Abstract
Ubiquitination-mediated degradation of the RelA subunit of nuclear factor-κB (NF-κB) is critical for the termination of NF-κB activation. However, the precise mechanism for the ubiquitination of RelA is still not fully understood. Here we report that tumor necrosis factor-α (TNFα) induces RelA polyubiquitination at the lysine 195 residue, and this ubiquitination event is critical for the degradation of RelA and termination of TNFα-mediated NF-κB activation. Overexpression of a RelA mutant with an arginine substitution for the lysine 195 residue dramatically inhibits RelA polyubiquitination and induces a stronger NF-κB activation compared with the wild type. Reconstitution of RelA-deficient mouse embryo fibroblast cells with wild-type RelA or RelA containing a K195R mutation revealed the importance of this site in TNFα-mediated RelA polyubiquitination, degradation, and attenuation of NF-κB activation. Our finding is the first report that substitution of a key RelA lysine residue with arginine inhibits TNFα-induced RelA ubiquitination and enhances TNFα-induced NF-κB activation.
Nuclear factor-κB (NF-κB)2 represents a family of dimeric transcription factors comprising RelA (p65), RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52) in mammals (1, 2). NF-κB proteins form various homodimers and heterodimers via their N-terminal Rel homology domains and mediate cellular responses in inflammation, immunity, development, cell proliferation, and apoptosis (3, 4). Tumor necrosis factor-α (TNFα) is a proinflammatory cytokine that exerts its function through activation of NF-κB along with other transcription factors upon binding to its receptor (2). In unstimulated cells, NF-κB dimers are sequestered in the cytoplasm as latent complexes by physical association of their Rel homology domains with NF-κB inhibitory proteins, IκBs (5). Binding of TNFα to its receptor on the cell surface induces activation of IκB kinase (IKK). The activated IKK phosphorylates two serine residues located in an IκB regulatory domain, and this phosphorylation event leads to IκB ubiquitination and subsequent degradation by the proteasome. Degradation of the IκB proteins frees the NF-κB complex to enter the nucleus and activates NF-κB-dependent gene expression (6, 7).
Recently, ubiquitination and degradation of the RelA subunit of NF-κB in the nucleus have been suggested to be involved in the negative regulation of NF-κB activation (8–12). Furthermore, RelA ubiquitination has been shown to be controlled by various factors, including SOCS1 (suppressor of cytokine signaling 1), PDLIM2 (PDZ and LIM domain 2), COMMD1 (COMM domain-containing 1), and GCN5 (8, 12–14). Interestingly, the ORF73 protein encoded by murid herpesvirus 4 inhibits NF-κB transcriptional activity through polyubiquitination and nuclear degradation of RelA (15).
RelA Ser-536 is a target for IKKα-dependent phosphorylation and mediates lipopolysaccharide-induced RelA turnover (16). Phosphorylation of RelA at Ser-468 controls its COMMD1-dependent ubiquitination and degradation (17). However, the mechanism and role of RelA ubiquitination in TNFα-induced NF-κB activation have not been fully characterized.
In this study, we used molecular and biochemical approaches to identify a potential ubiquitin lysine acceptor site(s) in the RelA protein. We present evidence that Lys-195 of the RelA protein is the key ubiquitin acceptor site responsible for mediating TNFα-induced RelA ubiquitination and degradation as well as attenuation of TNFα-mediated NF-κB activation.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK-293T cells and RelA-deficient MEF cells (18) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. For transfection, HEK-293T cells and RelA-deficient MEF cells were transiently transfected with expression plasmids by using FuGENE 6 (Roche Applied Science) and Lipofectamine 2000 (Invitrogen), respectively.
Antibodies and Reagents
Mouse monoclonal anti-RelA (sc-8008), anti-Myc (sc-40), anti-HA (sc-7392), anti-Ub (sc-8017), and anti-proliferating cell nuclear antigen (sc-56) antibodies and protein A-agarose were obtained from Santa Cruz Biotechnology. Polyclonal rabbit anti-phospho-IKKα/β (2681), anti-IKKβ (2684), and anti-IκBα (9242) antibodies and secondary antibodies conjugated to horseradish peroxidase were obtained from Cell Signaling. Mouse monoclonal anti-actin (A2228) and anti-FLAG (F3165) antibodies were from Sigma. Recombinant human and mouse TNFα were from R&D Systems. MG132 was from Calbiochem.
