Abstract
ATP-dependent chromatin remodeling complexes, such as SWI/SNF, are required for transcriptional activation of specific genes and are believed to be recruited to gene promoters by direct interaction with DNA binding transcription factors. However, we report here that recruitment of SWI/SNF to target genes of estrogen receptor α (ERα) requires the previously described nuclear receptor coactivator protein Flightless-I (Fli-I). Fli-I can bind directly to both ER and BAF53, an actin-related component of the SWI/SNF complex, suggesting that Fli-I may recruit SWI/SNF to ER target genes via interaction with BAF53. Point mutations in Fli-I that disrupt binding to ER or BAF53 compromised the ability of Fli-I to enhance ER-mediated activation of a transiently transfected reporter gene. Depletion of endogenous Fli-I or BAF53 inhibited estrogen-responsive expression of endogenous target genes of ER, indicating a critical role for Fli-I and BAF53. Moreover, depletion of endogenous Fli-I or BAF53 specifically eliminated part of the complex cyclical pattern of recruitment of SWI/SNF to estrogen-responsive promoters in a way that indicates multiple roles and multiple mechanisms of recruitment for SWI/SNF in estrogen-dependent target gene expression. These results begin to establish the functional relationships and interdependencies that coordinate the actions of the many coactivators participating in the transcriptional activation process.
Chromatin remodeling for transcriptional regulation involves protein complexes with two distinct types of enzymatic activities, i.e. ATP-dependent chromatin remodeling and posttranslational histone modification. The four major classes of ATP-dependent chromatin remodeling complexes, SWI/SNF, ISWI, Mi-2/NuRD, and INO80, are characterized by the identity of their ATPase subunit (1–3). Human SWI/SNF complexes contain either human Brahma or Brahma-related gene 1 (BRG1)2 protein as the catalytic ATPase subunit along with ∼10–12 BRG1-associated factors (BAFs), the composition of which varies (4). The nature of the remodeled chromatin state generated by SWI/SNF complexes has been extensively studied in vitro and is characterized by increased mobility of nucleosomes on DNA templates and altered degree of association of DNA with histones within nucleosomes (2, 5).
The role of SWI/SNF complexes in nuclear receptor (NR)-mediated transcriptional activation was first studied for the glucocorticoid receptor in a heterologous yeast model system and later in mammalian cells (3, 6, 7). SWI/SNF complexes play critical roles in establishing hypersensitive chromatin architecture associated with steroid hormone receptor binding sites on DNA and are recruited to these sites in a hormone-dependent manner (4, 8, 9).
Including the SWI/SNF complex, more than 300 coactivators and corepressors for NRs have been identified, but their mechanistic contributions to transcriptional activation and the mechanisms that coordinate the activities of multiple coactivator complexes on promoters of NR target genes are mostly unknown (10–13). Recent studies identified Flightless-I (Fli-I) as a transcriptional coactivator for NRs, including estrogen receptor α (ERα) and thyroid hormone receptor, in transient reporter gene assays (14). Fli-I has a leucine-rich repeat (LRR) region at its N terminus and a C-terminal gelsolin-like domain (Fig. 1A) which places it in the gelsolin protein family (14). Like other members of the gelsolin family, Fli-I binds to actin and actin-related proteins (15, 16). Also like gelsolin, the C-terminal gelsolin-like domain of Fli-I consists of two large tandem repeats, each of which contains three smaller repeats (Fig. 1A); in gelsolin, each small repeat folds as an independent structural module (17). Although we previously showed that Fli-I is required for activation of a transient reporter gene by steroid hormones (14), the mechanism by which Fli-I contributes to transcriptional activation of NR target genes is unknown. Our finding that Fli-I interacts directly with NRs and BAF53 (14), which is an actin-related protein component of SWI/SNF complexes (4, 18), suggests a possible functional relationship between Fli-I and SWI/SNF. This led us to propose that Fli-I might be responsible for recruitment of the SWI/SNF complex to steroid hormone-responsive promoters (14). In the current study we, therefore, tested whether depletion of Fli-I by siRNA affected the estrogen-dependent expression of endogenous ER target genes and recruitment of SWI/SNF to the corresponding promoters of ER target genes. Our findings indicate an important role for Fli-I in the transcription initiation process and the function of SWI/SNF and support a model for multiple roles of SWI/SNF complexes in estrogen-dependent transcription of ER target genes.
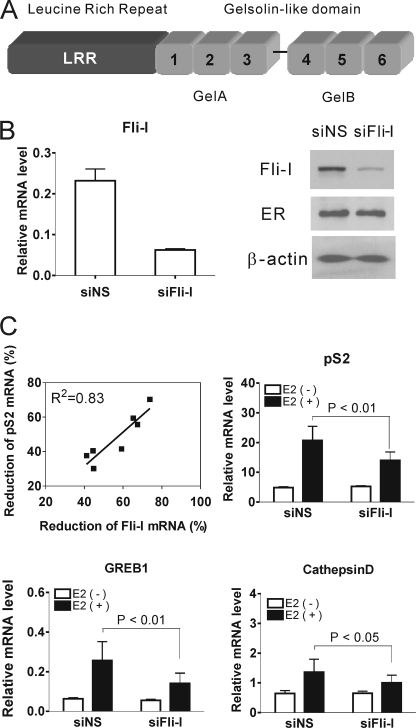
FIGURE 1.
Reduction of endogenous Fli-I attenuated the expression of ER target genes. A, domains of Fli-I. GelA and GelB, the gelsolin-like domain consisting of two large tandem repeats. The six numbered regions represent small repeated modules homologous to the actin binding motifs of gelsolin. B, depletion of Fli-I mRNA and protein by siRNA transfection. MCF7 cells were transfected with siRNA specific for Fli-I (siFli-I) or siNS and grown in hormone-free media for 72 h. Total RNA was analyzed for Fli-I mRNA by qRT-PCR and normalized to the level of GAPDH mRNA. Protein levels of Fli-I, ER, and β-actin were assessed by immunoblot. C, effect of reduced Fli-I on expression of estrogen-responsive genes. MCF7 cells were transfected with siFli-I or siNS and grown in hormone free-media. After 72 h cells were treated with 100 nm E2 or vehicle for 24 h. Total RNA was analyzed by qRT-PCR. The levels of pS2, GREB1, and cathepsin D mRNAs were normalized to that of GAPDH mRNA. Results shown in the bar graphs are the means and S.D. of data from seven independent experiments. p values were determined by paired, two-tailed t tests from the seven independent experiments, as indicated by the brackets in the figures. Results of all seven independent experiments are shown in supplemental Table S1. The line graph shows the correlation between reduction of Fli-I and pS2 mRNA levels. From the seven independent experiments in supplemental Table S1, % reduction of pS2 mRNA was plotted against % reduction of Fli-I mRNA, and the R2 value was calculated.
