Abstract
The c-Myb transcription factor is required for normal adult hematopoiesis. However, the embryonic lethality of Myb null mutations has been an impediment to identifying roles for c-Myb during lymphocyte development. We have used tissue-specific inactivation of the Myb locus in early progenitor cells to demonstrate that c-Myb is absolutely required for the differentiation of CD19+ B-lineage cells and B cell differentiation is profoundly blocked beyond the pre-pro-B cell stage in Mybf/f Mb1-cre mice. We demonstrate that c-Myb is required for the intrinsic survival of CD19+ pro-B cells as well as the proper expression of the α-chain of the IL-7 receptor (CD127) and Ebf1. However, survival of c-Myb deficient CD19+ pro-B cells cannot be rescued by transduction with CD127 producing retrovirus suggesting that c-Myb controls a survival pathway independent of CD127. Furthermore, c-Myb deficient progenitor cells inefficiently generate CD19+ B-lineage cells during stromal cell culture but this process can be partially rescued with exogenous Ebf1. Thus, c-Myb does not appear to be required for commitment to B cell differentiation but is crucial for B cell differentiation to the CD19+ pro-B cell stage as well as survival of CD19+ pro-B cells. Surprisingly, forced c-Myb expression in LMPPs favors differentiation toward the myeloid lineage, suggesting that proper c-Myb expression is crucial for B-lineage development.
Introduction
B cell development is characterized by the sequential expression of cell surface markers and the ordered rearrangement of immunoglobulin heavy and light chain gene segments (1). Hematopoietic stem cells give rise to progenitor B cells (pro–B cells) that undergo rearrangement of the immunoglobulin heavy–chain (H–chain) locus. Successful H–chain rearrangement leads to expression of μH–chain, which associates with surrogate light chain and the signaling molecules Ig–α and Ig–β to form the pre–BCR complex. Signaling through the pre–BCR results in clonal expansion of cytoplasmic μ+ cells and differentiation to the small pre–B cell stage of differentiation. Small pre–B cells undergo rearrangement at the immunoglobulin light chain (L–chain) loci. Pre–B cells that produce L–chain, paired with the μ heavy chain, express membrane bound IgM (mIgM) and are referred to as immature B cells.
The pathway from HSC to committed B cell progenitor involves the sequential generation of multipotent progenitor cells (MPP) that can give rise to each hematopoietic lineage and lymphoid-primed multipotent progenitors (LMPPs) (2). LMPPs can give rise to lymphocytes, granulocytes and macrophage but have generally lost the ability to generate erythrocytes and megakaryocytes (3,4). HSCs, MPPs and LMPPs are distinguished by increasing expression of Flt3 (3). LMPPs serve as precursors for common lymphoid progenitors (CLPs), which express the interleukin-7 receptor (IL-7R) and retain broad differentiation potential for the lymphocytic lineage but not granulocytes or macrophage (5,6). CLPs give rise to a population of cells referred to as pre-pro-B cells, Fraction A or CLP2 cells that express B220 on the surface and represent the first clear B-lineage progenitor (7-10). Expression of the transcription factors PU.1 and Ikaros in HSC direct development away from the myeloid lineage and favors lymphocyte development (11,12). Deficiency in either PU.1 or Ikaros results in a block to B cell development during transition from the MPP to CLP stage as well as the lack of expression of B-lineage associated genes. Ikaros is crucial for expression of Flt3 in LMPPs (13-15) and likely plays a further role in differentiation of LMPPs and subsequent specification of the B cell fate (16). The transcription factors E2A, Ebf1 and Pax5 are required for specification of the B cell fate and transition from the CLP stage to the B-lineage committed CD19+ pro-B cell (17,18). Signals through Flt3 and PU.1 appear to upregulate expression of the IL7R (19,20). Though signaling through the IL-7R is not strictly required for formation of CLPs or pre-pro-B cells (21,22) it appears to be required for maintaining B-lineage potential in CLPs and pre-pro-B cells augmenting expression of Ebf1 (22-24). E2A and Ebf1 lead to induction of Pax-5, which is required for expression of CD19 and maintaining commitment to the B cell lineage (25-28).
The Myb protooncogene (c-Myb) encodes a nuclear, DNA-binding protein that functions as both a transcription activator and repressor (29,30). Expression of c-Myb has been primarily associated with hematopoietic tissue, although expression of Myb mRNA has been reported in other tissues (30). Hematopoietic stem cells (HSC) and progenitors of each hematopoietic lineage express c–Myb and the down–regulation of Myb expression is associated with hematopoietic maturation (31,32). The pattern of Myb expression suggested that it plays a significant role in regulating hematopoiesis and this has been supported experimentally. Myb null embryos develop normally to day fourteen after which they die with severely disrupted patterns of erythroid and myeloid development (33). However, gaining insight into lineage specific roles for c-Myb has been difficult due to the embryonic lethality of null Myb alleles. Normal B cell progenitors and mature B cells contain Myb mRNA and c–Myb protein suggesting that c-Myb may play a role in B cell differentiation (34,35). A role for c–Myb during B cell development was further supported by experiments that demonstrated Myb null embryonic stem cells were unable to give rise to B–lineage cells in the Rag1−/− blastocyst complementation system (36). In addition, several hypomorphic mutants have been reported with reduced numbers of peripheral B cells and an apparent block in transition from the pro-B cell to the pre-B cell stage of differentiation (37-39). However, defective B cell development in these models could be due to defects in HSC or very early progenitor cells.
We have previously used conditional inactivation of the Myb locus to demonstrate that c-Myb is crucial for transition from the pro-B cell to the pre-B cell stage of development and the maintenance of the pre-B cell compartment (34). Inactivation of the Myb locus in this study took place in pro-B cells, making clear that c-Myb was important in B-lineage precursors. The pro-B to pre-B cell transition is particularly sensitive to the amount of c-Myb produced as Myb+/− heterozygotes are partially blocked at this stage of differentiation (40). Furthermore, pro-B cell to pre-B cell transition is disrupted by transgenic expression of the MiR-150 miRNA, which targets the Myb mRNA (40). Thus, different stages of B cell development appear to be exquisitely sensitive to differences in the amount of c-Myb produced.
We have now used cell specific inactivation of the Myb locus in very early B-lineage progenitors using the Mb1-cre allele (41). The Mb1-cre allele leads to inactivation of a loxP targeted Myb locus (Mybf) beginning in the pre-pro-B cell stage and is essentially complete in CD19+ pro-B cells. We demonstrate that c-Myb is absolutely required for B cell development and Mybf/f Mb1-cre progenitors fail to give rise to CD19+ pro-B cells. We demonstrate that c-Myb plays a critical role in the survival of CD19+ pro-B cells as well as proper expression of the α-chain of the IL-7R (CD127) and Ebf1. However, the survival defect in c-Myb deficient CD19+ pro-B cells appears to be independent of altered CD127 expression. Furthermore, development of CD19+ B-lineage cells can be partially rescued from Mybf/f Mb1-cre LMPPs by exogenously supplied Ebf1, suggesting that c-Myb regulates B cell development in part by controlling IL-7R driven expression of Ebf1 as well as the survival of CD19+ pro-B cells. Finally, we find that increased c-Myb expression in normal LMPPs can skew differentiation away from the B cell lineage toward development of myeloid cells.
Materials and Methods
Mice
Mybf/f, Mybf/+, Myb+/−, CD19-cre and Mb1-cre mice have been described (41-43). Rag2−/− mice (Taconic) were bred at the University of Virginia. Mice were 8 – 12 weeks old when used for experiments and were housed in a barrier mouse facility at the University of Virginia.
Antibodies and Flow cytometry
Single-cell suspensions were prepared from 8 - 12-week-old mice and 2×106 cells were stained with optimal amounts of fluorochrome-conjugated antibodies as previously described (34). Cells were subsequently analyzed on a FACSCalibur (BD Immunocytometry Systems, San Jose, CA) or CyAn ADP (DakoCytomation, Glostrup, Denmark). For intracellular staining, cell surface staining was performed first, followed by fixation and permeabilization (BD Pharmigen) and intracellular staining. For cell cycle analysis, cell surface staining was performed first followed by incubation with DRAQ5 (Biostatus Limited, Shepshed, Leicestershire, UK) for 20 minutes at 37°C. Flow cytometric data were analyzed using FlowJo (Tree Star, Ashland, OR). Cell sorting was performed on a FACSVantage SE Turbo Sorter with DIVA option (BD Immunocytometry Systems). Antibodies and reagents were purchased as follows. CalTag (Burlingame, CA): anti-CD25-PE (PC61 5.3). BD Pharmigen (San Diego, CA): anti-B220-biotin (RA3-6B2), anti-B220-PE-Texas Red (RA3-6B2), anti-CD3e-biotin (145-2C11), anti-CD5-PE (53-7.3), anti-CD11b-biotin (M1/70), anti-CD43-PE (S7), anti-IgM-FITC (R6-60.2), anti-Gr1-biotin (RB6-8C5), anti-Ly6C-biotin (AL-21) and anti-Ter119-biotin (Ter-119). eBioscience (San Diego, CA): anti-B220-APC (RA3-6B2), anti-B220-FITC (RA3-6B2), anti-CD3e-FITC (145-2C11), anti-CD4-biotin (GK1.5), anti-CD4-PE (GK1.5, L3T4), anti-CD8-biotin (53-6.7), anti-CD8-FITC (53-6.7), anti-CD11b-FITC (M1/70), anti-CD19-FITC (MB19-1), anti-CD19-PECy5.5 (6D5), anti-CD24-FITC (30-F1), anti-CD44-FITC (1M7), anti-CD117-APC (2B8), anti-CD117-APC-AlexaFluor750 (2B8), anti-CD127-PE (A7R34), anti-CD127-APC-AlexaFluor750 (A7R34), anti-CD135-biotin (A2F10), anti-CD135-PE (A2F10), anti-Ly6C-FITC (Al-21), anti-Gr1-FITC (RB6-8C5), anti-NK1.1-PECy7 (PK136), anti-PanNK-biotin (DX5), anti-Sca1-PECy5.5 (D7), anti-Ter119-FITC (TER-119), streptavidin-APC, streptavidin-APCCy7, and streptavidin-PECy7. Cell Signaling (Danvers, MA): rabbit anti-mouse Bcl-xL (54H6). Rockland (Gilbertsville, PA): rabbit anti-mouse Mcl1. Jackson ImmunoResearch (West Grove, PA): anti-rabbit IgG-APC. Sigma-Aldrich (St. Louis, MO): 4′,6-diamidino-2-phenylindole (DAPI) and propidium iodide (PI): Invitrogen/Molecular Probes (Carlsbad, CA): 7-amino-actinomycin D (7-AAD).
