Abstract
The negative selection of T cells in the thymus is necessary for the maintenance of self tolerance. Thymic medullary epithelial cells have a key function in this process as they express a large number of tissue-specific self antigens that are presented to developing T cells. Mutations in the transcriptional regulator AIRE cause a breakdown of central tolerance associated with decreased expression of self antigens in the thymus. In this Review, we discuss the role of AIRE in the thymus and recent advances in our understanding of how AIRE might function to regulate gene expression.
Autoimmunity is caused by the breakdown of mechanisms that maintain immune tolerance to self tissues. Most self-reactive T cells are deleted in the thymus, resulting in central tolerance [G], which is further supported by regulatory mechanisms outside of primary lymphoid tissues, which are collectively known as peripheral tolerance. As the main mechanism of central tolerance, the negative selection [G] of self-reactive thymocytes occurs mainly in the medullary compartment of the thymus 1, 2. The medullary thymic epithelial cells (mTECs) express a large number of genes, including tissue-specific antigens (TSAs, also named tissue-restricted antigens or peripheral tissue antigens) that are normally present only in specialized peripheral organs and are apparently not required for the direct function of mTECs 3, 4. During negative selection, these encoded TSAs are presented by mTECs or dendritic cells to differentiating thymocytes as self antigens5, 6, leading to the induction of tolerance either by clonal deletion, functional inactivation of self-reactive T cells or clonal deviation 7-9.
The autoimmune regulator (AIRE) protein has been found to be important for the maintenance of self tolerance 10, 11. Mutations in the AIRE gene cause autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED) (BOX 1), a syndrome that is characterized by the presence of autoantibodies specific for multiple self antigens, leading to lymphocytic infiltration of endocrine glands and respective autoimmune disorders 12, 13. Mice with mutations in the Aire gene have pathological autoimmune features similar to those of patients with APECED, with multiorgan lymphocytic infiltration and autoantibody production. Many studies of recent years have shown that AIRE is a crucial factor for the promiscuous expression of TSAs in the thymus, and that mutations in this gene lead to the escape of self-reactive T cells from the thymus, which consequently results in autoimmunity.
BOX 1. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED).
APECED (also known as autoimmune polyendocrine syndrome type 1, APS1) is a rare monogenic autoimmune syndrome (http://www.apeced.net) caused by mutations in autoimmune regulator (AIRE). The disease is more common among certain populations, such as Finnish (1: 25000) and Sardinian (1:14000) populations. Most commonly, patients with APECED have chronic mucocutaneous candidiasis, hypoparathyroidism and Addison’s disease. The spectrum of clinical features is broad and variable and includes several endocrine autoimmune disorders such as premature gonadal failure, hypothyroidism, hypophysitis, pernicious anaemia, type 1 diabetes, autoimmune hepatitis and gastritis, as well as autoimmune skin diseases such as alopecia and vitiligo (for a Review, see REF 12). Patients with APECED characteristically have serum autoantibodies specific for self antigens that often have structural or functional similarities. The self antigens are usually enzymes that are expressed tissue-specifically — for example, steroidogenic P450 enzymes that are expressed in the adrenal cortex and gonads, insulin expressed in pancreatic islets or the recently described NALP5 expressed in parathyroid glands 124 (for a Review, see REF 13). The presence of specific autoantibodies correlates with or can even predict the outcome of disease. For example, autoantibodies specific for type I interferons are so prevalent and specific to patients with APECED that they can be used as diagnostic markers for the disease 125. These antibodies result in a decrease in the expression of interferon-stimulated genes in blood cells 126, which resembles the autoimmune pathogenesis in thymomas 127. Currently, approximately 60 different mutations in the AIRE gene have been reported in patients with APECED, of which the two most common mutations are R257X in exon 6 and a 13-base-pair deletion in exon 8. The mechanistic explanation for the association of the clinically important condition chronic mucocutaneous candidiasis with APECED is unknown. Unfortunately, AIRE-deficient mice seem not to be overtly susceptible to candidiasis (Hubert FX and Scott HS, personal communication), which might result from differences between mouse and human immune systems 128. This underlines the importance of studies with samples from human patients with APECED.
In this Review, we provide an overview of our current knowledge about the function of AIRE in directing the promiscuous expression of TSAs in the thymic medulla to control autoimmunity. In light of recent findings, including the involvement of AIRE in transcription elongation and binding to chromatin, we discuss the molecular mechanisms by which AIRE might regulate gene expression and central tolerance.
AIRE deficiency results in autoimmunity
Similar to patients with APECED, AIRE-deficient mice develop tissue infiltrations of mononuclear cells and autoantibodies specific for multiple peripheral tissues 14-19. Most commonly, the affected tissues are salivary glands, eye, stomach and liver. Recent studies have shown that T cells, in particular T helper 1 (TH1)-polarized CD4+ T cells, are a crucial component of the tissue infiltrations in AIRE-deficient mice, whereas B cells have more limited function but are still required for early and severe autoimmunity 20, 21. However, the autoimmune phenotype varies greatly with different genetic backgrounds 22, which indicates that other genes are important for the overall penetrance [G] of the phenotype and organ specificity. It should be noted that AIRE-deficient mice and patients with APECED have several differences in their target autoantigens and disease manifestations 23, 24. Despite this phenotypic discrepancy, which could be due to different environmental or genetic influences in the two species 24, AIRE-deficient mice still provide a good mechanistic model for human AIRE function and APECED-related autoimmunity.
Importantly, mTECs from AIRE-deficient mice have a profound decrease in expression of multiple transcripts encoding peripheral TSAs 3, 19, including the insulin 2 (Ins2) and salivary protein 1 (Spt1) genes, which seem to be under the direct control of AIRE 25. However, not all transcripts encoding peripherally expressed antigens (such as glutamic acid decarboxylase 67 and C-reactive protein) are downregulated in AIRE-deficient thymus 3, 19, which indicates that other factors are necessary for the efficient expression of these genes.
Experimental evidence for the function of AIRE in negative selection was obtained using mice expressing transgenic T-cell receptors (TCRs) specific for neo-self antigens. AIRE-dependent negative selection of specific TCR-transgenic T cells was observed in transgenic mice expressing hen egg lysozyme 26, 27 or ovalbumin 28 as neo-self antigens from the rat insulin promoter, which drives the expression of these antigens in mTECs in addition to pancreatic beta cells. These data provided compelling evidence that AIRE is essential for efficient thymic T-cell deletion. Importantly, AIRE-deficient double transgenic mice expressing hen egg lysozome 26 or ovalbumin 28 antigens from the rat insulin promoter, together with the respective transgenic TCR, developed aggressive diabetes at birth, showing the functional autoimmune capacity of T cells that escape negative selection.
So, the consequence of AIRE deficiency is thought to be disturbed negative selection of T cells specific for self antigens, which could be caused by defective expression of self antigens by thymic epithelial cells 29. In support of this idea, AIRE-deficient mice spontaneously develop autoimmune uveitis and autoimmune gastritis as a result of autoimmunity against interphotoreceptor retinoid-binding protein (IRBP; also known as retinoid binding protein 3, RBP3) 30 and mucin 6 (MUC6) antigens 31, respectively, the expression of which is regulated by AIRE in mTECs. In humans, differences in AIRE expression level, together with genetic variances in insulin and acetylcholine receptor genes, influence the expression levels of these two established self antigens of type 1 diabetes and autoimmune myasthenia gravis, respectively 32, 33.
Evidence is accumulating, however, that the activation of expression of tissue-specific genes by mTECs is not the only AIRE-associated mechanism involved in negative selection. Strikingly, AIRE-deficient mice also develop autoimmunity to endogenous self antigens whose expression in mTECs is not regulated by AIRE. For example, the autoimmune reaction against AIRE-independently expressed α-fodrin (spectrin-α non-erythrocytic 1, SPTAN1) causes Sjögren-syndrome-like [G] pathology in AIRE-deficient mice 14, and autoimmunity against pancreas-specific protein disulfide isomerase A2 (PDIA2; which is also expressed by mTECs independent of AIRE) causes autoimmune destruction of the exocrine pancreas in AIRE-deficient NOD mice 16. It is possible that the immune reaction to these two antigens is a result of epitope spreading and the initiating autoantigen remains to be identified. However, these observations indicate that AIRE might have roles in negative selection other than activating the expression of peripheral TSAs 18. These mechanisms could be associated with chemokine expression or antigen presentation, as several genes involved in these processes are downregulated in AIRE-deficient mTECs. Indeed, the capacity of AIRE-deficient mTECs to present endogenously synthesized or exogenously added ovalbumin peptide to ovalbumin-reactive T cells is decreased compared with control mTECs 28. Intriguingly, AIRE deficiency was recently reported to cause a developmental block of late-stage CD4+ single-positive thymocyte differentiation, leading to the accumulation of immature CD4+ thymocytes from the thymus 34. This result indicates that in addition to the induction of TSA expression, AIRE might also affect the final maturation of thymocytes.
