Abstract
Human CD38 is a pleiotropic glycoprotein belonging to a family of genes coding for enzymes/receptors involved in the catabolism of extracellular nucleotides. CD38 receptor activities are regulated through binding to the non-substrate ligand CD31. CD38 expression above a critical threshold represents a negative prognostic marker for CLL patients. Activation of CD38 by means of agonistic monoclonal antibodies or the CD31 ligand induces proliferation and immunoblast differentiation of CLL cells. Here we define the genetic signature that follows long-term in vitro interactions between CD38+ CLL lymphocytes and CD31+ cells. The emerging profile confirms that the CD31/CD38 axis activates genetic programs relevant for proliferative responses. It also indicates a contribution of this pathway to the processes mediating migration and homing. These results further support the notion that the CD31/CD38 axis is part of a network of accessory signals that modify the microenvironment, favoring localization of leukemic cells to growth-permissive sites.
Introduction
Human CD38 is a pleiotropic glycoprotein belonging to a complex family of genes coding for enzymes of the cell surface involved in the catabolism of extracellular nucleotides (1). During evolution, CD38 acquired the ability to mediate cell-cell interactions, acting as a receptor and binding the non-substrate ligand CD31. CD31/CD38 interactions drive activation and proliferation of distinct lymphocyte populations (2).
Along with the absence of mutations in the immunoglobulin variable region heavy chain gene (IgVH) (3) and expression of the cytoplasmic kinase ZAP-70 (4), CD38 expression above a critical threshold represents a reliable negative prognostic marker for chronic lymphocytic leukemia (CLL) patients (5). The working hypothesis of our group is that CD38 is not a mere a marker in CLL, but a cell surface receptor and adhesion molecule directly involved in the delivery of growth signals (6). Several pieces of evidence support this view. First, CD38 expression is higher within the bone marrow and the lymph nodes, where the proliferative core of the disease resides (7). Moreover, within each CLL clone, cells expressing CD38 are enriched in expression of Ki-67, suggesting that CD38+ cells represent a cycling subset (8). The CD38+ subset is also characterized by a specific genetic profile, showing up-regulation of proliferation/survival pathways (9).
We showed that activation of CD38 induces proliferation and immunoblast differentiation of CLL cells (10). These signals are regulated by physical localization of the CD38 receptor within lipid micro-domains and act in synergy with the B cell receptor (BCR)/CD19 complex (11). The initial observations were obtained using agonistic monoclonal antibodies (mAbs) and then extended to a model closely resembling the physiological environment where CLL cells grow and expand. Indeed, CD38+ CLL cells can bind to murine fibroblasts transfected with the CD31 ligand with resulting increased growth and survival (12). In vivo, several CD31+ cells can be found in lymphoid organs, often in close contact with CD38+ CLL cells (13).
This work was undertaken in order to obtain a genome-wide signature of the transcriptional events that follow activation of the CD31/CD38 axis. It also provides some answers to the issues raised by a recent report, where the relevance of CD31/CD38 interactions in the context of CLL was called into question (14). The emerging genetic profile indicates that CD31/CD38 interactions are indeed followed by activation of genetic programs relevant not only for proliferative responses, as could be anticipated on the basis of in vitro experiments, but also for migration and homing. This latter point deserves further attention and points to a potential role for CD38 in directing CLL cells to specific microenvironments.
Materials and Methods
Patients and cells
10 samples from CLL patients presenting with typical morphology and immunophenotype were obtained after written informed consent in accordance with Institutional Guidelines and Declaration of Helsinki. Analyses included CD38 and ZAP-70 stainings [cut-off positivity values of ≥20% and ≥10%, respectively (15)], IgVH mutational status (unmutated if ≥98% homology to the germline gene) and fluorescence in situ hybridization (FISH) for chromosomes 11, 12, 13 and 17 (Table 1). B cells were purified from PBMC by negative selection (15).
Table 1.
Clinical and biological features of the CLL patients
| Patient No. | Age (yr) | Sex | Stage | YD | TX | IgVH (%)* | CD38 (%) | ZAP-70 (%) | FISH |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 60 | F | 0/A | 10 | T | 100 | 54 | 2 | 11q- |
| 2 | 65 | F | 2/A | 7 | T | 100 | 35 | 12 | 11q-,13q- |
| 3 | 72 | F | 0/A | 7 | NT | - | 70 | 11 | +12,17p- |
| 4 | 47 | M | 2/A | 21 | T | 91.2 | 75 | 0 | ND |
| 5 | 57 | M | 4/C | 7 | T | 97 | 79 | 9 | ND |
| 6 | 83 | M | 2/A | 5 | T | 99.7 | 66 | 23 | +12 |
| 7 | 75 | F | 1/B | 6 | NT | 99 | 42 | 12 | 17p- |
| 8 | 61 | M | 4/A | 8 | NT | 99.3 | 78 | 21 | ND |
| 9 | 48 | M | 2/B | 8 | T | 99.1 | 34 | 35 | 13q- |
| 10 | 67 | M | 2/A | 13 | T | 99 | 58 | 14 | +12 |
Stage: defined according to Rai and Binet diagnostic criteria. YD: years since diagnosis. TX: therapy; NT: untreated.
Shown as percentage similarity to the closest germline gene.
-: missing data;
ND: none detected.
Culture conditions
Mouse L-cells transfected with human CD31 (L-CD31+) or with the empty plasmid (L-mock) were obtained as described (16). Freshly purified CLL cells (1.5x106) were plated on mitomycin C-inactivated (0.5 mg/ml, 20 min at 37 °C) L-CD31+ or L-mock fibroblasts (2x105/well in a 24w plate) for 5 days.
Gene profiling
Data collection. RNA was obtained from freshly purified CLL cells (“basal”) or after culture on L-CD31+ (“L-CD31+” profile) or L-mock fibroblasts (L-mock” profile). Amplified and labeled specimens from each sample (15) were combined with the Human Universal Reference Total RNA (Clontech, Saint-Germain-en-Laye, France), fragmented and hybridized to oligonucleotide glass arrays representing 41,000 human unique genes and transcripts (Human Whole Genome Oligo Microarray Array, Agilent Technologies, Cernusco, Italy). A dye-swap replicate was performed for each sample.
Data analysis. Data were pre-processed using the Rosetta Resolver SE software (Rosetta Biosoftware, Seattle, WA) (15). Error-weighted Student two-sample paired t-test with Benjamini and Hochberg p-value adjustment was applied to obtain differentially expressed sequences in the “L-CD31+ vs L-mock, “L-CD31+ vs basal” and “L-mock vs basal” class comparisons, after a sequence pre-filtering step (average expression ratio p-value respect to the Universal Reference less than 0.01 in at least 50% of the patients of each class comparison). Venn diagrams were then created to visualize intersections between class comparison results and to select the sequences of interest.
Pathway analysis. Significant differential expression at the level of a collection of pre-defined pathways from the KEGG database was identified using PLAGE (http://dulci.org/pathways/) (17).
Hierarchical clustering. Results were represented using Cluster 3.0 and TreeView (http://rana.lbl.gov/EisenSoftware.htm) and applying hierarchical clustering to the pathway matrix output given by PLAGE.
The data are deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) and are accessible through the GEO series GSE14063.
Results and Discussion
CD38 receptor activities are regulated through binding to CD31, a non-substrate ligand surrogated by ligation with agonistic mAbs [reviewed in (1)]. In vitro experiments showed that these interactions induce marked proliferation and immunoblast transformation of a fraction of the leukemic clone. We asked whether these results could find a confirmation using a different technological approach, needed also because the relevance of the CD31/CD38 axis in CLL has recently been called into question (14). This issue was answered by obtaining a genome-wide signature of the events taking place after CD31/CD38 interactions. To this aim, CLL cells from 10 CD38+ patients were used immediately (“basal” profile) or cultured for 5 days in the presence of L-CD31+ transfectants (“L-CD31+” profile) or of mock-transfected cells (“L-mock” profile), used as controls. The choice of the 5-days time point was based on previous results indicating that proliferation and survival are highest after this incubation time (12). Moreover, this approach rests on physiological considerations in that CD31/CD38 static interactions are likely to take place within lymphoid organs, where leukemic cells lie in close and prolonged contacts with CD31+ residential elements (13).
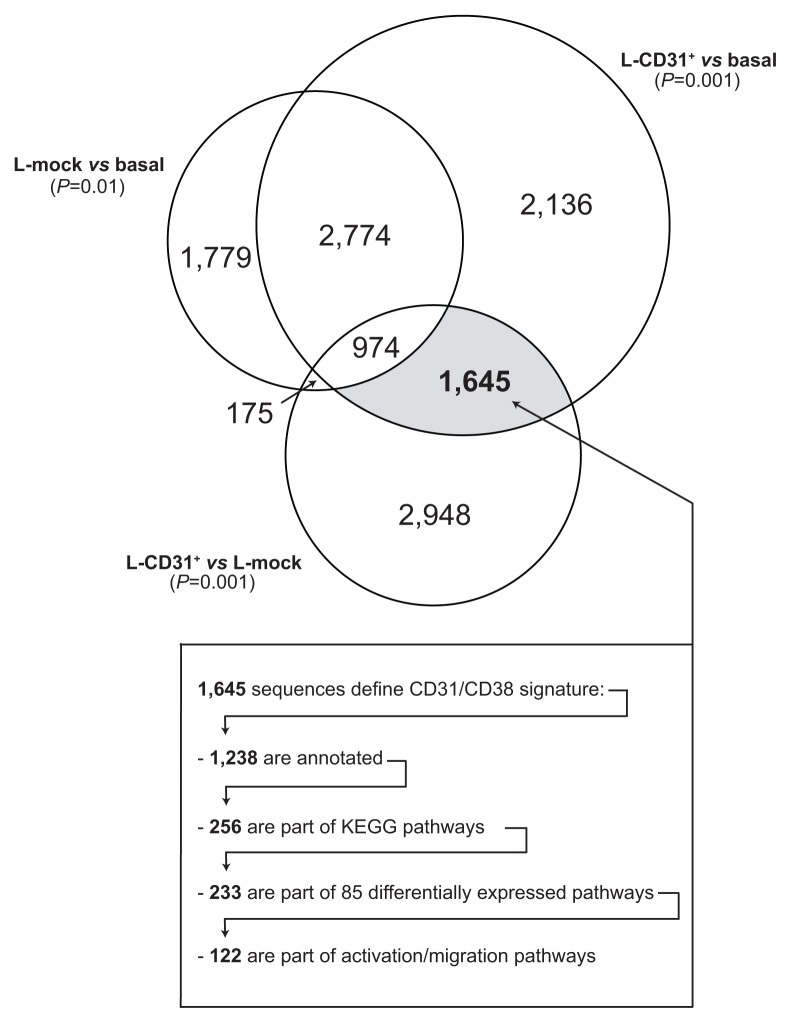
Sample and Reference RNAs were co-hybridized to arrays representing 41,000 human unique genes and transcripts. The strategy selected to identify the signature in response to CD31/CD38 interactions relied upon i) identification of genes differentially expressed in the “L-CD31+ vs L-mock” comparison, and ii) subsequent exclusion of genes modulated in the “L-mock vs basal” comparison. Results are represented in the Venn diagram of Figure 1. Two p-value thresholds were adopted to i) exclude genes that changed expression simply as a result of co-culture (“L-mock” vs “basal”, p-value 0.01), and to ii) select sequences with a more significant level of modulation (“L-CD31+” vs “L-mock” and “L-CD31+” vs “basal”, p-value 0.001) among the ones differentially expressed as a direct consequence of co-culture on L-CD31+ transfectants.
Figure 1. CD38 engagement by the CD31 ligand triggers genetic pathways leading to cell proliferation and movement.
Venn diagram showing the strategy selected to identify the signature in response to CD31/CD38 interactions. Results were obtained by applying error-weighted Student two-sample paired t-test with Benjamini and Hochberg p-value adjustment to “L-CD31+ vs L-mock”, “L-CD31+ vs basal” and “L-mock vs basal” class comparisons. Circle radii are proportional to the number of transcripts differentially expressed in each condition. The grey area defines the CD31/CD38 signature. A cut-off of 0.01 for the adjusted p-value was applied to exclude genes that changed expression simply as a result of co-culture (“L-mock vs basal”), while a more stringent threshold (adjusted P<0.001) was applied to the other two comparisons. The resulting 1,645 sequences are involved in 85 differentially regulated pathways identified by PLAGE. 30% of these pathways are implicated in lymphocyte adhesion/movement and/or signalling.
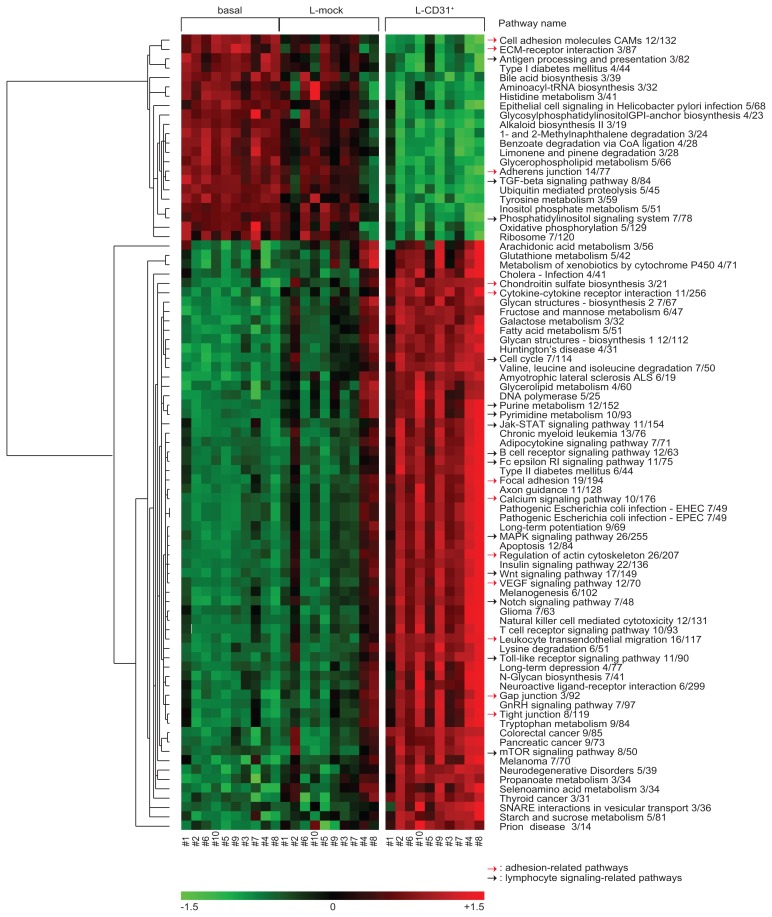
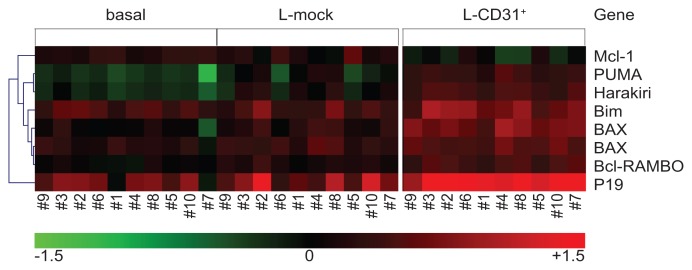
The result was the identification of 1,645 sequences modulated after CD31/CD38 interactions. The list of genes contained into the pathways of the Kyoto Encyclopedia of Genes and Genomes (KEGG) with significant enrichment score (adjusted p-value <0.01) is available as Supplementary Table 1. Pathway Level Analysis of Gene Expression (PLAGE) of the sequences identified 85 differentially regulated pathways (Figure 2 and Supplementary Table 2). Of these, 14 pathways (16%) are involved in lymphocyte signaling (black arrows in Figure 2), while 12 pathways (14%) are implicated in cell adhesion/movement phenomena (red arrows in Figure 2). Relevant examples of up-modulated genes regulating activation/proliferation pertain to the BCR, the MAPK and the TLR signaling pathways (Figure 2 and Supplementary Table 2). Specific attention was dedicated to the panel of 30 apoptosis-regulating genes found unmodulated by Tonino and colleagues (14). In our series, 7/30 genes are differentially regulated upon CD31/CD38 interactions, including P19, Bax, Bcl-RAMBO, Bim, PUMA, Harakiri and Mcl-1 (Figure 3). All of these genes, with the exception of Mcl-1, are up-regulated upon CD31/CD38 interactions, suggesting that de-regulation of apoptosis is a component of this signature. The apparent contrast of these results with those of Tonino et al. could be explained on the basis of experimental differences (3T3 cells versus L-fibroblasts, 72 versus 120 hrs incubation) and/or on the basis of characteristics intrinsic to the patients selected.
Figure 2. Representation of the expression pattern of 85 pathways differentially expressed as a consequence of CD31/CD38 interactions.
Hierarchical clustering was applied to the matrix of the activity levels of the 85 KEGG pathways found as differentially expressed in the L-CD31+ samples as compared to the basal and L-mock ones by PLAGE analysis.
Figure 3. Representation of the 7 apoptosis-related genes differentially expressed upon CD31/CD38 engagement.
Expression pattern of 7 genes of the 1,645 gene signature that are also part of the 30 apoptosis-regulating genes reported in (14). Red squares refer to up-regulation in the sample with respect to the Universal Reference, while green squares to down-regulation. Expression ratios are represented in a log10 scale. All the genes but Mcl-1 increase their expression as a consequence of CD31/CD38 interactions. Two different Agilent probes specific for Bax are over-expressed in the L-CD31+ condition.
In the context of the control of cell movement, relevant examples are up-modulation of genes ruling leukocyte transendothelial migration, actin cytoskeleton and focal adhesion on the one hand, and down-regulation of genes influencing interactions with the extracellular matrix and coding for cell adhesion molecules on the other. This genetic signature implies that long-term CD31/CD38 interactions modulate the competence of CLL cells to grow and progress in response to microenvironmental signals and conditions. Independent immunohistochemical studies on lymph node sections revealed a direct association between the number of endothelial cells (CD31+) and the level of CD38 expression by CLL cells (13), indirectly confirming that CD31/CD38 static interactions occur in vivo. Moreover, the percentage of CD38+ CLL cells (8, 18) and the density of tumor vessels (19, 20) correlate with neoplastic proliferation and disease aggressiveness.
In conclusion, these results provide further support to the hypothesis that CLL maintenance and progression derive not only from the essential drive provided by the antigen but also from a number of accessory signals that modify the microenvironment favoring localization of neoplastic cells to growth-permissive sites. The CD31/CD38 axis is part of this network.
Supplementary Material
| Accession # | Name | Description | L-CD31+ vs L-mock | p value |
|---|---|---|---|---|
| NM_199203 | Kua-UEV | Ubiquitin-conjugating enzyme E2 variant 1 | −12.01382 | 7.62476E-07 |
| NM_004218 | RAB11B | RAB11B, member RAS oncogene family | 12.37772 | 5.9108E-07 |
| NM_006544 | EXOC5 | Exocyst complex component 5 | 10.28434 | 2.8321E-06 |
| NM_212472 | PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 16.0055 | 6.41336E-08 |
| NM_003934 | FUBP3 | Far upstream element (FUSE) binding protein 3 | 10.04849 | 3.43697E-06 |
| NM_153693 | HOXC6 | Homeobox C4 | 10.30985 | 2.77405E-06 |
| NM_022491 | SUDS3 | Suppressor of defective silencing 3 homolog (S. cerevisiae) | −11.04648 | 1.55368E-06 |
| NM_015330 | SPECC1L | SPECC1-like | −10.25039 | 2.91144E-06 |
| NM_001429 | EP300 | E1A binding protein p300 | −11.65812 | 9.84564E-07 |
| BX090412 | BX090412 | Transcribed locus | −14.57157 | 1.45057E-07 |
| NM_001261 | CDK9 | Cyclin-dependent kinase 9 (CDC2-related kinase) | 9.86276 | 4.01395E-06 |
| NM_002661 | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | −11.67385 | 9.73359E-07 |
| NM_000945 | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | 9.70376 | 4.59333E-06 |
| NM_177965 | C8orf37 | Chromosome 8 open reading frame 37 | 12.04077 | 7.48054E-07 |
| NM_024325 | ZNF343 | Zinc finger protein 343 | −9.47095 | 5.61486E-06 |
| NM_025136 | OPA3 | Optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) | 9.4887 | 5.52874E-06 |
| NM_201266 | NRP2 | Neuropilin 2 | 10.06726 | 3.38394E-06 |
| NM_019028 | ZDHHC13 | Zinc finger, DHHC-type containing 13 | 9.82075 | 4.15875E-06 |
| THC2304728 | THC2304728 | N1L4_HUMAN (Q99733) Nucleosome assembly protein 1-like 4 (Nucleosome assembly protein 2) (NAP2), partial (59%) [THC2304728] | 9.63357 | 4.87791E-06 |
| NM_000123 | ERCC5 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 (xeroderma pigmentosum, complementation group G (Cockayne syndrome)) | −10.55662 | 2.27557E-06 |
| NM_007363 | NONO | Non-POU domain containing, octamer-binding | −10.45614 | 2.4655E-06 |
| NM_006595 | API5 | Apoptosis inhibitor 5 | 10.44438 | 2.48886E-06 |
| AA832084 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 9.99145 | 3.60381E-06 |
| NM_020640 | DCUN1D1 | DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) | 10.73637 | 1.97476E-06 |
| NM_016040 | TMED5 | Transmembrane emp24 protein transport domain containing 5 | 11.90305 | 8.25054E-07 |
| NM_182540 | DDX26B | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B | −10.73728 | 1.97335E-06 |
| NM_003893 | LDB1 | LIM domain binding 1 | 10.7935 | 1.88856E-06 |
| NM_016545 | IER5 | Immediate early response 5 | 10.53902 | 2.30764E-06 |
| NM_006805 | HNRPA0 | Heterogeneous nuclear ribonucleoprotein A0 | 9.18978 | 7.19589E-06 |
| NM_001229 | CASP9 | Caspase 9, apoptosis-related cysteine peptidase | −12.50316 | 5.42256E-07 |
| NM_004614 | TK2 | Thymidine kinase 2, mitochondrial | 8.99903 | 8.54562E-06 |
| NM_014876 | JOSD1 | Josephin domain containing 1 | 8.61553 | 0.00001 |
| NM_006459 | SPFH1 | SPFH domain family, member 1 | 9.04944 | 8.16368E-06 |
| THC2400714 | THC2400714 | Unknown | −9.97506 | 3.65339E-06 |
| NM_152734 | C6orf89 | Chromosome 6 open reading frame 89 | 12.84733 | 4.29726E-07 |
| NM_012426 | SF3B3 | Splicing factor 3b, subunit 3, 130kDa | 9.53653 | 5.30387E-06 |
| A_23_P207143 | A_23_P207143 | Unknown | 8.51395 | 0.00001 |
| NM_144726 | FLJ31951 | hypothetical protein FLJ31951 | 12.17957 | 6.7841E-07 |
| NM_005109 | OXSR1 | Oxidative-stress responsive 1 | 9.0603 | 8.08387E-06 |
| Y10152 | CRHR2 | Corticotropin releasing hormone receptor 2 | 10.38139 | 2.61814E-06 |
| NM_014554 | SENP1 | SUMO1/sentrin specific peptidase 1 | 8.65206 | 0.00001 |
| NM_022171 | TCTA | T-cell leukemia translocation altered gene | 8.52684 | 0.00001 |
| NM_006565 | CTCF | CCCTC-binding factor (zinc finger protein) | 8.69879 | 0.00001 |
| A_24_P290214 | A_24_P290214 | Unknown | 8.54654 | 0.00001 |
| NM_052987 | CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | 9.19897 | 7.13708E-06 |
| NM_016506 | KBTBD4 | Mitochondrial carrier homolog 2 (C. elegans) | −9.39145 | 6.01911E-06 |
| NM_005026 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | 8.93923 | 9.02436E-06 |
| NM_020462 | ERGIC1 | Endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 | 12.75714 | 4.5648E-07 |
| NM_003005 | SELP | Selectin P (granule membrane protein 140kDa, antigen CD62) | −10.38446 | 2.61168E-06 |
| NM_007126 | VCP | Valosin-containing protein | 8.20306 | 0.00002 |
| NM_015349 | KIAA0240 | KIAA0240 | −8.93919 | 9.02463E-06 |
| NM_177966 | 2'-PDE | 2'-phosphodiesterase | 9.44127 | 5.76213E-06 |
| NM_015184 | PLCL2 | Phospholipase C-like 2 | −9.67039 | 4.72625E-06 |
| NM_013236 | ATXN10 | Ataxin 10 | 9.30208 | 6.51217E-06 |
| A_24_P581311 | A_24_P581311 | Unknown | 8.04345 | 0.00002 |
| A_24_P400616 | A_24_P400616 | Unknown | 7.90396 | 0.00002 |
| NM_174907 | PPP4R2 | Protein phosphatase 4, regulatory subunit 2 | 8.18701 | 0.00002 |
| NM_017897 | OXSM | 3-oxoacyl-ACP synthase, mitochondrial | −10.2951 | 2.80746E-06 |
| NM_054027 | ANKH | Ankylosis, progressive homolog (mouse) | 9.32545 | 6.37908E-06 |
| NM_005498 | AP1M2 | Adaptor-related protein complex 1, mu 2 subunit | 8.86146 | 9.69158E-06 |
| NM_005986 | SOX1 | SRY (sex determining region Y)-box 1 | 13.28268 | 3.22755E-07 |
| NM_007236 | CHP | calcium binding protein P22 | 14.02006 | 2.02561E-07 |
| BC034752 | C9orf62 | Chromosome 9 open reading frame 62 | 10.18258 | 3.07735E-06 |
| NM_022067 | C14orf133 | Chromosome 14 open reading frame 133 | −8.90563 | 9.30619E-06 |
| NM_002709 | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform | 8.0112 | 0.00002 |
| NM_016312 | WBP11 | WW domain binding protein 11 | −10.32371 | 2.74306E-06 |
| NM_006391 | IPO7 | Importin 7 | 9.46839 | 5.62738E-06 |
| NM_022107 | GPSM3 | G-protein signalling modulator 3 (AGS3-like, C. elegans) | 9.33884 | 6.30415E-06 |
| NM_001190 | BCAT2 | Branched chain aminotransferase 2, mitochondrial | 8.2365 | 0.00002 |
| NM_031892 | SH3KBP1 | SH3-domain kinase binding protein 1 | 8.18059 | 0.00002 |
| BC004815 | MGC5139 | hypothetical protein MGC5139 | −11.00369 | 1.60539E-06 |
| NM_001417 | EIF4B | Eukaryotic translation initiation factor 4B | 11.08825 | 1.50497E-06 |
| NM_017615 | NSMCE4A | Non-SMC element 4 homolog A (S. cerevisiae) | 8.20619 | 0.00002 |
| NM_153335 | LYK5 | protein kinase LYK5 | −14.13835 | 1.88372E-07 |
| AK074764 | SDHALP2 | Succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2 | 7.93021 | 0.00002 |
| NM_005298 | GPR25 | G protein-coupled receptor 25 | 9.62722 | 4.90459E-06 |
| NM_033547 | INTS4 | Integrator complex subunit 4 | 9.65114 | 4.8049E-06 |
| A_24_P780709 | A_24_P780709 | Unknown | 9.37541 | 6.10447E-06 |
| NM_005163 | AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 8.33875 | 0.00002 |
| NM_012316 | KPNA6 | Karyopherin alpha 6 (importin alpha 7) | −11.38421 | 1.20449E-06 |
| A_24_P144556 | A_24_P144556 | Unknown | 8.55649 | 0.00001 |
| NM_001128 | AP1G1 | Adaptor-related protein complex 1, gamma 1 subunit | −11.10174 | 1.4896E-06 |
| NM_001039651 | C6orf26 | MutS homolog 5 (E. coli) | −9.24198 | 6.86871E-06 |
| NM_130791 | WWOX | WW domain containing oxidoreductase | 9.20432 | 7.10308E-06 |
| NM_004082 | DCTN1 | Dynactin 1 (p150, glued homolog, Drosophila) | 10.47834 | 2.42208E-06 |
| NM_031454 | SELO | selenoprotein O | 10.02399 | 3.50756E-06 |
| NM_006544 | EXOC5 | Exocyst complex component 5 | 8.34375 | 0.00002 |
| NM_020960 | GPR107 | G protein-coupled receptor 107 | −10.56817 | 2.25479E-06 |
| NM_203364 | GPIAP1 | GPI-anchored membrane protein 1 | 7.64062 | 0.00003 |
| NM_144576 | COQ10A | Coenzyme Q10 homolog A (S. cerevisiae) | −8.26821 | 0.00002 |
| NM_003089 | SNRP70 | Small nuclear ribonucleoprotein 70kDa polypeptide (RNP antigen) | 8.47566 | 0.00001 |
| NM_005839 | SRRM1 | Serine/arginine repetitive matrix 1 | −10.29528 | 2.80705E-06 |
| NM_016604 | JMJD1B | Jumonji domain containing 1B | −10.08354 | 3.3387E-06 |
| NM_178438 | LCE5A | Late cornified envelope 5A | 9.60463 | 5.00087E-06 |
| AK074764 | SDHALP2 | Succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2 | 8.66586 | 0.00001 |
| NM_002109 | HARS | Histidyl-tRNA synthetase | −9.09351 | 7.84517E-06 |
| NM_152219 | GJC1 | Gap junction protein, chi 1, 31.9kDa (connexin 31.9) | 10.85341 | 1.8026E-06 |
| NM_032322 | RNF135 | Ring finger protein 135 | −9.9372 | 3.77077E-06 |
| A_23_P369758 | A_23_P369758 | Unknown | 10.8002 | 1.87873E-06 |
| NM_001933 | DLST | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 7.44271 | 0.00004 |
| NM_015600 | ABHD12 | Abhydrolase domain containing 12 | 7.97343 | 0.00002 |
| NM_012215 | MGEA5 | Meningioma expressed antigen 5 (hyaluronidase) | −9.01908 | 8.39141E-06 |
| NM_004831 | CRSP7 | Cofactor required for Sp1 transcriptional activation, subunit 7, 70kDa | 8.94066 | 9.01256E-06 |
| NM_016143 | NSFL1C | NSFL1 (p97) cofactor (p47) | −7.96774 | 0.00002 |
| NM_139013 | MAPK14 | Mitogen-activated protein kinase 14 | 8.46192 | 0.00001 |
| CR621132 | NSUN7 | Full-length cDNA clone CS0DC015YK09 of Neuroblastoma Cot 25-normalized of Homo sapiens (human) | −7.48544 | 0.00004 |
| NM_001005404 | YPEL2 | Yippee-like 2 (Drosophila) | −8.59126 | 0.00001 |
| NM_153334 | SCARF2 | Scavenger receptor class F, member 2 | 7.52965 | 0.00004 |
| NM_018416 | FOXJ2 | ForkheadJ2 box | −8.811 | 0.00001 |
| NM_014607 | UBXD2 | UBX domain containing 2 | 8.99319 | 8.59109E-06 |
| NM_025263 | PRR3 | Major histocompatibility complex, class I, E | 8.13755 | 0.00002 |
| NM_052853 | ADCK2 | AarF domain containing kinase 2 | −9.20982 | 7.06829E-06 |
| THC2316116 | THC2316116 | Unknown | −9.80843 | 4.20233E-06 |
| AK023797 | KIAA0672 | KIAA0672 gene product | −12.98441 | 3.92319E-07 |
| NM_025207 | FLAD1 | FAD1 flavin adenine dinucleotide synthetase homolog (S. cerevisiae) | 8.22176 | 0.00002 |
| BQ548663 | ALOX5 | Arachidonate 5-lipoxygenase | −8.8633 | 9.67523E-06 |
| A_24_P101493 | A_24_P101493 | Unknown | 8.46769 | 0.00001 |
| NM_018144 | SEC61A2 | Sec61 alpha 2 subunit (S. cerevisiae) | 8.43895 | 0.00001 |
| NM_002593 | PCOLCE | Procollagen C-endopeptidase enhancer | 7.63508 | 0.00003 |
| NM_024545 | SAP130 | Sin3A-associated protein, 130kDa | −8.3626 | 0.00002 |
| NM_005744 | ARIH1 | Ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) | 14.02495 | 2.01951E-07 |
| CR627148 | LOC728649 | hypothetical protein LOC728649 | −12.98113 | 3.93171E-07 |
| NM_173833 | SCARA5 | Scavenger receptor class A, member 5 (putative) | 10.19322 | 3.05064E-06 |
| NM_014337 | PPIL2 | Peptidylprolyl isomerase (cyclophilin)-like 2 | 10.82689 | 1.84012E-06 |
| NM_003791 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | −13.00009 | 3.88276E-07 |
| NM_182543 | NSUN6 | NOL1/NOP2/Sun domain family, member 6 | −7.91983 | 0.00002 |
| NM_197957 | MAX | MYC associated factor X | 8.86102 | 9.69551E-06 |
| NM_014669 | NUP93 | Nucleoporin 93kDa | 7.4057 | 0.00004 |
| NM_006522 | WNT6 | Wingless-type MMTV integration site family, member 6 | 8.10676 | 0.00002 |
| NM_170662 | CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | −7.59917 | 0.00003 |
| NM_014800 | ELMO1 | Engulfment and cell motility 1 | −8.86216 | 9.68535E-06 |
| NM_014932 | NLGN1 | Neuroligin 1 | 11.85202 | 8.55769E-07 |
| NM_000401 | EXT2 | Exostoses (multiple) 2 | −11.00481 | 0.000001604 |
| NM_000439 | PCSK1 | Proprotein convertase subtilisin/kexin type 1 | 10.68261 | 2.05985E-06 |
| NM_032228 | MLSTD2 | Male sterility domain containing 2 | 7.87725 | 0.00003 |
| NM_015575 | TNRC15 | Trinucleotide repeat containing 15 | 10.81159 | 1.86214E-06 |
| NM_032383 | HPS3 | Hermansky-Pudlak syndrome 3 | −7.7146 | 0.00003 |
| NM_014303 | PES1 | Pescadillo homolog 1, containing BRCT domain (zebrafish) | 11.27587 | 1.30602E-06 |
| NM_138811 | C7orf31 | Chromosome 7 open reading frame 31 | −7.84466 | 0.00003 |
| THC2306788 | THC2306788 | ALU1_HUMAN (P39188) Alu subfamily J sequence contamination warning entry, partial (10%) [THC2306788] | 8.36187 | 0.00002 |
| NM_001077207 | SEC31A | SEC31 homolog A (S. cerevisiae) | −10.01301 | 3.53971E-06 |
| NM_021030 | ZNF14 | Zinc finger protein 14 | −9.40692 | 5.93797E-06 |
| NM_001002259 | C1QDC1 | C1q domain containing 1 | −7.34532 | 0.00004 |
| NM_005370 | RAB8A | RAB8A, member RAS oncogene family | 8.28127 | 0.00002 |
| AK131269 | MBOAT1 | Membrane bound O-acyltransferase domain containing 1 | −9.47467 | 5.5967E-06 |
| NM_005827 | SLC35B1 | Solute carrier family 35, member B1 | 8.26348 | 0.00002 |
| A_24_P332754 | A_24_P332754 | Unknown | 7.25643 | 0.00005 |
| NM_031992 | EIF4H | Eukaryotic translation initiation factor 4H | 8.331 | 0.00002 |
| NM_001011658 | TRAPPC2 | Trafficking protein particle complex 2 | −9.81802 | 4.16839E-06 |
| NM_198563 | TMEM110 | Transmembrane protein 110 | 7.20495 | 0.00005 |
| NM_024419 | PGS1 | Phosphatidylglycerophosphate synthase 1 | 11.5625 | 1.05585E-06 |
| THC2284389 | THC2284389 | C40201 artifact-warning sequence (translated ALU class C) - human {Homo sapiens;} , partial (6%) [THC2284389] | −8.27074 | 0.00002 |
| NR_002144 | LOC407835 | Homo sapiens mitogen-activated protein kinase kinase 2 pseudogene (LOC407835) on chromosome 7. | 7.15658 | 0.00005 |
| NM_012179 | FBXO7 | F-box protein 7 | −8.73795 | 0.00001 |
| NM_016076 | C1orf121 | Chromosome 1 open reading frame 121 | 11.06158 | 1.53587E-06 |
| A_24_P878366 | A_24_P878366 | Unknown | 13.11769 | 3.59374E-07 |
| NM_032336 | GINS4 | Golgi autoantigen, golgin subfamily a, 7 | 7.52996 | 0.00004 |
| NM_004530 | MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 12.50249 | 5.42507E-07 |
| A_32_P83520 | A_32_P83520 | Unknown | 8.84018 | 9.8835E-06 |
| NM_198976 | TH1L | TH1-like (Drosophila) | 6.989 | 0.00006 |
| NM_017668 | NDE1 | NudE nuclear distribution gene E homolog 1 (A. nidulans) | 7.69641 | 0.00003 |
| NM_198963 | DHX57 | DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 | −10.92259 | 1.70868E-06 |
| NM_006101 | KNTC2 | Kinetochore associated 2 | 7.11802 | 0.00006 |
| NM_002967 | SAFB | Scaffold attachment factor B | 7.17136 | 0.00005 |
| NM_017745 | BCOR | BCL6 co-repressor | 8.15186 | 0.00002 |
| NM_006711 | RNPS1 | RNA binding protein S1, serine-rich domain | 7.2874 | 0.00005 |
| NM_014268 | MAPRE2 | Microtubule-associated protein, RP/EB family, member 2 | 7.51812 | 0.00004 |
| BC057844 | BC057844 | CDNA clone MGC:71846 IMAGE:5313492 | 10.04434 | 3.44883E-06 |
| NM_002525 | NRD1 | Nardilysin (N-arginine dibasic convertase) | −9.26143 | 0.000006751 |
| NM_014089 | NUPL1 | Nucleoporin like 1 | 7.10305 | 0.00006 |
| NM_007131 | ZNF75 | Zinc finger protein 75 (D8C6) | −8.06537 | 0.00002 |
| NM_014877 | HELZ | Helicase with zinc finger | −10.7689 | 1.92516E-06 |
| NM_012478 | WBP2 | WW domain binding protein 2 | 8.60644 | 0.00001 |
| NM_001620 | AHNAK | AHNAK nucleoprotein (desmoyokin) | 7.85031 | 0.00003 |
| NM_018195 | C11orf57 | Chromosome 11 open reading frame 57 | −8.5476 | 0.00001 |
| NM_024541 | C10orf76 | Chromosome 10 open reading frame 76 | 9.53683 | 5.30248E-06 |
| NM_016290 | UIMC1 | Ubiquitin interaction motif containing 1 | −7.07217 | 0.00006 |
| NM_015005 | KIAA0284 | KIAA0284 | 9.65504 | 4.78885E-06 |
| BC002431 | B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 | 10.07832 | 3.35313E-06 |
| NM_014760 | TATDN2 | TatD DNase domain containing 2 | −7.94034 | 0.00002 |
| NM_024881 | SLC35E1 | Solute carrier family 35, member E1 | 8.64898 | 0.00001 |
| NM_003111 | SP3 | Sp3 transcription factor | 10.18982 | 3.05914E-06 |
| NM_019024 | KIAA1414 | KIAA1414 protein | −10.63453 | 2.13938E-06 |
| NM_138707 | BCL7B | B-cell CLL/lymphoma 7B | 7.05423 | 0.00006 |
| NM_016824 | ADD3 | Adducin 3 (gamma) | −7.17037 | 0.00005 |
| NM_004309 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 8.3467 | 0.00002 |
| THC2314371 | THC2314371 | ALU8_HUMAN (P39195) Alu subfamily SX sequence contamination warning entry, partial (10%) [THC2314371] | −8.75011 | 0.00001 |
| NM_016079 | VPS24 | Vacuolar protein sorting 24 homolog (S. cerevisiae) | 10.94974 | 1.67331E-06 |
| NM_013282 | UHRF1 | Ubiquitin-like, containing PHD and RING finger domains, 1 | 7.78277 | 0.00003 |
| NM_032775 | KLHL22 | Kelch-like 22 (Drosophila) | 7.71952 | 0.00003 |
| NM_020648 | TWSG1 | Twisted gastrulation homolog 1 (Drosophila) | 7.84782 | 0.00003 |
| THC2444634 | THC2444634 | AF195969 rho GTPase activating protein 8 isoform 2 {Homo sapiens;} , partial (8%) [THC2444634] | 8.87065 | 9.60999E-06 |
| AK056150 | LOC149478 | hypothetical protein LOC149478 | 7.34421 | 0.00004 |
| A_24_P101181 | A_24_P101181 | Unknown | 7.72485 | 0.00003 |
| NM_014657 | KIAA0406 | KIAA0406 | 13.42386 | 2.94683E-07 |
| NM_004808 | NMT2 | N-myristoyltransferase 2 | −6.83368 | 0.00008 |
| NM_004249 | RAB28 | RAB28, member RAS oncogene family | 7.06441 | 0.00006 |
| NM_018121 | C10orf6 | Chromosome 10 open reading frame 6 | −9.42738 | 5.83253E-06 |
| NM_012395 | PFTK1 | PFTAIRE protein kinase 1 | −7.45833 | 0.00004 |
| NM_080596 | HIST1H2AH | Histone cluster 1, H2ai | 7.1406 | 0.00005 |
| NM_003806 | HRK | Harakiri, BCL2 interacting protein (contains only BH3 domain) | 6.88927 | 0.00007 |
| NM_032438 | L3MBTL3 | L(3)mbt-like 3 (Drosophila) | −7.00205 | 0.00006 |
| NM_006290 | TNFAIP3 | Tumor necrosis factor, alpha-induced protein 3 | 8.65812 | 0.00001 |
| NM_015061 | JMJD2C | Jumonji domain containing 2C | −10.21014 | 3.00869E-06 |
| NM_153813 | ZFPM1 | Zinc finger protein, multitype 1 | 8.03175 | 0.00002 |
| BC040982 | PIK3C3 | Phosphoinositide-3-kinase, class 3 | −8.2424 | 0.00002 |
| NM_001012651 | NKTR | Natural killer-tumor recognition sequence | −8.32262 | 0.00002 |
| BC028192 | LOC161635 | similar to casein kinase I alpha | 6.79522 | 0.00008 |
| AK002023 | FLJ25715 | CDNA FLJ11161 fis, clone PLACE1007021 | −9.2345 | 6.9146E-06 |
| NM_001684 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | 9.10183 | 7.78657E-06 |
| NM_004171 | SLC1A2 | Homo sapiens solute carrier family 1 (glial high affinity glutamate transporter), member 2 (SLC1A2), mRNA. | 7.93271 | 0.00002 |
| NM_006766 | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 | −8.77256 | 0.00001 |
| NM_018622 | PARL | Presenilin associated, rhomboid-like | 8.68819 | 0.00001 |
| NM_006224 | PITPNA | Phosphatidylinositol transfer protein, alpha | 6.70394 | 0.00009 |
| NM_002873 | RAD17 | RAD17 homolog (S. pombe) | −8.20583 | 0.00002 |
| THC2280638 | THC2280638 | RL2A_HUMAN (P46776) 60S ribosomal protein L27a, partial (24%) [THC2280638] | −7.53358 | 0.00004 |
| BX427435 | BX427435 | Transcribed locus | 7.05574 | 0.00006 |
| NM_001017395 | TMCC1 | Transmembrane and coiled-coil domain family 1 | 7.44023 | 0.00004 |
| A_24_P502660 | A_24_P502660 | Unknown | 8.49614 | 0.00001 |
| NM_181838 | UBE2D2 | Ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | −14.33002 | 1.67656E-07 |
| NM_002468 | MYD88 | Myeloid differentiation primary response gene (88) | 7.41285 | 0.00004 |
| NM_022487 | DCLRE1C | DNA cross-link repair 1C (PSO2 homolog, S. cerevisiae) | −8.35217 | 0.00002 |
| NM_032364 | DNAJC14 | DnaJ (Hsp40) homolog, subfamily C, member 14 | −11.12091 | 1.46806E-06 |
| NM_002737 | PRKCA | Protein kinase C, alpha | 11.80087 | 8.87837E-07 |
| BQ317309 | GCSH | Glycine cleavage system protein H (aminomethyl carrier) | 6.8177 | 0.00008 |
| NM_014360 | NKX2-8 | NK2 transcription factor related, locus 8 (Drosophila) | 7.31477 | 0.00004 |
| AJ002425 | HSAJ2425 | p65 protein | 8.11665 | 0.00002 |
| NM_006763 | BTG2 | BTG family, member 2 | −7.82584 | 0.00003 |
| A_24_P118721 | A_24_P118721 | Unknown | 9.15287 | 7.43754E-06 |
| NM_138763 | BAX | BCL2-associated X protein | 9.65811 | 4.77628E-06 |
| NM_005819 | STX6 | Syntaxin 6 | 7.83975 | 0.00003 |
| NM_017730 | QRICH1 | Glutamine-rich 1 | −10.66821 | 2.08332E-06 |
| NM_006827 | TMED10 | Transmembrane emp24-like trafficking protein 10 (yeast) | 7.98498 | 0.00002 |
| NM_012118 | CCRN4L | CCR4 carbon catabolite repression 4-like (S. cerevisiae) | 7.30251 | 0.00005 |
| NM_033532 | CDC2L2 | Cell division cycle 2-like 2 (PITSLRE proteins) | 6.8601 | 0.00007 |
| NM_177966 | 2'-PDE | 2'-phosphodiesterase | 8.47363 | 0.00001 |
| NM_002103 | GYS1 | Glycogen synthase 1 (muscle) | −8.92352 | 9.15489E-06 |
| NM_182547 | TMED4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 56 | 7.46187 | 0.00004 |
| NM_021149 | COTL1 | Coactosin-like 1 (Dictyostelium) | −7.01049 | 0.00006 |
| NM_002145 | HOXB2 | Homeobox B2 | 8.19278 | 0.00002 |
| NM_001002909 | GPATCH8 | G patch domain containing 8 | −9.04408 | 8.20341E-06 |
| NM_021649 | TICAM2 | Toll-like receptor adaptor molecule 2 | 6.59711 | 0.0001 |
| NM_022558 | GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) | 8.05993 | 0.00002 |
| NM_194247 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 7.43335 | 0.00004 |
| NM_014153 | ZC3H7A | Zinc finger CCCH-type containing 7A | 7.05942 | 0.00006 |
| NM_153045 | C9orf91 | Chromosome 9 open reading frame 91 | −7.73743 | 0.00003 |
| ENST00000312847 | ENST00000312847 | Unknown | 6.86646 | 0.00007 |
| NM_018013 | FLJ10159 | hypothetical protein FLJ10159 | −7.60685 | 0.00003 |
| X16323 | HGF | Human mRNA for hepatocyte growth factor (HGF). | 9.5554 | 5.21792E-06 |
| A_32_P331700 | A_32_P331700 | Unknown | 6.7671 | 0.00008 |
| AK095600 | ZNF709 | Homo sapiens cDNA FLJ38281 fis, clone FCBBF3005729, moderately similar to Homo sapiens GIOT-4 mRNA for gonadotropin inducible transcription repressor-4. | −7.53387 | 0.00004 |
| NM_001031628 | LOC57228 | small trans-membrane and glycosylated protein | 10.54713 | 2.29279E-06 |
| NM_015001 | SPEN | Spen homolog, transcriptional regulator (Drosophila) | −10.27584 | 2.85175E-06 |
| NM_002229 | JUNB | Jun B proto-oncogene | 12.09317 | 7.20878E-07 |
| BX647217 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 8.416 | 0.00001 |
| NM_014384 | ACAD8 | Acyl-Coenzyme A dehydrogenase family, member 8 | −7.18087 | 0.00005 |
| NM_138575 | PGAM5 | Phosphoglycerate mutase family member 5 | 7.49043 | 0.00004 |
| NM_003884 | PCAF | P300/CBP-associated factor | −8.25455 | 0.00002 |
| AK122976 | ZNF275 | Zinc finger protein 275 | −8.96519 | 8.81293E-06 |
| NM_017911 | FAM118A | Family with sequence similarity 118, member A | 11.18404 | 1.39951E-06 |
| NM_003791 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | −11.73778 | 9.29231E-07 |
| AK096643 | FLJ22184 | hypothetical protein FLJ22184 | 10.23909 | 2.9384E-06 |
| NM_017651 | AHI1 | Abelson helper integration site 1 | −8.65797 | 0.00001 |
| NM_002541 | OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | 7.30081 | 0.00005 |
| NM_005561 | LAMP1 | Lysosomal-associated membrane protein 1 | 7.79006 | 0.00003 |
| NM_015476 | C18orf10 | Chromosome 18 open reading frame 10 | 7.4762 | 0.00004 |
| NM_006380 | APPBP2 | Amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 8.89445 | 9.40209E-06 |
| BC007879 | FAM120A | Family with sequence similarity 120A | 7.06812 | 0.00006 |
| NM_018221 | MOBK1B | MOB1, Mps One Binder kinase activator-like 1B (yeast) | 7.46482 | 0.00004 |
| NM_147148 | GSTM4 | Glutathione S-transferase M4 | 8.61646 | 0.00001 |
| AA451906 | BIN1 | Bridging integrator 1 | 7.4277 | 0.00004 |
| NM_032383 | HPS3 | Hermansky-Pudlak syndrome 3 | −6.97134 | 0.00007 |
| A_24_P808838 | A_24_P808838 | Unknown | 6.73859 | 0.00008 |
| NM_001042537 | SLC9A6 | Solute carrier family 9 (sodium/hydrogen exchanger), member 6 | −7.75826 | 0.00003 |
| NM_031434 | TMUB1 | Transmembrane and ubiquitin-like domain containing 1 | 12.22266 | 6.5827E-07 |
| AB020648 | KIAA0841 | KIAA0841 | −8.89967 | 9.35718E-06 |
| NM_015120 | ALMS1 | Alstrom syndrome 1 | −8.99658 | 8.5646E-06 |
| NM_015401 | HDAC7A | Histone deacetylase 7A | −6.57001 | 0.0001 |
| NM_000189 | HK2 | Hexokinase 2 | 6.72803 | 0.00009 |
| NM_021149 | COTL1 | Coactosin-like 1 (Dictyostelium) | −6.86527 | 0.00007 |
| CR603450 | TRIM52 | Tripartite motif-containing 52 | −7.76938 | 0.00003 |
| NM_175863 | ARID1B | AT rich interactive domain 1B (SWI1-like) | 12.1051 | 7.14841E-07 |
| THC2295157 | THC2295157 | Unknown | −7.06649 | 0.00006 |
| AJ008005 | PSEN1 | Presenilin 1 (Alzheimer disease 3) | 9.19485 | 7.16338E-06 |
| NM_007326 | CYB5R3 | Cytochrome b5 reductase 3 | 6.89825 | 0.00007 |
| AK096104 | LOC727820 | hypothetical protein LOC727820 | −9.71542 | 4.54783E-06 |
| NM_000319 | PEX5 | Peroxisomal biogenesis factor 5 | 10.03095 | 3.48736E-06 |
| BF895757 | TRIM28 | Tripartite motif-containing 28 | 6.34251 | 0.00013 |
| BM509577 | THC2281731 | Transcribed locus, strongly similar to XP_001152474.1 hypothetical protein [Pan troglodytes] | 10.69767 | 2.03562E-06 |
| NM_021188 | ZNF410 | Zinc finger protein 410 | −12.20709 | 6.65471E-07 |
| NM_001681 | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 6.35897 | 0.00013 |
| THC2377679 | THC2377679 | Unknown | −10.62465 | 2.15615E-06 |
| NM_198494 | ZNF642 | Zinc finger protein 642 | −7.31971 | 0.00004 |
| NM_017706 | WDR55 | WD repeat domain 55 | 7.59863 | 0.00003 |
| NM_024757 | EHMT1 | Euchromatic histone-lysine N-methyltransferase 1 | 8.32535 | 0.00002 |
| A_24_P340966 | A_24_P340966 | Unknown | 7.90415 | 0.00002 |
| ENST00000380831 | ENST00000380831 | LPXK_RHIME (Q92RP7) Tetraacyldisaccharide 4'-kinase (Lipid A 4'-kinase) , partial (5%) [THC2374771] | −6.42385 | 0.00012 |
| NM_013446 | MKRN1 | Makorin, ring finger protein, 1 | −6.90506 | 0.00007 |
| XM_928285 | ENST00000375923 | PREDICTED: Homo sapiens similar to protein expressed in prostate, ovary, testis, and placenta 15, transcript variant 1 (LOC653239), mRNA [XM_928285] | 6.42221 | 0.00012 |
| BX648945 | EYA3 | Eyes absent homolog 3 (Drosophila) | −8.35174 | 0.00002 |
| NM_152367 | C1orf161 | Chromosome 1 open reading frame 161 | 8.07991 | 0.00002 |
| NM_015361 | R3HDM1 | R3H domain containing 1 | −8.80786 | 0.00001 |
| NM_152896 | UHRF2 | Ubiquitin-like, containing PHD and RING finger domains, 2 | −6.81973 | 0.00008 |
| NM_001001396 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | −6.32724 | 0.00014 |
| NM_020847 | TNRC6A | Homo sapiens trinucleotide repeat containing 6A (TNRC6A), mRNA. | 7.32895 | 0.00004 |
| AK074590 | SFRS16 | Splicing factor, arginine/serine-rich 16 | 8.55204 | 0.00001 |
| NM_001018052 | POLR3H | Polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) | 6.76245 | 0.00008 |
| NM_012256 | ZNF212 | Zinc finger protein 398 | −7.93151 | 0.00002 |
| NM_004850 | ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | −8.12669 | 0.00002 |
| NM_053041 | COMMD7 | COMM domain containing 7 | 6.43078 | 0.00012 |
| NM_015382 | HECTD1 | HECT domain containing 1 | −9.02879 | 8.3178E-06 |
| NM_033116 | NEK9 | NIMA (never in mitosis gene a)- related kinase 9 | 6.35421 | 0.00013 |
| NM_002886 | RAP2B | RAP2B, member of RAS oncogene family | 9.69267 | 4.63706E-06 |
| NM_014846 | KIAA0196 | KIAA0196 | −6.66666 | 0.00009 |
| NM_022658 | HOXC8 | Homeobox C4 | 6.23915 | 0.00015 |
| NM_004788 | UBE4A | Ubiquitination factor E4A (UFD2 homolog, yeast) | −7.44899 | 0.00004 |
| NM_022776 | OSBPL11 | Oxysterol binding protein-like 11 | −7.44389 | 0.00004 |
| BC018035 | IMP5 | intramembrane protease 5 | 10.04526 | 3.4462E-06 |
| NM_001007559 | SS18 | Synovial sarcoma translocation, chromosome 18 | 13.94595 | 2.12052E-07 |
| AK000675 | 1-Mar | Membrane-associated ring finger (C3HC4) 1 | −6.55294 | 0.0001 |
| NM_004710 | SYNGR2 | Synaptogyrin 2 | 6.65626 | 0.00009 |
| NM_014749 | KIAA0586 | KIAA0586 | −6.46727 | 0.00012 |
| NM_014925 | R3HDM2 | Homo sapiens R3H domain containing 2 (R3HDM2), mRNA. | −8.92219 | 9.16608E-06 |
| NM_030567 | PRR7 | Proline rich 7 (synaptic) | 7.36757 | 0.00004 |
| NM_173514 | FLJ90709 | hypothetical protein FLJ90709 | −7.07812 | 0.00006 |
| NM_144653 | BTBD14A | BTB (POZ) domain containing 14A | 9.27991 | 6.64126E-06 |
| BC063441 | BC063441 | Homo sapiens cDNA clone IMAGE:5527329, partial cds. | 7.14285 | 0.00005 |
| NM_000379 | XDH | Xanthine dehydrogenase | 6.77939 | 0.00008 |
| BX648958 | BX648958 | Shinc-4 mRNA, partial sequence | −8.74774 | 0.00001 |
| NM_007275 | TUSC2 | Tumor suppressor candidate 2 | 8.78322 | 0.00001 |
| NM_005487 | HMG2L1 | High-mobility group protein 2-like 1 | −7.02563 | 0.00006 |
| NM_020245 | TULP4 | Tubby like protein 4 | −8.25514 | 0.00002 |
| NM_032788 | ZNF514 | Mitochondrial ribosomal protein S5 | −8.39584 | 0.00002 |
| ENST00000331736 | ENST00000331736 | Q8NH99 (Q8NH99) Seven transmembrane helix receptor, partial (77%) [THC2437350] | 8.07755 | 0.00002 |
| NM_182314 | COVA1 | Cytosolic ovarian carcinoma antigen 1 | −10.02086 | 3.51669E-06 |
| NM_015330 | SPECC1L | SPECC1-like | −9.41199 | 5.91167E-06 |
| NM_014649 | SAFB2 | Small nuclear ribonucleoprotein polypeptide E | 7.43943 | 0.00004 |
| NM_007253 | CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | 8.22057 | 0.00002 |
| AK130071 | ZBTB44 | Zinc finger and BTB domain containing 44 | −6.57362 | 0.0001 |
| NM_145006 | SUSD3 | Sushi domain containing 3 | −7.18067 | 0.00005 |
| NM_001394 | DUSP4 | Dual specificity phosphatase 4 | 7.89185 | 0.00002 |
| NM_007175 | SPFH2 | SPFH domain family, member 2 | −8.77846 | 0.00001 |
| NM_080611 | DUSP15 | Dual specificity phosphatase 15 | 6.86459 | 0.00007 |
| NM_033296 | MRFAP1 | Mof4 family associated protein 1 | 7.20985 | 0.00005 |
| NM_006037 | HDAC4 | Histone deacetylase 4 | 6.91398 | 0.00007 |
| NM_001033858 | DCLRE1C | DNA cross-link repair 1C (PSO2 homolog, S. cerevisiae) | −6.19183 | 0.00016 |
| NM_012181 | FKBP8 | FK506 binding protein 8, 38kDa | 10.34449 | 2.69733E-06 |
| AK055921 | MST101 | MSTP101 | −10.08009 | 3.34822E-06 |
| AK057884 | COG3 | Component of oligomeric golgi complex 3 | 6.83815 | 0.00008 |
| NM_015448 | RP11-529I10.4 | deleted in a mouse model of primary ciliary dyskinesia | 7.35999 | 0.00004 |
| NM_205861 | DHDDS | Dehydrodolichyl diphosphate synthase | 9.46651 | 5.63664E-06 |
