casein) were found in datasets from a specific subset of labs (5 found albumin, 5 casein, and 3 both proteins; albumin was incorrectly reported as human when in fact it was bovine, and both bovine serum albumin and casein are likely abundant proteins used in these labs for standardization); and d) that autolytic trypsin peptides resulted from added trypsin. Excluding the contaminants introduced by the labs, 94% of the tandem mass spectra were accounted for by the 20 recombinant proteins, and the remaining tandem MS were assigned to the E. coli proteins (Fig 1a right side). False negatives (one or more of the 20 recombinant proteins not detected) were likely a consequence of variability in trypsin digestion and the stochastic sampling of the mass spectrometry analysis.
Laboratories that used exclusively liquid phase separations in general had fewer spectra that could be assigned to epidermal keratins than laboratories that used a combination of protein separation by gel electrophoresis followed by in-gel digestion, peptide extraction and HPLC peptide separation prior to MS/MS analysis (Supplementary Fig. 11). This trend is probably caused by the fact that each gel slice was exposed to the environment individually, effectively increasing the load of environmental contaminants. The number of spectra that could be assigned to keratins was also broadly correlated with the identification of low-concentration sample source contaminants (E. coli proteins) and reagent proteins (trypsin), suggesting that in most cases these proteins were present at significantly lower concentrations than the 20 test sample proteins (Supplementary Table 15).
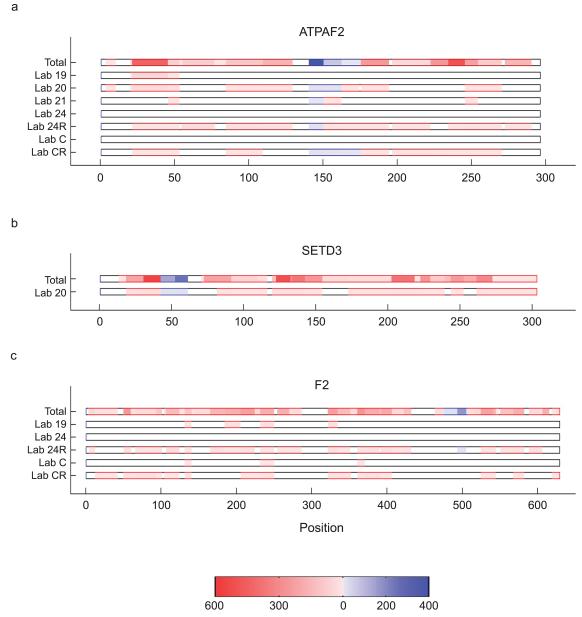
Our centralized analysis confirmed that raw data initially reported by 4 labs were incomplete (Supplementary Table 15). Repeat analysis by these labs generated sufficient data to identify the 20 proteins. As seen in Fig. 2, no tandem mass spectra were initially observed for the ATAF2 protein by labs 24 and C, but in a repeat analysis, these labs generated sufficient tandem mass spectra (marked as 24R and CR) to characterize the protein as well as the 1250-Da peptide. However, labs 19, 20 and 21 generated sufficient tandem mass spectra for protein ATPAF2, lab 20 generated sufficient tandem mass spectra for protein SETD3 and labs 19 and C generated sufficient tandem mass spectra for protein F2, but still did not initially report the identification of these proteins. We determined that lab 20 had a database problem for protein SETD3 and lab 19 had an acrylamide modification problem for protein F2. Lab 24 had a trypsinization problem for protein F2, which was fixed upon repeat analysis (24R). Although lab C initially reported a trypsinization problem for the F2 protein, the raw data proved otherwise. Lab C’s repeat analysis (CR) revealed more tandem mass spectra assigned to protein F2 but insufficient data for the peptide of mass 1250 Da. Detailed central analysis of each lab’s data submitted to Tranche justified the removal of results of lab 24 (but not of this lab’s repeat analysis, 24R) from the heat map shown in Fig 1b. Inspection of the results for lab 24 (Supplementary Table 13) revealed that ~95% of the tandem mass spectra were assigned to peptides with cyclized N-terminal glutamine amino acid (pyroGln) which is not typical for analysis of tryptic peptides. Further in-depth analysis of the raw data failed to identify tandem mass spectra; aberrant chemically induced modifications may have been introduced.
Figure 2. Discrepancies between reported data and centralized analysis identify erroneous reporting.