AIRE expression in mTECs
AIRE is expressed in the thymus by a mTEC subpopulation that is positive for the co-stimulatory markers CD80, CD86 and CD40 and that has high levels of MHC class II expression 35-37. In addition, AIRE expression has been detected by rare cells in the periphery (BOX 2). There are phenotypic and functional differences between mTEC subpopulations and at least two subsets of mTECs can be detected in the thymic medulla, consisting of CD80lowMHC class IIlow and CD80hiMHC classIIhi cells, which have provided a basis for two models of mTEC development (for a review, see REFS 2, 4). According to the terminal-differentiation model, AIRE expression activates the promiscuous expression of TSAs in late-stage mature mTECs (CD80hiMHC classIIhi cells) with efficient antigen-presentation capacity. Another, developmental model proposes that heterogeneity between mTEC populations and the pattern of TSA expression is more compatible with developmental regulation of mTECs by AIRE 38, 39. In this model, AIRE is seen as a transcription factor that directs mTEC differentiation 40. The requirement of AIRE for mTEC differentiation or in forming a tolerogenic environment by regulating the function of neighbouring AIRE-negative mTECs has also been proposed by others 41, 42.
BOX 2. AIRE expression in the periphery.
In addition to AIRE expression in mTECs, AIRE expression has been reported in peripheral lymphoid tissues, such as lymph nodes and spleen, and in other tissues. Most of these reports have described AIRE mRNA in monocytes or dendritic cells (DCs) 35, 121, 129, 130, lymphocytes 131 or spermatogonia 132; however, the expression of AIRE in peripheral tissues at the protein level has been controversial and, for example, a recent study failed to detect AIRE protein in various subsets of DCs 133. Nevertheless, there are several indications of AIRE involvement outside of the thymus as AIRE-deficient mice have increased numbers of blood monocytes and an increased risk of marginal-zone B-cell lymphoma, which might indicate a defect in the early differentiation of myeloid-lineage monocyte and metallophilic macrophage cells, which therefore modifies their phenotype in the periphery 134. In support of AIRE expression by the monocyte-DC lineage, AIRE-deficient DCs have increased ability to activate naïve T cells 135 and have increased expression of VCAM1 135, 136. The expression of AIRE in the periphery is, however, not limited to monocytes and DCs. Recent studies have identified a rare stromal type of Aire-expressing cells in lymph nodes and spleen 122, 137. These cells are negative for the DC marker CD11c and fibroblastic reticular cell markers but express the epithelial marker EpCam, and MHC class II molecules and programmed death-1 ligand 1 (PDL1), but somewhat surprisingly do not express the costimulatory molecules CD80 and CD86. Most importantly, similarly to thymic AIRE-expressing cells, these AIRE+ peripheral cells, which are generally confined to the T-cell zones of the secondary lymphoid organs, can express endogenously expressed TSAs and promote the deletion of self-reactive T cells. As the characterization of the extrathymic AIRE-expressing cell phenotype was made using transgenic mice expressing green fluorescent protein from the AIRE promoter, further studies are required to prove the existence of these cells in more physiological settings and to confirm the presence of AIRE-expressing cells in human secondary lymphoid tissues. The finding that AIRE can regulate the expression of TSAs in peripheral lymphoid tissues lends support to the idea that peripheral expression of self antigens serves as a complement for AIRE-mediated negative selection of self-reactive T cells in the thymus.
The expression of a large pool of TSAs is not a universal property of mTECs as a homogeneous population; rather, the expression of AIRE and individual antigens is restricted to a subset of mTECs, which indicates that there is high heterogeneity between mTEC subpopulations 39, 43, 44. As one factor of heterogeneity, some mTECs proliferate extensively during periods of thymic growth: for example, 2–6% of mTECs divide mitotically with a turnover period of approximately 10–14 days 45-47. AIRE+MHC class II+ mTECs have been shown to differentiate from mTEC progenitors expressing the tight-junction [G] components claudins and occludin 48. Two recent reports showed that AIRE+ MHC class IIhi mTECs are almost entirely postmitotic cells and are derived from mitotic AIRE- MHC class IIhi cells 46, 47. These data support a linear series of differentiation events (from CD80lowAIRE- to CD80hiAIRE- to CD80hiAIRE+ cells); however, other differentiation schemes remain possible 49.
Several studies have addressed the question of the specific signals that are necessary for AIRE expression during mTEC differentiation. A correct three-dimensional medullary microarchitecture and proper signaling is required as mTECs lose AIRE expression when cultured ex vivo 25, 50. In keeping with these requirements, AIRE expression is decreased under conditions where normal thymic architecture is disturbed 51, including defects in components of the alternative NF-κB pathway such as RelB 36, 52, NIK 53 NF-κB2 54 and IKKα 55. Recently, several studies identified RANK signal as an important factor for the differentiation of mTECs from AIRE- cells to AIRE+ cells in the embryonic thymus 50, 56 as well as the role of RANK and CD40 signals in AIRE+ mTEC differentiation in postnatal thymus 57-59.
Structural and functional characteristics of AIRE
Although the role of AIRE in the regulation of promiscuous gene expression is well established, the molecular mechanisms underlying the actions of AIRE are not well understood. It has been unclear how AIRE directs the transcription of TSAs at the molecular level and what are the interacting proteins involved in this process. Several studies using in vitro and in vivo approaches have now revealed new insights into the molecular mechanisms of AIRE function.
Functional domains
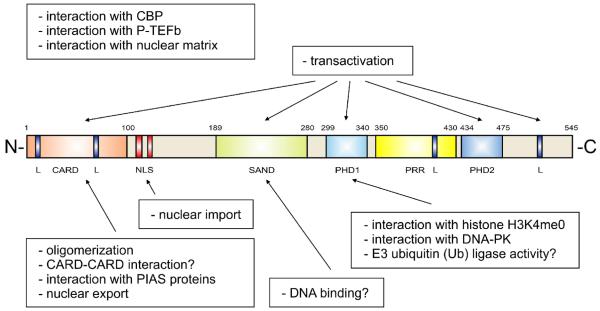
AIRE has a unique combination of domains characteristic of transcriptional regulators and chromatin-binding proteins (Figure 1). It has the closest structural similarity to Sp100 (Speckled protein 100 kDa)-family members, which are involved in transcriptional activation and repression mechanisms 60. Although it is mainly described as a nuclear protein, transfected AIRE is also seen in fibrillar structures in the cytoplasm. In accord with this, AIRE has both a functional nuclear-import signal for the classical importin-α/β pathway 61 and a CRM1-dependent nuclear-export signal in its amino-terminal region 62. The N-terminal region of AIRE was initially described as a HSR (homogenously staining region) domain 63; however, the recent structural reanalysis has revealed a six-helix structure with high similarity to a caspase-recruitment domain (CARD) [G] 64, 65. The CARD has been implicated in oligomerization of AIRE 62, 66 and in dimerization of other proteins that function in inflammation or apoptosis 67.
Figure 1. Schematic representation of human AIRE protein.
The domains and functional elements of the AIRE protein are shown in different colors; CARD (caspase-recruitment domain), SAND (Sp100, AIRE-1, NucP41/P75 and Drosophila DEAF-1), PHD (plant homeodomain), PRR (proline-rich region), L (LXXLL motive), NLS (nuclear-localization signal). Functions of AIRE are shown in boxes and indicated by arrows pointing to the corresponding domain, if known. The CARD (caspase-recruitment domain) has been implicated in homo-oligomerization and is required for nuclear-dot formation and for the heterodimerization of CARD proteins that function in inflammation or apoptosis. Interaction of the CARD with PIAS1 (Protein Inhibitors of Activated STAT) proteins influences the transactivation ability of AIRE. SAND is a putative DNA-binding domain. The first PHD finger has been shown to interact with histone H3 unmethylated at lysine 4 and DNA-PK (DNA protein kinase). Both of these interactions are required for proper transactivation. In addition, PHD1 has been shown to have E3 ubiquitin ligase activity, a mechanism by which a ubiquitin molecule is covalently attached to the target protein. AIRE has also been shown to interact with the transcriptional coactivator CBP (CREB binding protein) and positive transcription elongation factor b (P-TEFb). The latter interaction indicates that AIRE might function in elongation rather than the initiation of transcription. In addition, AIRE has shown to bind the nuclear matrix.
