Abstract
Huntingtin (Htt) is a widely expressed protein that causes tissue-specific degeneration when mutated to contain an expanded polyglutamine (poly(Q)) domain. Although Htt is large, 350 kDa, the appearance of amino-terminal fragments of Htt in extracts of postmortem brain tissue from patients with Huntington disease (HD), and the fact that an amino-terminal fragment, Htt exon 1 protein (Httex1p), is sufficient to cause disease in models of HD, points to the importance of the amino-terminal region of Htt in the disease process. The first exon of Htt encodes 17 amino acids followed by a poly(Q) repeat of variable length and culminating with a proline-rich domain of 50 amino acids. Because modifications to this fragment have the potential to directly affect pathogenesis in several ways, we have surveyed this fragment for potential post-translational modifications that might affect Htt behavior and detected several modifications of Httex1p. Here we report that the most prevalent modifications of Httex1p are NH2-terminal acetylation and phosphorylation of threonine 3 (pThr-3). We demonstrate that pThr-3 occurs on full-length Htt in vivo, and that this modification affects the aggregation and pathogenic properties of Htt. Thus, therapeutic strategies that modulate these events could in turn affect Htt pathogenesis.
Aberrant behavior of mutant Huntingtin protein (Htt),2 caused by an expansion of the CAG triplet repeat sequence within the first exon of the huntingtin (IT15) gene, results in neurodegeneration and leads to Huntington disease (HD) (1). Full-length Htt protein is 350 kDa in size, but a truncated form of Htt (Httex1p), which includes the expanded polyglutamine region, is sufficient to cause pathology in animal models (2–4). Moreover, an amino-terminal fragment of Htt is detected in nuclear extracts from patient brain and is not detected in control cortex samples (5). In fact, recent studies suggest that production of truncated fragments is essential for disease (6, 7).
The first 17 amino acids of Htt, MATLEKLMKAFESLKSF, are highly conserved throughout mammalian evolution (8, 9), suggesting an important function for these residues. It is well established that post-translational modifications of a protein can affect activity state, intracellular localization, turnover rate, and protein-protein interactions. Several modifications of Htt, without the addition of exogenous modifiers, have been identified (10–18) and implicated in HD (18, 19), but to date, none of these occur within the pathogenic Httex1p fragment. Given that this domain is sufficient to cause HD-like phenotypes, modifications that occur within this pathologic fragment may directly affect either its biophysical properties or its interaction with cellular components that affect pathology. Within the first 17 amino acids of Httex1p, there are several candidate amino acids for post-translational modification. Whereas genetic mutation of the lysines in this region alters HD pathology (20, 21), direct evidence for modifications of the amino-terminal fragment, e.g. by mass spectrometry, and identification of the modified residues, remains undocumented.
In addition to affecting interactions with cellular components, recent reports indicate that mutations in the first 17 amino acids can alter the intrinsic structure of the peptide and modulate the propensity of Htt to aggregate (8, 22). The role of Htt-containing aggregates in HD remains unclear, with recent studies suggesting that visible aggregates may be protective and function as a coping response to toxic mutant Htt (22, 23). An increasingly popular notion is that oligomer/protofibrillar soluble intermediates formed during the aggregation process are the pathogenic structures (24). Post-translational modification of the first 17 amino acids could influence Httex1p aggregation behavior by changing the properties of the modified residue much like the amino acid substitutions reported (8, 22).
In this study, we use mass spectrometry to present the first direct physical evidence for post-translational modification of the pathogenic exon 1 fragment of Htt without overexpressing modifying moieties or enzymes. We find that Htt is modified by the native cellular machinery and that the most common modifications of Httex1p are amino (NH2)-terminal acetylation and phosphorylation of threonine 3 (Thr3). Furthermore, we show that Thr-3 phosphorylation occurs in vivo on full-length, endogenous Htt, that the length of the poly(Q) tract affects the relative abundance of this modification, and that Thr-3 phosphorylation affects HD pathology and the propensity for Htt aggregation in vitro and in vivo.
EXPERIMENTAL PROCEDURES
Cell Culture Plasmid Constructs
Constructs encoding the first exon of Htt, containing 25, 46, or 97 Qs were previously cloned into pcDNA 3.1 (21). The sequence for the HBH tag (25) was inserted, in-frame, 3′ of the Httex1p sequence between the BamHI and XbaI restriction sites. Thr-3 mutant constructs were generated by inserting double-stranded oligonucleotides between restriction site sequences for XhoI and StuI, 5′ of and within, the Httex1p sequence, using pBluescript SK(−) (Stratagene) as an intermediate vector during cloning.
Cell Culture
ST14A (26) and HeLa cells were cultured as previously described (21). We used Lipofectamine 2000 (Invitrogen) to transiently transfect cells, according to the manufacturer's instructions.
Protein Purification and Mass Spectrometric Analysis
ST14A and HeLa cells were transiently transfected with Httex1p containing an unexpanded (25Q) or expanded (46Q) poly(Q) tract fused to a COOH-terminal HBH tag (25) in 10-cm dishes. Cells reached confluence 48 h following transfection and were incubated with 10 nm calyculin A (Calbiochem) for 30 min. Calyculin A treatment was used in all cell culture experiments, to retain phosphorylation modifications (27), when probing for phosphorylation by mass spectrometry or immunoblot (Figs. 1, and 3, A and B; also Fig. 3, C and D, described below). Following phosphatase inhibitor pretreatment, cells were washed with cold 1× phosphate-buffered saline, and then each plate was lysed and harvested in 1 ml of denaturing lysis buffer (8 m urea, 500 mm NaCl, 50 mm NaH2PO4, 10 mm imidazole, 0.5% Triton X-100, 50 mm Tris-HCl, pH 8.0 + complete mini protease inhibitor pellet (Roche)). Passing 20 times through a 20-gauge needle furthered the cell lysis and sheared the DNA. Cells were then centrifuged to remove cellular debris. The supernatant was incubated with 25 μl of a nickel-Sepharose 6 Fast Flow (Amersham Biosciences) bead slurry 3 h to overnight, at room temperature. Protein-bound beads were then washed 4 times with wash buffer (8 m urea, 500 mm NaCl, 50 mm NaH2PO4, 20 mm imidazole, 0.5% Triton X-100, 50 mm Tris-HCl, pH 6.3). For mass spectrometric analysis, beads from 20 plates of the same condition were combined and the urea buffer was replaced with 50 mm NH4CO3.
FIGURE 1.
Httex1p is post-translationally modified. The following ESI-MS/MS spectra were obtained after chymotryptic digestion and collision induced dissociation (CID) of nickel-purified Httex1p HBH: A, NH2-terminally acetylated Htt peptide Ac-ATLEKLMKAFESLKSF, [MH3]3+ at m/z 629.09 (MH+ = 1885.27 Da); B, NH2-terminally acetylated and mono-phosphorylated peptide Ac-ApT3LEKLMKAF, [MH2]2+ at m/z 637.32 (MH+ = 1273.64 Da). b/yn* denotes the b/yn ion with the neutral loss of H3PO4.
FIGURE 3.
