Abstract
Purpose of review
Cystic fibrosis (CF) is a common, life-limiting monogenic disease, which typically manifests as progressive bronchiectasis, exocrine pancreatic dysfunction, and recurrent sinopulmonary infections. Although the gene responsible for CF (CFTR) was described in 1989, it has become increasingly evident that modifier genes and environmental factors play substantial roles in determining the severity of disease, particularly lung disease. Identifying these factors is crucial in devising therapies and other interventions to decrease the morbidity and mortality associated with this disorder.
Recent findings
Although many genes have been proposed as potential modifiers of CF, only a handful have withstood the test of replication. Several of the replicated findings reveal that genes affecting inflammation and infection response play a key role in modifying CF lung disease severity. Interactions between CFTR genotype, modifier genes, and environmental factors have been documented to influence lung function measures and infection status in CF patients.
Summary
Several genes have been demonstrated to affect disease severity in CF. Furthermore, it is likely that gene–gene and gene–environment interactions can explain a substantial portion of the variation of lung disease. Ongoing genome-wide studies are likely to identify novel genetic modifiers. Continued exploration of the role of genetic and nongenetic modifiers of CF is likely to yield new options for combating this debilitating disease.
Keywords: cystic fibrosis, genome-wide, interaction, modifier gene
Introduction
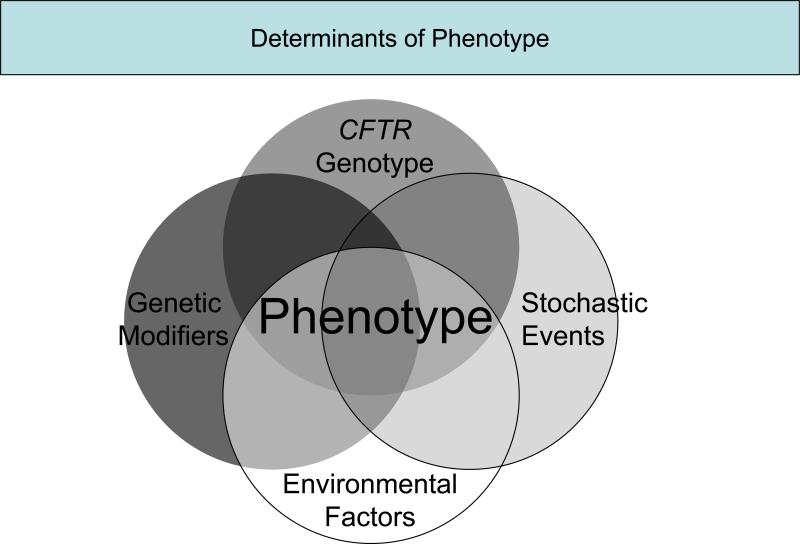
Cystic fibrosis (CF) is an autosomal recessive disorder affecting over 70 000 individuals worldwide and leads to significant morbidity and mortality with a median predicted age of survival of 37 years in 2006 (www.cff.org). The gene responsible for CF, CFTR, was identified in 1989 [1,2]. Phenotypic variation in a monogenic disorder, such as CF, can be attributed to several sources, including different mutations in the disease-causing gene, genetic modifiers, environmental factors, random events, and interactions among and between any of these sources (Fig. 1 Fig. 1). For some manifestations of CF, such as exocrine pancreatic insufficiency, CFTR genotype correlates with variation in organ function. However for other manifestations of CF, phenotypic variation remains substantial, even among patients with identical mutations in CFTR. The latter is demonstrated by the wide variation in lung function observed in patients carrying the same CFTR mutations (ΔF508 homozygotes) [3–5]. Identifying sources of disease variation is crucial to improving care for individuals with CF. This knowledge may improve the efficacy of current symptomatic therapies as well as identifying patients at risk for poor outcomes. New therapeutic targets may be discovered by this approach and some of these targets may be more accessible than CFTR. An additional impetus for elucidating the modifiers of a monogenic disease, such as CF, is the potential insight into factors that underlie variation in lung function in polygenic disorders, such as asthma. Often the first step in distilling the sources of disease variation is determining the relative contribution of genes (i.e., heritability) versus environmental factors. Classically, twin and sibling studies have been widely used to assess the degree of heritability of various phenotypes. In CF, twin studies have demonstrated that there is a genetic contribution to lung disease (heritability estimates: 0.54−1.0) [6,7•], diabetes (∼1.0) [8], and meconium ileus (∼1.0) [9], but not distal intestinal obstruction syndrome [9]. Not only can heritability studies justify the search for genetic modifiers, particularly for lung disease, they also provide a quantitative estimate of the effect of environmental modifiers and gene–environment interactions.
000 individuals worldwide and leads to significant morbidity and mortality with a median predicted age of survival of 37 years in 2006 (www.cff.org). The gene responsible for CF, CFTR, was identified in 1989 [1,2]. Phenotypic variation in a monogenic disorder, such as CF, can be attributed to several sources, including different mutations in the disease-causing gene, genetic modifiers, environmental factors, random events, and interactions among and between any of these sources (Fig. 1 Fig. 1). For some manifestations of CF, such as exocrine pancreatic insufficiency, CFTR genotype correlates with variation in organ function. However for other manifestations of CF, phenotypic variation remains substantial, even among patients with identical mutations in CFTR. The latter is demonstrated by the wide variation in lung function observed in patients carrying the same CFTR mutations (ΔF508 homozygotes) [3–5]. Identifying sources of disease variation is crucial to improving care for individuals with CF. This knowledge may improve the efficacy of current symptomatic therapies as well as identifying patients at risk for poor outcomes. New therapeutic targets may be discovered by this approach and some of these targets may be more accessible than CFTR. An additional impetus for elucidating the modifiers of a monogenic disease, such as CF, is the potential insight into factors that underlie variation in lung function in polygenic disorders, such as asthma. Often the first step in distilling the sources of disease variation is determining the relative contribution of genes (i.e., heritability) versus environmental factors. Classically, twin and sibling studies have been widely used to assess the degree of heritability of various phenotypes. In CF, twin studies have demonstrated that there is a genetic contribution to lung disease (heritability estimates: 0.54−1.0) [6,7•], diabetes (∼1.0) [8], and meconium ileus (∼1.0) [9], but not distal intestinal obstruction syndrome [9]. Not only can heritability studies justify the search for genetic modifiers, particularly for lung disease, they also provide a quantitative estimate of the effect of environmental modifiers and gene–environment interactions.
