Abstract
Initially identified in Chlamydomonas, RSP3 (radial spoke protein 3) is 1 of more than 20 identified radial spoke structural components of motile cilia and is required for axonemal sliding and flagellar motility. The mammalian orthologs for this and other radial spoke proteins, however, remain to be characterized. We found mammalian RSP3 to bind to the MAPK ERK2 through a yeast two-hybrid screen designed to identify interacting proteins that have a higher affinity for the phosphorylated, active form of the protein kinase. Consistent with the screening result, the human homolog, RSPH3, interacts with and is a substrate for ERK1/2. Moreover, RSPH3 is a protein kinase A-anchoring protein (AKAP) that scaffolds the cAMP-dependent protein kinase holoenzyme. The binding of RSPH3 to the regulatory subunits of cAMP-dependent protein kinase, RIIα and RIIβ, is regulated by ERK1/2 activity and phosphorylation. Here we describe an ERK1/2-interacting AKAP and suggest a mechanism by which cAMP-dependent protein kinase-AKAP binding can be modulated by the activity of other enzymes.
A persistent question in understanding cellular networks is how different input signals converge on MAPK3 signaling cascades to elicit multiple and often divergent responses. More specifically, how do different MAPK modules integrate various input stimuli and ultimately cause selective activation and actions of these cascades at distinct sites within the cell? Within this context, scaffolding and anchoring proteins are important regulators in coordinating ERK1/2 activity through a variety of mechanisms, including confining and/or dynamically distributing these kinases to distinct sites of action in the cell and integrating multiple inputs into the ERK1/2 MAPK cascade from other signaling modalities to allow for the divergent responses of the ERK1/2 pathway (1). Scaffolds and anchoring proteins may also affect the magnitude and duration of signals through the pathway, mediate cross-talk with other pathways, and serve in inhibitory roles, preventing inappropriate action of the kinases in certain cellular compartments (2). Scaffolds themselves are often regulated and may be subject to multiple post-translational modifications, e.g. phosphorylation, sumoylation, and ubiquitylation. These modifications may regulate the stability of the scaffolds as well as their roles in interconnecting pathways in a manner dependent on the particular stimulus or change in cell context (3, 4).
One particularly well characterized class of scaffolding proteins are the protein kinase A-anchoring proteins (AKAPs) that bind to the cAMP-dependent protein kinase (PKA) holoenzyme (5–8). PKA is a ubiquitous protein kinase involved in numerous regulatory events in the cell and is composed of two catalytic subunits and two regulatory subunits. The PKA holoenzyme is classified as type I or type II, based on the regulatory subunits, RIα/β or RIIα/β, present in the heterotetrameric complex (7, 9, 10). AKAPs concentrate PKA holoenzymes to defined locations within the cell, where the catalytic subunits can phosphorylate surrounding substrates in response to increased concentrations of cAMP, thus providing spatiotemporal specificity to PKA activity and cAMP-dependent signaling. AKAPs bind to hydrophobic residues of the N-terminal dimerization interface of the regulatory subunits via a conserved 14–18-residue amphipathic helical region (8). More than 50 AKAPs have been identified, many with multivalent binding capacity, interacting not only with PKA but also with components of often disparate signaling pathways. AKAPs have the ability to integrate multiple inputs and facilitate cross-talk of pathways as a function of these specific interactions, resulting in unique, localized outcomes within the cell.
RSP3 (radial spoke protein 3) is one of at least 23 known radial spoke proteins identified in Chlamydomonas reinhardtii and is assembled in a large multisubunit complex required for flagellar motility (11). Radial spoke proteins are thought to be important in transducing signals from the inner pair of microtubules to the outer doublets in the flagellar axoneme, regulating dynein-mediated axonemal sliding and subsequent flagellar motility. Genetic analysis of RSP3 function in Chlamydomonas indicates that flagella are paralyzed, and radial spokes are not assembled in the absence of RSP3 (12, 13). Additionally, biochemical studies of Chlamydomonas RSP3 show that it is an AKAP, and loss of its ability to bind to PKA also results in abnormal flagellar motility and paralyzed flagella (14, 15). More recently, RSP3 has been shown to form a homodimer within the radial spoke structure. This dimer is proposed to provide the base for radial spoke assembly (16).
Through proteomic analysis of human bronchial epithelial cells and immunofluorescence staining of mouse tracheal epithelial cells, RSP3 has been found in motile cilia in mammals (16, 17). Mammals contain one RSP3 gene (mapped to chromosomal locus 6q25.3), composed of eight exons and seven introns. The gene is believed to contain alternative start sites that generate two transcripts to produce a long and a short form. The short form, annotated as RSP3, is made up of 418 amino acids, whereas the 560-amino acid-long form, extended by 142 amino acids at the N terminus, is referred to as RSPH3 (radial spoke protein homolog 3). Human and mouse RSP3 are ∼84% similar at the amino acid level and share 67% similarity within the radial spoke domain to Chlamydomonas RSP3. The radial spoke domain and the AKAP domain of RSP3 are conserved among a variety of species. The mammalian orthologs for this and other radial spoke proteins, however, have not been characterized. Here we describe the interaction of mammalian RSP3/RSPH3 with ERK1/2 and PKA and describe some features of its regulation. This work identifies the only AKAP thus far known to interact with components of the ERK1/2 kinase cascade.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Harvest
HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 1% l-glutamine at 5% CO2. Generally, cells were reverse-transfected using FuGENE 6 according to the manufacturer's protocol. 1.5 μg of plasmid(s) was used in transfections, and cells were harvested 36–40 h post-transfection. After indicated treatments as described under “Results” and figure legends, cells were washed twice with iced phosphate-buffered saline and lysed on ice in 50 mm HEPES, pH 7.5, 150 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 0.2 mm Na3VO4, 100 mm NaF, 50 mm β-glycerophosphate, 10% glycerol, 0.1% Triton X-100, 1.6 μg/ml aprotinin, 0.1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each of Nα-p-tosyl-l-lysine chloromethyl ketone, Nα-p-tosyl-l-arginine methyl ester, pepstatin A, and leupeptin. After 15 min on ice, lysates were frozen in N2 (liquid) and thawed on ice, followed by centrifugation for 15 min at 16,000 × g in a microcentrifuge at 4 °C. Supernatants were stored at −80 °C until further analysis.
