Abstract
Certain prokaryotic transport proteins similar to the lactose permease of Escherichia coli (LacY) have been identified by BLAST searches from available genomic databanks. These proteins exhibit conservation of amino acid residues that participate in sugar binding and H+ translocation in LacY. Homology threading of prokaryotic transporters based on the X-ray structure of LacY (PDB ID: 1PV7) and sequence similarities reveals a common overall fold for sugar transporters belonging to the Major Facilitator Superfamily (MFS) and suggest new targets for study. Evolution-based searches for sequence similarities also identify eukaryotic proteins bearing striking resemblance to MFS sugar transporters. Like LacY, the eukaryotic proteins are predicted to have 12 transmembrane domains (TMDs), and many of the irreplaceable residues for sugar binding and H+ translocation in LacY appear to be largely conserved. The overall size of the eukaryotic homologs is about twice that of prokaryotic permeases with longer N and C termini and loops between TMDs III–IV and VI–VII. The human gene encoding protein FLJ20160 (NM_017694) consists of six exons located on more than 60,000 bp of DNA sequences and requires splicing to produce mature mRNA. Cellular localization predictions suggest membrane insertion with possible proteolysis at the N terminus, and expression studies with the human protein FJL20160 demonstrate membrane insertion in both E. coli and Pichia pastoris. Widespread expression of the eukaryotic sugar transport candidates suggests an important role in cellular metabolism, particularly in brain and tumors. Homology is observed in the TMDs of both the eukaryotic and prokaryotic proteins that contain residues involved in sugar binding and H+ translocation in LacY.
Keywords: membrane transporters, structure, homology threading, sugar binding, proton translocation
Introduction
Membrane proteins mediate a huge variety of physiological processes, and as many as 20% of all cellular proteins are polytopic membrane proteins, as judged from genome sequences.1 However, structures of this class of proteins comprise only a fraction of known protein structures.
The Major Facilitator Superfamily (MFS) of transport proteins (permeases) catalyze transport of a diverse range of substrates against concentration gradients by using the energy stored in electro-chemical ion gradients across membranes.2 Compounds transported by MFS permeases include simple sugars, oligosaccharides, nucleosides, drugs, amino acids, and various organic and inorganic anions and cations. The MFS family appears to be evolutionarily related and found in membranes from archaea to the mammalian central nervous system.
MFS proteins likely have similar secondary structures, as predicted by sequence alignments and hydropathy profiling.3 All permeases in the MFS possess either 12 or 14 transmembrane helices, which likely arose from tandem gene duplications. Studying the structure and mechanism of membrane proteins in general and MFS transporters in particular is thorny, since hydrophobic integral membrane proteins are notoriously difficult to handle by using routine biochemical techniques designed for water-soluble proteins. Nevertheless, information on this class of proteins is essential, since MFS transporters are ubiquitous in living cells and are involved in a variety of physiological processes.2,4
The hydrophobic, conformationally flexible nature of the MFS transporters makes them metastable and difficult to crystallize. However, crystal structures have been obtained recently for the lactose permease (LacY),5 which catalyzes the stoichiometric symport of galactoside and a H+, and the glycerol-3-phosphate/phosphate anti-porter (GlpT),6 which catalyzes the antiport of cytoplasmic phosphate for glycerol-3-phosphate in the external medium. LacY was the first membrane protein to be sequenced,7 purified to homogeneity and reconstituted into proteoliposomes in a fully functional state.8,9 As summarized in a recent review,10 a large amount of biochemical and biophysical data have been obtained for LacY over the last 20 years, much of which is confirmed by the crystal structure.5
LacY has been used to model tertiary structures of different MFS transporters,11–16 but significant homology of amino acid residues involved in substrate binding and H+ translocation is apparent for only a few prokaryotic transporters,17–21 The availability of a LacY X-ray structure allows homology threading of MFS sugar transporters that have amino acid sequences similar to LacY. The process yields a predicted low resolution structure22 in which accuracy depends upon the level of homology between proteins, and together with sequence alignment, may provide a powerful tool for predicting amino acid residues that may be mechanistically important.
Here, a number of eukaryotic proteins that exhibit significant homology to LacY-type MFS sugar transport proteins are identified by employing an evolution-based BLAST search. These putative transporters are present in eukaryotes from insects to mammals, appear to be twice as big as LacY, but show dramatic conservation of structural and functional elements located in the suggested transmembrane domains. Moreover, dozens of new MFS homologs are also identified, many of which are from pathogenic bacteria. In the accompanying paper,23 it is shown that despite low homology with LacY, not only is the overall fold of sucrose permease (CscB) similar to that of LacY, but specific conserved residues obligatory for LacY activity are also essential for the activity of CscB.
Results and Discussion
Prokaryotic homologs
LacY homologs and threading
More than a dozen LacY homologs were identified from BLAST searches by using either the Swissprot Database or by searching for DNA sequences encoding open reading frames in genomic databases starting with the amino acid sequence of LacY. The proteins suitable for initial threading were selected according to level of homology, generally have from 20 to more than 60% identity with LacY (Table 1) and are members of the MFS sub-family termed Oligosaccharide/H+ Symporters.3,4 Secondary structure predictions for these membrane proteins24–27 suggest 12 transmembrane domains (TMDs) connected by short loops with a longer loop between helices VI and VII. It is likely that these proteins have a topology similar to LacY with the N and C termini inside the cell and are appropriate for homology threading.
Table 1.