EXPERIMENTAL PROCEDURES
Plasmids
The following plasmids were described previously (14): pSG5-ER(DBD-AF2), human Fli-I expression vectors pSG5-FLAG-LRR and pSG5-FLAG-GelA, and glutathione S-transferase (GST) fusion protein expression vector pGEX4T1-ER(LBD). pSG5-HA-BAF53 contains the BAF53 coding region inserted into XhoI and BglII sites of pSG5.HA (19). pSG5-FLAG (14) expression vectors encoding the following proteins were constructed by inserting the appropriate PCR-amplified cDNA coding regions into EcoRI and XhoI sites: pSG5-FLAG-G1 (Fli-I amino acids 495–597), pSG5-FLAG-G2 (amino acids 598–709), pSG5-FLAG-G3 (amino acids 710–822), and pSG5-FLAG-GelB (amino acids 825–1269). GST protein expression vectors encoding the following proteins were constructed by inserting the appropriate cDNA coding region into EcoRI and XhoI sites of the vector: pGEX4T1-LRR (amino acids 1–494), pGEX4T1-GelA (amino acids 495–822), pGEX4T1-GelB (amino acids 825–1269), pGEX4T1-G1 (amino acids 495–597), pGEX4T1-G3 (amino acids 710–822). His-tagged full-length Fli-I (pTriEX4-Fli-I) was created by PCR amplification of pCDNA-Fli-I and subcloning into NcoI and XhoI sites of pTriEX4 vector (Novagen). pTriEX4-Fli-I-ΔG1 lacking amino acids 495–597 was generated by two-step PCR amplification and insertion. The first PCR product comprised of the first 1410 bp of Fli-I cDNA was inserted into NcoI and BamHI sites of pTriEX4, and the second PCR fragment comprised of bp 1719–3801 was inserted into the XmaI site within the previous insert and the XhoI site in pTriEX4 vector. Point mutations in Fli-I, GelA, and G1 were introduced by site-directed mutagenesis using the QuikChange II site-directed mutagenesis kit (Stratagene). BRG1 expression plasmid was kindly provided by Dr. Anthony N. Imbalzano (University of Massachusetts Medical School).
Protein Interaction Assays and Immunoblot
The procedure for GST pulldown assays was described previously (14). GST fusion proteins were expressed in Escherichia coli BL21(DE3) strain and purified by incubation with glutathione-Sepharose beads and washing with NETN buffer (300 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl (pH 8.0), and 0.01% Nonidet P-40). His-tagged BAF53 and Fli-I were also expressed in E. coli BL21(DE3) strain and purified with nickel-nitrilotriacetic acid-agarose beads (Qiagen). FLAG-tagged Fli-I fragments were synthesized by transcription and translation in vitro using the TNT-Quick-coupled reticulocyte lysate system (Promega) according to the manufacturer's protocol. For coimmunoprecipitation assay, 293T cells were plated at 1.5 × 106 cells per 10-cm dish and transiently transfected using Lipofectamine 2000 (Invitrogen) and the indicated amount of plasmids. At 48 h after transfection, cell extracts were prepared in 1.0 ml of radioimmune precipitation assay buffer (50 mm Tris-Cl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 2 mm EDTA). Immunoblotting was performed as described previously (14) using the following antibodies: anti-His, anti-ERα, anti-Fli-I, anti-BRG1, anti-BAF53, anti-β-actin, anti-GAL4DBD, and normal mouse or rabbit IgG (Santa Cruz Biotechnology) and anti-HA (Roche Applied Science); anti-FLAG (Sigma Aldrich).
Chromatin Immunoprecipitation Assay (ChIP)
ChIP assays were performed according to previously described protocols (14). Briefly, MCF7 cells were transfected with siRNA and then cultured for 3 days in phenol red-free Dulbecco's modified Eagle's medium supplemented with 5% charcoal-dextran-stripped fetal bovine serum. At ∼90% confluency cells were treated with 100 nm estradiol (E2) or vehicle for the indicated time. After cross-linking with formaldehyde, cell extracts were prepared from control and E2-treated MCF7 cells. Immunoprecipitation of sonicated chromatin solutions was conducted by overnight incubation at 4 °C with normal mouse or rabbit IgG, anti-ERα, anti-Fli-I, anti-BRG1, anti-TRAP220 (Santa Cruz Biotechnology) or anti-RNA polymerase II (Millipore). Cross-linking was reversed by heating, and immunoprecipitated DNA was purified by phenol-chloroform extraction and ethanol precipitation. The purified DNA was dissolved in 100 μl of TE buffer (10 mm Tris-HCl (pH 8.0), 1 mm EDTA) and analyzed by quantitative PCR using the Stratagene Mx3000P system with SYBR Green dye. The primers used were: pS2 promoter region, 5′-GGCAGGCTCTGTTTGCTTAAAGAGCG-3′ (forward) and 5′-GGCCATCTCTCACTATGAATCACTTCTGC-3′ (reverse); pS2 coding region, 5′-TGCCAGCTGTGGGGAGCTGAATAACTT-3′ (forward) and 5′-CAGTTCGTTCTGTACACCGAGGCCACT-3′ (reverse); GREB1 promoter region, 5′-GTGGCAACTGGGTCATTCTGA-3′ (forward) and 5′-CGACCCACAGAAATGAAAAGG-3′ (reverse).
RNA Interference and qRT-PCR
Small interfering RNA experiments were performed according to previously published methods (14). The sequences of siRNA used were: siFli-I, 5′-CAACCUGACCACGCUUCAUdTdT-3′ (sense) and 5′-AUGAAGCGUGGUCAGGUUGdTdT-3′ (antisense); siFli-I(2), 5′-GCUGGAACACUUGUCUGUGdTdT-3′ (sense) and 5′-CACAGACAAGUGUUCCAGCdTdT-3′ (antisense);siBAF53, 5′-GCUUUCCUUGAAAUGCACUdTdT-3′ (sense) and 5′-AGUGCAUUUCAAGGAAAGCdTdT-3′ (antisense); siBAF53(2), 5′-GGUACUUCAAGUGUCAGAUdTdT-3′ (sense) and 5′-AUCUGACACUUGAAGUACCdTdT-3′ (antisense); siBRG1, 5′-CAUGCACCAGAUGCACAAGdTdT-3′ (sense) and 5′-CUUGUGCAUCUGGUGCAUGdTdT-3′ (antisense); siBRG1(2), 5′-CCGUGGACUUCAAGAAGAUdTdT-3′ (sense) and 5′-AUCUUCUUGAAGUCCACGGdTdT-3′ (antisense); nonspecific siRNA (siNS), 5′-UUCUCCGAACGUGUCACGUdTdT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAAdTdT-3′ (antisense) (20). Transfections in MCF7 cells were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Total RNA was isolated from MCF7 cells with Trizol (Invitrogen) after hormone treatment as indicated and subjected to reverse transcription by the iScript cDNA synthesis kit (Bio-Rad). 5 μl of reverse transcription product was used for qPCR analysis with the following primers: pS2, 5′-GAACAAGGTGATCTGCG-3′ (forward) and 5′-TGGTATTAGGATAGAAGCACCA-3′ (reverse); cathepsin D, 5′-GTACATGATCCCCTGTGAGAAGGT-3′ (forward) and 5′-GTCACCGGAGTCCATCACGATG-3′ (reverse) (21); GREB1, 5′-CAAAGAATAACCTGTTGGCCCTGC-3′ (forward) and 5′-GACATGCCTGCGCTCTCATACTTA-3′ (reverse) (22); BRG1, 5′-CATCATCGTGCCTCTCTCAAC-3′ (forward) and 5′-ACACGCACCTCGTTCTGCTG-3′ (reverse); BAF53, 5′-GAGTTCCCAAGCTTCTACCTTCCT-3′ (forward) and 5′-CATTCTACAAAAGATGGTCATTCTTTTC-3′ (reverse); β-actin, 5′-ACCCCATCGAGCACGGCATCG-3′ (forward) and 5′-GTCACCGGAGTCCATCACGATG-3′ (reverse) (23); GAPDH, 5′-TCTGGTAAAGTGGATATTGTTG-3′ (forward) and 5′-GATGGTGATGGGATTTCC-3′ (reverse) (24). Relative expression levels were normalized to GAPDH mRNA levels.