| NM_003255 | TIMP2 | TIMP metallopeptidase inhibitor 2 | 9.53344 | 5.31808E-06 |
| AJ420589 | FAM120B | Family with sequence similarity 120B | −8.32685 | 0.00002 |
| NM_017851 | PARP16 | Poly (ADP-ribose) polymerase family, member 16 | −8.30035 | 0.00002 |
| NM_001002021 | PFKL | Phosphofructokinase, liver | 6.55024 | 0.00011 |
| NM_004491 | GRLF1 | Glucocorticoid receptor DNA binding factor 1 | 6.56125 | 0.0001 |
| NM_022457 | RFWD2 | Ring finger and WD repeat domain 2 | −6.49304 | 0.00011 |
| NM_006555 | YKT6 | YKT6 v-SNARE homolog (S. cerevisiae) | 6.62617 | 0.0001 |
| NM_018452 | C6orf35 | Chromosome 6 open reading frame 35 | 6.82308 | 0.00008 |
| NM_006464 | TGOLN2 | Trans-golgi network protein 2 | −7.19236 | 0.00005 |
| NM_002860 | ALDH18A1 | Aldehyde dehydrogenase 18 family, member A1 | −10.20157 | 3.02985E-06 |
| NM_145232 | ATPBD3 | ATP binding domain 3 | 6.34185 | 0.00013 |
| NM_001233 | CAV2 | Caveolin 2 | 10.02277 | 3.51112E-06 |
| A_24_P322908 | A_24_P322908 | Unknown | −7.38585 | 0.00004 |
| NM_139246 | C9orf97 | Chromosome 9 open reading frame 97 | 7.67513 | 0.00003 |
| NM_025154 | UNC84A | Unc-84 homolog A (C. elegans) | −7.71957 | 0.00003 |
| NM_015655 | ZNF337 | Zinc finger protein 337 | −6.34247 | 0.00013 |
| NM_033083 | EAF1 | ELL associated factor 1 | 6.56433 | 0.0001 |
| NM_021096 | CACNA1I | Calcium channel, voltage-dependent, alpha 1I subunit | 8.34352 | 0.00002 |
| NM_014423 | AFF4 | AF4/FMR2 family, member 4 | −6.759 | 0.00008 |
| BC010538 | C18orf18 | Hypothetical protein LOC339290 | −7.97833 | 0.00002 |
| NM_002541 | OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | 8.14865 | 0.00002 |
| NM_174887 | IFT20 | Intraflagellar transport 20 homolog (Chlamydomonas) | 9.26877 | 6.70721E-06 |
| XM_936535 | NDOR1 | PREDICTED: Homo sapiens hypothetical protein LOC648245, transcript variant 1 (LOC648245), mRNA [XM_936535] | 7.74239 | 0.00003 |
| NM_019896 | POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | 8.36258 | 0.00002 |
| NM_016154 | RAB4B | RAB4B, member RAS oncogene family | 6.27108 | 0.00015 |
| NM_033129 | SCRT2 | Scratch homolog 2, zinc finger protein (Drosophila) | 6.66725 | 0.00009 |
| NM_001012614 | CTBP1 | C-terminal binding protein 1 | 7.38024 | 0.00004 |
| BC045778 | XYLT1 | Xylosyltransferase I | −6.16707 | 0.00017 |
| NM_012208 | HARSL | Histidyl-tRNA synthetase-like | −8.54862 | 0.00001 |
| NM_014411 | NAG8 | nasopharyngeal carcinoma associated gene protein-8 | −6.87141 | 0.00007 |
| NM_022893 | BCL11A | B-cell CLL/lymphoma 11A (zinc finger protein) | −7.86838 | 0.00003 |
| NM_000019 | ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 6.17359 | 0.00016 |
| NM_014802 | KIAA0528 | KIAA0528 | −7.27048 | 0.00005 |
| NM_014912 | CPEB3 | Cytoplasmic polyadenylation element binding protein 3 | −9.89204 | 3.91629E-06 |
| NM_152285 | ARRDC1 | Arrestin domain containing 1 | 6.40343 | 0.00012 |
| NM_020438 | DOLPP1 | Dolichyl pyrophosphate phosphatase 1 | 6.43503 | 0.00012 |
| AK090478 | TMC8 | Transmembrane channel-like 8 | −7.59888 | 0.00003 |
| NM_014329 | EDC4 | Enhancer of mRNA decapping 4 | 6.31661 | 0.00014 |
| NM_015229 | KIAA0664 | KIAA0664 | 6.8895 | 0.00007 |
| NM_018360 | CXorf15 | Chromosome X open reading frame 15 | 9.35232 | 6.2297E-06 |
| AK026760 | C20orf119 | Chromosome 20 open reading frame 119 | −7.30047 | 0.00005 |
| NM_048368 | CTDP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 | 6.38575 | 0.00013 |
| NM_174891 | C14orf79 | Chromosome 14 open reading frame 79 | 8.13124 | 0.00002 |
| AK056484 | LOC441204 | hypothetical locus LOC441204 | 8.75709 | 0.00001 |
| NM_020711 | KIAA1189 | KIAA1189 | −7.43592 | 0.00004 |
| NM_024891 | FLJ11783 | hypothetical protein FLJ11783 | −14.18157 | 1.83464E-07 |
| S72422 | DLSTP | E2k=alpha-ketoglutarate dehydrogenase complex dihydrolipoyl succinyltransferase [human, fetal brain, mRNA, 2987 nt]. | 6.15369 | 0.00017 |
| AB018323 | JMJD2C | Jumonji domain containing 2C | −7.69576 | 0.00003 |
| NM_006696 | BRD8 | Bromodomain containing 8 | −8.73726 | 0.00001 |
| NM_177439 | FTSJ1 | FtsJ homolog 1 (E. coli) | 6.55266 | 0.0001 |
| THC2392085 | THC2392085 | Unknown | −7.57601 | 0.00003 |
| NM_005645 | TAF13 | TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa | 6.49776 | 0.00011 |
| NM_182612 | PDDC1 | Parkinson disease 7 domain containing 1 | 10.24294 | 2.92916E-06 |
| NM_004284 | CHD1L | Chromodomain helicase DNA binding protein 1-like | −8.18555 | 0.00002 |
| NM_032354 | TMEM107 | Transmembrane protein 107 | −9.96453 | 3.68563E-06 |
| NM_002182 | IL1RAP | Interleukin 1 receptor accessory protein | −6.23672 | 0.00015 |
| NM_001007071 | RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | 6.0887 | 0.00018 |
| NM_032349 | NUDT16L1 | Nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 | 6.94114 | 0.00007 |
| NM_017951 | SMPD4 | Sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) | 5.9566 | 0.00021 |
| NM_032042 | C5orf21 | Chromosome 5 open reading frame 21 | −5.99255 | 0.0002 |
| AI090937 | LOC729616 | hypothetical protein LOC729616 | −7.02475 | 0.00006 |
| NM_022764 | MTHFSD | Methenyltetrahydrofolate synthetase domain containing | −9.35301 | 6.22594E-06 |
| NM_001315 | MAPK14 | Mitogen-activated protein kinase 14 | −9.49739 | 5.48709E-06 |
| NM_198867 | ALKBH6 | AlkB, alkylation repair homolog 6 (E. coli) | 5.99375 | 0.0002 |
| NM_004155 | SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 8.93566 | 9.05384E-06 |
| NM_198970 | AES | Amino-terminal enhancer of split | 7.18158 | 0.00005 |
| NM_007353 | GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | 7.51614 | 0.00004 |
| BC087847 | ENST00000322446 | CDNA clone IMAGE:30389268 | 8.47062 | 0.00001 |
| NM_145869 | ANXA11 | Annexin A11 | −9.25282 | 6.80287E-06 |
| AB014574 | FKBP15 | FK506 binding protein 15, 133kDa | −13.99188 | 2.06113E-07 |
| NM_033426 | KIAA1737 | KIAA1737 | −7.68569 | 0.00003 |
| BQ425510 | ADNP | Transcribed locus | −8.23074 | 0.00002 |
| NM_016287 | HP1BP3 | Heterochromatin protein 1, binding protein 3 | 8.19698 | 0.00002 |
| NM_016255 | FAM8A1 | Family with sequence similarity 8, member A1 | −6.9287 | 0.00007 |
| NM_004637 | RAB7 | RAB7, member RAS oncogene family | 6.02105 | 0.0002 |
| NM_144982 | CCDC131 | Coiled-coil domain containing 131 | −6.4492 | 0.00012 |
| THC2365247 | THC2365247 | Q6QAQ1 (Q6QAQ1) Cytoskeletal beta actin (Fragment), partial (76%) [THC2365247] | 5.97469 | 0.00021 |
| NM_014719 | KIAA0738 | KIAA0738 gene product | −10.86208 | 1.79053E-06 |
| NM_016028 | SUV420H1 | Suppressor of variegation 4-20 homolog 1 (Drosophila) | 8.9239 | 9.15176E-06 |
| NM_033274 | ADAM19 | ADAM metallopeptidase domain 19 (meltrin beta) | −7.76503 | 0.00003 |
| AF086790 | ACON | Homo sapiens aconitase precursor (ACON) mRNA, nuclear gene encoding mitochondrial protein, partial cds. | 11.59634 | 1.02999E-06 |
| NM_032236 | USP48 | Ubiquitin specific peptidase 48 | −6.86748 | 0.00007 |
| NM_001125 | ADPRH | ADP-ribosylarginine hydrolase | 6.64904 | 0.00009 |
| NM_178508 | C6orf1 | Chromosome 6 open reading frame 1 | 8.21324 | 0.00002 |
| NM_015691 | WWC3 | WWC family member 3 | −9.22295 | 6.98604E-06 |
| NM_032353 | VPS25 | Vacuolar protein sorting 25 homolog (S. cerevisiae) | 7.29826 | 0.00005 |
| NM_016372 | GPR175 | G protein-coupled receptor 175 | 6.49813 | 0.00011 |
| AL359584 | BXDC5 | Brix domain containing 5 | −6.78264 | 0.00008 |
| NM_001009955 | SSBP3 | Single stranded DNA binding protein 3 | 8.57491 | 0.00001 |
| NM_201649 | SLC6A9 | Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 7.77425 | 0.00003 |
| AB075502 | C6orf62 | Chromosome 6 open reading frame 62 | −6.64251 | 0.00009 |
| NM_173587 | RCOR2 | REST corepressor 2 | 7.89684 | 0.00002 |
| NM_001006634 | ARHGAP17 | Rho GTPase activating protein 17 | −9.86551 | 4.00466E-06 |
| NM_003656 | CAMK1 | Calcium/calmodulin-dependent protein kinase I | 7.09836 | 0.00006 |
| NM_017640 | LRRC16 | Leucine rich repeat containing 16 | −6.80494 | 0.00008 |
| CB987747 | CB987747 | Transcribed locus | −7.90385 | 0.00002 |
| NM_052957 | ACRC | Acidic repeat containing | −7.3354 | 0.00004 |
| NM_005634 | SOX3 | SRY (sex determining region Y)-box 3 | 8.01509 | 0.00002 |
| NM_138348 | FAM105B | Family with sequence similarity 105, member B | 7.37875 | 0.00004 |
| NM_017813 | IMPAD1 | Inositol monophosphatase domain containing 1 | 7.59455 | 0.00003 |
| NM_030576 | LIMD2 | LIM domain containing 2 | 12.89038 | 4.17569E-07 |
| NM_000466 | PEX1 | Peroxisome biogenesis factor 1 | −10.02315 | 3.51001E-06 |
| NM_001521 | GTF3C2 | General transcription factor IIIC, polypeptide 2, beta 110kDa | 8.99983 | 8.5394E-06 |
| AK125038 | KIAA0500 | KIAA0500 protein | −5.89084 | 0.00023 |
| NM_015367 | BCL2L13 | BCL2-like 13 (apoptosis facilitator) | 6.02735 | 0.0002 |
| NM_015635 | GAPVD1 | GTPase activating protein and VPS9 domains 1 | 6.35776 | 0.00013 |
| NM_201589 | MAFA | V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian) | 7.51391 | 0.00004 |
| NM_017742 | ZCCHC2 | Zinc finger, CCHC domain containing 2 | −6.89791 | 0.00007 |
| NM_015138 | RTF1 | Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | −7.75704 | 0.00003 |
| NM_030802 | FAM117A | Family with sequence similarity 117, member A | −6.45889 | 0.00012 |
| NM_017580 | ZRANB1 | Zinc finger, RAN-binding domain containing 1 | 7.16513 | 0.00005 |
| CR624826 | WASF2 | CDNA clone IMAGE:3030163 | −7.8809 | 0.00002 |
| NM_004310 | RHOH | Ras homolog gene family, member H | −8.98267 | 8.67368E-06 |
| NM_014827 | ZC3H11A | Zinc finger CCCH-type containing 11A | −9.52615 | 5.35178E-06 |
| NM_021960 | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | −9.30554 | 6.4923E-06 |
| THC2275506 | THC2275506 | ALU4_HUMAN (P39191) Alu subfamily SB2 sequence contamination warning entry, partial (24%) [THC2275506] | −7.19705 | 0.00005 |
| NM_022066 | UBE2O | Ubiquitin-conjugating enzyme E2O | −8.02194 | 0.00002 |
| NM_015100 | POGZ | Pogo transposable element with ZNF domain | −9.44018 | 5.76767E-06 |
| NM_032982 | CASP2 | Caspase 2, apoptosis-related cysteine peptidase (neural precursor cell expressed, developmentally down-regulated 2) | −6.4344 | 0.00012 |
| NM_015568 | PPP1R16B | Protein phosphatase 1, regulatory (inhibitor) subunit 16B | 6.70901 | 0.00009 |
| NM_001017992 | DKFZp686D0972 | similar to RIKEN cDNA 4732495G21 gene | 7.41188 | 0.00004 |
| NM_002073 | GNAZ | Guanine nucleotide binding protein (G protein), alpha z polypeptide | 8.64948 | 0.00001 |
| A_24_P264416 | A_24_P264416 | Unknown | 6.15843 | 0.00017 |
| NM_152682 | RWDD4A | RWD domain containing 4A | −7.11982 | 0.00006 |
| NM_024833 | ZNF671 | Zinc finger protein 671 | −8.82104 | 0.00001 |
| A_24_P410070 | A_24_P410070 | Unknown | −7.09055 | 0.00006 |
| NM_018210 | FLJ10769 | hypothetical protein FLJ10769 | −10.36037 | 2.66292E-06 |
| NM_003194 | TBP | TATA box binding protein | −7.06616 | 0.00006 |
| THC2306239 | THC2306239 | HUMPKD1G08 polycystic kidney disease 1 protein {Homo sapiens;} , partial (12%) [THC2306239] | 6.39854 | 0.00013 |
| NM_021158 | TRIB3 | Tribbles homolog 3 (Drosophila) | 6.25082 | 0.00015 |
| NM_020771 | HACE1 | HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 | −6.76349 | 0.00008 |
| NM_030939 | C6orf62 | Chromosome 6 open reading frame 62 | −6.49244 | 0.00011 |
| ENST00000357697 | ENST00000357697 | Unknown | 7.83769 | 0.00003 |
| NM_021145 | DMTF1 | Cyclin D binding myb-like transcription factor 1 | −7.75897 | 0.00003 |
| NM_014912 | CPEB3 | Cytoplasmic polyadenylation element binding protein 3 | −8.90297 | 9.32893E-06 |
| NM_030569 | ITIH5 | Inter-alpha (globulin) inhibitor H5 | 9.22611 | 6.96638E-06 |
| NM_031858 | NBR1 | Neighbor of BRCA1 gene 1 | −11.10553 | 1.48531E-06 |
| NM_024050 | C19orf58 | Chromosome 19 open reading frame 58 | 7.04054 | 0.00006 |
| NM_014943 | ZHX2 | Zinc fingers and homeoboxes 2 | −8.10382 | 0.00002 |
| NM_173728 | ARHGEF15 | Rho guanine nucleotide exchange factor (GEF) 15 | 6.53943 | 0.00011 |
| NM_014710 | GPRASP1 | G protein-coupled receptor associated sorting protein 2 | −6.26554 | 0.00015 |
| NM_024580 | EFTUD1 | Elongation factor Tu GTP binding domain containing 1 | −6.34432 | 0.00013 |
| AK127374 | AK127374 | CDNA FLJ45450 fis, clone BRSTN2002691 | −8.71223 | 0.00001 |
| BC062753 | MCART1 | Mitochondrial carrier triple repeat 1 | −9.73391 | 4.47672E-06 |
| NM_003640 | IKBKAP | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | −7.39824 | 0.00004 |
| NM_007375 | TARDBP | TAR DNA binding protein | −6.66957 | 0.00009 |
| CR594281 | METTL3 | Methyltransferase like 3 | −8.8068 | 0.00001 |
| NM_002014 | FKBP4 | FK506 binding protein 4, 59kDa | 6.81268 | 0.00008 |
| NM_145279 | MOBKL2C | MOB1, Mps One Binder kinase activator-like 2C (yeast) | 10.59534 | 2.20673E-06 |
| BC043212 | EFHB | EF-hand domain family, member B | 6.97051 | 0.00007 |
| NM_003146 | SSRP1 | Structure specific recognition protein 1 | −6.13864 | 0.00017 |
| NM_015107 | PHF8 | PHD finger protein 8 | −7.1863 | 0.00005 |
| BC051311 | DKFZp547E087 | hypothetical gene LOC283846 | 9.24017 | 6.87976E-06 |
| NM_018688 | BIN3 | Bridging integrator 3 | 5.69995 | 0.00029 |
| NM_024620 | ZNF329 | Zinc finger protein 329 | −7.79871 | 0.00003 |
| NM_015001 | SPEN | Spen homolog, transcriptional regulator (Drosophila) | −8.49844 | 0.00001 |
| NM_014659 | HISPPD2A | Histidine acid phosphatase domain containing 2A | −6.14796 | 0.00017 |
| NM_016598 | ZDHHC3 | Zinc finger, DHHC-type containing 3 | 6.91463 | 0.00007 |
| NM_018353 | C14orf106 | Chromosome 14 open reading frame 106 | −6.98428 | 0.00006 |
| NM_006861 | RAB35 | RAB35, member RAS oncogene family | 11.1048 | 1.48613E-06 |
| NM_018474 | C20orf19 | Homo sapiens chromosome 20 open reading frame 19 (C20orf19), mRNA. | −6.2157 | 0.00016 |
| AB037855 | KIAA1434 | hypothetical protein KIAA1434 | −7.0343 | 0.00006 |
| AL834448 | DKFZp547C195 | hypothetical protein DKFZp547C195 | −11.98823 | 7.7645E-07 |
| NM_004629 | FANCG | Fanconi anemia, complementation group G | −6.22857 | 0.00015 |
| NM_033089 | ZCCHC3 | Zinc finger, CCHC domain containing 3 | −6.36039 | 0.00013 |
| THC2301362 | THC2301362 | Unknown | −7.58392 | 0.00003 |
| AK026896 | STK24 | Serine/threonine kinase 24 (STE20 homolog, yeast) | −5.95955 | 0.00021 |
| NM_032438 | L3MBTL3 | L(3)mbt-like 3 (Drosophila) | −6.4802 | 0.00011 |
| THC2378263 | THC2378263 | Unknown | −6.63826 | 0.0001 |
| NM_032560 | SMEK1 | SMEK homolog 1, suppressor of mek1 (Dictyostelium) | −7.87743 | 0.00003 |
| NM_030915 | LBH | Limb bud and heart development homolog (mouse) | 8.06636 | 0.00002 |
| NM_004364 | CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha | 9.5187 | 5.3865E-06 |
| NM_006278 | ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | 5.76877 | 0.00027 |
| NM_018996 | TNRC6C | Trinucleotide repeat containing 6C | 6.87855 | 0.00007 |
| NM_016479 | SCOTIN | scotin | −10.49939 | 2.38168E-06 |
| NM_019555 | ARHGEF3 | Rho guanine nucleotide exchange factor (GEF) 3 | −10.73596 | 1.9754E-06 |
| NM_207660 | ZC3H14 | Zinc finger CCCH-type containing 14 | 8.23037 | 0.00002 |
| AY726570 | ENST00000367696 | Clone TESTIS-724 mRNA sequence | −6.55525 | 0.0001 |
| NM_012239 | SIRT3 | Sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) | −7.27221 | 0.00005 |
| NM_139321 | ATRN | Attractin | −11.14924 | 1.43685E-06 |
| NM_006868 | RAB31 | RAB31, member RAS oncogene family | −8.70562 | 0.00001 |
| NM_003730 | RNASET2 | Ribonuclease T2 | −6.26012 | 0.00015 |
| BC045677 | FAM123A | Family with sequence similarity 123A | 7.88642 | 0.00002 |
| NM_005626 | SFRS4 | Splicing factor, arginine/serine-rich 4 | 6.47874 | 0.00011 |
| NM_003510 | HIST1H2AK | Histone cluster 1, H2ak | 6.79094 | 0.00008 |
| NM_006814 | PSMF1 | Proteasome (prosome, macropain) inhibitor subunit 1 (PI31) | −6.85967 | 0.00007 |
| NR_002939 | RUNDC2C | Homo sapiens RUN domain containing 2C (RUNDC2C) on chromosome 16. | 10.2493 | 2.91402E-06 |
| NM_005946 | MT1A | Metallothionein 1A (functional) | 5.81991 | 0.00025 |
| NM_145698 | ACBD5 | Acyl-Coenzyme A binding domain containing 5 | 8.7742 | 0.00001 |
| AY629351 | ZCCHC4 | Zinc finger, CCHC domain containing 4 | 7.46566 | 0.00004 |
| NM_145071 | CISH | Cytokine inducible SH2-containing protein | 6.338 | 0.00013 |
| NM_016072 | GOLT1B | Golgi transport 1 homolog B (S. cerevisiae) | 8.13565 | 0.00002 |
| AK056744 | VPS13C | Vacuolar protein sorting 13 homolog C (S. cerevisiae) | −6.21249 | 0.00016 |
| AF174606 | SHFM3P1 | Breakpoint cluster region | 6.2552 | 0.00015 |
| NM_023923 | PHACTR4 | Phosphatase and actin regulator 4 | −9.54987 | 5.24294E-06 |
| CR591805 | TMEM183A | Chromosome 1 open reading frame 37 | −7.74218 | 0.00003 |
| AY163812 | C17orf80 | Chromosome 17 open reading frame 80 | −6.70328 | 0.00009 |
| NM_001127 | AP1B1 | Adaptor-related protein complex 1, beta 1 subunit | 6.3445 | 0.00013 |
| AK024489 | PPP1R3E | Protein phosphatase 1, regulatory (inhibitor) subunit 3E | −5.77543 | 0.00027 |
| NM_014827 | ZC3H11A | Zinc finger CCCH-type containing 11A | −9.04607 | 8.18865E-06 |
| NM_020651 | PELI1 | Pellino homolog 1 (Drosophila) | −10.29063 | 2.81767E-06 |
| NM_001004351 | MGC57359 | similar to Williams Beuren syndrome chromosome region 19 | 9.7492 | 4.41886E-06 |
| A_23_P140454 | A_23_P140454 | Unknown | 7.70324 | 0.00003 |
| NM_021639 | GPBP1L1 | GC-rich promoter binding protein 1-like 1 | −9.69439 | 4.63023E-06 |
| THC2437906 | THC2437906 | Unknown | −7.29071 | 0.00005 |
| NM_014751 | MTSS1 | Metastasis suppressor 1 | −6.83835 | 0.00008 |
| THC2274685 | THC2274685 | AF326941 5H1 {Caenorhabditis elegans;} , partial (16%) [THC2274685] | −5.8588 | 0.00024 |
| NM_006994 | BTN3A3 | Butyrophilin, subfamily 3, member A3 | −6.7053 | 0.00009 |
| NM_194302 | CCDC108 | Coiled-coil domain containing 108 | 8.27613 | 0.00002 |
| NM_018475 | TMEM165 | Transmembrane protein 165 | 6.52923 | 0.00011 |
| NM_018355 | ZNF415 | Zinc finger protein 415 | −6.32122 | 0.00014 |
| AW948903 | ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | 10.10496 | 3.28015E-06 |
| NM_017733 | PIGG | Phosphatidylinositol glycan anchor biosynthesis, class G | −8.82095 | 0.00001 |
| NM_013433 | TNPO2 | Transportin 2 (importin 3, karyopherin beta 2b) | 5.7888 | 0.00026 |
| NM_003379 | VIL2 | Villin 2 (ezrin) | 7.14114 | 0.00005 |
| AK093641 | FAM120AOS | Family with sequence similarity 120A opposite strand | −8.05047 | 0.00002 |
| NM_023015 | INTS3 | Integrator complex subunit 3 | −6.0334 | 0.00019 |
| NM_002017 | FLI1 | Friend leukemia virus integration 1 | −6.68628 | 0.00009 |
| NM_145808 | MTPN | Myotrophin | 8.14242 | 0.00002 |
| A_24_P118591 | A_24_P118591 | Unknown | −7.11249 | 0.00006 |
| BG620990 | LOC339352 | similar to ATP binding domain 3 | 5.91871 | 0.00022 |
| NM_002862 | PYGB | Phosphorylase, glycogen; brain | 6.26247 | 0.00015 |
| XM_291857 | A_24_P499282 | Unknown | 8.70354 | 0.00001 |
| NM_022040 | LAT2 | Linker for activation of T cells family, member 2 | 5.99705 | 0.0002 |
| AL122093 | ZP4 | Zona pellucida glycoprotein 4 | 6.09319 | 0.00018 |
| NM_012448 | STAT5B | Signal transducer and activator of transcription 5B | 7.84235 | 0.00003 |
| NM_017777 | MKS1 | Meckel syndrome, type 1 | 6.36619 | 0.00013 |
| NM_052831 | C6orf192 | Chromosome 6 open reading frame 192 | −5.92287 | 0.00022 |
| NM_207336 | ZNF467 | Zinc finger protein 467 | 9.10574 | 7.75922E-06 |
| NM_005550 | KIFC3 | Kinesin family member C3 | 7.12481 | 0.00006 |
| NM_001312 | CRIP2 | Cysteine-rich protein 2 | 5.77985 | 0.00027 |
| XM_372970 | ENST00000312710 | Unknown | 6.04444 | 0.00019 |
| NM_015135 | NUP205 | Nucleoporin 205kDa | −9.28016 | 6.63981E-06 |
| NM_005800 | USPL1 | Ubiquitin specific peptidase like 1 | −6.5711 | 0.0001 |
| THC2284174 | THC2284174 | Unknown | −8.71174 | 0.00001 |
| A_24_P213175 | A_24_P213175 | Unknown | −5.91113 | 0.00023 |
| NM_000873 | ICAM2 | Intercellular adhesion molecule 2 | −6.44068 | 0.00012 |
| NM_014742 | TM9SF4 | Transmembrane 9 superfamily protein member 4 | −9.05755 | 8.10401E-06 |
| NM_003858 | CCNK | Cyclin K | −6.93939 | 0.00007 |
| NM_144498 | OSBPL2 | Oxysterol binding protein-like 2 | −7.43179 | 0.00004 |
| NM_032819 | ZNF341 | Zinc finger protein 341 | 8.05677 | 0.00002 |
| A_24_P273004 | A_24_P273004 | Unknown | −11.34914 | 1.23637E-06 |
| NM_178170 | NEK8 | NIMA (never in mitosis gene a)- related kinase 8 | 15.27718 | 9.62147E-08 |
| NM_003791 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | 6.5654 | 0.0001 |
| XM_497271 | A_24_P761490 | Unknown | 7.0537 | 0.00006 |
| NM_020800 | IFT80 | Intraflagellar transport 80 homolog (Chlamydomonas) | −5.90569 | 0.00023 |
| NM_015412 | C3orf17 | Chromosome 3 open reading frame 17 | −7.11308 | 0.00006 |
| NM_021064 | HIST1H2AG | Histone cluster 1, H2ag | 6.92417 | 0.00007 |
| NM_015893 | PRLH | Prolactin releasing hormone | 8.0239 | 0.00002 |
| NM_024040 | CUEDC2 | CUE domain containing 2 | 6.71805 | 0.00009 |
| A_24_P340916 | A_24_P340916 | Unknown | 5.93574 | 0.00022 |
| NM_006738 | AKAP13 | A kinase (PRKA) anchor protein 13 | 9.53395 | 5.31574E-06 |
| NM_014587 | SOX8 | SRY (sex determining region Y)-box 8 | 6.76814 | 0.00008 |
| NM_001126 | ADSS | Adenylosuccinate synthase | 6.04218 | 0.00019 |
| NM_014189 | ADD1 | Adducin 1 (alpha) | 6.99685 | 0.00006 |
| NM_001024847 | TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | 5.75954 | 0.00027 |
| NM_014708 | KNTC1 | Kinetochore associated 1 | −6.31481 | 0.00014 |
| NM_033402 | LRRCC1 | Leucine rich repeat and coiled-coil domain containing 1 | −7.93516 | 0.00002 |
| NM_006129 | BMP1 | Bone morphogenetic protein 1 | 8.37174 | 0.00002 |
| NM_007260 | LYPLA2 | Lysophospholipase II | 5.78566 | 0.00026 |
| AL834499 | LOC90113 | hypothetical protein BC009862 | 8.85453 | 9.75368E-06 |
| BX640624 | IGHA1 | Immunoglobulin heavy constant alpha 1 | 5.77955 | 0.00027 |
| NM_058187 | C21orf63 | Chromosome 21 open reading frame 63 | 6.98823 | 0.00006 |
| AK055501 | RPL15 | Ribosomal protein L15 | −6.50653 | 0.00011 |
| AF039564 | RBBP9 | Homo sapiens retinoblastoma binding protein (RBBP9) mRNA, complete cds. | 7.94915 | 0.00002 |
| BC041387 | MCTP2 | Multiple C2 domains, transmembrane 2 | −6.79143 | 0.00008 |
| A_23_P252501 | A_23_P252501 | Unknown | 10.69754 | 2.03582E-06 |
| NM_014023 | WDR37 | WD repeat domain 37 | −6.4766 | 0.00011 |
| NM_172037 | RDH10 | Retinol dehydrogenase 10 (all-trans) | 8.73814 | 0.00001 |
| A_24_P106166 | A_24_P106166 | Unknown | −6.28848 | 0.00014 |
| NM_178313 | SPTBN1 | Spectrin, beta, non-erythrocytic 1 | 5.45395 | 0.0004 |
| THC2403165 | THC2403165 | Q6TXI7 (Q6TXI7) LRRGT00012, partial (4%) [THC2403165] | −7.92713 | 0.00002 |
| NM_080660 | ZC3HAV1L | Zinc finger CCCH-type, antiviral 1-like | 8.99264 | 8.59541E-06 |
| AK123844 | C9orf45 | Spermatid perinuclear RNA binding protein | −6.84307 | 0.00008 |
| NM_207581 | DUOXA2 | Dual oxidase maturation factor 2 | 7.20138 | 0.00005 |
| AK128714 | LOC647121 | similar to embigin homolog | −6.07806 | 0.00018 |
| NM_152906 | C22orf25 | Chromosome 22 open reading frame 25 | 6.93325 | 0.00007 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −6.65631 | 0.00009 |
| NM_133330 | WHSC1 | Wolf-Hirschhorn syndrome candidate 1 | −10.55755 | 2.27388E-06 |
| NM_003636 | KCNAB2 | Potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 8.84297 | 9.85807E-06 |
| NM_033064 | ATCAY | Ataxia, cerebellar, Cayman type (caytaxin) | 9.55873 | 5.20293E-06 |
| THC2427841 | THC2427841 | Q9JM93 (Q9JM93) SRp25 nuclear protein (ADP-ribosylation factor-like 6 interacting protein 4), partial (7%) [THC2427841] | −7.12454 | 0.00006 |
| NM_012115 | CASP8AP2 | CASP8 associated protein 2 | −7.48469 | 0.00004 |
| XM_496950 | LOC441320 | PREDICTED: Homo sapiens similar to seven transmembrane helix receptor (LOC441320), mRNA [XM_496950] | 7.82806 | 0.00003 |
| NM_006521 | TFE3 | Transcription factor binding to IGHM enhancer 3 | −9.40066 | 5.97063E-06 |
| NM_032813 | TMTC4 | Transmembrane and tetratricopeptide repeat containing 4 | −7.55316 | 0.00003 |
| XM_929854 | LOC646890 | PREDICTED: Homo sapiens hypothetical protein LOC646890 (LOC646890), mRNA [XM_929854] | −5.8798 | 0.00023 |
| NM_172193 | KLHDC1 | Kelch domain containing 1 | 5.70628 | 0.00029 |
| NM_006289 | TLN1 | Talin1 | 5.85014 | 0.00024 |
| NM_004609 | TCF15 | Transcription factor 15 (basic helix-loop-helix) | 6.93939 | 0.00007 |
| NM_018442 | IQWD1 | IQ motif and WD repeats 1 | −11.63764 | 9.99374E-07 |
| NM_003110 | SP2 | Sp2 transcription factor | −7.66008 | 0.00003 |
| NM_016089 | ZNF589 | Zinc finger protein 589 | −7.66171 | 0.00003 |
| THC2378504 | THC2378504 | Unknown | −5.67893 | 0.0003 |
| NM_018117 | BRWD2 | Bromodomain and WD repeat domain containing 2 | −6.96869 | 0.00007 |
| NM_024676 | C1orf113 | Chromosome 1 open reading frame 113 | 5.59764 | 0.00034 |
| AK098835 | ZFHX1B | Zinc finger homeobox 1b | −5.92034 | 0.00022 |
| NM_000235 | LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | −6.63589 | 0.0001 |
| NM_001852 | COL9A2 | Collagen, type IX, alpha 2 | −7.20238 | 0.00005 |
| NM_021170 | HES4 | Hairy and enhancer of split 4 (Drosophila) | 9.59815 | 5.02884E-06 |
| NM_032340 | C6orf125 | Chromosome 6 open reading frame 125 | 12.24403 | 6.48527E-07 |
| NM_012407 | PICK1 | Protein interacting with PRKCA 1 | 6.45076 | 0.00012 |
| NM_005343 | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | 5.45148 | 0.0004 |
| AK130576 | ARHGAP24 | Rho GTPase activating protein 24 | −5.8887 | 0.00023 |
| AL831999 | SUGT1 | MRNA; cDNA DKFZp451K063 (from clone DKFZp451K063) | −7.49324 | 0.00004 |
| DQ287932 | DPY19L1 | Dpy-19-like 1 (C. elegans) | 7.50563 | 0.00004 |
| AK098256 | LOC728723 | hypothetical protein LOC728723 | 7.87079 | 0.00003 |
| THC2441708 | THC2441708 | Unknown | 9.15445 | 0.000007427 |
| NM_004649 | C21orf33 | Chromosome 21 open reading frame 33 | −7.56222 | 0.00003 |
| NM_021003 | PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | −6.37986 | 0.00013 |
| AF307332 | MGEA5 | Meningioma expressed antigen 5 (hyaluronidase) | −6.55078 | 0.00011 |
| NM_005159 | ACTC1 | Actin, alpha, cardiac muscle 1 | 5.65957 | 0.00031 |
| NM_014824 | FCHSD2 | FCH and double SH3 domains 2 | −7.13614 | 0.00005 |
| NM_001397 | ECE1 | Endothelin converting enzyme 1 | 5.45317 | 0.0004 |
| NM_019042 | PUS7 | Pseudouridylate synthase 7 homolog (S. cerevisiae) | 8.95172 | 8.92198E-06 |
| NM_004096 | EIF4EBP2 | Eukaryotic translation initiation factor 4E binding protein 2 | −6.01179 | 0.0002 |
| A_32_P119165 | A_32_P119165 | Unknown | −6.04598 | 0.00019 |
| NM_013398 | ZNF224 | Zinc finger protein 224 | −6.49534 | 0.00011 |
| NM_001039888 | ANKRD34 | Ankyrin repeat domain 34 | −5.80244 | 0.00026 |
| A_24_P67378 | A_24_P67378 | Unknown | 6.82367 | 0.00008 |
| NM_152549 | CCDC112 | Coiled-coil domain containing 112 | −6.31102 | 0.00014 |
| AK092824 | AMN | Amnionless homolog (mouse) | −5.76454 | 0.00027 |
| NM_194247 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 7.60297 | 0.00003 |
| NM_006569 | CGREF1 | Cell growth regulator with EF-hand domain 1 | 8.72836 | 0.00001 |
| NM_152687 | C5orf29 | Chromosome 5 open reading frame 29 | −5.52685 | 0.00037 |
| NM_005436 | CCDC6 | Coiled-coil domain containing 6 | −7.57251 | 0.00003 |
| NM_017723 | FLJ20245 | hypothetical protein FLJ20245 | −8.75065 | 0.00001 |
| NM_020145 | SH3GLB2 | Sec23 homolog B (S. cerevisiae) | 6.66636 | 0.00009 |
| A_24_P358205 | A_24_P358205 | Unknown | −7.12448 | 0.00006 |
| NM_001104 | ACTN3 | Actinin, alpha 3 | 5.36436 | 0.00045 |
| NM_001111 | ADAR | Adenosine deaminase, RNA-specific | 6.46515 | 0.00012 |
| NM_024778 | LONRF3 | LON peptidase N-terminal domain and ring finger 3 | 6.13423 | 0.00017 |
| NM_014670 | BZW1 | Basic leucine zipper and W2 domains 1 | 6.26341 | 0.00015 |
| NM_172020 | POM121 | POM121 membrane glycoprotein (rat) | −8.87468 | 9.57445E-06 |
| NM_000539 | RHO | Rhodopsin (opsin 2, rod pigment) (retinitis pigmentosa 4, autosomal dominant) | 8.27908 | 0.00002 |
| NM_020998 | MST1 | Macrophage stimulating 1 (hepatocyte growth factor-like) | 8.08571 | 0.00002 |
| NM_014417 | BBC3 | BCL2 binding component 3 | 6.68822 | 0.00009 |
| NM_005796 | NUTF2 | Nuclear transport factor 2 | 5.44463 | 0.00041 |
| NM_153342 | TMEM150 | Transmembrane protein 150 | 5.64117 | 0.00032 |
| AK091057 | LOC285535 | hypothetical protein LOC285535 | −8.33776 | 0.00002 |
| NM_032932 | RAB11FIP4 | RAB11 family interacting protein 4 (class II) | −7.08778 | 0.00006 |
| A_32_P167723 | A_32_P167723 | Unknown | 6.59109 | 0.0001 |
| NM_001007793 | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 5.60451 | 0.00033 |
| ENST00000372821 | ENST00000372821 | Q6DEY7 (Q6DEY7) MGC89376 protein, partial (38%) [THC2400949] | −7.5636 | 0.00003 |
| NM_032580 | HES7 | Hairy and enhancer of split 7 (Drosophila) | 8.7194 | 0.00001 |
| NM_144596 | TTC8 | Tetratricopeptide repeat domain 8 | −6.49123 | 0.00011 |
| NM_001024457 | RGPD1 | Plasminogen-like B1 | −5.37308 | 0.00045 |
| NM_014157 | CCDC113 | Coiled-coil domain containing 113 | 7.65554 | 0.00003 |
| NM_152262 | ZNF439 | Zinc finger protein 439 | −5.84169 | 0.00025 |
| NM_015358 | MORC3 | MORC family CW-type zinc finger 3 | −6.20742 | 0.00016 |
| NM_197941 | ADAMTS6 | ADAM metallopeptidase with thrombospondin type 1 motif, 6 | −6.43052 | 0.00012 |
| NM_146421 | GSTM1 | Glutathione S-transferase M1 | 5.20983 | 0.00056 |
| NM_052861 | MGC21675 | hypothetical protein MGC21675 | −6.36104 | 0.00013 |
| NM_000182 | HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 8.29677 | 0.00002 |
| NM_017443 | POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | −6.66635 | 0.00009 |
| NM_018254 | RCOR3 | REST corepressor 3 | −6.75332 | 0.00008 |
| NM_153002 | GPR156 | G protein-coupled receptor 156 | 7.48264 | 0.00004 |
| NM_001010919 | LOC441168 | hypothetical protein LOC441168 | −8.95495 | 8.8957E-06 |
| NM_005452 | WDR46 | WD repeat domain 46 | 5.35346 | 0.00046 |
| NM_004290 | RNF14 | Ring finger protein 14 | 5.61613 | 0.00033 |
| AK094269 | LOC285813 | hypothetical protein LOC285813 | −6.16194 | 0.00017 |
| NM_014641 | MDC1 | Mediator of DNA damage checkpoint 1 | −10.5734 | 2.24545E-06 |
| NM_018084 | KIAA1212 | KIAA1212 | −5.64438 | 0.00032 |
| NM_001008735 | HMG1L1 | High-mobility group (nonhistone chromosomal) protein 1-like 1 | 6.98657 | 0.00006 |
| NM_001012398 | FTS | Fused toes homolog (mouse) | 5.31553 | 0.00048 |
| NM_016546 | C1RL | Complement component 1, r subcomponent-like | −9.50091 | 5.47037E-06 |
| NM_003153 | STAT6 | Signal transducer and activator of transcription 6, interleukin-4 induced | −6.43102 | 0.00012 |
| NM_001074 | UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | 7.85504 | 0.00003 |
| NM_013328 | PYCR2 | Pyrroline-5-carboxylate reductase family, member 2 | 5.42514 | 0.00042 |
| NM_015897 | PIAS4 | Protein inhibitor of activated STAT, 4 | 6.55103 | 0.00011 |
| NM_001283 | AP1S1 | Adaptor-related protein complex 1, sigma 1 subunit | 5.21037 | 0.00056 |
| NM_000923 | PDE4C | Phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | 7.48989 | 0.00004 |
| NM_018676 | THSD1 | Thrombospondin, type I, domain containing 1 | −7.01524 | 0.00006 |
| NM_007042 | RPP14 | Ribonuclease P 14kDa subunit | 5.88159 | 0.00023 |
| NM_015926 | TEX264 | Testis expressed sequence 264 | 6.03192 | 0.00019 |
| NM_021216 | ZNF71 | Zinc finger protein 71 | −7.39256 | 0.00004 |
| NM_001659 | ARF3 | ADP-ribosylation factor 3 | 6.08657 | 0.00018 |
| THC2435127 | THC2435127 | Unknown | −7.00523 | 0.00006 |
| NM_015153 | PHF3 | PHD finger protein 3 | −7.08168 | 0.00006 |
| THC2407434 | THC2407434 | ALU1_HUMAN (P39188) Alu subfamily J sequence contamination warning entry, partial (20%) [THC2407434] | −9.80581 | 4.21166E-06 |
| NM_018051 | WDR60 | WD repeat domain 60 | −5.79231 | 0.00026 |
| NM_177987 | TUBB8 | tubulin, beta 8 | 5.56498 | 0.00035 |
| BC037535 | BC037535 | CDNA clone IMAGE:5263531 | −5.96076 | 0.00021 |
| NM_015568 | PPP1R16B | Protein phosphatase 1, regulatory (inhibitor) subunit 16B | −7.15947 | 0.00005 |
| NM_021813 | BACH2 | BTB and CNC homology 1, basic leucine zipper transcription factor 2 | −5.18425 | 0.00058 |
| THC2345721 | THC2345721 | Unknown | −5.77505 | 0.00027 |
| NM_015291 | DNAJC16 | DnaJ (Hsp40) homolog, subfamily C, member 16 | −5.67192 | 0.0003 |
| THC2437034 | THC2437034 | Q9N083 (Q9N083) Unnamed portein product, partial (43%) [THC2437034] | −5.95354 | 0.00021 |
| NM_004804 | CIAO1 | Cytosolic iron-sulfur protein assembly 1 homolog (S. cerevisiae) | 9.32183 | 6.39948E-06 |
| NM_001558 | IL10RA | Interleukin 10 receptor, alpha | −7.16204 | 0.00005 |
| NM_006016 | CD164 | CD164 molecule, sialomucin | 9.32468 | 6.38342E-06 |
| NM_003581 | NCK2 | NCK adaptor protein 2 | 7.177 | 0.00005 |
| NM_020432 | PHTF2 | Putative homeodomain transcription factor 2 | −7.55837 | 0.00003 |
| NM_016263 | FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | 5.79913 | 0.00026 |
| NM_006901 | MYO9A | Myosin IXA | −6.32218 | 0.00014 |
| BU661610 | BU661610 | Transcribed locus | −7.93911 | 0.00002 |
| NM_001310 | CREBL2 | CAMP responsive element binding protein-like 2 | −8.63283 | 0.00001 |
| NM_015187 | KIAA0746 | KIAA0746 protein | −7.19882 | 0.00005 |
| NM_014319 | LEMD3 | LEM domain containing 3 | −5.23419 | 0.00054 |
| NM_021008 | DEAF1 | Deformed epidermal autoregulatory factor 1 (Drosophila) | 7.79753 | 0.00003 |
| NM_015336 | ZDHHC17 | Zinc finger, DHHC-type containing 17 | −6.20521 | 0.00016 |
| NM_006888 | CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | −6.26682 | 0.00015 |
| NM_018198 | DNAJC11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | 5.29002 | 0.0005 |
| AF277175 | KIAA0265 | KIAA0265 protein | −6.54094 | 0.00011 |
| NM_198974 | TWF1 | Twinfilin, actin-binding protein, homolog 1 (Drosophila) | 9.75026 | 4.41488E-06 |
| NM_005370 | RAB8A | RAB8A, member RAS oncogene family | 5.7961 | 0.00026 |
| BG695979 | BG695979 | Transcribedlocus | −6.26052 | 0.00015 |
| NM_019044 | CCDC93 | Coiled-coil domain containing 93 | −7.08537 | 0.00006 |
| NM_032377 | ELOF1 | Elongation factor 1 homolog (S. cerevisiae) | 5.33163 | 0.00047 |
| NM_020336 | KIAA1219 | KIAA1219 | −9.56667 | 5.16733E-06 |
| NM_017704 | ANKRD49 | Ankyrin repeat domain 49 | −6.59777 | 0.0001 |
| BU618279 | PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | 8.48486 | 0.00001 |
| NM_014730 | KIAA0152 | KIAA0152 | 5.53054 | 0.00037 |
| NM_005153 | USP10 | Ubiquitin specific peptidase 10 | 6.93512 | 0.00007 |
| NM_001001433 | STX16 | Syntaxin 16 | 8.14081 | 0.00002 |
| BX098637 | RAD50 | Transcribed locus, weakly similar to NP_689672.2 protein LOC146556 [Homo sapiens] | −7.28539 | 0.00005 |
| NM_001885 | CRYAB | Crystallin, alpha B | 7.84386 | 0.00003 |
| NR_002797 | LOC255783 | hypothetical protein LOC255783 | 7.45402 | 0.00004 |
| NM_031219 | HDHD3 | Haloacid dehalogenase-like hydrolase domain containing 3 | 6.5466 | 0.00011 |
| NM_019002 | ETAA1 | Ewing's tumor-associated antigen 1 | −6.00953 | 0.0002 |
| THC2250585 | THC2250585 | Unknown | −10.02693 | 3.49902E-06 |
| NM_002999 | SDC4 | Syndecan 4 (amphiglycan, ryudocan) | 5.27453 | 0.00051 |
| CR620831 | C12orf47 | Chromosome 12 open reading frame 47 | −7.40664 | 0.00004 |
| NM_005500 | SAE1 | SUMO1 activating enzyme subunit 1 | −7.04103 | 0.00006 |
| NM_014045 | LRP10 | Low density lipoprotein receptor-related protein 10 | 5.77774 | 0.00027 |
| NM_173622 | CDRT4 | CMT1A duplicated region transcript 4 | −6.39054 | 0.00013 |
| NM_000594 | TNF | Tumor necrosis factor (TNF superfamily, member 2) | 5.25446 | 0.00052 |
| NM_001822 | CHN1 | Chimerin (chimaerin) 1 | 6.92225 | 0.00007 |
| A_24_P872359 | A_24_P872359 | Unknown | 6.07579 | 0.00018 |
| NM_016219 | MAN1B1 | Mannosidase, alpha, class 1B, member 1 | 6.64725 | 0.00009 |
| NM_173607 | C14orf24 | Chromosome 14 open reading frame 24 | 7.36156 | 0.00004 |
| THC2349560 | THC2349560 | Unknown | 6.3882 | 0.00013 |
| AK126092 | LOC221442 | hypothetical LOC221442 | −6.7597 | 0.00008 |
| NM_006804 | STARD3 | START domain containing 3 | 6.72679 | 0.00009 |
| NM_000848 | GSTM2 | Glutathione S-transferase M2 (muscle) | 5.42214 | 0.00042 |
| NM_005399 | PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | −5.63095 | 0.00032 |
| NM_012296 | GAB2 | GRB2-associated binding protein 2 | −6.86247 | 0.00007 |
| NM_024707 | GEMIN7 | Gem (nuclear organelle) associated protein 7 | −5.73351 | 0.00028 |
| NM_004162 | RAB5A | RAB5A, member RAS oncogene family | 5.07256 | 0.00067 |
| NM_199072 | MDFIC | MyoD family inhibitor domain containing | 5.16111 | 0.00059 |
| NM_016454 | TMEM85 | Transmembrane protein 85 | 6.36911 | 0.00013 |
| NM_024295 | DERL1 | Der1-like domain family, member 1 | 6.49264 | 0.00011 |
| NM_017679 | BCAS3 | Breast carcinoma amplified sequence 3 | 7.15775 | 0.00005 |
| NM_152574 | C9orf52 | Chromosome 9 open reading frame 52 | −5.48097 | 0.00039 |
| NM_147190 | LASS5 | LAG1 homolog, ceramide synthase 5 (S. cerevisiae) | 7.77914 | 0.00003 |
| A_32_P65793 | A_32_P65793 | Unknown | 7.34402 | 0.00004 |
| NM_006923 | SDF2 | Stromal cell-derived factor 2 | 5.09884 | 0.00065 |
| NM_001012423 | GOLGA8E | Golgi autoantigen, golgin subfamily a, 8E | 7.02657 | 0.00006 |
| A_24_P824578 | A_24_P824578 | Unknown | 5.25718 | 0.00052 |
| NM_015550 | OSBPL3 | Oxysterol binding protein-like 3 | −7.36736 | 0.00004 |
| NM_024653 | PRKRIP1 | Postmeiotic segregation increased 2-like 3 | 5.39368 | 0.00044 |
| NM_133636 | HEL308 | DNA helicase HEL308 | −6.10973 | 0.00018 |
| A_32_P11425 | A_32_P11425 | Unknown | 7.51075 | 0.00004 |
| THC2279840 | THC2279840 | Unknown | −5.90895 | 0.00023 |
| CR623166 | TTC14 | Tetratricopeptide repeat domain 14 | −6.56973 | 0.0001 |
| NM_014868 | RNF10 | Ring finger protein 10 | −6.69199 | 0.00009 |
| NM_207443 | FLJ45244 | FLJ45244 protein | −6.99635 | 0.00006 |
| NM_012218 | ILF3 | Interleukin enhancer binding factor 3, 90kDa | −9.40237 | 5.96169E-06 |
| NM_199345 | LOC375133 | similar to phosphatidylinositol 4-kinase alpha | 6.26274 | 0.00015 |
| NM_017727 | FLJ20254 | hypothetical protein FLJ20254 | 6.90032 | 0.00007 |
| A_32_P1291 | A_32_P1291 | Unknown | 7.28232 | 0.00005 |
| BC020095 | MGC17403 | hypothetical protein MGC17403 | −6.5012 | 0.00011 |
| NM_002729 | HHEX | Homeobox, hematopoietically expressed | −7.00539 | 0.00006 |
| A_32_P112100 | A_32_P112100 | Unknown | 5.97742 | 0.00021 |
| XM_941086 | LOC651831 | similar to CG31232-PA, isoform A | 5.07231 | 0.00067 |
| AK024092 | CHD2 | Chromodomain helicase DNA binding protein 2 | −5.56197 | 0.00035 |
| AJ227863 | MBNL1 | Muscleblind-like (Drosophila) | −6.04839 | 0.00019 |
| NM_015678 | NBEA | Neurobeachin | −5.67699 | 0.0003 |
| NM_003611 | OFD1 | Oral-facial-digital syndrome 1 | −6.13772 | 0.00017 |
| NM_021144 | PSIP1 | PC4 and SFRS1 interacting protein 1 | −5.05928 | 0.00068 |
| THC2441367 | THC2441367 | Unknown | −6.27525 | 0.00015 |
| AJ001898 | GDNF | Glial cell derived neurotrophic factor | 6.43449 | 0.00012 |
| NM_014781 | RB1CC1 | RB1-inducible coiled-coil 1 | −6.24829 | 0.00015 |
| BC022830 | LOC642852 | hypothetical LOC642852 | 6.3943 | 0.00013 |
| BX337332 | BX337332 | Transcribed locus, moderately similar to XP_001136080.1 hypothetical protein [Pan troglodytes] | −6.2502 | 0.00015 |
| NM_001006634 | ARHGAP17 | Rho GTPase activating protein 17 | −8.33571 | 0.00002 |
| NM_006761 | YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 4.97347 | 0.00077 |
| NM_002498 | NEK3 | NIMA (never in mitosis gene a)-related kinase 3 | −5.97924 | 0.00021 |
| CR605444 | FBXL4 | F-box and leucine-rich repeat protein 4 | −5.49736 | 0.00038 |
| NM_017991 | FLJ10081 | hypothetical protein FLJ10081 | −8.26415 | 0.00002 |
| NM_000428 | LTBP2 | Latent transforming growth factor beta binding protein 2 | 7.35375 | 0.00004 |
| NM_133439 | TADA2L | Transcriptional adaptor 2 (ADA2 homolog, yeast)-like | 5.53759 | 0.00036 |
| NM_005718 | ARPC4 | Actin related protein 2/3 complex, subunit 4, 20kDa | 5.39072 | 0.00044 |
| A_24_P238819 | A_24_P238819 | Unknown | 7.58575 | 0.00003 |
| NM_024069 | C19orf50 | Chromosome 19 open reading frame 50 | −8.00106 | 0.00002 |
| NM_005665 | EVI5 | Ecotropic viral integration site 5 | −6.01249 | 0.0002 |
| NM_002211 | ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | −6.426 | 0.00012 |
| NM_022362 | MMS19L | MMS19-like (MET18 homolog, S. cerevisiae) | −7.9471 | 0.00002 |
| AK022140 | FLJ12078 | hypothetical protein FLJ12078 | −5.85668 | 0.00024 |
| NM_017730 | QRICH1 | Glutamine-rich 1 | −7.12784 | 0.00005 |
| NM_001989 | EVX1 | Eve, even-skipped homeobox homolog 1 (Drosophila) | 7.23511 | 0.00005 |
| NM_018405 | C17orf79 | Chromosome 17 open reading frame 79 | 5.68363 | 0.0003 |
| A_32_P167212 | A_32_P167212 | Unknown | −5.12463 | 0.00062 |
| NM_006711 | RNPS1 | RNA binding protein S1, serine-rich domain | 6.2515 | 0.00015 |
| DB341590 | LOC728499 | similar to Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145) | −5.18063 | 0.00058 |
| BE535679 | BE535679 | Transcribed locus | −6.17163 | 0.00016 |
| A_32_P327750 | A_32_P327750 | Unknown | 5.15782 | 0.0006 |
| NM_018035 | FLJ10241 | hypothetical protein FLJ10241 | 5.1834 | 0.00058 |
| NM_021727 | FADS3 | Fatty acid desaturase 3 | 5.3224 | 0.00048 |
| NM_181607 | KRTAP19-1 | Keratin associated protein 19-1 | 5.70077 | 0.00029 |
| NM_003199 | TCF4 | Transcription factor 4 | −5.32466 | 0.00048 |
| AK024898 | AK024898 | CDNA: FLJ21245 fis, clone COL01184 | −7.51329 | 0.00004 |
| NM_001018055 | BRCC3 | BRCA1/BRCA2-containing complex, subunit 3 | −5.30636 | 0.00049 |
| NM_016238 | ANAPC7 | Anaphase promoting complex subunit 7 | 5.43935 | 0.00041 |
| NM_013259 | TAGLN3 | Transgelin 3 | 6.8389 | 0.00008 |
| NM_001010983 | GLT8D1 | Glycosyltransferase 8 domain containing 1 | −5.69977 | 0.00029 |
| NM_006390 | IPO8 | Importin 8 | 6.90698 | 0.00007 |
| NM_005520 | HNRPH1 | Heterogeneous nuclear ribonucleoprotein H1 (H) | −8.24555 | 0.00002 |
| NM_145233 | ZNF625 | Zinc finger protein 20 | 6.01856 | 0.0002 |
| NM_020805 | KLHL14 | Kelch-like 14 (Drosophila) | −8.24609 | 0.00002 |
| AV698092 | C6orf62 | Transcribed locus | 5.96382 | 0.00021 |
| NM_002945 | RPA1 | Replication protein A1, 70kDa | −7.5748 | 0.00003 |
| NM_182691 | SRPK2 | SFRS protein kinase 2 | 5.04526 | 0.00069 |
| NM_138706 | B3GNT6 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) | 7.94837 | 0.00002 |
| NM_007172 | NUP50 | Nucleoporin 50kDa | 5.09318 | 0.00065 |
| AJ404330 | TBC1D12 | TBC1 domain family, member 12 | −8.08182 | 0.00002 |
| THC2312756 | THC2312756 | O60448 (O60448) Neuronal thread protein AD7c-NTP, partial (9%) [THC2312756] | −6.28489 | 0.00014 |
| NM_004897 | MINPP1 | Multiple inositol polyphosphate histidine phosphatase, 1 | 4.91264 | 0.00083 |
| NM_013335 | GMPPA | GDP-mannose pyrophosphorylase A | 5.16344 | 0.00059 |
| THC2377877 | THC2377877 | Q87WI3 (Q87WI3) Dipeptide ABC transporter, permease protein, partial (5%) [THC2377877] | −5.53889 | 0.00036 |
| A_24_P256050 | A_24_P256050 | Unknown | 5.35666 | 0.00046 |
| A_32_P12282 | A_32_P12282 | Unknown | 8.58347 | 0.00001 |
| NM_005056 | JARID1A | Jumonji, AT rich interactive domain 1A | −4.92183 | 0.00082 |
| NM_020230 | PPAN | Peter pan homolog (Drosophila) | 5.57565 | 0.00034 |
| NM_032558 | HIATL1 | Hippocampus abundant transcript-like 1 | 7.72907 | 0.00003 |
| NM_030672 | ARHGAP28 | Rho GTPase activating protein 28 | 7.72512 | 0.00003 |
| NM_001005386 | ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | 6.62652 | 0.0001 |
| NM_001012968 | LOC139886 | hypothetical protein LOC139886 | −5.45114 | 0.00041 |
| NM_183001 | SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | 8.174 | 0.00002 |
| CR620532 | RASA2 | RAS p21 protein activator 2 | −6.29865 | 0.00014 |
| NM_177543 | PPAP2C | Phosphatidic acid phosphatase type 2C | 8.2774 | 0.00002 |
| NM_018381 | FLJ11286 | hypothetical protein FLJ11286 | −9.16765 | 7.33973E-06 |
| XM_497140 | A_24_P135921 | Unknown | 4.9042 | 0.00084 |
| NM_000819 | GART | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 5.26257 | 0.00052 |
| NM_002009 | FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | 4.87946 | 0.00087 |
| NM_003316 | TTC3 | Tetratricopeptide repeat domain 3 | −5.03045 | 0.00071 |
| NM_013326 | C18orf8 | Chromosome 18 open reading frame 8 | −6.43782 | 0.00012 |
| AF419616 | NBPF10 | Neuroblastoma breakpoint family, member 10 | −6.22377 | 0.00015 |
| NM_004329 | BMPR1A | Bone morphogenetic protein receptor, type IA | 6.20251 | 0.00016 |