Properties of selected bacterial MFS transporters compared to LacY
| Protein | Source | Length (amino acids) | Assigned function | Homology threading | Identical amino acids (%) | Sequence homology (%) |
|---|---|---|---|---|---|---|
| P02920—LacY | Escherichia coli | 417 | Lactose transport | 1PV7.pdb | 100 | 100 |
| P47234–LacY | Citrobacter freundii | 416 | Lactose transport | Yes | 88 | 95 |
| AAM86136–LacY | Yersinia pestis | 414 | Galactoside transport | Yes | 63 | 78 |
| P18817–LacY | Klebsiella oxytoca | 416 | Lactose transport | Yes | 59 | 80 |
| BAA19154–MelY | Enterobacter cloacae | 425 | Galactoside transport | Yes | 57 | 75 |
| P16552–RafB | E. coli | 425 | Raffinose transport | Yes | 55 | 75 |
| AAK79497–RafB | Clostridium acetobutilicum | 411 | Raffinose transport | Yes | 40 | 60 |
| AAM19070–FruP | Bacillus megaterium | 413 | Fructose transport? | Yes | 39 | 61 |
| P30000–CscB | E. coli | 415 | Sucrose transport | Yes | 28 | 51 |
| CAD26969–CscB | Bifidobacterium animalis | 441 | Sucrose transport | Yes | 26 | 49 |
| BAB05995 – MaltP | Bacillus halodurans | 392 | Maltose transport? | Yes | 17 | 35 |
| P76417 – YegT | E. coli | 425 | Nucleoside transport | Partial | 16 | 31 |
| P44629 – PhenP | Haemophilus influenza | 388 | Phenylpropionic acid transport | Partial | 16 | 34 |
| P02982–Tcr1 | E. coli | 399 | Antibiotic resistance | Partial | 16 | 32 |
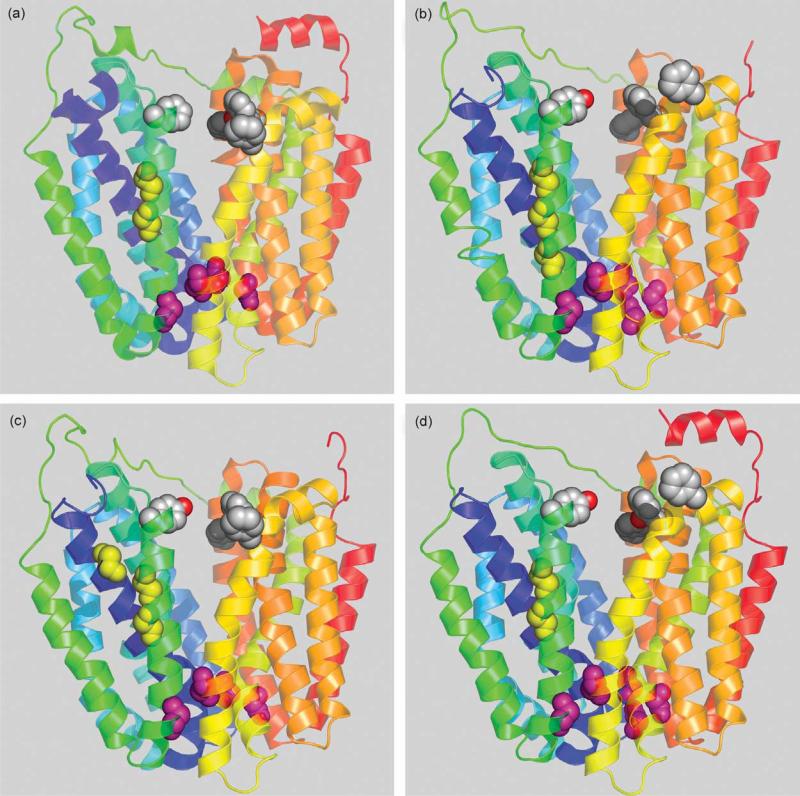
Figure 1 shows a side view of LacY and three other known (CscB, RafB) or putative (FruP) sugar transport proteins threaded into the LacY structure. Clearly, in each instance, 12 transmembrane helices are organized into two pseudo-symmetrical six-helix bundles tightly closed on the periplasmic side with a large cavity open to cytoplasm representing the inward-facing conformation.
Figure 1.
Homologous bacterial MFS transporters threaded into LacY structure. Transmembrane helices are depicted as ribbons colored from blue (helix I) to red (helix XII). The cytoplasmic side is at the top. Conserved Gly residues in helices I and V are shown as yellow spheres. Conserved Gly residues on the extracellular surface between the N and C-terminal six-helix bundles are shown as pink spheres. Conserved aromatic residues facing hydrophilic cavity are shown as grey spheres. (a) LacY (PDB code 1PV7); (b) E. coli sucrose permease CscB; (c) E. coli raffinose permease RafB; (d) Bacillus megaterium fructose permease FruP.
Residues involved in substrate binding
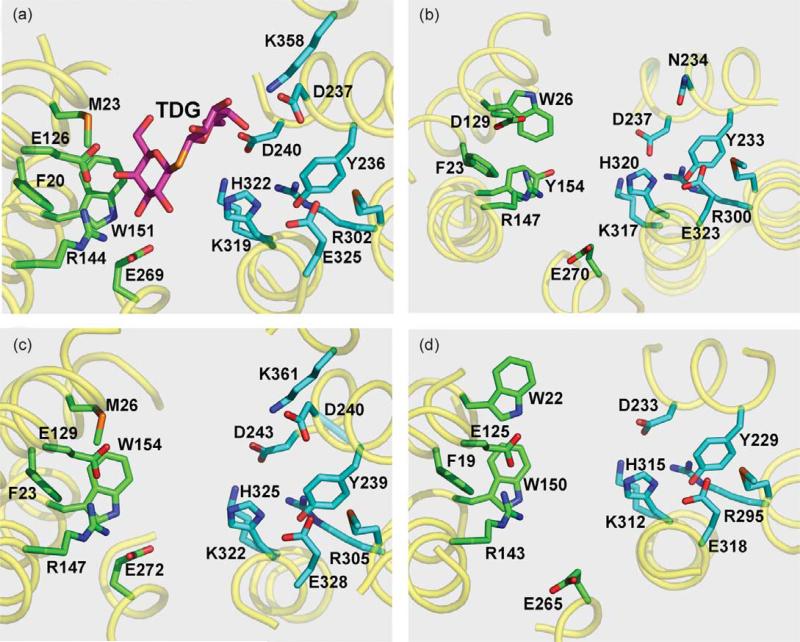
There is also a remarkable similarity in the structural arrangement of side-chains that form the sugar-binding site in LacY and the homologs (Figure 2). All of the substrate specificity of LacY is directed towards the galactopyranosyl moiety of its substrates, and substituents on the anomeric position increase affinity by non-specific interactions.28 The side-chains involved in LacY specificity are located in N-terminal six-helix bundle in helices I, IV, and V (Figure 2(a)), as identified by site-directed and Cys-scanning mutagenesis29 and by the X-ray structure.5 Arg144 (helix V) forms a bidentate H-bond with the C4–OH and C3–OH of the galactopyranosyl ring. Another essential amino acid, Glu126 (helix IV), is in close proximity to Arg144 and may interact with the C5–OH and/or C6–OH via a water molecule, and Glu269 (helix VIII) may interact weakly with the C3–OH of the sugar.30 Trp151 (helix V) stacks hydrophobically with the galactopyranosyl ring,5,31,32 as observed in a number of sugar binding proteins.33–36 As indicated by the X-ray structure of LacY, Met23 (helix I) is close to the C6 of the galactopyranosyl ring, and Phe20 is in close proximity to Trp151.5 Although the F20A mutant exhibits a 50-fold decrease in affinity, as judged by sugar protection against alkylation of Cys148, replacement of Met23 with Ala has no effect on affinity.23
Figure 2.
