Abstract
Mitochondria play a central role during neurogenesis by providing energy in the form of ATP for cytoskeletal remodelling, outgrowth of neuronal processes, growth cone activity and synaptic activity. However, the fundamental question of how differentiating neurons control mitochondrial biogenesis remains vastly unexplored. Since our previous studies have shown that the neurogenic bHLH (basic helix–loop–helix) transcription factor NeuroD6 is sufficient to induce differentiation of the neuronal progenitor-like PC12 cells and that it triggers expression of mitochondrial-related genes, we investigated whether NeuroD6 could modulate the mitochondrial biomass using our PC12-ND6 cellular paradigm. Using a combination of flow cytometry, confocal microscopy and mitochondrial fractionation, we demonstrate that NeuroD6 stimulates maximal mitochondrial mass at the lamellipodia stage, thus preceding axonal growth. NeuroD6 triggers remodelling of the actin and microtubule networks in conjunction with increased expression of the motor protein KIF5B, thus promoting mitochondrial movement in developing neurites with accumulation in growth cones. Maintenance of the NeuroD6-induced mitochondrial mass requires an intact cytoskeletal network, as its disruption severely reduces mitochondrial mass. The present study provides the first evidence that NeuroD6 plays an integrative role in co-ordinating increase in mitochondrial mass with cytoskeletal remodelling, suggestive of a role of this transcription factor as a co-regulator of neuronal differentiation and energy metabolism.
Keywords: basic helix–loop–helix transcription factor, cytoskeletal remodelling, mitochondrial biogenesis, NeuroD family, neuronal differentiation
Abbreviations: bHLH, basic helix–loop–helix; COX, cytochrome c oxidase; E, embryonic day; ESC, embryonic stem cell; F-actin, filamentous actin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MAP, microtubule-associated protein; MMP, mitochondrial membrane potential; mtDNA, mitochondrial DNA; MTG, MitoTracker® Green; MTR, MitoTracker® Red; NGF, nerve growth factor; NRF, nuclear respiratory factor; PDL, poly-d-lysine; PGC-1, peroxisome-proliferator-activated receptor-γ co-activator-1; SOD2, superoxide dismutase 2; WGA, wheat germ agglutinin
INTRODUCTION
Differentiation of progenitor cells into neurons requires energy in the form of ATP produced in mitochondria via oxidative phosphorylation for cytoskeletal assembly, axonal and dendritic growth, growth cone activity and organelle transport (Bernstein and Bamburg, 2003). Early studies have shown that developing neurons exhibit a biphasic increase in total mitochondrial proteins, suggestive of augmented mitochondrial biogenesis during early neuronal differentiation and maturation corresponding to growth cone formation and establishment of the neuronal network with synaptic activity respectively (Cordeau-Lossouarn et al., 1991; Vayssiere et al., 1992). Although mitochondrial trafficking during neuronal differentiation has been well studied (Hollenbeck and Saxton, 2005; Chan, 2006; Chang and Reynolds, 2006), little emphasis has been placed on mitochondrial biogenesis and its transcriptional regulation. The fact that mitochondrial dysfunctions are often associated with neurodevelopmental disorders, such as mitochondrial encephalopathies and axonal Charcot–Marie–Tooth neuropathy (Darin et al., 2001; Zeviani and Di Donato, 2004) raises the fundamental question of how mitochondrial biogenesis is controlled in differentiating neuronal cells.
Mitochondrial biogenesis is a complex process, requiring bi-genomic co-ordination between the nuclear and mitochondrial genomes, with more than 95% of the genes being nuclear-encoded, as the mitochondrial genome encodes a small number (13) of protein subunits of the respiratory complexes I, III, IV and V, along with the necessary tRNAs and rRNAs for their translation (reviewed by Wallace, 2005, Bonawitz et al., 2006; Scarpulla, 2008). To add to the complexity, nuclear-encoded genes are expressed in both a pan- and neural-specific manner, suggesting that mitochondria may also be ‘micro-regulated' in a neuronal-specific fashion.
Since most mitochondrial-related transcriptional studies have been performed within the context of adipocyte and muscle cell differentiation (Carmona et al., 2002; Wu et al., 2002; Wilson-Fritch et al., 2003; Handschin et al., 2003; Wu et al., 2006; Franko et al., 2008), there is a gap in our knowledge regarding the identity of neuronal-specific transcription factors regulating mitochondrial biogenesis in response to energy needs during neurogenesis. Thus far, only PGC-1α (peroxisome-proliferator-activated receptor-γ co-activator 1α), the original member of the PGC-1 family of transcriptional co-activators, has been well established as a master regulator of mitochondrial biogenesis in adipocyte and muscle cell differentiation (Puigserver et al., 1998; Wu et al., 1999; Lehman et al., 2000). It functions as an integrator of physiological and environmental cues to implement a specific transcriptional programme, of which NRF (nuclear respiratory factor)-1 and -2 are key transcriptional regulators co-ordinating expression of several nuclear-encoded subunits of the respiratory complexes and mitochondrial transcription factors (reviewed by Scarpulla, 2008). It is only recently that the expression and activity of NRF-1 and -2 have been correlated with activity of visual cortical neurons (Liang et al., 2006; Yang et al., 2006; Dhar et al., 2008; Dhar and Wong-Riley, 2009).
Therefore the question of whether neurogenic transcription factors are capable of stimulating mitochondrial biogenesis directly while executing terminal neuronal differentiation of progenitor cells remains largely unexplored. Members of the bHLH (basic helix–loop–helix) NeuroD family are potential candidates, being key transcriptional regulators of terminal differentiation during neurogenesis (Guillemot, 2007), at a time when increase in total mitochondrial proteins was observed (Cordeau-Lossouarn et al., 1991). We focused on NeuroD6 (Nex1/MATH-2) whose expression is triggered at E11 (where E is embryonic day), coinciding with neuronal progenitor cells undergoing cell-cycle arrest and terminal differentiation, and further peaks during the first postnatal week, when neuritogenesis and synaptogenesis are highly active (Schwab et al., 1998, 2000</citref>; Schuurmans et al., 2004). Our in vitro functional studies have shown that NeuroD6 by itself initiates and executes differentiation of neuronal-like progenitor PC12 cells, through several interconnected but distinct transcriptional programmes linking neuritogenesis, cell-cycle withdrawal and neuronal survival via the mitochondrial pathway (Uittenbogaard and Chiaramello, 2002, 2004, 2005). Using a genome-wide microarray approach, we found that overexpression of NeuroD6 resulted in a co-ordinated increase in the expression of cytoskeletal elements and mitochondrial-related genes (Uittenbogaard et al., 2009).
The goal of the present study was to investigate whether NeuroD6 could induce and co-ordinate mitochondrial mass with neuronal differentiation using our PC12-ND6 cellular paradigm, which constitutively overexpresses NeuroD6 at levels comparable with that of NGF (nerve growth factor) exposure (Uittenbogaard and Chiaramello, 2002). Using a combination of flow cytometry, confocal microscopy and mitochondrial fractionation, we found that NeuroD6 induced maximal increase of mitochondrial mass at the very first stage of neuronal differentiation, the lamellipodia stage. NeuroD6 also increased expression of the motor protein KIF5B, while generating a permissive gradient of microtubule-associated proteins for proper mitochondrial movement in developing neurites with accumulation in the growth cones. Finally, maintenance of the NeuroD6-induced mitochondrial mass required an intact cytoskeletal network, as its disruption severely reduced mitochondrial mass. Collectively, the present study provides the first evidence that NeuroD6 plays an integrative role in co-ordinating increase in mitochondrial mass with cytoskeletal remodelling in the very early stages of neuronal differentiation, suggestive of a potential role as a co-regulator of neuronal differentiation and energy metabolism.
MATERIALS AND METHODS
Cell culture and pharmacological treatments
Rat pheochromocytoma PC12 cells and PC12-ND6 cells (previously called PC12-Nex1, medium-expressing clones) were grown on collagen I-coated plates (Becton Dickinson Labware) as described by Uittenbogaard and Chiaramello (2002). When indicated, cells were differentiated in the presence of NGF (100 ng/ml) (Roche Molecular Biochemicals). When necessary, cells were grown on PDL (poly-d-lysine)-coated coverslips. To disrupt microtubules or actin polymerization, PC12-ND6 cells were grown for a period of 10 h in the presence of 10 μg/ml nocodazole (Sigma) diluted from a 0.4 mg/ml stock in DMSO or 15 μM latrunculin B diluted from a 5.0 mM stock in DMSO. Control cells were cultured in the presence of 2.5% DMSO. For the recovery period, DMSO or drug-containing medium was removed by washing the cells three times with drug-free medium.
Flow cytometry
Cells were grown on collagen I-coated plates to 80% confluence before staining with either 50 nM MTR (MitoTracker® Red) (chloromethyl-X-rosamine; Molecular Probes) or 70 nM MTG (MitoTracker® Green) (Molecular Probes), for 30 min at 37°C. Cells were then washed in PBS, trypsizined, collected by centrifugation at 228 g for 5 min, and analysed with a FACScalibur flow cytometer (BD Bioscience). Data were collected from 20000 cells for each sample and analysed using the CellQuant software (BD Biosciences).
Immunocytochemistry and confocal microscopy
For immunocytochemistry labelling, cells were seeded on to PDL-coated coverslips. Mitochondria and plasma membrane were labelled by incubating cells in the presence of 500 nM MTR and 1.0 μM Alexa Fluor® 488-conjugated WGA (wheat germ agglutinin) (Molecular Probes) diluted in growth medium for 10 min at 37°C. All samples were fixed in 4.0% (w/v) paraformaldehyde for 5 min and permeabilized in 0.2% Triton X-100 for 5 min. To label polymerized actin filaments, cells were incubated with Alexa Fluor® 488–phalloidin diluted to a final concentration of 82.5 nM in PBS for 20 min at room temperature (25°C) following fixation and permeabilization. Cells were blocked in 10% goat serum (Invitrogen) for 1 h at room temperature. All primary and secondary antibody incubations were performed at room temperature as follows: 1 h of incubation with diluted primary antibodies, followed by 1 h of incubation with the appropriate diluted Alexa Fluor®-conjugated secondary antibodies (Table 1). When indicated, cells were incubated with the nuclear counterstain TO-PRO®-3 (Molecular Probes) diluted 1:500 in PBS for 15 min at room temperature. All samples were mounted with Fluoromount G (Electron Microscopy Sciences). Images were acquired using a Zeiss LSM 510 confocal system equipped with a 63× Plan-Apochromat® (numerical aperture 1.4) objective and Zen software.
