Abstract
The existence of cytotoxic T cells (CTL) cross-reacting with the human major histocompatibility antigens HLA-B14 and HLA-B27 suggests that their alloreactivity could be due to presentation of shared peptides in similar binding modes by these molecules. We therefore determined the crystal structures of the subtypes HLA-B*1402, HLA-B*2705, and HLA-B*2709 in complex with a proven self-ligand, pCatA (peptide with the sequence IRAAPPPLF derived from cathepsin A (residues 2–10)), and of HLA-B*1402 in complex with a viral peptide, pLMP2 (RRRWRRLTV, derived from latent membrane protein 2 (residues 236–244) of Epstein-Barr virus). Despite the exchange of 18 residues within the binding grooves of HLA-B*1402 and HLA-B*2705 or HLA-B*2709, the pCatA peptide is presented in nearly identical conformations. However, pLMP2 is displayed by HLA-B*1402 in a conformation distinct from those previously found in the two HLA-B27 subtypes. In addition, the complexes of HLA-B*1402 with the two peptides reveal a nonstandard, tetragonal mode of the peptide N terminus anchoring in the binding groove because of the exchange of the common Tyr-171 by His-171 of the HLA-B*1402 heavy chain. This exchange appears also responsible for reduced stability of HLA-B14-peptide complexes in vivo and slow assembly in vitro. The studies with the pCatA peptide uncover that CTL cross-reactive between HLA-B14 and HLA-B27 might primarily recognize the common structural features of the bound peptide, thus neglecting amino acid replacements within the rim of the binding grooves. In contrast, structural alterations between the three complexes with the pLMP2 peptide indicate how heavy chain polymorphisms can influence peptide display and prevent CTL cross-reactivity between HLA-B14 and HLA-B27 antigens.
T cells possessing the ability to recognize major histocompatibility complex (MHC)2 molecules from another individual of the same species, also termed alloreactive T cells, may constitute up to 10% of the T cell pool of an individual, and their precursor frequency can be 100–1,000-fold higher than that of self-restricted T cells directed against a foreign peptide (1, 2). The ability of alloreactive T cells to cross-react with nonself-MHC molecules is a major obstacle preventing successful organ transplantations (3–5). Two mechanisms, direct or indirect allorecognition, can be responsible for the rejection of a transplant by alloreactive T cells (6). In the first case, donor cells expressing MHC molecules are directly recognized by host T cells (7), whereas indirect allorecognition involves the presentation of peptides derived from donor proteins by MHC molecules of the host, followed by the detection of the complexes by the host T cells (8). However, although alloreactive T cells are very common and of great clinical importance, neither the primary basis for their existence nor the reasons underlying their cross-reactivity are sufficiently understood to draw general conclusions (9–11). Only very few studies have addressed the structural basis for the recognition of distinct MHC antigens by cross-reactive T cells (12–18). One of the most important questions regards the individual contribution of the bound peptide and binding groove residues of the heavy chain (HC) of MHC class I antigens to the interaction with T cell receptors (TCR).
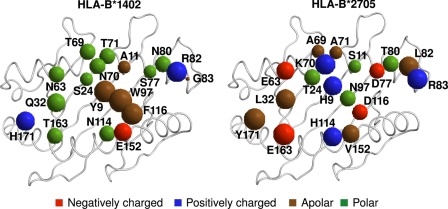
Here we analyze an HLA-B14 subtype, HLA-B*1402 (named B*1402), as well as two HLA-B27 subtypes, HLA-B*2705 and HLA-B*2709 (named B*2705 and B*2709), to shed light on the structural basis of peptide presentation and T cell alloreactivity among these HLA-B molecules. The amino acid sequences of B*1402 and B*2705 HC differ from each other at 18 positions, all of which are part of the peptide-binding groove (Fig. 1). These amino acid exchanges result in different repertoires of bound peptides; B*1402 and B*2705 share only about 4% of their peptides (19), whereas this value rises to 88% for the B*2705 and B*2709 subtypes (20), which are distinguished only by a single residue at the floor of the binding groove (B*2705, Asp-116; B*2709, His-116). The structural similarities between the two HLA-B27 subtypes (21–27) permit extensive cross-reactivity (up to 90%) of cytotoxic T cells (CTL) (28), whereas CTL alloreactivity between B*1402 and B*2705 is drastically reduced (to about 3%) (19), in line with the very limited overlap of their peptide repertoires.
FIGURE 1.
Amino acid sequence differences among B*1402 and B*2705 HC. The 18 residues distinguishing the two subtypes are all located in or in the immediate vicinity of the peptide-binding groove. B*2705 differs from B*2709 only by a D116H exchange (not shown). The residues are indicated by spheres with volumes roughly proportional to the volumes of the respective amino acid side chain in solution (77). The spheres are colored according to the biochemical properties of the respective amino acids, as indicated at the bottom of the image.
The HLA-B14 and HLA-B27 subtypes are distinguished from most other HLA class I molecules in their requirement for an arginine at anchor position 2 of the bound peptide (p2) (20, 29, 30). This preference is nearly absolute in B*2705 and B*2709 (31), whereas B*1402 tolerates also glutamine, glutamate, and proline as p2 anchors (19, 29). Statistically significant differences between B*1402 and B*2705 are also found at several other peptide positions (19). Previous structural and cellular studies of the HLA-B27 subtypes have suggested that molecular mimicry between the viral peptide pLMP2 (RRRWRRLTV, derived from Epstein-Barr virus latent membrane protein 2, residues 236–244) and the self-peptide pVIPR (RRKWRRWHL, derived from vasoactive intestinal peptide type 1 receptor, residues 400–408), when bound to B*2705, serves as an example of how a cellular immune response could be triggered that might contribute to the onset of ankylosing spondylitis (AS) through an autoimmune mechanism (22, 24). CTL that recognize the B*2705 and the B*2709 subtypes in complex with the self-peptide pVIPR (22) exemplify alloreactivity in this system, although the D116H micropolymorphism is deeply buried and not directly accessible to a TCR.
Alloreactive T cells are known to recognize a very diverse array of alloantigen-bound peptides (32, 33), so that virtually each T cell clone can be assumed to be specific for a distinct peptide. For this reason, the substantial correlation found in previous studies between peptide and the alloreactive T cell epitope sharing among HLA-B27 (reviewed in Ref. 34) or HLA-B14 subtypes (only 28.4% partial or full cross-reactivity, similar to peptide overlapping between the subtypes B*1402 and B*1403, see Ref. 19) supports a prominent role of peptides in determining alloreactive T cell cross-reaction, and it suggests that many shared ligands adopt antigenically similar conformations when bound to distinct HLA-B molecules. On the other hand, the results reported by Merino et al. (19) also demonstrate that the few CTL that cross-react with B*1402 and B*2705 did not exhibit cross-reactivity with B*1403, which is distinguished from B*1402 only by a single amino acid exchange in the α2-helix. Furthermore, they show that alloreactive CTL from various donors directed against B*2705 did not lyse cells expressing either B*1402 or B*1403, although the number of CTL tested might not have been high enough to detect a presumably low degree of cross-reactivity. Without structural data from HLA-B14 subtypes, however, these results are difficult to interpret.
The pCatA peptide (IRAAPPPLF, derived from the signal sequence of cathepsin A, residues 2–10) is among the very few known common ligands of B*1402, B*2705 (19), and B*27093 and can thus serve to study how a very different (B*1402) and two very similar subtypes (B*2705 and B*2709) handle a common ligand. On the other hand, the pLMP2 peptide is a proven natural ligand only of B*2705, whose possible presentation in vivo by B*2709 and HLA-B14 is not yet known, although this peptide can be complexed in vitro with B*2709 (24) and also with B*1402 (35). From previous crystallographic studies, it was known that pLMP2 is presented by the two HLA-B27 antigens in very different conformations (24). We expected that the pronounced sequence differences between B*1402 and the HLA-B27 alloantigens (Fig. 1) might even enhance the conformational dissimilarities that are observed when two very closely related subtypes such as B*2705 and B*2709 are compared. Discrepancies in peptide display could reasonably be expected to prevent CTL cross-reaction, so that pLMP2 might be considered as a representative of the vast majority of HLA-B14- and HLA-B27-presented ligands that must be responsible for the low degree of CTL cross-reactivity between these alloantigens. Despite these presumed differences between pCatA and pLMP2, both peptides may be seen as examples of ligands that could principally allow direct allorecognition.
