Abstract
Epidermal fatty acid-binding protein (E-FABP), a member of the family of FABPs, exhibits a robust expression in neurons during axonal growth in development and in nerve regeneration following nerve injury. This study examines the impact of E-FABP expression in normal neurite extension in differentiating pheochromocytoma cell (PC12) cultures supplemented with selected long chain free fatty acids (LCFFA). We found that E-FABP binds to a broad range of saturated and unsaturated LCFFAs, including those with potential interest for neuronal differentiation and axonal growth such as C22:6n-3 docosahexaenoic acid (DHA), C20:5n-3 eicosapentaenoic acid (EPA), and C20:4n-6 arachidonic acid (ARA). PC12 cells exposed to nerve growth factor (NGFDPC12) exhibit high E-FABP expression that is blocked by mitogen-activated protein kinase kinase (MEK) inhibitor U0126. Nerve growth factor-differentiated pheochromocytoma cells (NGFDPC12) antisense clones (NGFDPC12-AS) which exhibit low E-FABP expression have fewer/shorter neurites than cells transfected with vector only or NGFDPC12 sense cells (NGFDPC12-S). Replenishing NGFDPC12-AS cells with biotinylated recombinant E-FABP (biotin-E-FABP) protein restores normal neurite outgrowth. Cellular localization of biotin-E-FABP in NGFDPC12 was detected mostly in the cytoplasm and in the nuclear region. Treatment of NGFDPC12 with DHA, EPA, or ARA further enhances neurite length but it does not trigger further induction of TrkA or MEK phosphorylation or E-FABP mRNA observed in differentiating PC12 cells without LCFFA supplementation. Significantly, DHA and EPA neurite stimulating effects are higher in NGFDPC12-S than in NGFDPC12-AS cells. These findings are consistent with the scenario that neurite extension of differentiating PC12 cells, including further stimulation by DHA and EPA, requires sufficient cellular levels of E-FABP.
Keywords: C20:5n-3 eicosapentaenoic acid, C22:6n-3 docosahexaenoic acid, epidermal fatty acid-binding protein, fatty acid binding, n-3/n-6 polyunsaturated fatty acids, neuronal differentiation
Axon growth during neuronal differentiation and nerve regeneration may require the contribution of key lipids and growth associated proteins to develop normal synapse formation and maturation (Martin et al. 2000; Darios and Davletov 2006; Davletov et al. 2007). Neuronal membranes are specially rich in long chain polyunsaturated fatty acids (LCPUFAs) like C22:6n-3 docosahexaenoic acid (DHA) and C20:4n-6 arachidonic acid (ARA) (Jump 2002; Barcelo-Coblijn et al. 2003) that may play a role in maintaining proper membrane fluidity that facilitates plasticity and signaling during axon growth. Deficiency of n-3 LCPUFA can lead to cognitive abnormalities, reduced neuroplasticity, memory loss, learning disabilities, and neurodegeneration (Connor et al. 1992; Bazan 2006; Cole and Frautschy 2006; Carlson and Kingston 2007; Hichami et al. 2007). Additional reports have shown that n-6 LCPUFA deficit can also result in impaired learning and memory (Roegge et al. 2005). Deficiency of n-3 LCPUFA results in fewer isoforms of glucose transporter (Pifferi et al. 2005), reduction in G protein-coupled signaling efficiency (Niu et al. 2004), and shrinkage of neurons (Ahmad et al. 2002). These findings raise the importance to study proteins associated with the shuttling of these LCPUFAs within the cells to evaluate whether they might play a role in normal neuronal axon growth.
Intracellular fatty acid-binding proteins (FABPs) are a 15-kDa cytosolic family of proteins that can have high affinity to long chain free fatty acids (LCFFA). Several members of the FABP family have been identified in the nervous system with a distinct spatial and temporal distribution, suggesting that they may have different roles (for review, see Veerkamp and Zimmerman 2001). Myelin-FABP (P2 myelin protein) is located in the PNS and is important in the metabolism of peripheral myelin lipids (Polverini et al. 2006). Brain FABP is expressed in glial cells and astrocytes and has been associated with differentiation of glial cells (Kurtz et al. 1994; Owada et al. 1996b). Heart FABP is detected in postnatal neurons and participates in neurite formation and synapse maturation (Sellner et al. 1995). Epidermal FABP (E-FABP/FABP5) is induced following various types of nerve injuries (De Leon et al. 1996; Owada et al. 1996a) and has been proposed to play a role during neuronal differentiation and axonal growth in nervous system. During development the expression of E-FABP mRNA and protein is robust in neurons in the retina, hippocampus, cerebellum, and cerebral cortex as well as motor neurons in the spinal cord (Liu et al. 1997, 2000; Allen et al. 2001). In the rat, E-FABP immunoreactivity co-localizes with growth-associated protein 43 in the cell body and axons of retinal ganglion cells and its up-regulation by embryonic day 14 and 15 timely correlates with their terminal differentiation and axonal extension (Allen et al. 2001).
Fatty acid-binding proteins are believed to bind hydrophobic ligands (i.e., LCFFA) and shuttle them to specific intracellular organelles (For review, see Veerkamp and Zimmerman 2001). Understanding the functions of FABPs such as E-FABP in neurons may require characterization of its binding characteristics and identifying cellular processes that rely on both E-FABP and its LCFFA ligands to succeed. The pheochromocytoma cell line (PC12) is an excellent model to explore this question because it has been well characterized for studies in neuronal differentiation and exhibits robust and quantifiable neurite formation. Nerve growth factor (NGF) is prominent in its ability to regulate neuronal differentiation and induce neuronal-like differentiation in PC12 cells. Further, NGF is a potent inducer of E-FABP expression and PC12 clones defective in the expression of E-FABP show shorter and fewer neurites (Allen et al. 2000). The present study examines connections between E-FABP levels and its endogenous ligands [DHA, C20:5n-3 eicosapentaenoic acid (EPA), C18:1n-9 (oleic acid) (OA), and ARA)] with regard to neurite extension in NGF-differentiated PC12 (NGFDPC12) cells.
Materials and methods
PC12 cell culture
Pheochromocytoma cells were cultured in Dulbecco’s Modified Eagle’s Medium (Mediatech Inc., Herndon, VA, USA) with 10% horse serum (Atlantic Biological, Lawrenceville, GA, USA), 5% fetal bovine serum (FBS, Atlantic Biological), and penicillin (100 units/mL)/streptomycin (100 µg/mL). To induce differentiation, the cells were changed to Dulbecco’s Modified Eagle’s Medium containing reduced serum (1% FBS) and 50 ng/mL of NGF (Alomone Labs, Jerusalem, Israel). All cultures were maintained in a humidified 5% CO2 atmosphere at 37°C.
For the signaling pathway experiment, mitogen-activated protein kinase kinase (MEK) inhibitor U0126 (Promega, Madison, WI, USA) was first prepared as a 10 mM stock in dimethylsulfoxide and diluted to 30 µM in reduced serum medium with or without NGF. PC12 cells were cultured in test medium for 3 days before RNA analysis. For ligand-supplemented experiments, DHA, EPA, ARA, and OA were purchased from Sigma (St Louis, MO, USA) and 150 mM stock solutions in 100% ethanol were prepared. The free fatty acid (FFA) stocks were diluted into 1% FBS medium containing 150 µM of fatty acid-free bovine serum albumin (BSA) (EMD Biosciences, La Jolla, CA, USA) to a final concentration of 60 µM. FFA/BSA mixture was used to ensure stable unbound FFA concentration in the medium during the course of experiment. Because FFA/BSA ratio determines the concentration of unbound FFA, the 60 µM/150 µM complex (0.4 : 1) we used in this study is similar to that reported by others (Marszalek et al. 2004). Reduced serum medium (1% FBS) with 150 µM BSA and 0.04% ethanol was used as the control. Cells were cultured in test medium for up to 6 days before RNA or morphology analysis.
Construction of PC12-AS, PC12-S, and PC12-CMV cells
To generate E-FABP antisense clones, rat E-FABP cDNA was amplified by PCR with primers 5′-GGTCTAGATTGCTGCTTTTGTGCTCTCC-3′ and 5′-GGAAGCTTTTCTGTACGACCTGCTCATT-3′. To generate E-FABP sense clones, E-FABP cDNA was amplified by PCR with primers 5′-GGAAGCTTTTGCTGCTTTTGTGCTCTCC-3′ and 5′-GGTCTAGATTCTGTACGACCTGCTCATT-3′. The PCR products were then cloned into the pRc/cytomegalovirus (CMV) vector (Invitrogen, Carlsbad, CA, USA) using XbaI and HindIII sites. After confirming the sequence, the E-FABP antisense vector was used to transfect PC12 cells to create PC12 cells expressing a reduced level of E-FABP (PC12-AS). The E-FABP sense clone was used to create PC12 cells expressing an elevated level of E-FABP (PC12-S). PC12 cells with vector only (PC12-CMV) served as the control. The transfected cells were selected by G418 (500 µg/mL). Five clonal lines were established for all three types of transfected PC12 cells; clone 4 was selected for use from each of the three types of transfected PC12 cells, i.e., PC12-AS4, PC12-S4, and PC12-CMV4.
Production of recombinant rat E-FABP
Recombinant rat E-FABP was produced and purified by the IMPAC T7 system (New England Biolabs, Beverly, MA, USA). First, rat E-FABP cDNA was cloned into the pTYB1 vector, then Escherchia coli host strain ER2566 was used to produce a fusion protein of E-FABP and intein by induction with isopropyl-β-D-thiagaloctoside at a cool temperature (15°C) to reduce the formation of inclusion bodies. The E. coli were centrifuged and the cell pellet was resuspended in 20 mM Tris–HCl (pH 8.0) with 500 mM NaCl, 0.1 mM EDTA, and 0.1% Triton X-100, then lysed with lysozyme (0.6 mg/mL) for 10 min, followed by sonication. The following purification procedures were carried out at 4°C. After centrifugation, clarified cell extract was loaded onto a chitin column and washed with washing buffer (20 mM Tris–HCl, pH8.0 with 1.5 M NaCl, 0.1 mM EDTA, and 0.25% Triton X-100). After washing, the column was flushed with cleavage buffer (20 mM Tris–HCl, pH 8.0 with 50 mM NaCl, 0.1 mM EDTA, and 30 mM 2-mercaptoethanol) for 15 min, the flow was stopped and cleavage was induced at 4°C overnight. On the next day, recombinant E-FABP (rE-FABP) was eluted with cleavage buffer without 2-mercaptoethanol. Small fraction of samples from each purification step was saved and verified on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis followed by Coomassie blue staining or western blot analysis. The purified rE-FABP was further verified by liquid chromatography/masspectrometry or mass spectrometry (Q-TOF, Waters, Manchester, UK). The rE-FABP was delipidated before performing fatty acid binding studies. First, rE-FABP was concentrated and the buffer was changed to 10 mM potassium phosphate (pH 7.4) using a Centricon YM-10 (Millipore, Bedford, MA, USA). A Lipidex 1000 column (Hydroxyalkoxypropyl dextran, type IX, Sigma) was equilibrated with the same buffer at 37°C. rE-FABP was loaded onto the column at the flow rate of 1 mL/10 min. Void volume fraction, which contained delipidated rE-FABP, was collected and the protein concentration was estimated by Bradford protein assay and by spectrophotometry, using an extinction coefficient ε278 = 1.55 × 104/cm/M (Kane et al. 1996). The concentration determined by protein assay agreed within 10% with the concentration determined by absorbance.
Production of biotinylated E-FABP
Biotin (Long Arm) NHS (Vector Laboratories, Burlingame, CA, USA) was dissolved in dimethylsulfoxide (20 mg/mL). rE-FABP was concentrated to 2 mg/mL in 100 mM HEPES, pH 8.5. One millilitre of E-FABP solution was mixed with 20 µL of biotin solution and incubated at 25°C for 2 h. Ten mg of glycine was added to the mixture to stop the reaction. The unbound biotin was removed by dialysis of the reaction mixture against three changes of two-liters of buffer containing 10 mM potassium phophate, pH 7.4 and 40 mM potassium chloride. Lastly, the biotin-E-FABP was delipidated by Lipidex 1000 as described above.
