Abstract
Growth-blocking peptide (GBP) is a hormone-like peptide that suppresses the growth of the host armyworm. Although the 23-amino acid GBP (1–23 GBP) is expressed in nonparasitized armyworm plasma, the parasitization by wasp produces the 28-amino acid GBP (1–28 GBP) through an elongation of the C-terminal amino acid sequence. In this study, we characterized the GBP variants, which consist of various lengths of the C-terminal region, by comparing their biological activities and three-dimensional structures. The results of an injection study indicate that 1–28 GBP most strongly suppresses larval growth. NMR analysis shows that these peptides have basically the same tertiary structures and that the extension of the C-terminal region is disordered. However, the C-terminal region of 1–28 GBP undergoes a conformational transition from a random coiled state to an α-helical state in the presence of dodecylphosphocholine micelles. This suggests that binding of the C-terminal region would affect larval growth activity.
Growth-blocking peptide (GBP)2 was initially identified from the hemolymph of armyworm Pseudaletia separata as a 25-amino acid peptide (1–25 GBP) that prevents the onset of pupation of the host by parasitization of wasp Cotesia kariyai (1–4). Injection of GBP into nonparasitized armyworm larvae early in the last instar delays larval growth and retards pupation for more than a few days. Our previous studies showed that GBP is a hormone-like biogenic peptide of the host armyworm (5, 6). In nonparasitized larvae, the concentrations of GBP were much higher in the early larval stages than in the latter ones. However, parasitization by wasp induces an elevation of GBP in the last larval stages. This elevation was shown to lead to growth retardation via repression of juvenile hormone esterase activity (7–9). Interestingly, a cDNA analysis indicated that the cDNA encodes a 23-amino acid GBP (1–23 GBP), although GBP purified from parasitized armyworm plasma consists of 25 amino acid residues. GBP was expressed as a 23-residue peptide (1–23 GBP) in nonparasitized armyworm larvae, whereas 1–25 GBP, containing Tyr24 and Gln25, was purified from the parasitized larvae. Moreover, the preliminary peptide sequencing of GBP prepared from parasitized larval hemolymph showed the 26th and 27th residues on rare occasions (Leu and Ile, respectively) (6). On the basis of these results, we concluded that the TAG stop codon for the 24th amino acid was unusually decoded as Tyr upon parasitization by parasitoid wasps (10) and predicted that an intact and mature GBP synthesized in the parasitized armyworm larvae would consist of 28 amino acid residues (1–28 GBP).
GBP has multiple functions: adhesion and spreading of a specific class of immune cells (plasmatocytes), proliferation of various cultured cells, and induction of larval paralysis (11–13). More than 10 GBP homologous peptides have been identified in Lepidopteran insects, and based on their N-terminal consensus sequences (Glu1-Asn2-Phe3), they have been categorized as the ENF peptide family (14). The tertiary structure of 1–25 GBP consists of a disordered N-terminal region (residues Glu1–Gly6), a well ordered core region (residues Cys7–Thr22) stabilized by a disulfide bond and a short antiparallel β-sheet, and a short unstructured C-terminal region (Phe23–Glu25) (15). Because no GBP receptor or its gene has been isolated yet, the nature of either of them at the cellular and molecular levels is poorly understood at present. In contrast, the relationship between the structure and activity of GBP has been well studied by analyzing the biological activities of several variants of GBP and plasmatocyte-spreading peptide (one of the ENF family peptides). Especially, extensive studies on the N termini (residues 1–6) of GBP and plasmatocyte-spreading peptide demonstrated the importance of Phe3 for exerting their hemocyte stimulating activity, thereby suggesting a possible mechanism for receptor activation that requires binding of the aromatic ring of Phe3 and a closely spaced primary amine with receptor activating properties (16–19).
In contrast, the C termini of GBP and other ENF peptides have received less attention, because of the weak secondary structure predictions. Therefore, in this study we focused on the C terminus region of GBP and analyzed its contribution to the expression of some biological activities and to the tertiary structure of this peptide. Especially, we prepared GBP with 28 amino acids and characterized the C-terminal region of 1–28 GBP (residues Phe23–Thr28), because we knew that GBP is present as a 23-amino acid peptide in nonparasitized healthy larvae and that GBP with 28 amino acids has been found only in parasitized host larvae. Our results suggest that the elongation of the C-terminal region of Phe23–Thr28 greatly reinforced GBP binding with the membrane. Further, the elongation increased GBP inhibition of larval growth.
EXPERIMENTAL PROCEDURES
Animals
P. separata was reared on an artificial diet at 25 ± 1 °C with a photoperiod of 16 h of light and 8 h of dark (1). The penultimate instar larvae undergoing ecdysis between 2 and 2.5 h after lights on were designated as day 0 last instar larvae (1).
Preparation and Identification of GBPs
All of the GBPs used in this study were prepared by the Escherichia coli expression system. GBPs (Table 1) were expressed and purified using our previous procedure (20). A cDNA encoding the entire GBP sequence was used as a template, and polymerase chain reactions were performed with each GBP primer. GBPs were expressed as thioredoxin fusion proteins and purified with nickel-nitrilotriacetic acid-agarose (Qiagen). 15N labeling was achieved by growing the bacteria in minimal medium containing 15NH4Cl as the sole nitrogen source. The purified proteins were cleaved by enterokinase enzyme (Invitrogen) and further purified by reverse phase high performance liquid chromatography (RP-HPLC). The peptides after HPLC purification were judged to be homogeneous after a second analysis by RP-HPLC, which found that the purity of each analogue was above 95%. The purified peptides were further confirmed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS).
TABLE 1.
Sequences of 1–23, 1–25, and 1–28 GBP
| 1–23 GBP | ENFSG GCVAG YMRTP DGRCK PTF |
| 1–25 GBP | ENFSG GCVAG YMRTP DGRCK PTFYQ |
| 1–28 GBP | ENFSG GCVAG YMRTP DGRCK PTFYQ LIT |
Peptide Degradation Assay
GBPs were dissolved at a concentration of 0.4 mm in 60 μl of 50 mm phosphate buffer at pH 6.6. These solutions were incubated in 100 μl of the plasma isolated from insect hemolymph at 25 °C. After incubation, the peptide samples were submitted to RP-HPLC analysis on a 5C18-AR-300 column (Nacalai Tesque; 250 × 10 mm) using a linear gradient of acetonitrile/trifluoroacetic acid (99.9/0.1 v/v). Fractions corresponding to each peak were collected and subjected to MALDI-TOF MS analysis.
