Abstract
Cells exposed to environmental stress rapidly activate the MAPK cascade (MKKK/MKK/MAPK). The transient nature of stress signaling is a consequence of negative feedback signals that lead to kinase dephosphorylation, degradation, and sequestration, which have not been fully elucidated for MKK family members. Here, we investigated the signals that negatively regulate MKK4/SEK1, an upstream activator of the MAPKs JNK and p38/HOG1. Following exposure of cells to sorbitol, MKK4 underwent ubiquitination and degradation in a proteasome-dependent manner. MKK4 ubiquitination required JNK kinase activity. The JNK substrate Itch (a HECT domain-containing Nedd4-like ubiquitin protein ligase) bound to MKK4, ubiquitinated lysines 140 and 143, and promoted MKK4 degradation. Other E3 ligases within the MAPK modular complex did not ubiquitinate MKK4. These data suggest that MKK4 is negatively regulated through a feedback loop involving the E3 ubiquitin ligase Itch, which has a fundamental role in the mechanism that controls MKK4 protein levels.
Cellular responses to environmental stress are regulated by an intracellular phosphorelay system involving at least four groups of MAPKs,2 including ERK1/2, JNK1/2/3, p38α/β/γ/δ, and ERK5, which are substrates for specific MAP2K proteins (MKKs); ERK1 and ERK2 are substrates for MKK1/2, p38 for MKK3/6, JNK for MKK4/7, and ERK5 for MKK5 (1, 2). Each MKK is regulated by multiple MAP3K proteins (MKKKs), of which there are more than a dozen mammalian family members, increasing the complexity and diversity of MAPK signaling (3–5). The specificity of these protein interactions is preserved by scaffolding proteins that organize into a single module composed of the three components of a MAPK cascade and its upstream activators (1, 6).
Cells exposed in vitro to stress typically exhibit rapid activation and decay of MAPK activity (2). The degradation of the MAPK signal is a consequence of negative feedback loops that regulate kinase activity, abundance, and localization through changes in kinase phosphorylation and ubiquitination (3, 6, 7). For example, kinase phosphorylation regulates interactions with E3 ligases, which transfer polyubiquitin chains onto lysine residues, by regulating the subcellular location of the kinase or by creating phosphodegrons, which are recognition signals for specific E3 ligases (8–10). The transfer of ubiquitin occurs passively in the case of RING (really interesting new gene) finger-containing E3 ligases, which function as adaptor molecules between the E2 ubiquitin-conjugating enzyme and substrate, or actively in the case of HECT (homologous to the E6-associated protein C terminus) domain-containing E3 ligases, which serve as a catalytic intermediate in the transfer process (11, 12). Through their interactions with E3 ligases, certain MKKKs (MEKK1 and MEKK2) (13, 14) and MAPKs (ERK2 and ERK7) (15, 16) undergo ubiquitination, which marks these proteins for degradation by the 26 S proteasome, thereby attenuating MAPK signaling. In contrast, less progress has been made in understanding how mammalian MKK family members are negatively regulated. Given that MKK homologs in yeast (17) and Dictyostelium (18) are ubiquitinated following prolonged stimulation, we postulated that, like MKKKs and MAPKs, mammalian MKKs are regulated through protein ubiquitination, a reasonable postulate given the modular organization and coordinate regulation of kinases within the MAPK cascade.
In this study, we found that MKKs undergo ubiquitination and proteasomal degradation in response to environmental stress. MKK4 associates with the ubiquitin ligase Itch (which belongs to the HECT domain-containing Nedd4-like E3 family), is an important regulator of murine epithelial and hematopoietic cell function (19), and is absent in 18H (Itchy) mice, which exhibit profound immune defects (20). Itch binds to and ubiquitinates MKK4 and mediates MKK4 protein degradation. Notably, stress-induced MKK4 degradation is dependent upon activation of the MKK4 substrate JNK, which phosphorylates and activates Itch. We conclude that MKK4 protein stability is regulated through a negative feedback loop involving Itch.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies against MKK4, phospho-MKK4, phospho-JNK, c-Jun, phospho-c-Jun, ERK1/2, and Myc (Cell Signaling Technologies); JNK, MEKK1, Itch, HA, GST, and His (Santa Cruz Biotechnology, Inc.); and FLAG and actin (Sigma) were purchased as indicated. Sorbitol was from Sigma, and SP600125 and clasto-lactacystin β-lactone were from Calbiochem.