Western blots
The polyclonal antibodies used for E-FABP western blots were generated in rabbits against rE-FABP produced in the laboratory. Phospho-TrkA and Phospho-MEK1/2 antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). Protein extracts of PC12 cells were resolved on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane. After blocking with 7.5% milk in Tris-buffered saline with 0.05% Tween 20, pH 7.4 (TTBS) for 1 h, the membrane was incubated with E-FABP antiserum (1 : 1000) in 5% milk TTBS, phospho-TrkA, or phospho-MEK1/2 antibodies (1 : 500) in 5% BSA-TTBS 4°C overnight. Subsequently, the membrane was washed three times with TTBS and incubated with horseradish peroxidase-goat anti-rabbit IgG (1 : 1000, GE Healthcare Bio-Science, Piscataway, NJ, USA) for 1 h, followed by three washes with TTBS. The signal was then detected by enhanced chemiluminescence-plus (GE Healthcare Bio-Science). The membrane was reblocked and analyzed for β-actin to verify loading.
Fatty acid binding assay
Free fatty acids were purchased from Sigma. OA, C18:0 (stearic acid), C16:0 (palmitic acid), and C14:0 (myristic acid) were purchased as the sodium salt form. Stock solutions (100 mM) were prepared in 0.01 N NaOH. The rest of FFA used in the binding assay was purchased as the FFA form. Stock solutions (100 mM) were prepared in 100% ethanol then diluted with 0.01 N NaOH for the binding assay. A fluorescent probe 1-anilinonaphthalene-8-sulfonic acid (ANS) was purchased from Molecular Probes (Eugene, OR, USA) and an 8 mM stock solution in 100% ethanol was prepared.
The fatty acid-binding assay was carried out at 25°C with a Shimadzu RF1501 spectrofluorophotometer (Shimadzu Corp., Tokyo, Japan). rE-FABP (or biotin-E-FABP) and ANS were diluted with 10 mM potassium phosphate (pH 7.4) with 40 mM KCl.
To determine the binding affinity of rE-FABP to ANS, increasing concentration of rE-FABP was mixed with 500 nM ANS for 2 min in dark, followed by measuring the fluorescence (excitation/emission at 370 nm/480 nm) of the mixture. For fatty acid displacement assay, 2000-fold stock solutions of fatty acids were prepared in 0.01 N NaOH and kept at 50°C, except that C18:0 (stearic acid) and C16:0 (palmitic acid) stock solutions were kept at 85°C. Fatty acid was added to the binding assay solution containing 0.4 µM of recombinant protein and 4 µM ANS in an increment of 0.5 or 1 µM. Mixture was kept in dark for 2 min before fluorescence measurement.
The binding affinity or the apparent dissociation constant (Kd) of E-FABP to ANS was calculated using the Graphpad Prism software (version 4.0a; GraphPad Software Inc., La Jolla, CA, USA) with non-linear regression one-site binding analysis. The Kd of E-FABP to selected fatty acids was calculated using the following equations (Widstrom et al. 2001):
Kd = 1/Ka. ‘F’ is the measured fluorescence, F0 is the fluorescence in the absence of fatty acid, PT is the total protein concentration (0.4 µM in our case), LT is the fatty acid concentration, Ka is the apparent association constant for the titrant fatty acid, and Fmax is the fluorescence emission after complete saturation of E-FABP with ligand fatty acid. Non-linear regression analysis was carried out with Graphpad Prism.
Delivery of biotin-E-FABP to PC12 cells
BioPORTER reagent (Gene Therapy Systems, San Diego, CA, USA) was dissolved in 250 µL of chloroform and vortexed for 20 s. Then, the desired amount of BioPORTER was transferred into 1.5-mL vial and the solvent was evaporated in a tissue culture hood for 3–4 h. The vials were stored at −20°C for future use. Delipidated biotin-E-FABP was diluted in 10 mM HEPES, 150 mM NaCl, pH 7.0 and hydrated with the dried BioPORTER reagent in the vials. The solution was incubated at 25°C for 5 min. Serum-free medium was added to the complex solution to make the final volume (250 µL for 24-well plate).
Pheochromocytoma cells were plated on Biocoat collagen I 4-well culture slides (Becton Dickinson, Bedford, MA, USA) in full serum medium. After attachment, the cells were changed to 1% FBS medium and cultured overnight. Subsequently, the medium was aspirated and the transfection solution (biotin-E-FABP/BioPORTER complex from above) was added to the wells. The cells were incubated with the transfection solution for 3–4 h and full serum medium was added to the wells for 4 h. The transfected PC12 cells were then differentiated with 50 ng/mL of NGF in 1% FBS medium for 3 days.
Fluorescent staining
The biotin-E-FABP transfected PC12 cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min. After rinsing with PBS, the fixed cells were blocked with 3% BSA and 0.1% Triton X-100 in PBS for 30 min. Afterward, fluorescein avidin DCS (cell sorter grade) (Vector Laboratories), which has a high affinity to biotin-E-FABP, was incubated with the cells at 20 µg/mL for 1 h. Cells were washed with 0.1% Tween-20 in PBS and blocked again with 1.5% BSA in PBS for 30 min, followed by counter-staining with Texas red-phalloidin (Molecular Probes) for 30 min and washing. Then, the slide was mounted with Vectashield (Vector Laboratories) and checked under a fluorescent microscope (Olympus BX50, Olympus Optical Co., Tokyo, Japan).
Morphometric analysis
Pheochromocytoma cells having at least one neurite that is longer than one cell body width were considered differentiated. Percent differentiation was calculated by number of differentiated cells divided by total number of cells in a random field containing at least 50 cells. The length of the longest neurite of each differentiated cell was determined using NIH Image 1.63 software (National Institutes of Health, Bethesda, MD, USA). The same images used in determination of percent differentiation were employed for neurite length measurements.
Northern blots
Northern blots were performed as previously described (Allen et al. 2000). Briefly, total RNA from PC12 cells was purified by TRI REAGENT® (Molecular Research Center, Cincinnati, OH, USA), separated by 1% formaldehyde gel and transferred to a nylon membrane. After pre-hybridization, the membrane was hybridized with 32P-labeled E-FABP cDNA probe. Afterward, the blot was stripped and reprobed with 32P-labeled cyclophilin cDNA probe.
Two step quantitative RT-PCR
RNA samples were first reversed transcribed to cDNA using iSCRIPT cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA, USA), followed by real-time PCR using iQ Sybr Green supermix (Bio-Rad). The primers used for E-FABP were 5′-TTACCCTCGACGGCARACARA-3′ and 5′-CCATCAGCTGTGGTTTCATCA-3′. The primers used for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were 5′-CARACTACATGGTCTACATGTTC-3′ and 5′-CTCGCTCCTGGARAGATG-3′. Reactions were performed in a 25 µL mixture in three replicates using iCycler (Bio-Rad). To exclude the contamination of non-specific PCR products such as primer dimers, melt curve analysis was applied to all final PCR products. Also, PCR reactions without cDNA were performed as the negative control.
As the PCR efficiency of E-FABP and GAPDH (internal reference) was approximately equal, the amount of E-FABP mRNA in the experimental group relative to the amount E-FABP mRNA in the control group was determined by comparative CT (threshold cycle) method using arithmetic formulas: 2−ΔΔCT. ΔCT is the average CT value for E-FABP subtracted by the average CT value for GAPDH. ΔΔCT is the ΔCT for the experimental group subtracted by the ΔCT for the control group.
Statistical analysis
All the experiments were repeated at least three times. Values represent means ± SE. Statistical comparisons were made using one-way or two-way anova adjusted with least significant differences. Significance was accepted at p < 0.05.
Results
Regulation of E-FABP in PC12 cells
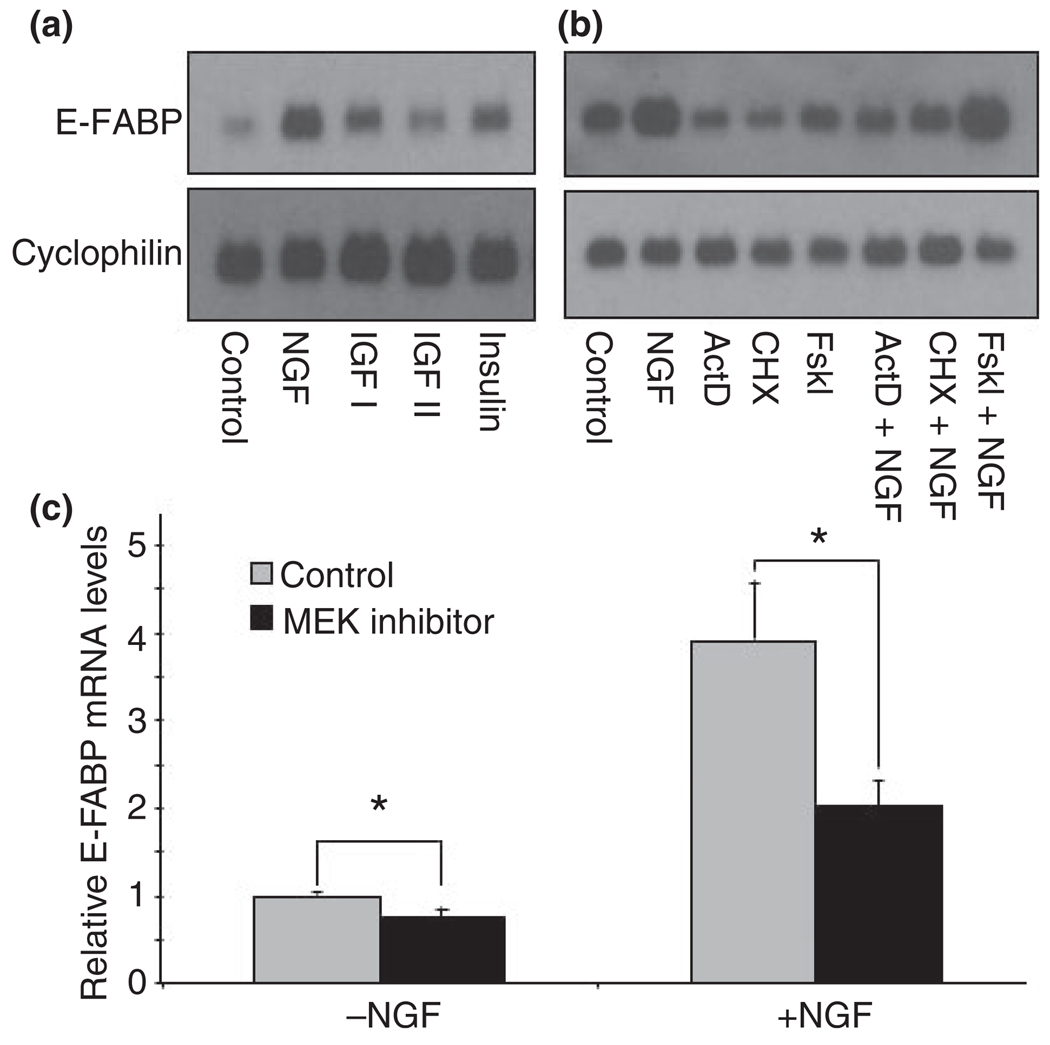
Epidermal-fatty acid-binding protein mRNA levels were compared following exposure to NGF, insulin, insulin-like growth factor (IGF)-I and IGF-II (Fig. 1a). NGF elicited the strongest induction of E-FABP mRNA expression, followed by IGF-I and insulin. IGF-II triggered only a small elevation of E-FABP mRNA level. Figure 1b shows the effects of forskolin (adenylyl cyclase activator), actinomycin D (DNA synthesis inhibitor), and cyclohexamide (protein synthesis inhibitor) on NGF regulation of E-FABP mRNA. Forskolin enhanced the NGF effect while actinomycin D and cycloheximide inhibited both the basal and NGF-induced levels of E-FABP mRNA. NGF phosphorylates the TrkA receptor and activates signaling pathways for the appearance of a differentiated phenotype in PC12 cells, including the formation of neurites (Huang and Reichardt 2003). A major downstream signaling cascade stimulated by NGF is the mitogen-activated protein (MAP) kinase pathway. We used U0126, a MEK inhibitor, to test whether this pathway mediates the signal transduction of NGF-regulated expression of E-FABP. Similar to previous findings (Das et al. 2004), PC12 cells exposed to NGF in the presence of U0126 showed inhibition of neurite formation. As expected, quantitative RT-PCR showed that U0126 inhibits the already low basal levels of E-FABP mRNA by 25% and triggers a twofold repression in the NGF-stimulated expression of E-FABP (Fig. 1c). These results suggest that both the basal and NGF-stimulated expression of E-FABP is regulated by the MAP kinase pathway.