Table 1. List of antibodies used for the immunofluorescence studies and immunoblot analyses.
IB, immunoblot analysis; ICC, immunocytochemistry; mAb, monoclonal antibody; pAb, polyclonal antibody.
| Antibody | Type | Source (catalogue number) | Dilution | Application |
| Zyxin | Mouse mAb | Abcam (ab50391) | 1:500 | IB, ICC |
| Gelsolin | Mouse mAb | BD Bioscience (610412) | 1:200 | IB, ICC |
| MAP-1A | Mouse mAb | Chemicon (MAB362) | 1:500 | ICC |
| MAP-2 | Mouse mAb | Chemicon (MAB364) | 1:250 | ICC |
| Total tau | Mouse mAb | Millipore (MAB361) | 1:500 | ICC |
| Unphosphorylated tau | Mouse mAb | Chemicon (MAB3420) | 1:200 | ICC |
| KIF5B | Mouse mAb | Abcam (ab28060) | 1:100 | IB, ICC |
| Neurofilament M | Rabbit pAb | Chemicon (AB1987) | 1:200 | IB, ICC |
| β-III-tubulin | Mouse mAb | Convance (MMS-435P) | 1:500 | ICC |
| β-III-tubulin | Rabbit pAb | Abcam (ab18207) | 1:500 | ICC |
| SOD2 | Rabbit pAb | Abcam (ab13534) | 1:500 | ICC |
| COX III | Mouse mAb | MitoSciences (MS304) | 1:1000 | ICC, IB |
| KIF5B | Mouse mAb | Abcam (ab28060) | 1:100 | ICC, IB |
| GAPDH | Mouse mAb | Aplied Biosystems | 1:2000 | IB |
| Actin | Mouse mAb | Chemicon (MAB15010) | 1:2000 | IB |
| Alexa Fluor® 488-conjugated anti-rabbit IgG | Goat pAb | Invitrogen (A11034) | 1:1000 | ICC |
| Alexa Fluor® 488-conjugated anti-mouse IgG | Goat pAb | Invitrogen (A11029) | 1:1000 | ICC |
| Alexa Fluor® 488-conjugated anti-goat IgG | Donkey pAb | Invitrogen (A11055) | 1:1000 | ICC |
| Alexa Fluor® 568-conjugated anti-rabbit IgG | Goat pAb | Invitrogen (A11036) | 1:1000 | ICC |
| Alexa Fluor® 568-conjugated anti-mouse IgG | Goat pAb | Invitrogen(A11031) | 1:1000 | ICC |
| Alexa Fluor® 647-conjugated anti-rabbit IgG | Goat pAb | Invitrogen (A21245) | 1:1000 | ICC |
| Alexa Fluor® 647-conjugated anti-mouse IgG | Goat pAb | Invitrogen (A21236) | 1:1000 | ICC |
Live-cell imaging
PC12 and PC12-ND6 cells were grown on PDL-coated glass-bottom dishes (Warner Instruments), labelled with 100 nM MTR for 1 h at 37°C and imaged in MTR-free medium. Cells were visualized with an oil-immersion objective 63× on a Zeiss LSM 510 confocal microscope equipped with a TempModuleS and CO2 Module (Zeiss) to maintain the temperature and CO2 levels at 37°C and 5.0%.
Measurement of mitochondrial area
The mitochondrial area was measured using the histogram function of the Image J software (http://rsb.info.nih.gov/ij/) by setting a threshold value excluding 95% of the background staining, as determined by the pixel values from the nuclear area (Liang et al., 2007). To delineate the cytoplasm, we used either the combination of Alexa Fluor® 488-conjugated WGA and TO-PRO®-3 staining or the phalloidin and β-III-tubulin labelling.
ATP bioluminescence assay
ATP content was assessed by the luciferin/luciferase-based CellTiter-Glo™ luminescent assay (Promega). Briefly, PC12 and PC12-ND6 cells were grown in triplicate in white opaque-walled 96-well collagen I-coated plates (Becton Dickinson) at a density of 2.5×104 cells/well, and 100 μl of the CellTiter-Glo® reagent was added to each well. Cells were lysed and incubated for 10 min at room temperature. An ATP standard curve was generated to calculate the amount in fmol of ATP per cell. Chemiluminescence was quantified in triplicate from three independent experiments using the LMax microplate luminometer (Molecular Devices).
Immunoblot analysis
Whole-cell extracts from PC12 and PC12-ND6 cells were prepared as described by Uittenbogaard et al. (2009). Proteins (40 μg) were resolved on a 10% NuPAGE Bis-Tris gel (Invitrogen) and transferred on to a nitrocellulose membrane, which was probed with specific antibodies (Table 1) and corresponding secondary horseradish peroxidase-conjugated antibodies (Pierce). The antigen–antibody complex was detected using the Supersignal West Pico Chemiluminescent Substrate kit (Pierce). Blots were then stripped using Restore™ Western blot stripping buffer (Pierce) and re-probed with the anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) antibody to confirm equal protein loading.
Mitochondrial fractionation
Mitochondria-enriched fractions were isolated using a mitochondrial/cytosol fractionation kit (Sigma) according to the manufacturer's instructions. PC12 and PC12-ND6 cells were grown to 90% confluence and counted using a haematocytometer. Cells were trypsinized, centrifuged at 600 g for 5 min, washed twice with PBS and resuspended in an extraction buffer in the presence of protease inhibitors. Cells were gently homogenized using a Dounce homogenizer, and the degree of homogenization was visualized by Trypan Blue staining. The homogenate was centrifuged at 1000 g for 10 min at 4°C. The supernatant was centrifuged again at 3500 g for 10 min at 4°C, and the pelleted mitochondria were lysed in the lysis buffer provided in the presence of protease inhibitors. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories).
Statistical analysis
Statistical analyses were performed using the unpaired Student's t test, and results are expressed as means±S.D.
RESULTS
PC12-ND6 cells recapitulate the early stages of neuronal differentiation
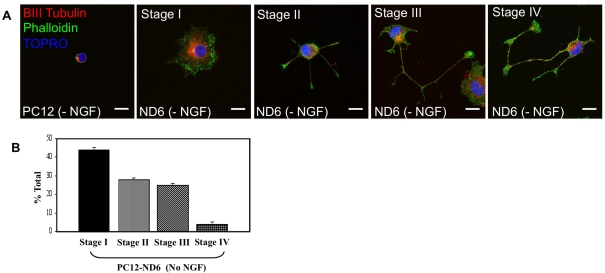
We initially confirmed that PC12-ND6 cells recapitulated the early stages of neuronal differentiation, as defined by Dotti et al. (1988). The first three stages of neuronal differentiation, with stage I being defined as the lamellipodia stage, stage II as outgrowth of neuritic process, and stage III as axonal outgrowth, occur very rapidly within 36 h of in vitro culture, while stage IV, during which dendritic growth is triggered, begins 2–3 days later, with their subsequent maturation (stage V) occurring beyond 1 week of culture.
Untreated PC12 and PC12-ND6 cells were labelled with the fluorescent F-actin (filamentous actin) marker, Alexa Fluor® 488–phalloidin and the nuclear counterstain TO-PRO®-3, and analysed by confocal fluorescence microscopy. PC12-ND6 cells spontaneously recapitulated the first four stages of neuronal differentiation within the first 48 h after plating on coverglass in the absence of NGF (Figure 1A), with 44% at stage I, 28 and 25% at stages II and III respectively, and 3% at stage IV (Figure 1B). By day 9, most of the PC12-ND6 cells reached stage III/IV, when left unperturbed. As expected, untreated PC12 cells, which do not express endogenous NeuroD6, displayed a morphology characteristic of progenitor-like neuronal cells, small size without an extensive lamellipodium, as observed with stage I PC12-ND6 cells (Figure 1A). Thus the PC12-ND6 cell system provides an unlimited supply of differentiating neuronal cells, making it a suitable and instructive paradigm to investigate NeuroD6's effect on mitochondrial mass throughout neuronal differentiation.
Figure 1. PC12-ND6 cells recapitulate the first four stages of neuronal differentiation.
(A) Morphological characteristics of PC12-ND6 cells. Untreated PC12 and PC12-ND6 (referred to as ND6) cells were labelled with the F-actin marker Alexa Fluor 488®–phalloidin (green), anti-β-III-tubulin antibody (red) and the nuclear marker TO-PRO®-3 (blue) and analysed by confocal microscopy. Scale bar, 10 μm. (B) Quantification of the distinct neuronal differentiation stages in cultured PC12-ND6 cells labelled with phalloidin 48 h after seeding. Results are means±S.D. for three independent experiments, with n = 200 cells per experiment.
NeuroD6 increases mitochondrial mass in PC12-ND6 cells in a manner similar to that of NGF treatment
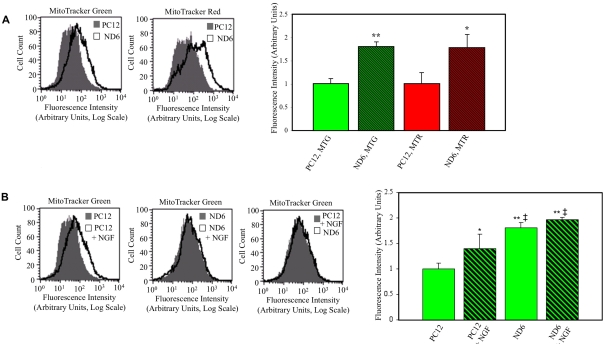
On the basis of the correlation between NeuroD6 and mitochondrial-related gene expression highlighted by our genome-wide microarray study (Uittenbogaard et al., 2009), we hypothesized that NeuroD6 could stimulate mitochondrial mass. We assessed mitochondrial mass by flow cytometry using PC12 and PC12-ND6 cells stained with MTG or MTR. Fluorescence intensity emitted by MTG has been used as an estimate of overall mitochondrial mass, as it accumulates in the lipid compartment of the mitochondria in an MMP (mitochondrial membrane potential)-independent manner, whereas fluorescence intensity emitted by MTR assesses the content of energized mitochondria, as its accumulation and retention has been shown to be MMP-dependent (Pendergrass et al., 2004). Quantitative flow cytometry revealed that PC12-ND6 cells displayed 1.8-fold increased fluorescence emission of each dye, compared with control PC12 cells (Figure 2A), indicative of increased mitochondrial mass upon NeuroD6 expression.
