Abstract
Several recent reports have suggested that microRNAs (miRNAs) might play critical roles in acute myocardial infarction (AMI). However, the miRNA expression signature in the early phase of AMI has not been identified. In this study, the miRNA expression signature was investigated in rat hearts 6 h after AMI. Compared with the expression signature in the noninfarcted areas, 38 miRNAs were differentially expressed in infarcted areas and 33 miRNAs were aberrantly expressed in the border areas. Remarkably, miR-21 expression was significantly down-regulated in infarcted areas, but was up-regulated in border areas. The down-regulation of miR-21 in the infarcted areas was inhibited by ischemic preconditioning, a known cardiac protective method. Overexpression of miR-21 via adenovirus expressing miR-21 (Ad-miR-21) decreased myocardial infarct size by 29% at 24 h and decreased the dimension of left ventricles at 2 weeks after AMI. Using both gain-of-function and loss-of-function approaches in cultured cardiac myocytes, we identified that miR-21 had a protective effect on ischemia-induced cell apoptosis that was associated with its target gene programmed cell death 4 and activator protein 1 pathway. The protective effect of miR-21 against ischemia-induced cardiac myocyte damage was further confirmed in vivo by decreased cell apoptosis in the border and infarcted areas of the infarcted rat hearts after treatment with Ad-miR-21. The results suggest that miRNAs such as miR-21 may play critical roles in the early phase of AMI.
MicroRNAs (miRNAs)3 are endogenous, noncoding, single-stranded RNAs of ∼22 nucleotides and constitute a novel class of gene regulators (1–3). Analogous to the first RNA revolution in the 1980s, when Zaug and Cech (4) discovered the enzymatic activity of RNA, the more recent discoveries of RNA interference and miRNA may represent the second RNA revolution (5). Although the first miRNA, lin-4, was discovered in 1993 (6, 7), their presence in vertebrates was confirmed only in 2001 (8). miRNAs are initially transcribed in the nucleus by RNA polymerase II or III to form large pri-miRNA transcripts (9). These pri-miRNAs are then processed by the RNase III enzymes, Drosha, Pasha, and Dicer, to generate 18- to 24-nucleotide mature miRNAs. In addition to this miRNA biogenesis pathway, some miRNA precursors are able to bypass Drosha processing to produce miRNAs via Dicer, possibly representing an alternative pathway for miRNA biogenesis (10, 11). The mature miRNAs bind to the 3′-untranslated region of their mRNA targets and negatively regulate gene expression via degradation or translational inhibition.
Currently, about 600 miRNAs have been cloned and sequenced in humans, and the estimated number of miRNA genes is as high as 1,000 in the human genome (12, 13). Functionally, an individual miRNA is as important as a transcription factor because it is able to regulate the expression of its multiple target genes. As a group, miRNAs are estimated to regulate over 30% of the genes in a cell (14). It is thus not surprising that miRNAs are involved in the regulation of almost all major cellular functions including apoptosis and necrosis, which are two key cellular events in acute myocardial infarction (AMI).
AMI has long been the leading cause of death in developed countries. Several recent reports have suggested that miRNAs might play critical roles in the pathophysiology of AMI (15–19). Yang et al. (15) have found that the expression of a cardiac arrhythmia-related miRNA, miR-1, is increased in human hearts with coronary heart disease and in rat hearts with AMI. The results of miR-1 expression change in human hearts with coronary artery disease are still controversial, because another recent study has demonstrated that the miR-1 expression tends to be down-regulated in human hearts with coronary artery disease (16). The potential involvement of miRNAs in AMI is also suggested in a study using miR-126 null mice, in which Wang et al. (17) have found that the survival rate in miR-126-deficient mice following AMI is significantly reduced compared with that in wild-type mice. The expression signature in the late phase of AMI (3 and 14 days after AMI) has just been identified by an excellent study reported by van Rooij et al. (18). These investigators found that miR-29 plays an important role in cardiac fibrosis during the repair process after AMI. During manuscript preparation, another excellent study was reported by Kukreja's group (19). In an in vitro ischemia/reperfusion injury model, they have found that, in mouse hearts preinjected with heat shock-induced miRNAs including miR-21, myocardial infarct size after ischemia/reperfusion injury in vitro is reduced. Still, the miRNA expression signature in the early phase of AMI has not been identified. Moreover, the potential effects of miRNA treatment on myocardial infarct size in an in vivo AMI model have not been investigated. The objective of the current study was to determine the expression signatures of different areas in infarcted rat hearts at 6 h after AMI and to investigate the role of an aberrantly expressed miRNA, miR-21, in AMI and its potential cellular and molecular mechanisms.
EXPERIMENTAL PROCEDURES
AMI and Ischemic Preconditioning (IP) Animal Models
To determine the miRNA expression changes in infarcted hearts, we applied a well established rat AMI model using left coronary artery ligation as described (20). In brief, 10-week-old male Sprague-Dawley rats (weighing 250–300 g) were anesthetized with ketamine (80 mg/kg intraperitoneally) and xylazine (5 mg/kg intraperitoneally). Under sterile conditions, an anterior transmural AMI was created by occlusion of the left anterior descending coronary artery with a silk suture. Sham-operated rats served as controls. Sham operation involved an identical procedure, except the suture was passed around the vessel without left anterior descending coronary artery occlusion. IP was achieved via four cycles of 5-min coronary occlusion/5-min reperfusion cycles of the left anterior descending coronary artery as described (21). All protocols were approved by the Institutional Animal Care and Use Committee at the University of Medicine and Dentistry of New Jersey-New Jersey Medical School, and were consistent with the Guide for the Care and Use of Laboratory Animals (National Institutes of Health publication 85-23).
Measurement of Infarct Size and Determination of Infarcted, Border, and Noninfarcted Areas
At 6 and 24 h after the occlusion of the coronary artery, the rats were anesthetized and 6 ml of 1% Evans blue dye was injected into the vena cava to delineate the noninfarcted portion of the heart. The myocardial ischemic area at risk was identified as the region lacking blue staining. The ventricles of the hearts were sliced transversely into 2-mm thick slices. The slices were incubated in 1% triphenyltetrazolium chloride (TTC) at 37 °C for 30 min to identify the noninfarcted and infarcted areas. Infarct size was expressed as a percentage of the ischemic area at risk. In myocardial slices, noninfarcted area was defined as the Evans blue-stained area. The infarcted area was displayed as the TTC unstained area. The border area was identified as Evans blue unstained and TTC-stained areas.
Measurement of Dimensions of the Left Ventricles (LV)
Two weeks after AMI, the anesthetized rat hearts were arrested in diastole by an intravenous injection of saturated potassium chloride solution via the left jugular vein. The dimensions of the LV were measured in transverse slices at the level of papillary muscle as described (22, 23).
miRNA Expression Signature Array
miRNAs were isolated from the infarcted, border, and noninfarcted areas of rat left ventricles at 6 h after left anterior descending coronary artery ligation using the mirVana miRNA isolation kit (Ambion, Inc.). Left ventricular miRNAs isolated from the sham group were used as the sham controls. In addition, one group of miRNAs were isolated from rat left ventricles at 6 h after IP. Each group had six rats, and miRNA expression profiling was done by miRNA microarray analysis using a chip containing 341 mature miRNAs (Chip ID miRRat 12.0 version; LC Sciences) (24, 25). The miRNA expression was demonstrated by the mean of the 6 biological replicates. Proprietary “spike-in” controls were used at each step of the process.
