Abstract
The cannabinoid receptor 1 (CB1) and CB2 cannabinoid receptors, associated with drugs of abuse, may provide a means to treat pain, mood, and addiction disorders affecting widespread segments of society. Whether the orphan G-protein coupled receptor GPR55 is also a cannabinoid receptor remains unclear as a result of conflicting pharmacological studies. GPR55 has been reported to be activated by exogenous and endogenous cannabinoid compounds but surprisingly also by the endogenous non-cannabinoid mediator lysophosphatidylinositol (LPI). We examined the effects of a representative panel of cannabinoid ligands and LPI on GPR55 using a β-arrestin-green fluorescent protein biosensor as a direct readout of agonist-mediated receptor activation. Our data demonstrate that AM251 and SR141716A (rimonabant), which are cannabinoid antagonists, and the lipid LPI, which is not a cannabinoid receptor ligand, are GPR55 agonists. They possess comparable efficacy in inducing β-arrestin trafficking and, moreover, activate the G-protein-dependent signaling of protein kinase CβII. Conversely, the potent synthetic cannabinoid agonist CP55,940 acts as a GPR55 antagonist/partial agonist. CP55,940 blocks GPR55 internalization, the formation of β-arrestin GPR55 complexes, and the phosphorylation of ERK1/2; CP55,940 produces only a slight amount of protein kinase CβII membrane recruitment but does not stimulate membrane remodeling like LPI, AM251, or rimonabant. Our studies provide a paradigm for measuring the responsiveness of GPR55 to a variety of ligand scaffolds comprising cannabinoid and novel compounds and suggest that at best GPR55 is an atypical cannabinoid responder. The activation of GPR55 by rimonabant may be responsible for some of the off-target effects that led to its removal as a potential obesity therapy.
The CB12 and CB2 cannabinoid receptors comprise a two-member subfamily of G-protein-coupled receptors (GPCRs) that are notable as the targets of the tetrahydrocannabinol (THC) derivatives found in marijuana. More recently CB1 receptors along with other GPCRs have been promoted as therapeutic pharmacological targets in the billion dollar weight loss market for controversial drugs such as rimonabant (SR141716A) and Fen-phen. Thus, an important utility of cannabinoid family receptors to society appears to arise from their role in regulating a broad spectrum of addiction-based behaviors, and the addition of new members to the cannabinoid receptor family may have social and economic implications that reach far beyond the initial scientific discovery. As a consequence, the re-classification of an orphan GPCR as a cannabinoid family member should be done with caution requiring strict criteria of receptor activation by THC derivatives or endogenous cannabinoid compounds and a widespread agreement of the results by the scientific community.
Marijuana, one of the most widely abused substances (1), mediates many of its psychotropic effects by targeting CB1 receptors in the central nervous system, but studies with CB1 and CB2 knock-out mice indicate that the complex pharmacological properties on pain, mood, and memory exhibited by exogenous cannabinoids and the endogenous arachidonic acid-based endo-cannabinoids, including anandamide and 2-arachidonoylglycerol (2-AG), are not fully explained by their activation of CB1 and CB2 (2–4). The CB1 and CB2 receptors are 44% identical and signal through Gi/o-mediated pathways. Activation of either receptor is inhibitory for cAMP production via adenylyl cyclase and stimulatory for mitogen-activated protein kinase (MAPK) (extracellular-regulated protein kinase 1/2 (ERK1/2)) activation (5). However, the failure of these two receptors to account for the full complement of physiological effects observed with cannabinoid ligands has led to the hypothesis that additional cannabinoid-like receptors exist.
The orphan GPCR, GPR55, which exhibits only 10–15% homology to the two human cannabinoid receptors (6), is one of a number of plausible cannabinoid family member candidates (7). GPR55 was first identified and mapped to human chromosome 2q37 a decade ago (8). In the human central nervous system, it is predominantly localized to the caudate, putamen, and striatum (8), coupling to Gα13 (9, 10), Gα12, or Gαq (11).
GPR55 has been tested against a number of cannabinoid ligands with mixed results. Observations using a GTPγS functional assay indicate that GPR55 is activated by nanomolar concentrations of the endocannabinoids 2-AG, virodhamine, noladin ether, and palmitoylethanolamine (10) and the atypical cannabinoids Abn-CBD and O-1602 (12) as well as by the drugs CP55,950, HU210, and Δ9-THC (11). Exposure of GPR55 to the cannabinoids THC and JWH015 in dorsal root ganglion neurons and in receptor-transfected HEK293 cells correlates with increases of intracellular Ca2+ (11). In contrast, GPR55 is insensitive to the CB1 inverse agonist AM281 and the potent cannabinoid agonist WIN55212-2 but is antagonized by the marijuana constituent CBD (9, 10). However, Oka et al. (13) reported that GPR55 is not a typical cannabinoid receptor, as numerous endogenous and synthetic cannabinoids, including many mentioned above, had no effect on GPR55 activity. They present compelling data suggesting that the endogenous lipid LPI and its 2-arachidonyl analogs are agonists at GPR55 as a result of their abilities to phosphorylate extracellular-regulated kinase and induce calcium signaling (13, 14). Further studies indicate that LPI and the rimonabant-like CB1 inverse agonist AM251 induce oscillatory Ca2+ release through Gα13 and RhoA (9). These reports were all performed in HEK 293 cells, yet each documented a distinct and conflicting chemical space of agonists that recognized GPR55. To resolve these inconsistencies in classification, an alternative approach for identifying GPR55 ligands that is insensitive to the endogenous complement of cellular receptors could circumvent many of the challenges that have arisen in the measurements of G-protein signaling.
β-Arrestins are intracellular proteins that bind and desensitize activated GPCRs and in the process form stable receptor/arrestin signaling complexes (15, 16). β-Arrestin redistribution to the activated membrane-bound receptor represents one of the early intracellular events provoked by agonist binding and, consequently, is less prone to a false positive or negative readout as compared with studying a downstream signaling event as a readout of receptor activation. β-arrestin-green fluorescent chimeras can make this process attractive to monitor by forming remarkably sensitive and specific probes of GPCR activation that are independent of downstream G-protein-mediated signaling (17–19). We have determined GPR55 responsiveness to a representative panel of cannabinoid ligands and LPI in the presence (and absence) of a β-arrestin2-green fluorescent protein (βarr2-GFP) biosensor. Our data demonstrate that LPI, the CB1 inverse agonist/antagonists SR141716A, and AM251 are GPR55 agonists, and the CB1 agonist CP55940 is a GPR55 antagonist/partial agonist. These data together with our inability to observe activation of GPR55 by Δ9-THC and endocannabinoids indicate that GPR55 should be classified as an atypical cannabinoid receptor at best.