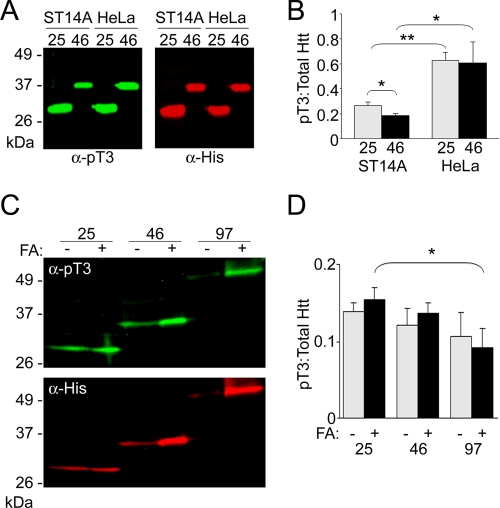
Htt Thr-3 phosphorylation is affected by polyglutamine length and cell type. Unexpanded and expanded Htt were nickel-purified from ST14A (left two lanes of both panels) or HeLa (right two lanes of both panels) cells and analyzed for relative pThr-3 content by immunoblot (A). Thr-3 phosphorylation was detected using anti-pThr-3 (left panels) and total Htt, including modified and unmodified Htt, was determined using anti-His (right panels). pThr-3 and His signals were detected and quantified using LI-COR. pThr-3:total Htt ratios were calculated for three independent experiments in B. Whole cell extracts from ST14A cells expressing Httex1p with 25Q, 46Q, or 97Q were left untreated (−) or were treated with 100% formic acid (+) to solubilize aggregated material, and then analyzed by immunoblot using pThr-3 (top panel) and His (bottom panel) antibodies (C). pThr-3:total Htt ratios were calculated for 25Q (top bar graph), 46Q (middle bar graph), and 97Q (bottom bar graph), from three independent experiments, to determine the relative amount of pThr-3 in soluble versus total (soluble plus insoluble) Httex1p (D). wt, wild-type; FA, formic acid. Error bars are the S.D. from three independent experiments. *, p ≤ 0.05; **, p ≤ 0.01.
Nickel bead-bound protein was digested with chymotrypsin (2% by weight) overnight at 37 °C. The resultant peptides were extracted from the beads using 25% acetonitrile, 0.1% formic acid 3 times and pooled. The samples were then concentrated by SpeedVac and acidified using 0.1% formic acid prior to mass spectrometric analysis. These samples were analyzed by reverse phase liquid chromatography coupled to tandem mass spectrometry (MS/MS) on a quadropole orthogonal time-of-flight tandem (QSTAR XL) mass spectrometer (Applied Biosystems/PE Sciex) (28).
Extraction of the monoisotopc masses (m/z) of parent ions, their charge states, and their corresponding fragment ions, was performed automatically using Analyst software (Applied Biosystems). These data were then submitted for automated data base searching for protein identification using Batch-tag and MS-tag search programs in Protein Prospector (University of California, San Francisco) (29). All post-translational modifications were confirmed by manual inspection of the MS/MS spectra.
Primary Antibodies
The specific Htt phosphothreonine 3 antibody (anti-pThr-3; 1:1,000) was raised against the following peptide: Ac-A(pT)LEKLMKAFESC, and double affinity purified to yield a phosphospecific antibody (New England peptide). Other primary antibodies used were: rabbit anti-actin (1:1,000; Sigma), rabbit anti-COOH-terminal Htt HF-1 (1:1000; a gift from M. MacDonald, Mass General Hospital), mouse anti-penta-His (1:200; Qiagen), sheep anti-Htt S830 (1:1,000; a gift from G. Bates, Kings College), and rat anti-Elav (1:200 dilution, Iowa Hybridoma Bank).
Immunoblotting
To determine the specificity of the Htt-pThr-3 antibody, unmodified (Ac-ATLEKLMKAFESC) and Thr-3-phosphorylated (Ac-A(pT)LEKLMKAFESC) peptides were dissolved in water and directly spotted, in 10-fold increments between 1 ng and 10 μg in equal volumes, on nitrocellulose. The membrane was air dried for 30 min at room temperature. The dry membrane was stained with Ponceau to confirm similar loading for the two peptides, and then probed with anti-pThr-3 and rabbit anti-horseradish peroxidase (1:5,000; Thermo). For cell extracts, cells were lysed under denaturing conditions as described for protein purification for mass spectrometry. Protein was then nickel-purified as described above or precipitated using 20% trichloroacetic acid, washed with acetone, and resuspended in 2× SDS gel loading buffer. Fly head extracts were prepared as described by Wolfgang et al. (30). Protein concentration was measured by the Bradford assay, and 50 μg of protein (approximately equivalent to 5 fly heads) was loaded per lane. For mouse brain extracts, the striatum and cortex were harvested from 3 wild-type C57BL/6J mice and 5 CAG140 mice. The genotypes and CAG repeat numbers were determined by Laragen and described by Simmons et al. (31). Tissue was homogenized in T-per extraction buffer (Piercenet) supplemented with Complete protease inhibitor mixture (Roche) at 150 mg/ml with a Downs homogenizer. Following homogenization, extracts were centrifuged at 15,000 × g for 15 min at 4 °C, and the supernatants were collected as the soluble fraction (32). The protein concentration of the soluble fraction was determined by Bradford assay, and 10 μg of protein was loaded per lane. Protein extracts from cells, flies, and mice were resolved by SDS-PAGE and transferred to polyvinylidene fluoride or nitrocellulose membranes. The filter retardation assay was conducted as described by Green et al. (32), using 5 fly head equivalents (∼50 μg) per spot. Membrane-bound proteins were detected using one of the primary antibodies listed above. Secondary antibodies used for immunoblotting were goat anti-rabbit horseradish peroxidase (1:5,000; Thermo), Alexa Fluor® 680-conjugated donkey anti-sheep (1:1,000; Molecular Probes), IRDye® 800CW-conjugated goat anti-mouse (1:15,000; LI-COR), IRDye® 680-conjugated goat anti-mouse (1:15,000; LI-COR), IRDye 800CW-conjugated goat anti-rabbit (1:15,000; LI-COR), and IRDye 680-conjugated goat anti-rabbit (1:15,000; LI-COR). Fluorophore-conjugated antibodies were detected and quantified using an Odyssey® Imager (LI-COR). Horseradish peroxidase-conjugated antibodies were detected using SuperSignal West Pico (Thermo). The immunoblot signal was detected using film for horseradish peroxidase-conjugated secondary antibodies and the LI-COR was used to detect and quantify fluorophore-conjugated antibodies. All experiments quantified by the LI-COR were carried out in triplicate, using independently generated samples, and the averages and standard deviations were calculated for the three independent experiments.