| A_24_P915883 | A_24_P915883 | Unknown | 5.59313 | 0.00034 |
| NM_018486 | HDAC8 | Histone deacetylase 8 | 5.80887 | 0.00026 |
| THC2364821 | THC2364821 | Unknown | 6.33357 | 0.00014 |
| NM_002540 | ODF2 | Outer dense fiber of sperm tails 2 | −8.03397 | 0.00002 |
| NM_199437 | PRDM10 | PR domain containing 10 | −5.28301 | 0.00051 |
| NM_032179 | CPSF3L | Homo sapiens cleavage and polyadenylation specific factor 3-like (CPSF3L), transcript variant 2, mRNA [NM_032179] | 5.25928 | 0.00052 |
| NM_016598 | ZDHHC3 | Zinc finger, DHHC-type containing 3 | −7.1702 | 0.00005 |
| BU852331 | BU852331 | Transcribed locus | 8.69121 | 0.00001 |
| NM_004882 | CIR | CBF1 interacting corepressor | −5.75692 | 0.00027 |
| NM_000425 | L1CAM | L1 cell adhesion molecule | 6.26131 | 0.00015 |
| NM_022757 | CCDC14 | Coiled-coil domain containing 14 | −5.98571 | 0.00021 |
| A_24_P204454 | A_24_P204454 | Unknown | −5.98313 | 0.00021 |
| NM_015020 | PHLPPL | PH domain and leucine rich repeat protein phosphatase-like | −6.317 | 0.00014 |
| A_24_P170309 | A_24_P170309 | Unknown | 5.14466 | 0.00061 |
| A_24_P392099 | A_24_P392099 | Unknown | 5.32541 | 0.00048 |
| AK027248 | LOC56757 | hypothetical protein LOC56757 | −8.86084 | 9.69717E-06 |
| NM_170607 | MLX | MAX-like protein X | 6.44866 | 0.00012 |
| BC037281 | LOC439951 | hypothetical LOC439951 protein | 8.02017 | 0.00002 |
| NM_003800 | RNGTT | RNA guanylyltransferase and 5'-phosphatase | −7.68253 | 0.00003 |
| NM_198829 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 6.91893 | 0.00007 |
| THC2282321 | THC2282321 | AF235005 suppression of tumorigenicity 16 protein {Homo sapiens;} , partial (13%) [THC2282321] | −4.99222 | 0.00075 |
| NM_014828 | KIAA0737 | KIAA0737 | −7.25431 | 0.00005 |
| X82545 | PRNP | Prion protein (p27–30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 9.03795 | 8.24904E-06 |
| NM_017634 | KCTD9 | Potassium channel tetramerisation domain containing 9 | −7.29441 | 0.00005 |
| NM_012268 | PLD3 | Phospholipase D family, member 3 | 7.3033 | 0.00005 |
| NM_017633 | FAM46A | Family with sequence similarity 46, member A | −5.89253 | 0.00023 |
| NM_016353 | ZDHHC2 | Zinc finger, DHHC-type containing 2 | 6.58499 | 0.0001 |
| NM_005845 | ABCC4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 5.25204 | 0.00053 |
| THC2279840 | THC2279840 | Unknown | −6.05565 | 0.00019 |
| ENST00000361859 | ENST00000361859 | Unknown | 5.9073 | 0.00023 |
| BX538248 | BX538248 | CDNA clone IMAGE:30408657 | −5.68515 | 0.0003 |
| AK095904 | MIA3 | Melanoma inhibitory activity family, member 3 | −7.19205 | 0.00005 |
| NM_003040 | SLC4A2 | Solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) | 5.0469 | 0.00069 |
| NM_138819 | FAM122C | Family with sequence similarity 122C | −6.11779 | 0.00018 |
| NM_015442 | CNOT10 | CCR4-NOT transcription complex, subunit 10 | −6.29023 | 0.00014 |
| NM_152270 | SLFN11 | Schlafen family member 11 | −6.48172 | 0.00011 |
| AF085995 | LOC645513 | similar to septin 7 | −6.71126 | 0.00009 |
| NM_144566 | ZNF700 | Zinc finger protein 700 | −7.22033 | 0.00005 |
| NM_002570 | PCSK6 | Proprotein convertase subtilisin/kexin type 6 | 7.32909 | 0.00004 |
| NM_014827 | ZC3H11A | Zinc finger CCCH-type containing 11A | −7.8499 | 0.00003 |
| NM_000234 | LIG1 | Ligase I, DNA, ATP-dependent | −5.36772 | 0.00045 |
| NM_052928 | SMYD4 | SET and MYND domain containing 4 | −5.63457 | 0.00032 |
| THC2386194 | THC2386194 | Unknown | −5.35511 | 0.00046 |
| NM_005358 | LMO7 | LIM domain 7 | −7.08854 | 0.00006 |
| NM_022833 | FAM129B | Family with sequence similarity 129, member B | 7.60785 | 0.00003 |
| NM_181831 | NF2 | Neurofibromin 2 (bilateral acoustic neuroma) | 5.7013 | 0.00029 |
| NM_016551 | TM7SF3 | Transmembrane 7 superfamily member 3 | −5.82276 | 0.00025 |
| NM_057169 | GIT2 | G protein-coupled receptor kinase interactor 2 | −6.32361 | 0.00014 |
| AK000923 | SENP3 | Homo sapiens cDNA FLJ10061 fis, clone HEMBA1001413. | −5.87671 | 0.00024 |
| NM_033551 | LARP1 | La ribonucleoprotein domain family, member 1 | −7.90965 | 0.00002 |
| NM_015051 | TXNDC4 | Thioredoxin domain containing 4 (endoplasmic reticulum) | −6.64755 | 0.00009 |
| THC2339776 | THC2339776 | Unknown | −8.41951 | 0.00001 |
| AK024900 | AK024900 | CDNA: FLJ21247 fis, clone COL01205 | −7.56648 | 0.00003 |
| A_24_P799580 | A_24_P799580 | Unknown | 6.60497 | 0.0001 |
| ENST00000360523 | ENST00000360523 | GB|AB065479.1|BAC05733.1 seven transmembrane helix receptor [Homo sapiens] [NP511107] | 6.64141 | 0.00009 |
| A_24_P32836 | A_24_P32836 | Unknown | −6.19519 | 0.00016 |
| AB020633 | FRYL | FRY-like | −6.38433 | 0.00013 |
| NM_002118 | HLA-DMB | Major histocompatibility complex, class II, DM beta | −5.54567 | 0.00036 |
| THC2408277 | THC2408277 | Unknown | −5.38709 | 0.00044 |
| NM_017994 | C7orf42 | Chromosome 7 open reading frame 42 | −6.74705 | 0.00008 |
| NM_001659 | ARF3 | ADP-ribosylation factor 3 | 5.1421 | 0.00061 |
| NM_001752 | CAT | Catalase | −5.64093 | 0.00032 |
| NM_020657 | ZNF304 | Zinc finger protein 304 | −6.87846 | 0.00007 |
| BC056896 | HSPC157 | HSPC157 protein | −5.44409 | 0.00041 |
| NM_014859 | KIAA0672 | KIAA0672 gene product | −7.21036 | 0.00005 |
| NM_178520 | TMEM105 | Transmembrane protein 105 | 6.20265 | 0.00016 |
| NM_001011538 | LOC402176 | similar to 60S ribosomal protein L21 | −7.23041 | 0.00005 |
| NM_133446 | CTGLF1 | Centaurin, gamma-like family, member 1 | −7.90796 | 0.00002 |
| NM_194314 | ZBTB41 | Zinc finger and BTB domain containing 41 | −7.18381 | 0.00005 |
| NM_015609 | C1orf144 | Chromosome 1 open reading frame 144 | 5.0769 | 0.00067 |
| BC043279 | BC043279 | CDNA clone IMAGE:5297259 | −6.22554 | 0.00015 |
| NM_053042 | KIAA1729 | KIAA1729 protein | −7.36048 | 0.00004 |
| AK096054 | SLC6A19 | Solute carrier family 6 (neutral amino acid transporter), member 19 | 6.73775 | 0.00008 |
| NM_014990 | GARNL1 | GTPase activating Rap/RanGAP domain-like 1 | −8.21092 | 0.00002 |
| NM_014362 | HIBCH | 3-hydroxyisobutyryl-Coenzyme A hydrolase | −5.28173 | 0.00051 |
| NM_006804 | STARD3 | START domain containing 3 | 6.12902 | 0.00017 |
| NM_001042 | SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | 8.12027 | 0.00002 |
| NM_001040440 | CCDC112 | Coiled-coil domain containing 112 | −4.8855 | 0.00086 |
| NM_020918 | GPAM | Glycerol-3-phosphate acyltransferase, mitochondrial | −6.05652 | 0.00019 |
| NM_020381 | PDSS2 | Prenyl (decaprenyl) diphosphate synthase, subunit 2 | −5.38002 | 0.00044 |
| NM_001029857 | LOC158381 | hypothetical protein LOC158381 | 6.13599 | 0.00017 |
| BX641669 | ACOX1 | Acyl-Coenzyme A oxidase 1, palmitoyl | −6.4566 | 0.00012 |
| NM_033161 | SURF4 | Surfeit 4 | 6.55595 | 0.0001 |
| NM_144673 | CMTM2 | CKLF-like MARVEL transmembrane domain containing 2 | 5.76847 | 0.00027 |
| NM_015659 | RSL1D1 | Ribosomal L1 domain containing 1 | 5.34644 | 0.00046 |
| AK021842 | FLJ25476 | FLJ25476 protein | 6.14491 | 0.00017 |
| NM_183233 | SLC22A18 | Solute carrier family 22 (organic cation transporter), member 18 | 5.61224 | 0.00033 |
| NM_024649 | BBS1 | Bardet-Biedl syndrome 1 | −6.4087 | 0.00012 |
| ENST00000373264 | ENST00000373264 | Unknown | 5.31863 | 0.00048 |
| NM_025228 | TRAF3IP3 | TRAF3 interacting protein 3 | −5.6509 | 0.00031 |
| NM_005718 | ARPC4 | Actin related protein 2/3 complex, subunit 4, 20kDa | 5.28128 | 0.00051 |
| NM_007234 | DCTN3 | Dynactin 3 (p22) | 5.24163 | 0.00053 |
| NM_014949 | KIAA0907 | KIAA0907 | 7.69915 | 0.00003 |
| NM_001024847 | TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | −7.62359 | 0.00003 |
| A_24_P399341 | A_24_P399341 | Unknown | −6.21519 | 0.00016 |
| AK098491 | OR5T2 | RPA interacting protein | −5.42913 | 0.00042 |
| BC039296 | C20orf19 | Chromosome 20 open reading frame 19 | −5.18365 | 0.00058 |
| NM_005026 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | 6.21873 | 0.00016 |
| NM_005119 | THRAP3 | Thyroid hormone receptor associated protein 3 | 8.12691 | 0.00002 |
| AB051518 | KIAA1731 | KIAA1731 | −5.99995 | 0.0002 |
| BC012484 | TNRC15 | Homo sapiens trinucleotide repeat containing 15, mRNA (cDNA clone IMAGE:4472100), partial cds. | −6.86227 | 0.00007 |
| NM_001119 | ADD1 | Adducin 1 (alpha) | 6.32625 | 0.00014 |
| NM_173511 | ALS2CR13 | Amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 13 | −4.78976 | 0.00099 |
| NM_014813 | LRIG2 | Leucine-rich repeats and immunoglobulin-like domains 2 | −6.24565 | 0.00015 |
| NM_015143 | METAP1 | Methionyl aminopeptidase 1 | 6.82305 | 0.00008 |
| NM_206962 | PRMT2 | Protein arginine methyltransferase 2 | 5.94335 | 0.00022 |
| AK001118 | UBR2 | Ubiquitin protein ligase E3 component n-recognin 2 | −4.83769 | 0.00092 |
| THC2314600 | THC2314600 | Unknown | −7.0493 | 0.00006 |
| NM_002950 | RPN1 | Ribophorin I | −7.71446 | 0.00003 |
| NM_014969 | WDR47 | WD repeat domain 47 | −4.99127 | 0.00075 |
| AK130862 | FLJ27352 | hypothetical LOC145788 | −6.98877 | 0.00006 |
| NM_003104 | SORD | Sorbitol dehydrogenase | −5.18411 | 0.00058 |
| NM_033017 | TRIM4 | Tripartite motif-containing 4 | 5.35891 | 0.00046 |
| AK025221 | RAD50 | RAD50 homolog (S. cerevisiae) | −7.71671 | 0.00003 |
| NM_144664 | FAM76B | Family with sequence similarity 76, member B | −5.5059 | 0.00038 |
| NM_012219 | MRAS | Muscle RAS oncogene homolog | 6.83581 | 0.00008 |
| THC2437476 | THC2437476 | Unknown | −7.20877 | 0.00005 |
| BX116997 | BX116997 | Transcribed locus | −6.06866 | 0.00019 |
| NM_016272 | TOB2 | Transducer of ERBB2, 2 | −5.96088 | 0.00021 |
| NM_023111 | FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 5.0047 | 0.00073 |
| NM_152756 | RICTOR | rapamycin-insensitive companion of mTOR | −5.79934 | 0.00026 |
| CR590163 | KIAA2018 | KIAA2018 | −5.82281 | 0.00025 |
| NM_006141 | DYNC1LI2 | Dynein, cytoplasmic 1, light intermediate chain 2 | −5.10652 | 0.00064 |
| NM_005088 | CXYorf3 | Chromosome X and Y open reading frame 3 | −5.75059 | 0.00028 |
| NM_003309 | TSPYL1 | TSPY-like 1 | −6.02861 | 0.0002 |
| NM_004891 | MRPL33 | Mitochondrial ribosomal protein L33 | −7.30793 | 0.00005 |
| NM_033406 | FBXO3 | F-box protein 3 | 6.64954 | 0.00009 |
| NM_001037533 | GON4L | YY1 associated protein 1 | −7.07118 | 0.00006 |
| NM_004323 | BAG1 | BCL2-associated athanogene | −4.94334 | 0.0008 |
| NM_198567 | C5orf25 | Chromosome 5 open reading frame 25 | −6.25358 | 0.00015 |
| NM_006709 | EHMT2 | Euchromatic histone-lysine N-methyltransferase 2 | 5.73631 | 0.00028 |
| NM_004375 | COX11 | COX11 homolog, cytochrome c oxidase assembly protein (yeast) | −4.9474 | 0.00079 |
| AK074291 | HIPK2 | Homeodomain interacting protein kinase 2 | −7.86645 | 0.00003 |
| NM_001032289 | SLC35A2 | Solute carrier family 35 (UDP-galactose transporter), member A2 | −8.23296 | 0.00002 |
| NM_002778 | PSAP | Prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy) | 4.87526 | 0.00088 |
| NM_012330 | MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | −6.78062 | 0.00008 |
| AW518445 | RPS23 | Ribosomal protein S23 | −6.93887 | 0.00007 |
| NM_021260 | ZFYVE1 | Zinc finger, FYVE domain containing 1 | −5.06342 | 0.00068 |
| NM_005052 | RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | 7.20917 | 0.00005 |
| A_24_P24790 | A_24_P24790 | Unknown | −5.64628 | 0.00032 |
| A_32_P122285 | A_32_P122285 | Unknown | −5.35697 | 0.00046 |
| NM_006885 | ATBF1 | AT-binding transcription factor 1 | 7.13739 | 0.00005 |
| NM_020382 | SETD8 | SET domain containing (lysine methyltransferase) 8 | 5.29077 | 0.0005 |
| NM_000202 | IDS | Iduronate 2-sulfatase (Hunter syndrome) | −5.14859 | 0.0006 |
| A_24_P93184 | A_24_P93184 | Unknown | 5.20135 | 0.00056 |
| NM_002557 | OVGP1 | Oviductal glycoprotein 1, 120kDa (mucin 9, oviductin) | −6.11482 | 0.00018 |
| U68019 | SMAD3 | SMAD family member 3 | −5.73809 | 0.00028 |
| NM_001120 | TETRAN | tetracycline transporter-like protein | 5.16055 | 0.00059 |
| A_24_P298616 | A_24_P298616 | Unknown | −5.85459 | 0.00024 |
| NM_199341 | LOC374920 | hypothetical protein LOC374920 | −6.70474 | 0.00009 |
| AK025695 | TMED8 | Glutathione transferase zeta 1 (maleylacetoacetate isomerase) | −6.84167 | 0.00008 |
| NM_002721 | PPP6C | Protein phosphatase 6, catalytic subunit | −6.83703 | 0.00008 |
| NM_001229 | CASP9 | Caspase 9, apoptosis-related cysteine peptidase | −7.15685 | 0.00005 |
| NM_017541 | CRYGS | Crystallin, gamma S | −6.28412 | 0.00014 |
| NM_001013406 | KRIT1 | KRIT1, ankyrin repeat containing | 8.09633 | 0.00002 |
| NM_005867 | DSCR4 | Down syndrome critical region gene 4 | 4.79505 | 0.00098 |
| NM_004187 | JARID1C | Jumonji, AT rich interactive domain 1C | −4.8437 | 0.00092 |
| A_32_P32905 | A_32_P32905 | Unknown | −5.02028 | 0.00072 |
| XM_934270 | LOC647065 | hypothetical LOC647065 | −5.78325 | 0.00026 |
| AK097149 | SPPL3 | signal peptide peptidase 3 | −5.07639 | 0.00067 |
| NM_002004 | FDPS | Farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | −7.93324 | 0.00002 |
| A_24_P109962 | A_24_P109962 | Unknown | 6.63781 | 0.0001 |
| AK092875 | AK092875 | CDNA FLJ35556 fis, clone SPLEN2004844 | 6.52427 | 0.00011 |
| NM_014847 | UBAP2L | Ubiquitin associated protein 2-like | 7.49946 | 0.00004 |
| AA902595 | IQGAP1 | IQ motif containing GTPase activating protein 1 | −7.31533 | 0.00004 |
| AK125206 | LOC149448 | hypothetical protein LOC149448 | −7.45844 | 0.00004 |
| A_24_P812265 | A_24_P812265 | Unknown | 5.37664 | 0.00045 |
| A_32_P171984 | A_32_P171984 | Unknown | 5.94952 | 0.00022 |
| XM_373367 | A_24_P401150 | Unknown | 4.88814 | 0.00086 |
| NM_030809 | C12orf22 | Chromosome 12 open reading frame 22 | −6.26667 | 0.00015 |
| NM_001014436 | DBNL | Drebrin-like | 5.82723 | 0.00025 |
| NM_014774 | KIAA0494 | ATP synthase mitochondrial F1 complex assembly factor 1 | 6.08021 | 0.00018 |
| NM_005482 | PIGK | Phosphatidylinositol glycan anchor biosynthesis, class K | −6.21898 | 0.00016 |
| NM_174898 | LOC129530 | hypothetical protein LOC129530 | −6.1107 | 0.00018 |
| NM_201434 | RAB5C | RAB5C, member RAS oncogene family | 5.60187 | 0.00033 |
| AK095151 | EDD1 | E3 ubiquitin protein ligase, HECT domain containing, 1 | −5.90406 | 0.00023 |
| NM_152409 | C5orf24 | Chromosome 5 open reading frame 24 | 5.18137 | 0.00058 |
| A_24_P626812 | A_24_P626812 | Unknown | 5.48701 | 0.00039 |
| NM_001521 | GTF3C2 | General transcription factor IIIC, polypeptide 2, beta 110kDa | −5.31194 | 0.00049 |
| NM_018116 | MSTO1 | Death associated protein 3 | 5.70862 | 0.00029 |
| NM_031483 | ITCH | Itchy homolog E3 ubiquitin protein ligase (mouse) | −5.65809 | 0.00031 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −6.76771 | 0.00008 |
| NM_005692 | ABCF2 | ATP-binding cassette, sub-family F (GCN20), member 2 | 4.85315 | 0.0009 |
| NM_007249 | KLF12 | Kruppel-like factor 12 | −5.04433 | 0.0007 |
| AK125099 | AK125099 | CDNA FLJ43109 fis, clone CTONG2025516, moderately similar to Homo sapiens general transcription factor II, i (GTF2I) | 6.01691 | 0.0002 |
| NM_203447 | DOCK8 | Dedicator of cytokinesis 8 | −7.32816 | 0.00004 |
| BC020955 | TBC1D23 | TBC1 domain family, member 23 | −6.35938 | 0.00013 |
| BC045820 | C21orf51 | Chromosome 21 open reading frame 51 | −5.01304 | 0.00073 |
| AY831680 | HC1 | HC1 (HC1) | −6.02551 | 0.0002 |
| NM_022894 | PAPOLG | Poly(A) polymerase gamma | −7.18187 | 0.00005 |
| NM_018371 | ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | 5.05306 | 0.00069 |
| A_32_P211048 | A_32_P211048 | Unknown | −5.23129 | 0.00054 |
| NM_020235 | BBX | Bobby sox homolog (Drosophila) | 5.65165 | 0.00031 |
| AL117511 | C14orf125 | Homo sapiens mRNA; cDNA DKFZp434I1735 (from clone DKFZp434I1735); partial cds. | −5.13994 | 0.00061 |
| AK024925 | FLJ21272 | hypothetical protein FLJ21272 | −5.11626 | 0.00063 |
| NM_021100 | NFS1 | NFS1 nitrogen fixation 1 homolog (S. cerevisiae) | −6.4712 | 0.00012 |
| NM_006133 | C11orf11 | Chromosome 11 open reading frame 11 | 6.22281 | 0.00015 |
| AL832861 | MTERF | Mitochondrial transcription termination factor | −6.82522 | 0.00008 |
| NM_199320 | PHF17 | PHD finger protein 17 | −5.41371 | 0.00043 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −5.8857 | 0.00023 |
| NM_145233 | ZNF625 | Zinc finger protein 20 | 5.42341 | 0.00042 |
| NM_032782 | HAVCR2 | Hepatitis A virus cellular receptor 2 | −5.54466 | 0.00036 |
| XM_497894 | A_24_P24912 | Unknown | 5.83076 | 0.00025 |
| NM_015530 | GORASP2 | Golgi reassembly stacking protein 2, 55kDa | 5.85766 | 0.00024 |
| NM_172241 | CTAGE1 | Cutaneous T-cell lymphoma-associated antigen 1 | 7.17695 | 0.00005 |
| AK001173 | AQR | Aquarius homolog (mouse) | −7.77232 | 0.00003 |
| NM_001299 | CNN1 | Calponin 1, basic, smooth muscle | 5.51348 | 0.00037 |
| AB058774 | ZNF594 | Zinc finger protein 594 | −5.7672 | 0.00027 |
| NM_001607 | ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | 5.0508 | 0.00069 |
| NM_001005526 | SF3B1 | Splicing factor 3b, subunit 1, 155kDa | 5.20139 | 0.00056 |
| NM_006241 | PPP1R2 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 | −5.25608 | 0.00052 |
| W18193 | THC2304444 | IMAGE:20064 Soares infant brain 1NIB Homo sapiens cDNA clone IMAGE:20064, mRNA sequence. | −5.51323 | 0.00037 |
| AK125922 | CSNK2A2 | Casein kinase 2, alpha prime polypeptide | −7.4646 | 0.00004 |
| BU567832 | BU567832 | Transcribed locus | −4.93845 | 0.0008 |
| NM_000791 | DHFR | Dihydrofolate reductase | 4.95027 | 0.00079 |
| BC104478 | RPL21 | Ribosomal protein L21 | −6.41461 | 0.00012 |
| NM_001040431 | CCDC56 | Coiled-coil domain containing 56 | 5.15456 | 0.0006 |
| AJ420510 | PHF10 | PHD finger protein 10 | −6.30643 | 0.00014 |
| BC011660 | BC011660 | Homo sapiens cDNA clone IMAGE:4120690, **** WARNING: chimeric clone ****. | 5.70061 | 0.00029 |
| NM_025164 | KIAA0999 | KIAA0999 protein | −6.08738 | 0.00018 |
| NM_006195 | PBX3 | Pre-B-cell leukemia transcription factor 3 | −6.5323 | 0.00011 |
| NM_014149 | HSPC049 | HSPC049 protein | −5.65533 | 0.00031 |
| NM_018029 | FLJ10213 | hypothetical protein FLJ10213 | −5.53682 | 0.00036 |
| XM_373538 | ENST00000344556 | Unknown | 7.28826 | 0.00005 |
| THC2316878 | THC2316878 | Unknown | −5.50095 | 0.00038 |
| NM_003634 | NIPSNAP1 | Nipsnap homolog 1 (C. elegans) | 5.96498 | 0.00021 |
| NM_153631 | HOXA3 | Homeobox A5 | 7.53786 | 0.00004 |
| THC2404993 | THC2404993 | Unknown | −4.88629 | 0.00086 |
| NM_001008709 | PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | 4.95731 | 0.00078 |
| NM_000889 | ITGB7 | Integrin, beta 7 | −5.0143 | 0.00072 |
| NM_014033 | METTL7A | LETM1 domain containing 1 | −5.10185 | 0.00064 |
| NM_023926 | ZNF447 | Zinc finger protein 447 | 7.08335 | 0.00006 |
| NM_007139 | ZNF92 | Zinc finger protein 92 | −5.33052 | 0.00047 |
| NM_032645 | RAPSN | Receptor-associated protein of the synapse, 43kD | 7.31724 | 0.00004 |
| AI299143 | LOC51255 | hypothetical protein LOC51255 | −6.62138 | 0.0001 |
| NM_014624 | S100A6 | S100 calcium binding protein A6 | 5.69194 | 0.0003 |
| NM_032837 | FAM104A | Family with sequence similarity 104, member A | 7.10417 | 0.00006 |
| NM_005292 | GPR18 | G protein-coupled receptor 18 | −5.39734 | 0.00043 |
| AK001179 | ANKIB1 | Ankyrin repeat and IBR domain containing 1 | 6.78686 | 0.00008 |
| AK097258 | DOK3 | Docking protein 3 | −6.21585 | 0.00016 |
| NM_175852 | TXLNA | Taxilin alpha | 5.07701 | 0.00067 |
| NM_006231 | POLE | Polymerase (DNA directed), epsilon | 5.3557 | 0.00046 |
| NM_015216 | HISPPD1 | Histidine acid phosphatase domain containing 1 | −5.75216 | 0.00028 |
| NM_020400 | GPR92 | G protein-coupled receptor 92 | −6.72524 | 0.00009 |
| NM_020781 | ZNF398 | Zinc finger protein 398 | −6.47114 | 0.00012 |
| A_24_P632160 | A_24_P632160 | Unknown | 5.12173 | 0.00063 |
| NM_014224 | PGA5 | Pepsinogen 5, group I (pepsinogen A) | 5.1415 | 0.00061 |
| NM_014824 | FCHSD2 | FCH and double SH3 domains 2 | 7.01313 | 0.00006 |
| NM_015723 | PNPLA8 | Patatin-like phospholipase domain containing 8 | 7.24769 | 0.00005 |
| AK001179 | ANKIB1 | Ankyrin repeat and IBR domain containing 1 | −5.9491 | 0.00022 |
| AA420998 | LOC286076 | hypothetical protein LOC286076 | 5.31666 | 0.00048 |
| NM_033184 | KRTAP2-4 | Keratin associated protein 2-4 | 6.81809 | 0.00008 |
| NM_033481 | FBXO9 | F-box protein 9 | −5.53612 | 0.00036 |
| NM_020964 | KIAA1632 | KIAA1632 | −5.70799 | 0.00029 |
| NM_000405 | GM2A | GM2 ganglioside activator | 4.81139 | 0.00096 |
| NM_024713 | C15orf29 | Chromosome 15 open reading frame 29 | 6.26529 | 0.00015 |
| NM_006620 | HBS1L | HBS1-like (S. cerevisiae) | −5.65177 | 0.00031 |
| NM_153353 | LRRC34 | Leucine rich repeat containing 34 | −5.18522 | 0.00058 |
| NM_032271 | TRAF7 | TNF receptor-associated factor 7 | 5.42648 | 0.00042 |
| NM_002498 | NEK3 | NIMA (never in mitosis gene a)-related kinase 3 | −6.56948 | 0.0001 |
| BQ188033 | EPB41L4A | Erythrocyte membrane protein band 4.1 like 4A | −7.30113 | 0.00005 |
| AK025056 | ZYG11B | Zyg-11 homolog B (C. elegans) | −6.13158 | 0.00017 |
| AK126501 | CDC2L2 | Cell division cycle 2-like 2 (PITSLRE proteins) | −7.34404 | 0.00004 |
| NM_001945 | HBEGF | Heparin-binding EGF-like growth factor | 5.29197 | 0.0005 |
| AK093505 | CXorf18 | SPANX family, member A2 | 7.21242 | 0.00005 |
| NM_022838 | ARMCX5 | G protein-coupled receptor associated sorting protein 2 | −6.83633 | 0.00008 |
| NM_002883 | RANGAP1 | Ran GTPase activating protein 1 | 5.21738 | 0.00055 |
| NM_018448 | CAND1 | Cullin-associated and neddylation-dissociated 1 | 6.43506 | 0.00012 |
| NM_172193 | KLHDC1 | Kelch domain containing 1 | −4.97574 | 0.00076 |
| NM_002874 | RAD23B | RAD23 homolog B (S. cerevisiae) | 5.10293 | 0.00064 |
| NM_000402 | G6PD | Glucose-6-phosphate dehydrogenase | 4.90285 | 0.00084 |
| NM_004665 | VNN2 | Vanin 2 | −5.49775 | 0.00038 |
| AF111848 | PRO0529 | Homo sapiens PRO0529 mRNA, complete cds. | 7.2649 | 0.00005 |
| A_32_P83174 | A_32_P83174 | Unknown | −6.08174 | 0.00018 |
| BE044472 | C22orf27 | Chromosome 22 open reading frame 27 | −6.12607 | 0.00017 |
| NM_001012754 | LOC387921 | similar to RIKEN cDNA 8030451K01 | −6.13156 | 0.00017 |
| NM_020382 | SETD8 | SET domain containing (lysine methyltransferase) 8 | 6.25922 | 0.00015 |
| NM_006313 | USP15 | Ubiquitin specific peptidase 15 | −5.27031 | 0.00051 |
| NM_004727 | SLC24A1 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | −5.58374 | 0.00034 |
| XM_371160 | ENST00000310822 | PREDICTED: Homo sapiens similar to ribosomal protein L21 isoform 1 (LOC388532), mRNA [XM_371160] | −6.21526 | 0.00016 |
| NM_004879 | EI24 | Etoposide induced 2.4 mRNA | 4.81487 | 0.00095 |
| NM_018660 | ZNF395 | Zinc finger protein 395 | −6.76571 | 0.00008 |
| NM_005880 | DNAJA2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 5.86414 | 0.00024 |
| NM_014017 | MAPBPIP | mitogen-activated protein-binding protein-interacting protein | 6.46094 | 0.00012 |
| NM_013339 | ALG6 | Asparagine-linked glycosylation 6 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | −5.2637 | 0.00052 |
| THC2375818 | THC2375818 | Q15464 (Q15464) Shb, partial (4%) [THC2375818] | 5.43053 | 0.00042 |
| NM_015227 | POFUT2 | Protein O-fucosyltransferase 2 | −5.82663 | 0.00025 |
| NM_018367 | PHCA | Phytoceramidase, alkaline | 5.4057 | 0.00043 |
| AK124515 | AK124515 | CDNA FLJ30141 fis, clone BRACE2000148 | −5.80037 | 0.00026 |
| AK025335 | AP1GBP1 | AP1 gamma subunit binding protein 1 | −6.17463 | 0.00016 |
| NM_145867 | LTC4S | Leukotriene C4 synthase | 5.71862 | 0.00029 |
| A_24_P50707 | A_24_P50707 | Unknown | −7.21616 | 0.00005 |
| NM_014990 | GARNL1 | GTPase activating Rap/RanGAP domain-like 1 | −6.79782 | 0.00008 |
| NM_005803 | FLOT1 | Flotillin 1 | 4.88546 | 0.00086 |
| NM_032230 | C12orf26 | Chromosome 12 open reading frame 26 | −5.72089 | 0.00029 |
| NM_000632 | ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | −6.173 | 0.00016 |
| AK125015 | MTHFSD | Homo sapiens cDNA FLJ43025 fis, clone BRTHA2018707. | −6.19194 | 0.00016 |
| NM_003705 | SLC25A12 | Solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | 5.56608 | 0.00035 |
| NM_032632 | PAPOLA | Poly(A) polymerase alpha | −6.93832 | 0.00007 |
| NM_007331 | WHSC1 | Wolf-Hirschhorn syndrome candidate 1 | −5.54554 | 0.00036 |
| BM711907 | THC2373429 | Transcribed locus | −5.5243 | 0.00037 |
| A_24_P264413 | A_24_P264413 | Unknown | 4.86773 | 0.00089 |
| NM_032778 | MINA | MYC induced nuclear antigen | 5.1964 | 0.00057 |
| AK095260 | FLJ10769 | hypothetical protein FLJ10769 | 6.78238 | 0.00008 |
| NM_004930 | CAPZB | Capping protein (actin filament) muscle Z-line, beta | 5.14986 | 0.0006 |
| NM_003951 | SLC25A14 | Solute carrier family 25 (mitochondrial carrier, brain), member 14 | −5.04853 | 0.00069 |
| NM_012108 | BRDG1 | BCR downstream signaling 1 | −5.17296 | 0.00058 |
| NM_000788 | DCK | Deoxycytidine kinase | −6.25805 | 0.00015 |
| NM_019015 | CSGlcA-T | chondroitin sulfate glucuronyltransferase | 5.53232 | 0.00036 |
| NM_001040150 | C15orf28 | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | −5.31078 | 0.00049 |
| NM_052839 | PANX2 | Pannexin 2 | 5.81643 | 0.00025 |
| NM_001024732 | MECR | Mitochondrial trans-2-enoyl-CoA reductase | 6.46046 | 0.00012 |
| NM_018949 | UTS2R | Urotensin 2 receptor | 6.14462 | 0.00017 |
| NM_000786 | CYP51A1 | Cytochrome P450, family 51, subfamily A, polypeptide 1 | −5.98488 | 0.00021 |
| NM_001361 | DHODH | Dihydroorotate dehydrogenase | −4.81064 | 0.00096 |
| THC2442489 | THC2442489 | Unknown | −5.7987 | 0.00026 |
| AK024144 | FLJ14082 | hypothetical protein FLJ14082 | 5.0193 | 0.00072 |
| THC2315966 | THC2315966 | Unknown | 5.44628 | 0.00041 |
| THC2279305 | THC2279305 | Unknown | −6.62236 | 0.0001 |
| BG768878 | THC2312236 | 602743557F1 NIH_MGC_49 Homo sapiens cDNA clone IMAGE:4872878 5', mRNA sequence. | 5.64061 | 0.00032 |
| NM_001031735 | C19orf36 | Chromosome 19 open reading frame 36 | 5.29118 | 0.0005 |
| NM_001329 | CTBP2 | C-terminal binding protein 2 | 6.02499 | 0.0002 |
| NM_007372 | DDX42 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | 5.48554 | 0.00039 |
| AF343078 | ATAD3B | Homo sapiens TOB3 mRNA, complete cds. | 5.06251 | 0.00068 |
| NM_001013842 | C8orf58 | Chromosome 8 open reading frame 58 | −5.24475 | 0.00053 |
| AV739735 | HNRPC | Transcribed locus, moderately similar to NP_001020804.1 nuclear ribonucleoprotein C [Rattus norvegicus] | 5.62056 | 0.00033 |
| NM_002641 | PIGA | Phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) | −5.71759 | 0.00029 |
| NM_018047 | RBM22 | RNA binding motif protein 22 | 6.25385 | 0.00015 |
| NM_153187 | SLC22A1 | Solute carrier family 22 (organic cation transporter), member 1 | 5.05565 | 0.00069 |
| AK127081 | USP54 | Ubiquitin specific peptidase 54 | −6.40172 | 0.00013 |
| AK125832 | TBC1D3C | TBC1 domain family, member 3C | −5.35704 | 0.00046 |
| CR619110 | LOC401588 | hypothetical LOC401588 | −5.72382 | 0.00029 |
| NM_003861 | WDR22 | WD repeat domain 22 | −6.2627 | 0.00015 |
| NM_138363 | CCDC45 | Coiled-coil domain containing 45 | −4.87186 | 0.00088 |
| A_32_P55799 | A_32_P55799 | Unknown | −5.05573 | 0.00068 |
| NM_014940 | MON1B | MON1 homolog B (yeast) | 7.0631 | 0.00006 |
| BX648857 | SP3 | Sp3 transcription factor | −5.68022 | 0.0003 |
| NM_006120 | HLA-DMA | Major histocompatibility complex, class II, DM beta | −5.36711 | 0.00045 |
| NM_006669 | LILRB1 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | −6.90789 | 0.00007 |
| BQ189538 | THC2316936 | Transcribedlocus | −5.01081 | 0.00073 |
| NM_015205 | ATP11A | ATPase, Class VI, type 11A | −6.56291 | 0.0001 |
| NM_138364 | LOC90826 | hypothetical protein BC004337 | −5.01733 | 0.00072 |
| BC089156 | BC089156 | Homo sapiens, clone IMAGE:4346533, mRNA | −5.40338 | 0.00043 |
| A_24_P16273 | A_24_P16273 | Unknown | 5.78523 | 0.00026 |
| CR606044 | LOC93622 | hypothetical protein BC006130 | −4.92763 | 0.00082 |
| NM_199044 | NSUN4 | NOL1/NOP2/Sun domain family, member 4 | 5.527 | 0.00037 |
| NM_000455 | STK11 | Serine/threonine kinase 11 | 6.37056 | 0.00013 |
| NM_173694 | ATP11C | ATPase, Class VI, type 11C | −6.79799 | 0.00008 |
| NM_006962 | ZNF182 | Zinc finger protein 182 | −5.38748 | 0.00044 |
| BM680823 | BM680823 | Transcribed locus | −5.0325 | 0.00071 |
| BC053353 | PRPF38B | CDNA clone IMAGE:5246408 | −5.95255 | 0.00021 |
| AB014514 | FLJ30092 | AF-1 specific protein phosphatase | 6.40387 | 0.00012 |
| NM_001152 | SLC25A5 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 4.99296 | 0.00075 |
| BF213738 | CLEC2B | C-type lectin domain family 2, member B | −6.15737 | 0.00017 |
| NM_025231 | ZNF435 | Zinc finger protein 435 | −6.70982 | 0.00009 |
| NM_001076786 | QSER1 | Glutamine and serine rich 1 | −5.86969 | 0.00024 |
| NM_019007 | ARMCX6 | Armadillo repeat containing, X-linked 6 | −5.98809 | 0.00021 |
| NM_145911 | ZNF23 | Ribosomal protein L27 | −6.73374 | 0.00009 |
| NM_024819 | DCAKD | Dephospho-CoA kinase domain containing | −4.9368 | 0.00081 |
| NM_005207 | CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | 5.38662 | 0.00044 |
| NM_175736 | FMNL3 | Formin-like 3 | 5.60824 | 0.00033 |
| NM_017794 | KIAA1797 | KIAA1797 | −5.35919 | 0.00046 |
| NM_004074 | COX8A | Cytochrome c oxidase subunit 8A (ubiquitous) | 6.05767 | 0.00019 |
| AF144054 | AF144054 | Apoptosis related protein APR-4 | 6.06285 | 0.00019 |
| THC2344420 | THC2344420 | ALU7_HUMAN (P39194) Alu subfamily SQ sequence contamination warning entry, partial (21%) [THC2344420] | −5.13064 | 0.00062 |
| AF229166 | LOC220077 | hypothetical gene supported by AF229166 | 5.41636 | 0.00042 |
| AB014550 | SMCHD1 | Structural maintenance of chromosomes flexible hinge domain containing 1 | −6.90178 | 0.00007 |
| NM_024738 | C12orf49 | Chromosome 12 open reading frame 49 | 5.44473 | 0.00041 |
| NM_153649 | TPM3 | Tropomyosin 3 | 5.58209 | 0.00034 |
| NM_012247 | SEPHS1 | Selenophosphate synthetase 1 | 4.8008 | 0.00097 |
| NM_014339 | IL17RA | Interleukin 17 receptor A | −4.86703 | 0.00089 |
| NM_000534 | PMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) | −5.93393 | 0.00022 |
| AL832783 | LMLN | Leishmanolysin-like (metallopeptidase M8 family) | −4.79121 | 0.00099 |
| NM_015525 | IBTK | Inhibitor of Bruton agammaglobulinemia tyrosine kinase | −6.3151 | 0.00014 |
| A_32_P77421 | A_32_P77421 | Unknown | −6.30639 | 0.00014 |
| NM_019843 | EIF4ENIF1 | Eukaryotic translation initiation factor 4E nuclear import factor 1 | −6.83768 | 0.00008 |
| NM_024898 | DENND1C | DENN/MADD domain containing 1C | −6.91959 | 0.00007 |
| NM_172217 | IL16 | Interleukin 16 (lymphocyte chemoattractant factor) | −5.28949 | 0.0005 |
| NM_001192 | TNFRSF17 | Tumor necrosis factor receptor superfamily, member 17 | −6.95798 | 0.00007 |
| NM_001010853 | ACY1L2 | Aminoacylase 1-like 2 | −6.81869 | 0.00008 |
| THC2428671 | THC2428671 | Unknown | −6.52112 | 0.00011 |
| NM_004423 | DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | 5.46922 | 0.0004 |
| BC040695 | WBSCR16 | Williams-Beuren syndrome chromosome region 16 | −5.70567 | 0.00029 |
| A_24_P418590 | A_24_P418590 | Unknown | −5.20758 | 0.00056 |
| NM_001018062 | LOC51149 | hypothetical LOC51149 | −4.88888 | 0.00086 |
| NM_017869 | BANP | BTG3 associated nuclear protein | 5.46398 | 0.0004 |
| D86980 | TTC9 | Tetratricopeptide repeat domain 9 | −5.47605 | 0.00039 |
| A_24_P333077 | A_24_P333077 | Unknown | 5.92063 | 0.00022 |
| NM_001556 | IKBKB | Golgi autoantigen, golgin subfamily a, 7 | −6.58675 | 0.0001 |
| NM_001008225 | CNOT4 | CCR4-NOT transcription complex, subunit 4 | 4.9542 | 0.00079 |
| NM_021173 | POLD4 | Polymerase (DNA-directed), delta 4 | 5.57566 | 0.00034 |
| NM_031303 | KATNAL2 | Katanin p60 subunit A-like 2 | −6.56838 | 0.0001 |
| NM_007234 | DCTN3 | Dynactin 3 (p22) | 5.07507 | 0.00067 |
| NM_022845 | CBFB | Core-binding factor, beta subunit | 6.17287 | 0.00016 |
| XM_167254 | LOC402643 | PREDICTED: Homo sapiens similar to tropomyosin 3 isoform 2 (LOC402643), mRNA [XM_167254] | 4.89622 | 0.00085 |
| NM_032868 | FLJ14981 | hypothetical protein FLJ14981 | 5.68109 | 0.0003 |
| NM_013296 | GPSM2 | G-protein signalling modulator 2 (AGS3-like, C. elegans) | 5.72586 | 0.00028 |
| NM_006134 | TMEM50B | Transmembrane protein 50B | 6.77942 | 0.00008 |
| NM_001781 | CD69 | CD69molecule | −5.92149 | 0.00022 |
| NM_021818 | SAV1 | Salvador homolog 1 (Drosophila) | −5.9986 | 0.0002 |
| NM_174913 | C14orf21 | Cell death-inducing DFFA-like effector b | 6.12658 | 0.00017 |
| NM_013943 | CLIC4 | Chloride intracellular channel 4 | 5.03541 | 0.0007 |
| NM_003161 | RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | −5.92216 | 0.00022 |
| NM_153649 | TPM3 | Tropomyosin 3 | −6.5978 | 0.0001 |
| NM_198040 | PHC2 | Polyhomeotic homolog 2 (Drosophila) | 6.49653 | 0.00011 |
| NM_014766 | SCRN1 | Secernin 1 | −5.9542 | 0.00021 |
| X78926 | ZNF268 | Phosphatidylinositol glycan anchor biosynthesis, class F | −6.26836 | 0.00015 |
| NM_005051 | QARS | Glutaminyl-tRNA synthetase | 5.02506 | 0.00071 |
| NM_032272 | MAF1 | MAF1 homolog (S. cerevisiae) | 5.03895 | 0.0007 |
| NM_178862 | STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | 5.53837 | 0.00036 |
| NM_031157 | HNRPA1 | Heterogeneous nuclear ribonucleoprotein A1 | −5.34903 | 0.00046 |
| BC018676 | LOC730846 | similar to S-adenosylmethionine decarboxylase proenzyme 2 (AdoMetDC 2) (SamDC 2) | 6.02167 | 0.0002 |
| NM_015251 | ASCIZ | ATM/ATR-Substrate Chk2-Interacting Zn2+-finger protein | −4.91078 | 0.00084 |
| NM_024557 | RIC3 | Resistance to inhibitors of cholinesterase 3 homolog (C. elegans) | −5.18745 | 0.00057 |
| NM_005389 | PCMT1 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase | 4.80631 | 0.00097 |
| AK056402 | TDRKH | Tudor and KH domain containing | −5.98927 | 0.00021 |
| BX113895 | BX113895 | Transcribed locus | −5.24899 | 0.00053 |
| NM_199418 | PRCP | Prolylcarboxypeptidase (angiotensinase C) | −6.17688 | 0.00016 |
| NM_022074 | FAM111A | Family with sequence similarity 111, member A | −6.19018 | 0.00016 |
| NM_032826 | SLC35B4 | Solute carrier family 35, member B4 | 5.93208 | 0.00022 |
| AK024473 | LOC92017 | similar to RIKEN cDNA 4933437K13 | −6.33059 | 0.00014 |
| NM_002766 | PRPSAP1 | Phosphoribosyl pyrophosphate synthetase-associated protein 1 | −5.41712 | 0.00042 |
| NM_006052 | DSCR3 | Down syndrome critical region gene 3 | 5.6984 | 0.00029 |
| CR603982 | SESN3 | MRNA; cDNA DKFZp313B1017 (from clone DKFZp313B1017) | −5.53487 | 0.00036 |
| NM_015509 | NECAP1 | NECAP endocytosis associated 1 | 5.46732 | 0.0004 |
| BC047412 | ZNF433 | Zinc finger protein 433 | −6.47114 | 0.00012 |
| NM_198829 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 5.72007 | 0.00029 |
| NM_018191 | RCBTB1 | Regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 | −6.58896 | 0.0001 |
| NM_002075 | GNB3 | Guanine nucleotide binding protein (G protein), beta polypeptide 3 | 5.91288 | 0.00023 |
| NM_012164 | FBXW2 | Homo sapiens F-box and WD repeat domain containing 2 (FBXW2), mRNA. | 4.83277 | 0.00093 |
| A_32_P163472 | A_32_P163472 | Unknown | −6.16132 | 0.00017 |
| NM_015365 | AMMECR1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region, gene 1 | −5.71868 | 0.00029 |
| AI887037 | AI887037 | Transcribed locus, weakly similar to NP_114185.2 Y-box protein YB2 [Rattus norvegicus] | 5.73536 | 0.00028 |
| NM_003913 | PRPF4B | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | −6.5001 | 0.00011 |
| NM_018981 | DNAJC10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | −5.57747 | 0.00034 |
| NM_000904 | NQO2 | NAD(P)H dehydrogenase, quinone 2 | 5.15535 | 0.0006 |
| AK123940 | FAM76A | CDNA FLJ41946 fis, clone PLACE6019701 | −5.47561 | 0.00039 |
| NM_005254 | GABPB2 | GA binding protein transcription factor, beta subunit 2 | −5.57953 | 0.00034 |
| A_23_P21804 | A_23_P21804 | Unknown | −5.00709 | 0.00073 |
| BC071966 | FAM60A | Family with sequence similarity 60, member A | 5.73366 | 0.00028 |
| U52054 | EMB | S6 H-8 mRNA expressed in chromosome 6-suppressed melanoma cells | −6.50457 | 0.00011 |
| NM_005463 | HNRPDL | Homo sapiens heterogeneous nuclear ribonucleoprotein D-like (HNRPDL), transcript variant 1, mRNA [NM_005463] | −6.07265 | 0.00019 |
| NM_152729 | NT5DC1 | 5'-nucleotidase domain containing 1 | −5.41419 | 0.00043 |
| NM_032775 | KLHL22 | Kelch-like 22 (Drosophila) | 5.32193 | 0.00048 |
| A_32_P177097 | A_32_P177097 | Unknown | −5.55856 | 0.00035 |
| THC2400533 | THC2400533 | Unknown | −6.2533 | 0.00015 |
| NM_005877 | SF3A1 | Splicing factor 3a, subunit 1, 120kDa | −4.80231 | 0.00097 |
| NM_001008392 | CTDSPL | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like | 5.07055 | 0.00067 |
| NM_015914 | TXNDC11 | Thioredoxin domain containing 11 | −5.91849 | 0.00022 |
| NM_025040 | ZNF614 | Zinc finger protein 614 | 5.42227 | 0.00042 |
| AF067420 | IGHA1 | Immunoglobulin heavy constant alpha 1 | 5.36829 | 0.00045 |
| BF592096 | RPL22 | Ribosomal protein L22 | −4.87536 | 0.00088 |
| XM_931434 | LOC400027 | hypothetical gene supported by BC047417 | −5.55031 | 0.00036 |
| NM_004124 | GMFB | Glia maturation factor, beta | 5.74047 | 0.00028 |
| NM_003932 | ST13 | Suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 6.04059 | 0.00019 |
| NM_014341 | MTCH1 | Mitochondrial carrier homolog 1 (C. elegans) | −6.36315 | 0.00013 |
| NM_002494 | NDUFC1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa | −5.97566 | 0.00021 |
| NM_194324 | MGC39900 | hypothetical protein MGC39900 | −5.2886 | 0.0005 |
| BX118285 | LOC729717 | hypothetical protein LOC729717 | −6.00778 | 0.0002 |
| NM_017615 | NSMCE4A | Non-SMC element 4 homolog A (S. cerevisiae) | 5.78582 | 0.00026 |
| NM_178354 | LCE1F | Late cornified envelope 1F | 5.95595 | 0.00021 |
| NM_006699 | MAN1A2 | Mannosidase, alpha, class 1A, member 2 | 5.92092 | 0.00022 |
| NM_015245 | ANKS1A | Ankyrin repeat and sterile alpha motif domain containing 1A | −5.50607 | 0.00038 |
| NM_018714 | COG1 | Component of oligomeric golgi complex 1 | −5.45574 | 0.0004 |
| NM_032730 | RTN4IP1 | Reticulon 4 interacting protein 1 | −6.07719 | 0.00018 |
| BX647358 | LOC283970 | hypothetical protein LOC283970 | −6.36374 | 0.00013 |
| NM_005171 | ATF1 | Activating transcription factor 1 | 4.79615 | 0.00098 |
| NM_006537 | USP3 | Ubiquitin specific peptidase 3 | 5.28511 | 0.0005 |
| NM_020764 | CASKIN1 | CASK interacting protein 1 | 5.90091 | 0.00023 |
| XM_929707 | LOC646756 | similar to Hermansky-Pudlak syndrome 1 protein isoform b | 6.01252 | 0.0002 |
| NM_016025 | METTL9 | Methyltransferase like 9 | −5.30304 | 0.00049 |
| NM_014718 | CLSTN3 | Calsyntenin 3 | 5.55953 | 0.00035 |
| NM_016220 | ZNF588 | Zinc finger protein 588 | −6.08105 | 0.00018 |
| NM_152391 | PQLC3 | PQ loop repeat containing 3 | −5.42606 | 0.00042 |
| NM_005093 | CBFA2T2 | Core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | −4.93458 | 0.00081 |
| NM_003132 | SRM | Spermidine synthase | 6.26425 | 0.00015 |
| AB075859 | ZNF525 | Zinc finger protein 525 | −6.0377 | 0.00019 |
| NM_012133 | COPG2 | Coatomer protein complex, subunit gamma 2 | −5.60169 | 0.00033 |
| NM_015391 | ANAPC13 | Anaphase promoting complex subunit 13 | 5.88569 | 0.00023 |
| NM_007279 | U2AF2 | U2 small nuclear RNA auxiliary factor 2 | 6.28623 | 0.00014 |
| NM_181657 | LTB4R | Leukotriene B4 receptor | 6.31112 | 0.00014 |
| BC049371 | HERPUD2 | HERPUD family member 2 | −5.4236 | 0.00042 |
| NM_024865 | NANOG | Nanog homeobox | −4.86843 | 0.00089 |
| NM_015366 | PRR5 | Proline rich 5 (renal) | 5.66776 | 0.00031 |
| NM_003161 | RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | −5.22779 | 0.00054 |
| NM_007186 | CEP250 | Centrosomal protein 250kDa | −5.15592 | 0.0006 |
| AI241366 | AI241366 | Transcribed locus | −5.77357 | 0.00027 |
| NM_018660 | ZNF395 | Zinc finger protein 395 | −5.96506 | 0.00021 |
| NM_006352 | ZNF238 | Zinc finger protein 238 | 5.02507 | 0.00071 |
| NM_032199 | ARID5B | AT rich interactive domain 5B (MRF1-like) | −5.49226 | 0.00038 |
| NM_001184 | ATR | Ataxia telangiectasia and Rad3 related | −6.35054 | 0.00013 |
| NM_203448 | MGC21881 | hypothetical protein MGC21881 | −5.36318 | 0.00045 |
| NM_015584 | POLDIP2 | Polymerase (DNA-directed), delta interacting protein 2 | −5.75051 | 0.00028 |
| A_24_P315644 | A_24_P315644 | Unknown | 5.15013 | 0.0006 |
| NM_014679 | CEP57 | Centrosomal protein 57kDa | −6.20966 | 0.00016 |
| NM_030776 | ZBP1 | Z-DNA binding protein 1 | −5.73526 | 0.00028 |
| A_24_P684274 | A_24_P684274 | Unknown | 6.05786 | 0.00019 |
| NM_013347 | RPA4 | Diaphanous homolog 2 (Drosophila) | −5.34338 | 0.00047 |
| NM_000028 | AGL | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (glycogen debranching enzyme, glycogen storage disease type III) | −5.88724 | 0.00023 |
| NM_005544 | IRS1 | Insulin receptor substrate 1 | −5.99343 | 0.0002 |
| NM_017890 | VPS13B | Vacuolar protein sorting 13B (yeast) | −5.26169 | 0.00052 |
| NM_003925 | MBD4 | Methyl-CpG binding domain protein 4 | −6.08677 | 0.00018 |
| NM_194278 | C14orf43 | Chromosome 14 open reading frame 43 | −6.1667 | 0.00017 |
| NM_013450 | BAZ2B | Bromodomain adjacent to zinc finger domain, 2B | −6.24334 | 0.00015 |
| NM_002120 | HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | −5.22613 | 0.00054 |
| AK075364 | SLC10A7 | Chromosome 4 open reading frame 13 | −6.08302 | 0.00018 |
| AL390214 | TRIO | Triple functional domain (PTPRF interacting) | −5.41553 | 0.00042 |
| NM_033535 | FBXL5 | F-box and leucine-rich repeat protein 5 | −5.47671 | 0.00039 |
| A_24_P234871 | A_24_P234871 | Unknown | −5.18812 | 0.00057 |
| NM_017846 | TRSPAP1 | TRNA selenocysteine associated protein 1 | −4.89339 | 0.00086 |
| NM_203304 | RKHD1 | Ring finger and KH domain containing 1 | 5.65882 | 0.00031 |
| NM_016824 | ADD3 | Adducin 3 (gamma) | −6.2579 | 0.00015 |
| NM_012089 | ABCB10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | −5.17797 | 0.00058 |
| NM_054028 | AMAC1L2 | Acyl-malonyl condensing enzyme 1-like 2 | −6.13479 | 0.00017 |
| THC2405550 | THC2405550 | Unknown | −5.82626 | 0.00025 |
| NM_013314 | BLNK | B-cell linker | −6.15231 | 0.00017 |
| NM_001424 | EMP2 | Epithelial membrane protein 2 | 5.01766 | 0.00072 |
| NM_032932 | RAB11FIP4 | RAB11 family interacting protein 4 (class II) | −5.21617 | 0.00055 |
| BC036626 | SENP5 | SUMO1/sentrin specific peptidase 5 | −6.1731 | 0.00016 |
| NM_014300 | SEC11A | SEC11 homolog A (S. cerevisiae) | −5.51732 | 0.00037 |
| NM_032444 | BTBD12 | BTB (POZ) domain containing 12 | −5.94585 | 0.00022 |
| NM_002795 | PSMB3 | Proteasome (prosome, macropain) subunit, beta type, 3 | 5.01349 | 0.00073 |
| A_24_P609932 | A_24_P609932 | Unknown | 5.95203 | 0.00021 |
| NM_198868 | TBC1D9B | TBC1 domain family, member 9B (with GRAM domain) | −5.14293 | 0.00061 |
| NM_015272 | KIAA1005 | KIAA1005 protein | −5.38264 | 0.00044 |
| XM_496158 | A_24_P341126 | Unknown | 4.80149 | 0.00097 |
| NM_006099 | PIAS3 | Protein inhibitor of activated STAT, 3 | 5.57718 | 0.00034 |
| A_24_P126902 | A_24_P126902 | Unknown | −5.90443 | 0.00023 |
| XM_930744 | LOC653056 | PREDICTED: Homo sapiens similar to hypothetical gene supported by AK024248; AL137733, transcript variant 12 (LOC653056), mRNA [XM_930744] | 5.61483 | 0.00033 |
| NM_032199 | ARID5B | AT rich interactive domain 5B (MRF1-like) | 5.09208 | 0.00065 |
| AF318367 | C9orf86 | Homo sapiens pp8875 mRNA, complete cds. | 4.89882 | 0.00085 |
| NM_001969 | EIF5 | Eukaryotic translation initiation factor 5 | −6.21026 | 0.00016 |
| CN404647 | CCDC18 | Coiled-coil domain containing 18 | −5.25271 | 0.00053 |
| NM_004894 | C14orf2 | Chromosome 14 open reading frame 2 | −5.8179 | 0.00025 |
| NM_152458 | ZNF785 | Zinc finger protein 785 | −6.19706 | 0.00016 |
| NM_153816 | SNX14 | Sorting nexin 14 | −4.95674 | 0.00078 |
| NM_024783 | AGBL2 | ATP/GTP binding protein-like 2 | −5.29806 | 0.0005 |
| NM_003342 | UBE2G1 | Ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | 5.20823 | 0.00056 |
| AK129584 | AK129584 | Full-length cDNA clone CS0DI007YH13 of Placenta Cot 25-normalized of Homo sapiens (human) | 5.90722 | 0.00023 |
| NM_012089 | ABCB10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | −5.38255 | 0.00044 |
| NM_052873 | C14orf179 | Chromosome 14 open reading frame 179 | 5.93542 | 0.00022 |
| NM_152500 | CCDC17 | Coiled-coil domain containing 17 | −5.39547 | 0.00044 |
| NM_001040138 | CKLF | Chemokine-like factor | −5.93626 | 0.00022 |
| AK022223 | DCAKD | Dephospho-CoA kinase domain containing | −5.27457 | 0.00051 |
| NM_014314 | DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −6.10636 | 0.00018 |
| NM_003938 | AP3D1 | Adaptor-related protein complex 3, delta 1 subunit | 6.20295 | 0.00016 |