Cytoplasmic view of substrate binding and H+ translocation sites of LacY and homologs. Amino acid residues known (a), or predicted ((b)–(d)) to be involved in sugar binding (green) or H+ translocation (cyan) are shown as sticks. Essential residues are numbered according to protein sequences deposited in SwissProt Databank. (a) X-ray structure of LacY (PDB code 1PV7) with bound sugar β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG); (b) E. coli sucrose permease CscB; (c) E. coli raffinose permease RafB; (d) B. megaterium fructose permease FruP.
Homology alignment and threading of other protein sequences into LacY reveal conservation of Trp, Tyr or Phe at the position corresponding to Trp151 in LacY (Figure 2). The sucrose transporter CscB (Figure 2(b)) has a Tyr at position 154, which is homologous to Trp151 in LacY, and Trp26 (homologous to Met23 in LacY) in close proximity to Tyr154. The organization of the residues putatively involved in RafB sugar binding (Figure 2(c)) is virtually identical with that of LacY. FruP, a putative transport protein (Figure 2(d)), has a Trp at position 150 homologous to Trp151 in LacY and an additional aromatic residue, Trp22, in place of Met23 in LacY. All of the proteins shown in Figure 2 have a conserved Phe at the position corresponding to Phe20 in LacY. In addition, several proteins listed in Table 1 have either Trp or Tyr in positions homologous to Met23 in LacY. It is likely that these aromatic amino acids in helix I play a role in substrate binding and specificity.
An Arg residue at a position corresponding to 144 in LacY is strictly conserved in all proteins listed in Table 1. This Arg residue together with a Glu or Asp, homologous to Glu126 in LacY, are positioned almost identically in the threaded structures (Figure 2) and likely form a charge pair in the absence of substrate.37,38 In several bacterial sugar transporters (e.g. CscB from Escherichia coli (Figure 2(b)) and MaltP from Bacillus halodurans (not shown)), an Asp residue is present in a position homologous to Glu126 (helix IV) in LacY (Table 1). When this is the case, a Glu residue corresponding to Glu269 is shifted one turn of helix VIII towards the cytoplasmic surface and is in a position homologous to Asn272 in LacY. RafB and FruP (Figure 2(c) and (d)) have Glu residues homologous to Glu126 and Glu269 in LacY. Glu269 is in close proximity to Trp151 in LacYand may be the key residue in coupling sugar and H+ translocation.5,38 With the exception of replacement with Asp, which significantly compromises binding and transport,39,40 all other replacements for Glu269 are completely defective with respect to substrate binding and all translocation reactions catalyzed by LacY.
Residues involved in H+ translocation
The structural relationship between residues thought to be involved in H+ translocation is also significantly conserved (Figure 2). Three amino acid residues in helices IX and X (homologous to irreplaceable Arg302, His322 and Glu325 in LacY) are positioned close to each other and highly conserved in each protein (R300, H320 and E323 in CscB; R305, H325 and E328 in RafB; R295, H315 and E318 in FruP). The hydroxyl group of Tyr236 in LacY is also in close proximity to these three residues and may participate in a salt-bridge/H-bond network5 that is critical for active transport.41 The distances estimated from the X-ray structure of LacY between the OH of Tyr236 and Arg302, His322 or Glu325 are within 3–4 Å. Although replacement of Tyr236 with Phe abolishes active transport,41 replacement with Cys decreases lactose accumulation by only threefold.42 Possibly, the Y236F mutation places a hydrophobic insulator between Arg302, His322 and Glu325, thereby disrupting important H-bonds that can be partially replaced by H2O when Cys is in this position. The importance of the Tyr residue homologous to Tyr236 in LacY in other bacterial MFS sugar transporters (Figure 2) is suggested by conservation of Tyr at the analogous position in the other proteins studied (Tyr233 in CscB, Tyr239 in RafB and Tyr229 in FruP).
Biochemical studies indicate that His322 in LacY may be in close proximity to Glu26943,44 and interaction between the two residues may play an important role in sugar-coupled H+ translocation.10 In the inward-facing conformation,5,38 Glu269 is close to Trp151. Similar positioning of a Glu residue is observed in all homologs shown (Figure 2(b)–(d)), suggesting that this residue may play a similar role in other MFS transporters.
Charge paired residues between helices VII and X or XI
Asp240 (helix VII) and Lys319 (helix X) in LacY are salt-bridged,45 but unlike the charge pair between Asp237 (helix VII) and Lys358 (helix XI), Asp 240 and Lys319 can not be interchanged. In the threaded structures (Figure 2(b)–(d)), homologous Asp and Lys residues occupy similar positions as Asp240-Lys319 in LacY and likely form a salt-bridge connecting helices VII and X (Asp237-Lys317 in CscB, Asp243-Lys322 in RafB, and Asp233-Lys312 in FruP). Charge pair Asp237/Lys358 is important for membrane insertion of LacY and also participates non-specifically in binding of the substituent at the anomeric position of galactosidic substrates.5 The pair is present in RafB (Figure 2(c)), but is much less conserved in other members of the Oligosaccharide/H+ Symport subfamily. Possibly, this charge pair contributes to sugar affinity, but not specificity.5 Thus, CscB may not bind sugars in an extended conformation because there is no interaction with the C-terminal six-helix bundle as in LacY. Engineering the charge pair into CscB (mutant N234D/S356K) has been shown18 to increase membrane expression of the protein.
Gly residues may stabilize the inward-facing conformation
Sequence alignment and threading also reveals the location of conserved Gly residues at positions homologous to positions 46, 159, 262 and 370 in LacY (Figure 1; pink spheres). Gly residues may facilitate close contact between helices,46,47 and this is the case with helices II and XI (Gly46 and Gly370) and helices V and VIII (Gly159 and Gly262) in LacY. It is possible that the positions of these pairs of Gly residues facilitate tight closure at the periplasmic side of the molecule, thereby stabilizing the inward-facing conformation. It is also noteworthy that another 12-helix MFS transporter, GlpT,6 also crystallizes in an inward-facing conformation. In GlpT, Gly residues are also found in close proximity at the periplasmic ends of helices II and XI (Gly63, Gly66 and Gly405), as well as helix V (Gly169 and Gly170). Also, in the proposed outward-facing conformation of LacY5 Gly or amino acids with short side-chains are located in the postulated zone of contact between the N and C-terminal six-helix bundles on the cytoplasmic site.
Comparative sequence alignments with LacY and homology threading reveal that Gly residues 147 and 150 positioned on one side of helix V in the approximate middle of the membrane are highly conserved (Figure 1; yellow spheres) and are across from bulky side-chains in helix I. Therefore, it seems reasonable to suggest that conserved Gly residues on one side of helix V allow close contact with helix I, which is important for transport,48–50 and may be another general feature of MFS family members.