Experimental Injection of GBP Variants into Larvae
The experimental injection method was a modification of the previously published method of Hayakawa and Ohnishi (11). Day 0 last instar larvae (0.17–0.18 g) of armyworm were injected with the indicated amounts of GBPs early in the last larval instar and weighed at 24 h after injection.
NMR Spectroscopy
To elucidate the structure, each GBP was dissolved in 250 μl of 90% H2O, 10% D2O to give a final concentration of 3 mm (pH 4.4 and 6.8). NMR samples of 1–28 GBP bound to DPC micelles were prepared under the same conditions containing 200 mm d38-DPC. The NMR experiments were carried out on either a JEOL ECA 600 MHz or a Bruker DRX 500 MHz spectrometer. All of the NMR spectra were recorded at 10, 20, or 30 °C. The chemical shifts were measured from the internal standard of sodium 2,2-dimethyl-2-silapentane-5-sulfonate (The chemical shifts have been submitted to the Biological Magnetic Resonance Bank (BMRB) under accession numbers 11068, 11069, and 11070.). At various peptide concentrations (0.1–3 mm) and pH levels (pH 4.4 and 6.8), there were no meaningful changes in chemical shifts or peak line widths of 1H one-dimensional NMR spectra. Double quantum filtered correlation spectroscopy spectra (21), total correlation spectroscopy spectra (22), and NOE spectroscopy spectra (23) with mixing times of 100–300 ms were recorded during suppression of the water signal by a water gate pulse (24). The NMR spectra of the 15N-labeled sample of 1–28 GBP were recorded under the same conditions as nonlabeled samples except for the peptide concentration of 1 mm using 1H-15N heteronuclear single quantum coherence (HSQC) (25), 15N-edited HSQC-total correlation spectroscopy (26), and 15N-edited HSQC-NOE spectroscopy (26). The data for the 1–28 GBP bound to DPC micelles were recorded under the same conditions containing 100 mm d38-DPC. The data were processed using NMRpipe 4.1 and NMRDraw 2.3 (27) and analyzed using Sparky 3.111 software (28). The average chemical shift differences in the amide proton and nitrogen resonances between 1–28 GBP in water and those in DPC micelles were calculated using the following equation (29),
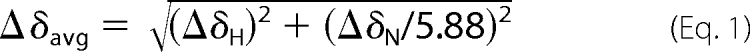 |
where ΔδH and ΔδN are the chemical shift differences of the amide proton and nitrogen resonances between 1–28 GBP in water and those in DPC micelles.
15N-{1H} NOE values were measured for each of the 15N amides using standard pulse sequences with a gradient at 500 MHz. 15N-Labeled 1–28 GBP was dissolved in 250 μl of 90% H2O, 10% D2O (in the absence or presence of 100 mm d38-DPC) to give a final concentration of 1 mm (pH 4.4). 15N-{1H} NOE values are reported as peak intensity ratios obtained with and without 1H saturation.
Structure Calculations
The three-dimensional structures were calculated in the program ARIA 1.2 (30) implemented in CNS 1.1 (31). Angle restraints were derived from an analysis of the 3JNHα coupling constant, and the minimum range employed for ϕ torsion angle restraints was ±30°. The hydrogen bonding pattern was included as a constraint for secondary structure elements, where the proximity and orientation between amide protons and carbonyl groups could be determined. The 20 refined structures with the lowest energy contributions from intramolecular interactions were chosen to represent the NMR ensemble. These final ensembles of accepted structures satisfy the following criteria: no NOE violations greater than 0.5 Å and no RMSD for angle deviations from the ideal greater than 5°. The quality of the final structures was assessed using the program PROCHECK-NMR (32). The conformers, including calculation of root mean square deviation values, were analyzed by the MOLMOL program (33).
Microfiltration Assay
GBPs were mixed with dodecylphosphocholine micelles to a final concentration of 150 μm GBP and 50 mm DPC micelles in 10 mm phosphate buffer (pH 6.8). Samples (400 μl) were incubated at room temperature for 30–60 min, then layered on filters (10,000 molecular weight cut-off), and spun at 2000 × g in a microcentrifuge for 30 min. The concentrations of filtrated GBPs were determined by measuring the absorbance at 280 nm and compared with those of GBPs not exposed to DPC micelles.
Surface Plasmon Resonance Experiments
Small unilamellar vesicles of 1 mm dimyristoylphosphatidylcholine were prepared in 20 mm phosphate buffer (pH 6.8) by sonication. Biosensor experiments were carried out with a BIACORE X analytical system (Biacore AB, Uppsala, Sweden) using an L1 sensor chip. The running buffer used for all of the experiments was 20 mm phosphate buffer. The surface was washed by injecting 25 μl of 40 mm N-octyl β-d-glucopyranoside at a flow rate of 5 μl/min. Small unilamellar vesicles (80 μl) were immediately applied to the sensor chip surface at a flow rate of 5 μl/min. The multilamellar structures were removed from the lipid surface by injecting 30 μl of 10 mm sodium hydroxide at a flow rate of 50 μl/min, in a stable base line. The negative control bovine serum albumin (10 μl, 0.1 mg/ml in 20 mm phosphate buffer) was injected to confirm the complete coverage of the chip surface with lipid by the absence of nonspecific binding. Peptide solutions (25 μl, 150 μm in 20 mm phosphate buffer) were injected for 300 s at a flow rate of 5 μl/min. Upon completion of injection, buffer flow was continued for 600 s to allow for dissociation. The operating temperature was 25 °C.
Hydrogen-Deuterium (H/D) Exchange
To measure the H/D exchange of the amide protons, an ∼1 mm 15N-labeled 1–28 GBP sample was dissolved in 100% D2O in the presence of 100 mm d38-DPC, and 1H-15N HSQC spectra were recorded at different time intervals. HSQC peaks were monitored for disappearance at 15, 25, 35, and 45 min upon the addition of the solvent.
Spin Label Experiments
The DPC-bound and water-accessible residues of 1–28 GBP in DPC micelles were identified by measuring the effect of 5-doxyl stearic acid (5-DSA) on the intensities of peaks in 1H one-dimensional spectra or 1H-15N HSQC spectra. In 100 μl of 30% DPC solution, 1.5 mg of 5-DSA was solubilized. The 5-DSA solution was titrated to 1 mm unlabeled (in 100% D2O) or 15N-labeled 1–28 GBP (in 90% H2O/10% D2O) in the presence of 100 mm d38-DPC. The one-dimensional spectra and HSQC spectra were collected for each point, and the intensities were compared. The intensity reductions of HSQC spectra caused by close proximity to the spin label were calculated as the ratios of peak intensities in spectra from the sample containing the respective spin labels to the peak intensities derived from the same sample before adding the probes. The 5-DSA did not alter the protein structure, as evidenced by the absence of chemical shift perturbations.
