Abstract
The large differences in redox potentials between the HiPIPs and ferredoxins are generally attributed to hydrogen bonds and electrostatic effects from the protein and solvent. Recent ligand K-edge X-ray absorption studies by Solomon and co-workers show that the Fe-S covalencies of [4Fe-4S] clusters in the two proteins differ considerably apparently because of hydrogen bonds from water, indicating electronic effects may be important. However, combined density function theory (DFT) and photoelectron spectroscopy studies by our group and Wang and co-workers indicate that hydrogen bonds tune the potential of [4Fe-4S] clusters by mainly electrostatics. The DFT studies here rationalize both results, namely that the observed change in the Fe-S covalency is due to differences in ligand conformation between the two proteins rather than hydrogen bonds. Moreover, the ligand conformation affects the calculated potentials by ~100 mV, and thus is a heretofore unconsidered means of tuning the potential.
A classical question about oxidation-reduction potentials in proteins is why the [4Fe-4S] clusters in the high potential iron-sulfur proteins (HiPIPs) have a 1-/2- reduction at 100 to 450 mV vs. NHE, while the ferredoxins (Fds) have a 2-/3- reduction at −100 to −645 mV.1 The differences are generally attributed to hydrogen bonding and electrostatic effects from the surrounding protein and solvent.2–5 Recent ligand K-edge X-ray absorption spectroscopy (XAS) experiments find large differences in the Fe-S covalency between HiPIPs and Fds apparently due to hydration,6 and attribute the redox potential differences to electronic effects of water hydrogen bonds based on a correlation between electrochemical redox potentials and metal-ligand bond covalencies in iron-sulfur complexes.7 However, our combined density functional theory (DFT) and photoelectron spectroscopy (PES) studies indicate hydrogen bond effects are actually primarily electrostatic since the electronic structure and bonding of the clusters are not significantly affected.8 Here, we present a new mechanism for redox tuning, namely the ligand conformation, which reconciles the seemingly conflicting experimental results.
Due to the complexity of the protein environment, the effects of hydration and ligand conformation on Fe-S covalencies and redox properties are separated using broken symmetry DFT (BS-DFT)9 calculations of the analog [Fe4S4(SEt)4]2− (Et = ethyl), which is a good model for the [Fe4S4(Cys)4]n− found in proteins.8 Redox energies in the gas phase were calculated at the B3LYP/6-31(++)SG**//B3LYP/6-31G** level and Fe-L covalencies as measured by the percent ligand (%L) character mixing in the Fe 3d orbitals were obtained from natural bond orbital (NBO) analysis10 of geometries at the B3LYP/6-31G** level. Thus, the calculations focus on a single effect at a time without approximations for the environment and take advantage of our previous calibrations of a variety of clusters against experiment.11,12 Moreover, using these methods, vertical detachment energies (VDE) correlate well with total %L character for a series of [Fe4S4L4]2− clusters in the gas phase with different ligands12, in agreement with the above mentioned XAS experiments.6,7 However, the vertical reduction energies (VRE), which are defined similarly to the VDE as the energy to add an electron without allowing relaxation, have a more complicated dependence on the total %L character.