AIRE and Sp100 proteins share a SAND (Sp100, AIRE1, NucP41/75, DEAF1) domain. As an indication of the functional importance of the SAND domain, a dominant-negative mutation in this domain (G228W) is found in patients with APECED. The G228W mutation disrupts the transactivation capacity 68 and alters the subcellular localization of the AIRE protein in vitro and in vivo 69, and it predisposes to autoimmune thyroiditis as a single-copy mutation in humans and mice 69, 70. In its carboxy-terminal region, AIRE contains two PHD (plant homeodomain)-type zinc fingers [G] (also present in Sp100 proteins) that adopt a globular fold in which two zinc atoms coordinate cysteine and histidine residues 71. Mutations in the PHD fingers markedly decrease the transcriptional-activation capacity of the protein 63, 72-74. Although one of the AIRE PHD fingers has been suggested to act as an E3 ubiquitin ligase 75, this has not been confirmed by others 71. In addition to these well-conserved domains, the AIRE protein has four nuclear-receptor-binding LXXLL motifs (where X is any amino acid), which also occur in transcriptional coactivator proteins such as CBP, p300 and SRC1 76. AIRE has also a proline-rich region that can be found in other proteins involved in transcription; for example, in the transcriptional activator protein TAp63, the proline-rich region is required for transcriptional activity and pro-apoptotic functions 77. All of the functional domains are highly conserved between mammalian AIRE homologues, and human AIRE has significant sequence identity with mouse, rat, bovine and dog AIRE proteins 78, 79.
Localization to nuclear bodies
In the cell nucleus, AIRE is located in discrete dot-like structures that resemble, but are distinct from, PML (promyelocytic leukemia) nuclear bodies [G]. Recently, these AIRE bodies were shown to be distributed in vivo immediately adjacent to nuclear speckles 69. The nuclear-body localization of AIRE depends on the CARD, as many mutations in the CARD block the formation of these bodies 62, 65, 74. However, the involvement of other AIRE domains in nuclear-body localization, such as the SAND domain, has also been reported 62, 65, 66, 80-82. The PML bodies contain many proteins including Sp100-family members and the AIRE-interacting protein CBP (see later), and they have been implicated in diverse cellular processes including cell-growth control, antiviral defence, tumour suppression and apoptosis 83. Recent studies have indicated that they have a role in the modulation of transcription at specific genomic loci. For example, PML bodies were shown to specifically associate with the MHC locus 84, 85. Nuclear bodies are tightly bound to the nuclear matrix [G], which provides a structural network for chromatin organization and gene regulation; consistent with AIRE localization to nuclear bodies, a large proportion of the AIRE protein is associated with the nuclear matrix 86, 87. It should be noted that AIRE nuclear structures (similar to PML bodies) do not contain chromatin or RNA polymerase II 88 and, therefore, are not sites of transcription. Nevertheless, AIRE nuclear bodies might influence the nuclear organization of chromatin by indirect contacts. As exemplified in studies with PML bodies, it has been proposed that AIRE regulates gene expression by recruiting a transcriptional-activation complex to specific regions of the genome through interactions with the nuclear matrix 86, 87. It should be noted, however, that despite AIRE localization to nuclear bodies, a significant amount of AIRE protein is also detectable in the nucleoplasm 86.
AIRE as a transcriptional activator
Even without functional experiments, the presence of domains that are characteristic of a transcriptional protein, together with its localization to nuclear bodies, indicate a role for AIRE in the regulation of gene expression. Many studies of transfected AIRE in tissue culture cells have confirmed the function of AIRE as a potent transcriptional activator of various reporter and endogenous promoters 62, 63, 65, 66, 68, 73, 74, 89-91. The transcriptional-activation region of AIRE was initially mapped to the two C-terminal PHD fingers, as mutations in these domains markedly decreased transactivation capacity 63, 72. More recent reports have shown that several APECED-causing mutations in the CARD or SAND domain of AIRE also decrease its transactivation ability, indicating that all of the main AIRE domains participate in transcriptional activation, although the extent of the effect varies between the mutations 65, 74, 88.
Most likely, AIRE acts as a coactivator in a large transcriptional complex. Although in vitro DNA-binding experiments and chromatin-immunoprecipitation experiments in vivo indicate that AIRE might ‘prefer’ certain DNA sequences 81, 92, 93, conclusive data showing that AIRE binds to defined cis-acting sequences are still lacking. The crystal structure of the Sp100B and GMEB (glucocorticoid modulatory element binding protein) SAND domains has been solved 94, 95. Both of these proteins have been shown to bind DNA through a K(D/N)WK motif, which is replaced with a KNKA motif in the AIRE protein, indicating that there are differences in the binding characteristics between these proteins. Considering the large number and chromosomal clustering 96 of AIRE-regulated genes, AIRE does not seem to act as a specific DNA-binding transcription factor, but rather has a more general function in transcription.
In the context of transcriptional activation, several protein partners with seemingly diverse functions have been identified for AIRE. The first protein found to bind directly to AIRE was CBP63, a common transcriptional coactivator that is localized to PML bodies. CBP and AIRE preferentially colocalize to nuclear bodies in cells that lack or have relatively low expression level of the PML protein 63. CBP and AIRE coactivate the transcription of various reporter promoters and endogenous genes in tissue culture cells 65, 86, 88. The relevance of CBP for AIRE activity seems to be integrated with mTEC differentiation, as signaling through RANK (which induces the differentiation of mTECs from AIRE- to AIRE+ cells), but not LTBR, results in the translocation of CBP from the cytoplasm to the nucleus and the coaccumulation of AIRE and CBP in mTEC nuclear dots 65. So, before the expression of AIRE protein, the immature (CD80low) mTECs might lack the intranuclear architecture required to trigger nuclear localization of CBP, resulting in failure of this transcriptional complex to reach its nuclear targets 65. As CBP typically functions in transcription initiation, as well as in the acetylation of histones and non-histone proteins 97, it is possible that the collaborative action of AIRE and CBP results in histone acetylation, the recruitment of chromatin-remodeling and general transcription factors and promotion of transcription of the target genes.
In addition to CBP, the DNA-dependent protein kinase (DNA-PK) complex and SP-RING domain protein inhibitor of activated STAT 1 (PIAS1) have been identified as protein partners of AIRE 89, 98. Both of these proteins are known to bind nuclear-matrix-associated DNA sequences 98-100. Earlier studies have shown that the nuclear matrix is attached to nuclear bodies and can function as a platform for the assembly of macromolecular structures including chromatin 101, 102. In agreement with this, matrix-associated DNA regions frequently colocalize with enhancer [G] and insulator [G] regions. It is therefore conceivable that the interaction of AIRE with DNA-PK and PIAS1 is important for the formation of AIRE-associated nuclear structures or in the regulation of AIRE-mediated gene expression. For example, AIRE expression increases the formation of PIAS1-containing nuclear bodies, which are distinct from but localized adjacent to AIRE-containing bodies. This indicates that AIRE and PIAS1 might associate with a common complex through other components of the nuclear matrix 98. Simultaneous expression of PIAS1 and AIRE resulted in the coactivation of an insulin-promoter-driven reporter and this coactivation was dependent on the SP-RING domain of PIAS1 98. By contrast, AIRE acted as a transcriptional repressor on the promoter of the housekeeping gene encoding cystatin B. These results indicate that the effects of AIRE on different gene promoters are modulated by interactions with other transcriptional regulators such as PIAS1, but might also depend on DNA elements or the chromatin structure of the specific promoter. Another AIRE-interacting and nuclear-matrix-associated protein, the DNA-PK complex, has an important role in DNA repair and in phosphorylation of many proteins involved in transcription and cell cycling. DNA-PK mainly phosophorylates target proteins that are attached to the same DNA molecule, and indeed, the in vitro phosphorylation of two residues (Thr68 and Ser156) in the N-terminal region of AIRE by DNA-PK depends on the presence of DNA. The interaction with DNA-PK also influences AIRE-mediated transcriptional regulation, as mutations in the putative DNA-PK phosphorylation sites in AIRE markedly decrease its transcriptional-activation activity 89. Although the functional details of the interaction of AIRE with these proteins remain to be studied, both DNA-PK and PIAS1 interactions implicate AIRE in nuclear-matrix-mediated transcriptional regulation.
Transcriptional elongation and histone binding
Although there is general agreement about the role of AIRE in transcriptional regulation, the molecular mechanisms underlying this function are not well understood. How does AIRE influence the expression of so many genes? Recent studies associate the function of AIRE with basic, but apparently separate, aspects of transcription, providing novel insights into these mechanisms.