Here we report the crystal structures of B*1402·pCatA, B*2705·pCatA, B*2709·pCatA, and B*1402·pLMP2, and we compare them with each other and with the previously reported structures of B*2705·pLMP2 and B*2709·pLMP2 (24).
MATERIALS AND METHODS
Protein Expression and Purification
The HC of B*1402 (19), B*2705, and B*2709 (21), as well as β2-microglobulin (β2m) were expressed separately in Escherichia coli as inclusion bodies, purified and dissolved in 50% (w/v) urea solution. The chemically synthesized nona-peptides pCatA (IRAAPPPLF) and pLMP2 (RRRWRRLTV) were purchased from EZBiolab, Westfield, IN. The HLA-B-peptide complexes were prepared by reconstituting a given HC, β2m, and the respective peptide according to protocols described previously (21, 35, 36). Briefly, the reconstitution mixture of HC, β2m, and a peptide (3 mg of HC, 2.5 mg of β2m, and 1 mg of peptide in a 100-ml solution containing 400 mm arginine-HCl, 2 mm EDTA, 5 mm reduced glutathione, 0.5 mm oxidized glutathione, and 100 mm Tris-HCl, pH 7.5) was incubated at 277 K for 8–12 weeks (B*1402) or 7–15 days (HLA-B27 subtypes), concentrated using Amicon Ultra-15 concentrators, and purified by size-exclusion chromatography using Sephadex 75 columns (GE Healthcare). The purified protein complexes were electrophoretically checked for purity, concentrated to ∼15 mg/ml, and used for crystallization trials.
Crystallization, X-ray Data Collection, and Structure Determination
The crystallization of the B*1402·pCatA and the B*1402·pLMP2 complexes have already been described in detail (35). Crystals of the B*2705·pCatA and the B*2709·pCatA complexes were obtained in a hanging-drop vapor diffusion setup using a reservoir solution containing 20–24% polyethylene glycol 8000, 100 mm Tris-HCl, pH 7.5 or 8.0, and a drop containing 1.1 μl of protein solution and 1.1 μl of reservoir solution. The crystals of all four complexes were soaked for 20–30 s in cryoprotectant solutions containing the respective reservoir solution and glycerol. Optimal concentrations of glycerol were found to be 20% (w/v) for B*1402·pLMP2, 15% (w/v) for B*1402·pCatA, 17% (w/v) for B*2705·pCatA, and 15% (w/v) for B*2709·pCatA crystals.
X-ray diffraction data sets of all four complexes were collected at 100 K on Protein Structure Factory beamline 14.2 at the BESSY II synchrotron facility in Berlin, Germany, using a wavelength of 0.91841 Å. Crystals of the B*1402·pLMP2 complex diffracted to 2.55 Å resolution, whereas the crystals of the other three complexes diffracted to about 1.8 Å resolution. The collected data sets were integrated and scaled using the HKL2000 suite (37). The B*1402·pLMP2 crystal has two complexes per crystal asymmetric unit, whereas the other three crystals contain only one complex per asymmetric unit. Data collection and refinement statistics are listed in Table 1. The data collection statistics for the two HLA-B14 complexes have already been provided by Kumar et al. (35) and are only shown here for comparison with the HLA-B27 structures.
TABLE 1.
Data collection and refinement statistics
A Values in parentheses refer to the highest resolution shell.
B Rsym = ΣhΣi|Ih,i − 〈Ih〉|/ΣhΣiIh,i.
C Rcryst = Σh|Fo − Fc|/ΣFo (working set, no σ cutoff applied).
D Rfree is the same as Rcryst but calculated on 5% of the data excluded from refinement.
E r.m.s.d. is from target geometries.
The CCP4 (38) suite programs MolRep (39) and PHASER (40) were used for the localization of the HLA-B molecules in crystal unit cells by molecular replacement. The atomic coordinates of HC and β2m obtained from the structures of B*2705·m9 (Protein Data Bank accession code 1JGE) (21) and B*2709·pLMP2 (Protein Data Bank accession code 1UXW) (24) were employed as a search model for the localization of HC and β2m of the B*2709-pCatA and the B*1402·pLMP2 complexes, respectively, in their unit cells. The atomic coordinates of HC and β2m obtained from the refined models of B*2709·pCatA and B*1402·pLMP2 were then used as search models for the localization of HC and β2m of the B*2705·pCatA and the B*1402·pCatA complexes, respectively, in their unit cells by molecular replacement. The obtained models were subjected to iterative cycles of restrained maximum likelihood refinement, including isotropic temperature factor adjustment using CNS (41) for the B*1402·pLMP2 structure and REFMAC (42) for the other three structures, followed by manual rebuilding using COOT (43). Water molecules were positioned using CNS for the B*1402·pLMP2 structure and Arp/Warp (44) for the other three structures. All contacts were calculated with the CONTACT program of the CCP4 suite. All superpositions and root mean square deviation (r.m.s.d.) calculations were performed with LSQKAB (45). Calculations of the electrostatic surface potentials were performed by employing the PDB2PQR web server (46) and the Adaptive Poisson-Boltzmann Solver program (47). Unless otherwise mentioned, the Cα atoms of HC peptide-binding groove residues (1–180 of a HC) were overlaid during comparisons.
The two-dimensional figures showing structural details were prepared using PyMOL (48). The fully interactive three-dimensional Fig. 3 (A and B) is embedded into conventional two-dimensional images and was created using the Adobe Acrobat 3D Toolkit and Adobe Acrobat software. We have recently described the procedure to generate such images (49, 50), starting from raw Protein Data Bank files (3BXN, 3BVN, 3BP4, 1UXS, 3BP7, and 1UXW).
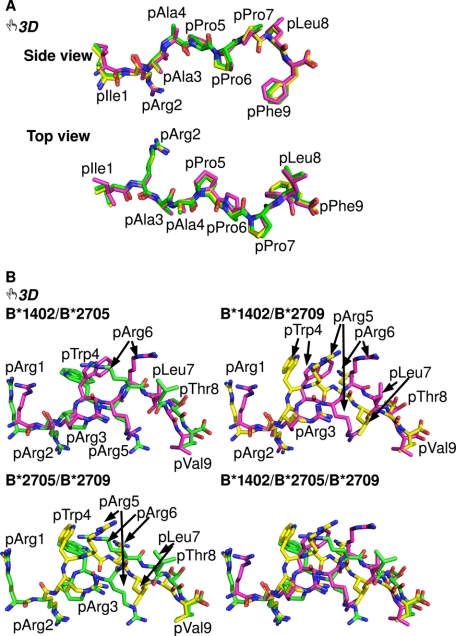
FIGURE 3.
Interactive three-dimensional comparison of the pCatA and pLMP2 peptides bound to three HLA-B molecules. An overlay of the conformations of pCatA in B*1402 (violet), B*2705 (green), and B*2709 (yellow) is provided in A and for pLMP2 in B. By clicking on each of the two-dimensional images in the PDF version of the article, the three-dimensional functions for a given peptide become available (they can be terminated by right-clicking on the three-dimensional display and choosing the “Disable three-dimensional” function). In each case, the model tree icon allows the view of individual components (with their designations shown on the left) of the three-dimensional model, and offers a preselected “tour” of the model. During the tour (with 6 and 10 images for B*1402 or B*2705, respectively), each of the models can be manipulated individually using the mouse (the tools to rotate, pan, or zoom an image can be selected through the toolbar or the contextual menu). A better understanding of many structural characteristics of the models can be obtained by “playing” with the structures. For example, the different contacts made by pArg5 in the binding grooves of B*1402 and B*2705, respectively, are easily compared with each other, as Asp-74 and the polymorphic residue 116 (Phe in B*1402, Asp in B*2705, and His in B*2709) are shown, along with residue 97 (views 6–10). Colors of the peptides as in Fig. 2.
RESULTS
Reconstitution of the HLA-B14 and HLA-B27 Complexes in Vitro
Reconstitution of the HLA-B14 complexes required a much longer time than in the case of the HLA-B27 complexes. Whereas complexes of B*2705 and B*2709 could be purified from the reconstitution mixture, although in low yield, already after 7 days of incubation, the B*1402 complexes required a minimum of 3 weeks of incubation for complex formation. The yield of reconstitution of the B*1402·pCatA complex was about 30 times lower than that of the B*2709·pCatA molecule. In general, the yields following reconstitution of B*1402 complexes were >10 times lower than the yields obtained after refolding the HLA-B27 complexes. Attempts to optimize the yield of the B*1402 complexes by varying the relative amounts of HC, β2m, and peptide, by changing the concentrations of oxidized and reduced glutathione or by adding glycerol (51), failed.