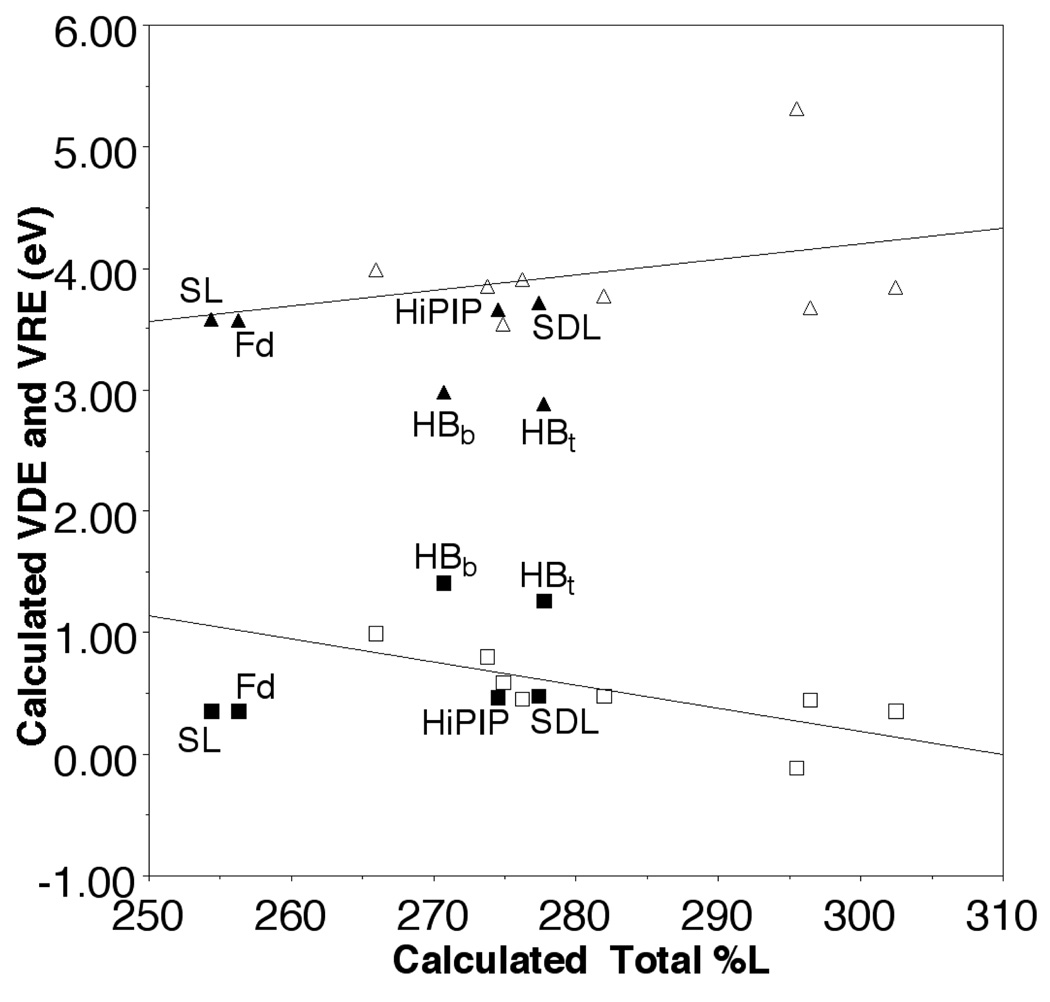
To elucidate hydration effects, [Fe4S4(SEt)4]2− was compared to hydrated complexes in which a water molecule was placed near each terminal or bridging sulfur ([Fe4S4(SEt)4]2−-HB(t) and [Fe4S4(SEt)4]2−-HB(b), respectively), resulting in a total of four water molecules per cluster. In the optimized structures, each water forms two hydrogen bonds, with [Fe4S4(SEt)4]2−-HB(t) slightly more favorable by ~0.9 eV. Although VDE and VRE change by ~0.7 eV upon hydration, the change in %L character is small and accounts for less than ~0.1 eV of the VDE and VRE differences based on the correlation for other ligands (Figure 1). Thus, the major contribution to the redox potential differences appears to be the electrostatic effect of the hydrogen bonds, consistent with PES/DFT studies8 and protein calculations.13–15
Figure 1.
The correlation of the calculated total Fe-S/L covalency (%L) with VDE (open square) and VRE (open triangle) of [Fe4S4L4]2− (left to right, L=Cl, SH, S-t-butyl, SCH3, SeCH3, H, PH2, and P(CH3)2 and with VDE (solid square) and VRE (solid triangle) of [Fe4S4(SEt)4]2− for the spin-delocalized state (SDL), the spin-localized state (SL), with waters hydrogen bonded to the bridging (HBb) and terminal (HBt) sulfurs, and in the Fd- and HiPIP-like conformations.