PCR
HSC, LMPP, CLP, pre-pro-B cells, pro-B cells, pre-B cells, immature B cells, and mature B cells from bone marrow as well as DN, DP, CD4 SP, and CD8 SP thymocytes, and CD4+ T-cells, CD8+ T-cells, and CD19+ B-cells from spleen were electronically sorted and genomic DNA was prepared using the PicoPure DNA Extraction Kit (Arcturus, Mt. View, CA). PCR to determine deletion at the Myb locus was performed as previously described (42).
Semiquantitative and Quantitative RT-PCR
HSC, LMPP, CLP, pre-pro-B cells, pro-B cells, pre-B cells, immature B cells, and mature B cells from C57BL/6 mice were electronically sorted and total cellular RNA was isolated using RNeasy Mini Kits (Qiagen, Valencia, CA). Mybf/f Rag2−/− CD19+ pro-B cells transduced with MSCV-IRES-tNGFR and MSCV-Cre-IRES-tNGFR were positively selected using anti-tNGFR-biotin followed by streptavidin magnetic beads (Miltenyi) and total cellular RNA was isolated using the PicoPure RNA Extraction Kit (Arcturus). Contaminating genomic DNA was removed by treatment with RNase-free DNase I, and cDNA was prepared with SuperScript II First-Strand Synthesis System (Invitrogen). For semiquantitative RT-PCR, each sample was normalized by PCR for ActB mRNA content, and analysis of Ebf1 and Il7r (CD127) mRNA was performed on serially diluted cDNA (1:3) as previously described (44). RT-PCR products were fractionated on agarose gels and quantified using ImageQuant TL 2005 software. Semiquantitative RT-PCR to measure expression of Bim, Bcl2, BclxL, and Mcl1 mRNA was performed on serially diluted cDNA (1:3) as previously described (22,45,46). RT-PCR products were fractionated on agarose gels and visualized by ethidium bromide staining. Quantitative RT-PCR was performed on cDNA with Titanium Taq Polymerase (BD/Clonetech, Palo Alto, CA) with 1X SYBR Green (Molecular Probes) and 0.4 μM of the primer set of interest in 25 μl reaction mixtures in an OptiCon DNA Engine (MJ Research, Waltham, MA). Conditions for quantitative RT-PCR were as follows: 95°C for 3 minutes, then 40 cycles of 95°C for 40 seconds, 66°C for 20 seconds, and 72°C for 30 seconds, followed by an extension 72°C for 1 minute. Melting curve analysis was then performed to ensure equivalent and appropriate melting temperatures. Each sample was normalized relative to the expression of Hprt1 (encoding hypoxanthine guanine phosphoribosyl transferase). Primers used were as followed: Myb (forward, 5’-AACGACCTGAAGGGACAGCA-3′; reverse, 5′-TGGCATGGTGTTCTCCCAAA-3′) and Hprt1 (forward, 5′-TGCCGAGGATTTGGAAAAAGTG-3′; reverse, 5′-CACAGAGGGCCACAATGTGATG-3′).
Retroviral vectors
The retroviral vectors pMIG-R1, pMIG-EBF, pMIG-IL7Rα and pMSCV-IRES-tNGFR have been previously described (44,47,48). The retroviral vector pMIG-BclxL was a kind gift from Dr. Thomas J. Braciale (University of Virginia, Charlottesville, VA). To generate pMIG-cMYB a cDNA encoding a murine c-Myb with an influenza hemaglutanin tag at the amino terminal end was cloned into the BamHI/BglII site of pMIG-R1 (supplemental Fig. S1). To generate pMIG-Cre, Cre cDNA was isolated from pPGK-Cre-bpA and cloned into the XhoI/EcoRI site of pMIG-R1. To generate ptNGFR-Cre, Cre cDNA was cloned into the BglII/EcoRI site of pMSCV-IRES-tNGFR. Retroviral supernatants were generated by transient transfection of HEK-293T cells and titered on NIH-3T3 cells by flow cytometry as previously described (49).
Cell culture
Pro-B cells from Myb+/+Rag2−/−, Myb+/− Rag2-/-, and Mybf/f CD19-cre Rag2−/− mice were positively selected from bone marrow using CD19-labelled magnetic beads (Miltenyi). Cells were cultured in Opti-MEM supplemented with 10% (vol/vol) FBS (Gibco), penicillin-streptomycin (100 U/ml), L-glutamine (2 mM) and 2-mercaptoethanol (50 μM) in flat bottom 96-well plates, and total cells per well were analyzed 24 hours, 48 hours, and 72 hours later by trypan blue exclusion. For transduction of Rag2−/− CD19+ pro-B cells, cells were selected on CD19-labelled magnetic beads, cultured for 24 hours in Opti-MEM supplemented with 15% (vol/vol) FBS (Gibco), recombinant IL-7 (5 ng/ml, Peprotech), penicillin-streptomycin (100 U/ml), L-glutamine (2 mM) and 2-mercaptoethanol (50 μM) and transduced with retroviral vectors as previously described (50). Following transduction, pro-B cells were cultured in Opti-MEM with 10% (vol/vol) FBS (Gibco), penicillin-streptomycin (100 U/ml), L-glutamine (2 mM) and 2-mercaptoethanol (50 μM) with or without IL-7 (5 ng/ml) and were analyzed 24 hours, 36 hours, and 48 hours later by flow cytometry.
For transduction of LMPPs, LMPPs, defined as Lin- CD117+ Sca1+ CD135hi, were electronically sorted from lineage-depleted bone marrow from Mybf/f and Mybf/f Mb1-cre mice and transduced as previously described (16). Following two successive rounds of retroviral transduction, cells were placed on OP9 stromal cells in Opti-MEM with 2.5% (vol/vol) FBS, penicillin-streptomycin (100 U/ml), L-glutamine (2 mM) and 2-mercaptoethanol (50 μM) supplemented with SCF (10 ng/ml, Peprotech), Flt3L (10 ng/ml, Peprotech), and IL-7 (5 ng/ml, Peprotech) and analyzed 10-14 days later by flow cytometry. Day 14.5 fetal liver progenitors were isolated following anti-CD24 complement-mediated lysis (Cedarlane, Hornby, Ontario, Canada) as previously described (51). Following isolation, fetal liver progenitors were transduced and analyzed as described above.
Results
Expression of c-Myb mRNA in bone marrow B-lineage progenitor cells
We have previously used Mybf/f CD19-cre mice to demonstrate that c-Myb is crucial for B cell development during transition from the pro-B cell to pre-B cell stage as well as for the maintenance of the pre-B cell compartment (34). However, while pre-B cells contain the greatest amount of Myb mRNA among bone marrow B-lineage subsets, HSC and each progenitor stage of B cell development contain 10-fold more Myb mRNA than peripheral B cells suggesting that c-Myb may be important for B-lymphopoiesis prior to the pro-B to pre-B cell transition (Fig. 1). CD19 expression initiates in Fraction B pro-B cells but deletion efficiency mediated by CD19-cre is incomplete in pro-B cells and is not detectable at earlier stages of B cell development limiting its utility to assess c-Myb function earlier during B cell development (34,41) (T. Bender, unpublished data). To circumvent this problem, we crossed Mybf/f mice to the Mb1-cre mouse strain to produce Mybf/+ Mb1-cre mice. The Mb1-cre allele was created by replacing exons 2 and 3 of the Cd79a (Mb1) locus with a mammalian codon optimized Cre recombinase encoding cDNA, which has been reported to efficiently delete loxP targeted DNA in CD19+ bone marrow B cells (41). To determine if Mb1-cre also is able to delete in B-lineage progenitors prior to the pro-B cell stage, we electronically sorted HSC, LMPP, CLP and pre-pro-B cells as well as pro-B, pre-B, immature B and recirculating B cells from Mybf/+ Mb1-cre mice and assessed the deletion efficiency of the Mybf allele by PCR (Fig. 2). Cre mediated deletion of the floxed Myb locus was barely detectable in HSC, LMPP and CLPs. However, deletion of the floxed Myb allele was approximately 40% in pre-pro-B cells and was essentially complete in CD19+ pro-B cells and all subsequent stages of B cell development. Comparable results were obtained when the Mb1-cre allele was crossed onto the ROSA-floxed EYFP reporter mouse (not shown). Thus, Mb1-cre provides earlier and more complete deletion of the Mybf allele than CD19-cre.