Construction of the Adenovirus Expressing miR-21, Programmed Cell Death 4 (PDCD4), or Control Adenovirus Expressing GFP
The adenovirus expressing miR-21 (Ad-miR-21), PDCD4 (Ad-PDCD4), or control adenovirus expressing GFP (Ad-GFP) were generated using the Adeno-XTM Expression Systems 2 kit (Clontech) according to the manufacturer's protocols. These adenoviruses were purified by cesium chloride gradient ultracentrifugation and titrated using a standard plaque assay.
Adenovirus-mediated miR-21 Gene Transfer in Vivo
Ad-miR-21 or Ad-GFP were delivered into the rat hearts 3 days before AMI as described (26). Briefly, rats were anesthetized with ketamine (80 mg/kg intraperitoneal) and xylazine (5 mg/kg intraperitoneally). The pericardium was opened via the third intercostal space. The aorta and pulmonary artery were identified. A 23-gauge catheter containing 200 μl of adenovirus was advanced from the apex of the left ventricle to the aortic root. The aorta and pulmonary arteries were clamped distal to the site of the catheter and the solution was injected. The clamp was maintained for 10 s when the heart pumped against a closed system (isovolumically). This procedure allows the solution that contains the adenovirus to circulate down the coronary arteries and perfuse the heart. After 10 s, clamps on the aorta and pulmonary artery were released and the chest was closed.
Cardiac Myocyte Culture and Cell Ischemia Injury Model
Primary cultures of neonatal rat cardiac ventricular myocytes were performed as described previously (12). In brief, hearts from 1–2-day-old Sprague-Dawley rats were placed in ice-cold 1× phosphate-buffered saline solution. After repeated rinsing, the atria were cut off, and the ventricles were minced with scissors. The minced tissue and ventricular cells were dispersed by digestion with collagenase type IV (0.45 mg/ml), 0.1% trypsin, and 15 μg/ml DNase I. Cardiomyocytes (0.33 × 106 cells/ml) were cultured in cardiac myocyte culture medium containing Dulbecco's modified Eagle's medium/F-12 supplemented with 5% horse serum, 4 μg/ml of transferrin, 0.7 ng/ml of sodium selenite, 2 g/liter of bovine serum albumin, 3 mmol/liter of pyruvic acid, 15 mmol/liter of HEPES, 100 μmol/liter of ascorbic acid, 100 μg/ml of ampicillin, 5 μg/ml of linoleic acid, 1% penicillin, 1% streptomycin, and 100 μmol/liter 5-bromo-2′-deoxyuridine and seeded into six-well plates.
Cell ischemic injury, induced by hypoxia in a serum- and glucose-free medium, and reoxygenation (H/R) was performed as described (27). Hypoxia was achieved by placing the cells in a hypoxia chamber filled with 5% CO2 and 95% N2 at 37 °C for 4 h. Following hypoxia exposure, the cells were reoxygenated with 5% CO2 and 95% O2 for 3 h in Dulbecco's modified Eagle's medium containing 5% serum and normal glucose.
Oligo Transfection, miR-21 Knockdown, miR-21 Overexpression, and PDCD4 Gene Up-regulation in Cultured Cardiac Myocytes
Oligo transfection was performed according to an established protocol (24, 25). For the miR-21 knockdown, miR-21 inhibitor (LNA-anti-miR-21) (Exiqon, Inc.) was added to the culture medium at a final oligonucleotide concentration of 30 nm. For miR-21 up-regulation, pre-miR-21 (Ambion, Inc.) was added directly to the complexes at a final oligonucleotide concentration of 30 nm. PDCD4 gene up-regulation was performed by Ad-PDCD4 (30 m.o.i.). The transfection medium was replaced 4 h post-transfection by regular culture medium. Vehicle control, oligo controls for LNA-anti-miR-21 and pre-miR-21 (Ambion, Inc.), and adenovirus control (Ad-GFP) were applied.
Quantitative Reverse Transcriptase-PCR (qRT-PCR)
Some aberrantly expressed miRNAs in infarcted hearts were further confirmed by qRT-PCR (24, 25). Moreover, as miR-21 is the selected miRNA for the study, its expression in sham-opened hearts and in different areas of the infarcted hearts was determined at 6 and 24 h after AMI. In addition, qRT-PCR was also used to determine the miR-21 levels in cultured cells. qRT-PCR was performed on cDNA generated from 50 ng of total RNA using the protocol of the mirVana qRT-PCR miRNA Detection Kit (Ambion, Inc). Amplification and detection of specific products were performed with a Roche Lightcycler 480 Detection System. As an internal control, U6 was used for template normalization. The primers used were provided by Ambion, Inc. Fluorescent signals were normalized to an internal reference, and the threshold cycle (Ct) was set within the exponential phase of the PCR. The relative gene expression was calculated by comparing cycle times for each target PCR. The target PCR Ct values were normalized by subtracting the U6 Ct value, which provided the ΔCt value. The relative expression level between treatments was then calculated using the following equation: relative gene expression = 2−(ΔCt,sample-ΔCt,control).
Western Blot Analysis
Proteins isolated from cultured cardiac myocytes were determined by Western blot analysis. Equal amounts of protein were subjected to SDS-PAGE. A standard Western blot analysis was conducted using PDCD4 antibody (Santa Cruz Biotechnology). Glyceraldehyde-3-phosphate dehydrogenase antibody (1:5000 dilution; Cell Signaling) was used as the loading control.
Luciferase Assay
Activator protein 1 (AP-1) activity was measured using luciferase assay as described (28). Briefly, adenoviral vector (Ad-AP1-Luc) controlled by a synthetic promoter with direct repeats of the transcription recognition sequences for AP-1 was purchased from Vector Biolabs. Cultured cardiac myocytes pretreated with vehicle, control oligos (30 nm), LNA-anti-miR-21 (30 nm), pre-miR-21 (30 nm), Ad-GFP (30 m.o.i.), or Ad-PDCD4 (30 m.o.i.) for 4 h were transfected with Ad-AP1-Luc for 5 h with 10 pfu/cell. Luciferase activity was measured after 24 h using a scintillation counter and normalized to protein levels.
Detection of Apoptosis
Cultured cardiac myocyte apoptosis and apoptosis in heart sections were measured by terminal deoxynucleotide transferase dUTP nick end labeling (TUNEL) staining as described previously (24, 25). Briefly, cardiac myocytes cultured on coverslips in 24-well plates were fixed in 4% paraformaldehyde. In heart tissue, TUNEL staining was performed in frozen heart sections (8 μm). The TUNEL staining was done using the in situ cell death detection kit (Roche) according to the manufacturer's protocol. The numbers of TUNEL-positive cells and the total cells in infarcted and border areas were counted under a fluorescence microscope. The infarcted, border, and noninfarcted areas were identified by histology in hematoxylin and eosin-stained heart sections.