Formic Acid Dissolution of Aggregates
ST14A cells were plated in 6-well plates and transfected with Httex1p25Q-HBH, Httex1p46Q-HBH, or Httex1p97Q-HBH. The cells reached confluence 48 h after transfection, and the cells were treated with 20 nm calyculin A (Biomol) for 10 min prior to harvest (see Fig. 3, C and D). Cells were resuspended in denaturing lysis buffer (the same buffer used for protein purification) and homogenized by passing 20 times through a 20-gauge needle. An equal volume of cold 20% trichloroacetic acid was added to the lysates and incubated on ice for 30 min. Precipitated protein was pelleted in a microcentrifuge at 14,000 × g. Pellets were washed with acetone. Following the wash, the pellets were resuspended in 60 μl of 2× SDS gel loading buffer (control) or 60 μl of 100% formic acid. Formic acid-treated samples were incubated at 37 °C for 1 h, dried by vacuum centrifugation, and resuspended in 60 μl of 2× SDS gel loading buffer (procedure modified from Hazeki et al. (33)). 5 μl of 1 m Tris-HCl, pH 8.0, was added to control and formic acid-treated samples to neutralize the pH prior to Western analysis.
Immunofluorescence of Cells
ST14A cells were grown on coverslips (4 coverslips for each condition) and transiently transfected with Httex1p46Q-HBH, Httex1p46QT3A-HBH, or Httex1p46QT3D-HBH using Lipofectamine 2000. 48 h following transfection, cells were fixed with 4% paraformaldehyde, permeablized with 0.1% Triton X-100, and blocked with 5% bovine serum albumin. Sheep anti-Htt (S830) was used at 1:200. A fluorescein isothiocyanate-conjugated donkey anti-sheep secondary antibody (1:250 Jackson ImmunoResearch, Inc.) was used for visualization. 4′,6-Diamidino-2-phenylindole counterstained the cell nuclei. Images were collected on a confocal microscope using a ×20 objective. Non-overlapping pictures containing a total of ∼300 transfected cells per coverslip were taken. The number of cells containing aggregates, divided by the total number of transfected cells, was calculated for each coverslip. Averages and standard deviations were calculated from the four coverslips per condition.
Drosophila Constructs, Stocks, and Crosses
Httex1p97Q, -97QT3A, and -97QT3D were cloned into the pUASTattB vector and this construct was injected into approximately one-half h old embryos of the genotype yw hs-flp; AttP-zh86Fb; vas-phi-zh 102D. This method enables site-specific integration into the same chromosomal position (in this case in 86F on the third chromosome) using the Phi-C31-mediated integration system (34). Several homozygous lines were established for each construct using appropriate balancers. For Httex1p expression, male Htt transgenic flies were mated with females homozygous for the pan-neuronal elav driver w; P{w+mW.hs = GawB}elavC155 (Bloomington Drosophila Stock Center). The un-injected stock flies, with an identical genotype as the transgenic flies without the transgene (yw hs-flp; AttP-zh86Fb; vas-phi-zh 102D), were also crossed to elavC155 virgins to generate wild-type controls. Cultures were raised at 25 °C.
Immunofluorescence of Eye Imaginal Discs
Eye imaginal discs were dissected from 3rd instar larvae, fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with anti-Elav (1:200) and anti-Htt (S830; 1:1000) in phosphate-buffered saline, 0.1% Tween 20 (PBST) overnight at 4 °C. Secondary antibodies used were Cy3-conjugated donkey anti-rat (1:250; Jackson ImmunoResearch) and fluorescein isothiocyanate-conjugated donkey anti-sheep (1:250; Jackson ImmunoResearch, Inc.). Z-stack images were taken and merged using a Zeiss LSM510-META 2-photon confocal microscope. For quantification of aggregation, merged Z-stack images were cropped to equal size at the start of and parallel to Elav expression. Fluorescence from aggregated Htt was measured using Scion imaging software (Scion Corporation) after subjecting the Htt channel to grayscale and threshold commands to remove the signal from soluble Htt. Pixels containing fluorescent signal from aggregates were counted for each disc and the averages and standard deviations were calculated for at least 4 discs per transgene.
Fly Survival
Male flies homozygous for the Htt transgenes were mated to double-balanced virgin females (TM3/TM6). F1 males, containing the Htt transgene over a TM3 balancer (Htt/TM3), were crossed to elavC155 virgin female flies. Cultures were raised at 25 °C, and survival was calculated as the ratio of enclosed adults expressing the transgene to those carrying the balancer.
Pseudopupil Analysis
4- and 11-day-old female flies were decapitated and subjected to pseudopupil analysis to count the number of surviving photoreceptor neurons as previously described (35). Female flies were used as opposed to male flies because of the low survival of males. A minimum of 6 flies per genotype were used for pseudopupil analysis.
Statistics
Unpaired Student's t tests were used to calculate the significance of the differences between measurements: * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001.
RESULTS
Httex1p Is Subject to Post-translational Modification
To determine whether Httex1p is post-translationally modified by endogenous protein activities, we transiently transfected striatal progenitor cells, ST14A, which have properties similar to the medium spiny neurons most overtly affected in HD (26, 36), and non-neuronal HeLa cells with Httex1p containing a polyglutamine tract of 25 or 46Qs, fused to an HBH tag (25) at the carboxyl terminus. The HBH tag contains two His6 tags, enabling purification via nickel chromatography under strong denaturing conditions (8 m urea). To reduce the loss of phosphorylation, transfected cells were pre-treated with the Ser/Thr phosphatase inhibitor, calyculin A prior to purification (27). Nickel-purified Httex1p25Q-HBH and Httex1p46Q-HBH proteins were subjected to on-bead chymotryptic digestion, and the resultant peptides were separated and analyzed by reverse phase liquid chromatography coupled to tandem mass spectrometry (LC MS/MS). The MS/MS spectra were submitted to Protein Prospector (29) for automated data base searching to determine peptide sequences and post-translational modifications. All spectra were examined manually to confirm the Protein Prospector results.
NH2-terminal acetylation and phosphorylation of threonine 3 (Fig. 1), among other modifications (supplemental Fig. S1), were identified in both ST14A and HeLa cell types. Pretreatment with the phosphatase inhibitor calyculin A enhanced the phosphorylation of threonine 3, but phosphorylation of this residue occurs without the addition of the inhibitor (data not shown, also see Fig. 2, B and C, and supplemental Fig. S2 as described below). NH2-terminal acetylation and threonine 3 phosphorylation were present in both cell types on expanded and unexpanded Httex1p, but the intensity of their signals varied. The theoretical monoisotopic mass for a peptide consisting of the first 17 amino acids of Htt is MH+ = 1974.04 Da. One peptide, detected by mass spectrometry, had a mass of 1885.27 Da (Fig. 1A). The difference between these two masses is equivalent to the loss of the NH2-terminal methionine and subsequent acetylation of the penultimate residue, alanine. The peptide sequence was determined by a series of y and b ions present in the MS/MS spectrum (Fig. 1A). Virtually all Htt peptides detected were processed such that the NH2-terminal methionine was removed and the alanine was NH2-terminally acetylated. This is consistent with the widespread removal of the NH2-terminal methionine and acetylation of most eukaryotic proteins but it has the potential to affect the physical properties of this quite critical portion of the Htt protein.
FIGURE 2.
