Abstract
Three new structures of Escherichia coli succinate-quinone oxidoreductase (SQR) have been solved. One with the specific quinone-binding site (Q-site) inhibitor carboxin present has been solved at 2.4 Å resolution and reveals how carboxin inhibits the Q-site. The other new structures are with the Q-site inhibitor pentachlorophenol and with an empty Q-site. These structures reveal important details unresolved in earlier structures. Comparison of the new SQR structures shows how subtle rearrangements of the quinone-binding site accommodate the different inhibitors. The position of conserved water molecules near the quinone binding pocket leads to a reassessment of possible water-mediated proton uptake networks that complete reduction of ubiquinone. The dicarboxylate-binding site in the soluble domain of SQR is highly similar to that seen in high resolution structures of avian SQR (PDB 2H88) and soluble flavocytochrome c (PDB 1QJD) showing mechanistically significant structural features conserved across prokaryotic and eukaryotic SQRs.
Succinate:quinone oxidoreductase (SQR,4 succinate dehydrogenase) and menaquinol:fumarate oxidoreductase (QFR, fumarate reductase), members of the Complex II family, are homologous integral membrane proteins which couple the interconversion of succinate and fumarate with quinone and quinol (1–4). SQR is a key enzyme in the Krebs cycle, oxidizing succinate to fumarate during aerobic growth and reducing quinone to quinol and, thus, acts as a direct link between the Krebs cycle and the respiratory chain. QFR is found in anaerobic or facultative bacteria and lower eukaryotes, where it couples the oxidation of reduced quinones to the reduction of fumarate (1, 4). Escherichia coli SQR has four subunits, two hydrophilic subunits exposed to the cytoplasm (SdhA and SdhB), which interact with two hydrophobic membrane-intrinsic subunits (SdhC and SdhD) (5). SdhA contains the dicarboxylate-binding site and a covalently bound FAD cofactor which cycles between FAD and FADH2 redox states during succinate oxidation (6). The electrons from succinate oxidation are sequentially transferred via a [2Fe-2S], a [4Fe-4S], and a [3Fe-4S] iron-sulfur cluster relay system in SdhB to a quinone-binding site (QP) located at the interface of the SdhB, SdhC, and SdhD subunits. SdhC and SdhD are both composed of three transmembrane helices and coordinate a low spin b-type heme via His residues contributed by each subunit (7, 8).
The first structural information about members of the Complex II family came from x-ray structures of the QFR enzymes from E. coli at 3.3 Å resolution (9) and Wolinella succinogenes at 2.2 Å resolution (10). These structures revealed details of the overall architecture of the subunits, the position of key redox cofactors, the electron transfer pathway, and the quinone-binding sites. At around the same time, the structures of soluble fumarate reductases found in anaerobic and microaerophilic bacteria and structurally homologous to the flavoprotein subunit of Complex II were solved by x-ray crystallography (1). Analysis of these soluble fumarate reductases has proven particularly informative in describing the mechanism of fumarate reduction and succinate oxidation at the dicarboxylate-binding site (11–14).
Structures of SQRs lagged behind those of the QFRs until the structure of the E. coli enzyme was solved at 2.6 Å (15). This structure, solved in space group R32, revealed that the E. coli enzyme is packed as a trimer. The structures of the SdhA and SdhB subunits were highly similar to those of E. coli and W. succinogenes QFRs, but the transmembrane SdhC and SdhD subunits showed differences compared with their QFR counterparts. The structure revealed the position of the redox sites and the dicarboxylate- and quinone-binding (Q) sites. The heme b molecule was shown to lie away from the electron transfer pathway, suggesting electrons are preferentially transferred from the [3Fe-4S] cluster to ubiquinone, on the grounds of the edge-to-edge distances and redox potentials of the relevant groups. The structure revealed density in the Q-site that was interpreted as ubiquinone, and the position of the binding site was confirmed by the structure of the E. coli enzyme co-crystallized with the Q-site inhibitor 2-(1-methyl-hexyl)-4,6-dinitrophenol (DNP-17, PDB code 1NEN (15)). The E. coli enzyme was subsequently co-crystallized with the Q-site inhibitor Atpenin A5 (AA5) (PDB code 2ACZ (16)). This inhibitor was bound deeper into the quinone-binding site than ubiquinone or DNP-17, suggesting that there are two binding positions for ubiquinone in its binding site. The structure also identified a water-mediated proton pathway, proposed to deliver protons to the quinone-binding site. The first structure of a mitochondrial SQR was from porcine heart at 2.4 Å resolution (PDB code 1ZOY (17). This structure revealed a monomer in the asymmetric unit, suggesting that mitochondrial SQRs were likely to function as monomers. Superposition of the porcine and E. coli SQR structures revealed the high structural similarity of the SdhA and SdhB subunits and the conservation in position of the redox cofactors. Larger divergences were observed in the transmembrane subunits.
Further structural information about SQRs was obtained by analysis of structures of avian SQR crystallized with oxaloacetate (2.2 Å resolution, PDB code 1YQ3), with 3-nitropropionate (2.4 Å resolution, PDB code 1YQ4), and with the Q-site inhibitor carboxin (2.1 Å resolution, PDB code 2FBW) (18). These structures revealed important differences in the position of key residues in the dicarboxylate-binding site compared with the E. coli and porcine structures. Arg-297 (equivalent to Arg-298 in porcine and Arg-286 in E. coli SQRs) was ideally located to act as a general base catalyst, accepting a proton during dehydrogenation of succinate, as in the soluble Shewanella flavocytochrome c3 (PDB code 1QJD) (11), suggesting conservation of mechanism between these distantly related enzymes. An unusual cis-serine peptide bond was proposed to position another arginine residue for binding dicarboxylates. Density for the dicarboxylate in 1YQ3 and 2FBW was shown to be distinctly non-planar and could be modeled by the “malate-like intermediate” seen in 1QJD. The nature of the ligand in the dicarboxylate site was further analyzed in a 1.74 Å resolution structure of avian SQR (PDB code 2H88), confirming the high structural similarity of the ligand and binding site residues in the SQR and flavocytochrome c3 structures (11, 12, 14).
Despite the structural information described above, there are still unresolved issues regarding the structure and function of SQRs and QFRs. These include the location of conserved waters, which may form a channel involved in protonation of quinone, and the ability of the Q-site to accommodate different quinones and inhibitors. To further address these issues, we pursued structure-function studies of E. coli SQR. We developed alternative crystallization conditions that provided crystals more reproducibly and diffracting to higher resolution. By exchanging the enzyme into decyl-β-d-maltoside (DM) during purification, it was possible to crystallize the enzyme in the orthorhombic P212121 space group. These crystals routinely diffracted in the 3–3.5 Å resolution range. Co-crystallization with the biochemically well characterized Q-site inhibitor carboxin improved diffraction to 2.1–2.8 Å. This structure shows new features related to the dicarboxylate-binding site of E. coli SQR including a rare cis-peptide bond in SdhA, as found in avian SQR (14), which helps shape the geometry of the active site. Comparisons of the structure with those of SQR binding PCP and SQR with an empty Q-site show how subtle rearrangements of the Q-site accommodate the different inhibitors. The orientation of carboxin in the Q-site differs with computational predictions (16) and with that seen in avian SQR (2FBW). The position of conserved water molecules around the Q-site suggests a new water-mediated proton uptake pathway consistent with recent mutational and biophysical studies (19).