General Features of the Structures
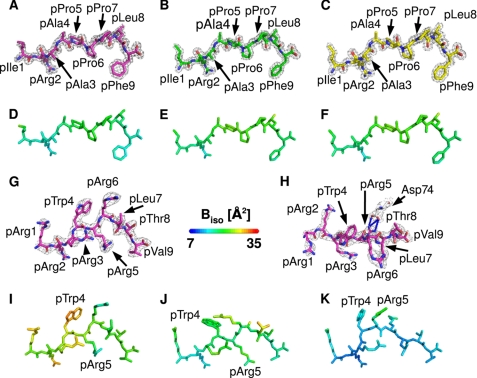
The complexes of the B*1402, B*2705, and B*2709 subtypes with the pCatA peptide crystallized isomorphously in the orthorhombic space group P212121 with one molecule per asymmetric unit, whereas the complex of B*1402 with pLMP2 crystallized in the monoclinic P21 space group with two molecules in the asymmetric unit. All four complexes exhibit the typical MHC class I architecture (52). The structures were refined to Rcryst and Rfree values of 16.9/21.9% (B*1402·pCatA), 17.1/22.1% (B*2705·pCatA), and 16.7/20.4% (B*2709·pCatA), all at about 1.8 Å resolution, and the B*1402·pLMP2 complex structure was refined to Rfree and Rcryst values of 24.0 and 26.8% at 2.55 Å resolution (Table 1). The complexes of B*2705 and B*2709 with pLMP2 crystallized in the monoclinic space group P21 as well, albeit under different crystallization conditions, and their crystals contained one molecule per asymmetric unit (24). The high quality electron density maps enabled us to model the molecules unambiguously (Fig. 2, A–C, G, and H).
FIGURE 2.
General properties of the pCatA and pLMP2 peptides bound to three HLA-B subtypes. A–C, final 2Fo − Fc electron density of pCatA conformations in B*1402 (A), B*2705 (B), and B*2709 (C), contoured at 1.5σ level. d–F, pCatA bound by B*1402 (D), B*2705 (E), and B*2709 (F) color-coded by isotropic B-factor. G and H, final 2Fo − Fc electron density of pLMP2 in B*1402 shown from the α2-helix (G) or the top of the binding groove (H), contoured at 1.5σ level. Electron density is depicted only for one molecule in the asymmetric unit. I–K, pLMP2 bound by B*1402 (I), B*2705 (J), and B*2709 (K) color-coded by isotropic B-factor. In all figures, unless mentioned otherwise, peptides bound to B*1402 are shown in violet, and peptides bound to B*2705 are shown in green, and peptides bound to B*2709 in are shown in yellow.
The peptide-binding grooves of B*1402 and B*2705 differ from each other at 18 positions (Fig. 1). These amino acid exchanges influence at least three features as follows: the general shape of the binding grooves, the interactions of several pocket-forming residues with atoms of the peptide, and the potential of residues of the α1- and α2-helices to interact with cell surface-expressed receptor molecules on effector cells. However, the overall structures of the HC and β2m of all complexes described here are very similar, with r.m.s.d. values for Cα overlay (HC/β2m) of 0.8/0.2 Å (B*1402·pCatA on B*1402·pLMP2), 0.3/0.2 Å (B*1402·pCatA on B*2705·pCatA or B*2709·pCatA), 0.7/0.3 Å (B*1402·pLMP2 on B*2705·pLMP2), and 0.6/0.3 Å (B*1402·pLMP2 on B*2709·pLMP2).
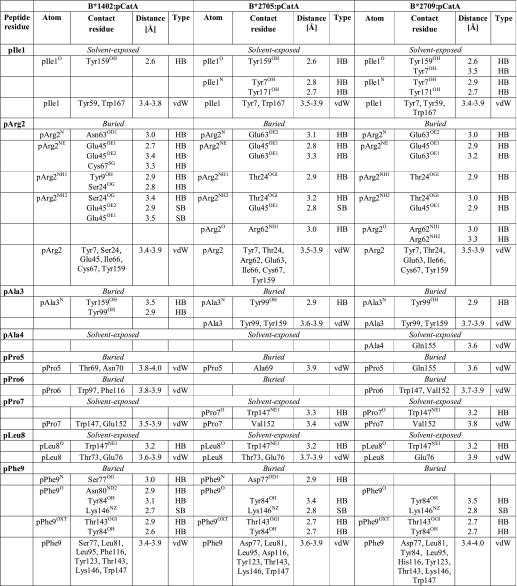
The pCatA peptide is displayed by the three HLA-B molecules in almost identical conventional orientations termed p4α (i.e. the main chain ϕ and ψ torsion angles are in α-helical conformation at p4, and all other ϕ/ψ torsion angles as in β-strands) (Fig. 2, A–C). In all cases, the primary anchor residue pArg2 and the secondary anchor residue pPhe9 are bound in nearly indistinguishable fashion in the B and F pockets, despite the presence of polymorphic HC residues that could influence peptide accommodation (Fig. 1). The interactions of other pCatA amino acids with residues of the binding groove are similar in the three complexes as well (Table 2). A comparison of the B-factor values shows that all atoms of pCatA exhibit low thermal vibration (Fig. 2, D–F).
TABLE 2.
Comparison of pCatA binding to the B*1402, B*2705, and B*2709 molecules
Only direct hydrogen bonds (HB, ≤3.5Å), salt bridges (SB, ≤3.5Å), and van der Waals interactions (vdW, ≤4.0Å) between peptide residues and HLA residues are listed. In case of peptide residues, van der Waals interactions between backbone atoms and between atoms of neighboring residues are not shown. Water-mediated interaction are not shown as well.
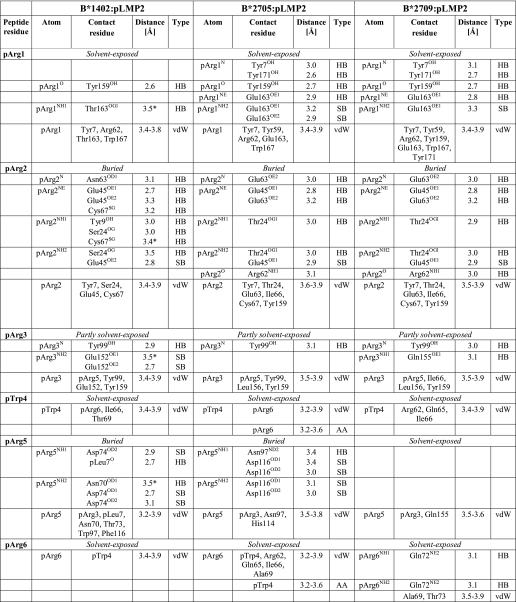
On the other hand, the pLMP2 peptide is presented by B*1402 in the noncanonical “p6α”-binding mode (i.e. the main chain ϕ and ψ torsion angles are in α-helical conformation at p6, and all other ϕ/ψ torsion angles as in β-strands), with the residue pArg5 anchored deep in the binding groove, where it forms salt bridges with Asp-74 and a very short intra-peptide H-bond with pLeu7O (Table 3 and Fig. 2, G and H) (24). These interactions are instrumental in holding the side chain of pArg5 firmly within the groove as indicated by low B-factor values (Fig. 2I). In contrast, high B-factor values are found for several pLMP2 residues, most notably pTrp4 but also in case of the arginine residues at p1, p2, and p3, when contrasted e.g. with pVal9 (Fig. 2I). Although the different resolutions preclude a direct comparison of B*1402 and of HLA-B27 with the pLMP2 peptide, the primary anchor residue pArg2 clearly exhibits a very low degree of flexibility when bound to B*2705 and B*2709 (Fig. 2, J and K), and this residue is also more fixed in the B*1402·pCatA complex (Fig. 2D).
TABLE 3.
Comparison of pLMP2 binding to B*1402, B*2705, and B*2709
Only direct hydrogen bonds (HB, ≤3.5Å), salt bridges (SB, ≤3.5Å), van der Waals interactions (vdW, ≤4.0Å), and amino aromatic interactions (AA) between peptide residues and HLA residues are listed. In case of peptide residues, van der Waals interactions between backbone atoms and between atoms of neighboring residues are not shown. Water-mediated interactions are not shown as well.