Figure 2. NeuroD6 overexpression results in increased mitochondrial mass in a manner similar to that of NGF treatment.
(A) Quantification of the fluorescence intensity of PC12 and PC12-ND6 labelled with mitochondria-specific dyes. Untreated control PC12 and untreated PC12-ND6 (ND6) cells were stained with MTG or MTR and analysed by flow cytometry. Flow cytometric profiles of PC12 and PC12-ND6 labelled with mitochondria-specific dyes (left-hand panel). Results are expressed as means±S.D. for five independent experiments (right-hand panel). *P<0.05, **P<0.0001 (Student's t test), compared with control PC12 cells. (B) Quantification of the fluorescence intensity of MTG-labelled untreated and NGF-treated PC12 and PC12-ND6 cells. Untreated and NGF-treated (100 ng/ml, 48 h) PC12 and PC12-ND6 cells were stained with MTG and analysed by flow cytometry. Flow cytometric profiles of PC12 and PC12-ND6 cells grown in the presence or absence of NGF (left-hand panel). Results are expressed as means±S.D. for five independent experiments (right-hand panel). *P<0.05 compared with untreated PC12 cells; **P<0.0001 compared with untreated PC12 cells; ‡P<0.05 compared with PC12 cells treated with NGF for 48 h.
As NeuroD6 is a critical effector of the NGF pathway (Uittenbogaard and Chiaramello, 2002), we asked whether NeuroD6 could mimic NGF treatment in terms of mitochondrial mass. PC12 and PC12-ND6 cells were differentiated for 2 days with NGF and stained with MTG for flow cytometry analysis. Although NGF-treated PC12 cells displayed a 1.4-fold increase in mitochondrial mass, NGF-treated PC12-ND6 cells failed to show any further increase in mitochondrial mass (Figure 2B), despite their more advanced degree of differentiation with a preponderance of stages III and IV (Uittenbogaard and Chiaramello, 2002). In contrast, most NGF-treated PC12 cells were differentiated at stages I and II (Uittenbogaard and Chiaramello, 2002). Interestingly, untreated PC12-ND6 cells exhibited a moderate but consistent increase in mitochondrial mass, compared with NGF-treated PC12 cells (Figure 2B). Collectively, these results suggest that increase in mitochondrial mass may occur in the very early stages of neuronal differentiation and precede axonal growth.
PC12-ND6 cells display maximal mitochondrial area at the lamellipodia stage of neuronal differentiation
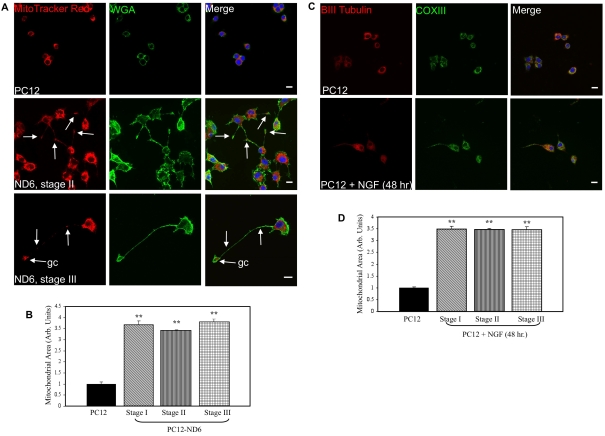
Since mitochondrial area is another reliable indicator of mitochondrial content, similar to mitochondrial mass (Berman et al., 2009), we measured mitochondrial area by confocal fluorescence microscopy using PC12 and PC12-ND6 cells labelled with the plasma membrane marker, Alexa Fluor® 488-conjugated WGA, the mitochondrial dye MTR, and the nuclear counterstain TO-PRO®-3 (Figure 3A). We confirmed that MTR labelled the entire population of mitochondria by co-staining cells with MTR and an antibody against the mitochondrial enzyme SOD2 (superoxide dismutase 2), an antioxidant protein known to localize exclusively in the mitochondrial matrix (see Supplementary Figure S1 at http://asnneuro.org/an/001/an001e016.add.htm).
Figure 3. PC12-ND6 cells display increased mitochondrial area.
(A) Staining of the mitochondrial compartment. PC12 and PC12-ND6 cells were stained with MTR (red) and the plasma membrane marker Alexa Fluor® 488-conjugated WGA (green) and TO-PRO®-3 (blue). White arrows indicate the presence of mitochondria within neurites or growth cones (gc). Scale bar, 10 μm. (B) Quantification of mitochondrial area in PC12 and PC12-ND6 cells at different stages of neuronal differentiation. Results are means±S.D. for three independent experiments, with a minimum of n = 150 cells for each cell type and a minimum of n = 50 cells per stage. **P<0.0001 (Student's t test). (C) Staining of the mitochondrial compartment in untreated and NGF-differentiated PC12 cells. Untreated and NGF-treated PC12 cells for 48 h (100 μg/ml) were stained with antibodies against β-III-tubulin (red), COX III (green) and subsequently counterstained with the TO-PRO®-3 (blue). Scale bar, 10 μm. (D) Quantification of mitochondrial area in untreated and NGF-treated PC12 cells. Results are means±S.D. for three independent experiments, with n = 150 cells per stage. **P<0.0001 (Student's t test).
Overexpression of NeuroD6 led not only to increased mitochondrial area, but also to differential distribution of mitochondria, when compared with control PC12 cells (Figure 3A). In PC12 cells, mitochondria were localized within the perinuclear region, whereas mitochondria in stage II PC12-ND6 cells were positioned in a row to migrate toward the neuritic tip (Figure 3A, middle row, white arrows). Similarly, in stage III PC12-ND6 cells, mitochondria were present throughout the developing neuronal process and accumulated in the growth cone (Figure 3A, bottom row, white arrows). During the course of this analysis, we also assessed mitochondrial morphology using fixed and live PC12 and PC12-ND6 cells stained with SOD2 and MTR respectively (see Supplementary Figure S2 at http://asnneuro.org/an/001/an001e016.add.htm). Mitochondria from control PC12 cells displayed a predominant short and punctate morphology (arrowheads), whereas stage I PC12-ND6 cells harboured a heterogeneous mixture of elongated tubular (arrows) and short (arrowheads) forms of mitochondria. In contrast, the majority of mitochondria in dividing stage I PC12-ND6 cells displayed a homogeneous short morphology. In stage III PC12-ND6 cells, mitochondria morphology varied depending on their subcellular localization: elongated in the soma, and short in the growth cones.
To quantify the mitochondrial area, the plasma membrane and nuclear stains were used to delineate the cytoplasm, while the mitochondrial area labelled with MTR was measured for each cell type using the Image J software. Since the results from flow cytometry were suggestive of increased mitochondrial mass during the early stages of neuronal differentiation, we quantified the mitochondrial area at each stage of neuronal differentiation (I–III) for PC12-ND6 cells. We observed a 3.6-fold increase in mitochondrial area (P<0.0001) upon NeuroD6 expression at the very first stage of neuronal differentiation, the lamellipodia stage, when compared with PC12 cells (Figure 3B). We found no statistically significant difference in the mitochondrial area of PC12-ND6 cells throughout stages I–III, consistent with the flow cytometry analysis. The timing of increased mitochondrial compartment during neuronal differentiation was confirmed further by quantifying the mitochondrial area at distinct stages of NGF-differentiated PC12 cells for 48 h, stained with an antibody against the core 2 protein of COX (cytochrome c oxidase) III instead of MTR (Figures 3C and 3D) to avoid any potential problems of MMP heterogeneity, as NGF signalling is known to modulate MMP (Verburg and Hollenbeck, 2008). Taken together, these results reveal that NeuroD6 triggers an increase in mitochondrial mass in a manner similar to that of NGF signalling, which essentially occurs during the transition from undifferentiated neuronal progenitor-like PC12 cells to stage I differentiating PC12-ND6 cells.
Overexpression of NeuroD6 results in increased total mitochondrial proteins and ATP levels
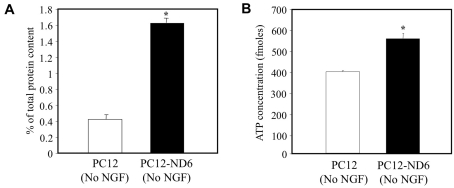
To confirm NeuroD6-increased mitochondrial mass, we isolated mitochondrial-enriched fractions from PC12 and PC12-ND6 cells and expressed the protein content of mitochondrial fractions as percent of total protein content of the corresponding whole-cell extract (Figure 4A). Total mitochondrial protein content increased 3.88-fold upon NeuroD6 expression, in keeping with the 3.6-fold increase in mitochondrial area measured by confocal microscopy (Figure 3B).
Figure 4. Overexpression of NeuroD6 increases the mitochondrial protein content and cellular ATP levels.
(A) Measurement of mitochondrial protein content in PC12 and PC12-ND6 cells. Mitochondrial-enriched fractions were isolated from PC12 and PC12-ND6 cells grown in the absence of NGF, and the protein content of mitochondrial fractions was expressed as percentage of total protein content of the corresponding whole-cell extract. Results are means±S.D. for three independent experiments. *P<0.001 (Student's t test). (B) Determination of cellular ATP levels. ATP levels were measured in triplicate from PC12 and PC12-ND6 cells grown in the absence of NGF using the CellTiter-Glo™ assay. Results are means±S.D. for three independent experiments. *P<0.001 (Student's t test).
We next measured total ATP levels from PC12 and PC12-ND6 cells, as described in the Materials and methods section, and found 1.56-fold increased ATP levels upon NeuroD6 overexpression (Figure 4B), which paralleled the 1.8-fold increase in mitochondrial mass measured by flow cytometry (Figure 2A) and the 2.2-fold increased expression of the δ subunit of the ATP synthase complex (Uittenbogaard and Chiaramello, 2004), which is essential for its assembly, being part of the stalk connecting the F1 and Fo moieties (Capaldi et al., 1994; Zhang et al., 1995). Collectively, these data strongly suggest that NeuroD6 overexpression results in increased mitochondrial content and ATP levels.