Figure 1.
Multiple factors interact to determine phenotype gr1
Candidate gene studies
Having identified manifestations of CF that are more prone to be influenced by genetic variation, such as lung function, the next step is to identify modifier genes. Candidates are generally selected on a mechanistic basis either because they function in key disease-related pathways or have been previously identified as modifiers in similar diseases. For example, TGFB1 has been implicated as a modifier of respiratory disease [10–16], and PPARγ as a modifier of diabetes risk [17]. Linkage studies can help to identify regions of interest within the genome associated with particular CF phenotypes in order to target searches for candidate genes [9,18] (L.L. Vanscoy, unpublished observation). Genetic modifiers have also been evaluated by association analysis using case–control or family-based study designs.
A number of candidate genes have been tested for association with different CF phenotypic manifestations. Published studies have examined mostly immune or inflammatory genes, including ACE [19,20], ADRβ2 [20–23], ATB0 [24], CAPN10 [25], ClCN2 [26], DEFβ4 [27], ENaC [28], FcβRII [29], GCLC [30], GSTM1 [20,31–35], GSTM3 [34], GSTP1 [20,33–35], GSTT1 [34], HFE [36,37], HLA-I [38], HLA-II [38–41], HLA-III [38], HSP70 [39], IFN-γ [19], IL1-β [39], IL6 [25], IL10 [19,20,42], IL18 [25], KCNJ11 [25], MBL2 [20,43–52,53••], MIF [54], NOS1 [55–57], NOS3 [20,58], MASP-2 [48,51], PPARβ [25], SERPINA1 [20,59–67], SERPINA3 [68], SFTPA1 [50], SFTPA2 [50], TLR4 [69], TGFB1 [19,20,52,53ββ,67,70–72••,73••], TNFα [19,20,25,32,52,74–76], TNFα-receptor [28], and TNFβ [39].
One of the advantages to candidate gene association studies is their less costly and less resource intensive means of obtaining data than genome-wide approaches. Additionally, studying candidates within known mechanistic pathways increases the likelihood that off-the-shelf therapies already exist to alter the pathway if an association is found. However, the candidate gene approach, being restricted to known pathways, does not allow for the discovery of novel pathways that affect disease. Furthermore, there exists a bias toward false-positive results as any positive association (even in underpowered studies) may be considered relevant as the candidate gene was selected from a pathway suspected of being linked to disease. Given the relatively low number of patients in most studies (<500) and the likely low magnitude of effect size for many modifier genes, replication of results in independent populations is essential. Of the aforementioned studies, only 13 candidate genes have been studied in two or more separate populations of individuals with CF. This review focuses on seven candidate genes in which at least three separate studies have examined the effect of a polymorphism on a particular phenotype (Table 1 Table 1); the majority of studies examine the phenotype of lung function.
Table 1.
Summary of replicated findings in candidate gene association studies
| Gene | Lung function | Infection | Nutritional | Survival |
|---|---|---|---|---|
| Acquisition/Prevalence | Status | |||
| TGFB1 | Probable effect | No effect | Likely no effect | |
| MBL2 | Probable effect | Possible effect | ||
| TNFα | No effect | Likely no effect | Likely no effect | Likely no effect |
| SERPINA1 | No effect | Likely no effect | ||
| GSTM1 | No effect | |||
| GSTP1 | No effect | |||
| ADRB2 | No effect |
Probable effect: association observed in three or more independent studies with ≥1000 participants in aggregate.
Possible effect: association observed in two or more independent studies with ≥500 participants in aggregate.
Likely no effect: no association observed in two or more independent studies with ≥500 participants in aggregate.
No effect: no association observed in three or more independent studies with ≥1000 participants in aggregate.
Transforming growth factor β1
TGFB1, a cytokine and mediator of fibrosis, has been studied in a number of lung diseases besides CF. Polymorphisms in TGFB1 regulate levels of transforming growth factor β1 (TGFβ1) production and have been shown to modify the development and/or severity of lung disease in fibrotic lung disease [77,78], asthma [10,13,15], and chronic obstructive pulmonary disease (COPD) [11,12,14]. At least three polymorphisms have been investigated for their role in CF, including one in the promoter region (−509C>T) and two in exon 1, Leu10Pro (c869C>T) and Arg25Pro (c915G>C). No associations have been observed between codon 25 variants and lung disease in several studies [19,20,70,71]. Polymorphisms at the −509 promoter site and codon 10 may affect lung function; however, neither variant has been observed to affect infection status. Studies by Drumm et al. [20] and Bremer et al. [73••] have demonstrated that the −509T allele is associated with improved lung function, whereas Brazova et al. [71] did not observe any association. Differing results with reference to lung phenotypes have been found with codon 10; better-powered studies suggest that the T allele correlated with higher lung function [20,73••]; however, other studies have reported lower function with the T/T genotype [70], a steeper decline with both the T/T and C/C genotypes [53••], and two studies reported no association between genotype and lung function [19,71]. Together, these studies suggest that TGFβ1 is a modifier of CF lung function, and the best-powered studies are consistent for association of the −509C allele and the codon 10 T allele with improved lung function.
Other manifestations that correlate with the severity of CF lung disease (e.g., colonization or the age of acquisition of Pseudomonas or Burkholderia cepacia) do not show associations with any of the three TGFB1 polymorphisms [19,53••,70,71]. More limited literature suggests that none of the polymorphisms affect survival [19,52,70]. Replication in some studies [73••], but not others, could be due to the differences of these studies to detect low effect variants. Additionally, other variables such as age or cohort effects may confound associations [53••,79•]. Finally, differing results could also be due to the presence of unaccounted for candidate gene interactions with CFTR, other genetic modifiers, or environmental factors. For TGFB1 codon 10 variants, interactions have been documented with MBL2 [53••], and for both the −509 promoter site and codon 10 interactions have been reported with CFTR genotype (ΔF508 homozygote versus not) [73••], and the environmental factor of secondhand smoke exposure in the home [72••]. In summary, polymorphisms of TGFB1 modify CF lung function and interactions with other genetic and environmental factors modulate the modifier effect of this gene.