Peptide heat map comparisons of the centralized analysis compiled for all 27 labs (Total), with the data from selected individual labs indicated below for the proteins (a) ATPAF2, (b) SETD3 and (c) F2. Blue, the 1250 Da peptides; red, all other tryptic peptides. Scale bar represents the number of redundant peptides. Missed cleavages account for the different degree of shading for peptides of mass 1250 Da.
Discussion
Our results demonstrate that, from a cross-section of 27 labs, only 7 labs were initially able to characterize an equimolar sample of 20 human proteins. However, our centralized analysis of the raw data demonstrates that each of the labs, with a few exceptions, had in fact generated mass spectrometry data of very high quality, more than sufficient to identify all 20 proteins and most of the 22 1250-Da peptides. This demonstrates the important need for education and training to properly apply such a complex technology.
Most notably, generic problems in databases were found to be the major hurdle for the correct characterization of proteins in the test sample. The search engines used here are currently unable to distinguish among different identifiers for the same protein, deriving from the way the databases are constructed. Indeed, the search engines used either for the centralized data analysis or by the individual labs suggest an erroneous confidence to the assignments of peptides and proteins. This erroneous confidence necessitates the use of manual verification of both the peptide assignments and protein assignments for low confidence identifications.
An extended standardized FASTA format (http://psidev.info/index.php?q=node/317) has been proposed by HUPO-PSI that would resolve the problem of standardized annotation. Currently, manual curation of tandem MS search results is needed for correct reporting. This includes the non-redundant assignments of tandem MS to overcome the common errors in the apparent characterization of different proteins that are one and the same. We have observed that algorithms used by different search engines to calculate molecular weight are variable (data not shown). It is therefore reasonable to suggest that a common method for calculating molecular weight be chosen and used throughout the community. Additionally, the automatic matching of tandem mass spectra of high quality to a protein coding genome with a single representative protein for each gene could overcome several of the current errors in protein naming and redundancies.
A test sample containing 20 proteins at 5 pmol equimolar abundance is not representative of a proteomics study with complex mixtures. However, a routine 100% success rate of protein and 1250-Da peptide identification of such a test sample could be implemented as a standard, as well as the routine deposition of raw data into Tranche. This would enable a greater degree of trust in the conclusions deduced for proteomics studies in general. A limited number of the 20 test sample protein mixtures have been prepared and are available by contacting the lead author (AWB). These samples, however, are stored in 7.5 M urea, which leads to variable carbamylation and this may affect trypsinization as well as data analysis. Such test samples should be helpful as a benchmarking tool for labs embarking on a proteomics study with complex mixtures. At the least, their abilities to collect sufficient data for unambiguous identification of 20 human proteins and 22 1250-Da peptides can be assessed. A peptide by peptide comparison for any individual lab with those from a centralized analysis of the data should be informative to the inability of any lab to detect proteins or specific peptides. For any large-scale, multi-laboratory proteomics effort, we recommend the use of a centralized analysis, especially if data is generated on more than one platform, location or collected over time.
Our study has allowed us to deduce a number of guidelines for performing any proteomics experiment. Sources of laboratory derived contamination need to be identified and monitored closely, with the two major sources being environmental contamination carried over from prior experiments, and keratins (largely from gel-based analysis). The use of target-decoy search strategies should be made mandatory, and false discovery rates should be reported. The monitoring of unique peptides and unique tandem mass spectra is needed to ensure that the minimum list of protein identifications is reported, in order to address the issue of redundant identifications (sequence variants of the same protein). A gene-centric database could ensure that only a single descriptive name would be assigned to each protein sequence, eliminating aliases. The creation of tools for transforming data (raw data, peak lists, peptide lists, and protein lists) into standardized formats would aid the ease of submission to repositories such as Tranche. The distribution of all data deposited in Tranche to the community, via PRIDE, Human ProteinPedia, PeptideAtlas and GPM, would facilitate centralized data analysis which may help lead to new insights in proteomics experiments.
In summary, our analysis shows that even with a sample consisting of highly purified human proteins, many participating laboratories had difficulties in reporting data correctly. However, the majority of the participants deposited raw data where each had more than sufficient coverage of the 20 proteins. Thus a major contributing factor to erroneous reporting resides at the level of database and search engines used and once corrected for, provided an almost perfect score for most participants. Therefore, we expect that once databases and search engines have been improved and made compatible with MS-based proteomics, the accuracy of data reporting will increase and along with it, the fidelity of proteomics.