Plasmids and Site-directed Mutagenesis
Plasmids containing MAP2K4 and ERK2 were gifts from Dr. Jia Le Dai (M. D. Anderson Cancer Center) and Dr. Elizabeth J. Goldsmith (University of Texas at Southwestern Medical Center), respectively. These cDNAs were FLAG-tagged at the N termini and inserted into the pLHCX vector (Clontech). Plasmids containing MEKK1 (wild-type, D1369A, and C433A/C478A), HA-ubiquitin, and HA-JNKK2-JNK1 were gifts from Dr. Zhimin Lu (M. D. Anderson Cancer Center), Dr. Edward Yeh (M. D. Anderson Cancer Center), and Dr. Anning Lin (University of Alabama at Birmingham), respectively. FLAG-MAP2K3 (plasmid 14671), FLAG-MAP2K6 (plasmid 13517), FLAG-MAP2K7b1 (plasmid 14622), Myc-POSH (plasmid 15908), Myc-ITCH (wild-type, plasmid 11427), Myc-ITCH-C822S (plasmid 11428), HA-MEKK4α (wild-type, plasmid 12187), and HA-MEKK4α-K1361M (plasmid 12188) were purchased from Addgene. For protein purification, MAP2K4 and the N-terminal C2 domain-deleted ITCH (ΔC2, Asn236–Glu865) were inserted into the pET15b (His tag; Novagen) and pGEX-4T-1 (GST tag; Amersham Biosciences) vectors, respectively. To construct MAP2K4 mutants, a PCR-based site-directed mutagenesis strategy (21) was carried out using pLHCX-FLAG-MAP2K4 as a backbone.
Cell Culture and Transfection
293T and CHO cells were maintained in Dulbecco's modified Eagle's medium (Mediatech) supplemented with 10% fetal bovine serum (Sigma). MEKK1 wild-type and null mouse embryonic fibroblasts were gifts from Dr. Gary L. Johnson (University of North Carolina School of Medicine) and were cultured in Iscove's modified Dulbecco's medium (Invitrogen) with 10% fetal bovine serum. Transient and stable transfections were performed using Lipofectamine PlusTM (Invitrogen). Small interfering RNA against Itch (Dharmacon) was cotransfected with other plasmids using DharmaFECT® Duo (Dharmacon). Cells were treated with sorbitol (500 mm) or UV light (250 J/m2) to induce ubiquitination and degradation of MKK4.
Western Blotting and Immunoprecipitation
Cells were lysed in 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and protease/phosphatase inhibitor mixture (Sigma). Cell lysates were used for Western blotting or were immunoprecipitated with anti-FLAG M2 affinity gel (Sigma) to isolate FLAG-MKK4 or with anti-Myc antibody and protein G-Sepharose (Amersham Biosciences) to isolate Myc-Itch. Interactions between endogenous MKK4 and Itch were examined by performing anti-MKK4 immunoprecipitation followed by Western blotting with anti-Itch antibody.
In Vitro Binding Assay
His-MKK4 and GST-ItchΔC2 were purified using nickel-Sepharose and GSH-Sepharose (Amersham Biosciences), respectively, according to the manufacturer's protocol. Purified proteins were incubated in binding buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, and protease inhibitors), and the protein complex was pulled down with GSH-Sepharose. After SDS-PAGE, MKK4 bound to Itch was detected by Western blotting with anti-His antibody. GST (Sigma) was used as a negative control.
Ubiquitination Assay
293T cells were transfected with FLAG-MKK and HA-ubiquitin, lysed, and subjected to immunoprecipitation with anti-FLAG M2 affinity gel. After SDS-PAGE, polyubiquitinated MKKs were detected by Western blotting with anti-HA antibody. A ubiquitination kit (BIOMOL) was used for in vitro ubiquitination assay with purified His-MKK4 or immunoprecipitated FLAG-MKK4 (target protein), GST-ItchΔC2 (E3 ligase), and UbcH6 as the E2 ubiquitin-conjugating enzyme.