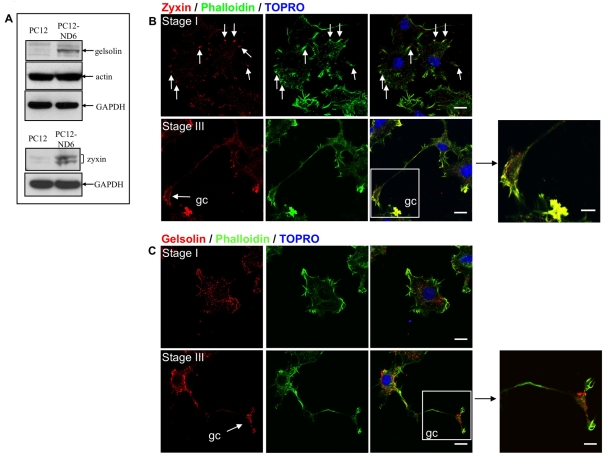
NeuroD6 promotes rearrangement of actin in conjunction with the actin regulatory proteins, zyxin and gelsolin
Actin remodelling plays a critical role in localized and retrograde movement of mitochondria within axons (Morris and Hollenbeck, 1995; Ligon and Steward, 2000a; Chada and Hollenbeck, 2004). The fact that phalloidin labelling revealed differential distribution of the actin cytoskeleton at distinct neuronal stages of PC12-ND6 cells (Figure 1A) combined with 5.4- and 2.3-fold increased mRNA levels of the actin-remodelling proteins zyxin and gelsolin respectively (Uittenbogaard et al., 2009) prompted us to assess the spatio-temporal co-localization of actin, zyxin and gelsolin in PC12-ND6 cells at distinct stages of neuronal differentiation using confocal microscopy. We initially confirmed increased zyxin and gelsolin expression at the protein level by immunoblot analysis (Figure 5A), with zyxin migrating as a doublet, consistent with its identity as a phosphoprotein (Crawford and Beckerle, 1991).
Figure 5. Reorganization of actin and the actin-binding proteins zyxin and gelsolin.
(A) Immunoblot analysis of actin, gelsolin and zyxin proteins in PC12 and PC12-ND6 cells. Immunoblot analysis was performed using whole-cell extracts from PC12 and PC12-ND6 cells using antibodies against gelsolin, zyxin and actin. Equal loading of protein was verified using an anti-GAPDH antibody. Results shown are representative of at least three independent experiments. (B) Stage-specific rearrangement of zyxin. PC12-ND6 cells were labelled with Alexa Fluor® 488–phalloidin (green), anti-zyxin antibody (red) and TO-PRO®-3 (blue). Stage I PC12-ND6 cells (top row) display accumulation of zyxin labelling at the focal adhesion points of the lamellipodium (arrows). In stage III PC12-ND6 cells (bottom row), zyxin labelling has been redistributed to the growth cone (gc, arrow). Scale bar, 10 μm. In the right-hand panel, a high magnification of the growth cone is shown. Scale bar, 5 μm. (C) Stage-specific rearrangement of gelsolin. PC12-ND6 cells were labelled with Alexa Fluor® 488–phalloidin (green), anti-gelsolin antibody (red) and TO-PRO®-3 (blue). In stage I PC12-ND6 cells (top row), gelsolin labelling is predominantly found at the edge of the lamellipodium. In stage III PC12-ND6 cells (bottom row), a portion of the gelsolin labelling is found within the growth cone (gc, arrow). Scale bar, 10 μm. In the right-hand panel, a high magnification of the growth cone is shown. Scale bar, 5 μm.
Zyxin co-localized with actin in clusters at the leading edge of the lamellipodia and focal adhesions of stage I PC12-ND6 cells and in the growth cones of stage III PC12-ND6 cells (Figure 5B, white arrows), consistent with its known ability to direct cytoskeletal reorganization at the leading edge of the lamellipodium and growth cones (Beckerle, 1986, 1997; Crawford and Beckerle, 1991; Drees et al., 1999). Gelsolin co-localized with actin filaments in growing neurites of stage II PC12-ND6 cells with enrichment at the tip, whereas PC12-ND6 cells with stage III morphology exhibit concentrated expression of gelsolin in the growth cones where it is expressed in the central domain as well as the leading edge of the lamellipodia and filopodia (Figure 5C). These results indicate that NeuroD6 promotes actin remodelling by simultaneously increasing the expression levels of key actin-remodelling proteins and promoting their appropriate spatio-temporal regulation.
NeuroD6 promotes rearrangement of microtubules and microtubule-binding proteins
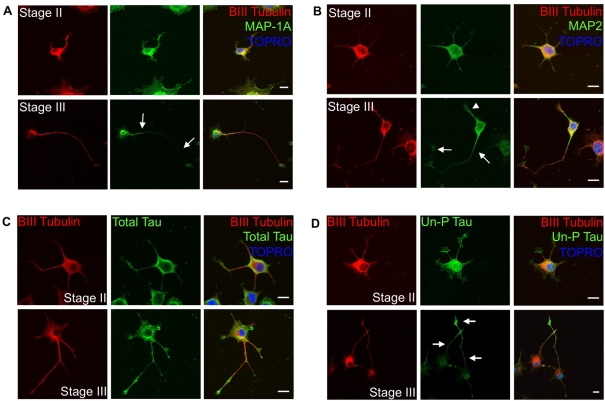
We next focused on the microtubule network, as it plays a critical role in long distance trafficking of mitochondria in the developing axon toward the growth cone (Morris and Hollenbeck 1993, 1995; Hollenbeck 1996; Ruthel and Hollenbeck, 2003). The fact that NeuroD6 increases the expression of β-III-tubulin protein in the absence of NGF (Uittenbogaard and Chiaramello, 2004), and promotes its spatial redistribution throughout the early stages of neuronal differentiation (Figure 1A), enhances further its potential contribution to co-ordinate an increase in mitochondrial mass with movement. On the basis that a proper gradient of MAPs (microtubule-associated proteins) is critical for mitochondrial movement in developing axons (Ebneth et al., 1998; Trinczek et al., 1999; Seitz et al., 2002; Mandelkow et al., 2003; Jimenez-Mateos et al., 2006; Dixit et al., 2008), we assessed the expression pattern of MAP-1A, MAP-2 and Tau in PC12-ND6 cells at different stages of neuronal differentiation, all of them being up-regulated by NeuroD6 (Uittenbogaard and Chiaramello, 2004). Stage II PC12-ND6 cells displayed equal distribution of MAP-1A among all growing neurites, whereas stage III PC12-ND6 cells showed that MAP-1A expression was excluded from the distal portion of nascent axons (Figure 6A). In contrast, decreased MAP-2 expression was observed in developing axons of stage III PC12-ND6 cells, with a drastic reduction at their distal portions (Figure 6B). Thus constitutive NeuroD6 expression led to proper redistribution of MAP-1A and MAP-2 at distinct stages of neuronal differentiation, as observed in cultured embryonic hippocampal neurons (Cãceres et al., 1986; Dotti et al., 1988; Pennypacker et al., 1991; Ulloa et al., 1994; Díez-Guerra and Avila, 1995).
Figure 6. Reorganization of microtubules and corresponding microtubule-binding proteins in PC12-ND6 cells.
(A) Stage-specific redistribution of MAP-1A. PC12-ND6 cells were labelled with anti-β-III-tubulin antibody (red), anti-MAP-1A antibody (green) and TO-PRO®-3 (blue). In stage II PC12-ND6 cells (top row), MAP-1A is evenly distributed throughout the cell. In stage III PC12-ND6 cells (bottom row), a MAP-1A gradient has been established with MAP-1A labelling excluded from the nascent axon, as indicated by white arrows. Scale bar, 10 μm. (B) Stage-specific redistribution of MAP-2. PC12-ND6 cells were labelled with anti-β-III-tubulin antibody (red), anti-MAP-2 antibody (green) and TO-PRO®-3 (blue). In stage II PC12-ND6 cells (top row), MAP-2 is evenly distributed throughout the cell. In stage III PC12-ND6 cells (bottom row), a gradient of MAP-2 has been established, with MAP-2 labelling excluded from the growing axon (white arrows), but present within developing dendrites (white arrowhead). Scale bar, 10 μm. (C) Total tau is evenly distributed in PC12-ND6 cells at stages II and III. PC12-ND6 cells were labelled with anti-β-III-tubulin antibody (red), an antibody recognizing all forms of tau (total tau, green) and TO-PRO®-3 (blue). Scale bar, 10 μm. (D) Expression of unphosphorylated tau is enriched in the distal portion of growing axons in stage III PC12-ND6 cells. PC12-ND6 cells were labelled with anti-β-III-tubulin antibody (red), antibody recognizing only the unphosphorylated form of tau (Un-P Tau, green) and TO-PRO®-3 (blue). In stage II PC12-ND6 cells (top row), unphosphorylated tau is evenly distributed throughout the soma and neurites. In contrast, stage III PC12-ND6 cells (bottom row) display unphosphorylated tau enriched within the distal portion of the growing axon, as indicated by arrows. Scale bar, 10 μm.
We next examined the distribution pattern of the unphosphorylated and phosphorylated forms of tau, since their gradients of expression influence microtubule organization and mitochondrial movement during axonal outgrowth (Cãceres and Kosik, 1990; Cãceres et al., 1991; Dixit et al., 2008). We used two distinct anti-tau antibodies, one specific for the unphosphorylated tau form (at the Tau-1 epitope) and the other one raised against total tau, to perform immunocytochemistry on PC12-ND6 cells at different stages of differentiation. Unphosphorylated tau was evenly distributed among growing neurites of stage II PC12-ND6 cells, whereas unphosphorylated tau was enriched in the distal portion of the developing axon of stage III PC12-ND6 cells (Figure 6C). Such a proximo-distal gradient was maintained in stage IV PC12-ND6 cells (data not shown). In contrast, total tau was evenly distributed in growing processes of PC12-ND6 cells at all stages (Figure 6D). These results are consistent with the tau expression pattern observed in cultured embryonic hippocampal neurons (Mandell and Banker, 1996). Taken together, our results indicate that PC12-ND6 cells recapitulate the spatial and temporal expression patterns of key players compatible with cytoskeletal rearrangement and mitochondrial trafficking during early neuronal differentiation.