RESULTS
Reduction of Endogenous Fli-I Attenuated the Expression of ER Target Genes
We previously showed that Fli-I enhances and is required for efficient NR mediated transcription of transiently transfected reporter genes (14). Here we examined endogenous ER target genes in MCF7 cells. Compared with siNS, siRNA specific for Fli-I (siFli-I) effectively reduced Fli-I mRNA and protein levels (Fig. 1B). Fli-I siRNA also significantly inhibited E2-dependent expression of the endogenous pS2, GREB1, and cathepsin D genes (Fig. 1C, showing the mean and S.D. from the results of seven independent experiments). For the seven independent experiments, the reduction of Fli-I mRNA levels was 57 ± 13%, and the -fold induction of ER target gene expression by E2 was reduced by 48 ± 14% for pS2 mRNA (p < 0.01 by paired, two-tailed t test), 47 ± 8% for GREB1 mRNA (p < 0.01), and 49 ± 21% for cathepsin D mRNA (p < 0.05) (Fig. 1C and supplemental Table S1). The percentages given above for the effect of siRNA on ER target gene expression represent the % reduction in the E2-induced mRNA level after subtracting out the basal mRNA level (as basal expression was not significantly affected by the Fli-I-specific siRNA); see supplemental Table S1 for the formula used in the calculation. Although knockdown efficiency of Fli-I varied from experiment to experiment, there was a strong linear correlation between reduction of Fli-I mRNA and reduction of E2-induced levels of pS2 mRNA with an R2 correlation coefficient of 0.83 (Fig. 1C). The reduction of Fli-I mRNA level by another siRNA targeting a different part of the coding region of Fli-I caused a similar inhibition of ER target gene expression (supplemental Fig. S1). Thus, Fli-I is required for hormonal induction of ER target genes in MCF7 cells.
Mapping the Interaction Sites of Fli-I with ER
We previously showed that Fli-I binds to full-length ER (14). To further explore possible functional relationships, we mapped the ER binding site on Fli-I. ER-ligand binding domain (LBD) fused to GST bound in vitro in an E2-dependent manner to the GelA domain (amino acids 495–822) of Fli-I but not to the LRR domain (amino acids 1–494) or the GelB domain (amino acids 825–1269) (Fig. 2A). In similar GST pulldown assays with each of the three repetitive subregions of GelA (Fig. 1A), only the third repeat (G3, amino acids 710–822) bound ER-LBD, and binding was E2-dependent, indicating that G3 is the major binding site for ER-LBD (Fig. 2B, upper panel). Conversely, GST-tagged G3 bound an in vitro translated ER fragment consisting of the DNA binding domain (DBD) and LBD in a hormone-dependent manner (Fig. 2B, lower panel).
FIGURE 2.
Interaction of Fli-I with ER. A, GST pulldown assays were performed with in vitro translated FLAG-tagged LRR, GelA, or GelB incubated in the absence or presence of E2 with GST-fused ER-LBD bound to glutathione-Sepharose beads. Bound proteins were analyzed by immunoblot with anti-FLAG antibody. B, GST pulldown assays were performed with in vitro translated small repeating modules (G1, G2, and G3) of Fli-I incubated with GST-fused ER-LBD in the absence or presence of E2 (upper panel). In vitro translated ER fragment containing DBD and LBD (AF2) was incubated with GST-fused G3 fragment in the absence or presence of E2 (lower panel). C and D, the leucine-rich motif in G3 is involved in hormone-dependent interaction with ER LBD. GST pulldown assays were performed with in vitro translated GelA or GelA(LL/AA) mutant (mt) protein (C) or with E. coli-expressed His-tagged full-length Fli-I or Fli-I(LL/AA) mutant protein (D) incubated with GST-ER LBD in the absence or presence of E2. wt, wild type.
Interestingly, the G3 fragment contains an LXXLL motif (where L represents leucine, and X denotes any amino acid) which is observed in many coactivators that interact with hormone-activated NR. To determine whether the LXXLL motif in G3 is required for the hormone-dependent interaction with ERα, a GST pulldown assay was performed using a mutant GelA fragment with Leu-769 and Leu-770 changed to Ala (i.e. LXXLL to LXXAA). This mutation eliminated binding of GelA to hormone-activated ERα (Fig. 2C). Furthermore, the same mutation in His-tagged full-length Fli-I almost completely eliminated binding to ERα LBD compared with wild type Fli-I (Fig. 2D), suggesting that the ER-Fli-I interaction is direct, that the G3 gelsolin repeat is the primary site in Fli-I for binding to ER LBD, and that the LXXLL motif is critical for this interaction.
Fli-I Interacts Directly with BAF53
We previously demonstrated by coimmunoprecipitation that the GelA fragment of Fli-I binds BAF53, an actin-like component of the SWI/SNF complex (14). In further mapping studies, deletion of the G1 module (amino acids 495–597) from His-tagged full-length Fli-I substantially decreased co-immunoprecipitation of HA-tagged BAF53 by anti-His-tag antibodies from total cell extracts of transiently transfected 293T cells (Fig. 3A). Thus, the G1 fragment is required for binding of BAF53 to full-length Fli-I. Furthermore, purified recombinant His-tagged BAF53 was bound by GST-tagged GelA, but not by GST-LRR or GST-GelB, indicating that the GelA-BAF53 interaction is direct (Fig. 3B). The G1 fragment of GelA was sufficient to bind BAF53 (Fig. 3C).
FIGURE 3.
Fli-I interacts with BAF53 directly. A and D, coimmunoprecipitation was performed with 1 ml of total cell extracts from 293T cells (100-mm dish) transfected with pSG5-HA-BAF53 (2 μg) and pTriEX4-Fli-I, pTriEX4-Fli-IΔG1 (lacking amino acids 495–597) or pTriEX4-Fli-I(E/K) (2 μg). Fli-I fragments were immunoprecipitated (IP) with 1 μg of anti-His-tag antibody or normal mouse IgG, and BAF53 was detected in immunoblots (IB) with anti-HA antibody. B, GST pulldown assays were performed with bacterially expressed His-tagged BAF53 incubated with GST-fused Fli-I fragments. Bound protein was analyzed by immunoblot with anti-His-tag antibody (upper panel). GST proteins eluted from the beads were visualized by Coomassie Blue staining (lower panel). C, GST pulldown assay performed with bacterially expressed His-tagged BAF53 incubated with GST-fused G1 fragment of Fli-I. E, GST pulldown assays were performed with in vitro translated HA-tagged BAF53 incubated with GST-fused ER-LBD in the absence or presence of E2. No bound protein was detected by immunoblot with anti-HA antibody.
A Glu-to-Lys mutation in the first actin binding motif of the protein gelsolin reduces binding of gelsolin to actin (25). We previously showed that the analogous E585K mutation in the G1 motif of Fli-I reduced binding of the GelA fragment of Fli-I to BAF53 (14). The same mutation in full-length Fli-I substantially reduced binding to BAF53 (Fig. 3D). This result confirmed that G1 is the primary BAF53 binding site in Fli-I and that Glu585 in full-length Fli-I is critically involved in the interaction with BAF53. In contrast, purified recombinant BAF53 did not bind to GST-ER-LBD (Fig. 3E); thus, any association between BAF53 and ER would have to be indirect and could potentially be mediated by Fli-I.