Quantitative Reverse Transcription-PCR
Total RNA was isolated from small interfering RNA-transfected 293T cells using TRIzol® (Invitrogen). After reverse transcription, quantitative PCR assays were performed using a SYBR® Green-based system (Applied Biosystems) to detect Itch mRNA levels (forward primer, 5′-ACCCTGAGGACAGCCTCATA-3′; and reverse primer, 5′-AAATGTGGCAAAAGTCCCTG-3′). Ribosomal protein L32 (forward primer, 5′-CCTTGTGAAGCCCAAGATCG-3′; and reverse primer, 5′-TGCCGGATGAACTTCTTGGT-3′) was used as a control.
RESULTS AND DISCUSSION
MKKs Are Ubiquitinated and Negatively Regulated by Stress
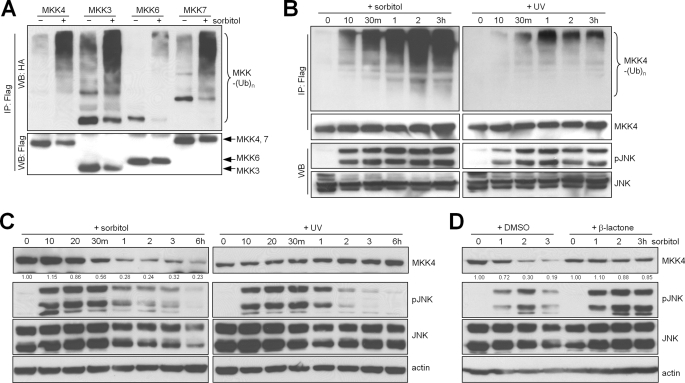
We first examined the effects of stress on MKK4 ubiquitination by treating MKKs with sorbitol or UV light. Treatment with 500 mm sorbitol, which creates a hyperosmotic condition, increased the ubiquitination of MKK3, MKK4, MKK6, and MKK7 (Fig. 1A). Sorbitol-induced MKK ubiquitination was detectable as early as 10 min after treatment initiation, coinciding with a prominent increase in JNK phosphorylation (Fig. 1B), and was followed by a reduction in MKK protein levels (Fig. 1C), which was reversed by treatment with the proteasome inhibitor clasto-lactacystin β-lactone (Fig. 1D). In contrast, the ubiquitination and abundance of MKK4 minimally changed following treatment with UV light (Fig. 1, B and C). Collectively, these findings indicate that MKK is ubiquitinated and degraded through the proteasomal pathway following treatment with sorbitol but not UV light, which is a pattern of sensitivity to stress inducers similar to that of ERK1/2 (15).
FIGURE 1.
Sorbitol treatment increases MKK4 ubiquitination and degradation. A, Western blotting (WB) to detect ubiquitinated (upper panel) and total (lower panel) exogenous protein levels in 293T cells transfected with FLAG-MAP2K and HA-ubiquitin (Ub), treated with sorbitol (500 mm) for 3 h, and subjected to immunoprecipitation (IP) with anti-FLAG M2-agarose. Arrows point to specific bands of interest. B, immunoprecipitation/Western blotting to detect ubiquitinated MKK4 (upper panels), total MKK4 (second panels), phosphorylated JNK (pJNK; third panels), and total JNK (lower panels) in transfected 293T cells. m, minutes. C, Western blot of CHO cells treated for the indicated durations with sorbitol (500 mm) or with a pulse of UV light (250 J/m2) and then incubated for the indicated times. D, Western blot of CHO cells pretreated with the proteasome inhibitor clasto-lactacystin β-lactone (β-lactone; 10 μm) followed by sorbitol. Densitometric values are located under the bands and are expressed relative to those of untreated cells, which were set at 1.0. Actin was used as a loading control. DMSO, dimethyl sulfoxide.