Statistics
All data are presented as mean ± S.E. For relative gene expression, the mean value of the vehicle control group was defined as 100% or 1. Two-tailed unpaired Student's t tests and analysis of variance were used for statistical evaluation of the data. The Sigma stat statistical analysis program was used for data analysis. A p value <0.05 was considered significant.
RESULTS
miRNA Expression Signatures in Different Areas of Infarcted Rat Hearts in the Early Phase of AMI
Compared with those in the noninfarcted areas, miRNAs were aberrantly expressed in both infarcted (Table 1) and border areas (Table 2) of the infarcted hearts at 6 h after AMI. Among the 341 arrayed miRNAs, 38 miRNAs were differentially expressed (21 up and 17 down) in the infarcted areas with over 30% change and p value <0.01. In the border areas, 33 miRNAs were differentially expressed (19 up and 14 down). Although most of the miRNAs had a similar expression in the noninfarcted area of the infarcted hearts compared with the sham-opened controls, the expression of some miRNAs in the two groups was still different. Among them, miR-21, miR-27a, miR-27b, miR-30b-5p, miR-30c, miR-125a-5p, miR-126, mir-150, miR-26a, and miR-26b were up-regulated, but miR-107, miR-130a, miR-145, miR-16, and miR-22 were down-regulated in the noninfarcted areas of the infarcted hearts.
TABLE 1.
Aberrant expression of miRNAs in the infarcted area of rat hearts at 6 h after AMI
| miRNAs | Expression in noninfarcted area |
Expression in infarcted area |
Noninfarcted value | p value | ||
|---|---|---|---|---|---|---|
| Mean | S.E. | Mean | S.E. | |||
| % | ||||||
| miR-30e | 1,143 | 130 | 424 | 80 | 37.11 | 3.78E-09 |
| miR-451 | 738 | 105 | 291 | 94 | 39.48 | 1.51E-05 |
| miR-29c | 288 | 53 | 126 | 26 | 43.63 | 2.17E-07 |
| miR-99a | 414 | 84 | 183 | 43 | 44.27 | 1.46E-06 |
| miR-499 | 672 | 100 | 355 | 63 | 52.83 | 7.96E-07 |
| miR-199a-3p | 1,024 | 162 | 544 | 82 | 53.11 | 5.41E-07 |
| miR-100 | 252 | 54 | 142 | 24 | 56.33 | 2.45E-05 |
| miR-25 | 445 | 36 | 253 | 54 | 56.95 | 1.94E-05 |
| miR-16 | 4,111 | 387 | 2,361 | 252 | 57.44 | 1.01E-08 |
| miR-92a | 314 | 45 | 181 | 34 | 57.82 | 3.48E-06 |
| miR-143 | 3,069 | 261 | 1,811 | 162 | 59.01 | 1.75E-09 |
| miR-24 | 5,312 | 376 | 3,337 | 402 | 62.83 | 3.56E-07 |
| miR-1 | 33,598 | 1,320 | 21,430 | 1,108 | 63.79 | 1.22E-08 |
| miR-195 | 1,920 | 132 | 1,249 | 164 | 65.05 | 1.67E-06 |
| miR-126 | 11,620 | 721 | 7,908 | 916 | 68.06 | 1.57E-06 |
| miR-21 | 1,125 | 103 | 448 | 56 | 39.80 | 3.74E-03 |
| miR-125b-5p | 3,540 | 313 | 2,474 | 267 | 69.89 | 2.31E-06 |
| miR-133b | 6,347 | 299 | 8,488 | 1,099 | 133.72 | 1.80E-04 |
| miR-378 | 1,230 | 146 | 1,688 | 220 | 137.18 | 8.59E-05 |
| miR-145 | 4,466 | 254 | 6,771 | 484 | 151.62 | 2.02E-09 |
| let-7e | 1,399 | 186 | 2,387 | 625 | 170.64 | 1.46E-04 |
| let-7d | 5,471 | 564 | 9,463 | 532 | 172.96 | 2.09E-07 |
| miR-328 | 61 | 25 | 107 | 17 | 176.18 | 8.06E-03 |
| miR-181a | 737 | 94 | 1,300 | 145 | 176.28 | 6.95E-08 |
| miR-26b | 884 | 148 | 1,632 | 308 | 184.63 | 2.89E-06 |
| miR-320 | 778 | 110 | 1,448 | 171 | 185.98 | 4.05E-08 |
| miR-214 | 889 | 135 | 1,709 | 210 | 192.29 | 3.80E-08 |
| miR-146a | 537 | 60 | 1,096 | 123 | 204.03 | 1.37E-09 |
| miR-181c | 45 | 15 | 102 | 16 | 225.33 | 8.18E-05 |
| miR-674-5p | 67 | 18 | 199 | 36 | 295.12 | 9.90E-08 |
| miR-223 | 56 | 23 | 210 | 64 | 373.66 | 2.69E-06 |
| miR-206 | 85 | 23 | 429 | 88 | 505.56 | 7.44E-09 |
| miR-352 | 118 | 40 | 611 | 204 | 515.90 | 4.50E-08 |
| miR-672 | 96 | 35 | 720 | 234 | 749.53 | 8.90E-08 |
| miR-466b | 16 | 9 | 189 | 55 | 1208.08 | 1.54E-05 |
| miR-188 | 24 | 7 | 343 | 48 | 1455.51 | 5.43E-11 |
| miR-327 | 11 | 3 | 183 | 45 | 1728.84 | 1.31E-03 |
| miR-290 | 16 | 7 | 1,880 | 120 | 11899.49 | 2.56E-04 |
TABLE 2.