Thr-3 is phosphorylated on endogenous Htt in vivo. A, specificity of rabbit polyclonal antibody against phospho-T3 Htt (pT3) as determined by dot blot assay. Increasing amounts of phospho-T3 peptide and unmodified peptide were spotted onto a nitrocellulose membrane and probed with the pThr-3 antibody. Corresponding extracts from the striata (B) and cortices (C) of 3 wild-type C57BL/6 mice (1–3) and 5 knock-in CAG140 mice (1–5) were analyzed by immunoblot using the pThr-3 antibody (upper panel), the Htt carboxyl-terminal HF-1 antibody (1981–2580 amino acids) (middle panel), and actin antibody as a loading control (bottom panel).
Phosphorylation of threonine 3 (pThr-3) was the most frequent side chain modification detected. Thr-3-phosphorylated peptides were detected on expanded and unexpanded forms of Httex1p isolated from both ST14A and HeLa cells. Fig. 1B illustrates the MS/MS spectrum of a chymotryptic peptide ([MH2]2+ 637.32). A fragment ion ([MH2]2+ 588.32), having lost H3PO4 (−98 Da) from the parent ion ([MH2]2+ 637.32), was observed, indicating that the detected peptide was phosphorylated. Based on the series of b and y ions, the peptide was determined to be the phosphorylated form of the peptide Ac-ATLEKLMKAF, matching the NH2-terminal sequence of Httex1p. In this sequence, Thr-3 is the only potential phosphorylation site. Observation of the non-phosphorylated forms of y1–y8 ions, and detection of phosphorylated forms of y9 and b2 ∼ b9 ions and their corresponding neutral loss fragment ions further confirmed that Thr-3 is the phosphorylated residue in this peptide. Several mass spectrometric analyses of Httex1p post-translational modification were performed, and the pThr-3 modification was consistently detected, leading us to further investigate the biological role of Thr-3 phosphorylation in HD pathology.
T3 Phosphorylation Occurs in Vivo
To further explore the consequences of phosphorylation of Htt Thr-3, a phosphospecific antibody was generated against the Htt-pThr-3 peptide. Specificity of the anti-pThr-3 antiserum was demonstrated by its ability to detect the Thr-3-phosphorylated peptide but not recognize the unmodified peptide by dot blot (Fig. 2A).
To eliminate the possibility that Thr-3 phosphorylation occurs only on an overexpressed and/or truncated fragment of Htt, we asked whether endogenous, full-length Htt is phosphorylated at Thr-3 in vivo in tissues relevant to HD. We utilized wild-type C57BL/6 mice and a previously described HD mouse model, the CAG140 mouse, in which human htt exon 1 containing 140 CAG repeats was inserted into the native murine htt locus (hdh) (37). The mouse Hdh and human Htt proteins are 91% identical (38), with no deviations in the first 17 amino acids, permitting detection of Thr-3 phosphorylation of both the wild-type mouse Htt and the human-mouse hybrid Htt in knock-in mice. Because most of the neuron loss occurs in the striatum and cortex of HD patients, independent extracts from the striatum and cortex of five 6-month-old CAG140 mice and three wild-type littermates were analyzed for Thr-3 phosphorylation. The phosphospecific antibody detected Htt Thr-3 phosphorylation in the striatum and cortex of both the wild-type and mutant mice at molecular weights corresponding to the full-length forms (Fig. 2, B and C). The pThr-3-positive bands were also recognized by an antibody that recognizes the COOH terminus of Htt (HF-1) (39), confirming that full-length Htt is phosphorylated on Thr-3 in vivo. In all samples from the CAG140 mice, a reduced amount of Htt protein was observed, as is typically seen due to the redistribution of soluble expanded Htt into insoluble aggregates (see Fig. 3C, lower panel). The parallel changes in signal intensity of HF-1 and anti-pThr-3 in the striatum illustrate that the pThr-3 antibody is indeed recognizing a modified form of Htt and not a nonspecific protein of coincidentally the same size (Fig. 2B). We conclude from these data that Thr-3 phosphorylation occurs on full-length, endogenous Htt in brain tissue.
Polyglutamine Length Affects Thr-3 Phosphorylation
After detecting phosphorylation of Thr-3 in vivo on full-length Htt, we asked whether Thr-3 phosphorylation is differentially sensitive to polyglutamine expansion. Levels of Thr-3 phosphorylation on soluble Htt in ST14A striatal progenitor cells using nickel-purified, expanded (46Q) and unexpanded (25Q) HBH-tagged Httex1p were compared by quantitative Western blot using an Odyssey Imager (LI-COR). This instrument has a wide linear range for accurate quantification, especially of weak bands (40). In ST14A cells, unexpanded Httex1p exhibited a higher fraction of pThr-3 phosphorylation than expanded Httex1p (Fig. 3, A, left two lanes of each panel, quantified in B). Because expanded Htt is prone to inclusion body formation, we compared the fractions of Thr-3 phosphorylation in untreated whole cell extracts with those treated with formic acid to dissociate insoluble Htt-containing material, yielding monomeric Htt (33). Formic acid treatment increased the amount of soluble expanded Htt, but did not alter the ratio of phosphorylated Thr-3 (Fig. 3, C, quantified in D).
The same method was used to determine the ratio of pThr-3:total Htt for expanded and unexpanded Htt in non-neuronal HeLa cells. A statistically significant increase in the proportion of Thr-3-phosphorylated Htt was observed in both expanded and unexpanded Httex1p (Fig. 3, A, right two lanes of each panel, quantified in B). Interestingly, there was no difference observed in the Thr-3 phosphorylation of expanded versus unexpanded Htt in the HeLa cell type. These results suggest that a higher ratio of pThr-3:total Htt is inversely correlated with disease and susceptibility of the cell to Htt challenge.
T3 Phosphomimetic Enhances Htt Aggregation in Cell Culture
The results above raise the question of how pThr-3 affects HD phenotypes. Because the first 17 amino acids are known to modulate Httex1p aggregation (8, 9, 22), we generated phosphomimetic and nonphosphorylatable mutants of Httex1p in which Thr-3 was converted to either aspartic acid (T3D) or alanine (T3A), respectively. These mutants were generated in the moderately expanded Httex1p (46Q) rather than Httex1p with a larger expansion (97Q) to increase the sensitivity of our experiment, because Httex1p46Q exhibits much less aggregation than the Httex1p97Q protein and thus may be more sensitive to modulatory effects. Aggregation of wild-type expanded Httex1p (46Q) was compared with that of 46QT3D or 46QT3A forms of Httex1p in ST14A cells (Figs. 4, A and B). Cells expressing 46QT3A displayed slightly decreased aggregation compared with those expressing the wild-type sequence (30 versus 35% of cells contain aggregates; Fig. 4, A, left and center panels, quantified in B). On the other hand, expression of the 46QT3D enhanced Htt aggregation (54% of cells contain aggregates) (Fig. 4, A, right panel, quantified in B). The absence of aggregates in ST14A cells expressing unexpanded Htt (25Q) was unaffected by either the T3D or T3A mutation (data not shown). These data are consistent with the possibility that Thr-3 phosphorylation enhances Htt aggregation.
FIGURE 4.