Online Methods
Test sample generation and distribution
As more completely described in the Supplementary Methods, all test sample proteins were cloned23 and expressed24 in E. coli, purified from inclusion bodies under denaturing conditions, and mixed in equimolar (5 pmol) amounts. A committee made up of funding agency representatives (NIH, CIHR), journal editors and the HUPO Executive Committee proposed a list of 55 laboratories. Invitations were extended to 41 laboratories and 24 accepted to participate. Further, 6 mass spectrometer vendors were selected by the HUPO Industrial Advisory Board (IAB) and all agreed to participate but only 3 provided results. The 27 laboratories that participated are indicated here as co-authors. Dried samples containing 5 picomoles of each protein were shipped on dry-ice, along with detailed examples of LC-MS proteomics analyses (http://www.invitrogen.com/etc/medialib/en/filelibrary/pdf.Par.72904.File.dat/HumanProteinStandardsforMassSpectrometry.pdf). Samples were shipped from Invitrogen (Carlsbad, California) and deliveries were overnight by DHL (www.dhl-usa.com/) in the USA and DHL International or FEDEX International (http://fedex.com/us/) express overseas (1 to 3 business day delivery). Delivery to Australia was delayed on 2 occasions due to incomplete Customs-related documentation that resulted in the samples attaining ambient temperatures and hence their replacement. A further 2 samples were received at the recipient institutes but failed to arrive at the host laboratory. One vial was reported to be empty as negligible signal was observed by Coomassie blue staining of a 2D gel. In all cases, more material was supplied. Participants were instructed to use a specified NCBInr database (http://portal.proteomics.mcgill.ca:8080/hupo-standards/nr_human_20061127_v2.fasta.), to report details of methodologies employed and proteins identified, and to deposit raw data and reports to Tranche (http://tranche.proteomecommons.org/) (Supplementary Note online).
Instructions to laboratories and vendors
Test Samples were distributed to participating laboratories, who were instructed to i) identify the 20 human proteins, ii) report the details of the identifications (protein name, NCBI gi number, sequence coverage, number of peptides, and number of tandem MS) following the criteria of Carr et al.25, and iii) report the details of methodology. The following description of the sample was supplied: The sample is an equimolar mixture (5 picomoles) of 20 human proteins that were expressed in E. coli under conditions to maximize inclusion body formation. The expression system results in an N-terminal extension of 7 amino acids (sequence MYKKAGT) followed by the encoded initiator methionine. The 20 proteins were purified by preparative SDS PAGE or 2D-LC (anion exchange and reversed phase) to > 95% purity. Trypsin digestion of the purified constructs results in the generation of a tripeptide (MYK) plus free K or a tetrapeptide (MYKK) resulting from 1 missed cleavage and an N-terminal extension of 3 (AGT) or 4 (KAGT, 1 missed cleavage) amino acids. Contaminants do not exceed 1% in the final mixture. Details regarding the proteomics MS analysis as well as the selection and purification of the Test Sample proteins by Invitrogen were also supplied (poster presentation (http://www.invitrogen.com/etc/medialib/en/filelibrary/pdf.Par.72904.File.dat/HumanProteinStandardsforMassSpectrometry.pdf) that was presented at the HUPO 5th Annual World Congress (Long Beach, California)).
Protein identification reports were scored based on acceptable names as found in the specified database. For reassessment, each lab was instructed to make corrections based on naming, redundant, false positive and contaminant identifications, and acrylamide alkylation of cysteines. Labs that failed to achieve 100% after reassessment were requested to repeat the analysis of a fresh sample.
Reporting of peptides of mass 1250 ± 5 Da was requested, with reassessment as above, and reports were scored two-fold, for analysis and reporting completeness.
Database Selection
To limit variation in data evaluation, a single database, the NCBInr human protein database of November 27, 2006, was selected. The NCBInr database contained all 20 test proteins with their exact matches represented.
Previous efforts to benchmark proteomics through test samples have usually allowed participating laboratories to choose whatever database they felt might be the most appropriate to match their tandem mass spectra. As we have argued elsewhere6,26, most databases are still in a constant flux changing from one release to another. These changes lead to increased variation in data evaluation. Here, we compared the predicted amino acid sequence of the 20 test proteins selected as identified above with the NCBI non-redundant database, the Universal Protein Resource (UniProt) and the International Protein Index (IPI) databases. Comparisons were made by employing blastp (http://www.ncbi.nim.nih.gov/BLAST/). The reciprocal matching (database to ORF and ORF to database) process revealed differences in protein length as well as amino acid substitutions, most of which occurred in the IPI database and are likely to be related to the specific assembly process of the IPI27. Longer (blue shading) or shorter (pink shading) sequences in the database indicate extensions or truncations and/or differences in editing (removal of potential introns) the predicted DNA sequences. Amino acid substitutions are indicated by orange and green shading. An exact match is indicated by 100% identity in both directions. From this database assessment only the NCBI nr database had all recombinant proteins with their exact matches represented.