MKK4 Ubiquitination Is Dependent upon MEKK1/JNK Activity
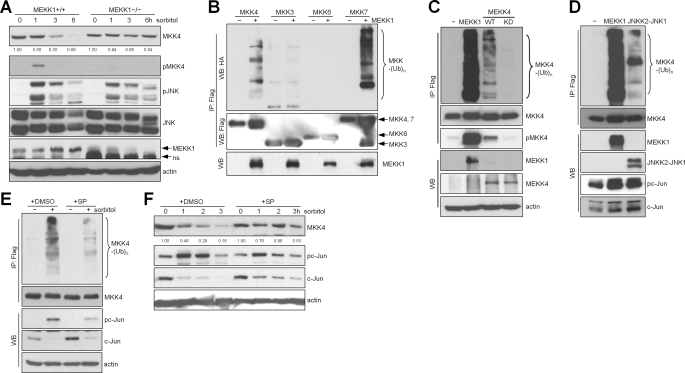
We next examined whether MKK ubiquitination is part of a negative feedback loop activated by stress kinases. Indeed, sorbitol treatment decreased MKK4 levels in MEKK1 wild-type but not null mouse embryo fibroblasts (Fig. 2A) (22), suggesting that an upstream activator of MKK4 is required. Supporting this possibility, MKK4 ubiquitination increased following forced expression of the MKK4 kinases MEKK1 and MEKK4 (Fig. 2, B and C). Interestingly, MEKK1 transfection increased the ubiquitination of MKK4 and MKK7 but not MKK3 or MKK6 (Fig. 2B), suggesting that, of these MKK family members, only MEKK1 substrates are ubiquitinated (23). We next examined whether this effect of MEKK1 requires activation of an MKK4 substrate. Indeed, transfection of an activated MKK7(JNKK2)-JNK fusion construct (24) increased MKK4 ubiquitination (Fig. 2D), and treatment with SP600125, a JNK small molecule inhibitor, abrogated sorbitol-induced MKK4 ubiquitination and degradation (Fig. 2, E and F). We conclude that MKK4 ubiquitination is dependent upon JNK activity.
FIGURE 2.
MKK4 ubiquitination is dependent on the MEKK1/JNK signaling pathway. A, Western blot of MEKK1 wild-type and null mouse embryonic fibroblasts treated with sorbitol (500 mm) to detect the indicated total and phosphorylated (p) protein levels. ns, nonspecific band. B–E, immunoprecipitation (IP)/Western blot (WB) of cotransfected 293T cells to detect ubiquitinated (Ub; upper panels) and total or phosphorylated protein levels (lower panels). WT, wild-type; KD, kinase-dead (K1361M). JNKK2-JNK1 is a constitutively active fusion construct. DMSO, dimethyl sulfoxide. F, Western blot of CHO cells treated with the JNK inhibitor SP600125 (SP; 50 μm). Densitometric values are located under the bands and are expressed relative to those of untreated cells, which were set at 1.0. Actin was used as a loading control.
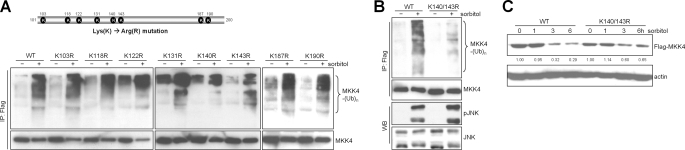
MKK4 Ubiquitination at Lys140 and Lys143
MKK4 is ubiquitinated within the kinase domain,3 but it is unclear which of the lysines within this region are ubiquitinated. All eight lysines within the kinase domain were mutated to arginine (Fig. 3A), and the ubiquitination of these mutant proteins was compared with that of wild-type MKK4. Following transfection of these constructs into cells and treatment of the transfectants with sorbitol, MKK4-K140R and MKK4-K143R exhibited attenuated ubiquitination, whereas the ubiquitination of the other mutants was indistinguishable from that of the wild type (Fig. 3A). A double mutant (K140R/K143R) exhibited strikingly attenuated ubiquitination (Fig. 3B) and greater stability (Fig. 3C) compared with wild-type MKK4 (35% versus 71% reductions in abundance, respectively) following treatment of the transfectants with sorbitol. Although other ubiquitination sites probably exist due to the small, albeit detectable, degradation of the K140R/K143R mutant, we conclude that Lys140 and Lys143 are the primary ubiquitination sites required for the proteolytic effect of sorbitol.
FIGURE 3.
Lys140 and Lys143 are primary MKK4 ubiquitination sites. A: upper panel, locations of lysines within a 100-amino acid region of the MKK4 kinase domain required for ubiquitination. These eight lysines were individually mutated to arginines. Lower panel, immunoprecipitation (IP)/Western blotting of 293T cells transfected with the designated mutants and treated with sorbitol to detect ubiquitinated (Ub; upper) and total (lower) MKK4. B: immunoprecipitation/Western blotting (WB) of 293T cells transfected with wild-type (WT) MAP2K4 or the double mutant K140R/K143R to detect ubiquitinated (upper panel) and phosphorylated (p) and total proteins (lower panels). C: Western blot of CHO cells transfected with wild-type MAP2K4 or the double mutant K140R/K143R. Actin was used as a loading control. Densitometric values are located under the bands and are expressed relative to those of untreated cells, which were set at 1.0.