EXPERIMENTAL PROCEDURES
Carboxin (5,6-dihydro-2-methyl-1,4-oxathiin-3-carboxanilide), >99% pure, was obtained from Duchefa Biochemie, Haarlem, The Netherlands. PCP, 98% pure, was obtained from Aldrich.
Strains, Plasmids, and Mutagenesis
E. coli strain DW35 (ΔfrdABCD, sdhC::kan) was used as the host for expression of SQR as described (20). Plasmid pFAS (sdhC+D+A+B+) was used for expression of wild type and SQR mutants. Mutations were introduced using the QuikChange (Stratagene, La Jolla, CA) mutagenesis kit as described (20).
Assay for Stability in Detergents
The SQR enzyme was purified as previously described (21, 22). Protein was diluted 50-fold to a final concentration of 1 mg/ml in buffers containing 20 mm Tris, pH 7.6, and a range of detergents at a concentration of twice the critical micelle concentration. The UV-visible spectrum (250–750 nm) was measured for each sample (Beckman DU Series 700 UV-visible scanning spectrophotometer), and samples were then incubated at 20 °C. At regular time intervals, the samples were centrifuged at high speed to pellet aggregates (100,000 x g, 30 min, Beckman TLA120.1 rotor). The supernatant was recovered, and the UV-visible spectrum was measured. Incubation was then continued at 20 °C.
Protein Expression and Purification
As a result of the detergent stability assays, protein for crystallization was purified using a modification of earlier procedures (21, 22). Briefly, membranes containing 600–800 mg of total protein were resuspended in buffer A (20 mm Tris pH 7.4, 0.2 mm EDTA, Roche Applied Science Complete protease inhibitor tablets) to a concentration of about 10 mg of total protein/ml. The detergent C12E9 (Anatrace) was added to give a final concentration of 2% (w/v). The suspension was stirred briefly at 4 °C and then centrifuged at 100,000 x g for 40 min. The reddish-brown supernatant was filtered through a 0.2-μm filter (Millipore) before being applied to a Q-Sepharose Fast Flow 26/10 column (GE Healthcare, bed volume 50 ml) equilibrated with buffer B (20 mm Tris, pH 7.4, 0.2 mm EDTA, 0.05% (w/v) C12E9). After washing with two bed volumes of buffer B and two bed volumes of buffer B with 100 mm NaCl, the protein was eluted with a linear gradient of buffer B with 100–350 mm NaCl over 6 bed volumes at 2.5 ml/min. Fractions containing SQR were selected on the basis of their A407/A280 ratio, pooled, and concentrated using a 30-kDa cutoff membrane in an Amicon Stirred Cell Concentrator. The concentrated sample was diluted in buffer A before being applied to a Poros 50HQ column (bed volume, 25 ml, Perseptive Biosystems, Framingham, MA) equilibrated with buffer B. The loading and running of the column was identical to that of the Q-Sepharose column, except that the flow rate was increased to 3.5 ml/min. The pooled fractions were concentrated to 2 ml as described above. The sample was applied to a Sephacryl S-300 26/60 column (GE Healthcare), equilibrated with buffer C (20 mm Tris, pH 7.6, 0.2% (w/v) DM (Anatrace)), and run at 0.25 ml/min. SQR eluted as a single peak, and pooled fractions were concentrated to 20 mg/ml with an Amicon Centricon with a 100-kDa cutoff membrane. The sample was filtered with a 0.1-μm filter before crystallization.
Protein Crystallization
Initial crystal hits were found using commercially available screens (Molecular Dimensions Memstart/Memsys screen). Crystals suitable for data collection were obtained using the hanging-drop vapor diffusion technique. SQR was mixed in a 1:10 (v/v) ratio with the inhibitors carboxin (from a 10 mm stock solution in ethanol) or PCP (from a 5 mm stock solution in ethanol). For crystals of SQR binding carboxin, 1 μl of inhibited enzyme was mixed with 1 μl of reservoir solution (0.1 m Tris, pH 8.5, 0.1 m Li2SO4, 0.1 m NaCl, 0.009% dodecyl-β-d-maltoside, 11% (w/v) polyethylene glycol 4000). For crystals of SQR binding PCP, the reservoir solution was modified to contain 12% (w/v) polyethylene glycol 4000. Crystals of SQR with an empty Q-site came from an unsuccessful co-crystallization trial with the inhibitor flutolanil and were grown using a reservoir solution containing 0.1 m Tris, pH 8.5, 0.1 m MgSO4, 10% polyethylene glycol 4000. In all cases crystals appeared overnight and were frozen within 4 days of setup using 30% glycerol as the cryoprotectant and mounted mesh LithoLoopsTM (Molecular Dimensions Ltd, UK) to support the large plate crystals.
Data Collection
X-ray data were collected from frozen crystals at 100 K at the European Synchrotron Radiation Facility beamline ID23–1. The SQR/carboxin dataset was collected from a single position on the crystal using a 100 × 100-μm beam. The SQR/PCP and empty Q-site datasets were collected from three different positions on the respective crystals, to minimize the effects of radiation damage and increase the resolution of the complete dataset, using a 50 × 50-μm beam (PCP) or 100 × 100-μm beam (empty Q-site). Intensities were integrated using the program MOSFLM (23) from the CCP4 suite (24) or XDS (25) and merged using SCALA (26, 27). For all datasets, the space group was P212121, with a trimer of SQR molecules in the asymmetric unit. Data collection statistics are shown in supplemental Table S1.
Structure Determination and Refinement
For determination of the SQR/carboxin structure, molecular replacement was done using the program PHASER (28) and the SQR coordinates in the R32 crystal form at 2.6 Å resolution (PDB code 1NEK) as a search model. Non-protein atoms were omitted from the search model and initial phase calculation. Density for carboxin was clearly visible in the maps calculated from these initial phases, giving us confidence in the molecular replacement solution and the quality of the phasing. Some flexible loops of the model, particularly in the SdhA subunit, did not fit the density maps well. Automated model building was, therefore, attempted using the programs RESOLVE in the PHENIX suite (29, 30) and ARP/wARP (31). Although neither program successfully built the whole SQR trimer in the asymmetric unit, output from the programs helped correct regions of the initial model. Rotamers were checked using the side chains option of ARP/wARP, and many side-chain conformations were rebuilt. Manual model building was performed using the programs Coot (32) and O (33), and refinement was done using phenix. refine (34) and Refmac (35). Strict NCS restraints were applied to the protein part of the model during refinement. Coordinates and restraints for carboxin were downloaded from the PRODRG server (36). Ideal coordinates for TEO were obtained from the HIC-Up data base (37) and input to the PRODRG server to generate final coordinate and restraint files for modeling. The ligand building procedure within ARP/wARP (38) was used to fit both ligands into the density maps. Water molecules were fitted using phenix.refine and Coot and validated using Coot. The model was refined to Rcryst = 19.25% and Rfree = 22.09%. TLS refinement (39) was implemented in the final stages of refinement, defining each chain in the asymmetric unit as a separate TLS group. This produced a model with a final Rcryst = 17.1% and Rfree = 20.0%. The TLS model was validated using TLSANL (40). The final model consists of residues 1–588 of SdhA (chains A, E, and I and FADs), residues 1–238 of SdhB (chains B, F, and J and FeS clusters), residues 8–128 of SdhC (chains C, G, and K), residues 11–115 of SdhD (chains D, H, and L), 3 hemes, 3 carboxin molecules, 3 TEO molecules, 3 Na+ ions, 3 sulfates, and 1116 water molecules. The side chain of SdhD Trp 113 was truncated at the Cβ atom as density for the side chain was poor, and it was not possible to model it as a common rotamer without introducing clashes with symmetry-related side chains.