Fig. 1.
Selected factors affecting the expression of epidermal fatty acid-binding protein (E-FABP) in pheochromocytoma cells (PC12). (a) PC12 cells were treated with nerve growth factor (NGF, 50 ng/mL), insulin-like growth factor I (IGF I, 20 ng/mL), IGF II (20 ng/mL) or insulin (10 ng/mL) for 2 days. E-FABP mRNA was examined by Northern blot analysis. A representative blot of at least three independent experiments is shown. (b) PC12 cells were incubated with actinomycin D (ActD, 0.5 µg/mL), cycloheximide (CHX, 1 µg/mL), or forskolin (Fskl, 20 µM) in the presence or absence of NGF (50 ng/mL), for 24 h. E-FABP mRNA was evaluated by Northern blot analysis. A representative blot of at least three independent experiments is shown. (c) PC12 cells were treated with NGF for 3 days in the presence or absence of 30 µM U0126, mitogen-activated protein kinase kinase (MEK) inhibitor and the levels of E-FABP mRNA were analyzed by quantitative RT-PCR. Error bars represent the SEM of three independent experiments. Statistical significance between control and U0126-treated groups was determined by one-way anova. *p < 0.05.
Linkage between E-FABP levels in cytoplasm and normal neurite formation in NGFDPC12-CMV, NGFDPC12-AS, and NGFDPC12-S cells
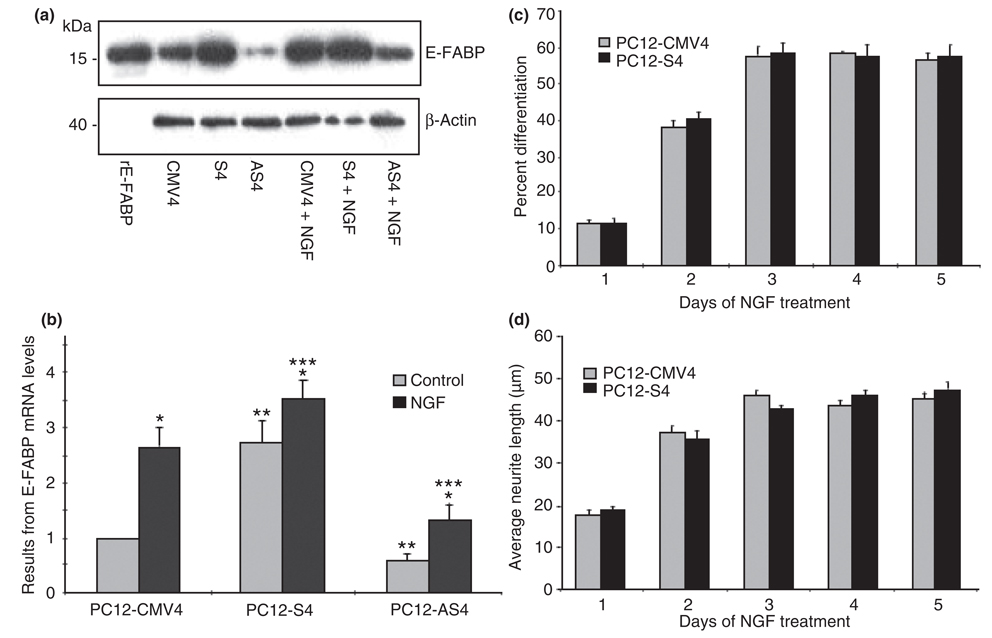
Nerve growth factor-differentiated PC12-AS4 cells with depleted intracellular levels of E-FABP exhibit fewer and shorter neurites (Allen et al. 2000). However, the question always remains whether this effect on neurite formation can be indisputably attributed to E-FABP or to another secondary effect of the stable antisense transfection. The next series of experiments examined the neurite phenotype of stable transfected sense and antisense E-FABP PC12 cells clones following exposure to NGF. Figure 2a and b demonstrate that PC12 cells transfected with a pRc/CMV plasmid containing a rat E-FABP cDNA in the sense orientation downstream to the CMV promoter (PC12-S4) exhibited more than a twofold increase in the levels of E-FABP mRNA and protein than clones transfected with plasmid alone (PC12-CMV4). Further, PC12 cells transfected with E-FABP cDNA in the antisense orientation (PC12-AS4) showed a 40% reduction of the already low basal levels of E-FABP when compared with PC12-CMV4. NGF exposure increased E-FABP expression in all the transfected PC12 cells clones. However, differentiated PC12-MCV4 (NGFDPC12-CMV) and differentiated PC12-AS4 (NGFDPC12-AS) cells showed more than a twofold induction, while E-FABP levels in differentiated PC12-S4 (NGFDPC12-S) cells increased only 25%. Despite increasing E-FABP expression following NGF exposure, NGFDPC12-AS still showed a significant deficit in cellular E-FABP levels when compared with NGFDPC12-CMV (Fig. 2a and b). Morphologically, NGFDPC12-AS cells had shorter neurites and less percent differentiation compared with NGFDPC12-CMV (Allen et al. 2000). Undifferentiated PC12-S4 had comparable levels of E-FABP as NGFDPC12-CMV, but did not exhibit the neurite phenotype in the absence of NGF (data not shown). Moreover, NGFDPC12-S did not show additional stimulation of neurite extension (Fig. 2c) or neurite length (Fig. 2d) when compared with NGFDPC12-CMV.
Fig. 2.
Nerve growth factor (NGF) regulation of epidermal fatty acid-binding protein (E-FABP) expression and neurite outgrowth in pheochromocytoma cells (PC12)-CMV4, PC12-S4, and PC12-AS4 cells. (a) E-FABP protein analysis by western blots. Recombinant rat E-FABP (rE-FABP) was used as a standard. (b) E-FABP mRNA analysis by QTRTPCR. The data represent mean ± SE of three experiments. PC12-CMV4 = PC12 cells line transfected with CMV vector only; PC12-S4 = PC12 cells line transfected with E-FABP cDNA in the sense orientation; PC12-AS4 = PC12 cells with E-FABP cDNA in the antisense direction. Statistical significance was determined by two-way anova. *p < 0.05 for all three cell types compare NGF-treated cells with control cells. **p < 0.05 for cells without NGF, compare S4 and AS4 cells with CMV4 cells. ***p < 0.05 for cells with NGF, compare S4 and AS4 cells to CMV4 cells. (c) Quantitative analysis of percent differentiation after NGF treatment. (d) Quantitative analysis of average neurite length after NGF treatment. The data represent mean ± SE of three experiments. There is no statistical significance between PC12-CMV4 and PC12-S4 groups.
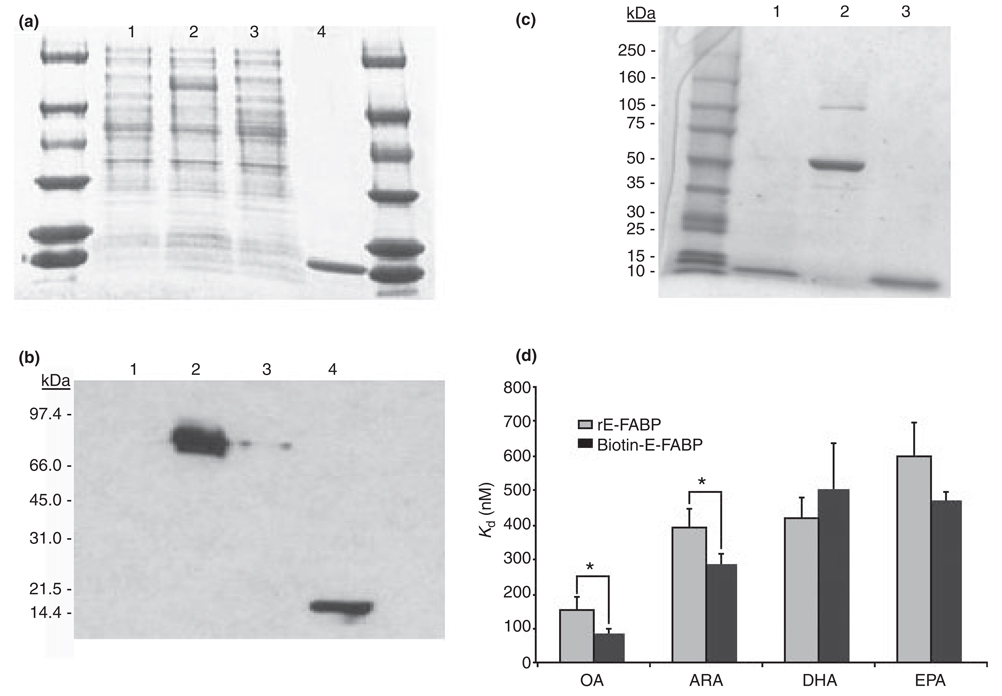
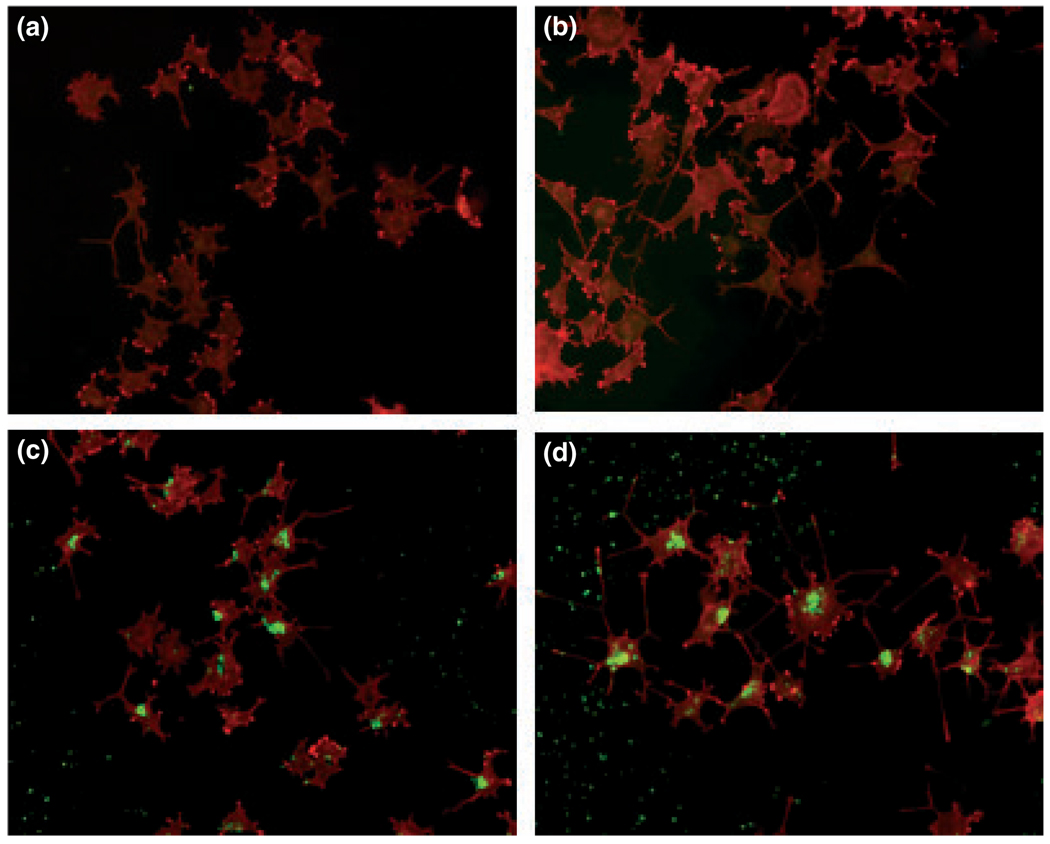
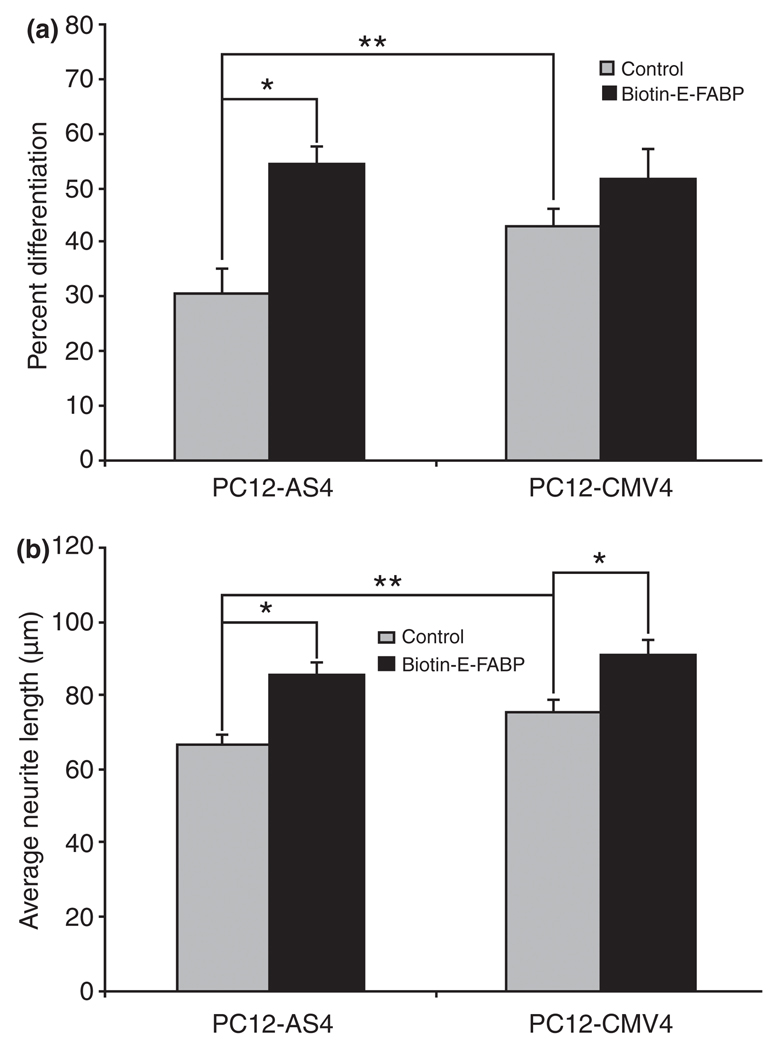
To further examine the potential effects of E-FABP in the formation of neurites following NGF treatment, we used rE-FABP to replenish NGFDPC12-AS clones. The purity and the immunoreactivity of the rE-FABP are shown in Fig. 3a and b. To trace the exogenous E-FABP in the cells, rE-FABP was modified by biotinylation (Fig. 3c). The biotinylated rE-FABP probe (biotin-E-FABP) and wild-type rE-FABP had similar fatty acid binding affinity to DHA and EPA, but biotin-E-FABP had higher affinity to OA and ARA (Fig. 3d). Next, biotin-E-FABP was delivered to PC12-AS4 and PC12-CMV4 cultures using ‘BioPORTER’ reagent before NGF treatment to study whether replenishing NGFDPC12-AS clones with E-FABP protein can restore normal neurite phenotype. As shown in Fig. 4, the incorporation of biotin-E-FABP protein was uneven in the treated cells, but more than 80% of the cells treated with biotin-E-FABP showed significant levels of fluorescent staining, predominantly in the cytoplasm and nuclear region. Figure 4c is a representative field of a culture treated with biotin-E-FABPP showing an