Co-localization of mitochondria, microtubules and microtubule-based motor protein KIF5B in PC12-ND6 cells
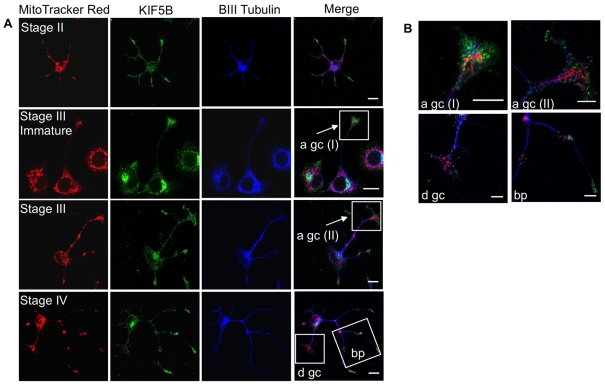
Since proper transport and intracellular distribution of mitochondria are vital for proper neuronal differentiation to insure high metabolic demands associated with active growth cones and branching points (Povlishock, 1976; Morris and Hollenbeck, 1993; Ruthel and Hollenbeck, 2003), we examined using confocal microscopy the spatio-temporal expression pattern of the motor protein KIF5B (also called kinesin-1 heavy chain, KHC) at distinct stages of neuronal differentiation of untreated PC12-ND6 cells. KIF5B, a member of the kinesin superfamily, is an essential microtubule-based motor protein for anterograde mitochondrial transport, as a body of studies has shown that inhibition of KIF5B, targeted disruption of kif5b or mutations of the Milton and Miro proteins necessary to mediate linkage of KIF5B to mitochondria drastically reduce anterograde mitochondrial transport (Nangaku et al., 1994; Hollenbeck, 1996; Hurd and Saxton, 1996; Hurd et al., 1996; Pereira et al., 1997; Tanaka et al., 1998; Goldstein and Yang, 2000; Stowers et al., 2002; Górska-Andrzejak et al., 2003; Hollenbeck and Saxton, 2005; Glater et al., 2006; Pilling et al., 2006).
Figure 7 clearly illustrates a spatio-temporal pattern of KIF5B expression compatible with its function in mitochondrial transport and subcellular distribution associated with distinct stages of neuronal differentiation. Stage II PC12-ND6 cells displayed a uniform distribution of KIF5B throughout the cell body and growing neurites with an overlapping expression pattern with MTR and β-III-tubulin, and enriched KIF5B expression at the tip of growing neurites with high density of mitochondria (Figure 7A). During the early phase of stage II–III transition (referred to as immature stage III), KIF5B immunoreactivity remained enriched in growth cones, when compared with the soma (Figure 7A). However, stage III PC12-ND6 cells no longer displayed such enrichment of KIF5B (Figure 7A). High magnification revealed a differential spatial distribution of KIF5B protein in the growth cones depending on the degree of differentiation of stage III PC12-ND6 cells, with accumulation of KIF5B in the central domain of growth cones of immature stage III PC12-ND6 cells and dispersion of KIF5B protein at the leading edge of growth cones of mature stage III PC12-ND6 cells (Figure 7B), which is in keeping with the well-documented interactions between KIF5B and axonal synaptic vesicles (Cai et al., 2007; Cai and Sheng, 2009).
Figure 7. Subcellular distribution of the motor protein KIF5B and mitochondria in PC12-ND6 cells at distinct stages of neuronal differentiation.
(A) Mitochondria, KIF5B and tubulin closely overlap throughout the early stages of neuronal differentiation. PC12-ND6 cells were labelled with MTR and antibodies against KIF5B (green) and β-III-tubulin (blue). Scale bar, 10 μm. (B) Co-localization of KIF5B and mitochondria within growth cones and branching points. High-magnification images of the boxed regions reveal that mitochondria are located in axonal (a) and dendritic (d) growth cones (gc), whereas KIF5B protein is detected at the leading edge of the growth cones. Mitochondria also accumulate in the branching points (bp) of growing processes. Scale bar, 5 μm.
Finally, stage IV PC12-ND6 cells showed a uniform distribution of KIF5B in the soma, axon and dendrites, with the exception of future branching points and axonal growth cones, where clusters of KIF5B were observed (Figure 7A). High magnification of the future branching points showed co-localization of mitochondria and KIF5B in the absence or low levels of β-III-tubulin (Figure 7B). It is worth noting the lack of KIF5B accumulation in dendritic growth cones when compared with axonal growth cones (Figure 7B). This is most likely to be the result of differences in the polarity of the microtubules, with axonal microtubules uniformly oriented with their plus-ends distal to the cell body and dendritic microtubules displaying a mixture of plus-end and minus-end orientations (Heidemann, 1996; Baas, 2002), as KIF5B functions as a plus-end-directed motor protein. Taken together, our results provide the first demonstration that NeuroD6 promotes proper mitochondrial subcellular distribution through the expression and appropriate stage-specific redistribution of key cytoskeletal elements, cytoskeletal-associated proteins and the motor protein KIF5B during the early stages of neuronal differentiation.
Depolymerization of the actin or microtubule network results in decreased mitochondrial area despite constitutive NeuroD6 expression
We next asked the question of whether NeuroD6-mediated increase in mitochondrial mass could be sustained upon depolymerization of the actin or microtubule network. Since the microtubule and actin networks have distinct functions in mitochondrial trafficking (Morris and Hollenbeck, 1995; Ligon and Steward, 2000a; Chada and Hollenbeck, 2004), we elected to disrupt each cytoskeletal component separately using either nocodazole to disrupt the microtubule network or latrunculin B to inhibit actin polymerization. PC12-ND6 cells were treated with 10 μg/ml nocodazole or 15 μM latrunculin B for 10 h, followed by a 30-h-long recovery period, during which we monitored the integrity of actin or microtubule network in conjunction with mitochondrial area, as described in the Materials and methods section.
As expected, incubation with nocodazole or latrunculin B led to substantial retraction of neuritic processes, resulting in decreased frequency of stage II and III PC12-ND6 cells, while vehicle (DMSO)-only treated PC12-ND6 cells retained their original frequency (see Supplementary Figure S3A at http://asnneuro.org/an/001/an001e016.add.htm). The immunohistochemistry results confirmed that nocodazole treatment selectively disrupted the microtubule network and latrunculin B selectively depolarized actin filaments (see Supplementary Figures S3B and S3C). Disruption of the actin or microtubule network had a severe impact on the mitochondrial area of treated PC12-ND6 cells, as compared with vehicle-only treated PC12-ND6 cells. Interestingly, it differentially affected the distribution pattern of mitochondria, with latrunculin B-treated PC12-ND6 cells displaying mitochondria essentially aggregated around the MTOC (microtubule-organizing centre) (see Supplementary Figure S3B) and nocodazole-treated PC12-ND6 cells harbouring mitochondria evenly localized to the perinuclear region (see Supplementary Figure S3C).
We next investigated whether the decline in mitochondrial content was reversible upon re-establishment of proper cytoskeletal reorganization by switching to drug-free medium over a period of 30 h. PC12-ND6 cells regained their original morphology, characteristic of stages II and II within a 30-h period, with mitochondrial area being restored to original levels in a time-dependent manner peaking after a 15-h-long recovery (see Supplementary Figures S3B–S3D). Although phalloidin and α-tubulin labelling revealed recovery of the microtubule and actin networks within the first 2 h following drug removal, recovery of mitochondrial content was significantly delayed, with partial restoration of the mitochondrial compartment after 6 h of recovery, which was stably maintained after 30 h, suggesting that the observed decrease in mitochondrial content was not a by-product of cytotoxicity from nocodazole or latrunculin B exposure, but was most likely to be due to disruption of the cytoskeletal architecture (see Supplementary Figure S3D).
DISCUSSION
In the present paper, we report a novel role for NeuroD6 in increasing mitochondrial mass. First, we have demonstrated that the increase in mitochondrial mass occurs during the very first step of neuronal differentiation, the lamellipodia stage I, thus preceding neurite outgrowth. Secondly, we have shown that NeuroD6 concurrently co-ordinates mitochondrial mass increase with cytoskeletal remodelling, resulting in mitochondrial distribution in developing neuronal processes with accumulation in growth cones (Figure 8). Thirdly, such co-ordination is required to sustain mitochondrial mass during neuronal differentiation. Thus the present study provides the first evidence of a neurogenic-specific regulation of mitochondrial mass in the context of neuronal differentiation, as most studies have essentially revealed ubiquitously expressed transcription factors modulating mitochondrial biogenesis in the context of physiological adaptations and differentiation of cellular systems, such as adipocytes and muscle cells.
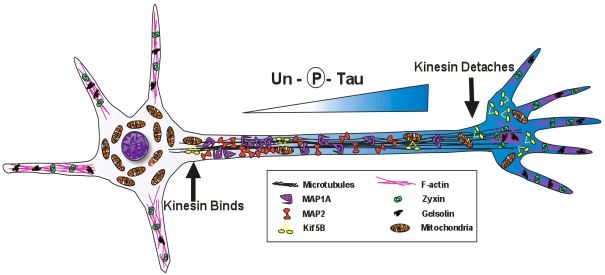
Figure 8. Working model of NeuroD6-mediated co-ordination of mitochondrial mass with cytoskeletal rearrangement in PC12-ND6 cells.
NeuroD6 induces cytoskeletal remodelling, appropriate stage-specific redistribution of MAP-1A and MAP-2, as well as a gradient of unphosphorylated tau in developing processes of stage III PC12-ND6 cells, which facilitate efficient binding of KIF5B with mitochondria in the proximal portion of the developing axon and the release of KIF5B with its cargo at the distal portion of the developing axon.
The lack of a suitable cellular paradigm and/or animal models has hampered studies on mitochondrial biogenesis at the early stages of neurogenesis. Knockout of key transcriptional regulators for mitochondrial biogenesis, such as the mitochondrial-specific polymerase γ PolG and Tfam, results in early embryonic lethality, at E8.5 (Hance et al., 2005) and E10.5 (Larsson et al., 1998) respectively. Although cultured embryonic E18 hippocampal neurons are widely used to investigate the mechanisms regulating mitochondrial trafficking, they are not amenable to investigate mitochondrial biogenesis and its neuronal regulators during the early stages of neuronal differentiation, since the first three stages occur within an extremely narrow timeframe of 36 h after plating (Dotti et al., 1988).