EXPERIMENTAL PROCEDURES
Materials
Dulbecco's modified Eagle's medium, Hanks' balanced salt solution, and fetal bovine serum were purchased from Cellgro, Mediatech, Inc. and Hyclone (Fisher). LPI, cannabidiol, poly-d-lysine, and anti-phospho-ERK antibodies were purchased from Sigma. WIN55212-2, CP55,940, O-1602, JWH015, and AM281 were obtained from Tocris (Ellsville, MO). Anandamide, methanandamide, 2-AG, AM251, O-1918, and abnormal cannabidiol (Abn-CBD) were purchased from Cayman Chemicals (Ann Arbor, MI). SR141716A and SR144528 were obtained from the National Institute on Drug Abuse drug supply program at the Research Triangle Institute). HU210 was a generous gift from Dr. R. Mechoulam (Hebrew University). Anti-HA mouse monoclonal antibody was purchased from Covance (Emeryville, CA). Actin monoclonal antibody was purchased from MP Biomedicals (Aurora, OH). Alexa Fluor 568 goat anti-mouse antibody, zeocin, SlowFade Gold, and Lipofectamine 2000 were purchased from Invitrogen. IRDye 800-conjugated anti-mouse IgG was from LI-COR. Protease inhibitors were from Roche Applied Science. G418 was purchased from A. G. Scientific (San Diego, CA). HEK293 cells were from American Type Culture Collection (Manassas, VA), and HA-GPR55E and CB1RE plasmids and cell lines were provided by the Duke University GPCR Assay Bank. All other reagents were obtained from Sigma or other standard sources.
Plasmids, Transfection, and Cell Culture
βarr2-GFP is described in Barak (17). Protein kinase CβII (PKCβII)-GFP is described in Feng et al. (20). Human GPR55 in the vector pCMV-sport6 (NIH Image Consortium) were transiently transfected in HEK293 cells with (5:1 ratio) and without βarr2-GFP or PKCβII-GFP plasmids using Lipofectamine 2000 as specified by the manufacturer or coprecipitation with calcium phosphate as previously described (17). The human N-terminal HA-tagged GPR55E receptor in pCDNA3.1zeo(−) was constructed from GPR55 by inserting the HA sequence YPYDVPDYA after the start Met of the receptor and modifying its C terminus by replacing the terminal GPR55 amino acid sequence HRPSRVQLVLQDTTISRG by the four-amino acid linker CAAA containing a putative cysteine palmitoylation site followed by the human vasopressin2 receptor terminal tail sequence RGRTPPSLGPQDESCTTASSSLAKDTSS (21). A U2OS cell line stably expressing GPR55E and βarr2-GFP (Renilla) was engineered using 0.4 mg/ml zeocin and 0.4 mg/ml G418 selection and maintained in 100 μg/ml G418 and 50 μg/ml zeocin in a 37 °C, 5% CO2 incubator. The plasmid CB1RE was constructed by replacing the human cannabinoid receptor C-tail, the segment after the amino acid sequence FPSC, by the linker AAA and the human substance P receptor tail PFISAGDYEGLEMKSTRYLQTQGSVYKVSRLETTISTVVGAHEEEPEDGPKATPSSLDLTSNCSSRSDSKTMTESFSFSSNVLS. The CB1-RE cell line was constructed as above.
β-Arrestin Assay for Determining Receptor Responsiveness
HEK 293 cells transiently expressing GPR55 receptors and βarr2-GFP were utilized 48 h after transfection. U2OS cells permanently expressing HA-GPR55E and βarr2-GFP were plated onto coverslips that were placed in 24-well plates which had been pretreated for 1 h with 0.02 mg/ml poly-d-lysine. Cells were maintained at 37 °C in 5% CO2 until ready for experiments (80–85% confluent) and washed once with Hanks' balanced salt solution before drug application. Agonist-stimulated redistribution of βarr2-GFP was assessed after drug treatment for 40 min. To measure the effects of antagonists, both agonist and antagonist were co-applied. Cells were then fixed with 4% paraformaldehyde for 20 min at room temperature followed by three washes with Hanks' balanced salt solution. Glass coverslips were mounted on slides in SlowFade Gold mounting media and were imaged on a (Nikon E800) fluorescence microscope using a 40× oil objective and 488-nm excitation for GFP and 568-nm excitation for Alexa Fluor 568 antibody. Confocal images were acquired with Leica TCS SP5 and Zeiss LSM-510 microscopes.
Internalization Assay, Immunocytochemistry
GPR55-expressing cells grown on coverslips were incubated over ice for 40 min with a 1:500 dilution of mouse monoclonal anti-HA antibody in blocking buffer (3% bovine serum albumin in phosphate-buffered saline). This was followed by appropriate washes and a 40-min incubation with 1:1500 dilution of Alexa Fluor 568 goat anti-mouse secondary antibody. Antibody-labeled cells were treated with agonist alone or in combination with antagonist for 40 min at 37 °C. Cells were imaged as described above.
On-cell Western Blot for Quantification of Receptor Internalization
GPR55E-expressing cells were grown until 90% confluent in poly-d-lysine-treated 96-well glass-bottom plates (BD Falcon). Cells were incubated with a mouse monoclonal anti-HA antibody at a 1:100 dilution for 45-min at 37 °C. Cells were washed once with phosphate-buffered saline before drug treatment, fixed in paraformaldehyde as described, and washed 3 times with phosphate-buffered saline for 5 min each. Cells were then treated with LI-COR Odyssey blocking buffer for 45 min at room temperature and then incubated in the dark with the secondary IRDye 800-conjugated anti-mouse IgG antibody diluted 1:1000 in LI-COR blocking buffer for 1 h at room temperature. Cells were then washed 5 times in TBST (137 mm NaCl, 10 mm Tris with 0.05% Tween 20) and scanned on the LI-COR Odyssey Infrared Imager set at 169 μm resolution, 4 focus offset, and 4.5–6 intensity. Data were analyzed using Excel and Prism 4.0 software.
Western Analysis for Determination of ERK Activity
GPR55E-expressing U2OS cells were grown to sub-confluence in 60-mm plates and serum-starved overnight before assay. After drug treatment the cells were disrupted in a lysis buffer (50 mm Hepes, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 10% glycerol, 1% Triton X-100, 10 μm MgCl2, 20 mm p-nitrophenyl phosphate, 1 mm Na3VO4, 25 mm NaF, and a protease inhibitor mixture (1:25, pH 7.5). Lysates were immediately placed on ice for 10 min and then centrifuged at 16,000 × g for 30 min at 4 °C. Supernatants, corresponding to the cytosolic fraction, were collected, and protein concentrations were determined by the Bradford assay (Bio-Rad) using bovine serum albumin as a standard. Cytosolic fractions (20 μg) were separated on a 10% gel by SDS-PAGE followed by immunoblotting (22). Antibodies against double-phosphorylated ERK1/2 (1:5000) were detected using a Fuji imager LAS-1000 (Fujifilm Life Science, Woodbridge, CT). A monoclonal antibody against actin (1:10,000) was used to confirm equal protein loading. Densitometric analysis was performed using ImageJ software (rsb.nih.gov/ij). The value obtained for both ERK1 and ERK2 was normalized to anti-actin levels. The data were normalized to control and presented as percentage stimulation.
PKCβII Assay for Determining Receptor Responsiveness
HEK 293 cells plated in 35-mm glass well Matek plastic dishes were transiently transfected with 175 μl of solution containing 1.5 μg/ml PKCβII-GFP cDNA or the PKC plasmid and 5 μg/ml human GPR55 cDNA in pCMV-Sport6 (Open Biosystems, Huntsville, Al) using a standard calcium phosphate protocol. Cells expressing GPR55 and PKCβII-GFP were utilized 24 h after transfection. Cells were washed with warm minimum Eagle's medium and maintained at 37 °C in 5% CO2 for 30–45 min after drug application. Agonist stimulated redistribution of PKCβII-GFP was assessed after drug treatment at room temperature.