Plasmids and Antibodies
Human RSPH3 in a pSPORT6 vector was obtained from ATCC and cloned into pCMV7.1 N-terminal 3xFLAG vector for mammalian expression. Site-directed mutagenesis was performed to generate RSPH3 mutants in ERK1/2 phosphorylation sites and the AKAP domain. 3xFLAG-RSPH3 truncation mutants were also generated. Normal mouse or rabbit control IgG antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Plasmids encoding RIIα, RIIβ, and RIα were gifts from G. S. McKnight (University of Washington). Mouse monoclonal antibodies against PKA RIIα or RIIβ were obtained from BD Transduction Laboratories. Rabbit anti-PRKAR2A/PKA-RIIα antibody was obtained from Bethyl Laboratories, Inc. (Montgomery, TX). Anti-Myc antibody was obtained from the National Cell Culture Center. Mouse monoclonal anti-activated MAPK (diphosphorylated ERK1/2) and mouse monoclonal anti-FLAG M2 were obtained from Sigma. Antibodies to ERK1/2 (Y691, X837), MEK1 (A2227), and Rsk (S732) were as described (18–20). The MEK inhibitor, U0126, was obtained from Promega (Madison, WI).
Immunoprecipitation
For immunoprecipitations, cell extracts containing 1 mg of total protein were pre-cleared with 30 μl of protein A-Sepharose beads for 1 h at 4 °C prior to incubation with the indicated primary antibodies (1:250 dilution) at 4 °C overnight. Then 30 μl of protein A-Sepharose beads was incubated with the lysate for 2 h at 4 °C. The beads were centrifuged at 5000 × g for 3 min prior to three 10-min washes with 10 mm Tris, pH 7.4, 5 mm MgCl2, 500 mm NaCl. The beads were subsequently boiled in Laemmli sample buffer (2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromphenol blue, 50 mm Tris-HCl) and subjected to SDS-PAGE. Proteins were electrotransferred to nitrocellulose membranes (Millipore, Billerica, MA). Membranes were blocked with 5% nonfat powdered milk in 20 mm Tris, pH 7.5, 0.15 m NaCl (TBS) for 2 h at 4 °C, incubated with the primary antibody overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibody diluted 1:2000 in 5% milk in TBS for 1 h at room temperature. Proteins were detected by enhanced chemiluminescence. In some cases, immunoblots were analyzed by densitometry using Image J.
Immunoprecipitation Kinase Assays
After washing the immunoprecipitates as above, protein A-Sepharose beads were washed with 10 mm HEPES, pH 8.0, 10 mm MgCl2. Beads were subsequently resuspended in 10 mm HEPES, pH 8.0, 10 mm MgCl2, 1 mm 1,4-dithiothreitol, and 1 mm benzamidine. Recombinant active phosphorylated ERK2 (pERK2) and the purified catalytic subunit of PKA, each prepared as described (21, 22), were used as enzymes. Immunoprecipitation kinase assays were performed at 30 °C for 10 min. Reactions contained 10 μm ATP in a 30-μl total reaction volume (60–70 cpm/fmol [γ-32P]ATP). Reactions were quenched with Laemmli sample buffer and heated at 100 °C for 5 min. Samples were resolved on 12% polyacrylamide gels in SDS, stained with Coomassie Blue, dried, and analyzed using autoradiography.
Phosphoamino Acid Analysis
One μg of His6-RSP3 phosphorylated in vitro with pERK2 was hydrolyzed with 6 n HCl, and the products were resolved by electrophoresis on a thin layer plate using a Hunter apparatus as described previously (23, 24). The migration of phosphorylated residues from the samples was compared with migration of phosphoamino acid standards visualized with ninhydrin.
Quantification and Statistical Analyses
Results are expressed as means determined from at least three independent experiments. Bars represent S.E. Statistical significance was calculated using paired or unpaired one- or two-tail t tests or nonparametric Kruskal-Wallis analysis as indicated in the figure legends. Densitometric quantification of immunoblots was performed using ImageJ. Scanned images of film exposed with intensities of bands in the linear range as determined by exposure of a sensitometer imprinting an intensity gradient on the film were used for all quantification analyses.
RESULTS
RSPH3 Binds to ERK1/2
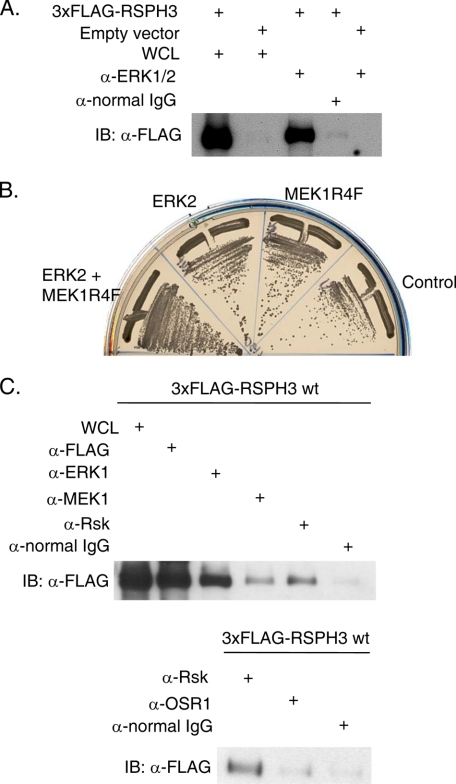
The interaction between ERK2 and RSP3 was found through a yeast two-hybrid screen designed to identify interacting proteins that have a higher affinity for the phosphorylated active form of ERK2. Yeast were transformed with bait vectors containing an ERK2 and active MEK1 (MEK1R4F) co-expression plasmid (25), ERK2 alone, or MEK1R4F alone, along with a fetal mouse brain cDNA library as prey. Growth on restrictive medium and β-galactosidase activity were used as readouts for an interaction. RSP3 was identified and bound 2.5-fold more strongly to ERK2 co-expressed with active MEK1 than to ERK2 alone, based on β-galactosidase activity. To test the yeast two-hybrid finding, 3xFLAG-tagged RSPH3 or empty vector was expressed in HEK293 cells, and endogenous ERK1/2 was immunoprecipitated. 3xFLAG-RSPH3 was readily detected in the immunoprecipitate (Fig. 1A). 3xFLAG-RSPH3 also co-immunoprecipitated with Myc-tagged ERK2 (data not shown), consistent with the two-hybrid result suggesting that phosphorylation and activation of the kinase are not required. Despite a theoretical molecular mass of 67 kDa with the 3xFLAG epitope, 3xFLAG-RSPH3 migrates between 75 and 90 kDa on a 12% polyacrylamide gel. This slower migration is characteristic of several AKAPs and has been attributed to the exposed charge of the amphipathic helical domain.
