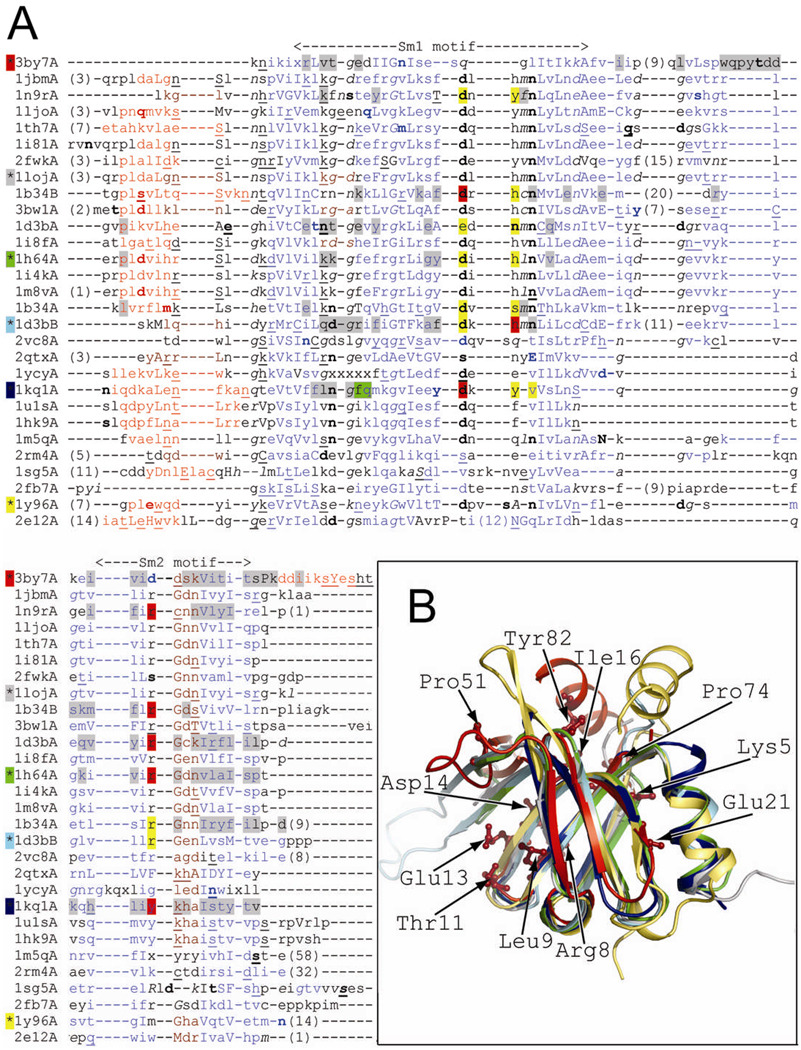
Figure 2.
(A) Structure-based sequence alignment. The structural features are annotated with JOY software 53 (solvent inaccessible, uppercase; solvent accessible, lowercase; positive φ, italic; hydrogen bond to main-chain amide, bold; hydrogen bond to main-chain carbonyl, underlined; α-helix, red; β-strand, blue; 310-helix, maroon; unknown, x). Residues involved in RNA or protein binding are highlighted (binding both RNA and adjacent subunit, red; RNA binding, yellow; adjacent subunit binding, gray; adjacent hexamer binding, green) according to PDBsum server (PDB code 3by7) or annotations from CDD database 5 (PDB codes: 1loj, 1h64, 1d3b, 1kq1, and 1y96). The background color of the asterisk preceding the PDB code corresponds to the color of the cartoon representation in Fig. 2B. The Sm1 and Sm2 motifs correspond to previously characterized structures, but not PDB code 3by7. (B) Structure superposition of ECX21941 (PDB code 3by7A, red) and structurally diverse representatives of proteins with Sm-like fold: archaeal protein SmAP1 from M. thermoautotrophicum (the most structurally similar protein; PDB code 1loj, gray), archaeal Sm1 from P. abyssi (PDB code 1h64A, green), human small nuclear ribonucleoprotein (snRNP)-associated protein B (SmB, PDB code 1d3bB, cyan), bacterial translational regulator Hfq from S. aureus (PDB code 1kq1A, navy blue), and gemin6 from the human SMN complex (PDB code 1y96A, yellow). Residues conserved among ECX21941 homologs (Lys5, Arg8, Leu9, Thr11, Glu13, Asp14, Ile16, Glu21, Pro51, Pro74, and Tyr82; PDB code 3by7) are shown by a stick representation.