increased number of cells exhibiting neurites in the NGFDPC12-AS group when compared with NGFDPC12- AS cultures treated with only the ‘bioPORTER’ reagent (Fig. 4a). This result was quantified as shown in Fig. 5a and b, demonstrating that NGFDPC12-AS cells had less percent differentiation and shorter average neurite length compared with NGFDPC12-CMV cells. In response to biotin-E-FABP treatment, these differentiating phenotypes were significantly improved in NGFDPC12-AS cells. Adding biotin-E-FABP to NGFDPC12-CMV did not further enhance the percentage of cells exhibiting neurites but increased the average neurite length. Treating PC12-CMV4 cultures with the biotin-E-FABP without NGF did not elicit neurite formation (data not shown). Thus, E-FABP protein is up-regulated by NGF and positively correlates with neurite formation, but is not self-sufficient for neurite formation or differentiation.
Fig. 3.
Rat recombinant E-FABP (rE-FABP) production and comparison the fatty acid binding affinity of rE-FABP and biotin-E-FABP. Rat rE-FABP was produced using the IMPACT system, by which method a fusion protein of epidermal fatty acid-binding protein (E-FABP)-intein was first produced in an Escherchia coli transformant and purified on a chitin column. The rE-FABP was cleaved and eluted from the chitin column. (a) Coomassie blue stained gel analysis 0.2% aliquot of 1 L E. coli culture. Lane1, cell extact from uninduced E. coli transformant; lane2, cell extract from induced E. coli transformant, showing the induction of E-FABP-intein fusion protein (70 kDa); lane3, chitin column flow through; lane4, eluted rE-FABP fraction (15 kDa). (b) Western blots analysis for E-FABP on a duplicate gel with one-tenth sample loading. (c) Coomassie blue stained gel analysis of purified biotin-E-FABP. Lane1, rE-FABP; lane 2, bovine serum albumin; Lane 3: biotin-E-FABP. (d) The binding affinity of rE-FABP and biotin-E-FABP to C18:1n-9 oleic acid (OA), C20:4n-6 arachidonic acid (ARA), C22:6n-3 docosahexaenoic acid (DHA), and C20:5n-3 eicosapentaenoic acid (EPA) was determined using 1-anilinonaphthalene-8-sulfonic acid method. Kd values are presented as mean ± SD of three separate measurements. The data used for rE-FABP here are also used in Table 1. Difference of Kd between rE-FABP and biotin-E-FABP was analyzed by one-way anova. *p < 0.05.
Fig. 4.
Localization of biotin-E-FABP in nerve growth factor-differentiated pheochromocytoma cells (NGFDPC12). Representative images of PC12-AS4 and PC12-CMV4 cells treated with biotin-E-FABP protein followed by nerve growth factor (NGF) treatment. (a) Control NGFDPC12-AS cells treated with BioPORTER only; (b) Control NGFDPC12-CMV cells treated with BioPORTER only; (c) NGFDPC12-AS cells treated with biotin- E-FABP/BioPORTER complex; (d) NGFDPC12-CMV cells treated with biotin-E-FABP/BioPORTER complex. Biotin-E-FABP was detected by fluorescein avidin DCS (green staining) and cell morphology was visualized by Texas red-phalloidin. E-FABP, epidermal fatty acid-binding protein.
Fig. 5.
Biotin-E-FABP increases neurite outgrowth and neurite length of nerve growth factor-differentiated pheochromocytoma cells (NGFDPC12). (a) Quantitative analysis of percent differentiation. For biotin-E-FABP groups, the percent differentiation was determined only among biotin-E-FABP-containing cells (green-staining cells). (b) Quantitative analysis of average neurite length. Again, only greenstaining cells were included for biotin-E-FABP groups. Error bars represent the SEM of three experiments. Statistical significance between groups was determined by two-way anova. *p < 0.01; **p < 0.05. E-FABP, epidermal fatty acid-binding protein.
Characterization of binding affinity of selected saturated and unsaturated LCFFAs to rE-FABP
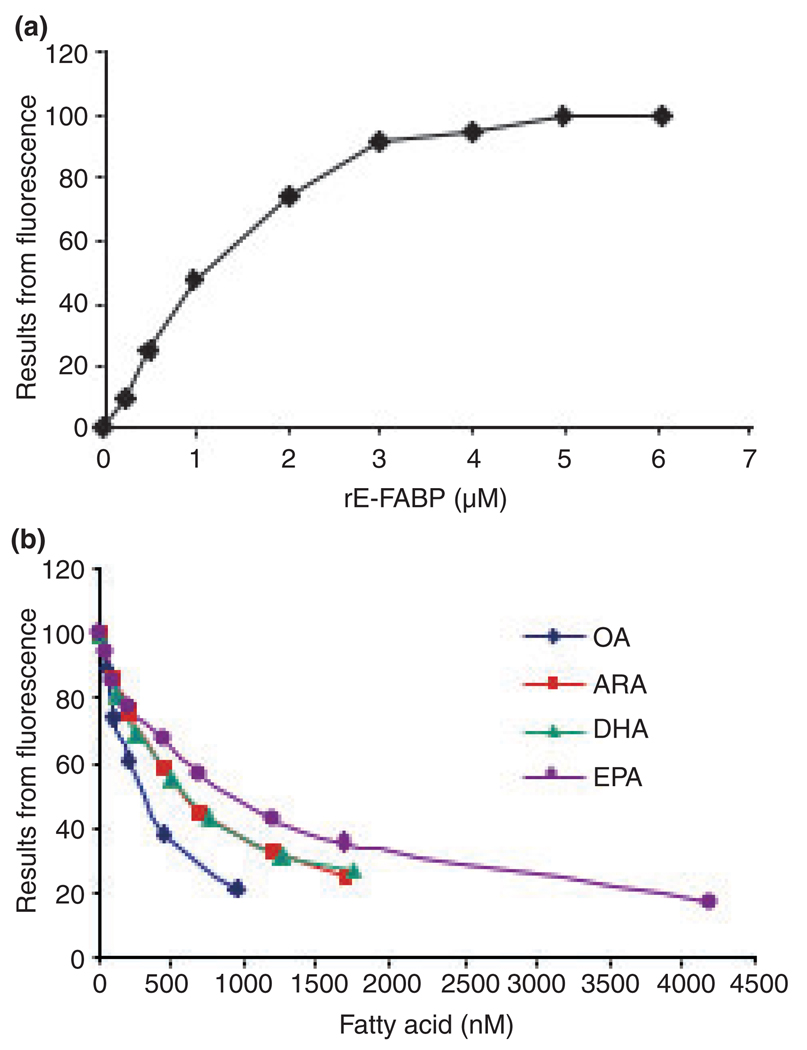
Epidermal-fatty acid-binding protein belongs to a family of fatty acid binding proteins in the nervous system (Veerkamp and Zimmerman 2001). Presumably, a postulated neuronal differentiation/neurite formation role of E-FABP following exposure to NGF might include binding LCFFA necessary for neurite extension during neuronal differentiation. We chose the ANS methods described elsewhere (Kane and Bernlohr 1996; Kirk et al. 1996) to characterize the binding properties of rE-FABP to LCFFA ligands. A representative binding saturation curve is shown in Fig. 6a and the Kd value yields from three measurements is 1.58 ± 0.29 µM, which is higher than that reported for murine E-FABP (0.53 ± 0.14 µM, Kane and Bernlohr 1996). The high affinity of ANS to E-FABP was more than adequate to carry out fatty acid displacement experiments (Fig. 6b) to accurately measure the affinity (Kd) of rE-FABP to selected fatty acids listed in Table 1. The data show that OA and C18:0 (stearic acid) possess the highest binding affinity (Kd < 200 nM) to rE-FABP. ARA, DHA, C16:0 (palmitic acid), C18:2n-6 (linoleic acid), and EPA showed a high moderate binding affinity (200 nM < Kd < 800 nM), while C17:0 (heptadecanoic acid), C22:0 (behenic acid), C18:3n-3 (linolenic acid), and C20:0 (arachidic acid) showed a low moderate binding affinity (800 nM < Kd < 1200 nM). The rE-FABP had a weak affinity to C15:0 (pentadecanoic acid), C14:0 (myristic acid), C24:1n-9 (nervonic acid), and C12:0 (lauric acid) (1200 nM < Kd < 3000 nM) and little affinity to all-trans retinoic acid (Kd > 4000 nM). These data demonstrated that E-FABP has the ability to bind a wide range of saturated and unsaturated LCFFAs and show that LCPUFA (DHA, EPA, and ARA) are ligands of E-FABP.
Fig. 6.