Plasmids
Human RelA cDNA was inserted into the expression vector pcDNA3.1 with a FLAG or Myc tag at its N terminus. Point mutations of each lysine were generated by site-directed mutagenesis. Retroviral wild-type RelA and K195R mutant constructs were generated using the pBabe-puro vector. The NF-κB-dependent firefly luciferase reporter and pCMV promoter-dependent Renilla luciferase reporter plasmids were purchased from Clontech.
Establishment of RelA Stable MEF Cell Lines
For retroviral infection, the pBabe empty vector pBabe-RelA wild type, and K195R mutant were cotransfected with retrovirus packing vector Pegpam 3e and pLC-ECO in HEK-293T cells to obtain retroviral supernatants. Viral supernatants were collected after 48 and 72 h. RelA-deficient MEF cells were incubated with retroviral supernatant in the presence of 4 μg/ml Polybrene. Stable cell lines were established after 10 days of puromycin (3 μg/ml) selection.
Preparation of Cytoplasmic and Nuclear Extracts
Nuclear and cytosolic extracts were obtained as described (19). Cells were lysed on ice for 30 min with hypotonic buffer (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 20 mm glycerophosphate, 0.1 mm sodium orthovanadate, 10 μg/ml aprotinin, and 10 μg/ml leupeptin). After centrifugation of samples at 15,000 × g for 15 min, supernatants were collected and were used as cytoplasmic fractions. Pellets were then lysed for 30 min on ice in hypertonic buffer (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 20 mm glycerophosphate, 1 mm sodium orthovanadate, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) with brief vortexing. After centrifugation at 15,000 × g for 15 min, supernatants were collected as nuclear fractions.
Immunoprecipitation and Immunoblotting
For immunoprecipitation, cells were washed three times with ice-cold phosphate-buffered saline on ice, and then whole cell extracts were prepared by lysing cells in lysis buffer (25 mm HEPES, pH 7.7, 135 mm NaCl, 3 mm EDTA, 1% Triton X-100, 25 mm β-glycerophosphate, 0.1 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm benzamidine, 20 mm disodium p-nitrophenyl phosphate, and phosphatase inhibitor mixture A and B (Sigma)). After collecting the lysate by centrifugation at 15,000 × g for 15 min at 4 °C, primary antibodies were added to the supernatant and incubated with rotation for 3 h at 4 °C. After adding a protein A-agarose bead suspension, the mixture was further incubated with rotation for 3 h at 4 °C. The precipitates were washed three times using pre-cold washing buffer (20 mm HEPES, pH 7.7, 50 mm NaCl, 2.5 mm MgCl2, 0.1 mm EDTA, and 0.05% Triton X-100), and then the beads were resuspended in Laemmli sample buffer and boiled for 10 min. The immunoprecipitates or the whole cell lysates were resolved by SDS-PAGE and transferred to nitrocellulose membranes (Bio-Rad). The membranes were probed with appropriate antibodies. Horseradish peroxidase-conjugated IgG antibodies were used as the secondary antibodies. The proteins were detected using the ECL Plus Western blotting detection system.
Electrophoretic Mobility Shift Assay
An electrophoretic mobility shift assay was performed with Gel Shift assay systems (Promega). Briefly, MEF cells (1 × 106) were stimulated for the indicated time points. Nuclear extracts isolated from these cells were then incubated with 32P-labeled probes in binding buffer containing 0.5 mm EDTA, 0.5 mm dithiothreitol, 4% glycerol, 1 mm MgCl2, 50 mm NaCl, 10 mm Tris-HCl, pH 7.5, and 0.05 mg/ml poly(dI-dC) for 20 min at room temperature. DNA-protein complexes were resolved at 200 V for 2 h by using native 5% polyacrylamide gels. The gels were transferred to Whatman No. 3MM paper, dried, and exposed to x-ray film overnight at −80 °C.
Reverse Transcription and Quantitative Real-time PCR
Total cellular RNA was isolated by using TRIzol reagent (Invitrogen), and cDNA was prepared using SuperScript III gene expression tools (Invitrogen) according to the manufacturer's protocol. One μg of RNA was used for cDNA synthesis from oligo(dT) primers using the SuperScript first-strand synthesis system (Invitrogen). Quantitative real-time PCR was performed using specific primers described previously (20) and SYBR Green ROX mix (Qiagen), and reactions were analyzed using an Applied Biosystems 7300 real-time PCR system. Data were normalized to the housekeeping glyceraldehyde-3-phosphate dehydrogenase gene, and the relative abundance of transcripts was calculated by using Ct models.