RESULTS AND DISCUSSION
Peptide Degradation Assay
GBP has been purified for the first time from the hemolymph of the host armyworm. Follow-up studies demonstrated that GBP is synthesized as a propeptide precursor that is processed by processing enzyme(s) in hemolymph. After processing, the matured GBP must be exposed to various endogenous proteases in vivo. We expected to find differences in the half-lives of the GBPs (1–23 GBP, 1–25 GBP, and 1–28 GBP). The difference in the degradation kinetics of GBP in insect hemolymph may be involved with their growth inhibitory activities. For this reason, we analyzed their half-lives by RP-HPLC of the peptides after incubating them with plasma fractions prepared from the armyworm larvae.
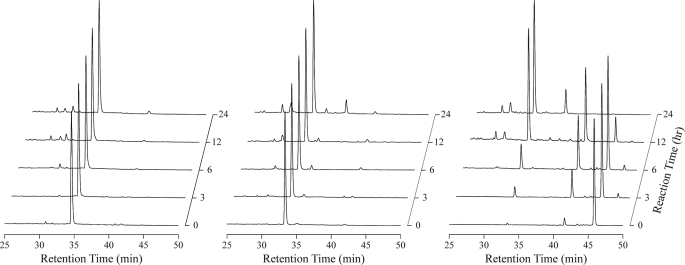
Fig. 1 shows chromatograms after incubation of recombinant GBP peptides with the plasma preparation without blood cells in vitro. The retention times of 1–23 GBP, 1–25 GBP, and 1–28 GBP were 34, 33, and 45 min, respectively. The chromatograms of 1–23 GBP and 1–25 GBP show that these peptides are hardly degraded in plasma for 24 h. In contrast, 1–28 GBP was degraded immediately. The degradation products were observed at the retention times of 35 and 40 min. The fractions corresponding to each peak were 1–25 GBP and 1–26 GBP by MALDI-TOF MS analysis. These results indicate that the elongated C-terminal regions were highly susceptible to degradation in insect plasma.
FIGURE 1.
Peptide degradation assay. RP-HPLC chromatograms of 1–23 GBP (left panel), 1–25 GBP (middle panel), and 1–28 GBP (right panel) dissolved in insect plasma. Chromatograms were acquired 0–24 h after incubation.
Our previous studies suggested that the 24th stop codon would be decoded as Tyr unconventionally by polydnavirus infection in the same manner as tobacco mosaic virus infection (34, 35) and that the mature GBP in armyworm plasma would consist of 28 amino acids in the parasitized larvae (6). However, GBP purified from the plasma of the parasitized larvae was dominantly 1–25 GBP. Judging from the results of the present study, 1–25 GBP in parasitoid armyworm plasma would be the degradation product of 1–28 GBP. The Leu26-Ile27-Thr28 residues of 1–28 GBP are expected to be immediately degraded to 1–25 GBP by a protease in the hemolymph of the larvae.
Larval Growth Activity of GBP Variants
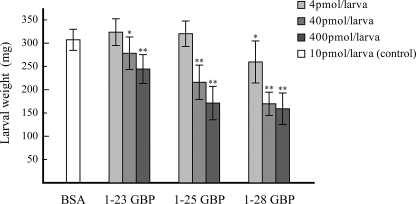
To reveal the effect of the elongated C-terminal region on the biological activities of GBP, we compared three peptide variants in terms of larval growth retardation. In the previous study, the injection of 1–25 GBP into the armyworm P. separata larvae dose-dependently reduced larval growth rates (1, 2). Therefore, we compared the weights of GBP-injected larvae with those of control larvae injected with bovine serum albumin. Larval growth was clearly inhibited by 40 or 400 pmol of 1–25 GBP, whereas larvae injected with 4 pmol exhibited no reduction in weight gain relative to control larvae (Fig. 2). On the other hand, larvae injected with 40 or 400 pmol of 1–23 GBP also gained less weight than control larvae but gained more weight than larvae injected with 1–25 GBP. Interestingly, larvae injected with 1–28 GBP even at 4 pmol gained significantly less weight than control larvae. The injection with 40 or 400 pmol of 1–28 GBP completely inhibited larval growth for at least 24 h after injection. Therefore, 1–28 GBP exhibited growth blocking activity 100-fold greater than 1–23 GBP, although the C-terminal region of 1–28 GBP was readily degraded in insect plasma.
FIGURE 2.
Biological activities of GBPs. The effect of each GBP on weight gain in P. separata is shown. Larvae injected with 10 pmol of bovine serum albumin (BSA) served as negative controls. Each bar indicates the mean percentage ± S.D. of weight gain of the treated larva (n = 14–17). The asterisks indicate significant differences between GBP-injected larvae and control (bovine serum albumin-injected) larvae (t test). *, p < 0.01; **, p < 0.001.
NMR Analysis of GBPs in Aqueous Solution
The NMR structure of 1–25 GBP in aqueous solution was previously reported (15). The core structure of 1–25 GBP consists of a short segment of double stranded β-sheet in the region of Tyr11–Arg13 and Cys19–Pro21, a type II β-turn in the region of Val8–Tyr11, and a type I β-turn in the region of Thr14–Gly17. To investigate the structural features of the C-terminal region, we assigned the chemical shifts of 1–23 GBP and 1–28 GBP. The NMR signals of the backbone protons for 1–23 GBP and 1–28 GBP were completely assigned.
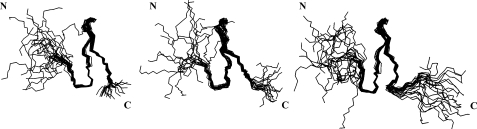
A structural calculation based on NOE data, reported in Table 2, was performed. Superpositions of the backbones of the 20 lowest energy structures of GBPs are shown in Fig. 3. The secondary structures of 1–23 GBP and 1–28 GBP each consist of an antiparallel β-sheet in the regions of Tyr11–Arg13 and Cys19–Pro21. Although there are differences in the lengths of the C termini of the GBPs, the core structures of these three peptides are extremely similar. The C-terminal region of GBP seems to have an insignificant effect on the structure in the core region because the elongated C-terminal region of 1–28 GBP is unstructured in water. Moreover, this structural information suggested that the C-terminal region of 1–28 GBP will be easily degraded by the various proteases in armyworm plasma.
TABLE 2.