Data Reporting
The number of proteins reported and number correct are indicated as are the number of false positive (proteins identified by shared peptides) and contaminant (proteins not in the sample) identifications and those proteins identified more than once but reported as separate proteins (redundant). Subsequent to the initial reporting by the 27 labs (numbers and letters are used to identify academic labs and vendors, respectively), one of us (AWB) discussed with each lab problems associated with providing non-descriptive names (e.g. hypothetical protein, ORF), and also the reporting of redundant identifications, and false positive and contaminating proteins. Problems associated with spurious alkylation of cysteine residues by acrylamide during preparative electrophoresis were also discussed. Participants were requested to reassess search results and to submit updated final reports. A scoring system was devised to take into account incomplete reporting as well as erroneous identifications. The score (Table 1) was calculated as follows: score = fraction identified (number correct ÷ 20) × accuracy (number correct ÷ number reported) × 100. For Table 1, details for the proteomics analyses on a lab-by-lab basis including protein separation, mass spectrometer, peaklist software, and database search engine as well as turn-around-time (time from the lab receiving the sample until results were submitted by email (average 67 days)) are indicated. All laboratories employed trypsin. Mass spectrometers employed included: ion trap (IT); QToF (QT); hybrid (H) including LTQ-FT or LTQ-Orbitrap; and ToFToF (TT). Peaklists were generated by employing the following software: Bioworks Browser (Thermo Electron) (B); Data Analysis mzXML (D); Distiller (Matrix Science) (Di); DTA Supercharge (DTA); Extract_msn (Thermo Electron) (E); Explorer (Applied Biosystems) (Ex); Masslynx (Waters) (M); ProteinLynx Global Server (Waters) (P); Protein Pilot (Applied Biosystems) (PP); Spectrum Mill (Agilent) (Sp); X! Tandem (X); and Xcaliber (Thermo Electron) (Xc) and all labs employed default parameter with lab 5 including total ion current (TIC) threshold of 100 and a minimum of 10 peaks; and lab 7 including correlation threshold (CT) of 0.7, signal to noise ratio (SNR) of 20, reject width outliers and baseline correction. Database search engines included: Mascot (Matrix Science) (M); Sequest (Thermo Electron) (S); Spectrum Mill (Sp); and other (O) that include IdentityE (PLGS, http://www.waters.com), ProteinPilot (Applied Biosystems) or X!Tandem. All procedures used are reported in Tranche (Supplementary Table 14).
The methodology, the peak lists, the peptide statistics, and protein identifications were transferred to Tranche, a repository for raw data. Detailed instructions (see Supplementary Note online) were provided to each participating laboratory with regards to the preparation and transferring of supporting data and information to Tranche (http://www.proteomecommons.org/dev/dfs/examples/hupo-2007/Tranche-HUPO.jsp.). All problems in the transfer of data from host laboratories to Tranche (e.g. CD disk and courier transmission, firewall problems, unresponsive servers) were overcome. The successful transfer of data culminated with the generation of a Tranche hash and passphrase codes that are returned by email to the submitter and to one of us (AWB). The final set of codes is appended (Supplementary Table 14).
Transferring of peaklists, search results, peptide statistics, and protein identifications from Tranche to PRIDE by the PRIDE personnel has led to the successful transfer of 29 datasets (accession numbers: 8130-8158, inclusive). The data can be accessed by these accession numbers or by project name (HUPO test samples) from the ‘Browse experiments’ portal at PRIDE. The information in PRIDE comprises protein identifications and spectra from all the groups involved, and all the associated metadata.
Centralized Analysis of the Collective Data
To provide an independent assessment of all individual analyses, we reanalyzed all data collectively by using a uniform protocol of searching with X!Tandem21 and post-processing with the Trans Proteomic Pipeline22 to assign probabilities to all identifications and global false discovery rates.