Enzyme-linked Immunosorbent Assay
To measure levels of mouse IL-6, RelA-deficient and wild-type RelA- and K195R mutant-reconstituted MEF cells were treated with or without mouse TNFα (2 ng/ml), and the supernatants were collected at different time points. Mouse IL-6 concentrations in the supernatants were determined by enzyme-linked immunosorbent assay using a BD Biosciences kit according to the manufacturer's instructions.
Luciferase Reporter Assay
Targeted cells were seeded at 3 × 105 cells/well and cultured overnight in 6-well plates. The cells were transfected with NF-κB-dependent firefly luciferase construct, Renilla luciferase, and expression vectors. Luciferase activity was measured according to the manufacturer's protocol (Promega). The relative luciferase activity was calculated by dividing the firefly luciferase activity by the Renilla luciferase activity. Data represent three independent experiments performed in triplicate.
RESULTS
Lys-195 Is Required for RelA Overexpression-mediated Polyubiquitination
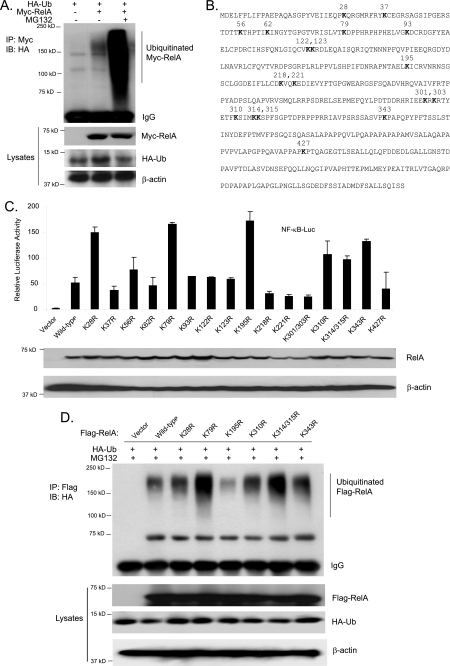
To determine whether RelA is ubiquitinated, HA-ubiquitin expression vectors were cotransfected with vector control or Myc-RelA expression vectors into HEK-293T cells. Cells were lysed after treatment with or without MG132, and Myc-RelA proteins were immunoprecipitated and immunoblotted with anti-HA antibodies. In this assay, we found that overexpressed Myc-RelA proteins were partially ubiquitinated, and MG132 treatment dramatically increased the level of ubiquitinated Myc-RelA (Fig. 1A).
FIGURE 1.
Lys-195 is required for RelA overexpression-mediated polyubiquitination. A, overexpression of RelA induces its polyubiquitination. HEK-293T cells were transfected with control vectors or expression vectors encoding Myc-RelA along with HA-Ub. Cells were lysed after treatment with MG132 for 3 h. Myc-RelA proteins in the cell lysates were immunoprecipitated (IP) with anti-Myc antibodies and immunoblotted (IB) with anti-HA antibodies. B, RelA primary sequence, with the lysine residues indicated. C, effects of overexpression of RelA lysine-to-arginine mutants on RelA overexpression-induced NF-κB activation. NF-κB luciferase (NF-κB-Luc) reporter and control Renilla luciferase reporter vectors were cotransfected into RelA-deficient MEF cells with empty vector or expression vectors encoding wild-type RelA or lysine-to-arginine mutants, respectively. The relative luciferase activity was measured 48 h later and normalized with the Renilla activity. Error bars indicate standard deviations from triplicate experiments. D, effects of overexpression of RelA lysine-to-arginine mutants on RelA overexpression-induced RelA polyubiquitination. Expression vectors encoding FLAG-tagged wild-type RelA or lysine-to-arginine mutants were individually transfected into HEK-293T cells with expression vectors encoding HA-Ub. FLAG-RelA proteins in the cell lysates were immunoprecipitated with anti-FLAG antibodies and immunoblotted with anti-HA antibodies.
