Abstract
The glycosylation state of envelope glycoproteins in Human and Simian Immunodeficiency Viruses (HIV/SIV) is critical to viral infectivity and tropism, viral protein processing, and in virus evasion of the immune system. Using a rapid fluorescent two-dimensional gel based method coupled with enzymatic pre-treatment of virus with PNGase F (Peptide: N-Glycosidase F) and fluorescent 2D gels or 2D gel Western blotting, we show significant differences in the glycosylation patterns of two SIV strains widely used in animal models of HIV disease and vaccine studies. We also demonstrate the modification of a host protein important in HIV biology (HLA-DR) by O-GlcNAc. Further, this experimental pipeline allows for the identification of the modified protein and the site of N-linked glycosylation by fluorescent two-dimensional gel electrophoresis coupled with mass spectrometry (MS) and the qualitative and semi-quantitative assessment of viral glycosylation. The method is fully compatible with downstream glycomics analysis. This approach will permit correlation of virus glycosylation status with pathological severity and may serve as a rapid screen of viruses from physiological samples for further study by more advanced MS methodology.
Keywords: HIV-1, SIV, Glycosylation, O-GlcNAc, N-Linked Glycans, Mass Spectrometry, Proteomics, Two-Dimensional Gels, DIGE, Fluorescence
Introduction
Glycosylation of HIV and SIV proteins plays central roles during multiple stages of the infectivity cycle. During infection, viral glycoproteins influence binding of viral surface proteins gp120 and gp41 to host cell CD4 and co-receptor [1] and can enhance the interactions of HIV and SIV with different cell types, including dendritic cells [2]. Following infection, glycosylation is required for cleavage of the envelope precursor protein (gp160) into gp120 (ENV) and gp41 (transmembrane protein) [3]. Upon release of the virus, glycosylation remains vital to immune evasion, since changes in envelope glycosylation allow the virus to evade immune responses [4, 5]. Glycosylation of viral proteins has thus been an important consideration in vaccine design [6, 7]. Despite the recognition of the importance of glycosylation by the field, to date, methods to assess glycosylation rapidly and globally remain limited. Existing approaches have relied on the exogenous expression of large quantities of viral proteins in non-physiological expression systems and require extensive efforts by mass spectrometry to thoroughly identify which oligosaccharide modifications are found at which residue(s) [8-10]. A rapid, sensitive method to assess glycosylation globally would assist in the characterization of strains exhibiting differential pathogenicity and, ultimately, in the development of effective vaccines.
The experimental pipeline presented here addresses current methodological shortcomings and is intended as a complement to existing mass spectrometry approaches as a rapid, initial qualitative and semi-quantitative screen of HIV/SIV glycosylation. The approach combines a differential in-gel electrophoresis (DIGE) technique [11] with one-dimensional and two-dimensional gel electrophoresis (1DE, 2DE), coupled with enzymatic removal of N-linked glycans [12] for direct analysis or with immunological detection of specific targets. In this manner, the degree of modifications of viral proteins with N-linked glycans can be determined by looking for spots that disappear following enzymatic treatment or by direct detection of specific oligosaccharides by Western blotting (Supplemental Figure 1 (S1)). This pipeline is also compatible with direct carbohydrate extraction from gels for glycomics studies[13] (Figure S1 shaded area). Although success was limited by protein abundance, we were able to map five glycosylation sites out of the 20 possible sites in HIV ENV. Of significance, we found dramatic differences in gp120 glycosylation patterns between two SIV strains widely used in animal models of HIV disease and vaccine studies and show for the first time by fluorescent Western blotting the O-GlcNAc modification of virus-associated HLA-DR. These findings have the potential to enhance understanding of viral pathogenicity to allow for better structural modeling of viral proteins, and to aid in understanding vaccine trial results and designing better vaccines.
METHODS
Virus production
Viruses were purified as previously reported [14]. Briefly, viruses were isolated from clarified cell culture supernatants by ultracentrifugation. The first round of ultracentrifugation was through 25 to 50% sucrose gradients (Beckman CFTU rotor, Beckman, Carlsbad, CA). Virus was identified by UV absorption at 254 and 280 nm. Peak UV fractions were pooled, diluted to below 20% sucrose with TNE buffer (0.01 M Tris-HCl (pH 7.2), 0.1 M NaCl, and 1 mM EDTA in double deionized water), ultracentrifuged to a pellet (Beckman SW-28 Rotor, 100,000 X G for 1hour), and resuspended in TNE buffer. Samples were stored at −80°C.
PNGase F treatment
PNGase F was obtained from New England Biolabs (Ipswich, MA) and used according to the manufacturer’s instructions. Briefly, 1/10 volume of 10X lysis buffer (5% SDS, 10% beta-Mercaptoethanol) was added to virus, and the solution was boiled for 5 min. NP40 was added to a final concentration of 1%, and the solution was adjusted to 50mM Sodium Phosphate, pH 7.5, for the reaction to proceed. The sample was split into half and PNGase F (2U per 10ug of protein) was added to one of the solutions. Both solutions were incubated at 37 degrees for 1 hour (PNGase F treated and control). Samples were then precipitated with trichloroacetic acid (TCA)/acetone. TFA was added to a final concentration of 15% and incubated for 16 hrs at 4°C. The sample was spun for 15 minutes at 15 000 X g in a microfuge (Beckman) and washed with 15% ice cold TCA for 1 minute. It was repelleted as above, then washed twice with−20°C acetone. Samples were dried using a SpeedVAC (Thermo). Samples were resuspended in buffer for fluorescent labeling as described below.
2DE and fluorescent labeling
2DE analysis of HIV and SIV was performed as previously described [15], except fluorescent labeling of samples was done (according to manufacturer’s instructions, GE Healthcare, Piscataway NJ). Briefly, protein samples (at 2mg/ml to ensure consistency) were prepared in 8M urea, 2M thiourea, 4% ASB-14, 50mM tris pH8.0, and labeled with the indicated Cy Dye (GE Healthcare, Piscataway NJ) at a dye ration of 400pmol:50ug of protein. The reaction was kept on ice for 30 minutes in the dark. Samples were quenched with 2.5× v/v of dye with 1M free lysine for 10 min at room temperature in the dark after vortexing and brief centrifugation (1s pulse). Samples were isoelectrically focused in sample buffer containing 8M urea, 2M thiourea, 4% CHAPS, 1% w/v DTT, 1.5% v/v HED and 0.5% ampholytes at the indicated pH range of the strip (see figure legends). Voltage steps were: overnight rehydration at 50V; 150V, 1hour; 300V, 1 hour; 600V 1hour; linear ramp to 1000V in 1 hour; 1000V for 1 hour; linear ramp to 10000V in 1 hour; 10000V for 35 000 KVh (18 cm strips) or at 8 000V for 8 000KVh (7cm strips). Strips were reduced and alkylated as previously described[15] during equilibration in a 4% SDS solution and transferred to the second dimension using in-house bis-tris gels or precast 4-12% gels (Novex, Invitrogen). Gels were either silver stained and scanned as previously described [15] or the fluorescent gels were imaged using a Typhoon 9400 (GE Healthcare) at 50 microns for 10cm gels, and 100 microns for 20 cm gels. PMT voltages were adjusted to ensure that intensities were within the linear range of the instrument and to correct for any minor errors in protein loading as per the manufacturer’s instructions (intensity of capsid protein was compared).
2D Gel Analysis
Gel images were processed by Progenesis ™ Workstation 2005 (V2005, Nonlinear Dynamics, Newcastle, UK) automatic analysis. Following the automated analysis, the images were manually evaluated for boundary and match correction. The volumes of the identified spots were modified using Progenesis ™ backgound subtraction and normalized by total spot volume. Proteins of interest were identified by tandem MS. Regulated proteins with identical identifications and spot patterns indicative of PTMs (horizontal trail) had their cumulative detected values for volume summed and compared between sample conditions.