Given the ability of NeuroD6 to promote neuritogenesis via increased expression of cytoskeletal genes (Uittenbogaard and Chiaramello, 2002, 2004; Uittenbogaard et al., 2009) and the recently identified NeuroD6 target gene, PRG-1, also called plasticity-related gene-1, a member of the LPP (lipid phosphate phosphatase) family facilitating axonal elongation during development (Bräuer et al., 2003; Bräuer and Nitsch, 2008; Yamada et al., 2008), the PC12-ND6 cells spontaneously recapitulate the first four stages of neuronal differentiation, thus providing a limitless source of differentiating neuronal cells, making them an instructive in vitro neuronal system to investigate mitochondrial biogenesis and its transcriptional regulation. Furthermore, the PC12-ND6 cellular paradigm allows investigating the intrinsic properties of NeuroD6, independently of any extrinsic differentiation signalling and the functional redundancy between members of the NeuroD family. Since the untreated PC12 cells share characteristics with neuroprogenitors (Leonard et al., 1987; Greene et al., 1998; Angelastro et al., 2000, 2002), our current experimental paradigm has enabled us to investigate the dynamic modulation of mitochondrial mass during the early stages of transition between neuroprogenitor-like cells to differentiating neuronal cells. Therefore it is an advantageous in vitro cellular system to initiate studies of the transcription pathways controlling mitochondrial biogenesis at the outset of neuronal differentiation, which can then be tested further in more challenging in vivo systems. However, the PC12-ND6 cellular paradigm has inherent limitations, such as the lack of diversity in terms of lineages and response to diverse signalling pathways, which prevent addressing the question of how mitochondrial biogenesis is tailored to specific needs of neuronal populations upon cell fate commitment and differentiation.
Using a combination of flow cytometry, confocal microscopy and mitochondrial protein fractionation, we have demonstrated that constitutive expression of NeuroD6 results in increased mitochondrial content, coinciding with cytoplasmic expansion associated with the lamellipodia formation. Although the overall cell volume of PC12-ND6 cells is greater than that of the progenitor-like PC12 cells, the results of mitochondrial protein fractionation normalized against total protein content show a significant 3.66-fold increase in mitochondrial content, even when taking into account the increased cellular volume. Our findings are in keeping with the emerging link between mitochondrial expansion and spontaneous differentiation of mouse and human ESCs (embryonic stem cells) (St John et al., 2005; Cho et al., 2006). Undifferentiated and proliferative D3 mouse ESCs contain few and immature mitochondria, whereas loss of ESC pluripotency coupled with onset of cellular differentiation is accompanied by a gradual increase in mtDNA (mitochondrial DNA) replication (Facucho-Oliveira et al., 2007).
Whereas our results demonstrate that NeuroD6 increases the mitochondrial content, the underlying mechanisms by which NeuroD6 triggers mitochondrial biogenesis remain to be elucidated. It could involve shifting the balance from fusion to fission, increasing mtDNA replication, stimulating the mitochondrial biogenic transcriptional cascade, diminishing mitochondria turnover or any combination of these regulatory pathways depending on the stage of neuronal differentiation.
Since mitochondrial biogenesis is not suspected to occur de novo (Detmer and Chan, 2007), an increase in mitochondrial mass may in part result from increased rate of mitochondrial fission at the expense of fusion (Nunnari et al., 1997; Mozdy and Shaw, 2003; Scott et al., 2003; Karbowski and Youle, 2003). Mitochondrial fission is mediated by three proteins, Fis1, Drp-1/DLP1/Dmn1 and the recently identified Mff protein (Mozdy et al., 2000; Jakobs et al., 2003; James et al., 2003; Yoon et al., 2003; Schauss et al., 2006; Gandre-Babbe and van der Bliek, 2008), whereas mitochondrial fusion is under the control of the Mitofusin-1 and -2 proteins (Mfn1 and Mfn2), and the optic atrophy-1 protein (OPA1) (Santel and Fuller, 2001; Olichon et al., 2002; Ishihara et al., 2003; Satoh et al., 2003; Cerveny et al., 2007). Future studies are required to directly assess the link between NeuroD6 expression and the rate of fusion and fission using a mitochondrial matrix-targeted photoactivable green fluorescent protein (Patterson and Lippincott-Schwartz, 2002; Karbowski et al., 2004).
The fact that NeuroD6 increases the expression of mitochondrial-related genes, involved in several aspects of mitochondrial function, including mitochondrial bioenergetics, protein folding and oxidative metabolism (Uittenbogaard et al., 2009), strongly supports the notion of NeuroD6 promoting mitochondrial biogenesis. It is conceivable that NeuroD6 may regulate the mitochondrial biogenic transcriptional pathway to stimulate mtDNA replication and transcription, via the well-established regulator PGC-1α. Although the role of PGC-1α in mitochondrial biogenesis has not been addressed in a neuronal context, recent studies suggest a potential role in neuronal function (Lin et al., 2004; Leone et al., 2005; Liang and Wong-Riley, 2006; Cowell et al., 2007), which is in keeping with increased neuronal expression of NRF-1 and NRF-2, both under PGC-1α control to promote the expression of nuclear- and mitochondrial-encoded subunits of COX IV, essential for mitochondrial biogenesis (Ongwijitwat and Wong-Riley, 2005; Ongwijitwat et al., 2006; Meng et al., 2007; Dhar and Wong-Riley, 2009).
The finding that constitutive NeuroD6 expression is sufficient to induce cytoskeletal remodelling has important functional implications, since cytoskeleton plays a cardinal role in both the morphological changes upon differentiation from a precursor to a neuron and mitochondrial trafficking (reviewed by Matus, 1988; Hollenbeck and Saxton, 2005). In keeping with our previous findings of increased expression of microtubules and MAPs (Uittenbogaard and Chiaramello, 2004), we showed that the actin and microtubule cytoskeletons and their associated binding proteins are appropriately remodelled in PC12-ND6 cells at each stage of neuronal differentiation. Actin remodelling is particularly relevant, as maximal increase in mitochondrial mass occurs at stage I, during which the actin network is extensively remodelled for lamellipodia and filopodia formation (reviewed by da Silva and Dotti, 2002). This concerted action is critical, as reorganization of actin filaments and microtubules must be co-ordinated during neuritogenesis (reviewed by Dehmelt and Halpain, 2004; Dent et al., 2007).
NeuroD6 by itself is sufficient to induce a gradient of MAP-1A and MAP-2 expression as well as a proximo-distal gradient of unphosphorylated tau, similar to that observed in developing cultured hippocampal neurons, with MAP-1A and MAP-2 predominantly expressed in the proximal segment of nascent axons and enriched expression of unphosphorylated tau (at the Tau-1 epitope) in the distal portion of developing axons. Such gradients are critical not only to regulate microtubule dynamics in developing axons and dendrites, but also to promote mitochondrial transport, as microtubules serve as tracks for anterograde mitochondrial trafficking (Nangaku et al., 1994; Morris and Hollenbeck, 1995; Rickard and Kreis, 1996) in conjunction with the motor protein KIF5B (Cai et al., 2005, 2007; Hollenbeck and Saxton, 2005; Pilling et al., 2006; Glater et al., 2006). MAPs inhibit kinesin- and dynein-dependent mitochondrial movement along microtubules, by competing for binding to the microtubule surface (Ebneth et al., 1998; Trinczek et al., 1999; Seitz et al., 2002; Mandelkow et al., 2003; Jimenez-Mateos et al., 2006). The correlation between the proximo-distal gradient of phosphorylated tau, mitochondria labelling, and KIF5B pattern of distribution in PC12-ND6 cells (Figure 8), is consistent with the notion that phosphorylated tau permits anterograde movement of mitochondria by allowing the binding of KIF5B coupled to mitochondria on to microtubules (Doble and Woodgett, 2003; Tatebayashi et al., 2004; Mazanetz and Fischer, 2007). The preponderance of unphosphorylated tau in the distal portion of the developing axons of stage III PC12-ND6 cells would favour the release of KIF5B from microtubules, resulting in increase of free (unbound) KIF5B and dispersion of mitochondria, a mechanism that could deliver mitochondria to regions with high-energy demands, such as growth cones (Figure 8).
The fact that maximal increase in mitochondrial mass coincides with lamellipodia formation adds support to the notion that actin plays a critical role in local trafficking of mitochondria. Although most mitochondrial movement in neurons is microtubule-dependent (Ligon and Steward, 2000b), a growing body of evidence has highlighted actin-based mitochondrial local transport in growth cones and branching points, where microtubules are scarce (Morris and Hollenbeck, 1995, Ligon and Steward, 2000a; Langford 2002; Bridgman 2004; Hollenbeck and Saxton, 2005; Frederick and Shaw, 2007). Actin filaments are also necessary for the NGF-induced docking of mitochondria along axons of cultured sympathetic neurons (Chada and Hollenbeck, 2003, 2004). In the unicellular budding yeast system, proper mitochondrial inheritance along the mother-bud axis is ensured by actin-mediated motility (reviewed by Boldogh and Pon, 2006).
In conclusion, the present study documents for the first time that the neurogenic transcription factor NeuroD6 co-ordinates increase in mitochondrial mass concurrently with cytoskeletal remodelling at the very early stages of neuronal differentiation. Most importantly, our results shed some light on the transcriptional cues controlling the delicate balance between generating and maintaining the mitochondrial biomass, which is tailored to meet the specific energetic needs of differentiating neuronal cells. It is also conceivable that defects in acquiring proper mitochondrial mass at the outset of neural development may play a role in development of neurodegenerative diseases later in life.
Online data
ACKNOWLEDGEMENTS
We extend our special thanks to Dr Vittorio Gallo for his insightful comments on the manuscript. We thank Dr Anastas Popratiloff for his expertise in confocal fluorescence microscopy and Teresa Hawley for her technical advice on flow cytometry. This work will be part of a dissertation presented to the Graduate Program of Molecular Medicine, George Washington University Institute for Biomedical Sciences, in partial fulfilment of the requirements for the Ph.D. degree.
Footnotes
This work was supported by the National Institutes of Health, National Institutes of Neurological Disorders and Stroke [grant number R01-NS041391] to A.C. and by National Institute of Child Health and Development [grant number P30HD40677].