To address the effect of polyubiquitination of RelA on its activation, it is critical to map the Ub lysine acceptor site(s) of RelA and characterize the functional effects of eliminating the site(s) on NF-κB activation. To determine the location of the Ub lysine acceptor site(s) of RelA, we systematically replaced each of the RelA Lys residues with an Arg, which would maintain the positive charge but would not serve as an acceptor site for Ub modification (Fig. 1B). To determine the functional effects of these mutants of RelA on its transactivity, the expression vectors encoding wild-type RelA and mutants with different Lys-to-Arg mutations were cotransfected along with NF-κB luciferase reporter constructs in the RelA-deficient MEF cells. As shown in Fig. 1C, overexpression of RelA K28R, K79R, K195R, K310R, K314R/K315R, and K343R mutants induced a dramatically increased NF-κB-dependent luciferase reporter gene expression compared with wild-type RelA, whereas other mutants showed a decreased reporter gene expression. This result suggests that Lys-to-Arg mutations at these seven sites may abolish RelA polyubiquitination and degradation and result in an increased RelA transactivity. Therefore, one or more of these seven Lys residues are potential Ub acceptor site(s) to mediate RelA polyubiquitination.
To further map the Ub lysine acceptor site(s) for mediation of RelA polyubiquitination, we cotransfected FLAG-tagged wild-type RelA and the seven Lys-to-Arg mutants described above along with HA-ubiquitin into HEK-293T cells. FLAG-RelA proteins were immunoprecipitated and immunoblotted with anti-HA antibodies. In this assay, we found that only the K195R mutant showed dramatically decreased polyubiquitination (Fig. 1D).
Interestingly, we found that RelA K79R and K314R/K315R overexpression induced a higher NF-κB activation in the reporter assay, whereas these two RelA mutants had a slightly higher level of polyubiquitination compared with the wild type (Fig. 1, C and D). One of the explanations for this result is that RelA K79R and K314R/K315R mutants have a decreased binding affinity for IκBα. To confirm this hypothesis, FLAG-tagged wild-type RelA and K195R, K79R, and K314R/K315R mutants overexpressed in the HEK-293T cells were immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-IκBα antibody. In this assay, we found that RelA K79R and K314R/K315R mutants had a decreased binding affinity for endogenous IκBα compared with the wild type and K195R (data not shown). Together, these results strongly suggest that the RelA Lys-195 residue is the Ub acceptor site that mediates RelA polyubiquitination.
RelA Lys-195 Mediates TNFα-induced RelA Polyubiquitination and Degradation
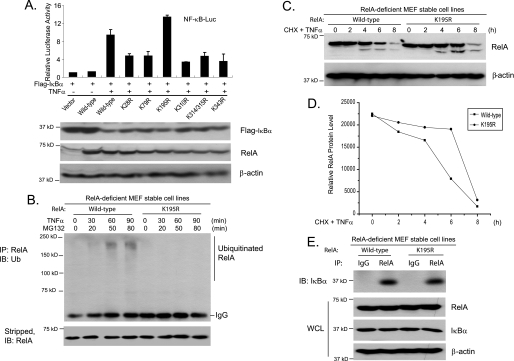
To further determine whether the RelA Lys-195 residue is the predominant Ub acceptor site that mediates TNFα-induced RelA ubiquitination and degradation, wild-type RelA and the seven Lys-to-Arg RelA mutants described above were cotransfected along with IκBα and NF-κB luciferase reporter constructs into RelA-deficient MEF cells. In this assay, we could examine the effect of a RelA Lys-to-Arg mutation on TNFα-induced NF-κB activation because RelA overexpression-induced NF-κB activation would be blocked by the overexpressed IκBα in the cells. As shown in Fig. 2A, TNFα induced higher NF-κB-dependent luciferase reporter gene expression only in RelA K195R mutant-transfected cells compared with wild-type RelA, whereas other RelA Lys-to-Arg mutants partially inhibited TNFα-induced NF-κB reporter activities.
FIGURE 2.
RelA Lys-195 mediates TNFα-induced RelA polyubiquitination. A, effects of overexpression of RelA lysine-to-arginine mutants on TNFα-induced NF-κB activation. Expression vectors encoding IκBα, NF-κB luciferase (NF-κB-Luc) reporter, and Renilla luciferase plasmids were cotransfected into RelA-deficient MEF cells along with wild-type RelA and lysine-to-arginine mutants, and then the cells were either untreated or treated with TNFα (1 ng/ml) for 6 h. The relative luciferase activity was measured and normalized with the Renilla activity. Error bars indicate standard deviations from triplicate experiments. B, TNFα induces RelA polyubiquitination at Lys-195. RelA-deficient MEF cells with stable expression of wild-type RelA or the K195R mutant were either untreated or treated with TNFα (10 ng/ml) for the times indicated in the presence of MG132 and subsequently lysed. RelA proteins in the cell lysates were immunoprecipitated (IP) with anti-RelA antibodies and immunoblotted (IB) with anti-Ub antibodies to detect the presence of ubiquitinated RelA. C, RelA Lys-195 is required for TNFα-induced RelA degradation. RelA-deficient MEF cells with stable expression of wild-type RelA or the K195R mutant were either untreated or treated with TNFα (10 ng/ml) for the time points indicated in the presence of cycloheximide (CHX) and subsequently lysed. The whole cell lysates (WCL) were immunoblotted with anti-RelA antibodies to detect the stability of RelA. D, quantification of results from C. E, Wild-type RelA and K195R have comparable binding affinities for IκBα. Wild-type RelA and K195R proteins in the reconstituted MEF cells were immunoprecipitated with anti-RelA antibodies and immunoblotted with anti-IκBα antibodies to detect the presence of IκBα in the immunoprecipitates.