E3 Ubiquitin Ligase Itch Controls MKK4 Stability
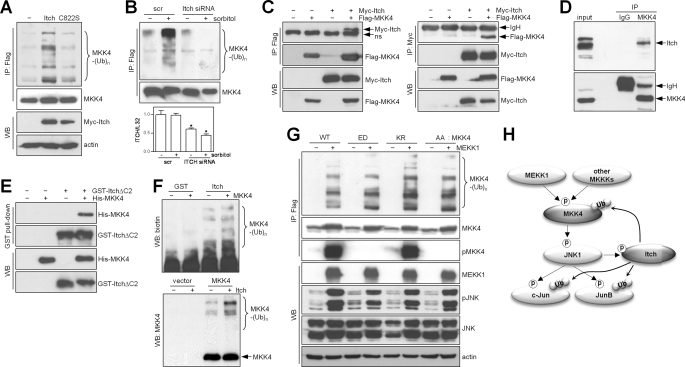
Given our finding that MKK4 was ubiquitinated in a feedback manner by JNK activation, we reasoned that the E3 ligase for MKK4 would be either a component of the MEKK1-MKK4-JNK modular complex that has E3 ligase activity, including tumor necrosis factor receptor-associated factors (25), MEKK1 (15, 26), or POSH (plenty of SH−3) (27), or the JNK substrate Itch (28, 29), which is a member of the HECT domain-containing E3 ligase family (11, 19). Arguing against the former possibility, MKK4 ubiquitination increased similarly in cells transfected with wild-type or E3 ligase-deficient MEKK1 (supplemental Fig. S1, A and B), and MKK4 ubiquitination was suppressed, not enhanced, by forced expression of POSH (supplemental Fig. S1, C and D), suggesting that the POSH scaffolding protein competes with E3 ligases for binding to MKK4. In contrast, basal ubiquitination of MKK4 was enhanced by forced expression of Itch (Fig. 4A), an effect that required Itch E3 ligase activity, and sorbitol-induced MKK4 ubiquitination was abolished in cells transfected with Itch small interfering RNA (Fig. 4B). Itch co-immunoprecipitated with MKK4 (Fig. 4, C and D) and directly bound to MKK4 in an in vitro binding assay using recombinant Itch and MKK4 proteins (Fig. 4E). Itch ubiquitinated MKK4 in an in vitro ubiquitination assay (Fig. 4F). To determine whether the MEKK1 phosphorylation sites on MKK4 (Ser257 and Thr261) are phosphodegrons for Itch, we performed transfections in cells to examine the ability of MEKK1 to ubiquitinate MKK4 with mutations at Ser257 and Thr261 that render MKK4 catalytically inactive (S257A/T261A) or active (S257E/T261D) and a third mutant that is inactive due to a mutation in the ATP-binding site (K131R). In fact, the ubiquitination of wild-type MKK4 was indistinguishable from that of these mutants (Fig. 4G), suggesting that neither the phosphorylation status nor the catalytic activity of MKK4 regulates its ubiquitination. The persistent JNK phosphorylation in cells transfected with the K131R mutant (Fig. 4G) reflects either an incomplete blockade of endogenous MKK4 activity or maintenance of JNK phosphorylation through MKK4-independent mechanisms, such as MKK7.
FIGURE 4.