One study showed that AIRE promotes transcriptional elongation by binding and recruiting the positive transcription elongation factor b (P-TEFb) complex to RNA polymerase II to target gene promoters 91. Mammalian gene transcription is initiated by binding of RNA polymerase II to gene promoters, thereby ensuring correct formation of the preinitiation complex for transcription. This complex initiates transcription but does not continue to the elongation process unless P-TEFb forms a complex with RNA polymerase II. Recruitment of P-TEFb removes the negative elongation factor (N-TEF), mediates the phosphorylation of RNA polymerase II and thereby allows transcription to proceed to the elongation phase. Active P-TEFb is a heterodimer of a C-type cyclin (CYCT1, CYCT2 or CYCK) and cyclin-dependent kinase 9 (CDK9) 103. Both CDK9 and CYCT1 were shown to interact with AIRE in cell lysates and CYCT1 was seen to partially co-localize with AIRE in the same nuclear structures. Furthermore, it was found that CYCT1 and CDK9 are present on target gene (such as Ins2 and Spt1) promoters and coding regions only in the presence of AIRE. Moreover, mutations in the mouse CycT1 gene correlated with decreased thymic expression of the Ins2 and Spt1 genes, and these mice had lymphocytic infiltrations in their lacrimal glands 91. These findings link CycT1 and Cdk9 genes to autoimmune pathogenesis, and more importantly, indicate a role for AIRE as a global transcriptional activator affecting different target genes through the stimulation of transcriptional elongation.
Two other studies stem from the recent findings implicating PHD fingers as interaction domains for histone H3, which is a core structural unit of nucleosomes in chromatin 104. Interactions between chromatin-binding proteins and histones are subject to subtle changes as a result of epigenetic modifications of histones 105. These modifications greatly influence chromatin accessibility and the epigenetic control of transcribed genes. In particular, trimethylation of histone H3 lysine 4 (H3K4me3) is highly associated with actively transcribed gene promoters, whereas lack of H3 lysine 4 methylation (H3K4me0) is associated with silenced genes. Two studies showed that the first PHD finger of AIRE preferentially binds to the N-terminal tail of histone H3 when lysine 4 is unmethylated, which indicates a possible role for AIRE in the epigenetic control of target genes 106, 107. Consistent with this hypothesis, AIRE can activate promoters with low levels of H3K4me3 in a cell-culture model 106. So, it is proposed that outside of their normal functional cell environment, for example in mTECs, TSA genes are usually not highly expressed and therefore are unmethylated or monomethylated at histone H3 lysine 4 106. After the expression of AIRE by CD80hiMHC class IIhi mTECs, AIRE functions as a transcriptional activator and can ‘read’ the unmethylated H3K4 to initiate gene expression. Genome-wide studies of chromatin modifications in pluripotent and differentiated cells have divided genes into three distinct groups based on the abundance of transcriptionally active H3K4me3 and silent H3K27me3 modifications 108. As expected, genes containing only H3K4me3 are typically active and genes containing both modifications are poised for transcription. A third set of genes that lacks both modifications in embryonic stem cells is mainly expressed in differentiated tissues and only obtains significant H3K4 methylation when the cells are committed to specific lineages 109. This would indicate that TSA genes lacking H3K4me3 on their promoters in mTECs would be preferential targets for AIRE-mediated transcriptional control. In light of the function of AIRE in enhancing the transcription-elongation process and the finding that AIRE preferentially binds to particular histone modifications that are often found in transcriptionally silent promoters, it can be proposed that AIRE recruits elongation and co-transcriptional mRNA-processing factors to TSA gene promoters that are otherwise inactive (Figure 2). As a result of AIRE recruitment to such promoters of TSA genes, expression of a wide range of TSAs would be activated in mTECs. It remains to be determined whether the recruitment of AIRE to target promoters correlates with the epigenetic changes of histone modifications.
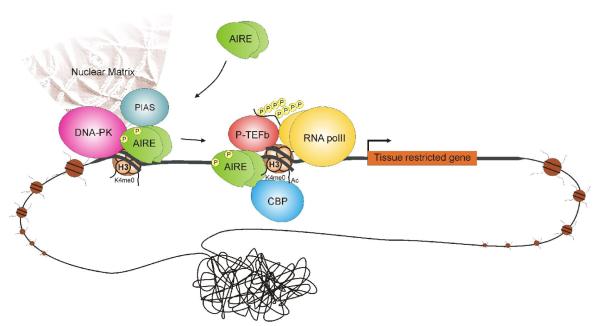
Figure 2. Proposed mechanism of AIRE-mediated gene activation.
AIRE is preferentially recruited to promoters with low levels of histone H3K4me3 (histone H3 trimethylated at lysine 4). In addition to histone binding, a direct interaction with DNA might be necessary to guide AIRE to specific regions. On target gene regulatory regions, AIRE recruits a P-TEFb complex that phosporylates serine residues of RNA polymerase II. This phosphorylation converts the polymerase from a paused to elongating form and results in the activation of gene expression. AIRE also interacts with CBP, which acetylates histones allowing further access to chromatin and DNA. Interaction of AIRE with DNA-PK and PIAS proteins further modulates transcriptional activation. DNA-PK can phosphorylate AIRE and, through the interaction with nuclear matrix, collaborate with AIRE in the formation of chromatin loops. Because of the adjacent localization of PIAS1- and AIRE-containing nuclear bodies, PIAS1 might also function in the nuclear organization of chromatin.
Stochastic gene regulation?
Analysis of gene-expression patterns in AIRE-deficient mice and in transfected cells indicates that AIRE might function not only as an activator but also as a repressor of multiple genes 87, 96, 98, 110. Although it cannot be excluded that the repressive effects of AIRE are indirect, this bidirectional nature of transcriptional regulation and the large number of regulated genes indicates that AIRE has a more complicated function than simple transcription-factor activity.
As the AIRE-regulated genes tend to occur in clusters, it has been proposed that AIRE functions as a regulator of genomic clusters 87, 96, 111. Co-regulation of clustered genes is a common phenomenon in the eukaryotic genome and is found in many cell types. According to this view, AIRE is recruited to discrete locations in chromatin together with other components of the transcriptional machinery 87, possibly through the interaction with the nuclear matrix. As a result, the expression of certain genes within the cluster is activated, whereas transcription of other genes is repressed (Figure 3). So far, however, there is no evidence that AIRE binds to any specific regulatory element or somehow changes chromatin structure within the gene clusters. The observed clustering of AIRE target genes and their expression by mTECs could be due to the accessibility of open chromatin regions, independent of the tissue specificity of the genes or their requirement for thymic negative selection 3. In this light, it is conceivable that AIRE functions in protein complexes of transcriptional regulators that access specific chromatin regions.
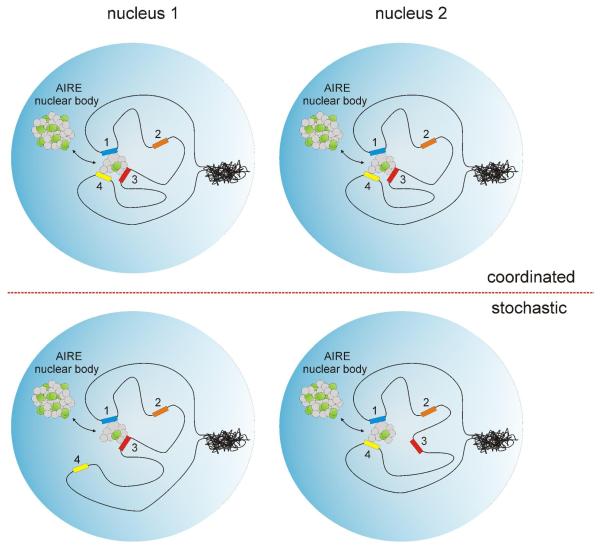
Figure 3. Stochastic and coordinated regulation of target genes.
In the nucleus AIRE (green circles) is seen in nuclear bodies or recruited to target gene regions. A genomic cluster with four genes is depicted. Genes 1, 3, and 4 are regulated by AIRE; gene 2 is an AIRE-independent gene, which can be active or inactive. In the case of coordinated regulation (upper part), tight transcriptional control is used and the same genes are expressed in nucleus 1 and nucleus 2. Stochastic expression (lower part) leads to fluctuations in the genes that are activated in different nuclei, so that various combinations of clustered genes are expressed in each single cell. In both cases, all three AIRE-regulated cluster genes are expressed when analyzed on a population level. The expression of gene 2 might or might not be influenced by indirect changes in chromatin structure. For example, chromatin reorganization by AIRE might lead to mild repression of gene 2.