Tandem MS
Tandem MS was performed as previously described [16]. Briefly, samples were excised from 2D gels and digested as previously described with sequencing grade modified trypsin, as per the manufacturer’s instructions (Promega, Madison, WI) at an enzyme-to-substrate ratio of 1:50 and at 37 °C overnight (>12 h). 10% trifluoroacetic acid was added to stop the digestion. ESI-MS/MS of tryptic peptides was performed either on an LTQ-ion trap-MS/MS instrument (Thermo Finnigan, San Jose, CA) with an Eskigent nano-liquid chromatography system (Eksigent, Dublin, CA) at The Technical Implementation and Coordination Core, Johns Hopkins University, Baltimore, MD, or on an Orbitrab-LTQ ion trap-MS/MS instrument (Thermo Finnigan, San Jose, CA) with an Agilent nano-liquid chromatography system in the authors’ laboratory (Agilent. Santa Clara, CA). Peptides were separated with a 1% / minute AB linear gradient over 60 minutes as previously described [16].
Database Searching
For the experiments performed on the LTQ instrument, tandem mass spectra were extracted and deisotoped by Mascot Distiller version 2.0. (Matrix Science, www.matrixscience.com, London, UK). Charge state deconvolution was not performed. Mascot (Matrix Science; version 2.0.01) was used to first identify peptides against the NCBInr release 20060312 for human-specific searches or against TrEMBL release 20060207 for virus-specific searches using trypsin as the enzyme specified for digestion, allowing for up to one missed cleavage. For MASCOT, parent mass tolerance was set to 1.5, and MS/MS tolerance to 0.8. Instrument settings were set to ESI-TRAP. All MS/MS samples were then analyzed using X! Tandem (www.thegpm.org; version 2006.04.01.2). X! Tandem was set up to search subset databases of all of the original hits identified by MASCOT, also assuming trypsin digestion and allowing for up to 1 missed cleavage. Deamidation of asparagine (deglycosylation N->D) and oxidation of methionine were specified in X! Tandem as variable modifications. Oxidation of methionine and iodoacetamide derivatives of cysteine were specified in Mascot as variable modifications. For the experiments using the Orbitrap-MS, tandem mass spectra were extracted, charge state deconvoluted and deisotoped by BioWorks version 3.3. All MS/MS samples were analyzed using Sequest (ThermoFinnigan, San Jose, CA; version v.27, rev. 11) and X! Tandem (www.thegpm.org ; version 2007.01.01.2). X! Tandem was set up to search a subset of the siv_and_hiv_uniprot_trembl database assuming the digestion enzyme trypsin. Sequest was set up to search the siv_and_hiv_uniprot_trembl database (extracted from TREMBLE / SWISSPROT 20060207, 152212 entries) assuming the digestion enzyme trypsin. Sequest was searched with a fragment ion mass tolerance of 0 Da and a parent ion tolerance of 1.8 Da. X! Tandem was searched with a fragment ion mass tolerance of 0.50 Da and a parent ion tolerance of 1.8 Da. Deamidation of asparagine (deglycosylation N->D) and oxidation of methionine were specified in Sequest as variable modifications. Deamidation of asparagine (deglycosylation N->D) and oxidation of methionine were specified in X! Tandem as variable modifications.
Criteria for protein identification
Scaffold (version Scaffold-01_06_19, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm [17]. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm [18]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony (Table 1, Supplemental tables 1, 2 and 4). The theoretical pI and mass values were recorded from the National Center for Biotechnology Information (NCBI) entry. To validate potential N to D conversions (from PNGase treatment), sequences with potential N to D conversion spectra were manually validated to ensure that there was at least one b or y ion (major fragmentation ions) on either side of the modified amino acid in the fragmentation spectrum, given the higher mass resolution for the fragmentation spectra (in the LTQ). With data generated from the Orbitrap showing a deamidation of the parent ion, we allowed for substation of minor ions in manual validation. Data were processed in an unbiased fashion. Once a potential modification was identified, we examined the sequence manually to ensure that a consensus N-linked glycosylation sequon was utilized (Asn-X-Ser/Thr, where X is not Pro), and assigned a hit only if the sequence existed. We cannot exclude the possibility that there are non-canonical sequences that may have N-linked modifications. Identifications based on one peptide have not been reported, and any peptides associated with Keratin have not been reported (supplemental tables 1, 2 and 4).
Table 1. gp120 residues modified by N-Linked glycosylation.
The mass spectrometry results (Supplemental Table 4) were searched for putative modifications and the corresponding spectra were manually validated (see methods). The spectra corresponding to the sequence (column 2) and spectrum identifier (column 6) are presented in supplemental figures 8-11.
| Virus Strain (PNGaseF Treated) |
Peptide sequence |
Sequence position in gp120 (V- variable; C- constant; I- intervening Region) |
Sequence Alignment to gp120 (HIV-HXB2) (http://www.hiv.lanl.gov/content/hiv-db/EPILIGN/EPI.html) |
Protein accession numbers |
Spectrum Identifier (Supp. Figures 8-11) |
|---|---|---|---|---|---|
| HIV-1/H9 | LRKqFGEnR | 356 (I2) | 349-357 | Q8AGB3_9HIV1 | 0234.0234.3 |
| HIV-1/H9 | AEGnGTITLPCR | 411 (V3 region) | 408-419 | Q6BC31_9HIV1 | 0772.0772.2 |
| SIV/MNE | HFHnRTIVF | 355 (I2) | 352-361 | Q5UB55_9HIV1 | 0710.0710.2 |
| SIV/MNE | LKEHFPnKTIIK | 355 (I2) | 349-361 | Q6TCN3_9HIV1 | 1135.1135.3 |
OGlcNAc blotting
OGlcNAc Western blots were done as previously described [19], with modifications for fluorescent western blotting (below). Briefly, HIV was fluorescently labeled and run on 7cm 3-11 IPG strips and then on 4-12% Bis-Tris gels. The gels were then transferred to low background PVDF (Millipore), blocked overnight with 3% milk in Tris-buffered saline/0.1% Tween 20 at 4°C, and probed as described [19]. We modified the protocol by probing the anti-OGlcNAc IgM with a Cy5-labeled secondary antibody (false coloured blue during scan; Jackson Immunoresearch, West Grove, PA) and scanned the images using a BioRad Molecular Imager Fx Pro Fluorescent Imager. PNGase-treated samples at higher loads (250 ug each) were run as described above on large-format (25 cm) 2D gels. Proteins spots were mapped to these gels, excised, digested and identified by mass spectrometry as described above.
Results
Differential display of laboratory-adapted SIV and HIV strains show remarkable differences in their protein compositions and glycosylation status
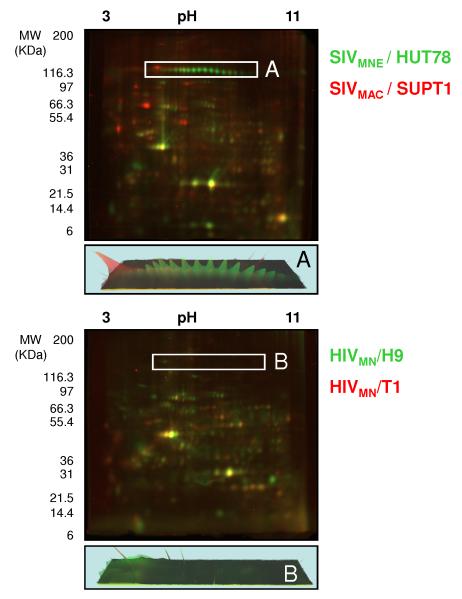
To demonstrate both qualitative and semi-quantitative differences between various laboratory-adapted strains of SIV and HIV (see methods), viruses normalized based on the quantity of the structural protein capsid (CA, p24 for HIV and p27 for SIV were used as an internal standard to normalize by particle number by ELISA, not shown) prior to analysis by two-dimensional electrophoresis (2DE) (Figure 1), were assessed by dual-in-gel-electrophoresis (DIGE) technique, where two viruses are simultaneously analyzed in the same gel using different fluorescent labels (Figure 1, Top panel SIV, Bottom, HIV). The two SIV strains were selected as they are widely used in animal models of HIV disease and vaccine studies[20-27]. SIVMAC 239 (MAC) was originally isolated from Rhesus Macaques (Macaque mulatta)[20] and SIVMNE (MNE) was isolated from Southern Pig-tailed Macaques (Macaca nemestrina)[28] and SIVMNE ClE11s was a subclone of SIVMNE derived from HUT78 cells[25]. These non-human primate animal models represent the fundamental backbone of HIV pathogenesis and vaccine research. Similarly, the HIV strains were selected as these laboratory-adapted strains are used in invitro studies showing significant differences in the induction of anergy and apoptosis in patient-derived PBMCs, putatively by a mechanism of IFN-α secretion and induction of TRAIL (T-Cell Related Apoptosis Inducing Ligand) expression in cells [29, 30]. The SIV and HIV viruses have been propagated in different T-cell lines (H9, HUT78 or SUPT1) or a T-cell B-cell hybrid line (CEMX174) which is indicated in the nomenclature for the HIV and SIV strains (HIVMN / T1; HIVMN/H9 and SIVMNE / HUT78 andSIVMAC / SUPT1 ). As shown in Figure 1, the protein composition of the virus preparations with regard to one another varied dramatically. It is difficult to make direct conclusions of the glycosylation status of host-derived proteins directly packaged into viruses, since the presence of microvesicles in these virus preparations [23] means that they may or may not be virion-associated. However, since these preparations are used in animal challenge experiments (SIV) or in vitro pathogenesis studies (HIV) in the direct form shown here, including microvesicle contamination, qualitative assessment of all protein differences may be extremely useful in determining differences in responses attributable to the host proteins regardless of whether they are virus associated or microvesicle associated. Differences in viral proteins can be assessed without ambiguity, since viral proteins are excluded from microvesicles [31]. This allows the determination of the level of post-translational modifications of viral proteins, since virus input is normalized by particle number (CA, above; see boxes in Figure 1 for selective differences in the viral surface glycoprotein, gp120 between each virus).