To confirm the above results, we stably reintroduced wild-type RelA and the K195R mutant into RelA-deficient MEF cells and treated these two cell lines with TNFα in the presence of MG132 at the time points indicated. In this assay, we found that TNFα failed to induce obvious RelA ubiquitination in the MEF cells reconstituted with the K195R mutant compared with the cells reconstituted with wild-type RelA (Fig. 2B). These results indicate that RelA Lys-195 is the predominant Ub acceptor site that mediates TNFα-induced RelA polyubiquitination and degradation.
To determine whether the RelA Lys-195 residue mediates TNFα-induced RelA degradation, we treated wild-type RelA- and K195R mutant-reconstituted MEF cells with TNFα at the time points indicated in the presence of cycloheximide, an inhibitor of protein biosynthesis. Consistent with the above results, we found that TNFα-induced RelA degradation was inhibited in RelA K195R-reconstituted MEF cells compared with the RelA wild type (Fig. 2, C and D). To rule out the possibility that the RelA K195R mutant has a decreased binding affinity for IκBα compared with the wild type, RelA in the MEF cells reconstituted with wild-type RelA or the K195R mutant were immunoprecipitated with anti-RelA antibodies and immunoblotted with anti-IκBα antibody. As shown in Fig. 2E, both wild-type RelA and the K195R mutant had a comparable binding affinity for IκBα. Taken together, these results suggest that the RelA Lys-195 residue mediates TNFα-induced RelA polyubiquitination and degradation.
RelA Lys-195 Is Required for Down-regulation of TNFα-induced NF-κB Activation
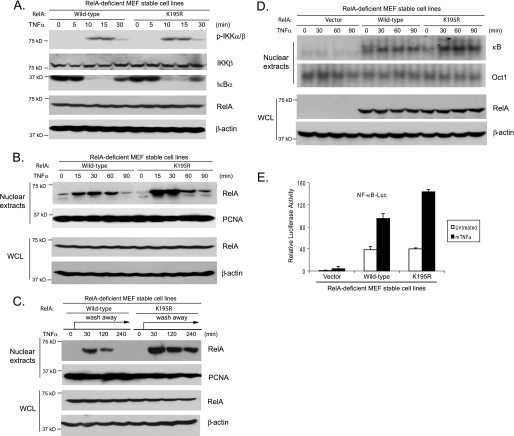
To characterize the physiological role of the RelA Lys-195 site in the TNFα-mediated NF-κB pathway, MEF cell lines expressing wild-type RelA or the K195R mutant were treated with TNFα for the time points indicated, and then the cell lysates were immunoblotted with the indicated antibodies to examine the TNFα-induced IKK phosphorylation, IκBα degradation, and NF-κB/RelA nuclear translocation. In this assay, TNFα induced similar levels of IKK phosphorylation and IκBα degradation in both wild-type RelA- and K195R-reconstituted MEF cells (Fig. 3A). However, a TNFα-induced NF-κB nuclear presence was much higher and lasted much longer in the RelA K195R mutant-reconstituted cells compared with the wild type (Fig. 3, B and C). We also found that TNFα induced a higher and a longer NF-κB activation in the K195R-reconstituted cells than the wild type in an electrophoretic mobility shift assay (Fig. 3D).
FIGURE 3.