Aberrant expression of miRNAs in border area of the rat hearts at 6 h after AMI
| miRNAs | Expression in noninfarcted area |
Expression in border area |
Noninfarcted value | p value | ||
|---|---|---|---|---|---|---|
| Mean | S.E. | Mean | S.E. | |||
| % | ||||||
| miR-130a | 403 | 84 | 128 | 30 | 31.72 | 9.44E-08 |
| miR-22 | 1,782 | 181 | 730 | 59 | 40.97 | 2.81E-12 |
| miR-214 | 889 | 135 | 483 | 52 | 54.41 | 1.52E-07 |
| miR-107 | 591 | 22 | 332 | 21 | 56.20 | 2.51E-11 |
| miR-143 | 3,069 | 261 | 1,741 | 146 | 56.73 | 4.74E-10 |
| miR-1 | 33,598 | 1,320 | 19,244 | 853 | 57.28 | 2.92E-11 |
| miR-181a | 737 | 94 | 424 | 109 | 57.49 | 1.75E-04 |
| miR-378 | 1,230 | 146 | 723 | 35 | 58.77 | 2.47E-07 |
| miR-103 | 639 | 66 | 382 | 25 | 59.76 | 1.17E-08 |
| miR-199a-3p | 1,024 | 162 | 669 | 40 | 65.34 | 4.20E-05 |
| miR-99a | 414 | 84 | 283 | 28 | 68.19 | 8.88E-04 |
| miR-99b | 390 | 74 | 268 | 36 | 68.69 | 4.76E-04 |
| miR-24 | 5,312 | 376 | 3,656 | 240 | 68.83 | 6.70E-09 |
| let-7c | 12,210 | 1,106 | 8,452 | 489 | 69.22 | 3.53E-04 |
| miR-27b | 1,623 | 161 | 2,171 | 197 | 133.75 | 3.03E-05 |
| miR-30a | 1,443 | 102 | 1,922 | 141 | 133.13 | 4.96E-07 |
| miR-146a | 537 | 60 | 818 | 58 | 152.26 | 4.58E-07 |
| miR-27a | 1,180 | 113 | 1,935 | 129 | 163.95 | 4.13E-09 |
| miR-150 | 1,199 | 101 | 1,982 | 120 | 165.34 | 8.09E-10 |
| miR-92b | 75 | 19 | 131 | 17 | 174.24 | 1.30E-04 |
| miR-21 | 1125 | 119 | 1,976 | 176 | 175.69 | 3.35E-09 |
| miR-146b | 57 | 11 | 127 | 21 | 222.11 | 2.45E-07 |
| miR-322 | 161 | 50 | 374 | 29 | 232.40 | 6.41E-05 |
| miR-29c | 288 | 53 | 677 | 69 | 234.64 | 3.37E-08 |
| miR-30b-5p | 1,534 | 185 | 3,664 | 351 | 238.80 | 4.20E-11 |
| miR-125a-5p | 500 | 77 | 1,231 | 84 | 246.18 | 3.10E-08 |
| miR-26b | 884 | 148 | 2,641 | 248 | 298.85 | 1.08E-09 |
| miR-499 | 672 | 100 | 2,038 | 131 | 303.29 | 3.14E-09 |
| miR-352 | 118 | 40 | 551 | 41 | 465.17 | 9.47E-07 |
| miR-223 | 56 | 23 | 502 | 108 | 892.02 | 1.24E-08 |
| miR-122 | 15 | 3 | 223 | 9 | 1497.62 | 3.45E-03 |
| miR-290 | 16 | 7 | 304 | 64 | 1923.27 | 2.00E-03 |
Verification of Some Aberrantly Expressed miRNAs in Infarcted Areas of the Hearts by qRT-PCR
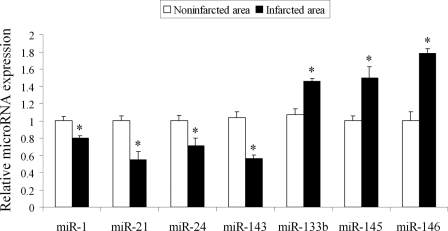
Some aberrantly expressed miRNAs in infarcted areas were further verified by qRT-PCR (Fig. 1). Remarkably, miR-21 expression in the infarcted areas was significantly decreased compared with that in noninfarcted area.
FIGURE 1.
Verification of some aberrantly expressed miRNAs in infarcted areas of the hearts by qRT-PCR. miRNAs were isolated in the noninfarcted area and the infarcted area of the infarcted hearts. The expression of some miRNAs that were aberrantly expressed in the infarcted area was verified by qRT-PCR. Note: Data presented as mean ± S.E. (error bars), n = 5, *, p < 0.05 compared with the noninfarcted control.
miR-21 Expression in the Different Areas of the Infarcted Hearts in the Early Phase of AMI
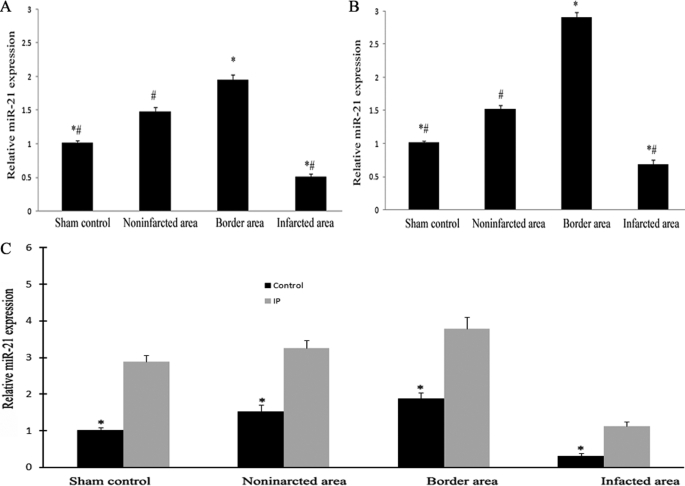
To further determine the expression changes of miR-21 in the different areas of the infarcted hearts and its time course response in the early phase of AMI, miRNAs were isolated in the different areas of the infarcted hearts as well as in sham-opened hearts at 6 and 24 h after AMI. As shown in Fig. 2, miR-21 expression was down-regulated in the infarcted areas at both 6 (Fig. 2A) and 24 h (Fig. 2B) after AMI compared with that in other areas and in sham-opened hearts. Interestingly, compared with that in other areas and in sham-opened hearts, miR-21 expression in border areas was significantly increased. In addition, miR-21 expression in noninfarcted areas of the infarcted hearts was also higher than that in sham-opened hearts.
FIGURE 2.
miR-21 expression in different areas of the infarcted hearts and the effect of IP on the expression in the early phase of AMI. miRNAs were isolated in different areas of the infarcted hearts as well as in sham-opened hearts at 6 (A) and 24 h (B) after AMI. The expression of miR-21 was determined by qRT-PCR. miR-21 expression was down-regulated in the infarcted area at both 6 and 24 h after AMI compared with that in other areas and in sham-opened hearts. Notably, compared with that in other areas and sham-opened hearts, miR-21 expression in the border area was significantly increased. C, the effect of IP on miR-21 expression at 6 h after AMI. The IP was performed 6 h before the AMI. Note: Data presented as mean ± S.E. (error bars), n = 6, *, p < 0.05 compared with the noninfarcted control in A and C, and with the IP group in C. #, p < 0.05 compared with that in the border area.
The Effects of IP on miRNA Expression
IP is a well established method to protect the heart against myocardial infarction. To further explore the pathological involvement of miR-21 in AMI, the effect of IP on miR-21 was investigated. As shown in Fig. 2C, 6 h after IP, the expression in non-infarcted rat hearts was increased. In IP-pretreated infarcted rat hearts, the down-regulated expression of miR-21 in infarcted areas was reversed. In border and noninfarcted areas, IP resulted in an additional increase in miR-21 expression in hearts at 6 h after AMI (Fig. 2C). The effects of IP on the expression of miRNAs that were aberrantly expressed in the infarcted area of the rat hearts determined by microarray analysis are listed in Table 3.
TABLE 3.