Thr-3 mutant aggregation in striatal progenitor cells. ST14A cells expressing 46Q, 46QT3A, or 46QT3D were immunostained with anti-Htt (S830; green) and nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue) (A). Images were taken using a ×20 confocal objective (A; top row). The bottom row depicts enlarged regions (×2) of the boxed regions in the top row. The number of transfected cells containing at least 1 aggregate were counted and normalized to total number of transfected cells (B). The scale bar equals 100 μm in A. *, p ≤ 0.05; ***, p ≤ 0.001. Error bars are the S.D. from four independent coverslips.
T3 Mutation Affects Httex1p Aggregation in Vivo
Given the impact of Thr-3 modification on Htt aggregation in cultured cells, we sought to confirm the influential role of Thr-3 phosphorylation in in vivo protein aggregation. Drosophila models of Huntington disease, expressing expanded Httex1p, exhibit several phenotypes including reduced viability and lifespan, neurodegeneration, and protein aggregation (3, 35, 41). In addition, the human protein is phosphorylated by the Drosophila cellular machinery (supplemental Fig. S2). We therefore generated transgenic Drosophila that ectopically express expanded (97Q) wild-type, phosphomimetic (97QT3D), or non-phosphorylatable (97QT3A) Httex1p to compare the pathogenicity of these proteins. To allow for direct comparison of multiple transgenes, without the effects of different integration sites (42–44), the transgenes were inserted into a defined site and orientation by targeted insertion (34), and expressed in neurons using the elav promoter and the UAS-Gal4 system.
Quantitative reverse transcriptase-PCR demonstrated that fly lines expressing Httex1p97Q, Httex1p97QT3A, and Httex1p97QT3D produce similar levels of transgene expression (data not shown). Extracts from these Htt-expressing flies were also assayed for soluble protein content by quantitative Western blot (Fig. 5, A, quantified in B) and aggregation by filter retardation assay (Fig. 5, C, quantified in D). All lines contained similar levels of soluble Htt (Fig. 5, A and B), whereas 97QT3D mutant flies exhibited a marked increase in insoluble Httex1p compared with Httex1p97Q or -97QT3A (Fig. 5, C and D). This is consistent with the aggregation behavior of Thr-3 mutants observed in cell culture.
FIGURE 5.
The effect of Thr-3 mutation on Httex1p aggregation in vivo. Flies expressing 97Q, 97QT3A, or 97QT3D were assayed for soluble Htt (anti-Htt; S830; top panel) and actin (bottom panel) levels using quantitative immunoblot (A). The Htt signal was quantified and normalized to the actin signal (LI-COR) (B). Insoluble material from Htt-expressing flies was measured by filter retardation assay (C). Equal amounts of fly extract were spotted onto a nitrocellulose membrane, and stained with Ponceau for total protein (top panel) and then probed with anti-Htt (S830) to detect the amount of insoluble Htt in 97Q versus 97QT3A versus 97QT3D (bottom panel). The Htt signal was measured by LI-COR and quantified in D. Eye imaginal discs from 3rd instar larvae were stained for Elav (red, first column) and Htt (S830; green, middle column) (E). Boxes drawn in the merged (last) column, in E, identify the area of the Htt signal that was converted to grayscale (F) and analyzed for total aggregate load using Scion Imaging software as described under “Experimental Procedures” (G). The scale bars equal 50 μm in E and F. wt, wild-type. Error bars are the S.D. from four or more imaginal discs for each genotype (E and F) and three independent dot blots (C and D). *, p ≤ 0.05; **, p ≤ 0.01.
Httex1p aggregate formation was also analyzed using Drosophila larval tissue. During development, a morphogenetic furrow travels across the eye-imaginal disc of 3rd instar larvae, initiating neurogenesis and expression from the pan-neuronal elav promoter. Elav protein and Httex1p expression, under the regulation of elav, commences immediately posterior to this furrow with each row of photoreceptor neurons toward the posterior having expressed the proteins for 2 h longer. This induction of protein expression over time permits visualization of the time course of Httex1p expression and aggregate accumulation. We dissected and fixed eye-imaginal discs from 3rd instar larvae expressing Httex1p97Q, -97QT3A, and -97QT3D under the regulations of elav-Gal4 and stained for Httex1p and Elav (Fig. 5E). The presence of Elav protein defines cells that are expressing Httex1p (Fig. 5E, left column in red). To determine the effect of the Thr-3 mutation on aggregate formation, we measured the total aggregate load generated from expression of the different transgenes. To this end, boxes of identical size were drawn with one edge placed at the leading edge of Elav expression parallel to the wave of Htt expression (Fig. 5E, right column), and the fluorescent signal from aggregated, but not soluble material, within the boxes (Fig. 5F) was analyzed to quantify the total aggregate load (Fig. 5G). Expression of Httex1p97Q results in protein aggregates in fly larval eye-imaginal discs (Fig. 5, E, top row, quantified in G). Mutation of Thr-3 to alanine (Ala) results in reduced Httex1p aggregation (Fig. 5E, middle row, and quantified in G), whereas the T3D mutant yields an increase in aggregation within the defined area (Fig. 5, E, bottom row, quantified in F). The opposing aggregation behavior of the T3A and T3D mutants in the eye imaginal discs was consistent with what was seen in cell culture and with insoluble Httex1p levels from adult fly extracts.
T3 Mutation Rescues Htt-induced Lethality and Neurodegeneration in Vivo
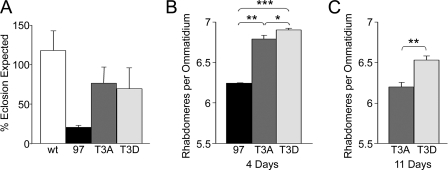
In addition to protein aggregation, expression of expanded Httex1p results in reduced viability and neurodegeneration in Drosophila (3, 35, 41). We have compared the viability and neurodegeneration of wild-type versus the T3A and T3D mutants in our site-directed lines. Percent viability was measured by taking a ratio of Htt-expressing flies to non-expressing siblings that emerged as adults. When expanded Htt expression was induced by the X-chromosomal elav-Gal4 driver, male flies exhibited reduced viability: males expressing Httex1p97Q exhibited 80% lethality; whereas those expressing T3A or T3D, exhibited about 25 and 30% lethality, respectively (Fig. 6A). Females from all three lines, reared at 25 °C, did not exhibit reduced viability (data not shown). The difference in male to female viability is that the females are expressing approximately half as much protein as the males because they are heterozygous for the X-chromosomal driver. In addition to assaying viability, we analyzed the degeneration of photoreceptor neurons upon expression of the transgenes in the adult eye using the pseudopupil assay (3, 41, 45). Mutation of Thr-3 to Asp or Ala significantly reduces the photoreceptor loss caused by Httex1p97Q in female flies at 4 days post-eclosion (Fig. 6B), although rescue from T3D was greater than from the T3A form. This difference was even more significant at 11 days post-eclosion (Fig. 6C). Although T3A and T3D mutations have differing effects on expanded Htt aggregation, they both significantly reduce the neurodegeneration caused by expanded Htt.
FIGURE 6.