Raw data and supporting documentation as deposited by each lab to Tranche were downloaded by employing Tranche hash and passphrase codes (Supplementary Table 14 online). For labs 01-05, 07, 09-14, 15_1, 16-21, 23R, 24, 24R and A, raw mass spectrometer output files were deposited in the native instrument vendor format. These files were transformed into the open XML format mzXML28. Labs 06, 08, 15_2, 22R, and B did not provide mass spectrometer output files, and in these cases the text-format peak list files were used in the centralized analysis. For labs C and CR, mzData files were submitted and used for the analysis. Lab A data were acquired in MSe29 mode that include low energy (MS scans) and high energy (fragmentation scans) scans without peptide ion selection. Standard processing techniques cannot be applied to the output MSe spectra because co-eluting peptide ions are fragmented simultaneously. For the centalized analysis, Lab A provided PKL files with time-deconvolved peaklists. These PKL files were converted to mzXML and processed in the same manner as the others. For Lab 7, the conversion from vendor format to mzXML did not sum consecutive scans, which would have resulted in approximately twice as many identified spectra. For this reason, the MGF files provided by the lab that already contained summed scans were used for the analysis.
All of the datasets were subjected to a uniform processing and validation in order to provide a homogeneous analysis environment in an attempt to minimize data processing differences among the groups. The tandem mass spectra were searched against a reference database constructed from a) the human IPI 3.50 protein list (www.ebi.ac.uk/IPI/); b) the non-redundant E. coli database distributed by NCI ABCC dated 2008-02-06 (ftp://ftp.ncifcrf.gov/pub/nonredun/); c) the cRAP set of common contaminant proteins from the Global Proteome Machine data base (GPMDB) dated 2008-10-01 (http://www.thegpm.org/cRAP/index.html); d) the 20 recombinant proteins present in the test samples with the vector-derived N-terminal extension of 7 amino acids; and e) finally an appended set of decoy proteins derived by scrambling all tryptic peptides in the target sequences described above. A copy of this constructed database is available at http://www.peptideatlas.org/tmp/HsIPI3.50_Ec_cRAP_20_TargetDecoy.fasta. The spectra were searched using the X! Tandem search engine21 with the K-score plugin30.
The search parameter files used for each experiment are available in the centalized reanalysis Tranche project file (Supplementary Table 14 online). In general, the search parameters were: 2 allowed missed cleavages, precursor m/z tolerance from −2.1 to +4.1, fragment m/z tolerance 0.4. Searches were performed with variable methionine oxidation, pyroGlu formation (from N-terminal Glu and Gln), and variable iodoacetamide and acrylamide modifications on cysteine, or iTRAQ modifications if appropriate. If the native data contained charge state information, it was used; when charge state information was not available, either +1 or both +2, +3 were searched. Consideration for potential ion pairs that might degrade MS-analysis (i.e., Glu and Asp residues in carboxylate form and ion-paired with Na+ or K+) revealed a negligible contribution, and these ion pairs were not included.
Validation of the search results was performed using the Trans Proteomic Pipeline (TPP) software suite22. The TPP tool PeptideProphet31 modeled the correct and incorrect spectrum assignments, calculating a probability of being correct to each match based on the models. The ProteinProphet tool32 was then used to adjust the identification probabilities based on corroborating evidence of other identifications that include tandem MS of similar matching characteristics but of lower quality within each dataset, and importantly, perform a protein-inference step that coalesces the identifications that map to multiple proteins into single consensus identifications. This processing and validation produced a high-quality set of identifications for each lab. A final centralized processing of all PeptideProphet results through a single ProteinProphet run yields a global picture of all proteins detected by the 27 labs in the mass spectrometry analyses.