Fli-I Binding to ER and to the SWI/SNF Complex Is Important for ER-mediated Transcription
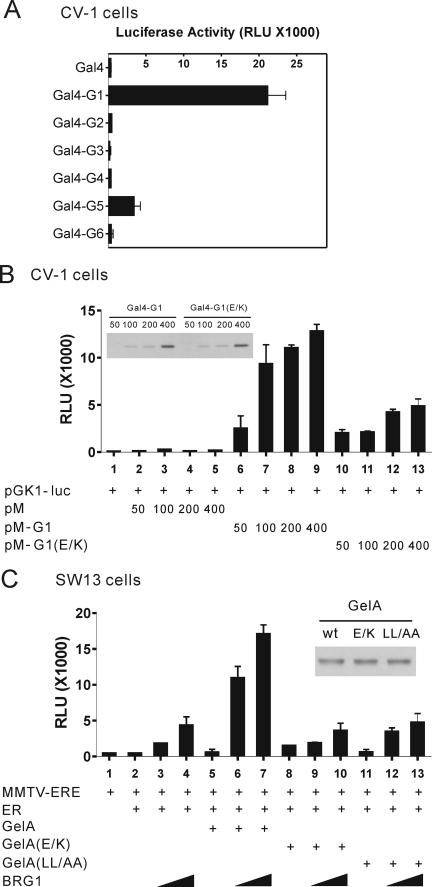
Components of the SWI/SNF complex form a very strong protein complex which is stable even in 3 m urea, and most SWI/SNF subunits (e.g. BRG1, human Brahma, and BAFs) exist in complexes in vivo (26). We failed to observe direct interaction of BRG1 with Fli-I (data not shown), but we found that the GelA fragment of Fli-I binds to BAF53 directly (Fig. 3B). Therefore, we speculated that Fli-I recruits the SWI/SNF complex via interaction with BAF53. Among the three actin binding motifs in the GelA region, the G1 fragment showed strong autonomous transcriptional activation when fused to Gal4 DBD (Fig. 4A), and we also found that the G1 fragment is sufficient to interact with BAF53 (Fig. 3C), suggesting that the recruitment of the SWI/SNF complex by G1 may account for the transcriptional activation. The observation that the G1(E585K) mutant protein, which does not bind to BAF53, showed much weaker transcriptional activation (Fig. 4B) supports our hypothesis that Fli-I recruits the SWI/SNF complex via the interaction with BAF53 to activate ER-mediated transcription.
FIGURE 4.
Fli-I binding to ER and to the SWI/SNF complex is important for ER-mediated transcription. A and B, autonomous activation function in the G1 motif of Fli-I. CV-1 cells were transfected with pGK1-luc reporter plasmid (200 ng) controlled by Gal4-responsive elements and expression plasmids encoding Gal4 DBD or Gal4 DBD fused to each gelsolin-like motif (G1 through G6) or to the E585K mutant of the G1 fragment (200 ng). Transfected cells were grown for 2 days and harvested for luciferase assays (bar graphs) and immunoblots using antibodies against Gal4 (upper panel of B). C, SW13 cells were transfected with murine mammary tumor virus-estrogen response element (MMTV-ERE) luciferase reporter plasmid (200 ng) and expression plasmids encoding ERα (0.1 ng), BRG1 (10 ng or 50 ng), and GelA, GelA(E585K) or GelA(LL/AA) (100 ng) as indicated. Transfected cells were grown with E2 for 48 h, and luciferase activities of the transfected-cell extracts were determined by luminometer (bar graph). Expression of wild type (wt) and mutant GelA was monitored by immunoblot using antibodies against the FLAG epitope (inset). RLU, relative luciferase units.
To assess whether Fli-I requires interaction with ER or the SWI/SNF chromatin remodeling complex for its coactivator function, we performed ER-mediated reporter gene assays in SW13 cells. The SW13 cell line contains no detectable BRG1 or human Brahma proteins (the ATPase core subunits of SWI/SNF) but nonetheless contains all the BAF subunits of the SWI/SNF complex (6, 27). ER-mediated activation of estrogen-responsive genes was diminished in these cells, but reintroduction of the BRG1 protein induces formation of a functional SWI/SNF complex and restores estrogen stimulated gene expression, indicating that SWI/SNF activity is essential for estrogen receptor-mediated transcription. In our study wild type GelA, GelA(E/K) or GelA(LL/AA) were transiently expressed with ERα in the absence or presence of co-expressed BRG1. In the absence of BRG1, ERα failed to activate transcription efficiently in the presence of E2 (Fig. 4C, lanes 1–2). However, expression of BRG1 in SW13 cells by transient transfection restored hormone-dependent activation of the reporter gene (lanes 3–4). Interestingly, the level of transcription was synergistically enhanced by co-expression of BRG1 with GelA (lanes 6–7), which can bind to ER through the G3 motif and to BAF53 in the SWI/SNF complex through the G1 motif. However, GelA alone did not activate reporter gene expression in the absence of BRG1 (lane 5). In contrast, the GelA(E585K) mutant, which does not bind to BAF53, and the GelA(LL/AA) mutant, which does not bind to ERα, failed to cooperate functionally with BRG1 and ERα (Fig. 4C, lanes 8–13). These results further support the ability of Fli-I to recruit the SWI/SNF complex to ER target promoters by the interaction of GelA with ERα through the leucine-rich motif (LXXLL) in G3 and by the interaction of GelA with BAF53 through the G1 motif.
Fli-I Is Required for Recruitment of SWI/SNF Chromatin Remodeling Complex to the Promoter of an ER Target Gene, pS2
Results from our protein interaction and transient reporter gene assays (Figs. 2–4) indicated that the interaction of Fli-I with ER and with the SWI/SNF subunit BAF53 are critical for the coactivator function of Fli-I. Therefore, we tested whether Fli-I is involved in recruitment of SWI/SNF to the promoter region of an endogenous ER target gene. E2 treatment results in cyclical recruitment of ER and various coregulators to the pS2 promoter in MCF7 cells (28–30). ERα and Fli-I were associated with the pS2 promoter at 30 and 60 min after E2 treatment of MCF7 cells (Fig. 5A), but Fli-I was not recruited to the pS2 coding region (Fig. 5B), demonstrating regional specificity of Fli-I recruitment. A more detailed time course of Fli-I occupancy on the pS2 promoter during E2 treatment found that there were two peaks of Fli-I occupancy around 20 and 80 min of E2 treatment (supplemental Fig. S2). E2 also induced occupancy of BRG1 on the pS2 promoter at 30 min; BRG1 occupancy was reduced to the uninduced level at 60 min (Fig. 5A), consistent with the different timing previously observed for recruitment of BRG1 versus ER (30).
FIGURE 5.
Recruitment of ER, Fli-I, and BRG1 to the pS2 promoter in MCF7 cells. Chromatin immunoprecipitation assays were performed with MCF7 cells in 150-mm dishes treated with 100 nm E2 or vehicle for 30 or 60 min. After immunoprecipitation of cross-linked chromatin fragments with the indicated antibody, the amount of pS2 promoter region (A) or coding region (B) present was determined by qPCR. The data are plotted as the percentage of total input before immunoprecipitation and are from a single experiment which is representative of at least two independent experiments. Error bars represent the range of variation of duplicate PCR reactions.