Aromatic side-chains and the inward-closed conformation
Current models for transport by MFS family members are consistent with alternating accessibility of substrate-binding sites to each side of the membrane.5,6,51 Since LacY and GlpT crystallize in an inward-facing conformation, the outward-facing conformation may represent a transition-state stabilized by interaction of amino acid side-chains. LacY in the inward-facing conformation exhibits three aromatic side-chains positioned in the same plane on the cytoplasmic side of the molecule (Phe140, Phe334 and Tyr350; shown as grey spheres on Figure 1(a)). These side-chains protrude into hydrophilic cavity above the sugar-binding site, but the distances between them are too great to make contact in the postulated inward-facing conformation. Nevertheless, close interaction of these amino acid residues was demonstrated by cross-linking experiments.52 Significant conservation of aromatic residues in other sugar transporters corresponding to Phe140, Phe334 and Tyr350 in LacY (Figure 1(b)–(d); grey spheres) suggests an important role in the mechanism.
Conserved Phe residues 277 and 278 in trans-membrane helix VIII of LacY are also positioned in the same plane as Phe140, Phe334 and Tyr350 close to the cytoplasmic cavity, but are directed away from the hydrophilic interior of the cavity. Glu269 is also located in helix VIII, but close to the middle of the molecule. It has been suggested that Glu269 is in close proximity to His322 in one conformation, but interacts with Arg144 and Trp151 when sugar occupies the binding site.5,43,44 Several amino acid residues in helix VIII of LacY exhibit altered accessibility to N-ethylmaleimide in the presence of sugar.37,50,53 In addition, a Cys at position 278 cross-links with a Cys placed at either position 140 or 143 in the presence of o-phenylenedimaleimide.52 Areasonable explanation for the observations is that helix VIII rotates, as in the crystal structure of LacY, Phe278 is directed towards the outside of the protein and cross-linking is unlikely without rotation. Rotation of helix VIII may bring Phe277 and Phe278 in close contact with Phe140, further facilitating closure of the cytoplasmic cavity.
Eukaryotic homologs
LacY homologs in eukaryotes
LacY is arguably the best studied of the MFS transport proteins and has been used as a model for structure/function predictions for other transporters.11–15,54,55 Nevertheless, significant sequence homology with LacY has been found with only a few bacterial transporters grouped in the Oligosaccharide/H+ Symport subfamily.3 A BLAST search was performed using the translated genomic databases starting from the amino acid sequence of the maltose permease (MaltP) homolog from the deep sea alkalophile B. halodurans,56 a homolog of LacY with conservation of the most essential amino acid residues (17% identity, see Table 1). The search exhibits significant similarities to more than 100 proteins of both bacterial and eukaryotic origin, including several from nerve tissue and tumors. One of the most intriguing proteins, human FLJ20160, was originally predicted to have nine TMDs and less than 500 amino acid residues (AAH50537). However, it was found recently that the number of amino acid residues is 791 (NM_017694). Human FLJ20160 is thought to be encoded by six exons located in a sequence of more than 60,000 bp of chromosome 2. The identification of the correct amino acid sequence of FLJ20160 is facilitated by available partial sequences in cDNA libraries that represent mature mRNA.
A total of 12 transmembrane helices were predicted24–27 from amino acid sequences of eukaryotic proteins similar to LacY. Secondary structure predictions allow alignment of these eukaryotic proteins with their prokaryotic homologs. Table 2 shows 12 eukaryotic and 28 prokaryotic proteins aligned over entire sequences with regions around the amino acid residues essential for substrate binding (helices IV–V, VIII) and H+ translocation (helices IX–X) in LacY. Although the function of these eukaryotic proteins and their location in the cell is unknown, significant similarities in the structural organization of the TMDs to MFS bacterial sugar transporters suggests that they may function in sugar binding or transport. Prediction of the cellular location for human protein FLJ20160 using the PSORT web-based program57 suggests localization in the cytoplasmic membrane with a possible post-translational modification of the N terminus by proteolysis. Additional searches in eukaryotic genomic databases starting from the orphan human protein FLJ20160 uncover a number of novel eukaryotic proteins with similarity to LacY in insects, worms, fish, birds and mammals, although significant differences are found in the overall lengths of the proteins. There are some uncertainties in the amino acid sequences of these proteins deduced by identifying possible splicing signals, which are not well defined. Nevertheless, using human protein FLJ20160 as a template facilitates the analysis of the translated sequences bringing the overall length of the eukaryotic homologs to about 800 amino acid residues.
Table 2.
Selected sequence alignment around regions essential for substrate binding and proton translocation for 40 eukaryotic and prokaryotic sugar MFS transporters similar to LacY
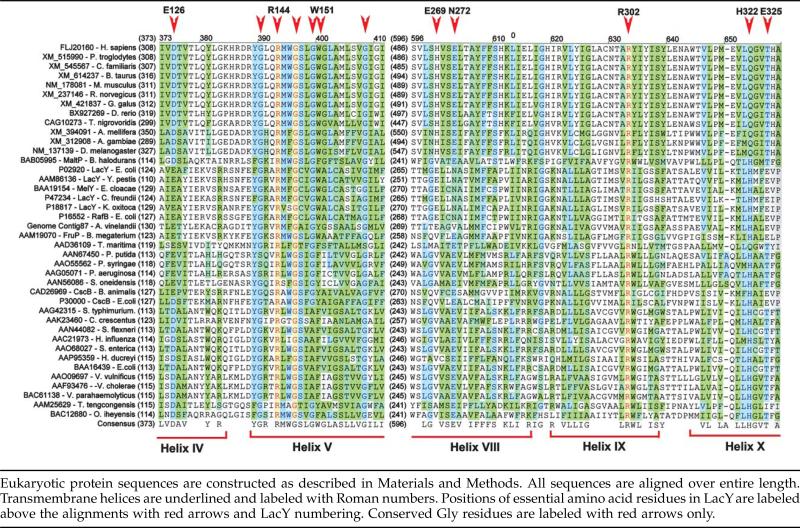 |
Putative eukaryotic homologs of LacY generally have longer N and C termini, longer cytoplasmic loops between helices VI and VII and a long loop between helices III and IV facing the outside of the cell. There is no homology between the eukaryotic proteins and their bacterial counterparts in these segments. On the other hand, very significant homology is found in the amino acid residues of the putativ TMDs (Table 2). All of the mammalian proteins have practically identical sequences in the TMDs. Most differences are observed in the N and C termini, as well as in loop III/IV. Loop III/IV, which is presumably located on the outer face of the membrane, has several predicted glycosylation sites. The most dramatic results from aligning the bacterial and eukaryotic proteins are in the conservation of amino acid residues essential for the structure and function of LacY and in all likelihood certain other MFS sugar transporters.