A recent single-cell PCR analysis of genes localized to the casein locus, which is one of the gene clusters regulated by AIRE, showed that random combinations of casein locus genes were expressed by single mTECs rather than the uniform coexpression of all genes in the locus by every cell, as occurs in mammary epithelium 44. These findings were also supported by another study analysing S100 gene cluster 112. Although the approach has its limitation by providing a static picture of an otherwise very dynamic process, it might be anticipated that this phenomenon applies to other gene clusters that are regulated by AIRE. This indicates that AIRE might activate genes in a stochastic manner (Figure 3). Recent studies have shown that identical cell clones show variations in the expression of proteins in an individual cell. For example, a transcriptional regulator that is equally present in two identical cells might be bound to a promoter in one cell and unbound in another cell, resulting in unequal amounts of expressed protein per cell 113. The stochastic nature of gene expression is influenced by fluctuations in several factors, including varying amounts of gene regulators, availability of chromosomal positions or posttranscriptional regulation events such as mRNA processing and stability 114-116. The emerging view is that the assembly of large macromolecular transcriptional complexes on chromatin occurs through the short-period stochastic recruitment of soluble preformed subcomplexes. According to this so-called self-organization model, transcriptional protein subcomplexes freely diffuse in the nucleoplasm, scan for specific binding sites, and stochastically assemble around the genes forming transcriptional centers for a short period of time 117. Furthermore, AIRE-regulated genes in mTECs seem to be expressed at relatively low levels 19, 44(P.P., T.O. and A.R., unpublished observations). This observation might be seen as additional evidence for stochastic gene expression of AIRE regulated genes, as genes with a low expression level tend to have more fluctuations in their expression and are more susceptible to stochastic transcriptional changes 118, 119.
The stochastic model of AIRE action is not in conflict with the proposed role of AIRE in the regulation of genomic clusters. Large chromatin loops containing gene clusters are formed in the nucleus and placed into distinct nuclear environments to optimize their activity. The spatial distribution and transcriptional activation of clusters is driven by stochastic self-organization in the nucleus that might contribute to various gene-expression patterns 120. Accordingly, the selection and the control of AIRE-regulated genes would be determined by the epigenetic or transcriptional status of the target genes and by other transcriptional proteins, which could vary between specific cellular environments. In support of this, there is little overlap of gene expression between mTECs, extrathymic AIRE-expressing cells in lymph nodes and various cell lines with overexpression of AIRE 19, 121, 122 (P.P., T.O. and A.R., unpublished observations). In conclusion, the role of AIRE in the regulation of gene clusters, presumably acting in a stochastic manner, and its function in transcriptional elongation and histone binding makes AIRE an interesting model protein, not only for studies of the mechanisms of immune tolerance but also for research into the transcriptional regulation of complex higher-order chromatin structures.
Future directions
Unravelling the mechanisms of negative selection of self-reactive T cells in the thymus is important to improve our knowledge of the development of autoimmune pathogenesis. Clearly, AIRE is one of the key regulators of promiscuous expression and is central to the thymic-selection process, as a considerable amount of data accumulated from mouse studies have shown its role in the deletion of self-reactive T cells. Recent studies, however, challenge the view that AIRE-mediated self-antigen expression is the only reason for defective negative selection in Aire-deficient thymus (for review 18, 123). In this context, it will be important to clarify whether and how AIRE contributes to other mechanisms in the thymus, for example by promoting the expression of chemokines, increasing antigen-processing and/or -presentation capacity of mTECs or modulating thymic differentiation programs. Obviously, our understanding of cellular events needs to be integrated with the information about the molecular mechanisms of AIRE action, and therefore, the interacting partners and role of AIRE in transcription, chromatin organization and epigenetic regulation remain important topics for further studies. Finally, we have just begun to understand the specific signals and factors that determine the expression of AIRE in mTECs and further analysis of these mechanisms should help us to modulate the expression of TSAs (for example insulin) and hopefully to develop better treatments to prevent autoimmunity.
Acknowledgements
The authors would like to thank all laboratory members and many colleagues including M. Anderson, G. Holländer, K. Krohn, O. Kämpe, B. Kyewski, A. Marx, M. Matsumoto, G. Musco, T. Rich, H. Scott, I. Ulmanen and N. Willcox and useful discussions. This work was supported by EU FP6 project Thymaide, The Wellcome Trust, European Regional Development Fund and Estonian Science Foundation.
Glossary
- Negative selection
The deletion of self-reactive thymocytes in the thymus. Thymocytes expressing T-cell receptors that strongly recognize self-peptide bound to self-MHC molecules undergo apoptosis in response to the signalling generated by high-affinity binding of their target antigen presented by cells in the thymic medulla.
- Central tolerance
The lack of self-responsiveness that occurs during lymphocyte development in the central lymphoid organs. B-cell progenitors in the bone marrow and T-cell progenitors in the thymus that strongly recognize self antigen either undergo further rearrangement of antigen-receptor genes to avoid reactivity to self or face deletion by apoptosis.
- Non-obese diabetic (NOD) mice
NOD mice develop spontaneous diabetes, including the development of islet-specific autoantibodies and inflammation of the pancreatic islets.
- Penetrance
The proportion of affected individuals among carriers of a particular genotype. If all individuals with a disease genotype show the disease phenotype, then the disease is said to be completely penetrant.
- Sjögren syndrome
An organ-specific autoimmune disorder characterized by lymphocytic infiltrates, tissue destruction and functional decline of salivary or lacrimal glands, and the systemic production of autoantibodies to ribonucleoproteins.
- Tight junction
A belt-like region of adhesion between adjacent epithelial or endothelial cells that regulates paracellular flux. Tight-junction proteins include the integral membrane proteins occludin and claudin, in association with cytoplasmic zonula occludens proteins.
- Lymphoid-tissue inducer cells
A cell that is present in developing lymph nodes, Peyer’s patches and nasopharynx-associated lymphoid tissue (NALT). Lymphoid-tissue inducer cells are required for the development of these lymphoid organs. The inductive capacity of these cells for the generation of Peyer’s patches and NALT has been shown by adoptive transfer, and it is generally assumed that they have a similar function in the formation of lymph nodes.
- Caspase-recruitment domain
(CARD). A protein domain that is found in certain initiator caspases (for example, mammalian caspase-9) and their adaptor proteins (for example, apoptotic-protease-activating factor 1, APAF1) that function in inflammation and apoptosis. This domain mediates protein-protein interaction.
- PHD zinc-finger domains
These comprise about 60 amino acids and are characterized by a C4HC3 (four cysteines, one histidine, three cysteines) signature that binds two zinc ions. They have been shown recently to bind to histone H3 modified or unmodified at specific residues.
- Nuclear bodies
Structurally and functionally heterogenous subnuclear structures (0.2–1μm) present in most mammalian cells that have been implicated in cellular senescence, apoptosis, proliferation and maintenance of genomic stability through transcriptional repression, transcriptional activation and protein degradation.
- Nuclear matrix
A network of nuclear proteins that provides a structural framework for chromatin organization.
- Nuclear speckles
Dynamic subnuclear structures that contain pre-messenger RNA splicing factors and other proteins that are involved in transcription, 3′- end RNA-processing and reversible protein phosphorylation.
- Insulator
A boundary element in genome that may block activity of nearby enhancer regions or function as barrier against heterochromatin spreading into active chromatin area.
- Enhancer
A region of DNA that can bind activator proteins, which can initiate the transcription of a gene promoter that is a long distance away from the enhancer.
Footnotes
- AIRE is mainly expressed by mature thymic medullary epithelial cells, where it promotes the promiscuous expression of many tissue-specific antigens (TSAs).
- TSAs that are upregulated by AIRE are presented to developing thymocytes, and this is required for efficient negative selection. Aberrant negative selection in the absence of AIRE leads to the escape of self-reactive thymocytes to the periphery and subsequent autoimmunity.
- AIRE contains several domains that are characteristic of transcriptional regulators and chromatin-binding proteins, such as a CARD, a SAND domain and PHD fingers. Concordantly, many studies have confirmed AIRE’s function as a potent transcriptional activator.
- Several proteins have been found to interact with AIRE, including CBP, PIAS1, DNA-PK, histone H3 with an unmethylated N-terminus (lysine 4) and P-TEFb.
- Interaction with histone H3 that is unmethylated at lysine 4 allows AIRE to bind to certain chromatin regions. At target gene promoters, AIRE promotes transcriptional elongation by binding and recruiting the positive transcription elongation factor b (P-TEFb) complex to RNA polymerase II.
- AIRE-regulated genes tend to cluster in the genome; however, recent findings indicate that at a single-cell level, the expression of TSAs varies, which indicates that gene activation by AIRE can be a stochastic event.