Phases for the wild type/PCP and empty Q-site structures were initially determined by rigid-body refinement in REFMAC of the protein part of the wild type/carboxin model. This was followed by positional refinement in REFMAC with tight NCS restraints. For the PCP dataset, difference density for the inhibitor was clearly visible in the Q-site. Ideal coordinates for PCP were obtained from the HIC-UP data base, and restraints were generated using the SKETCHER interface to LIBCHECK. PCP was built into the density map using the ligand building procedure in ARP/wARP. The model was refined to an Rcryst = 22.2% and Rfree = 25.8%. TLS refinement, with each chain in the asymmetric unit as a separate TLS group, reduced both R factors to final values of Rcryst = 19.7% and Rfree = 22.8%. The final model consists of residues 1–588 of SdhA (chains A, E, and I and FADs), residues 1–238 of SdhB (chains B, F, and J and FeS clusters), residues 8–129 of SdhC (chains C, G, and K), residues 11–115 of SdhD (chains D, H, and L), 3 hemes, 3 PCP molecules, 3 TEO molecules, and 3 Na+ ions. For the empty Q-site structure, positional refinement with tight NCS restraints reduced Rcryst to 26.5% and Rfree to 30.3%. TLS refinement, with one TLS group per chain, produced a final model with Rcryst = 20.5% and Rfree = 23.3%. The mFo − DFc map after refinement shows a small residual density peak in the Q-site, but this cannot accommodate an inhibitor or native ubiquinone; hence, we consider this structure to show an “empty” Q-site. The final model consists of residues 1–588 of SdhA (chains A, E, and I and FADs), residues 1–238 of SdhB (chains B, F, and J and FeS clusters), residues 8–128 of SdhC (chains C, G, and K), residues 11–115 of SdhD (chains D, H, and L), 3 hemes, 3 TEO molecules, and 3 Na+ ions.
Molprobity (41) was used to validate the final models and check for Asn/Gln/His flips. Refinement statistics for all datasets are shown in supplemental Table S1). Crystal contacts and residues involved in trimer formation were analyzed using the CCP4 program AREAIMOL. The root mean square deviation of superposed structures was determined using the CCP4 program LSQKAB. Figs. 1–5 and supplemental Figs. S2, S4, S6, and S7 were prepared using PyMOL (42). Supplemental Figs. S3 and S5 were prepared using CCP4 mg (43).
FIGURE 1.
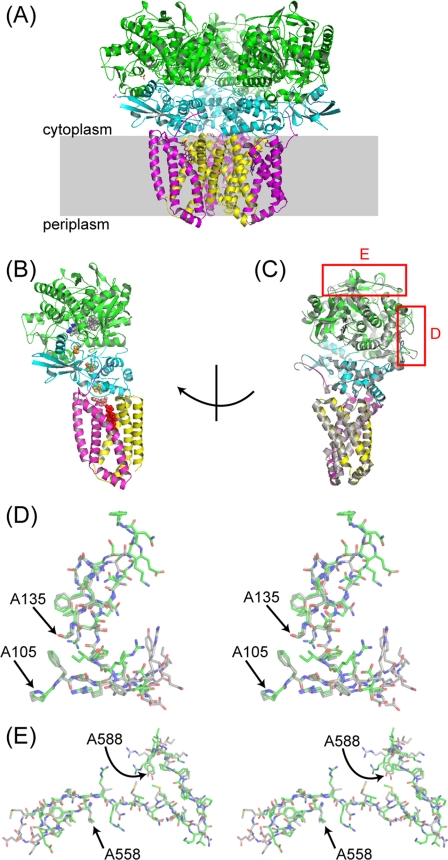
Overall structure of E. coli SQR in the P212121 crystals and differences in cytoplasmic loops in the SdhA subunit. The asymmetric unit contains a trimer of SQR molecules (A), with SdhA subunits colored green, SdhB colored cyan, SdhC colored magenta, and SdhD colored yellow. A shows a view approximately parallel to the membrane, with the membrane bilayer shown as a gray box. An SQR monomer (chains A–D) is shown in B with chains colored as in A and with FAD atoms shown as gray spheres, TEO as blue spheres, carboxin as salmon spheres, heme as red spheres, and FeS clusters as orange and yellow spheres. In C, the SQR monomer has been rotated and is shown with 1NEK (in gray, aligned by superposing the SdhA subunits in Coot (34)). Red boxes highlight significant differences in the position of cytoplasmic loops in the two structures. These differences are shown as stereo figures in D for the region around A110-A130 and E for the region around A560–A588. Carbon atoms from 1NEK are displayed in gray, and carbons from the P212121 structure are in green, whereas oxygen, nitrogen, and sulfur atoms are shown in red, blue, and yellow to assist side-chain identification.
FIGURE 2.
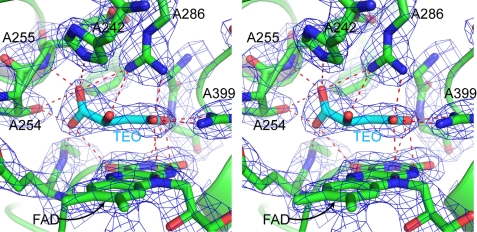
Architecture of the dicarboxylate-binding site and ligand in the carboxin-inhibited structure. The stereo figure shows residues within 4.0 Å of the ligand TEO, a malate-like intermediate. Interactions between residues and the ligand are shown as red dotted lines. The density, shown as a blue mesh, is a 2mFo − DFc map, contoured at 1.8 σ. Residues 251–253 and 354 of SdhA have been removed to enable a clearer view of the binding site.
FIGURE 3.
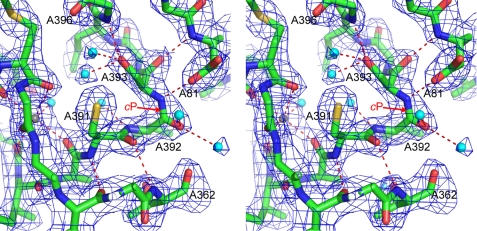
An unusual cis-peptide bond between Val-A392 and Ser-A393. The stereo figure shows residues around the cis-peptide (labeled cP). Polar contacts between residues are shown as red dotted lines, water molecules as cyan spheres, and a metal ion as a gray sphere. The density, shown as a blue mesh, is a 2mFo − DFc map, contoured at 1.3 σ.
FIGURE 4.
Views of the Q-site in the carboxin-inhibited, PCP-inhibited, and empty states. A stereo view of the Q-site in carboxin-inhibited SQR is shown in A. B shows a stereo view of the Q-site in PCP-inhibited SQR. In A and B, residues within 4 Å of the inhibitor are shown in stick representation, and polar contacts are shown as red dotted lines. The density, shown as a blue mesh, is a 2mFo − DFc map, contoured at 1.3 σ. Pro-B60 and the amide N of Ser-B61 have been removed to allow a clear view of the binding site. C shows a superposition of the carboxin-bound (colored) and empty Q-site (in gray) structures; the view of the Q-site is the same as in A. D shows a superposition of the PCP-bound (colored) and empty Q-site (in gray) structures; the view of the Q-site is as in B. In C and D, the structures were superimposed by minimizing the root mean square deviations of residues within the Q-site (B164, B207, B209, C20, C31, D82, and D83) using the CCP4 program LSQKAB.
