Abstract
DNA-protein cross-links (DPCs) are unique among DNA lesions in their unusually bulky nature. The steric hindrance imposed by cross-linked proteins (CLPs) will hamper DNA transactions, such as replication and transcription, posing an enormous threat to cells. In bacteria, DPCs with small CLPs are eliminated by nucleotide excision repair (NER), whereas oversized DPCs are processed exclusively by RecBCD-dependent homologous recombination (HR). Here we have assessed the roles of NER and HR for DPCs in mammalian cells. We show that the upper size limit of CLPs amenable to mammalian NER is relatively small (8–10 kDa) so that NER cannot participate in the repair of chromosomal DPCs in mammalian cells. Moreover, CLPs are not polyubiquitinated and hence are not subjected to proteasomal degradation prior to NER. In contrast, HR constitutes the major pathway in tolerance of DPCs as judged from cell survival and RAD51 and γ-H2AX nuclear foci formation. Induction of DPCs results in the accumulation of DNA double strand breaks in HR-deficient but not HR-proficient cells, suggesting that fork breakage at the DPC site initiates HR and reactivates the stalled fork. DPCs activate both ATR and ATM damage response pathways, but there is a time lag between two responses. These results highlight the differential involvement of NER in the repair of DPCs in bacterial and mammalian cells and demonstrate the versatile and conserved role of HR in tolerance of DPCs among species.
The chromosomal DNA of living organisms continuously suffers from a variety of lesions induced by endogenous and environmental agents. DNA-protein cross-links (DPCs)4 account for a class of the most ubiquitous DNA lesions and are known to be produced by chemical agents, such as formaldehyde (FA) and transition metals, and by physical agents, such as ionizing radiation and UV light (1). DPCs are also produced by anticancer drugs, such as 5-aza-2′-deoxycytidine (azadC) and cisplatin (1, 2). Although some classes of DPCs contain a flanking strand break (e.g. covalently trapped topoisomerases) (3), typical (and probably the most common) DPCs contain proteins irreversibly trapped on the uninterrupted DNA strand. It is readily inferred from the unusually bulky nature of cross-linked proteins (CLPs) that steric hindrance imposed by CLPs on proteins involved in DNA transactions would hamper replication, transcription, and repair. Consistent with this notion, DPCs incorporated into oligonucleotides and plasmid DNA block DNA replication in vitro (4, 5) and in vivo (6, 7), respectively. Moreover, CLPs attenuate the binding of the damage recognition protein (UvrB) involved in bacterial nucleotide excision repair (NER) in a size-dependent manner (7).
Conversely, it has been largely elusive how cells circumvent the genotoxic effects of DPCs. We recently showed that NER and homologous recombination (HR) play pivotal roles in mitigating the genotoxic effects of DPCs in bacteria (7). Interestingly, the two pathways contribute differentially to the tolerance of DPCs. In NER catalyzed by UvrABC, the excision efficiency for DPCs varies dramatically with the size of CLPs both in vitro and in vivo and is attenuated by steric hindrance of CLPs. The upper size limit of CLPs amenable to NER in vitro was around 16 kDa, but the biologically relevant size limit was lower in vivo, at around 11 kDa. DPCs with oversized CLPs are processed exclusively by RecB-dependent HR. Given that HR can also process DPCs containing small CLPs, HR is a versatile mechanism for DPC tolerance, whereas NER makes a limited but still significant contribution to the repair of DPCs in bacterial cells. Although the study provides a paradigm for the repair of DPCs in cells, it remains to be seen whether NER and HR are also closely coordinated to deal with unusually bulky DNA lesions, such as DPCs, in higher organisms.
As for bacterial NER (8, 9), it has been reported that the in vitro activity of mammalian NER for DPCs is dependent on the size of CLPs. Mammalian cell-free extracts (CFEs) make efficient damage-specific incisions for DPCs containing short peptides comprising 4 or 12 amino acids (0.57 and 1.5 kDa) but not for those containing 16-kDa T4 endonuclease V, 22-kDa histone H1, and 37-kDa HhaI DNA cytosine methyltransferase (DNMT) (5, 10, 11). The damage-specific incision for short peptide adducts was absent with CFEs from NER-deficient cells. Although these data indicate that the mammalian NER system is sensitive to the size of CLPs, it remains to be clarified whether NER participates in the repair of DPCs in mammalian cells as in bacterial cells. In addition to direct repair of DPCs by NER, an alternative repair model of DPCs has also been proposed, in which CLPs are initially degraded to short peptides by the proteasome, and the resulting DNA-peptide cross-links are removed by NER (3, 9–11). Again, the validity of this alternative model also remains to be examined in vivo.