References
- 1.Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–35. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- 2.Anderson G, Lane PJ, Jenkinson EJ. Generating intrathymic microenvironments to establish T-cell tolerance. Nat Rev Immunol. 2007;7:954–63. doi: 10.1038/nri2187. [DOI] [PubMed] [Google Scholar]
- 3.Derbinski J, et al. Promiscuous gene expression in thymic epithelial cells is regulated at multiple levels. J Exp Med. 2005;202:33–45. doi: 10.1084/jem.20050471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kyewski B, Klein L. A central role for central tolerance. Annu Rev Immunol. 2006;24:571–606. doi: 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- 5.Gallegos AM, Bevan MJ. Central tolerance to tissue-specific antigens mediated by direct and indirect antigen presentation. J Exp Med. 2004;200:1039–49. doi: 10.1084/jem.20041457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aschenbrenner K, et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol. 2007;8:351–8. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 7.Palmer E. Negative selection--clearing out the bad apples from the T-cell repertoire. Nat Rev Immunol. 2003;3:383–91. doi: 10.1038/nri1085. [DOI] [PubMed] [Google Scholar]
- 8.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol. 2005;5:772–82. doi: 10.1038/nri1707. [DOI] [PubMed] [Google Scholar]
- 9.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 10.Nagamine K, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–8. doi: 10.1038/ng1297-393. [DOI] [PubMed] [Google Scholar]
- 11.An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. doi: 10.1038/ng1297-399. [DOI] [PubMed] [Google Scholar]
- 12.Perheentupa J. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab. 2006;91:2843–50. doi: 10.1210/jc.2005-2611. [DOI] [PubMed] [Google Scholar]
- 13.Peterson P, Peltonen L. Autoimmune polyendocrinopathy syndrome type 1 (APS1) and AIRE gene: new views on molecular basis of autoimmunity. J Autoimmun. 2005;25(Suppl):49–55. doi: 10.1016/j.jaut.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 14.Kuroda N, et al. Development of autoimmunity against transcriptionally unrepressed target antigen in the thymus of Aire-deficient mice. J Immunol. 2005;174:1862–70. doi: 10.4049/jimmunol.174.4.1862. [DOI] [PubMed] [Google Scholar]
- 15.Ramsey C, et al. Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet. 2002;11:397–409. doi: 10.1093/hmg/11.4.397. [DOI] [PubMed] [Google Scholar]
- 16.Niki S, et al. Alteration of intra-pancreatic target-organ specificity by abrogation of Aire in NOD mice. J Clin Invest. 2006;116:1292–301. doi: 10.1172/JCI26971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang W, Anderson MS, Bronson R, Mathis D, Benoist C. Modifier loci condition autoimmunity provoked by Aire deficiency. J Exp Med. 2005;202:805–15. doi: 10.1084/jem.20050693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathis D, Benoist C. A decade of AIRE. Nat Rev Immunol. 2007;7:645–50. doi: 10.1038/nri2136. [DOI] [PubMed] [Google Scholar]
- 19.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 20.Gavanescu I, Benoist C, Mathis D. B cells are required for Aire-deficient mice to develop multiorgan autoinflammation: A therapeutic approach for APECED patients. Proc Natl Acad Sci U S A. 2008;105:13009–14. doi: 10.1073/pnas.0806874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devoss JJ, et al. Effector mechanisms of the autoimmune syndrome in the murine model of autoimmune polyglandular syndrome type 1. J Immunol. 2008;181:4072–9. doi: 10.4049/jimmunol.181.6.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venanzi ES, Melamed R, Mathis D, Benoist C. The variable immunological self: Genetic variation and nongenetic noise in Aire-regulated transcription. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0808070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontynen N, et al. Aire deficient mice do not develop the same profile of tissue-specific autoantibodies as APECED patients. J Autoimmun. 2006;27:96–104. doi: 10.1016/j.jaut.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Kekalainen E, Miettinen A, Arstila TP. Does the deficiency of Aire in mice really resemble human APECED? Nat Rev Immunol. 2007;7:1. doi: 10.1038/nri2136-c1. [DOI] [PubMed] [Google Scholar]
- 25.Kont V, et al. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol. 2008;45:25–33. doi: 10.1016/j.molimm.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liston A, et al. Gene dosage--limiting role of Aire in thymic expression, clonal deletion, and organ-specific autoimmunity. J Exp Med. 2004;200:1015–26. doi: 10.1084/jem.20040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003;4:350–4. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 28.Anderson MS, et al. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–39. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Villasenor J, Benoist C, Mathis D. AIRE and APECED: molecular insights into an autoimmune disease. Immunol Rev. 2005;204:156–64. doi: 10.1111/j.0105-2896.2005.00246.x. [DOI] [PubMed] [Google Scholar]
- 30.DeVoss J, et al. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med. 2006;203:2727–35. doi: 10.1084/jem.20061864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavanescu I, Kessler B, Ploegh H, Benoist C, Mathis D. Loss of Aire-dependent thymic expression of a peripheral tissue antigen renders it a target of autoimmunity. Proc Natl Acad Sci U S A. 2007;104:4583–7. doi: 10.1073/pnas.0700259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabater L, et al. Insulin alleles and autoimmune regulator (AIRE) gene expression both influence insulin expression in the thymus. J Autoimmun. 2005;25:312–8. doi: 10.1016/j.jaut.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 33.Giraud M, et al. An IRF8-binding promoter variant and AIRE control CHRNA1 promiscuous expression in thymus. Nature. 2007;448:934–7. doi: 10.1038/nature06066. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Developmental pathway of CD4+CD8- medullary thymocytes during mouse ontogeny and its defect in Aire-/- mice. Proc Natl Acad Sci U S A. 2007;104:18175–80. doi: 10.1073/pnas.0708884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heino M, et al. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem Biophys Res Commun. 1999;257:821–5. doi: 10.1006/bbrc.1999.0308. [DOI] [PubMed] [Google Scholar]
- 36.Zuklys S, et al. Normal thymic architecture and negative selection are associated with Aire expression, the gene defective in the autoimmune-polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) J Immunol. 2000;165:1976–83. doi: 10.4049/jimmunol.165.4.1976. [DOI] [PubMed] [Google Scholar]
- 37.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001;2:1032–9. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 38.Gillard GO, Farr AG. Contrasting models of promiscuous gene expression by thymic epithelium. J Exp Med. 2005;202:15–9. doi: 10.1084/jem.20050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gillard GO, Farr AG. Features of medullary thymic epithelium implicate postnatal development in maintaining epithelial heterogeneity and tissue-restricted antigen expression. J Immunol. 2006;176:5815–24. doi: 10.4049/jimmunol.176.10.5815. [DOI] [PubMed] [Google Scholar]
- 40.Gillard GO, Dooley J, Erickson M, Peltonen L, Farr AG. Aire-dependent alterations in medullary thymic epithelium indicate a role for Aire in thymic epithelial differentiation. J Immunol. 2007;178:3007–15. doi: 10.4049/jimmunol.178.5.3007. [DOI] [PubMed] [Google Scholar]
- 41.Matsumoto M. Transcriptional regulation in thymic epithelial cells for the establishment of self tolerance. Arch Immunol Ther Exp (Warsz) 2007;55:27–34. doi: 10.1007/s00005-007-0007-9. [DOI] [PubMed] [Google Scholar]
- 42.Meager A, Peterson P, Willcox N. Hypothetical review: thymic aberrations and type-I interferons; attempts to deduce autoimmunizing mechanisms from unexpected clues in monogenic and paraneoplastic syndromes. Clin Exp Immunol. 2008;154:141–51. doi: 10.1111/j.1365-2249.2008.03739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanahan D. Peripheral-antigen-expressing cells in thymic medulla: factors in self-tolerance and autoimmunity. Curr Opin Immunol. 1998;10:656–62. doi: 10.1016/s0952-7915(98)80085-x. [DOI] [PubMed] [Google Scholar]
- 44.Derbinski J, Pinto S, Rosch S, Hexel K, Kyewski B. Promiscuous gene expression patterns in single medullary thymic epithelial cells argue for a stochastic mechanism. Proc Natl Acad Sci U S A. 2008;105:657–62. doi: 10.1073/pnas.0707486105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gray DH, et al. Developmental kinetics, turnover, and stimulatory capacity of thymic epithelial cells. Blood. 2006;108:3777–85. doi: 10.1182/blood-2006-02-004531. [DOI] [PubMed] [Google Scholar]