REFERENCES
- Angelastro JM, Klimaschewski L, Tang S, Vitolo OV, Weissman TA, Donlin LT, Shelanski ML, Greene LA. Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc Natl Acad Sci USA. 2000;97:10424–10429. doi: 10.1073/pnas.97.19.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelastro JM, Torocsik B, Greene LA. Nerve growth factor selectively regulates expression of transcripts encoding ribosomal proteins. BMC Neurosci. 2002;3:3. doi: 10.1186/1471-2202-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas PW. Neuronal polarity: microtubules strike back. Nat Cell Biol. 2002;4:E194–E195. doi: 10.1038/ncb0802-e194. [DOI] [PubMed] [Google Scholar]
- Beckerle MC. Identification of a new protein localized at sites of cell–substrate adhesion. J Cell Biol. 1986;103:1679–1687. doi: 10.1083/jcb.103.5.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckerle MC. Zyxin: zinc fingers at sites of cell adhesion. BioEssays. 1997;19:949–957. doi: 10.1002/bies.950191104. [DOI] [PubMed] [Google Scholar]
- Berman SB, Chen YB, Qi B, McCaffery JM, Rucker EB, 3rd, Goebbels S, Nave KA, Arnold BA, Jonas EA, Pineda FJ, Hardwick JM. Bcl-xL increases mitochondrial fission, fusion, and biomass in neurons. J Cell Biol. 2009;184:707–719. doi: 10.1083/jcb.200809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Actin–ATP hydrolysis is a major energy drain for neurons. J Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Bräuer AU, Nitsch R. Plasticity-related genes (PRGs/LRPs): a brain-specific class of lysophospholipid-modifying proteins. Biochim Biophys Acta. 2008;1781:595–600. doi: 10.1016/j.bbalip.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Bräuer AU, Savaskan NE, Kuhn H, Prehn S, Ninnemann O, Nitsch R. A new phospholipid phosphatase, PRG-1, is involved in axon growth and regenerative sprouting. Nat Neurosci. 2003;6:572–578. doi: 10.1038/nn1052. [DOI] [PubMed] [Google Scholar]
- Bridgman PC. Myosin-dependent transport in neurons. J Neurobiol. 2004;58:164–174. doi: 10.1002/neu.10320. [DOI] [PubMed] [Google Scholar]
- Cãceres A, Kosik KS. Inhibition of neurite polarity by tau antisense oligonucleotides in primary cerebellar neurons. Nature. 1990;343:461–463. doi: 10.1038/343461a0. [DOI] [PubMed] [Google Scholar]
- Cãceres A, Banker GA, Binder L. Immunocytochemical localization of tubulin and microtubule-associated protein 2 during the development of hippocampal neurons in culture. J Neurosci. 1986;6:714–722. doi: 10.1523/JNEUROSCI.06-03-00714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cãceres A, Potrebic S, Kosik KS. The effect of tau antisense oligonucleotides on neurite formation of cultured cerebellar macroneurons. J Neurosci. 1991;11:1515–1523. doi: 10.1523/JNEUROSCI.11-06-01515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Sheng ZH. Molecular motors and synaptic assembly. Neuroscientist. 2009;15:78–89. doi: 10.1177/1073858408329511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Gerwin C, Sheng ZH. Syntabulin-mediated anterograde transport of mitochondria along neuronal processes. J Cell Biol. 2005;170:959–969. doi: 10.1083/jcb.200506042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Pan PY, Sheng ZH. Syntabulin–kinesin-1 family member 5B-mediated axonal transport contributes to activity-dependent presynaptic assembly. J Neurosci. 2007;27:7284–7296. doi: 10.1523/JNEUROSCI.0731-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi RA, Aggeler R, Turina P, Wilkens S. Coupling between catalytic sites and the proton channel in F1Fo-type ATPases. Trends Biochem Sci. 1994;19:284–289. doi: 10.1016/0968-0004(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Carmona MC, Iglesias R, Obregon MJ, Darlington GJ, Villarroya F, Giralt M. Mitochondrial biogenesis and thyroid status maturation in brown fat require CCAAT/enhancer-binding protein alpha. J Biol Chem. 2002;277:21489–21498. doi: 10.1074/jbc.M201710200. [DOI] [PubMed] [Google Scholar]
- Cerveny KL, Studer SL, Jensen RE, Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev Cell. 2007;12:363–375. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Mitochondrial movement and positioning in axons: the role of growth factor signaling. J Exp Biol. 2003;206:1985–1992. doi: 10.1242/jeb.00263. [DOI] [PubMed] [Google Scholar]
- Chada SR, Hollenbeck PJ. Nerve growth factor signaling regulates motility and docking of axonal mitochondria. Curr Biol. 2004;14:1272–1276. doi: 10.1016/j.cub.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chang DT, Reynolds IJ. Mitochondrial trafficking and morphology in healthy and injured neurons. Prog Neurobiol. 2006;80:241–268. doi: 10.1016/j.pneurobio.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park DJ, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348:1472–1478. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Cordeau-Lossouarn L, Vayssiere JL, Larcher JC, Gros F, Croizat B. Mitochondrial maturation during neuronal differentiation in vivo and in vitro. Biol Cell. 1991;71:57–65. doi: 10.1016/0248-4900(91)90051-n. [DOI] [PubMed] [Google Scholar]
- Cowell RM, Blake KR, Russell JW. Localization of the transcriptional coactivator PGC-1α to GABAergic neurons during maturation of the rat brain. J Comp Neurol. 2007;502:1–18. doi: 10.1002/cne.21211. [DOI] [PubMed] [Google Scholar]
- Crawford AW, Beckerle MC. Purification and characterization of zyxin, an 82,000-dalton component of adherens junctions. J Biol Chem. 1991;266:5847–5853. [PubMed] [Google Scholar]
- da Silva JS, Dotti CG. Breaking the neuronal sphere: Regulation of the actin cytoskeleton in neuritogenesis. Nat Rev Neurosci. 2002;3:694–704. doi: 10.1038/nrn918. [DOI] [PubMed] [Google Scholar]
- Darin N, Oldfors A, Moslemi AR, Holme E, Tulinius M. The incidence of mitochondrial encephalomyopathies in childhood: clinical features and morphological, biochemical, and DNA anbormalities. Ann Neurol. 2001;49:377–383. [PubMed] [Google Scholar]
- Dehmelt L, Halpain S. Actin and microtubules in neurite initiation: are MAPs the missing link? J Neurobiol. 2004;58:18–33. doi: 10.1002/neu.10284. [DOI] [PubMed] [Google Scholar]
- Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, Gupton S, Van Veen JE, Furman C, Zhang J, Alberts AS, Mori S, Gertler FB. Filopodia are required for cortical neurite initiation. Nat Cell Biol. 2007;9:1347–1359. doi: 10.1038/ncb1654. [DOI] [PubMed] [Google Scholar]
- Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díez-Guerra FJ, Avila J. An increase in phosphorylation of microtubule-associated protein 2 accompanies dendrite extension during the differentiation of cultured hippocampal neurones. Eur J Biochem. 1995;227:68–77. doi: 10.1111/j.1432-1033.1995.tb20360.x. [DOI] [PubMed] [Google Scholar]
- Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA. The establishment of polarity by hippocampal neurons in culture. J Neurosci. 1988;8:1454–1468. doi: 10.1523/JNEUROSCI.08-04-01454.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees BE, Andrews KM, Beckerle MC. Molecular dissection of zyxin function reveals its involvement in cell motility. J Cell Biol. 1999;147:1549–1560. doi: 10.1083/jcb.147.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E. Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer's disease. J Cell Biol. 1998;143:777–794. doi: 10.1083/jcb.143.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120:4025–4034. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- Franko A, Mayer S, Thiel G, Mercy L, Arnould T, Hornig-Do HT, Wiesner RJ, Goffart S. CREB-1α is recruited to and mediates upregulation of the cytochrome c promoter during enhanced mitochondrial biogenesis accompanying skeletal muscle differentiation. Mol Cell Biol. 2008;28:2446–2459. doi: 10.1128/MCB.00980-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: The roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- Górska-Andrzejak J, Stowers RS, Borycz J, Kostyleva R, Schwarz TL, Meinertzhagen IA. Mitochondria are redistributed in Drosophila photoreceptors lacking milton, a kinesin-associated protein. J Comp Neurol. 2003;463:372–388. doi: 10.1002/cne.10750. [DOI] [PubMed] [Google Scholar]
- Greene LA, Farinelli SE, Cunningham ME, Park DS. Culture and experimental use of the PC12 rat pheochromocytoma cell line. In: Goslin K, Banker G, editors. Culturing Nerve Cells, 2nd edn. Cambridge, MA, U.S.A: MIT Press; 1998. pp. 161–187. [Google Scholar]
- Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- Hance N, Ekstrand MI, Trifunovic A. Mitochondrial DNA polymerase γ is essential for mammalian embryogenesis. Hum Mol Genet. 2005;14:1775–1783. doi: 10.1093/hmg/ddi184. [DOI] [PubMed] [Google Scholar]
- Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc Natl Acad Sci USA. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidemann SR. Cytoplasmic mechanisms of axonal and dendritic growth in neurons. Int Rev Cytol. 1996;165:235–296. doi: 10.1016/s0074-7696(08)62224-x. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. The pattern and mechanism of mitochondrial transport in axons. Front Biosci. 1996;1:d91–d102. doi: 10.2741/a118. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd DD, Stern M, Saxton WM. Mutation of the axonal transport motor kinesin enhances paralytic and suppresses shaker in Drosophila. Genetics. 1996;142:195–204. doi: 10.1093/genetics/142.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Jofuku A, Eura Y, Mihara K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem Biophys Res Commun. 2003;301:891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- Jakobs S, Martini N, Schauss AC, Egner A, Westermann B, Hell SW. Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p. J Cell Sci. 2003;116:2005–2014. doi: 10.1242/jcs.00423. [DOI] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- Jimenez-Mateos EM, Gonzalez-Billault C, Dawson HN, Vitek MP, Avila J. Role of MAP1B in axonal retrograde transport of mitochondria. Biochem J. 2006;397:53–59. doi: 10.1042/BJ20060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- Karbowski M, Arnoult D, Chen H, Chan DC, Smith CL, Youle RJ. Quantitation of mitochondrial dynamics by photolabeling of individual organelles shows that mitochondrial fusion is blocked during the bax activation phase of apoptosis. J Cell Biol. 2004;164:493–499. doi: 10.1083/jcb.200309082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford GM. Myosin-V, a versatile motor for short-range vesicle transport. Traffic. 2002;3:859–865. doi: 10.1034/j.1600-0854.2002.31202.x. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard DG, Ziff EB, Greene LA. Identification and characterization of mRNAs regulated by nerve growth factor in PC12 cells. Mol Cell Biol. 1987;7:3156–3167. doi: 10.1128/mcb.7.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CL, Wang TT, Luby-Phelps K, German DC. Mitochondria mass is low in mouse substantia nigra dopamine neurons: implications for Parkinson's disease. Exp Neurol. 2007;203:370–380. doi: 10.1016/j.expneurol.2006.08.015. [DOI] [PubMed] [Google Scholar]