Supplementary Material
Acknowledgements
Supported in part by Canadian Institutes of Health Research to the Human Proteome Organisation (HUPO) Head Quarters (S. Ouellette) for coordination of this HUPO test sample initiative. A. Bell and C. Au were supported by Genome Quebec and McGill University. We thank D. Juncker (McGill Univeristy, Montreal), G. Temple (NHGRI, NIH, Bethesda), J. van Oostrum (Chair, HUPO Industrial Advisory Board), G. Omenn (University of Michigan), K. Colwill (Mount Sinai Hospital, Toronto) and M. Hallett (McGill University) for their comments on the manuscript. We also thank Dr. Dominic M. Desiderio, University of Tennessee, for helpful comments on the manuscript. This test sample effort builds on pioneering efforts from several other groups and especially Association of Biomolecular Resource Facilities. This study is a HUPO test sample initiative and HUPO welcomes collaborative efforts to benefit proteomics. The following sources of grant support are acknowledged: E. W. Deutsch is supported by the National Heart, Lung, and Blood Institute, National Institutes of Health (NIH), under contract No. N01-HV-28179. The University of California, Los Angeles-Burnham Institute for Medical Research-NIH grant number RR020843; University of California, Los Angeles (NHLBI P01-008111); University College Dublin; access and use of The University College Dublin Conway Mass Spectrometry Resource instrumentation is acknowledged, supported by Science Foundation, Ireland Grant No. 04/RPI/B499; University of Michigan-NIH P41RR018627; PRIDE- J. A. Vizcaíno is a Postdoctoral Fellow of the “Especialización en Organismos Internacionales” program from the Spanish Ministry of Education and Science. L. Martens is supported by the “ProDaC” grant LSHG-CT-2006-036814 of the European Union. Samuel Lunenfeld Research Institute, Mount Sinai, Toronto is supported by Genome Canada through Ontario Genomics Institute. J. A. Vizcaíno and L. Martens would like to thank H. Hermjakob and R. Apweiler for their support. Finally, A. W. Bell thanks L. Roy (McGill University and Génome Québec Innovation Centre, Montreal), and Z. Bencsath-Makkai (McGill University) for help in data submission and analysis.
Appendix
A complete list of authors appears at the end of this paper:
HUPO Test Sample Working Group
Thomas A. Beardslee1, Thomas Chappell2, Gavin Meredith3, Peter Sheffield4, Phillip Gray5, Mahbod Hajivandi3, Marshall Pope3, Paul Predki3, Majlinda Kullolli6, Marina Hincapie6, William S. Hancock6, Wei Jia7, Lina Song7, Lei Li7, Junying Wei7, Bing Yang7, Jinglan Wang7, Wantao Ying7, Yangjun Zhang7, Yun Cai7, Xiaohong Qian7, Fuchu He7, Helmut E. Meyer8, Christian Stephan8, Martin Eisenacher8, Katrin Marcus8, Elmar Langenfeld8, Caroline May8, Steve Carr9, Rushdy Ahmad9, Wenhong Zhu10, Jeffrey W. Smith10, Samir M. Hanash11, Jason J. Struthers11, Hong Wang11, Qing Zhang11, Yanming An12, Radoslav Goldman12, Elisabet Carlsohn13, Sjoerd van der Post13, Kenneth E. Hung14, David A. Sarracino15, Kenneth Parker14, Bryan Krastins15, Raju Kucherlapati14, Sylvie Bourassa16, Guy G. Poirier17, Eugene Kapp18, Heather Patsiouras18, Robert Moritz18, Richard Simpson18, Benoit Houle19, Sylvie LaBoissiere20, Pavel Metalnikov21, Vivian Nguyen22, Tony Pawson22, Catherine C. L. Wong23, Daniel Cociorva23, John R. Yates III23, Michael J. Ellison24, Ana Lopez-Campistrous24, Paul Semchuk24, Yueju Wang25, Peipei Ping25, Giuliano Elia26, Michael J. Dunn26, Kieran Wynne26, Angela K. Walker27, John R. Strahler27, Philip C. Andrews27, Brian L. Hood28,29, William L. Bigbee28,30, Thomas P. Conrads28,29, Derek Smith31, Christoph H. Borchers31, Gilles A. Lajoie32, Sean C. Bendall32, Kaye D. Speicher33, David W. Speicher33, Masanori Fujimoto34, Kazuyuki Nakamura34, Young-Ki Paik35, Sang Yun Cho35, Min-Seok Kwon35, Hyoung-Joo Lee35, Seul-Ki Jeong35, An Sung Chung35, Christine A. Miller36, Rudolf Grimm36, Katy Williams37, Craig Dorschel38, Jayson A. Falkner39, Lennart Martens40, Juan Antonio Vizcaíno40