Data Analysis
βarr2-GFP aggregates were identified using a wavelet-based Microsoft Windows compatible computer program written in the MatLab programming environment. A program algorithm extracts from two-dimensional images those pixels that generate objects of interest that fall within a predetermined range of sizes and intensities and that are embedded among widely varying local backgrounds (L. Barak, available from the Duke University GPCR Assay Bank). Concentration-effect curves for agonist-mediated receptor activation and competition-inhibition curves for antagonist studies were analyzed by nonlinear regression techniques using GraphPad Prism 4.0 software (GraphPad, San Diego, CA), and data were fitted to sigmoidal dose-response curves to obtain EC50 or IC50 values. The KI (apparent) value of the antagonist was calculated using the Cheng-Prusoff equation, KI = IC50/(1 + [L]/EC50) (23), where [L] is the agonist concentration. Statistical analysis was performed using one-way analysis of variance followed by Dunnett's post-test or two-tailed unpaired Student's t test. p values of <0.05 were considered significant.
RESULTS
GPR55 and HA-GPR55 Recruit βarr2-GFP in Response to LPI
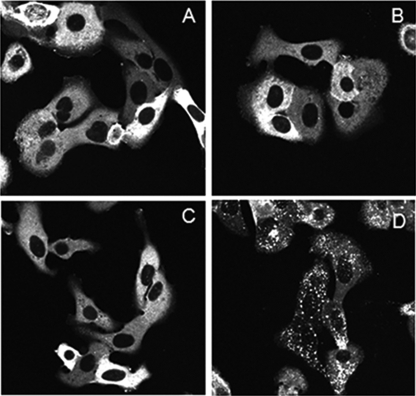
Arrestin proteins mediate GPCR desensitization by binding to activated GPCRs, and in the process the arrestins relocate from the cytosol to form a complex with the membrane receptor. The strength of this association determines the subsequent fate of the complex, with weaker β-arrestin-receptor complexes dissociating at the plasma membrane while in clathrin-coated pits and more stable ones internalizing and concentrating in cytosolic endosomes (18, 21). In the absence of agonist, β-arrestin-GFP is uniformly distributed in the cytoplasm with no apparent compartmentalization at the plasma membrane or nucleus (Fig. 1, A and D). Using β-arrestin-GFP as a biosensor of receptor activation, the relatively weak β-arrestin-receptor complexes are observed as membrane-associated fluorescence aggregates and the more stable ones as brighter intracellular objects. HEK293 cells transiently transfected with human GPR55 and βarr2-GFP develop membrane aggregates when treated with 10 μm LPI (Fig. 1, A and B). The addition of serine phosphorylation sites to a GPCR C-tail can increase receptor affinity for arrestin without changing its response profile to different ligands (21, 24). Therefore, to increase GPR55 assay sensitivity, we employed an HA-epitope-tagged variant of GPR55 with a serine enhanced C terminus (HA-GPR55E). This resulted in a much more robust βarr2-GFP response in HEK-293 cells when HA-GPR55E was exposed to 10 μm LPI (Fig. 1C). A time-course analysis of ligand treatment demonstrated robust agonist-mediated βarr2-GFP trafficking at 40-min ligand treatment (data not shown). Consequently, we used U2OS cells stably transfected with HA-GPR55E and βarr2-GFP for subsequent ligand characterization. Fig. 1D shows the pattern of βarr2-GFP expression in U2OS cells in the absence of agonist. No βarr2-GFP fluorescence was observed at the plasma membrane. Immunofluorescence images of anti-HA antibody labeling followed by Alexa Fluor secondary antibody verify the plasma membrane expression of HA-tagged GPR55E (Fig. 1E), with no membrane fluorescence visible in the absence of primary HA-antibody treatment (Fig. 1G).
FIGURE 1.
Confocal microscopy of co-transfected βarr2-GFP and GPR55. A, transiently transfected βarr2-GFP (in cytoplasm, see the arrow) and GPR55 in untreated HEK293 cells. B and C, 10 μm LPI induces redistribution of βarr2-GFP in HEK 293 cells expressing GPR55 (B) and HA-GPR55E (C). D and E, corresponding GFP and Alexa Fluor 568 fluorescence channels of a field of U2OS cells stably transfected with βarr2-GFP and HA-GPR55E. D, under basal conditions the βarr2-GFP is uniformly distributed in the cell cytoplasm. Panel E shows the plasma membrane staining of HA-GPR55E in these cells using anti-HA epitope mouse primary antibody and goat anti-mouse Alexa Fluor 568 secondary antibody. F depicts the overlay of panel D and E. G, cells as in E, in which treatment with primary anti-HA antibody is omitted. No membrane staining of receptor is observed in the Alexa Fluor 568 channel.
Agonist-stimulated βarr2-GFP Trafficking in GPR55E U2OS Cells
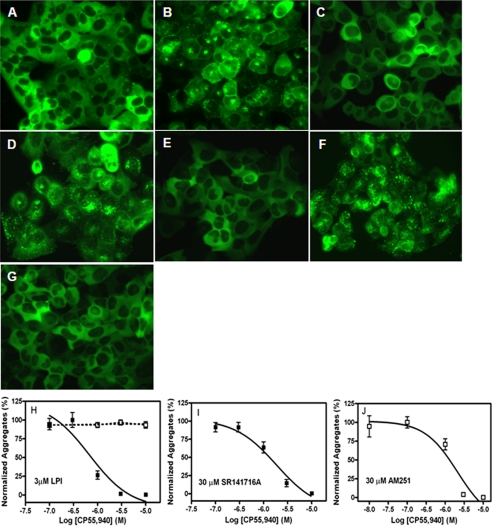
Using real-time confocal microscopy in live U2OS cells stably coexpressed with GPR55E and βarr2-GFP, we observed that 3 μm LPI induced a rapid relocation (60–90s time scale) of βarr2-GFP complex en masse to plasma membrane-bound GPR55E with a concomitant depletion of cytosolic fluorescence (see the supplemental videos). We next investigated the concentration dependence of ligands to induce internalization of GPR55·βarr2-GFP complexes. This LPI mediated-response of βarr2-GFP occurs with an EC50 of 1.2 μm (Figs. 2, A and B), whereas the CB1 receptor inverse agonist/antagonists SR141716A and AM251 produce recruitment of βarr2-GFP to GPR55E receptors with EC50 values of 3.9 and 9.6 μm, respectively (Fig. 2, C–F). Thus, these ligands act as GPR55E agonists and have efficacies similar to LPI (Fig. 2H). We also treated the GPR55E/βarr-GFP U2OS cells with a group of 15 structurally diverse cannabinoid ligands comprised of classic, non-classic, and endogenous agonists and antagonists (Table 1). None of the compounds at concentrations upward of 10–30 μm activated GPR55E to produce a distribution of βarr2-GFP different from the basal state or that observed in vehicle-treated cells. As a control, U2OS cells expressing CB1 receptors were exposed to vehicle, LPI, and SR141716A, and no βarr2-GFP trafficking was observed (Fig. 3, A–C), whereas CP55,940, a potent CB1 receptor agonist, activated βarr2-GFP trafficking in these cells (Fig. 3D).
FIGURE 2.