The effects of IP on the expression of miRNAs that are aberrantly expressed in the infarcted area of the rat hearts
| miRNAs | Expression in sham-opened hearts |
Expression in IP hearts |
Sham control | p value | ||
|---|---|---|---|---|---|---|
| Mean | S.E. | Mean | S.E. | |||
| % | ||||||
| miR-21 | 1,602 | 226 | 5,812 | 263 | 362.86 | 0.00E+00 |
| miR-16 | 2,491 | 247 | 4,203 | 266 | 168.69 | 8.44E-13 |
| miR-26a | 23,394 | 1,393 | 16,064 | 788 | 68.67 | 3.44E-11 |
| miR-352 | 815 | 295 | 316 | 93 | 38.76 | 8.32E-08 |
| miR-320 | 1,078 | 158 | 2,333 | 141 | 216.31 | 0.00E+00 |
| let-7e | 2,703 | 416 | 4,211 | 521 | 155.79 | 5.98E-13 |
| miR-214 | 414 | 86 | 812 | 88 | 195.95 | 3.20E-10 |
| miR-126 | 27,090 | 1,357 | 18,420 | 1,286 | 67.99 | 5.34E-12 |
| miR-499 | 3,496 | 768 | 913 | 99 | 26.11 | 3.33E-16 |
The Effect of Adenovirus-mediated miR-21 Gene Transfer on Myocardial Infarct Size
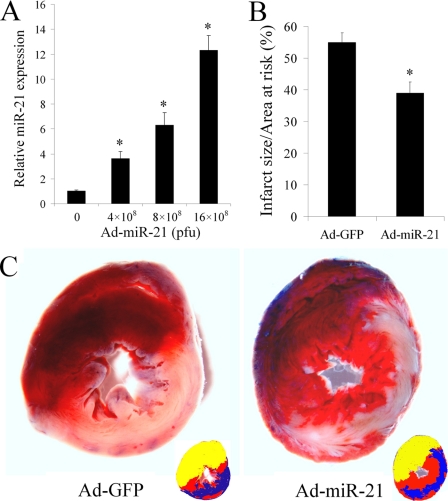
As shown in Fig. 3A, at 3 days after administration, Ad-miR-21 increased miR-21 expression in rat hearts in a dose-dependent manner. Based on the dose response of Ad-miR-21, we injected 8 × 108 pfu/rat into the animals using our delivery method at 3 days before AMI. As shown in Fig. 3B, compared with control adenovirus (Ad-GFP)-treated rats, Ad-miR-21 reduced myocardial infarct size by 29 ± 3.32% at 24 h after AMI. Representative TTC-stained heart slices from rats treated with Ad-miR-21 or Ad-GFP are shown in Fig. 3C.
FIGURE 3.
The effect of adenovirus-mediated miR-21 gene transfer on myocardial infarct size. A, Ad-miR-21 increased miR-21 expression in rat hearts in a dose-dependent manner. Ad-miR-21 was delivered into the rat hearts using the local delivery method at doses: 0 (Ad-GFP control), 4 × 108, 8 × 108, or 12 × 108 pfu/rat. Three days later, rat hearts were isolated for miR-21 expression assay by qRT-PCR. Note: Data presented as mean ± S.E. (error bars), n = 5, *, p < 0.05 compared with 0 group. B, Ad-miR-21 (8 × 108 pfu) reduced infarct size in rat hearts at 24 h after AMI. Note: Data presented as mean ± S.E. (error bars), n = 10; *, p < 0.05 compared with Ad-GFP control. C, representative TTC-stained heart slices from rats treated with control adenovirus, Ad-GFP, or Ad-miR-21. The infarcted area in the Ad-GFP-treated image was 56% and the infarcted area in the Ad-21-treated image was 37%. Corresponding color drawings (right corner) show the risk area (red and blue), infracted area (blue), and the Evans blue-stained area (yellow).
The Effect of miR-21 on Ischemia-related Cell Injury in Vitro
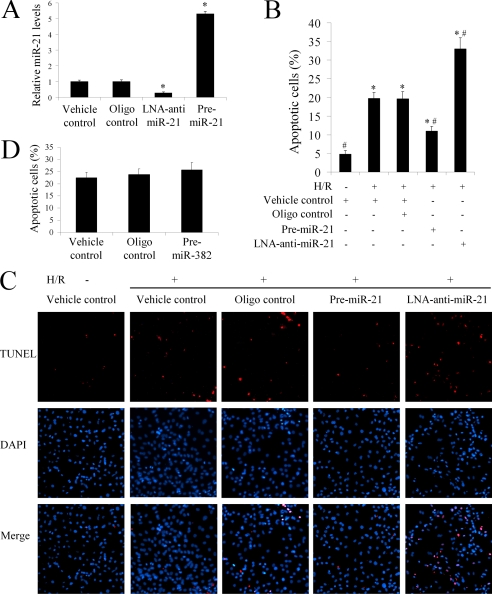
To determine the potential cellular mechanism behind miR-21-mediated protective effects during myocardial infarction, an ischemia-related cell injury model was applied in which injury to the cultured cells was induced by hypoxia in a serum- and glucose-free medium, followed by reoxygenation in normal culture medium. We found that the miR-21 inhibitor, LNA-anti-miR-21 (30 nm), decreased, but pre-miR-21 increased miR-21 expression in cultured cardiac cells (Fig. 4A). As expected, hypoxia/reoxygenation resulted in an increase in apoptosis. Notably, pre-miR-21 decreased hypoxia/reoxygenation-induced cardiac myocyte apoptosis (Fig. 4B). In contrast, cardiac myocyte apoptosis was exacerbated after treatment with LNA-anti-miR-21 (Fig. 4B). Representative TUNEL-stained photomicrographs from uninjured cardiac myocytes and injured cardiac myocytes treated with vehicle, control oligos, pre-miR-21, or LNA-anti-miR-21 are shown in Fig. 4C. The results indicate that miR-21 has a protective effect against ischemia-induced cardiac myocyte apoptosis in vitro. To confirm that the cell protective effect is miR-21 specific, we also determined the effect of unrelated miR-382, which has very low expression in the heart and has no significant expression during the AMI on cell injury response. As shown in Fig. 4D, miR-382 had no effect on hypoxia/reoxygenation-induced cardiac myocyte apoptosis.
FIGURE 4.
The effect of miR-21 on ischemia-related cell injury in vitro. Cultured cardiac cell injury was induced by hypoxia for 4 h in a serum- and glucose-free medium followed by reoxygenation (H/R) for 3 h in normal culture medium. Cell apoptosis was determined by TUNEL staining. A, miR-21 inhibitor, LNA-anti-miR-21 (30 nm) decreased, but pre-miR-21 (30 nm) increased miR-21 expression in cultured cardiac cells. Note: Data presented as mean ± S.E. (error bars), n = 5, *, p < 0.05 compared with the vehicle control. B, hypoxia/reoxygenation resulted in an increase in apoptosis. Pre-miR-21 decreased the cardiac myocyte apoptosis. In contrast, cardiac myocyte apoptosis was exacerbated after treatment with LNA-anti-miR-21. Note: n = 5, *, p < 0.05 compared with the noninjured (without H/R) control. #, p < 0.05 compared with H/R treated with the vehicle control. C, representative TUNEL-stained photomicrographs from uninjured cells, and cardiac myocytes treated with vehicle, control oligos, pre-miR-21, or LNA-anti-miR-21. Note: red is TUNEL staining representing apoptotic cells; blue is the cell nucleus stained by 4′,6-diamidino-2-phenylindole (DAPI). D, the effect of miR-382 on H/R-mediated cell apoptosis.