1-anilinonaphthalene-8-sulfonic acid (ANS) binding assays. (a) Increasing concentration of recombinant E-FABP (rE-FABP) was incubated with 500 nM ANS and the intensity of fluorescence recorded at excitation/emission 370 nm/480 nm. A representative measurement of three independent experiments is shown. (b) Competitive displacement of ANS from rE-FABP by free fatty acids. Representative experiments of C18:1n-9 oleic acid (OA), C20:4n-6 arachidonic acid (ARA), C22:6n-3 docosahexaenoic acid (DHA), and C20:5n-3 eicosapentaenoic acid (EPA) are shown. The binding affinity (Kd) of rE-FABP to fatty acid was calculated based on the reduction of fluorescence (see Material and Methods). E-FABP, epidermal fatty acid-binding protein.
Table 1.
Binding affinities of rE-FABP to selected fatty acids
| Fatty acid | Kd (nM) |
|---|---|
| C18:1n-9 (Oleic acid) | 154.6 ± 35.3 |
| C18:0 (Stearic acid) | 168.1 ± 38.1 |
| C20:4n-6 (Arachidonic acid) | 390.9 ± 54.2 |
| C22:6n-3 (Docosahexaenoic acid) | 422.2 ± 58.1 |
| C16:0 (Palmitic acid) | 478.0 ± 76.7 |
| C18:2n-6 (Linoleic acid) | 512.0 ± 34.5 |
| C20:5n-3 (Eicosapentaenoic acid) | 598.4 ± 100.5 |
| C17:0 (Heptadecanoic acid) | 973.6 ± 89.3 |
| C22:0 (Behenic acid) | 1021.5 ± 225.7 |
| C18:3n-3 (Linolenic acid) | 1027.7 ± 63.7 |
| C20:0 (Arachidic acid) | 1043.5 ± 140.8 |
| C15:0 (Pentadecanoic acid) | 1381.0 ± 310.5 |
| C14:0 (Myristic acid) | 1438.6 ± 461.9 |
| C24:1n-9 (Nervonic acid) | 1983.6 ± 545.6 |
| C12:0 (Lauric acid) | 2460.5 ± 532.1 |
| All-trans retinoic acid | 4539.6 ± 712.5 |
The binding affinity was measured using the ANS method as described in Material and Methods. Kd values are presented as mean ± SD of three separate measurements. ANS, 1-anilinonaphthalene-8-sulfonic acid; rE-FABP, recombinant epidermal fatty acid-binding protein.
LCPUFA enhancement of NGF-induced neurite formation is regulated by intracellular levels of E-FABP
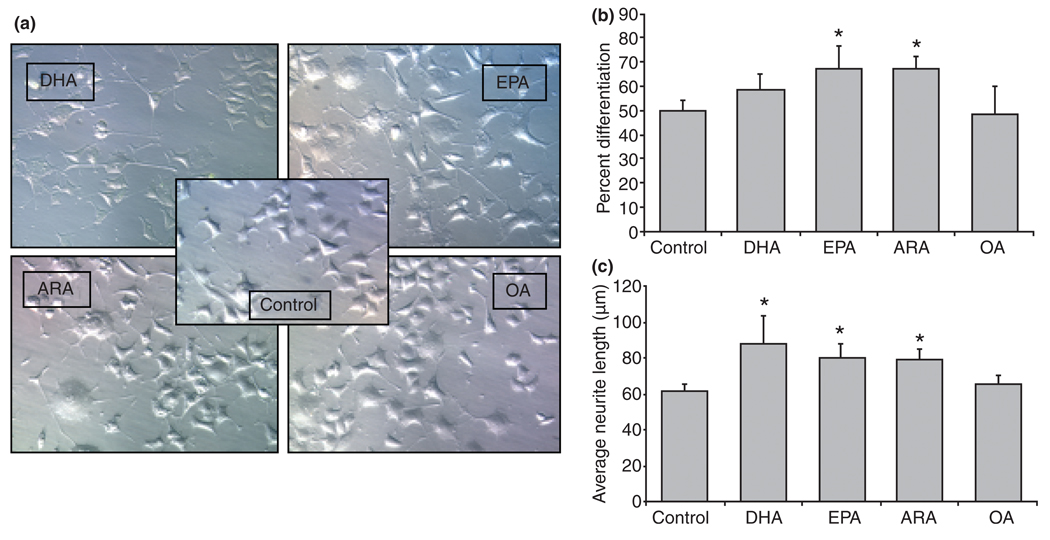
Fatty acid-binding proteins are believed to function as intracellular transporters of LCFFAs (Chmurzynska 2006). This hypothesis predicts that cellular actions of biologically active LCFFA may require appropriate levels of E-FABPs in the cytosol to shuttle them to their final destination site and execute their specific function. Considering that LCPUFA have been shown to facilitate neuronal differentiation and neurite outgrowth (Calderon and Kim 2004; Marszalek et al. 2004), we asked whether intracellular levels of E-FABPs influence the potential differentiation role of these FFAs. PC12 cells were grown in tissue culture conditions enriched with n-3 LCPUFA (such as DHA and EPA), n-6 LCPUFA (such as ARA), or monounsaturated LCFFA (such as OA) in the presence of NGF. PC12 cells supplemented with EPA, DHA, ARA, but not OA, expressed longer neurite formation than cultures treated with NGF alone (Fig. 7a). Quantitative analysis (Fig. 7b and c) showed that EPA and ARA significantly increased both percent differentiation and average neurite length in the presence of NGF. DHA supplementation in the presence of NGF significantly increased the average neurite length but not percentage of differentiation. Under these conditions, OA did not show a significant effect on either percent differentiation or average neurite length above the stimulation observed with NGF alone.
Fig. 7.
The effects of unsaturated long chain free fatty acid (LCFFA) on neurite extension in nerve growth factor-differentiated pheochromocytoma cells (NGFDPC12). Pheochromocytoma cells (PC12) were differentiated with 50 ng/mL of nerve growth factor (NGF) for 6 days with the supplement of 60 µM C22:6n-3 docosahexaenoic acid (DHA), C20:5n-3 eicosapentaenoic acid (EPA), C20:4n-6 arachidonic acid (ARA), or C18:1n-9 oleic acid (OA), which were complexed with 150 µM bovine serum albumin. Cells were then fixed and Hoffman modulation contrast images were taken. (a) Representative images. (b) Quantitative analysis of percent differentiation. (c) Quantitative analysis of average neurite length. The data represent mean ± SE of three experiments. Statistical significance between LCFFA treated groups and control groups were determined by one-way anova. *p < 0.01.
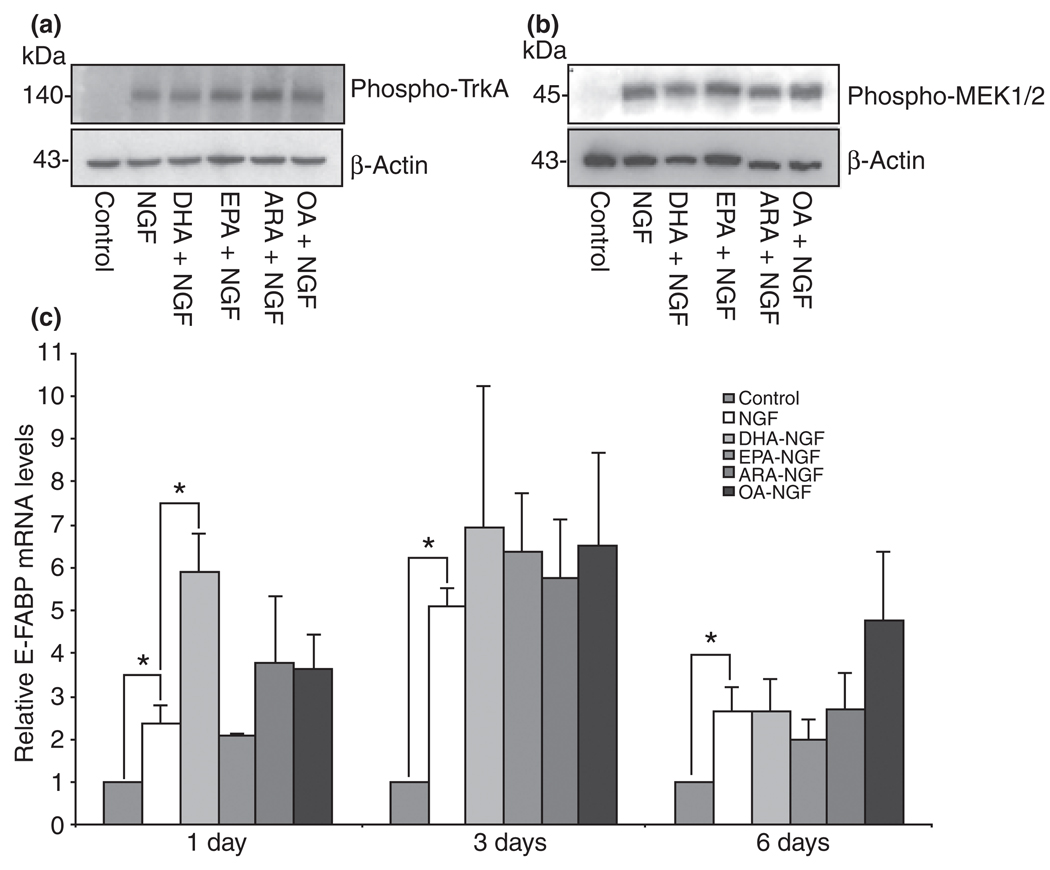
One possible mechanism by which the LCPUFA stimulate neurite extension is by increasing the phosphorylation of TrKA or other kinases associated with the NGF signaling pathway. PC12 cells were treated with DHA, EPA, ARA, or OA for 24 h following a 5 min treatment with NGF. Cells were harvested and the TrKA and MEK phosphorylation levels were analyzed using western blots as described in the Materials and methods. None of these fatty acids stimulated TrkA or MEK1/2 phosphorylation above NGF-induced levels (Fig. 8a and b). Next, we asked whether the neurite stimulating effects observed after treatment with these FFAs required an increase in E-FABP expression above the induced levels triggered by NGF. Cultures supplemented with DHA in the presence of NGF triggered a transient increase of E-FABP mRNA after 24 h above NGF induced levels (Fig. 8c). Treatment of NGFDPC12 with EPA, ARA, or OA did not significantly enhance E-FABP mRNA above the induced level seen in the presence of NGF alone.
Fig. 8.
The effects of unsaturated long chain free fatty acid on nerve growth factor (NGF)-induced phosphorylated TrkA and mitogen-activated protein kinase kinase (MEK1/2) and expression of epidermal fatty acid-binding protein (E-FABP) in pheochromocytoma cells (PC12) cells. PC12 cells were treated with 150 µM bovine serum albumin (BSA) plus 60 µM of C22:6n-3 docosahexaenoic acid (DHA), C20:5n-3 eicosapentaenoic acid (EPA), C20:4n-6 arachidonic acid (ARA), or C18:1n-9 oleic acid (OA) for 24 h and then stimulated with NGF for 5 min. Cells were washed and lysed with sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris–HCl, pH 6.8, 2% SDS, 10% glycerol, 50 mM dichlorodiphenyltrichloroethane, and 0.01% bromophenol blue). Western blots analysis of (a) phosphorylated TrkA and (b) phosphorylated MEK 1/2 were performed as described in Materials and methods. The experiment was repeated at least three times and a representative blot is shown. (c) PC12 cells were treated with NGF in the presence of 60 µM of DHA, EPA, ARA, or OA (complexed with 150 µM BSA) for 1, 3, and 6 days. The level of E-FABP mRNA was determined by QTRTPCR. The data represent mean ± SE of three or four experiments. Statistical significances between groups were determined by two-way anova. *p < 0.05.