Two groups of prokaryotic and eukaryotic proteins can be identified based on aligning amino acid residues required for sugar binding in LacY (Table 2). Both groups have an absolutely conserved Arg at a position homologous to Arg144 in LacY. Several bacterial homologs have a Glu residue at position 126 in helix IV (LacY numbering), Glu269 in helix VIII and an Asn at position 272. Another group of transporters, which includes CscB from E. coli, MaltP and the putative eukaryotic homologs, have an Asp residue at a position homologous to 126 in LacY. As discussed above, when an Asp residue is found in place of Glu at this position, it is associated with a shift of Glu269 to a position homologous to Asn272 in LacY. Such a shift may lead to a change in sugar specificity. In all homologs, an aromatic residue is found at a position corresponding to Trp151 in LacY.
Homology threading of human FLJ20160 protein
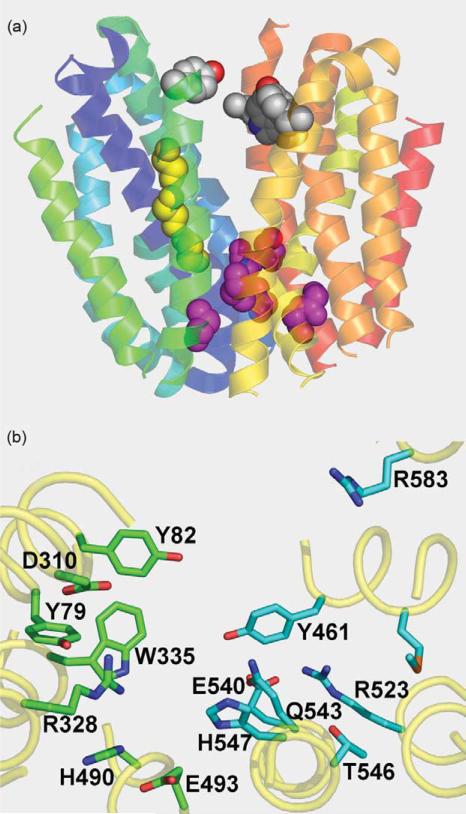
The homology observed in the TMDs of LacY and the eukaryotic homologs (Table 2) allows 3D modeling of TMDs in human protein FLJ20160. The organization of the 12 TMDs suggests that the structural features of bacterial sugar transporters (Figure 1) may be present in this human protein (Figure 3(a)). The exterior side of the protein appears to be closed, and conserved Gly residues provide close contact between helices V–VIII and II–XI (pink spheres). Gly residues that are conserved in the alignments (Table 2) are positioned in helix V facing helix I (Figure 3(a); yellow spheres). Aromatic residues facing hydrophilic cavity at the cytoplasmic opening are also conserved (grey spheres).
Figure 3.
Predicted structure of human FLJ20160 protein transmembrane domains threaded into LacY structure. (a) Organization of transmembrane helices with cytoplasmic side at the top. Connecting loops, and N and C termini are removed. Transmembrane helices are depicted as ribbons colored from blue (helix I) to red (helix XII). Conserved Gly residues in helices I and V are shown as yellow spheres. Conserved Gly residues on the extracellular surface between the N and C-terminal six-helix bundles are shown as pink spheres. Conserved aromatic residues facing hydrophilic cavity are shown as grey spheres. (b) Positions of residues that may be involved in sugar binding and/or H+ translocation viewed from the cytoplasmic side of the hydrophilic cavity. Amino acid residues with possible importance for sugar binding (green) or H+ translocation (cyan) are shown as sticks.
Homology threading also shows similarity of the putative sugar-binding site to the known binding site in LacY (Figures 2(a) and 3(b)). Trp335 in the human protein aligns nicely with Trp151 in LacY. Also, strictly conserved Arg328 lies just above Trp335, and Asp310 lies in close proximity in analogy to Glu126 and Arg144 in LacY. Glu493, which corresponds to Glu269 in LacY, is also close to Trp335, as in LacY or CscB. There are two Tyr residues at positions 79 and 82 located on the same side of the helix I in FLJ20160 that are homologous to Phe20 and Met23 in LacY.
Amino acid residues involved in H+ translocation in LacY also generally appear to be in a similar location in the eukaryotic homologs (Figures 2(a) and 3(b)). Strictly conserved Arg523 (Arg302 in LacY) is surrounded by Tyr461 (Tyr236 in LacY), Gln543 (His322 in LacY), Glu540 (Lys319 in LacY), Thr546 and His547 (Glu325 in LacY). The precise positions of the amino acid side-chains in the eukaryotic homologs that are involved in H+ translocation in LacY cannot be modeled because of the uncertainty of homology threading. However, the amino acid residues lying in close proximity to Arg523 are either the same or similar to those in LacY that are involved in H+ translocation. Unfortunately, none of these eukaryotic homologs has a known function. Therefore, any conclusions regarding either sugar binding and/or transport or sugar-coupled H+ translocation are highly speculative at this time.
Human and mouse homologs were initially identified from cDNA libraries constructed from brain mRNA. The expression level of human protein FLJ20160 is based on analyses of novel expressed sequence tag (EST) counts in the UniGene Database.58–60 The highest levels are found in brain although the ESTs are also found in many other tissues, including tumors. Since nerve cells, as well as tumors, use glucose as a primary energy source, it is enticing to speculate that the eukaryotic homologs may represent a class of sugar binding/transport proteins.
Cloning and expression of human protein FLJ20160
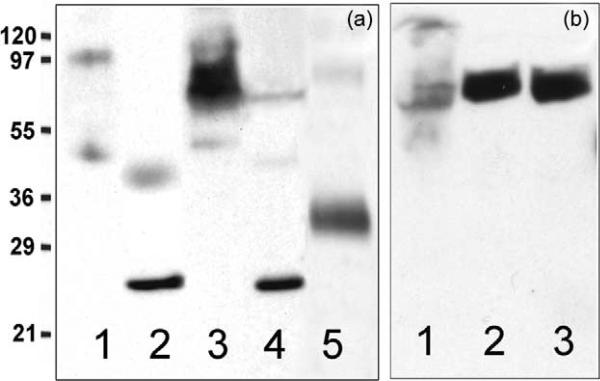
Putative eukaryotic homologs of LacY should be inserted into membranes. Evidence supporting this notion was obtained by cloning the FLJ20160 gene into bacterial and yeast expression vectors. A construct encoding full-length FLJ20160 protein under the LacZ promoter/operator was used to test expression in E. coli XL1-Blue (Figure 4(a)). Clearly, FLJ20160 protein is expressed (lane 3), and there is essentially no protein in either the membrane or cytoplasmic fractions from control untransformed cells (lanes 1 and 2). The cytoplasmic fraction (lane 4) from the same cells harboring the plasmid also does not contain FLJ20160 protein after induction. Rather, FLJ20160 is found exclusively in the membrane fraction and appears to be expressed at a reasonably high level. In addition, treatment of the membrane fraction with 5 M urea for 30 min does not remove human FLJ20160 protein from the membrane, indicating that the protein is trans-membrane (data not shown). The apparent molecular mass of FLJ20160 is about double that of LacY on SDS/polyacrylamide gel electrophoresis (compare lines 3 and 5 in Figure 4(a)). Like LacY (45.5 kDa from DNA sequence) and other hydro-phobic membrane proteins, FLJ20160 (89 kDa from DNA sequence) migrates at a lower apparent molecular mass.