Mannose binding lectin 2
Mannose binding lectin 2 (MBL2), which codes for the collectin MBL, plays an important role in innate immunity by promoting phagocytosis of infectious organisms [80]. Polymorphisms in MBL2 are associated with differing levels of MBL production [81], and deficiencies in MBL production have been linked to an increased incidence of infection, including pneumonia and sepsis, even in patients with no defects in adaptive immunity [80]. The polymorphisms most frequently studied include three amino acid substitutions in exon 1 (52, 54, 57) and one in the promoter (−221G>C). By convention, an exon 1 haplotype is classified as ‘A’ if all three codon variants are wild-type and ‘O’ if any one of the three variants is not wild-type. O variants typically lead to decreased or deficient production. The −221 promoter variant (X/Y) is often analyzed in the context of the exon 1 haplotype, with genetic combinations of the promoter and exon 1 variants frequently being categorized into three groups: low or deficient producers, intermediate producers, and high producers. Only one study [48] has been published on the role of the −550G>C promoter variant (H/L) in CF.
MBL-deficient states in CF patients have been associated with reduced survival, most likely secondary to more severe lung disease, but not necessarily due to increased susceptibility to infection. Two studies examining survival [44,52] found that it was decreased in patients with insufficient MBL states as defined by exon 1 genotypes. The relative deficiency of serum MBL has been shown to be associated with accelerated lung function decline in individuals with CF [82]. The majority of candidate gene studies [43,44,46,47,49,53••] have documented worse lung disease with insufficient MBL2 genotypes; however, a few studies [20,48,50] have seen no effect, and one study [51] saw worse lung function with high or intermediate producers. Most studies [43,44,46,47,51] have seen no effect of MBL2 variation on colonization or the age of acquisition of Pseudomonas; however, other studies [49,53••] have observed earlier acquisition of Pseudomonas with deficient states or conversely with sufficient states [48]. MBL2 genotype was found not to affect B. cepacia colonization in two studies [46,51], but colonization was found to be more likely with A/O or O/O genotypes in another study [44]. In summary, genotypes linked with decreased MBL production appear associated with lower lung function that likely is the cause of the observed increased mortality.
Tumor necrosis factor α
Polymorphisms of tumor necrosis factor α (TNFα), an inflammatory cytokine, have been associated with the development of asthma [83] and COPD [84] in meta-analyses. These polymorphisms are associated with differing levels of TNFα production, and in individuals with CF, higher sputum TNFα levels have been associated with decreased lung function [85]. Several polymorphisms of TNFα have been investigated as potential modifiers of CF including three within the promoter region, −851C>T [74], −308G>A [19,20,32,52,74–76,86], and −238G>A [52,74], and 1 in intron 1, +691G ins/del [74]. However, only −308G>A has been studied multiple times. Published studies suggest that this polymorphism does not affect lung function, infection, diabetes, or survival, though results are mixed for nutritional status. Most studies [19,20,74,76] have seen no effect of the −308G>A polymorphism on lung disease, although one study [32] did report worse function with the G/A genotype. Similarly, most studies [19,32,74,76] have not seen any genotype effect on the acquisition or colonization of Pseudomonas, although one study [75] has reported that earlier acquisition and colonization with Pseudomonas is more likely with the G allele. More limited literature suggests that the −308G>A polymorphism does not affect survival [19,52] or the prevalence of diabetes [19,76,86]. Although two studies [74,76] have reported no effect of the polymorphism on weight or BMI, another study [32] reported that the G/A genotype was associated with a decreased weight z-score. In summary, the TNFα −308G>A polymorphism does not appear to modify some of the more common manifestations of CF; however, the role of other variants of TNFα is uncertain due to lack of replication.
α1-Antitrypsin
Over 100 mutations have been identified in α1-antitrypsin (SERPINA1) [87], a gene coding for the proteinase inhibitor α1-antitrypsin. Three commonly studied mutations include the S mutation in exon 3 (Glu264Val or c2313T>A) and the Z mutation in exon 5 (Glu342Lys or c4627G>A), which result reduced plasma levels of α1-antitrypsin, and +1237G>A in the 3′-untranslated region, which may result in a decrease in acute phase levels [60]. Deficiency of α1-antitrypsin is associated with COPD. Available literature suggests that the Z, S, or +1237G>A polymorphisms do not affect lung disease or nutritional status and have mixed results regarding infection status and liver disease. Most studies [20,59,62,64,65] have seen no effect of these polymorphisms on lung function; however, one study [60] reported better function with an S or Z mutation, and another study [66] reported worse function with the G/G genotype at position +1237.
Most studies [60,63,64] have reported no association between the age of acquisition of or colonization with Pseudomonas with genotypic variants of the +1237 position, although one study [62] reported colonization was more likely with the G/G genotype. Two studies [59,64] have reported that earlier acquisition and higher prevalence of Pseudomonas are seen with the S and Z mutations, other studies [60,63] have not observed similar findings. Although two studies [60,62] did not see any effect on the prevalence of liver disease, a more recent study [67] found an increased prevalence of the Z mutation in CF patients with liver disease. More limited literature suggests that polymorphisms in SERPINA1 do not affect nutritional status [64,66]. In summary, mutations in SERPINA1 seem unlikely to affect the more common manifestations of CF, but may alter the susceptibility to the development of hepatobiliary complications.
Glutathione S-transferase M1 and glutathione S-transferase P1
The null mutation in glutathione S-transferase M1 (GSTM1), an antioxidant gene, results in no protein production and may lead to decreased lung function growth in children [88]. Polymorphisms in glutathione S-transferase P1 (GSTP1), another antioxidant gene in the glutathione-S-transferase family, have been associated with susceptibility to asthma [89–91]. The four studies that have examined the role of GSTM1 (null deletion) and GSTP1 (Ile105Val (c1375A>G) polymorphism) in CF suggest that the null deletion does not affect lung function, infection status, or nutritional status, and the polymorphism c1375A>G does not affect lung function. Three studies [20,32,34] demonstrated no effect on lung function for both genes; one study [35] reported an interaction between GSTM1 wild type and the GSTP1 Ile105Val genotypes, but it is unclear whether this genetic combination is overrepresented in a group of individuals with severe CF lung disease for protective or detrimental reasons. More limited literature suggests that the null deletion does not affect the prevalence of colonization with Pseudomonas or nutritional status as measured by weight or BMI z-scores [32,34].