* This bond exists in only one of the two molecules in the asymmetric unit.
An overlay of the bound peptides (Fig. 3, A and B) reveals that the distances between the Cα atoms of the N- and C-terminal residues in the two B*1402 complexes are smaller than in case of the HLA-B27-peptide complexes as follows: B*1402 in comparison with either B*2705 or B*2709, 0.3 Å for complexes with bound pCatA, 1.2 Å for B*1402/B*2705·pLMP2, and 0.7 Å for B*1402/B*2709-pLMP2.
To clarify differences in the conformations of the peptides in the three HLA-B complexes, we compared interactive three-dimensional models (49, 50) of pCatA and pLMP2 in B*1402 and in the two HLA-B27 subtypes (Fig. 3, A and B). Access to the three-dimensional feature is obtained by clicking on any part of the conventional two-dimensional figures in the PDF. A better understanding of many structural features is thus provided, and it is particularly helpful to be able to compare the peptide conformations by zooming in and by rotating the structures. The freely available Adobe Reader (version 9) is required as well as any Windows-based computer with a modern graphic card (128+ MB memory) and sufficient RAM (1024+ MB). The “Help” option within the program provides an introduction into the possibilities that are offered. A further useful feature of the three-dimensional images is the possibility to hide certain structural elements such as β2m, HC, or any of the peptide conformations. A “tour” of the molecule resembling a pre-rendered video file of the structures is available through the pre-set views as well. At any stage, however, full interaction with the structures remains possible.
In the case of Fig. 3A, an overlay of the three pCatA structures allows us to view the peptide conformations from the side of the α2-helix (view 1), then from the top (view 2), and finally in the context of the whole HLA-B complex (views 3–6). The pCatA conformations are extremely similar when superimposed, because noticeable differences are only found for the N- and C-terminal residues. In contrast, the noncanonical p6α conformation of pLMP2 in B*1402 bears resemblance only to the noncanonical binding mode observed for pLMP2 in complex with B*2705 (Fig. 3B and views 1–10). The peptide backbone has very similar conformations in the two structures, and with the exception of pTrp4 and pArg6, all peptide residue side chains have comparable orientations. However, the precise location of similarly oriented side chains can differ considerably, e.g. in case of pArg1, pArg5, and pVal9 (Table 3). The three-dimensional images show these features very clearly, and they reveal also the distinct coordination of pArg5, either with Asp-74 in B*1402 (view 6) or with Asp-116 in B*2705 (view 7). As expected from the previously described differences between the B*2705 and B*2709 subtypes in complex with pLMP2 (24), the orientations of this peptide bound to B*1402 and B*2709, where the latter occupies the conventional “p4α” binding mode, have very little in common, in contrast to B*1402 and B*2705 (Fig. 3B, views 9 and 10). With the exception of pArg2, the side chains of all other peptide residues occupy distinct and in most cases totally different locations, whereas a comparison of the pLMP2-binding modes in B*2705 and B*2709 shows that despite the distinct peptide orientations at least pArg1, pArg2, pThr8, and pVal9 are bound in a nearly identical manner (Fig. 3B and Table 3) (24).
Consequences of Differential A Pocket Architectures
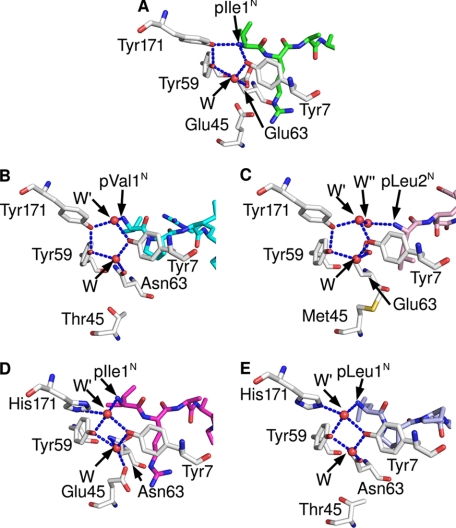
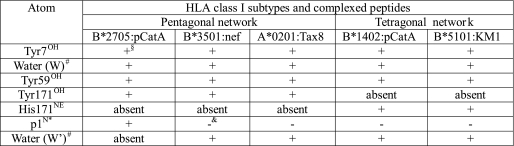
The N termini of peptides are anchored through a pentagonal network of hydrogen bonds in nearly all known MHC class I structures (52), including the four HLA-B27 structures compared here. This network additionally encompasses Tyr-7, Tyr-59, Tyr-171, and a water molecule termed “W” in Fig. 4 and Table 4. This water molecule is also contacted by residue 63 (Asn in B*1402 and Glu in B*2705 and B*2709), as shown by the B*2705·pCatA complex (Fig. 4A). In exceptional cases, here exemplified by B*3501 in complex with the octameric peptide nef (Fig. 4B) (53) and A*0201 presenting a peptide without its N-terminal amino acid (Fig. 4C) (54), a water molecule (W′) takes the place of p1N, thus retaining the pentagonal network, although the peptide N terminus is no longer part of it. However, in the B*1402·pCatA complex, p1N is connected to a tetragonal network of hydrogen bonds formed by Tyr-7, Tyr-59, and two water molecules (W and W′) (Fig. 4D and Table 4). The two water molecules serve as hydrogen bonding partners of p1N and His-171NE, as well as Glu-45 and Asn-63, respectively. The N terminus of pLMP2 in the B*1402 complex is anchored in both molecules of the asymmetric unit in precisely the same way, with all the elements of the tetragonal network identically positioned, including the water molecule (W′) at the position where it is located in B*1402·pCatA. However, the electron density map of B*1402·pLMP2 permits the placement of W′ only in one complex within the asymmetric unit. Since the tetragonal network engages also Asn-63 and Glu-45, two HC residues that possess identical conformations, and because the electron density map contoured at 0.8σ level shows some electron density at the position where W′ is located in the B*1402·pCatA complex, it is likely that the tetragonal network is present also in the other molecule of B*1402·pLMP2 within the asymmetric unit.
FIGURE 4.
HLA-B subtype-dependent anchoring of peptide N termini. The crucial role of the Y171H exchange for the anchoring of peptide N termini is revealed by comparing the A pockets of B*2705·pCatA (A), B*3501 bound to the octameric nef peptide (B), A*0201 complexed with an N-terminally truncated peptide (C), B*1402·pCatA (D), and B*5101·KM1 (E). Whereas the first three structures (all with Tyr-171) show a pentagonal hydrogen bonding network, the subtypes B*1402 and B*5101 anchor the peptide N terminus only indirectly, via a water molecule (W′) which is also contacted by His-171NE, resulting in the formation of a tetragonal network of H-bonds. HC residues are colored gray, and peptide residues are in different colors.
TABLE 4.
Compilation of atoms comprising the pentagonal or tetragonal network of hydrogen bonds within the A pocket of various HLA class I molecules
§ The atom is part of the network.
# For designations of the water atoms, please refer to Fig. 5.
* N terminus of the peptide is shown.
& The atom is not part of the network.
The B*5101 subtype is one of the few HLA-B molecules that possess His-171. There are two structures available for this subtype, both in complex with immunodominant peptides as follows: KM1 (LPPVVAKEI) and KM2 (TAFTIPSI) of human immunodeficiency virus, type 1 (55). The N terminus of the KM1 peptide is displayed in the 2.2 Å resolution structure of the B*5101-KM1 complex in the same nonconventional mode of N-terminal anchoring that has been observed for the B*1402 subtype (Fig. 4, D and E). The binding mode of the N terminus of the KM2 peptide to B*5101 is somewhat different from that resulting in the tetragonal network (data not shown), very likely because of the lower resolution (3.0 Å) of this structure, which may not have permitted unambiguous positioning of all atoms participating in anchoring of the KM2 N terminus.