The miR-21-mediated Protective Effect against Myocardial Infarction Is Related to Its Anti-apoptotic Effect on Cardiac Cells in Vivo
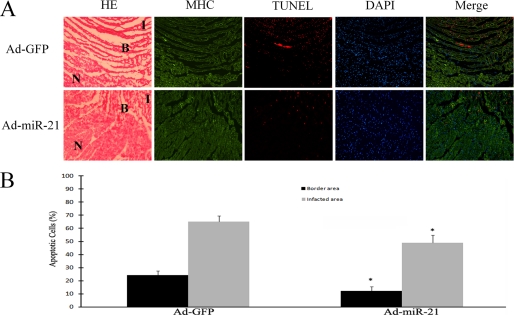
To verify the cellular mechanism involved in the miR-21-mediated protective effect against myocardial infarction in vivo, apoptosis was determined in infarcted heart sections by immunofluorescence with TUNEL staining. The different areas were further identified by histology in hematoxylin and eosin-stained sections as shown in Fig. 5A and the myocytes were identified by immunofluorescence with myosin heavy chain antibody (Fig. 5A). Representative TUNEL-stained photomicrographs in heart sections from rats treated with Ad-GFP or Ad-miR-21 are displayed in Fig. 5A. The images reveal that, at 24 h after AMI, there were many apoptotic cells in both infarcted and border areas. As shown in Fig. 5B, compared with the Ad-GFP-treated group, apoptosis of the cardiac cells in both the border and infarcted areas was significantly decreased in the Ad-miR-21-treated group. The results indicate that the miR-21-mediated protective effect against myocardial infarction is related to its anti-apoptotic effect on cardiac cells in vivo.
FIGURE 5.
The effect of miR-21 overexpression on cardiac cell apoptosis in the border and infarcted areas of rat hearts. A, representative hematoxylin and eosin (HE), myosin heavy chain (MHC), and TUNEL-stained photomicrographs from cardiac myocytes in heart sections from rats treated with Ad-GFP or Ad-miR-21 (8 × 108 per rat). Note: I, infarcted area; B, border area; and n, non-infarcted area. Red is TUNEL staining representing apoptotic cells; blue is the cell nucleus stained by 4′,6-diamidino-2-phenylindole (DAPI). B, quantitative analysis of the apoptotic cells in heart sections. Note: Data presented as mean ± S.E. (error bars), n = 6, *, p < 0.05 compared with the Ad-GFP control.
The Effect of Adenovirus-mediated miR-21 Gene Transfer on LV Dimensions
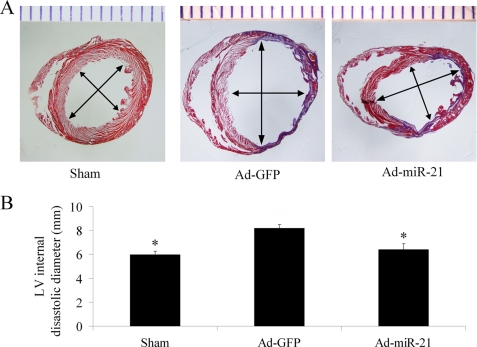
The reduced myocardial size and heart cell apoptosis should have functional results on LV remodeling such as change in LV dimensions. To test these results, the infarcted hearts from both Ad-GFP and Ad-miR-21 pre-treated rats were isolated at 2 weeks after AMI. As shown in Fig. 6, the LV internal diastolic diameter in Ad-miR-21-treated animals (6.4 ± 0.41 mm) was significantly smaller than that in Ad-GFP-treated animals (8.2 ± 0.29 mm).
FIGURE 6.
The effect of adenovirus-mediated miR-21 gene transfer on LV dimensions. A, representative heart sections from sham-opened, Ad-GFP, or Ad-miR-21-treated rats at 2 weeks after AMI. B, Ad-miR-21 (8 × 108 pfu) reduced internal diastolic diameter of left ventricles in rat hearts at 2 weeks after AMI. Note: Data presented as mean ± S.E. (error bars), n = 8; *, p < 0.05 compared with Ad-GFP control.
PDCD4 Is a miR-21 Target Gene That Is Involved in miR-21-mediated Anti-apoptotic Effects on Cardiac Cells That May Be Related to Its Downstream Signaling Molecule, AP-1
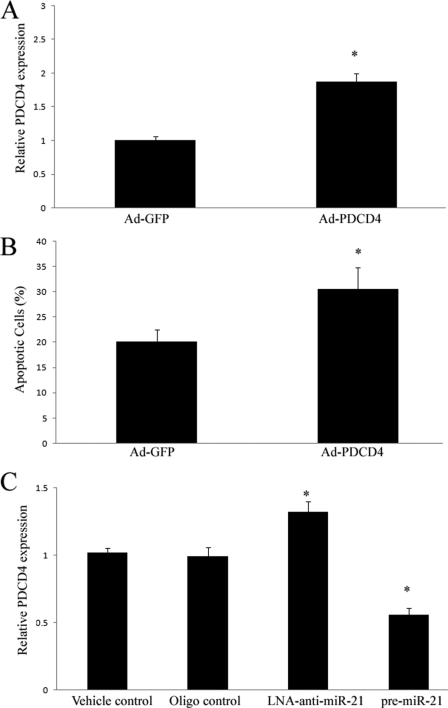
Computational analysis indicates that PDCD4 is a potential target gene of miR-21 as described in our recent study (24). The role of PDCD4 in cardiac cell apoptosis is currently unclear. To test this analysis, we overexpressed PDCD4 by Ad-PDCD4 in cultured cardiac myocytes. As shown in Fig. 7A, PDCD4 expression was increased by Ad-PDCD4 in these cardiac cells. Consistent with the PDCD4 expression, Ad-PDCD4 increased cardiac myocyte apoptosis induced by hypoxia/reoxygenation (Fig. 7B). In addition, PDCD4 expression in cardiac cells was up-regulated by LNA-anti-miR-21 (30 nm), but down-regulated by pre-miR-21 (Fig. 7C). The results suggest that PDCD4 is a miR-21 target gene that is involved in miR-21-mediated anti-apoptotic effects on cardiac cells.
FIGURE 7.
PDCD4 is a miR-21 target gene that is involved in miR-21-mediated anti-apoptotic effect on cardiac cells. A, overexpression of Ad-PDCD4 by Ad-PDCD4 (30 m.o.i.) in cultured cardiac myocytes. Note: n = 3, *, p < 0.05 compared with the Ad-GFP control. B, Ad-PDCD4 (30 m.o.i.) increased cardiac myocyte apoptosis induced by hypoxia/reoxygenation as determined by TUNEL staining. Note: n = 5, *, p < 0.05 compared with the Ad-GFP control. C, modulation of PDCD4 expression in cardiac myocytes by LNA-anti-miR-21 (30 nm) and pre-miR-21 (30 nm). Note: Data presented as mean ± S.E. (error bars), n = 6, *, p < 0.05 compared with the vehicle control.