β-Adrenergic receptor
β-Adrenergic receptor (ADRβ2) codes for an airway receptor that modulates reactivity; its pharmogenetic effects in asthma are still unclear [92]. The four studies that have examined the role of ADRβ2 in CF have focused on the Arg16Gly and Gln27Glu polymorphisms; one study [21] has examined the Thr164Ile polymorphism. In regard to clinical phenotypes, only effects on lung disease were investigated by more than one study. Three studies [20,22,23] saw no association of lung disease with the codon 16 or codon 27 polymorphisms, although one study [21] reported worse function and decline in individuals who carried a Gly/Gly or Arg/Gly genotype at codon 16 or a Glu/Glu or Gln/Glu genotype at codon 27.
Interactions
No gene operates in a vacuum, and although several CF modifier genes have been identified by association and will continued to be identified through genome-wide studies, it is likely that most modifier genes in isolation confer small effects upon phenotypic variation, analogous to the effect of common variants in common diseases. However, genetic modifiers in concert with other factors such as CFTR genotype, variants in other modifier genes, or environmental exposures are likely to have more substantial and therefore detectable effects. Several interactions have been reported to date and two have been replicated in independent populations: an MBL2–CFTR interaction and an MBL2–Pseudomonas interaction.
Gene–gene interactions
To date several gene–gene interactions have been reported between CFTR and modifier genes, including GCLC [30], MBL2 [49,50], FCγRII [29], NOS1 [55], and TGFβ1 [19,73••]. Most of these reported gene–CFTR interactions demonstrate the presence of or increased variant polymorphism effects in the context of ΔF508 or other severe mutation homozygosity, excepting the GCLC–CFTR interaction and the TGFB1–CFTR interaction as reported by Bremer et al. Other reported gene–gene interactions in CF include MBL2–TGFB1 [53••], TNF•-8.1 ancestral MHC genotype [75], and GSTM1–GSTP1 [35].
With the candidate gene approach, testing for gene–gene interactions is achievable as most modifier gene studies have typed only CFTR and a few other select polymorphisms. However, with the new technology to obtain genome-wide data, the number of genes in a study that uses a genome-wide approach will have increased exponentially. One current challenge in the field of modifier gene studies is developing analytic tools to determine which of potentially millions of permutations of gene–gene combinations result in biologically significant interactions.
Gene–environment interactions
Identifying gene–environment interactions in CF could prove to be clinically valuable as environmental factors may be more amenable to interventions than altering the functions of genetic modifiers. However, accurately describing and quantifying environmental exposures is more challenging and more time-consuming than genotyping. Furthermore, environmental exposures generally occur along a continuum, and often for the simplicity of analysis, these truly continuous variables are divided into discrete variables with the potential introduction of bias to simplification of a continuous variable. Also, the analytic tools for identifying gene–environment interactions using genome-wide data are still in their infancy.
It can be hypothesized that adverse effects seen with particular variant polymorphisms are amplified in the presence of adverse environmental stimuli. To date, only a handful of gene–environment interactions have been identified; these include interactions between MBL2 and Pseudomonas [44,49], and MBL2 and Staphylococcus aureus [48] leading to additional decreases in lung function. Other than infections, published data on interactions with known environmental modifiers of CF, for example, the effect of air pollution on CF lung function [93], is virtually nonexistent. However, recently interactions between secondhand smoke exposure in the home and TGFB1, as well as CFTR, have been identified [72••].
The few CF studies that have considered environmental exposures in studying interactions have focused on risk factors. In the future, it is expected that studies will consider treatment modalities as environmental exposures. Identification of genotypes that respond to particular interventions may lead to improvements in customizing therapies for individuals. Asthma research has already identified a number of genetic polymorphisms that modulate the response of individuals to specific treatment modalities, including β2-agonists, glucocorticosteroids, and leukotriene modifiers [94]. As a similar example in CF, VX-770, which is in phase II trials as of early 2008, is being tested on individuals with the G551D mutation in CFTR.
Genome-wide studies
As genetic technology has progressed, the more sophisticated tools of genome-wide approaches are now available for studying genetic variation. With the decreasing costs of genotyping assays, it is likely that the genome-wide approach will supersede the candidate approach. However, due to statistical probabilities, it is possible that thousands of SNPs will be falsely associated with specific clinical phenotypes. Although the use of a much lower P value than the commonly accepted value of 0.05 can reduce the number of falsely positive SNPs that are identified; however, detailed evaluations of identified SNPs will be essential. Evaluations must include serious discussion of plausible biological mechanisms, linkage studies, haplotype studies, and replications in independently collected populations, in order to determine which statistically significant SNPs are truly biologically significant.
In 2007, three North American groups studying genetic modifiers in CF formed a consortium to sequence the genomes of participants within their study populations (each numbering over a thousand participants) and to pool the resulting information. It is hoped that such an effort with such detailed genetic information and increased power through collaboration will lead to new insights concerning genetic modifiers into CF. With this information in hand, the subsequent challenge will be translating these discoveries to continue the fight to decrease the morbidity and mortality of this devastating disorder.
Conclusion
Although therapeutic advancements have led to decreased morbidity and improved survival in individuals with CF, the median predicted age of survival in 2006 is approximately half that of the general population. Further advances in care will come from understanding the genetic and environmental modifiers of CF disease to target existing therapies more efficiently and to develop novel therapies based on newly discovered modifiers. Recent studies have revealed several genetic modifiers and imminent genome-wide studies are likely to uncover additional genes that could be targets for therapy for this life-limiting disease.
References
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 000−000).