Structural statistics for the final 20 structures of GBP without micelles
| 1–23 GBP | 1–28 GBP | |
|---|---|---|
| Number of experimental restraints | ||
| Total unambiguous NOEs | 326 | 543 |
| Intraresidue | 139 | 214 |
| Sequential | 90 | 159 |
| Medium range | 31 | 65 |
| Long range | 66 | 105 |
| Total hydrogen bond restraints | 18 | 14 |
| Dihedral angle restraints | ||
| ϕ | 15 | 20 |
| Average pairwise RMSD (Å) | ||
| All backbone atoms (residues 7–22) (Å) | 0.53 ± 0.16 | 0.55 ± 0.19 |
| All heavy atoms (residues 7–22) (Å) | 1.08 ± 0.24 | 1.36 ± 0.28 |
| RMSD from experimental data | ||
| Distances (Å) | 0.015 ± 0.003 | 0.017 ± 0.003 |
| Dihedral angles (°) | 0.770 ± 0.212 | 0.367 ± 0.177 |
| Most favored regions (%) | 81.7 | 73.8 |
| Additional allowed regions (%) | 17.7 | 25.5 |
| Generously allowed regions (%) | 0.7 | 0.8 |
| Disallowed regions (%) | 0.0 | 0.0 |
FIGURE 3.
Solution structures of GBPs. The 20 best energy-minimized conformers representing the solution structures of 1–23 GBP (left panel), 1–25 GBP (middle panel), and 1–28 GBP (right panel). The backbone atoms of residues 7–22 were used for the superimposition. The drawing of 1–25 GBP was made using the atomic coordinates obtained from the Protein Data Bank (code 1BQF).
Structural Changes in 1–28 GBP in Membrane Mimetic Environment
Despite the rapid degradation of the elongated C terminus in insect plasma, 1–28 GBP strongly inhibited the armyworm larval growth. What part does the C-terminal region of 1–28 GBP play in the growth inhibitory activity? Gololobov et al. (36) studied a membrane-binding peptide, vasoactive intestinal peptide, and proposed that the interactions of peptides with membrane components protect the peptides from degradation in vivo. This result leads us to hypothesize that the elongation of the C terminus of the 1–28 GBP interacts with insect membrane components and is protected from degradation.
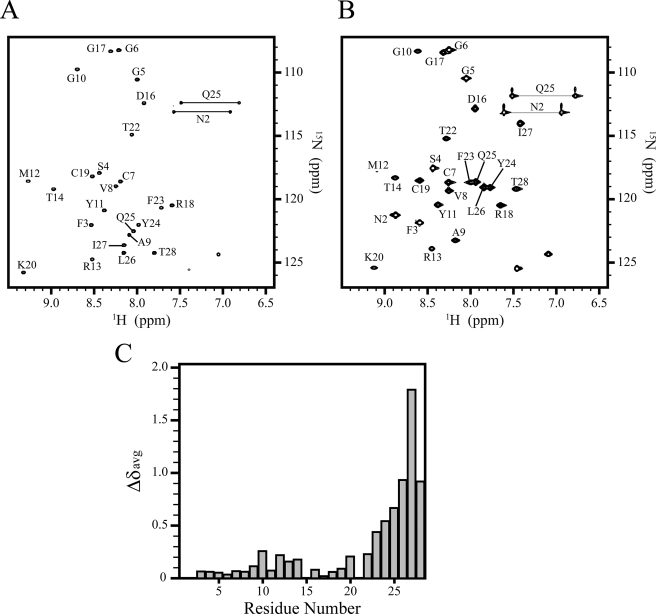
To verify this hypothesis, we tested the GBPs for the ability to interact with dodecylphosphocholine (DPC) micelles. DPC is a well characterized model membrane system used frequently in solution NMR studies of peptide-membrane interactions (37). DPC has a neutral head group and mimics the anisotropic environment of a lipid membrane. DPC forms a stable micelle with a molecular mass of ∼20 kDa, resulting in reasonable correlation time and manageable line width for studies utilizing solution NMR. However, compared with a planar membrane, a micelle exhibits a rather different, highly curved surface. Fig. 4 (A and B) shows the 1H-15N HSQC spectrum of 1–28 GBP in the absence or presence of DPC micelles at 20 °C. Broader cross-peaks were observed in the C-terminal region in DPC micelles. The chemical shift differences of the backbone amide proton and nitrogen resonances between aqueous solution and DPC micelles were calculated (Fig. 4C). Although the chemical shift differences of the N-terminal and core residues hardly changed, the C-terminal residues represented large chemical shift perturbations.
FIGURE 4.
NMR spectra and chemical shift differences of 1–28 GBP. A and B, 1H-15N HSQC spectra in aqueous solution, pH 4.4, 20 °C (A) and in the presence of DPC, pH 4.4, 20 °C (B). Spectral cross-peaks are labeled by a one-letter amino acid code and residue number. The NMR signal of Asn2 of 1–28 GBP in water was too broad because of the amide proton exchange in this condition. C, the weighted average 1H/15N backbone amide chemical shift changes of 1–28 GBP upon binding to DPC micelles.
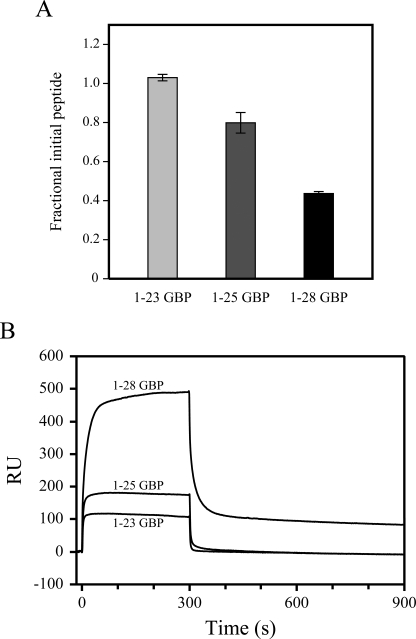
To confirm the interaction between GBPs and DPC micelles, we performed microfiltration assays (38). The relative ratios of the filtrate concentrations of GBP variants in the absence and presence of DPC micelles are shown in Fig. 5A. In this experiment, free GBPs (<3,000 Da) passed through an ultra-filtration membrane (10,000 molecular weight cut-off), whereas GBPs bound to DPC micelles (>20,000 Da) could not pass. The filtrate concentrations of 1–23 GBP are almost the same in the absence of DPC micelles as in their presence. In contrast, the concentration of filtrated 1–28 GBP was significantly decreased in the presence of DPC micelles. This shows the strong binding between DPC micelles and 1–28 GBP. Similarly, the filtrate concentration of 1–25 GBP was reduced in the presence of DPC micelles. 1–25 GBP exhibits a lower affinity for DPC micelles than 1–28 GBP. These results suggest that the binding affinities of GBPs for membrane were enhanced according to the length of the C terminus.