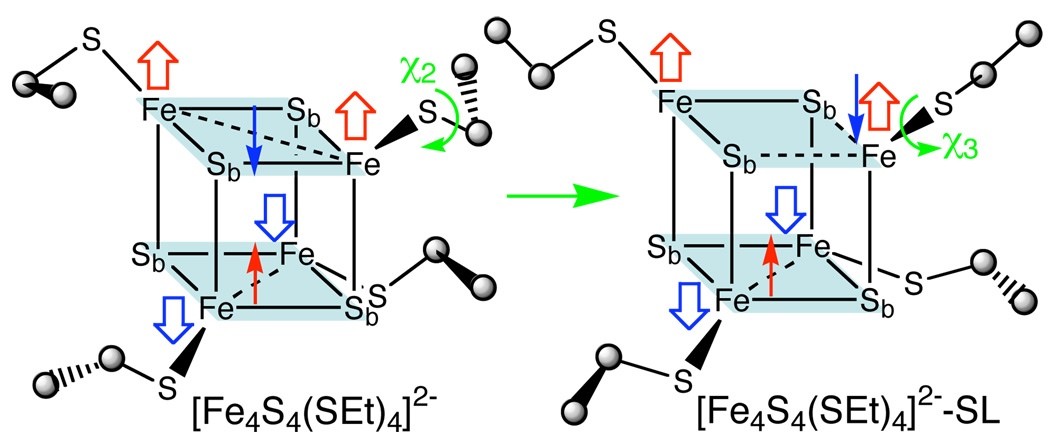
However, the %L character of the HiPIPs and the Fd are significantly different in the XAS studies, which must have an underlying cause. A possibility is suggested by another conformation of [Fe4S4(SEt)4]2− with different χ3 (C-St-Fe–Sb, where Sb is the bridging sulfur on the opposite layer and St is the terminal sulfur ligand) torsions (Figure 2) that has very similar VDE and VRE but only 92% of calculated total %L character of [Fe4S4(SEt)4]2− (Figure 1), apparently due to the minority spin exchange states of the two antiferromagnetically coupled layers that comprise the cubane.9 Specifically, [Fe4S4(SEt)4]2−, which is like experimental conformational structure,16 has all four χ3 ≈ 50° and the layers each have a minority spin delocalized to create a symmetric Fe2.5+-Fe2.5+ pair. On the other hand, the other conformer is characterized by two χ3 ≈ 85° and two χ3 ≈ 60° and has one layer in which the minority spin becomes localized to create a Fe3+-Fe2+ pair, henceforth referred to as [Fe4S4(SEt)4]2−-SL (Figure 2). Similar spin state have been found in the fission transition state of [4Fe-4S] clusters17 and in the hetero-ligand [Fe4S4(SCH3)2L2]2− (L = Cl, H).18
Figure 2.
The schematic spin-delocalized [Fe4S4(SEt)4]2− and spin-localized [Fe4S4(SEt)4]2−-SL structures.
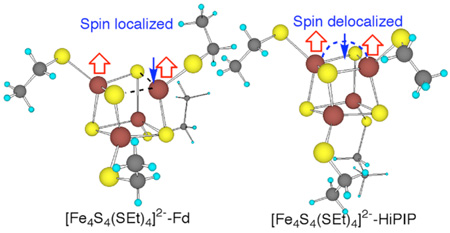
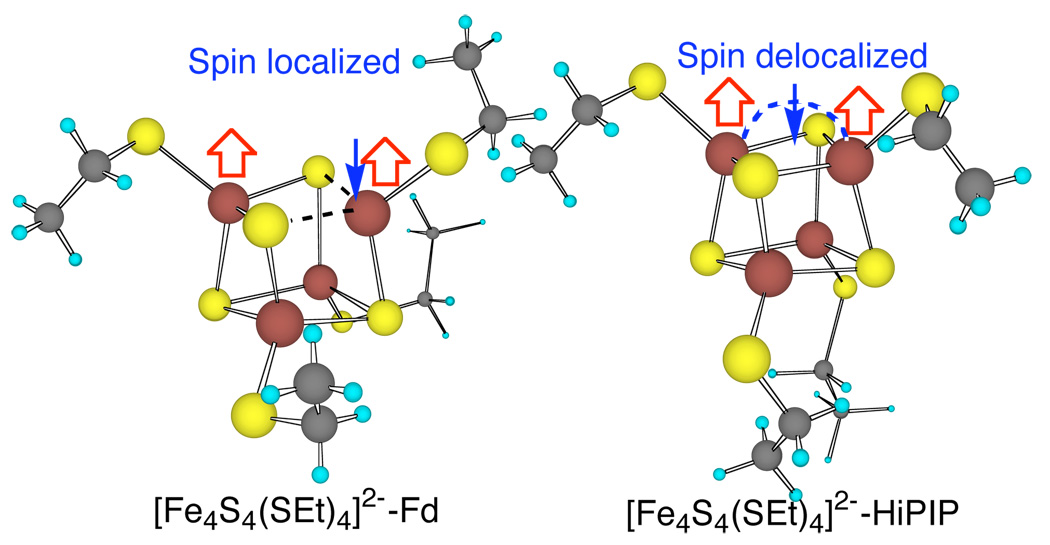
Two more conformations of [Fe4S4(SEt)4]2− (Figure 3) were optimized from X-ray structures of redox sites of Thermochromatium tepidum (Tt)19 HiPIP at 0.80 Å resolution ([Fe4S4(SEt)4]2−-HiPIP), and Bacillus thermoproteolyticus (Bt)20 Fd at 0.92 Å resolution, ([Fe4S4(SEt)4]2−-Fd). Interestingly, [Fe4S4(SEt)4]2−-HiPIP has two Fe2.5+-Fe2.5+ layers with χ3 = −65 and +68° for one and χ3 = −153 and 172° for the other, and its cubane structure, spin state, and redox properties are similar to those of [Fe4S4(SEt)4]2−, in very good agreement with Mössbauer of analogs and HiPIPs.21 On the other hand, [Fe4S4(SEt)4]2−-Fd has one Fe3+-Fe2+ layer with χ3 ≈ 84 and 74° and one Fe2.5+-Fe2.5+ layer with χ3 ≈ −76 and +66° and its cubane structure, spin state, and redox properties are similar to those of [Fe4S4(SEt)4]2−-SL. The Fe2.5+ in Fd versus the Fe3+ in HiPIP also appears consistent with Mössbauer results in which the [Fe4S4(Cys)4]2− cluster in Fd has less quadrupole splitting (ΔEQ) than in HiPIP.21,22 Remarkably, the calculated total Fe-S covalencies of [Fe4S4(SEt)4]2−-Fd and [Fe4S4(SEt)4]2−-HiPIP are 92% and 99% of [Fe4S4(SEt)4]2− values, respectively, in good agreement with the experimental results that Bt Fd and Chromatium vinosum HiPIP are 85% and 96% of [Fe4S4(SEt)4]2−.6
Figure 3.
The calculated Fd- and HiPIP-like [Fe4S4(SEt)4]2− structures.
In summary, BS-B3LYP calculations show dramatic changes in redox energies of [Fe4S4(SEt)4]2− due to hydration with only slight changes in the Fe-S covalency. On the other hand, the conformational changes in thiolate ligands of [Fe4S4(SEt)4]2−-SL and [Fe4S4(SEt)4]2−-Fd induce minority spin localization, leading to large decreases in Fe-S covalency with only small changes in redox energies relative to [Fe4S4(SEt)4]2− and [Fe4S4(SEt)4]2−-HiPIP. These findings suggest that the large change in the Fe-S covalency between Fd and HiPIP observed by XAS is due differences in the conformations of the cysteinyl ligands of the cluster. Furthermore, the resulting differences in redox energies of ~100 mV are significant; thus, ligand conformation is important to consider in understanding redox properties of iron-sulfur proteins.
Supplementary Material
The hydrogen bonding of terminal and bridging ligands of [Fe4S4(SEt)4]2−, [Fe4S4(SEt)4]2−-HB(t) and [Fe4S4(SEt)4]2−-HB(b), (Figure S1), the B3LYP/6–31G** optimized geometry parameters of spin-delocalized, spinlocalized, Fd-like and HiPIP-like [Fe4S4(SEt)4]2− structures (Table S1). These materials are available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
This work was supported by a grant from the National Institutes of Health (GM-45303). The calculations were performed at the Environmental Molecular Sciences Laboratory, a national user facility sponsored by the U.S. DOE’s Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, operated for DOE by Battelle, under the grant GC3565 and GC20901. Additional computational resources were provided by the William G. McGowan Foundation.