FIGURE 1.
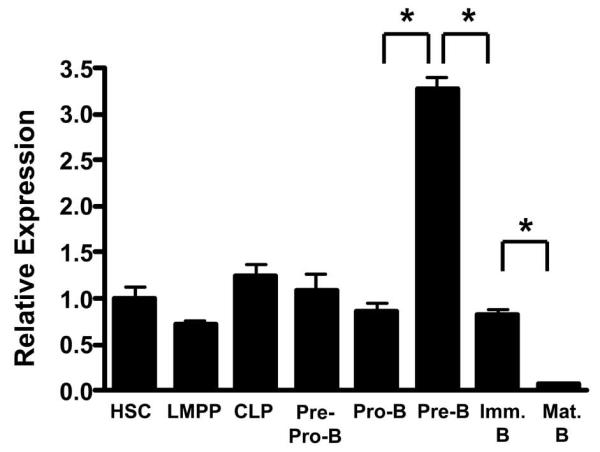
Expression of Myb mRNA in bone marrow B-lineage progenitors. Total cellular RNA was isolated from HSC (Lin− CD117+ Sca1+ CD135−), LMPP (Lin− CD117+ Sca1+ CD135hi), CLP (Lin− CD19− B220− CD127+ CD135+), pre-pro-B cell (Ly6C− NK1.1− B220+ CD43+ CD19−), pro-B cell (Ly6C− NK1.1− B220+ CD43+ CD19+), pre-B cell (B220+ CD43−), immature B cell (B220+ IgM+), and mature B cell (B220hi IgM+) subsets isolated from C57BL/6J bone marrow. Myb expression was analyzed by quantitative RT-PCR and normalized to the expression of HPRT. * p<0.01
FIGURE 2.
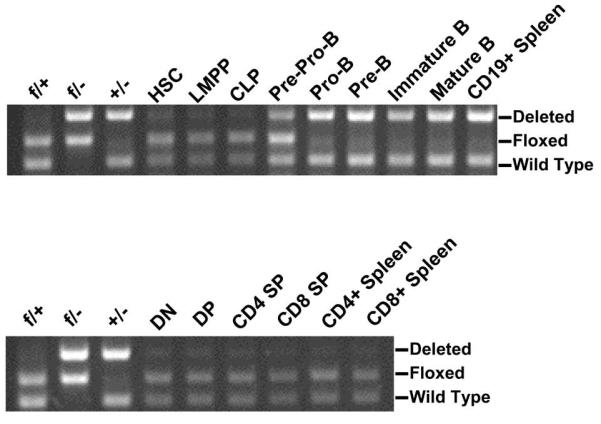
Deletion efficiency of the floxed Myb allele in Mybf/+ Mb1-cre mice. Deletion at the Myb locus was analyzed in B-lineage subsets from the bone marrow as described in Figure 1 as well as CD19+ splenocytes (top panel) and thymic DN (CD4− CD8−), DP (CD4+ CD8+), CD4 SP (CD4+ CD8−), and CD8 SP (CD4− CD8+) subsets as well as CD4+ and CD8+ splenocytes (bottom panel). Control PCR from Mybf/+ (f/+), Mybf/− (f/−) and Myb+/− (+/−) splenocytes was included as markers.
c-Myb is required for the accumulation of CD19+ B cells in peripheral lymphoid tissue and bone marrow
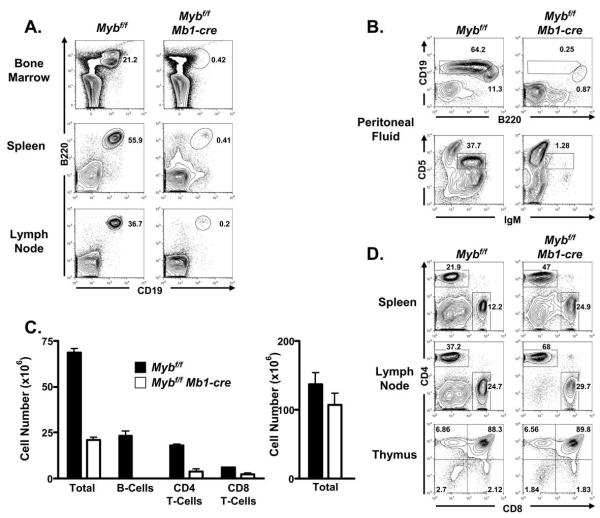
Mb1-cre mice were crossed with Mybf/f to produce Mybf/f Mb1-cre mice. We initially compared the proportion of CD19+ B cells in the spleen, inguinal lymph nodes and bone marrow of 8 – 12 week old Mybf/f Mb1-cre and control mice (controls are both Mybf/f and Myb+/+ Mb1-cre mice) by flow cytometry and found few, if any, B-lineage cells in each compartment (Fig. 3A). Similarly, the proportion of B1 B cells was severely reduced in peritoneal fluid from Mybf/f Mb1-cre compared to controls (Fig. 3B). The absolute number of B cells in the spleen was reduced >99% (Fig. 3C). In contrast, the proportion of CD4 and CD8 T cells in the spleen and inguinal lymph nodes was increased although the absolute number of splenic CD4 and CD8 T cells was reduced by approximately 70% and 50% respectively (Figs. 3C and 3D). However, the absolute number of thymocytes was not statistically different in thymi from Mybf/f Mb1-cre and control mice nor did we detect a difference in the distribution of CD4 and CD8 subsets in the thymi of mutant and control mice (Figs. 3C and 3D). Cre mediated deletion was extremely low in thymocytes (Fig. 2) and the reduced number of T cells detected in peripheral lymphoid tissues is consistent with the decreased number of peripheral T cells detected in other mouse models with severe defects in B-lymphopoiesis that do not alter thymocyte development (52). Thus, in the absence of c-Myb, CD19+ B cells fail to accumulate in peripheral lymphoid tissue.
FIGURE 3.
Lack of peripheral B cells in Mybf/f Mb1-cre mice. A, Bone marrow, spleen, and inguinal lymph node cells were analyzed for surface expression of B220 and CD19 by flow cytometry. Viable cells were identified as PI−. Number next to gates is percent of total PI− cells. Data is representative of at least 10 mutant and control mice. B, Peritoneal fluid cells were analyzed for surface expression of B220 and CD19 (top panel) or CD5 and IgM (bottom panel) to detect B1a and B1b or B1a B-cells defined as B220lo CD19+ and CD5+ IgM+, respectively. Viable cells were identified as PI−. Data is representative of 3 mutant and control mice of each strain. C, Absolute number of total nucleated cells, B220+ CD19+ B-cells, CD4+ T-cells, and CD8+ T-cells from Mybf/f and Mybf/f Mb1-cre spleen (left panel). Total splenic cellularity, CD4+ T-cells and CD8+ T-cells were significantly reduced in Mybf/f Mb1-cre mice compared to controls (p<0.05). No statistically significant difference in total thymic cellularity was detected between Mybf/f and Mybf/f Mb1-cre mice (right panel). D, Spleen, inguinal lymph node and thymus cells were analyzed for surface expression of CD4 and CD8 by flow cytometry. Viable cells were identified as PI−.
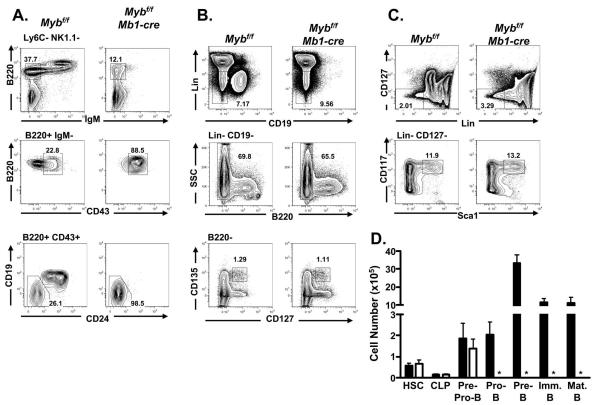
To better understand the drastic decrease between the number of peripheral B cells detected in Mybf/f Mb1-cre and control mice we examined the distribution of bone marrow B-lineage subsets by flow cytometry (Fig. 4A). Few, if any, B220+ IgM+ B cells were detected in the bone marrow of Mybf/f Mb1-cre mice compared to controls. We previously identified a partial block during the pro-B to pre-B cell transition in Mybf/f CD19-cre mice that was manifested in part by a decreased number of Fraction C’ pro-B cells as defined by Hardy and colleagues (34,53). However, when we gated on B220+ IgM- cells and examined the distribution of CD43 surface expression on cells from Mybf/f Mb1-cre and control bone marrow we detected few, if any, B220+ mIgM- CD43- pre-B cells in Mybf/f Mb1-cre bone marrow. Furthermore, we failed to detect CD19+ pro-B cells in Mybf/f Mb1-cre bone marrow (Fig. 4A). In contrast, we did not detect a statistically significant difference between the number of pre-pro-B cells (defined as B220+ CD43+ Ly6C- Nk1.1- CD19- CD24-) in bone marrow from Mybf/f Mb1-cre compared to controls (Figs. 4A and 4D). No difference was detected between the number of HSCs, LMPPs or CLPs in bone marrow from Mybf/f Mb1-cre mice compared to controls (Figs. 4B, 4C and 4D) , which is in agreement with the lack of deletion detected at the floxed Myb locus in these cells (Fig. 2). Taken together, these results suggest that c-Myb is required for differentiation to the CD19+ pro-B cell compartment, survival of the CD19+ pro-B cell compartment or both.