FIGURE 5.
Microfiltration assay and surface plasmon resonance sensorgrams. A, binding of GBP variants to DPC micelles in PBS as measured by microfiltration assay. The peptide concentrations of the filtration samples were determined by measuring the absorbance at 280 nm and compared with that of GBP variants not exposed to DPC micelles. B, representative surface plasmon sensorgrams of 150 μm solutions of GBP variants, pH 6.8, 20 mm phosphate buffer at 25 °C on a dimyristoylphosphatidylcholine-coated surface.
We utilized surface plasmon resonance experiments to confirm the binding of GBP variants to gain a better understanding of the membrane system. Fig. 5B shows the comparative sensorgram obtained for each peptide with immobilized lipids. The sensorgram for 1–28 GBP showed markedly higher response levels than that for 1–23 or 1–25 GBP. As reported by other groups, sensorgram of peptides bound to the lipid surface generally show complex biphasic patterns for both binding and dissociation because the peptides first bind to the lipid head groups and then become inserted further into the hydrocarbon region of the lipid bilayer (39–41). The initial association of the membrane-binding peptide to the immobilized lipid surface begins as a very fast process. Then the peptide continues to accumulate on the lipid surface during the entire injection phase without reaching the equilibrium level. At the end of the peptide injection, the signal decreases very quickly in the initial dissociation phase because the running buffer flow removes a large amount of free or weakly bound peptide. The dissociation is followed by a much slower step. Typically, peptide sensorgrams do not return to the base line within the dissociation phase, indicating that a proportion of the peptide remains inserted into the inner surface of the immobilized lipid bilayer (39). In our surface plasmon resonance experiments, this was the case for 1–28 GBP-lipid interaction. In contrast, the sensorgrams of 1–23 GBP and 1–25 GBP were simple and clearly returned to zero. A comparison of the results of GBP variants suggests that the elongation C-terminal region allows it to bind more strongly to immobilized lipid, enhancing the subsequent hydrophobic binding and that the absence of C-terminal residues results in low binding affinities of these peptides with lipid.
To obtain structural information of 1–28 GBP in DPC micelles, distance restraints were obtained from the 1H NOE spectra recorded at 30 °C (Table 3). As with the solution structure, the long range NOEs between Tyr11–Arg13 and Cys19–Pro21 were also observed in the micelle environment. These data indicate that 1–28 GBP in water and that in DPC micelles have very similar core structures. On the other hand, several contiguous helix-indicative NOEs were observed in the C-terminal region when compared with aqueous solution. Furthermore, α-protons of C-terminal residues show the same trend with the upfield shift. These results suggest the possibility that the C-terminal region forms an α-helix upon DPC micelle binding. Following these results, structural calculation was performed in the expectation of conformational changes of the C-terminal region in DPC micelles. The superposition of the NMR ensemble of 1–28 GBP bound to DPC micelles is displayed in Fig. 6. The structure calculation clearly indicated the presence of a helical region in the C terminus between Phe23 and Ile27.
TABLE 3.
Structural statistics for the final 20 structures of 1–28 GBP with DPC micelles
| Number of experimental restraints | |
| Total unambiguous NOEs | 558 |
| Intraresidue | 204 |
| Sequential | 168 |
| Medium range | 82 |
| Long range | 104 |
| Total hydrogen bonds restraints | 20 |
| Dihedral angle restraints | |
| ϕ | 20 |
| Average pairwise RMSD (Å) | |
| All backbone atoms (residues 7–22) (Å) | 0.80 ± 0.22 |
| All heavy atoms (residues 7–22) (Å) | 1.55 ± 0.28 |
| RMSD from experimental data | |
| Distances (Å) | 0.020 ± 0.002 |
| Dihedral angles (°) | 0.543 ± 0.211 |
| Ramachandran plot analysis (from PROCHECK) | |
| Most favored regions (%) | 85.8 |
| Additional allowed regions (%) | 12.5 |
| Generously allowed regions (%) | 1.2 |
| Disallowed regions (%) | 0.5 |
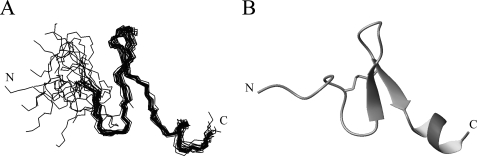
FIGURE 6.
Three-dimensional structures of 1–28 GBP bound to DPC micelles. A, ensemble of the 20 lowest energy structures of micelle-bound 1–28 GBP. The backbone atoms of residues 7–27 were used for the superimposition. B, secondary structure of micelle-bound 1–28 GBP of a representative conformer.
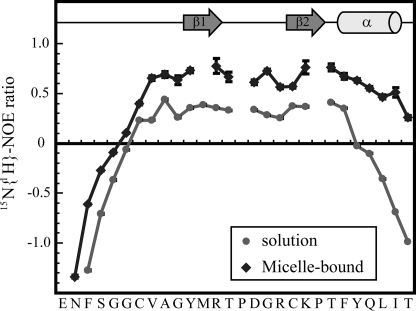
To characterize the dynamics of 1–28 GBP, we have also recorded 15N relaxation data for 1–28 GBP. A comparison of the 15N-{1H} NOE ratio of 1–28 GBP in solution with that in the micelle-bound state is shown in Fig. 7. 15N-{1H} NOE data have been used to characterize the backbone dynamics of proteins. Low 15N-{1H} NOE values represent the highly dynamic nature of the backbone, and stably folded regions in proteins generally exhibit NOE ratios above 0.5. In the C-terminal region, the flexibility of the backbone amide bond vectors continuously increased from Phe23 to Thr28 in aqueous solution. This indicates that the C-terminal region of 1–28 GBP has a highly dynamic nature in aqueous solution. On the other hand, the backbone is stabilized in the micelle-bound state, because is supported by the structure calculation in which a helix is formed in the C-terminal region. We concluded that the elongated C-terminal region undergoes a conformational transition from a random coil state to an α-helical state in the presence of micelles.
FIGURE 7.
15N relaxation data for 1–28 GBP in the absence or presence of DPC micelles. 15N-{1H} NOE values for 1–28 GBP in the absence (●) or presence (♦) of DPC micelles at 20 °C. Relaxation data were obtained at 500 MHz. The error bars indicate the standard deviation of the NOE intensity ratio determined from measured background noise levels. Residues with missing relaxation data include prolines and amides that display significantly weakened signals in the 1H-15N HSQC spectrum. The positions of the secondary structural elements are shown for the micelle-bound 1–28 GBP.