| BC035854 | DSCR1L2 | Down syndrome critical region gene 1-like 2 | −5.19219 | 0.00057 |
| NM_015342 | PPWD1 | Peptidylprolyl isomerase domain and WD repeat containing 1 | −5.06668 | 0.00067 |
| NM_175876 | EXOC8 | Exocyst complex component 8 | −5.67475 | 0.0003 |
| NM_033557 | YIF1B | Yip1 interacting factor homolog B (S. cerevisiae) | 4.88311 | 0.00087 |
| NM_002470 | MYH3 | Myosin, heavy chain 3, skeletal muscle, embryonic | −5.68484 | 0.0003 |
| NM_178126 | LOC162427 | hypothetical protein LOC162427 | 5.77981 | 0.00027 |
| BG007597 | ALKBH5 | Transcribed locus, strongly similar to NP_060228.2 protein LOC54890 [Homo sapiens] | 5.35565 | 0.00046 |
| NM_003982 | SLC7A7 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | −5.53607 | 0.00036 |
| NM_001326 | CSTF3 | Cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa | −6.14277 | 0.00017 |
| U97060 | SNAI2 | Snail homolog 2 (Drosophila) | 5.73829 | 0.00028 |
| NM_006640 | 9-Sep | Septin 9 | −5.35521 | 0.00046 |
| NM_025139 | ARMC9 | Armadillo repeat containing 9 | 5.18952 | 0.00057 |
| NM_006996 | SLC19A2 | Solute carrier family 19 (thiamine transporter), member 2 | −5.95859 | 0.00021 |
| NM_014501 | UBE2S | Ubiquitin-conjugating enzyme E2S | 5.46013 | 0.0004 |
| NM_016184 | CLEC4A | C-type lectin domain family 4, member A | −6.07896 | 0.00018 |
| NM_020470 | YIF1A | Yip1 interacting factor homolog A (S. cerevisiae) | 5.81837 | 0.00025 |
| NM_005922 | MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | −5.64318 | 0.00032 |
| S74620 | PRKCH | Protein kinase C, eta | 5.30026 | 0.00049 |
| A_32_P62137 | A_32_P62137 | Unknown | 5.50854 | 0.00038 |
| L08436 | COTL1 | Coactosin-like 1 (Dictyostelium) | −6.03708 | 0.00019 |
| NM_172345 | SPAG9 | Sperm associated antigen 9 | 5.40408 | 0.00043 |
| NM_003119 | SPG7 | Spastic paraplegia 7, paraplegin (pure and complicated autosomal recessive) | 4.91896 | 0.00083 |
| AK128396 | TRAF6 | CDNA FLJ46539 fis, clone THYMU3037836 | −4.9648 | 0.00078 |
| NM_014480 | ZNF544 | Zinc finger protein 544 | −5.52426 | 0.00037 |
| NM_024050 | C19orf58 | Chromosome 19 open reading frame 58 | 5.81018 | 0.00026 |
| NM_014017 | MAPBPIP | mitogen-activated protein-binding protein-interacting protein | 5.65193 | 0.00031 |
| NM_018952 | HOXB6 | Homeobox B5 | 4.78253 | 0.001 |
| AL832683 | PTAR1 | Protein prenyltransferase alpha subunit repeat containing 1 | −5.18786 | 0.00057 |
| NM_024812 | BAALC | Brain and acute leukemia, cytoplasmic | 5.29334 | 0.0005 |
| XM_496043 | A_24_P761457 | Unknown | 4.99333 | 0.00075 |
| BX538300 | MRPS30 | Full-length cDNA clone CS0DK012YA15 of HeLa cells Cot 25-normalized of Homo sapiens (human) | 5.38356 | 0.00044 |
| NM_145309 | LRRC51 | Leucine rich repeat containing 51 | 5.71612 | 0.00029 |
| NM_005647 | TBL1X | Transducin (beta)-like 1X-linked | −5.4274 | 0.00042 |
| NM_006788 | RALBP1 | RalA binding protein 1 | 5.44876 | 0.00041 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −4.85896 | 0.0009 |
| CR590530 | PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | −5.90778 | 0.00023 |
| NM_006135 | CAPZA1 | Capping protein (actin filament) muscle Z-line, alpha 1 | 5.48417 | 0.00039 |
| NM_198490 | RAB43 | RAB43, member RAS oncogene family | −5.83049 | 0.00025 |
| BM461836 | BM461836 | Transcribed locus | −6.01488 | 0.0002 |
| AK022479 | HDHD1A | Haloacid dehalogenase-like hydrolase domain containing 1A | −5.47807 | 0.00039 |
| AL832806 | HDAC4 | Histone deacetylase 4 | −5.80606 | 0.00026 |
| BC021898 | AP1S3 | Homo sapiens adaptor-related protein complex 1, sigma 3 subunit, mRNA (cDNA clone MGC:17284 IMAGE:4340257), complete cds. | 5.33898 | 0.00047 |
| NM_001991 | EZH1 | Enhancer of zeste homolog 1 (Drosophila) | −5.85304 | 0.00024 |
| NM_033222 | PSIP1 | PC4 and SFRS1 interacting protein 1 | −5.29831 | 0.00049 |
| NM_182538 | MGC29671 | hypothetical protein MGC29671 | 5.11574 | 0.00063 |
| NM_002055 | GFAP | Glial fibrillary acidic protein | 5.4633 | 0.0004 |
| NM_004552 | NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) | −5.31081 | 0.00049 |
| AB011149 | KIAA0577 | Homo sapiens mRNA for KIAA0577 protein, partial cds. | −5.56005 | 0.00035 |
| AK098220 | LOC728555 | hypothetical protein LOC728555 | −5.66434 | 0.00031 |
| NM_199168 | CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 5.36752 | 0.00045 |
| XM_001134097 | LOC731989 | similar to protein tyrosine phosphatase 4a1 | 5.88768 | 0.00023 |
| NM_021140 | UTX | Ubiquitously transcribed tetratricopeptide repeat, X chromosome | −5.36566 | 0.00045 |
| NM_001032410 | USP16 | Ubiquitin specific peptidase 16 | −5.39423 | 0.00044 |
| NM_031206 | LAS1L | LAS1-like (S. cerevisiae) | −5.31002 | 0.00049 |
| NM_032750 | ABHD14B | Abhydrolase domain containing 14B | 4.95275 | 0.00079 |
| NM_013375 | ABT1 | Activator of basal transcription 1 | −5.94292 | 0.00022 |
| NM_178831 | GATS | opposite strand transcription unit to STAG3 | 5.62774 | 0.00032 |
| NM_138426 | GLCCI1 | Glucocorticoid induced transcript 1 | −5.09956 | 0.00065 |
| NM_025160 | WDR26 | WD repeat domain 26 | −4.99453 | 0.00074 |
| NM_017914 | C19orf24 | Chromosome 19 open reading frame 24 | −5.7746 | 0.00027 |
| NM_019053 | EXOC6 | Exocyst complex component 6 | −5.5546 | 0.00035 |
| NM_012249 | RHOQ | Ras homolog gene family, member Q | 5.05453 | 0.00069 |
| NM_032823 | C9orf3 | Chromosome 9 open reading frame 3 | 5.65131 | 0.00031 |
| NM_014388 | C1orf107 | Chromosome 1 open reading frame 107 | −5.4832 | 0.00039 |
| BC054050 | THOC2 | THO complex 2 | −5.54513 | 0.00036 |
| NM_001017915 | INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | −5.83365 | 0.00025 |
| NM_032380 | GFM2 | G elongation factor, mitochondrial 2 | −4.82169 | 0.00094 |
| NM_025054 | VCPIP1 | Valosin containing protein (p97)/p47 complex interacting protein 1 | −5.25903 | 0.00052 |
| NM_006454 | MXD4 | MAX dimerization protein 4 | 5.83018 | 0.00025 |
| THC2412623 | THC2412623 | Unknown | 5.5052 | 0.00038 |
| NM_131916 | PPIL3 | Peptidylprolyl isomerase (cyclophilin)-like 3 | −5.7303 | 0.00028 |
| NM_005730 | CTDSP2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 | −4.87806 | 0.00087 |
| NM_001000 | RPL39 | Ribosomal protein L39 | −5.27874 | 0.00051 |
| L06133 | ATP7A | ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome) | −5.18321 | 0.00058 |
| NM_145059 | FUK | Fucokinase | −5.02457 | 0.00071 |
| BC043173 | BC043173 | CDNA clone IMAGE:5287121 | −4.87502 | 0.00088 |
| NM_015261 | NCAPD3 | Non-SMC condensin II complex, subunit D3 | −5.01908 | 0.00072 |
| NM_175709 | CBX7 | Chromobox homolog 7 | −5.37048 | 0.00045 |
| NM_138764 | BAX | BCL2-associated X protein | 4.98972 | 0.00075 |
| NM_001002261 | ZFYVE27 | Zinc finger, FYVE domain containing 27 | −5.04773 | 0.00069 |
| BX538256 | ENST00000369183 | MRNA; cDNA DKFZp686C1384 (from clone DKFZp686C1384) | −5.41008 | 0.00043 |
| NM_139015 | SPPL3 | signal peptide peptidase 3 | −5.26749 | 0.00052 |
| NM_176818 | 15E1.2 | hypothetical protein LOC283459 | 4.87135 | 0.00088 |
| BX648207 | BX648207 | CDNA FLJ40891 fis, clone UTERU2001110 | −5.16386 | 0.00059 |
| A_32_P19616 | A_32_P19616 | Unknown | 5.60695 | 0.00033 |
| NM_014829 | DDX46 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 | −5.52869 | 0.00037 |
| AK023831 | FLJ13769 | hypothetical protein FLJ13769 | −5.65982 | 0.00031 |
| A_24_P298835 | A_24_P298835 | Unknown | −4.94729 | 0.00079 |
| NM_052862 | RCSD1 | RCSD domain containing 1 | −4.94157 | 0.0008 |
| XM_496145 | LOC440361 | PREDICTED: Homo sapiens similar to Ig heavy chain V-III region VH26 precursor (LOC440361), mRNA [XM_496145] | 4.79274 | 0.00098 |
| NM_014649 | SAFB2 | Small nuclear ribonucleoprotein polypeptide E | −4.92345 | 0.00082 |
| NM_152604 | ZNF383 | Zinc finger protein 383 | −4.95902 | 0.00078 |
| NM_022748 | TNS3 | Tensin 3 | −5.23702 | 0.00054 |
| BC062636 | RHBDD1 | Rhomboid domain containing 1 | −5.31878 | 0.00048 |
| NM_006591 | POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | −5.68071 | 0.0003 |
| NM_024329 | EFHD2 | EF-hand domain family, member D2 | 5.37781 | 0.00045 |
| NM_015485 | RWDD3 | RWD domain containing 3 | −5.33594 | 0.00047 |
| AK094181 | TTC15 | Tetratricopeptide repeat domain 15 | −5.28622 | 0.0005 |
| NM_017613 | DONSON | Downstream neighbor of SON | −4.89151 | 0.00086 |
| BF476310 | PGPEP1 | Pyroglutamyl-peptidase I | 5.57075 | 0.00035 |
| AK022016 | MYO1E | Myosin IE | −5.28289 | 0.00051 |
| NM_018602 | DNAJA4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 5.08766 | 0.00066 |
| THC2405710 | THC2405710 | Unknown | −5.43828 | 0.00041 |
| NM_016227 | C1orf9 | Chromosome 1 open reading frame 9 | −5.11283 | 0.00063 |
| NM_006568 | CGRRF1 | Cell growth regulator with ring finger domain 1 | −5.30073 | 0.00049 |
| XM_497928 | A_24_P409891 | Unknown | −5.10488 | 0.00064 |
| NM_032358 | CCDC77 | Coiled-coil domain containing 77 | −4.97185 | 0.00077 |
| NM_020807 | ZNF319 | Zinc finger protein 319 | −4.89333 | 0.00086 |
| A_32_P145302 | A_32_P145302 | Unknown | 5.14089 | 0.00061 |
| NM_001007467 | SFI1 | Sfi1 homolog, spindle assembly associated (yeast) | −5.24419 | 0.00053 |
| NM_021167 | GATAD1 | GATA zinc finger domain containing 1 | 5.4081 | 0.00043 |
| A_24_P616082 | A_24_P616082 | Unknown | 5.48231 | 0.00039 |
| AK000103 | LOC153277 | hypothetical protein LOC153277 | −5.60166 | 0.00033 |
| AK096109 | LOC339483 | hypothetical LOC339483 | −5.45198 | 0.0004 |
| NM_138621 | BCL2L11 | BCL2-like 11 (apoptosis facilitator) | 5.11569 | 0.00063 |
| NM_016563 | RASL12 | RAS-like, family 12 | 5.14387 | 0.00061 |
| NM_004551 | NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) | 5.3438 | 0.00047 |
| NM_006618 | JARID1B | Jumonji, AT rich interactive domain 1B | −5.52211 | 0.00037 |
| NM_015503 | SH2B1 | SH2B adaptor protein 1 | 5.12367 | 0.00062 |
| NM_020062 | SLC2A4RG | SLC2A4 regulator | 5.48553 | 0.00039 |
| NM_006321 | ARIH2 | Ariadne homolog 2 (Drosophila) | 4.82099 | 0.00095 |
| NM_020235 | BBX | Bobby sox homolog (Drosophila) | −5.22583 | 0.00054 |
| NM_021167 | GATAD1 | GATA zinc finger domain containing 1 | −4.7975 | 0.00098 |
| NM_000255 | MUT | Methylmalonyl Coenzyme A mutase | −5.32572 | 0.00048 |
| NM_014636 | RALGPS1 | Ral GEF with PH domain and SH3 binding motif 1 | −5.3981 | 0.00043 |
| NM_003899 | ARHGEF7 | Rho guanine nucleotide exchange factor (GEF) 7 | −4.98475 | 0.00075 |
| BC032369 | HNRPUL2 | Heterogeneous nuclear ribonucleoprotein U-like 2 | 4.95545 | 0.00079 |
| NM_014739 | BCLAF1 | Homo sapiens BCL2-associated transcription factor 1 (BCLAF1), transcript variant 1, mRNA. | −4.93999 | 0.0008 |
| NM_000364 | TNNT2 | Troponin T type 2 (cardiac) | 4.96733 | 0.00077 |
| NM_030639 | BHLHB9 | Basic helix-loop-helix domain containing, class B, 9 | −4.9057 | 0.00084 |
| NM_013430 | GGT1 | Gamma-glutamyltransferase 1 | 4.78165 | 0.001 |
| NM_006594 | AP4B1 | Adaptor-related protein complex 4, beta 1 subunit | −5.45928 | 0.0004 |
| NM_003188 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | −5.50014 | 0.00038 |
| THC2415754 | THC2415754 | Q9Y6H2 (Q9Y6H2) Ku70-binding protein (Fragment), partial (41%) [THC2415754] | −4.97453 | 0.00077 |
| XM_496741 | LOC441066 | PREDICTED: Homo sapiens similar to Beta-glucuronidase precursor (LOC441066), mRNA [XM_496741] | 4.89513 | 0.00085 |
| NM_012430 | SEC22A | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) | −5.0367 | 0.0007 |
| NM_014805 | EPM2AIP1 | EPM2A (laforin) interacting protein 1 | −5.02229 | 0.00072 |
| BC045718 | AMZ2 | archaemetzincins-2 | −4.86184 | 0.00089 |
| AK023737 | AK023737 | Homo sapiens cDNA FLJ13675 fis, clone PLACE1011875, highly similar to Homo sapiens mRNA for KIAA0580 protein. | −5.39305 | 0.00044 |
| A_24_P110521 | A_24_P110521 | Unknown | −5.43875 | 0.00041 |
| NM_015355 | SUZ12 | Suppressor of zeste 12 homolog (Drosophila) | −5.08232 | 0.00066 |
| THC2320257 | THC2320257 | Unknown | −5.26085 | 0.00052 |
| BM069797 | C18orf17 | Transcribed locus, strongly similar to XP_512065.1 similar to squamous cell carcinoma-related protein-1 [Pan troglodytes] | −5.06886 | 0.00067 |
| THC2373083 | THC2373083 | Unknown | −5.22908 | 0.00054 |
| NM_001439 | EXTL2 | Exostoses (multiple)-like 2 | −5.22046 | 0.00055 |
| THC2340322 | THC2340322 | Q5T931 (Q5T931) Sorbin and SH3 domain containing 1, partial (3%) [THC2340322] | −5.24933 | 0.00053 |
| NM_018297 | NGLY1 | N-glycanase 1 | −5.39139 | 0.00044 |
| A_32_P55438 | A_32_P55438 | Unknown | −5.09221 | 0.00065 |
| NM_019593 | KIAA1434 | hypothetical protein KIAA1434 | −4.86596 | 0.00089 |
| NM_005813 | PRKD3 | Protein kinase D3 | −5.19393 | 0.00057 |
| NM_001750 | CAST | Calpastatin | −5.38217 | 0.00044 |
| THC2266506 | THC2266506 | RS24_HUMAN (P62847) 40S ribosomal protein S24, complete [THC2266506] | −5.34445 | 0.00047 |
| NM_001029 | RPS26 | Ribosomal protein S26 | −5.39967 | 0.00043 |
| NM_147171 | AKAP9 | A kinase (PRKA) anchor protein (yotiao) 9 | −5.10376 | 0.00064 |
| THC2671898 | THC2671898 | Q3WYF8_9ACTN (Q3WYF8) Sulfotransferase, partial (6%) [THC2671898] | 4.9822 | 0.00076 |
| NM_005773 | ZNF256 | Zinc finger protein 418 | −5.08148 | 0.00066 |
| A_32_P8971 | A_32_P8971 | Unknown | −4.99489 | 0.00074 |
| NM_173854 | SLC41A1 | Solute carrier family 41, member 1 | −5.30434 | 0.00049 |
| NM_016467 | ORMDL1 | ORM1-like 1 (S. cerevisiae) | −5.13111 | 0.00062 |
| THC2437881 | THC2437881 | Q9NBA9 (Q9NBA9) Stretchin-MLCK (Fragment), partial (5%) [THC2437881] | −5.17389 | 0.00058 |
| NM_004487 | GOLGB1 | Golgi autoantigen, golgin subfamily b, macrogolgin (with transmembrane signal), 1 | −5.26839 | 0.00051 |
| NM_013444 | UBQLN2 | Ubiquilin 2 | −4.98166 | 0.00076 |
| NM_001011667 | CHCHD7 | Coiled-coil-helix-coiled-coil-helix domain containing 7 | −5.02654 | 0.00071 |
| NM_020666 | CLK4 | CDC-like kinase 4 | −5.18565 | 0.00057 |
| NM_006221 | PIN1 | Protein (peptidylprolyl cis/trans isomerase) NIMA-interacting 1 | 5.0031 | 0.00074 |
| NM_001029 | RPS26 | Ribosomal protein S26 | −5.3064 | 0.00049 |
| THC2280343 | THC2280343 | ALU6_HUMAN (P39193) Alu subfamily SP sequence contamination warning entry, partial (9%) [THC2280343] | −4.93191 | 0.00081 |
| AK023675 | UTRN | Utrophin (homologous to dystrophin) | −5.03783 | 0.0007 |
| NM_024065 | PDCL3 | Phosducin-like 3 | −4.79228 | 0.00098 |
| NM_182663 | RASSF5 | Ras association (RalGDS/AF-6) domain family 5 | 4.79344 | 0.00098 |
| NM_018456 | EAF2 | ELL associated factor 2 | −5.1398 | 0.00061 |
| A_23_P158868 | A_23_P158868 | Unknown | 5.19558 | 0.00057 |
| XM_940047 | LOC654152 | PREDICTED: Homo sapiens similar to ribosomal protein L21 isoform 1 (LOC654152), mRNA [XM_940047] | −5.06821 | 0.00067 |
| NM_003264 | TLR2 | Toll-like receptor 2 | −5.20009 | 0.00056 |
| NM_015117 | ZC3H3 | Zinc finger CCCH-type containing 3 | 5.05631 | 0.00068 |
| NM_024112 | C9orf16 | Chromosome 9 open reading frame 16 | 5.0859 | 0.00066 |
| NM_006051 | APBB3 | Amyloid beta (A4) precursor protein-binding, family B, member 3 | −4.83419 | 0.00093 |
| NM_032714 | C14orf151 | Chromosome 14 open reading frame 151 | 5.05654 | 0.00068 |
| NM_015911 | ZNF691 | Zinc finger protein 691 | −4.89911 | 0.00085 |
| THC2306096 | THC2306096 | Unknown | −5.15365 | 0.0006 |
| NM_015088 | TNRC6B | Trinucleotide repeat containing 6B | −4.78159 | 0.001 |
| A_32_P158543 | A_32_P158543 | Unknown | 4.83166 | 0.00093 |
| NM_004278 | PIGL | Phosphatidylinositol glycan anchor biosynthesis, class L | 5.03195 | 0.00071 |
| NM_024313 | NOL12 | Nucleolar protein 12 | 4.80569 | 0.00097 |
| NM_014238 | KSR1 | Kinase suppressor of ras 1 | 4.99409 | 0.00074 |
| NM_015412 | C3orf17 | Chromosome 3 open reading frame 17 | −5.07804 | 0.00066 |
| NM_018255 | STATIP1 | Signal transducer and activator of transcription 3 interacting protein 1 | −4.88966 | 0.00086 |
| A_32_P180185 | A_32_P180185 | Unknown | −4.90553 | 0.00084 |
| L31408 | FGF2 | Fibroblast growth factor 2 (basic) | 4.99531 | 0.00074 |
| NM_014608 | CYFIP1 | Cytoplasmic FMR1 interacting protein 1 | −4.97789 | 0.00076 |
| NM_000655 | SELL | Selectin L (lymphocyte adhesion molecule 1) | −4.93527 | 0.00081 |
| NM_153240 | NPHP3 | Nephronophthisis 3 (adolescent) | −4.85518 | 0.0009 |
| NM_031444 | C22orf13 | Chromosome 22 open reading frame 13 | 4.95359 | 0.00079 |
| NM_003811 | TNFSF9 | Tumor necrosis factor (ligand) superfamily, member 9 | 4.87787 | 0.00087 |
| NM_016310 | POLR3K | Polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa | 4.78534 | 0.00099 |
| NM_021149 | COTL1 | Coactosin-like 1 (Dictyostelium) | 4.83716 | 0.00092 |
| THC2409283 | THC2409283 | Unknown | −4.98158 | 0.00076 |
| X81724 | CPVL | Carboxypeptidase, vitellogenic-like | 4.78924 | 0.00099 |
| NM_001039360 | ZBTB7C | Zinc finger and BTB domain containing 7C | 4.94196 | 0.0008 |
| NM_003387 | WIPF1 | WAS/WASL interacting protein family, member 1 | −4.91746 | 0.00083 |
| BE774743 | THC2363476 | CDNA FLJ13267 fis, clone OVARC1000964 | 4.92569 | 0.00082 |
| NM_032330 | CAPNS2 | Calpain, small subunit 2 | 4.85241 | 0.00091 |
| NM_015540 | RPAP1 | RNA polymerase II associated protein 1 | −4.89166 | 0.00086 |
| XM_376787 | RPS26P10 | Ribosomal protein S26 pseudogene 10 | −4.90436 | 0.00084 |
| NM_018125 | ARHGEF10L | Rho guanine nucleotide exchange factor (GEF) 10-like | 4.88064 | 0.00087 |
| NM_017644 | KLHL24 | Kelch-like 24 (Drosophila) | −4.8297 | 0.00093 |
| NM_015132 | SNX13 | Sorting nexin 13 | −4.82155 | 0.00094 |
| NM_153219 | ZNF524 | Zinc finger protein 524 | −4.82122 | 0.00095 |
| A_23_P206568 | A_23_P206568 | Unknown | 4.81298 | 0.00096 |
| Pathway | Gene | Description |
|---|---|---|
| Cell adhesion molecules (CAMs) | HLA-DMB | Major histocompatibility complex, class II, DM beta |
| ITGB7 | Integrin, beta 7 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| HLA-DMA | Major histocompatibility complex, class II, DM beta | |
| SDC4 | Syndecan 4 (amphiglycan, ryudocan) | |
| HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | |
| SELP | Selectin P (granule membrane protein 140kDa, antigen CD62) | |
| NLGN1 | Neuroligin 1 | |
| SELL | Selectin L (lymphocyte adhesion molecule 1) | |
| L1CAM | L1 cell adhesion molecule | |
| ICAM2 | Intercellular adhesion molecule 2 | |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | |
| ECM-receptor interaction | ITGB7 | Integrin, beta 7 |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| SDC4 | Syndecan 4 (amphiglycan, ryudocan) | |
| Antigen processing and presentation | HLA-DMB | Major histocompatibility complex, class II, DM beta |
| HLA-DMA | Major histocompatibility complex, class II, DM beta | |
| HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | |
| Type I diabetes mellitus | HLA-DMB | Major histocompatibility complex, class II, DM beta |
| HLA-DMA | Major histocompatibility complex, class II, DM beta | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | |
| Bile acid biosynthesis | ACAD8 | Acyl-Coenzyme A dehydrogenase family, member 8 |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | |
| ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | |
| Aminoacyl-tRNA biosynthesis | QARS | Glutaminyl-tRNA synthetase |
| HARSL | Histidyl-tRNA synthetase-like | |
| HARS | Histidyl-tRNA synthetase | |
| Histidine metabolism | PRMT2 | Protein arginine methyltransferase 2 |
| HARS | Histidyl-tRNA synthetase | |
| HARSL | Histidyl-tRNA synthetase-like | |
| Epithelial cell signaling in Helicobacter pylori infection | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| HBEGF | Heparin-binding EGF-like growth factor | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | PIGA | Phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) |
| PIGK | Phosphatidylinositol glycan anchor biosynthesis, class K | |
| PIGG | Phosphatidylinositol glycan anchor biosynthesis, class G | |
| PIGL | Phosphatidylinositol glycan anchor biosynthesis, class L | |
| Alkaloid biosynthesis II | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | |
| 1- and 2-Methylnaphthalene degradation | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| ACAD8 | Acyl-Coenzyme A dehydrogenase family, member 8 | |
| Benzoate degradation via CoA ligation | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| Limonene and pinene degradation | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| Glycerophospholipid metabolism | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| GPAM | Glycerol-3-phosphate acyltransferase, mitochondrial | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| LYPLA2 | Lysophospholipase II | |
| PPAP2C | Phosphatidic acid phosphatase type 2C | |
| Adherens junction | ACTC1 | Actin, alpha, cardiac muscle 1 |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | |
| ACTN3 | Actinin, alpha 3 | |
| EP300 | E1A binding protein p300 | |
| WASF2 | CDNA clone IMAGE:3030163 | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| SNAI2 | Snail homolog 2 (Drosophila) | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| SMAD3 | SMAD family member 3 | |
| LMO7 | LIM domain 7 | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| TGF-beta signaling pathway | ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | |
| EP300 | E1A binding protein p300 | |
| BMPR1A | Bone morphogenetic protein receptor, type IA | |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| SMAD3 | SMAD family member 3 | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Ubiquitin mediated proteolysis | ITCH | Itchy homolog E3 ubiquitin protein ligase (mouse) |
| ANAPC7 | Anaphase promoting complex subunit 7 | |
| EDD1 | E3 ubiquitin protein ligase, HECT domain containing, 1 | |
| UBE2D2 | Ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | |
| FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | |
| Tyrosine metabolism | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| PRMT2 | Protein arginine methyltransferase 2 | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| Inositol phosphate metabolism | PIK3C3 | Phosphoinositide-3-kinase, class 3 |
| MINPP1 | Multiple inositol polyphosphate histidine phosphatase, 1 | |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Phosphatidylinositol signaling system | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| PIK3C3 | Phosphoinositide-3-kinase, class 3 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| PRKCA | Protein kinase C, alpha | |
| PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Oxidative phosphorylation | NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| NDUFC1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa | |
| COX11 | COX11 homolog, cytochrome c oxidase assembly protein (yeast) | |
| NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) | |
| COX8A | Cytochrome c oxidase subunit 8A (ubiquitous) | |
| Ribosome | RPL39 | Ribosomal protein L39 |
| RPL21 | Ribosomal protein L21 | |
| RPL22 | Ribosomal protein L22 | |
| RPS26P10 | Ribosomal protein S26 pseudogene 10 | |
| RPS26 | Ribosomal protein S26 | |
| RPL15 | Ribosomal protein L15 | |
| RPS23 | Ribosomal protein S23 | |
| Arachidonic acid metabolism | GGT1 | Gamma-glutamyltransferase 1 |
| LTC4S | Leukotriene C4 synthase | |
| ALOX5 | Arachidonate 5-lipoxygenase | |
| Glutathione metabolism | GSTM2 | Glutathione S-transferase M2 (muscle) |
| GSTM1 | Glutathione S-transferase M1 | |
| GSTM4 | Glutathione S-transferase M4 | |
| G6PD | Glucose-6-phosphate dehydrogenase | |
| GGT1 | Gamma-glutamyltransferase 1 | |
| Metabolism of xenobiotics by cytochrome P450 | GSTM4 | Glutathione S-transferase M4 |
| GSTM2 | Glutathione S-transferase M2 (muscle) | |
| GSTM1 | Glutathione S-transferase M1 | |
| UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | |
| Cholera - Infection | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| PRKCA | Protein kinase C, alpha | |
| SEC61A2 | Sec61 alpha 2 subunit (S. cerevisiae) | |
| ARF3 | ADP-ribosylation factor 3 | |
| Chondroitin sulfate biosynthesis | CSGlcA-T | chondroitin sulfate glucuronyltransferase |
| XYLT1 | Xylosyltransferase I | |
| ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | |
| Cytokine-cytokine receptor interaction | TNF | Tumor necrosis factor (TNF superfamily, member 2) |
| GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) | |
| IL1RAP | Interleukin 1 receptor accessory protein | |
| BMPR1A | Bone morphogenetic protein receptor, type IA | |
| TNFRSF1 | 7Tumor necrosis factor receptor superfamily, member 17 | |
| IL10RA | Interleukin 10 receptor, alpha | |
| HGF | Human mRNA for hepatocyte growth factor (HGF). | |
| TNFSF9 | Tumor necrosis factor (ligand) superfamily, member 9 | |
| IL17RA | Interleukin 17 receptor A | |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Glycan structures - biosynthesis 2 | B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | |
| PIGA | Phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) | |
| PIGK | Phosphatidylinositol glycan anchor biosynthesis, class K | |
| PIGG | Phosphatidylinositol glycan anchor biosynthesis, class G | |
| ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | |
| PIGL | Phosphatidylinositol glycan anchor biosynthesis, class L | |
| Fructose and mannose metabolism | PFKL | Phosphofructokinase, liver |
| GMPPA | GDP-mannose pyrophosphorylase A | |
| STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | |
| SORD | Sorbitol dehydrogenase | |
| FUK | Fucokinase | |
| HK2 | Hexokinase 2 | |
| Galactose metabolism | PFKL | Phosphofructokinase, liver |
| B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 | |
| HK2 | Hexokinase 2 | |
| Fatty acid metabolism | ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) |
| ACOX1 | Acyl-Coenzyme A oxidase 1, palmitoyl | |
| ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | |
| Glycan structures - biosynthesis 1 | B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| ALG6 | Asparagine-linked glycosylation 6 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | |
| ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | |
| XYLT1 | Xylosyltransferase I | |
| EXT2 | Exostoses (multiple) 2 | |
| B3GNT6 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) | |
| RPN1 | Ribophorin I | |
| CSGlcA-T | chondroitin sulfate glucuronyltransferase | |
| ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | |
| EXTL2 | Exostoses (multiple)-like 2 | |
| MAN1B1 | Mannosidase, alpha, class 1B, member 1 | |
| MAN1A2 | Mannosidase, alpha, class 1A, member 2 | |
| Huntington's disease | DCTN1 | Dynactin 1 (p150, glued homolog, Drosophila) |
| BAX | BCL2-associated X protein | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| EP300 | E1A binding protein p300 | |
| Cell cycle | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) |
| ANAPC7 | Anaphase promoting complex subunit 7 | |
| EP300 | E1A binding protein p300 | |
| YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | |
| ATR | Ataxia telangiectasia and Rad3 related | |
| SMAD3 | SMAD family member 3 | |
| FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | |
| Valine, leucine and isoleucine degradation | ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) |
| MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 | |
| ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | |
| BCAT2 | Branched chain aminotransferase 2, mitochondrial | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| MUT | Methylmalonyl Coenzyme A mutase | |
| Amyotrophic lateral sclerosis (ALS) | RAB5A | RAB5A, member RAS oncogene family |
| BAX | BCL2-associated X protein | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CAT | Catalase | |
| SLC1A2 | Homo sapiens solute carrier family 1 (glial high affinity glutamate transporter), member 2 (SLC1A2), mRNA. | |
| Glycerolipid metabolism | GPAM | Glycerol-3-phosphate acyltransferase, mitochondrial |
| STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | |
| PPAP2C | Phosphatidic acid phosphatase type 2C | |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | |
| DNA polymerase | POLD4 | Polymerase (DNA-directed), delta 4 |
| POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | |
| POLE | Polymerase (DNA directed), epsilon | |
| Purine metabolism | ADSS | Adenylosuccinate synthase |
| PDE4C | Phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | |
| XDH | Xanthine dehydrogenase | |
| POLR3H | Polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) | |
| POLR3K | Polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa | |
| POLD4 | Polymerase (DNA-directed), delta 4 | |
| POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| DCK | Deoxycytidine kinase | |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | |
| POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | |
| GART | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | |
| POLE | Polymerase (DNA directed), epsilon | |
| Pyrimidine metabolism | POLR3H | Polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) |
| POLD4 | Polymerase (DNA-directed), delta 4 | |
| POLR3K | Polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa | |
| DCK | Deoxycytidine kinase | |
| POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | |
| DHODH | Dihydroorotate dehydrogenase | |
| POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | |
| TK2 | Thymidine kinase 2, mitochondrial | |
| POLE | Polymerase (DNA directed), epsilon | |
| Jak-STAT signaling pathway | GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| STAT6 | Signal transducer and activator of transcription 6, interleukin-4 induced | |
| CISH | Cytokine inducible SH2-containing protein | |
| EP300 | E1A binding protein p300 | |
| PIAS4 | Protein inhibitor of activated STAT, 4 | |
| PIAS3 | Protein inhibitor of activated STAT, 3 | |
| IL10RA | Interleukin 10 receptor, alpha | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| STAT5B | Signal transducer and activator of transcription 5B | |
| Chronic myeloid leukemia | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| CTBP1 | C-terminal binding protein 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| GAB2 | GRB2-associated binding protein 2 | |
| CTBP2 | C-terminal binding protein 2 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| SMAD3 | SMAD family member 3 | |
| STAT5B | Signal transducer and activator of transcription 5B | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Adipocytokine signaling pathway | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| STK11 | Serine/threonine kinase 11 | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | |
| IRS1 | Insulin receptor substrate 1 | |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | |
| B cell receptor signaling pathway | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| CHP | calcium binding protein P22 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| BLNK | B-cell linker | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| Fc epsilon RI signaling pathway | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| GAB2 | GRB2-associated binding protein 2 | |
| PRKCA | Protein kinase C, alpha | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Type II diabetes mellitus | MAFA | V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian) |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| IRS1 | Insulin receptor substrate 1 | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | |
| Focal adhesion | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| ACTN3 | Actinin, alpha 3 | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| TLN1 | Talin 1 | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| PRKCA | Protein kinase C, alpha | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| CAV2 | Caveolin 2 | |
| ITGB7 | Integrin, beta 7 | |
| HGF | Human mRNA for hepatocyte growth factor (HGF). | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| GRLF1 | Glucocorticoid receptor DNA binding factor 1 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| Axon guidance | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| CHP | calcium binding protein P22 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| NCK2 | NCK adaptor protein 2 | |
| L1CAM | L1 cell adhesion molecule | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | |
| Calcium signaling pathway | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| CHP | calcium binding protein P22 | |
| CACNA1I | Calcium channel, voltage-dependent, alpha 1I subunit | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| PRKCA | Protein kinase C, alpha | |
| SLC25A5 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | |
| ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | |
| Pathogenic Escherichia coli infection - EHEC | TUBB8 | tubulin, beta 8 |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| VIL2 | Villin 2 (ezrin) | |
| PRKCA | Protein kinase C, alpha | |
| NCK2 | NCK adaptor protein 2 | |
| Long-term potentiation | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| CHP | calcium binding protein P22 | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| EP300 | E1A binding protein p300 | |
| PRKCA | Protein kinase C, alpha | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| MAPK signaling pathway | DUSP4 | Dual specificity phosphatase 4 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| RASA2 | RAS p21 protein activator 2 | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| MAX | MYC associated factor X | |
| CHP | calcium binding protein P22 | |
| PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| PRKCA | Protein kinase C, alpha | |
| CACNA1I | Calcium channel, voltage-dependent, alpha 1I subunit | |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| TRAF6 | CDNA FLJ46539 fis, clone THYMU3037836 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| FGF2 | Fibroblast growth factor 2 (basic) | |
| MRAS | Muscle RAS oncogene homolog | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Apoptosis | PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| BAX | BCL2-associated X protein | |
| MYD88 | Myeloid differentiation primary response gene (88) | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| IL1RAP | Interleukin 1 receptor accessory protein | |
| CHP | calcium binding protein P22 | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Regulation of actin cytoskeleton | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| ARPC4 | Actin related protein 2/3 complex, subunit 4, 20kDa | |
| FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| ACTN3 | Actinin, alpha 3 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| VIL2 | Villin 2 (ezrin) | |
| WASF2 | CDNA clone IMAGE:3030163 | |
| ITGB7 | Integrin, beta 7 | |
| PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CYFIP1 | Cytoplasmic FMR1 interacting protein 1 | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| ARHGEF7 | Rho guanine nucleotide exchange factor (GEF) 7 | |
| GRLF1 | Glucocorticoid receptor DNA binding factor 1 | |
| FGF2 | Fibroblast growth factor 2 (basic) | |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | |
| MRAS | Muscle RAS oncogene homolog | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| Insulin signaling pathway | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| PFKL | Phosphofructokinase, liver | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| RHOQ | Ras homolog gene family, member Q | |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | |
| FLOT1 | Flotillin 1 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| IRS1 | Insulin receptor substrate 1 | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | |
| GYS1 | Glycogen synthase 1 (muscle) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| PYGB | Phosphorylase, glycogen; brain | |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| Wnt signaling pathway | CTBP1 | C-terminal binding protein 1 |
| WNT6 | Wingless-type MMTV integration site family, member 6 | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| EP300 | E1A binding protein p300 | |
| CHP | calcium binding protein P22 | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| CTBP2 | C-terminal binding protein 2 | |
| PSEN1 | Presenilin 1 (Alzheimer disease 3) | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| TBL1X | Transducin (beta)-like 1X-linked | |
| PRKCA | Protein kinase C, alpha | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| SMAD3 | SMAD family member 3 | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| VEGF signaling pathway | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| CHP | calcium binding protein P22 | |
| PRKCA | Protein kinase C, alpha | |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Melanogenesis | CALM1 | Calmodulin 1 (phosphorylase kinase, delta) |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| WNT6 | Wingless-type MMTV integration site family, member 6 | |
| EP300 | E1A binding protein p300 | |
| PRKCA | Protein kinase C, alpha | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| Notch signaling pathway | CIR | CBF1 interacting corepressor |
| CTBP1 | C-terminal binding protein 1 | |
| PCAF | P300/CBP-associated factor | |
| EP300 | E1A binding protein p300 | |
| PSEN1 | Presenilin 1 (Alzheimer disease 3) | |
| CTBP2 | C-terminal binding protein 2 | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| Glioma | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| PRKCA | Protein kinase C, alpha | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Natural killer cell mediated cytotoxicity | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| CHP | calcium binding protein P22 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| PRKCA | Protein kinase C, alpha | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| ICAM2 | Intercellular adhesion molecule 2 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| T cell receptor signaling pathway | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) |
| CHP | calcium binding protein P22 | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| NCK2 | NCK adaptor protein 2 | |
| Leukocyte transendothelial migration | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| ACTN3 | Actinin, alpha 3 | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| VIL2 | Villin 2 (ezrin) | |
| PRKCA | Protein kinase C, alpha | |
| RASSF5 | Ras association (RalGDS/AF-6) domain family 5 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| RHOH | Ras homolog gene family, member H | |
| GRLF1 | Glucocorticoid receptor DNA binding factor 1 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Lysine degradation | DLST | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | |
| EHMT2 | Euchromatic histone-lysine N-methyltransferase 2 | |
| EHMT1 | Euchromatic histone-lysine N-methyltransferase 1 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| Toll-like receptor signaling pathway | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| MYD88 | Myeloid differentiation primary response gene (88) | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| TRAF6 | CDNA FLJ46539 fis, clone THYMU3037836 | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| TLR2 | Toll-like receptor 2 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| TICAM2 | Toll-like receptor adaptor molecule 2 | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Long-term depression | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| GNAZ | Guanine nucleotide binding protein (G protein), alpha z polypeptide | |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | |
| PRKCA | Protein kinase C, alpha | |
| N-Glycan biosynthesis | DHDDS | Dehydrodolichyl diphosphate synthase |
| B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 | |
| MAN1B1 | Mannosidase, alpha, class 1B, member 1 | |
| ALG6 | Asparagine-linked glycosylation 6 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | |
| MAN1A2 | Mannosidase, alpha, class 1A, member 2 | |
| RPN1 | Ribophorin I | |
| DOLPP1 | Dolichyl pyrophosphate phosphatase 1 | |
| Neuroactive ligand-receptor interaction | GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) |
| GPR156 | G protein-coupled receptor 156 | |
| CRHR2 | Corticotropin releasing hormone receptor 2 | |
| UTS2R | Urotensin 2 receptor | |
| PRLH | Prolactin releasing hormone | |
| LTB4R | Leukotriene B4 receptor | |
| Gap junction | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| TUBB8 | tubulin, beta 8 | |
| PRKCA | Protein kinase C, alpha | |
| GnRH signaling pathway | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | |
| HBEGF | Heparin-binding EGF-like growth factor | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| PRKCA | Protein kinase C, alpha | |
| MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Tight junction | ACTN3 | Actinin, alpha 3 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| PRKCA | Protein kinase C, alpha | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| MRAS | Muscle RAS oncogene homolog | |
| PRKCH | Protein kinase C, eta | |
| Tryptophan metabolism | TRAF7 | TNF receptor-associated factor 7 |
| OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| UHRF1 | Ubiquitin-like, containing PHD and RING finger domains, 1 | |
| CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | |
| CAT | Catalase | |
| PRMT2 | Protein arginine methyltransferase 2 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| CNOT4 | CCR4-NOT transcription complex, subunit 4 | |
| Colorectal cancer | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| BAX | BCL2-associated X protein | |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| SMAD3 | SMAD family member 3 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Pancreatic cancer | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| RALBP1 | RalA binding protein 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| SMAD3 | SMAD family member 3 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| mTOR signaling pathway | RICTOR | rapamycin-insensitive companion of mTOR |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| EIF4B | Eukaryotic translation initiation factor 4B | |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| LYK5 | protein kinase LYK5 | |
| STK11 | Serine/threonine kinase 11 | |
| Melanoma | FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| HGF | Human mRNA for hepatocyte growth factor (HGF). | |
| FGF2 | Fibroblast growth factor 2 (basic) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Neurodegenerative Disorders | BAX | BCL2-associated X protein |
| PRNP | Prion protein (p27–30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | |
| PSEN1 | Presenilin 1 (Alzheimer disease 3) | |
| EP300 | E1A binding protein p300 | |
| GFAP | Glial fibrillary acidic protein | |
| Propanoate metabolism | MUT | Methylmalonyl Coenzyme A mutase |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| Selenoamino acid metabolism | SEPHS1 | Selenophosphate synthetase 1 |
| PRMT2 | Protein arginine methyltransferase 2 | |
| GGT1 | Gamma-glutamyltransferase 1 | |
| Thyroid cancer | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| CCDC6 | Coiled-coil domain containing 6 | |
| TPM3 | Tropomyosin 3 | |
| SNARE interactions in vesicular transport | STX6 | Syntaxin 6 |
| YKT6 | YKT6 v-SNARE homolog (S. cerevisiae) | |
| STX16 | Syntaxin 16 | |
| Starch and sucrose metabolism | HK2 | Hexokinase 2 |
| PYGB | Phosphorylase, glycogen; brain | |
| AGL | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (glycogen debranching enzyme, glycogen storage disease type III) | |
| GYS1 | Glycogen synthase 1 (muscle) | |
| UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | |
| Prion disease | PRNP | Prion protein (p27–30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| GFAP | Glial fibrillary acidic protein |
Supplemental Data
Acknowledgements
The authors would like to thank the qualified technical assistance of Francesca Cottino and Katiuscia Gizzi. This work was supported by the Associazione Italiana Ricerca Cancro (AIRC; S.D.), the CLL Global Research Foundation (S.D.), the Compagnia SanPaolo (S.D., M.M.G. and G.C.), the Progetti Ricerca Interesse Nationale (PRIN; F.M.), the University of Turin (S.D. and F.M.), the Fondazione “Guido Berlucchi” (F.M.) and the Regione Piemonte (S.D., T.V. and F.M.). The Fondazione Internazionale Ricerche Medicina Sperimentale (FIRMS) provided financial and administrative assistance. S.A. is a student of the Ph.D. program in Advanced Techniques in the Localization of Human Tumors (University of Turin).