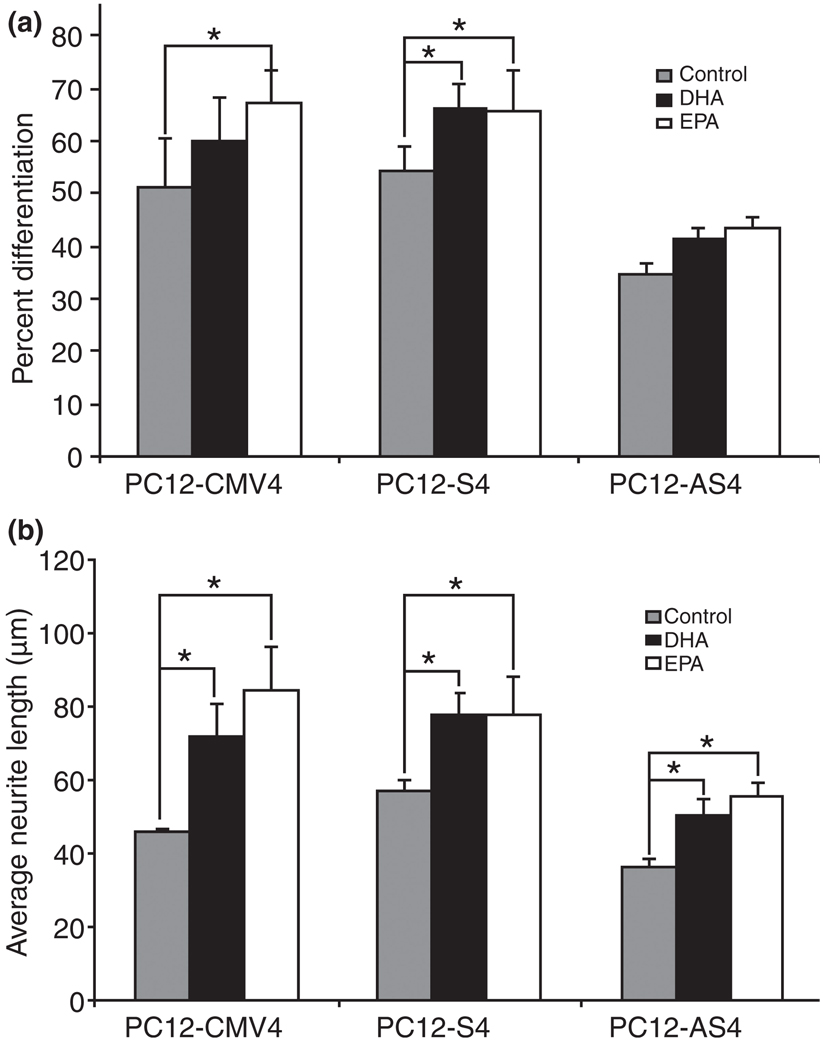
Considering that nerve growth factor-differentiated pheochromocytoma cells (NGFDPC12) already have induced levels of E-FABP, we examined whether DHA and EPA neurite stimulating effects were also observed after knocking down E-FABP mRNA and protein levels. We compared this effect in three different PC12 cell clonal lines that vary in the expression of E-FABP: NGFDPC12-CMV, NGFDPC12-S, and NGFDPC12-AS cells. Figure 9a and b show that EPA increased both the percent differentiation and average neurite length on NGFDPC12-CMV and NGFDPC12-S cells, but only increased average neurite length in NGFDPC12-AS cells that had a deficit in E-FABP expression (see Fig. 2a). DHA increased both the percent differentiation and average neurite length on NGFDPC12-S cells, while showing lower stimulation (only increased average neurite length) in NGFDPC12-CMV and NGFDPC12-AS cells. In addition, DHA and EPA supplementation did not improve average neurite length in NGFDPC12-AS to the levels seen in NGFDPC12-S and NGFDPC12-CMV cells.
Fig. 9.
Phenotype comparison of PC12-CMV4, PC12-S4, and PC12-AS4 cells following nerve growth factor (NGF) treatment in the presence of C22:6n-3 docosahexaenoic acid (DHA), or C20:5n-3 eicosapentaenoic acid (EPA). PC12-CMV4, PC12-S4, and PC12-AS4 cells were treated for 7 days with NGF combined with DHA/bovine serum albumin (BSA) or EPA/BSA at the concentration of 60 µM/150 µM. Cells were fixed and Hoffman modulation contrast images were taken. (a) Quantitative analysis of percent differentiation. (b) Quantitative analysis of average neurite length. The data represent SEM of three experiments. Statistical significance between groups was determined by two-way anova. *p < 0.05.
Discussion
The present study examined the effects E-FABP expression on neurite extension of differentiating PC12 cells with or without supplementation with LCFFA. The major findings are: (i) E-FABP expression is regulated by NGF and is needed to have normal neurite formation during differentiation; (ii) E-FABP alone is not sufficient to induce neurites in PC12 cells in the absence of NGF; (iii) E-FABP protein has high affinity to a broad range of saturated and unsaturated LCFFA including fatty acids that are needed for normal axonal growth; (iv) DHA, EPA, and ARA stimulate neurite outgrowth in differentiating PC12 cells following exposure to NGF; (v) neurite stimulation by DHA, EPA, and ARA does not require additional up-regulation of E-FABP to that triggered by NGF; (vi) enhancement of neurite extension in differentiating PC12 cells by DHA and EPA is impaired in PC12 cells antisense clones exhibiting repressed levels of E-FABP mRNA and protein; (vii) phenotypic impairment of neurite extension in PC12 antisense clones is overcome by treatment with recombinant biotin-E-FABP protein. Overall, the data suggest that normal neurite extension in differentiating PC12 cells requires adequate levels of LCPUFA and high expression levels of E-FABP.
The structure of E-FABP is similar to other members of the FABP family, which includes two α helices and 10 β-strands (β-barrel) that forms the fatty acid binding cavity (Gutierrez-Gonzalez et al. 2002). The binding stochiometry is one fatty acid molecule to one E-FABP molecule. Our results, the first report for the rat E-FABP, show that E-FABP binds to fatty acids with 16 to 22 carbons, i.e., long-chain fatty acids, and the affinity for a particular fatty acid varies significantly. This finding is consistent with the common binding properties reported for other FABPs (Zimmerman and Veerkamp 2002). Throughout the literature, Kd values of FFA to FABP analyzed by different binding methods are not always comparable. The Kd values reported here (using the ANS method) are in the nM range, which are modestly higher than those of FABPs using the AcryloDated Intestine Fatty Acid Binding Protein (ADIFAB) method (in low nM range) (Richieri et al. 1994, 2000), but lower than those of FABPs using the Lipidex method (in µM range) (Zimmerman and Veerkamp 2001; Zimmerman et al. 2001). E-FABP showed the highest affinity to two C18 fatty acids, OA and C18:0 (stearic acid), and did not appear to bind retinoic acid. This result agrees with the findings reported for E-FABP from mouse and human (Siegenthaler et al. 1994; Kane et al. 1996). Thus, our own findings and reports by others indicate that E-FABP has specific affinity to both saturated and unsaturated LCFFA. Brain-FABP preferentially binds n-3 LCPUFA (DHA) but shows no specific affinity to C16:0 (palmitic acid) (Feng et al. 1994; Xu et al. 1996) and heart-FABP shows higher affinity to n-6 LCPUFA (ARA) (Paulussen et al. 1988). These diverse binding characteristics for different FABPs are intriguing but may help in elucidating their particular function(s) in neurons. The broad range of ligands for E-FABP suggests that its binding characteristic is rather promiscuous suggesting a diverse role in the cell.
Our findings show that DHA, EPA, and ARA, but not OA, were capable of stimulating neurite extension in differentiating PC12 cells. These results are consistent with observations made by others (Calderon and Kim 2004; Marszalek et al. 2004). It is well established that the composition of phospholipids in the cell membrane is a major factor that influences membrane fluidity, which consequently affects essential neuronal functions, such as binding of hormone and growth factor receptors, activity of membrane-bound enzymes, transport of ions, and release and uptake neurotransmitters (Stubbs and Smith 1984; Daveloose et al. 1993; Zimmer et al. 2000; Nishio et al. 2004). LCPUFA, i.e., n-3 and n-6 fatty acids increase membrane fluidity because of their double bond-rich structure affecting a number of physiological processes (Valentine and Valentine 2004). An extra flexibility is needed in neuronal membranes because of their particular morphological properties, such as folded membranes in the disks of photoreceptors and highly curved membranes in synapses. In fact, about 50% of all acyl chains of phospholipids in the rod outer segment of the retina are DHA, mainly in phosphatidylethanolamine, phosphatidylserine (PS), and phosphatidylcholine, while minor lipids-like diacylglycerol, phosphatidylinositol, and phosphatidic acid contain predominantly ARA (Stinson et al. 1991). Deficiencies of n-3 fatty acids during development lead to decreased DHA accumulation in the brain lipids, particular in growth cone (Auestad and Innis 2000). This deficiency affects brain levels of DHA-containing PS (Hamilton et al. 2000), which is well known for its role in neuronal survival via phosphotidylinositol 3-kinase/Akt signaling pathway (Akbar et al. 2005). As a precursor of DHA, the effect of EPA may be mediated on its own or by converting to DHA. For instance, during phospholipid synthesis, PS is synthesized from preexisting PC or phosphatidylethanolamine by serine base exchange. DHA at the sn-2 position on PC is more favorably converted to PS that is rather abundant in nerve cells membranes (Kim et al. 2004). Interestingly, PC12 cells treated with [14C]-ARA, DHA, or OA, show more exogenous ARA and DHA incorporated into phospholipids than OA (Marszalek et al. 2005), which agrees with our finding that OA did not promote neurite extension in PC12 cells under the conditions used in this study. These data suggest that merely increasing the levels of a high affinity E-FABP ligand such as OA may not guarantee neurite stimulation if the ligand does not participate directly in that specific role.
Our finding that ARA supplementation of NGFDPC12 cultures resulted in longer neurites is consistent with the importance of this LCPUFA in neuronal differentiation. NGF-induced expressed levels of E-FABP seemed to be adequate to support this neurite stimulation. ARA is well known for its role as an intracellular second messenger in signal transduction. Upon stimulation, ARA is preferentially cleaved from the sn-2 position of phospholipids by Ca2+-dependent cytosolic phospholipase A2 and can be subsequently metabolized to leukotrienes by the action of lipoxygenase and to prostaglandins (PGs) by cyclooxygenase (COX). Both ARA and its metabolites regulate neuronal functions such as modulation of ion channels and neurotransmitter release (Ray et al. 1993; Williams et al. 1994), and increase long-term synaptic plasticity (Williams et al. 1989; Feinmark et al. 2003). For example, in PC12 cells, cytosolic phospholipase A2 and COX-1 are up-regulated after NGF treatment (Kaplan et al. 1997; Akiyama et al. 2004). COX products of ARA, especially prostaglandin PGJ2, increase neurite outgrowth in NGF-differentiated PC12 cells by activation of p38 MAP kinase and transcription factor activator protein-1 (Satoh et al. 1999; Jung et al. 2003). Further, Ca2+-independent PLA2 is found to be responsible for releasing DHA from the brain phospholipids (Strokin et al. 2003).
Stimulation of neurite length by DHA and EPA were more pronounced in PC12 cells with elevated levels of E-FABP (NGFDPC12-S) compared with PC12 cells with reduced levels of E-FABP (NGFDPC12-AS). More importantly, DHA and EPA improved the average neurite length of NGFDPC12-AS, yet their neurites were still significantly shorter than the ones exhibited by NGFDPC12-CMV and NGFDPC12-S cells. Darios and Davletov (2006) have demonstrated that ARA as well as other n-3 and n-6 LCPUFAs could act on syntaxsin 3, one of the proteins in lipid rafts in the growth cone plasma membrane, to stimulate soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex assembly. SNARE formation is critical for vesicle-associated membrane fusion during neurite outgrowth. This result suggests that LCPUFAs might directly stimulate membrane expending/neurite extension in addition to being incorporated into expanding membrane. Previous reports from our laboratory showed that E-FABP is present in neurites, axons, and growth cones (Liu et al. 1997; Allen et al. 2001). We speculate that E-FABP might play a role transporting/enriching LCPUFAs to the site of SNARE assembly, considering that syntaxin 3 itself exhibits relatively weak affinity to LCPUFAs (Darios and Davletov 2006).