RelA Lys-195 mediates down-regulation of TNFα-induced NF-κB activation. A, RelA Lys-195 is not involved in TNFα-induced IKK activation and IκBα degradation. RelA-deficient MEF cells with stable expression of wild-type RelA or the K195R mutant were either untreated or treated with TNFα (10 ng/ml) for the time points indicated and subsequently lysed. The whole cell lysates were immunoblotted with the indicated antibodies to detect IKK phosphorylation and IκBα degradation. B and C, RelA Lys-195 is required for TNFα-induced NF-κB degradation. RelA-deficient MEF cells with stable expression of wild-type RelA or the K195R mutant were either untreated or treated with TNFα (10 ng/ml) for the time points indicated (B), or the TNFα was washed away after treatment for 10 min (C), and the cells were subsequently harvested. The nuclear extracts were prepared and subjected to SDS-PAGE. Nuclear NF-κB RelA was determined by immunoblotting with an anti-RelA antibody, and proliferating cell nuclear antigen (PCNA) was detected as a loading control. D, vector control or wild-type RelA- or K195R-reconstituted MEF cells were either untreated or treated with TNFα for the time points indicated and subsequently harvested. Nuclear extracts were prepared and subjected to an electrophoretic mobility shift assay with 32P-labeled NF-κB and Oct-1 probes. Whole cell lysates (WCL) were subjected to SDS-PAGE and immunoblotting with the antibodies indicated. β-Actin was detected as a loading control for whole cell lysates. E, RelA Lys-195 is required for down-regulation of TNFα-induced NF-κB activation. NF-κB luciferase (NF-κB-Luc) reporter and Renilla luciferase plasmids were cotransfected into wild-type RelA- or K195R mutant-reconstituted MEF cells, and then the cells were either untreated or treated with TNFα (2 ng/ml) for 6 h. The relative luciferase activity was measured and normalized with the Renilla activity. Error bars indicate standard deviations from triplicate experiments.
Consistent with the above results, TNFα induced a much higher NF-κB-dependent luciferase activity in MEF cells expressing the RelA K195R mutant than in those expressing the wild type (Fig. 3E). Taken together, these results indicate that ubiquitination of RelA at the Lys-195 site is required for preventing excessive TNFα-induced NF-κB activation.
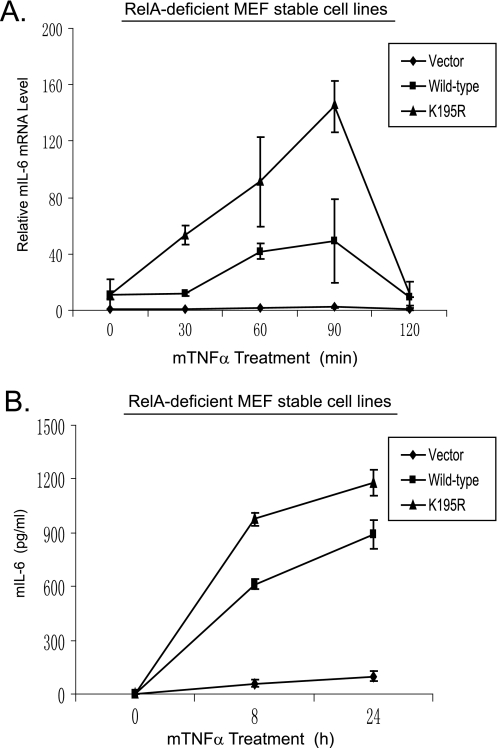
RelA Polyubiquitination at Lys-195 Attenuates TNFα-induced IL-6 Gene Expression
TNFα induces IL-6 gene expression via NF-κB activation (21–23). To determine the role of RelA ubiquitination at Lys-195 in TNFα-induced IL-6 gene expression, total RNA was isolated from the wild-type RelA and K195R mutant stable MEF cell lines treated with or without TNFα. Quantitative reverse transcription-PCR analysis showed that TNFα induced a higher level of IL-6 gene expression in the RelA K195R mutant-reconstituted cells compared with the RelA wild type (Fig. 4A). Consistent with this result, TNFα also induced a higher level of IL-6 in the medium from cells with RelA K195R compared with RelA wild type (Fig. 4B). These results demonstrate that ubiquitination of RelA at Lys-195 is required for preventing excessive TNFα-induced NF-κB activation and cellular responses.
FIGURE 4.
RelA Lys-195 mediates down-regulation of TNFα-induced NF-κB-dependent IL-6 expression. A, the RelA K195R mutation enhances the TNFα-induced NF-κB-dependent IL-6 gene expression. Wild-type RelA- or K195R mutant-reconstituted MEF cells were either untreated or treated with TNFα (2 ng/ml) for the time points indicated. Total RNA from these cells was harvested. IL-6 transcript levels in the cells were measured using quantitative reverse transcription-PCR normalized to glyceraldehyde-3-phosphate dehydrogenase levels. The data are presented as averages of three separate experiments with standard deviations. B, the RelA K195R mutation enhances the TNFα-induced IL-6 production. Wild-type RelA- and K195R mutant-reconstituted MEF cells were either untreated or treated with TNFα (2 ng/ml) for the time points indicated. The supernatants from these cell cultures were collected and subjected to a mouse IL-6 (mIL-6) enzyme-linked immunosorbent assay analysis according to the manufacturer's instructions.