Figure 4.
Insertion of human FLJ20160 protein into membranes of E. coli or P. pastoris. Membrane fractions were separated from cytoplasm by centrifugation and samples containing 200 μg (a) or 50 μg (b) of protein were subjected to SDS/polyacrylamide gel (12%) electrophoresis. The gel was electroblotted to PVDF membrane and probed with HRP-conjugated antibody directed against His-tag. Molecular weight ladder is shown in kDa. (a) Membrane (lane 1) or cytoplasm (lane 2) fractions isolated from control untransformed E. coli XL1-Blue cells. Membrane (lane 3) or cytoplasm (lane 4) fractions isolated from E. coli XL1-Blue cells harboring pSP72 plasmid encoding human FLJ20160 protein. Purified LacY/His10 (0.2 μg; lane 5) is shown for comparison. (b) Membrane fractions isolated from different P. pastoris strains harboring pPICZ-A plasmid encoding human FLJ20160 protein after induction with methanol. KM71H (lane 1), X-33 (lane 2), and GS115 (lane 3).
The yeast vector pPICZ-A from the Invitrogen EasySelect Pichia expression kit was used to clone the complete gene encoding FLJ20160 into the KpnI/XhoI sites in-frame with sequences encoding the Myc-epitope and a 6-His-tag. Several Pichia pastoris strains were transformed with linearized plasmid, selected for multiple-integrants with high Zeocin concentrations and used to test for expression (Figure 4(b)). As shown, although expression of FLJ20160 in membranes from P. pastoris KM71H is low, good expression is apparent in the membranes of the wild-type strain X-33 (lane 2), as well as strain GS115 (lane 3).
Materials and Methods
Analysis of sequence homology
Sequence homology searches were carried out by using an on-line search (Vector NTI 9.1 Suit, Invitrogen, Carlsbad, CA,) starting from protein sequences and using either protein databases (BLAST-p) or translated open reading frames in DNA databases (BLAST-tn). Resulting protein and DNA sequences were directly downloaded to Vector NTI, and sequence alignments were done by using the AlignX module of the same program. Secondary structure and transmembrane regions from protein sequences were predicted by using Predict Protein Server.24–27
A BLAST-tn search was performed from translated genomic databases using putative maltose permease (MaltP) from B. halodurans56 as a starting point. The human FLJ20160 amino acid sequence resulting from the search was used to find other homologous eukaryotic proteins from a second BLAST-tn search. Protein sequences from genomic databases were aligned with the human and mouse proteins with relatively reliable sequences as judged by cDNA sequences. The nucleotide sequences were analyzed for the protein coding sequences by using the web-based GeneBuilder program.61 Strong splicing signals were used to identify the correct sequences of the genes, and human protein FLJ20160 was selected as the template. Those amino acid sequences that were misaligned relative to the human protein were examined for possible splicing signals in the corresponding DNA sequences and appropriate corrections were made.
Homology threading and tertiary structure analysis
Homology threading was done for selected proteins with high sequence similarity. All proteins were threaded into the LacY structure by using the X-ray coordinates (PDB code 1PV7) as a template on the web-based SWISSPROT modeling server.22 All homology threading was done without manual optimization using default mode in order to eliminate human bias. 3D-structures obtained by homology threading were displayed using Pymol 0.97 (DeLano Scientific, LLC) and compared to the LacY structure. Homology threading of the human protein FLJ20160 was done by removing the long N and C-terminal sequences in addition to the longer connecting loops, so that the overall length was similar to LacY.
Cloning end expression of human FLJ20160 gene
DNA clone ID 6204874 from the MGC full-length (IRAT) collection encoding part of the human FLJ20160 protein (amino acid residues 247–791) in pCMV-Sport6 vector and BAC clone RP11-647K16 (encoding amino acid residues 1–511) in pBAC 3.6 vector were purchased from Invitrogen (Carlsbad, CA). PCR primers were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). Gene fragment no. 1 encoding the N terminus of FLJ20160 was amplified by PCR from the BAC clone using a direct primer with an engineered KpnI unique restriction site (CTTGCTGATGGTGGTGGTACCCCATGGCAGATGATAAAG) and a reverse primer (GCTTCAACTTCTTGTTGATCATAAACAAGCATG) containing a BclI restriction site. Gene fragment no. 2 encoding the second part of FLJ20160 was amplified by PCR from cDNA using a direct primer (CATGCTTGTTTATGATCAACAAGAAGTTGAAGC) containing restriction site BclI and a reverse primer that introduced a unique XhoI restriction site (GAGATGAGCAGGATGCCCTCGAGTGTCCTCCCGCGGCCAGC). Fragment no. 1 digested with KpnI and BclI and fragment no. 2 digested with BclI and XhoI were purified on agarose gels and sequentially ligated into plasmid pSP72, which confers ampicillin resistance, and contains KpnI, BclI and XhoI restriction sites followed by an in-frame 6-His tag. The final construct contained the full-length human gene encoding FLJ20160 under the LacZ promoter/operator and used for expression experiments. A Complete EasySelect Pichia expression kit was purchased from Invitrogen (Carlsbad, CA). The human gene isolated with KpnI/XhoI restriction enzymes from pSP72 plasmid harboring the FLJ20160 gene was ligated into the KpnI/XhoI sites of the yeast expression vector pPICZ-A in-frame with a Myc-epitope and a 6-His-tag. All plasmids were sequenced over the entire gene sequence before use in expression studies.
E. coli XL1-Blue cells (Stratagene, La Jolla, CA) transformed with pSP72 plasmid encoding the FLJ20160 gene were grown in LB medium at 37 °C with 100 mg/l ampicillin. Cells were induced with 0.3 mM IPTG, harvested after 4 h by centrifugation and disrupted by using an Emulsiflex (Avestin International, Canada). Membranes were separated from cytoplasm by centrifugation and subjected to SDS/polyacrylamide gel electrophoresis and Western blot analysis using His-tag directed HRP-conjugated antibody (Qiagen, Valencia, CA), as described.49 Protein concentrations were measured by using the Micro BCA method (Pierce, Rockford, IL).