Footnotes
Disclosure
The authors reported no potential conflict of interest.
Reference list
- 1.Malavasi F, Deaglio S, Funaro A, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev. 2008;88:841–886. doi: 10.1152/physrev.00035.2007. [DOI] [PubMed] [Google Scholar]
- 2.Deaglio S, Mallone R, Baj G, et al. CD38/CD31, a receptor/ligand system ruling adhesion and signaling in human leukocytes. Chem Immunol. 2000;75:99–120. [PubMed] [Google Scholar]
- 3.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 4.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 5.Matrai Z. CD38 as a prognostic marker in CLL. Hematology. 2005;10:39–46. doi: 10.1080/10245330400020470. [DOI] [PubMed] [Google Scholar]
- 6.Deaglio S, Vaisitti T, Aydin S, Ferrero E, Malavasi F. In-tandem insight from basic science combined with clinical research: CD38 as both marker and key component of the pathogenetic network underlying chronic lymphocytic leukemia. Blood. 2006;108:1135–1144. doi: 10.1182/blood-2006-01-013003. [DOI] [PubMed] [Google Scholar]
- 7.Jaksic O, Paro MM, Kardum Skelin I, Kusec R, Pejsa V, Jaksic B. CD38 on B-cell chronic lymphocytic leukemia cells has higher expression in lymph nodes than in peripheral blood or bone marrow. Blood. 2004;103:1968–1969. doi: 10.1182/blood-2003-11-3890. [DOI] [PubMed] [Google Scholar]
- 8.Damle RN, Temburni S, Calissano C, et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood. 2007;110:3352–3359. doi: 10.1182/blood-2007-04-083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepper C, Ward R, Lin TT, et al. Highly purified CD38(+) and CD38(−) sub-clones derived from the same chronic lymphocytic leukemia patient have distinct gene expression signatures despite their monoclonal origin. Leukemia. 2007 doi: 10.1038/sj.leu.2404587. [DOI] [PubMed] [Google Scholar]
- 10.Deaglio S, Capobianco A, Bergui L, et al. CD38 is a signaling molecule in B-cell chronic lymphocytic leukemia cells. Blood. 2003;102:2146–2155. doi: 10.1182/blood-2003-03-0989. [DOI] [PubMed] [Google Scholar]
- 11.Deaglio S, Vaisitti T, Billington R, et al. CD38/CD19: a lipid raft dependent signaling complex in human B cells. Blood. 2007;109:5390–5398. doi: 10.1182/blood-2006-12-061812. [DOI] [PubMed] [Google Scholar]
- 12.Deaglio S, Vaisitti T, Bergui L, et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood. 2005;105:3042–3050. doi: 10.1182/blood-2004-10-3873. [DOI] [PubMed] [Google Scholar]
- 13.Patten PE, Buggins AG, Richards J, et al. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood. 2008;111:5173–5181. doi: 10.1182/blood-2007-08-108605. [DOI] [PubMed] [Google Scholar]
- 14.Tonino SH, Spijker R, Luijks DM, van Oers MH, Kater AP. No convincing evidence for a role of CD31–CD38 interactions in the pathogenesis of chronic lymphocytic leukemia. Blood. 2008;112:840–843. doi: 10.1182/blood-2008-03-144576. [DOI] [PubMed] [Google Scholar]
- 15.Deaglio S, Vaisitti T, Aydin S, et al. CD38 and ZAP-70 are functionally linked and mark CLL cells with high migratory potential. Blood. 2007;110:4012–4021. doi: 10.1182/blood-2007-06-094029. [DOI] [PubMed] [Google Scholar]
- 16.Deaglio S, Morra M, Mallone R, et al. Human CD38 (ADP-ribosyl cyclase) is a counter-receptor of CD31, an Ig superfamily member. J Immunol. 1998;160:395–402. [PubMed] [Google Scholar]
- 17.Tomfohr J, Lu J, Kepler TB. Pathway level analysis of gene expression using singular value decomposition. BMC Bioinformatics. 2005;6:225. doi: 10.1186/1471-2105-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin TT, Hewamana S, Ward R, et al. Highly purified CD38 sub-populations show no evidence of preferential clonal evolution despite having increased proliferative activity when compared with CD38 sub-populations derived from the same chronic lymphocytic leukaemia patient. Br J Haematol. 2008;142:595–605. doi: 10.1111/j.1365-2141.2008.07236.x. [DOI] [PubMed] [Google Scholar]
- 19.Letilovic T, Vrhovac R, Verstovsek S, Jaksic B, Ferrajoli A. Role of angiogenesis in chronic lymphocytic leukemia. Cancer. 2006;107:925–934. doi: 10.1002/cncr.22086. [DOI] [PubMed] [Google Scholar]
- 20.Shanafelt TD, Kay NE. The clinical and biologic importance of neovascularization and angiogenic signaling pathways in chronic lymphocytic leukemia. Semin Oncol. 2006;33:174–185. doi: 10.1053/j.seminoncol.2006.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
| Accession # | Name | Description | L-CD31+ vs L-mock | p value |
|---|---|---|---|---|
| NM_199203 | Kua-UEV | Ubiquitin-conjugating enzyme E2 variant 1 | −12.01382 | 7.62476E-07 |
| NM_004218 | RAB11B | RAB11B, member RAS oncogene family | 12.37772 | 5.9108E-07 |
| NM_006544 | EXOC5 | Exocyst complex component 5 | 10.28434 | 2.8321E-06 |
| NM_212472 | PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | 16.0055 | 6.41336E-08 |
| NM_003934 | FUBP3 | Far upstream element (FUSE) binding protein 3 | 10.04849 | 3.43697E-06 |
| NM_153693 | HOXC6 | Homeobox C4 | 10.30985 | 2.77405E-06 |
| NM_022491 | SUDS3 | Suppressor of defective silencing 3 homolog (S. cerevisiae) | −11.04648 | 1.55368E-06 |
| NM_015330 | SPECC1L | SPECC1-like | −10.25039 | 2.91144E-06 |
| NM_001429 | EP300 | E1A binding protein p300 | −11.65812 | 9.84564E-07 |
| BX090412 | BX090412 | Transcribed locus | −14.57157 | 1.45057E-07 |
| NM_001261 | CDK9 | Cyclin-dependent kinase 9 (CDC2-related kinase) | 9.86276 | 4.01395E-06 |
| NM_002661 | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | −11.67385 | 9.73359E-07 |
| NM_000945 | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | 9.70376 | 4.59333E-06 |
| NM_177965 | C8orf37 | Chromosome 8 open reading frame 37 | 12.04077 | 7.48054E-07 |
| NM_024325 | ZNF343 | Zinc finger protein 343 | −9.47095 | 5.61486E-06 |
| NM_025136 | OPA3 | Optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) | 9.4887 | 5.52874E-06 |
| NM_201266 | NRP2 | Neuropilin 2 | 10.06726 | 3.38394E-06 |
| NM_019028 | ZDHHC13 | Zinc finger, DHHC-type containing 13 | 9.82075 | 4.15875E-06 |
| THC2304728 | THC2304728 | N1L4_HUMAN (Q99733) Nucleosome assembly protein 1-like 4 (Nucleosome assembly protein 2) (NAP2), partial (59%) [THC2304728] | 9.63357 | 4.87791E-06 |
| NM_000123 | ERCC5 | Excision repair cross-complementing rodent repair deficiency, complementation group 5 (xeroderma pigmentosum, complementation group G (Cockayne syndrome)) | −10.55662 | 2.27557E-06 |
| NM_007363 | NONO | Non-POU domain containing, octamer-binding | −10.45614 | 2.4655E-06 |
| NM_006595 | API5 | Apoptosis inhibitor 5 | 10.44438 | 2.48886E-06 |
| AA832084 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 9.99145 | 3.60381E-06 |
| NM_020640 | DCUN1D1 | DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) | 10.73637 | 1.97476E-06 |
| NM_016040 | TMED5 | Transmembrane emp24 protein transport domain containing 5 | 11.90305 | 8.25054E-07 |
| NM_182540 | DDX26B | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B | −10.73728 | 1.97335E-06 |
| NM_003893 | LDB1 | LIM domain binding 1 | 10.7935 | 1.88856E-06 |
| NM_016545 | IER5 | Immediate early response 5 | 10.53902 | 2.30764E-06 |
| NM_006805 | HNRPA0 | Heterogeneous nuclear ribonucleoprotein A0 | 9.18978 | 7.19589E-06 |
| NM_001229 | CASP9 | Caspase 9, apoptosis-related cysteine peptidase | −12.50316 | 5.42256E-07 |
| NM_004614 | TK2 | Thymidine kinase 2, mitochondrial | 8.99903 | 8.54562E-06 |
| NM_014876 | JOSD1 | Josephin domain containing 1 | 8.61553 | 0.00001 |
| NM_006459 | SPFH1 | SPFH domain family, member 1 | 9.04944 | 8.16368E-06 |
| THC2400714 | THC2400714 | Unknown | −9.97506 | 3.65339E-06 |
| NM_152734 | C6orf89 | Chromosome 6 open reading frame 89 | 12.84733 | 4.29726E-07 |
| NM_012426 | SF3B3 | Splicing factor 3b, subunit 3, 130kDa | 9.53653 | 5.30387E-06 |
| A_23_P207143 | A_23_P207143 | Unknown | 8.51395 | 0.00001 |
| NM_144726 | FLJ31951 | hypothetical protein FLJ31951 | 12.17957 | 6.7841E-07 |
| NM_005109 | OXSR1 | Oxidative-stress responsive 1 | 9.0603 | 8.08387E-06 |
| Y10152 | CRHR2 | Corticotropin releasing hormone receptor 2 | 10.38139 | 2.61814E-06 |
| NM_014554 | SENP1 | SUMO1/sentrin specific peptidase 1 | 8.65206 | 0.00001 |
| NM_022171 | TCTA | T-cell leukemia translocation altered gene | 8.52684 | 0.00001 |
| NM_006565 | CTCF | CCCTC-binding factor (zinc finger protein) | 8.69879 | 0.00001 |
| A_24_P290214 | A_24_P290214 | Unknown | 8.54654 | 0.00001 |
| NM_052987 | CDK10 | Cyclin-dependent kinase (CDC2-like) 10 | 9.19897 | 7.13708E-06 |
| NM_016506 | KBTBD4 | Mitochondrial carrier homolog 2 (C. elegans) | −9.39145 | 6.01911E-06 |
| NM_005026 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | 8.93923 | 9.02436E-06 |
| NM_020462 | ERGIC1 | Endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 | 12.75714 | 4.5648E-07 |
| NM_003005 | SELP | Selectin P (granule membrane protein 140kDa, antigen CD62) | −10.38446 | 2.61168E-06 |
| NM_007126 | VCP | Valosin-containing protein | 8.20306 | 0.00002 |
| NM_015349 | KIAA0240 | KIAA0240 | −8.93919 | 9.02463E-06 |
| NM_177966 | 2'-PDE | 2'-phosphodiesterase | 9.44127 | 5.76213E-06 |
| NM_015184 | PLCL2 | Phospholipase C-like 2 | −9.67039 | 4.72625E-06 |
| NM_013236 | ATXN10 | Ataxin 10 | 9.30208 | 6.51217E-06 |
| A_24_P581311 | A_24_P581311 | Unknown | 8.04345 | 0.00002 |
| A_24_P400616 | A_24_P400616 | Unknown | 7.90396 | 0.00002 |
| NM_174907 | PPP4R2 | Protein phosphatase 4, regulatory subunit 2 | 8.18701 | 0.00002 |
| NM_017897 | OXSM | 3-oxoacyl-ACP synthase, mitochondrial | −10.2951 | 2.80746E-06 |
| NM_054027 | ANKH | Ankylosis, progressive homolog (mouse) | 9.32545 | 6.37908E-06 |
| NM_005498 | AP1M2 | Adaptor-related protein complex 1, mu 2 subunit | 8.86146 | 9.69158E-06 |
| NM_005986 | SOX1 | SRY (sex determining region Y)-box 1 | 13.28268 | 3.22755E-07 |
| NM_007236 | CHP | calcium binding protein P22 | 14.02006 | 2.02561E-07 |
| BC034752 | C9orf62 | Chromosome 9 open reading frame 62 | 10.18258 | 3.07735E-06 |
| NM_022067 | C14orf133 | Chromosome 14 open reading frame 133 | −8.90563 | 9.30619E-06 |
| NM_002709 | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform | 8.0112 | 0.00002 |
| NM_016312 | WBP11 | WW domain binding protein 11 | −10.32371 | 2.74306E-06 |
| NM_006391 | IPO7 | Importin 7 | 9.46839 | 5.62738E-06 |
| NM_022107 | GPSM3 | G-protein signalling modulator 3 (AGS3-like, C. elegans) | 9.33884 | 6.30415E-06 |
| NM_001190 | BCAT2 | Branched chain aminotransferase 2, mitochondrial | 8.2365 | 0.00002 |
| NM_031892 | SH3KBP1 | SH3-domain kinase binding protein 1 | 8.18059 | 0.00002 |
| BC004815 | MGC5139 | hypothetical protein MGC5139 | −11.00369 | 1.60539E-06 |
| NM_001417 | EIF4B | Eukaryotic translation initiation factor 4B | 11.08825 | 1.50497E-06 |
| NM_017615 | NSMCE4A | Non-SMC element 4 homolog A (S. cerevisiae) | 8.20619 | 0.00002 |
| NM_153335 | LYK5 | protein kinase LYK5 | −14.13835 | 1.88372E-07 |
| AK074764 | SDHALP2 | Succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2 | 7.93021 | 0.00002 |
| NM_005298 | GPR25 | G protein-coupled receptor 25 | 9.62722 | 4.90459E-06 |
| NM_033547 | INTS4 | Integrator complex subunit 4 | 9.65114 | 4.8049E-06 |
| A_24_P780709 | A_24_P780709 | Unknown | 9.37541 | 6.10447E-06 |
| NM_005163 | AKT1 | V-akt murine thymoma viral oncogene homolog 1 | 8.33875 | 0.00002 |
| NM_012316 | KPNA6 | Karyopherin alpha 6 (importin alpha 7) | −11.38421 | 1.20449E-06 |
| A_24_P144556 | A_24_P144556 | Unknown | 8.55649 | 0.00001 |
| NM_001128 | AP1G1 | Adaptor-related protein complex 1, gamma 1 subunit | −11.10174 | 1.4896E-06 |
| NM_001039651 | C6orf26 | MutS homolog 5 (E. coli) | −9.24198 | 6.86871E-06 |
| NM_130791 | WWOX | WW domain containing oxidoreductase | 9.20432 | 7.10308E-06 |
| NM_004082 | DCTN1 | Dynactin 1 (p150, glued homolog, Drosophila) | 10.47834 | 2.42208E-06 |
| NM_031454 | SELO | selenoprotein O | 10.02399 | 3.50756E-06 |
| NM_006544 | EXOC5 | Exocyst complex component 5 | 8.34375 | 0.00002 |
| NM_020960 | GPR107 | G protein-coupled receptor 107 | −10.56817 | 2.25479E-06 |
| NM_203364 | GPIAP1 | GPI-anchored membrane protein 1 | 7.64062 | 0.00003 |
| NM_144576 | COQ10A | Coenzyme Q10 homolog A (S. cerevisiae) | −8.26821 | 0.00002 |
| NM_003089 | SNRP70 | Small nuclear ribonucleoprotein 70kDa polypeptide (RNP antigen) | 8.47566 | 0.00001 |
| NM_005839 | SRRM1 | Serine/arginine repetitive matrix 1 | −10.29528 | 2.80705E-06 |
| NM_016604 | JMJD1B | Jumonji domain containing 1B | −10.08354 | 3.3387E-06 |
| NM_178438 | LCE5A | Late cornified envelope 5A | 9.60463 | 5.00087E-06 |
| AK074764 | SDHALP2 | Succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2 | 8.66586 | 0.00001 |
| NM_002109 | HARS | Histidyl-tRNA synthetase | −9.09351 | 7.84517E-06 |
| NM_152219 | GJC1 | Gap junction protein, chi 1, 31.9kDa (connexin 31.9) | 10.85341 | 1.8026E-06 |
| NM_032322 | RNF135 | Ring finger protein 135 | −9.9372 | 3.77077E-06 |
| A_23_P369758 | A_23_P369758 | Unknown | 10.8002 | 1.87873E-06 |
| NM_001933 | DLST | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) | 7.44271 | 0.00004 |
| NM_015600 | ABHD12 | Abhydrolase domain containing 12 | 7.97343 | 0.00002 |
| NM_012215 | MGEA5 | Meningioma expressed antigen 5 (hyaluronidase) | −9.01908 | 8.39141E-06 |
| NM_004831 | CRSP7 | Cofactor required for Sp1 transcriptional activation, subunit 7, 70kDa | 8.94066 | 9.01256E-06 |
| NM_016143 | NSFL1C | NSFL1 (p97) cofactor (p47) | −7.96774 | 0.00002 |
| NM_139013 | MAPK14 | Mitogen-activated protein kinase 14 | 8.46192 | 0.00001 |
| CR621132 | NSUN7 | Full-length cDNA clone CS0DC015YK09 of Neuroblastoma Cot 25-normalized of Homo sapiens (human) | −7.48544 | 0.00004 |
| NM_001005404 | YPEL2 | Yippee-like 2 (Drosophila) | −8.59126 | 0.00001 |
| NM_153334 | SCARF2 | Scavenger receptor class F, member 2 | 7.52965 | 0.00004 |
| NM_018416 | FOXJ2 | ForkheadJ2 box | −8.811 | 0.00001 |
| NM_014607 | UBXD2 | UBX domain containing 2 | 8.99319 | 8.59109E-06 |
| NM_025263 | PRR3 | Major histocompatibility complex, class I, E | 8.13755 | 0.00002 |
| NM_052853 | ADCK2 | AarF domain containing kinase 2 | −9.20982 | 7.06829E-06 |
| THC2316116 | THC2316116 | Unknown | −9.80843 | 4.20233E-06 |
| AK023797 | KIAA0672 | KIAA0672 gene product | −12.98441 | 3.92319E-07 |
| NM_025207 | FLAD1 | FAD1 flavin adenine dinucleotide synthetase homolog (S. cerevisiae) | 8.22176 | 0.00002 |
| BQ548663 | ALOX5 | Arachidonate 5-lipoxygenase | −8.8633 | 9.67523E-06 |
| A_24_P101493 | A_24_P101493 | Unknown | 8.46769 | 0.00001 |
| NM_018144 | SEC61A2 | Sec61 alpha 2 subunit (S. cerevisiae) | 8.43895 | 0.00001 |
| NM_002593 | PCOLCE | Procollagen C-endopeptidase enhancer | 7.63508 | 0.00003 |
| NM_024545 | SAP130 | Sin3A-associated protein, 130kDa | −8.3626 | 0.00002 |
| NM_005744 | ARIH1 | Ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 (Drosophila) | 14.02495 | 2.01951E-07 |
| CR627148 | LOC728649 | hypothetical protein LOC728649 | −12.98113 | 3.93171E-07 |
| NM_173833 | SCARA5 | Scavenger receptor class A, member 5 (putative) | 10.19322 | 3.05064E-06 |
| NM_014337 | PPIL2 | Peptidylprolyl isomerase (cyclophilin)-like 2 | 10.82689 | 1.84012E-06 |
| NM_003791 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | −13.00009 | 3.88276E-07 |
| NM_182543 | NSUN6 | NOL1/NOP2/Sun domain family, member 6 | −7.91983 | 0.00002 |
| NM_197957 | MAX | MYC associated factor X | 8.86102 | 9.69551E-06 |
| NM_014669 | NUP93 | Nucleoporin 93kDa | 7.4057 | 0.00004 |
| NM_006522 | WNT6 | Wingless-type MMTV integration site family, member 6 | 8.10676 | 0.00002 |
| NM_170662 | CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | −7.59917 | 0.00003 |
| NM_014800 | ELMO1 | Engulfment and cell motility 1 | −8.86216 | 9.68535E-06 |
| NM_014932 | NLGN1 | Neuroligin 1 | 11.85202 | 8.55769E-07 |
| NM_000401 | EXT2 | Exostoses (multiple) 2 | −11.00481 | 0.000001604 |
| NM_000439 | PCSK1 | Proprotein convertase subtilisin/kexin type 1 | 10.68261 | 2.05985E-06 |
| NM_032228 | MLSTD2 | Male sterility domain containing 2 | 7.87725 | 0.00003 |
| NM_015575 | TNRC15 | Trinucleotide repeat containing 15 | 10.81159 | 1.86214E-06 |
| NM_032383 | HPS3 | Hermansky-Pudlak syndrome 3 | −7.7146 | 0.00003 |
| NM_014303 | PES1 | Pescadillo homolog 1, containing BRCT domain (zebrafish) | 11.27587 | 1.30602E-06 |
| NM_138811 | C7orf31 | Chromosome 7 open reading frame 31 | −7.84466 | 0.00003 |
| THC2306788 | THC2306788 | ALU1_HUMAN (P39188) Alu subfamily J sequence contamination warning entry, partial (10%) [THC2306788] | 8.36187 | 0.00002 |
| NM_001077207 | SEC31A | SEC31 homolog A (S. cerevisiae) | −10.01301 | 3.53971E-06 |
| NM_021030 | ZNF14 | Zinc finger protein 14 | −9.40692 | 5.93797E-06 |
| NM_001002259 | C1QDC1 | C1q domain containing 1 | −7.34532 | 0.00004 |
| NM_005370 | RAB8A | RAB8A, member RAS oncogene family | 8.28127 | 0.00002 |
| AK131269 | MBOAT1 | Membrane bound O-acyltransferase domain containing 1 | −9.47467 | 5.5967E-06 |
| NM_005827 | SLC35B1 | Solute carrier family 35, member B1 | 8.26348 | 0.00002 |
| A_24_P332754 | A_24_P332754 | Unknown | 7.25643 | 0.00005 |
| NM_031992 | EIF4H | Eukaryotic translation initiation factor 4H | 8.331 | 0.00002 |
| NM_001011658 | TRAPPC2 | Trafficking protein particle complex 2 | −9.81802 | 4.16839E-06 |
| NM_198563 | TMEM110 | Transmembrane protein 110 | 7.20495 | 0.00005 |
| NM_024419 | PGS1 | Phosphatidylglycerophosphate synthase 1 | 11.5625 | 1.05585E-06 |
| THC2284389 | THC2284389 | C40201 artifact-warning sequence (translated ALU class C) - human {Homo sapiens;} , partial (6%) [THC2284389] | −8.27074 | 0.00002 |
| NR_002144 | LOC407835 | Homo sapiens mitogen-activated protein kinase kinase 2 pseudogene (LOC407835) on chromosome 7. | 7.15658 | 0.00005 |
| NM_012179 | FBXO7 | F-box protein 7 | −8.73795 | 0.00001 |
| NM_016076 | C1orf121 | Chromosome 1 open reading frame 121 | 11.06158 | 1.53587E-06 |
| A_24_P878366 | A_24_P878366 | Unknown | 13.11769 | 3.59374E-07 |
| NM_032336 | GINS4 | Golgi autoantigen, golgin subfamily a, 7 | 7.52996 | 0.00004 |
| NM_004530 | MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | 12.50249 | 5.42507E-07 |
| A_32_P83520 | A_32_P83520 | Unknown | 8.84018 | 9.8835E-06 |
| NM_198976 | TH1L | TH1-like (Drosophila) | 6.989 | 0.00006 |
| NM_017668 | NDE1 | NudE nuclear distribution gene E homolog 1 (A. nidulans) | 7.69641 | 0.00003 |
| NM_198963 | DHX57 | DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 | −10.92259 | 1.70868E-06 |
| NM_006101 | KNTC2 | Kinetochore associated 2 | 7.11802 | 0.00006 |
| NM_002967 | SAFB | Scaffold attachment factor B | 7.17136 | 0.00005 |
| NM_017745 | BCOR | BCL6 co-repressor | 8.15186 | 0.00002 |
| NM_006711 | RNPS1 | RNA binding protein S1, serine-rich domain | 7.2874 | 0.00005 |
| NM_014268 | MAPRE2 | Microtubule-associated protein, RP/EB family, member 2 | 7.51812 | 0.00004 |
| BC057844 | BC057844 | CDNA clone MGC:71846 IMAGE:5313492 | 10.04434 | 3.44883E-06 |
| NM_002525 | NRD1 | Nardilysin (N-arginine dibasic convertase) | −9.26143 | 0.000006751 |
| NM_014089 | NUPL1 | Nucleoporin like 1 | 7.10305 | 0.00006 |
| NM_007131 | ZNF75 | Zinc finger protein 75 (D8C6) | −8.06537 | 0.00002 |
| NM_014877 | HELZ | Helicase with zinc finger | −10.7689 | 1.92516E-06 |
| NM_012478 | WBP2 | WW domain binding protein 2 | 8.60644 | 0.00001 |
| NM_001620 | AHNAK | AHNAK nucleoprotein (desmoyokin) | 7.85031 | 0.00003 |
| NM_018195 | C11orf57 | Chromosome 11 open reading frame 57 | −8.5476 | 0.00001 |
| NM_024541 | C10orf76 | Chromosome 10 open reading frame 76 | 9.53683 | 5.30248E-06 |
| NM_016290 | UIMC1 | Ubiquitin interaction motif containing 1 | −7.07217 | 0.00006 |
| NM_015005 | KIAA0284 | KIAA0284 | 9.65504 | 4.78885E-06 |
| BC002431 | B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 | 10.07832 | 3.35313E-06 |
| NM_014760 | TATDN2 | TatD DNase domain containing 2 | −7.94034 | 0.00002 |
| NM_024881 | SLC35E1 | Solute carrier family 35, member E1 | 8.64898 | 0.00001 |
| NM_003111 | SP3 | Sp3 transcription factor | 10.18982 | 3.05914E-06 |
| NM_019024 | KIAA1414 | KIAA1414 protein | −10.63453 | 2.13938E-06 |
| NM_138707 | BCL7B | B-cell CLL/lymphoma 7B | 7.05423 | 0.00006 |
| NM_016824 | ADD3 | Adducin 3 (gamma) | −7.17037 | 0.00005 |
| NM_004309 | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 8.3467 | 0.00002 |
| THC2314371 | THC2314371 | ALU8_HUMAN (P39195) Alu subfamily SX sequence contamination warning entry, partial (10%) [THC2314371] | −8.75011 | 0.00001 |
| NM_016079 | VPS24 | Vacuolar protein sorting 24 homolog (S. cerevisiae) | 10.94974 | 1.67331E-06 |
| NM_013282 | UHRF1 | Ubiquitin-like, containing PHD and RING finger domains, 1 | 7.78277 | 0.00003 |
| NM_032775 | KLHL22 | Kelch-like 22 (Drosophila) | 7.71952 | 0.00003 |
| NM_020648 | TWSG1 | Twisted gastrulation homolog 1 (Drosophila) | 7.84782 | 0.00003 |
| THC2444634 | THC2444634 | AF195969 rho GTPase activating protein 8 isoform 2 {Homo sapiens;} , partial (8%) [THC2444634] | 8.87065 | 9.60999E-06 |
| AK056150 | LOC149478 | hypothetical protein LOC149478 | 7.34421 | 0.00004 |
| A_24_P101181 | A_24_P101181 | Unknown | 7.72485 | 0.00003 |
| NM_014657 | KIAA0406 | KIAA0406 | 13.42386 | 2.94683E-07 |
| NM_004808 | NMT2 | N-myristoyltransferase 2 | −6.83368 | 0.00008 |
| NM_004249 | RAB28 | RAB28, member RAS oncogene family | 7.06441 | 0.00006 |
| NM_018121 | C10orf6 | Chromosome 10 open reading frame 6 | −9.42738 | 5.83253E-06 |
| NM_012395 | PFTK1 | PFTAIRE protein kinase 1 | −7.45833 | 0.00004 |
| NM_080596 | HIST1H2AH | Histone cluster 1, H2ai | 7.1406 | 0.00005 |
| NM_003806 | HRK | Harakiri, BCL2 interacting protein (contains only BH3 domain) | 6.88927 | 0.00007 |
| NM_032438 | L3MBTL3 | L(3)mbt-like 3 (Drosophila) | −7.00205 | 0.00006 |
| NM_006290 | TNFAIP3 | Tumor necrosis factor, alpha-induced protein 3 | 8.65812 | 0.00001 |
| NM_015061 | JMJD2C | Jumonji domain containing 2C | −10.21014 | 3.00869E-06 |
| NM_153813 | ZFPM1 | Zinc finger protein, multitype 1 | 8.03175 | 0.00002 |
| BC040982 | PIK3C3 | Phosphoinositide-3-kinase, class 3 | −8.2424 | 0.00002 |
| NM_001012651 | NKTR | Natural killer-tumor recognition sequence | −8.32262 | 0.00002 |
| BC028192 | LOC161635 | similar to casein kinase I alpha | 6.79522 | 0.00008 |
| AK002023 | FLJ25715 | CDNA FLJ11161 fis, clone PLACE1007021 | −9.2345 | 6.9146E-06 |
| NM_001684 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | 9.10183 | 7.78657E-06 |
| NM_004171 | SLC1A2 | Homo sapiens solute carrier family 1 (glial high affinity glutamate transporter), member 2 (SLC1A2), mRNA. | 7.93271 | 0.00002 |
| NM_006766 | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 | −8.77256 | 0.00001 |
| NM_018622 | PARL | Presenilin associated, rhomboid-like | 8.68819 | 0.00001 |
| NM_006224 | PITPNA | Phosphatidylinositol transfer protein, alpha | 6.70394 | 0.00009 |
| NM_002873 | RAD17 | RAD17 homolog (S. pombe) | −8.20583 | 0.00002 |
| THC2280638 | THC2280638 | RL2A_HUMAN (P46776) 60S ribosomal protein L27a, partial (24%) [THC2280638] | −7.53358 | 0.00004 |
| BX427435 | BX427435 | Transcribed locus | 7.05574 | 0.00006 |
| NM_001017395 | TMCC1 | Transmembrane and coiled-coil domain family 1 | 7.44023 | 0.00004 |
| A_24_P502660 | A_24_P502660 | Unknown | 8.49614 | 0.00001 |
| NM_181838 | UBE2D2 | Ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | −14.33002 | 1.67656E-07 |
| NM_002468 | MYD88 | Myeloid differentiation primary response gene (88) | 7.41285 | 0.00004 |
| NM_022487 | DCLRE1C | DNA cross-link repair 1C (PSO2 homolog, S. cerevisiae) | −8.35217 | 0.00002 |
| NM_032364 | DNAJC14 | DnaJ (Hsp40) homolog, subfamily C, member 14 | −11.12091 | 1.46806E-06 |
| NM_002737 | PRKCA | Protein kinase C, alpha | 11.80087 | 8.87837E-07 |
| BQ317309 | GCSH | Glycine cleavage system protein H (aminomethyl carrier) | 6.8177 | 0.00008 |
| NM_014360 | NKX2-8 | NK2 transcription factor related, locus 8 (Drosophila) | 7.31477 | 0.00004 |
| AJ002425 | HSAJ2425 | p65 protein | 8.11665 | 0.00002 |
| NM_006763 | BTG2 | BTG family, member 2 | −7.82584 | 0.00003 |
| A_24_P118721 | A_24_P118721 | Unknown | 9.15287 | 7.43754E-06 |
| NM_138763 | BAX | BCL2-associated X protein | 9.65811 | 4.77628E-06 |
| NM_005819 | STX6 | Syntaxin 6 | 7.83975 | 0.00003 |
| NM_017730 | QRICH1 | Glutamine-rich 1 | −10.66821 | 2.08332E-06 |
| NM_006827 | TMED10 | Transmembrane emp24-like trafficking protein 10 (yeast) | 7.98498 | 0.00002 |
| NM_012118 | CCRN4L | CCR4 carbon catabolite repression 4-like (S. cerevisiae) | 7.30251 | 0.00005 |
| NM_033532 | CDC2L2 | Cell division cycle 2-like 2 (PITSLRE proteins) | 6.8601 | 0.00007 |
| NM_177966 | 2'-PDE | 2'-phosphodiesterase | 8.47363 | 0.00001 |
| NM_002103 | GYS1 | Glycogen synthase 1 (muscle) | −8.92352 | 9.15489E-06 |
| NM_182547 | TMED4 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 56 | 7.46187 | 0.00004 |
| NM_021149 | COTL1 | Coactosin-like 1 (Dictyostelium) | −7.01049 | 0.00006 |
| NM_002145 | HOXB2 | Homeobox B2 | 8.19278 | 0.00002 |
| NM_001002909 | GPATCH8 | G patch domain containing 8 | −9.04408 | 8.20341E-06 |
| NM_021649 | TICAM2 | Toll-like receptor adaptor molecule 2 | 6.59711 | 0.0001 |
| NM_022558 | GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) | 8.05993 | 0.00002 |
| NM_194247 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 7.43335 | 0.00004 |
| NM_014153 | ZC3H7A | Zinc finger CCCH-type containing 7A | 7.05942 | 0.00006 |
| NM_153045 | C9orf91 | Chromosome 9 open reading frame 91 | −7.73743 | 0.00003 |
| ENST00000312847 | ENST00000312847 | Unknown | 6.86646 | 0.00007 |
| NM_018013 | FLJ10159 | hypothetical protein FLJ10159 | −7.60685 | 0.00003 |
| X16323 | HGF | Human mRNA for hepatocyte growth factor (HGF). | 9.5554 | 5.21792E-06 |
| A_32_P331700 | A_32_P331700 | Unknown | 6.7671 | 0.00008 |
| AK095600 | ZNF709 | Homo sapiens cDNA FLJ38281 fis, clone FCBBF3005729, moderately similar to Homo sapiens GIOT-4 mRNA for gonadotropin inducible transcription repressor-4. | −7.53387 | 0.00004 |
| NM_001031628 | LOC57228 | small trans-membrane and glycosylated protein | 10.54713 | 2.29279E-06 |
| NM_015001 | SPEN | Spen homolog, transcriptional regulator (Drosophila) | −10.27584 | 2.85175E-06 |
| NM_002229 | JUNB | Jun B proto-oncogene | 12.09317 | 7.20878E-07 |
| BX647217 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 8.416 | 0.00001 |
| NM_014384 | ACAD8 | Acyl-Coenzyme A dehydrogenase family, member 8 | −7.18087 | 0.00005 |
| NM_138575 | PGAM5 | Phosphoglycerate mutase family member 5 | 7.49043 | 0.00004 |
| NM_003884 | PCAF | P300/CBP-associated factor | −8.25455 | 0.00002 |
| AK122976 | ZNF275 | Zinc finger protein 275 | −8.96519 | 8.81293E-06 |
| NM_017911 | FAM118A | Family with sequence similarity 118, member A | 11.18404 | 1.39951E-06 |
| NM_003791 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | −11.73778 | 9.29231E-07 |
| AK096643 | FLJ22184 | hypothetical protein FLJ22184 | 10.23909 | 2.9384E-06 |
| NM_017651 | AHI1 | Abelson helper integration site 1 | −8.65797 | 0.00001 |
| NM_002541 | OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | 7.30081 | 0.00005 |
| NM_005561 | LAMP1 | Lysosomal-associated membrane protein 1 | 7.79006 | 0.00003 |
| NM_015476 | C18orf10 | Chromosome 18 open reading frame 10 | 7.4762 | 0.00004 |
| NM_006380 | APPBP2 | Amyloid beta precursor protein (cytoplasmic tail) binding protein 2 | 8.89445 | 9.40209E-06 |
| BC007879 | FAM120A | Family with sequence similarity 120A | 7.06812 | 0.00006 |
| NM_018221 | MOBK1B | MOB1, Mps One Binder kinase activator-like 1B (yeast) | 7.46482 | 0.00004 |
| NM_147148 | GSTM4 | Glutathione S-transferase M4 | 8.61646 | 0.00001 |
| AA451906 | BIN1 | Bridging integrator 1 | 7.4277 | 0.00004 |
| NM_032383 | HPS3 | Hermansky-Pudlak syndrome 3 | −6.97134 | 0.00007 |
| A_24_P808838 | A_24_P808838 | Unknown | 6.73859 | 0.00008 |
| NM_001042537 | SLC9A6 | Solute carrier family 9 (sodium/hydrogen exchanger), member 6 | −7.75826 | 0.00003 |
| NM_031434 | TMUB1 | Transmembrane and ubiquitin-like domain containing 1 | 12.22266 | 6.5827E-07 |
| AB020648 | KIAA0841 | KIAA0841 | −8.89967 | 9.35718E-06 |
| NM_015120 | ALMS1 | Alstrom syndrome 1 | −8.99658 | 8.5646E-06 |
| NM_015401 | HDAC7A | Histone deacetylase 7A | −6.57001 | 0.0001 |
| NM_000189 | HK2 | Hexokinase 2 | 6.72803 | 0.00009 |
| NM_021149 | COTL1 | Coactosin-like 1 (Dictyostelium) | −6.86527 | 0.00007 |
| CR603450 | TRIM52 | Tripartite motif-containing 52 | −7.76938 | 0.00003 |
| NM_175863 | ARID1B | AT rich interactive domain 1B (SWI1-like) | 12.1051 | 7.14841E-07 |
| THC2295157 | THC2295157 | Unknown | −7.06649 | 0.00006 |
| AJ008005 | PSEN1 | Presenilin 1 (Alzheimer disease 3) | 9.19485 | 7.16338E-06 |
| NM_007326 | CYB5R3 | Cytochrome b5 reductase 3 | 6.89825 | 0.00007 |
| AK096104 | LOC727820 | hypothetical protein LOC727820 | −9.71542 | 4.54783E-06 |
| NM_000319 | PEX5 | Peroxisomal biogenesis factor 5 | 10.03095 | 3.48736E-06 |
| BF895757 | TRIM28 | Tripartite motif-containing 28 | 6.34251 | 0.00013 |
| BM509577 | THC2281731 | Transcribed locus, strongly similar to XP_001152474.1 hypothetical protein [Pan troglodytes] | 10.69767 | 2.03562E-06 |
| NM_021188 | ZNF410 | Zinc finger protein 410 | −12.20709 | 6.65471E-07 |
| NM_001681 | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 6.35897 | 0.00013 |
| THC2377679 | THC2377679 | Unknown | −10.62465 | 2.15615E-06 |
| NM_198494 | ZNF642 | Zinc finger protein 642 | −7.31971 | 0.00004 |
| NM_017706 | WDR55 | WD repeat domain 55 | 7.59863 | 0.00003 |
| NM_024757 | EHMT1 | Euchromatic histone-lysine N-methyltransferase 1 | 8.32535 | 0.00002 |
| A_24_P340966 | A_24_P340966 | Unknown | 7.90415 | 0.00002 |
| ENST00000380831 | ENST00000380831 | LPXK_RHIME (Q92RP7) Tetraacyldisaccharide 4'-kinase (Lipid A 4'-kinase) , partial (5%) [THC2374771] | −6.42385 | 0.00012 |
| NM_013446 | MKRN1 | Makorin, ring finger protein, 1 | −6.90506 | 0.00007 |
| XM_928285 | ENST00000375923 | PREDICTED: Homo sapiens similar to protein expressed in prostate, ovary, testis, and placenta 15, transcript variant 1 (LOC653239), mRNA [XM_928285] | 6.42221 | 0.00012 |
| BX648945 | EYA3 | Eyes absent homolog 3 (Drosophila) | −8.35174 | 0.00002 |
| NM_152367 | C1orf161 | Chromosome 1 open reading frame 161 | 8.07991 | 0.00002 |
| NM_015361 | R3HDM1 | R3H domain containing 1 | −8.80786 | 0.00001 |
| NM_152896 | UHRF2 | Ubiquitin-like, containing PHD and RING finger domains, 2 | −6.81973 | 0.00008 |
| NM_001001396 | ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | −6.32724 | 0.00014 |
| NM_020847 | TNRC6A | Homo sapiens trinucleotide repeat containing 6A (TNRC6A), mRNA. | 7.32895 | 0.00004 |
| AK074590 | SFRS16 | Splicing factor, arginine/serine-rich 16 | 8.55204 | 0.00001 |
| NM_001018052 | POLR3H | Polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) | 6.76245 | 0.00008 |
| NM_012256 | ZNF212 | Zinc finger protein 398 | −7.93151 | 0.00002 |
| NM_004850 | ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | −8.12669 | 0.00002 |
| NM_053041 | COMMD7 | COMM domain containing 7 | 6.43078 | 0.00012 |
| NM_015382 | HECTD1 | HECT domain containing 1 | −9.02879 | 8.3178E-06 |
| NM_033116 | NEK9 | NIMA (never in mitosis gene a)- related kinase 9 | 6.35421 | 0.00013 |
| NM_002886 | RAP2B | RAP2B, member of RAS oncogene family | 9.69267 | 4.63706E-06 |
| NM_014846 | KIAA0196 | KIAA0196 | −6.66666 | 0.00009 |
| NM_022658 | HOXC8 | Homeobox C4 | 6.23915 | 0.00015 |
| NM_004788 | UBE4A | Ubiquitination factor E4A (UFD2 homolog, yeast) | −7.44899 | 0.00004 |
| NM_022776 | OSBPL11 | Oxysterol binding protein-like 11 | −7.44389 | 0.00004 |
| BC018035 | IMP5 | intramembrane protease 5 | 10.04526 | 3.4462E-06 |
| NM_001007559 | SS18 | Synovial sarcoma translocation, chromosome 18 | 13.94595 | 2.12052E-07 |
| AK000675 | 1-Mar | Membrane-associated ring finger (C3HC4) 1 | −6.55294 | 0.0001 |
| NM_004710 | SYNGR2 | Synaptogyrin 2 | 6.65626 | 0.00009 |
| NM_014749 | KIAA0586 | KIAA0586 | −6.46727 | 0.00012 |
| NM_014925 | R3HDM2 | Homo sapiens R3H domain containing 2 (R3HDM2), mRNA. | −8.92219 | 9.16608E-06 |
| NM_030567 | PRR7 | Proline rich 7 (synaptic) | 7.36757 | 0.00004 |
| NM_173514 | FLJ90709 | hypothetical protein FLJ90709 | −7.07812 | 0.00006 |
| NM_144653 | BTBD14A | BTB (POZ) domain containing 14A | 9.27991 | 6.64126E-06 |
| BC063441 | BC063441 | Homo sapiens cDNA clone IMAGE:5527329, partial cds. | 7.14285 | 0.00005 |
| NM_000379 | XDH | Xanthine dehydrogenase | 6.77939 | 0.00008 |
| BX648958 | BX648958 | Shinc-4 mRNA, partial sequence | −8.74774 | 0.00001 |
| NM_007275 | TUSC2 | Tumor suppressor candidate 2 | 8.78322 | 0.00001 |
| NM_005487 | HMG2L1 | High-mobility group protein 2-like 1 | −7.02563 | 0.00006 |
| NM_020245 | TULP4 | Tubby like protein 4 | −8.25514 | 0.00002 |
| NM_032788 | ZNF514 | Mitochondrial ribosomal protein S5 | −8.39584 | 0.00002 |
| ENST00000331736 | ENST00000331736 | Q8NH99 (Q8NH99) Seven transmembrane helix receptor, partial (77%) [THC2437350] | 8.07755 | 0.00002 |
| NM_182314 | COVA1 | Cytosolic ovarian carcinoma antigen 1 | −10.02086 | 3.51669E-06 |
| NM_015330 | SPECC1L | SPECC1-like | −9.41199 | 5.91167E-06 |
| NM_014649 | SAFB2 | Small nuclear ribonucleoprotein polypeptide E | 7.43943 | 0.00004 |
| NM_007253 | CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | 8.22057 | 0.00002 |
| AK130071 | ZBTB44 | Zinc finger and BTB domain containing 44 | −6.57362 | 0.0001 |
| NM_145006 | SUSD3 | Sushi domain containing 3 | −7.18067 | 0.00005 |
| NM_001394 | DUSP4 | Dual specificity phosphatase 4 | 7.89185 | 0.00002 |
| NM_007175 | SPFH2 | SPFH domain family, member 2 | −8.77846 | 0.00001 |
| NM_080611 | DUSP15 | Dual specificity phosphatase 15 | 6.86459 | 0.00007 |
| NM_033296 | MRFAP1 | Mof4 family associated protein 1 | 7.20985 | 0.00005 |
| NM_006037 | HDAC4 | Histone deacetylase 4 | 6.91398 | 0.00007 |
| NM_001033858 | DCLRE1C | DNA cross-link repair 1C (PSO2 homolog, S. cerevisiae) | −6.19183 | 0.00016 |
| NM_012181 | FKBP8 | FK506 binding protein 8, 38kDa | 10.34449 | 2.69733E-06 |
| AK055921 | MST101 | MSTP101 | −10.08009 | 3.34822E-06 |
| AK057884 | COG3 | Component of oligomeric golgi complex 3 | 6.83815 | 0.00008 |
| NM_015448 | RP11-529I10.4 | deleted in a mouse model of primary ciliary dyskinesia | 7.35999 | 0.00004 |
| NM_205861 | DHDDS | Dehydrodolichyl diphosphate synthase | 9.46651 | 5.63664E-06 |
| NM_003255 | TIMP2 | TIMP metallopeptidase inhibitor 2 | 9.53344 | 5.31808E-06 |
| AJ420589 | FAM120B | Family with sequence similarity 120B | −8.32685 | 0.00002 |
| NM_017851 | PARP16 | Poly (ADP-ribose) polymerase family, member 16 | −8.30035 | 0.00002 |
| NM_001002021 | PFKL | Phosphofructokinase, liver | 6.55024 | 0.00011 |
| NM_004491 | GRLF1 | Glucocorticoid receptor DNA binding factor 1 | 6.56125 | 0.0001 |
| NM_022457 | RFWD2 | Ring finger and WD repeat domain 2 | −6.49304 | 0.00011 |
| NM_006555 | YKT6 | YKT6 v-SNARE homolog (S. cerevisiae) | 6.62617 | 0.0001 |
| NM_018452 | C6orf35 | Chromosome 6 open reading frame 35 | 6.82308 | 0.00008 |
| NM_006464 | TGOLN2 | Trans-golgi network protein 2 | −7.19236 | 0.00005 |
| NM_002860 | ALDH18A1 | Aldehyde dehydrogenase 18 family, member A1 | −10.20157 | 3.02985E-06 |
| NM_145232 | ATPBD3 | ATP binding domain 3 | 6.34185 | 0.00013 |
| NM_001233 | CAV2 | Caveolin 2 | 10.02277 | 3.51112E-06 |
| A_24_P322908 | A_24_P322908 | Unknown | −7.38585 | 0.00004 |
| NM_139246 | C9orf97 | Chromosome 9 open reading frame 97 | 7.67513 | 0.00003 |
| NM_025154 | UNC84A | Unc-84 homolog A (C. elegans) | −7.71957 | 0.00003 |
| NM_015655 | ZNF337 | Zinc finger protein 337 | −6.34247 | 0.00013 |
| NM_033083 | EAF1 | ELL associated factor 1 | 6.56433 | 0.0001 |
| NM_021096 | CACNA1I | Calcium channel, voltage-dependent, alpha 1I subunit | 8.34352 | 0.00002 |
| NM_014423 | AFF4 | AF4/FMR2 family, member 4 | −6.759 | 0.00008 |
| BC010538 | C18orf18 | Hypothetical protein LOC339290 | −7.97833 | 0.00002 |
| NM_002541 | OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | 8.14865 | 0.00002 |
| NM_174887 | IFT20 | Intraflagellar transport 20 homolog (Chlamydomonas) | 9.26877 | 6.70721E-06 |
| XM_936535 | NDOR1 | PREDICTED: Homo sapiens hypothetical protein LOC648245, transcript variant 1 (LOC648245), mRNA [XM_936535] | 7.74239 | 0.00003 |
| NM_019896 | POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | 8.36258 | 0.00002 |
| NM_016154 | RAB4B | RAB4B, member RAS oncogene family | 6.27108 | 0.00015 |
| NM_033129 | SCRT2 | Scratch homolog 2, zinc finger protein (Drosophila) | 6.66725 | 0.00009 |
| NM_001012614 | CTBP1 | C-terminal binding protein 1 | 7.38024 | 0.00004 |
| BC045778 | XYLT1 | Xylosyltransferase I | −6.16707 | 0.00017 |
| NM_012208 | HARSL | Histidyl-tRNA synthetase-like | −8.54862 | 0.00001 |
| NM_014411 | NAG8 | nasopharyngeal carcinoma associated gene protein-8 | −6.87141 | 0.00007 |
| NM_022893 | BCL11A | B-cell CLL/lymphoma 11A (zinc finger protein) | −7.86838 | 0.00003 |
| NM_000019 | ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | 6.17359 | 0.00016 |
| NM_014802 | KIAA0528 | KIAA0528 | −7.27048 | 0.00005 |
| NM_014912 | CPEB3 | Cytoplasmic polyadenylation element binding protein 3 | −9.89204 | 3.91629E-06 |
| NM_152285 | ARRDC1 | Arrestin domain containing 1 | 6.40343 | 0.00012 |
| NM_020438 | DOLPP1 | Dolichyl pyrophosphate phosphatase 1 | 6.43503 | 0.00012 |
| AK090478 | TMC8 | Transmembrane channel-like 8 | −7.59888 | 0.00003 |
| NM_014329 | EDC4 | Enhancer of mRNA decapping 4 | 6.31661 | 0.00014 |
| NM_015229 | KIAA0664 | KIAA0664 | 6.8895 | 0.00007 |
| NM_018360 | CXorf15 | Chromosome X open reading frame 15 | 9.35232 | 6.2297E-06 |
| AK026760 | C20orf119 | Chromosome 20 open reading frame 119 | −7.30047 | 0.00005 |
| NM_048368 | CTDP1 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 | 6.38575 | 0.00013 |
| NM_174891 | C14orf79 | Chromosome 14 open reading frame 79 | 8.13124 | 0.00002 |
| AK056484 | LOC441204 | hypothetical locus LOC441204 | 8.75709 | 0.00001 |
| NM_020711 | KIAA1189 | KIAA1189 | −7.43592 | 0.00004 |
| NM_024891 | FLJ11783 | hypothetical protein FLJ11783 | −14.18157 | 1.83464E-07 |
| S72422 | DLSTP | E2k=alpha-ketoglutarate dehydrogenase complex dihydrolipoyl succinyltransferase [human, fetal brain, mRNA, 2987 nt]. | 6.15369 | 0.00017 |
| AB018323 | JMJD2C | Jumonji domain containing 2C | −7.69576 | 0.00003 |
| NM_006696 | BRD8 | Bromodomain containing 8 | −8.73726 | 0.00001 |
| NM_177439 | FTSJ1 | FtsJ homolog 1 (E. coli) | 6.55266 | 0.0001 |
| THC2392085 | THC2392085 | Unknown | −7.57601 | 0.00003 |
| NM_005645 | TAF13 | TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa | 6.49776 | 0.00011 |
| NM_182612 | PDDC1 | Parkinson disease 7 domain containing 1 | 10.24294 | 2.92916E-06 |
| NM_004284 | CHD1L | Chromodomain helicase DNA binding protein 1-like | −8.18555 | 0.00002 |
| NM_032354 | TMEM107 | Transmembrane protein 107 | −9.96453 | 3.68563E-06 |
| NM_002182 | IL1RAP | Interleukin 1 receptor accessory protein | −6.23672 | 0.00015 |
| NM_001007071 | RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | 6.0887 | 0.00018 |
| NM_032349 | NUDT16L1 | Nudix (nucleoside diphosphate linked moiety X)-type motif 16-like 1 | 6.94114 | 0.00007 |
| NM_017951 | SMPD4 | Sphingomyelin phosphodiesterase 4, neutral membrane (neutral sphingomyelinase-3) | 5.9566 | 0.00021 |
| NM_032042 | C5orf21 | Chromosome 5 open reading frame 21 | −5.99255 | 0.0002 |
| AI090937 | LOC729616 | hypothetical protein LOC729616 | −7.02475 | 0.00006 |
| NM_022764 | MTHFSD | Methenyltetrahydrofolate synthetase domain containing | −9.35301 | 6.22594E-06 |
| NM_001315 | MAPK14 | Mitogen-activated protein kinase 14 | −9.49739 | 5.48709E-06 |
| NM_198867 | ALKBH6 | AlkB, alkylation repair homolog 6 (E. coli) | 5.99375 | 0.0002 |
| NM_004155 | SERPINB9 | Serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 8.93566 | 9.05384E-06 |
| NM_198970 | AES | Amino-terminal enhancer of split | 7.18158 | 0.00005 |
| NM_007353 | GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | 7.51614 | 0.00004 |
| BC087847 | ENST00000322446 | CDNA clone IMAGE:30389268 | 8.47062 | 0.00001 |
| NM_145869 | ANXA11 | Annexin A11 | −9.25282 | 6.80287E-06 |
| AB014574 | FKBP15 | FK506 binding protein 15, 133kDa | −13.99188 | 2.06113E-07 |
| NM_033426 | KIAA1737 | KIAA1737 | −7.68569 | 0.00003 |
| BQ425510 | ADNP | Transcribed locus | −8.23074 | 0.00002 |
| NM_016287 | HP1BP3 | Heterochromatin protein 1, binding protein 3 | 8.19698 | 0.00002 |
| NM_016255 | FAM8A1 | Family with sequence similarity 8, member A1 | −6.9287 | 0.00007 |
| NM_004637 | RAB7 | RAB7, member RAS oncogene family | 6.02105 | 0.0002 |
| NM_144982 | CCDC131 | Coiled-coil domain containing 131 | −6.4492 | 0.00012 |
| THC2365247 | THC2365247 | Q6QAQ1 (Q6QAQ1) Cytoskeletal beta actin (Fragment), partial (76%) [THC2365247] | 5.97469 | 0.00021 |
| NM_014719 | KIAA0738 | KIAA0738 gene product | −10.86208 | 1.79053E-06 |
| NM_016028 | SUV420H1 | Suppressor of variegation 4-20 homolog 1 (Drosophila) | 8.9239 | 9.15176E-06 |
| NM_033274 | ADAM19 | ADAM metallopeptidase domain 19 (meltrin beta) | −7.76503 | 0.00003 |
| AF086790 | ACON | Homo sapiens aconitase precursor (ACON) mRNA, nuclear gene encoding mitochondrial protein, partial cds. | 11.59634 | 1.02999E-06 |
| NM_032236 | USP48 | Ubiquitin specific peptidase 48 | −6.86748 | 0.00007 |
| NM_001125 | ADPRH | ADP-ribosylarginine hydrolase | 6.64904 | 0.00009 |
| NM_178508 | C6orf1 | Chromosome 6 open reading frame 1 | 8.21324 | 0.00002 |
| NM_015691 | WWC3 | WWC family member 3 | −9.22295 | 6.98604E-06 |
| NM_032353 | VPS25 | Vacuolar protein sorting 25 homolog (S. cerevisiae) | 7.29826 | 0.00005 |
| NM_016372 | GPR175 | G protein-coupled receptor 175 | 6.49813 | 0.00011 |
| AL359584 | BXDC5 | Brix domain containing 5 | −6.78264 | 0.00008 |
| NM_001009955 | SSBP3 | Single stranded DNA binding protein 3 | 8.57491 | 0.00001 |
| NM_201649 | SLC6A9 | Solute carrier family 6 (neurotransmitter transporter, glycine), member 9 | 7.77425 | 0.00003 |
| AB075502 | C6orf62 | Chromosome 6 open reading frame 62 | −6.64251 | 0.00009 |
| NM_173587 | RCOR2 | REST corepressor 2 | 7.89684 | 0.00002 |
| NM_001006634 | ARHGAP17 | Rho GTPase activating protein 17 | −9.86551 | 4.00466E-06 |
| NM_003656 | CAMK1 | Calcium/calmodulin-dependent protein kinase I | 7.09836 | 0.00006 |
| NM_017640 | LRRC16 | Leucine rich repeat containing 16 | −6.80494 | 0.00008 |
| CB987747 | CB987747 | Transcribed locus | −7.90385 | 0.00002 |
| NM_052957 | ACRC | Acidic repeat containing | −7.3354 | 0.00004 |
| NM_005634 | SOX3 | SRY (sex determining region Y)-box 3 | 8.01509 | 0.00002 |