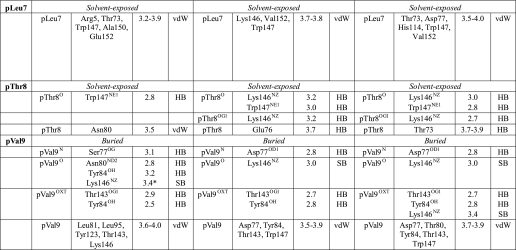
As a consequence of the nonconventional location, the N termini of both B*1402-bound peptides in the crystal asymmetric unit are anchored rather shallowly and are shifted “upward” by 1.6–1.7 Å in comparison with the HLA-B27-bound peptides (Fig. 3, A and B). Furthermore, the location of the methyl group of the pIle1 side chain of the pCatA peptide differs between the B*1402 and B*2705 complexes, and also the apex of the pArg1 side chain of the pLMP2 peptide is separated by 3.37 Å between these HLA-B molecules (Fig. 3, A and B). Residue 163 (Thr in B*1402 and Glu in B*2705 and B*2709) contributes to fixing the side chain of pArg1 through direct contacts in all three structures. Thus, although stabilization of the pArg1 side chain, a predominant residue among HLA-B27 ligands, is dependent on Glu-163, substitution of this residue by Thr still allows hydrogen bonding with pArg1. Furthermore, whereas Arg-62 and Trp-167 also have important roles in anchoring an N-terminal arginine in HLA-B27 molecules (56), hydrophobic interactions with these two residues are largely abrogated in the B*1402·pLMP2 structure (Fig. 5, D–F).
FIGURE 5.
Molecular surfaces of the pCatA and pLMP2 peptides bound to three HLA-B subtypes. Shown are molecular surface representations of B*1402 (A and D), B*2705 (B and E), and B*2709 (C and F) complexed with the pCatA peptide (A–C) or the pLMP2 peptide (D–F), respectively, as viewed by an approaching TCR. Colors of the peptides are as in Fig. 2. In each image, electrostatic surfaces are shown only for the respective HC, with red indicating a negative and blue a positive charge. Gray areas are uncharged. A number of crucial peptide or HC residues (see text) are indicated.
Pockets for Primary and Secondary Peptide Anchors
The B pocket of the peptide-binding groove, which accommodates the primary anchor of the peptide (p2) in both the B*1402 and the HLA-B27 subtypes, exhibits four substitutions in B*1402, relative to B*2705 or B*2709 as follows: Y9H, S24T, N63E, and N70K. These exchanges, however, do not result in a reorientation of the pArg2 side chain in the two complexes with B*1402. Despite a number of distinct contacts, this amino acid is accommodated in nearly identical conformations in all six structures compared here (Tables 2 and 3). In neither of the two B*2705 complexes does Lys-70 form water-mediated hydrogen bonds with pArg2 or pAla/Arg3, whereas Asn-70 forms water-mediated hydrogen bonds with both of these peptide residues in the B*1402·pCatA structure. The apparent absence of these water-mediated contacts in the B*1402·pLMP2 complex could be due to the lower resolution of this structure.
A surface representation of the three HLA-B molecules (Fig. 5) demonstrates that in the HLA-B27-peptide complexes, a bridge-like structure arches over the N-terminal half of a peptide, connecting Arg-62 and Glu-163 via a water molecule (Fig. 5, B, C, E, and F), as described previously in detail for the structure of B*2709 complexed to the s10R peptide that contains, like pLMP2, an arginine at p1 (56). With a glycine at this position, as in case of the m9 peptide (21), the side chain of Arg-62 assumes a distinct orientation that results in loss of the “bridge.” Similarly, the E163T exchange between HLA-B27 and B*1402 alters the orientation of the Arg-62 side chain, leading to an abrogation of the Arg-62 to Glu-163 contact between the two α-helices (Fig. 5, A and D).
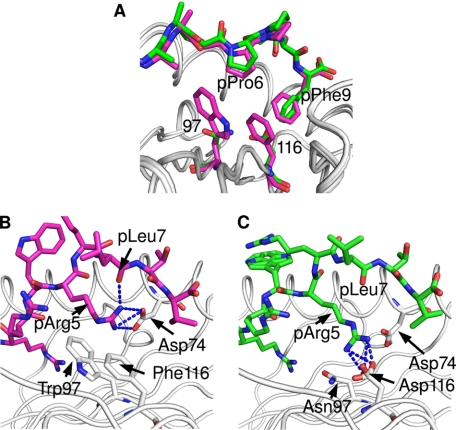
Second only to the B pocket, the F pocket also fulfills an important role in both B*1402 and HLA-B27 (19, 29, 30). It exhibits differences at positions S77D, N80T, and F116D between B*1402 and B*2705 (Fig. 1). Apart from a difference in charge, the exchange of residue 116 leads also to a higher occupation of the F pocket by more atoms in B*1402, to which the bulky Trp-97 (instead of Asn in B*2705) is also likely to contribute. Although no difference between the coordinations of pPhe9 in B*1402 and the HLA-B27 structures can be observed (Table 2), apart from the replacement of a direct pPhe9O → Asn-80ND2 contact in B*1402 by a water-mediated interaction between the pPhe9O and Thr-80OG1 in B*2705 and B*2709 (not shown in Table 2), the aromatic ring of pPhe9 in the pCatA peptide is oriented at an angle of ∼60° in the B*1402 complex in comparison with the two HLA-B27 complexes (Fig. 6A). This peculiar orientation facilitates an optimal face-to-edge stacking (C-H-π bonding) between the aromatic rings of pPhe9 and Phe-116. Comparable interactions are frequently found in protein structures (57) and allow a better sharing of π electrons. The C-terminal residue of the pCatA peptide is also stabilized by C-H-π bonding to the nonpolymorphic residue Tyr-123 in all three structures.
FIGURE 6.
Differential anchoring of the middle of the pCatA and pLMP2 peptides. The bulky residues at HC positions 97 (Trp) and 116 (Phe) in the B*1402 subtype lead to differential contacts when compared with B*2705 (Asn-97 and Asp-116). The structures with pCatA (violet) (A) and pLMP2 (B) (green, A and C) are shown. Whereas pPro6 of pCatA is shifted only slightly into the groove in case of B*1402, the smaller side chains of residues 97 and 116 do not contact pPro6. The pArg5 residue of the pLMP2 peptide forms a bidentate salt bridge with Asp-74 and a hydrogen bond with pLeu7 in B*1402, whereas salt bridges with Asp-116 characterize the noncanonical pLMP2 conformation in case of B*2705.
Anchoring of the Middle of the Peptides
In contrast, striking HLA-B subtype-dependent changes are observed when residues in the middle of the peptides are considered, in particular pTrp4, pArg5, and pArg6 in the case of pLMP2. However, conformational differences are also found between the three structures with pCatA as follows: the presence of Phe-116 (B*1402), but not Asp-116 (B*2705) or His-116 (B*2709), permits van der Waals contacts with pPro6, leading to a shift (0.3 Å) of this residue into the binding groove (Fig. 6A). This location of pPro6 is also stabilized through van der Waals interactions with Trp-97 (B*1402), which cannot form with the much smaller Asn-97 that is found in the two HLA-B27 molecules (Fig. 6A). These differences do not, however, lead to a very pronounced displacement of the pCatA peptide between B*1402 and B*2705/B*2709 (Fig. 3A).
Prolines in native proteins are always observed in either completely cis or trans form, because the relative ease of flipping between these forms is overwhelmed by favorable interactions with proximal groups (58). All three prolines of the pCatA peptide are in trans conformation in the three structures. We built trans-Pro-cis-Pro variant models of the pCatA peptide, disregarding electron density maps and keeping the Cα1–Cα9 distance and the orientation of the residues p1–p3 and p9 the same as in the B*1402·pCatA structure. All models with one to three cis-Pro were found to contain at least two and a maximum of five residues lying in disallowed regions of the Ramachandran plot, suggesting that the all-trans model that is observed in the three structures is sterically the most favored.
In contrast to pCatA, drastic conformational changes are obvious with regard to the pLMP2 peptide (Fig. 3B). As mentioned previously, these are primarily due to the different orientations of pArg5 in the three structures, and although this residue is solvent-exposed in B*2709 (Fig. 2K), it contacts Asp-74 (B*1402) or Asp-116 (B*2705) within the binding groove through salt bridges (interactive Fig. 3B and Fig. 6, B and C). Whereas these distinct anchoring modes of pArg5 are responsible for the differences found in the pLMP2-binding modes, it is more difficult to explain the considerable changes between B*1402 and B*2705 that characterize the conformations of other peptide side chains (Fig. 3B). Apart from alterations that are a consequence of A pocket polymorphisms (Fig. 4), the short salt bridge between pArg3 and Glu-152 (Table 3) could influence the insertion of pLMP2 into the B*1402-binding groove. In both HLA-B27 subtypes, pArg3 forms an intra-peptide hydrogen bond with pTrp4O instead. pArg6 and pTrp4 contact each other by van der Waals bonds both in B*1402 and B*2705, despite the distinct orientations of the side chains of both residues (Fig. 2, I and J).