DISCUSSION
Except for IκBα-mediated nuclear export of NF-κB, ubiquitin- and proteasome-dependent degradation of the nuclear RelA subunit of NF-κB has also been suggested to be a critical mechanism of terminating NF-κB activation and preventing excessive TNFα-mediated cellular responses. Until now, the mechanism and role of NF-κB ubiquitination in the negative regulation of the TNFα-induced NF-κB activation had not been completely defined. In this investigation, we further examined the mechanism of RelA polyubiquitination in the attenuation of TNFα-mediated NF-κB activation and gained novel insights into the regulatory role of polyubiquitination in NF-κB function. Here we show that the RelA subunit of NF-κB is polyubiquitinated at the Lys-195 residue in response to TNFα stimulation and that RelA polyubiquitination at the Lys-195 residue is a critical mechanism for termination of immune and inflammatory responses mediated by RelA-containing NF-κB. Our studies suggest that ubiquitination-dependent degradation of the RelA subunit of NF-κB plays an important role in maintaining a delicate balance in TNFα-mediated NF-κB-dependent cellular responses.
In this study, we found that TNFα induces polyubiquitination of only one single lysine residue, Lys-195, on RelA to mediate proteasome-dependent RelA degradation and termination of NF-κB activation. Previously, both the Ser-536 and Ser-468 residues were shown to be involved in the regulation of TNFα-induced RelA turnover (16, 17). It is highly likely that phosphorylation of one or both of these serine residues mediates TNFα-induced polyubiquitination of RelA at the Lys-195 residue. However, future studies are needed to determine whether TNFα-induced phosphorylation of other serine/threonine sites on RelA is involved in the mediation of RelA polyubiquitination and degradation.
Previously, Rodrigues et al. (15) reported that the ORF73 protein encoded by murid herpesvirus 4 acts as an E3 ligase to mediate RelA polyubiquitination and degradation by targeting the Rel homology domain of RelA. Consistent with this report, we found that the RelA Lys-195 residue within its Rel homology domain is the ubiquitin acceptor site for mediating RelA polyubiquitination and degradation. Furthermore, several mammalian E3 ligases have been reported to be involved in the regulation of RelA polyubiquitination and degradation (8, 12, 13). However, further studies are required to determine their specificity in mediating TNFα-induced RelA polyubiquitination and degradation.
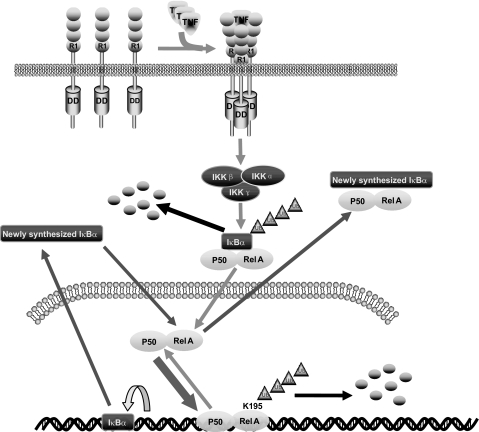
In summary, our study revealed that polyubiquitination of the Lys-195 residue of RelA is involved in the negative regulation of TNFα-induced NF-κB activation. Importantly, our findings in this study are the first report that substitution of a key ubiquitin acceptor lysine residue of RelA with an arginine residue extends the retention time of RelA in the nucleus and increases the NF-κB-dependent gene expression in response to TNFα stimulation. In view of the data presented here and in previous reports, we propose a working model (Fig. 5) in which, upon TNFα binding to its receptor, TNFα receptor-mediated signaling events lead to RelA phosphorylation, nuclear translocation, and binding to the NF-κB-dependent gene promoter. After transactivation, the promoter-bound RelA subunit of NF-κB is polyubiquitinated at Lys-195 and degraded in the nucleus to terminate NF-κB-mediated gene expression.
FIGURE 5.