Figure 1. Comparison of SIV and HIV strains, illustrating the differences in viral and host protein composition.
Two-dimensional gel electrophoresis with fluorescent labeling of 50μg of capsid protein equivalents for each virus was used to compare SIVMNE /HUT78, Cy3-labeled (green), and SIVMAC / SUPT1, Cy5-labeled (red) (top panel) and HIVMN / H9, Cy3-labeled (green), and HIVMN / T1, Cy5-labeled (red) (bottom panel). The region containing gp120 in SIV and HIV is indicated as box 1 and 2 respectively, and the representative three-dimensional protein abundance plot is presented for each region below. Gels were visualized by fluorescent scanning.
Differences were expected between the SIV strains as they have different amino acid sequences for the ENV protein, and SIVMNE has a truncated transmembrane protein which can result in different glycosylation levels [32]. However, the differences in ENV glycosylation that were observed were surprising, both in the extent of glycosylation but also in the degree of isoelectric point shift in the glycosylation patterns for SIVMNE, suggesting the differential incorporation of a negatively-charged oligosaccharide [33, 34]. The level of gp120 incorporation into the ENV in the HIV strains (molecular clones of each other but isolated from different cell types) was near the limit of detection, so no obvious differences were observed. The following three sections of the experimental pipeline (Supplemental Figure 1) are used to identify those proteins with N- and then O-linked glycosylation; to determine the degree of glycosylation in a semi-quantitative manner; and to identify the modified residue(s) by MS.
Carbohydrate staining of glycosylated residues in HIV and SIV laboratory-adapted strains, and 2DE confirms that the different isoelectric point isoforms of gp120 are glycosylated
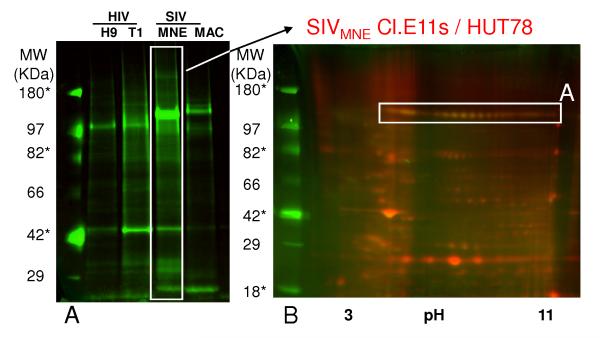
To demonstrate the extent of viral envelope protein glycosylation, a general carbohydrate stain (ProQ Emerald, GE Healthcare) was used on 1 dimensional SDS-PAGE gels of HIV and SIV, where viral input was normalized to the capsid protein. Figure 2 confirms glycosylation of the ENV protein (compare signal from the sample to signal-to-noise of glycosylated markers indicated with an * to unglycosylated markers), and suggests that significant differences can be attributed to the amount of envelope and the extent of glycosylation. SIVMNE was the most highly glycosylated of these viruses. In fact, different isoforms of ENV were glycosylated when observed using a combined staining technique with fluorescently-labeled virus in a 2D gel (Figure 2B). We were unable to detect glycosylation in the other HIV and SIV viruses with confidence using this method due to limitations in the sensitivity of the assay, leading us to the next step in the experimental pipeline, wherein we use enzymatic cleavage of N-linked glycans.
Figure 2. Pro-Q emerald staining (general carbohydrate stain) demonstrating the degree of glycosylation in HIV and SIV by 1 and 2-dimensional gel electrophoresis.
Virus was normalized to capsid protein, and subjected to one-dimensional electrophoresis (left panel, approximately 10 μg of total protein each per lane), followed by staining with Pro-Q emerald to show differences in glycosylation. Notation is truncated. In the right panel, 15 μg of SIVMNE was labeled with Cy5, mixed with 35μg of unlabelled sample and subjected to two-dimensional gel electrophoresis followed by staining with Pro-Q emerald. Gels were visualized by fluorescent scanning. Region A corresponds to region A in Figure 1. * - represents molecular weight markers that are glycosylated.
PNGase F treatment in combination with Cy dye labeling reveals the complexity associated with glycosylation, and concentrates gp120 to detectible levels in HIV by 2DE
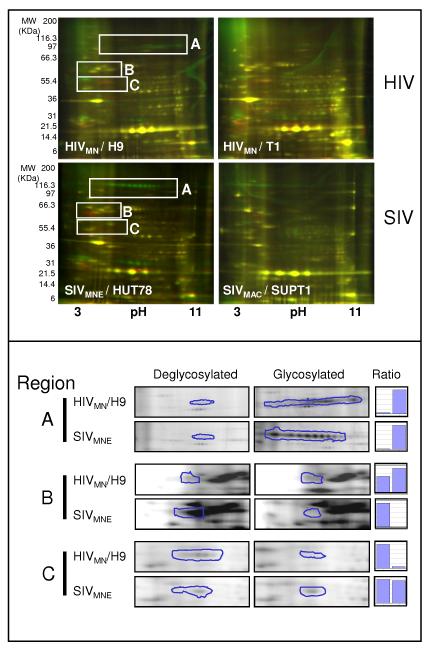
To determine the extent of N-linked glycosylation of ENV, virion preparations were treated with PNGase F, an enzyme that specifically removes the N-linked sugar from the modified asparagine residue (Supplemental Figure 2, mechanism of enzyme action). Samples were labeled Cy3 (control, green) or Cy5 (PNGase F-treated, red), prior to 2DE and were normalized based on the quantity of the structural protein capsid (CA) prior to analysis by two-dimensional electrophoresis (2DE). As illustrated in Supplemental Figure 2, during the reaction with PNGase F, the asparagine residue is converted to an aspartic acid residue, resulting in an acidic shift and molecular weight reduction of the treated sample. For example, a collapsing of the glycosylated charge trains (Cy3, green) occurs with appearance of red spots (cy5) that are of a lower molecular weight and are shifted to the acidic side of the IPG strip (Figure 3, top panel). Comparison of the two different HIV and SIV strains illustrates the differences in both the degree of glycosylation and the degree of incorporation of some glycosylated host proteins (Figure 3, top panel). Since relative quantitation of gp120 is often an issue due to low incorporation of the proteins into the virion, deglycosylation has the additional benefit of concentrating these proteins at a single isoelectric point and molecular mass, allowing for potentially more accurate quantitation than obtained by other techniques like HPLC, where changes in glycosylation can cause spreading of retention times [35]. The enzymatic removal is efficient and complete as illustrated in Figure 3. Region A demonstrates the near-complete disappearance of signal following PNGase F treatment of either SIV or HIV. In addition, this method allows the separation and quantification of gp120 from SIV and HIV: the virus-specific proteins migrate at approximately the same apparent molecular mass and isoelectric point prior to N-linked deglycosylation, but not after deglycosylation (Figure 3, Region B shows the appearance of the deglycosylated gp120 in SIV, but not HIV compare circled regions and bar graphs comparing ratios— whereas region C shows the appearance of gp120 in HIV, but not in SIV).