FIGURE 4.
B-cell development in Mybf/f Mb1-cre mice. A, Bone marrow from Mybf/f Mb1-cre and Mybf/f control mice was stained for surface expression of Ly6C, NK1.1, B220, IgM, CD43, CD19, and CD24. Viable cells were identified as DAPI−. Top tier represents B220 versus IgM after gating out Ly6C+ and NK1.1+ cells. Mature B-cells were defined as Ly6C− NK1.1− B220hi IgM+ and immature B-cells were defined as Ly6C− NK1.1− B220+ IgM+. Middle tier represents B220 versus CD43 through a B220+ IgM− gate. Pre-B cells were defined as Ly6C− NK1.1− B220+ IgM− CD43−. Bottom tier represents CD19 versus CD24 through B220+ CD43+ gate. Pro-B cells were defined as Ly6C− NK1.1− B220+ IgM− CD43+ CD19+ CD24+ and pre-pro-B cells were defined as Ly6C− NK1.1− B220+ IgM− CD43+ CD19− CD24−. Data is representative of at least 10 mutant and control mice. B, Bone marrow was stained for surface expression of lineage markers [CD3e, CD11b, Ly6C, Ter119], CD19, B220, CD135, and CD127. Viable cells were identified as DAPI−. CLPs were defined as Lin− CD19− B220− CD127+ CD135+. Data is representative of three mutant and three control mice. C, Bone marrow was stained for surface expression of lineage markers [B220, CD3e, CD11b, Ly6C, Ter119], CD127, CD117, and Sca1. Viable cells were identified as DAPI−. HSCs were defined as Lin− CD127− CD117+ Sca1+. Data is representative of three mutant and three control mice. D, Absolute number of HSCs, CLPs, pre-pro-B cells, pro-B cells, pre-B cells, immature B cells, and mature B cells detected in Mybf/f Mb1-cre and Mybf/f mice. Data is compiled from three mutant and control mice. No statistically significant difference in HSCs, CLPs, and pre-pro-B cells between Mybf/f Mb1-cre mice and LMC. Asterisks (*) mark populations that are severely reduced in Mybf/f Mb1-cre mice.
c-Myb is critical for the survival of CD19+ pro-B cells
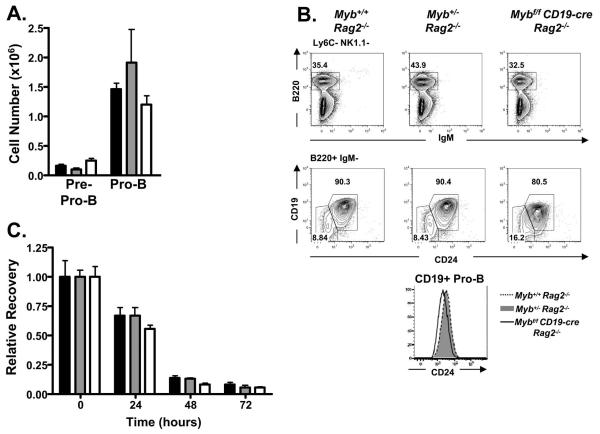
We have reported that Mybf/f CD19-cre and Myb+/− mice are impaired in pro-B to pre-B cell transition yet the absolute number of pro-B cells is not decreased in these mice compared to Mybf/f mice, suggesting that c-Myb is not important for the maintenance of the pro-B cell compartment (34,40). However, interpretation of these models is complicated by ongoing selection at the pre-Bcr checkpoint. To separate maintenance of the pro-B cell compartment from events associated with selection at the pre-Bcr checkpoint we bred Mybf/f CD19-cre Rag2−/−, CD19-cre Rag2−/− and Myb+/− Rag2−/− mice and characterized the CD19+ pro-B cell compartment in each mouse strain. We detect no difference in the absolute number of pre-pro-B cells or CD19+ pro-B cells between Mybf/f CD19-cre Rag2−/−, Myb+/− Rag2−/− and control mice (Fig. 5A). However, surface expression of CD19 and CD24 was reduced on CD19+ pro-B cells from Mybf/f CD19-cre Rag2−/− mice compared to Myb+/− Rag2−/− or Myb+/+ Rag2−/− CD19+ pro-B cells (Fig 5B). The CD19-cre allele was made by replacing CD19 coding sequences with Cre, which results in a null allele (43). Thus, mice that carry a single CD19-cre allele have reduced expression of CD19 on all CD19+ cells. However, reduced expression of CD24 is consistent with the notion that c-Myb deficient pro-B cells either fail to CD24 expression or fail to survive.
FIGURE 5.
B-cell development in Myb+/+ Rag2−/−, Myb+/− Rag2−/−, and Mybf/f CD19-cre Rag2−/− mice. A, Absolute number of pre-pro-B cells and pro-B cells in Myb+/+ Rag2−/− (black bar), Myb+/− Rag2−/− (grey bar), and Mybf/f CD19-cre Rag2−/− (white bar) mice. None are statistically significant. B, Bone marrow was stained for surface expression of Ly6C, NK1.1, B220, IgM, CD24, and CD19. Top tier, B220 versus IgM after gating out Ly6C+ and NK1.1+ cells. Bottom tier, CD19 versus CD24 through the B220+ IgM− gate. Pro-B cells were defined as Ly6C− NK1.1− B220+ IgM− CD19+ CD24+ and pre-pro-B cells were defined as Ly6C− NK1.1− B220+ IgM− CD19− CD24−. Histogram represents surface expression of CD24 on CD19+ pro-B cells, defined as Ly6C− NK1.1− B220+ IgM− CD19+, from Myb+/+ Rag2−/−, Myb+/− Rag2−/−, and Mybf/f CD19-cre Rag2−/− mice. C, Relative recovery of CD19+ pro-B cells isolated from Myb+/+ Rag2−/−, Myb+/− Rag2−/−, and Mybf/f CD19-cre Rag2−/− mice after 24, 48, and 72 hours in liquid culture without exogenous IL-7. Viable cell counts were determined by trypan blue exclusion. Relative recovery was determined by normalization to the total number of cells plated.
To initially compare survival between Mybf/f CD19-cre Rag2−/−, Myb+/− Rag2−/− and control Myb+/+ Rag2−/− CD19+ pro-B cells, we purified CD19+ pro-B cells from each strain on anti-CD19 coated magnetic beads and placed them in liquid culture without an exogenous source of IL-7. Pro-B cells were harvested and viable cells counted by trypan dye exclusion (Fig. 5C) or by flow cytometry (not shown) after 24, 48 and 72 hours of culture. We did not detect a statistically significant difference in the number of viable pro-B cells between strains at any time point during the experiment. However, we did detect a consistent decrease in the number of viable Mybf/f CD19-cre Rag2−/− pro-B cells after 24 and 48 hours of culture compared to the other strains although this did not reach statistical significance. To better assess the survival of c-Myb deficient CD19+ pro-B cells, CD19+ pro-B cells were isolated from Mybf/f Rag2−/− mice on anti-CD19 coated magnetic beads and placed in liquid culture supplemented with IL-7 for 24 hours. Subsequently, the pro-B cells were washed and transduced with retroviruses that produce Cre recombinase and GFP (MIG-Cre) or GFP alone (MIG-R1). After retrovirus transduction, the pro-B cells were placed back in liquid culture without exogenous IL-7 for 24, 48 and 72 hours (Fig. 6A). Twenty-four hours after transduction, approximately 40% of the cells that were transduced with either virus were GFP+ as measured by flow cytometry. The proportion of GFP+ and GFP- cells remained constant in pro-B cell cultures that were transduced with GFP alone over 72 hours suggesting that neither the transduced or untransduced cells had a growth advantage. In contrast, after transduction with MIG-Cre, only 10% of the viable pro-B cells were GFP+ by 48 hours after transduction, suggesting that the cells transduced with MIG-Cre failed to survive as well as the cells transduced with the MIG-R1 GFP only producing virus. When the absolute number of viable cells was calculated, we found that the number of viable cells that were transduced with MIG-Cre rapidly decreased in culture compared to the number of cells transduced with MIG-R1 (Fig. 6A). Importantly, the number of viable Myb+/+ Rag2−/− CD19+ pro-B cells transduced with MIG-Cre did not decrease more rapidly in culture than untransduced cells (Fig. 6B). Thus, c-Myb is critical for the survival of CD19+ pro-B cells.
FIGURE 6.