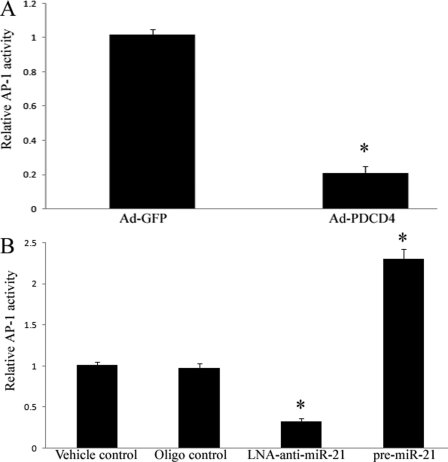
AP-1 might be a downstream signaling molecule of PDCD4 that is associated with the PDCD4-mediated effect on cell apoptosis (27). To determine the potential involvement of AP-1 in miR-21-induced effects on cardiac cells, AP-1 activity was determined in cultured cardiac cells. As shown in Fig. 8A, overexpression of PDCD4 by Ad-PDCD4 inhibited AP-1 activity. In addition, increasing PDCD4 expression via LNA-anti-miR-21 (Fig. 7C) resulted in a decrease in AP-1 activity (Fig. 8B). In contrast, decreasing PDCD4 expression via pre-miR-21 (Fig. 7C) resulted in an increase in AP-1 activity (Fig. 8B). These results suggest that AP-1 may be a downstream signaling molecule of PDCD4 that is related to the miR-21-mediated effect on cardiac myocytes.
FIGURE 8.
AP-1 is a downstream signaling molecule of PDCD4 that may be involved in miR-21-mediated effect on cardiac myocytes. A, overexpression of PDCD4 by Ad-PDCD4 (30 m.o.i.) inhibited AP-1 activity. Note: n = 5, *, p < 0.05 compared with the Ad-GFP control. B, modulation of AP-1 activity in cardiac myocytes by LNA-anti-miR-21 (30 nm) and pre-miR-21 (30 nm). Note: Data presented as mean ± S.E. (error bars), n = 5, *, p < 0.05 compared with the vehicle control.
DISCUSSION
It is well established that AMI is a complex process in which multiple genes have been found to be dysregulated (29). miRNAs are a novel layer of gene regulators that regulate over 30% of genes in a cell. It is therefore reasonable to hypothesize that miRNAs could be involved in AMI. The current study demonstrated for the first time that multiple miRNAs were aberrantly expressed in the early phase (6 h) of AMI both in the border and infarcted areas. The multiple dysregulated miRNAs found in this study match the complexity of AMI.
Pathophysiological changes after occlusion of the coronary artery include early myocyte ischemia followed by injury and infarction within the area of its blood supply (8). Even after infarction, however, there are still some ischemic and injured cells near the infarction zone (border area). The border area could therefore provide an excellent therapeutic window for AMI. One important finding in the current study is that the miRNA signature in the border area is much different from that of the infarcted area of the infarcted hearts. We think that some of the aberrantly expressed miRNAs might have defensive effects under AMI conditions. Another finding in the study is that the expression of some miRNAs in the non-infarcted area at 6 h after AMI is also different from that in sham-opened hearts. This result is consistent with a recent report by van Rooij et al. (18), in which these investigators also found that the expression of some miRNAs in the non-infarcted area of hearts in the late phase of AMI (3 and 14 days after AMI) is different from that in sham-opened control hearts. These results suggest that some miRNAs in the non-infarcted area might also participate in the pathophysiologic response to AMI.
One recent report has indicated that miR-1 is up-regulated in ischemic heart tissue (15). However, another recent study has demonstrated that miR-1 expression tends to be down-regulated in human hearts with coronary heart disease (16). In the current study, we found that miR-1 is significantly down-regulated in the infarcted areas at 6 h after AMI. The reasons for the discrepancies in miR-1 expression among these studies are unclear. However, the variations in disease state, sample localization, and time points for sample isolations might be responsible for the discrepancies. Indeed, we have found that the expression of multiple miRNAs is different in the different areas of the infarcted hearts at 6 h after AMI. In addition, the loading controls used in the miRNA array might also affect the results of miRNA expression. For example, total RNA is often used as the loading control in the miRNA microarray, whereas U6 is typically used as the loading control in qRT-PCR. However, under some pathological conditions, the rate of degradation of total RNA is not consistent with that of U6. Our data indicate that after AMI, the degradation of U6 is relatively slower than that of total RNA.4 Thus, selection of a correct loading control under different pathological conditions is still an unsolved issue in miRNA assay.
In the current study, one remarkable result is that miR-21 is significantly increased in the border area of the infarcted hearts at both 6 and 24 h after AMI. In contrast, its expression is down-regulated in the infarcted area. Moreover, IP, a well established cardiac protective procedure, is able to reverse down-regulation of miR-21 in the infarcted area. To further determine the effects of IP on cardiac miRNA expression, a miRNA microarray was performed in rat hearts at 6 h after IP. Interestingly, compared with expression changes in the infarcted areas, IP elicited an opposite change in the expression of miR-21, mir-16, miR-26a, and miR-352. In contrast, IP elicited a similar change in the expression of miR-320, miR-7e, miR-214, miR-126, and miR-499, compared with that in the infarcted areas (see Tables 1 and 3). The results indicated that these aberrantly expressed miRNAs in infarcted hearts may have either anti- or pro-injury effects on cardiac cells under ischemic conditions. Our recent studies (24, 25) have revealed that miR-21 has a strong anti-apoptotic effect on vascular smooth muscle cells and in cultured heart cells. To test the potential role of miR-21 in AMI, adenovirus-mediated miR-21 gene transfer was applied. We found that overexpression of miR-21 significantly decreases the myocardial infarct size in the rat model. Although the extent of the decrease in myocardial infarct size is modest, the modulation of one single miRNA that caused such a reduction in myocardial infarct size indicates that miRNAs may play an important role in the pathophysiology of AMI.
To further determine the cellular mechanism in miR-21-mediated effects on AMI, an ischemia-injury cardiac cell model was applied. Using both gain-of-function and loss-of-function approaches, we have identified that miR-21 has an anti-apoptotic effect on cardiac cells under ischemia-related injury in vitro. Moreover, the anti-apoptotic effect of miR-21 on cardiac cells has been confirmed in vivo in the infarcted heart slices from rats treated with Ad-miR-21. The reduced myocardial size and cell apoptosis following Ad-miR-21 treatment resulted in the long-term remodeling of the hearts as shown by the protective effect on LV internal dimension enlargement. Cardiac function should be measured in future studies to evaluate the miR-21-mediated functional outcome. Lack of functional data is a limitation of the current study.
During manuscript preparation, we were happy to see another independent group report their findings about the role of miR-21 on myocardial infarction in an in vitro model (19). Our study used an in vivo AMI model that is different from their study. In addition, we have identified the early expression signature of miRNAs in the different zones of the infarcted hearts. Moreover, we have demonstrated that a novel signaling pathway, miR-21/PDCD4/AP-1, may be involved in miR-21-mediated cardiac protection. Thus, our study has provided further information in this new research area. More importantly, the reporting of a protective effect of miR-21 on cardiac injury from our two independent groups indicated that miR-21 is indeed a critical miRNA related to cardiac protection.
miRNAs modulate biological functions via their multiple target gene mRNAs. Although their potential gene targets can be predicted by computational analysis, these targets must be experimentally verified in investigatory cells because miRNA-mediated effects on gene expression and cellular functions are cell specific. Computational analysis suggests that PDCD4 may be a miR-21 target. In the current study, we have experimentally verified that PDCD4 is a target gene in cardiac cells. Moreover, we have identified, for the first time, that PDCD4 has a pro-apoptotic effect on cardiac cells. It is well established that AP-1 is a key signaling molecule that determines life or death cell fates in response to extracellular stimuli (30). In the current study, we have identified that AP-1 is associated with PDCD4, which may be involved in miR-21-mediated effects on cardiac cells.