FIGURE 1.
RSPH3 binds to ERK1/2 and other cascade components. A, endogenous ERK1/2 were immunoprecipitated from HEK293 cell lysates expressing 3xFLAG-RSPH3 or empty vector (negative control) and immunoblotted (IB) with the anti-FLAG antibody. Lanes labeled whole cell lysate (WCL) contain 10% of the input for immunoprecipitation. Immunoprecipitation with normal rabbit IgG was used as a negative control. B, yeast were co-transformed with plasmids containing mouse RSP3 and ERK2, MEK1R4F, a co-expression plasmid containing ERK2 plus MEK1R4F, or empty vector as a control. C, endogenous ERK1/2, MEK1, and Rsk (top) or Rsk and OSR1 (bottom) were immunoprecipitated from cells transfected with 3xFLAG-RSPH3. Precipitates were immunoblotted with anti-FLAG antibodies. Normal IgG was immunoprecipitated as control. The whole cell lysate lane contains 5% of the input for immunoprecipitation. Experiments in A–C were repeated 10, 2, and 3 times.
RSPH3 Coordinates a MAPK Signaling Module
The two-hybrid screen that identified RSP3 used a bait that also contained MEK1. Thus, we determined whether RSP3 also had the ability to bind to the upstream protein kinase MEK1. In a two-hybrid test, RSP3 interacted most strongly with ERK2 co-expressed with MEK1R4F; weaker binding was also detected with ERK2 alone, as noted earlier (Fig. 1B). RSP3 also bound to MEK1R4F alone but less so to wild-type MEK1 (see Fig. 2E), suggesting that RSP3 binds directly not only to ERK1/2 but also to active MEK1. Subsequently, endogenous ERK1/2, MEK1, and the ERK1/2 substrate protein kinase Rsk were immunoprecipitated from HEK293 cells expressing 3xFLAG-RSPH3. Tagged RSPH3 co-precipitated with ERK1/2, MEK1, and Rsk but not with an unrelated protein kinase OSR1 (Fig. 1C, upper and lower panels). Co-precipitation with MEK1 was weaker than with Rsk or ERK1/2.
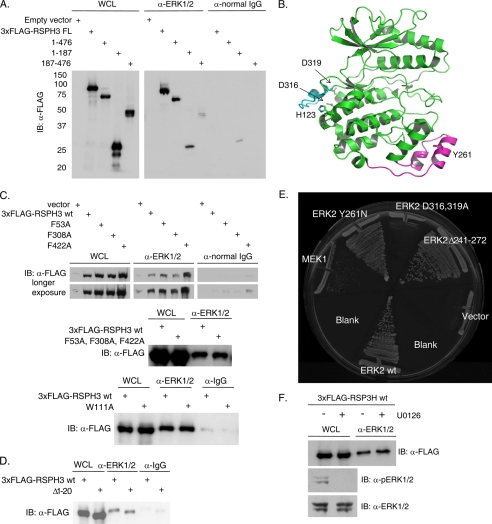
FIGURE 2.
ERK CD region is involved in the RSPH3 interaction. A, endogenous ERK1/2 was immunoprecipitated from HEK293 cells expressing 3xFLAG-tagged RSPH3 truncation mutants, residues 1–476, 1–187, and 187–476. Immunoprecipitation with normal rabbit IgG was used as a negative control. Precipitates were immunoblotted (IB) with anti-FLAG antibody. The whole cell lysate (WCL) lane contains 5% of the input for the immunoprecipitations. B, ribbon representation of ERK2 (green) bound via its CD domain to a peptide derived from the hematopoietic tyrosine phosphatase HePTP (cyan) (Protein Data Bank code 2gph). The MAPK insert is highlighted in magenta, with Tyr-261 indicated. C, three putative FXF motifs within 3xFLAG-RSPH3 were mutated singly (top panel) or in combination (middle panel), and a WXY motif within 3xFLAG-RSPH3 was mutated (bottom), and all were tested for ERK1/2 binding. Endogenous ERK1/2 immunoprecipitates were immunoblotted with anti-FLAG. Immunoprecipitation from cells transfected with empty vector or with normal rabbit IgG were used as negative controls. The whole cell lysate lane contains 5% of the input for immunoprecipitation. D, experiment as in C with a truncation of the first 20 residues of 3xFLAG-RSPH3. E, pairwise yeast two-hybrid analysis of mouse RSP3 interaction with wild-type (wt) ERK2 or ERK2 mutants. Yeast containing RSP3 were co-transformed with wild-type MEK1 or empty vector as controls. F, HEK293 cells expressing 3xFLAG-RSPH3 were treated with 10 μm U0126 or diluent for 10 min prior to lysis. Endogenous ERK1/2 were immunoprecipitated, and the precipitates were immunoblotted (IB) with anti-FLAG. The whole cell lysate lane contains 5% of the input. Phosphorylated and total ERK1/2 were immunoblotted in the lysates. n = 3. Experiments in each panel were performed three times, except the two-hybrid test which was performed twice.