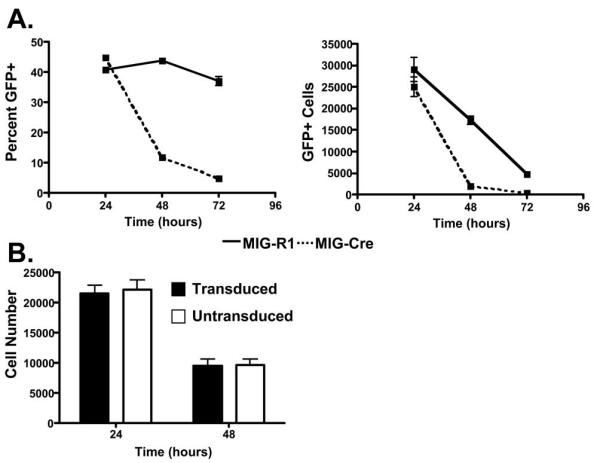
c-Myb-deficient CD19+ pro-B cells have an intrinsic survival defect. A, CD19+ pro-B cells isolated from Mybf/f Rag2−/− mice were transduced with MIG-R1 or MIG-Cre and cultured without exogenous IL-7. Cells were analyzed 24, 48, and 72 hours post-transduction for expression of GFP and total cells per well. B, CD19+ pro-B cells isolated from Myb+/+ Rag2−/− mice were transduced with MIG-Cre and cultured without exogenous IL-7. Cells were analyzed 24 and 48 hours post-transduction for the expression of GFP and total cells per well.
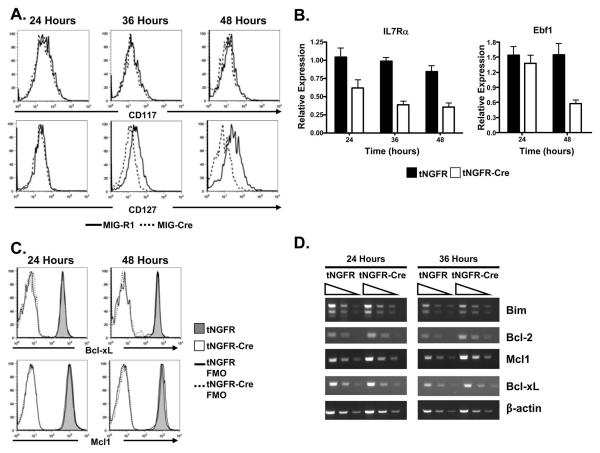
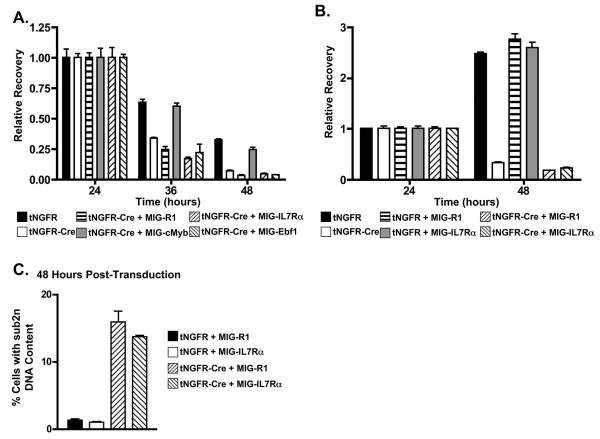
To begin to gain insight into the basis for decreased pro-B cell survival in the absence of c-Myb we initially assessed the surface expression of CD117 and CD127, which are cytokine receptors that are involved in maintaining the survival of pro-B cells, by flow cytometry. In particular, CD117 has been reported to be a target of c-Myb activity (54,55). However, we did not detect a change in CD117 expression on Mybf/f Rag2 −/− CD19+ pro-B cells after transduction with a Cre/GFP producing retrovirus (Fig. 7A). In contrast, surface expression of CD127 was decreased on Mybf/f Rag2−/− CD19+ pro-B cells following transduction with Cre/GFP producing retrovirus, suggesting that signaling through the IL-7R may be compromised. In addition, we found that mRNAs encoding CD127 and Ebf1 were decreased in c-Myb deficient pro-B cells (Fig. 7B) but detected no change in expression of Bcl-xL or Mcl-1 as measured by flow cytometry (Fig. 7C) or RT-PCR (Fig. 7D). Furthermore, we did not detect decreased expression of Bcl-2 or Bim encoding mRNA (Fig. 7D). Decreased expression of CD127 in c-Myb deficient pro-B cells may result in decreased survival signals. To test this notion, we isolated CD19+ pro-B cells from Mybf/f Rag2−/− mice and transduced them with retroviruses that encode IL7Rα/GFP (MIG-IL7Rα), Ebf1/GFP (MIG-Ebf1) or c-Myb (MIG-cMYB) (Fig. 8A). The transduced cells were then cultured in IL-7 for 24 hours and subsequently transduced with a second virus the encodes Cre and a truncated form of the nerve growth factor receptor that is unable to signal (tNGFR-Cre). The cells were then placed in liquid culture with or without IL-7 and survival of the cotransduced cells (identified by coexpression of GFP and tNGFR using flow cytometry) was assessed after 24 and 48 hours of culture. c-Myb was able to rescue the survival of CD19+ cells in liquid culture lacking IL-7 (Fig. 8A). However, survival was not rescued with exogenous CD127 or Ebf1. Signaling through the IL-7 receptor provides cues for survival and proliferation to pro-B cells in liquid culture. When CD19+ pro-B cells from Mybf/f Rag2−/− mice were put in liquid culture supplemented with IL-7 after transduction with tNGFR, tNGFR plus MIG-R1 or tNGFR plus MIG-IL7Rα, the relative recovery of CD19+ pro-B cells more than doubled between 24 and 48 of culture (Fig. 8B). In contrast, the recovery of CD19+ pro-B cells transduced with tNGRF-cre or tNGRF-cre plus MIG-R1 decreased by approximately 65% between 24 and 48 hours of culture. Similarly, the recovery of CD19+ pro-B cells from Mybf/f Rag2−/− mice transduced with tNGFR-cre plus MIG-IL7Rα also decreased by approximately 65% between 24 and 48 hours of liquid culture supplemented in the presence of IL-7, demonstrating that c-Myb deficient CD19+ pro-B cells fail to survive in the presence of IL-7 even when provided with exogenously supplied IL7Rα. In addition, CD19+ pro-B cells transduced with control virus (tNGFR) or a Cre producing virus (tNGFR-Cre) plus MIG-R1 or MIG-IL7Rα were stained with DRAQ5 and the DNA content measured by flow cytometry (Fig. 8C). Populations of cells that were transduced with tNGRF-Cre plus MIG-R1 or MIG-IL7Rα contained 5 to 7-fold more cells with a <2n DNA content than cells transduced with tNGRF plus MIG-R1 or MIG-IL7Rα. Thus, decreased viability of c-Myb deficient pro-B cells is not simply due to reduced expression of CD127 or Ebf1.
FIGURE 7.
Reduced expression of Il7r and Ebf1 mRNA in the absence of c-Myb. A, Surface expression of CD117 (c-kit) and CD127 (IL-7Rα) on Mybf/f Rag2−/− CD19+ pro-B cells transduced with MIG-R1 or MIG-Cre and analyzed 24, 36, and 48 hours post-transduction. Viable cells were identified as 7AAD−. B, Relative expression of Il7r and Ebf1 mRNA in Mybf/f Rag2−/− CD19+ pro-B cells transduced with tNGFR or tNGFR-Cre was measured by semi-quantitative RT-PCR 24, 36, and 48 hours post-transduction and normalized to the expression of ActB. Transduced cells were isolated after treatment with biotinylated anti-tNGFR antibody and streptavidin coated magnetic beads prior to making RNA. C, Intracellular expression of Bcl-xL and Mcl1 in Mybff Rag2−/− CD19+ pro-B cells transduced with tNGFR or tNGFR-Cre was measured by flow cytometry 24 and 48 hours post-transduction. D, Semi-quantitative RT-PCR analysis of Bcl2, Bcl-xL, Mcl1, and Bim performed on 3-fold serial dilutions of cDNA prepared from Mybf/f Rag2−/− CD19+ pro-B cells transduced with tNGFR or tNGFR-Cre 24 and 36 hours post-transduction. ActB serves as a loading control.
FIGURE 8.
Overexpression of IL-7Rα or EBF is unable to rescue the survival defect in Myb-deficient CD19+ pro-B cells. A, CD19+ pro-B cells from Mybf/f Rag2−/− mice were cotransduced with tNGFR-Cre and MIG-R1, MIG-cMyb, MIG-IL7Rα, or MIG-Ebf1, cultured in the absence of exogenous IL-7, and analyzed 24, 36, and 48 hours post-transduction for expression of tNGFR and GFP and total cells per well. Relative recovery was determined by normalization to the number of cotransduced cells 24 hours post-transduction. B, CD19+ pro-B cells from Mybf/f Rag2−/− mice were cotransduced with tNGFR or tNGFR-Cre and MIG-R1 or MIG-IL7Rα, cultured in the presence of exogenous IL-7, and analyzed 24 and 48 hours post-transduction for expression of tNGFR and GFP and total cells per well. C, Sub2n DNA content of Mybf/f Rag2−/− CD19+ pro-B cells cotransduced with tNGFR or tNGFR-Cre and MIG-R1 or MIG-IL7Rα was determined by flow cytometry 48 hours post-transduction.