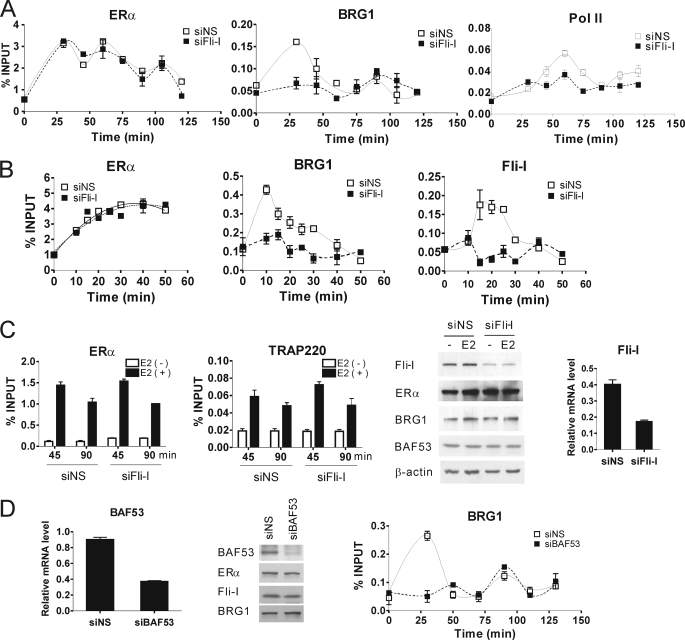
Next, transfection with siRNA was used to test whether Fli-I is required for recruitment of BRG1 to the pS2 promoter in response to E2. The siRNA against Fli-I reduced the endogenous cellular level of Fli-I protein and mRNA but had no effect on the level of ERα, BRG1, BAF53, or actin protein (Fig. 6C, two right panels). Using MCF7 cells transfected with nonspecific or Fli-I-specific siRNA, we analyzed the occupancy of ER and selected coregulators and RNA polymerase II on the pS2 promoter at time points between 0 and 120 min after adding E2. Although the peaks are small, we consistently observed three distinct cycles of ERα recruitment to the pS2 promoter during 120 min of E2 treatment, consistent with previous reports (30); moreover, this pattern of ERα occupancy of the pS2 promoter was the same in cells transfected with nonspecific siRNA or siRNA against Fli-I (Fig. 6A, left panel). We observed a large, early peak of BRG1 recruitment to the pS2 promoter at or before 30 min of E2 treatment and a second smaller peak at ∼90 min; this was also consistent with a previous finding that the first peak of BRG1 recruitment overlaps with but reaches its maximum before the initial peak of ER recruitment and that subsequent peaks of BRG1 recruitment were out of phase with peaks of ER recruitment (30). When endogenous Fli-I protein level was reduced, the early peak of BRG1 recruitment was eliminated, but the later peak was not (Fig. 6A, middle panel). The recruitment of RNA polymerase II reached a peak at 60 min after the beginning of hormone treatment in siNS-transfected MCF7 cells and reached another peak at 120 min or later (right panel). The two peaks of RNA polymerase II recruitment correspond to the second and third peaks of ERα recruitment observed here and previously (30). Depletion of endogenous Fli-I substantially compromised the hormone-dependent recruitment of RNA polymerase II at both 60 and 120 min (Fig. 6A, right panel).
FIGURE 6.
Role of Fli-I and BAF53 in cyclical, estrogen-dependent recruitment of SWI/SNF complex to the pS2 promoter. A and B, chromatin immunoprecipitation assays were performed with MCF7 cells as in Fig. 5 after transfection with siRNA against Fli-I (siFli-I) or siNS and treatment with E2 for the indicated times. Results shown are from a single experiment and are representative of at least two independent experiments. C, ChIP assays were performed in MCF7 cells transfected 72 h previously with siFli-I or siNS. E2 or vehicle was added 45 or 90 min before cross linking of chromatin, ChIP was performed with the indicated antibodies, and the presence of the pS2 promoter region in the immunoprecipitates was determined by qPCR (first two panels). Fli-I, ERα, BRG1, BAF53, and β-actin protein levels from total cell lysates of MCF7 cells were determined by immunoblot (third panel). Fli-I mRNA levels were determined by qRT-PCR (fourth panel). D, ChIP assays were performed as in A, but siRNA against BAF53 was substituted for siRNA against Fli-I (third panel). BAF53 mRNA levels were determined by qRT-PCR (first panel). BAF53, ERα, Fli-I, and BRG1 protein levels from total cell lysate of MCF7 cells were determined by immunoblot (middle panel).
Although these results were reproducible in multiple independent experiments, the small number of time points in the first peak of BRG1 recruitment led us to examine in more detail the early time period after E2 addition. We observed a broad peak of ER occupancy on the pS2 promoter with a maximum value at ∼40 min, whereas Fli-I and BRG1 occupancy peaked at ∼10–15 min (Fig. 6B). Transfection with siRNA against Fli-I had no effect on recruitment of ER, but the early peaks of Fli- I and BRG1 recruitment were eliminated. The reduction of Fli-I level by another siRNA targeting a different part of the coding region of Fli-I caused a similar inhibition of BRG1 occupancy on the pS2 promoter (supplemental Fig. S3A). Thus, endogenous Fli-I is required for the initial peak of recruitment of BRG1 and presumably the SWI/SNF complex to the pS2 promoter in response to E2.
In contrast to the effect on BRG1 recruitment, reduction of endogenous Fli-I had no effect on the E2-dependent recruitment of Med1/TRAP220, a component of the Mediator complex, to the pS2 promoter (Fig. 6C), demonstrating a specific role for Fli-I in recruitment of BRG1. The Mediator complex is involved in recruitment of RNA polymerase II to many promoters.
Although the ability of Fli-I to bind BAF53 suggests a possible mechanism for recruitment of SWI/SNF, Fli-I could potentially also interact with other components of the SWI/SNF complex, such as actin, to which it is known to bind (15, 16), or Fli-I could influence SWI/SNF recruitment by an indirect mechanism. To explore the role of BAF53 in Fli-I mediated recruitment of SWI/SNF, additional ChIP assays were performed in MCF7 cells with reduced levels of BAF53. As when Fli-I levels were reduced, depletion of BAF53 by siRNA eliminated the initial E2-induced peak of BRG1 recruitment to the pS2 promoter but did not affect the second smaller peak of BRG1 recruitment (Fig. 6D). Thus, BAF53 as well as Fli-I is required for the initial peak of BRG1 recruitment, consistent with a mechanism of SWI/SNF recruitment by Fli-I which involves interaction of Fli-I with BAF53.
Role of Fli-I in E2-dependent Recruitment of BRG1 to the GREB1 Promoter
We examined another ER target gene, GREB1, which has three putative estrogen response elements. The most robust recruitment of ERα occurs at a site 1.6 kilobases upstream from the transcription initiation site, and recruitment of ER occurs there in a cyclical fashion, as with the pS2 promoter (23). We observed a cyclical recruitment of BRG1 to the GREB1 promoter, with peaks at ∼15 and 90 min, similar to what we observed on the pS2 promoter, but in contrast to the pS2 promoter, the second peak of BRG1 recruitment was higher than the first peak on the GREB1 promoter (compare Fig. 7A with Fig. 6, A and D). As on the pS2 promoter, depletion of Fli-I by siRNA transfection eliminated the first peak of BRG1 recruitment but had no effect on the second peak (Fig. 7A). A more detailed analysis of the first 60 min of E2 treatment confirmed that depletion of Fli-I had no effect on ER recruitment but eliminated the first peak of BRG1 recruitment (Fig. 7B). Similar results were obtained with another siRNA targeting a different part of the coding region of Fli-I (supplemental Fig. S3B). Thus, Fli-I is required for recruitment of BRG1 to the GREB1 promoter as well as the pS2 promoter, suggesting a broader role for Fli-I in estrogen regulation of various ER target genes.
FIGURE 7.
Role of Fli-I in cyclical, estrogen-dependent recruitment of SWI/SNF complex to the GREB1 promoter. A and B, recruitment of ER, BRG1, and Fli-I to the GREB1 promoter was determined by ChIP assay as in Fig. 6 using specific primers for GREB1 promoter region. ERE, estrogen response element; kb, kilobases.