Topology of the Peptide-Micelle Interface
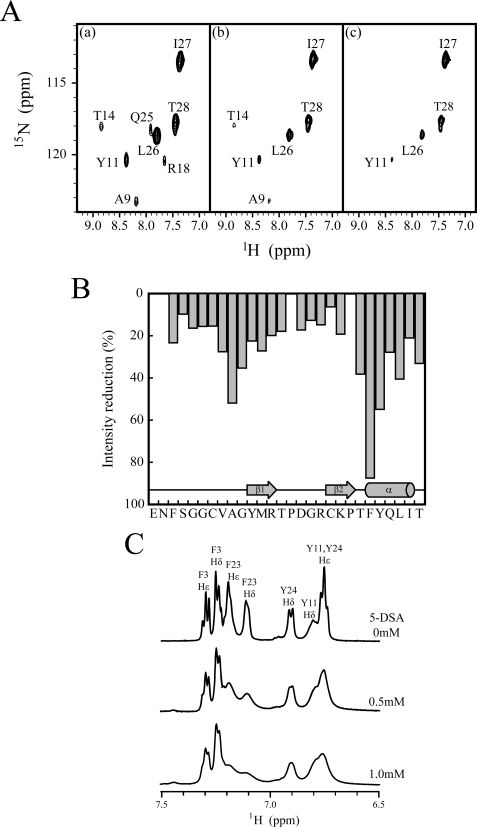
To investigate the details of 1–28 GBP association with DPC micelles, we performed H/D exchange experiments. These experiments have long been used to reveal that amide protons are involved in hydrogen bonds or are protected to a large extent from solvent access (42). Furthermore, these experiments have been recognized as a useful tool to determine the residues involved in the binding of peptides to DPC micelles. Amide protons from residues at the interface are protected from solvent and display reduced exchange rates. Fig. 8A shows the change in 1H-15N HSQC spectra in the presence of DPC micelles in 100% D2O. Nearly all peaks had vanished after 35 min, with the exception of the signals residues Ala9, Tyr11, The14, Leu26, Ile27, and Thr28. Residues Tyr11 and Thr14 formed hydrogen bonds to Val8 and Arg18 and exhibited slow H/D exchange rates because of hydrogen bond formation even in aqueous solution. The signals remaining after 45 min corresponded to residues Leu26, Ile27, and Thr28. These signals were not protected at all in aqueous solution. Interestingly, Ala9 is relatively inaccessible to D2O solvent in DPC micelles, although the amide proton of Ala9 is not involved in the hydrogen bond and rapidly decreases its signal intensity in water. These results suggest the direct interaction between DPC micelles and the core region (around Ala9) as well as the C-terminal region.
FIGURE 8.
H/D exchange experiments and spin label experiments of 1–28 GBP. A, representative 1H-15N HSQC spectra of 15N-labeled 1–28 GBP in 100 mm DPC. Spectra were acquired 25 min (panel a), 35 min (panel b), and 45 min (panel c) after the addition of D2O to a lyophilized sample. B, paramagnetic broadening effects of amide cross-peaks of 1H-15N HSQC spectra obtained for 1 mm 15N-labeled 1–28 GBP in 100 mm DPC micelles using 5-DSA. Intensity retention plot for 1–28 GBP in the presence of 1.5 mm of 5-DSA. C, paramagnetic line broadening effects in aromatic region of 1H one-dimensional spectra obtained for 1 mm unlabeled 1–28 GBP in 100 mm DPC micelles using 5-DSA.
To further define the membrane association of the 1–28 GBP with DPC micelles, we utilized the distance-dependent broadening effect of paramagnetic agents. The micelle-integrating spin label 5-DSA induces paramagnetic relaxation enhancements and causes the line broadening of NMR resonances of nuclei located in the proximity of the micelle surface. The broadening was detected as signal intensity changes of amide cross-peaks in 1H-15N HSQC spectra or aromatic cross-peaks in 1H one-dimensional spectra upon the stepwise addition of 5-DSA solution. The relative ratios of cross-peak intensities of 1–28 GBP in the absence or presence of the spin label are shown in Fig. 8B. When the 5-DSA was titrated to the 15N-labeled 1–28 GBP containing DPC micelles, the Ala9, Gly10, Thr22, Phe23, Tyr24, and Leu26 amide signal intensities were considerably reduced. This also shows the presence of the contacts between the core region (around Ala9) and DPC micelles. Fig. 8C shows the 1H one-dimensional spectra of the aromatic region of 1–28 GBP in the absence or presence of 5-DSA. Compared with Tyr11 and Tyr24, pronounced signal reductions were observed for Phe23 in the C-terminal helix, indicating that the aromatic side chain of Phe23 becomes integrated with the micelle interior. In addition, only weak signal reductions were observed for Phe3 in the disordered N terminus, which supports the view that the N-terminal region diffuses freely in solution.
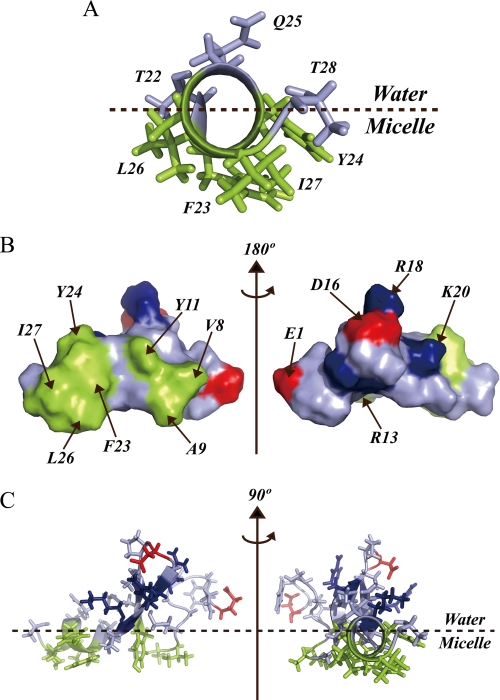
As shown, the conformational properties of 1–28 GBP are strongly influenced by mimics of the cell membrane. A structural model of micelle-bound 1–28 GBP is displayed in Fig. 9. The C-terminal amphipathic helix from residues Phe23 to Ile27 is placed at the micelle-water interface with hydrophobic side chains of the residues toward the micelle interior (Fig. 9A). There is good agreement between this model and the data on 1H one-dimensional spectra in spin label experiments. In addition to the C-terminal region, the side chains of residues Val8, Ala9, and Tyr11 form a hydrophobic patch. As the spin label experiments revealed, this region will make additional contacts with DPC micelles. Consequently, once the hydrophobic surface of the C-terminal helix comes in contact with the membrane, the formation of this hydrophobic patch is expected to enhance the peptide-micelle interaction (Fig. 9, B and C).