DISCUSSION
The present crystallographic study was primarily undertaken to provide a better understanding for the observation that alloreactive CTL can be generated against cells expressing B*1402 that cross-react with HLA-B27 molecules, despite the considerable exchanges that characterize the peptide-binding grooves of the two antigens (Fig. 1) (19). Most of these differences affect the α1-helix and the floor of the binding groove with eight and seven exchanges, respectively, whereas there are only three α2-helical exchanges. We analyzed a proven shared ligand of B*1402 and HLA-B27 antigens, the self-peptide pCatA, as well as a nonself-ligand, the viral peptide pLMP2. The overall structures of the B*1402 and HLA-B27 complexes with both peptides are related, but a detailed comparison of the two peptides reveals that they are bound either very similarly (pCatA) or in distinct conformations (pLMP2) (Tables 2 and 3; Figs. 2 and 3).
The anchoring of the N termini of bound peptides through a pentagonal hydrogen bond network within the A pocket (Fig. 4; Table 4) is known to be one of the most concordant features of peptide presentation by MHC molecules (52). Furthermore, the structure of B*3501 in complex with an octameric peptide reveals that the pentagonal network can also exist in a slightly modified form in which the N terminus of the peptide is shifted upwards, preventing its direct participation but permitting the establishment of a connection to the pentagonal network through a water molecule (53), instead of forming one of the five corners of the pentagonal network. The latter represents the “normal” situation and can be found in HLA-B35 molecules presenting nonameric peptides (15, 16, 59), in all other published structures of “classical” MHC class I molecules, or in the structures of the HLA-B27 subtypes with the pCatA peptide described here. A comparison of the binding of the nonameric Tax peptide (Tax9) and an N-terminally truncated octamer (Tax8) to HLA-A2 shows that the removal of p1, a residue essential for formation of the pentagonal network, significantly reduces the affinity of the interaction between the ligand and the A pocket (54), but it does not eliminate the pentagonal network that is present in the Tax9 as well as the Tax8 peptide structures, with positions of the p1 atoms crucial for connecting the peptide with the ring formed by water molecules in the Tax8 structure.
However, a drastically different coordination of the peptide N terminus is triggered by the presence of His-171, as in B*1402. It results in the formation of a tetragonal network of hydrogen bonds. The water molecule (W′ in Fig. 4), which “replaces” the hydroxyl group of Tyr-171, is also contacted by the peptide N terminus. In contrast, the involvement of the other three members of both the tetragonal and the pentagonal networks (Tyr-7, the water molecule W, and Tyr-59) remains unchanged. The causative role of His-171 in implementing the formation of a tetragonal network is supported by the only other high resolution structure in which the MHC molecule exhibits this exchange at residue 171, B*5101 in complex with the KM1 peptide (55). A comparison of the diverse contacts formed by the pentagonal and tetragonal networks in the different structures (Fig. 4; Table 4) reveals that it is in fact the exchange at residue 171, and not that of the polymorphic A pocket residue Asp/Asn-63 (Fig. 1), that is responsible for the formation of the unusual network in B*1402 and B*5101; Asn-63 occurs also, for example, in B*3501 and B*3508, both of which bind the N terminus of nonoctameric peptides via a pentagonal network (15, 16, 59). A further and very divergent solution for the N-terminal anchoring of a ligand is realized in case of the mouse H-2M3 antigen. It binds only peptides with an N-formylated methionine at the N terminus. A number of crucial A pocket polymorphisms are responsible for a unique architecture that permits this molecule to adjust to the specialized binding requirements of ligands derived from the N terminus of proteins produced by bacteria and mitochondria (60).
The presence of the tetragonal network only in the B*1402 and the B*5101 antigens strongly suggests that their exceptional reconstitution behavior in vitro is a consequence of this structural alteration; complexes of both HLA-B molecules need 2–3 months to reconstitute (35, 55), whereas HLA-B27 molecules require 2–3 weeks (21), and the majority of other MHC class I molecules only a few days (36). HLA-B14-peptide complexes also exhibit a high maturation rate and a reduced stability in vivo when compared with B*2705, as well as a substantial rate of dissociation in post-endoplasmic reticulum compartments. These distinguishing features are possibly due to a less optimized peptide repertoire (61). A reduced stability of HLA-B14 antigens might additionally be ascribed to the absence of the water-mediated contact between Arg-62 and Glu-163 that characterizes all HLA-B27-peptide complexes except those with pGly1 (see Refs. 21–26, 62 and this study). This bridge-like structural feature contributes to keeping the peptide in place; it cannot form in B*1402 because of the presence of Thr-163 (Fig. 5). Nevertheless, the B*1402 subtype is widely distributed in various human populations, suggesting that the seemingly disadvantageous structural and biochemical features that are due to its unorthodox A pocket are no impediment for being a “successful” HLA class I antigen. However, the biological advantage conferred by expressing HLA-B14 molecules or of possessing HLA-B14-carrying haplotypes (63), if any, remains currently unclear.
Possibly the most conspicuous biochemical feature that the HLA-B14 and the HLA-B27 antigens have in common is the preference (HLA-B14) (19, 29) or nearly absolute requirement (HLA-B27) (20, 31) for pArg2 of a bound peptide, despite the many amino acid exchanges that characterize the binding grooves of these HLA-B molecules (Fig. 1). The capacity to bind peptides with pArg2 is a feature of a relatively small number of MHC class I molecules, many of which seem to predispose to a development of spondyloarthropathies. Besides the well known and very strong association of HLA-B27 with AS (30, 64–66), B*3901 is associated with spondyloarthropathies in Japanese individuals (67, 68). This class I molecule as well as the very closely related B*3909 subtype present peptides with pArg2 (69, 70), and B*3901 binds also HLA-B27 ligands in vitro (68). Similarly, although carriers of the B*1402 allele are not at risk to develop AS, the very closely related B*1403 allele (distinguished only by an L156R exchange) is strikingly associated with AS in sub-Saharan populations from Togo (71) and Zambia (72), where AS is very infrequently encountered. B*1403 shares the preference for pArg2 of a bound peptide with B*1402 (19). Our finding that the pArg2 residues of the pCatA and pLMP2 peptides are similarly accommodated by the HLA-B14 and HLA-B27 B pockets (Figs. 2 and 3) provides a strong argument in favor of a role for this pocket in AS pathogenesis (30).
Further support for this contention can be gained from studies in gorillas with spondyloarthropathies. Gogo-B*0101 is an allele of an MHC class I molecule that binds peptides containing pArg2 (73) and is very frequently observed in affected animals. Residues belonging to the B pocket of this molecule exhibit a number of alterations in comparison with the AS-associated HLA-B27 alleles, most notably an exchange of Glu-45 (HLA-B27) to Met-45. Although this drastic mutation, when introduced into HLA-B27 molecules, abrogates the capacity of the mutant to bind peptides with pArg2 (74), it does not affect the B pocket preference for pArg2 in Gogo-B*0101 due to compensatory changes, mainly H9D (73). Thus, a variety of B pockets are able to bind pArg2 either with a highly conserved binding mode, as illustrated in this study for B*1402, B*2705, and B*2709, or in a different way, as in the Gogo-B*0101 antigen. However, neither the structure of the B pocket per se nor the capacity to present peptides with Arg-2 is sufficient to predispose to AS, as is obvious from the lack of association of B*2709 and B*1402 with this disease.
Anchoring of pArg5 to the floor of the peptide-binding groove is believed to be the defining feature for the accommodation of a peptide in a noncanonical binding mode by HLA-B27 molecules (22, 24, 26). In all the cases known so far, residue 116 forms a salt bridge (pArg5) or a hydrogen bond (citrulline at p5) with the side chain of the residue in the middle of a peptide, thus providing an additional anchor for the peptide and permitting it to adopt the noncanonical conformation. The B*1402·pLMP2 complex, however, exploits a further, and to our knowledge novel, mode of pArg5 anchoring to the groove; due to the exchange of Asp-116 by Phe-116, pArg5 forms a salt bridge with Asp-74. This binding mode appears also to be the result of steric hindrance because of the bulky residues on the floor of the peptide-binding groove (Figs. 1, 3, and 6), and it explains the observation that the B*1402 and in particular the B*1403 antigens show a marked preference for peptides with pArg5 (19). Because this preference is much higher than in case of B*2705, it is likely that the contact between pArg5 and Asp-74 in HLA-B14 molecules, despite the seemingly disadvantageous geometry of the curved pArg5 side chain, has a higher stabilizing effect on bound peptides than the interaction between pArg5 and Asp-116 in B*2705 (Figs. 3 and 6).