FIGURE 5.
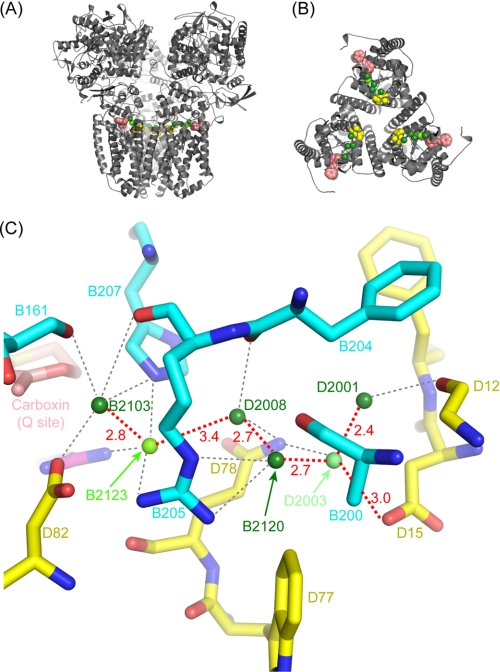
Possible water-mediated proton transfer network. Views of the E. coli SQR trimer are shown parallel to the membrane (A) and perpendicular to the membrane, showing the SdhC and SdhD subunits (B). The protein is shown in gray in schematic representation, carboxin (sitting in the Q-site) is shown as salmon spheres, waters are shown as green spheres, and Sdh D15 is shown as yellow spheres. C shows a detailed view of the pathway, including residues located within 4 Å of the waters. Carbon atoms from SdhB are shown in cyan, carbons from SdhD are shown in yellow, carbons from carboxin are shown in salmon, whereas oxygens are shown in red, and nitrogens are in blue. Water molecules are shown as green spheres, shaded according to their degree of conservation in the three NCS copies: dark green (100%), chartreuse (66%), and lime green (33%). Distances between water molecules on the pathway are shown in red (in Å). Possible hydrogen-bonds between the waters and protein residues are shown as gray dotted lines.
Enzyme Activity
Succinate oxidation with the artificial electron acceptor phenazine methosulfate and the ubiquinone analogue UQ1 was coupled to reduction of dichlorophenol indophenol as previously described (20).
RESULTS AND DISCUSSION
A New Crystal Form of E. coli SQR
One of the key factors influencing membrane protein crystallization is the choice of detergent, which may affect protein stability and crystal packing (44, 45). We, therefore, developed a simple assay described under “Experimental Procedures” to test the stability of SQR in a range of detergents. Representative experimental results are shown in supplemental Fig. S1. This assay revealed that E. coli SQR is about as stable in the maltoside detergents (dodecyl-β-d-maltoside, DM, and nonyl-β-d-maltoside) as it is in C12E9, a polyoxyethylene detergent that has been used extensively in SQR studies. As expected, E. coli SQR was slightly less stable in nonyl-β-d-maltoside compared with dodecyl-β-d-maltoside because of the shorter alkyl chain length of nonyl-β-d-maltoside. Interestingly, E. coli SQR was unstable in octyl-β-d-glucoside, aggregating quickly in this detergent even when used at low concentrations as a detergent additive in crystallization trials. This contrasts with the behavior of avian SQR in which the final stage before crystallization was a detergent exchange into octyl-β-d-glucoside (46).
Screening for new crystallization conditions identified several new hits (see “Experimental Procedures”). The best of these produced crystals diffracting routinely in the 3.0–3.5 Å range; however, co-crystallization with the Q-site inhibitor carboxin increased the resolution of the crystals to 2.1–2.8 Å. Datasets were collected for SQR inhibited by carboxin to 2.4 Å, SQR inhibited by PCP to 3.2 Å, and SQR with an empty Q-site to 3.2 Å. All datasets were in the orthorhombic P212121 space group, with slightly variable unit cell parameters.
Comparison of Orthorhombic and Trigonal Crystal Forms; Crystal Packing and B-factors
The overall structure of the E. coli enzyme is generally confirmed by the higher resolution structure in the orthorhombic crystal. The Cα atoms of the SdhA subunit can be superimposed with a root mean square deviation of 2.7 Å. Excluding 47 residues with the greatest differences (residues 110–130 and 561–588; see below) allows the SdhA subunits to be superimposed with a root mean square deviation of 0.5 Å for the Cα atoms. The other subunits can be superimposed with root mean square deviations for Cα atoms of 0.4 Å for SdhB, 0.3 Å for SdhC, and 0.4 Å for SdhD.
The new orthorhombic crystals contain a trimer of SQR molecules in the asymmetric unit (Fig. 1A and supplemental Fig. S2). This trimer of SQR molecules was first seen in the trigonal R32 crystal form (1NEK), where the monomers were related by a crystallographic 3-fold axis. The tight packing of the monomers in the R32 crystal led to the suggestion that the trimer association is physiological. The conservation of the trimer in the new orthorhombic crystals, after exchange into a new detergent during purification, is further support for its physiological relevance. Similar tight packing of the monomers is seen, with a monomer burying 1598 Å2 of surface area upon trimer formation.
The crystal packing in the two different crystal forms is compared in supplemental Fig. S2. In the R32 crystals, crystal contacts are made by the large hydrophilic SdhA subunits, but there are additional hydrophobic interactions between SdhC subunits. The hydrophobic interactions result in the three-dimensional crystal being composed of stacked two dimensional crystals, typically observed in a type I membrane protein crystal (47). In contrast, residues in the SdhA subunit dominate crystal contacts in the orthorhombic crystals. They interact predominantly with other SdhA subunits, but there are a few interactions with transmembrane SdhC/D subunits in neighboring molecules (supplemental Fig. S3). The transmembrane subunits are not tightly packed together, and there is room to accommodate a detergent micelle around the transmembrane helices. This is typically observed in a type II membrane protein crystal (47).
Although the average temperature factor (B-factor) is comparable for carboxin-inhibited SQR in the orthorhombic crystal and 1NEK, the B-factor distribution across an SQR monomer is strikingly different in the two crystal forms (supplemental Fig. S4). The orthorhombic crystal has a lower average B-factor for the SdhA subunit but higher B-factors for the transmembrane subunits compared with 1NEK. This is most likely because of the lack of crystal packing interactions between transmembrane subunits in the orthorhombic crystal. Unlike 1NEK, we do not find density that can be attributed to lipid molecules in the new density maps. It is possible that lipid molecules help stabilize the transmembrane region in 1NEK, reducing the B-factors for SdhC and SdhD.