FIGURE 9.
Interface between 1–28 GBP and DPC micelles. A, helical region (Thr22–Thr28) is viewed along the helical axis. B, view of the surface model. C, the average structure of membrane-bound 1–28 GBP. Hydrophobic patch (Val8, Ala9, Tyr11, Phe23, Tyr24, Leu26, and Ile27), acidic (Glu1 and Asp16), and basic (Arg13, Arg18, and Lys20) amino acid residues are colored green, red, and blue, respectively.
Effects of Interaction between 1–28 GBP and Membrane on Growth Activity
Although the C-terminal region from residues Leu26 to Thr28 of 1–28 GBP is readily degraded in a peptide degradation assay in vitro, the growth inhibitory activity of 1–28 GBP on larvae was more effective than that of 1–25 GBP. In this study, we showed that the elongated C-terminal region of 1–28 GBP bound to membrane mimetic compounds and caused the formation of the α-helical domain from residues Phe23 to Ile27. In addition, the relative affinities of GBPs for membrane mimetic components agree better with their larval growth inhibitory activity. Our results suggest that the interaction between the C-terminal region of 1–28 GBP and the membrane would be involved in larval growth activity.
There are two possible explanations for why the affinity for membrane correlates with the ability of GBP to retard growth. One possibility is that membrane interaction retards the degradation of the 1–28 GBP C-terminal region elongation in vivo. An insect cell membrane will confer a helical characteristic on the C-terminal region of 1–28 GBP as well as a membrane mimetic component, DPC or dimyristoylphosphatidylcholine, and will trap 1–28 GBP molecules via this interaction. Molecular interaction between the C-terminal region of 1–28 GBP and the membrane may play a role in stabilizing 1–28 GBP from degradation. In the case of the purification of GBP from parasitized larvae, it is likely that 1–28 GBP cannot be purified from hemolymph because of membrane trapping.
Previous studies revealed that the GBP-binding protein, GBPBP, is released through hemocyte lysis and immediately scavenges free GBP molecules in hemocoel to prevent GBP from excessively stimulating the hemocytes (43). Membrane interaction induces the stabilization of GBP and is likely to protect GBP molecules from GBPBP and endogenous protease components. Moreover, if the hydrophobic patch in the core region of GBP is required for its efficient binding by GBPBP, the stable conformation in the presence of membrane components will limit GBPBP recognition of GBP. It can be hypothesized, therefore, that the interaction with the cell membrane contributes to the increase in the longevity of circulating 1–28 GBP as well as in the maintenance of biologically effective concentrations of 1–28 GBP or its degraded product, 1–25 GBP.
Another possibility is that 1–28 GBP membrane interaction precedes receptor binding as proposed by the membrane compartment model (44, 45). Kaiser and Kezdy (46, 47) studied biologically active peptides that act at the membrane and lipid-water interface and demonstrated the role of amphipathic cell membrane in influencing the structures of peptides. Schwyzer and co-workers (44, 45) developed the concepts of Kaiser and Kezdy into the membrane compartments model. Their model proposes that membrane interaction is an important event preceding receptor binding. Peptide-membrane interaction would induce an increase in concentration in the vicinity of the receptor and a reduction of the search for the receptor from three dimensions to two dimensions. Moreover, this interaction would induce a conformation more closely related to the active one, leading to reduced losses in entropy upon receptor binding. In the case of GBP, biological data in the larval growth assay are better correlated to trends observed in micelle binding affinities. Therefore, it is possible that the recognition of 1–28 GBP by the GBP receptor is enhanced because of the interaction between 1–28 GBP and the cell membrane. Although the GBP receptor is still unclear (48), GBP may target membrane-embedded receptors such as peptidergic G protein-coupled receptors, whose extracellular loops recognize the endogenous peptide.
In summary, the elongation of the C-terminal region of GBP is involved in larval growth inhibition. We determined the three-dimensional structures of 1–23 GBP and 1–28 GBP in aqueous solution and that of 1–28 GBP in DPC micelles. NMR analysis has suggested a structural explanation for the differences in the activities of GBP variants.
Acknowledgment
We thank the staff of the High Resolution NMR Laboratory of the Graduate School of Science of Hokkaido University for the NMR measurements.
This work was supported in part by KAKENHI Grant 19770123 and the Program for the Promotion of Basic Research Activities for Innovative Biosciences.
The atomic coordinates and structure factors (codes 2EQH, 2EQQ, and 2EQT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GBP
- growth-blocking peptide
- GBPBP
- GBP-binding protein
- DPC
- dodecylphosphocholine
- DSA
- doxylstearic acid
- RP-HPLC
- reverse phase high performance liquid chromatography
- NOE
- nuclear Overhauser effect
- HSQC
- heteronuclear single quantum coherence
- RMSD
- root mean square deviation
- MALDI-TOF MS
- matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- H/D
- hydrogen-deuterium.