In summary, miRNA signatures in the early phase of AMI have revealed that multiple miRNAs are aberrantly expressed. Among them, miR-21 has a protective effect on myocardial infarction by reducing cardiac cell apoptosis via its target gene PDCD4. The results suggest that miRNAs may play important roles in the pathophysiology of AMI. Some miRNAs may be considered new therapeutic targets or even biomarkers for ischemic heart disease such as AMI.
Acknowledgment
We thank Dr. Sheldon Goldstein in our department for editing assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant HL080133 and American Heart Association Grant 09GRNT2250567.
S. Dong, Y. Cheng, J. Yang, J. Li, X. Liu, X. Wang, D. Wang, T. J. Krall, E. S. Delphin, and C. Zhang, unpublished data.
- miRNA
- microRNA
- TTC
- triphenyltetrazolium chloride
- m.o.i.
- multiplicity of infection
- qRT
- quantitative reverse transcriptase
- pfu
- plaque-forming unit
- TUNEL
- terminal deoxynucleotide transferase dUTP nick end labeling
- AMI
- acute myocardial infarction
- IP
- ischemic preconditioning
- PDCD4
- programmed cell death 4
- Ad
- adenovirus
- GFP
- green fluorescent protein
- LV
- left ventricle
- H/R
- hypoxia/reoxygenation
- LNA
- lacked nucleic acid.
REFERENCES
- 1.Ambros V. (2003) Cell 113, 673–676 [DOI] [PubMed] [Google Scholar]
- 2.Farh K. K., Grimson A., Jan C., Lewis B. P., Johnston W. K., Lim L. P., Burge C. B., Bartel D. P. (2005) Science 310, 1817–1821 [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli A. E., Hunter S., Bracht J. (2005) Curr. Opin. Genet. Dev. 15, 200–205 [DOI] [PubMed] [Google Scholar]
- 4.Zaug A. J., Cech T. R. (1986) Science 231, 470–475 [DOI] [PubMed] [Google Scholar]
- 5.Kong Y., Han J. H. (2005) Genomics Proteomics Bioinformatics 3, 62–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee R. C., Feinbaum R. L., Ambros V. (1993) Cell 75, 843–854 [DOI] [PubMed] [Google Scholar]
- 7.Wightman B., Ha I., Ruvkun G. (1993) Cell 75, 855–862 [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. (2001) Science 294, 853–858 [DOI] [PubMed] [Google Scholar]
- 9.Kim V. N. (2005) Nat. Rev. Mol. Cell Biol. 6, 376–385 [DOI] [PubMed] [Google Scholar]
- 10.Kim Y. K., Kim V. N. (2007) EMBO J. 26, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruby J. G., Jan C. H., Bartel D. P. (2007) Nature 448, 83–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentwich I., Avniel A., Karov Y., Aharonov R., Gilad S., Barad O., Barzilai A., Einat P., Einav U., Meiri E., Sharon E., Spector Y., Bentwich Z. (2005) Nat. Genet. 37, 766–770 [DOI] [PubMed] [Google Scholar]
- 13.Berezikov E., Guryev V., van de Belt J., Wienholds E., Plasterk R. H., Cuppen E. (2005) Cell 120, 21–24 [DOI] [PubMed] [Google Scholar]
- 14.Lewis B. P., Burge C. B., Bartel D. P. (2005) Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 15.Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B., Zhang Y., Xu C., Bai Y., Wang H., Chen G., Wang Z. (2007) Nat. Med. 13, 486–491 [DOI] [PubMed] [Google Scholar]
- 16.Ikeda S., Kong S. W., Lu J., Bisping E., Zhang H., Allen P. D., Golub T. R., Pieske B., Pu W. T. (2007) Physiol. Genomics 31, 367–373 [DOI] [PubMed] [Google Scholar]
- 17.Wang S., Aurora A. B., Johnson B. A., Qi X., McAnally J., Hill J. A., Richardson J. A., Bassel-Duby R., Olson E. N. (2008) Dev. Cell 15, 261–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Rooij E., Sutherland L. B., Thatcher J. E., DiMaio J. M., Naseem R. H., Marshall W. S., Hill J. A., Olson E. N. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13027–13032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin C., Wang X., Kukreja R. C. (2008) FEBS Lett. 582, 4137–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckle T., Grenz A., Köhler D., Redel A., Falk M., Rolauffs B., Osswald H., Kehl F., Eltzschig H. K. (2006) Am. J. Physiol. Heart Circ. Physiol. 291, H2533–H2540 [DOI] [PubMed] [Google Scholar]
- 21.Tang X. L., Sato H., Tiwari S., Dawn B., Bi Q., Li Q., Shirk G., Bolli R. (2006) Am. J. Physiol. Heart Circ. Physiol. 291, H2308–H2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolakowski S., Jr., Berry M. F., Atluri P., Grand T., Fisher O., Moise M. A., Cohen J., Hsu V., Woo Y. J. (2006) J. Card. Surg. 21, 559–564 [DOI] [PubMed] [Google Scholar]
- 23.Zheng B., Cao L. S., Zeng Q. T., Wang X., Li D. Z., Liao Y. H. (2004) Basic Res. Cardiol. 99, 264–271 [DOI] [PubMed] [Google Scholar]
- 24.Cheng Y., Liu X., Zhang S., Lin Y., Yang J., Zhang C. (2009) J. Mol. Cell. Cardiol. 47, 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji R., Cheng Y., Yue J., Yang J., Liu X., Chen H., Dean D. B., Zhang C. (2007) Circ. Res. 100, 1579–1588 [DOI] [PubMed] [Google Scholar]
- 26.Hajjar R. J., Schmidt U., Matsui T., Guerrero J. L., Lee K. H., Gwathmey J. K., Dec G. W., Semigran M. J., Rosenzweig A. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5251–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho M. A., Lee M. K., Nam K. H., Chung W. Y., Park C. S., Lee J. H., Noh T., Yang W. I., Rhee Y., Lim S. K., Lee H. C., Lee E. J. (2007) J. Endocrinol. 195, 255–263 [DOI] [PubMed] [Google Scholar]
- 28.Hwang S. K., Jin H., Kwon J. T., Chang S. H., Kim T. H., Cho C. S., Lee K. H., Young M. R., Colburn N. H., Beck G. R., Jr., Yang H. S., Cho M. H. (2007) Gene Ther. 14, 1353–1361 [DOI] [PubMed] [Google Scholar]
- 29.Burke A. P., Virmani R. (2007) Med. Clin. North Am. 91, 553–572 [DOI] [PubMed] [Google Scholar]
- 30.Shaulian E., Karin M. (2001) Oncogene 20, 2390–2400 [DOI] [PubMed] [Google Scholar]