The lower resolution PCP and empty Q-site datasets have slightly larger unit-cell parameters than the carboxin dataset, and this seems to be partly because of looser packing of the monomers within the trimers. In the PCP and empty Q-site structures, we see differences between the B-factors of NCS-related chains after TLS refinement. In the P212121 crystals the different monomers in the trimer are involved in different numbers of crystal contacts, with chains A-D having more contacts than chains E-H and chains I-L having the least. As a consequence, the refined B-factors are lowest for chains A-D and highest for chains I-L. This effect is more extreme for the lower resolution datasets, with chains I-L of the empty Q-site structure making fewer crystal contacts than is seen in the carboxin structure and far fewer contacts than chains A-D (compare supplemental Figs. S3A and S5A). As a consequence, chains I–L show greater anisotropic disorder than chains A–D (supplemental Fig. S5B). For the empty Q-site structure, this is reflected in poorer density, higher total B-factors (including residual and TLS components), and increased anisotropic atomic displacement parameters for chains I-L after TLS refinement. We have selected chains A–D, with the lowest B-factors, from each structure for the descriptions and comparisons in this paper.
Altered Conformation of Loop Regions in SdhA
Two loop regions (residues 110–130 and 560–588) in SdhA show significantly different conformations in 1NEK and the new P212121 crystal form (Fig. 1, C–E). In the new crystal form the region between residues 110 and 130 forms a tight loop between Leu-A109 and Ile-A114 and a more extended loop between Gln-A122 and Ala-A131. Loop region 110–130 is not involved in crystal contacts in either 1NEK or the new orthorhombic crystals, so the difference in conformation in this region cannot be attributed to crystal packing.
The region between 560 and 588 forms a tight turn between Ser-A563 and Glu-A564 and a β-strand between Glu-A564 and Arg-A569, which interacts with a β-strand running from His-A556 to Leu-A560. This loop is involved in crystal packing in both crystal forms. Residues 562, 567, and 568 make crystal contacts in 1NEK. In the orthorhombic crystals NCS-related chains within the asymmetric unit make different crystal contacts. For chain A, residues 567, 568, 582–585, and 587–588 make crystal contacts. For chain I, only residue 574 is involved in crystal contacts. Because chains A and I both show the same position for loop 560–588, in good density (for the carboxin dataset), crystal contacts are highly unlikely to be responsible for the different position of this loop.
The differences are unlikely to be because of functionally significant conformational changes and, as discussed above, are unlikely to be induced by crystal packing contacts. Rather, the differences are attributable to the regions being disordered in 1NEK. Our new structure provides a more accurate position of these two loops.
Comparison of E. coli and Eukaryotic SQR Structures
The SdhA and SdhB subunits of E. coli SQR (1NEK) and porcine SQR (1ZOY) superimpose with a low root mean square deviation (0.97 and 0.72 Å, respectively for the backbone atoms), but there are larger deviations in the transmembrane SdhC and SdhD subunits (17). However, differences were noted in the SdhA subunit, including an insertion from residues 135–143 (porcine sequence) and a deletion at residues 122–123 (porcine sequence). The new E. coli structures reveal that these differences are no longer present given the new position of the 110–130 loop. The new position agrees well with the equivalent positions in both the avian (highest resolution structure 2H88) and porcine SQRs (1ZOY). Cα atoms of residues 110–127 can be superimposed with equivalent residues from the avian and porcine structures with root mean square deviations of 0.3 and 0.4 Å, respectively. Residues 128–130 adopt a slightly different conformation in the structures. Both eukaryotic structures show that these SQRs contain a large loop inserted between residues 551–552 in SdhA (E. coli numbering). This remains the most significant difference in the structure of SdhA in E. coli versus eukaryotic SQRs.
Architecture of the Dicarboxylate-binding Site
Key active-site residues are absolutely conserved between E. coli SQR, QFR, eukaryotic SQRs, and Shewanella flavocytochrome c, and the new structure reveals that these residues now superimpose very well (Fig. 2). Clear density is present within the binding site for a dicarboxylate but is not well modeled by oxaloacetate as positioned in 1NEK. The density is distinctly non-planar and fits the malate-like intermediate which was termed TEO from its three-letter ID assignment in the PDB data base (14) and is seen in 1QJD and in the avian SQR structures (11, 14, 18). The exact chemical identity of the dicarboxylate termed TEO in the avian SQR structures is still a matter of debate (14, 18). It has been suggested that it could be either the keto or enol form of oxaloacetate, with a strained C1 carboxylate, or possibly oxaloacetate reduced to malate by radiation damage during data collection (18). The former is consistent with previous studies showing that SQR oxidizes malate to the enol form of oxaloacetate and that keto-enol tautomerization may be responsible for the strong oxaloacetate inhibition of oxidized SQR (48). Given the resolution of the E. coli SQR structure binding carboxin (2.4 Å), we have conservatively used “ideal” bond lengths and angles in our model of TEO (generated by the PRODRG server (36) from coordinates in the HIC-Up data base (37)), including an sp3-hybrdized C2. A higher resolution structure of avian SQR has enabled a free atom refinement of the geometry of TEO, and the resulting bond lengths and angles are the most accurate description of TEO bound to SQR currently available (14). Compared with our model of TEO, the higher resolution structure shows small differences in bond lengths and angles, including sp2 hybridization of C2, which also fits the planar nature of our density well.
In Fig. 2 the four-carbon backbone of the dicarboxylate, the oxygens of the C4 carboxylate, and the oxygen attached to C2 lie almost co-planar, sitting about 3 Å above and parallel to the isoalloxazine ring of the FAD. The C1 carboxylate group is twisted out of the plane defined by the four-carbon backbone and attached oxygens. TEO interacts with a large number of residues within the dicarboxylate-binding site. The C1 carboxylate forms hydrogen-bonds with five different residues; the O1B atom hydrogen-bonds to His-A242, Glu-A255 (to amide N), Arg-A286, and Thr-A254, and the O1A atom hydrogen-bonds to Thr-A254 and Gly-A51 (to amide N). The O1A atom lies only 2.5 Å from the oxygen of Thr-A254, and bridging electron density is seen between the atoms. As seen in avian SQR (18), the C1 carboxylate is sandwiched between two hydrophobic residues, Phe-A119 and Leu-A252. In Shewanella these residues are replaced by methionines. The large number of hydrogen bonds from the C1 carboxylate to residues in the binding site coupled with the steric hindrance provided by the hydrophobic residues (11) cause the C1 carboxylate to be twisted out of the flat plane formed by the other atoms in TEO. The oxygen attached to C2 forms hydrogen-bonds to His-A354 and His-A242. The C4 carboxylate forms hydrogen-bonds to Arg-A399 and Gly-A402 (amide N) via the O4A atom and to His-A354, Arg-A399, and Arg-A286 via the O4B atom. The O4A and O4B atoms can also make polar contacts to nitrogen atoms in the FAD isoalloxazine ring. The oxygen atoms of the C4 carboxylates interact particularly tightly with the nitrogens of Arg-A399, lying about 2.6 Å from them, and bridging electron density is seen between these residues.
In 1NEK, Arg-A286 was built as an unusual rotamer, pointing away from the dicarboxylate-binding site and with a water molecule between its ϵ nitrogen and oxaloacetate. The equivalent residue for porcine SQR was modeled in the same way in 1ZOY (18). In our high resolution structure, Arg-A286 adopts a common rotamer and points directly at TEO, forming hydrogen bonds to both the C1 and C4 carboxylates. The position of the catalytically critical Arg-A286 in the new structures superimposes very well with the equivalent arginine residues in the avian SQR structures and 1QJD. This arginine residue is ideally positioned to act as a general base catalyst. One of its terminal nitrogen atoms is placed 2.9 Å from C3 and 3.4 Å from C2. The other nitrogen atoms hydrogen-bond to carboxylates (the C4 carboxylate of TEO, Gln-A240 and Glu-A255), hold the guanidine group in place, and make Arg-A286 a stronger base.