1CODA Genomics, 26061 Merit Circle #101, Laguna Hills, CA 92653 2BioGrammatics, Inc., 2705 Glasgow Drive, Carlsbad, CA 92010 3Invitrogen Corporation, 1600 Faraday Avenue, PO Box 6482, Carlsbad, California 92008 4Allergan, 2525 Dupont Drive, Irvine, CA 92612 5Ambry Genetics, 100 Columbia #200, Aliso Viejo, CA 92656 6Barnett Institute and Department of Chemistry and Chemical Biology, Northeastern University, Boston, MA 02115, USA 733 Life Park Road, State Key Laboratory of Proteomics, Beijing Proteome Research Center Changping District, Beijing, 102206, P. R. China 8Bochum University, Ruhr-Universitaet Bochum, ZKF E.143, Universitaetsstrasse 150, Bochum, D-44801, Germany 9Proteomics, Broad Institute of MIT and Harvard, Cambridge, MA 02142-2025 10Burnham Institute for Medical Research, 10901 N. Torrey Pines Rd., La Jolla, CA 92037 11Fred Hutchinson Cancer Research Center, 1100 Fairview Ave. North, Seattle, WA 98109 12Georgetown University, Department of Oncology, 3970 Reservoir Rd NW, Washington, DC 20057 13Göteborg Proteomics Centre: The Proteomics Core Facility, Sahlgrenska Academy at the University of Göteborg, Göteborg, Sweden 14Harvard Partners Center for Genetics and Genomics, 65 Landsdowne Street, Cambridge, MA 02139 15Thermo-Fisher BRIMS Center, 790 Memorial Drive, Cambridge, MA 02139 16Proteomics Platform, Quebec Genomic Center, Laval University Medical Research Center, CHUQ, 2705 Boulevard Laurier, Quebec, Canada G1V 4G2 17Health and Environment Unit, Laval University Medical Research Center, CHUQ, 2705 Boulevard Laurier, Quebec, Canada G1V 4G2 18Joint ProteomicS Laboratory, Ludwig Institute For Cancer Research and The Walter & Eliza Hall Institute For Medical Research, Parkville, Victoria, Australia 3050 19Genizon BioSciences Inc., 880 McCaffrey Street, St. Laurent, Quebec Canada H4T 2C7 20McGill University and Genome Quebec Innovation Centre, 740, Dr Penfield Avenue, Room 5202, Montréal (Québec) Canada H3A 1A4 21Ontario Cancer Biomarker Network, MaRS Centre, 101 College Street, suite 200 Toronto, ON, M5G 1L7 22Samuel Lunenfeld Research Institute, Mount Sinai Hospital, 600 University Avenue, Toronto, Ontario M5G 1X5 23The Scripps Research Institute, Department of Chemical Physiology, 10550 N Torrey Pines Road, SR-11, La Jolla, CA 92037 24Department of Biochemistry, University of Alberta. Edmonton, Alberta 25Departments of Physiology, Medicine/Division of Cardiology, David Geffen School of Medicine, University of California, Los Angeles, CA 90095 26Proteome Research Centre, UCD Conway Institute of Biomolecular and Biomedical Research, University College Dublin, Dublin, Ireland 27Department of Biological Chemistry, University of Michigan, 300 N. Ingalls St., Room 1100, Ann Arbor Michigan 48109-0404v 28Clinical Proteomics Facility, University of Pittsburgh Cancer Institute 29Department of Pharmacology & Chemical Biology, University of Pittsburgh School of Medicine 30Department of Pathology, University of Pittsburgh School of Medicine; Magee-Womens Research Institute, 204 Craft Avenue, Suite B401, Pittsburgh, PA, 15213, USA 31University of Victoria, 4464 Markham St., Victoria, BC, Canada, V8Z 7X8 32Department of Biochemistry, University of Western Ontario, London, ON, N6A 5C1 33The Wistar Institute, Room 151, 3601 Spruce Street, Philadelphia, PA 19104 34Department of Biochemistry and Functional Proteomics, Yamaguchi University Graduate School of Medicine, 1-1-1 Minamikogushi, Ube, Yamaguchi 755-8505, Japan 35Yonsei Proteome Research Center, Industry-University Bldg, Yonsei University, 134 Shinchon-dong, Sudaemoon-ku, Seoul, 120-749 Korea 36Agilent Technologies Inc., 5301 Stevens Creek Blvd., Santa Clara, California 95051-8059 37Applied Biosystems, Foster City, CA 38Waters Corporation, 34 Maple Street, Milford, MA 01757 39Department of Biological Chemistry, University of Michigan, Ann Arbor, MI, USA 40EMBL Outstation, European Bioinformatics Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, UK
References
- 1.de Godoy LM, et al. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455:1251–4. doi: 10.1038/nature07341. [DOI] [PubMed] [Google Scholar]
- 2.Turck CW, et al. The Association of Biomolecular Resource Facilities Proteomics Research Group 2006 study: relative protein quantitation. Mol. Cell. Proteomics. 2007;6:1291–8. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Boutilier K, et al. Comparison of different search engines using validated MS/MS test datasets. Anal. Chim. Acta. 2005;534:11–20. [Google Scholar]
- 4.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods. 2005;2:667–75. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 5.Kapp EA, et al. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics. 2005;5:3475–90. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