- 46.Gray D, Abramson J, Benoist C, Mathis D. Proliferative arrest and rapid turnover of thymic epithelial cells expressing Aire. J Exp Med. 2007;204:2521–8. doi: 10.1084/jem.20070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabler J, Arnold J, Kyewski B. Promiscuous gene expression and the developmental dynamics of medullary thymic epithelial cells. Eur J Immunol. 2007;37:3363–72. doi: 10.1002/eji.200737131. [DOI] [PubMed] [Google Scholar]
- 48.Hamazaki Y, et al. Medullary thymic epithelial cells expressing Aire represent a unique lineage derived from cells expressing claudin. Nat Immunol. 2007;8:304–11. doi: 10.1038/ni1438. [DOI] [PubMed] [Google Scholar]
- 49.Dooley J, Erickson M, Farr AG. Alterations of the medullary epithelial compartment in the Aire-deficient thymus: implications for programs of thymic epithelial differentiation. J Immunol. 2008;181:5225–32. doi: 10.4049/jimmunol.181.8.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossi SW, et al. RANK signals from CD4(+)3(-) inducer cells regulate development of Aire-expressing epithelial cells in the thymic medulla. J Exp Med. 2007;204:1267–72. doi: 10.1084/jem.20062497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akiyama T, et al. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science. 2005;308:248–51. doi: 10.1126/science.1105677. [DOI] [PubMed] [Google Scholar]
- 52.Heino M, et al. RNA and protein expression of the murine autoimmune regulator gene (Aire) in normal, RelB-deficient and in NOD mouse. Eur J Immunol. 2000;30:1884–93. doi: 10.1002/1521-4141(200007)30:7<1884::AID-IMMU1884>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 53.Kajiura F, et al. NF-kappa B-inducing kinase establishes self-tolerance in a thymic stroma-dependent manner. J Immunol. 2004;172:2067–75. doi: 10.4049/jimmunol.172.4.2067. [DOI] [PubMed] [Google Scholar]
- 54.Zhu M, et al. NF-kappaB2 is required for the establishment of central tolerance through an Aire-dependent pathway. J Clin Invest. 2006;116:2964–71. doi: 10.1172/JCI28326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kinoshita D, et al. Essential role of IkappaB kinase alpha in thymic organogenesis required for the establishment of self-tolerance. J Immunol. 2006;176:3995–4002. doi: 10.4049/jimmunol.176.7.3995. [DOI] [PubMed] [Google Scholar]
- 56.Hikosaka Y, et al. The cytokine RANKL produced by positively selected thymocytes fosters medullary thymic epithelial cells that express autoimmune regulator. Immunity. 2008;29:438–50. doi: 10.1016/j.immuni.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 57.White AJ, et al. Sequential phases in the development of Aire-expressing medullary thymic epithelial cells involve distinct cellular input. Eur J Immunol. 2008;38:942–7. doi: 10.1002/eji.200738052. [DOI] [PubMed] [Google Scholar]
- 58.Akiyama T, et al. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity. 2008;29:423–37. doi: 10.1016/j.immuni.2008.06.015. [DOI] [PubMed] [Google Scholar]
- 59.Irla M, et al. Autoantigen-specific interactions with CD4+ thymocytes control mature medullary thymic epithelial cell cellularity. Immunity. 2008;29:451–63. doi: 10.1016/j.immuni.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 60.Bloch DB, et al. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol Cell Biol. 2000;20:6138–46. doi: 10.1128/mcb.20.16.6138-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ilmarinen T, et al. The monopartite nuclear localization signal of autoimmune regulator mediates its nuclear import and interaction with multiple importin alpha molecules. Febs J. 2006;273:315–24. doi: 10.1111/j.1742-4658.2005.05065.x. [DOI] [PubMed] [Google Scholar]
- 62.Pitkanen J, Vahamurto P, Krohn K, Peterson P. Subcellular localization of the autoimmune regulator protein. characterization of nuclear targeting and transcriptional activation domain. J Biol Chem. 2001;276:19597–602. doi: 10.1074/jbc.M008322200. [DOI] [PubMed] [Google Scholar]
- 63.Pitkanen J, et al. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J Biol Chem. 2000;275:16802–9. doi: 10.1074/jbc.M908944199. [DOI] [PubMed] [Google Scholar]
- 64.Sanchez-Pulido L, Valencia A, Rojas AM. Are promyelocytic leukaemia protein nuclear bodies a scaffold for caspase-2 programmed cell death? Trends Biochem Sci. 2007;32:400–6. doi: 10.1016/j.tibs.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 65.Ferguson BJ, et al. AIRE’s CARD revealed, a new structure for central tolerance provokes transcriptional plasticity. J Biol Chem. 2008;283:1723–31. doi: 10.1074/jbc.M707211200. [DOI] [PubMed] [Google Scholar]
- 66.Ramsey C, Bukrinsky A, Peltonen L. Systematic mutagenesis of the functional domains of AIRE reveals their role in intracellular targeting. Hum Mol Genet. 2002;11:3299–308. doi: 10.1093/hmg/11.26.3299. [DOI] [PubMed] [Google Scholar]
- 67.Park HH, et al. The death domain superfamily in intracellular signaling of apoptosis and inflammation. Annu Rev Immunol. 2007;25:561–86. doi: 10.1146/annurev.immunol.25.022106.141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ilmarinen T, et al. Functional analysis of SAND mutations in AIRE supports dominant inheritance of the G228W mutation. Hum Mutat. 2005;26:322–31. doi: 10.1002/humu.20224. [DOI] [PubMed] [Google Scholar]
- 69.Su MA, et al. Mechanisms of an autoimmunity syndrome in mice caused by a dominant mutation in Aire. J Clin Invest. 2008;118:1712–26. doi: 10.1172/JCI34523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cetani F, et al. A novel mutation of the autoimmune regulator gene in an Italian kindred with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy, acting in a dominant fashion and strongly cosegregating with hypothyroid autoimmune thyroiditis. J Clin Endocrinol Metab. 2001;86:4747–52. doi: 10.1210/jcem.86.10.7884. [DOI] [PubMed] [Google Scholar]
- 71.Bottomley MJ, et al. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J Biol Chem. 2005;280:11505–12. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 72.Bjorses P, et al. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am J Hum Genet. 2000;66:378–92. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meloni A, Incani F, Corda D, Cao A, Rosatelli MC. Role of PHD fingers and COOH-terminal 30 amino acids in AIRE transactivation activity. Mol Immunol. 2008;45:805–9. doi: 10.1016/j.molimm.2007.06.156. [DOI] [PubMed] [Google Scholar]
- 74.Halonen M, et al. APECED-causing mutations in AIRE reveal the functional domains of the protein. Hum Mutat. 2004;23:245–57. doi: 10.1002/humu.20003. [DOI] [PubMed] [Google Scholar]
- 75.Uchida D, et al. AIRE functions as an E3 ubiquitin ligase. J Exp Med. 2004;199:167–72. doi: 10.1084/jem.20031291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Savkur RS, Burris TP. The coactivator LXXLL nuclear receptor recognition motif. J Pept Res. 2004;63:207–12. doi: 10.1111/j.1399-3011.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- 77.Helton ES, Zhang J, Chen X. The proline-rich domain in p63 is necessary for the transcriptional and apoptosis-inducing activities of TAp63. Oncogene. 2008;27:2843–50. doi: 10.1038/sj.onc.1210948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mittaz L, et al. Isolation and characterization of the mouse Aire gene. Biochem Biophys Res Commun. 1999;255:483–90. doi: 10.1006/bbrc.1999.0223. [DOI] [PubMed] [Google Scholar]
- 79.Saltis M, et al. Evolutionarily conserved and divergent regions of the autoimmune regulator (Aire) gene: a comparative analysis. Immunogenetics. 2008;60:105–14. doi: 10.1007/s00251-007-0268-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bjorses P, et al. Localization of the APECED protein in distinct nuclear structures. Hum Mol Genet. 1999;8:259–66. doi: 10.1093/hmg/8.2.259. [DOI] [PubMed] [Google Scholar]
- 81.Kumar PG, et al. The autoimmune regulator (AIRE) is a DNA-binding protein. J Biol Chem. 2001;276:41357–64. doi: 10.1074/jbc.M104898200. [DOI] [PubMed] [Google Scholar]
- 82.Rinderle C, Christensen HM, Schweiger S, Lehrach H, Yaspo ML. AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered sub-cellular distribution of mutants lacking the PHD zinc fingers. Hum Mol Genet. 1999;8:277–90. doi: 10.1093/hmg/8.2.277. [DOI] [PubMed] [Google Scholar]
- 83.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–16. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 84.Shiels C, et al. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J Cell Sci. 2001;114:3705–16. doi: 10.1242/jcs.114.20.3705. [DOI] [PubMed] [Google Scholar]
- 85.Kumar PP, et al. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat Cell Biol. 2007;9:45–56. doi: 10.1038/ncb1516. [DOI] [PubMed] [Google Scholar]
- 86.Akiyoshi H, et al. Subcellular expression of autoimmune regulator is organized in a spatiotemporal manner. J Biol Chem. 2004;279:33984–91. doi: 10.1074/jbc.M400702200. [DOI] [PubMed] [Google Scholar]
- 87.Tao Y, et al. AIRE recruits multiple transcriptional components to specific genomic regions through tethering to nuclear matrix. Mol Immunol. 2006;43:335–45. doi: 10.1016/j.molimm.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 88.Pitkanen J, et al. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem Biophys Res Commun. 2005;333:944–53. doi: 10.1016/j.bbrc.2005.05.187. [DOI] [PubMed] [Google Scholar]