The effect of Thr-3 mutation on Httex1p-induced lethality and neurodegeneration. The numbers of male flies expressing no transgene (wild-type, wt; see “Experimental Procedures”), 97Q, 97QT3A, or 97T3D were taken as a percentage of non-expressing siblings carrying a balancer (A). The average rhabdomeres (photoreceptor neurons) per ommatidium (individual eye of a compound eye) were calculated for female flies expressing no transgene (wt), 97Q, 97QT3A, and 97QT3D at 4 days post-eclosion (B). The neurodegeneration was assessed at 11 days post-eclosion for 97QT3A and 97QT3D; there were no 97Q survivors at this time point (C). Error bars are the S.D. from four independent vials (A) or six or more flies per genotype (B and C). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
DISCUSSION
Poly(Q) Toxicity and Protein Context
CAG repeat expansion is responsible for a growing number of “triplet repeat” neurodegenerative diseases, including HD, spinal and bulbar muscular atrophy (Kennedy disease), dentatorubral-pallidoluysian atrophy, and spinocerebellar ataxias (SCA1, -2, -3, -7, and -17) (46). Expression of an expanded polyglutamine peptide alone is sufficient to cause neurodegenerative phenotypes (2, 47). However, the expanded polyglutamine tract seems not to be the sole determinant of disease, because different subsets of neurons are affected in each of the diseases despite ubiquitous expression throughout the brain and other tissues (46). This implies that the protein context, surrounding the poly(Q) repeat, is important for modulating the neurodegeneration caused in these specific diseases. This protein context can be influenced by any post-translational modification of the flanking sequences. Indeed in the case of spinocerebellar ataxias 1, post-translational modification has a role in pathogenesis (48). Because Htt exon 1 protein (Httex1p) is sufficient to cause pathology and contains several modifiable amino acids within its protein sequence, discovering modifications in this region of Httex1p is of particular interest and may reveal key modulators of HD pathology.
Httex1p Is NH2-terminally Acetylated
In this study, we provide the first direct identification of post-translational modifications in the first 17 amino acids of Httex1p, using mass spectrometry, in cells not overexpressing modifying enzymes or moieties. NH2-terminal modification is among these modifications. In most eukaryotic proteins, the NH2-terminal methionine is removed and the second amino acid is acetylated on its NH2-terminal amine group, especially if the penultimate residue has a radius of gyration of 1.29 Å or less (i.e. glycine, alanine, serine, cysteine, threonine, proline, and valine) (49, 50). The second residue of Htt is alanine and our detection of NH2-terminal acetylation following the cleavage of methionine on virtually all Httex1p peptides is consistent with expectation.
Htt Is Phosphorylated, and This Modification Is Affected by Poly(Q) Expansion and Cell Type
In addition to NH2-terminal acetylation, we find compelling evidence that full-length Htt and Httex1p can be phosphorylated on Thr-3 (see Figs. 1–3). Protein activity and subcellular localization are known to be regulated by phosphorylation (51–54). We have repeatedly detected Htt Thr-3 phosphorylation by mass spectrometry and immunoblot analysis using a phospho-T3 specific antibody. In cell culture, the prevalence of Thr-3 phosphorylation is reduced upon poly(Q) expansion, especifically in HD-susceptible cells, whereas pThr-3 is elevated in cells resistant to poly(Q) toxicity (i.e. HeLa) (see Fig. 3). Additionally, in vivo, we find that whereas Thr-3 is clearly phosphorylated in both the cortex and striatum of wild-type mice, in mice expressing expanded poly(Q) Htt, the relative levels of pThr-3 Htt appear to be greater in cortical versus striatal tissues (Fig. 2, B and C). Interestingly, CAG140 mice experience marked striatal neuron loss, whereas the cortex and other regions of the brain are much less affected (55). This difference was observed in all five of the independently prepared striatal extracts and their corresponding cortical extracts (Fig. 2, B and C). The apparently reduced amount of Htt in the CAG140 brain extracts is likely due to the loss of soluble Htt into insoluble aggregates during sample preparation, which is typical for both tissues, at this age (37). The inverse correlation between neurodegeneration and Thr-3 phosphorylation warrants further investigation, but these data are consistent with Thr-3 phosphorylation serving a protective role.
Mutations of the Thr-3 Phosphorylation Site May Alter the Intrinsic Biophysical Properties of Htt
Mutating Thr-3 to either Asp or Ala results in reduced lethality and neurodegeneration in our fly model of HD (see Fig. 6), although mutation to Asp leads to greater rescue than Ala. This could be because there is a modification of Thr-3, other than phosphorylation that makes 97Q more toxic than the Thr-3 mutants (e.g. glycosylation) that was undetected in our mass spectrometric studies, or because there is some intrinsic property of the threonine residue itself that leads to the toxicity seen with the wild-type sequence. The latter seems more likely because although both T3D and T3A mutations reduce the lethality and neurodegeneration caused by expanded Httex1p, they oppositely affect Httex1p aggregation behavior, suggesting different underlying mechanisms for the rescues observed.
Aggregation is a multistep process involving monomers, intermediate oligomeric and protofibrillar structures, and large fibrillar aggregates (56–58). There is a growing consensus that the intermediate oligomeric structures may be the toxic form of expanded Htt (24). If intermediate oligomers actually represent the toxic species and large inclusions are neutral or protective (24, 25), then neuronal protection and the appearance of macroscopic inclusions are separable processes as we observe here. Protection with accompanying aggregates that might occur are either by 1) preventing the initial oligomer formation step or 2) accelerating the aggregation process and sequestering the oligomers into biologically neutral macroscopic inclusions.
Htt aggregation and subcellular localization are modulated by the first 17 amino acids of Htt, and mutations within this region alter cytotoxicity (8, 9, 21, 22, 59). Amino acid substitution within the first 17 amino acids can alter the inherent structural properties of this peptide in a manner that can, directly (by changing the misfolding pathway or kinetics) or indirectly (by affecting membrane localization), affect cellular aggregation (8, 22). The finding that T3D or Ala are both neuroprotective while they result in opposite effects on aggregate formation is consistent with the emerging view that microscopic aggregates do not represent the toxic species (23). Thr-3 resides in what is predicted to be the hydrophobic face of a putative amphipathic α-helix structure (8, 9). By adding a negatively charged phosphate group to the hydrophobic face, phosphorylation of Thr-3 may disrupt this structure, thereby reducing its membrane association and increasing its availability in the cytoplasm for aggregation (8). Mutating Thr-3 to Ala may stabilize the α-helical structure by preventing the phosphorylation of Thr-3, thereby maintaining its membrane association and preventing aggregation.
A second complementary scenario is also possible. NMR structural analysis of the first 17 amino acids indicate that this region exists in a collapsed, compact coil state that resists aggregation and contains little secondary structure, and adopts an α-helical structure only when complexed with binding partners; the only detectable secondary structure within the first 17 amino acids, when in isolation, occurs between the Thr-3 and glutamate 5 residues (22). This collapsed, aggregate-resistant structure of the first 17 residues is maintained in the presence of unexpanded poly(Q), but is disrupted upon expansion of the polyglutamine tract, resulting in a more extended conformation and a corresponding accelerated association of the NH2-terminal 17 amino acids into oligomeric structures (22). When free in solution (i.e. not membrane-bound), it is possible that increasing the hydrophobic nature of Thr-3 (T3A) stabilizes the collapsed coil conformation of amino acids 1–17, retarding oligomer formation and reducing Httex1p pathology. Alternatively, introducing a negative charge at Thr-3 (pThr-3 or T3D) would impede the interaction of Thr-3 with Glu-5, generating a more extended conformation of amino acids 1–17, which might accelerate the seeding process of macroscopic poly(Q) aggregates and thus speed the conversion of oligomers to inert forms. Once seeded, both phosphorylated and non-phosphorylated Htt forms are recruited to the growing aggregates (see Fig. 3, C and D). This is consistent with the association of macroscopic aggregates with reduced neuronal toxicity as previously reported (23).