Working model for the role of Lys-195 in TNFα-induced RelA polyubiquitination and degradation. TNFα induces IKKβ-mediated IκBα degradation and NF-κB nuclear translocation. Once bound to the NF-κB-binding site on the promoter, the RelA subunit of NF-κB is polyubiquitinated at Lys-195 and subsequently degraded to attenuate TNFα-induced NF-κB activation. R1, TNFα receptor 1; DD, death domain.
Acknowledgment
We thank Paul Chiao for providing RelA-deficient MEFs.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R21CA106513-01A2. This work was also supported by American Cancer Society Grant RSG-06-070-01-TBE (to J. Y.), the Virginia and L. E. Simmons Family Foundation (to J. Y.), and the Fleming and Davenport Award (to H. Z.).
- NF-κB
- nuclear factor-κB
- TNFα
- tumor necrosis factor-α
- IKK
- IκB kinase
- MEF
- mouse embryo fibroblast
- IL-6
- interleukin-6
- HA
- hemagglutinin
- Ub
- ubiquitin.
REFERENCES
- 1.Sun S. C., Ley S. C. (2008) Trends Immunol. 29, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden M. S., Ghosh S. (2004) Genes Dev. 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 3.Baldwin A. S., Jr. (1996) Annu. Rev. Immunol. 14, 649–683 [DOI] [PubMed] [Google Scholar]
- 4.May M. J., Ghosh S. (1998) Immunol. Today 19, 80–88 [DOI] [PubMed] [Google Scholar]
- 5.Rothwarf D. M., Karin M. (1999) Sci. STKE 1999, RE1. [DOI] [PubMed] [Google Scholar]
- 6.Beg A. A., Baldwin A. S., Jr. (1993) Genes Dev. 7, 2064–2070 [DOI] [PubMed] [Google Scholar]
- 7.Verma I. M., Stevenson J. K., Schwarz E. M., Van Antwerp D., Miyamoto S. (1995) Genes Dev. 9, 2723–2735 [DOI] [PubMed] [Google Scholar]
- 8.Ryo A., Suizu F., Yoshida Y., Perrem K., Liou Y. C., Wulf G., Rottapel R., Yamaoka S., Lu K. P. (2003) Mol. Cell 12, 1413–1426 [DOI] [PubMed] [Google Scholar]
- 9.Saccani S., Marazzi I., Beg A. A., Natoli G. (2004) J. Exp. Med. 200, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmody R. J., Ruan Q., Palmer S., Hilliard B., Chen Y. H. (2007) Science 317, 675–678 [DOI] [PubMed] [Google Scholar]
- 11.Natoli G., Chiocca S. (2008) Sci. Signal. 1, pe1. [DOI] [PubMed] [Google Scholar]
- 12.Maine G. N., Mao X., Komarck C. M., Burstein E. (2007) EMBO J. 26, 436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka T., Grusby M. J., Kaisho T. (2007) Nat. Immunol. 8, 584–591 [DOI] [PubMed] [Google Scholar]
- 14.Mao X., Gluck N., Li D., Maine G. N., Li H., Zaidi I. W., Repaka A., Mayo M. W., Burstein E. (2009) Genes Dev. 23, 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigues L., Filipe J., Seldon M. P., Fonseca L., Anrather J., Soares M. P., Simas J. P. (2009) EMBO J. 28, 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence T., Bebien M., Liu G. Y., Nizet V., Karin M. (2005) Nature 434, 1138–1143 [DOI] [PubMed] [Google Scholar]
- 17.Geng H., Wittwer T., Dittrich-Breiholz O., Kracht M., Schmitz M. L. (2009) EMBO Rep. 10, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beg A. A., Sha W. C., Bronson R. T., Ghosh S., Baltimore D. (1995) Nature 376, 167–170 [DOI] [PubMed] [Google Scholar]
- 19.Sun W., Yu Y., Dotti G., Shen T., Tan X., Savoldo B., Pass A. K., Chu M., Zhang D., Lu X., Fu S., Lin X., Yang J. (2009) Cell. Signal. 21, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y., Ge N., Xie M., Sun W., Burlingame S., Pass A. K., Nuchtern J. G., Zhang D., Fu S., Schneider M. D., Fan J., Yang J. (2008) J. Biol. Chem. 283, 24497–24505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Libermann T. A., Baltimore D. (1990) Mol. Cell. Biol. 10, 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu H., Mitomo K., Watanabe T., Okamoto S., Yamamoto K. (1990) Mol. Cell. Biol. 10, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y. H., Lin J. X., Vilcek J. (1990) Mol. Cell. Biol. 10, 3818–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]