BAF53 and BRG1 as Well as Fli-I Are Required for Expression of ER Target Genes
The requirement of both Fli-I and BAF53 for estrogen-dependent recruitment of BRG1 to E2-responsive promoters leads us to propose that Fli-I recruits SWI/SNF through the interaction of Fli-I with BAF53. According to this hypothesis, BAF53 and BRG1 as well as Fli-I should be required for E2-dependent expression of ER target genes. Furthermore, because Fli-I and BAF53 are required for the first but not subsequent peaks of BRG1 recruitment to ER target genes, reduction of endogenous Fli-I or BAF53 might affect the kinetics of ER target gene expression in response to E2. We, therefore, performed a 48-h time course of ER target gene expression after adding E2 to siRNA-transfected MCF7 cells. Fli-I and BAF53 mRNA levels were effectively reduced in MCF7 cells by the appropriate siRNA (Fig. 8A). Depletion of either Fli-I or BAF53 compromised the E2-induced increase in mRNA levels of three ER target genes (pS2, GREB1, and cathepsin D) but had no effect on the level of β-actin mRNA relative to GAPDH mRNA (Fig. 8A). The reduction of target gene expression was consistent throughout the 48-h time period examined. The reduction of Fli-I or BAF53 level by another siRNA targeting a different part of the coding region of Fli-I or BAF53 caused a similar inhibition of ER target gene expression (supplemental Figs. S1 and S4). Furthermore, depletion of BRG1 by siRNA caused a similar reduction in E2-induced expression of ER target genes (Fig. 8B) as expected, suggesting that loss of the initial peak of BRG1 recruitment caused a sustained deficiency in E2-dependent transcription on these promoters. A similar result was observed with a second siRNA targeting a different part of the coding region of BRG1 (supplemental Fig. S5). Thus, both Fli-I and BAF53 are required for optimal recruitment of BRG1 to E2-responsive promoters, and all three of these proteins are required for E2-enhanced expression of ER target genes.
FIGURE 8.
Fli-I, BAF53, and BRG1 are required for estrogen-dependent expression of ER target genes. A, endogenous Fli-I or BAF53 mRNA levels in MCF7 cells were determined from total RNA preparations by qRT-PCR after transient transfection with the indicated siRNAs. MCF7 cells transfected with nonspecific siRNA or siRNA against Fli-I or BAF53 were treated with 100 nm E2 for the indicated times, and mRNA levels for the indicated ER target genes or β-actin gene were analyzed by qRT-PCR and normalized to the level of GAPDH mRNA. The means and ranges of variation for duplicate PCR reactions are shown. B, MCF7 cells were transfected with nonspecific siRNA or siRNA against BRG1, and mRNA levels for the indicated ER target genes were determined at various times after the beginning of E2 treatment as in (A). C, proposed model for recruitment of SWI/SNF complex by Fli-I. Fli-I is recruited to ER target genes in response to estradiol via binding to ER-LBD (via the G3 module in the gelsolin-like domain of Fli-I) or binding to the N-terminal region of GRIP1 or another p160 coactivator (via the leucine-rich repeat region in the N-terminal domain of Fli-I). Fli-I recruits the SWI/SNF complex by binding to BAF53 (via the G1 module in the gelsolin-like domain of Fli-I). Fli-I is required only for the initial peak of recruitment of SWI/SNF after adding estradiol. Subsequent recruitment of SWI/SNF is mediated by interactions of SWI/SNF with other undefined components of the transcription complex. Thus, upon hormone treatment Fli-I mediates the initial recruitment of SWI/SNF to facilitate remodeling of chromatin structure. Pol II, polymerase II; TBP, TATA box-binding protein.
DISCUSSION
Role of Fli-I in Recruitment of SWI/SNF Chromatin Remodeling Complexes to Estrogen-responsive Gene Promoters
Although Fli-I was previously shown to enhance transient reporter gene activation by nuclear receptors and was required for hormone dependent expression of the transient reporter gene (14), we show here that Fli-I is required for efficient hormonal induction of endogenous ER target genes (Figs. 1 and 8). In multiple independent experiments there was a strong linear correlation between the degree of reduction in Fli-I mRNA and the degree of reduction in target gene expression. Furthermore, the slope for the plot of % reduction of pS2 mRNA versus % reduction of Fli-I mRNA was close to 1 (Fig. 1C), suggesting that Fli-I has a very important role in hormonal activation of ER target genes.
SWI/SNF chromatin remodeling complexes are required for ER-mediated transcriptional activity (31, 32). Guided by the findings that Fli-I binds actin and actin-related SWI/SNF component BAF53 (15, 16) and that Fli-I requires the SWI/SNF complex to activate ER-mediated transcription in transient transfection assays (Fig. 4), we demonstrated that Fli-I plays an important role in recruitment of BRG1-containing SWI/SNF complexes to two different ER target genes (Figs. 6 and 7). Our results, therefore, suggest a model whereby SWI/SNF is recruited to estrogen-responsive promoters by the interaction of its BAF53 subunit with Fli-I (Fig. 8C). This result was unexpected given the previous demonstrations that various subunits of the SWI/SNF complex can interact with ERα and with the glucocorticoid receptor (6, 7, 33–36) and the implicit assumption that these interactions were responsible for recruitment of SWI/SNF to steroid hormone responsive promoters. Although our results demonstrate that Fli-I is required (presumably by bridging ER or ER-associated coactivators to BAF53) for recruitment of SWI/SNF to estrogen-responsive promoters, it is possible that interactions of ER with additional subunits of SWI/SNF also contribute to SWI/SNF recruitment, as discussed further below.
Fli-I, through the G3 module of its gelsolin-like domain, binds directly to the LBD of ER in a hormone-dependent manner (Fig. 2), and the Fli-I G1 module binds directly to BAF53 (Fig. 3). These findings support a model for Fli-I to recruit the SWI/SNF complex to DNA-bound ER (Fig. 8C) and also provide an explanation for the previously described ability of the GelA fragment of Fli-I to enhance transcriptional activation by ERα in transient reporter gene assays (14) (Fig. 4C). Indeed, the G1 module within the GelA fragment of Fli-I has a strong autonomous activation domain when fused to Gal4 DBD (Fig. 4A), and this activity may be because of its ability to recruit BAF53 and the associated SWI/SNF complex, as the E585K mutation in Fli-I eliminated binding to BAF53 (Fig. 3D) and most of the transactivation activity of G1 (Fig. 4B). In contrast to CV-1 cells (14), GelA did not show coactivator activity in SW13 cells, which lack BRG1 and human Brahma. However, upon reintroduction of BRG1 by transient transfection, GelA regained its coactivator function in a BRG1-dependent manner, suggesting that Fli-I requires the SWI/SNF complex for its coactivator function. Moreover, we found that the interactions with BAF53 and ER are critical for GelA to activate ER-mediated transcription (Fig. 4C). Neither the GelA(E585K) nor the GelA(LL/AA) mutant protein was able to activate transcription as efficiently as wild type GelA in SW13 cells transfected with BRG1. However, in addition to ER and BAF53, Fli-I can also bind to the p160 coactivator GRIP1 and to the protein methyltransferase CARM1 (14); which of these interactions is responsible for recruitment of Fli-I to estrogen-responsive promoters still remains to be determined. Multiple contacts with ER and other coactivators may contribute to stable recruitment of Fli-I to the promoter.