- 1.Riordan JR, Rommens JM, Kerem B, et al. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 2.Kerem B, Rommens JM, Buchanan JA, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 3.Koch C, Cuppens H, Rainisio M, et al. European Epidemiologic Registry of Cystic Fibrosis (ERCF): comparison of major disease manifestations between patients with different classes of mutations. Pediatr Pulmonol. 2001;31:1–12. doi: 10.1002/1099-0496(200101)31:1<1::aid-ppul1000>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 4.Kerem E, Corey M, Kerem B-S, et al. The relation between genotype and phenotype in cystic fibrosis – analysis of the most common mutation (deltaF508) N Engl J Med. 1990;323:1517–1522. doi: 10.1056/NEJM199011293232203. [DOI] [PubMed] [Google Scholar]
- 5.Cystic Fibrosis Genotype/Phenotype Consortium Correlation between genotype and phenotype in cystic fibrosis. N Engl J Med. 1993;329:1308–1313. doi: 10.1056/NEJM199310283291804. [DOI] [PubMed] [Google Scholar]
- 6.Mekus F, Ballmann M, Bronsveld I, et al. Categories of deltaF508 homozygous cystic fibrosis twin and sibling pairs with distinct phenotypic characteristics. Twin Res. 2000;3:277–293. doi: 10.1375/136905200320565256. [DOI] [PubMed] [Google Scholar]
- 7•.Vanscoy LL, Blackman SM, Collaco JM, et al. Heritability of lung disease severity in cystic fibrosis. Am J Respir Crit Care Med. 2007;175:1036–1043. doi: 10.1164/rccm.200608-1164OC. [• of special interestHeritability paper establishing the relative contribution of modifier genes to CF lung disease] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blackman SM, Hsu S, Ritter SE, et al. TCF7L2, A risk gene for type 2 diabetes, is associated with cystic fibrosis-related diabetes. Pediatr Pulmonol Suppl. 2007;30:265. [Google Scholar]
- 9.Blackman SM, Deering-Brose R, McWilliams R, et al. Relative contribution of genetic and nongenetic modifiers to intestinal obstruction in cystic fibrosis. Gastroenterology. 2006;131:1030–1039. doi: 10.1053/j.gastro.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, Romieu I, Wu H, et al. Genetic polymorphisms in transforming growth factor beta-1 (TGFB1) and childhood asthma and atopy. Hum Genet. 2007;121:529–538. doi: 10.1007/s00439-007-0337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Su ZG, Wen FQ, Feng YL, et al. Transforming growth factor-beta1 gene polymorphisms associated with chronic obstructive pulmonary disease in Chinese population. Acta Pharmacol Sin. 2005;26:714–720. [PubMed] [Google Scholar]
- 12.Wu L, Chau J, Young RP, et al. Transforming growth factor-beta1 genotype and susceptibility to chronic obstructive pulmonary disease. Thorax. 2004;59:126–129. doi: 10.1136/thorax.2003.005769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mak JC, Leung HC, Ho SP, et al. Analysis of TGF-beta(1) gene polymorphisms in Hong Kong Chinese patients with asthma. J Allergy Clin Immunol. 2006;117:92–96. doi: 10.1016/j.jaci.2005.08.049. [DOI] [PubMed] [Google Scholar]
- 14.Celedon JC, Lange C, Raby BA, et al. The transforming growth factor-beta1 (TGFB1) gene is associated with chronic obstructive pulmonary disease (COPD) Hum Mol Genet. 2004;13:1649–1656. doi: 10.1093/hmg/ddh171. [DOI] [PubMed] [Google Scholar]
- 15.Pulleyn LJ, Newton R, Adcock IM, Barnes PJ. TGFbeta1 allele association with asthma severity. Hum Genet. 2001;109:623–627. doi: 10.1007/s00439-001-0617-y. [DOI] [PubMed] [Google Scholar]
- 16.Silverman ES, Palmer LJ, Subramaniam V, et al. Transforming growth factor-beta1 promoter polymorphism C-509T is associated with asthma. Am J Respir Crit Care Med. 2004;169:214–219. doi: 10.1164/rccm.200307-973OC. [DOI] [PubMed] [Google Scholar]
- 17.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 18.Zielenski J, Markiewicz D, Yuan X, et al. Analysis of gene candidates for cystic fibrosis modifier 1 (CFM1) associated with meconium ileus in cystic fibrosis patients. Pediatr Pulmonol Suppl. 2002;24:226. [Google Scholar]
- 19.Arkwright PD, Pravica V, Geraghty PJ, et al. End-organ dysfunction in cystic fibrosis: association with angiotensin I converting enzyme and cytokine gene polymorphisms. Am J Respir Crit Care Med. 2003;167:384–389. doi: 10.1164/rccm.200204-364OC. [DOI] [PubMed] [Google Scholar]
- 20.Drumm ML, Konstan MW, Schluchter MD, et al. Genetic modifiers of lung disease in cystic fibrosis. N Engl J Med. 2005;353:1443–1453. doi: 10.1056/NEJMoa051469. [DOI] [PubMed] [Google Scholar]
- 21.Buscher R, Eilmes KJ, Grasemann H, et al. beta2 adrenoceptor gene polymorphisms in cystic fibrosis lung disease. Pharmacogenetics. 2002;12:347–353. doi: 10.1097/00008571-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Hart MA, Konstan MW, Darrah RJ, et al. Beta 2 adrenergic receptor polymorphisms in cystic fibrosis. Pediatr Pulmonol. 2005;39:544–550. doi: 10.1002/ppul.20210. [DOI] [PubMed] [Google Scholar]
- 23.Steagall WK, Barrow BJ, Glasgow CG, et al. Beta-2-adrenergic receptor polymorphisms in cystic fibrosis. Pharmacogenet Genomics. 2007;17:425–430. doi: 10.1097/FPC.0b013e3280119349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larriba S, Sumoy L, Ramos MD, et al. ATB(0)/SLC1A5 gene. Fine localisation and exclusion of association with the intestinal phenotype of cystic fibrosis. Eur J Hum Genet. 2001;9:860–866. doi: 10.1038/sj.ejhg.5200726. [DOI] [PubMed] [Google Scholar]