Figure 3. PNGase F treatment of HIV and SIV and assessment by fluorescent 2-dimensional gel electrophoresis.
Equal amounts of either HIV or SIV (strain indicated in white inlaid) were first treated with Peptide N-Glycosidase F (PNGase F) or with control buffers, labeled with either Cy3-control (green) or Cy5-PNGase F (red), and then subjected to two-dimensional electrophoresis (15 μg of each, for a total of 30μg per gel). Deglycosylation by PNGase F results in a decrease in molecular mass, and an acidic shift if the N-linked glycan is neutral (conversion of asparagines to aspartic acid). Gels were visualized by fluorescent scanning. In the bottom panel, regions identified as gp120 were analyzed by PROGENESIS software (see methods) and relative ratios of signal between glycosylated and deglycosylated gels were compared.
To test the applicability of this method for relative quantitation of gp120, we assessed the normalized spot volume for gp120 in the PNGase-treated gels for HIVMN / H9 and SIV MNE / HUT78. Although the levels of gp120 for SIVMAC were below the limits of detection by this assay, which was not surprising given previous findings showing low levels of gp120 incorporation in this strain [36], it was possible to compare the amount of signal contributed by gp120 in the two HIV strains and SIVMNE. These results show that, by assessing only the amount of gp120 by fluorescent methods and examining the fully glycosylated protein, SIVMNE contained approximately 4.3 fold greater amounts of gp120 than the HIV strains (Supplemental Table 3: “gp120 quantitation”). In fact, the amount of gp120 signal detected in de-glycosylated samples gels, SIVMNE was shown to contain approximately 1.4 fold more gp120 than the HIV strains (Supplemental Table 3 “gp120 quantitation”). This is in agreement with recent findings by Zhu and colleagues, who calculated an approximate 5.7 fold increase in the number of SIV trimers in the mutant virus used in this experiment as compared to HIV [32]. It has been suggested that a truncated transmembrane protein in SIVMNE is responsible for the increased incorporation of gp120 [36]. The quantitation results, based upon the signal for deglycosylated virus, indicate that findings based upon glycosylation may somewhat overestimate the amount of gp120 in highly glycosylated viruses, and that the levels of gp120 in HIV are potentially slightly higher in comparison with the mutant strain of SIV than previously reported [32, 37]. These results are only an estimate of abundance, as multiple replicates containing an internal standard were not used to obtain statistics or variability associated with this measure.
Identification of proteins with putative N-Linked glycosylation modifications and the sites of N-linked glycosylation in gp120 by tandem mass spectrometry
Thoroughly mapping all gp120 glycosylation sites was beyond the scope of our aims and would require extensive high resolution mass spectrometry and highly purified recombinant proteins as previously reported [9]. Rather, we wished to demonstrate the rapid applicability of a 2D gel-based approach for profiling viruses derived from infected cells. Therefore, we identified the spots corresponding to a deglycosylated protein by lower resolution MS methods (LTQ-MS) and selectively performed high resolution MS (LTQ-Orbitrap) on one HIV and SIV strain to demonstrate the utility of the method for determining site-specific usage of N-Linked glycosylation in gp120 as previously reported [9]. We identified spots demonstrating above a 10 fold change in abundance from Progenesis analysis by mass spectrometry using large-format gels (18 cm gels - See Supplemental Figures 3-6; Supplemental table 1 “viral peptides -LTQ” and supplemental table 2 “host peptides –LTQ.” The spot numbers corresponding to the identified proteins are indicated in the gel images and protein tables). Of the host proteins identified, several have been previously shown to be incorporated into virions [37, 38]; however, caution should be used in the interpretation of these data, as many of these proteins could be contaminants from microvesicles or highly abundant serum proteins that have adsorbed to the virus. Further, since there is no way to ensure normalization of host proteins (there may be varying amounts of contaminants), we have not presented any quantitative data on host proteins but only for viral proteins where viral particles have been carefully normalized for capsid protein.
The site-specific usage of N-linked glycoslyation in gp120 was determined by using 1D gels of PNGase F-treated and control HIV samples (Supplemental Figure 7). MS/MS data (ThermoFinnigan LTQ or LTQ-Orbitrap with nano-electrospray source) was analyzed using Mascot or Sequest software followed with a second round of searching using the X-TANDEM algorithm (see methods) to identify the deamination of ASN to ASP shift upon deglycosylation by PNGase F (see methods). Table 1 summarizes the modified N-linked residues of gp120 determined from the MS/MS spectra analysis (see Supplemental figures 8-11 for spectra). We identified two sites in HIV and 1 site in SIV for gp120 glycosylation that showed a potential N-linked glycosylation sequon (Asn-X-Ser/Thr, where X is any amino acid except proline), which we deemed necessary to call a “hit” (see methods). The peptides were then aligned to a reference HIV sequence by using the online tool EpiAlign (available at http://www.hiv.lanl.gov/content/hivdb/EPILIGN/EPI.html). To determine the structural regions in gp120 where these sites occurred, the residues were manually matched to the regions reported by Modrow and colleagues [39]. As shown in Table 1, modified residues were found in variable region (V) V3 (residue 411) in HIV. This critical antigenic region of gp120 often masks neutralization epitopes [40]. Two of these sites were previously observed in HIV-1 produced from CHO cells, and agree with our findings in that they were also found to be PNGase F sensitive [41]. Sites at less critical regions were also found, including intervening (I) region 2 (residues 355 and 356).
Presence of O-GlcNAc of viral-associated proteins
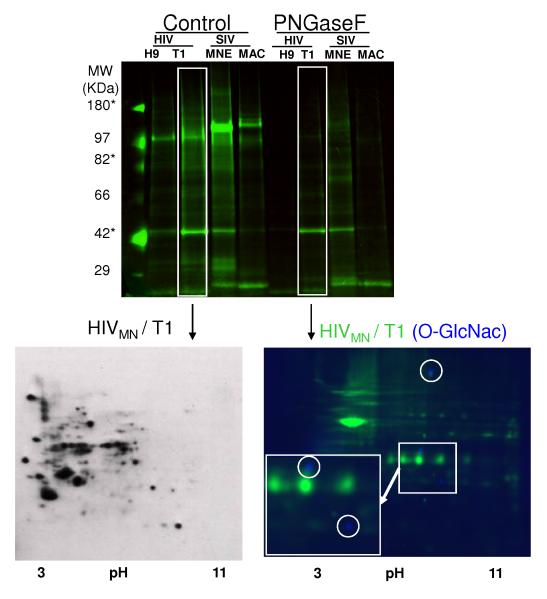
O-linked glycosylation could be contributing to the residual signal of the generalcarbohydrate staining in the samples following PNGase F treatment (Figure 2). Therefore, fluorescent Western blotting was carried out using an antibody specific to the O-GlcNAc moiety (see methods) on control virus (Figure 4, bottom left panel; PNGase F-treated sample was labeled with Cy3 (green); O-GlcNAc antibody (blue)). Of note, the antibody binding is out-competed with 100mM free GlcNAc (see Supplemental Figure 12), confirming the specificity of this antibody. Three distinct protein spots were shown to be specifically O-GlcNAc modified (Figure 4), which, although much less sensitive than enzymatic detection reactions (compare bottom right panel to bottom left panel), allowed easy matching of protein spots for identification by mass spectrometry. The spots containing O-GlcNAc corresponded to a mixture of peptides for moesin, erythrocyte membrane protein band 4.1-like 2, both ERM (eosin-radexin-moesin) family members (spot 45, Supplemental Figure 6, Supplemental Table 2); HLA-DR and Pan-leukocyte antigen (CD53) (spot 32, Supplemental Figure 5, Supplemental Table 2); polyubiquitin C and polymerase (Spots 20, Supplemental Figure 6, Supplemental Table 2). All identifications matched the observed molecular mass. Since O-GlcNAc is a labile modification by the tandem MS methods used in this study, we were unable to map the site of usage to verify which peptide and position was used.