- Liang HL, Wong-Riley MT. Activity-dependent regulation of nuclear respiratory factor-1, nuclear respiratory factor-2, and peroxisome proliferator-activated receptor γ coactivator-1 in neurons. NeuroReport. 2006;17:401–405. doi: 10.1097/01.wnr.0000204980.98876.11. [DOI] [PubMed] [Google Scholar]
- Liang HL, Ongwijitwat S, Wong-Riley MT. Bigenomic functional regulation of all 13 cytochrome c oxidase subunit transcripts in rat neurons in vitro and in vivo. Neuroscience. 2006;140:177–190. doi: 10.1016/j.neuroscience.2006.01.056. [DOI] [PubMed] [Google Scholar]
- Ligon LA, Steward O. Movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J Comp Neurol. 2000a;427:340–350. doi: 10.1002/1096-9861(20001120)427:3<340::aid-cne2>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Ligon LA, Steward O. Role of microtubules and actin filaments in the movement of mitochondria in the axons and dendrites of cultured hippocampal neurons. J Comp Neurol. 2000b;427:351–361. doi: 10.1002/1096-9861(20001120)427:3<351::aid-cne3>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Stamer K, Vogel R, Thies E, Mandelkow E. Clogging of axons by tau, inhibition of axonal traffic and starvation of synapses. Neurobiol Aging. 2003;24:1079–1085. doi: 10.1016/j.neurobiolaging.2003.04.007. [DOI] [PubMed] [Google Scholar]
- Mandell JW, Banker GA. A spatial gradient of tau protein phosphorylation in nascent axons. J Neurosci. 1996;16:5727–5740. doi: 10.1523/JNEUROSCI.16-18-05727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus A. Microtubule-associated proteins: their potential role in determining neuronal morphology. Annu Rev Neurosci. 1988;11:29–44. doi: 10.1146/annurev.ne.11.030188.000333. [DOI] [PubMed] [Google Scholar]
- Mazanetz MP, Fischer PM. Untangling tau hyperphosphorylation in drug design for neurodegenerative diseases. Nat Rev Drug Discov. 2007;6:464–479. doi: 10.1038/nrd2111. [DOI] [PubMed] [Google Scholar]
- Meng H, Liang HL, Wong-Riley M. Quantitative immuno-electron microscopic analysis of depolarization-induced expression of PGC-1α in cultured rat visual cortical neurons. Brain Res. 2007;1175:10–16. doi: 10.1016/j.brainres.2007.07.063. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. The regulation of bidirectional mitochondrial transport is coordinated with axonal outgrowth. J Cell Sci. 1993;104:917–927. doi: 10.1242/jcs.104.3.917. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozdy AD, Shaw JM. A fuzzy mitochondrial fusion apparatus comes into focus. Nat Rev Mol Cell Biol. 2003;4:468–478. doi: 10.1038/nrm1125. [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Marshall WF, Straight A, Murray A, Sedat JW, Walter P. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Mol Biol Cell. 1997;8:1233–1242. doi: 10.1091/mbc.8.7.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, Emorine LJ, Descoins E, Pelloquin L, Brichese L, Gas N, Guillou E, Delettre C, Valette A, Hamel CP, Ducommun B, Lenaers G, Belenguer P. The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 2002;523:171–176. doi: 10.1016/s0014-5793(02)02985-x. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Pendergrass W, Wolf N, Poot M. Efficacy of MitoTracker green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 2004;61:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- Pennypacker K, Fischer I, Levitt P. Early in vitro genesis and differentiation of axons and dendrites by hippocampal neurons analysed quantitatively with neurofilament-H and microtubule-associated protein 2 antibodies. Exp Neurol. 1991;111:25–35. doi: 10.1016/0014-4886(91)90047-g. [DOI] [PubMed] [Google Scholar]
- Pereira AJ, Dalby B, Stewart RJ, Doxsey SJ, Goldstein LS. Mitochondrial association of a plus end-directed microtubule motor expressed during mitosis in Drosophila. J Cell Biol. 1997;136:1081–1090. doi: 10.1083/jcb.136.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povlishock JT. The fine structure of the axons and growth cones of the human fetal cerebral cortex. Brain Res. 1976;114:379–389. doi: 10.1016/0006-8993(76)90961-6. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rickard JE, Kreis TE. CLIPs for organelle-microtubule interactions. Trends Cell Biol. 1996;6:178–183. doi: 10.1016/0962-8924(96)10017-9. [DOI] [PubMed] [Google Scholar]
- Ruthel G, Hollenbeck PJ. Response of mitochondrial traffic to axon determination and differential branch growth. J Neurosci. 2003;23:8618–8624. doi: 10.1523/JNEUROSCI.23-24-08618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Satoh M, Hamamoto T, Seo N, Kagawa Y, Endo H. Differential sublocalization of the dynamin-related protein OPA1 isoforms in mitochondria. Biochem Biophys Res Commun. 2003;300:482–493. doi: 10.1016/s0006-291x(02)02874-7. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schauss AC, Bewersdorf J, Jakobs S. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J Cell Sci. 2006;119:3098–3106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab MH, Druffel-Augustin S, Gass P, Jung M, Klugmann M, Bartholomae A, Rossner MJ, Nave KA. Neuronal basic helix–loop–helix proteins (NEX, neuroD, NDRF): spatiotemporal expression and targeted disruption of the NEX gene in transgenic mice. J Neurosci. 1998;18:1408–1418. doi: 10.1523/JNEUROSCI.18-04-01408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab MH, Bartholomae A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, Nave KA. Neuronal basic helix–loop–helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Cassidy-Stone A, Meeusen SL, Nunnari J. Staying in aerobic shape: how the structural integrity of mitochondria and mitochondrial DNA is maintained. Curr Opin Cell Biol. 2003;15:482–488. doi: 10.1016/s0955-0674(03)00070-x. [DOI] [PubMed] [Google Scholar]
- Seitz A, Kojima H, Oiwa K, Mandelkow EM, Song YH, Mandelkow E. Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J. 2002;21:4896–4905. doi: 10.1093/emboj/cdf503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John JC, Ramalho-Santos J, Gray HL, Petrosko P, Rawe VY, Navara CS, Simerly CR, Schatten GP. The expression of mitochondrial DNA transcription factors during early cardiomyocyte in vitro differentiation from human embryonic stem cells. Cloning Stem Cells. 2005;7:141–153. doi: 10.1089/clo.2005.7.141. [DOI] [PubMed] [Google Scholar]
- Stowers RS, Megeath LJ, Gorska-Andrzejak J, Meinertzhagen IA, Schwarz TL. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron. 2002;36:1063–1077. doi: 10.1016/s0896-6273(02)01094-2. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Tatebayashi Y, Haque N, Tung YC, Iqbal K, Grundke-Iqbal I. Role of tau phosphorylation by glycogen synthase kinase-3β in the regulation of organelle transport. J Cell Sci. 2004;117:1653–1663. doi: 10.1242/jcs.01018. [DOI] [PubMed] [Google Scholar]
- Trinczek B, Ebneth A, Mandelkow EM, Mandelkow E. Tau regulates the attachment/detachment but not the speed of motors in microtubule-dependent transport of single vesicles and organelles. J Cell Sci. 1999;112:2355–2367. doi: 10.1242/jcs.112.14.2355. [DOI] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. Constitutive overexpression of the basic helix–loop–helix Nex1/MATH-2 transcription factor promotes neuronal differentiation of PC12 cells and neurite regeneration. J Neurosci Res. 2002;67:235–245. doi: 10.1002/jnr.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. Expression profiling upon Nex1/MATH-2-mediated neuritogenesis in PC12 cells and its implication in regeneration. J Neurochem. 2004;91:1332–1343. doi: 10.1111/j.1471-4159.2004.02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. The basic helix–loop–helix transcription factor nex-1/Math-2 promotes neuronal survival of PC12 cells by modulating the dynamic expression of anti-apoptotic and cell cycle regulators. J Neurochem. 2005;92:585–596. doi: 10.1111/j.1471-4159.2004.02886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Baxter KK, Chiaramello A. NeuroD6 genomic signature bridging neuronal differentiation to survival via the molecular chaperone network. J Neurosci Res. 2009 doi: 10.1002/jnr.22182. doi: 10.1002/jnr.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa L, Diez-Guerra FJ, Avila J, Diaz-Nido J. Localization of differentially phosphorylated isoforms of microtubule-associated protein 1B in cultured rat hippocampal neurons. Neuroscience. 1994;61:211–223. doi: 10.1016/0306-4522(94)90225-9. [DOI] [PubMed] [Google Scholar]
- Vayssiere JL, Cordeau-Lossouarn L, Larcher JC, Basseville M, Gros F, Croizat B. Participation of the mitochondrial genome in the differentiation of neuroblastoma cells. In Vitro Cell Dev Biol. 1992;28A:763–772. doi: 10.1007/BF02631065. [DOI] [PubMed] [Google Scholar]
- Verburg J, Hollenbeck PJ. Mitochondrial membrane potential in axons increases with local nerve growth factor or semaphorin signaling. J Neurosci. 2008;28:8306–8315. doi: 10.1523/JNEUROSCI.2614-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1α transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci USA. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Shida Y, Takahashi K, Tanioka T, Nakano Y, Tobe T, Yamada M. Prg1 is regulated by the basic helix–loop–helix transcription factor Math2. J Neurochem. 2008;106:2375–2384. doi: 10.1111/j.1471-4159.2008.05579.x. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Liang HL, Wong-Riley MT. Activity-dependent transcriptional regulation of nuclear respiratory factor-1 in cultured rat visual cortical neurons. Neuroscience. 2006;141:1181–1192. doi: 10.1016/j.neuroscience.2006.04.063. [DOI] [PubMed] [Google Scholar]
- Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–5420. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–2172. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- Zhang FX, Pan W, Hutchins JB. Phosphorylation of F1Fo ATPase δ-subunit is regulated by platelet-derived growth factor in mouse cortical neurons in vitro. J Neurochem. 1995;65:2812–2815. doi: 10.1046/j.1471-4159.1995.65062812.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.