- 25.Derbel S, Doumaguet C, Hubert D, et al. Calpain 10 and development of diabetes mellitus in cystic fibrosis. J Cyst Fibros. 2006;5:47–51. doi: 10.1016/j.jcf.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Blaisdell CJ, Howard TD, Stern A, et al. CLC-2 single nucleotide polymorphisms (SNPs) as potential modifiers of cystic fibrosis disease severity. BMC Med Genet. 2004;5:26. doi: 10.1186/1471-2350-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollox EJ, Davies J, Griesenbach U, et al. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J Negat Results Biomed. 2005;4:9. doi: 10.1186/1477-5751-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanke F, Becker T, Cuppens H, et al. The TNFalpha receptor TNFRSF1A and genes encoding the amiloride-sensitive sodium channel ENaC as modulators in cystic fibrosis 1. Hum Genet. 2006;119:331–343. doi: 10.1007/s00439-006-0140-2. [DOI] [PubMed] [Google Scholar]
- 29.De R V, Arduino C, Cappello N, et al. Fcgamma receptor IIA genotype and susceptibility to P. aeruginosa infection in patients with cystic fibrosis. Eur J Hum Genet. 2005;13:96–101. doi: 10.1038/sj.ejhg.5201285. [DOI] [PubMed] [Google Scholar]
- 30.McKone EF, Shao J, Frangolias DD, et al. Variants in the glutamate-cysteine-ligase gene are associated with cystic fibrosis lung disease. Am J Respir Crit Care Med. 2006;174:415–419. doi: 10.1164/rccm.200508-1281OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baranov VS, Ivaschenko T, Bakay B, et al. Proportion of the GSTM1 0/0 genotype in some Slavic populations and its correlation with cystic fibrosis and some multifactorial diseases. Hum Genet. 1996;97:516–520. doi: 10.1007/BF02267078. [DOI] [PubMed] [Google Scholar]
- 32.Hull J, Thomson AH. Contribution of genetic factors other than CFTR to disease severity in cystic fibrosis. Thorax. 1998;53:1018–1021. doi: 10.1136/thx.53.12.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henrion-Caude A, Flamant C, Roussey M, et al. Liver disease in pediatric patients with cystic fibrosis is associated with glutathione S-transferase P1 polymorphism. Hepatology. 2002;36:913–917. doi: 10.1053/jhep.2002.35534. [DOI] [PubMed] [Google Scholar]
- 34.Flamant C, Henrion-Caude A, Boelle PY, et al. Glutathione-S-transferase M1, M3, P1 and T1 polymorphisms and severity of lung disease in children with cystic fibrosis. Pharmacogenetics. 2004;14:295–301. doi: 10.1097/00008571-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Korytina GF, Iaibaeva DG, Viktorova TV. Polymorphism of glutathione-S-transferase M1 and P1 genes in patients with cystic fibrosis and chronic respiratory tract diseases. Genetika. 2004;40:401–408. [PubMed] [Google Scholar]
- 36.Rohlfs EM, Shaheen NJ, Silverman LM. Is the hemochromatosis gene a modifier locus for cystic fibrosis? Genet Test. 1998;2:85–88. doi: 10.1089/gte.1998.2.85. [DOI] [PubMed] [Google Scholar]
- 37.Devaney J, Maher M, Smith T, et al. HFE alleles in an Irish cystic fibrosis population. Genet Test. 2003;7:155–158. doi: 10.1089/109065703322146876. [DOI] [PubMed] [Google Scholar]
- 38.Duthie A, Doherty DG, Donaldson PT, et al. The major histocompatibility complex influences the development of chronic liver disease in male children and young adults with cystic fibrosis. J Hepatol. 1995;23:532–537. doi: 10.1016/0168-8278(95)80058-1. [DOI] [PubMed] [Google Scholar]
- 39.Lanng S, Thorsteinsson B, Pociot F, et al. Diabetes mellitus in cystic fibrosis: genetic and immunological markers. Acta Paediatr. 1993;82:150–154. doi: 10.1111/j.1651-2227.1993.tb12628.x. [DOI] [PubMed] [Google Scholar]
- 40.Carrington M, Krueger LJ, Holsclaw DSJ, Jr, et al. Cystic fibrosis-related diabetes is associated with HLA DQB1 alleles encoding Asp-57-molecules. J Clin Immunol. 1994;14:353–358. doi: 10.1007/BF01546319. [DOI] [PubMed] [Google Scholar]
- 41.Aron Y, Polla BS, Bienvenu T, et al. HLA class II polymorphism in cystic fibrosis. Am J Resp Crit Care Med. 1999;159:1464–1468. doi: 10.1164/ajrccm.159.5.9807046. [DOI] [PubMed] [Google Scholar]
- 42.Brouard J, Knauer N, Boelle PY, et al. Influence of interleukin-10 on Aspergillus fumigatus infection in patients with cystic fibrosis. J Infect Dis. 2005;191:1988–1991. doi: 10.1086/429964. [DOI] [PubMed] [Google Scholar]
- 43.Gabolde M, Guilloud-Bataille M, Feingold J, Besmond C. Association of variant alleles of mannose binding lectin with severity of pulmonary disease in cystic fibrosis: cohort study. Br Med J. 1999;319:1166–1167. doi: 10.1136/bmj.319.7218.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garred P, Pressler T, Madsen HO, et al. Association of mannose-binding lectin gene heterogeneity with severity of lung disease and survival in cystic fibrosis. J Clin Invest. 1999;104:431–437. doi: 10.1172/JCI6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabolde M, Hubert D, Guilloud-Bataille M, et al. The mannose binding lectin gene influences the severity of chronic liver disease in cystic fibrosis. J Med Genet. 2001;38:310–311. doi: 10.1136/jmg.38.5.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies JC, Turner MW, Klein N. Impaired pulmonary status in cystic fibrosis adults with two mutated MBL-2 alleles. Eur Respir J. 2004;24:798–804. doi: 10.1183/09031936.04.00055404. [DOI] [PubMed] [Google Scholar]
- 47.Yarden J, Radojkovic D, De Boeck K, et al. Polymorphisms in the mannose binding lectin gene affect the cystic fibrosis pulmonary phenotype. J Med Genet. 2004;41:629–633. doi: 10.1136/jmg.2003.017947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlsson M, Sjoholm AG, Eriksson L, et al. Deficiency of the mannan-binding lectin pathway of complement and poor outcome in cystic fibrosis: bacterial colonization may be decisive for a relationship. Clin Exp Immunol. 2005;139:306–313. doi: 10.1111/j.1365-2249.2004.02690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trevisiol C, Boniotto M, Giglio L, et al. MBL2 polymorphisms screening in a regional Italian CF center. J Cyst Fibros. 2005;4:189–191. doi: 10.1016/j.jcf.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 50.Choi EH, Ehrmantraut M, Foster CB, et al. Association of common haplotypes of surfactant protein A1 and A2 (SFTPA1 and SFTPA2) genes with severity of lung disease in cystic fibrosis. Pediatr Pulmonol. 2006;41:255–262. doi: 10.1002/ppul.20361. [DOI] [PubMed] [Google Scholar]