Figure 4. PNGase F removal of N-Linked Glycans and subsequent visualization of O-GlcNAc modified proteins in HIVMN/T1 by standard and fluorescent Western Blotting.
The results from figure 2 are shown with the addition of results for PNGase F treatment of the same samples (PNGase F). Control virus (HIVMN/T1, 50 μg) was subjected to two-dimensional electrophoresis and transferred to PVDF. The blot was then probed with antibodies specific to O-GlcNAc and developed with an anti-mouse IgM-HRP conjugate (bottom left panel). Alternatively, following removal of N-linked glycans, HIVMN/T1 was Cy3-labeled (Green, 50 μg) and subjected to two-dimensional electrophoresis and then transferred to low-background PVDF. The blot was then probed with antibodies specific to OGlcNAc and subsequently with Cy2-labeled (blue) anti-mouse IgM. The blot was visualized by fluorescent scanning (bottom right panel).
The observations that there are significant differences at the level of glycosylation (and host protein incorporation) between different viruses (Figures 1 and 3) supports the proposal that this platform should be used to observe virus isolated from individual patient cell types and at different time points. This will provide increased understanding at the clinical level of the extent or level of glycosylation. An obvious limitation to this approach is virus production. From a patient standpoint, cells would have to be isolated, any drug therapies washed out, and virus cultured and expanded for approximately two weeks to obtain sufficient virus to do these assays; however, these methods are already well established [42]. By increasing the amount of fluorescent label, it is possible to perform an entire pipeline using less than 5 ug of total protein, which is approximately equivalent to 500 nanograms of p24 equivalents or less (See Supplemental Figure 13 demonstrating resolution of gp120). Although currently this amount of protein pushes the limits for identifications by mass spectrometry, this limitation can be addressed by preparing detailed 2D maps of viruses and/or Western blotting of the patient-derived virus following our 2D analysis to confirm protein identification by antibodies with high specificity to viral proteins.
Discussion
Glycosylation is a fundamentally important process in HIV and SIV biology. It is one of the major mediators of virus binding [1], is required for proper processing of the viral envelope proteins [3] and is key in immunologic escape [4, 5, 43, 44]’. From a vaccination standpoint, differences in glycosylation can change how antigen presenting cells interact with the vaccine [45] , and obviously change the nature of the immune responses.
The SIV strains chosen for this study have been widely utilized in the most relevant animal models for HIV pathogenesis and vaccine research [20, 22-28]. Therefore an understanding of the fundamental differences between these viruses may help to explain some of the differences in disease progression observed between the non-human primate models of HIV disease[46-51]. Regarding proteins differentially expressed in these preparations, although we cannot attribute the presence of absence of these proteins to virions or microvesicles, it is important to note that these proteins themselves can elicit immune responses. For example it has been shown that animals immunized with HLA-DR can confer resistance against challenge with SIV[52]. Therefore even the basic information obtained from a 2DE-DIGE experiment (Figure 1) may be revealing.
Although our success in identifying glycosylation site usage is limited in comparison to more recent studies using recombinant sources of ENV protein [9], we have shown glycosylation site usage in regions of the virus used in immune evasion and CD4 binding (V3) in physiologically relevant sample sources and did obtain high sequence coverage for gp120 (Supplemental Tables 1, 2 and 4). We were not surprised at the limited observation of gp120 glycosylation sites from our 1D gel experiments since we were only able to load 25ug per lane or 10 fold less gp120 than on a 2D gel. Direct assessment of spots from a 2D gel using MALDI-TOF/TOF or FT-ICR instrument and direct comparison of fully glycosylated and de-glycosylated spectra and the use of 018 labeled water to minimize the likelihood of false positives would likely increase the identification of putative glycosylation sites as recently reported [53]. The low extraction efficiency associated with gel-based methods is also likely responsible for the low number of modified peptides observed. It is therefore likely that if HPLC methods were used to first purify gp120 from the virus preparations [36] then much higher sequence coverage could be obtained; however, information on post-translational modifications imparted by different oligosaccharides would be lost, as these are best visualized by a 2D gel method. Further this method is extremely rapid as results may be interpreted directly at the 2D gel level.
Although many studies have used site directed mutagenesis to change potential glycosylation sites of HIV and SIV [54] , there is a paucity of findings of actual in vivo glycosylation sites due to the very large amounts of virus required to make this determination. Our experimental pipeline allows the determination of the actual usage of glycosylation sites in complex mixtures of virions. Furthermore, we can also address the issues of relative ENV incorporation and ENV heterogeneity in a sample. For instance, it has been estimated that HIV virions contain 7-10 trimers of gp120 based upon the amount of ENV protein [36]. However, is this ENV homogenous or heterogeneous with regard to glycosylation? The data presented in this study show that the heterogeneity observed with regard to isoelectric point is likely due to compositional changes in the N-linked carbohydrates rather than site utilization given the relative uniformity of molecular masses observed. Further studies will be required to determine the exact oligosaccharide composition for this viruses used in this study.
Finally, the role of O-GlcNAcylation in cell biology has only begun to be recognized [55]. Our results show that HLA-DR is O-GlcNAc modified (Figure 4). Many reports in the literature describe the specific incorporation of HLA-DR into HIV [38], but the specifics of this mechanism are still unknown. Although the role of the invariant chain in HLA-DR shuttling to endosomes has been well described [56], it is unclear what properties O-GlcNAc modifications of the cytoplasmic tail (O-GlcNAc transferase is localized to the nucleus and cytoplasm) of HLA-DR might confer. The immunological implications of our findings may potentially shed light on the specific nature of HLA incorporation into viral particles. These findings again raise the question of the significance of host protein incorporation into virion particles [57]. Using this methodology, one can track the presence and absence of different host proteins at a global level, and determine if there are differentially glycosylated forms of host proteins that are more highly enriched than in the cell, and potentially have different functions than their unmodified counterparts. The pilot results presented here suggest there will be dramatic differences in host protein composition depending on what cell type the virus is isolated from and the status of the cell at that time (Figure 1).
Collectively, these data will hopefully help to overcome some specific challenges facing vaccine design regarding antigen quality control, and help to provide an alternative method of validation for studies examining viral sequences and correlation of viral sequence with pathogenicity [58]. This is important since examination of the nucleic acid sequence information of virions alone cannot consider differences present at the level of glycosylation (or other post translational modifications observable by 2DE) between different infected target cells (Figure 3). Since glycosylation status can change based upon the state of the host cell [59], be induced by inflammation [60], or be unique to different cell types [33, 34, 61], such studies would overlook differences at the level of this important post-translational modification. Due to this limitation, even very well controlled studies that conclude that the ENV glycoprotein is not responsible for changes in pathogenicity may have overlooked the possibility that differences in glycosylation in ENV may be responsible for the effect [29, 30]. By applying our methods retrospectively to viruses that have been extensively studied and used in vaccine studies, like the SIV strains used in this study [21, 62], we may be able to better understand the types of results that were obtained, and potentially understand why certain vaccine trials failed to protect.
Conclusion
From a broad standpoint we present a combined approach for assessing the state of glycosylation globally, and also a method of identifying the specific modified amino acid residues that are utilized in vivo (see Supplemental Figure 1, “Workflow for assessing glycosylation”). From a clinical standpoint, it can be expected that by understanding glycosylation we could potentially make immediate impacts in the clinic. It has already been shown that general inhibitors of glycosylation, or natural compounds that bind to glycosylated proteins, can prevent or attenuate virus pathology [63, 64]. The identification of specific carbohydrate modifications that correlate to pathogenicity will hopefully allow for more specific targeting of pathways or specific small molecular interfering substances that will neutralize the activity of the identified carbohydrate moieties. By combining the techniques presented here with emerging methods in glycobiology [65] we hope to shorten the time from discovery to clinical translation.
Supplementary Material
Supplemental Figure 1: Shematic of experimental pipeline. The basic principles of how to approach potentially glycosylated samples is illustrated. The first step involves using general carbohydrate staining to assess the general state of glycosylation and the use of PNGase F to determine residual signal to other potential glycans (O-linked). The next step involves either quantitation and site specific determination (presented in this manuscript) by using a combination of DIGE and tandem mass spectrometry, or determination of carbohydrate structure by glycobiology methods like Fluorescent Assisted Carbohydrate Electrophoresis (*FACE, not shown) or mass spectrometry.