Itch is an E3 ligase for MKK4. A, immunoprecipitation (IP)/Western blot (WB) of 293T cells transfected with wild-type Itch or an inactive E3 ligase mutant (C822S) to detect ubiquitinated MKK4. Actin was used as a loading control. B, immunoprecipitation/Western blotting to detect ubiquitinated (upper panel) or total (middle panel) MKK4 in 293T cells transfected with scrambled (scr) control or Itch small interfering RNA (siRNA), which effectively knocked down Itch on the basis of quantitative reverse transcription-PCR analysis. In the bar graph (lower panel), values were normalized on the basis of ribosomal protein L32. *, p < 0.05 scrambled versus Itch (n = three per group). C, co-immunoprecipitation of Itch and MKK4 from 293T cells cotransfected with Myc-Itch and FLAG-MKK4. Binding was analyzed by performing anti-FLAG immunoprecipitation/anti-Myc Western blotting (left panels) or anti-Myc immunoprecipitation/anti-FLAG Western blotting (right panels). Arrows point to specific bands of interest. ns, nonspecific band. D, co-immunoprecipitation of endogenous Itch and MKK4 in 293T cells by anti-MKK4 immunoprecipitation/anti-Itch Western blotting. E, in vitro binding assay using GST-ItchΔC2 and His-MKK4 proteins purified from Escherichia coli. F, in vitro ubiquitination assay with purified His-MKK4 from E. coli (upper panel) and immunoprecipitated FLAG-MKK4 from 293T cells (lower panel), GST-ItchΔC2, and UbcH6 as an E2 ubiquitin-conjugating enzyme. Ubiquitinated MKK4 was detected by Western blotting using anti-biotin (upper panel) and anti-MKK4 (lower panel) antibodies. G, immunoprecipitation/Western blotting to detect MKK4 ubiquitination (upper panel) and phosphorylated (p) and total proteins (lower panels) in 293T cells cotransfected with MEKK1 and MKK4 (wild-type (WT) or mutant). ED, constitutively active S257E/T261D mutant; KR, kinase-dead K131R mutant; AA, kinase-dead S257A/T261A mutant. H, diagrammatic illustration of a negative feedback loop involving MKK4/JNK/Itch and other substrates of Itch and JNK (c-Jun and JunB). Arrows indicate directional phosphorylation (p) and ubiquitination (Ub).
Conclusions
Itch substrates include, among others, transcription factors (p63, p73, c-Jun, and JunB) (28, 30, 31), cytosolic kinases (phospholipase Cγ1 and protein kinase Cθ) (32), and transmembrane receptors (Notch1, ErbB4, and CXCR4) (33–35), suggesting that Itch participates in diverse biological functions, but its precise role in stress signaling has not been fully defined. The findings presented here add an important piece to the puzzle, given that MKK4 is an upstream activator of two other Itch substrates, c-Jun and JunB (28), which, like Itch (28, 29), are JNK substrates (Fig. 4H). Thus, Itch substrates cluster around MAPKs and their transcriptional mediators. Given that Itch undergoes substrate recruitment and catalytic activation following phosphorylation by JNK (28, 29), these findings suggest that Itch is part of a negative feedback loop that inhibits MAPK signaling at multiple levels of the cascade. Equally intriguing is our recent finding that most of the tumor-associated somatic mutations in MAP2K4 are loss-of-function mutations, encode C-terminally truncated proteins that are highly unstable, and enhance the invasive properties of tumor cells.3 In this setting, the E3 ligases that degrade MKK4 (such as Itch) may play an important role in tumorigenesis, a point made more compelling by evidence that other Itch substrates, such as p63 and p73 (30, 31), have tumor suppressor properties. Taken together, these data support the hypothesis that Itch plays an important physiologic role as a negative regulator of stress signaling through its effects on multiple substrates, and further studies are warranted to determine whether Itch is oncogenic in permissive cellular and genetic contexts.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA105155 from NCI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Y.-H. Ahn, Y. Yang, D. L. Gibbons, W. Lin, N. Thilaganathan, C. J. Creighton, E. J. Goldsmith, and J. M. Curie, unpublished data.
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- MKK
- MAPK kinase
- MKKK
- MAPK kinase kinase
- MEKK
- MAPK/ERK kinase kinase
- HA
- hemagglutinin
- GST
- glutathione S-transferase
- CHO
- Chinese hamster ovary.