Basis for RSP3-ERK1/2 Interaction
Truncation mutants of RSPH3 were generated to identify the minimal ERK1/2-interacting region within RSPH3. Residues 187–476 represent the conserved radial spoke 3 domain as defined for the Chlamydomonas protein. FLAG-tagged truncation mutants of RSPH3, residues 1–476 and 1–187, readily co-immunoprecipitated with endogenous ERK1/2. Binding of residues 187–476 to ERK1/2, although detectable, was diminished (Fig. 2A). These results are consistent with binding of ERK1/2 to the first 187 amino acids of RSPH3 with a possible contribution from more C-terminal residues. The two best described motifs that bind ERK1/2 are the FXF motif and the docking or D motif. Hydrophobic FXF motifs bind to ERK2 near the MAPK insert and underlying structures (26, 27) (Fig. 2B) and are thought to interact preferentially with pERK1/2, although interactions with the unphosphorylated proteins have been found (28, 29). D motifs, characterized by a basic/hydrophobic consensus sequence, interact in the common docking (CD) region of ERK1/2 and are thought to bind preferentially to unphosphorylated ERK1/2 (30–33) (Fig. 2B). Because RSPH3 binds better to pERK1/2, we first mutated the three FXF motifs in RSPH3, singly or together to test for loss of RSPH3-ERK1/2 binding. The four resulting mutants were able to co-immunoprecipitate with ERK1/2 in a manner indistinguishable from wild-type RSPH3 (Fig. 2C). A recent analysis using peptides suggested that tryptophan and tyrosine could replace phenylalanine in this type of motif (34). Thus, a fourth motif, including Trp-111, was mutated that also did not appear to disrupt the interaction with ERK1/2 (Fig. 2C). RSPH3 contains no strongly predicted D motifs (35), but near its N terminus is a sequence (LAKRR) identical to the ERK-binding sequence from the protein kinases Msk and Mnk that are ERK1/2 substrates (36). To test its potential role, the first 20 amino acids of RSPH3 were deleted, but the interaction was retained (Fig. 2D).
Because these motifs were apparently not important for ERK1/2 binding, we used an unbiased approach to elucidate the nature of the interaction. A two-hybrid library of ERK2 mutants (37) was screened with RSPH3 to find ERK2 binding-deficient mutants. Through this screen ERK2 H123Y was found to have impaired binding to RSPH3 (data not shown). This residue lies on the side opposite of the ERK2 active site near the CD region that binds D motifs (Fig. 2B). Thus, we compared RSP3 binding using ERK2 mutants defective in D motif and FXF interactions by pairwise yeast two-hybrid tests (Fig. 2E). Mutations within the MAPK insert (Y261N) and the MAPK insert deletion (Δ241–272), which disrupt FXF-mediated interactions, bound to RSP3 as well as did wild-type ERK2, consistent with the co-immunoprecipitation studies with RSPH3 FXF mutants. On the other hand, mutations in the CD region (D316A and D319A) showed reduced binding to RSP3. This suggests that as yet unidentified basic/hydrophobic D-like motifs within RSPH3 mediate its interaction with ERK1/2.
To test whether ERK1/2 activity affected the interaction with RSPH3 in a mammalian cell system, HEK293 cells expressing 3xFLAG-RSPH3 were treated with the MEK1/2 inhibitor, U0126, which inhibits the subsequent activation of ERK1/2, or DMSO as a control, for 10 min prior to immunoprecipitation of endogenous ERK1/2. Under these conditions, no apparent difference in co-immunoprecipitation of overexpressed RSPH3 with ERK1/2 was observed between the control- and U0126-treated samples (Fig. 2F).
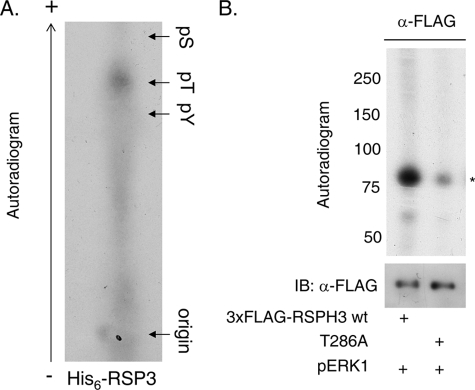
RSP3 Is an ERK1/2 Substrate
In vitro kinase assays performed with recombinant pERK1 and immunoprecipitated RSPH3 from HEK293 lysates or recombinant His6-RSP3 showed that RSP3 and RSPH3 are indeed substrates (Fig. 3, A and B). Recombinant His6-RSP3 was also used in in vitro kinase assays for phospho-residue identification (Fig. 3A). Phosphoamino acid analysis indicates that pERK1 predominantly phosphorylates threonine in RSP3/RSPH3. In silico prediction of consensus ERK1/2 phosphorylation motifs suggested the major site to be threonine 286 (35). Mutation of this residue to alanine in RSPH3 (T286A) attenuated phosphorylation by ERK1 by ∼70% in immunoprecipitation kinase assays (Fig. 3B).
FIGURE 3.
ERK1/2 phosphorylate RSPH3. A, phosphoamino acid analysis of His6-RSP3 phosphorylated in vitro with pERK1. An autoradiogram is shown. B, 3xFLAG-RSP3H wild-type or T286A was immunoprecipitated from HEK293 cells and used as substrate for an in vitro kinase reaction with purified, recombinant pERK1. Mutating T286A decreased 32P incorporation by ∼70% (n = 6). pS, phosphoserine; pT, phosphothreonine; pY, phosphotyrosine.