Supplemental Figure 2: Mechanism of PNGase F removal. The amino acid backbone is represented by the symbol X. During the process of PNGase F removal, the amino acid Asparagine is deamidated and converted into the amino acid Aspartate.
Supplemental Figure 3: Spot-map of HIVMN/H9 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 4: Spot-map of HIVMN/T1 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 5: Spot-map of PNGase F treated HIVMN/H9 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 6: Spot-map of PNGase F treated HIVMN/T1 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 7: 1D gel of PNGase F and Control virus and the corresponding bands cut for MS identification. PNGase F treated (Cy5, Red) or Control virus (Cy3, Green) were prepared and run on a 20cm SDS-page gel (1 μg each, with 50 μg of unlabelled PNGase F treated virus). The gels were subsequently silver-stained (see methods, not shown) and bands corresponding to the molecular mass of deglycosylated gp120 and exhibiting signal from Cy5 were cut and digested by trpysin for MS/MS identification (see methods).
Supplemental Figures 8-11: Spectra corresponding to Table 1. For each site of deamidation identified in Table 1, the corresponding spectrum, fragmentation table and spectrum model error is presented. The top panel for each figure illustrates the annotated spectrum (colored sequence indicate a definite amino acid identification as determined by flanking b or y ions – see fragmentation table). The middle panel represents all of the fragmentation ions observed (fragmentation table), and the bottom panel represents the error associated with each fragmentation ion (+/− 0.5 DA).
Supplemental Figure 12: O-GlcNAc western blotting of HIVMN/T1. 2×50 ug of HIV was subjected to two-dimensional gel electrophoresis and transferred to PVDF. The membranes were probed with an anti-O-GlcNAc antibody in the absence or presence of 100mM GlcNAc to demonstrate specificity of the primary antibody. The primary antibody was visualized with a secondary HRP-coupled antibody to mouse IgM.
Supplemental Figure 13: Visualization of patient-derived HIVNL4-3 from primary monocytes. 200 ng of patient derived virus was labeled at a ratio of 800 pmol of Cy3 (Green) dye to 50 ug of protein or the same volume/volume amount of control material at the same density of OptiPrep and labeled at the same ration with Cy5 (red) and subjected to two-dimensional gel electrophoresis on 24 cm IEF strips. The strips were cut into 3 7cm pieces and ran on 3 7 cm SDS-PAGE gels. The gels were visualized with a fluorescent scanner.
Supplemental Table 1: Viral Peptides-LTQ. Data was exported from Scaffold and organized by the Gel description and spot number of each gel as described in Supplemental Figures 3-6 into this table. Redundant header information was truncated (described in methods). To determine the identity of the proteins that are presented in the publication, use the sample description and spot number as key and compare to Supplemental figures 3-6. Data is presented in a format to comply with the guidelines for publications of proteomic data as outlined in the instructions to authors.
Supplemental Table 2: Host Peptides-LTQ. Data was exported from Scaffold and organized by the Gel description and spot number of each gel as described in Supplemental Figures 3-6 into this table. Redundant header information was truncated (described in methods). To determine the identity of the proteins that are presented in the publication, use the sample description and spot number as key and compare to Supplemental figures 3-6. Data is presented in a format to comply with the guidelines for publications of proteomic data as outlined in the instructions to authors.
Supplemental Table 3: Data used to calculate gp120 levels as presented in the text. Spot abundance information was obtained in Progenesis software as describe in the methods for each spot corresponding to gp120. The data was exported from Progenesis into this table.
Supplemental Table 4: Viral Peptides-Orbitrap. Data was exported from Scaffold and organized by the Gel description and spot number of each gel as described in Supplemental Figures 3-6 into this table. Redundant header information was truncated (described in methods). To determine the identity of the proteins that are presented in the publication, use the sample description and spot number as key and compare to Supplemental figures 3-6. Data is presented in a format to comply with the guidelines for publications of proteomic data as outlined in the instructions to authors.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the Johns Hopkins Technology Implementation and Co-ordination Center for performing the LTQ-MS in this study; Drs. Caroline Gilbert and Michel Tremblay at Laval University, Quebec, Canada, for providing the virus used in Supplemental Figure 20 and Dr. Jeff Lifson and Julian Bess at the AIDS vaccine program, SAIC-NCI-FCRDC, Frederick, Maryland, USA, for the HIV and SIV strains used in this study. This work was supported by the Dean’s Office of John Hopkins University (startup funds to D. G.) and by the National Heart Lung Blood Institute Proteomic Initiative (contract NO-HV-28120).
Abbreviations
- ENV
Envelope
- HIV-1
Human Immunodeficiency Virus Type 1
- PNGase F
Peptide: N-glycosidase F
- SIV
Simian Immunodeficiency Virus
- TRAIL
T-Cell Related Apoptosis Inducing Ligand
REFERENCES
- [1].Chen M, Shi C, Kalia V, Tencza SB, et al. HIV gp120 V(1)/V(2) and C(2)-V(3) domains glycoprotein compatibility is required for viral replication. Virus Res. 2001;79:91–101. doi: 10.1016/s0168-1702(01)00322-7. [DOI] [PubMed] [Google Scholar]
- [2].Geijtenbeek TB, van Kooyk Y. DC-SIGN: a novel HIV receptor on DCs that mediates HIV-1 transmission. Curr Top Microbiol Immunol. 2003;276:31–54. doi: 10.1007/978-3-662-06508-2_2. [DOI] [PubMed] [Google Scholar]
- [3].Land A, Braakman I. Folding of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum. Biochimie. 2001;83:783–790. doi: 10.1016/s0300-9084(01)01314-1. [DOI] [PubMed] [Google Scholar]
- [4].Kwong PD, Doyle ML, Casper DJ, Cicala C, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- [5].Shi Y, Brandin E, Vincic E, Jansson M, et al. Evolution of human immunodeficiency virus type 2 coreceptor usage, autologous neutralization, envelope sequence and glycosylation. J Gen Virol. 2005;86:3385–3396. doi: 10.1099/vir.0.81259-0. [DOI] [PubMed] [Google Scholar]
- [6].Mullins JI, Nickle DC, Heath L, Rodrigo AG, Learn GH. Immunogen sequence: the fourth tier of AIDS vaccine design. Expert Rev Vaccines. 2004;3:S151–159. doi: 10.1586/14760584.3.4.s151. [DOI] [PubMed] [Google Scholar]
- [7].Farrar WL, Harel-Bellan A, Ferris DK. Characterization of CD4 glycoprotein determinant-HIV envelope protein interactions: perspectives for analog and vaccine development. Crit Rev Immunol. 1988;8:315–339. [PubMed] [Google Scholar]
- [8].Gram GJ, Bolmstedt A, Schonning K, Biller M, et al. Detection of orientation-specific anti-gp120 antibodies by a new N-glycanase protection assay. Apmis. 2002;110:123–131. doi: 10.1034/j.1600-0463.2002.100202.x. [DOI] [PubMed] [Google Scholar]
- [9].Go EP, Irungu J, Zhang Y, Dalpathado DS, et al. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes’ accessibility. J Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Renouf DV, Hounsell EF. Molecular modelling of glycoproteins by homology with non-glycosylated protein domains, computer simulated glycosylation and molecular dynamics. Adv Exp Med Biol. 1995;376:37–45. doi: 10.1007/978-1-4615-1885-3_4. [DOI] [PubMed] [Google Scholar]
- [11].Unlu M, Morgan ME, Minden JS. Difference gel electrophoresis: a single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- [12].Packer NH, Lawson MA, Jardine DR, Sanchez JC, Gooley AA. Analyzing glycoproteins separated by two-dimensional gel electrophoresis. Electrophoresis. 1998;19:981–988. doi: 10.1002/elps.1150190613. [DOI] [PubMed] [Google Scholar]
- [13].Kuster B, Wheeler SF, Hunter AP, Dwek RA, Harvey DJ. Sequencing of N-linked oligosaccharides directly from protein gels: in-gel deglycosylation followed by matrix-assisted laser desorption/ionization mass spectrometry and normal-phase high-performance liquid chromatography. Anal Biochem. 1997;250:82–101. doi: 10.1006/abio.1997.2199. [DOI] [PubMed] [Google Scholar]