References
- 1.Cammack R. Adv. Inorg. Chem. 1992;38:281. [Google Scholar]
- 2.Stephens PJ, Jollie DR, Warshel A. Chem. Rev. 1996;96:2491. doi: 10.1021/cr950045w. [DOI] [PubMed] [Google Scholar]
- 3.Capozzi F, Ciurli S, Luchinat C. Metal Sites in Proteins and Models. 1998;90:127. [Google Scholar]
- 4.Adman ET, Watenpaugh KD, Jensen LH. Proc. Natl. Acad. Sci. U. S. A. 1975;72:4854. doi: 10.1073/pnas.72.12.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backes G, Mino Y, Loehr TM, Meyer TE, Cusanovich MA, Sweeny WV, Adman ET, Sanders-Loehr J. J. Am. Chem. Soc. 1991;113:2055. [Google Scholar]
- 6.Dey A, Francis EJ, Adams MWW, Babini E, Takahashi Y, Fukuyama K, Hodgson KO, Hedman B, Solomon EI. Science. 2007;318:1464. doi: 10.1126/science.1147753. [DOI] [PubMed] [Google Scholar]
- 7.Solomon EI, Hedman B, Hodgson KO, Dey A, Szilagyi RK. Coord. Chem. Rev. 2005;249:97. [Google Scholar]
- 8.Yang X, Niu S, Ichiye T, Wang L-S. J. Am. Chem. Soc. 2004;126:15790. doi: 10.1021/ja045709a. [DOI] [PubMed] [Google Scholar]
- 9.Noodleman L, Peng CY, Case DA, Mouesca JM. Coord. Chem. Rev. 1995;144:199. [Google Scholar]
- 10.All calculations were performed using the NWChem program package: Straatsma TP, et al. NWChem, A Computational Chemistry Package for Parallel Computers, Version 4.6`. Richland, Washington 99352-0999, USA: Pacific Northwest National Laboratory; 2004. .
- 11.Wang X-B, Niu S-Q, Yang X, Ibrahim SK, Pickett CJ, Ichiye T, Wang L-S. J. Am. Chem. Soc. 2003;125:14072. doi: 10.1021/ja036831x. [DOI] [PubMed] [Google Scholar]
- 12.Niu S-Q, Ichiye T. Unpublished work. 2008 [Google Scholar]
- 13.Jensen GM, Warshel A, Stephens PJ. Biochem. 1994;33:10911. doi: 10.1021/bi00202a010. [DOI] [PubMed] [Google Scholar]
- 14.Beck BW, Xie Q, Ichiye T. Biophys. J. 2001;81:601. doi: 10.1016/s0006-3495(01)75726-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagen KS, Watson AD, Holm RH. Inorg. Chem. 1984;23:2984. [Google Scholar]
- 17.Niu S-Q, Wang X-B, Yang X, Wang L-S, Ichiye T. J. Phys. Chem. A. 2004;108:6750. [Google Scholar]
- 18.Niu S-Q, Ichiye T. Unpublished work. 2008 [Google Scholar]
- 19.Liu LJ, Nogi T, Kobayashi M, Nozawa T, Miki K. Acta Crystallographica Section D-Biological Crystallography. 2002;58:1085. doi: 10.1107/s0907444902006261. [DOI] [PubMed] [Google Scholar]
- 20.Fukuyama K, Okada T, Kakuta Y, Takahashi Y. J. Mol. Biol. 2002;315:1155. doi: 10.1006/jmbi.2001.5292. [DOI] [PubMed] [Google Scholar]
- 21.Beinert H, Kennedy MC, Stout CD. Chem. Rev. 1996;96:2335. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- 22.Mouesca JM, Lamotte B. Coord. Chem. Rev. 1998;180:1573. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The hydrogen bonding of terminal and bridging ligands of [Fe4S4(SEt)4]2−, [Fe4S4(SEt)4]2−-HB(t) and [Fe4S4(SEt)4]2−-HB(b), (Figure S1), the B3LYP/6–31G** optimized geometry parameters of spin-delocalized, spinlocalized, Fd-like and HiPIP-like [Fe4S4(SEt)4]2− structures (Table S1). These materials are available free of charge via the Internet at http://pubs.acs.org.