REFERENCES
- 1.Chang L., Karin M. (2001) Nature 410, 37–40 [DOI] [PubMed] [Google Scholar]
- 2.Raman M., Chen W., Cobb M. H. (2007) Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 3.Johnson G. L., Dohlman H. G., Graves L. M. (2005) Curr. Opin. Chem. Biol. 9, 325–331 [DOI] [PubMed] [Google Scholar]
- 4.Chen W., White M. A., Cobb M. H. (2002) J. Biol. Chem. 277, 49105–49110 [DOI] [PubMed] [Google Scholar]
- 5.Deacon K., Blank J. L. (1997) J. Biol. Chem. 272, 14489–14496 [DOI] [PubMed] [Google Scholar]
- 6.Morrison D. K., Davis R. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 7.Owens D. M., Keyse S. M. (2007) Oncogene 26, 3203–3213 [DOI] [PubMed] [Google Scholar]
- 8.Hunter T. (2007) Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 9.Lu Z., Hunter T. (2009) Annu. Rev. Biochem. 78, 435–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laine A., Ronai Z. (2005) Sci. STKE. 2005, re5. [DOI] [PubMed] [Google Scholar]
- 11.Bernassola F., Karin M., Ciechanover A., Melino G. (2008) Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 12.Nakayama K. I., Nakayama K. (2006) Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 13.Liu W. H., Lai M. Z. (2005) Mol. Cell. Biol. 25, 1367–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita M., Ying S. X., Zhang G. M., Li C., Cheng S. Y., Deng C. X., Zhang Y. E. (2005) Cell 121, 101–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Z., Xu S., Joazeiro C., Cobb M. H., Hunter T. (2002) Mol. Cell 9, 945–956 [DOI] [PubMed] [Google Scholar]
- 16.Kuo W. L., Duke C. J., Abe M. K., Kaplan E. L., Gomes S., Rosner M. R. (2004) J. Biol. Chem. 279, 23073–23081 [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Ge Q., Houston D., Thorner J., Errede B., Dohlman H. G. (2003) J. Biol. Chem. 278, 22284–22289 [DOI] [PubMed] [Google Scholar]
- 18.Sobko A., Ma H., Firtel R. A. (2002) Dev. Cell 2, 745–756 [DOI] [PubMed] [Google Scholar]
- 19.Melino G., Gallagher E., Aqeilan R. I., Knight R., Peschiaroli A., Rossi M., Scialpi F., Malatesta M., Zocchi L., Browne G., Ciechanover A., Bernassola F. (2008) Cell Death Differ. 15, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 20.Perry W. L., Hustad C. M., Swing D. A., O'Sullivan T. N., Jenkins N. A., Copeland N. G. (1998) Nat. Genet. 18, 143–146 [DOI] [PubMed] [Google Scholar]
- 21.Meza R., Nuñez-Valdez M. E., Sanchez J., Bravo A. (1996) FEMS Microbiol. Lett. 145, 333–339 [DOI] [PubMed] [Google Scholar]
- 22.Yujiri T., Ware M., Widmann C., Oyer R., Russell D., Chan E., Zaitsu Y., Clarke P., Tyler K., Oka Y., Fanger G. R., Henson P., Johnson G. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7272–7277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia Y., Wu Z., Su B., Murray B., Karin M. (1998) Genes Dev. 12, 3369–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng C., Xiang J., Hunter T., Lin A. (1999) J. Biol. Chem. 274, 28966–28971 [DOI] [PubMed] [Google Scholar]
- 25.Matsuzawa A., Tseng P. H., Vallabhapurapu S., Luo J. L., Zhang W., Wang H., Vignali D. A., Gallagher E., Karin M. (2008) Science 321, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia Y., Wang J., Xu S., Johnson G. L., Hunter T., Lu Z. (2007) Mol. Cell. Biol. 27, 510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z., Kukekov N. V., Greene L. A. (2003) EMBO J. 22, 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., Karin M. (2004) Science 306, 271–275 [DOI] [PubMed] [Google Scholar]
- 29.Gallagher E., Gao M., Liu Y. C., Karin M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi M., Aqeilan R. I., Neale M., Candi E., Salomoni P., Knight R. A., Croce C. M., Melino G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi M., De Laurenzi V., Munarriz E., Green D. R., Liu Y. C., Vousden K. H., Cesareni G., Melino G. (2005) EMBO J. 24, 836–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heissmeyer V., Macián F., Im S. H., Varma R., Feske S., Venuprasad K., Gu H., Liu Y. C., Dustin M. L., Rao A. (2004) Nat. Immunol. 5, 255–265 [DOI] [PubMed] [Google Scholar]
- 33.Qiu L., Joazeiro C., Fang N., Wang H. Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y. C. (2000) J. Biol. Chem. 275, 35734–35737 [DOI] [PubMed] [Google Scholar]
- 34.Sundvall M., Korhonen A., Paatero I., Gaudio E., Melino G., Croce C. M., Aqeilan R. I., Elenius K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchese A., Raiborg C., Santini F., Keen J. H., Stenmark H., Benovic J. L. (2003) Dev. Cell 5, 709–722 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.