Ebf1 can partially rescue production of CD19+ cells from Mybf/f Mb1-cre progenitor cells
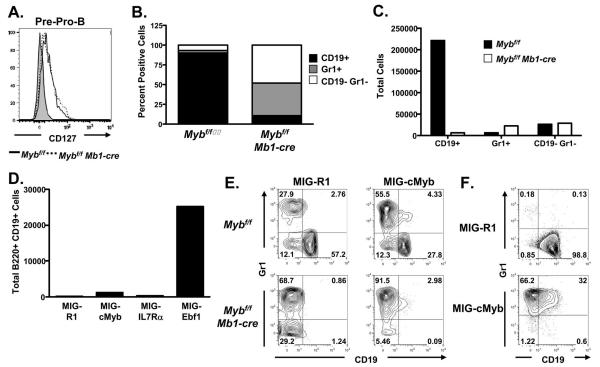
Transition from the pre-pro-B cell to the CD19+ pro-B cell compartment is dependent on signals through the IL-7R, which leads to increased expression of Ebf1 and differentiation to the CD19+ pro-B cell stage (22-24). Decreased expression of CD127 and Ebf1 after deletion of the floxed c-Myb alleles suggests that c-Myb may also be important for transition from the pre-pro-B cell to the CD19+ pro-B cell compartment in addition to survival of CD19+ pro-B cells. However, we did not detect a difference in expression of CD127 between Mybf/f Mb1-cre and control pre-pro-B cells (Fig. 9A), which could have been due to incomplete deletion at the Myb locus in pre-pro-B cells from Mybf/f Mb1-cre mice (Fig. 2) or that c-Myb deficient pre-pro-B cells have poor viability and die in vivo before we could detect a decrease in CD127 expression. LMPPs from Mybf/f Mb1-cre and control mice were electronically sorted and plated onto OP-9 stromal cell cultures supplemented with SCL, Flt3L and IL-7 to determine if they could generate CD19+ cells (Figs. 9B and 9C). The vast majority of cells that grew in cultures derived from control LMPPs were CD19+ B-lineage cells while very few CD19+ B-lineage cells grew from Mybf/f Mb1-cre LMPPs (Fig. 9C). While very few CD19+ cells grow out of the Mybf/f Mb1-cre LMPPs in these cultures compared to control LMPPs, there is not an appreciable increase in the number of Gr1+ or CD19- Gr1- cells that grow out of these cultures. Thus, the change in proportion of cells that grow out of Mybf/f Mb1-cre LMPPs compared to controls is primarily due to the vastly decreased growth of CD19+ cells that grow from Mybf/f Mb1-cre LMPPs. This result closely resembles the phenotype of B cell development in Mybf/f Mb1-cre mice compared to controls. To determine if decreased signals through the IL-7R could explain the lack of CD19+ pro-B cells in Mybf/f Mb1-cre mice, LMPPs were electronically sorted from Mybf/f Mb1-cre bone marrow, transduced with MIG-R1, MIG-cMYB, MIG-IL7Rα or MIG-Ebf1 and subsequently cultured for 14 days on OP-9 stromal cells as described above. Few CD19+ B-lineage cells grew out of the cultures transduced with MIG-R1, MIG-cMYB or MIG-IL7Rα producing retroviruses (Fig. 9D). Furthermore, transducing Mybf/f Mb1-cre LMPPs with a Bcl-xL producing retrovirus also was not able to rescue differentiation of CD19+ pro-B cells (supplemental Fig. 2) suggesting that survival is not the sole basis for the severe deficit in accumulation of CD19+ pro-B cells in Mybf/f Mb1-cre mice. However, LMPPs transduced with MIG-Ebf1 produced an increased number of CD19+ B-lineage cells, representing a >20-fold stimulation over cultures transduced with GFP or IL7Rα/GFP, suggesting that bypassing the need for signals through the IL-7R by overexpression of Ebf1 can at least in part overcome c-Myb deficiency to promote transition from the pre-pro-B cell to the CD19+ pro-B cell compartment. However, the number of CD19+ pro-B cells that grow out in OP-9 stromal cell culture from MIG-Ebf1 transduced Mybf/f Mb1-cre LMPPs is still only 10 – 15% the number CD19+ cells that grow from control LMPPs (compare Figs. 9C and 9D), suggesting that Ebf1 can only provide a partial rescue of B-lineage development. This is similar to the finding that Ebf1 can rescue differentiation of IL-7R or E2A deficient B-lineage progenitors but not proliferative expansion (22,56,57).
FIGURE 9.
Ebf1 can partially rescue B-lineage development from Mybf/f Mb1-cre LMPPs. A. Surface expression of CD127 on pre-pro-B cells, defined as Ly6C− NK1.1− B220+ CD43+ CD19− CD24−, from Mybf/f Mb1-cre and Mybf/f mice compared to FMO control. B, Five thousand LMPPs from Mybf/f and Mybf/f Mb1-cre mice were seeded on OP-9 stromal cells in the presence of SCF, Flt3L, and IL-7 and analyzed 14 days later by flow cytometry for surface expression of CD19 and Gr1. Bars represent percentage of positive cells from each culture. Data are representative of 4 independent experiments. C, Total number of CD19+, Gr1+, and CD19− Gr1− cells that grew out of OP-9 stromal cell coculture with Mybf/f and Mybf/f Mb1-cre LMPPs. Data are representative of 4 independent experiments. D, LMPPs were isolated from Mybf/f Mb1-cre bone marrow, seeded at 5,000 cells per well on OP-9 stromal cells and transduced with MIG-R1, MIG-cMyb, MIG-IL7Rα, or MIG-Ebf1. LMPPs were cultured in the presence of SCF, Flt3L, and IL-7 and analyzed 14 days later by flow cytometry for expression of B220 and CD19 and the number of GFP+ B220+ CD19+ cells were determined. Data are representative of 4 independent experiments. E, Mybf/f and Mybf/f Mb1-cre LMPPs were transduced with MIG-R1 or MIG-cMyb and cultured on OP9 stromal cells in the presence of SCF, Flt3L, and IL-7. Cultures were analyzed 14 days later by flow cytometry for the surface expression of CD19 and Gr1. Number in quadrants indicates percent cells in each well. Data are representative of four independent experiments. F, Day 14.5 fetal liver progenitors from Myb+/+ embryos were transduced with MIG-R1 or MIG-cMyb and cultured on OP-9 stromal cells in the presence of SCF, Flt3L, and IL-7. Cultures were analyzed 14 days later by flow cytometry for the surface expression of CD19 and Gr1. Number in quadrants indicates percent cells in each.
Surprisingly, c-Myb and CD127 failed to rescue production of CD19+ cells from Mybf/f Mb1-cre LMPPs. When we more closely examined the cells that grew out from Mybf/f Mb1-cre or control LMPPs transduced with MIG-cMYB we found that in both cases c-Myb appeared to favor the production of Gr1 rather than CD19 producing cells (Fig. 9E), suggesting that increased c-Myb expression in LMPPs may block differentiation to the B cell lineage or favor the growth of myeloid cells. We similarly tried to rescue CD19+ B-lineage cell development from Myb−/− fetal liver progenitor cells. In this case, we were unable to rescue growth of CD19+ or Gr1+ cells with MIG-R1, MIG-cMYB, MIG-IL7Rα or MIG-Ebf1 (data not shown). However, transduction of control fetal liver progenitors with MIG-cMYB resulted in the outgrowth of almost entirely Gr1+ cells (Fig. 9F). Thus, proper control of c-Myb expression appears to be crucial in determining the growth of myeloid versus B-lineage growth from LMPPs.
Discussion
Several studies have suggested that c-Myb plays an important role during B-lymphopoiesis. First, B cells failed to develop in Rag1−/− blastocyst chimeras made with Myb−/− mouse embryonic stem cells (36). Second, a reduced number of peripheral B cells has been identified in several hypomorphic c-Myb mutants (37-39). However, in each case the decrease in peripheral B cell number could be related to defects in HSC or early progenitors. We recently used Mybf/f CD19-cre mice to identify a partial block in B cell development during the pro-B to pre-B cell transition (34). In addition, peripheral B cells in these mice were hyporesponsive to BLyS. CD19-cre directs deletion of floxed Myb alleles relatively late in the pro-B cell compartment making it difficult to determine if c-Myb is important at earlier stages of B cell development (34,41,43). Most recently, we found that forced expression of the MiR150 miRNA, which targets the Myb mRNA, resulted in a partial block to B cell development during the pro-B to pre-B cell transition that was associated with cell death when B-lineage cells were grown in stromal cell cultures supplemented with IL-7 (40). This study made clear that relatively small differences in c-Myb expression can have a significant impact on B cell development. However, by breeding Mybf/f Mb1-cre mice, where inactivation of the Myb locus occurs earlier than in CD19-cre mice but specifically in the B-lineage, we have demonstrated that c-Myb is absolutely required for B cell development. The severe deficit in B cell development detected in Mybf/f Mb1-cre mice demonstrates that c-Myb must be considered along with E2A, Ebf1 and Pax5 as a transcription factor that is critical for B cell development and included in the regulatory network that controls B cell development.