The yeast expression plasmid was linearized with SacI and used for transformation of P. pastoris strains X-33, GS115 or KM71H, as described in the EasySelect Pichia expression kit (Invitrogen, Carlsbad, CA). Yeast cells were selected for multiple copy gene expression with increasing Zeocin concentrations and used for expression trials. Cells were grown and induced according to the manufacturer's recommendations. Harvested cells were disrupted with glass beads, membranes were separated by centrifugation and subjected to SDS/polyacrylamide gel electrophoresis and Western blot analysis as described above.
Acknowledgements
We thank Dr Viveka Vadyvaloo for excellent assistance in cloning of the human FLJ20160 gene into pSP72 plasmid, Junichi Sugihara for technical help with protein expression experiments and Dr Jun-Yong Choe for critically reading the manuscript. This work was supported by NIH grants DK06946301 and DK051131 to H.R.K. As well as the NSF grant 0450970.
Abbreviations used
- MFS
Major Facilitator Super-family
- TMD
transmembrane domain
References
- 1.Fleming KG. Riding the wave: structural and energetic principles of helical membrane proteins. Curr. Opin. Biotechnol. 2000;11:67–71. doi: 10.1016/s0958-1669(99)00056-7. [DOI] [PubMed] [Google Scholar]
- 2.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–32. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saier MH., Jr Families of transmembrane sugar transport proteins. Mol. Microbiol. 2000;35:699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 4.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, et al. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 5.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 7.Büchel DE, Gronenborn B, Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980;283:541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- 8.Newman MJ, Foster DL, Wilson TH, Kaback HR. Purification and reconstitution of functional lactose carrier from Escherichia coli. J. Biol. Chem. 1981;256:11804–11808. [PubMed] [Google Scholar]
- 9.Viitanen P, Garcia ML, Kaback HR. Purified reconstituted lac carrier protein from Escherichia coli is fully functional. Proc. Natl Acad. Sci. USA. 1984;81:1629–1633. doi: 10.1073/pnas.81.6.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaback HR, Sahin-Toth M, Weinglass AB. The Kamikaze approach to membrane transport. Nature Rev. Mol. Cell. Biol. 2001;2:610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 11.Zuniga FA, Shi G, Haller JF, Rubashkin A, Flynn DR, Iserovich P, Fischbarg J. A three-dimensional model of the human facilitative glucose transporter Glut1. J. Biol. Chem. 2001;276:44970–44975. doi: 10.1074/jbc.M107350200. [DOI] [PubMed] [Google Scholar]
- 12.Hirai T, Heymann JA, Maloney PC, Subramaniam S. Structural model for 12-helix transporters belonging to the major facilitator superfamily. J. Bacteriol. 2003;185:1712–1718. doi: 10.1128/JB.185.5.1712-1718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood JM, Culham DE, Hillar A, Vernikovska YI, Liu F, Boggs JM, Keates RA. A structural model for the osmosensor, transporter, and osmoregulator ProP of Escherichia coli. Biochemistry. 2005;44:5634–5646. doi: 10.1021/bi047383o. [DOI] [PubMed] [Google Scholar]
- 14.Popp C, Gorboulev V, Muller TD, Gorbunov D, Shatskaya N, Koepsell H. Amino acids critical for substrate affinity of rat organic cation transporter 1 line the substrate binding region in a model derived from the tertiary structure of lactose permease. Mol. Pharmacol. 2005;67:1600–1611. doi: 10.1124/mol.104.008839. [DOI] [PubMed] [Google Scholar]
- 15.Vardy E, Arkin IT, Gottschalk KE, Kaback HR, Schuldiner S. Structural conservation in the major facilitator superfamily as revealed by comparative modeling. Protein Sci. 2004;13:1832–1840. doi: 10.1110/ps.04657704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sigal N, Vardy E, Molshanski-Mor S, Eitan A, Pilpel Y, Schuldiner S, Bibi E. 3D model of the Escherichia coli multidrug transporter MdfA reveals an essential membrane-embedded positive charge. Biochemistry. 2005;44:14870–14880. doi: 10.1021/bi051574p. [DOI] [PubMed] [Google Scholar]
- 17.Aslanidis C, Schmid K, Schmitt R. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J. Bacteriol. 1989;171:6753–6763. doi: 10.1128/jb.171.12.6753-6763.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frillingos S, Sahin-Tóth M, Lengeler JW, Kaback HR. Helix packing in the sucrose permease of Escherichia coli: properties of engineered charge pairs between helices VII and XI. Biochemistry. 1995;34:9368–9373. doi: 10.1021/bi00029a012. [DOI] [PubMed] [Google Scholar]
- 19.Sahin-Tóth M, Kaback HR. Functional conservation in the putative substrate binding site of the sucrose permease from Escherichia coli. Biochemistry. 2000;39:6170–6175. doi: 10.1021/bi000125g. [DOI] [PubMed] [Google Scholar]
- 20.Shinnick SG, Perez SA, Varela MF. Altered substrate selection of the melibiose transporter (MelY) of Enterobacter cloacae involving point mutations in Leu-88. Leu-91, and Ala-182 that confer enhanced maltose transport. J. Bacteriol. 2003;185:3672–3677. doi: 10.1128/JB.185.12.3672-3677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiou CY, Wang HH, Shaw GC. Identification and characterization of the non-PTS fru locus of Bacillus megaterium ATCC 14581. Mol. Genet. Genomics. 2002;268:240–248. doi: 10.1007/s00438-002-0741-y. [DOI] [PubMed] [Google Scholar]
- 22.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucl. Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vadyvaloo V, Smirnova IN, Kasho VN, Kaback HR. Conservation of residues involved in sugar/H+ symport by the sucrose permease of Escherichia coli relative to lactose permease. J. Mol. Biol. 2006 doi: 10.1016/j.jmb.2006.02.050. In the press (accomp. paper) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins: Struct. Funct. Genet. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 25.Rost B, Fariselli P, Casadio R. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 28.Sahin-Tóth M, Gunawan P, Lawrence MC, Toyokuni T, Kaback HR. Binding of hydrophobic d-galactopyranosides to the lactose permease of Escherichia coli. Biochemistry. 2002;41:13039–13045. doi: 10.1021/bi0203076. [DOI] [PubMed] [Google Scholar]
- 29.Frillingos S, Sahin-Toth M, Wu J, Kaback HR. Cys-scanning mutagenesis: a novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 30.Weinglass AB, Whitelegge JP, Hu Y, Verner GE, Faull KF, Kaback HR. Elucidation of substrate binding interactions in a membrane transport protein by mass spectrometry. EMBO J. 2003;22:1467–1477. doi: 10.1093/emboj/cdg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan L, Hu Y, Kaback HR. Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry. 2003;42:1377–1382. doi: 10.1021/bi027152m. [DOI] [PubMed] [Google Scholar]