- [14].Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. Cholesterol depletion of human immunodeficiency virus type 1 and simian immunodeficiency virus with beta-cyclodextrin inactivates and permeabilizes the virions: evidence for virion-associated lipid rafts. J Virol. 2003;77:8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Graham DR, Garnham CP, Fu Q, Robbins J, Van Eyk JE. Improvements in two-dimensional gel electrophoresis by utilizing a low cost “in-house” neutral pH sodium dodecyl sulfate-polyacrylamide gel electrophoresis system. Proteomics. 2005;5:2309–2314. doi: 10.1002/pmic.200401249. [DOI] [PubMed] [Google Scholar]
- [16].Sheng S, Chen D, Van Eyk JE. Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol Cell Proteomics. 2006;5:26–34. doi: 10.1074/mcp.T500019-MCP200. [DOI] [PubMed] [Google Scholar]
- [17].Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- [18].Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- [19].Akimoto Y, Kreppel LK, Hirano H, Hart GW. Increased O-GlcNAc transferase in pancreas of rats with streptozotocin-induced diabetes. Diabetologia. 2000;43:1239–1247. doi: 10.1007/s001250051519. [DOI] [PubMed] [Google Scholar]
- [20].Chakrabarti L, Guyader M, Alizon M, Daniel MD, et al. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature. 1987;328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- [21].Lifson JD, Rossio JL, Piatak M, Jr., Bess J, Jr., et al. Evaluation of the safety, immunogenicity, and protective efficacy of whole inactivated simian immunodeficiency virus (SIV) vaccines with conformationally and functionally intact envelope glycoproteins. AIDS Res Hum Retroviruses. 2004;20:772–787. doi: 10.1089/0889222041524661. [DOI] [PubMed] [Google Scholar]
- [22].Kestler HW, 3rd, Li Y, Naidu YM, Butler CV, et al. Comparison of simian immunodeficiency virus isolates. Nature. 1988;331:619–622. doi: 10.1038/331619a0. [DOI] [PubMed] [Google Scholar]
- [23].Naidu YM, Kestler HW, 3rd, Li Y, Butler CV, et al. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J Virol. 1988;62:4691–4696. doi: 10.1128/jvi.62.12.4691-4696.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kuller L, Benveniste RE, Tsai CC, Clark EA, et al. Intrarectal inoculation of macaques by the simian immunodeficiency virus, SIVmne E11S: CD4+ depletion and AIDS. J Med Primatol. 1994;23:397–409. doi: 10.1111/j.1600-0684.1994.tb00127.x. [DOI] [PubMed] [Google Scholar]
- [25].Benveniste RE, Roodman ST, Hill RW, Knott WB, et al. Infectivity of titered doses of simian immunodeficiency virus clone E11S inoculated intravenously into rhesus macaques (Macaca mulatta) J Med Primatol. 1994;23:83–88. doi: 10.1111/j.1600-0684.1994.tb00106.x. [DOI] [PubMed] [Google Scholar]
- [26].Marthas ML, Ramos RA, Lohman BL, Van Rompay KK, et al. Viral determinants of simian immunodeficiency virus (SIV) virulence in rhesus macaques assessed by using attenuated and pathogenic molecular clones of SIVmac. J Virol. 1993;67:6047–6055. doi: 10.1128/jvi.67.10.6047-6055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Tsai CC, Follis KE, Grant RF, Nolte RE, et al. Infectivity and pathogenesis of titered dosages of simian immunodeficiency virus experimentally inoculated into longtailed macaques (Macaca fascicularis) Lab Anim Sci. 1993;43:411–416. [PubMed] [Google Scholar]
- [28].Benveniste RE, Morton WR, Clark EA, Tsai CC, et al. Inoculation of baboons and macaques with simian immunodeficiency virus/Mne, a primate lentivirus closely related to human immunodeficiency virus type 2. J Virol. 1988;62:2091–2101. doi: 10.1128/jvi.62.6.2091-2101.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Esser MT, Bess JW, Jr., Suryanarayana K, Chertova E, et al. Partial activation and induction of apoptosis in CD4(+) and CD8(+) T lymphocytes by conformationally authentic noninfectious human immunodeficiency virus type 1. J Virol. 2001;75:1152–1164. doi: 10.1128/JVI.75.3.1152-1164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Herbeuval JP, Grivel JC, Boasso A, Hardy AW, et al. CD4+ T-cell death induced by infectious and noninfectious HIV-1: role of type 1 interferon-dependent, TRAIL/DR5-mediated apoptosis. Blood. 2005;106:3524–3531. doi: 10.1182/blood-2005-03-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bess JW, Jr., Gorelick RJ, Bosche WJ, Henderson LE, Arthur LO. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- [32].Zhu P, Liu J, Bess J, Jr., Chertova E, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- [33].Mizuochi T, Spellman MW, Larkin M, Solomon J, et al. Structural characterization by chromatographic profiling of the oligosaccharides of human immunodeficiency virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese hamster ovary cells. Biomed Chromatogr. 1988;2:260–270. doi: 10.1002/bmc.1130020608. [DOI] [PubMed] [Google Scholar]
- [34].Mizuochi T, Spellman MW, Larkin M, Solomon J, et al. Carbohydrate structures of the human-immunodeficiency-virus (HIV) recombinant envelope glycoprotein gp120 produced in Chinese-hamster ovary cells. Biochem J. 1988;254:599–603. doi: 10.1042/bj2540599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Che FY, Song JF, Shao XX, Wang KY, Xia QC. Comparative study on the distribution of ovalbumin glycoforms by capillary electrophoresis. J Chromatogr A. 1999;849:599–608. doi: 10.1016/s0021-9673(99)00521-x. [DOI] [PubMed] [Google Scholar]
- [36].Chertova E, Bess JW, Jr, Crise BJ, Sowder IR, et al. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), Is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 2002;76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. Jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chertova E, Chertov O, Coren LV, Roser JD, et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006;80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ott DE. Cellular proteins in HIV virions. Rev Med Virol. 1997;7:167–180. doi: 10.1002/(sici)1099-1654(199709)7:3<167::aid-rmv199>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- [39].Modrow S, Hahn BH, Shaw GM, Gallo RC, et al. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J Virol. 1987;61:570–578. doi: 10.1128/jvi.61.2.570-578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wyatt R, Kwong PD, Desjardins E, Sweet RW, et al. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- [41].Leonard CK, Spellman MW, Riddle L, Harris RJ, et al. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- [42].Shen L, Hober D, Benyoucef S, Ajana F, et al. A study of two procedures of HIV-1 isolation from whole blood cultures. Microbiol Immunol. 1996;40:195–200. doi: 10.1111/j.1348-0421.1996.tb03334.x. [DOI] [PubMed] [Google Scholar]
- [43].Chackerian B, Rudensey LM, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wei X, Decker JM, Wang S, Hui H, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- [45].Pohlmann S, Baribaud F, Lee B, Leslie GJ, et al. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gardner MB, Luciw PA. Macaque models of human infectious disease. Ilar J. 2008;49:220–255. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zink MC, Laast VA, Helke KL, Brice AK, et al. From mice to macaques--animal models of HIV nervous system disease. Curr HIV Res. 2006;4:293–305. doi: 10.2174/157016206777709410. [DOI] [PubMed] [Google Scholar]
- [48].Kimata JT. HIV-1 fitness and disease progression: insights from the SIV-macaque model. Curr HIV Res. 2006;4:65–77. doi: 10.2174/157016206775197628. [DOI] [PubMed] [Google Scholar]
- [49].Joag SV. Primate models of AIDS. Microbes Infect. 2000;2:223–229. doi: 10.1016/s1286-4579(00)00266-5. [DOI] [PubMed] [Google Scholar]
- [50].Hulskotte EG, Geretti AM, Osterhaus AD. Towards an HIV-1 vaccine: lessons from studies in macaque models. Vaccine. 1998;16:904–915. doi: 10.1016/s0264-410x(97)00292-2. [DOI] [PubMed] [Google Scholar]
- [51].Anderson DM, Agy MB, Bowden D, Morton WR, Liggitt HD. HIV infection in non-human primates: the Macaca nemestrina model. Virus Res. 1994;32:269–282. doi: 10.1016/0168-1702(94)90045-0. [DOI] [PubMed] [Google Scholar]