The new structure shows that water is excluded from the dicarboxylate-binding site. The closest water molecule (A2103) lies about 6 Å from TEO and is separated from it by the backbone of Glu-A255. As observed in the avian SQR structure, the exclusion of water from the dicarboxylate-binding site contrasts with the rest of the SdhA subunit, which is well hydrated (14). A large number of these water molecules are conserved between our structure and the highest resolution avian structure (2H88). For example, the positions of water molecules A2111, A2161, A2102, A2106, A2105, and A2103 are well conserved, and these waters are involved in hydrogen-bond networks that help shape the geometry of the binding site. A similar conservation of water-molecules is seen around other key structural features of SdhA, such as the metal ion binding sites (see below).
A Conserved cis-Serine peptide in the Complex II Superfamily
The new structure reveals an unusual cis-peptide bond between Val-A392 and Ser-A393 (Fig. 3). The quality of the density in this region of the map gives us confidence that the peptide is now correctly modeled in the cis conformation. Furthermore, when refined in the trans conformation, analysis by Molprobity (41) shows a high Cβ deviation for Val-A392 and Ser-A393 and bond angle outliers for Ser-A393. These geometry distortions strongly indicate that the trans-conformation is inappropriate when refined against the experimental data (49).
The avian SQR structures also show a cis-peptide in an equivalent position, between Ala-401 and Ser-402 (18). It appears that this cis-peptide is a conserved feature across the SQR family. The cis-peptide is stabilized by a strong hydrogen-bond between the amide nitrogen of Ser-A393 and Asp-A81.
In the avian structures the cis-peptide occurs in a turn, which includes one of the key residues in the dicarboxylate-binding site and also a metal ion binding site (18). The new E. coli structure reveals the same features. The cis-peptide forms a tight turn that leads toward Arg-A399, which interacts directly with TEO. Furthermore, it lies very close to the metal ion-binding site (A1590).
Metal Ion Binding Sites
The high resolution E. coli SQR structure reveals the position of a metal ion-binding site. The metal ion site A1590 lies in the same position as the K1 site in avian SQR (18). A1590 shows approximately octahedral coordination, interacting with the carbonyl backbone of residues Met-A356, Met-A357, Gly-A358, Glu-A388, and Ala-A390. A water molecule (A2137) was modeled in the sixth co-ordination position, accounting for a residual peak in mFo − DFc maps. We have modeled the metal ion as Na+. Na+ accounts for the density after refinement, is consistent with the 2.5 Å distance between the metal and the carbonyl of Glu-A388, and is present during purification and crystallization. We do not find convincing evidence for a metal ion in the same position as the K2 site of avian SQR and have instead modeled a water molecule there.
Role of Cardiolipin in Maintaining SQR Structure
1NEK revealed two lipid molecules tightly bound to SQR; that is, a phosphatidylethanolamine and a cardiolipin (15). The cardiolipin is particularly interesting because it forms several close interactions with the protein, and two of its acyl chains sit between the SdhC and SdhD subunits, forming a plug beneath the heme b. This suggested that cardiolipin could have a role in stabilizing the interface between SdhC and SdhD and the position of the heme. We cannot find convincing density for lipid molecules in our 2.4 Å map. In particular, we do not see clear density for cardiolipin, although there is some residual density close to heme b and the position of the cardiolipin acyl chains in 1NEK. Our new structure has higher B-factors in the transmembrane region than 1NEK. This indicates that the cardiolipin molecule may stabilize the transmembrane helices but is not essential for the structural integrity of heme or SdhC and -D.
Inhibition of the Quinone-binding Site by Carboxin and PCP
The high resolution dataset from the P212121 crystals comes from a co-crystallization experiment with the Q-site inhibitor carboxin. Clear density for the inhibitor is present in the Q-site, allowing carboxin to be modeled into the density (Fig. 4A). Contouring 2mFo − DFc maps at high σ levels show a clear peak associated with the sulfur atom, the most electron dense atom in the methyl-oxathiin ring (supplemental Fig. S6A). Carboxin interacts via hydrogen-bonds to two residues within the Q-site, Trp-B164 (2.9 Å away) and Tyr-D83 (2.6 Å away). The structure of carboxin-inhibited avian SQR has also been determined (PDB code 2FBW) (18). Interestingly, the position of the methyl-oxathiin ring differs in 2FBW compared with our structure. Difference maps (mFo − DFc) calculated from the coordinates of 2FBW and the structure factors deposited at the PDB suggest that the orientation of the methyl-oxathiin ring that we see in our structure would fit the density better (supplemental Fig. S6B). Repositioning carboxin in 2FBW according to our model removes the difference map peaks. For these reasons, our new carboxin model in the Q-site may be valid not only for E. coli SQR but also for avian and other SQRs. It should be noted, however, that mammalian (bovine) and E. coli SQR show some differences in the kinetic parameters for Q-site inhibitors of quinone reduction (4). There is a 1 order magnitude of difference in the Ki for carboxin, PCP, and theonyl-trifluoroacetone inhibition, with the mammalian enzyme being more sensitive to the inhibitors. Thus, there may be subtle differences in how the Q-sites of eukaryotic and prokaryotic SQRs accommodate the inhibitors.
A theoretical model of carboxin binding to E. coli SQR (PDB code 2AD0) (16) did not predict the interaction of the carbonyl group of the inhibitor with Trp-B164 and Tyr-D83. The inhibitor was predicted to interact with the protein via its methyl-oxathiin ring, with the sulfur atom forming a hydrogen-bond to Ser-C27 and the oxygen atom forming a hydrogen-bond to His-B207, whose imidazole ring has been flipped to enable this interaction. Although the experimental structure shows different interactions between carboxin and the protein, the potential of a flip of His-B207 is interesting, as it has been proposed to play a role in the mechanism of ubiquinone reduction (Ref. 16 and see below). In the 2.4 Å x-ray structure, it is not possible to unambiguously assign the orientations of His rings. However, the hydrogen-bonding arrangements involving the ring provide indirect evidence for its orientation. For this reason we have not flipped the ring of His-B207, as the Nδ atom forms a strong hydrogen-bond to one of the heme propionates, whereas the Nϵ hydrogen-bonds to one of the water molecules on the putative alternative proton transfer wire (see below).
We have also refined the structures of SQR inhibited by PCP and SQR with an empty Q-site. Clear density for PCP was found in the Q-site after co-crystallization experiments with this inhibitor (Fig. 4B). The hydroxyl group of PCP interacts with the same Q-site residues as carboxin, lying 2.7 Å from Trp-B164 and 2.6 Å from Tyr-D83. This is similar to the interactions seen in PCP bound to Thermus thermophilus polysulfide reductase, where the hydroxyl oxygen lies 2.9 Å from a His residue and 2.3 Å from a Tyr residue (50).