Mb1-cre mediated deletion is initiated in pre-pro-B cells. Thus, the failure to produce CD19+ pro-B cells could be due to a failure of developing B cells to survive, failure to differentiate beyond the pre-pro-B cell stage or both. We have previously reported that B cell development is partially blocked during transition from the pro-B cell to the pre-B cell compartment (34). Furthermore, mice that are transgenic for the MiR150 miRNA, which targets c-Myb mRNA, have a similar block during B cell differentiation (40). Enriched B-lineage cells from MiR150 transgenic mice that are grown in stromal cell culture with IL-7 accumulate 2 to 3-fold more dead cells than cells from control mice suggesting that proliferating pro-B or pre-B cells undergo apoptotic cell death. Cell death in this experiment could have been due to events associated with pre-BCR selection. Although it seemed unlikely that pre-BCR mediated events could explain the severe deficit of CD19+ B cells in Mybf/f Mb1-cre mice we crossed Myb+/− and Mybf/f CD19-cre mice with Rag2−/− mice to separate events associated with V(D)J recombination and the pre-BCR checkpoint from the intrinsic survival of pro-B cells. We did not detect a difference in the number of pre-pro-B cells or CD19+ pro-B cells in Myb+/−Rag2−/− or Mybf/f CD19-cre Rag2−/− compared to control mice nor did we detect a difference in intrinsic survival between pro-B cells from these mice in liquid culture. However, surface expression of CD24 was consistently lower on pro-B cells from Mybf/f CD19-cre Rag2−/− mice compared to Myb+/− or Myb+/+ Rag2−/− pro-B cells, suggesting that the late inactivation of the Myb locus in these mice might limit differentiation or survival. When we isolated CD19+ pro-B cells from Mybf/f Rag2−/− mice and used retrovirus mediated transduction to introduce Cre into pro-B cells, we found that c-Myb deficient pro-B cells rapidly died in liquid culture without exogenous IL-7. Thus, c-Myb is critical for the intrinsic survival of CD19+ pro-B cells prior to the pre-BCR checkpoint.
To gain insight into how c-Myb might mediate survival we assessed expression of several genes that are associated with survival in pro-B cells including Il7r and Mcl1. Mice deficient for Il7r and Mcl1 have blocks to B cell development that are very similar to Mybf/f Mb1-cre mice (22,24,58,59). Inactivation of the Myb locus did not result in decreased expression of CD117, which is thought to be a direct c-Myb target in some systems (55,60), Bcl-xL, Mcl-1, Bcl-2 or Bim. Both CD127 surface expression and mRNA were decreased after Cre mediated inactivation of the Myb locus in CD19+ pro-B cells suggesting that c-Myb might control pro-B cell viability by regulating expression of CD127. However, transduction of CD19+ pro-B cells with CD127 prior to inactivation of the Myb locus did not rescue survival of the pro-B cells. Thus, c-Myb dependent survival of CD19+ pro-B cells appears to be mediated independent of CD127 expression. How c-Myb is involved in regulating expression of CD127 remains unknown. The CD127 promoter region contains two potential c-Myb binding sites but we have been unable to demonstrate direct binding of c-Myb to the CD127 promoter using a chromatin immunoprecipitation assay (unpublished data). Thus, c-Myb may be involved in regulating CD127 expression by an indirect mechanism. It remains possible that c-Myb may interact with unidentified regulatory regions that control expression of CD127 or by interacting with other proteins that may tether c-Myb to the CD127 promoter and in this respect, c-Myb has been reported to regulate transcription of the vascular endothelial growth factor promoter in a DNA-binding domain independent fashion (61). Further work is required to understand how c-Myb regulates expression of CD127.
Inactivation of the Myb locus in CD19+ pro-B cells resulted in decreased surface expression of CD127 as well as decreased Il7r and Ebf1 mRNA. Specification of B cell fate in CLPs and transition from the pre-pro-B cell stage to the CD19+ pro-B cell stage is dependent on signals through the IL-7R that result in increased expression of Ebf1 (22-24). Thus, c-Myb may be required for proper CD127 expression prior to the CD19+ pro-B cell stage. LMPPs isolated from Mybf/f Mb1-cre mice produced <0.5% the number of CD19+ cells produced by LMPPs from control mice when grown on OP-9 stromal cells. Thus, the phenotype of Mybf/f Mb1-cre mice could be replicated in vitro. To determine if c-Myb, CD127 or Ebf1 could rescue development to the CD19+ pro-B cell stage, we transduced LMPPs from Mybf/f Mb1-cre mice with retroviruses encoding c-Myb/GFP, CD127/GFP, Ebf1/GFP or GFP alone. Surprisingly, we found that neither c-Myb nor CD127 could rescue differentiation of Mybf/f Mb1-cre LMPPs. Proper levels of c-Myb expression have been found to be a crucial determinant of hematopoietic differentiation in a variety of normal and leukemic models of hematopoiesis (37,62-64). One study that used a tetracycline inducible c-Myb allele reported that overexpression of c-Myb could block differentiation of erythroid, megakaryocytic and B-lymphocyte development while myeloid development was not inhibited (62). We found that transduction of Mybf/f LMPPs or Myb+/+ fetal liver progenitors with c-Myb/GFP appeared to skew differentiation of normal LMPPs away from B cell development in OP-9 stromal cell cultures and toward myeloid development. It is likely that expression of exogenous c-Myb in cells that already produce normal amounts of c-Myb directs differentiation away from or blocks B-lineage development but allows myeloid development to progress. Similarly, overexpression of CD127 has previously been reported to block B cell development though the basis for this is not understood (65).
Transduction of Mybf/f Mb1-cre LMPPs with MIG-Ebf1 resulted in a 20-fold increase in the number of CD19+ cells produced compared to LMPPs transduced with MIG-R1. This represented a 20-fold increase over Mybf/f Mb1-cre LMPPs transducd with GFP alone, suggesting that c-Myb may be important for expression of CD127 prior to the CD19+ pro-B cell stage, which in turn is needed for increased expression of Ebf1 and transition to the CD19+ pro-B cell compartment. The finding that Ebf1 can partially rescue differentiation of Mybf/f Mb1-cre pro-B cells to CD19+ cells suggests that c-Myb is not required for commitment to the B cell lineage. Thus, c-Myb appears to play at least two important roles during B cell differentiation. First, c-Myb appears to be involved directly or indirectly in regulating expression of CD127, which is required for proper expression of Ebf1 and differentiation to the CD19+ pro-B cell compartment. Second, c-Myb is critical for survival of CD19+ pro-B cells prior to the pre-BCR checkpoint and this event is independent of CD127 expression. Finally, we find that a single functional Myb allele is sufficient to drive B cell development to the pro-B cell stage and for the survival of pro-B cells. However, Myb+/− mice are partially blocked in transition from the pro-B cell to the pre-B cell stage of differentiation, and c-Myb expression is greatest in the pre-B cell compartment, suggesting that two Myb alleles are required at this stage of B cell differentiation. It will be important to identify direct targets of c-Myb activity at different stages of B cell differentiation to understand the basis for c-Myb activity during B-lymphopoiesis.
Supplementary Material
Acknowledgements
The authors thank Professor Klaus Rajewsky for his advice and support. The authors are grateful to the Flow Cytometry Core Facility at the University of Virginia and in particular thank Ms. Joanne Lannigan and Mr. Michael Solga for their expert help and advice.
Footnotes
This work was supported by grant AI059294 from the National Institutes of Health (to TPB).