- [52].Arthur LO, Bess JW, Jr., Urban RG, Strominger JL, et al. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. J Virol. 1995;69:3117–3124. doi: 10.1128/jvi.69.5.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Irungu J, Go EP, Zhang Y, Dalpathado DS, et al. Comparison of HPLC/ESI-FTICR MS Versus MALDI-TOF/TOF MS for Glycopeptide Analysis of a Highly Glycosylated HIV Envelope Glycoprotein. J Am Soc Mass Spectrom. 2008 doi: 10.1016/j.jasms.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Johnson WE, Sauvron JM, Desrosiers RC. Conserved, N-linked carbohydrates of human immunodeficiency virus type 1 gp41 are largely dispensable for viral replication. J Virol. 2001;75:11426–11436. doi: 10.1128/JVI.75.23.11426-11436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem. 2006;97:71–83. doi: 10.1002/jcb.20676. [DOI] [PubMed] [Google Scholar]
- [56].Bertolino P, Rabourdin-Combe C. The MHC class II-associated invariant chain: a molecule with multiple roles in MHC class II biosynthesis and antigen presentation to CD4+ T cells. Crit Rev Immunol. 1996;16:359–379. [PubMed] [Google Scholar]
- [57].Ott DE. Potential roles of cellular proteins in HIV-1. Rev Med Virol. 2002;12:359–374. doi: 10.1002/rmv.367. [DOI] [PubMed] [Google Scholar]
- [58].Luciw PA, Shaw KE, Unger RE, Planelles V, et al. Genetic and biological comparisons of pathogenic and nonpathogenic molecular clones of simian immunodeficiency virus (SIVmac) AIDS Res Hum Retroviruses. 1992;8:395–402. doi: 10.1089/aid.1992.8.395. [DOI] [PubMed] [Google Scholar]
- [59].Franz S, Frey B, Sheriff A, Gaipl US, et al. Lectins detect changes of the glycosylation status of plasma membrane constituents during late apoptosis. Cytometry A. 2006;69:230–239. doi: 10.1002/cyto.a.20206. [DOI] [PubMed] [Google Scholar]
- [60].Kleinberg ME, Rotrosen D, Malech HL. Asparagine-linked glycosylation of cytochrome b558 large subunit varies in different human phagocytic cells. J Immunol. 1989;143:4152–4157. [PubMed] [Google Scholar]
- [61].Mizuochi T, Matthews TJ, Kato M, Hamako J, et al. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J Biol Chem. 1990;265:8519–8524. [PubMed] [Google Scholar]
- [62].Evans DT, Bricker JE, Sanford HB, Lang S, et al. Immunization of macaques with single-cycle simian immunodeficiency virus (SIV) stimulates diverse virus-specific immune responses and reduces viral loads after challenge with SIVmac239. J Virol. 2005;79:7707–7720. doi: 10.1128/JVI.79.12.7707-7720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Montefiori DC, Robinson WE, Jr., Mitchell WM. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1988;85:9248–9252. doi: 10.1073/pnas.85.23.9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mori T, Boyd MR. Cyanovirin-N, a potent human immunodeficiency virus-inactivating protein, blocks both CD4-dependent and CD4-independent binding of soluble gp120 (sgp120) to target cells, inhibits sCD4-induced binding of sgp120 to cell-associated CXCR4, and dissociates bound sgp120 from target cells. Antimicrob Agents Chemother. 2001;45:664–672. doi: 10.1128/AAC.45.3.664-672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Morelle W, Michalski JC. Glycomics and mass spectrometry. Curr Pharm Des. 2005;11:2615–2645. doi: 10.2174/1381612054546897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Shematic of experimental pipeline. The basic principles of how to approach potentially glycosylated samples is illustrated. The first step involves using general carbohydrate staining to assess the general state of glycosylation and the use of PNGase F to determine residual signal to other potential glycans (O-linked). The next step involves either quantitation and site specific determination (presented in this manuscript) by using a combination of DIGE and tandem mass spectrometry, or determination of carbohydrate structure by glycobiology methods like Fluorescent Assisted Carbohydrate Electrophoresis (*FACE, not shown) or mass spectrometry.
Supplemental Figure 2: Mechanism of PNGase F removal. The amino acid backbone is represented by the symbol X. During the process of PNGase F removal, the amino acid Asparagine is deamidated and converted into the amino acid Aspartate.
Supplemental Figure 3: Spot-map of HIVMN/H9 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 4: Spot-map of HIVMN/T1 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 5: Spot-map of PNGase F treated HIVMN/H9 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 6: Spot-map of PNGase F treated HIVMN/T1 used for protein identification. 250ug of HIV was subjected to two-dimensional SDS-PAGE electrophoresis and visualized by silver staining. The spots were excised and prepared for mass spectrometry as described in the materials and methods.
Supplemental Figure 7: 1D gel of PNGase F and Control virus and the corresponding bands cut for MS identification. PNGase F treated (Cy5, Red) or Control virus (Cy3, Green) were prepared and run on a 20cm SDS-page gel (1 μg each, with 50 μg of unlabelled PNGase F treated virus). The gels were subsequently silver-stained (see methods, not shown) and bands corresponding to the molecular mass of deglycosylated gp120 and exhibiting signal from Cy5 were cut and digested by trpysin for MS/MS identification (see methods).
Supplemental Figures 8-11: Spectra corresponding to Table 1. For each site of deamidation identified in Table 1, the corresponding spectrum, fragmentation table and spectrum model error is presented. The top panel for each figure illustrates the annotated spectrum (colored sequence indicate a definite amino acid identification as determined by flanking b or y ions – see fragmentation table). The middle panel represents all of the fragmentation ions observed (fragmentation table), and the bottom panel represents the error associated with each fragmentation ion (+/− 0.5 DA).
Supplemental Figure 12: O-GlcNAc western blotting of HIVMN/T1. 2×50 ug of HIV was subjected to two-dimensional gel electrophoresis and transferred to PVDF. The membranes were probed with an anti-O-GlcNAc antibody in the absence or presence of 100mM GlcNAc to demonstrate specificity of the primary antibody. The primary antibody was visualized with a secondary HRP-coupled antibody to mouse IgM.
Supplemental Figure 13: Visualization of patient-derived HIVNL4-3 from primary monocytes. 200 ng of patient derived virus was labeled at a ratio of 800 pmol of Cy3 (Green) dye to 50 ug of protein or the same volume/volume amount of control material at the same density of OptiPrep and labeled at the same ration with Cy5 (red) and subjected to two-dimensional gel electrophoresis on 24 cm IEF strips. The strips were cut into 3 7cm pieces and ran on 3 7 cm SDS-PAGE gels. The gels were visualized with a fluorescent scanner.
Supplemental Table 1: Viral Peptides-LTQ. Data was exported from Scaffold and organized by the Gel description and spot number of each gel as described in Supplemental Figures 3-6 into this table. Redundant header information was truncated (described in methods). To determine the identity of the proteins that are presented in the publication, use the sample description and spot number as key and compare to Supplemental figures 3-6. Data is presented in a format to comply with the guidelines for publications of proteomic data as outlined in the instructions to authors.
Supplemental Table 2: Host Peptides-LTQ. Data was exported from Scaffold and organized by the Gel description and spot number of each gel as described in Supplemental Figures 3-6 into this table. Redundant header information was truncated (described in methods). To determine the identity of the proteins that are presented in the publication, use the sample description and spot number as key and compare to Supplemental figures 3-6. Data is presented in a format to comply with the guidelines for publications of proteomic data as outlined in the instructions to authors.
Supplemental Table 3: Data used to calculate gp120 levels as presented in the text. Spot abundance information was obtained in Progenesis software as describe in the methods for each spot corresponding to gp120. The data was exported from Progenesis into this table.
Supplemental Table 4: Viral Peptides-Orbitrap. Data was exported from Scaffold and organized by the Gel description and spot number of each gel as described in Supplemental Figures 3-6 into this table. Redundant header information was truncated (described in methods). To determine the identity of the proteins that are presented in the publication, use the sample description and spot number as key and compare to Supplemental figures 3-6. Data is presented in a format to comply with the guidelines for publications of proteomic data as outlined in the instructions to authors.