| NM_138348 | FAM105B | Family with sequence similarity 105, member B | 7.37875 | 0.00004 |
| NM_017813 | IMPAD1 | Inositol monophosphatase domain containing 1 | 7.59455 | 0.00003 |
| NM_030576 | LIMD2 | LIM domain containing 2 | 12.89038 | 4.17569E-07 |
| NM_000466 | PEX1 | Peroxisome biogenesis factor 1 | −10.02315 | 3.51001E-06 |
| NM_001521 | GTF3C2 | General transcription factor IIIC, polypeptide 2, beta 110kDa | 8.99983 | 8.5394E-06 |
| AK125038 | KIAA0500 | KIAA0500 protein | −5.89084 | 0.00023 |
| NM_015367 | BCL2L13 | BCL2-like 13 (apoptosis facilitator) | 6.02735 | 0.0002 |
| NM_015635 | GAPVD1 | GTPase activating protein and VPS9 domains 1 | 6.35776 | 0.00013 |
| NM_201589 | MAFA | V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian) | 7.51391 | 0.00004 |
| NM_017742 | ZCCHC2 | Zinc finger, CCHC domain containing 2 | −6.89791 | 0.00007 |
| NM_015138 | RTF1 | Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | −7.75704 | 0.00003 |
| NM_030802 | FAM117A | Family with sequence similarity 117, member A | −6.45889 | 0.00012 |
| NM_017580 | ZRANB1 | Zinc finger, RAN-binding domain containing 1 | 7.16513 | 0.00005 |
| CR624826 | WASF2 | CDNA clone IMAGE:3030163 | −7.8809 | 0.00002 |
| NM_004310 | RHOH | Ras homolog gene family, member H | −8.98267 | 8.67368E-06 |
| NM_014827 | ZC3H11A | Zinc finger CCCH-type containing 11A | −9.52615 | 5.35178E-06 |
| NM_021960 | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | −9.30554 | 6.4923E-06 |
| THC2275506 | THC2275506 | ALU4_HUMAN (P39191) Alu subfamily SB2 sequence contamination warning entry, partial (24%) [THC2275506] | −7.19705 | 0.00005 |
| NM_022066 | UBE2O | Ubiquitin-conjugating enzyme E2O | −8.02194 | 0.00002 |
| NM_015100 | POGZ | Pogo transposable element with ZNF domain | −9.44018 | 5.76767E-06 |
| NM_032982 | CASP2 | Caspase 2, apoptosis-related cysteine peptidase (neural precursor cell expressed, developmentally down-regulated 2) | −6.4344 | 0.00012 |
| NM_015568 | PPP1R16B | Protein phosphatase 1, regulatory (inhibitor) subunit 16B | 6.70901 | 0.00009 |
| NM_001017992 | DKFZp686D0972 | similar to RIKEN cDNA 4732495G21 gene | 7.41188 | 0.00004 |
| NM_002073 | GNAZ | Guanine nucleotide binding protein (G protein), alpha z polypeptide | 8.64948 | 0.00001 |
| A_24_P264416 | A_24_P264416 | Unknown | 6.15843 | 0.00017 |
| NM_152682 | RWDD4A | RWD domain containing 4A | −7.11982 | 0.00006 |
| NM_024833 | ZNF671 | Zinc finger protein 671 | −8.82104 | 0.00001 |
| A_24_P410070 | A_24_P410070 | Unknown | −7.09055 | 0.00006 |
| NM_018210 | FLJ10769 | hypothetical protein FLJ10769 | −10.36037 | 2.66292E-06 |
| NM_003194 | TBP | TATA box binding protein | −7.06616 | 0.00006 |
| THC2306239 | THC2306239 | HUMPKD1G08 polycystic kidney disease 1 protein {Homo sapiens;} , partial (12%) [THC2306239] | 6.39854 | 0.00013 |
| NM_021158 | TRIB3 | Tribbles homolog 3 (Drosophila) | 6.25082 | 0.00015 |
| NM_020771 | HACE1 | HECT domain and ankyrin repeat containing, E3 ubiquitin protein ligase 1 | −6.76349 | 0.00008 |
| NM_030939 | C6orf62 | Chromosome 6 open reading frame 62 | −6.49244 | 0.00011 |
| ENST00000357697 | ENST00000357697 | Unknown | 7.83769 | 0.00003 |
| NM_021145 | DMTF1 | Cyclin D binding myb-like transcription factor 1 | −7.75897 | 0.00003 |
| NM_014912 | CPEB3 | Cytoplasmic polyadenylation element binding protein 3 | −8.90297 | 9.32893E-06 |
| NM_030569 | ITIH5 | Inter-alpha (globulin) inhibitor H5 | 9.22611 | 6.96638E-06 |
| NM_031858 | NBR1 | Neighbor of BRCA1 gene 1 | −11.10553 | 1.48531E-06 |
| NM_024050 | C19orf58 | Chromosome 19 open reading frame 58 | 7.04054 | 0.00006 |
| NM_014943 | ZHX2 | Zinc fingers and homeoboxes 2 | −8.10382 | 0.00002 |
| NM_173728 | ARHGEF15 | Rho guanine nucleotide exchange factor (GEF) 15 | 6.53943 | 0.00011 |
| NM_014710 | GPRASP1 | G protein-coupled receptor associated sorting protein 2 | −6.26554 | 0.00015 |
| NM_024580 | EFTUD1 | Elongation factor Tu GTP binding domain containing 1 | −6.34432 | 0.00013 |
| AK127374 | AK127374 | CDNA FLJ45450 fis, clone BRSTN2002691 | −8.71223 | 0.00001 |
| BC062753 | MCART1 | Mitochondrial carrier triple repeat 1 | −9.73391 | 4.47672E-06 |
| NM_003640 | IKBKAP | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein | −7.39824 | 0.00004 |
| NM_007375 | TARDBP | TAR DNA binding protein | −6.66957 | 0.00009 |
| CR594281 | METTL3 | Methyltransferase like 3 | −8.8068 | 0.00001 |
| NM_002014 | FKBP4 | FK506 binding protein 4, 59kDa | 6.81268 | 0.00008 |
| NM_145279 | MOBKL2C | MOB1, Mps One Binder kinase activator-like 2C (yeast) | 10.59534 | 2.20673E-06 |
| BC043212 | EFHB | EF-hand domain family, member B | 6.97051 | 0.00007 |
| NM_003146 | SSRP1 | Structure specific recognition protein 1 | −6.13864 | 0.00017 |
| NM_015107 | PHF8 | PHD finger protein 8 | −7.1863 | 0.00005 |
| BC051311 | DKFZp547E087 | hypothetical gene LOC283846 | 9.24017 | 6.87976E-06 |
| NM_018688 | BIN3 | Bridging integrator 3 | 5.69995 | 0.00029 |
| NM_024620 | ZNF329 | Zinc finger protein 329 | −7.79871 | 0.00003 |
| NM_015001 | SPEN | Spen homolog, transcriptional regulator (Drosophila) | −8.49844 | 0.00001 |
| NM_014659 | HISPPD2A | Histidine acid phosphatase domain containing 2A | −6.14796 | 0.00017 |
| NM_016598 | ZDHHC3 | Zinc finger, DHHC-type containing 3 | 6.91463 | 0.00007 |
| NM_018353 | C14orf106 | Chromosome 14 open reading frame 106 | −6.98428 | 0.00006 |
| NM_006861 | RAB35 | RAB35, member RAS oncogene family | 11.1048 | 1.48613E-06 |
| NM_018474 | C20orf19 | Homo sapiens chromosome 20 open reading frame 19 (C20orf19), mRNA. | −6.2157 | 0.00016 |
| AB037855 | KIAA1434 | hypothetical protein KIAA1434 | −7.0343 | 0.00006 |
| AL834448 | DKFZp547C195 | hypothetical protein DKFZp547C195 | −11.98823 | 7.7645E-07 |
| NM_004629 | FANCG | Fanconi anemia, complementation group G | −6.22857 | 0.00015 |
| NM_033089 | ZCCHC3 | Zinc finger, CCHC domain containing 3 | −6.36039 | 0.00013 |
| THC2301362 | THC2301362 | Unknown | −7.58392 | 0.00003 |
| AK026896 | STK24 | Serine/threonine kinase 24 (STE20 homolog, yeast) | −5.95955 | 0.00021 |
| NM_032438 | L3MBTL3 | L(3)mbt-like 3 (Drosophila) | −6.4802 | 0.00011 |
| THC2378263 | THC2378263 | Unknown | −6.63826 | 0.0001 |
| NM_032560 | SMEK1 | SMEK homolog 1, suppressor of mek1 (Dictyostelium) | −7.87743 | 0.00003 |
| NM_030915 | LBH | Limb bud and heart development homolog (mouse) | 8.06636 | 0.00002 |
| NM_004364 | CEBPA | CCAAT/enhancer binding protein (C/EBP), alpha | 9.5187 | 5.3865E-06 |
| NM_006278 | ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | 5.76877 | 0.00027 |
| NM_018996 | TNRC6C | Trinucleotide repeat containing 6C | 6.87855 | 0.00007 |
| NM_016479 | SCOTIN | scotin | −10.49939 | 2.38168E-06 |
| NM_019555 | ARHGEF3 | Rho guanine nucleotide exchange factor (GEF) 3 | −10.73596 | 1.9754E-06 |
| NM_207660 | ZC3H14 | Zinc finger CCCH-type containing 14 | 8.23037 | 0.00002 |
| AY726570 | ENST00000367696 | Clone TESTIS-724 mRNA sequence | −6.55525 | 0.0001 |
| NM_012239 | SIRT3 | Sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) | −7.27221 | 0.00005 |
| NM_139321 | ATRN | Attractin | −11.14924 | 1.43685E-06 |
| NM_006868 | RAB31 | RAB31, member RAS oncogene family | −8.70562 | 0.00001 |
| NM_003730 | RNASET2 | Ribonuclease T2 | −6.26012 | 0.00015 |
| BC045677 | FAM123A | Family with sequence similarity 123A | 7.88642 | 0.00002 |
| NM_005626 | SFRS4 | Splicing factor, arginine/serine-rich 4 | 6.47874 | 0.00011 |
| NM_003510 | HIST1H2AK | Histone cluster 1, H2ak | 6.79094 | 0.00008 |
| NM_006814 | PSMF1 | Proteasome (prosome, macropain) inhibitor subunit 1 (PI31) | −6.85967 | 0.00007 |
| NR_002939 | RUNDC2C | Homo sapiens RUN domain containing 2C (RUNDC2C) on chromosome 16. | 10.2493 | 2.91402E-06 |
| NM_005946 | MT1A | Metallothionein 1A (functional) | 5.81991 | 0.00025 |
| NM_145698 | ACBD5 | Acyl-Coenzyme A binding domain containing 5 | 8.7742 | 0.00001 |
| AY629351 | ZCCHC4 | Zinc finger, CCHC domain containing 4 | 7.46566 | 0.00004 |
| NM_145071 | CISH | Cytokine inducible SH2-containing protein | 6.338 | 0.00013 |
| NM_016072 | GOLT1B | Golgi transport 1 homolog B (S. cerevisiae) | 8.13565 | 0.00002 |
| AK056744 | VPS13C | Vacuolar protein sorting 13 homolog C (S. cerevisiae) | −6.21249 | 0.00016 |
| AF174606 | SHFM3P1 | Breakpoint cluster region | 6.2552 | 0.00015 |
| NM_023923 | PHACTR4 | Phosphatase and actin regulator 4 | −9.54987 | 5.24294E-06 |
| CR591805 | TMEM183A | Chromosome 1 open reading frame 37 | −7.74218 | 0.00003 |
| AY163812 | C17orf80 | Chromosome 17 open reading frame 80 | −6.70328 | 0.00009 |
| NM_001127 | AP1B1 | Adaptor-related protein complex 1, beta 1 subunit | 6.3445 | 0.00013 |
| AK024489 | PPP1R3E | Protein phosphatase 1, regulatory (inhibitor) subunit 3E | −5.77543 | 0.00027 |
| NM_014827 | ZC3H11A | Zinc finger CCCH-type containing 11A | −9.04607 | 8.18865E-06 |
| NM_020651 | PELI1 | Pellino homolog 1 (Drosophila) | −10.29063 | 2.81767E-06 |
| NM_001004351 | MGC57359 | similar to Williams Beuren syndrome chromosome region 19 | 9.7492 | 4.41886E-06 |
| A_23_P140454 | A_23_P140454 | Unknown | 7.70324 | 0.00003 |
| NM_021639 | GPBP1L1 | GC-rich promoter binding protein 1-like 1 | −9.69439 | 4.63023E-06 |
| THC2437906 | THC2437906 | Unknown | −7.29071 | 0.00005 |
| NM_014751 | MTSS1 | Metastasis suppressor 1 | −6.83835 | 0.00008 |
| THC2274685 | THC2274685 | AF326941 5H1 {Caenorhabditis elegans;} , partial (16%) [THC2274685] | −5.8588 | 0.00024 |
| NM_006994 | BTN3A3 | Butyrophilin, subfamily 3, member A3 | −6.7053 | 0.00009 |
| NM_194302 | CCDC108 | Coiled-coil domain containing 108 | 8.27613 | 0.00002 |
| NM_018475 | TMEM165 | Transmembrane protein 165 | 6.52923 | 0.00011 |
| NM_018355 | ZNF415 | Zinc finger protein 415 | −6.32122 | 0.00014 |
| AW948903 | ELK3 | ELK3, ETS-domain protein (SRF accessory protein 2) | 10.10496 | 3.28015E-06 |
| NM_017733 | PIGG | Phosphatidylinositol glycan anchor biosynthesis, class G | −8.82095 | 0.00001 |
| NM_013433 | TNPO2 | Transportin 2 (importin 3, karyopherin beta 2b) | 5.7888 | 0.00026 |
| NM_003379 | VIL2 | Villin 2 (ezrin) | 7.14114 | 0.00005 |
| AK093641 | FAM120AOS | Family with sequence similarity 120A opposite strand | −8.05047 | 0.00002 |
| NM_023015 | INTS3 | Integrator complex subunit 3 | −6.0334 | 0.00019 |
| NM_002017 | FLI1 | Friend leukemia virus integration 1 | −6.68628 | 0.00009 |
| NM_145808 | MTPN | Myotrophin | 8.14242 | 0.00002 |
| A_24_P118591 | A_24_P118591 | Unknown | −7.11249 | 0.00006 |
| BG620990 | LOC339352 | similar to ATP binding domain 3 | 5.91871 | 0.00022 |
| NM_002862 | PYGB | Phosphorylase, glycogen; brain | 6.26247 | 0.00015 |
| XM_291857 | A_24_P499282 | Unknown | 8.70354 | 0.00001 |
| NM_022040 | LAT2 | Linker for activation of T cells family, member 2 | 5.99705 | 0.0002 |
| AL122093 | ZP4 | Zona pellucida glycoprotein 4 | 6.09319 | 0.00018 |
| NM_012448 | STAT5B | Signal transducer and activator of transcription 5B | 7.84235 | 0.00003 |
| NM_017777 | MKS1 | Meckel syndrome, type 1 | 6.36619 | 0.00013 |
| NM_052831 | C6orf192 | Chromosome 6 open reading frame 192 | −5.92287 | 0.00022 |
| NM_207336 | ZNF467 | Zinc finger protein 467 | 9.10574 | 7.75922E-06 |
| NM_005550 | KIFC3 | Kinesin family member C3 | 7.12481 | 0.00006 |
| NM_001312 | CRIP2 | Cysteine-rich protein 2 | 5.77985 | 0.00027 |
| XM_372970 | ENST00000312710 | Unknown | 6.04444 | 0.00019 |
| NM_015135 | NUP205 | Nucleoporin 205kDa | −9.28016 | 6.63981E-06 |
| NM_005800 | USPL1 | Ubiquitin specific peptidase like 1 | −6.5711 | 0.0001 |
| THC2284174 | THC2284174 | Unknown | −8.71174 | 0.00001 |
| A_24_P213175 | A_24_P213175 | Unknown | −5.91113 | 0.00023 |
| NM_000873 | ICAM2 | Intercellular adhesion molecule 2 | −6.44068 | 0.00012 |
| NM_014742 | TM9SF4 | Transmembrane 9 superfamily protein member 4 | −9.05755 | 8.10401E-06 |
| NM_003858 | CCNK | Cyclin K | −6.93939 | 0.00007 |
| NM_144498 | OSBPL2 | Oxysterol binding protein-like 2 | −7.43179 | 0.00004 |
| NM_032819 | ZNF341 | Zinc finger protein 341 | 8.05677 | 0.00002 |
| A_24_P273004 | A_24_P273004 | Unknown | −11.34914 | 1.23637E-06 |
| NM_178170 | NEK8 | NIMA (never in mitosis gene a)- related kinase 8 | 15.27718 | 9.62147E-08 |
| NM_003791 | MBTPS1 | Membrane-bound transcription factor peptidase, site 1 | 6.5654 | 0.0001 |
| XM_497271 | A_24_P761490 | Unknown | 7.0537 | 0.00006 |
| NM_020800 | IFT80 | Intraflagellar transport 80 homolog (Chlamydomonas) | −5.90569 | 0.00023 |
| NM_015412 | C3orf17 | Chromosome 3 open reading frame 17 | −7.11308 | 0.00006 |
| NM_021064 | HIST1H2AG | Histone cluster 1, H2ag | 6.92417 | 0.00007 |
| NM_015893 | PRLH | Prolactin releasing hormone | 8.0239 | 0.00002 |
| NM_024040 | CUEDC2 | CUE domain containing 2 | 6.71805 | 0.00009 |
| A_24_P340916 | A_24_P340916 | Unknown | 5.93574 | 0.00022 |
| NM_006738 | AKAP13 | A kinase (PRKA) anchor protein 13 | 9.53395 | 5.31574E-06 |
| NM_014587 | SOX8 | SRY (sex determining region Y)-box 8 | 6.76814 | 0.00008 |
| NM_001126 | ADSS | Adenylosuccinate synthase | 6.04218 | 0.00019 |
| NM_014189 | ADD1 | Adducin 1 (alpha) | 6.99685 | 0.00006 |
| NM_001024847 | TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | 5.75954 | 0.00027 |
| NM_014708 | KNTC1 | Kinetochore associated 1 | −6.31481 | 0.00014 |
| NM_033402 | LRRCC1 | Leucine rich repeat and coiled-coil domain containing 1 | −7.93516 | 0.00002 |
| NM_006129 | BMP1 | Bone morphogenetic protein 1 | 8.37174 | 0.00002 |
| NM_007260 | LYPLA2 | Lysophospholipase II | 5.78566 | 0.00026 |
| AL834499 | LOC90113 | hypothetical protein BC009862 | 8.85453 | 9.75368E-06 |
| BX640624 | IGHA1 | Immunoglobulin heavy constant alpha 1 | 5.77955 | 0.00027 |
| NM_058187 | C21orf63 | Chromosome 21 open reading frame 63 | 6.98823 | 0.00006 |
| AK055501 | RPL15 | Ribosomal protein L15 | −6.50653 | 0.00011 |
| AF039564 | RBBP9 | Homo sapiens retinoblastoma binding protein (RBBP9) mRNA, complete cds. | 7.94915 | 0.00002 |
| BC041387 | MCTP2 | Multiple C2 domains, transmembrane 2 | −6.79143 | 0.00008 |
| A_23_P252501 | A_23_P252501 | Unknown | 10.69754 | 2.03582E-06 |
| NM_014023 | WDR37 | WD repeat domain 37 | −6.4766 | 0.00011 |
| NM_172037 | RDH10 | Retinol dehydrogenase 10 (all-trans) | 8.73814 | 0.00001 |
| A_24_P106166 | A_24_P106166 | Unknown | −6.28848 | 0.00014 |
| NM_178313 | SPTBN1 | Spectrin, beta, non-erythrocytic 1 | 5.45395 | 0.0004 |
| THC2403165 | THC2403165 | Q6TXI7 (Q6TXI7) LRRGT00012, partial (4%) [THC2403165] | −7.92713 | 0.00002 |
| NM_080660 | ZC3HAV1L | Zinc finger CCCH-type, antiviral 1-like | 8.99264 | 8.59541E-06 |
| AK123844 | C9orf45 | Spermatid perinuclear RNA binding protein | −6.84307 | 0.00008 |
| NM_207581 | DUOXA2 | Dual oxidase maturation factor 2 | 7.20138 | 0.00005 |
| AK128714 | LOC647121 | similar to embigin homolog | −6.07806 | 0.00018 |
| NM_152906 | C22orf25 | Chromosome 22 open reading frame 25 | 6.93325 | 0.00007 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −6.65631 | 0.00009 |
| NM_133330 | WHSC1 | Wolf-Hirschhorn syndrome candidate 1 | −10.55755 | 2.27388E-06 |
| NM_003636 | KCNAB2 | Potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 8.84297 | 9.85807E-06 |
| NM_033064 | ATCAY | Ataxia, cerebellar, Cayman type (caytaxin) | 9.55873 | 5.20293E-06 |
| THC2427841 | THC2427841 | Q9JM93 (Q9JM93) SRp25 nuclear protein (ADP-ribosylation factor-like 6 interacting protein 4), partial (7%) [THC2427841] | −7.12454 | 0.00006 |
| NM_012115 | CASP8AP2 | CASP8 associated protein 2 | −7.48469 | 0.00004 |
| XM_496950 | LOC441320 | PREDICTED: Homo sapiens similar to seven transmembrane helix receptor (LOC441320), mRNA [XM_496950] | 7.82806 | 0.00003 |
| NM_006521 | TFE3 | Transcription factor binding to IGHM enhancer 3 | −9.40066 | 5.97063E-06 |
| NM_032813 | TMTC4 | Transmembrane and tetratricopeptide repeat containing 4 | −7.55316 | 0.00003 |
| XM_929854 | LOC646890 | PREDICTED: Homo sapiens hypothetical protein LOC646890 (LOC646890), mRNA [XM_929854] | −5.8798 | 0.00023 |
| NM_172193 | KLHDC1 | Kelch domain containing 1 | 5.70628 | 0.00029 |
| NM_006289 | TLN1 | Talin1 | 5.85014 | 0.00024 |
| NM_004609 | TCF15 | Transcription factor 15 (basic helix-loop-helix) | 6.93939 | 0.00007 |
| NM_018442 | IQWD1 | IQ motif and WD repeats 1 | −11.63764 | 9.99374E-07 |
| NM_003110 | SP2 | Sp2 transcription factor | −7.66008 | 0.00003 |
| NM_016089 | ZNF589 | Zinc finger protein 589 | −7.66171 | 0.00003 |
| THC2378504 | THC2378504 | Unknown | −5.67893 | 0.0003 |
| NM_018117 | BRWD2 | Bromodomain and WD repeat domain containing 2 | −6.96869 | 0.00007 |
| NM_024676 | C1orf113 | Chromosome 1 open reading frame 113 | 5.59764 | 0.00034 |
| AK098835 | ZFHX1B | Zinc finger homeobox 1b | −5.92034 | 0.00022 |
| NM_000235 | LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | −6.63589 | 0.0001 |
| NM_001852 | COL9A2 | Collagen, type IX, alpha 2 | −7.20238 | 0.00005 |
| NM_021170 | HES4 | Hairy and enhancer of split 4 (Drosophila) | 9.59815 | 5.02884E-06 |
| NM_032340 | C6orf125 | Chromosome 6 open reading frame 125 | 12.24403 | 6.48527E-07 |
| NM_012407 | PICK1 | Protein interacting with PRKCA 1 | 6.45076 | 0.00012 |
| NM_005343 | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | 5.45148 | 0.0004 |
| AK130576 | ARHGAP24 | Rho GTPase activating protein 24 | −5.8887 | 0.00023 |
| AL831999 | SUGT1 | MRNA; cDNA DKFZp451K063 (from clone DKFZp451K063) | −7.49324 | 0.00004 |
| DQ287932 | DPY19L1 | Dpy-19-like 1 (C. elegans) | 7.50563 | 0.00004 |
| AK098256 | LOC728723 | hypothetical protein LOC728723 | 7.87079 | 0.00003 |
| THC2441708 | THC2441708 | Unknown | 9.15445 | 0.000007427 |
| NM_004649 | C21orf33 | Chromosome 21 open reading frame 33 | −7.56222 | 0.00003 |
| NM_021003 | PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | −6.37986 | 0.00013 |
| AF307332 | MGEA5 | Meningioma expressed antigen 5 (hyaluronidase) | −6.55078 | 0.00011 |
| NM_005159 | ACTC1 | Actin, alpha, cardiac muscle 1 | 5.65957 | 0.00031 |
| NM_014824 | FCHSD2 | FCH and double SH3 domains 2 | −7.13614 | 0.00005 |
| NM_001397 | ECE1 | Endothelin converting enzyme 1 | 5.45317 | 0.0004 |
| NM_019042 | PUS7 | Pseudouridylate synthase 7 homolog (S. cerevisiae) | 8.95172 | 8.92198E-06 |
| NM_004096 | EIF4EBP2 | Eukaryotic translation initiation factor 4E binding protein 2 | −6.01179 | 0.0002 |
| A_32_P119165 | A_32_P119165 | Unknown | −6.04598 | 0.00019 |
| NM_013398 | ZNF224 | Zinc finger protein 224 | −6.49534 | 0.00011 |
| NM_001039888 | ANKRD34 | Ankyrin repeat domain 34 | −5.80244 | 0.00026 |
| A_24_P67378 | A_24_P67378 | Unknown | 6.82367 | 0.00008 |
| NM_152549 | CCDC112 | Coiled-coil domain containing 112 | −6.31102 | 0.00014 |
| AK092824 | AMN | Amnionless homolog (mouse) | −5.76454 | 0.00027 |
| NM_194247 | HNRPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 7.60297 | 0.00003 |
| NM_006569 | CGREF1 | Cell growth regulator with EF-hand domain 1 | 8.72836 | 0.00001 |
| NM_152687 | C5orf29 | Chromosome 5 open reading frame 29 | −5.52685 | 0.00037 |
| NM_005436 | CCDC6 | Coiled-coil domain containing 6 | −7.57251 | 0.00003 |
| NM_017723 | FLJ20245 | hypothetical protein FLJ20245 | −8.75065 | 0.00001 |
| NM_020145 | SH3GLB2 | Sec23 homolog B (S. cerevisiae) | 6.66636 | 0.00009 |
| A_24_P358205 | A_24_P358205 | Unknown | −7.12448 | 0.00006 |
| NM_001104 | ACTN3 | Actinin, alpha 3 | 5.36436 | 0.00045 |
| NM_001111 | ADAR | Adenosine deaminase, RNA-specific | 6.46515 | 0.00012 |
| NM_024778 | LONRF3 | LON peptidase N-terminal domain and ring finger 3 | 6.13423 | 0.00017 |
| NM_014670 | BZW1 | Basic leucine zipper and W2 domains 1 | 6.26341 | 0.00015 |
| NM_172020 | POM121 | POM121 membrane glycoprotein (rat) | −8.87468 | 9.57445E-06 |
| NM_000539 | RHO | Rhodopsin (opsin 2, rod pigment) (retinitis pigmentosa 4, autosomal dominant) | 8.27908 | 0.00002 |
| NM_020998 | MST1 | Macrophage stimulating 1 (hepatocyte growth factor-like) | 8.08571 | 0.00002 |
| NM_014417 | BBC3 | BCL2 binding component 3 | 6.68822 | 0.00009 |
| NM_005796 | NUTF2 | Nuclear transport factor 2 | 5.44463 | 0.00041 |
| NM_153342 | TMEM150 | Transmembrane protein 150 | 5.64117 | 0.00032 |
| AK091057 | LOC285535 | hypothetical protein LOC285535 | −8.33776 | 0.00002 |
| NM_032932 | RAB11FIP4 | RAB11 family interacting protein 4 (class II) | −7.08778 | 0.00006 |
| A_32_P167723 | A_32_P167723 | Unknown | 6.59109 | 0.0001 |
| NM_001007793 | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 5.60451 | 0.00033 |
| ENST00000372821 | ENST00000372821 | Q6DEY7 (Q6DEY7) MGC89376 protein, partial (38%) [THC2400949] | −7.5636 | 0.00003 |
| NM_032580 | HES7 | Hairy and enhancer of split 7 (Drosophila) | 8.7194 | 0.00001 |
| NM_144596 | TTC8 | Tetratricopeptide repeat domain 8 | −6.49123 | 0.00011 |
| NM_001024457 | RGPD1 | Plasminogen-like B1 | −5.37308 | 0.00045 |
| NM_014157 | CCDC113 | Coiled-coil domain containing 113 | 7.65554 | 0.00003 |
| NM_152262 | ZNF439 | Zinc finger protein 439 | −5.84169 | 0.00025 |
| NM_015358 | MORC3 | MORC family CW-type zinc finger 3 | −6.20742 | 0.00016 |
| NM_197941 | ADAMTS6 | ADAM metallopeptidase with thrombospondin type 1 motif, 6 | −6.43052 | 0.00012 |
| NM_146421 | GSTM1 | Glutathione S-transferase M1 | 5.20983 | 0.00056 |
| NM_052861 | MGC21675 | hypothetical protein MGC21675 | −6.36104 | 0.00013 |
| NM_000182 | HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | 8.29677 | 0.00002 |
| NM_017443 | POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | −6.66635 | 0.00009 |
| NM_018254 | RCOR3 | REST corepressor 3 | −6.75332 | 0.00008 |
| NM_153002 | GPR156 | G protein-coupled receptor 156 | 7.48264 | 0.00004 |
| NM_001010919 | LOC441168 | hypothetical protein LOC441168 | −8.95495 | 8.8957E-06 |
| NM_005452 | WDR46 | WD repeat domain 46 | 5.35346 | 0.00046 |
| NM_004290 | RNF14 | Ring finger protein 14 | 5.61613 | 0.00033 |
| AK094269 | LOC285813 | hypothetical protein LOC285813 | −6.16194 | 0.00017 |
| NM_014641 | MDC1 | Mediator of DNA damage checkpoint 1 | −10.5734 | 2.24545E-06 |
| NM_018084 | KIAA1212 | KIAA1212 | −5.64438 | 0.00032 |
| NM_001008735 | HMG1L1 | High-mobility group (nonhistone chromosomal) protein 1-like 1 | 6.98657 | 0.00006 |
| NM_001012398 | FTS | Fused toes homolog (mouse) | 5.31553 | 0.00048 |
| NM_016546 | C1RL | Complement component 1, r subcomponent-like | −9.50091 | 5.47037E-06 |
| NM_003153 | STAT6 | Signal transducer and activator of transcription 6, interleukin-4 induced | −6.43102 | 0.00012 |
| NM_001074 | UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | 7.85504 | 0.00003 |
| NM_013328 | PYCR2 | Pyrroline-5-carboxylate reductase family, member 2 | 5.42514 | 0.00042 |
| NM_015897 | PIAS4 | Protein inhibitor of activated STAT, 4 | 6.55103 | 0.00011 |
| NM_001283 | AP1S1 | Adaptor-related protein complex 1, sigma 1 subunit | 5.21037 | 0.00056 |
| NM_000923 | PDE4C | Phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | 7.48989 | 0.00004 |
| NM_018676 | THSD1 | Thrombospondin, type I, domain containing 1 | −7.01524 | 0.00006 |
| NM_007042 | RPP14 | Ribonuclease P 14kDa subunit | 5.88159 | 0.00023 |
| NM_015926 | TEX264 | Testis expressed sequence 264 | 6.03192 | 0.00019 |
| NM_021216 | ZNF71 | Zinc finger protein 71 | −7.39256 | 0.00004 |
| NM_001659 | ARF3 | ADP-ribosylation factor 3 | 6.08657 | 0.00018 |
| THC2435127 | THC2435127 | Unknown | −7.00523 | 0.00006 |
| NM_015153 | PHF3 | PHD finger protein 3 | −7.08168 | 0.00006 |
| THC2407434 | THC2407434 | ALU1_HUMAN (P39188) Alu subfamily J sequence contamination warning entry, partial (20%) [THC2407434] | −9.80581 | 4.21166E-06 |
| NM_018051 | WDR60 | WD repeat domain 60 | −5.79231 | 0.00026 |
| NM_177987 | TUBB8 | tubulin, beta 8 | 5.56498 | 0.00035 |
| BC037535 | BC037535 | CDNA clone IMAGE:5263531 | −5.96076 | 0.00021 |
| NM_015568 | PPP1R16B | Protein phosphatase 1, regulatory (inhibitor) subunit 16B | −7.15947 | 0.00005 |
| NM_021813 | BACH2 | BTB and CNC homology 1, basic leucine zipper transcription factor 2 | −5.18425 | 0.00058 |
| THC2345721 | THC2345721 | Unknown | −5.77505 | 0.00027 |
| NM_015291 | DNAJC16 | DnaJ (Hsp40) homolog, subfamily C, member 16 | −5.67192 | 0.0003 |
| THC2437034 | THC2437034 | Q9N083 (Q9N083) Unnamed portein product, partial (43%) [THC2437034] | −5.95354 | 0.00021 |
| NM_004804 | CIAO1 | Cytosolic iron-sulfur protein assembly 1 homolog (S. cerevisiae) | 9.32183 | 6.39948E-06 |
| NM_001558 | IL10RA | Interleukin 10 receptor, alpha | −7.16204 | 0.00005 |
| NM_006016 | CD164 | CD164 molecule, sialomucin | 9.32468 | 6.38342E-06 |
| NM_003581 | NCK2 | NCK adaptor protein 2 | 7.177 | 0.00005 |
| NM_020432 | PHTF2 | Putative homeodomain transcription factor 2 | −7.55837 | 0.00003 |
| NM_016263 | FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | 5.79913 | 0.00026 |
| NM_006901 | MYO9A | Myosin IXA | −6.32218 | 0.00014 |
| BU661610 | BU661610 | Transcribed locus | −7.93911 | 0.00002 |
| NM_001310 | CREBL2 | CAMP responsive element binding protein-like 2 | −8.63283 | 0.00001 |
| NM_015187 | KIAA0746 | KIAA0746 protein | −7.19882 | 0.00005 |
| NM_014319 | LEMD3 | LEM domain containing 3 | −5.23419 | 0.00054 |
| NM_021008 | DEAF1 | Deformed epidermal autoregulatory factor 1 (Drosophila) | 7.79753 | 0.00003 |
| NM_015336 | ZDHHC17 | Zinc finger, DHHC-type containing 17 | −6.20521 | 0.00016 |
| NM_006888 | CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | −6.26682 | 0.00015 |
| NM_018198 | DNAJC11 | DnaJ (Hsp40) homolog, subfamily C, member 11 | 5.29002 | 0.0005 |
| AF277175 | KIAA0265 | KIAA0265 protein | −6.54094 | 0.00011 |
| NM_198974 | TWF1 | Twinfilin, actin-binding protein, homolog 1 (Drosophila) | 9.75026 | 4.41488E-06 |
| NM_005370 | RAB8A | RAB8A, member RAS oncogene family | 5.7961 | 0.00026 |
| BG695979 | BG695979 | Transcribedlocus | −6.26052 | 0.00015 |
| NM_019044 | CCDC93 | Coiled-coil domain containing 93 | −7.08537 | 0.00006 |
| NM_032377 | ELOF1 | Elongation factor 1 homolog (S. cerevisiae) | 5.33163 | 0.00047 |
| NM_020336 | KIAA1219 | KIAA1219 | −9.56667 | 5.16733E-06 |
| NM_017704 | ANKRD49 | Ankyrin repeat domain 49 | −6.59777 | 0.0001 |
| BU618279 | PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | 8.48486 | 0.00001 |
| NM_014730 | KIAA0152 | KIAA0152 | 5.53054 | 0.00037 |
| NM_005153 | USP10 | Ubiquitin specific peptidase 10 | 6.93512 | 0.00007 |
| NM_001001433 | STX16 | Syntaxin 16 | 8.14081 | 0.00002 |
| BX098637 | RAD50 | Transcribed locus, weakly similar to NP_689672.2 protein LOC146556 [Homo sapiens] | −7.28539 | 0.00005 |
| NM_001885 | CRYAB | Crystallin, alpha B | 7.84386 | 0.00003 |
| NR_002797 | LOC255783 | hypothetical protein LOC255783 | 7.45402 | 0.00004 |
| NM_031219 | HDHD3 | Haloacid dehalogenase-like hydrolase domain containing 3 | 6.5466 | 0.00011 |
| NM_019002 | ETAA1 | Ewing's tumor-associated antigen 1 | −6.00953 | 0.0002 |
| THC2250585 | THC2250585 | Unknown | −10.02693 | 3.49902E-06 |
| NM_002999 | SDC4 | Syndecan 4 (amphiglycan, ryudocan) | 5.27453 | 0.00051 |
| CR620831 | C12orf47 | Chromosome 12 open reading frame 47 | −7.40664 | 0.00004 |
| NM_005500 | SAE1 | SUMO1 activating enzyme subunit 1 | −7.04103 | 0.00006 |
| NM_014045 | LRP10 | Low density lipoprotein receptor-related protein 10 | 5.77774 | 0.00027 |
| NM_173622 | CDRT4 | CMT1A duplicated region transcript 4 | −6.39054 | 0.00013 |
| NM_000594 | TNF | Tumor necrosis factor (TNF superfamily, member 2) | 5.25446 | 0.00052 |
| NM_001822 | CHN1 | Chimerin (chimaerin) 1 | 6.92225 | 0.00007 |
| A_24_P872359 | A_24_P872359 | Unknown | 6.07579 | 0.00018 |
| NM_016219 | MAN1B1 | Mannosidase, alpha, class 1B, member 1 | 6.64725 | 0.00009 |
| NM_173607 | C14orf24 | Chromosome 14 open reading frame 24 | 7.36156 | 0.00004 |
| THC2349560 | THC2349560 | Unknown | 6.3882 | 0.00013 |
| AK126092 | LOC221442 | hypothetical LOC221442 | −6.7597 | 0.00008 |
| NM_006804 | STARD3 | START domain containing 3 | 6.72679 | 0.00009 |
| NM_000848 | GSTM2 | Glutathione S-transferase M2 (muscle) | 5.42214 | 0.00042 |
| NM_005399 | PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | −5.63095 | 0.00032 |
| NM_012296 | GAB2 | GRB2-associated binding protein 2 | −6.86247 | 0.00007 |
| NM_024707 | GEMIN7 | Gem (nuclear organelle) associated protein 7 | −5.73351 | 0.00028 |
| NM_004162 | RAB5A | RAB5A, member RAS oncogene family | 5.07256 | 0.00067 |
| NM_199072 | MDFIC | MyoD family inhibitor domain containing | 5.16111 | 0.00059 |
| NM_016454 | TMEM85 | Transmembrane protein 85 | 6.36911 | 0.00013 |
| NM_024295 | DERL1 | Der1-like domain family, member 1 | 6.49264 | 0.00011 |
| NM_017679 | BCAS3 | Breast carcinoma amplified sequence 3 | 7.15775 | 0.00005 |
| NM_152574 | C9orf52 | Chromosome 9 open reading frame 52 | −5.48097 | 0.00039 |
| NM_147190 | LASS5 | LAG1 homolog, ceramide synthase 5 (S. cerevisiae) | 7.77914 | 0.00003 |
| A_32_P65793 | A_32_P65793 | Unknown | 7.34402 | 0.00004 |
| NM_006923 | SDF2 | Stromal cell-derived factor 2 | 5.09884 | 0.00065 |
| NM_001012423 | GOLGA8E | Golgi autoantigen, golgin subfamily a, 8E | 7.02657 | 0.00006 |
| A_24_P824578 | A_24_P824578 | Unknown | 5.25718 | 0.00052 |
| NM_015550 | OSBPL3 | Oxysterol binding protein-like 3 | −7.36736 | 0.00004 |
| NM_024653 | PRKRIP1 | Postmeiotic segregation increased 2-like 3 | 5.39368 | 0.00044 |
| NM_133636 | HEL308 | DNA helicase HEL308 | −6.10973 | 0.00018 |
| A_32_P11425 | A_32_P11425 | Unknown | 7.51075 | 0.00004 |
| THC2279840 | THC2279840 | Unknown | −5.90895 | 0.00023 |
| CR623166 | TTC14 | Tetratricopeptide repeat domain 14 | −6.56973 | 0.0001 |
| NM_014868 | RNF10 | Ring finger protein 10 | −6.69199 | 0.00009 |
| NM_207443 | FLJ45244 | FLJ45244 protein | −6.99635 | 0.00006 |
| NM_012218 | ILF3 | Interleukin enhancer binding factor 3, 90kDa | −9.40237 | 5.96169E-06 |
| NM_199345 | LOC375133 | similar to phosphatidylinositol 4-kinase alpha | 6.26274 | 0.00015 |
| NM_017727 | FLJ20254 | hypothetical protein FLJ20254 | 6.90032 | 0.00007 |
| A_32_P1291 | A_32_P1291 | Unknown | 7.28232 | 0.00005 |
| BC020095 | MGC17403 | hypothetical protein MGC17403 | −6.5012 | 0.00011 |
| NM_002729 | HHEX | Homeobox, hematopoietically expressed | −7.00539 | 0.00006 |
| A_32_P112100 | A_32_P112100 | Unknown | 5.97742 | 0.00021 |
| XM_941086 | LOC651831 | similar to CG31232-PA, isoform A | 5.07231 | 0.00067 |
| AK024092 | CHD2 | Chromodomain helicase DNA binding protein 2 | −5.56197 | 0.00035 |
| AJ227863 | MBNL1 | Muscleblind-like (Drosophila) | −6.04839 | 0.00019 |
| NM_015678 | NBEA | Neurobeachin | −5.67699 | 0.0003 |
| NM_003611 | OFD1 | Oral-facial-digital syndrome 1 | −6.13772 | 0.00017 |
| NM_021144 | PSIP1 | PC4 and SFRS1 interacting protein 1 | −5.05928 | 0.00068 |
| THC2441367 | THC2441367 | Unknown | −6.27525 | 0.00015 |
| AJ001898 | GDNF | Glial cell derived neurotrophic factor | 6.43449 | 0.00012 |
| NM_014781 | RB1CC1 | RB1-inducible coiled-coil 1 | −6.24829 | 0.00015 |
| BC022830 | LOC642852 | hypothetical LOC642852 | 6.3943 | 0.00013 |
| BX337332 | BX337332 | Transcribed locus, moderately similar to XP_001136080.1 hypothetical protein [Pan troglodytes] | −6.2502 | 0.00015 |
| NM_001006634 | ARHGAP17 | Rho GTPase activating protein 17 | −8.33571 | 0.00002 |
| NM_006761 | YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | 4.97347 | 0.00077 |
| NM_002498 | NEK3 | NIMA (never in mitosis gene a)-related kinase 3 | −5.97924 | 0.00021 |
| CR605444 | FBXL4 | F-box and leucine-rich repeat protein 4 | −5.49736 | 0.00038 |
| NM_017991 | FLJ10081 | hypothetical protein FLJ10081 | −8.26415 | 0.00002 |
| NM_000428 | LTBP2 | Latent transforming growth factor beta binding protein 2 | 7.35375 | 0.00004 |
| NM_133439 | TADA2L | Transcriptional adaptor 2 (ADA2 homolog, yeast)-like | 5.53759 | 0.00036 |
| NM_005718 | ARPC4 | Actin related protein 2/3 complex, subunit 4, 20kDa | 5.39072 | 0.00044 |
| A_24_P238819 | A_24_P238819 | Unknown | 7.58575 | 0.00003 |
| NM_024069 | C19orf50 | Chromosome 19 open reading frame 50 | −8.00106 | 0.00002 |
| NM_005665 | EVI5 | Ecotropic viral integration site 5 | −6.01249 | 0.0002 |
| NM_002211 | ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | −6.426 | 0.00012 |
| NM_022362 | MMS19L | MMS19-like (MET18 homolog, S. cerevisiae) | −7.9471 | 0.00002 |
| AK022140 | FLJ12078 | hypothetical protein FLJ12078 | −5.85668 | 0.00024 |
| NM_017730 | QRICH1 | Glutamine-rich 1 | −7.12784 | 0.00005 |
| NM_001989 | EVX1 | Eve, even-skipped homeobox homolog 1 (Drosophila) | 7.23511 | 0.00005 |
| NM_018405 | C17orf79 | Chromosome 17 open reading frame 79 | 5.68363 | 0.0003 |
| A_32_P167212 | A_32_P167212 | Unknown | −5.12463 | 0.00062 |
| NM_006711 | RNPS1 | RNA binding protein S1, serine-rich domain | 6.2515 | 0.00015 |
| DB341590 | LOC728499 | similar to Nuclear envelope pore membrane protein POM 121 (Pore membrane protein of 121 kDa) (P145) | −5.18063 | 0.00058 |
| BE535679 | BE535679 | Transcribed locus | −6.17163 | 0.00016 |
| A_32_P327750 | A_32_P327750 | Unknown | 5.15782 | 0.0006 |
| NM_018035 | FLJ10241 | hypothetical protein FLJ10241 | 5.1834 | 0.00058 |
| NM_021727 | FADS3 | Fatty acid desaturase 3 | 5.3224 | 0.00048 |
| NM_181607 | KRTAP19-1 | Keratin associated protein 19-1 | 5.70077 | 0.00029 |
| NM_003199 | TCF4 | Transcription factor 4 | −5.32466 | 0.00048 |
| AK024898 | AK024898 | CDNA: FLJ21245 fis, clone COL01184 | −7.51329 | 0.00004 |
| NM_001018055 | BRCC3 | BRCA1/BRCA2-containing complex, subunit 3 | −5.30636 | 0.00049 |
| NM_016238 | ANAPC7 | Anaphase promoting complex subunit 7 | 5.43935 | 0.00041 |
| NM_013259 | TAGLN3 | Transgelin 3 | 6.8389 | 0.00008 |
| NM_001010983 | GLT8D1 | Glycosyltransferase 8 domain containing 1 | −5.69977 | 0.00029 |
| NM_006390 | IPO8 | Importin 8 | 6.90698 | 0.00007 |
| NM_005520 | HNRPH1 | Heterogeneous nuclear ribonucleoprotein H1 (H) | −8.24555 | 0.00002 |
| NM_145233 | ZNF625 | Zinc finger protein 20 | 6.01856 | 0.0002 |
| NM_020805 | KLHL14 | Kelch-like 14 (Drosophila) | −8.24609 | 0.00002 |
| AV698092 | C6orf62 | Transcribed locus | 5.96382 | 0.00021 |
| NM_002945 | RPA1 | Replication protein A1, 70kDa | −7.5748 | 0.00003 |
| NM_182691 | SRPK2 | SFRS protein kinase 2 | 5.04526 | 0.00069 |
| NM_138706 | B3GNT6 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) | 7.94837 | 0.00002 |
| NM_007172 | NUP50 | Nucleoporin 50kDa | 5.09318 | 0.00065 |
| AJ404330 | TBC1D12 | TBC1 domain family, member 12 | −8.08182 | 0.00002 |
| THC2312756 | THC2312756 | O60448 (O60448) Neuronal thread protein AD7c-NTP, partial (9%) [THC2312756] | −6.28489 | 0.00014 |
| NM_004897 | MINPP1 | Multiple inositol polyphosphate histidine phosphatase, 1 | 4.91264 | 0.00083 |
| NM_013335 | GMPPA | GDP-mannose pyrophosphorylase A | 5.16344 | 0.00059 |
| THC2377877 | THC2377877 | Q87WI3 (Q87WI3) Dipeptide ABC transporter, permease protein, partial (5%) [THC2377877] | −5.53889 | 0.00036 |
| A_24_P256050 | A_24_P256050 | Unknown | 5.35666 | 0.00046 |
| A_32_P12282 | A_32_P12282 | Unknown | 8.58347 | 0.00001 |
| NM_005056 | JARID1A | Jumonji, AT rich interactive domain 1A | −4.92183 | 0.00082 |
| NM_020230 | PPAN | Peter pan homolog (Drosophila) | 5.57565 | 0.00034 |
| NM_032558 | HIATL1 | Hippocampus abundant transcript-like 1 | 7.72907 | 0.00003 |
| NM_030672 | ARHGAP28 | Rho GTPase activating protein 28 | 7.72512 | 0.00003 |
| NM_001005386 | ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | 6.62652 | 0.0001 |
| NM_001012968 | LOC139886 | hypothetical protein LOC139886 | −5.45114 | 0.00041 |
| NM_183001 | SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | 8.174 | 0.00002 |
| CR620532 | RASA2 | RAS p21 protein activator 2 | −6.29865 | 0.00014 |
| NM_177543 | PPAP2C | Phosphatidic acid phosphatase type 2C | 8.2774 | 0.00002 |
| NM_018381 | FLJ11286 | hypothetical protein FLJ11286 | −9.16765 | 7.33973E-06 |
| XM_497140 | A_24_P135921 | Unknown | 4.9042 | 0.00084 |
| NM_000819 | GART | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | 5.26257 | 0.00052 |
| NM_002009 | FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | 4.87946 | 0.00087 |
| NM_003316 | TTC3 | Tetratricopeptide repeat domain 3 | −5.03045 | 0.00071 |
| NM_013326 | C18orf8 | Chromosome 18 open reading frame 8 | −6.43782 | 0.00012 |
| AF419616 | NBPF10 | Neuroblastoma breakpoint family, member 10 | −6.22377 | 0.00015 |
| NM_004329 | BMPR1A | Bone morphogenetic protein receptor, type IA | 6.20251 | 0.00016 |
| A_24_P915883 | A_24_P915883 | Unknown | 5.59313 | 0.00034 |
| NM_018486 | HDAC8 | Histone deacetylase 8 | 5.80887 | 0.00026 |
| THC2364821 | THC2364821 | Unknown | 6.33357 | 0.00014 |
| NM_002540 | ODF2 | Outer dense fiber of sperm tails 2 | −8.03397 | 0.00002 |
| NM_199437 | PRDM10 | PR domain containing 10 | −5.28301 | 0.00051 |
| NM_032179 | CPSF3L | Homo sapiens cleavage and polyadenylation specific factor 3-like (CPSF3L), transcript variant 2, mRNA [NM_032179] | 5.25928 | 0.00052 |
| NM_016598 | ZDHHC3 | Zinc finger, DHHC-type containing 3 | −7.1702 | 0.00005 |
| BU852331 | BU852331 | Transcribed locus | 8.69121 | 0.00001 |
| NM_004882 | CIR | CBF1 interacting corepressor | −5.75692 | 0.00027 |
| NM_000425 | L1CAM | L1 cell adhesion molecule | 6.26131 | 0.00015 |
| NM_022757 | CCDC14 | Coiled-coil domain containing 14 | −5.98571 | 0.00021 |
| A_24_P204454 | A_24_P204454 | Unknown | −5.98313 | 0.00021 |
| NM_015020 | PHLPPL | PH domain and leucine rich repeat protein phosphatase-like | −6.317 | 0.00014 |
| A_24_P170309 | A_24_P170309 | Unknown | 5.14466 | 0.00061 |
| A_24_P392099 | A_24_P392099 | Unknown | 5.32541 | 0.00048 |
| AK027248 | LOC56757 | hypothetical protein LOC56757 | −8.86084 | 9.69717E-06 |
| NM_170607 | MLX | MAX-like protein X | 6.44866 | 0.00012 |
| BC037281 | LOC439951 | hypothetical LOC439951 protein | 8.02017 | 0.00002 |
| NM_003800 | RNGTT | RNA guanylyltransferase and 5'-phosphatase | −7.68253 | 0.00003 |
| NM_198829 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 6.91893 | 0.00007 |
| THC2282321 | THC2282321 | AF235005 suppression of tumorigenicity 16 protein {Homo sapiens;} , partial (13%) [THC2282321] | −4.99222 | 0.00075 |
| NM_014828 | KIAA0737 | KIAA0737 | −7.25431 | 0.00005 |
| X82545 | PRNP | Prion protein (p27–30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | 9.03795 | 8.24904E-06 |
| NM_017634 | KCTD9 | Potassium channel tetramerisation domain containing 9 | −7.29441 | 0.00005 |
| NM_012268 | PLD3 | Phospholipase D family, member 3 | 7.3033 | 0.00005 |
| NM_017633 | FAM46A | Family with sequence similarity 46, member A | −5.89253 | 0.00023 |
| NM_016353 | ZDHHC2 | Zinc finger, DHHC-type containing 2 | 6.58499 | 0.0001 |
| NM_005845 | ABCC4 | ATP-binding cassette, sub-family C (CFTR/MRP), member 4 | 5.25204 | 0.00053 |
| THC2279840 | THC2279840 | Unknown | −6.05565 | 0.00019 |
| ENST00000361859 | ENST00000361859 | Unknown | 5.9073 | 0.00023 |
| BX538248 | BX538248 | CDNA clone IMAGE:30408657 | −5.68515 | 0.0003 |
| AK095904 | MIA3 | Melanoma inhibitory activity family, member 3 | −7.19205 | 0.00005 |
| NM_003040 | SLC4A2 | Solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) | 5.0469 | 0.00069 |
| NM_138819 | FAM122C | Family with sequence similarity 122C | −6.11779 | 0.00018 |
| NM_015442 | CNOT10 | CCR4-NOT transcription complex, subunit 10 | −6.29023 | 0.00014 |
| NM_152270 | SLFN11 | Schlafen family member 11 | −6.48172 | 0.00011 |
| AF085995 | LOC645513 | similar to septin 7 | −6.71126 | 0.00009 |
| NM_144566 | ZNF700 | Zinc finger protein 700 | −7.22033 | 0.00005 |
| NM_002570 | PCSK6 | Proprotein convertase subtilisin/kexin type 6 | 7.32909 | 0.00004 |
| NM_014827 | ZC3H11A | Zinc finger CCCH-type containing 11A | −7.8499 | 0.00003 |
| NM_000234 | LIG1 | Ligase I, DNA, ATP-dependent | −5.36772 | 0.00045 |
| NM_052928 | SMYD4 | SET and MYND domain containing 4 | −5.63457 | 0.00032 |
| THC2386194 | THC2386194 | Unknown | −5.35511 | 0.00046 |
| NM_005358 | LMO7 | LIM domain 7 | −7.08854 | 0.00006 |
| NM_022833 | FAM129B | Family with sequence similarity 129, member B | 7.60785 | 0.00003 |
| NM_181831 | NF2 | Neurofibromin 2 (bilateral acoustic neuroma) | 5.7013 | 0.00029 |
| NM_016551 | TM7SF3 | Transmembrane 7 superfamily member 3 | −5.82276 | 0.00025 |
| NM_057169 | GIT2 | G protein-coupled receptor kinase interactor 2 | −6.32361 | 0.00014 |
| AK000923 | SENP3 | Homo sapiens cDNA FLJ10061 fis, clone HEMBA1001413. | −5.87671 | 0.00024 |
| NM_033551 | LARP1 | La ribonucleoprotein domain family, member 1 | −7.90965 | 0.00002 |
| NM_015051 | TXNDC4 | Thioredoxin domain containing 4 (endoplasmic reticulum) | −6.64755 | 0.00009 |
| THC2339776 | THC2339776 | Unknown | −8.41951 | 0.00001 |
| AK024900 | AK024900 | CDNA: FLJ21247 fis, clone COL01205 | −7.56648 | 0.00003 |
| A_24_P799580 | A_24_P799580 | Unknown | 6.60497 | 0.0001 |
| ENST00000360523 | ENST00000360523 | GB|AB065479.1|BAC05733.1 seven transmembrane helix receptor [Homo sapiens] [NP511107] | 6.64141 | 0.00009 |
| A_24_P32836 | A_24_P32836 | Unknown | −6.19519 | 0.00016 |
| AB020633 | FRYL | FRY-like | −6.38433 | 0.00013 |
| NM_002118 | HLA-DMB | Major histocompatibility complex, class II, DM beta | −5.54567 | 0.00036 |
| THC2408277 | THC2408277 | Unknown | −5.38709 | 0.00044 |
| NM_017994 | C7orf42 | Chromosome 7 open reading frame 42 | −6.74705 | 0.00008 |
| NM_001659 | ARF3 | ADP-ribosylation factor 3 | 5.1421 | 0.00061 |
| NM_001752 | CAT | Catalase | −5.64093 | 0.00032 |
| NM_020657 | ZNF304 | Zinc finger protein 304 | −6.87846 | 0.00007 |
| BC056896 | HSPC157 | HSPC157 protein | −5.44409 | 0.00041 |
| NM_014859 | KIAA0672 | KIAA0672 gene product | −7.21036 | 0.00005 |
| NM_178520 | TMEM105 | Transmembrane protein 105 | 6.20265 | 0.00016 |
| NM_001011538 | LOC402176 | similar to 60S ribosomal protein L21 | −7.23041 | 0.00005 |
| NM_133446 | CTGLF1 | Centaurin, gamma-like family, member 1 | −7.90796 | 0.00002 |
| NM_194314 | ZBTB41 | Zinc finger and BTB domain containing 41 | −7.18381 | 0.00005 |
| NM_015609 | C1orf144 | Chromosome 1 open reading frame 144 | 5.0769 | 0.00067 |
| BC043279 | BC043279 | CDNA clone IMAGE:5297259 | −6.22554 | 0.00015 |
| NM_053042 | KIAA1729 | KIAA1729 protein | −7.36048 | 0.00004 |
| AK096054 | SLC6A19 | Solute carrier family 6 (neutral amino acid transporter), member 19 | 6.73775 | 0.00008 |
| NM_014990 | GARNL1 | GTPase activating Rap/RanGAP domain-like 1 | −8.21092 | 0.00002 |
| NM_014362 | HIBCH | 3-hydroxyisobutyryl-Coenzyme A hydrolase | −5.28173 | 0.00051 |
| NM_006804 | STARD3 | START domain containing 3 | 6.12902 | 0.00017 |
| NM_001042 | SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | 8.12027 | 0.00002 |