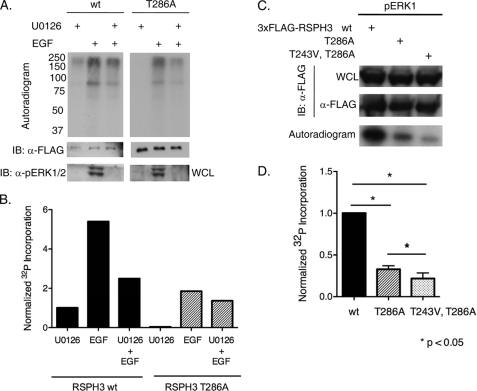
Labeling of cells with 32P indicated that RSPH3 phosphorylation is increased upon stimulation of cells with epidermal growth factor (EGF) (Fig. 4, A and B). Radiolabeled phosphate incorporation into RSPH3 was diminished by pretreatment with the MEK1/2 inhibitor U0126, suggesting that at least a portion of this phosphorylation is ERK1/-dependent. Furthermore, the extent of radiolabeled phosphate incorporation into RSPH3 T286A was much less than that into wild-type RSPH3, suggesting that threonine 286 is the major ERK1/2 phosphorylation site in cells. ERK2 also phosphorylates RSPH3 on threonine 243 to a lesser extent. Phosphorylation of the double mutant T243V/T286A RSPH3 was no more than 20% that of wild-type RSPH3 (Fig. 4, C and D).
FIGURE 4.
RSPH3 phosphorylation dependent on the ERK1/2 pathway. A, HEK293 expressing 3xFLAG-RSP3H wild-type (wt) or T286A were placed in serum-free medium for 20 h and then labeled with 1 mCi/ml [32P]orthophosphate with 10 μm U0126 or diluent for 60 min, and then the indicated groups were treated with 10 ng/ml EGF for 10 min. Autoradiograms of the 3xFLAG-RSP3H immunoprecipitates are shown. n = 3. IB, immunoblot; WCL, whole cell lysate. B, quantification of 32P incorporation into the RSP3H bands from one experiment. Incorporation was normalized to the U0126-treated RSPH3 wild-type sample and adjusted for differences in protein expression as assessed by immunoblotting and densitometry. C and D, 3xFLAG-RSP3H wild-type, T286A, or T243V/T286A was immunoprecipitated from HEK293 cells and used as substrate for in vitro kinase assays with pERK1. Graph represents fold 32P incorporation into RSP3H wild-type compared with the mutants (mean + S.E.). Protein amount in the immunoprecipitates was measured via densitometric analysis. Incorporation was normalized to that into the wild-type protein and corrected for differences in protein amount. n = 3. Mean values for T286A and T243V/T286A RSPH3 were 0.32 and 0.21, respectively. Statistical significance was evaluated with a nonparametric Kruskal-Wallis analysis.
Mammalian RSPH3 Is an AKAP and a Substrate for PKA
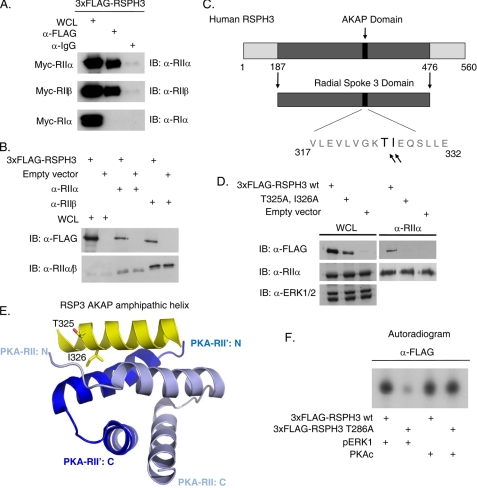
Chlamydomonas RSP3 has been shown to bind to the regulatory subunits of PKA in vitro in a similar manner to other known AKAPs. Orthologs of RSP3 exist in a variety of species, all of which contain a highly conserved similar sequence in the AKAP R-subunit binding domain (14). Thus, we tested whether the human isoform of RSPH3 has a similar AKAP function. 3xFLAG-RSPH3 and Myc-tagged RIIα, RIIβ, and RIα were co-expressed in HEK293 cells, and 3xFLAG-RSPH3 was immunoprecipitated. RIIα and RIIβ co-immunoprecipitated with RSPH3, whereas RIα did not, indicating that RSPH3 preferentially interacts with the type II PKA holoenzyme (Fig. 5A). Overexpressed RSPH3 also co-immunoprecipitated with endogenous RIIα and RIIβ, further confirming an interaction with the regulatory subunits of PKA (Fig. 5B). Mutation of residues threonine 325 and isoleucine 326 to alanines (T325A/I326A) within the amphipathic helix of RSPH3, which are critical for PKA regulatory subunit binding and helix folding, disrupted the co-immunoprecipitation interaction observed with overexpressed wild-type RSPH3 (Fig. 5, C–E). Ile-326 in RSPH3 is thought to bind within a hydrophobic groove created by R-subunit dimerization. In vitro kinase assays indicated that the purified catalytic subunit of PKA phosphorylates both wild-type and T286A mutant 3xFLAG-RSPH3 similarly (Fig. 5F).
FIGURE 5.
RSPH3 is AKAP. A, 3xFLAG-RSPH3 was co-expressed with Myc-tagged RIIα, RIIβ, or RIα PKA regulatory subunits in HEK293 cells. RSPH3 was immunoprecipitated with the anti-FLAG antibody and immunoblotted (IB) with antibodies against the regulatory subunits. n = 3. WCL, whole cell lysate. B, endogenous RIIα and RIIβ were immunoprecipitated from HEK293 cells expressing 3xFLAG-RSPH3. Co-immunoprecipitation was detected by immunoblotting with anti-FLAG antibodies. Empty vector transfected lysates were used as a negative control. The whole cell lysate lane contains 10% of the input. n = 4. C, schematic of human RSPH3 showing the RII-binding sequence of the AKAP domain, based on sequence similarity to the Chlamydomonas ortholog. D, endogenous RIIα was immunoprecipitated from the HEK293 cells expressing 3xFLAG-RSPH3 wild type (wt) or the AKAP-defective mutant, T325A/I326A. Samples were immunoblotted with the indicated antibodies. n = 4. E, ribbon diagram depicting the amphipathic AKAP helix in RSPH3 that mediates the interaction with the PKA regulatory subunit dimerization interface. F, overexpressed 3xFLAG-RSP3H wild type or T286A was immunoprecipitated from HEK293 cells and used as substrate for an in vitro kinase reaction with purified recombinant pERK1 or purified catalytic subunit of PKA (PKAc). n = 3.