Despite recent advances in the field of MHC molecules and their recognition by TCR (9, 10, 18, 75), there was no specific insight regarding the structural basis for similarities and differences in peptide presentation by the B*1402, B*2705, and B*2709 antigens. Our study now provides a better understanding of T cell alloreactivity among the HLA-B14 and the HLA-B27 antigens. The results with the pCatA peptide demonstrate that a proven self-ligand can be presented by the three alloantigens in nearly identical conformations (Fig. 5). A T cell receptor with a focus on the peptide (76) might thus be able to recognize all complexes, despite the dissimilarities between the three HC. The fact that a number of amino acids that are known to be crucial for the docking of many TCR (18) are unchanged between HLA-B14 and HLA-B27 (i.e. residues at positions 65 and 155 of an HC) permits us to predict that alloreactive T cells might exploit these similarities and “prefer” to neglect the many differences among potential contact residues.
On the other hand, the three distinct structures with the pLMP2 peptide (Fig. 5) show also how alloreactivity between these antigens can be prevented; in addition to the allelic differences, the display of the peptide is so divergent in the three structures that T cell cross-reactivity will almost certainly not occur.
It might be argued that the similar conformations adopted by pCatA bound to B*1402 and the HLA-B27 subtypes exemplify an exceptional situation favored by the presence of three consecutive proline residues in the peptide sequence, a feature that might limit the conformational flexibility of a bound ligand. However, the remarkable correlation between peptide overlap and alloreactive T cell epitope sharing found not only between B*1402 and B*2705 (19) but also very frequently among HLA-B27 subtypes (reviewed in Ref. 34) argues against this view. This correlation suggests that many shared ligands adopt antigenically similar conformations when bound to distinct MHC class I allotypes. Thus, the similar binding mode observed for pCatA in multiple contexts might not be unusual.
Acknowledgments
We thank Yvette Roske and Claudia Alings for help with the crystallization trials and the beamline staff at BESSY II, Berlin, Germany, for valuable assistance.
This work was supported by VolkswagenStiftung Grant I/79 983 (to A. Z.) and a stipend (to P. K.), Deutsche Forschungsgemeinschaft Grants SFB 449, TP B6, and Z3 (to W. S., B. U.-Z., and A. Z.), the Senate of Berlin NaFöG Fellowship (to P. K.), and Spanish Plan Nacional de I+D Grant SAF2008/00461 (to J. A. L. d. C.).
The atomic coordinates and structure factors (codes 3BXN, 3BVN, 3BP4, and 3BP7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
E. Merino and J. A. López de Castro, unpublished observations.
- MHC
- major histocompatibility complex
- AS
- ankylosing spondylitis
- CTL
- cytotoxic T lymphocytes
- HC
- heavy chain
- HLA
- human leukocyte antigen
- p1
- p2,…, peptide position 1, 2,…
- r.m.s.d.
- root mean square deviation
- TCR
- T cell receptor
- β2m
- β2-microglobulin.
REFERENCES
- 1.Lindahl K. F., Wilson D. B. (1977) J. Exp. Med. 145, 508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suchin E. J., Langmuir P. B., Palmer E., Sayegh M. H., Wells A. D., Turka L. A. (2001) J. Immunol. 166, 973–981 [DOI] [PubMed] [Google Scholar]
- 3.Afzali B., Lechler R. I., Hernandez-Fuentes M. P. (2007) Tissue Antigens 69, 545–556 [DOI] [PubMed] [Google Scholar]
- 4.Kamoun M., Holmes J. H., Israni A. K., Kearns J. D., Teal V., Yang W. P., Rosas S. E., Joffe M. M., Li H., Feldman H. I. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 18883–18888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris G. P., Allen P. M. (2009) J. Immunol. 182, 6639–6643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould D. S., Auchincloss H., Jr. (1999) Immunol. Today 20, 77–82 [DOI] [PubMed] [Google Scholar]
- 7.Bevan M. J. (1984) Immunol. Today 5, 128–130 [DOI] [PubMed] [Google Scholar]
- 8.Matzinger P., Bevan M. J. (1977) Cell Immunol. 29, 1–5 [DOI] [PubMed] [Google Scholar]
- 9.Felix N. J., Allen P. M. (2007) Nat. Rev. Immunol. 7, 942–953 [DOI] [PubMed] [Google Scholar]
- 10.Archbold J. K., Macdonald W. A., Burrows S. R., Rossjohn J., McCluskey J. (2008) Trends Immunol. 29, 220–226 [DOI] [PubMed] [Google Scholar]
- 11.Kazansky D. B. (2008) J. Immunotoxicol. 5, 369–384 [DOI] [PubMed] [Google Scholar]
- 12.Paradela A., García-Peydró M., Vázquez J., Rognan D., López de Castro J. A. (1998) J. Immunol. 161, 5481–5490 [PubMed] [Google Scholar]
- 13.Reiser J. B., Darnault C., Guimezanes A., Grégoire C., Mosser T., Schmitt-Verhulst A. M., Fontecilla-Camps J. C., Malissen B., Housset D., Mazza G. (2000) Nat. Immunol. 1, 291–297 [DOI] [PubMed] [Google Scholar]
- 14.Luz J. G., Huang M., Garcia K. C., Rudolph M. G., Apostolopoulos V., Teyton L., Wilson I. A. (2002) J. Exp. Med. 195, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tynan F. E., Elhassen D., Purcell A. W., Burrows J. M., Borg N. A., Miles J. J., Williamson N. A., Green K. J., Tellam J., Kjer-Nielsen L., McCluskey J., Rossjohn J., Burrows S. R. (2005) J. Exp. Med. 202, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hourigan C. S., Harkiolaki M., Peterson N. A., Bell J. I., Jones E. Y., O'Callaghan C. A. (2006) Eur. J. Immunol. 36, 3288–3293 [DOI] [PubMed] [Google Scholar]
- 17.Colf L. A., Bankovich A. J., Hanick N. A., Bowerman N. A., Jones L. L., Kranz D. M., Garcia K. C. (2007) Cell 129, 135–146 [DOI] [PubMed] [Google Scholar]
- 18.Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 19.Merino E., Montserrat V., Paradela A., López de Castro J. A. (2005) J. Biol. Chem. 280, 35868–35880 [DOI] [PubMed] [Google Scholar]
- 20.Ramos M., Paradela A., Vazquez M., Marina A., Vazquez J., Lopez de Castro J. A. (2002) J. Biol. Chem. 277, 28749–28756 [DOI] [PubMed] [Google Scholar]
- 21.Hülsmeyer M., Hillig R. C., Volz A., Rühl M., Schröder W., Saenger W., Ziegler A., Uchanska-Ziegler B. (2002) J. Biol. Chem. 277, 47844–47853 [DOI] [PubMed] [Google Scholar]
- 22.Hülsmeyer M., Fiorillo M. T., Bettosini F., Sorrentino R., Saenger W., Ziegler A., Uchanska-Ziegler B. (2004) J. Exp. Med. 199, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hülsmeyer M., Welfle K., Pöhlmann T., Misselwitz R., Alexiev U., Welfle H., Saenger W., Uchanska-Ziegler B., Ziegler A. (2005) J. Mol. Biol. 346, 1367–1379 [DOI] [PubMed] [Google Scholar]
- 24.Fiorillo M. T., Rückert C., Hülsmeyer M., Sorrentino R., Saenger W., Ziegler A., Uchanska-Ziegler B. (2005) J. Biol. Chem. 280, 2962–2971 [DOI] [PubMed] [Google Scholar]
- 25.Rückert C., Fiorillo M. T., Loll B., Moretti R., Biesiadka J., Saenger W., Ziegler A., Sorrentino R., Uchanska-Ziegler B. (2006) J. Biol. Chem. 281, 2306–2316 [DOI] [PubMed] [Google Scholar]