- 51.Olesen HV, Jensenius JC, Steffensen R, et al. The mannan-binding lectin pathway and lung disease in cystic fibrosis – disfunction of mannan-binding lectin-associated serine protease 2 (MASP-2) may be a major modifier. Clin Immunol. 2006;121:324–331. doi: 10.1016/j.clim.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Buranawuti K, Boyle MP, Cheng S, et al. Variants in mannose-binding lectin and tumor necrosis factor {alpha} affect survival in cystic fibrosis. J Med Genet. 2007;44:209–214. doi: 10.1136/jmg.2006.046318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.Dorfman R, Sandford A, Taylor C, et al. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J Clin Invest. 2008;118:1040–1049. doi: 10.1172/JCI33754. [•• of outstanding interestWell powered study demonstrating a gene–gene interaction between two modifier genes, MBL and TGFB1] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plant BJ, Gallagher CG, Bucala R, et al. Cystic fibrosis, disease severity, and a macrophage migration inhibitory factor polymorphism. Am J Respir Crit Care Med. 2005;172:1412–1415. doi: 10.1164/rccm.200412-1714OC. [DOI] [PubMed] [Google Scholar]
- 55.Grasemann H, Knauer N, Buscher R, et al. Airway nitric oxide levels in cystic fibrosis patients are related to a polymorphism in the neuronal nitric oxide synthase gene. Am J Respir Crit Care Med. 2000;162:2172–2176. doi: 10.1164/ajrccm.162.6.2003106. [DOI] [PubMed] [Google Scholar]
- 56.Grasemann H, Storm van's GK, Gartig S, et al. Nasal nitric oxide levels in cystic fibrosis patients are associated with a neuronal NO synthase (NOS1) gene polymorphism. Nitric Oxide. 2002;6:236–241. doi: 10.1006/niox.2001.0408. [DOI] [PubMed] [Google Scholar]
- 57.Texereau J, Marullo S, Hubert D, et al. Nitric oxide synthase 1 as a potential modifier gene of decline in lung function in patients with cystic fibrosis. Thorax. 2004;59:156–158. doi: 10.1136/thorax.2003.006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grasemann H, van's Gravesande KS, Buscher R, et al. Endothelial nitric oxide synthase variants in cystic fibrosis lung disease. Am J Respir Crit Care Med. 2003;167:390–394. doi: 10.1164/rccm.200202-155OC. [DOI] [PubMed] [Google Scholar]
- 59.Doring G, Krogh-Johansen H, Weidinger S, Hoiby N. Allotypes of alpha 1-antitrypsin in patients with cystic fibrosis, homozygous and heterozygous for deltaF508. Pediatr Pulmonol. 1994;18:3–7. doi: 10.1002/ppul.1950180104. [DOI] [PubMed] [Google Scholar]
- 60.Mahadeva R, Westerbeek RC, Perry DJ, et al. Alpha1-antitrypsin deficiency alleles and the Taq-I G→A allele in cystic fibrosis lung disease. Eur Respir J. 1998;11:873–879. doi: 10.1183/09031936.98.11040873. [DOI] [PubMed] [Google Scholar]
- 61.Mahadeva R, Stewart S, Bilton D, Lomas DA. Alpha-1 antitrypsin deficiency alleles and severe cystic fibrosis lung disease. Thorax. 1998;53:1022–1024. doi: 10.1136/thx.53.12.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henry MT, Cave S, Rendall J, et al. An alpha(1)-antitrypsin enhancer polymorphism is a genetic modifier of pulmonary outcome in cystic fibrosis. Eur J Hum Genet. 2001;9:273–278. doi: 10.1038/sj.ejhg.5200623. [DOI] [PubMed] [Google Scholar]
- 63.Meyer P, Braun A, Roscher AA. Analysis of the two common alpha-1-antitrypsin deficiency alleles PiMS and PiMZ as modifiers of Pseudomonas aeruginosa susceptibility in cystic fibrosis. Clin Genet. 2002;62:325–327. doi: 10.1034/j.1399-0004.2002.620413.x. [DOI] [PubMed] [Google Scholar]
- 64.Frangolias DD, Ruan J, Wilcox PJ, et al. Alpha 1-antitrypsin deficiency alleles in cystic fibrosis lung disease. Am J Respir Cell Mol Biol. 2003;29:390–396. doi: 10.1165/rcmb.2002-0271OC. [DOI] [PubMed] [Google Scholar]
- 65.de Faria EJ, de Faria IC, Alvarez AE, et al. Association between alpha 1 antitrypsin deficiency and cystic fibrosis severity. J Pediatr (Rio J) 2005;81:485–490. doi: 10.2223/JPED.1423. [DOI] [PubMed] [Google Scholar]
- 66.Courtney JM, Plant BJ, Morgan K, et al. Association of improved pulmonary phenotype in Irish cystic fibrosis patients with a 3′ enhancer polymorphism in alpha-1-antitrypsin. Pediatr Pulmonol. 2006;41:584–591. doi: 10.1002/ppul.20416. [DOI] [PubMed] [Google Scholar]
- 67.Stonebraker JR, Friedman KJ, Ling SC, et al. Genetic modifiers of severe liver disease in cystic fibrosis: a replication study. Pediatr Pulmonol Suppl. 2007;30:381. [Google Scholar]
- 68.Mahadeva R, Sharples L, Ross-Russell RI, et al. Association of alpha(1)-antichymotrypsin deficiency with milder lung disease in patients with cystic fibrosis. Thorax. 2001;56:53–58. doi: 10.1136/thorax.56.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Urquhart DS, Allen J, Elrayess M, et al. Modifier effect of the Toll-like receptor 4 D299G polymorphism in children with cystic fibrosis. Arch Immunol Ther Exp (Warsz) 2006;54:271–276. doi: 10.1007/s00005-006-0029-8. [DOI] [PubMed] [Google Scholar]