ERK1/2 Phosphorylation Regulates RSPH3 AKAP Activity
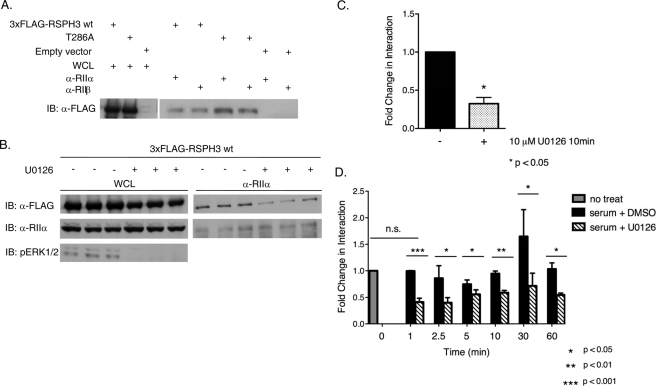
We next asked whether phosphorylation of RSPH3 by ERK1/2 has any consequence on RSPH3 AKAP function. We found that mutating the ERK1/2 phosphorylation site in RSPH3 affected its ability to co-immunoprecipitate with the regulatory subunits of PKA. Co-immunoprecipitation of the T286A mutant with endogenous RIIα or RIIβ was enhanced by ∼1.8-fold compared with wild-type overexpressed 3xFLAG-RSPH3 (Fig. 6A). Treating cells with the MEK1/2 inhibitor, U0126, which inhibits the subsequent activation of ERK1/2, diminished the interaction of RSPH3 with endogenous RIIα, however. HEK293 cells expressing 3xFLAG-RSPH3 were treated with fresh medium containing 10% fetal bovine serum and 10 μm U0126 or DMSO as a control for 10 min prior to immunoprecipitation of RIIα (Fig. 6, B and C). Stimulation with fresh serum-containing medium caused an expected increase in pERK1/2, and this activity was decreased in U0126-treated samples. U0126 also decreased RSPH3 in the RIIα immunoprecipitate by ∼70%, suggesting that inhibiting ERK1/2 activity decreases AKAP function of RSPH3. This was unexpected because of the enhanced interaction of the T286A mutant with RIIα. To examine the interaction between RSPH3 and RIIα further, a kinetic analysis of the effects of serum and U0126 upon the interaction was performed. The culture medium of RSPH3-transfected HEK293 cells was changed to fresh serum-containing medium supplemented with 10 μm U0126 or DMSO as a control for 1, 2.5, 5, 10, 30, and 60 min prior to lysis (Fig. 6D). As expected ERK1/2 activity was decreased by U0126. The difference in the amount of RSPH3 that co-immunoprecipitated with endogenous RIIα was negligible between untreated and samples treated with serum plus DMSO (control) for 1 min. Thereafter, the interaction between RIIα and RSPH3 was consistently diminished in the presence of U0126 over the time course of the experiment; the average reduction in RSPH3 pulled down with RIIα was 50% at all time points measured (Fig. 6D). Thus, inhibiting ERK1/2 activity appears to negatively regulate the AKAP function of RSPH3.
FIGURE 6.
ERK1/2 activity modulates the AKAP function of RSPH3. A, endogenous RIIα or RIIβ was immunoprecipitated from HEK293 cells expressing 3xFLAG-RSPH3 wild type (wt), T286A, or empty vector. Samples were immunoblotted (IB) with anti-FLAG antibody. Fold change in interaction (as described under “Results”) was determined using a ratiometric analysis of the intensity of the RSPH3 band relative to the whole cell lysate (WCL) for each protein. The experiment was performed three times each for RIIa and for RIIb. Statistical significance of the fold change in interaction was determined using a two-tailed t test, p value = 0.011. B, HEK293 cells expressing wild-type 3xFLAG-RSP3H were placed in serum-free medium for 12 h prior to stimulating with serum in the presence of 10 μm U0126 or diluent for 10 min. Endogenous RIIα was immunoprecipitated from the lysates, and the material was immunoblotted with the indicated antibodies. Data of B are shown in C as mean + S.E., n = 3. The fold change in interaction was based on the ratiometric analysis, and values were normalized to the DMSO-treated control. Statistical significance was analyzed using a two-tailed t test, p value = 0.0139. D, HEK293 cells expressing 3xFLAG-RSP3H were stimulated with serum with 10 μm U0126 or DMSO for the indicated times. RIIα was immunoprecipitated, and the precipitates were blotted with anti-FLAG. Fold change was determined as above. Samples were normalized to the untreated control. Data represent mean + S.E., n = 3. Statistical significance was analyzed using a two-tailed t test with p values as indicated.
DISCUSSION
Our findings reveal that mammalian RSPH3 is an ERK1/2-interacting AKAP; mutating residues within the amphipathic helix of RSPH3 (T325A/I326A) disrupted its interaction with RIIα, behavior typical of a bona fide AKAP. These findings on the human ortholog of RSP3 support previous work showing that the blue-green algae protein is an AKAP. Importantly, we show that RSPH3 binds multiple components of the ERK1/2 module, and at the interactions with ERK1/2 and MEK1 are most likely direct.
The yeast two-hybrid screen identified RSP3 as an interacting protein that has a higher affinity for the phosphorylated and active form of ERK2. Both two-hybrid and co-immunoprecipitation show that RSPH3 binds both pERK2 and ERK1/2. Consistent with weaker but significant binding to the unphosphorylated kinase, inhibiting ERK1/2 activation with U0126 did not disrupt the ability of RSPH3 to co-immunoprecipitate with endogenous ERK1/2. Corollary experiments using EGF and forskolin to activate the ERK1/2 and PKA signaling pathways, respectively, did not cause an apparent change in co-immunoprecipitation (data not shown).
Based on interactions with ERK2 mutants defective in binding to D or FXF motifs, RSPH3 most likely binds to ERK1/2, phosphorylated or not, through a D motif-CD region interaction. D motifs are not highly conserved sequences; consensus D motifs were not readily identified in RSPH3; as a result, we have not yet determined the relevant region of RSPH3 that binds to ERK1/2. Although the several FXF motifs on RSPH3 do not appear to contribute substantially to ERK1/2 binding, RSPH3, like other ERK1/2-binding proteins, may contain combinations of D and FXF that may function synergistically or contribute to differential interactions with the MAPKs depending on spatiotemporal or cellular context.