The present study did not evaluate whether E-FABP expression level affects fatty acid incorporation into the cell. However, there is evidence that FABPs, including E-FABP, participate in the intracellular transport of fatty acid ligands to key intracellular target(s) [for review, see Veerkamp and Zimmerman (2001)]. Veerkamp et al. (2000) showed that E-FABP as well as other FABPs in the nervous tissue, i.e., brain-FABP, heart-FABP, and myelin-FABP, increases the transfer of [14C] palmitic acid from immobilized liposomes to isolated mitochondria. Further, studies using heart-FABP knockout mice demonstrate a significant reduction in the incorporation of ARA into brain phospholipids, especially into phosphatidylcholine (Murphy et al. 2005). Of particular importance is the nuclear localization of E-FABP and other FABPs in the nervous system shown in this study and previous studies from our laboratory and others (Feng et al. 1994; Godbout et al. 1995; Allen et al. 2000; Liu et al. 2000). Nuclear localization of E-FABP suggests that this protein play a role with peroxisomes proliferators-activated receptors (PPARs). Previous studies have shown that E-FABP is associated with the actions of PPARs. PPARs are ligand-activated transcription factors that exist in different isoforms (α, β/δ, and γ) and tissues distribution in the nervous system (Moreno et al. 2004). They are dimerized with the retinoid × receptors and are involved in lipid metabolism as well as cell differentiation, apoptosis, inflammation, and neurodegeneration (Bordet et al. 2006; Heneka and Landreth 2007). LCPUFA activation of PPARs inhibit cell proliferation and induce differentiation in IMR-32 neuroblastoma cells suggesting that NGF, possibly through TrkA, is critical for this transcriptional stimulation (Fuenzalida et al. 2005). E-FABP, in response to ligand binding, is translocated to the nucleus and selectively enhances the activities of PPARβ. Interestingly, transfection of E-FABP antisense vector inhibits the PPARβ-mediated differentiation of primary keratinocyte cultures (Tan et al. 2002). Thus, we speculate that LCPUFAs such as DHA and ARA can be transported to the nucleus by E-FABP to activate PPARs and induce transcription of selected genes associated with neuronal differentiation and axonal growth. One target gene for this process is acyl-CoA synthetase 2 (ACS2), considering that increases of PPARβ/δ in cultured cortical neurons correlates with an increase in ACS2. ACS2 is an important enzyme that activates FFAs for further phospholipid synthesis needed for neurite extension (Cimini et al. 2005). Future studies are necessary to further evaluate these possibilities in differentiating NGFDPC12 cells.
Acknowledgements
We would like to acknowledge the technical assistance of Angie P. Mison. This work has been funded in part by awards NSF-9728662 and NIH 5P20MD001632, and 5R25GM060507.
Abbreviations used
- ACS2
acyl-CoA synthetase 2
- ANS
1-anilinonaphthalene-8-sulfonic acid
- ARA
C20:4n-6 arachidonic acid
- BSA
bovine serum albumin
- CMV
cytomegalovirus
- COX
cyclooxygenase
- DHA
C22:6n-3 docosahexaenoic acid
- E-FABP
epidermal fatty acid-binding protein
- EPA
C20:5n-3 eicosapentaenoic acid
- FABP
fatty acid-binding protein
- FBS
fetal bovine serum
- FFA
free fatty acid
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IGF
insulin-like growth factor
- LCFFA
long chain free fatty acid
- LCPUFA
long chain polyunsaturated fatty acid
- MAP
mitogen-activated protein
- MEK
mitogen-activated protein kinase kinase
- NGF
nerve growth factor
- NGFDPC12 cells
nerve growth factor-differentiated pheochromocytoma cells
- OA
C18:1n-9 oleic acid
- PBS
phosphate-buffered saline
- PC12 cells
pheochromocytoma cells
- PPAR
peroxisomes proliferators-activated receptor
- PS
phosphatidylserine
- rE-FABP
recombinant E-FABP
- SNARE
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- TTBS
Tris-buffered saline with 0.05% Tween 20
References
- Ahmad A, Murthy M, Greiner RS, Moriguchi T, Salem N., Jr A decrease in cell size accompanies a loss of docosahexaenoate in the rat hippocampus. Nutr. Neurosci. 2002;5:103–113. doi: 10.1080/10284150290018973. [DOI] [PubMed] [Google Scholar]
- Akbar M, Calderon F, Wen Z, Kim HY. Docosahexaenoic acid: a positive modulator of Akt signaling in neuronal survival. Proc. Natl Acad. Sci. USA. 2005;102:10858–10863. doi: 10.1073/pnas.0502903102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama N, Hatori Y, Takashiro Y, Hirabayashi T, Saito T, Murayama T. Nerve growth factor-induced up-regulation of cytosolic phospholipase A2 alpha level in rat PC12 cells. Neurosci. Lett. 2004;365:218–222. doi: 10.1016/j.neulet.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Allen GW, Liu JW, De Leon M. Depletion of a fatty acid-binding protein impairs neurite outgrowth in PC12 cells. Brain Res. Mol. Brain Res. 2000;76:315–324. doi: 10.1016/s0169-328x(00)00014-0. [DOI] [PubMed] [Google Scholar]
- Allen GW, Liu J, Kirby MA, De Leon M. Induction and axonal localization of epithelial/epidermal fatty acid-binding protein in retinal ganglion cells are associated with axon development and regeneration. J. Neurosci. Res. 2001;66:396–405. doi: 10.1002/jnr.1232. [DOI] [PubMed] [Google Scholar]
- Auestad N, Innis SM. Dietary n-3 fatty acid restriction during gestation in rats: neuronal cell body and growth-cone fatty acids. Am. J. Clin. Nutr. 2000;71 Suppl 1:312S–314S. doi: 10.1093/ajcn/71.1.312s. [DOI] [PubMed] [Google Scholar]
- Barcelo-Coblijn G, Kitajka K, Puskas LG, Hogyes E, Zvara A, Hackler L, Jr, Farkas T. Gene expression and molecular composition of phospholipids in rat brain in relation to dietary n-6 to n-3 fatty acid ratio. Biochim. Biophys. Acta. 2003;1632:72–79. doi: 10.1016/s1388-1981(03)00064-7. [DOI] [PubMed] [Google Scholar]
- Bazan NG. Cell survival matters: docosahexaenoic acid signaling, neuroprotection and photoreceptors. Trends Neurosci. 2006;29:263–271. doi: 10.1016/j.tins.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Bordet R, Ouk T, Petrault O, et al. PPAR: a new pharmacological target for neuroprotection in stroke and neurodegenerative diseases. Biochem. Soc. Trans. 2006;34:1341–1346. doi: 10.1042/BST0341341. [DOI] [PubMed] [Google Scholar]
- Calderon F, Kim HY. Docosahexaenoic acid promotes neurite growth in hippocampal neurons. J. Neurochem. 2004;90:979–988. doi: 10.1111/j.1471-4159.2004.02520.x. [DOI] [PubMed] [Google Scholar]
- Carlson BA, Kingston JD. Docosahexaenoic acid, the aquatic diet, and hominin encephalization: difficulties in establishing evolutionary links. Am. J. Hum. Biol. 2007;19:132–141. doi: 10.1002/ajhb.20579. [DOI] [PubMed] [Google Scholar]
- Chmurzynska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J. Appl. Genet. 2006;47:39–48. doi: 10.1007/BF03194597. [DOI] [PubMed] [Google Scholar]
- Cimini A, Benedetti E, Cristiano L, Sebastiani P, D’Amico MA, D’Angelo B, Di Loreto S. Expression of peroxisome proliferator-activated receptors (PPARs) and retinoic acid receptors (RXRs) in rat cortical neurons. Neuroscience. 2005;130:325–337. doi: 10.1016/j.neuroscience.2004.09.043. [DOI] [PubMed] [Google Scholar]
- Cole GM, Frautschy SA. Docosahexaenoic acid protects from amyloid and dendritic pathology in an Alzheimer’s disease mouse model. Nutr. Health. 2006;18:249–259. doi: 10.1177/026010600601800307. [DOI] [PubMed] [Google Scholar]
- Connor WE, Neuringer M, Reisbick S. Essential fatty acids: the importance of n-3 fatty acids in the retina and brain. Nutr. Rev. 1992;50:21–29. doi: 10.1111/j.1753-4887.1992.tb01286.x. [DOI] [PubMed] [Google Scholar]
- Darios F, Davletov B. Omega-3 and omega-6 fatty acids stimulate cell membrane expansion by acting on syntaxin 3. Nature. 2006;440:813–817. doi: 10.1038/nature04598. [DOI] [PubMed] [Google Scholar]
- Das KP, Freudenrich TM, Mundy WR. Assessment of PC12 cell differentiation and neurite growth: a comparison of morphological and neurochemical measures. Neurotoxicol. Teratol. 2004;26:397–406. doi: 10.1016/j.ntt.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Daveloose D, Linard A, Arfi T, Viret J, Christon R. Simultaneous changes in lipid composition, fluidity and enzyme activity in piglet intestinal brush border membrane as affected by dietary polyunsaturated fatty acid deficiency. Biochim. Biophys. Acta. 1993;1166:229–237. doi: 10.1016/0005-2760(93)90102-f. [DOI] [PubMed] [Google Scholar]
- Davletov B, Connell E, Darios F. Regulation of SNARE fusion machinery by fatty acids. Cell. Mol. Life Sci. 2007;64:1597–1608. doi: 10.1007/s00018-007-6557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon M, Welcher AA, Nahin RH, Liu Y, Ruda MA, Shooter EM, Molina CA. Fatty acid binding protein is induced in neurons of the dorsal root ganglia after peripheral nerve injury. J. Neurosci. Res. 1996;44:283–292. doi: 10.1002/(SICI)1097-4547(19960501)44:3<283::AID-JNR9>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Feinmark SJ, Begum R, Tsvetkov E, Goussakov I, Funk CD, Siegelbaum SA, Bolshakov VY. 12-lipoxygenase metabolites of arachidonic acid mediate metabotropic glutamate receptor-dependent long-term depression at hippocampal CA3-CA1 synapses. J. Neurosci. 2003;23:11427–11435. doi: 10.1523/JNEUROSCI.23-36-11427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Fuenzalida KM, Aguilera MC, Piderit DG, Ramos PC, Contador D, Quinones V, Rigotti A, Bronfman FC, Bronfman M. Peroxisome proliferator-activated receptor gamma is a novel target of the nerve growth factor signaling pathway in PC12 cells. J. Biol. Chem. 2005;280:9604–9609. doi: 10.1074/jbc.M409447200. [DOI] [PubMed] [Google Scholar]
- Godbout R, Marusyk H, Bisgrove D, Dabbagh L, Poppema S. Localization of a fatty acid binding protein and its transcript in the developing chick retina. Exp. Eye Res. 1995;60:645–657. doi: 10.1016/s0014-4835(05)80006-5. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Gonzalez LH, Ludwig C, Hohoff C, Rademacher M, Hanhoff T, Ruterjans H, Spener F, Lucke C. Solution structure and backbone dynamics of human epidermal-type fatty acid-binding protein (E-FABP) Biochem. J. 2002;364:725–737. doi: 10.1042/BJ20020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton L, Greiner R, Salem N, Jr, Kim HY. n-3 fatty acid deficiency decreases phosphatidylserine accumulation selectively in neuronal tissues. Lipids. 2000;35:863–869. doi: 10.1007/s11745-000-0595-x. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Landreth GE. PPARs in the brain. Biochim. Biophys. Acta. 2007;1771:1031–1045. doi: 10.1016/j.bbalip.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Hichami A, Datiche F, Ullah S, Lienard F, Chardigny JM, Cattarelli M, Khan NA. Olfactory discrimination ability and brain expression of c-fos, Gir and Glut1 mRNA are altered in n-3 fatty acid-depleted rats. Behav. Brain Res. 2007;184:1–10. doi: 10.1016/j.bbr.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu. Rev. Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- Jump DB. The biochemistry of n-3 polyunsaturated fatty acids. J. Biol. Chem. 2002;277:8755–8758. doi: 10.1074/jbc.R100062200. [DOI] [PubMed] [Google Scholar]