In the present study, we assessed the roles of NER and HR in tolerance of DPCs using a mammalian system. Several lines of in vitro and in vivo evidence indicate that NER alone or NER coupled with proteasomal degradation of CLPs does not contribute to the repair of DPCs, whereas HR initiated by fork breakage at DPCs plays a pivotal role in tolerance of DPCs in mammalian cells. These results highlight the differential involvement of NER in the repair of DPCs in bacterial and mammalian cells and demonstrate the versatile and conserved role of HR in tolerance of DPCs among species.
EXPERIMENTAL PROCEDURES
DNA, Proteins, and Cells
The 150-mer oligonucleotides containing oxanine (150OXA) or oxanine-protein cross-links (150OXA-DPC) were prepared as described in the supplemental materials. Preparation of 60-mer oligonucleotides containing oxanine (60OXA) or oxanine-protein cross-links (60OXA-DPC) were reported elsewhere (7). CLPs used in this study are listed in supplemental Table S1. 60OXA-DPC and 150OXA-DPC bearing a 5′-end 32P label or 3′-end [32P]dCMP were annealed to complementary strands and used for NER incision assays. Bacillus caldotenax UvrA and UvrB and Thermotoga maritima UvrC were purified as reported previously (12, 13). The cells used in the present study are listed in supplemental Table S2.
NER Incision Assays
NER incision assays with UvrABC (7, 14) or CFEs from HeLa, XPA, and XPF cells (14) were performed as reported previously. The sample was treated with proteinase K before PAGE analysis.
Fluorescence Analysis of CLPs
Chromosomal DNA was isolated by CsCl density gradient centrifugation, as described previously (7) with some modifications. Cells were grown in Dulbecco's modified Eagle's medium (DMEM; Nissui) supplemented with 10% fetal bovine serum (FBS) and kanamycin at 37 °C in a 5% CO2 atmosphere. Cells in the midlogarithmic phase were treated with 0.5 mm FA in FBS-free DMEM at 37 °C for 3 h and allowed to repair in fresh media for up to 4 h. Typically, cells from four 150-mm dishes were combined, lysed in PBS containing 1% Sarkosyl, and subjected to CsCl density gradient centrifugation. Fluorescence analysis of CLPs with fluorescein isothiocyanate (FITC; Dojindo) was performed as described previously (7). For analysis with 2-methoxy-2,4-diphenyl-3(2H)-furanone (MDPF; Tokyo Chemical Industry), DNA was heated at 70 °C for 6 h to release CLPs. The sample (150 μl) was dialyzed extensively against water using a microdialysis cup (Bio-Tech International) at 4 °C. Internal and external solutions of dialysis were recovered separately, and the volume was reduced to 150 μl by a centrifugation evaporator. The sample was incubated with 0.38 mm MDPF at room temperature for 1 h, and the fluorescence intensity was measured at 520 nm (FITC) or 485 nm (MDPF).
Analysis of Polyubiquitination of CLPs
DNA (50 μg) isolated from mock- and FA-treated WI38VA13 cells (with or without parallel treatment with MG132), ubiquitinated lysozyme (Hokudo), and lysozyme were vacuum slot-blotted on a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk; washed with Tris-buffered saline/Tween 20 (TBST) comprising 20 mm Tris-HCl (pH 7.6), 137 mm NaCl, and 0.1% Tween 20; incubated with the FK1 antibody conjugated with horseradish peroxidase (BIOMOL); and washed again with TBST. The chemiluminescence signal was detected with ECL Western blotting detection reagents (GE Healthcare) on a FAS1000 imaging analyzer (TOYOBO). Alternatively, DNA (160 μg, heated at 70 °C for 6 h to release CLPs) or control ubiquitinated lysozyme was mixed with an affinity matrix (40 μl) for ubiquitinated proteins (UbiQapture-Q kit; BIOMOL). The sample was mixed on a rotator at 4 °C overnight. The supernatant was removed (unbound fraction), and the matrix was washed twice with PBS (wash-1 and wash-2 fractions). Finally, bound proteins were eluted with SDS-loading buffer comprising 50 mm Tris-HCl (pH 6.8), 3% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 0.002% bromphenol blue (elute fraction). Proteins in each fraction were separated by 10% SDS-PAGE and stained with the Silver Stain-II kit (Daiich Pure Chemical).