REFERENCES
- 1.Hayakawa Y. (1990) J. Biol. Chem. 265, 10813–10816 [PubMed] [Google Scholar]
- 2.Hayakawa Y. (1991) J. Biol. Chem. 266, 7982–7984 [PubMed] [Google Scholar]
- 3.Hayakawa Y., Yasuhara Y. (1993) Insect Biochem. Mol. Biol. 23, 225–231 [Google Scholar]
- 4.Hayakawa Y. (1995) J. Insect Physiol. 41, 1–6 [Google Scholar]
- 5.Ohnishi A., Hayakawa Y., Matsuda Y., Kwon K. W., Takahashi T. A., Sekiguchi S. (1995) Insect Biochem. Mol. Biol. 25, 1121–1127 [DOI] [PubMed] [Google Scholar]
- 6.Hayakawa Y., Ohnishi A., Yamanaka A., Izumi S., Tomino S. (1995) FEBS Lett. 76, 185–189 [DOI] [PubMed] [Google Scholar]
- 7.Noguchi H., Hayakawa Y., Downer R. G. H. (1995) Insect Biochem. Mol. Biol. 25, 197–201 [Google Scholar]
- 8.Noguchi H., Hayakawa Y. (1996) Insect Biochem. Mol. Biol. 26, 659–665 [Google Scholar]
- 9.Hayakawa Y., Noguchi H. (1998) Eur. J. Biochem. 253, 810–816 [DOI] [PubMed] [Google Scholar]
- 10.Endo Y., Ohnishi A., Hayakawa Y. (1998) J. Insect Physiol. 44, 859–866 [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa Y., Ohnishi A. (1998) Biochem. Biophys. Res. Commun. 250, 194–199 [DOI] [PubMed] [Google Scholar]
- 12.Clark K. D., Pech L. L., Strand M. R. (1997) J. Biol. Chem. 272, 23440–23447 [DOI] [PubMed] [Google Scholar]
- 13.Skinner W. S., Dennis P. A., Li J. P., Summerfelt R. M., Carney R. L., Quistad G. B. (1991) J. Biol. Chem. 266, 12873–12877 [PubMed] [Google Scholar]
- 14.Strand M. R., Hayakawa Y., Clark K. D. (2000) J. Insect Physiol. 46, 817–824 [DOI] [PubMed] [Google Scholar]
- 15.Aizawa T., Fujitani N., Hayakawa Y., Ohnishi A., Ohkubo T., Kumaki Y., Kawano K., Hikichi K., Nitta K. (1999) J. Biol. Chem. 274, 1887–1890 [DOI] [PubMed] [Google Scholar]
- 16.Clark K. D., Volkman B. F., Thoetkiattikul H., Hayakawa Y., Strand M. R. (2001) J. Biol. Chem. 276, 37431–37435 [DOI] [PubMed] [Google Scholar]
- 17.Aizawa T., Hayakawa Y., Ohnishi A., Fujitani N., Clark K. D., Strand M. R., Miura K., Koganesawa N., Kumaki Y., Demura M., Nitta K., Kawano K. (2001) J. Biol. Chem. 276, 31813–31818 [DOI] [PubMed] [Google Scholar]
- 18.Tada M., Aizawa T., Shinohara Y., Matsubara K., Miura K., Yoshida M., Shitara K., Kouno T., Mizuguchi M., Nitta K., Hayakawa Y., Kawano K. (2003) J. Biol. Chem. 278, 10778–10783 [DOI] [PubMed] [Google Scholar]
- 19.Yoshida M., Aizawa T., Nakamura T., Shitara K., Hayakawa Y., Matsubara K., Miura K., Kouno T., Clark K. D., Strand M. R., Mizuguchi M., Demura M., Nitta K., Kawano K. (2004) J. Biol. Chem. 279, 51331–51337 [DOI] [PubMed] [Google Scholar]
- 20.Sekiya T. (1990) Tanpakushitsu Kakusan Koso. 35, 2977–2991 [PubMed] [Google Scholar]
- 21.Rance M., Sørensen O. W., Bodenhausen G., Wagner G., Ernst R. R., Wüthrich K. (1983) Biochem. Biophys. Res. Commun. 117, 479–485 [DOI] [PubMed] [Google Scholar]
- 22.Braunschweiler L., Ernst R. R. (1983) J. Magn. Reson. 53, 521–528 [Google Scholar]
- 23.Macura S., Huang Y., Suter D., Ernst R. R. (1981) J. Magn. Reson. 43, 259–281 [Google Scholar]
- 24.Piotto M., Saudek V., Sklenár V. (1992) J. Biomol. NMR 2, 661–665 [DOI] [PubMed] [Google Scholar]
- 25.Palmer A. G., 3rd, Cavanagh J., Wright P. E., Rance M. (1991) J. Magn. Reson. 93, 151–170 [Google Scholar]
- 26.Kay L. E., Marion D., Bax A. (1989) J. Magn. Reson. 84, 72–84 [Google Scholar]
- 27.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J Biomol NMR, 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 28.Goddard T. D., Kneller D. G. (2001) SPARKY, University of California, San Francisco [Google Scholar]
- 29.Farmer B. T., 2nd, Constantine K. L., Goldfarb V., Friedrichs M. S., Wittekind M., Yanchunas J., Jr., Robertson J. G., Mueller L. (1996) Nat. Struct. Biol. 3, 995–997 [DOI] [PubMed] [Google Scholar]
- 30.Linge J. P., Habeck M., Rieping W., Nilges M. (2003) Bioinformatics 19, 315–316 [DOI] [PubMed] [Google Scholar]
- 31.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 33.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 34.Beier H., Barciszewska M., Krupp G., Mitnacht R., Gross H. J. (1984) EMBO J. 3, 351–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beier H., Barciszewska M., Sickinger H. D. (1984) EMBO J. 3, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gololobov G., Noda Y., Sherman S., Rubinstein I., Baranowska-Kortylewicz J., Paul S. (1998) J. Pharmacol. Exp. Ther. 285, 753–758 [PubMed] [Google Scholar]
- 37.Lerch M., Mayrhofer M., Zerbe O. (2004) J. Mol. Biol. 339, 1153–1168 [DOI] [PubMed] [Google Scholar]
- 38.Haarer B. K., Petzold A. S., Brown S. S. (1993) Mol. Cell. Biol. 13, 7864–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Besenicar M., Macek P., Lakey J. H., Anderluh G. (2006) Chem. Phys. Lipids 141, 169–178 [DOI] [PubMed] [Google Scholar]
- 40.Lerch M., Kamimori H., Folkers G., Aguilar M. I., Beck-Sickinger A. G., Zerbe O. (2005) Biochemistry 44, 9255–9264 [DOI] [PubMed] [Google Scholar]
- 41.Mozsolits H., Wirth H. J., Werkmeister J., Aguilar M. I. (2001) Biochim. Biophys. Acta 1512, 64–76 [DOI] [PubMed] [Google Scholar]
- 42.Wagner G., Wüthrich K. (1982) J. Mol. Biol. 160, 343–361 [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto Y., Oda Y., Uryu M., Hayakawa Y. (2003) J. Biol. Chem. 278, 38579–38585 [DOI] [PubMed] [Google Scholar]
- 44.Sargent D. F., Schwyzer R. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 5774–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwyzer R. (1995) J. Mol. Recognit. 8, 3–8 [DOI] [PubMed] [Google Scholar]
- 46.Kaiser E. T., Kezdy F. J. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 1137–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaiser E. T., Kézdy F. J. (1984) Science 223, 249–255 [DOI] [PubMed] [Google Scholar]
- 48.Clark K. D., Garczynski S. F., Arora A., Crim J. W., Strand M. R. (2004) J. Biol. Chem. 279, 33246–33252 [DOI] [PubMed] [Google Scholar]