| NM_001040440 | CCDC112 | Coiled-coil domain containing 112 | −4.8855 | 0.00086 |
| NM_020918 | GPAM | Glycerol-3-phosphate acyltransferase, mitochondrial | −6.05652 | 0.00019 |
| NM_020381 | PDSS2 | Prenyl (decaprenyl) diphosphate synthase, subunit 2 | −5.38002 | 0.00044 |
| NM_001029857 | LOC158381 | hypothetical protein LOC158381 | 6.13599 | 0.00017 |
| BX641669 | ACOX1 | Acyl-Coenzyme A oxidase 1, palmitoyl | −6.4566 | 0.00012 |
| NM_033161 | SURF4 | Surfeit 4 | 6.55595 | 0.0001 |
| NM_144673 | CMTM2 | CKLF-like MARVEL transmembrane domain containing 2 | 5.76847 | 0.00027 |
| NM_015659 | RSL1D1 | Ribosomal L1 domain containing 1 | 5.34644 | 0.00046 |
| AK021842 | FLJ25476 | FLJ25476 protein | 6.14491 | 0.00017 |
| NM_183233 | SLC22A18 | Solute carrier family 22 (organic cation transporter), member 18 | 5.61224 | 0.00033 |
| NM_024649 | BBS1 | Bardet-Biedl syndrome 1 | −6.4087 | 0.00012 |
| ENST00000373264 | ENST00000373264 | Unknown | 5.31863 | 0.00048 |
| NM_025228 | TRAF3IP3 | TRAF3 interacting protein 3 | −5.6509 | 0.00031 |
| NM_005718 | ARPC4 | Actin related protein 2/3 complex, subunit 4, 20kDa | 5.28128 | 0.00051 |
| NM_007234 | DCTN3 | Dynactin 3 (p22) | 5.24163 | 0.00053 |
| NM_014949 | KIAA0907 | KIAA0907 | 7.69915 | 0.00003 |
| NM_001024847 | TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | −7.62359 | 0.00003 |
| A_24_P399341 | A_24_P399341 | Unknown | −6.21519 | 0.00016 |
| AK098491 | OR5T2 | RPA interacting protein | −5.42913 | 0.00042 |
| BC039296 | C20orf19 | Chromosome 20 open reading frame 19 | −5.18365 | 0.00058 |
| NM_005026 | PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | 6.21873 | 0.00016 |
| NM_005119 | THRAP3 | Thyroid hormone receptor associated protein 3 | 8.12691 | 0.00002 |
| AB051518 | KIAA1731 | KIAA1731 | −5.99995 | 0.0002 |
| BC012484 | TNRC15 | Homo sapiens trinucleotide repeat containing 15, mRNA (cDNA clone IMAGE:4472100), partial cds. | −6.86227 | 0.00007 |
| NM_001119 | ADD1 | Adducin 1 (alpha) | 6.32625 | 0.00014 |
| NM_173511 | ALS2CR13 | Amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 13 | −4.78976 | 0.00099 |
| NM_014813 | LRIG2 | Leucine-rich repeats and immunoglobulin-like domains 2 | −6.24565 | 0.00015 |
| NM_015143 | METAP1 | Methionyl aminopeptidase 1 | 6.82305 | 0.00008 |
| NM_206962 | PRMT2 | Protein arginine methyltransferase 2 | 5.94335 | 0.00022 |
| AK001118 | UBR2 | Ubiquitin protein ligase E3 component n-recognin 2 | −4.83769 | 0.00092 |
| THC2314600 | THC2314600 | Unknown | −7.0493 | 0.00006 |
| NM_002950 | RPN1 | Ribophorin I | −7.71446 | 0.00003 |
| NM_014969 | WDR47 | WD repeat domain 47 | −4.99127 | 0.00075 |
| AK130862 | FLJ27352 | hypothetical LOC145788 | −6.98877 | 0.00006 |
| NM_003104 | SORD | Sorbitol dehydrogenase | −5.18411 | 0.00058 |
| NM_033017 | TRIM4 | Tripartite motif-containing 4 | 5.35891 | 0.00046 |
| AK025221 | RAD50 | RAD50 homolog (S. cerevisiae) | −7.71671 | 0.00003 |
| NM_144664 | FAM76B | Family with sequence similarity 76, member B | −5.5059 | 0.00038 |
| NM_012219 | MRAS | Muscle RAS oncogene homolog | 6.83581 | 0.00008 |
| THC2437476 | THC2437476 | Unknown | −7.20877 | 0.00005 |
| BX116997 | BX116997 | Transcribed locus | −6.06866 | 0.00019 |
| NM_016272 | TOB2 | Transducer of ERBB2, 2 | −5.96088 | 0.00021 |
| NM_023111 | FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | 5.0047 | 0.00073 |
| NM_152756 | RICTOR | rapamycin-insensitive companion of mTOR | −5.79934 | 0.00026 |
| CR590163 | KIAA2018 | KIAA2018 | −5.82281 | 0.00025 |
| NM_006141 | DYNC1LI2 | Dynein, cytoplasmic 1, light intermediate chain 2 | −5.10652 | 0.00064 |
| NM_005088 | CXYorf3 | Chromosome X and Y open reading frame 3 | −5.75059 | 0.00028 |
| NM_003309 | TSPYL1 | TSPY-like 1 | −6.02861 | 0.0002 |
| NM_004891 | MRPL33 | Mitochondrial ribosomal protein L33 | −7.30793 | 0.00005 |
| NM_033406 | FBXO3 | F-box protein 3 | 6.64954 | 0.00009 |
| NM_001037533 | GON4L | YY1 associated protein 1 | −7.07118 | 0.00006 |
| NM_004323 | BAG1 | BCL2-associated athanogene | −4.94334 | 0.0008 |
| NM_198567 | C5orf25 | Chromosome 5 open reading frame 25 | −6.25358 | 0.00015 |
| NM_006709 | EHMT2 | Euchromatic histone-lysine N-methyltransferase 2 | 5.73631 | 0.00028 |
| NM_004375 | COX11 | COX11 homolog, cytochrome c oxidase assembly protein (yeast) | −4.9474 | 0.00079 |
| AK074291 | HIPK2 | Homeodomain interacting protein kinase 2 | −7.86645 | 0.00003 |
| NM_001032289 | SLC35A2 | Solute carrier family 35 (UDP-galactose transporter), member A2 | −8.23296 | 0.00002 |
| NM_002778 | PSAP | Prosaposin (variant Gaucher disease and variant metachromatic leukodystrophy) | 4.87526 | 0.00088 |
| NM_012330 | MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | −6.78062 | 0.00008 |
| AW518445 | RPS23 | Ribosomal protein S23 | −6.93887 | 0.00007 |
| NM_021260 | ZFYVE1 | Zinc finger, FYVE domain containing 1 | −5.06342 | 0.00068 |
| NM_005052 | RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | 7.20917 | 0.00005 |
| A_24_P24790 | A_24_P24790 | Unknown | −5.64628 | 0.00032 |
| A_32_P122285 | A_32_P122285 | Unknown | −5.35697 | 0.00046 |
| NM_006885 | ATBF1 | AT-binding transcription factor 1 | 7.13739 | 0.00005 |
| NM_020382 | SETD8 | SET domain containing (lysine methyltransferase) 8 | 5.29077 | 0.0005 |
| NM_000202 | IDS | Iduronate 2-sulfatase (Hunter syndrome) | −5.14859 | 0.0006 |
| A_24_P93184 | A_24_P93184 | Unknown | 5.20135 | 0.00056 |
| NM_002557 | OVGP1 | Oviductal glycoprotein 1, 120kDa (mucin 9, oviductin) | −6.11482 | 0.00018 |
| U68019 | SMAD3 | SMAD family member 3 | −5.73809 | 0.00028 |
| NM_001120 | TETRAN | tetracycline transporter-like protein | 5.16055 | 0.00059 |
| A_24_P298616 | A_24_P298616 | Unknown | −5.85459 | 0.00024 |
| NM_199341 | LOC374920 | hypothetical protein LOC374920 | −6.70474 | 0.00009 |
| AK025695 | TMED8 | Glutathione transferase zeta 1 (maleylacetoacetate isomerase) | −6.84167 | 0.00008 |
| NM_002721 | PPP6C | Protein phosphatase 6, catalytic subunit | −6.83703 | 0.00008 |
| NM_001229 | CASP9 | Caspase 9, apoptosis-related cysteine peptidase | −7.15685 | 0.00005 |
| NM_017541 | CRYGS | Crystallin, gamma S | −6.28412 | 0.00014 |
| NM_001013406 | KRIT1 | KRIT1, ankyrin repeat containing | 8.09633 | 0.00002 |
| NM_005867 | DSCR4 | Down syndrome critical region gene 4 | 4.79505 | 0.00098 |
| NM_004187 | JARID1C | Jumonji, AT rich interactive domain 1C | −4.8437 | 0.00092 |
| A_32_P32905 | A_32_P32905 | Unknown | −5.02028 | 0.00072 |
| XM_934270 | LOC647065 | hypothetical LOC647065 | −5.78325 | 0.00026 |
| AK097149 | SPPL3 | signal peptide peptidase 3 | −5.07639 | 0.00067 |
| NM_002004 | FDPS | Farnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) | −7.93324 | 0.00002 |
| A_24_P109962 | A_24_P109962 | Unknown | 6.63781 | 0.0001 |
| AK092875 | AK092875 | CDNA FLJ35556 fis, clone SPLEN2004844 | 6.52427 | 0.00011 |
| NM_014847 | UBAP2L | Ubiquitin associated protein 2-like | 7.49946 | 0.00004 |
| AA902595 | IQGAP1 | IQ motif containing GTPase activating protein 1 | −7.31533 | 0.00004 |
| AK125206 | LOC149448 | hypothetical protein LOC149448 | −7.45844 | 0.00004 |
| A_24_P812265 | A_24_P812265 | Unknown | 5.37664 | 0.00045 |
| A_32_P171984 | A_32_P171984 | Unknown | 5.94952 | 0.00022 |
| XM_373367 | A_24_P401150 | Unknown | 4.88814 | 0.00086 |
| NM_030809 | C12orf22 | Chromosome 12 open reading frame 22 | −6.26667 | 0.00015 |
| NM_001014436 | DBNL | Drebrin-like | 5.82723 | 0.00025 |
| NM_014774 | KIAA0494 | ATP synthase mitochondrial F1 complex assembly factor 1 | 6.08021 | 0.00018 |
| NM_005482 | PIGK | Phosphatidylinositol glycan anchor biosynthesis, class K | −6.21898 | 0.00016 |
| NM_174898 | LOC129530 | hypothetical protein LOC129530 | −6.1107 | 0.00018 |
| NM_201434 | RAB5C | RAB5C, member RAS oncogene family | 5.60187 | 0.00033 |
| AK095151 | EDD1 | E3 ubiquitin protein ligase, HECT domain containing, 1 | −5.90406 | 0.00023 |
| NM_152409 | C5orf24 | Chromosome 5 open reading frame 24 | 5.18137 | 0.00058 |
| A_24_P626812 | A_24_P626812 | Unknown | 5.48701 | 0.00039 |
| NM_001521 | GTF3C2 | General transcription factor IIIC, polypeptide 2, beta 110kDa | −5.31194 | 0.00049 |
| NM_018116 | MSTO1 | Death associated protein 3 | 5.70862 | 0.00029 |
| NM_031483 | ITCH | Itchy homolog E3 ubiquitin protein ligase (mouse) | −5.65809 | 0.00031 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −6.76771 | 0.00008 |
| NM_005692 | ABCF2 | ATP-binding cassette, sub-family F (GCN20), member 2 | 4.85315 | 0.0009 |
| NM_007249 | KLF12 | Kruppel-like factor 12 | −5.04433 | 0.0007 |
| AK125099 | AK125099 | CDNA FLJ43109 fis, clone CTONG2025516, moderately similar to Homo sapiens general transcription factor II, i (GTF2I) | 6.01691 | 0.0002 |
| NM_203447 | DOCK8 | Dedicator of cytokinesis 8 | −7.32816 | 0.00004 |
| BC020955 | TBC1D23 | TBC1 domain family, member 23 | −6.35938 | 0.00013 |
| BC045820 | C21orf51 | Chromosome 21 open reading frame 51 | −5.01304 | 0.00073 |
| AY831680 | HC1 | HC1 (HC1) | −6.02551 | 0.0002 |
| NM_022894 | PAPOLG | Poly(A) polymerase gamma | −7.18187 | 0.00005 |
| NM_018371 | ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | 5.05306 | 0.00069 |
| A_32_P211048 | A_32_P211048 | Unknown | −5.23129 | 0.00054 |
| NM_020235 | BBX | Bobby sox homolog (Drosophila) | 5.65165 | 0.00031 |
| AL117511 | C14orf125 | Homo sapiens mRNA; cDNA DKFZp434I1735 (from clone DKFZp434I1735); partial cds. | −5.13994 | 0.00061 |
| AK024925 | FLJ21272 | hypothetical protein FLJ21272 | −5.11626 | 0.00063 |
| NM_021100 | NFS1 | NFS1 nitrogen fixation 1 homolog (S. cerevisiae) | −6.4712 | 0.00012 |
| NM_006133 | C11orf11 | Chromosome 11 open reading frame 11 | 6.22281 | 0.00015 |
| AL832861 | MTERF | Mitochondrial transcription termination factor | −6.82522 | 0.00008 |
| NM_199320 | PHF17 | PHD finger protein 17 | −5.41371 | 0.00043 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −5.8857 | 0.00023 |
| NM_145233 | ZNF625 | Zinc finger protein 20 | 5.42341 | 0.00042 |
| NM_032782 | HAVCR2 | Hepatitis A virus cellular receptor 2 | −5.54466 | 0.00036 |
| XM_497894 | A_24_P24912 | Unknown | 5.83076 | 0.00025 |
| NM_015530 | GORASP2 | Golgi reassembly stacking protein 2, 55kDa | 5.85766 | 0.00024 |
| NM_172241 | CTAGE1 | Cutaneous T-cell lymphoma-associated antigen 1 | 7.17695 | 0.00005 |
| AK001173 | AQR | Aquarius homolog (mouse) | −7.77232 | 0.00003 |
| NM_001299 | CNN1 | Calponin 1, basic, smooth muscle | 5.51348 | 0.00037 |
| AB058774 | ZNF594 | Zinc finger protein 594 | −5.7672 | 0.00027 |
| NM_001607 | ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | 5.0508 | 0.00069 |
| NM_001005526 | SF3B1 | Splicing factor 3b, subunit 1, 155kDa | 5.20139 | 0.00056 |
| NM_006241 | PPP1R2 | Protein phosphatase 1, regulatory (inhibitor) subunit 2 | −5.25608 | 0.00052 |
| W18193 | THC2304444 | IMAGE:20064 Soares infant brain 1NIB Homo sapiens cDNA clone IMAGE:20064, mRNA sequence. | −5.51323 | 0.00037 |
| AK125922 | CSNK2A2 | Casein kinase 2, alpha prime polypeptide | −7.4646 | 0.00004 |
| BU567832 | BU567832 | Transcribed locus | −4.93845 | 0.0008 |
| NM_000791 | DHFR | Dihydrofolate reductase | 4.95027 | 0.00079 |
| BC104478 | RPL21 | Ribosomal protein L21 | −6.41461 | 0.00012 |
| NM_001040431 | CCDC56 | Coiled-coil domain containing 56 | 5.15456 | 0.0006 |
| AJ420510 | PHF10 | PHD finger protein 10 | −6.30643 | 0.00014 |
| BC011660 | BC011660 | Homo sapiens cDNA clone IMAGE:4120690, **** WARNING: chimeric clone ****. | 5.70061 | 0.00029 |
| NM_025164 | KIAA0999 | KIAA0999 protein | −6.08738 | 0.00018 |
| NM_006195 | PBX3 | Pre-B-cell leukemia transcription factor 3 | −6.5323 | 0.00011 |
| NM_014149 | HSPC049 | HSPC049 protein | −5.65533 | 0.00031 |
| NM_018029 | FLJ10213 | hypothetical protein FLJ10213 | −5.53682 | 0.00036 |
| XM_373538 | ENST00000344556 | Unknown | 7.28826 | 0.00005 |
| THC2316878 | THC2316878 | Unknown | −5.50095 | 0.00038 |
| NM_003634 | NIPSNAP1 | Nipsnap homolog 1 (C. elegans) | 5.96498 | 0.00021 |
| NM_153631 | HOXA3 | Homeobox A5 | 7.53786 | 0.00004 |
| THC2404993 | THC2404993 | Unknown | −4.88629 | 0.00086 |
| NM_001008709 | PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | 4.95731 | 0.00078 |
| NM_000889 | ITGB7 | Integrin, beta 7 | −5.0143 | 0.00072 |
| NM_014033 | METTL7A | LETM1 domain containing 1 | −5.10185 | 0.00064 |
| NM_023926 | ZNF447 | Zinc finger protein 447 | 7.08335 | 0.00006 |
| NM_007139 | ZNF92 | Zinc finger protein 92 | −5.33052 | 0.00047 |
| NM_032645 | RAPSN | Receptor-associated protein of the synapse, 43kD | 7.31724 | 0.00004 |
| AI299143 | LOC51255 | hypothetical protein LOC51255 | −6.62138 | 0.0001 |
| NM_014624 | S100A6 | S100 calcium binding protein A6 | 5.69194 | 0.0003 |
| NM_032837 | FAM104A | Family with sequence similarity 104, member A | 7.10417 | 0.00006 |
| NM_005292 | GPR18 | G protein-coupled receptor 18 | −5.39734 | 0.00043 |
| AK001179 | ANKIB1 | Ankyrin repeat and IBR domain containing 1 | 6.78686 | 0.00008 |
| AK097258 | DOK3 | Docking protein 3 | −6.21585 | 0.00016 |
| NM_175852 | TXLNA | Taxilin alpha | 5.07701 | 0.00067 |
| NM_006231 | POLE | Polymerase (DNA directed), epsilon | 5.3557 | 0.00046 |
| NM_015216 | HISPPD1 | Histidine acid phosphatase domain containing 1 | −5.75216 | 0.00028 |
| NM_020400 | GPR92 | G protein-coupled receptor 92 | −6.72524 | 0.00009 |
| NM_020781 | ZNF398 | Zinc finger protein 398 | −6.47114 | 0.00012 |
| A_24_P632160 | A_24_P632160 | Unknown | 5.12173 | 0.00063 |
| NM_014224 | PGA5 | Pepsinogen 5, group I (pepsinogen A) | 5.1415 | 0.00061 |
| NM_014824 | FCHSD2 | FCH and double SH3 domains 2 | 7.01313 | 0.00006 |
| NM_015723 | PNPLA8 | Patatin-like phospholipase domain containing 8 | 7.24769 | 0.00005 |
| AK001179 | ANKIB1 | Ankyrin repeat and IBR domain containing 1 | −5.9491 | 0.00022 |
| AA420998 | LOC286076 | hypothetical protein LOC286076 | 5.31666 | 0.00048 |
| NM_033184 | KRTAP2-4 | Keratin associated protein 2-4 | 6.81809 | 0.00008 |
| NM_033481 | FBXO9 | F-box protein 9 | −5.53612 | 0.00036 |
| NM_020964 | KIAA1632 | KIAA1632 | −5.70799 | 0.00029 |
| NM_000405 | GM2A | GM2 ganglioside activator | 4.81139 | 0.00096 |
| NM_024713 | C15orf29 | Chromosome 15 open reading frame 29 | 6.26529 | 0.00015 |
| NM_006620 | HBS1L | HBS1-like (S. cerevisiae) | −5.65177 | 0.00031 |
| NM_153353 | LRRC34 | Leucine rich repeat containing 34 | −5.18522 | 0.00058 |
| NM_032271 | TRAF7 | TNF receptor-associated factor 7 | 5.42648 | 0.00042 |
| NM_002498 | NEK3 | NIMA (never in mitosis gene a)-related kinase 3 | −6.56948 | 0.0001 |
| BQ188033 | EPB41L4A | Erythrocyte membrane protein band 4.1 like 4A | −7.30113 | 0.00005 |
| AK025056 | ZYG11B | Zyg-11 homolog B (C. elegans) | −6.13158 | 0.00017 |
| AK126501 | CDC2L2 | Cell division cycle 2-like 2 (PITSLRE proteins) | −7.34404 | 0.00004 |
| NM_001945 | HBEGF | Heparin-binding EGF-like growth factor | 5.29197 | 0.0005 |
| AK093505 | CXorf18 | SPANX family, member A2 | 7.21242 | 0.00005 |
| NM_022838 | ARMCX5 | G protein-coupled receptor associated sorting protein 2 | −6.83633 | 0.00008 |
| NM_002883 | RANGAP1 | Ran GTPase activating protein 1 | 5.21738 | 0.00055 |
| NM_018448 | CAND1 | Cullin-associated and neddylation-dissociated 1 | 6.43506 | 0.00012 |
| NM_172193 | KLHDC1 | Kelch domain containing 1 | −4.97574 | 0.00076 |
| NM_002874 | RAD23B | RAD23 homolog B (S. cerevisiae) | 5.10293 | 0.00064 |
| NM_000402 | G6PD | Glucose-6-phosphate dehydrogenase | 4.90285 | 0.00084 |
| NM_004665 | VNN2 | Vanin 2 | −5.49775 | 0.00038 |
| AF111848 | PRO0529 | Homo sapiens PRO0529 mRNA, complete cds. | 7.2649 | 0.00005 |
| A_32_P83174 | A_32_P83174 | Unknown | −6.08174 | 0.00018 |
| BE044472 | C22orf27 | Chromosome 22 open reading frame 27 | −6.12607 | 0.00017 |
| NM_001012754 | LOC387921 | similar to RIKEN cDNA 8030451K01 | −6.13156 | 0.00017 |
| NM_020382 | SETD8 | SET domain containing (lysine methyltransferase) 8 | 6.25922 | 0.00015 |
| NM_006313 | USP15 | Ubiquitin specific peptidase 15 | −5.27031 | 0.00051 |
| NM_004727 | SLC24A1 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 | −5.58374 | 0.00034 |
| XM_371160 | ENST00000310822 | PREDICTED: Homo sapiens similar to ribosomal protein L21 isoform 1 (LOC388532), mRNA [XM_371160] | −6.21526 | 0.00016 |
| NM_004879 | EI24 | Etoposide induced 2.4 mRNA | 4.81487 | 0.00095 |
| NM_018660 | ZNF395 | Zinc finger protein 395 | −6.76571 | 0.00008 |
| NM_005880 | DNAJA2 | DnaJ (Hsp40) homolog, subfamily A, member 2 | 5.86414 | 0.00024 |
| NM_014017 | MAPBPIP | mitogen-activated protein-binding protein-interacting protein | 6.46094 | 0.00012 |
| NM_013339 | ALG6 | Asparagine-linked glycosylation 6 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | −5.2637 | 0.00052 |
| THC2375818 | THC2375818 | Q15464 (Q15464) Shb, partial (4%) [THC2375818] | 5.43053 | 0.00042 |
| NM_015227 | POFUT2 | Protein O-fucosyltransferase 2 | −5.82663 | 0.00025 |
| NM_018367 | PHCA | Phytoceramidase, alkaline | 5.4057 | 0.00043 |
| AK124515 | AK124515 | CDNA FLJ30141 fis, clone BRACE2000148 | −5.80037 | 0.00026 |
| AK025335 | AP1GBP1 | AP1 gamma subunit binding protein 1 | −6.17463 | 0.00016 |
| NM_145867 | LTC4S | Leukotriene C4 synthase | 5.71862 | 0.00029 |
| A_24_P50707 | A_24_P50707 | Unknown | −7.21616 | 0.00005 |
| NM_014990 | GARNL1 | GTPase activating Rap/RanGAP domain-like 1 | −6.79782 | 0.00008 |
| NM_005803 | FLOT1 | Flotillin 1 | 4.88546 | 0.00086 |
| NM_032230 | C12orf26 | Chromosome 12 open reading frame 26 | −5.72089 | 0.00029 |
| NM_000632 | ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | −6.173 | 0.00016 |
| AK125015 | MTHFSD | Homo sapiens cDNA FLJ43025 fis, clone BRTHA2018707. | −6.19194 | 0.00016 |
| NM_003705 | SLC25A12 | Solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | 5.56608 | 0.00035 |
| NM_032632 | PAPOLA | Poly(A) polymerase alpha | −6.93832 | 0.00007 |
| NM_007331 | WHSC1 | Wolf-Hirschhorn syndrome candidate 1 | −5.54554 | 0.00036 |
| BM711907 | THC2373429 | Transcribed locus | −5.5243 | 0.00037 |
| A_24_P264413 | A_24_P264413 | Unknown | 4.86773 | 0.00089 |
| NM_032778 | MINA | MYC induced nuclear antigen | 5.1964 | 0.00057 |
| AK095260 | FLJ10769 | hypothetical protein FLJ10769 | 6.78238 | 0.00008 |
| NM_004930 | CAPZB | Capping protein (actin filament) muscle Z-line, beta | 5.14986 | 0.0006 |
| NM_003951 | SLC25A14 | Solute carrier family 25 (mitochondrial carrier, brain), member 14 | −5.04853 | 0.00069 |
| NM_012108 | BRDG1 | BCR downstream signaling 1 | −5.17296 | 0.00058 |
| NM_000788 | DCK | Deoxycytidine kinase | −6.25805 | 0.00015 |
| NM_019015 | CSGlcA-T | chondroitin sulfate glucuronyltransferase | 5.53232 | 0.00036 |
| NM_001040150 | C15orf28 | Acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | −5.31078 | 0.00049 |
| NM_052839 | PANX2 | Pannexin 2 | 5.81643 | 0.00025 |
| NM_001024732 | MECR | Mitochondrial trans-2-enoyl-CoA reductase | 6.46046 | 0.00012 |
| NM_018949 | UTS2R | Urotensin 2 receptor | 6.14462 | 0.00017 |
| NM_000786 | CYP51A1 | Cytochrome P450, family 51, subfamily A, polypeptide 1 | −5.98488 | 0.00021 |
| NM_001361 | DHODH | Dihydroorotate dehydrogenase | −4.81064 | 0.00096 |
| THC2442489 | THC2442489 | Unknown | −5.7987 | 0.00026 |
| AK024144 | FLJ14082 | hypothetical protein FLJ14082 | 5.0193 | 0.00072 |
| THC2315966 | THC2315966 | Unknown | 5.44628 | 0.00041 |
| THC2279305 | THC2279305 | Unknown | −6.62236 | 0.0001 |
| BG768878 | THC2312236 | 602743557F1 NIH_MGC_49 Homo sapiens cDNA clone IMAGE:4872878 5', mRNA sequence. | 5.64061 | 0.00032 |
| NM_001031735 | C19orf36 | Chromosome 19 open reading frame 36 | 5.29118 | 0.0005 |
| NM_001329 | CTBP2 | C-terminal binding protein 2 | 6.02499 | 0.0002 |
| NM_007372 | DDX42 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | 5.48554 | 0.00039 |
| AF343078 | ATAD3B | Homo sapiens TOB3 mRNA, complete cds. | 5.06251 | 0.00068 |
| NM_001013842 | C8orf58 | Chromosome 8 open reading frame 58 | −5.24475 | 0.00053 |
| AV739735 | HNRPC | Transcribed locus, moderately similar to NP_001020804.1 nuclear ribonucleoprotein C [Rattus norvegicus] | 5.62056 | 0.00033 |
| NM_002641 | PIGA | Phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) | −5.71759 | 0.00029 |
| NM_018047 | RBM22 | RNA binding motif protein 22 | 6.25385 | 0.00015 |
| NM_153187 | SLC22A1 | Solute carrier family 22 (organic cation transporter), member 1 | 5.05565 | 0.00069 |
| AK127081 | USP54 | Ubiquitin specific peptidase 54 | −6.40172 | 0.00013 |
| AK125832 | TBC1D3C | TBC1 domain family, member 3C | −5.35704 | 0.00046 |
| CR619110 | LOC401588 | hypothetical LOC401588 | −5.72382 | 0.00029 |
| NM_003861 | WDR22 | WD repeat domain 22 | −6.2627 | 0.00015 |
| NM_138363 | CCDC45 | Coiled-coil domain containing 45 | −4.87186 | 0.00088 |
| A_32_P55799 | A_32_P55799 | Unknown | −5.05573 | 0.00068 |
| NM_014940 | MON1B | MON1 homolog B (yeast) | 7.0631 | 0.00006 |
| BX648857 | SP3 | Sp3 transcription factor | −5.68022 | 0.0003 |
| NM_006120 | HLA-DMA | Major histocompatibility complex, class II, DM beta | −5.36711 | 0.00045 |
| NM_006669 | LILRB1 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | −6.90789 | 0.00007 |
| BQ189538 | THC2316936 | Transcribedlocus | −5.01081 | 0.00073 |
| NM_015205 | ATP11A | ATPase, Class VI, type 11A | −6.56291 | 0.0001 |
| NM_138364 | LOC90826 | hypothetical protein BC004337 | −5.01733 | 0.00072 |
| BC089156 | BC089156 | Homo sapiens, clone IMAGE:4346533, mRNA | −5.40338 | 0.00043 |
| A_24_P16273 | A_24_P16273 | Unknown | 5.78523 | 0.00026 |
| CR606044 | LOC93622 | hypothetical protein BC006130 | −4.92763 | 0.00082 |
| NM_199044 | NSUN4 | NOL1/NOP2/Sun domain family, member 4 | 5.527 | 0.00037 |
| NM_000455 | STK11 | Serine/threonine kinase 11 | 6.37056 | 0.00013 |
| NM_173694 | ATP11C | ATPase, Class VI, type 11C | −6.79799 | 0.00008 |
| NM_006962 | ZNF182 | Zinc finger protein 182 | −5.38748 | 0.00044 |
| BM680823 | BM680823 | Transcribed locus | −5.0325 | 0.00071 |
| BC053353 | PRPF38B | CDNA clone IMAGE:5246408 | −5.95255 | 0.00021 |
| AB014514 | FLJ30092 | AF-1 specific protein phosphatase | 6.40387 | 0.00012 |
| NM_001152 | SLC25A5 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | 4.99296 | 0.00075 |
| BF213738 | CLEC2B | C-type lectin domain family 2, member B | −6.15737 | 0.00017 |
| NM_025231 | ZNF435 | Zinc finger protein 435 | −6.70982 | 0.00009 |
| NM_001076786 | QSER1 | Glutamine and serine rich 1 | −5.86969 | 0.00024 |
| NM_019007 | ARMCX6 | Armadillo repeat containing, X-linked 6 | −5.98809 | 0.00021 |
| NM_145911 | ZNF23 | Ribosomal protein L27 | −6.73374 | 0.00009 |
| NM_024819 | DCAKD | Dephospho-CoA kinase domain containing | −4.9368 | 0.00081 |
| NM_005207 | CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | 5.38662 | 0.00044 |
| NM_175736 | FMNL3 | Formin-like 3 | 5.60824 | 0.00033 |
| NM_017794 | KIAA1797 | KIAA1797 | −5.35919 | 0.00046 |
| NM_004074 | COX8A | Cytochrome c oxidase subunit 8A (ubiquitous) | 6.05767 | 0.00019 |
| AF144054 | AF144054 | Apoptosis related protein APR-4 | 6.06285 | 0.00019 |
| THC2344420 | THC2344420 | ALU7_HUMAN (P39194) Alu subfamily SQ sequence contamination warning entry, partial (21%) [THC2344420] | −5.13064 | 0.00062 |
| AF229166 | LOC220077 | hypothetical gene supported by AF229166 | 5.41636 | 0.00042 |
| AB014550 | SMCHD1 | Structural maintenance of chromosomes flexible hinge domain containing 1 | −6.90178 | 0.00007 |
| NM_024738 | C12orf49 | Chromosome 12 open reading frame 49 | 5.44473 | 0.00041 |
| NM_153649 | TPM3 | Tropomyosin 3 | 5.58209 | 0.00034 |
| NM_012247 | SEPHS1 | Selenophosphate synthetase 1 | 4.8008 | 0.00097 |
| NM_014339 | IL17RA | Interleukin 17 receptor A | −4.86703 | 0.00089 |
| NM_000534 | PMS1 | PMS1 postmeiotic segregation increased 1 (S. cerevisiae) | −5.93393 | 0.00022 |
| AL832783 | LMLN | Leishmanolysin-like (metallopeptidase M8 family) | −4.79121 | 0.00099 |
| NM_015525 | IBTK | Inhibitor of Bruton agammaglobulinemia tyrosine kinase | −6.3151 | 0.00014 |
| A_32_P77421 | A_32_P77421 | Unknown | −6.30639 | 0.00014 |
| NM_019843 | EIF4ENIF1 | Eukaryotic translation initiation factor 4E nuclear import factor 1 | −6.83768 | 0.00008 |
| NM_024898 | DENND1C | DENN/MADD domain containing 1C | −6.91959 | 0.00007 |
| NM_172217 | IL16 | Interleukin 16 (lymphocyte chemoattractant factor) | −5.28949 | 0.0005 |
| NM_001192 | TNFRSF17 | Tumor necrosis factor receptor superfamily, member 17 | −6.95798 | 0.00007 |
| NM_001010853 | ACY1L2 | Aminoacylase 1-like 2 | −6.81869 | 0.00008 |
| THC2428671 | THC2428671 | Unknown | −6.52112 | 0.00011 |
| NM_004423 | DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | 5.46922 | 0.0004 |
| BC040695 | WBSCR16 | Williams-Beuren syndrome chromosome region 16 | −5.70567 | 0.00029 |
| A_24_P418590 | A_24_P418590 | Unknown | −5.20758 | 0.00056 |
| NM_001018062 | LOC51149 | hypothetical LOC51149 | −4.88888 | 0.00086 |
| NM_017869 | BANP | BTG3 associated nuclear protein | 5.46398 | 0.0004 |
| D86980 | TTC9 | Tetratricopeptide repeat domain 9 | −5.47605 | 0.00039 |
| A_24_P333077 | A_24_P333077 | Unknown | 5.92063 | 0.00022 |
| NM_001556 | IKBKB | Golgi autoantigen, golgin subfamily a, 7 | −6.58675 | 0.0001 |
| NM_001008225 | CNOT4 | CCR4-NOT transcription complex, subunit 4 | 4.9542 | 0.00079 |
| NM_021173 | POLD4 | Polymerase (DNA-directed), delta 4 | 5.57566 | 0.00034 |
| NM_031303 | KATNAL2 | Katanin p60 subunit A-like 2 | −6.56838 | 0.0001 |
| NM_007234 | DCTN3 | Dynactin 3 (p22) | 5.07507 | 0.00067 |
| NM_022845 | CBFB | Core-binding factor, beta subunit | 6.17287 | 0.00016 |
| XM_167254 | LOC402643 | PREDICTED: Homo sapiens similar to tropomyosin 3 isoform 2 (LOC402643), mRNA [XM_167254] | 4.89622 | 0.00085 |
| NM_032868 | FLJ14981 | hypothetical protein FLJ14981 | 5.68109 | 0.0003 |
| NM_013296 | GPSM2 | G-protein signalling modulator 2 (AGS3-like, C. elegans) | 5.72586 | 0.00028 |
| NM_006134 | TMEM50B | Transmembrane protein 50B | 6.77942 | 0.00008 |
| NM_001781 | CD69 | CD69molecule | −5.92149 | 0.00022 |
| NM_021818 | SAV1 | Salvador homolog 1 (Drosophila) | −5.9986 | 0.0002 |
| NM_174913 | C14orf21 | Cell death-inducing DFFA-like effector b | 6.12658 | 0.00017 |
| NM_013943 | CLIC4 | Chloride intracellular channel 4 | 5.03541 | 0.0007 |
| NM_003161 | RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | −5.92216 | 0.00022 |
| NM_153649 | TPM3 | Tropomyosin 3 | −6.5978 | 0.0001 |
| NM_198040 | PHC2 | Polyhomeotic homolog 2 (Drosophila) | 6.49653 | 0.00011 |
| NM_014766 | SCRN1 | Secernin 1 | −5.9542 | 0.00021 |
| X78926 | ZNF268 | Phosphatidylinositol glycan anchor biosynthesis, class F | −6.26836 | 0.00015 |
| NM_005051 | QARS | Glutaminyl-tRNA synthetase | 5.02506 | 0.00071 |
| NM_032272 | MAF1 | MAF1 homolog (S. cerevisiae) | 5.03895 | 0.0007 |
| NM_178862 | STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | 5.53837 | 0.00036 |
| NM_031157 | HNRPA1 | Heterogeneous nuclear ribonucleoprotein A1 | −5.34903 | 0.00046 |
| BC018676 | LOC730846 | similar to S-adenosylmethionine decarboxylase proenzyme 2 (AdoMetDC 2) (SamDC 2) | 6.02167 | 0.0002 |
| NM_015251 | ASCIZ | ATM/ATR-Substrate Chk2-Interacting Zn2+-finger protein | −4.91078 | 0.00084 |
| NM_024557 | RIC3 | Resistance to inhibitors of cholinesterase 3 homolog (C. elegans) | −5.18745 | 0.00057 |
| NM_005389 | PCMT1 | Protein-L-isoaspartate (D-aspartate) O-methyltransferase | 4.80631 | 0.00097 |
| AK056402 | TDRKH | Tudor and KH domain containing | −5.98927 | 0.00021 |
| BX113895 | BX113895 | Transcribed locus | −5.24899 | 0.00053 |
| NM_199418 | PRCP | Prolylcarboxypeptidase (angiotensinase C) | −6.17688 | 0.00016 |
| NM_022074 | FAM111A | Family with sequence similarity 111, member A | −6.19018 | 0.00016 |
| NM_032826 | SLC35B4 | Solute carrier family 35, member B4 | 5.93208 | 0.00022 |
| AK024473 | LOC92017 | similar to RIKEN cDNA 4933437K13 | −6.33059 | 0.00014 |
| NM_002766 | PRPSAP1 | Phosphoribosyl pyrophosphate synthetase-associated protein 1 | −5.41712 | 0.00042 |
| NM_006052 | DSCR3 | Down syndrome critical region gene 3 | 5.6984 | 0.00029 |
| CR603982 | SESN3 | MRNA; cDNA DKFZp313B1017 (from clone DKFZp313B1017) | −5.53487 | 0.00036 |
| NM_015509 | NECAP1 | NECAP endocytosis associated 1 | 5.46732 | 0.0004 |
| BC047412 | ZNF433 | Zinc finger protein 433 | −6.47114 | 0.00012 |
| NM_198829 | RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | 5.72007 | 0.00029 |
| NM_018191 | RCBTB1 | Regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 | −6.58896 | 0.0001 |
| NM_002075 | GNB3 | Guanine nucleotide binding protein (G protein), beta polypeptide 3 | 5.91288 | 0.00023 |
| NM_012164 | FBXW2 | Homo sapiens F-box and WD repeat domain containing 2 (FBXW2), mRNA. | 4.83277 | 0.00093 |
| A_32_P163472 | A_32_P163472 | Unknown | −6.16132 | 0.00017 |
| NM_015365 | AMMECR1 | Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region, gene 1 | −5.71868 | 0.00029 |
| AI887037 | AI887037 | Transcribed locus, weakly similar to NP_114185.2 Y-box protein YB2 [Rattus norvegicus] | 5.73536 | 0.00028 |
| NM_003913 | PRPF4B | PRP4 pre-mRNA processing factor 4 homolog B (yeast) | −6.5001 | 0.00011 |
| NM_018981 | DNAJC10 | DnaJ (Hsp40) homolog, subfamily C, member 10 | −5.57747 | 0.00034 |
| NM_000904 | NQO2 | NAD(P)H dehydrogenase, quinone 2 | 5.15535 | 0.0006 |
| AK123940 | FAM76A | CDNA FLJ41946 fis, clone PLACE6019701 | −5.47561 | 0.00039 |
| NM_005254 | GABPB2 | GA binding protein transcription factor, beta subunit 2 | −5.57953 | 0.00034 |
| A_23_P21804 | A_23_P21804 | Unknown | −5.00709 | 0.00073 |
| BC071966 | FAM60A | Family with sequence similarity 60, member A | 5.73366 | 0.00028 |
| U52054 | EMB | S6 H-8 mRNA expressed in chromosome 6-suppressed melanoma cells | −6.50457 | 0.00011 |
| NM_005463 | HNRPDL | Homo sapiens heterogeneous nuclear ribonucleoprotein D-like (HNRPDL), transcript variant 1, mRNA [NM_005463] | −6.07265 | 0.00019 |
| NM_152729 | NT5DC1 | 5'-nucleotidase domain containing 1 | −5.41419 | 0.00043 |
| NM_032775 | KLHL22 | Kelch-like 22 (Drosophila) | 5.32193 | 0.00048 |
| A_32_P177097 | A_32_P177097 | Unknown | −5.55856 | 0.00035 |
| THC2400533 | THC2400533 | Unknown | −6.2533 | 0.00015 |
| NM_005877 | SF3A1 | Splicing factor 3a, subunit 1, 120kDa | −4.80231 | 0.00097 |
| NM_001008392 | CTDSPL | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like | 5.07055 | 0.00067 |
| NM_015914 | TXNDC11 | Thioredoxin domain containing 11 | −5.91849 | 0.00022 |
| NM_025040 | ZNF614 | Zinc finger protein 614 | 5.42227 | 0.00042 |
| AF067420 | IGHA1 | Immunoglobulin heavy constant alpha 1 | 5.36829 | 0.00045 |
| BF592096 | RPL22 | Ribosomal protein L22 | −4.87536 | 0.00088 |
| XM_931434 | LOC400027 | hypothetical gene supported by BC047417 | −5.55031 | 0.00036 |
| NM_004124 | GMFB | Glia maturation factor, beta | 5.74047 | 0.00028 |
| NM_003932 | ST13 | Suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) | 6.04059 | 0.00019 |
| NM_014341 | MTCH1 | Mitochondrial carrier homolog 1 (C. elegans) | −6.36315 | 0.00013 |
| NM_002494 | NDUFC1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa | −5.97566 | 0.00021 |
| NM_194324 | MGC39900 | hypothetical protein MGC39900 | −5.2886 | 0.0005 |
| BX118285 | LOC729717 | hypothetical protein LOC729717 | −6.00778 | 0.0002 |
| NM_017615 | NSMCE4A | Non-SMC element 4 homolog A (S. cerevisiae) | 5.78582 | 0.00026 |
| NM_178354 | LCE1F | Late cornified envelope 1F | 5.95595 | 0.00021 |
| NM_006699 | MAN1A2 | Mannosidase, alpha, class 1A, member 2 | 5.92092 | 0.00022 |
| NM_015245 | ANKS1A | Ankyrin repeat and sterile alpha motif domain containing 1A | −5.50607 | 0.00038 |
| NM_018714 | COG1 | Component of oligomeric golgi complex 1 | −5.45574 | 0.0004 |
| NM_032730 | RTN4IP1 | Reticulon 4 interacting protein 1 | −6.07719 | 0.00018 |
| BX647358 | LOC283970 | hypothetical protein LOC283970 | −6.36374 | 0.00013 |
| NM_005171 | ATF1 | Activating transcription factor 1 | 4.79615 | 0.00098 |
| NM_006537 | USP3 | Ubiquitin specific peptidase 3 | 5.28511 | 0.0005 |
| NM_020764 | CASKIN1 | CASK interacting protein 1 | 5.90091 | 0.00023 |
| XM_929707 | LOC646756 | similar to Hermansky-Pudlak syndrome 1 protein isoform b | 6.01252 | 0.0002 |
| NM_016025 | METTL9 | Methyltransferase like 9 | −5.30304 | 0.00049 |
| NM_014718 | CLSTN3 | Calsyntenin 3 | 5.55953 | 0.00035 |
| NM_016220 | ZNF588 | Zinc finger protein 588 | −6.08105 | 0.00018 |
| NM_152391 | PQLC3 | PQ loop repeat containing 3 | −5.42606 | 0.00042 |
| NM_005093 | CBFA2T2 | Core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | −4.93458 | 0.00081 |
| NM_003132 | SRM | Spermidine synthase | 6.26425 | 0.00015 |
| AB075859 | ZNF525 | Zinc finger protein 525 | −6.0377 | 0.00019 |
| NM_012133 | COPG2 | Coatomer protein complex, subunit gamma 2 | −5.60169 | 0.00033 |
| NM_015391 | ANAPC13 | Anaphase promoting complex subunit 13 | 5.88569 | 0.00023 |
| NM_007279 | U2AF2 | U2 small nuclear RNA auxiliary factor 2 | 6.28623 | 0.00014 |
| NM_181657 | LTB4R | Leukotriene B4 receptor | 6.31112 | 0.00014 |
| BC049371 | HERPUD2 | HERPUD family member 2 | −5.4236 | 0.00042 |
| NM_024865 | NANOG | Nanog homeobox | −4.86843 | 0.00089 |
| NM_015366 | PRR5 | Proline rich 5 (renal) | 5.66776 | 0.00031 |
| NM_003161 | RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | −5.22779 | 0.00054 |
| NM_007186 | CEP250 | Centrosomal protein 250kDa | −5.15592 | 0.0006 |
| AI241366 | AI241366 | Transcribed locus | −5.77357 | 0.00027 |
| NM_018660 | ZNF395 | Zinc finger protein 395 | −5.96506 | 0.00021 |
| NM_006352 | ZNF238 | Zinc finger protein 238 | 5.02507 | 0.00071 |
| NM_032199 | ARID5B | AT rich interactive domain 5B (MRF1-like) | −5.49226 | 0.00038 |
| NM_001184 | ATR | Ataxia telangiectasia and Rad3 related | −6.35054 | 0.00013 |
| NM_203448 | MGC21881 | hypothetical protein MGC21881 | −5.36318 | 0.00045 |
| NM_015584 | POLDIP2 | Polymerase (DNA-directed), delta interacting protein 2 | −5.75051 | 0.00028 |
| A_24_P315644 | A_24_P315644 | Unknown | 5.15013 | 0.0006 |
| NM_014679 | CEP57 | Centrosomal protein 57kDa | −6.20966 | 0.00016 |
| NM_030776 | ZBP1 | Z-DNA binding protein 1 | −5.73526 | 0.00028 |
| A_24_P684274 | A_24_P684274 | Unknown | 6.05786 | 0.00019 |
| NM_013347 | RPA4 | Diaphanous homolog 2 (Drosophila) | −5.34338 | 0.00047 |
| NM_000028 | AGL | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (glycogen debranching enzyme, glycogen storage disease type III) | −5.88724 | 0.00023 |
| NM_005544 | IRS1 | Insulin receptor substrate 1 | −5.99343 | 0.0002 |
| NM_017890 | VPS13B | Vacuolar protein sorting 13B (yeast) | −5.26169 | 0.00052 |
| NM_003925 | MBD4 | Methyl-CpG binding domain protein 4 | −6.08677 | 0.00018 |
| NM_194278 | C14orf43 | Chromosome 14 open reading frame 43 | −6.1667 | 0.00017 |
| NM_013450 | BAZ2B | Bromodomain adjacent to zinc finger domain, 2B | −6.24334 | 0.00015 |
| NM_002120 | HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | −5.22613 | 0.00054 |
| AK075364 | SLC10A7 | Chromosome 4 open reading frame 13 | −6.08302 | 0.00018 |
| AL390214 | TRIO | Triple functional domain (PTPRF interacting) | −5.41553 | 0.00042 |
| NM_033535 | FBXL5 | F-box and leucine-rich repeat protein 5 | −5.47671 | 0.00039 |
| A_24_P234871 | A_24_P234871 | Unknown | −5.18812 | 0.00057 |
| NM_017846 | TRSPAP1 | TRNA selenocysteine associated protein 1 | −4.89339 | 0.00086 |
| NM_203304 | RKHD1 | Ring finger and KH domain containing 1 | 5.65882 | 0.00031 |
| NM_016824 | ADD3 | Adducin 3 (gamma) | −6.2579 | 0.00015 |
| NM_012089 | ABCB10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | −5.17797 | 0.00058 |
| NM_054028 | AMAC1L2 | Acyl-malonyl condensing enzyme 1-like 2 | −6.13479 | 0.00017 |
| THC2405550 | THC2405550 | Unknown | −5.82626 | 0.00025 |
| NM_013314 | BLNK | B-cell linker | −6.15231 | 0.00017 |
| NM_001424 | EMP2 | Epithelial membrane protein 2 | 5.01766 | 0.00072 |
| NM_032932 | RAB11FIP4 | RAB11 family interacting protein 4 (class II) | −5.21617 | 0.00055 |
| BC036626 | SENP5 | SUMO1/sentrin specific peptidase 5 | −6.1731 | 0.00016 |
| NM_014300 | SEC11A | SEC11 homolog A (S. cerevisiae) | −5.51732 | 0.00037 |
| NM_032444 | BTBD12 | BTB (POZ) domain containing 12 | −5.94585 | 0.00022 |
| NM_002795 | PSMB3 | Proteasome (prosome, macropain) subunit, beta type, 3 | 5.01349 | 0.00073 |
| A_24_P609932 | A_24_P609932 | Unknown | 5.95203 | 0.00021 |
| NM_198868 | TBC1D9B | TBC1 domain family, member 9B (with GRAM domain) | −5.14293 | 0.00061 |
| NM_015272 | KIAA1005 | KIAA1005 protein | −5.38264 | 0.00044 |
| XM_496158 | A_24_P341126 | Unknown | 4.80149 | 0.00097 |
| NM_006099 | PIAS3 | Protein inhibitor of activated STAT, 3 | 5.57718 | 0.00034 |
| A_24_P126902 | A_24_P126902 | Unknown | −5.90443 | 0.00023 |
| XM_930744 | LOC653056 | PREDICTED: Homo sapiens similar to hypothetical gene supported by AK024248; AL137733, transcript variant 12 (LOC653056), mRNA [XM_930744] | 5.61483 | 0.00033 |
| NM_032199 | ARID5B | AT rich interactive domain 5B (MRF1-like) | 5.09208 | 0.00065 |
| AF318367 | C9orf86 | Homo sapiens pp8875 mRNA, complete cds. | 4.89882 | 0.00085 |
| NM_001969 | EIF5 | Eukaryotic translation initiation factor 5 | −6.21026 | 0.00016 |
| CN404647 | CCDC18 | Coiled-coil domain containing 18 | −5.25271 | 0.00053 |
| NM_004894 | C14orf2 | Chromosome 14 open reading frame 2 | −5.8179 | 0.00025 |
| NM_152458 | ZNF785 | Zinc finger protein 785 | −6.19706 | 0.00016 |
| NM_153816 | SNX14 | Sorting nexin 14 | −4.95674 | 0.00078 |
| NM_024783 | AGBL2 | ATP/GTP binding protein-like 2 | −5.29806 | 0.0005 |
| NM_003342 | UBE2G1 | Ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | 5.20823 | 0.00056 |
| AK129584 | AK129584 | Full-length cDNA clone CS0DI007YH13 of Placenta Cot 25-normalized of Homo sapiens (human) | 5.90722 | 0.00023 |
| NM_012089 | ABCB10 | ATP-binding cassette, sub-family B (MDR/TAP), member 10 | −5.38255 | 0.00044 |
| NM_052873 | C14orf179 | Chromosome 14 open reading frame 179 | 5.93542 | 0.00022 |
| NM_152500 | CCDC17 | Coiled-coil domain containing 17 | −5.39547 | 0.00044 |
| NM_001040138 | CKLF | Chemokine-like factor | −5.93626 | 0.00022 |
| AK022223 | DCAKD | Dephospho-CoA kinase domain containing | −5.27457 | 0.00051 |
| NM_014314 | DDX58 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 | −6.10636 | 0.00018 |
| NM_003938 | AP3D1 | Adaptor-related protein complex 3, delta 1 subunit | 6.20295 | 0.00016 |
| BC035854 | DSCR1L2 | Down syndrome critical region gene 1-like 2 | −5.19219 | 0.00057 |
| NM_015342 | PPWD1 | Peptidylprolyl isomerase domain and WD repeat containing 1 | −5.06668 | 0.00067 |
| NM_175876 | EXOC8 | Exocyst complex component 8 | −5.67475 | 0.0003 |
| NM_033557 | YIF1B | Yip1 interacting factor homolog B (S. cerevisiae) | 4.88311 | 0.00087 |
| NM_002470 | MYH3 | Myosin, heavy chain 3, skeletal muscle, embryonic | −5.68484 | 0.0003 |
| NM_178126 | LOC162427 | hypothetical protein LOC162427 | 5.77981 | 0.00027 |
| BG007597 | ALKBH5 | Transcribed locus, strongly similar to NP_060228.2 protein LOC54890 [Homo sapiens] | 5.35565 | 0.00046 |
| NM_003982 | SLC7A7 | Solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 | −5.53607 | 0.00036 |
| NM_001326 | CSTF3 | Cleavage stimulation factor, 3' pre-RNA, subunit 3, 77kDa | −6.14277 | 0.00017 |
| U97060 | SNAI2 | Snail homolog 2 (Drosophila) | 5.73829 | 0.00028 |
| NM_006640 | 9-Sep | Septin 9 | −5.35521 | 0.00046 |
| NM_025139 | ARMC9 | Armadillo repeat containing 9 | 5.18952 | 0.00057 |
| NM_006996 | SLC19A2 | Solute carrier family 19 (thiamine transporter), member 2 | −5.95859 | 0.00021 |
| NM_014501 | UBE2S | Ubiquitin-conjugating enzyme E2S | 5.46013 | 0.0004 |
| NM_016184 | CLEC4A | C-type lectin domain family 4, member A | −6.07896 | 0.00018 |
| NM_020470 | YIF1A | Yip1 interacting factor homolog A (S. cerevisiae) | 5.81837 | 0.00025 |
| NM_005922 | MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | −5.64318 | 0.00032 |
| S74620 | PRKCH | Protein kinase C, eta | 5.30026 | 0.00049 |
| A_32_P62137 | A_32_P62137 | Unknown | 5.50854 | 0.00038 |
| L08436 | COTL1 | Coactosin-like 1 (Dictyostelium) | −6.03708 | 0.00019 |
| NM_172345 | SPAG9 | Sperm associated antigen 9 | 5.40408 | 0.00043 |
| NM_003119 | SPG7 | Spastic paraplegia 7, paraplegin (pure and complicated autosomal recessive) | 4.91896 | 0.00083 |
| AK128396 | TRAF6 | CDNA FLJ46539 fis, clone THYMU3037836 | −4.9648 | 0.00078 |
| NM_014480 | ZNF544 | Zinc finger protein 544 | −5.52426 | 0.00037 |
| NM_024050 | C19orf58 | Chromosome 19 open reading frame 58 | 5.81018 | 0.00026 |
| NM_014017 | MAPBPIP | mitogen-activated protein-binding protein-interacting protein | 5.65193 | 0.00031 |
| NM_018952 | HOXB6 | Homeobox B5 | 4.78253 | 0.001 |
| AL832683 | PTAR1 | Protein prenyltransferase alpha subunit repeat containing 1 | −5.18786 | 0.00057 |
| NM_024812 | BAALC | Brain and acute leukemia, cytoplasmic | 5.29334 | 0.0005 |
| XM_496043 | A_24_P761457 | Unknown | 4.99333 | 0.00075 |
| BX538300 | MRPS30 | Full-length cDNA clone CS0DK012YA15 of HeLa cells Cot 25-normalized of Homo sapiens (human) | 5.38356 | 0.00044 |
| NM_145309 | LRRC51 | Leucine rich repeat containing 51 | 5.71612 | 0.00029 |
| NM_005647 | TBL1X | Transducin (beta)-like 1X-linked | −5.4274 | 0.00042 |
| NM_006788 | RALBP1 | RalA binding protein 1 | 5.44876 | 0.00041 |
| NM_000982 | RPL21 | Ribosomal protein L21 | −4.85896 | 0.0009 |
| CR590530 | PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | −5.90778 | 0.00023 |
| NM_006135 | CAPZA1 | Capping protein (actin filament) muscle Z-line, alpha 1 | 5.48417 | 0.00039 |
| NM_198490 | RAB43 | RAB43, member RAS oncogene family | −5.83049 | 0.00025 |
| BM461836 | BM461836 | Transcribed locus | −6.01488 | 0.0002 |
| AK022479 | HDHD1A | Haloacid dehalogenase-like hydrolase domain containing 1A | −5.47807 | 0.00039 |
| AL832806 | HDAC4 | Histone deacetylase 4 | −5.80606 | 0.00026 |
| BC021898 | AP1S3 | Homo sapiens adaptor-related protein complex 1, sigma 3 subunit, mRNA (cDNA clone MGC:17284 IMAGE:4340257), complete cds. | 5.33898 | 0.00047 |
| NM_001991 | EZH1 | Enhancer of zeste homolog 1 (Drosophila) | −5.85304 | 0.00024 |
| NM_033222 | PSIP1 | PC4 and SFRS1 interacting protein 1 | −5.29831 | 0.00049 |
| NM_182538 | MGC29671 | hypothetical protein MGC29671 | 5.11574 | 0.00063 |
| NM_002055 | GFAP | Glial fibrillary acidic protein | 5.4633 | 0.0004 |
| NM_004552 | NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) | −5.31081 | 0.00049 |
| AB011149 | KIAA0577 | Homo sapiens mRNA for KIAA0577 protein, partial cds. | −5.56005 | 0.00035 |