Our observations indicate that the first 17 amino acids of Htt are subject to post-translational modification and that modification of this region affects the behavior of toxic expanded Htt and could also affect the behavior of unexpanded Htt. In particular, we demonstrate that Thr-3 can be phosphorylated and mutating this phosphorylation site significantly reduces expanded Htt toxicity. Furthermore, expansion of poly(Q) reduces Thr-3 phosphorylation in HD-susceptible cell types (e.g. striatal progenitor ST14A cells), whereas cells less sensitive to degeneration (e.g. non-neuronal HeLa cells) exhibit elevated levels of pThr-3. Therefore, the modification status of Thr-3 can play a central role in influencing HD pathogenesis. Because phosphorylation is considered to be a “druggable” target (60), identifying pathways that enhance this modification may provide a promising avenue for therapeutic development.
Supplementary Material
Acknowledgments
We thank Elizabeth Thomas (The Scripps Research Institute), Chunni Zu (University of California, Los Angeles), and Marie-Francoise Chesselet (University of California, Los Angeles) for help evaluating mouse tissues. We also thank Peter Kaiser (University of California, Irvine) for the gift of the HBH tag and Lee Bardwell (University of California, Irvine) for the use of the LI-COR instrument. We thank Gil Bates (King's College London School of Medicine) and Marcy MacDonald (Massachusetts General Hospital) for the S830 and HF-1 Htt antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grants NS52789 (to L. M. T. and J. L. M.), HD36081 (to J. L. M.), NS045283 (to J. L. M. and L. M. T.), T32GM0731130 (to C. T. A.), and GM-74830 (to L. H.). This work was also supported by Optical Biology Shared Resource of the Cancer Center Support Grant CA-62203 at the University of California, Irvine, and grants from the Hereditary Disease Foundation (to J. S. S., L. M. T., and J. L. M.), the Fox Family Foundation (to J. S. S.), the High Q Foundation (to J. S. S., L. M. T., and J. L. M.), and the Huntington's Disease Society of America Coalition for the Cure (to L. M. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- Htt
- Huntingtin
- HD
- Huntington disease
- Hdh
- Huntington disease homologue (murine Huntingtin)
- Httex1p
- Huntingtin exon 1 protein
- poly(Q)
- polyglutamine
- pThr-3
- phosphorylated threonine 3
- MS
- mass spectrometry
- UAS
- upstream activating sequence
- wt
- wild-type
- HBH
- His-biotin-His.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group (1993) Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2.Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 3.Steffan J. S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B. L., Kazantsev A., Schmidt E., Zhu Y. Z., Greenwald M., Kurokawa R., Housman D. E., Jackson G. R., Marsh J. L., Thompson L. M. (2001) Nature 413, 739–743 [DOI] [PubMed] [Google Scholar]
- 4.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 5.DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 6.Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. (2006) Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 7.Schilling G., Klevytska A., Tebbenkamp A. T., Juenemann K., Cooper J., Gonzales V., Slunt H., Poirer M., Ross C. A., Borchelt D. R. (2007) J. Neuropathol. Exp. Neurol. 66, 313–320 [DOI] [PubMed] [Google Scholar]
- 8.Atwal R. S., Xia J., Pinchev D., Taylor J., Epand R. M., Truant R. (2007) Hum. Mol. Genet. 16, 2600–2615 [DOI] [PubMed] [Google Scholar]
- 9.Rockabrand E., Slepko N., Pantalone A., Nukala V. N., Kazantsev A., Marsh J. L., Sullivan P. G., Steffan J. S., Sensi S. L., Thompson L. M. (2007) Hum. Mol. Genet. 16, 61–77 [DOI] [PubMed] [Google Scholar]
- 10.Gafni J., Ellerby L. M. (2002) J. Neurosci. 22, 4842–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermel E., Gafni J., Propp S. S., Leavitt B. R., Wellington C. L., Young J. E., Hackam A. S., Logvinova A. V., Peel A. L., Chen S. F., Hook V., Singaraja R., Krajewski S., Goldsmith P. C., Ellerby H. M., Hayden M. R., Bredesen D. E., Ellerby L. M. (2004) Cell Death Differ. 11, 424–438 [DOI] [PubMed] [Google Scholar]
- 12.Humbert S., Bryson E. A., Cordelières F. P., Connors N. C., Datta S. R., Finkbeiner S., Greenberg M. E., Saudou F. (2002) Dev. Cell 2, 831–837 [DOI] [PubMed] [Google Scholar]
- 13.Luo S., Vacher C., Davies J. E., Rubinsztein D. C. (2005) J. Cell Biol. 169, 647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pardo R., Colin E., Régulier E., Aebischer P., Déglon N., Humbert S., Saudou F. (2006) J. Neurosci. 26, 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schilling B., Gafni J., Torcassi C., Cong X., Row R. H., LaFevre-Bernt M. A., Cusack M. P., Ratovitski T., Hirschhorn R., Ross C. A., Gibson B. W., Ellerby L. M. (2006) J. Biol. Chem. 281, 23686–23697 [DOI] [PubMed] [Google Scholar]
- 16.Warby S. C., Chan E. Y., Metzler M., Gan L., Singaraja R. R., Crocker S. F., Robertson H. A., Hayden M. R. (2005) Hum. Mol. Genet. 14, 1569–1577 [DOI] [PubMed] [Google Scholar]
- 17.Yanai A., Huang K., Kang R., Singaraja R. R., Arstikaitis P., Gan L., Orban P. C., Mullard A., Cowan C. M., Raymond L. A., Drisdel R. C., Green W. N., Ravikumar B., Rubinsztein D. C., El-Husseini A., Hayden M. R. (2006) Nat. Neurosci. 9, 824–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeong H., Then F., Melia T. J., Jr., Mazzulli J. R., Cui L., Savas J. N, Voisine C., Paganetti P., Tanese N., Hart A. C., Yamamoto A., Krainc D. (2009) Cell 137, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ona V. O., Li M., Vonsattel J. P., Andrews L. J., Khan S. Q., Chung W. M., Frey A. S., Menon A. S., Li X. J., Stieg P. E., Yuan J., Penney J. B., Young A. B., Cha J. H., Friedlander R. M. (1999) Nature 399, 263–267 [DOI] [PubMed] [Google Scholar]
- 20.Kalchman M. A., Graham R. K., Xia G., Koide H. B., Hodgson J. G., Graham K. C., Goldberg Y. P., Gietz R. D., Pickart C. M., Hayden M. R. (1996) J. Biol. Chem. 271, 19385–19394 [DOI] [PubMed] [Google Scholar]