- Jung KM, Park KS, Oh JH, et al. Activation of p38 mitogen-activated protein kinase and activator protein-1 during the promotion of neurite extension of PC-12 cells by 15-deoxydelta12,14-prostaglandin J2. Mol. Pharmacol. 2003;63:607–616. doi: 10.1124/mol.63.3.607. [DOI] [PubMed] [Google Scholar]
- Kane CD, Bernlohr DA. A simple assay for intracellular lipid-binding proteins using displacement of 1-anilinonaphthalene-8-sulfonic acid. Anal. Biochem. 1996;233:197–204. doi: 10.1006/abio.1996.0028. [DOI] [PubMed] [Google Scholar]
- Kane CD, Coe NR, Vanlandingham B, Krieg P, Bernlohr DA. Expression, purification, and ligand-binding analysis of recombinant keratinocyte lipid-binding protein (MAL-1), an intracellular lipid-binding found overexpressed in neoplastic skin cells. Biochemistry. 1996;35:2894–2900. doi: 10.1021/bi952476e. [DOI] [PubMed] [Google Scholar]
- Kaplan MD, Olschowka JA, O’Banion MK. Cyclooxygenase-1 behaves as a delayed response gene in PC12 cells differentiated by nerve growth factor. J. Biol. Chem. 1997;272:18534–18537. doi: 10.1074/jbc.272.30.18534. [DOI] [PubMed] [Google Scholar]
- Kim HY, Bigelow J, Kevala JH. Substrate preference in phosphatidylserine biosynthesis for docosahexaenoic acid containing species. Biochemistry. 2004;43:1030–1036. doi: 10.1021/bi035197x. [DOI] [PubMed] [Google Scholar]
- Kirk WR, Kurian E, Prendergast FG. Characterization of the sources of protein-ligand affinity: 1-sulfonato-8-(1′) anilinonaphthalene binding to intestinal fatty acid binding protein. Biophys. J. 1996;70:69–83. doi: 10.1016/S0006-3495(96)79592-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz A, Zimmer A, Schnutgen F, Bruning G, Spener F, Muller T. The expression pattern of a novel gene encoding brain fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–2649. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- Liu Y, Molina CA, Welcher AA, Longo LD, De Leon M. Expression of DA11, a neuronal-injury-induced fatty acid binding protein, coincides with axon growth and neuronal differentiation during central nervous system development. J. Neurosci. Res. 1997;48:551–562. doi: 10.1002/(sici)1097-4547(19970615)48:6<551::aid-jnr8>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Liu Y, Longo LD, De Leon M. In situ and immunocytochemical localization of E-FABP mRNA and protein during neuronal migration and differentiation in the rat brain. Brain Res. 2000;852:16–27. doi: 10.1016/s0006-8993(99)02158-7. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Kitidis C, Dararutana A, Lodish HF. Acyl-CoA synthetase 2 overexpression enhances fatty acid internalization and neurite outgrowth. J. Biol. Chem. 2004;279:23882–23891. doi: 10.1074/jbc.M313460200. [DOI] [PubMed] [Google Scholar]
- Marszalek JR, Kitidis C, Dirusso CC, Lodish HF. Long-chain acyl-CoA synthetase 6 preferentially promotes DHA metabolism. J. Biol. Chem. 2005;280:10817–10826. doi: 10.1074/jbc.M411750200. [DOI] [PubMed] [Google Scholar]
- Martin RE, Wickham JQ, Om AS, Sanders J, Ceballos N. Uptake and incorporation of docosahexaenoic acid (DHA) into neuronal cell body and neurite/nerve growth cone lipids: evidence of compartmental DHA metabolism in nerve growth factor-differentiated PC12 cells. Neurochem. Res. 2000;25:715–723. doi: 10.1023/a:1007575406896. [DOI] [PubMed] [Google Scholar]
- Moreno S, Farioli-Vecchioli S, Ceru MP. Immunolocalization of peroxisome proliferator-activated receptors and retinoid × receptors in the adult rat CNS. Neuroscience. 2004;123:131–145. doi: 10.1016/j.neuroscience.2003.08.064. [DOI] [PubMed] [Google Scholar]
- Murphy EJ, Owada Y, Kitanaka N, Kondo H, Glatz JF. Brain arachidonic acid incorporation is decreased in heart fatty acid binding protein gene-ablated mice. Biochemistry. 2005;44:6350–6360. doi: 10.1021/bi047292r. [DOI] [PubMed] [Google Scholar]
- Nishio M, Fukumoto S, Furukawa K, Ichimura A, Miyazaki H, Kusunoki S, Urano T. Overexpressed GM1 suppresses nerve growth factor (NGF) signals by modulating the intracellular localization of NGF receptors and membrane fluidity in PC12 cells. J. Biol. Chem. 2004;279:33368–33378. doi: 10.1074/jbc.M403816200. [DOI] [PubMed] [Google Scholar]
- Niu SL, Mitchell DC, Lim SY, Wen ZM, Kim HY, Salem N, Jr, Litman BJ. Reduced G protein-coupled signaling efficiency in retinal rod outer segments in response to n-3 fatty acid deficiency. J. Biol. Chem. 2004;279:31098–31104. doi: 10.1074/jbc.M404376200. [DOI] [PubMed] [Google Scholar]
- Owada Y, Yoshimoto T, Kondo H. Increased expression of the mRNA for brain- and skin-type but not heart-type fatty acid binding proteins following kainic acid systemic administration in the hippocampal glia of adult rats. Brain Res. Mol. Brain Res. 1996a;42:156–160. doi: 10.1016/s0169-328x(96)00182-9. [DOI] [PubMed] [Google Scholar]
- Owada Y, Yoshimoto T, Kondo H. Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J. Chem. Neuroanat. 1996b;12:113–122. doi: 10.1016/s0891-0618(96)00192-5. [DOI] [PubMed] [Google Scholar]
- Paulussen RJ, van der Logt CP, Veerkamp JH. Characterization and binding properties of fatty acid-binding proteins from human, pig, and rat heart. Arch. Biochem. Biophys. 1988;264:533–545. doi: 10.1016/0003-9861(88)90319-0. [DOI] [PubMed] [Google Scholar]
- Pifferi F, Roux F, Langelier B, Alessandri JM, Vancassel S, Jouin M, Lavialle M, Guesnet P. (n-3) polyunsaturated fatty acid deficiency reduces the expression of both isoforms of the brain glucose transporter GLUT1 in rats. J. Nutr. 2005;135:2241–2246. doi: 10.1093/jn/135.9.2241. [DOI] [PubMed] [Google Scholar]
- Polverini E, Fornabaio M, Fasano A, Carlone G, Riccio P, Cavatorta P. The pH-dependent unfolding mechanism of P2 myelin protein: an experimental and computational study. J. Struct. Biol. 2006;153:253–263. doi: 10.1016/j.jsb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Ray P, Berman JD, Middleton W, Brendle J. Botulinum toxin inhibits arachidonic acid release associated with acetylcholine release from PC12 cells. J. Biol. Chem. 1993;268:11057–11064. [PubMed] [Google Scholar]
- Richieri GV, Ogata RT, Kleinfeld AM. Equilibrium constants for the binding of fatty acids with fatty acid-binding proteins from adipocyte, intestine, heart, and liver measured with the fluorescent probe ADIFAB. J. Biol. Chem. 1994;269:23918–23930. [PubMed] [Google Scholar]
- Richieri GV, Ogata RT, Zimmerman AW, Veerkamp JH, Kleinfeld AM. Fatty acid binding proteins from different tissues show distinct patterns of fatty acid interactions. Biochemistry. 2000;39:7197–7204. doi: 10.1021/bi000314z. [DOI] [PubMed] [Google Scholar]
- Roegge CS, Widholm JJ, Engeseth NJ, Wang X, Brosch KO, Seegal RF, Schantz SL. Delayed spatial alternation impairments in adult rats following dietary n-6 deficiency during development. Neurotoxicol. Teratol. 2005;27:485–495. doi: 10.1016/j.ntt.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Satoh T, Furuta K, Suzuki M, Watanabe Y. Prostaglandin J2 and its metabolites promote neurite outgrowth induced by nerve growth factor in PC12 cells. Biochem. Biophys. Res. Commun. 1999;258:50–53. doi: 10.1006/bbrc.1999.0587. [DOI] [PubMed] [Google Scholar]
- Sellner PA, Chu W, Glatz JF, Berman NE. Developmental role of fatty acid-binding proteins in mouse brain. Brain Res. Dev. Brain Res. 1995;89:33–46. doi: 10.1016/0165-3806(95)00099-y. [DOI] [PubMed] [Google Scholar]
- Siegenthaler G, Hotz R, Chatellard-Gruaz D, Didierjean L, Hellman U, Saurat JH. Purification and characterization of the human epidermal fatty acid-binding protein: localization during epidermal cell differentiation in vivo and in vitro. Biochem. J. 1994;302:363–371. doi: 10.1042/bj3020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinson AM, Wiegand RD, Anderson RE. Fatty acid and molecular species compositions of phospholipids and diacylglycerols from rat retinal membranes. Exp. Eye Res. 1991;52:213–218. doi: 10.1016/0014-4835(91)90261-c. [DOI] [PubMed] [Google Scholar]
- Strokin M, Sergeeva M, Reiser G. Docosahexaenoic acid and arachidonic acid release in rat brain astrocytes is mediated by two separate isoforms of phospholipase A2 and is differently regulated by cyclic AMP and Ca2+ Br. J. Pharmacol. 2003;139:1014–1022. doi: 10.1038/sj.bjp.0705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs CD, Smith AD. The modification of mammalian membrane polyunsaturated fatty acid composition in relation to membrane fluidity and function. Biochim. Biophys. Acta. 1984;779:89–137. doi: 10.1016/0304-4157(84)90005-4. [DOI] [PubMed] [Google Scholar]
- Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine RC, Valentine DL. Omega-3 fatty acids in cellular membranes: a unified concept. Prog. Lipid Res. 2004;43:383–402. doi: 10.1016/j.plipres.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Veerkamp JH, Zimmerman AW. Fatty acid-binding proteins of nervous tissue. J. Mol. Neurosci. 2001;16:133–142. doi: 10.1385/JMN:16:2-3:133. discussion 151–7. [DOI] [PubMed] [Google Scholar]
- Veerkamp JH, Van HtM, Zimmerman AW. Effect of fatty acid-binding proteins on intermembrane fatty acid transport studies on different types and mutant proteins. Eur. J. Biochem. 2000;267:5959–5966. doi: 10.1046/j.1432-1327.2000.01665.x. [DOI] [PubMed] [Google Scholar]
- Widstrom RL, Norris AW, Spector AA. Binding of cytochrome P450 monooxygenase and lipoxygenase pathway products by heart fatty acid-binding protein. Biochemistry. 2001;40:1070–1076. doi: 10.1021/bi001602y. [DOI] [PubMed] [Google Scholar]
- Williams JH, Errington ML, Lynch MA, Bliss TV. Arachidonic acid induces a long-term activity-dependent enhancement of synaptic transmission in the hippocampus. Nature. 1989;341:739–742. doi: 10.1038/341739a0. [DOI] [PubMed] [Google Scholar]
- Williams EJ, Walsh FS, Doherty P. The production of arachidonic acid can account for calcium channel activation in the second messenger pathway underlying neurite outgrowth stimulated by NCAM, N-cadherin, and L1. J. Neurochem. 1994;62:1231–1234. doi: 10.1046/j.1471-4159.1994.62031231.x. [DOI] [PubMed] [Google Scholar]
- Xu LZ, Sanchez R, Sali A, Heintz N. Ligand specificity of brain lipid-binding protein. J. Biol. Chem. 1996;271:24711–24719. doi: 10.1074/jbc.271.40.24711. [DOI] [PubMed] [Google Scholar]
- Zimmer L, Delpal S, Guilloteau D, Aioun J, Durand G, Chalon S. Chronic n-3 polyunsaturated fatty acid deficiency alters dopamine vesicle density in the rat frontal cortex. Neurosci. Lett. 2000;284:25–28. doi: 10.1016/s0304-3940(00)00950-2. [DOI] [PubMed] [Google Scholar]
- Zimmerman AW, Veerkamp JH. Fatty-acid-binding proteins do not protect against induced cytotoxicity in a kidney cell model. Biochem. J. 2001;360:159–165. doi: 10.1042/0264-6021:3600159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AW, Veerkamp JH. New insights into the structure and function of fatty acid-binding proteins. Cell. Mol. Life Sci. 2002;59:1096–1116. doi: 10.1007/s00018-002-8490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman AW, van Moerkerk HT, Veerkamp JH. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int. J. Biochem. Cell Biol. 2001;33:865–876. doi: 10.1016/s1357-2725(01)00070-x. [DOI] [PubMed] [Google Scholar]