- 32.Vazquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc. Natl Acad. Sci. USA. 2003;100:12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merritt EA, Sarfaty S, Feil IK, Hol WG. Structural foundation for the design of receptor antagonists targeting Escherichia coli heat-labile enter-otoxin. Structure. 1997;5:1485–1499. doi: 10.1016/s0969-2126(97)00298-0. [DOI] [PubMed] [Google Scholar]
- 34.Walser PJ, Haebel PW, Kunzler M, Sargent D, Kues U, Aebi M, Ban N. Structure and functional analysis of the fungal galectin CGL2. Structure (Camb) 2004;12:689–702. doi: 10.1016/j.str.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Fotinou C, Black I, Fairweather NF, Charles IG, Watts C, et al. The structures of the H(C) fragment of tetanus toxin with carbohydrate subunit complexes provide insight into ganglioside binding. J. Biol. Chem. 2000;275:8889–8894. doi: 10.1074/jbc.275.12.8889. [DOI] [PubMed] [Google Scholar]
- 36.Seetharaman J, Kanigsberg A, Slaaby R, Leffler H, Barondes SH, Rini JM. X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-Å resolution. J. Biol. Chem. 1998;273:13047–13052. doi: 10.1074/jbc.273.21.13047. [DOI] [PubMed] [Google Scholar]
- 37.Venkatesan P, Kaback HR. The substrate-binding site in the lactose permease of Escherichia coli. Proc. Natl Acad. Sci. USA. 1998;95:9802–9807. doi: 10.1073/pnas.95.17.9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ujwal ML, Sahin-Tóth M, Persson B, Kaback HR. Role of glutamate-269 in the lactose permease of Escherichia coli. Mol. Membr. Biol. 1994;11:9–16. doi: 10.3109/09687689409161024. [DOI] [PubMed] [Google Scholar]
- 40.Franco PJ, Brooker RJ. Functional roles of Glu-269 and Glu-325 within the lactose permease of Escherichia coli. J. Biol. Chem. 1994;269:7379–7386. [PubMed] [Google Scholar]
- 41.Roepe PD, Kaback HR. Site-directed mutagenesis of tyrosine residues in the lac permease of Escherichia coli. Biochemistry. 1989;28:6127–6132. doi: 10.1021/bi00440a060. [DOI] [PubMed] [Google Scholar]
- 42.Frillingos S, Sahin-Tóth M, Persson B, Kaback HR. Cysteine-scanning mutagenesis of putative helix VII in the lactose permease of Escherichia coli. Biochemistry. 1994;33:8074–8081. doi: 10.1021/bi00192a012. [DOI] [PubMed] [Google Scholar]
- 43.Jung K, Jung H, Wu J, Privé GG, Kaback HR. Use of site-directed fluorescence labeling to study proximity relationships in the lactose permease of Escherichia coli. Biochemistry. 1993;32:12273–12278. doi: 10.1021/bi00097a001. [DOI] [PubMed] [Google Scholar]
- 44.Jung K, Voss J, He M, Hubbell WL, Kaback HR. Engineering a metal binding site within a polytopic membrane protein, the lactose permease of Escherichia coli. Biochemistry. 1995;34:6272–6277. doi: 10.1021/bi00019a003. [DOI] [PubMed] [Google Scholar]
- 45.Sahin-Tóth M, Kaback HR. Properties of interacting aspartic acid and lysine residues in the lactose permease of Escherichia coli. Biochemistry. 1993;32:10027–10035. doi: 10.1021/bi00089a019. [DOI] [PubMed] [Google Scholar]
- 46.Senes A, Ubarretxena-Belandia I, Engelman DM. The Calpha···H···O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. Proc. Natl Acad. Sci. USA. 2001;98:9056–9061. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu W, Eilers M, Patel AB, Smith SO. Helix packing moments reveal diversity and conservation in membrane protein structure. J. Mol. Biol. 2004;337:713–729. doi: 10.1016/j.jmb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 48.Jung K, Jung H, Colacurcio P, Kaback HR. Role of glycine residues in the structure and function of lactose permease, an Escherichia coli membrane transport protein. Biochemistry. 1995;34:1030–1039. doi: 10.1021/bi00003a038. [DOI] [PubMed] [Google Scholar]
- 49.Smirnova IN, Kaback HR. A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry. 2003;42:3025–3031. doi: 10.1021/bi027329c. [DOI] [PubMed] [Google Scholar]
- 50.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Interhelical packing modulates conformational flexibility in the lactose permease of Escherichia coli. Biochemistry. 2005;44:7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 51.Guan L, Kaback HR. Binding affinity of lactose permease is not altered by the H+ electrochemical gradient. Proc. Natl Acad. Sci. USA. 2004;101:12148–12152. doi: 10.1073/pnas.0404936101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, Hardy D, Kaback HR. Tertiary contacts of helix V in the lactose permease determined by site-directed chemical crosslinking in situ. Biochemistry. 1999;38:2320–2325. doi: 10.1021/bi982288z. [DOI] [PubMed] [Google Scholar]
- 53.Frillingos S, Kaback HR. The role of helix VIII in the lactose permease of Escherichia coli. II. Site-directed sulfhydryl modification. Protein Sci. 1997;6:438–443. doi: 10.1002/pro.5560060221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gottschalk KE, Soskine M, Schuldiner S, Kessler H. A structural model of EmrE, a multi-drug transporter from Escherichia coli. Biophys. J. 2004;86:3335–3348. doi: 10.1529/biophysj.103.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patching SG, Baldwin SA, Baldwin AD, Young JD, Gallagher MP, Henderson PJ, Herbert RB. The nucleoside transport proteins, NupC and NupG, from Escherichia coli: specific structural motifs necessary for the binding of ligands. Org. Biomol. Chem. 2005;3:462–470. doi: 10.1039/b414739a. [DOI] [PubMed] [Google Scholar]
- 56.Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, et al. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucl. Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Horton P, Nakai K. Better prediction of protein cellular localization sites with the k nearest neighbors classifier. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 58.Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 59.Wheeler DL, Church DM, Edgar R, Federhen S, Helmberg W, Madden TL, et al. Database resources of the National Center for Biotechnology Information: update. Nucl. Acids Res. 2004;32:D35–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wheeler DL, Church DM, Federhen S, Lash AE, Madden TL, Pontius JU, et al. Database resources of the National Center for Biotechnology. Nucl. Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milanesi L, D'Angelo D, Rogozin IB. GeneBuilder: interactive in silico prediction of gene structure. Bioinformatics. 1999;15:612–621. doi: 10.1093/bioinformatics/15.7.612. [DOI] [PubMed] [Google Scholar]