- 21.Steffan J. S., Agrawal N., Pallos J., Rockabrand E., Trotman L. C., Slepko N., Illes K., Lukacsovich T., Zhu Y. Z., Cattaneo E., Pandolfi P. P., Thompson L. M., Marsh J. L. (2004) Science 304, 100–104 [DOI] [PubMed] [Google Scholar]
- 22.Thakur A. K., Jayaraman M., Mishra R., Thakur M., Chellgren V. M., Byeon I. J., Anjum D. H., Kodali R., Creamer T. P., Conway J. F., Gronenborn A. M., Wetzel R. (2009) Nat. Struct. Mol. Biol. 16, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arrasate M., Mitra S., Schweitzer E. S., Segal M. R., Finkbeiner S. (2004) Nature 431, 805–810 [DOI] [PubMed] [Google Scholar]
- 24.Caughey B., Lansbury P. T. (2003) Annu. Rev. Neurosci. 26, 267–298 [DOI] [PubMed] [Google Scholar]
- 25.Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 26.Ehrlich M. E., Conti L., Toselli M., Taglietti L., Fiorillo E., Taglietti V., Ivkovic S., Guinea B., Tranberg A., Sipione S., Rigamonti D., Cattaneo E. (2001) Exp. Neurol. 167, 215–226 [DOI] [PubMed] [Google Scholar]
- 27.Schroeder M. J., Webb D. J., Shabanowitz J., Horwitz A. F., Hunt D. F. (2005) J. Proteome Res. 4, 1832–1841 [DOI] [PubMed] [Google Scholar]
- 28.Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalkley R. J., Baker P. R., Huang L., Hansen K. C., Allen N. P., Rexach M., Burlingame A. L. (2005) Mol. Cell. Proteomics 4, 1194–1204 [DOI] [PubMed] [Google Scholar]
- 30.Wolfgang W. J., Miller T. W., Webster J. M., Huston J. S., Thompson L. M., Marsh J. L., Messer A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 11563–11568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simmons D. A., Rex C. S., Palmer L., Pandyarajan V., Fedulov V., Gall C. M., Lynch G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4906–4911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green K. N., Steffan J. S., Martinez-Coria H., Sun X., Schreiber S. S., Thompson L. M., LaFerla F. M. (2008) J. Neurosci. 28, 11500–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hazeki N., Tukamoto T., Goto J., Kanazawa I. (2000) Biochem. Biophys. Res. Commun. 277, 386–393 [DOI] [PubMed] [Google Scholar]
- 34.Bischof J., Maeda R. K., Hediger M., Karch F., Basler K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agrawal N., Pallos J., Slepko N., Apostol B. L., Bodai L., Chang L. W., Chiang A. S., Thompson L. M., Marsh J. L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 3777–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sipione S., Rigamonti D., Valenza M., Zuccato C., Conti L., Pritchard J., Kooperberg C., Olson J. M., Cattaneo E. (2002) Hum. Mol. Genet. 11, 1953–1965 [DOI] [PubMed] [Google Scholar]
- 37.Menalled L. B., Sison J. D., Dragatsis I., Zeitlin S., Chesselet M. F. (2003) J. Comp. Neurol. 465, 11–26 [DOI] [PubMed] [Google Scholar]
- 38.Barnes G., Duyao M., Ambrose C., McNeil S., Persichetti F., Srinidhi J., Gusella J., MacDonald M. (1994) Somatic Cell Mol. Genet. 20, 87–97 [DOI] [PubMed] [Google Scholar]
- 39.Persichetti F., Ambrose C. M., Ge P., McNeil S. M., Srinidhi J., Anderson M. A., Jenkins B., Barnes G. T., Duyao M. P., Kanaley L. (1995) Mol. Med. 1, 374–383 [PMC free article] [PubMed] [Google Scholar]
- 40.Schutz-Geschwender A., Zhang Y., Holt T., McDermitt D., Olive D. M. (2004) www.licor.com/bio/PDF/IRquant.pdf, LI-COR Biosciences, Lincoln, NE [Google Scholar]
- 41.Marsh J. L., Pallos J., Thompson L. M. (2003) Hum. Mol. Genet. 12, R187–R193 [DOI] [PubMed] [Google Scholar]
- 42.Bayer T. A., Campos-Ortega J. A. (1992) Development 115, 421–426 [DOI] [PubMed] [Google Scholar]
- 43.Jaenisch R., Jähner D., Nobis P., Simon I., Löhler J., Harbers K., Grotkopp D. (1981) Cell 24, 519–529 [DOI] [PubMed] [Google Scholar]
- 44.Lacy E., Roberts S., Evans E. P., Burtenshaw M. D., Costantini F. D. (1983) Cell 34, 343–358 [DOI] [PubMed] [Google Scholar]
- 45.Franceschini N. (1972) Pupil and Pseudopupil in the Compound Eye of Drosophila, Springer-Verlag, Berlin [Google Scholar]
- 46.Zoghbi H. Y., Orr H. T. (2000) Annu. Rev. Neurosci. 23, 217–247 [DOI] [PubMed] [Google Scholar]
- 47.Marsh J. L., Walker H., Theisen H., Zhu Y. Z., Fielder T., Purcell J., Thompson L. M. (2000) Hum. Mol. Genet. 9, 13–25 [DOI] [PubMed] [Google Scholar]
- 48.Emamian E. S., Kaytor M. D., Duvick L. A., Zu T., Tousey S. K., Zoghbi H. Y., Clark H. B., Orr H. T. (2003) Neuron 38, 375–387 [DOI] [PubMed] [Google Scholar]
- 49.Sherman F., Stewart J. W., Tsunasawa S. (1985) Bioessays 3, 27–31 [DOI] [PubMed] [Google Scholar]
- 50.Flinta C., Persson B., Jörnvall H., von Heijne G. (1986) Eur. J. Biochem. 154, 193–196 [DOI] [PubMed] [Google Scholar]
- 51.Petersen B. O., Lukas J., Sørensen C. S., Bartek J., Helin K. (1999) EMBO J. 18, 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quélo I., Gauthier C., St-Arnaud R. (2005) Gene Expr. 12, 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reimer T., Weiwad M., Schierhorn A., Ruecknagel P. K., Rahfeld J. U., Bayer P., Fischer G. (2003) J. Mol. Biol. 330, 955–966 [DOI] [PubMed] [Google Scholar]
- 54.Rothbächer U., Laurent M. N., Deardorff M. A., Klein P. S., Cho K. W., Fraser S. E. (2000) EMBO J. 19, 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hickey M. A., Kosmalska A., Enayati J., Cohen R., Zeitlin S., Levine M. S., Chesselet M. F. (2008) Neuroscience 157, 280–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G. P., Lehrach H., Wanker E. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4604–4609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poirier M. A., Li H., Macosko J., Cai S., Amzel M., Ross C. A. (2002) J. Biol. Chem. 277, 41032–41037 [DOI] [PubMed] [Google Scholar]
- 58.Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 59.Duennwald M. L., Jagadish S., Muchowski P. J., Lindquist S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 11045–11050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hopkins A. L., Groom C. R. (2002) Nat. Rev. Drug Discov. 1, 727–730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.