Agonist induced trafficking of βarr2-GFP in U2OS cells containing GPR55E. The redistribution of βarr2-GFP fluorescence was visualized after a 40-min treatment with 3 μm LPI (A), 30 μm SR141716A (C), and 30 μm AM251 (E). B, D, and F represent the concentration-response curves for LPI, SR141716A, and AM251, respectively. G shows the cytosolic distribution of βarr2-GFP in vehicle-treated cells. 4′,6-diamidino-2-phenylindole nuclear staining (arrows) shows nuclear exclusion of βarr2. H, the maximum number of β-arrestin aggregates (objects) was determined from the calculated plateaus of the concentration-response curves for each compound to determine relative efficacies. The data represent the mean ± S.E. from at least three independent experiments where triplicate images of fields containing multiple cells were analyzed. Representative images that were captured at 40× magnification are depicted.
TABLE 1.
β-Arrestin-dependent ligand-mediated activation of GPR55E
CI, confidence interval.
| Ligand | GPR55 EC50 (95% CI) | Relative efficacy ± S.E. | CB1 (KI)a or EC50 |
|---|---|---|---|
| LPI | 1.2 μm (0.3–4.0), agonist | 1.0 ± 0.13 | No response up to 30 μm |
| SR141716A | 3.9 μm (0.9–17), agonist | 0.85 ± 0.15 | (3.3 nm)a (31), inverse agonist |
| AM251 | 9.6 μm (3.6–26), agonist | 1.21 ± 0.13 | (7.5 nm)a (31), inverse agonist |
| AM281 | No response up to 30 μm | (21 nm)a (31), inverse agonist | |
| 2-AG | No response up to 30 μm | 30 nm (42), agonist | |
| Anandamide | No response up to 30 μm | 116 nm, agonistb | |
| Palmitoyl ethanolamide | No response up to 30 μm | ||
| Methanandamide | No response up to 30 μm | ||
| THC | No response up to 30 μm | 6 nm (10), agonist | |
| HU210 | No response up to 30 μm | 70.2 pm (31), agonist | |
| CP55,940 | No response up to 30 μm | 1.2 nm (31), agonist | |
| O-1602 | No response up to 30 μm | >30,000 nm (10), agonist | |
| O-1918 | No response up to 30 μm | No binding (43) | |
| Abn-CBD | No response up to 30 μm | No binding (43) | |
| CBD | No response up to 30 μm | >30,000 nm (10), agonist | |
| SR144528 | No response up to 30 μm | (0.7 nm)a (44), CB2 antagonist | |
| WIN55212-2 | No response up to 30 μm | 5.5 nm (31), agonist | |
| JWH-015 | No response up to 30 μm | (13.8 nm) (45), CB2 agonist |
Previously published.
A. Kapur, P. Zhao, H. Sharir, Y. Bai, M. G. Caron, L. S. Barak, and M. E. Abood, unpublished information.
FIGURE 3.
Ligand induced trafficking of βarr2-GFP in U2OS cells containing CB1RE, a receptor with a phosphorylation site-enhanced C-tail. Shown are representative live cell confocal images of a line of U2OS cells permanently transfected with CB1E receptors and β2-arrestin GFP. Cells were treated for 40 min at 37 °C with media containing vehicle (A), 10 μm LPI (B), 10 μm SR141716A (C), and 3 μm CP55,940 (D).
Antagonist Inhibition of βarr2-GFP Trafficking in GPR55E U2OS Cells
We next tested whether cannabinoid ligands that failed as GPR55 agonists could be GPR55 antagonists by measuring their ability to block receptor activation and βarr2-GFP redistribution. The non-classical cannabinoid agonist CP55940 produced a concentration-dependent inhibition of LPI-induced redistribution of βarr2-GFP with a KI of 194 nm (Fig. 4, B, C, and H, and Table 2). Likewise, CP55,940 antagonized the ability of the GPR55 agonists SR141716A and AM251 to recruit arrestins with KI values of 213 nm (Fig. 4, D, E, and I) and 540 nm (Fig. 4, F, G, and J), respectively. The remaining compounds from Table 1 failed to block LPI-mediated βarr2-GFP recruitment at concentrations of 10–30 μm (data not shown).
FIGURE 4.
CP55,940-mediated antagonism of βarr2-GFP in U2OS cells containing GPR55E. βarr2-GFP cytosolic fluorescence was visualized after a 40-min treatment of the GPR55E cells with the various combinations of ligands to assess the ability of CP55,940 to act as an antagonist. A, incubation with 3 μm CP55,940 alone. B, 100 nm CP55,940 was applied in combination with 3 μm LPI. C, 3 μm CP55,940 was applied along with 3 μm LPI. D, 100 nm CP55,940 was applied in combination with 30 μm AM251. E, 3 μm CP55,940 was applied with 30 μm AM251. F, 100 nm CP55,940 was applied along with 30 μm SR141716A. G, 3 μm CP55,940 was applied in combination with 30 μm SR141716A Images A-G were acquired 40× magnification. Panels H–I shows the dose-dependent CP55,940-mediated inhibition of β-arrestin redistribution in the absence (dotted line) or presence of 3 μm LPI (H), 30 μm SR141716A (I), and 30 μm AM251 (J). Each curve represents the analysis of at least three fields of cells at each set of ligand concentrations from three independent experiments ±S.E.
TABLE 2.
CP55,940-mediated inhibition of β−arrestin2 redistribution to GPR55
Log IC50 ± S.E. values were determined from concentration-effect curves. KI values were determined from IC50 values as described under “Experimental Procedures.”
| Competitive agonist | Log IC50 ± S.E. | IC50 | Calculated KI |
|---|---|---|---|
| m | nm | nm | |
| LPI | −6.17 ± 0.17 | 678 | 194 |
| LPI (On-Cell)a | −5.92 ± 0.15 | 1210 | 173 |
| SR141716A | −5.73 ± 0.15 | 1850 | 213 |
| AM251 | −5.56 ± 0.42 | 2750 | 542 |
Inhibition of LPI-mediated receptor internalization.
Agonist-induced HA-GPR55E Internalization in U2OS Cells
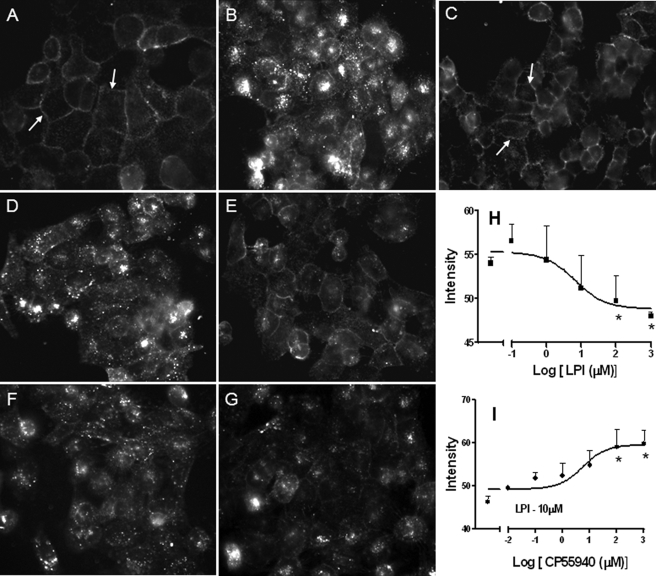
Many GPCRs undergo endocytosis from the plasma membrane by a clathrin/dynamin-mediated mechanism in response to agonist activation (25–27). Therefore, we examined whether LPI, SR141716A, and AM251 application could decrease the number of plasma membrane GPR55E receptors as an additional readout for receptor activation. Immunofluorescence labeling of live HA-GPR55E U2OS cells using anti HA monoclonal antibodies revealed a rich complement of membrane receptors (Fig. 5A). Exposing the cells to 3 μm LPI for 40 min resulted in a dramatic loss of membrane staining and the appearance of cytosolic fluorescent aggregates corresponding to internalized receptors (Fig. 5B). Treatment of cells with 30 μm SR141716A or AM251 also induced robust receptor-mediated receptor internalization (Fig. 5, D and F). As expected from previous results, CP55,940 at 3 μm inhibited LPI-, SR141716A-, or AM251-mediated HA-GPR55E internalization (Fig. 5, C, E, and G), as plasma membrane receptor fluorescence in cells with CP-55,940 was comparable with vehicle-treated cells.