| AK098220 | LOC728555 | hypothetical protein LOC728555 | −5.66434 | 0.00031 |
| NM_199168 | CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 5.36752 | 0.00045 |
| XM_001134097 | LOC731989 | similar to protein tyrosine phosphatase 4a1 | 5.88768 | 0.00023 |
| NM_021140 | UTX | Ubiquitously transcribed tetratricopeptide repeat, X chromosome | −5.36566 | 0.00045 |
| NM_001032410 | USP16 | Ubiquitin specific peptidase 16 | −5.39423 | 0.00044 |
| NM_031206 | LAS1L | LAS1-like (S. cerevisiae) | −5.31002 | 0.00049 |
| NM_032750 | ABHD14B | Abhydrolase domain containing 14B | 4.95275 | 0.00079 |
| NM_013375 | ABT1 | Activator of basal transcription 1 | −5.94292 | 0.00022 |
| NM_178831 | GATS | opposite strand transcription unit to STAG3 | 5.62774 | 0.00032 |
| NM_138426 | GLCCI1 | Glucocorticoid induced transcript 1 | −5.09956 | 0.00065 |
| NM_025160 | WDR26 | WD repeat domain 26 | −4.99453 | 0.00074 |
| NM_017914 | C19orf24 | Chromosome 19 open reading frame 24 | −5.7746 | 0.00027 |
| NM_019053 | EXOC6 | Exocyst complex component 6 | −5.5546 | 0.00035 |
| NM_012249 | RHOQ | Ras homolog gene family, member Q | 5.05453 | 0.00069 |
| NM_032823 | C9orf3 | Chromosome 9 open reading frame 3 | 5.65131 | 0.00031 |
| NM_014388 | C1orf107 | Chromosome 1 open reading frame 107 | −5.4832 | 0.00039 |
| BC054050 | THOC2 | THO complex 2 | −5.54513 | 0.00036 |
| NM_001017915 | INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | −5.83365 | 0.00025 |
| NM_032380 | GFM2 | G elongation factor, mitochondrial 2 | −4.82169 | 0.00094 |
| NM_025054 | VCPIP1 | Valosin containing protein (p97)/p47 complex interacting protein 1 | −5.25903 | 0.00052 |
| NM_006454 | MXD4 | MAX dimerization protein 4 | 5.83018 | 0.00025 |
| THC2412623 | THC2412623 | Unknown | 5.5052 | 0.00038 |
| NM_131916 | PPIL3 | Peptidylprolyl isomerase (cyclophilin)-like 3 | −5.7303 | 0.00028 |
| NM_005730 | CTDSP2 | CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 | −4.87806 | 0.00087 |
| NM_001000 | RPL39 | Ribosomal protein L39 | −5.27874 | 0.00051 |
| L06133 | ATP7A | ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome) | −5.18321 | 0.00058 |
| NM_145059 | FUK | Fucokinase | −5.02457 | 0.00071 |
| BC043173 | BC043173 | CDNA clone IMAGE:5287121 | −4.87502 | 0.00088 |
| NM_015261 | NCAPD3 | Non-SMC condensin II complex, subunit D3 | −5.01908 | 0.00072 |
| NM_175709 | CBX7 | Chromobox homolog 7 | −5.37048 | 0.00045 |
| NM_138764 | BAX | BCL2-associated X protein | 4.98972 | 0.00075 |
| NM_001002261 | ZFYVE27 | Zinc finger, FYVE domain containing 27 | −5.04773 | 0.00069 |
| BX538256 | ENST00000369183 | MRNA; cDNA DKFZp686C1384 (from clone DKFZp686C1384) | −5.41008 | 0.00043 |
| NM_139015 | SPPL3 | signal peptide peptidase 3 | −5.26749 | 0.00052 |
| NM_176818 | 15E1.2 | hypothetical protein LOC283459 | 4.87135 | 0.00088 |
| BX648207 | BX648207 | CDNA FLJ40891 fis, clone UTERU2001110 | −5.16386 | 0.00059 |
| A_32_P19616 | A_32_P19616 | Unknown | 5.60695 | 0.00033 |
| NM_014829 | DDX46 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 | −5.52869 | 0.00037 |
| AK023831 | FLJ13769 | hypothetical protein FLJ13769 | −5.65982 | 0.00031 |
| A_24_P298835 | A_24_P298835 | Unknown | −4.94729 | 0.00079 |
| NM_052862 | RCSD1 | RCSD domain containing 1 | −4.94157 | 0.0008 |
| XM_496145 | LOC440361 | PREDICTED: Homo sapiens similar to Ig heavy chain V-III region VH26 precursor (LOC440361), mRNA [XM_496145] | 4.79274 | 0.00098 |
| NM_014649 | SAFB2 | Small nuclear ribonucleoprotein polypeptide E | −4.92345 | 0.00082 |
| NM_152604 | ZNF383 | Zinc finger protein 383 | −4.95902 | 0.00078 |
| NM_022748 | TNS3 | Tensin 3 | −5.23702 | 0.00054 |
| BC062636 | RHBDD1 | Rhomboid domain containing 1 | −5.31878 | 0.00048 |
| NM_006591 | POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | −5.68071 | 0.0003 |
| NM_024329 | EFHD2 | EF-hand domain family, member D2 | 5.37781 | 0.00045 |
| NM_015485 | RWDD3 | RWD domain containing 3 | −5.33594 | 0.00047 |
| AK094181 | TTC15 | Tetratricopeptide repeat domain 15 | −5.28622 | 0.0005 |
| NM_017613 | DONSON | Downstream neighbor of SON | −4.89151 | 0.00086 |
| BF476310 | PGPEP1 | Pyroglutamyl-peptidase I | 5.57075 | 0.00035 |
| AK022016 | MYO1E | Myosin IE | −5.28289 | 0.00051 |
| NM_018602 | DNAJA4 | DnaJ (Hsp40) homolog, subfamily A, member 4 | 5.08766 | 0.00066 |
| THC2405710 | THC2405710 | Unknown | −5.43828 | 0.00041 |
| NM_016227 | C1orf9 | Chromosome 1 open reading frame 9 | −5.11283 | 0.00063 |
| NM_006568 | CGRRF1 | Cell growth regulator with ring finger domain 1 | −5.30073 | 0.00049 |
| XM_497928 | A_24_P409891 | Unknown | −5.10488 | 0.00064 |
| NM_032358 | CCDC77 | Coiled-coil domain containing 77 | −4.97185 | 0.00077 |
| NM_020807 | ZNF319 | Zinc finger protein 319 | −4.89333 | 0.00086 |
| A_32_P145302 | A_32_P145302 | Unknown | 5.14089 | 0.00061 |
| NM_001007467 | SFI1 | Sfi1 homolog, spindle assembly associated (yeast) | −5.24419 | 0.00053 |
| NM_021167 | GATAD1 | GATA zinc finger domain containing 1 | 5.4081 | 0.00043 |
| A_24_P616082 | A_24_P616082 | Unknown | 5.48231 | 0.00039 |
| AK000103 | LOC153277 | hypothetical protein LOC153277 | −5.60166 | 0.00033 |
| AK096109 | LOC339483 | hypothetical LOC339483 | −5.45198 | 0.0004 |
| NM_138621 | BCL2L11 | BCL2-like 11 (apoptosis facilitator) | 5.11569 | 0.00063 |
| NM_016563 | RASL12 | RAS-like, family 12 | 5.14387 | 0.00061 |
| NM_004551 | NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) | 5.3438 | 0.00047 |
| NM_006618 | JARID1B | Jumonji, AT rich interactive domain 1B | −5.52211 | 0.00037 |
| NM_015503 | SH2B1 | SH2B adaptor protein 1 | 5.12367 | 0.00062 |
| NM_020062 | SLC2A4RG | SLC2A4 regulator | 5.48553 | 0.00039 |
| NM_006321 | ARIH2 | Ariadne homolog 2 (Drosophila) | 4.82099 | 0.00095 |
| NM_020235 | BBX | Bobby sox homolog (Drosophila) | −5.22583 | 0.00054 |
| NM_021167 | GATAD1 | GATA zinc finger domain containing 1 | −4.7975 | 0.00098 |
| NM_000255 | MUT | Methylmalonyl Coenzyme A mutase | −5.32572 | 0.00048 |
| NM_014636 | RALGPS1 | Ral GEF with PH domain and SH3 binding motif 1 | −5.3981 | 0.00043 |
| NM_003899 | ARHGEF7 | Rho guanine nucleotide exchange factor (GEF) 7 | −4.98475 | 0.00075 |
| BC032369 | HNRPUL2 | Heterogeneous nuclear ribonucleoprotein U-like 2 | 4.95545 | 0.00079 |
| NM_014739 | BCLAF1 | Homo sapiens BCL2-associated transcription factor 1 (BCLAF1), transcript variant 1, mRNA. | −4.93999 | 0.0008 |
| NM_000364 | TNNT2 | Troponin T type 2 (cardiac) | 4.96733 | 0.00077 |
| NM_030639 | BHLHB9 | Basic helix-loop-helix domain containing, class B, 9 | −4.9057 | 0.00084 |
| NM_013430 | GGT1 | Gamma-glutamyltransferase 1 | 4.78165 | 0.001 |
| NM_006594 | AP4B1 | Adaptor-related protein complex 4, beta 1 subunit | −5.45928 | 0.0004 |
| NM_003188 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | −5.50014 | 0.00038 |
| THC2415754 | THC2415754 | Q9Y6H2 (Q9Y6H2) Ku70-binding protein (Fragment), partial (41%) [THC2415754] | −4.97453 | 0.00077 |
| XM_496741 | LOC441066 | PREDICTED: Homo sapiens similar to Beta-glucuronidase precursor (LOC441066), mRNA [XM_496741] | 4.89513 | 0.00085 |
| NM_012430 | SEC22A | SEC22 vesicle trafficking protein homolog A (S. cerevisiae) | −5.0367 | 0.0007 |
| NM_014805 | EPM2AIP1 | EPM2A (laforin) interacting protein 1 | −5.02229 | 0.00072 |
| BC045718 | AMZ2 | archaemetzincins-2 | −4.86184 | 0.00089 |
| AK023737 | AK023737 | Homo sapiens cDNA FLJ13675 fis, clone PLACE1011875, highly similar to Homo sapiens mRNA for KIAA0580 protein. | −5.39305 | 0.00044 |
| A_24_P110521 | A_24_P110521 | Unknown | −5.43875 | 0.00041 |
| NM_015355 | SUZ12 | Suppressor of zeste 12 homolog (Drosophila) | −5.08232 | 0.00066 |
| THC2320257 | THC2320257 | Unknown | −5.26085 | 0.00052 |
| BM069797 | C18orf17 | Transcribed locus, strongly similar to XP_512065.1 similar to squamous cell carcinoma-related protein-1 [Pan troglodytes] | −5.06886 | 0.00067 |
| THC2373083 | THC2373083 | Unknown | −5.22908 | 0.00054 |
| NM_001439 | EXTL2 | Exostoses (multiple)-like 2 | −5.22046 | 0.00055 |
| THC2340322 | THC2340322 | Q5T931 (Q5T931) Sorbin and SH3 domain containing 1, partial (3%) [THC2340322] | −5.24933 | 0.00053 |
| NM_018297 | NGLY1 | N-glycanase 1 | −5.39139 | 0.00044 |
| A_32_P55438 | A_32_P55438 | Unknown | −5.09221 | 0.00065 |
| NM_019593 | KIAA1434 | hypothetical protein KIAA1434 | −4.86596 | 0.00089 |
| NM_005813 | PRKD3 | Protein kinase D3 | −5.19393 | 0.00057 |
| NM_001750 | CAST | Calpastatin | −5.38217 | 0.00044 |
| THC2266506 | THC2266506 | RS24_HUMAN (P62847) 40S ribosomal protein S24, complete [THC2266506] | −5.34445 | 0.00047 |
| NM_001029 | RPS26 | Ribosomal protein S26 | −5.39967 | 0.00043 |
| NM_147171 | AKAP9 | A kinase (PRKA) anchor protein (yotiao) 9 | −5.10376 | 0.00064 |
| THC2671898 | THC2671898 | Q3WYF8_9ACTN (Q3WYF8) Sulfotransferase, partial (6%) [THC2671898] | 4.9822 | 0.00076 |
| NM_005773 | ZNF256 | Zinc finger protein 418 | −5.08148 | 0.00066 |
| A_32_P8971 | A_32_P8971 | Unknown | −4.99489 | 0.00074 |
| NM_173854 | SLC41A1 | Solute carrier family 41, member 1 | −5.30434 | 0.00049 |
| NM_016467 | ORMDL1 | ORM1-like 1 (S. cerevisiae) | −5.13111 | 0.00062 |
| THC2437881 | THC2437881 | Q9NBA9 (Q9NBA9) Stretchin-MLCK (Fragment), partial (5%) [THC2437881] | −5.17389 | 0.00058 |
| NM_004487 | GOLGB1 | Golgi autoantigen, golgin subfamily b, macrogolgin (with transmembrane signal), 1 | −5.26839 | 0.00051 |
| NM_013444 | UBQLN2 | Ubiquilin 2 | −4.98166 | 0.00076 |
| NM_001011667 | CHCHD7 | Coiled-coil-helix-coiled-coil-helix domain containing 7 | −5.02654 | 0.00071 |
| NM_020666 | CLK4 | CDC-like kinase 4 | −5.18565 | 0.00057 |
| NM_006221 | PIN1 | Protein (peptidylprolyl cis/trans isomerase) NIMA-interacting 1 | 5.0031 | 0.00074 |
| NM_001029 | RPS26 | Ribosomal protein S26 | −5.3064 | 0.00049 |
| THC2280343 | THC2280343 | ALU6_HUMAN (P39193) Alu subfamily SP sequence contamination warning entry, partial (9%) [THC2280343] | −4.93191 | 0.00081 |
| AK023675 | UTRN | Utrophin (homologous to dystrophin) | −5.03783 | 0.0007 |
| NM_024065 | PDCL3 | Phosducin-like 3 | −4.79228 | 0.00098 |
| NM_182663 | RASSF5 | Ras association (RalGDS/AF-6) domain family 5 | 4.79344 | 0.00098 |
| NM_018456 | EAF2 | ELL associated factor 2 | −5.1398 | 0.00061 |
| A_23_P158868 | A_23_P158868 | Unknown | 5.19558 | 0.00057 |
| XM_940047 | LOC654152 | PREDICTED: Homo sapiens similar to ribosomal protein L21 isoform 1 (LOC654152), mRNA [XM_940047] | −5.06821 | 0.00067 |
| NM_003264 | TLR2 | Toll-like receptor 2 | −5.20009 | 0.00056 |
| NM_015117 | ZC3H3 | Zinc finger CCCH-type containing 3 | 5.05631 | 0.00068 |
| NM_024112 | C9orf16 | Chromosome 9 open reading frame 16 | 5.0859 | 0.00066 |
| NM_006051 | APBB3 | Amyloid beta (A4) precursor protein-binding, family B, member 3 | −4.83419 | 0.00093 |
| NM_032714 | C14orf151 | Chromosome 14 open reading frame 151 | 5.05654 | 0.00068 |
| NM_015911 | ZNF691 | Zinc finger protein 691 | −4.89911 | 0.00085 |
| THC2306096 | THC2306096 | Unknown | −5.15365 | 0.0006 |
| NM_015088 | TNRC6B | Trinucleotide repeat containing 6B | −4.78159 | 0.001 |
| A_32_P158543 | A_32_P158543 | Unknown | 4.83166 | 0.00093 |
| NM_004278 | PIGL | Phosphatidylinositol glycan anchor biosynthesis, class L | 5.03195 | 0.00071 |
| NM_024313 | NOL12 | Nucleolar protein 12 | 4.80569 | 0.00097 |
| NM_014238 | KSR1 | Kinase suppressor of ras 1 | 4.99409 | 0.00074 |
| NM_015412 | C3orf17 | Chromosome 3 open reading frame 17 | −5.07804 | 0.00066 |
| NM_018255 | STATIP1 | Signal transducer and activator of transcription 3 interacting protein 1 | −4.88966 | 0.00086 |
| A_32_P180185 | A_32_P180185 | Unknown | −4.90553 | 0.00084 |
| L31408 | FGF2 | Fibroblast growth factor 2 (basic) | 4.99531 | 0.00074 |
| NM_014608 | CYFIP1 | Cytoplasmic FMR1 interacting protein 1 | −4.97789 | 0.00076 |
| NM_000655 | SELL | Selectin L (lymphocyte adhesion molecule 1) | −4.93527 | 0.00081 |
| NM_153240 | NPHP3 | Nephronophthisis 3 (adolescent) | −4.85518 | 0.0009 |
| NM_031444 | C22orf13 | Chromosome 22 open reading frame 13 | 4.95359 | 0.00079 |
| NM_003811 | TNFSF9 | Tumor necrosis factor (ligand) superfamily, member 9 | 4.87787 | 0.00087 |
| NM_016310 | POLR3K | Polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa | 4.78534 | 0.00099 |
| NM_021149 | COTL1 | Coactosin-like 1 (Dictyostelium) | 4.83716 | 0.00092 |
| THC2409283 | THC2409283 | Unknown | −4.98158 | 0.00076 |
| X81724 | CPVL | Carboxypeptidase, vitellogenic-like | 4.78924 | 0.00099 |
| NM_001039360 | ZBTB7C | Zinc finger and BTB domain containing 7C | 4.94196 | 0.0008 |
| NM_003387 | WIPF1 | WAS/WASL interacting protein family, member 1 | −4.91746 | 0.00083 |
| BE774743 | THC2363476 | CDNA FLJ13267 fis, clone OVARC1000964 | 4.92569 | 0.00082 |
| NM_032330 | CAPNS2 | Calpain, small subunit 2 | 4.85241 | 0.00091 |
| NM_015540 | RPAP1 | RNA polymerase II associated protein 1 | −4.89166 | 0.00086 |
| XM_376787 | RPS26P10 | Ribosomal protein S26 pseudogene 10 | −4.90436 | 0.00084 |
| NM_018125 | ARHGEF10L | Rho guanine nucleotide exchange factor (GEF) 10-like | 4.88064 | 0.00087 |
| NM_017644 | KLHL24 | Kelch-like 24 (Drosophila) | −4.8297 | 0.00093 |
| NM_015132 | SNX13 | Sorting nexin 13 | −4.82155 | 0.00094 |
| NM_153219 | ZNF524 | Zinc finger protein 524 | −4.82122 | 0.00095 |
| A_23_P206568 | A_23_P206568 | Unknown | 4.81298 | 0.00096 |
| Pathway | Gene | Description |
|---|---|---|
| Cell adhesion molecules (CAMs) | HLA-DMB | Major histocompatibility complex, class II, DM beta |
| ITGB7 | Integrin, beta 7 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| HLA-DMA | Major histocompatibility complex, class II, DM beta | |
| SDC4 | Syndecan 4 (amphiglycan, ryudocan) | |
| HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | |
| SELP | Selectin P (granule membrane protein 140kDa, antigen CD62) | |
| NLGN1 | Neuroligin 1 | |
| SELL | Selectin L (lymphocyte adhesion molecule 1) | |
| L1CAM | L1 cell adhesion molecule | |
| ICAM2 | Intercellular adhesion molecule 2 | |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | |
| ECM-receptor interaction | ITGB7 | Integrin, beta 7 |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| SDC4 | Syndecan 4 (amphiglycan, ryudocan) | |
| Antigen processing and presentation | HLA-DMB | Major histocompatibility complex, class II, DM beta |
| HLA-DMA | Major histocompatibility complex, class II, DM beta | |
| HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | |
| Type I diabetes mellitus | HLA-DMB | Major histocompatibility complex, class II, DM beta |
| HLA-DMA | Major histocompatibility complex, class II, DM beta | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| HLA-DOB | Homo sapiens major histocompatibility complex, class II, DO beta (HLA-DOB), mRNA. | |
| Bile acid biosynthesis | ACAD8 | Acyl-Coenzyme A dehydrogenase family, member 8 |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | |
| ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | |
| Aminoacyl-tRNA biosynthesis | QARS | Glutaminyl-tRNA synthetase |
| HARSL | Histidyl-tRNA synthetase-like | |
| HARS | Histidyl-tRNA synthetase | |
| Histidine metabolism | PRMT2 | Protein arginine methyltransferase 2 |
| HARS | Histidyl-tRNA synthetase | |
| HARSL | Histidyl-tRNA synthetase-like | |
| Epithelial cell signaling in Helicobacter pylori infection | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| HBEGF | Heparin-binding EGF-like growth factor | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | PIGA | Phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) |
| PIGK | Phosphatidylinositol glycan anchor biosynthesis, class K | |
| PIGG | Phosphatidylinositol glycan anchor biosynthesis, class G | |
| PIGL | Phosphatidylinositol glycan anchor biosynthesis, class L | |
| Alkaloid biosynthesis II | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | |
| 1- and 2-Methylnaphthalene degradation | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| ACAD8 | Acyl-Coenzyme A dehydrogenase family, member 8 | |
| Benzoate degradation via CoA ligation | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| Limonene and pinene degradation | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| Glycerophospholipid metabolism | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| GPAM | Glycerol-3-phosphate acyltransferase, mitochondrial | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| LYPLA2 | Lysophospholipase II | |
| PPAP2C | Phosphatidic acid phosphatase type 2C | |
| Adherens junction | ACTC1 | Actin, alpha, cardiac muscle 1 |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | |
| ACTN3 | Actinin, alpha 3 | |
| EP300 | E1A binding protein p300 | |
| WASF2 | CDNA clone IMAGE:3030163 | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| SNAI2 | Snail homolog 2 (Drosophila) | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| SMAD3 | SMAD family member 3 | |
| LMO7 | LIM domain 7 | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| TGF-beta signaling pathway | ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | |
| EP300 | E1A binding protein p300 | |
| BMPR1A | Bone morphogenetic protein receptor, type IA | |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| SMAD3 | SMAD family member 3 | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Ubiquitin mediated proteolysis | ITCH | Itchy homolog E3 ubiquitin protein ligase (mouse) |
| ANAPC7 | Anaphase promoting complex subunit 7 | |
| EDD1 | E3 ubiquitin protein ligase, HECT domain containing, 1 | |
| UBE2D2 | Ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | |
| FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | |
| Tyrosine metabolism | MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 |
| PRMT2 | Protein arginine methyltransferase 2 | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| Inositol phosphate metabolism | PIK3C3 | Phosphoinositide-3-kinase, class 3 |
| MINPP1 | Multiple inositol polyphosphate histidine phosphatase, 1 | |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Phosphatidylinositol signaling system | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| PIK3C3 | Phosphoinositide-3-kinase, class 3 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| PRKCA | Protein kinase C, alpha | |
| PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Oxidative phosphorylation | NDUFS3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) |
| NDUFC1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa | |
| COX11 | COX11 homolog, cytochrome c oxidase assembly protein (yeast) | |
| NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) | |
| COX8A | Cytochrome c oxidase subunit 8A (ubiquitous) | |
| Ribosome | RPL39 | Ribosomal protein L39 |
| RPL21 | Ribosomal protein L21 | |
| RPL22 | Ribosomal protein L22 | |
| RPS26P10 | Ribosomal protein S26 pseudogene 10 | |
| RPS26 | Ribosomal protein S26 | |
| RPL15 | Ribosomal protein L15 | |
| RPS23 | Ribosomal protein S23 | |
| Arachidonic acid metabolism | GGT1 | Gamma-glutamyltransferase 1 |
| LTC4S | Leukotriene C4 synthase | |
| ALOX5 | Arachidonate 5-lipoxygenase | |
| Glutathione metabolism | GSTM2 | Glutathione S-transferase M2 (muscle) |
| GSTM1 | Glutathione S-transferase M1 | |
| GSTM4 | Glutathione S-transferase M4 | |
| G6PD | Glucose-6-phosphate dehydrogenase | |
| GGT1 | Gamma-glutamyltransferase 1 | |
| Metabolism of xenobiotics by cytochrome P450 | GSTM4 | Glutathione S-transferase M4 |
| GSTM2 | Glutathione S-transferase M2 (muscle) | |
| GSTM1 | Glutathione S-transferase M1 | |
| UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | |
| Cholera - Infection | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| PRKCA | Protein kinase C, alpha | |
| SEC61A2 | Sec61 alpha 2 subunit (S. cerevisiae) | |
| ARF3 | ADP-ribosylation factor 3 | |
| Chondroitin sulfate biosynthesis | CSGlcA-T | chondroitin sulfate glucuronyltransferase |
| XYLT1 | Xylosyltransferase I | |
| ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | |
| Cytokine-cytokine receptor interaction | TNF | Tumor necrosis factor (TNF superfamily, member 2) |
| GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) | |
| IL1RAP | Interleukin 1 receptor accessory protein | |
| BMPR1A | Bone morphogenetic protein receptor, type IA | |
| TNFRSF1 | 7Tumor necrosis factor receptor superfamily, member 17 | |
| IL10RA | Interleukin 10 receptor, alpha | |
| HGF | Human mRNA for hepatocyte growth factor (HGF). | |
| TNFSF9 | Tumor necrosis factor (ligand) superfamily, member 9 | |
| IL17RA | Interleukin 17 receptor A | |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Glycan structures - biosynthesis 2 | B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | |
| PIGA | Phosphatidylinositol glycan anchor biosynthesis, class A (paroxysmal nocturnal hemoglobinuria) | |
| PIGK | Phosphatidylinositol glycan anchor biosynthesis, class K | |
| PIGG | Phosphatidylinositol glycan anchor biosynthesis, class G | |
| ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | |
| PIGL | Phosphatidylinositol glycan anchor biosynthesis, class L | |
| Fructose and mannose metabolism | PFKL | Phosphofructokinase, liver |
| GMPPA | GDP-mannose pyrophosphorylase A | |
| STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | |
| SORD | Sorbitol dehydrogenase | |
| FUK | Fucokinase | |
| HK2 | Hexokinase 2 | |
| Galactose metabolism | PFKL | Phosphofructokinase, liver |
| B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 | |
| HK2 | Hexokinase 2 | |
| Fatty acid metabolism | ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) |
| ACOX1 | Acyl-Coenzyme A oxidase 1, palmitoyl | |
| ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | |
| Glycan structures - biosynthesis 1 | B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| ALG6 | Asparagine-linked glycosylation 6 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | |
| ST3GAL4 | ST3 beta-galactoside alpha-2,3-sialyltransferase 4 | |
| XYLT1 | Xylosyltransferase I | |
| EXT2 | Exostoses (multiple) 2 | |
| B3GNT6 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) | |
| RPN1 | Ribophorin I | |
| CSGlcA-T | chondroitin sulfate glucuronyltransferase | |
| ChGn | chondroitin beta1,4 N-acetylgalactosaminyltransferase | |
| EXTL2 | Exostoses (multiple)-like 2 | |
| MAN1B1 | Mannosidase, alpha, class 1B, member 1 | |
| MAN1A2 | Mannosidase, alpha, class 1A, member 2 | |
| Huntington's disease | DCTN1 | Dynactin 1 (p150, glued homolog, Drosophila) |
| BAX | BCL2-associated X protein | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| EP300 | E1A binding protein p300 | |
| Cell cycle | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) |
| ANAPC7 | Anaphase promoting complex subunit 7 | |
| EP300 | E1A binding protein p300 | |
| YWHAE | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | |
| ATR | Ataxia telangiectasia and Rad3 related | |
| SMAD3 | SMAD family member 3 | |
| FZR1 | Fizzy/cell division cycle 20 related 1 (Drosophila) | |
| Valine, leucine and isoleucine degradation | ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) |
| MYST3 | MYST histone acetyltransferase (monocytic leukemia) 3 | |
| ACAA1 | Acetyl-Coenzyme A acyltransferase 1 (peroxisomal 3-oxoacyl-Coenzyme A thiolase) | |
| BCAT2 | Branched chain aminotransferase 2, mitochondrial | |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| MUT | Methylmalonyl Coenzyme A mutase | |
| Amyotrophic lateral sclerosis (ALS) | RAB5A | RAB5A, member RAS oncogene family |
| BAX | BCL2-associated X protein | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CAT | Catalase | |
| SLC1A2 | Homo sapiens solute carrier family 1 (glial high affinity glutamate transporter), member 2 (SLC1A2), mRNA. | |
| Glycerolipid metabolism | GPAM | Glycerol-3-phosphate acyltransferase, mitochondrial |
| STT3B | STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) | |
| PPAP2C | Phosphatidic acid phosphatase type 2C | |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase (Wolman disease) | |
| DNA polymerase | POLD4 | Polymerase (DNA-directed), delta 4 |
| POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | |
| POLE | Polymerase (DNA directed), epsilon | |
| Purine metabolism | ADSS | Adenylosuccinate synthase |
| PDE4C | Phosphodiesterase 4C, cAMP-specific (phosphodiesterase E1 dunce homolog, Drosophila) | |
| XDH | Xanthine dehydrogenase | |
| POLR3H | Polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) | |
| POLR3K | Polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa | |
| POLD4 | Polymerase (DNA-directed), delta 4 | |
| POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| DCK | Deoxycytidine kinase | |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | |
| POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | |
| GART | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase | |
| POLE | Polymerase (DNA directed), epsilon | |
| Pyrimidine metabolism | POLR3H | Polymerase (RNA) III (DNA directed) polypeptide H (22.9kD) |
| POLD4 | Polymerase (DNA-directed), delta 4 | |
| POLR3K | Polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa | |
| DCK | Deoxycytidine kinase | |
| POLE4 | Polymerase (DNA-directed), epsilon 4 (p12 subunit) | |
| POLE3 | Polymerase (DNA directed), epsilon 3 (p17 subunit) | |
| DHODH | Dihydroorotate dehydrogenase | |
| POLD3 | Polymerase (DNA-directed), delta 3, accessory subunit | |
| TK2 | Thymidine kinase 2, mitochondrial | |
| POLE | Polymerase (DNA directed), epsilon | |
| Jak-STAT signaling pathway | GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| STAT6 | Signal transducer and activator of transcription 6, interleukin-4 induced | |
| CISH | Cytokine inducible SH2-containing protein | |
| EP300 | E1A binding protein p300 | |
| PIAS4 | Protein inhibitor of activated STAT, 4 | |
| PIAS3 | Protein inhibitor of activated STAT, 3 | |
| IL10RA | Interleukin 10 receptor, alpha | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| STAT5B | Signal transducer and activator of transcription 5B | |
| Chronic myeloid leukemia | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| CTBP1 | C-terminal binding protein 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| GAB2 | GRB2-associated binding protein 2 | |
| CTBP2 | C-terminal binding protein 2 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| SMAD3 | SMAD family member 3 | |
| STAT5B | Signal transducer and activator of transcription 5B | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Adipocytokine signaling pathway | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| STK11 | Serine/threonine kinase 11 | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | |
| IRS1 | Insulin receptor substrate 1 | |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | |
| B cell receptor signaling pathway | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| CHP | calcium binding protein P22 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| BLNK | B-cell linker | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| Fc epsilon RI signaling pathway | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| GAB2 | GRB2-associated binding protein 2 | |
| PRKCA | Protein kinase C, alpha | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Type II diabetes mellitus | MAFA | V-maf musculoaponeurotic fibrosarcoma oncogene homolog A (avian) |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| IRS1 | Insulin receptor substrate 1 | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | |
| Focal adhesion | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| ACTN3 | Actinin, alpha 3 | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| TLN1 | Talin 1 | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| PRKCA | Protein kinase C, alpha | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| CAV2 | Caveolin 2 | |
| ITGB7 | Integrin, beta 7 | |
| HGF | Human mRNA for hepatocyte growth factor (HGF). | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| GRLF1 | Glucocorticoid receptor DNA binding factor 1 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| Axon guidance | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| CHP | calcium binding protein P22 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| NCK2 | NCK adaptor protein 2 | |
| L1CAM | L1 cell adhesion molecule | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | |
| Calcium signaling pathway | ATP2A2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| CHP | calcium binding protein P22 | |
| CACNA1I | Calcium channel, voltage-dependent, alpha 1I subunit | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| PRKCA | Protein kinase C, alpha | |
| SLC25A5 | Solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 | |
| ATP2B4 | ATPase, Ca++ transporting, plasma membrane 4 | |
| Pathogenic Escherichia coli infection - EHEC | TUBB8 | tubulin, beta 8 |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| VIL2 | Villin 2 (ezrin) | |
| PRKCA | Protein kinase C, alpha | |
| NCK2 | NCK adaptor protein 2 | |
| Long-term potentiation | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| CHP | calcium binding protein P22 | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| EP300 | E1A binding protein p300 | |
| PRKCA | Protein kinase C, alpha | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| MAPK signaling pathway | DUSP4 | Dual specificity phosphatase 4 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| RASA2 | RAS p21 protein activator 2 | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| MAX | MYC associated factor X | |
| CHP | calcium binding protein P22 | |
| PPM1A | Protein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| PRKCA | Protein kinase C, alpha | |
| CACNA1I | Calcium channel, voltage-dependent, alpha 1I subunit | |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| TRAF6 | CDNA FLJ46539 fis, clone THYMU3037836 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| FGF2 | Fibroblast growth factor 2 (basic) | |
| MRAS | Muscle RAS oncogene homolog | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Apoptosis | PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| BAX | BCL2-associated X protein | |
| MYD88 | Myeloid differentiation primary response gene (88) | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| IL1RAP | Interleukin 1 receptor accessory protein | |
| CHP | calcium binding protein P22 | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Regulation of actin cytoskeleton | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| ARPC4 | Actin related protein 2/3 complex, subunit 4, 20kDa | |
| FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| ACTN3 | Actinin, alpha 3 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| VIL2 | Villin 2 (ezrin) | |
| WASF2 | CDNA clone IMAGE:3030163 | |
| ITGB7 | Integrin, beta 7 | |
| PIP5K2B | Phosphatidylinositol-4-phosphate 5-kinase, type II, beta | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | |
| IQGAP1 | IQ motif containing GTPase activating protein 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| CYFIP1 | Cytoplasmic FMR1 interacting protein 1 | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| ARHGEF7 | Rho guanine nucleotide exchange factor (GEF) 7 | |
| GRLF1 | Glucocorticoid receptor DNA binding factor 1 | |
| FGF2 | Fibroblast growth factor 2 (basic) | |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | |
| MRAS | Muscle RAS oncogene homolog | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| Insulin signaling pathway | PPP1CB | Protein phosphatase 1, catalytic subunit, beta isoform |
| PFKL | Phosphofructokinase, liver | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| PRKAR1A | Protein kinase, cAMP-dependent, regulatory, type I, alpha (tissue specific extinguisher 1) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | |
| PPP1CA | Protein phosphatase 1, catalytic subunit, alpha isoform | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| RHOQ | Ras homolog gene family, member Q | |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | |
| FLOT1 | Flotillin 1 | |
| INPP5D | Inositol polyphosphate-5-phosphatase, 145kDa | |
| IRS1 | Insulin receptor substrate 1 | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| PRKAB2 | Protein kinase, AMP-activated, beta 2 non-catalytic subunit | |
| GYS1 | Glycogen synthase 1 (muscle) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| PYGB | Phosphorylase, glycogen; brain | |
| SLC2A4 | Solute carrier family 2 (facilitated glucose transporter), member 4 | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| CRKL | V-crk sarcoma virus CT10 oncogene homolog (avian)-like | |
| Wnt signaling pathway | CTBP1 | C-terminal binding protein 1 |
| WNT6 | Wingless-type MMTV integration site family, member 6 | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| EP300 | E1A binding protein p300 | |
| CHP | calcium binding protein P22 | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| CTBP2 | C-terminal binding protein 2 | |
| PSEN1 | Presenilin 1 (Alzheimer disease 3) | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| TBL1X | Transducin (beta)-like 1X-linked | |
| PRKCA | Protein kinase C, alpha | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| SMAD3 | SMAD family member 3 | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| VEGF signaling pathway | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) | |
| CHP | calcium binding protein P22 | |
| PRKCA | Protein kinase C, alpha | |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Melanogenesis | CALM1 | Calmodulin 1 (phosphorylase kinase, delta) |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| WNT6 | Wingless-type MMTV integration site family, member 6 | |
| EP300 | E1A binding protein p300 | |
| PRKCA | Protein kinase C, alpha | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| Notch signaling pathway | CIR | CBF1 interacting corepressor |
| CTBP1 | C-terminal binding protein 1 | |
| PCAF | P300/CBP-associated factor | |
| EP300 | E1A binding protein p300 | |
| PSEN1 | Presenilin 1 (Alzheimer disease 3) | |
| CTBP2 | C-terminal binding protein 2 | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| Glioma | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| PRKCA | Protein kinase C, alpha | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Natural killer cell mediated cytotoxicity | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) |
| PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) | |
| CHP | calcium binding protein P22 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| PRKCA | Protein kinase C, alpha | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| SHC1 | SHC (Src homology 2 domain containing) transforming protein 1 | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| ICAM2 | Intercellular adhesion molecule 2 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| T cell receptor signaling pathway | PPP3R1 | Protein phosphatase 3 (formerly 2B), regulatory subunit B, 19kDa, alpha isoform (calcineurin B, type I) |
| CHP | calcium binding protein P22 | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| CBLB | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| PPP3CA | Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform (calcineurin A alpha) | |
| NCK2 | NCK adaptor protein 2 | |
| Leukocyte transendothelial migration | PLCG2 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| ACTN3 | Actinin, alpha 3 | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | |
| ROCK2 | Rho-associated, coiled-coil containing protein kinase 2 | |
| VIL2 | Villin 2 (ezrin) | |
| PRKCA | Protein kinase C, alpha | |
| RASSF5 | Ras association (RalGDS/AF-6) domain family 5 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| RHOH | Ras homolog gene family, member H | |
| GRLF1 | Glucocorticoid receptor DNA binding factor 1 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | |
| CXCL12 | Chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Lysine degradation | DLST | Dihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | |
| EHMT2 | Euchromatic histone-lysine N-methyltransferase 2 | |
| EHMT1 | Euchromatic histone-lysine N-methyltransferase 1 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| Toll-like receptor signaling pathway | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| MYD88 | Myeloid differentiation primary response gene (88) | |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| TRAF6 | CDNA FLJ46539 fis, clone THYMU3037836 | |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| TLR2 | Toll-like receptor 2 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| TICAM2 | Toll-like receptor adaptor molecule 2 | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Long-term depression | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| GNAZ | Guanine nucleotide binding protein (G protein), alpha z polypeptide | |
| GNA12 | Guanine nucleotide binding protein (G protein) alpha 12 | |
| PRKCA | Protein kinase C, alpha | |
| N-Glycan biosynthesis | DHDDS | Dehydrodolichyl diphosphate synthase |
| B4GALT2 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 | |
| MAN1B1 | Mannosidase, alpha, class 1B, member 1 | |
| ALG6 | Asparagine-linked glycosylation 6 homolog (S. cerevisiae, alpha-1,3-glucosyltransferase) | |
| MAN1A2 | Mannosidase, alpha, class 1A, member 2 | |
| RPN1 | Ribophorin I | |
| DOLPP1 | Dolichyl pyrophosphate phosphatase 1 | |
| Neuroactive ligand-receptor interaction | GH2 | Chorionic somatomammotropin hormone 1 (placental lactogen) |
| GPR156 | G protein-coupled receptor 156 | |
| CRHR2 | Corticotropin releasing hormone receptor 2 | |
| UTS2R | Urotensin 2 receptor | |
| PRLH | Prolactin releasing hormone | |
| LTB4R | Leukotriene B4 receptor | |
| Gap junction | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| TUBB8 | tubulin, beta 8 | |
| PRKCA | Protein kinase C, alpha | |
| GnRH signaling pathway | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| MMP2 | Matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) | |
| HBEGF | Heparin-binding EGF-like growth factor | |
| CALM1 | Calmodulin 1 (phosphorylase kinase, delta) | |
| PRKCA | Protein kinase C, alpha | |
| MAP3K4 | Mitogen-activated protein kinase kinase kinase 4 | |
| MAPK14 | Mitogen-activated protein kinase 14 | |
| Tight junction | ACTN3 | Actinin, alpha 3 |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| PRKCA | Protein kinase C, alpha | |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| CSNK2A2 | Casein kinase 2, alpha prime polypeptide | |
| ACTC1 | Actin, alpha, cardiac muscle 1 | |
| MRAS | Muscle RAS oncogene homolog | |
| PRKCH | Protein kinase C, eta | |
| Tryptophan metabolism | TRAF7 | TNF receptor-associated factor 7 |
| OGDH | Oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) | |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| UHRF1 | Ubiquitin-like, containing PHD and RING finger domains, 1 | |
| CYP4F8 | Cytochrome P450, family 4, subfamily F, polypeptide 8 | |
| CAT | Catalase | |
| PRMT2 | Protein arginine methyltransferase 2 | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| CNOT4 | CCR4-NOT transcription complex, subunit 4 | |
| Colorectal cancer | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| BAX | BCL2-associated X protein | |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| SMAD3 | SMAD family member 3 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| DVL3 | Dishevelled, dsh homolog 3 (Drosophila) | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| Pancreatic cancer | AKT1 | V-akt murine thymoma viral oncogene homolog 1 |
| CASP9 | Caspase 9, apoptosis-related cysteine peptidase | |
| RAC3 | Ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | |
| RALBP1 | RalA binding protein 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| IKBKB | Golgi autoantigen, golgin subfamily a, 7 | |
| SMAD3 | SMAD family member 3 | |
| RAC1 | Ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) | |
| TGFBR2 | Transforming growth factor, beta receptor II (70/80kDa) | |
| mTOR signaling pathway | RICTOR | rapamycin-insensitive companion of mTOR |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| EIF4B | Eukaryotic translation initiation factor 4B | |
| RPS6KB2 | Ribosomal protein S6 kinase, 70kDa, polypeptide 2 | |
| RPS6KB1 | Ribosomal protein S6 kinase, 70kDa, polypeptide 1 | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| LYK5 | protein kinase LYK5 | |
| STK11 | Serine/threonine kinase 11 | |
| Melanoma | FGF7 | Fibroblast growth factor 7 (keratinocyte growth factor) |
| AKT1 | V-akt murine thymoma viral oncogene homolog 1 | |
| HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog | |
| FGFR1 | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | |
| HGF | Human mRNA for hepatocyte growth factor (HGF). | |
| FGF2 | Fibroblast growth factor 2 (basic) | |
| PIK3CD | Phosphoinositide-3-kinase, catalytic, delta polypeptide | |
| Neurodegenerative Disorders | BAX | BCL2-associated X protein |
| PRNP | Prion protein (p27–30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | |
| PSEN1 | Presenilin 1 (Alzheimer disease 3) | |
| EP300 | E1A binding protein p300 | |
| GFAP | Glial fibrillary acidic protein | |
| Propanoate metabolism | MUT | Methylmalonyl Coenzyme A mutase |
| ACAT1 | Acetyl-Coenzyme A acetyltransferase 1 (acetoacetyl Coenzyme A thiolase) | |
| HADHA | Hydroxyacyl-Coenzyme A dehydrogenase/3-ketoacyl-Coenzyme A thiolase/enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit | |
| Selenoamino acid metabolism | SEPHS1 | Selenophosphate synthetase 1 |
| PRMT2 | Protein arginine methyltransferase 2 | |
| GGT1 | Gamma-glutamyltransferase 1 | |
| Thyroid cancer | HRAS | V-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| CCDC6 | Coiled-coil domain containing 6 | |
| TPM3 | Tropomyosin 3 | |
| SNARE interactions in vesicular transport | STX6 | Syntaxin 6 |
| YKT6 | YKT6 v-SNARE homolog (S. cerevisiae) | |
| STX16 | Syntaxin 16 | |
| Starch and sucrose metabolism | HK2 | Hexokinase 2 |
| PYGB | Phosphorylase, glycogen; brain | |
| AGL | Amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (glycogen debranching enzyme, glycogen storage disease type III) | |
| GYS1 | Glycogen synthase 1 (muscle) | |
| UGT2B7 | UDP glucuronosyltransferase 2 family, polypeptide B7 | |
| Prion disease | PRNP | Prion protein (p27–30) (Creutzfeldt-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) |
| TNF | Tumor necrosis factor (TNF superfamily, member 2) | |
| GFAP | Glial fibrillary acidic protein |