The empty Q-site structure allows us to determine whether conformational changes occur in the Q-site after inhibitor binding. Structures have been superimposed by minimizing the root mean square deviation between residues forming the Q-site. Fig. 4C shows an overlay of the empty Q-site and carboxin-bound structures. Fig. 4D shows an overlay of the empty Q-site and PCP-bound structures. Carboxin binding does not involve significant conformational changes to residues in the Q-site, as has also been observed for the inhibitors DNP-17 (15) and Atpenin A5 (16). Surprisingly, PCP binding leads to small localized conformational changes involving residues Trp-B164 and His-B207. Negative peaks around the imidazole ring of His-B207 in the mFo − DFc map suggest that the ring is mobile. A second dataset collected from a crystal of PCP-inhibited SQR confirms the conformational changes shown in Fig. 4D.
A structural and computational study of the Q-site of E. coli SQR has led to the proposal of two binding positions for ubiquinone within the Q-site, called the Q1 and Q2 sites (16). The Q1-site is close to the cytoplasmic opening of the Q-site and allows the O1 carbonyl group of ubiquinone to hydrogen-bond to Trp-B164 and Tyr-D83. The presence of electrons in the redox cofactor chain is proposed to drive ubiquinone deeper into the Q-site, occupying the Q2 position and enabling the O4 carbonyl to interact with Ser-C27. The side chain of His-B207 is proposed to rotate and form a hydrogen bond to the O3 methoxy group of ubiquinone. Comparing the position of the carbonyl group of carboxin and the hydroxyl group of PCP interacting with Trp-B164 and Tyr-D83 with the structures of E. coli SQR binding ubiquinone (1NEK), DNP-17 (1NEN), and Atpenin A5 (2ACZ) reveals that PCP binds in the Q1-site, whereas carboxin binds deeper, in the Q2-site, but without triggering rotation of His-B207. The current structures provide experimental support for the proposal of two binding positions within the Q-site and show the plasticity of the Q-site in SQR.
Arrangement of Ordered Waters and a Pathway for Proton Transfer from the Cytoplasm to the Quinone-binding Site
Because SQR does not generate a protonmotive force during catalysis, the quinone reduction reaction must consume cytoplasmic protons, which are released during succinate oxidation. Based on the structure of 1NEK, a putative proton transfer pathway has been proposed involving 13 water molecules that cross the protein from Glu-C101 and Asp-C95 at the cytoplasmic surface through Lys-B230 to His-B207, Arg-C31, and Asp- D82 at the Q-site (16). Such key water molecules should be conserved in different crystal forms of the enzyme. The new P212121 structure, however, does not confirm significant conservation of the waters on the putative pathway but, rather, reveals ordered hydrogen-bonding waters that link residues at the cytoplasmic surface (Asp-D15) with those at the Q-site (His-B207 and Asp-D83) (Fig. 5, A and B). These water molecules have an average B-factor of 46.7 Å2, comparable with the average B-factor of the protein, showing that the water molecules are well ordered. A recent mutagenesis study also suggested a possible alternative entrance for protons involving Asp-D15 (19). In Fig. 5C the waters have been colored to reflect their degree of conservation within the three NCS copies of the monomer; dark green for those waters conserved in all three copies (B2103, D2008, B2120, and D2001), chartreuse for the one present in two copies (B2123), and lime green for the water present in one copy (D2003). Significantly, waters D2003 and D2008 hydrogen-bond to Gln-D78, which lies on the entrance to possible water channels identified in the porcine and avian SQR structures (19).
To probe both proposed water channels, we have mutated two residues. Lysine B230 was thought to be critical for the initially proposed channel (16), and Arg-B205 is involved in structural support of the water network by coordination of waters B2120 and B2123 (Fig. 5C). The data in Table 1 show that substitution of Arg-B205 induced a 1 order of magnitude increase in Km value and partial loss of quinone reductase activity by SQR as evidenced by the lower Q1/phenazine ethosulfate activity ratio. This suggests a structural rearrangement affecting the Q-site in this mutant. By contrast, mutation of Lys-B230 to Leu does not impose apparent structural perturbation of the Q-site. As seen in the table, the loss of enzyme activity in the SdhB K230L mutant is similar in either the phenazine ethosulfate-dichlorophenol indophenol or Q1-reductase assays, as is the binding of Q1. This suggests that Lys-B230 is not essential for protonation of quinone and is unlikely to be essential as a component of a water channel. The lowered activity of the SdhB substitution may be caused by changes in the redox properties of the [3Fe-4S] cluster rather than direct involvement of the Q-site (supplemental Fig. S7).5
TABLE 1.
Kinetic parameters of isolated SQR enzymes
Enzyme activity was measured at 30 °C after activation of SQR, and kinetic parameters were determined as described (20). Wild type and mutant enzymes were purified as previously described (20). DCIP, dichlorophenol indophenol; ND, not determined.
| Enzyme | Turnover |
Q1/PES | KM(Q1) | KI(PCP) | |
|---|---|---|---|---|---|
| PES-DCIP | Q1 | ||||
| s−1 | s−1 | μm | μm | ||
| Wild type SQR | 98 | 102 | 1.04 | 2.5 | 13 |
| SdhB R205S | 98 | 68 | 0.69 | 25.0 | ND |
| SdhB R205L | 95 | 52 | 0.55 | 22.5 | 6 |
| SdhB K230L | 58 | 50 | 0.86 | 5.0 | 12 |
The conservation of the waters shown in Fig. 5C and the similar location of possible water channels in eukaryotic SQRs (19) are consistent with some involvement of the water chain in protonation/deprotonation reactions of quinones. In E. coli the SQR trimer may also provide a water sink to provide bulk solvent water for these reactions. Other possibilities should be considered, however, as protein re-protonation may not absolutely require a structurally critical water channel but, rather, can be achieved by bulk solvent water near the Q-binding site. Evidence for this may be seen from the avian SQR structure (2H88) where a ligand-free quinone binding site accommodates several water molecules inside the ubiquinone binding cavity. Evidently further study is warranted.
Conclusion
In this paper we present three structures of E. coli SQR crystallized in the P212121 space group: SQR inhibited by carboxin at 2.4 Å, SQR inhibited by PCP at 3.2 Å, and SQR with an empty Q-site at 3.2 Å. These structures significantly improve our understanding of the E. coli SQR structure. The results show that the E. coli SdhA subunit is structurally highly similar to the equivalent subunit from eukaryotic SQR and the flavocytochrome c3 from Shewanella. The active site geometry and chemistry required for succinate oxidation and fumarate reduction has, therefore, been highly conserved through evolution.
The current structures provide experimental support for the proposal of two binding positions within the Q-site and show the plasticity of the Q-site in SQR. Although the structures indicate more perturbation of the Q-site after PCP binding, the different positions of the inhibitors in the Q-site may help explain their different kinetic behavior (51). The arrangement of ordered waters also supports the proposal of an alternative entrance for protons to quinone involving Asp-D15 (19).
Supplementary Material
Acknowledgments
J. Ruprecht acknowledges Drs. Jade Li, Gebhard Schertler, and Chris Tate from the MRC Laboratory of Molecular Biology, Cambridge, UK for advice on membrane protein crystallization and structure determination and Drs. Andrew McCarthy and Didier Nurizzo from the European Synchrotron Radiation Facility for support during data collection. We also thank Violetta Kotlyar for help.