- 70.Arkwright PD, Laurie S, Super M, et al. TGF-beta(1) genotype and accelerated decline in lung function of patients with cystic fibrosis. Thorax. 2000;55:459–462. doi: 10.1136/thorax.55.6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brazova J, Sismova K, Vavrova V, et al. Polymorphisms of TGF-beta1 in cystic fibrosis patients. Clin Immunol. 2006;121:350–357. doi: 10.1016/j.clim.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 72••.Collaco JM, Vanscoy L, Bremer L, et al. Interactions between secondhand smoke and genes that affect cystic fibrosis lung disease. JAMA. 2008;299:417–424. doi: 10.1001/jama.299.4.417. [•• of outstanding interestGene–environment study demonstrating an interaction between TGFB1 and secondhand smoke] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73••.Bremer LA, Blackman SM, Vanscoy LL, et al. Interaction between a novel TGFB1 haplotype and CFTR genotype is associated with improved lung function in cystic fibrosis. Hum Mol Genet. 2008;17:2228–2237. doi: 10.1093/hmg/ddn123. [•• of outstanding interestFamily-based study demonstrating a CFTR–TGFB1 interaction] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yarden J, Radojkovic D, De Boeck K, et al. Association of tumour necrosis factor alpha variants with the CF pulmonary phenotype. Thorax. 2005;60:320–325. doi: 10.1136/thx.2004.025262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Laki J, Laki I, Nemeth K, et al. The 8.1 ancestral MHC haplotype is associated with delayed onset of colonization in cystic fibrosis. Int Immunol. 2006;18:1585–1590. doi: 10.1093/intimm/dxl091. [DOI] [PubMed] [Google Scholar]
- 76.Schmitt-Grohe S, Stuber F, Book M, et al. TNF-alpha promoter polymorphism in relation to TNF-alpha production and clinical status in cystic fibrosis 1. Lung. 2006;184:99–104. doi: 10.1007/s00408-005-2568-x. [DOI] [PubMed] [Google Scholar]
- 77.Awad MR, El Gamel A, Hasleton P, et al. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation. 1998;66:1014–1020. doi: 10.1097/00007890-199810270-00009. [DOI] [PubMed] [Google Scholar]
- 78.Xaubet A, Marin-Arguedas A, Lario S, et al. Transforming growth factor-beta1 gene polymorphisms are associated with disease progression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2003;168:431–435. doi: 10.1164/rccm.200210-1165OC. [DOI] [PubMed] [Google Scholar]
- 79•.Lasky-Su J, Lyon HN, Emilsson V, et al. On the replication of genetic associations: timing can be everything! Am J Hum Genet. 2008;82:849–858. doi: 10.1016/j.ajhg.2008.01.018. [• of special interestA good review of challenges in gene association studies and demonstration of age as a confounding factor in such studies] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eisen DP, Minchinton RM. Impact of mannose-binding lectin on susceptibility to infectious diseases. Clin Infect Dis. 2003;37:1496–1505. doi: 10.1086/379324. [DOI] [PubMed] [Google Scholar]
- 81.Bouwman LH, Roep BO, Roos A. Mannose-binding lectin: clinical implications for infection, transplantation, and autoimmunity. Hum Immunol. 2006;67:247–256. doi: 10.1016/j.humimm.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 82.Muhlebach MS, MacDonald SL, Button B, et al. Association between mannan-binding lectin and impaired lung function in cystic fibrosis may be age-dependent. Clin Exp Immunol. 2006;145:302–307. doi: 10.1111/j.1365-2249.2006.03151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Castro-Giner F, Kogevinas M, Mächler M, et al. TNFA –308G>A in two international population-based cohorts and risk for asthma. Eur Respir J. 2008;32:350–361. doi: 10.1183/09031936.00155607. [DOI] [PubMed] [Google Scholar]
- 84.Hu GP, Peng GY, Hu JX, Ran PX. Association of tumor necrosis factor alpha 308 G/A gene promoter polymorphism with the presence of chronic obstructive pulmonary disease: a meta-analysis. Zhonghua Jie He He Hu Xi Za Zhi. 2007;30:588–594. [PubMed] [Google Scholar]
- 85.Greally P, Hussein MJ, Cook AJ, et al. Sputum tumour necrosis factor-alpha and leukotriene concentrations in cystic fibrosis. Arch Dis Child. 1993;68:389–392. doi: 10.1136/adc.68.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Derbel S, Doumaguet C, Hubert D, et al. Calpain 10 and development of diabetes mellitus in cystic fibrosis. J Cyst Fibros. 2006;5:47–51. doi: 10.1016/j.jcf.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 87.Fregonese L, Stolk J, Frants RR, Veldhuisen B. Alpha-1 antitrypsin null mutations and severity of emphysema. Respir Med. 2008;102:876–884. doi: 10.1016/j.rmed.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Gilliland FD, Gauderman WJ, Vora H, et al. Effects of glutathione-S-transferase M1, T1, and P1 on childhood lung function growth. Am J Respir Crit Care Med. 2002;166:710–716. doi: 10.1164/rccm.2112065. [DOI] [PubMed] [Google Scholar]
- 89.Kamada F, Mashimo Y, Inoue H, et al. The GSTP1 gene is a susceptibility gene for childhood asthma and the GSTM1 gene is a modifier of the GSTP1 gene. Int Arch Allergy Immunol. 2007;144:275–286. doi: 10.1159/000106316. [DOI] [PubMed] [Google Scholar]
- 90.Li YF, Gauderman WJ, Conti DV, et al. Glutathione S-transferase P1, maternal smoking, and asthma in children: a haplotype-based analysis. Environ Health Perspect. 2008;116:409–415. doi: 10.1289/ehp.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Imboden M, Rochat T, Brutsche M, et al. Glutathione S-transferase genotype increases risk of progression from bronchial hyperresponsiveness to asthma in adults. Thorax. 2008;63:322–328. doi: 10.1136/thx.2007.085555. [DOI] [PubMed] [Google Scholar]
- 92.Hawkins GA, Peters SP. Pharmacogenetics of asthma Methods. Mol Biol. 2008;448:359–378. doi: 10.1007/978-1-59745-205-2_11. [DOI] [PubMed] [Google Scholar]
- 93.Goss CH, Newsom SA, Schildcrout JS, et al. Effect of ambient air pollution on pulmonary exacerbations and lung function in cystic fibrosis. Am J Respir Crit Care Med. 2004;169:816–821. doi: 10.1164/rccm.200306-779OC. [DOI] [PubMed] [Google Scholar]
- 94.Morrow T. Implications of pharmacogenomics in the current and future treatment of asthma. J Manag Care Pharm. 2007;13:497–505. doi: 10.18553/jmcp.2007.13.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]