FIGURE 5.
Agonist-mediated GPR55E internalization in U2OS Cells. Live U2OS cells expressing HA-GPR55E prelabeled with anti-HA mouse antibody and Alexa Fluor 568 secondary antibody were treated with various compounds and the membrane staining imaged by fluorescence microscopy at 40X magnification. Receptor internalization resulted in a loss of cell surface immunofluorescence. A, vehicle-treated cells show predominantly plasma membrane receptor staining. B, D, and F, internalization of membrane-bound GPR55 occurs upon treatment with 3 μm LPI, 30 μm SR141716A, and 30 μm AM251, respectively. C, E, and G, the co-application of 3 μm CP55,940 attenuated receptor internalization and plasma membrane receptor staining in the presence of 3 μm LPI (C), 30 μm SR141716A (E), and 30 μm AM251 (G). On-Cell Western analysis is shown in H and I of the concentration-dependent LPI-induced loss of plasma membrane receptor (H) and concentration-dependent CP55,940-mediated antagonism of membrane receptor loss in the presence of 10 μm LPI(I). *, p < 0.05 was deemed statistically significant.
To determine ligand potency, an On-Cell Western analysis was performed to measure the concentration-dependent complement of cell surface receptors. LPI reduced HA-GPR55E membrane fluorescence, with an EC50 value of 1.6 ± 0.12 μm (Fig. 5H). In contrast, CP55,940 prevented the LPI-induced loss of plasma membrane HA-GPR55E, with a KI of 173 nm (Fig. 5I, Table 2).
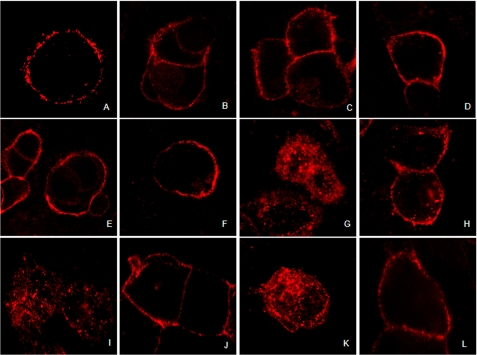
We next extended our GPR55 internalization studies (in the absence of βarr2-GFP) in an additional cell line, HEK293, to overcome any cellular bias. HEK293 cells transiently transfected with HA-tagged GPR55E were labeled and treated as described under “Materials and Methods.” Cell surface labeling was visualized using confocal microscopy after treatment with various ligands. Treatment with 30 μm 2-AG and anandamide (endocannabinoids), 30 μm THC (prototypical classic agonist), or 10 μm CP55,940 (non-classic agonist) had no effect on membrane-stained HA-GPR55 and resembled vehicle-treated cells (Fig. 6, A–F). As in U2OS cells (Fig. 5), treatment of cells with 3 μm LPI, 30 μm SR141716A, or AM251 resulted in loss of membrane staining (Fig. 6, G, I, and K), an effect that was reversed with antagonist treatment, CP55940 (Fig. 6, H, J, and L).
FIGURE 6.
Agonist-mediated GPR55E internalization in HEK293 cells transiently transfected with HA-GPR55E. Live HEK293 cells expressing HA-GPR55E prelabeled with anti-HA mouse antibody and Alexa Fluor 568 secondary antibody were treated with various compounds, and membrane staining was imaged by confocal microscopy at 63× magnification. Receptor internalization resulted in a loss of cell surface immunofluorescence. A, vehicle-treated cells show predominantly plasma membrane receptor staining. B–F, treatment with 30 μm 2-AG, 30 μm anandamide, 30 μm THC, 30 μm O-1602, and 10 μm CP55,940, respectively, resulted in no loss of receptor surface staining and resembled vehicle treated cells. G, I, and K, internalization of membrane-bound GPR55 occurs upon treatment with 3 μm LPI, 30 μm SR141716A, and 30 μm AM251, respectively. H, J, and L, the co-application of 3 μm CP55,940 in the presence of 3 μm LPI, 30 μm SR141716A, and 30 μm AM251, respectively, attenuated receptor internalization and restored plasma membrane receptor staining.
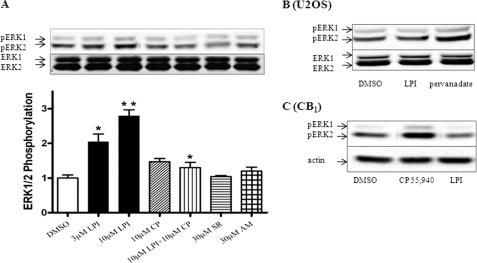
LPI Induced Phosphorylation of ERK1/2 in GPR55
A key consequence of GPCR activation is the induction of the ERK, MAPK signaling cascade (28). LPI has previously been shown to activate ERK in GPR55-transfected HEK293 cells (13). In Fig. 7A we show that LPI induced a significant activation of ERK1/2 in U2OS cells expressing HA-GPR55E. LPI-induced ERK1/2 phosphorylation did not occur in untransfected U2OS cells. LPI-dependent ERK1/2 phosphorylation was evident at 5 min and peaked at 10 min (data not shown). In contrast, AM251 and SR141716A did not induce any significant phosphorylation of ERK1/2 (pERK) in U2OS cells expressing GPR55. Likewise, the prototypical endocannabinoids anandamide and 2-AG did not activate ERK1/2 (data not shown). Co-application of CP55,940 with LPI inhibited LPI-mediated ERK activation. Furthermore, LPI-induced ERK activation was sensitive to inhibition by 3 μm MEK (ERK kinase) inhibitor U0126 (data not shown). The ERK signaling pathway was still intact in GPR55-deficient cells (U2OS cells) as treatment with 1 mm pervanadate, a potent inhibitor of tyrosine phosphatases and indirect activator of MEK (29) resulted in a marked increase of ERK1/2 phosphorylation (Fig. 7B). Moreover, LPI did not activate ERK1/2 in CB1E-transfected cells, in contrast to 10 μm CB1 agonist CP55,940, which caused a pronounced ERK1/2 phosphorylation as compared with vehicle-treated cells (Fig. 7C).
FIGURE 7.