The major site of ERK1/2 phosphorylation in RSPH3 is Thr-286 within a MAPK consensus motif. Radiolabeled phosphate incorporation into RSPH3 in response to EGF was not completely abolished by mutating Thr-286, or a second minor site Thr-243, or by blocking ERK1/2 activation. In addition to PKA and ERK1/2, our experiments suggest that there are additional EGF-sensitive, ERK1/2-independent protein kinases that also phosphorylate RSPH3. These phosphorylations may also influence interactions of RSPH3 with these signaling proteins as well as the resulting output.
Inhibiting ERK1/2 activation and subsequent activity decreased the amount of RSPH3 that co-immunoprecipitated with endogenous RIIα. Mutating the major ERK1/2 phosphorylation site in RSPH3 to alanine, however, increased the capacity of RSPH3 to co-immunoprecipitate with RIIα. Although mutation of a phosphorylation site to a small hydrophobic residue is generally expected to have an effect opposite to phosphorylation of that residue, several studies have found that removing the hydroxyl group alone, as in a Thr to Ala or Val mutation, is sufficient to mimic phosphorylation (38). Another possibility is that the mutation may cause a conformational change in RSPH3 that indirectly affects interactions; this has been noted in several transcription factors (39). Such a change might alter RIIα binding to RSPH3 relative to the unphosphorylated wild-type protein. It also must be pointed out that the decreasing interaction of RSPH3 with RIIα observed at early time points after addition of fresh serum-containing medium was consistent with the T286A mutant experiments, suggesting that there are multiple changes that take place upon ERK1/2 activation only one of which may be affected by phosphorylation of Thr-286. This idea is also suggested by the time course of binding in the presence of U0126, i.e. reduced RIIα binding at 1 and 2.5 min but a smaller effect at longer times of exposure.
In summary, we have identified RSPH3 as an AKAP that may serve as a point of convergence for ERK1/2 and PKA signaling in cilia. Proteomic analysis of human bronchial epithelial cells and immunofluorescence staining of mouse tracheal epithelial cells have shown that RSP3 is present in motile cilia of mammals (16, 17). Consistent with our findings, MEK1 and a phosphorylated form have also been found in cilia (40). Until this study, AKAPs have not been found directly associated with MAPK pathways. The muscle-specific AKAP mAKAP binds proteins involved in feedback loops with phosphodiesterase 4D3 and a cAMP-sensitive guanine nucleotide exchange factor, Epac1, regulating cAMP levels in cardiac myocytes (41). mAKAP coordinates PKA function with the ERK5 MAPK pathway. However, ERK5 binds indirectly to mAKAP through a direct interaction with the phosphodiesterase. MAP2 (microtubule-associated protein 2), an ERK1/2 substrate, is also an AKAP (42). ERK1/2 may facilitate regulation of microtubules in part through MAP2 interactions. Although stable binding of ERK1/2 to microtubules is well documented (43–46), it is not clear if MAP2 itself stably binds ERK1/2 to coordinate with PKA signaling. Thus, it is possible that RSPH3 stands alone as an ERK1/2-binding AKAP. The purpose of PKA and ERK1/2 binding to RSPH3 may be to orchestrate RSP3 actions in ciliary motility. The fact that the AKAP function of RSPH3 is regulated in an ERK1/2-dependent manner suggests that there may be additional ciliary functions associated with this ERK1/2-associated AKAP that remain to be determined.
Acknowledgments
We thank Maureen Wirschell and Winfield Sale (Emory University School of Medicine, Atlanta, GA) and Bill Snell (University of Texas Southwestern) for comments about the data and manuscript, Stan McKnight (University of Washington) for R subunit plasmids, and Dionne Ware for administrative assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant R37 DK34128. This work was also supported by a grant from the THECB Advanced Research Program and Grant I1243 from the Welch Foundation (to M. H. C.).
- MAPK
- mitogen-activated protein kinase
- AKAP
- protein kinase A-anchoring protein
- PKA
- cAMP-dependent protein kinase
- ERK
- extracellular signal-regulated protein kinase
- MEK
- MAPK/ERK kinase
- CD
- common docking
- D
- docking
- EGF
- epidermal growth factor.