- 26.Beltrami A., Rossmann M., Fiorillo M. T., Paladini F., Sorrentino R., Saenger W., Kumar P., Ziegler A., Uchanska-Ziegler B. (2008) J. Biol. Chem. 283, 27189–27199 [DOI] [PubMed] [Google Scholar]
- 27.Ziegler A., Loll B., Misselwitz R., Uchanska-Ziegler B. (2009) in Molecular Mechanisms of Spondyloarthropathies (López-Larrea C., Díaz-Peña R. eds) Vol. 649, pp. 177–195, Landes Bioscience, Austin, TX [Google Scholar]
- 28.García-Peydró M., Martí M., López de Castro J. A. (1999) J. Immunol. 163, 2299–2305 [PubMed] [Google Scholar]
- 29.DiBrino M., Parker K. C., Margulies D. H., Shiloach J., Turner R. V., Biddison W. E., Coligan J. E. (1994) J. Biol. Chem. 269, 32426–32434 [PubMed] [Google Scholar]
- 30.Ramos M., López de Castro J. A. (2002) Tissue Antigens 60, 191–205 [DOI] [PubMed] [Google Scholar]
- 31.Lopez de Castro J. A., Alvarez I., Marcilla M., Paradela A., Ramos M., Sesma L., Vázquez M. (2004) Tissue Antigens 63, 424–445 [DOI] [PubMed] [Google Scholar]
- 32.Heath W. R., Kane K. P., Mescher M. F., Sherman L. A. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W., Man S., Gulden P. H., Hunt D. F., Engelhard V. H. (1998) J. Immunol. 160, 1091–1097 [PubMed] [Google Scholar]
- 34.López de Castro J. A. (2009) in Molecular Mechanisms of Spondyloarthropathies (López-Larrea C., Díaz-Peña R. eds) Vol. 649, pp. 196–209 [Google Scholar]
- 35.Kumar P., Vahedi-Faridi A., Merino E., López de Castro J. A., Volz A., Ziegler A., Saenger W., Uchanska-Ziegler B. (2007) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 631–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garboczi D. N., Hung D. T., Wiley D. C. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 3429–3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 39.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 40.Storoni L. C., McCoy A. J., Read R. J. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 432–438 [DOI] [PubMed] [Google Scholar]
- 41.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 42.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 43.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 44.Morris R. J., Perrakis A., Lamzin V. S. (2003) Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W. (1976) Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 46.Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) Nucleic Acids Res. 35, W522–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 49.Kumar P., Ziegler A., Ziegler J., Uchanska-Ziegler B., Ziegler A. (2008) Trends Biochem. Sci. 33, 408–412 [DOI] [PubMed] [Google Scholar]
- 50.Kumar P., Vahedi-Faridi A., Saenger W., Ziegler A., Uchanska-Ziegler B. (2009) Protein Sci. 18, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loll B., Zawacka A., Biesiadka J., Petter C., Rückert C., Saenger W., Uchanska-Ziegler B., Ziegler A. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 939–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madden D. R. (1995) Annu. Rev. Immunol. 13, 587–622 [DOI] [PubMed] [Google Scholar]
- 53.Smith K. J., Reid S. W., Stuart D. I., McMichael A. J., Jones E. Y., Bell J. I. (1996) Immunity 4, 203–213 [DOI] [PubMed] [Google Scholar]
- 54.Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C. (2000) J. Immunol. 164, 6398–6405 [DOI] [PubMed] [Google Scholar]
- 55.Maenaka K., Maenaka T., Tomiyama H., Takiguchi M., Stuart D. I., Jones E. Y. (2000) J. Immunol. 165, 3260–3267 [DOI] [PubMed] [Google Scholar]
- 56.Hillig R. C., Hülsmeyer M., Saenger W., Welfle K., Misselwitz R., Welfle H., Kozerski C., Volz A., Uchanska-Ziegler B., Ziegler A. (2004) J. Biol. Chem. 279, 652–663 [DOI] [PubMed] [Google Scholar]
- 57.Waters M. L. (2002) Curr. Opin. Chem. Biol. 6, 736–741 [DOI] [PubMed] [Google Scholar]
- 58.Wedemeyer W. J., Welker E., Scheraga H. A. (2002) Biochemistry 41, 14637–14644 [DOI] [PubMed] [Google Scholar]
- 59.Menssen R., Orth P., Ziegler A., Saenger W. (1999) J. Mol. Biol. 285, 645–653 [DOI] [PubMed] [Google Scholar]
- 60.Wang C. R., Castaño A. R., Peterson P. A., Slaughter C., Lindahl K. F., Deisenhofer J. (1995) Cell 82, 655–664 [DOI] [PubMed] [Google Scholar]
- 61.Merino E., Galocha B., Vázquez M. N., López de Castro J. A. (2008) Arthritis Rheum. 58, 3693–3704 [DOI] [PubMed] [Google Scholar]
- 62.Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. (1992) Cell 70, 1035–1048 [DOI] [PubMed] [Google Scholar]
- 63.Li X., Ghandri N., Piancatelli D., Adams S., Chen D., Robbins F. M., Wang E., Monaco A., Selleri S., Bouaouina N., Stroncek D., Adorno D., Chouchane L., Marincola F. M. (2007) J. Transl. Med. 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khan M. A., Mathieu A., Sorrentino R., Akkoc N. (2007) Autoimmun. Rev. 6, 183–189 [DOI] [PubMed] [Google Scholar]
- 65.Brewerton D. A., Hart F. D., Nicholls A., Caffrey M., James D. C., Sturrock R. D. (1973) Lancet 1, 904–907 [DOI] [PubMed] [Google Scholar]
- 66.Schlosstein L., Terasaki P. I., Bluestone R., Pearson C. M. (1973) N. Engl. J. Med. 288, 704–706 [DOI] [PubMed] [Google Scholar]
- 67.Yamaguchi A., Tsuchiya N., Mitsui H., Shiota M., Ogawa A., Tokunaga K., Yoshinoya S., Juji T., Ito K. (1995) Arthritis Rheum. 38, 1672–1677 [DOI] [PubMed] [Google Scholar]
- 68.Sobao Y., Tsuchiya N., Takiguchi M., Tokunaga K. (1999) Arthritis Rheum. 42, 175–181 [DOI] [PubMed] [Google Scholar]
- 69.Falk K., Rötzschke O., Takiguchi M., Gnau V., Stevanoviæ S., Jung G., Rammensee H. G. (1995) Immunogenetics 41, 162–164 [DOI] [PubMed] [Google Scholar]
- 70.Yagüe J., Ramos M., Vázquez J., Marina A., Albar J. P., López de Castro J. A. (1999) Tissue Antigens 53, 227–236 [DOI] [PubMed] [Google Scholar]
- 71.López-Larrea C., Mijiyawa M., González S., Fernández-Morera J. L., Blanco-Gelaz M. A., Martínez-Borra J., López-Vázquez A. (2002) Arthritis Rheum. 46, 2968–2971 [DOI] [PubMed] [Google Scholar]
- 72.Díaz-Peña R., Blanco-Gelaz M. A., Njobvu P., López-Vazquez A., Suárez-Alvarez B., López-Larrea C. (2008) J. Rheumatol. 35, 2236–2240 [DOI] [PubMed] [Google Scholar]
- 73.Urvater J. A., Hickman H., Dzuris J. L., Prilliman K., Allen T. M., Schwartz K. J., Lorentzen D., Shufflebotham C., Collins E. J., Neiffer D. L., Raphael B., Hildebrand W., Sette A., Watkins D. I. (2001) J. Immunol. 166, 3334–3344 [DOI] [PubMed] [Google Scholar]
- 74.Villadangos J. A., Galocha B., García F., Albar J. P., López de Castro J. A. (1995) Eur. J. Immunol. 25, 2370–2377 [DOI] [PubMed] [Google Scholar]
- 75.Housset D., Malissen B. (2003) Trends Immunol. 24, 429–437 [DOI] [PubMed] [Google Scholar]
- 76.Tynan F. E., Burrows S. R., Buckle A. M., Clements C. S., Borg N. A., Miles J. J., Beddoe T., Whisstock J. C., Wilce M. C., Silins S. L., Burrows J. M., Kjer-Nielsen L., Kostenko L., Purcell A. W., McCluskey J., Rossjohn J. (2005) Nat. Immunol. 6, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 77.Zamyatnin A. A. (1972) Prog. Biophys. Mol. Biol. 24, 107–123 [DOI] [PubMed] [Google Scholar]