LPI mediated ERK1/2 phosphorylation in U2OS cells containing GPR55E. A, GPR55E cells treated for 10 min with 3 and 10 μm LPI demonstrated ERK1/2 activation significantly different from vehicle-treated cells (*, p < 0.05; **, p < 0.001). Co-application of 10 μm CP55,940 significantly inhibited 10 μm LPI-mediated ERK1/2 activation. In contrast, 10 μm CP55,940 (CP)-, 30 μm SR141716A (SR)-, and AM251 (AM)-mediated ERK activation was not different from vehicle treatment. The data represent the mean ± S.E. from at least three independent experiments performed in duplicate. B, 10 μm LPI fails to evoke ERK1/2 activation in untransfected U2OS cells, whereas activation of ERK occurs with 1 mm pervanadate treatment. C, in U2OS cells stably expressing the CB1RE, 10 μm LPI failed to activate ERK1/2, whereas treatment with 10 μm CB1 agonist, CP 55,940, resulted in a marked increase of ERK1/2 activation.
Likewise, in HEK293 cells transiently transfected with GPR55 (in the absence of βarr2-GFP), LPI induced a similar extent of ERK1/2 phosphorylation, whereas AM251, SR141716A, anandamide, and 2-AG did not produce an effect different from that observed in vehicle-treated cells (data not shown).
GPR55 Mediated PKCβII Activation
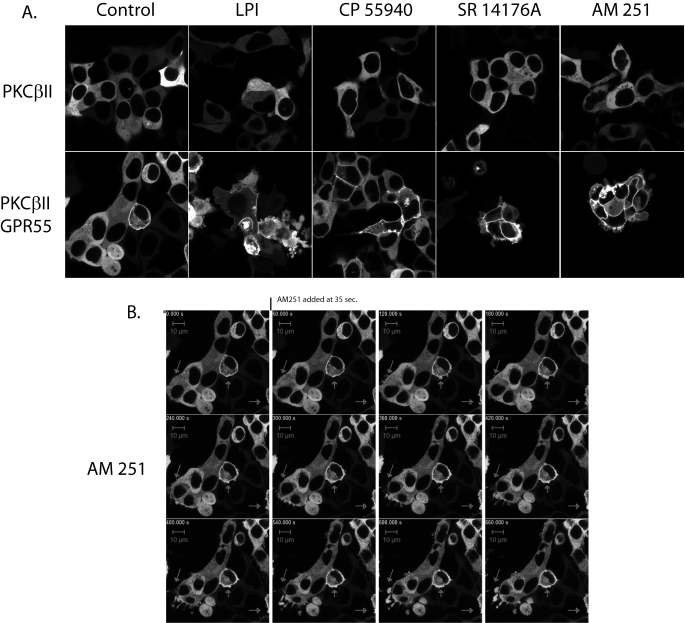
Activation of GPR55 has been shown to elicit release of intracellular calcium via activation of phospholipase C (9, 11). GPR55 has been suggested to couple through Gq, G12, and G13 (9, 11). Therefore, as a measure of GPR55-mediated G-protein signaling, we tested the ability of the agonists identified in this report, LPI, AM251, and rimonabant, to recruit PKCβII to the plasma membrane or to produce membrane remodeling or blebbing (20, 30). 10 μm concentrations of each drug were applied for 30–45 min to cells containing PKCβII-GFP with and without GPR55 expression. The live cells were then examined by confocal microscopy (Fig. 8A). No translocation of PKCβII-GFP was observed in the absence of co-transfection with GPR55 (Fig. 8A, upper and lower left panels). However, some constitutive recruitment of PKCβII-GFP to the plasma membrane along with an increased rounding of cells was observed in untreated GPR55-transfected cells. The addition of 10 μm CP55,940 produced a slight increase in PKC recruitment over controls but no increased membrane blebbing or remodeling (Fig. 8A, upper and lower panels, third from the left), suggesting that CP55,940 may only weakly activate G-proteins at GPR55. In contrast, AM251, LPI, and rimonabant (SR14176A) treatment of GPR55 cells produced not only increases in PKCβII-GFP recruitment to the plasma membrane but dramatic remodeling of the plasma membrane characterized by blebbing and the formation of large membrane protrusions (Fig. 8A, lower panels, second, fourth, and fifth from the left). AM251 appeared the most efficacious of the three agonists in inducing PKCβII-GFP activation, and an AM251 treatment time course of PKCβII-GFP recruitment showed that remodeling began within 45 s of compound addition (Fig. 8B; also see the supplemental videos).
FIGURE 8.
Agonist induced trafficking of PKCβII in HEK293 cells transiently transfected with PKCβII-GFP and GPR55. A, live HEK293 cells expressing PKCβII-GFP or PKCβII-GFP + GPR55 were treated with the indicated compounds, and membrane staining was imaged 30–45 min after drug application by confocal microscopy using a 40× oil objective. B, time series of live cell imaging of PKCβII-GFP recruitment after the addition of 10 μm AM251. Arrows indicate that within 60 s of the addition of AM251, membrane rearrangements begin to occur followed by protrusions and blebbing of the cell membranes.
DISCUSSION
In this study we have exploited the property that β-arrestins recognize the activated state of GPCRs to clarify a GPR55 pharmacology that has remained clouded by conflicting studies (10, 12, 13). We observed that of numerous cannabinoid compounds tested, only two unambiguously activated GPR55 in addition to the lysophospholipid LPI. These compounds, LPI, SR141716A, and AM251, have a rank order of potency of LPI > SR141716A > AM251 and comparable efficacies. Interestingly, SR141716A and AM251 are cannabinoid receptor inverse agonist/antagonists (31, 32), whereas the one GPR55 receptor antagonist we identified, CP55,940, is a cannabinoid receptor agonist. In contrast, AM281, which is structurally related to biarylpyrazole analogs (SR141716A and AM251), failed to activate GPR55. Such distinct pharmacological profiles of ligands at two receptors can provide valuable structure-activity information for the development of ligands with high specificity. Furthermore, our assays confirm that the endogenous compound LPI is unequivocally a GPR55 agonist at low micromolar concentrations, causing β-arrestin activation, receptor internalization, and ERK1/2 phosphorylation.
A consensus among our findings reported here and in previous studies (10–12) is that the aminoalkylindole, WIN55212-2, a potent CB1 and CB2 receptor agonist, does not activate GPR55. However, JWH015, a structurally related aminoalkylindole analog, has been shown to increase intracellular Ca2+ (11). In our hands, JWH015 (CB2 receptor agonist), SR144528 (CB2 receptor antagonist), the classical CB1 agonists (HU210 and THC), the endocannabinoids (anandamide and 2-AG), and cannabidiol had no effect on their own or on LPI-induced βarr2 trafficking in GPR55 U2OS cells. Our study also demonstrates that the endocannabinoids (anandamide and 2-AG) and atypical cannabinoids (Abn-CBD, O-1602, and O-1918) fail to evoke GPR55-modulated βarr2 redistribution. These compounds had previously been reported to activate GTPγS binding (10, 12), but to date there have been no additional reports confirming these data. The most parsimonious explanation of this apparent disagreement with previous reports (10–13) is that distinct conformations of the receptor resulting from the binding of different ligands might couple differentially and/or to multiple downstream effectors (6, 33). In addition, agonist-mediated receptor internalization in HEK293 cells (in the absence of βarr-GFP) are consistent with our β-arrestin recruitment studies that LPI, SR141716A, and AM251 are agonists at GPR55E and CP55,940 antagonizes their effects.