Abbreviations used in this paper: HSC, hematopoietic stem cell; MPP, mutipotent progenitor; LMPP, lymphoid-primed multipotent progenitor; CLP, common lymphoid progenitor; GFP, green fluorescent protein; tNGFR, truncated nerve growth factor receptor
References
- 1.Loffert D, Schaal S, Ehlich A, Hardy RR, Zou YR, Muller W, Rajewsky K. Early B-cell development in the mouse: insights from mutations introduced by gene targeting. Imm. Rev. 1994;137:135. doi: 10.1111/j.1600-065x.1994.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 2.Monroe JG, Dorshkind K. Fate Decisions Regulating Bone Marrow and Peripheral B Lymphocyte Development. In: Frederick WA, editor. Advances in Immunology. Academic Press; 2007. p. 1. [DOI] [PubMed] [Google Scholar]
- 3.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LAM, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3+ Lympho-Myeloid Stem Cells Lacking Erythro-Megakaryocytic Potential: A Revised Road Map for Adult Blood Lineage Commitment. Cell. 2005;121:295. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, Buza-Vidas N, Qian H, Soneji S, Enver T, Sigvardsson M, Jacobsen SE. Molecular Evidence for Hierarchical Transcriptional Lineage Priming in Fetal and Adult Stem Cells and Multipotent Progenitors. Immunity. 2007;26:407. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 6.Traver D, Akashi K. Lineage Commitment and Developmental Plasticity in Early Lymphoid Progenitor Subsets. In: Cantor L. G. a. F. Harvey., editor. Advances in Immunology T Cell Subsets: Cellular Selection, Commitment and Identity. Academic Press; 2004. p. 1. [DOI] [PubMed] [Google Scholar]
- 7.Gounari F, Aifantis I, Martin C, Fehling HJ, Hoeflinger S, Leder P, von BH, Reizis B. Tracing lymphopoiesis with the aid of a pTalpha-controlled reporter gene. Nat. Immunol. 2002;3:489. doi: 10.1038/ni778. [DOI] [PubMed] [Google Scholar]
- 8.Li YS, Hayakawa K, Hardy RR. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J. Exp. Med. 1993;178:951. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173:1213. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp. Med. 2006;203:675. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 12.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida T, Yao-Ming NS, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 2006;7:382. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in Hemopoietic Stem Cell Activity in Ikaros Mutant Mice. J. Exp. Med. 1999;190:1201. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynaud D, Demarco A, Reddy L, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 2008;9:927. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nutt SL, Kee BL. The Transcriptional Regulation of B Cell Lineage Commitment. Immunity. 2007;26:715. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Singh H, Pongubala JM. Gene regulatory networks and the determination of lymphoid cell fates. Curr. Opin. Immunol. 2006;18:116. doi: 10.1016/j.coi.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 19.DeKoter RP, Schweitzer BL, Kamath MB, Jones D, Tagoh H, Bonifer C, Hildeman DA, Huang KJ. Regulation of the Interleukin-7 Receptor {alpha} Promoter by the Ets Transcription Factors PU.1 and GA-binding Protein in Developing B Cells. J. Biol. Chem. 2007;282:14194. doi: 10.1074/jbc.M700377200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borge OJ, Adolfsson J, Jacobsen A. M. I.-L. M. a. S. Lymphoid-Restricted Development From Multipotent Candidate Murine Stem Cells: Distinct and Complimentary Functions of the c-kit and flt3-Ligands. Blood. 1999;94:3781. [PubMed] [Google Scholar]
- 21.Dias S, Silva H, Jr., Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 2005;201:971. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 2005;201:1197. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi K, Kasai H, Watanabe A, Lai AY, Kondo M. IL-7 Specifies B Cell Fate at the Common Lymphoid Progenitor to Pre-ProB Transition Stage by Maintaining Early B Cell Factor Expression. J. Immunol. 2008;181:383. doi: 10.4049/jimmunol.181.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller JP, Izon D, DeMuth W, Gerstein R, Bhandoola A, Allman D. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J. Exp. Med. 2002;196:705. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Riordan M, Grosschedl R. Coordinate Regulation of B Cell Differentiation by the Transcription Factors EBF and E2A. Immunity. 1999;11:21. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 26.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 27.Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 28.Mikkola I, Heavey B, Horcher M, Busslinger M. Reversion of B cell commitment upon loss of Pax5 expression. Science. 2002;297:110. doi: 10.1126/science.1067518. [DOI] [PubMed] [Google Scholar]
- 29.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Sem. Immunol. 2008;20:247. doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 31.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 32.Westin EH, Gallo RC, Arya SK, Eva A, Souza LM, Baluda MA, Aaronson SA, Wong-Staal F. Differential expression of the amv gene in human hematopoietic cells. Proc. Natl. Acad. Sci. USA. 1982;79:2194. doi: 10.1073/pnas.79.7.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr., Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 34.Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity. 2005;23:275. doi: 10.1016/j.immuni.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Golay J, Cusmano G, Introna M. Independent regulation of c-myc, B-myb, and c-myb gene expression by inducers and inhibitors of proliferation in human B lymphocytes. J. Immunol. 1992;149:300. [PubMed] [Google Scholar]
- 36.Allen RD, III, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes & Devel. 1999;13:1073. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emambokus N, Vegiopoulos A, Harman B, Jenkinson E, Anderson G, Frampton J. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carpinelli MR, Hilton DJ, Metcalf D, Antonchuk JL, Hyland CD, Mifsud SL, Di RL, Hilton AA, Willson TA, Roberts AW, Ramsay RG, Nicola NA, Alexander WS. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl.Acad. Sci. USA. 2004;101:6553. doi: 10.1073/pnas.0401496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandberg ML, Sutton SE, Pletcher MT, Wiltshire T, Tarantino LM, Hogenesch JB, Cooke MP. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Devel. Cell. 2005;8:153. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 40.Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell. 2007;131:146. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 41.Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, Pelanda R, Reth M. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 2006;103:13789. doi: 10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat. Immunol. 2004;5:721. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 43.Rickert RC, Rajewsky K, Roes J. Impairment of T-cell-dependent B-cell responses and B-1 cell development in CD19-deficient mice. Nature. 1995;376:352. doi: 10.1038/376352a0. [DOI] [PubMed] [Google Scholar]
- 44.DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- 45.Yin KJ, Hsu CY, Hu XY, Chen H, Chen SW, Xu J, Lee JM. Protein Phosphatase 2A Regulates bim Expression via the Akt/FKHRL1 Signaling Pathway in Amyloid-beta Peptide-Induced Cerebrovascular Endothelial Cell Death. J. Neurosci. 2006;26:2290. doi: 10.1523/JNEUROSCI.5103-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Exley GE, Tang C, McElhinny AS, Warner CM. Expression of Caspase and BCL-2 Apoptotic Family Members in Mouse Preimplantation Embryos. Biol. Reprod. 1999;61:231. doi: 10.1095/biolreprod61.1.231. [DOI] [PubMed] [Google Scholar]
- 47.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780. [PubMed] [Google Scholar]
- 48.Izon DJ, Punt JA, Xu L, Karnell FG, Allman D, Myung PS, Boerth NJ, Pui JC, Koretzky GA, Pear WS. Notch1 regulates maturation of CD4+ and CD8+ thymocytes by modulating TCR signal strength. Immunity. 2001;14:253. doi: 10.1016/s1074-7613(01)00107-8. [DOI] [PubMed] [Google Scholar]
- 49.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dengler HS, Baracho GV, Omori SA, Bruckner S, Arden KC, Castrillon DH, DePinho RA, Rickert RC. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 2008;9:1388. doi: 10.1038/ni.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carlyle JR, Michie AM, Cho SK, Zuniga-Pflucker JC. Natural killer cell development and function precede alpha beta T cell differentiation in mouse fetal thymic ontogeny. J. Immunol. 1998;160:744. [PubMed] [Google Scholar]
- 52.Kitamura D, Rajewsky K. Targeted disruption of mu chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356:154. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 53.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 1991;173:1213. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ratajczak MZ, Luger SM, DeRiel K, Abrahm J, Calabretta B, Gewirtz AM. Role of the KIT protooncogene in normal and malignant human hematopoiesis. Proc. Natl. Acad. Sci. USA. 1992;89:1710. doi: 10.1073/pnas.89.5.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogg A, Schirm S, Nakagoshi H, Bartley P, Ishii S, Bishop JM, Gonda TJ. Inactivation of a c-Myb/estrogen receptor fusion protein in transformed primary cells leads to granulocyte/macrophage differentiation and down regulation of c-kit but not c-myc or cdc2. Oncogene. 1997;15:2885. doi: 10.1038/sj.onc.1201472. [DOI] [PubMed] [Google Scholar]
- 56.Dias S, Silva H, Jr., Cumano A, Vieira P. Interleukin-7 is necessary to maintain the B cell potential in common lymphoid progenitors. J. Exp. Med. 2005;201:971. doi: 10.1084/jem.20042393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seet CS, Brumbaugh RL, Kee BL. Early B Cell Factor Promotes B Lymphopoiesis with Reduced Interleukin 7 Responsiveness in the Absence of E2A. J. Exp. Med. 2004;199:1689. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 59.Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 1994;180:1955. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ratajczak MZ, Perrotti D, Melotti P, Powzaniuk M, Calabretta B, Onodera K, Kregenow DA, Machalinski B, Gewirtz AM. Myb and ets proteins are candidate regulators of c-kit expression in human hematopoietic cells. Blood. 1998;91:1934. [PubMed] [Google Scholar]
- 61.Lutwyche JK, Keough RA, Hunter J, Coles LS, Gonda TJ. DNA binding-independent transcriptional activation of the vascular endothelial growth factor gene (VEGF) by the Myb oncoprotein. Bioch. Biophys. Res. Com. 2006;344:1300. doi: 10.1016/j.bbrc.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 62.Sakamoto H, Dai G, Tsujino K, Hashimoto K, Huang X, Fujimoto T, Mucenski M, Frampton J, Ogawa M. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood. 2006;108:896. doi: 10.1182/blood-2005-09-3846. [DOI] [PubMed] [Google Scholar]
- 63.Kumar A, Lee CM, Reddy EP. c-Myc is essential but not sufficient for c-Myb-mediated block of granulocytic differentiation. J. Biol. Chem. 2003;278:11480. doi: 10.1074/jbc.M300080200. [DOI] [PubMed] [Google Scholar]
- 64.Sumner R, Crawford A, Mucenski M, Frampton J. Initiation of adult myelopoiesis can occur in the absence of c-Myb whereas subsequent development is strictly dependent on the transcription factor. Oncogene. 2000;19:3335. doi: 10.1038/sj.onc.1203660. [DOI] [PubMed] [Google Scholar]
- 65.Purohit SJ, Stephan RP, Kim HG, Herrin BR, Gartland L, Klug CA. Determination of lymphoid cell fate is dependent on the expression status of the IL-7 receptor. EMBO J. 2003;22:5511. doi: 10.1093/emboj/cdg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.