This work was supported, in whole or in part, by National Institutes of Health Grant GM61606. This work was also supported by the Department of Veterans Affairs, Office of Research and Development, Biomedical Laboratory Research Division (to G. C.). The work in the UK was supported primarily by the Biotechnology and Biological Sciences Research Council (#B17935) with important contributions from the Human Receptor Crystallography Project, ERATO, Japan Science and Technology Agency.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S7.
The atomic coordinates and structure factors (codes 2WDQ, 2WDR, and 2WDV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
J. Ruprecht, V. Yankovskaya, E. Maklashina, S. Iwata, and G. Cecchini, manuscript in preparation.
- SQR
- succinate:quinone oxidoreductase
- QFR
- quinol:fumarate reductase
- PCP
- pentachlorophenol
- DM
- decyl-β-D-maltopyranoside
- NCS
- non-crystallographic symmetry
- Q-site
- ubiquinone-binding site
- Q1
- ubiquinol with one isoprenoid unit as the side chain
- TEO
- the malate-like intermediate
- TLS
- translation libration screw.
REFERENCES
- 1.Cecchini G. (2003) Annu. Rev. Biochem. 72, 77–109 [DOI] [PubMed] [Google Scholar]
- 2.Hägerhäll C. (1997) Biochim. Biophys. Acta 1320, 107–141 [DOI] [PubMed] [Google Scholar]
- 3.Lancaster C. R. D. (2002) Biochim. Biophys. Acta 1553, 1–6 [DOI] [PubMed] [Google Scholar]
- 4.Cecchini G., Schröder I., Gunsalus R. P., Maklashina E. (2002) Biochim. Biophys. Acta 1553, 140–157 [DOI] [PubMed] [Google Scholar]
- 5.Horsefield R., Iwata S., Byrne B. (2004) Curr. Protein Pept. Sci. 5, 107–118 [DOI] [PubMed] [Google Scholar]
- 6.Maklashina E., Iverson T. M., Sher Y., Kotlyar V., Andréll J., Mirza O., Hudson J. M., Armstrong F. A., Rothery R. A., Weiner J. H., Cecchini G. (2006) J. Biol. Chem. 281, 11357–11365 [DOI] [PubMed] [Google Scholar]
- 7.Vibat C. R., Cecchini G., Nakamura K., Kita K., Gennis R. B. (1998) Biochemistry 37, 4148–4159 [DOI] [PubMed] [Google Scholar]
- 8.Maklashina E., Rothery R. A., Weiner J. H., Cecchini G. (2001) J. Biol. Chem. 276, 18968–18976 [DOI] [PubMed] [Google Scholar]
- 9.Iverson T. M., Luna-Chavez C., Cecchini G., Rees D. C. (1999) Science 284, 1961–1966 [DOI] [PubMed] [Google Scholar]
- 10.Lancaster C. R., Kröger A., Auer M., Michel H. (1999) Nature 402, 377–385 [DOI] [PubMed] [Google Scholar]
- 11.Taylor P., Pealing S. L., Reid G. A., Chapman S. K., Walkinshaw M. D. (1999) Nat. Struct. Biol. 6, 1108–1112 [DOI] [PubMed] [Google Scholar]
- 12.Leys D., Tsapin A. S., Nealson K. H., Meyer T. E., Cusanovich M. A., Van Beeumen J. J. (1999) Nat. Struct. Biol. 6, 1113–1117 [DOI] [PubMed] [Google Scholar]
- 13.Bamford V., Dobbin P. S., Richardson D. J., Hemmings A. M. (1999) Nat. Struct. Biol. 6, 1104–1107 [DOI] [PubMed] [Google Scholar]
- 14.Huang L. S., Shen J. T., Wang A. C., Berry E. A. (2006) Biochim. Biophys. Acta 1757, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yankovskaya V., Horsefield R., Törnroth S., Luna-Chavez C., Miyoshi H., Léger C., Byrne B., Cecchini G., Iwata S. (2003) Science 299, 700–704 [DOI] [PubMed] [Google Scholar]
- 16.Horsefield R., Yankovskaya V., Sexton G., Whittingham W., Shiomi K., Omura S., Byrne B., Cecchini G., Iwata S. (2006) J. Biol. Chem. 281, 7309–7316 [DOI] [PubMed] [Google Scholar]
- 17.Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z. (2005) Cell 121, 1043–1057 [DOI] [PubMed] [Google Scholar]
- 18.Huang L. S., Sun G., Cobessi D., Wang A. C., Shen J. T., Tung E. Y., Anderson V. E., Berry E. A. (2006) J. Biol. Chem. 281, 5965–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng V. W., Johnson A., Rothery R. A., Weiner J. H. (2008) Biochemistry 47, 9107–9116 [DOI] [PubMed] [Google Scholar]
- 20.Maklashina E., Berthold D. A., Cecchini G. (1998) J. Bacteriol. 180, 5989–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Törnroth S., Yankovskaya V., Cecchini G., Iwata S. (2002) Biochim. Biophys Acta 1553, 171–176 [DOI] [PubMed] [Google Scholar]
- 22.Horsefield R., Yankovskaya V., Törnroth S., Luna-Chavez C., Stambouli E., Barber J., Byrne B., Cecchini G., Iwata S. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 600–602 [DOI] [PubMed] [Google Scholar]
- 23.Leslie A. G. W. (1992) Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography, No. 26 [Google Scholar]
- 24.The Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 25.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 26.Evans P. R. (1993) Proceedings of CCP4 Study Weekend on Data Collection and Processing (Sawyer L., Isaacs N., Bailey S. eds) pp. 114–122, CLRC Daresbury Laboratory, Warrington, UK [Google Scholar]
- 27.Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 28.McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 30.Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamzin V. S., Perrakis A., Wilson K. S. (2001) in International Tables for Crystallography, Volume F: Crystallography of Biological Macromolecules (Rossmann M. G., Arnold E. eds) pp. 720–722, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 32.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 34.Afonine P. V., Grosse-Kunstleve R. W., Adams P. D. (2005) CCP4 Newsletter 42, contribution 8, CLRC Daresbury Laboratory, Warrington, UK [Google Scholar]
- 35.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 36.Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 37.Kleywegt G. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 94–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evrard G. X., Langer G. G., Perrakis A., Lamzin V. S. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn M. D., Isupov M. N., Murshudov G. N. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 40.Howlin B., Butler S. A., Moss D. S., Harris G. W., Driessen H. P. C. (1993) J. Appl. Crystallogr. 26, 622–624 [Google Scholar]
- 41.Davies I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 43.Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 44.Kühlbrandt W. (1988) Q. Rev. Biophys. 21, 429–477 [DOI] [PubMed] [Google Scholar]
- 45.Wiener M. C. (2004) Methods 34, 364–372 [DOI] [PubMed] [Google Scholar]
- 46.Huang L. S., Borders T. M., Shen J. T., Wang C. J., Berry E. A. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel H. (1983) Trends Biochem. Sci. 8, 56–59 [Google Scholar]
- 48.Belikova Y. O., Kotlyar A. B., Vinogradov A. D. (1988) Biochim. Biophys. Acta 936, 1–9 [DOI] [PubMed] [Google Scholar]
- 49.Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 50.Jormakka M., Yokoyama K., Yano T., Tamakoshi M., Akimoto S., Shimamura T., Curmi P., Iwata S. (2008) Nat. Struct. Mol. Biol. 15, 730–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maklashina E., Cecchini G. (1999) Arch. Biochem. Biophys. 369, 223–232 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.