GPR55 has been shown to utilize Gq, G12, or G13 for signal transduction; RhoA and phospholipase C are activated (9, 11). This signaling mode is associated with temporal changes in cytoplasmic calcium, membrane-bound diacylglycerol, diacylglycerol, and plasma membrane topology. PKCβII is a conventional PKC isoform that transduces calcium- and diacylglycerol-dependent signals (34). Thus, PKCβII-GFP activation reflects intracellular calcium and diacylglycerol bioavailability, which may be sustained for extended periods after ligand stimulation (34). Our findings that LPI, SR141716A, and AM251 recruit PKCβII-GFP and cause widespread plasma membrane remodeling indicate that they mediate a GPR55 G-protein signaling pathway. Moreover, we found that these compounds are agonists for both β-arrestin-dependent and -independent signaling, suggesting that the most plausible explanation for the complement of our β-arrestin findings is that GPR55 signals differently from the CB1 cannabinoid receptors and that less common pharmacological explanations such as biased agonism are inconsistent with our findings (35).
Lauckner et al. (11) reported that SR141716A is a GPR55 antagonist at 2 μm in a calcium signaling assay. We clearly show that 10–30 μm SR141716A produces robust activation and internalization of GPR55. This discrepancy may be a reflection of the different range of doses and efficacy of the compounds that were utilized. That rimonabant (SR141716A) activates GPR55 at 10 μm concentrations may be clinically relevant. This drug had been marketed for the treatment of obesity, and off-target effects of this and related compounds may be manifest at GPR55.
Our observation that LPI alone induces a significant activation of ERK1/2 in cells expressing GPR55, whereas cannabinoids (AM251, SR141716A, anandamide, and 2-AG) fail to do so are in agreement with Oka et al. (13). This divergence in response suggests that LPI and cannabinoids display functional selectivity in modulating the MAPK signaling pathways in GPR55, a phenomenon widely known to occur in GPCRs (36).
The physiological relevance of GPR55 has been investigated in GPR55 knock-out mice (37). These GPR55−/− mice are resistant to mechanical hyperalgesia and have increased levels of anti-inflammatory cytokines as compared with wild-type animals. These data suggest that the manipulation of GPR55 may have therapeutic potential in the treatment of both inflammatory and neuropathic pain. It is tempting to speculate that some of the known antinociceptive effects of CP55,940 may also be mediated by action at GPR55 receptors.
Recently published studies characterizing GPR55 activation using a related technology (β-arrestin-PathHunter) or demonstrating that monitoring agonist-mediated receptor internalization and β-arrestin redistribution can recognize agonists, antagonists, and allosteric modulators support our findings (38, 39). The preponderance of data emerging from particularly this and other reports (9–11, 38) indicate that GPR55 is not activated by cannabinoid receptor ligands in a manner compatible with expected cannabinoid receptor pharmacology. Whether GPR55 can be categorized as a putative cannabinoid receptor in light of this and the finding that a large majority of the endogenous and synthetic cannabinoids tested failed to elicit β-arrestin trafficking, a key element of the GPCR signaling mechanism, is problematic. Of all the compounds tested by the various groups, LPI is the only endogenous one with the potential to signal through both G-protein and β-arrestin pathways (6, 33). This raises the questions of whether LPI is the “endogenous ligand” of GPR55 or whether more potent endogenous ligands remain to be found that might serve to counter the CB1 receptor and are chemically related to SR141716A. At best, GPR55 would certainly be an atypical representative of the cannabinoid receptor family. To provide a justification for classifying GPR55 as an additional cannabinoid receptor requires its further characterization by computational modeling and mutational studies to demonstrate a substantial relationship between its ligand binding motif and those of the CB1 and CB2 receptors (31, 40, 41).
Supplementary Material
Acknowledgment
We thank Dr. Robert Oakley for help in the development of many of the assays used in this work.
This work was supported, in whole or in part, by National Institutes of Health Grants DA023204 and DA05274 (to M. E. A.) and DA022950 (National Institute on Drug Abuse; to M. G. C. and L. S. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental videos.
- CB1
- cannabinoid receptor 1
- GPCR
- G-protein-coupled receptor
- LPI
- lysophosphatidylinositol
- CP55940
- (-)-3-[2-hydroxyl-4-(1,1-dimethylheptyl)phenyl]-4-[3-hydroxyl propyl] cyclohexan-1-ol
- HU210
- (−)-11-hydroxyl-Δ8-tetrahydrocannabinol-dimethylheptyl
- SR141716A
- N-(piperidin-1-yl)-5-(4-chlorophenyl)-1-(2,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carboxamide
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- WIN 5212-2
- (R)-(+)-[2,3-dihydro-5-methyl-3-[(4-morpholinyl)methyl]pyrrolo[1,2,3-de]-1,4-benzoxazin-6-yl](1-naphthalenyl)methanone
- JWH015
- (2-methyl-1-propyl-1H-3yl)-1-naphthalenyl-methanone
- 2-AG
- 2-arachidonoyl glycerol
- AM281
- 1-(2,4-dicholorophenyl)-5-(4-iodophenyl)-4-methyl-N-1-piperidinyl-1H-pyrazole-3-carboxamide
- AM251
- 1-(2,4-dicholorophenyl)-5-(4-iodophenyl)-4-morphoniyl-1H-pyrazole-3-carboxamide
- O-1602
- 5-methyl-4-[(1R,6R)-3-methyl-6-(1-methylenyl)-2-cyclohexen-1-yl]-1,3-benzenediol
- O-1918
- 1,3-dimethoxy-5-methyl-2-[(1R,6R)-3-methyl-6-(1-methylethenyl)-2-cyclohexen-1-yl]-benzene
- cannabidiol
- 2-[1R-3-methyl-6R-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-1,3-benzenediol
- Abn-CBD
- abnormal cannabidiol, 4-[3-methyl-6R-(1-methylethenyl)-2-cyclohexen-1-yl]-5-pentyl-1,3-benzenediol
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular-regulated protein kinase
- PKC
- protein kinase C
- THC
- tetrahydrocannabinol
- LPI
- lysophosphatidylinositol
- GFP
- green fluorescent protein
- βarr
- 2β-arrestin2
- HA
- hemagglutinin.