- 6.Bell AW, Nilsson T, Kearney RE, Bergeron JJ. The protein microscope: incorporating mass spectrometry into cell biology. Nat. Methods. 2007;4:783–4. doi: 10.1038/nmeth1007-783. [DOI] [PubMed] [Google Scholar]
- 7.Gilchrist A, et al. Quantitative proteomics analysis of the secretory pathway. Cell. 2006;127:1265–81. doi: 10.1016/j.cell.2006.10.036. [DOI] [PubMed] [Google Scholar]
- 8.Klie S, et al. Analyzing large-scale proteomics projects with latent semantic indexing. J. Proteome Res. 2008;7:182–91. doi: 10.1021/pr070461k. [DOI] [PubMed] [Google Scholar]
- 9.Zubarev R, Mann M. On the proper use of mass accuracy in proteomics. Mol Cell Proteomics. 2007;6:377–81. doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Cortez L. The implementation of accreditation in a chemical laboratory. Trends Analyt. Chem. 1999;18:638–43. [Google Scholar]
- 11.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 12.Yates JR, 3rd, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell. Biol. 2005;6:702–14. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 13.Shi L, Perkins RG, Fang H, Tong W. Reproducible and reliable microarray results through quality control: good laboratory proficiency and appropriate data analysis practices are essential. Curr. Opin. Biotechnol. 2008;19:10–8. doi: 10.1016/j.copbio.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Making the most of microarrays. Nat. Biotechnol. 2006;24:1039. doi: 10.1038/nbt0906-1039. [DOI] [PubMed] [Google Scholar]
- 15.Proteomics’ new order. Nature. 2005;437:169. doi: 10.1038/437169b. [DOI] [PubMed] [Google Scholar]
- 16.Domon B, Aebersold R. Challenges and opportunities in proteomics data analysis. Mol. Cell. Proteomics. 2006;5:1921–6. doi: 10.1074/mcp.R600012-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Falkner JA, Andrews PC. P6-T Tranche: Secure Decentralized Data Storage for the Proteomics Community. J. Biomol. Tech. 2007;18:3. [Google Scholar]
- 18.Martens L, et al. PRIDE: the proteomics identifications database. Proteomics. 2005;5:3537–45. doi: 10.1002/pmic.200401303. [DOI] [PubMed] [Google Scholar]
- 19.Liang F, et al. ORFDB: an information resource linking scientific content to a high-quality Open Reading Frame (ORF) collection. Nucleic Acids Res. 2004;32:D595–9. doi: 10.1093/nar/gkh118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strausberg RL, Feingold EA, Klausner RD, Collins FS. The mammalian gene collection. Science. 1999;286:455–7. doi: 10.1126/science.286.5439.455. [DOI] [PubMed] [Google Scholar]
- 21.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 22.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 2005;1:2005–0017. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan S, Hsu R, Jones A, Ross IL, Hart DN, Kato M. Identification of the dominant translation start site in the attB1 sequence of the pET-DEST42 Gateway vector. Protein Expr. Purif. 2006;49:102–7. doi: 10.1016/j.pep.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Fahnert B, Lilie H, Neubauer P. Inclusion Bodies: Formation and Utilisation. Adv. Biochem. Eng. Biotechnol. 2004;89:93–142. doi: 10.1007/b93995. [DOI] [PubMed] [Google Scholar]
- 25.Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol. Cell. Proteomics. 2004;3:531–3. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Au CE, Bell AW, Gilchrist A, Hiding J, Nilsson T, Bergeron JJ. Organellar proteomics to create the cell map. Curr. Opin. Cell. Biol. 2007;19:376–85. doi: 10.1016/j.ceb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–8. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 28.Pedrioli PG, et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 2004;22:1459–66. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- 29.Silva JC, et al. Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 2005;77:2187–200. doi: 10.1021/ac048455k. [DOI] [PubMed] [Google Scholar]
- 30.MacLean B, Eng JK, Beavis RC, McIntosh M. General framework for developing and evaluating database scoring algorithms using the TANDEM search engine. Bioinformatics. 2006;22:2830–2. doi: 10.1093/bioinformatics/btl379. [DOI] [PubMed] [Google Scholar]
- 31.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 32.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.