REFERENCES
- 1.Raman M., Chen W., Cobb M. H. (2007) Oncogene 26, 3100–3112 [DOI] [PubMed] [Google Scholar]
- 2.Elion E. A. (2001) J. Cell Sci. 114, 3967–3978 [DOI] [PubMed] [Google Scholar]
- 3.Kolch W. (2005) Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 4.Morrison D. K., Davis R. J. (2003) Annu. Rev. Cell Dev. Biol. 19, 91–118 [DOI] [PubMed] [Google Scholar]
- 5.Rubin C. S. (1994) Biochim. Biophys. Acta 1224, 467–479 [PubMed] [Google Scholar]
- 6.Gold M. G., Lygren B., Dokurno P., Hoshi N., McConnachie G., Taskén K., Carlson C. R., Scott J. D., Barford D. (2006) Mol. Cell 24, 383–395 [DOI] [PubMed] [Google Scholar]
- 7.Taylor S. S., Yang J., Wu J., Haste N. M., Radzio-Andzelm E., Anand G. (2004) Biochim. Biophys. Acta 1697, 259–269 [DOI] [PubMed] [Google Scholar]
- 8.Taskén K., Aandahl E. M. (2004) Physiol. Rev. 84, 137–167 [DOI] [PubMed] [Google Scholar]
- 9.Beavo J. A., Bechtel P. J., Krebs E. G. (1975) Adv. Cyclic Nucleotide Res. 5, 241–251 [PubMed] [Google Scholar]
- 10.Amieux P. S., McKnight G. S. (2002) Ann. N.Y. Acad. Sci. 968, 75–95 [DOI] [PubMed] [Google Scholar]
- 11.Yang P., Diener D. R., Yang C., Kohno T., Pazour G. J., Dienes J. M., Agrin N. S., King S. M., Sale W. S., Kamiya R., Rosenbaum J. L., Witman G. B. (2006) J. Cell Sci. 119, 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luck D., Piperno G., Ramanis Z., Huang B. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 3456–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piperno G., Huang B., Ramanis Z., Luck D. J. (1981) J. Cell Biol. 88, 73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaillard A. R., Diener D. R., Rosenbaum J. L., Sale W. S. (2001) J. Cell Biol. 153, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard A. R., Fox L. A., Rhea J. M., Craige B., Sale W. S. (2006) Mol. Biol. Cell 17, 2626–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wirschell M., Zhao F., Yang C., Yang P., Diener D., Gaillard A., Rosenbaum J. L., Sale W. S. (2008) Cell Motil. Cytoskeleton 65, 238–248 [DOI] [PubMed] [Google Scholar]
- 17.Ostrowski L. E., Blackburn K., Radde K. M., Moyer M. B., Schlatzer D. M., Moseley A., Boucher R. C. (2002) Mol. Cell. Proteomics 1, 451–465 [DOI] [PubMed] [Google Scholar]
- 18.Boulton T. G., Cobb M. H. (1991) Cell Regul. 2, 357–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S., Robbins D., Frost J., Dang A., Lange-Carter C., Cobb M. H. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6808–6812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory J. S., Boulton T. G., Sang B. C., Cobb M. H. (1989) J. Biol. Chem. 264, 18397–18401 [PubMed] [Google Scholar]
- 21.Robbins D. J., Zhen E., Owaki H., Vanderbilt C. A., Ebert D., Geppert T. D., Cobb M. H. (1993) J. Biol. Chem. 268, 5097–5106 [PubMed] [Google Scholar]
- 22.Reimann E. M., Beham R. A. (1983) Methods Enzymol. 99, 51–55 [DOI] [PubMed] [Google Scholar]
- 23.Hunter T., Sefton B. M. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 1311–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyle W. J., van der Geer P., Hunter T. (1991) Methods Enzymol. 201, 110–149 [DOI] [PubMed] [Google Scholar]
- 25.Robinson M. J., Stippec S. A., Goldsmith E., White M. A., Cobb M. H. (1998) Curr. Biol. 8, 1141–1150 [DOI] [PubMed] [Google Scholar]
- 26.Jacobs D., Glossip D., Xing H., Muslin A. J., Kornfeld K. (1999) Genes Dev. 13, 163–175 [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T., Hoofnagle A. N., Kabuyama Y., Stroud J., Min X., Goldsmith E. J., Chen L., Resing K. A., Ahn N. G. (2004) Mol. Cell 14, 43–55 [DOI] [PubMed] [Google Scholar]
- 28.Polychronopoulos S., Verykokakis M., Yazicioglu M. N., Sakarellos-Daitsiotis M., Cobb M. H., Mavrothalassitis G. (2006) J. Biol. Chem. 281, 25601–25611 [DOI] [PubMed] [Google Scholar]
- 29.Yazicioglu M. N., Goad D. L., Ranganathan A., Whitehurst A. W., Goldsmith E. J., Cobb M. H. (2007) J. Biol. Chem. 282, 28759–28767 [DOI] [PubMed] [Google Scholar]
- 30.Yang S. H., Yates P. R., Whitmarsh A. J., Davis R. J., Sharrocks A. D. (1998) Mol. Cell. Biol. 18, 710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu B., Wilsbacher J. L., Collisson T., Cobb M. H. (1999) J. Biol. Chem. 274, 34029–34035 [DOI] [PubMed] [Google Scholar]
- 32.Tanoue T., Adachi M., Moriguchi T., Nishida E. (2000) Nat. Cell Biol. 2, 110–116 [DOI] [PubMed] [Google Scholar]
- 33.Tanoue T., Maeda R., Adachi M., Nishida E. (2001) EMBO J. 20, 466–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheridan D. L., Kong Y., Parker S. A., Dalby K. N., Turk B. E. (2008) J. Biol. Chem. 283, 19511–19520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson G., Robinson F., Beers, Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 37.Robinson F. L., Whitehurst A. W., Raman M., Cobb M. H. (2002) J. Biol. Chem. 277, 14844–14852 [DOI] [PubMed] [Google Scholar]
- 38.Proft M., Struhl K. (2004) Cell 118, 351–361 [DOI] [PubMed] [Google Scholar]
- 39.Lee H. Y., Suh Y. A., Robinson M. J., Clifford J. L., Hong W. K., Woodgett J. R., Cobb M. H., Mangelsdorf D. J., Kurie J. M. (2000) J. Biol. Chem. 275, 32193–32199 [DOI] [PubMed] [Google Scholar]
- 40.Schneider L., Clement C. A., Teilmann S. C., Pazour G. J., Hoffmann E. K., Satir P., Christensen S. T. (2005) Curr. Biol. 15, 1861–1866 [DOI] [PubMed] [Google Scholar]
- 41.Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., Scott J. D. (2005) Nature 437, 574–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hausken Z. E., Dell'Acqua M. L., Coghlan V. M., Scott J. D. (1996) J. Biol. Chem. 271, 29016–29022 [DOI] [PubMed] [Google Scholar]
- 43.Reszka A. A., Seger R., Diltz C. D., Krebs E. G., Fischer E. H. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 8881–8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gotoh Y., Nishida E., Matsuda S., Shiina N., Kosako H., Shiokawa K., Akiyama T., Ohta K., Sakai H. (1991) Nature 349, 251–254 [DOI] [PubMed] [Google Scholar]
- 45.Shapiro P. S., Vaisberg E., Hunt A. J., Tolwinski N. S., Whalen A. M., McIntosh J. R., Ahn N. G. (1998) J. Cell Biol. 142, 1533–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zecevic M., Catling A. D., Eblen S. T., Renzi L., Hittle J. C., Yen T. J., Gorbsky G. J., Weber M. J. (1998) J. Cell Biol. 142, 1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]