- 89.Liiv I, et al. DNA-PK contributes to the phosphorylation of AIRE: importance in transcriptional activity. Biochim Biophys Acta. 2008;1783:74–83. doi: 10.1016/j.bbamcr.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Björses P, et al. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am J Hum Genet. 2000;66:378–92. doi: 10.1086/302765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oven I, et al. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol Cell Biol. 2007;27:8815–23. doi: 10.1128/MCB.01085-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purohit S, Kumar PG, Laloraya M, She JX. Mapping DNA-binding domains of the autoimmune regulator protein. Biochem Biophys Res Commun. 2005;327:939–44. doi: 10.1016/j.bbrc.2004.12.093. [DOI] [PubMed] [Google Scholar]
- 93.Ruan QG, et al. The autoimmune regulator directly controls the expression of genes critical for thymic epithelial function. J Immunol. 2007;178:7173–80. doi: 10.4049/jimmunol.178.11.7173. [DOI] [PubMed] [Google Scholar]
- 94.Surdo PL, Bottomley MJ, Sattler M, Scheffzek K. Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions. Mol Endocrinol. 2003;17:1283–95. doi: 10.1210/me.2002-0409. [DOI] [PubMed] [Google Scholar]
- 95.Bottomley MJ, et al. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol. 2001;8:626–33. doi: 10.1038/89675. [DOI] [PubMed] [Google Scholar]
- 96.Johnnidis JB, et al. Chromosomal clustering of genes controlled by the aire transcription factor. Proc Natl Acad Sci U S A. 2005;102:7233–8. doi: 10.1073/pnas.0502670102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem Pharmacol. 2004;68:1145–55. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 98.Ilmarinen T, et al. Functional interaction of AIRE with PIAS1 in transcriptional regulation. Mol Immunol. 2008;45:1847–62. doi: 10.1016/j.molimm.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 99.Mauldin SK, Getts RC, Liu W, Stamato TD. DNA-PK-dependent binding of DNA ends to plasmids containing nuclear matrix attachment region DNA sequences: evidence for assembly of a repair complex. Nucleic Acids Res. 2002;30:4075–87. doi: 10.1093/nar/gkf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okubo S, et al. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem. 2004;279:31455–61. doi: 10.1074/jbc.M403561200. [DOI] [PubMed] [Google Scholar]
- 101.Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr Opin Genet Dev. 2007;17:408–14. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 102.Razin SV, et al. Chromatin domains and regulation of transcription. J Mol Biol. 2007;369:597–607. doi: 10.1016/j.jmb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 103.Bres V, Yoh SM, Jones KA. The multi-tasking P-TEFb complex. Curr Opin Cell Biol. 2008;20:334–40. doi: 10.1016/j.ceb.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–4. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 105.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 106.Org T, et al. The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Rep. 2008;9:370–6. doi: 10.1038/embor.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Koh AS, et al. Aire employs a histone-binding module to mediate immunological tolerance, linking chromatin regulation with organ-specific autoimmunity. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0808470105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao XD, et al. Whole-genome mapping of histone h3 lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–98. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 110.Sato K, et al. Aire downregulates multiple molecules that have contradicting immune-enhancing and immune-suppressive functions. Biochem Biophys Res Commun. 2004;318:935–40. doi: 10.1016/j.bbrc.2004.04.116. [DOI] [PubMed] [Google Scholar]
- 111.Gotter J, Brors B, Hergenhahn M, Kyewski B. Medullary epithelial cells of the human thymus express a highly diverse selection of tissue-specific genes colocalized in chromosomal clusters. J Exp Med. 2004;199:155–66. doi: 10.1084/jem.20031677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Villasenor J, Besse W, Benoist C, Mathis D. Ectopic expression of peripheral-tissue antigens in the thymic epithelium: Probabilistic, monoallelic, misinitiated. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0808069105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kaufmann BB, van Oudenaarden A. Stochastic gene expression: from single molecules to the proteome. Curr Opin Genet Dev. 2007;17:107–12. doi: 10.1016/j.gde.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 114.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 115.Blake WJ, M KA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–7. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- 116.Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat Rev Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- 117.Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 118.Bar-Even A, et al. Noise in protein expression scales with natural protein abundance. Nat Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 119.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pombo A, Branco MR. Functional organisation of the genome during interphase. Curr Opin Genet Dev. 2007;17:451–5. doi: 10.1016/j.gde.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 121.Sillanpaa N, et al. Autoimmune regulator induced changes in the gene expression profile of human monocyte-dendritic cell-lineage. Mol Immunol. 2004;41:1185–98. doi: 10.1016/j.molimm.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 122.Gardner JM, et al. Deletional tolerance mediated by extrathymic Aire-expressing cells. Science. 2008;321:843–7. doi: 10.1126/science.1159407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheng MH, Shum AK, Anderson MS. What’s new in the Aire? Trends Immunol. 2007;28:321–7. doi: 10.1016/j.it.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 124.Alimohammadi M, et al. Autoimmune polyendocrine syndrome type 1 and NALP5, a parathyroid autoantigen. N Engl J Med. 2008;358:1018–28. doi: 10.1056/NEJMoa0706487. [DOI] [PubMed] [Google Scholar]
- 125.Meager A, et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med. 2006;3:e289. doi: 10.1371/journal.pmed.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kisand K, et al. Interferon autoantibodies associated with AIRE deficiency decrease the expression of IFN-stimulated genes. Blood. 2008;112:2657–66. doi: 10.1182/blood-2008-03-144634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Strobel P, et al. Deficiency of the autoimmune regulator AIRE in thymomas is insufficient to elicit autoimmune polyendocrinopathy syndrome type 1 (APS-1) J Pathol. 2007;211:563–71. doi: 10.1002/path.2141. [DOI] [PubMed] [Google Scholar]
- 128.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172:2731–8. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 129.Nagafuchi S, et al. Mitogen-activated protein kinase pathway controls autoimmune regulator (AIRE) gene expression in granulo-monocyte colony stimulating factor (GM-CSF)-stimulated myelomonocytic leukemia OTC-4 cells. Immunol Lett. 2005;99:130–5. doi: 10.1016/j.imlet.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 130.Kogawa K, et al. Expression of AIRE gene in peripheral monocyte/dendritic cell lineage. Immunol Lett. 2002;80:195–8. doi: 10.1016/s0165-2478(01)00314-5. [DOI] [PubMed] [Google Scholar]
- 131.Suzuki E, et al. Expression of AIRE in thymocytes and peripheral lymphocytes. Autoimmunity. 2008;41:133–9. doi: 10.1080/08916930701773941. [DOI] [PubMed] [Google Scholar]
- 132.Schaller CE, et al. Expression of Aire and the early wave of apoptosis in spermatogenesis. J Immunol. 2008;180:1338–43. doi: 10.4049/jimmunol.180.3.1338. [DOI] [PubMed] [Google Scholar]
- 133.Hubert FX, et al. A specific anti-Aire antibody reveals aire expression is restricted to medullary thymic epithelial cells and not expressed in periphery. J Immunol. 2008;180:3824–32. doi: 10.4049/jimmunol.180.6.3824. [DOI] [PubMed] [Google Scholar]
- 134.Hassler S, et al. Aire-deficient mice develop hematopoetic irregularities and marginal zone B-cell lymphoma. Blood. 2006;108:1941–8. doi: 10.1182/blood-2006-04-019679. [DOI] [PubMed] [Google Scholar]
- 135.Ramsey C, et al. Increased antigen presenting cell-mediated T cell activation in mice and patients without the autoimmune regulator. Eur J Immunol. 2006;36:305–17. doi: 10.1002/eji.200535240. [DOI] [PubMed] [Google Scholar]
- 136.Pontynen N, et al. Critical immunological pathways are downregulated in APECED patient dendritic cells. J Mol Med. 2008 doi: 10.1007/s00109-008-0374-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee JW, et al. Peripheral antigen display by lymph node stroma promotes T cell tolerance to intestinal self. Nat Immunol. 2007;8:181–90. doi: 10.1038/ni1427. [DOI] [PubMed] [Google Scholar]