REFERENCES
- 1.Murray R. M., Morrison P. D., Henquet C., Di Forti M. (2007) Nat. Rev. Neurosci. 8, 885–895 [DOI] [PubMed] [Google Scholar]
- 2.Breivogel C. S., Griffin G., Di Marzo V., Martin B. R. (2001) Mol. Pharmacol. 60, 155–163 [PubMed] [Google Scholar]
- 3.Hájos N., Freund T. F. (2002) Chem. Phys. Lipids 121, 73–82 [DOI] [PubMed] [Google Scholar]
- 4.Wagner J. A., Varga K., Járai Z., Kunos G. (1999) Hypertension 33, 429–434 [DOI] [PubMed] [Google Scholar]
- 5.Felder C. C., Joyce K. E., Briley E. M., Mansouri J., Mackie K., Blond O., Lai Y., Ma A. L., Mitchell R. L. (1995) Mol. Pharmacol. 48, 443–450 [PubMed] [Google Scholar]
- 6.Ross R. A. (2009) Trends Pharmacol. Sci. 30, 156–163 [DOI] [PubMed] [Google Scholar]
- 7.Baker D., Pryce G., Davies W. L., Hiley C. R. (2006) Trends Pharmacol. Sci. 27, 1–4 [DOI] [PubMed] [Google Scholar]
- 8.Sawzdargo M., Nguyen T., Lee D. K., Lynch K. R., Cheng R., Heng H. H., George S. R., O'Dowd B. F. (1999) Brain Res. Mol. Brain Res. 64, 193–198 [DOI] [PubMed] [Google Scholar]
- 9.Henstridge C. M., Balenga N. A., Ford L. A., Ross R. A., Waldhoer M., Irving A. J. (2009) FASEB J. 23, 183–193 [DOI] [PubMed] [Google Scholar]
- 10.Ryberg E., Larsson N., Sjögren S., Hjorth S., Hermansson N. O., Leonova J., Elebring T., Nilsson K., Drmota T., Greasley P. J. (2007) Br. J. Pharmacol. 152, 1092–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lauckner J. E., Jensen J. B., Chen H. Y., Lu H. C., Hille B., Mackie K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 2699–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johns D. G., Behm D. J., Walker D. J., Ao Z., Shapland E. M., Daniels D. A., Riddick M., Dowell S., Staton P. C., Green P., Shabon U., Bao W., Aiyar N., Yue T. L., Brown A. J., Morrison A. D., Douglas S. A. (2007) Br. J. Pharmacol. 152, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka S., Nakajima K., Yamashita A., Kishimoto S., Sugiura T. (2007) Biochem. Biophys. Res. Commun. 362, 928–934 [DOI] [PubMed] [Google Scholar]
- 14.Oka S., Toshida T., Maruyama K., Nakajima K., Yamashita A., Sugiura T. (2009) J. Biochem. 145, 13–20 [DOI] [PubMed] [Google Scholar]
- 15.Gurevich E. V., Gurevich V. V. (2006) Genome Biology 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenoy S. K., Lefkowitz R. J. (2005) Nat. Cell Biol. 7, 1159–1161 [DOI] [PubMed] [Google Scholar]
- 17.Barak L. S., Ferguson S. S., Zhang J., Caron M. G. (1997) J. Biol. Chem. 272, 27497–27500 [DOI] [PubMed] [Google Scholar]
- 18.Marion S., Oakley R. H., Kim K. M., Caron M. G., Barak L. S. (2006) J. Biol. Chem. 281, 2932–2938 [DOI] [PubMed] [Google Scholar]
- 19.McGuinness D., Malikzay A., Visconti R., Lin K., Bayne M., Monsma F., Lunn C. A. (2009) J. Biomol. Screen 14, 49–58 [DOI] [PubMed] [Google Scholar]
- 20.Feng X., Zhang J., Barak L. S., Meyer T., Caron M. G., Hannun Y. A. (1998) J. Biol. Chem. 273, 10755–10762 [DOI] [PubMed] [Google Scholar]
- 21.Oakley R. H., Laporte S. A., Holt J. A., Barak L. S., Caron M. G. (1999) J. Biol. Chem. 274, 32248–32257 [DOI] [PubMed] [Google Scholar]
- 22.Gallagher S., Winston S. E., Fuller S. A., Hurrell J. G. (2008) Curr. Protoc. Immunol. Chapter 8, Unit 8.10 [DOI] [PubMed] [Google Scholar]
- 23.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 24.Haasen D., Schnapp A., Valler M. J., Heilker R. (2006) Methods Enzymol. 414, 121–139 [DOI] [PubMed] [Google Scholar]
- 25.Goodman O. B., Jr., Krupnick J. G., Gurevich V. V., Benovic J. L., Keen J. H. (1997) J. Biol. Chem. 272, 15017–15022 [DOI] [PubMed] [Google Scholar]
- 26.Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 27.Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luttrell D. K., Luttrell L. M. (2003) Assay Drug Dev. Technol. 1, 327–338 [DOI] [PubMed] [Google Scholar]
- 29.Zhao Z., Tan Z., Diltz C. D., You M., Fischer E. H. (1996) J. Biol. Chem. 271, 22251–22255 [DOI] [PubMed] [Google Scholar]
- 30.Barak L. S., Warabi K., Feng X., Caron M. G., Kwatra M. M. (1999) J. Biol. Chem. 274, 7565–7569 [DOI] [PubMed] [Google Scholar]
- 31.Kapur A., Samaniego P., Thakur G. A., Makriyannis A., Abood M. E. (2008) J. Pharmacol. Exp. Ther. 325, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lan R., Liu Q., Fan P., Lin S., Fernando S. R., McCallion D., Pertwee R., Makriyannis A. (1999) J. Med. Chem. 42, 769–776 [DOI] [PubMed] [Google Scholar]
- 33.Violin J. D., Lefkowitz R. J. (2007) Trends Pharmacol. Sci. 28, 416–422 [DOI] [PubMed] [Google Scholar]
- 34.Newton A. C. (2009) J. Lipid Res. 50, (suppl.) 266–271 [Google Scholar]
- 35.Kenakin T. (2007) Mol. Pharmacol. 72, 1393–1401 [DOI] [PubMed] [Google Scholar]
- 36.González-Maeso J., Sealfon S. C. (2009) Curr. Med. Chem. 16, 1017–1027 [DOI] [PubMed] [Google Scholar]
- 37.Staton P. C., Hatcher J. P., Walker D. J., Morrison A. D., Shapland E. M., Hughes J. P., Chong E., Mander P. K., Green P. J., Billinton A., Fulleylove M., Lancaster H. C., Smith J. C., Bailey L. T., Wise A., Brown A. J., Richardson J. C., Chessell I. P. (2008) Pain 139, 225–236 [DOI] [PubMed] [Google Scholar]
- 38.Yin H., Chu A., Li W., Wang B., Shelton F., Otero F., Nguyen D. G., Caldwell J. S., Chen Y. A. (2009) J. Biol. Chem. 284, 12328–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Lee M. M., Blomenröhr M., van der Doelen A. A., Wat J. W., Smits N., Hanson B. J., van Koppen C. J., Zaman G. J. (2009) J. Biomol. Screen 14, 811–823 [DOI] [PubMed] [Google Scholar]
- 40.Hurst D. P., Lynch D. L., Barnett-Norris J., Hyatt S. M., Seltzman H. H., Zhong M., Song Z. H., Nie J., Lewis D., Reggio P. H. (2002) Mol. Pharmacol. 62, 1274–1287 [DOI] [PubMed] [Google Scholar]
- 41.McAllister S. D., Rizvi G., Anavi-Goffer S., Hurst D. P., Barnett-Norris J., Lynch D. L., Reggio P. H., Abood M. E. (2003) J. Med. Chem. 46, 5139–5152 [DOI] [PubMed] [Google Scholar]
- 42.Sugiura T., Waku K. (2000) Chem. Phys. Lipids 108, 89–106 [DOI] [PubMed] [Google Scholar]
- 43.Offertáler L., Mo F. M., Bátkai S., Liu J., Begg M., Razdan R. K., Martin B. R., Bukoski R. D., Kunos G. (2003) Mol. Pharmacol. 63, 699–705 [DOI] [PubMed] [Google Scholar]
- 44.Griffin G., Wray E. J., Tao Q., McAllister S. D., Rorrer W. K., Aung M. M., Martin B. R., Abood M. E. (1999) Eur. J. Pharmacol. 377, 117–125 [DOI] [PubMed] [Google Scholar]
- 45.Showalter V. M., Compton D. R., Martin B. R., Abood M. E. (1996) J. Pharmacol. Exp. Ther. 278, 989–999 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.