Roles of BAFs in Chromatin Remodeling Complexes
It has been proposed that β-actin and actin-related proteins are required for the maximum ATPase activity of SWI/SNF (37, 38) and for the stable association of chromatin remodeling complexes with chromatin (39). Recently, it has been demonstrated that BAF53b, a neuron-specific BAF53-related component of SWI/SNF, is specifically required for targeting SWI/SNF to the promoter of genes essential for dendritic growth (40). Our findings indicate an important role for BAF53 in E2-stimulated expression of ER target genes (Fig. 8). The essentially identical reductions in E2-dependent expression of ER target genes caused by reduction of BAF53 or Fli-I (Fig. 8) coupled with the direct binding of BAF53 to Fli-I in vitro (Fig. 3) further support the model that the interaction of Fli-I with BAF53 is responsible for recruitment of SWI/SNF to promoters of ER target genes. It was recently demonstrated that BAF53 is required for expression and inter-chromosomal interactions between two ER target genes in response to E2 (41). Our findings provide an apparent molecular mechanism for the involvement of BAF53 in the reorganization of chromatin domains by showing that Fli-I is responsible for linking BAF53 to the transcription complex formed by hormone-activated ER.
Other subunits of SWI/SNF complexes have previously been shown to bind steroid hormone receptors. BAF57 binds to ERα and to p160 coactivators and is required for E2-dependent expression of transiently transfected reporter genes and endogenous ER target genes (33, 34). Interaction of the glucocorticoid receptor with SWI/SNF was reported to be mediated by BAF60a in a ligand-independent manner (35) and by BAF250 in a ligand-dependent manner (36). Similarly, the interaction of SWI/SNF complexes with androgen receptor is mediated by direct interaction of androgen receptor with BAF57 and BAF155 (42, 43). However, whether any of these interactions is required for recruitment of SWI/SNF to steroid hormone-regulated target genes has not been determined. Because SWI/SNF complexes are very large (∼2 MDal), multiple protein-protein interactions might be involved in its association with the transcription initiation complex assembled by ER.
Role of Fli-I in the Complex Temporal Pattern of SWI/SNF Recruitment and Implications for the Mechanism of Transcriptional Activation
Chromatin immunoprecipitation studies defined an ordered, sequential, and cyclical pattern of steady-state level occupancy by ER, various coregulators, and various histone modifications on the pS2 promoter upon exposure of MCF7 cells to E2 (28–30). Multiple peaks of steady-state ER recruitment are observed, separated by intervals of ∼40 min. After the beginning of E2 treatment, BRG1 occupancy increases in parallel with ER recruitment but reaches its first peak of occupancy at 10 min, before the first ER peak of occupancy; subsequent peaks of BRG1 occupancy occur in between the peaks of ER occupancy and coincide with recruitment of histone deacetylase 1 and 7 (30). It was, therefore, proposed that the first peak of BRG1 occupancy helps to establish a chromatin conformation permissive for transcription initiation, whereas the subsequent peaks of BRG1 recruitment facilitate clearance of ER and the transcription complexes from the promoter.
We observed a similar E2-induced cyclical pattern of pS2 and GREB1 promoter occupancy by ER and BRG1 in MCF7 cells, with the first peak of BRG1 recruitment overlapping with but reaching a maximum before the first peak of ER occupancy and the second peak of BRG1 recruitment occurring between peaks of ER occupancy (Figs. 6 and 7). Remarkably, we observed that only the initial peak of recruitment of BRG1 was eliminated by reducing cellular levels of Fli-I or BAF53, whereas the second peak of BRG1 occupancy at 90 min still occurred normally (Figs. 6 and 7). These results suggest that BRG1 and SWI/SNF complexes are recruited by Fli-I interaction with BAF53 during the initial peak of BRG1 occupancy, but SWI/SNF uses a different mechanism of recruitment that does not require Fli-I or BAF53 in subsequent peaks of recruitment. This conclusion apparently fits well with the previous conclusion that the BRG1-based SWI/SNF complex is required for promoting transcription during its first peak of occupancy but is involved with clearing ER and the transcription complex from the promoter during subsequent peaks of recruitment (30). In any case, inhibition of only the initial, but not subsequent, peaks of BRG1 and SWI/SNF recruitment by depletion of Fli-I or BAF53 has a profound effect on the E2-induced expression of ER target genes (Fig. 8A). Although the actual peak of Fli-I occupancy occurs slightly after the first peak of BRG1 occupancy at 10 min, there may be sufficient Fli-I on the promoter at 10 min to recruit BRG1. Alternatively, Fli-I may facilitate BRG1 recruitment by an indirect mechanism that is not currently apparent. In any case, our data demonstrate that Fli-I is required for recruitment of SWI/SNF complexes to the promoter to help establish the initial transcription initiation complex (Fig. 8C), whereas subsequent rounds of promoter occupancy by SWI/SNF involve another mechanism of recruitment which is yet undefined.
Implications for Coordination of SWI/SNF and Mediator Functions
Whether chromatin remodeling by SWI/SNF-type complexes and histone modifying enzymes is required for recruitment of the Mediator complex (which is implicated in recruitment of RNA polymerase II) or vice versa has been a topic of considerable debate and may depend on promoter context (44–48). Sequential recruitment of the p160 coactivator complex (which contains several histone modifying enzymes), Mediator complexes, and SWI/SNF-type complexes has been proposed to account for their combinatorial and complementary actions in mediating transcriptional activation (29, 30, 49). Our observations indicate that the recruitment of Mediator complex to the pS2 promoter is not dependent on the initial recruitment of SWI/SNF, as depletion of Fli-I with the accompanying elimination of the first peak of SWI/SNF recruitment had no effect on recruitment of Mediator component Med1/TRAP220 (Fig. 6C). However, depletion of Fli-I compromised recruitment of RNA polymerase II in response to hormone (Fig. 6A), suggesting that both Fli-I and Mediator contribute to recruitment of RNA polymerase II to the pS2 promoter after hormone treatment. We also recently showed that another coactivator, CCAR1, is required for recruiting the Mediator complex to the pS2 promoter (50). Because both Fli-I (this study) and CCAR1 (50) can interact with ER and with components of the p160 complex, Fli-I and CCAR1 would appear to provide a possible mechanism for coordination of the complementary functions of three important coactivator complexes, i.e. p160, Mediator, and SWI/SNF. Thus, we have begun to establish the functional relationships and interdependencies among the many coactivators required for transcriptional activation.
Supplementary Material
Acknowledgments
We thank Dan Gerke and Kelly Chang (University of Southern California) for expert technical assistance, Dr. Geoffrey L. Greene (University of Chicago) for plasmid pGEX-ERα-LBD, Dr. Anthony N. Imbalzano (University of Massachusetts Medical School) for the plasmid expressing BRG1, and Dr. Trevor K. Archer (National Institutes of Health) for SW13 cells.
This work was supported, in whole or in part, by National Institutes of Health Grant DK43093 (to M. R. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S5.
- BRG1
- Brahma-related gene 1
- BAF
- BRG1-associated factor
- ER
- estrogen receptor
- LRR
- leucine-rich repeat
- HA
- hemagglutinin
- siRNA
- small interfering RNA
- NR
- nuclear receptor
- Fli-I
- Flightless-I
- ChIP
- chromatin immunoprecipitation
- qRT-PCR
- quantitative reverse transcription-PCR
- E2
- estradiol
- siNS
- nonspecific siRNA
- GST
- glutathione S-transferase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- LBD
- ligand binding domain
- DBD
- DNA binding domain.
REFERENCES
- 1.Peterson C. L., Workman J. L. (2000) Curr. Opin. Genet. Dev. 10, 187–192 [DOI] [PubMed] [Google Scholar]
- 2.Eberharter A., Becker P. B. (2004) J. Cell Sci. 117, 3707–3711 [DOI] [PubMed] [Google Scholar]
- 3.Trotter K. W., Archer T. K. (2007) Mol. Cell. Endocrinol. 265–266, 162–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trotter K. W., Archer T. K. (2008) Nucl. Recept. Signal 6, e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan H. Y., He X., Kingston R. E., Narlikar G. J. (2003) Mol. Cell 11, 1311–1322 [DOI] [PubMed] [Google Scholar]
- 6.Muchardt C., Yaniv M. (1993) EMBO J. 12, 4279–4290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshinaga S. K., Peterson C. L., Herskowitz I., Yamamoto K. R. (1992) Science 258, 1598–1604 [DOI] [PubMed] [Google Scholar]
- 8.Johnson T. A., Elbi C., Parekh B. S., Hager G. L., John S. (2008) Mol. Biol. Cell 19, 3308–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.John S., Sabo P. J., Johnson T. A., Sung M. H., Biddie S. C., Lightman S. L., Voss T. C., Davis S. R., Meltzer P. S., Stamatoyannopoulos J. A., Hager G. L. (2008) Mol. Cell 29, 611–624 [DOI] [PubMed] [Google Scholar]
- 10.Lonard D. M., O'Malley B. W. (2006) Cell 125, 411–414 [DOI] [PubMed] [Google Scholar]
- 11.Roeder R. G. (2005) FEBS Lett. 579, 909–915 [DOI] [PubMed] [Google Scholar]
- 12.Stallcup M. R., Kim J. H., Teyssier C., Lee Y. H., Ma H., Chen D. (2003) J. Steroid Biochem. Mol. Biol. 85, 139–145 [DOI] [PubMed] [Google Scholar]
- 13.Rosenfeld M. G., Lunyak V. V., Glass C. K. (2006) Genes Dev. 20, 1405–1428 [DOI] [PubMed] [Google Scholar]
- 14.Lee Y. H., Campbell H. D., Stallcup M. R. (2004) Mol. Cell. Biol. 24, 2103–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davy D. A., Ball E. E., Matthaei K. I., Campbell H. D., Crouch M. F. (2000) Immunol. Cell Biol. 78, 423–429 [DOI] [PubMed] [Google Scholar]
- 16.Davy D. A., Campbell H. D., Fountain S., de Jong D., Crouch M. F. (2001) J. Cell Sci. 114, 549–562 [DOI] [PubMed] [Google Scholar]
- 17.Urosev D., Ma Q., Tan A. L., Robinson R. C., Burtnick L. D. (2006) J. Mol. Biol. 357, 765–772 [DOI] [PubMed] [Google Scholar]
- 18.Goodson H. V., Hawse W. F. (2002) J. Cell Sci. 115, 2619–2622 [DOI] [PubMed] [Google Scholar]
- 19.Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 20.Takata H., Matsunaga S., Morimoto A., Ono-Maniwa R., Uchiyama S., Fukui K. (2007) FEBS Lett. 581, 3783–3788 [DOI] [PubMed] [Google Scholar]
- 21.Oxelmark E., Roth J. M., Brooks P. C., Braunstein S. E., Schneider R. J., Garabedian M. J. (2006) Mol. Cell. Biol. 26, 5205–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rae J. M., Johnson M. D., Scheys J. O., Cordero K. E., Larios J. M., Lippman M. E. (2005) Breast Cancer Res. Treat. 92, 141–149 [DOI] [PubMed] [Google Scholar]
- 23.Sun J., Nawaz Z., Slingerland J. M. (2007) Mol. Endocrinol. 21, 2651–2662 [DOI] [PubMed] [Google Scholar]
- 24.Chen Y. H., Kim J. H., Stallcup M. R. (2005) Mol. Cell. Biol. 25, 5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Way M., Pope B., Weeds A. G. (1992) J. Cell Biol. 116, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao K., Wang W., Rando O. J., Xue Y., Swiderek K., Kuo A., Crabtree G. R. (1998) Cell 95, 625–636 [DOI] [PubMed] [Google Scholar]
- 27.Dunaief J. L., Strober B. E., Guha S., Khavari P. A., Alin K., Luban J., Begemann M., Crabtree G. R., Goff S. P. (1994) Cell 79, 119–130 [DOI] [PubMed] [Google Scholar]
- 28.Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. (2000) Cell 103, 843–852 [DOI] [PubMed] [Google Scholar]
- 29.Burakov D., Crofts L. A., Chang C. P., Freedman L. P. (2002) J. Biol. Chem. 277, 14359–14362 [DOI] [PubMed] [Google Scholar]
- 30.Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Cell 115, 751–763 [DOI] [PubMed] [Google Scholar]
- 31.DiRenzo J., Shang Y., Phelan M., Sif S., Myers M., Kingston R., Brown M. (2000) Mol. Cell. Biol. 20, 7541–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiba H., Muramatsu M., Nomoto A., Kato H. (1994) Nucleic Acids Res. 22, 1815–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belandia B., Orford R. L., Hurst H. C., Parker M. G. (2002) EMBO J. 21, 4094–4103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.García-Pedrero J. M., Kiskinis E., Parker M. G., Belandia B. (2006) J. Biol. Chem. 281, 22656–22664 [DOI] [PubMed] [Google Scholar]
- 35.Hsiao P. W., Fryer C. J., Trotter K. W., Wang W., Archer T. K. (2003) Mol. Cell. Biol. 23, 6210–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nie Z., Xue Y., Yang D., Zhou S., Deroo B. J., Archer T. K., Wang W. (2000) Mol. Cell. Biol. 20, 8879–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen X., Ranallo R., Choi E., Wu C. (2003) Mol. Cell 12, 147–155 [DOI] [PubMed] [Google Scholar]
- 38.Rando O. J., Zhao K., Janmey P., Crabtree G. R. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olave I. A., Reck-Peterson S. L., Crabtree G. R. (2002) Annu. Rev. Biochem. 71, 755–781 [DOI] [PubMed] [Google Scholar]
- 40.Wu J. I., Lessard J., Olave I. A., Qiu Z., Ghosh A., Graef I. A., Crabtree G. R. (2007) Neuron 56, 94–108 [DOI] [PubMed] [Google Scholar]
- 41.Hu Q., Kwon Y. S., Nunez E., Cardamone M. D., Hutt K. R., Ohgi K. A., Garcia-Bassets I., Rose D. W., Glass C. K., Rosenfeld M. G., Fu X. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 19199–19204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hong C. Y., Suh J. H., Kim K., Gong E. Y., Jeon S. H., Ko M., Seong R. H., Kwon H. B., Lee K. (2005) Mol. Cell. Biol. 25, 4841–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Link K. A., Burd C. J., Williams E., Marshall T., Rosson G., Henry E., Weissman B., Knudsen K. E. (2005) Mol. Cell. Biol. 25, 2200–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He Q., Battistella L., Morse R. H. (2008) J. Biol. Chem. 283, 5276–5286 [DOI] [PubMed] [Google Scholar]
- 45.Yoon S., Qiu H., Swanson M. J., Hinnebusch A. G. (2003) Mol. Cell. Biol. 23, 8829–8845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Govind C. K., Yoon S., Qiu H., Govind S., Hinnebusch A. G. (2005) Mol. Cell. Biol. 25, 5626–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang G., Balamotis M. A., Stevens J. L., Yamaguchi Y., Handa H., Berk A. J. (2005) Mol. Cell 17, 683–694 [DOI] [PubMed] [Google Scholar]
- 48.Lemieux K., Gaudreau L. (2004) EMBO J. 23, 4040–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito M., Roeder R. G. (2001) Trends Endocrinol. Metab. 12, 127–134 [DOI] [PubMed] [Google Scholar]
- 50.Kim J. H., Yang C. K., Heo K., Roeder R. G., An W., Stallcup M. R. (2008) Mol. Cell 31, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.