Cell Survival
Human and Chinese hamster ovary (CHO) cells (supplemental Table S2) were grown in DMEM and Eagle's MEM (Nissui), respectively, supplemented with 10% FBS and kanamycin in a 5% CO2 atmosphere. Exponentially growing cells were seeded in 100-mm dishes and incubated for 12 h. For FA treatment, the medium was changed to FBS-free DMEM or Eagle's MEM containing FA (0–0.5 mm), and cells were incubated at 37 °C for 3 h. For azadC treatment, the medium was changed to DMEM or Eagle's MEM containing azadC (0–5 μm) and FBS, and cells were incubated at 37 °C for 24 h. Cells were washed with fresh media twice and allowed to form colonies. Colonies with more than 50 cells were counted after 7–12 days.
Immunofluorescence Staining
The formation of RAD51 and γ-H2AX nuclear foci was analyzed, as reported previously (15, 16). Briefly, MRC5SV cells were plated onto glass slides, treated with 0.1 mm FA or 1 μm azadC, and subjected to repair incubation as described under “Cell Survival.” Cells were fixed with cold methanol for 20 min, rinsed with cold acetone for several seconds, and then air-dried. The fixed cells were incubated with anti-RAD51 (BioAcademia) and anti-γ-H2AX (Upstate Biotechnology) antibodies and subsequently with secondary antibodies conjugated with Alexa 488 or Alexa 546 (Molecular Probes), mounted with fluorescent mounting medium (DAKO) containing 2 μg/ml 4′,6-diamidino-2-phenylindole. Stained cells were visualized on a fluorescence microscope.
Comet Assays
Cells were treated with FA (0.2 mm, 3 h) or azadC (1 μm, 24 h) as described above and further incubated for 6 h (FA) or 12 h (azadC) in fresh media without FA and azadC. Comet assays (single cell gel electrophoresis) were performed according to the published procedure (17). Aliquots (10 μl) of cell suspension (about 30,000 cells) were mixed with 0.5% low melting point agarose (120 μl) and added to microscope slides, which had been covered with a bottom layer of 1.5% agarose. Cell lysis and electrophoresis were performed at 4 °C. With FA, an additional proteinase K treatment was performed after the lysis of FA-treated cells (18). Cells were lysed at pH 10 overnight. Slides were kept in neutral or alkaline electrophoresis buffer for 20 min before electrophoresis. Electrophoresis was performed at 1 V/cm for 20 min (neutral) or 30 min (alkaline). Cells were stained with ethidium bromide, and their images were captured on a fluorescence microscope. Tail moments of 50–100 randomly selected cells were measured using Comet Assay IV software (Perceptive Instruments).
Phosphorylation of CHK1 and CHK2
WI38VA13 cells were treated with FA (0.4 mm) or azadC (2 μm) as described under “Cell Survival.” For control, cells were mock-treated or irradiated by UV light (50 J/m2). After appropriate periods of repair incubation (indicated in Fig. 6A), cells were collected and lysed with lysis buffer comprising 10 mm Tris-HCl (pH 8.0), 130 mm NaCl, 1% Triton X-100, 10 mm NaF, protease inhibitor mixture (Roche Applied Science), and phosphatase inhibitor mixture (Sigma). Proteins (150 μg) were separated by 10% SDS-PAGE, blotted on a nitrocellulose membrane, and probed with CHK1, CHK2, phospho-CHK1 (Ser317), and phospho-CHK2 (Ser19) antibodies (Cell Signaling Technology) or with the monoclonal actin antibody (Thermo Scientific). Horseradish peroxidase-linked anti-rabbit IgG antibodies (Cell Signaling Technology) or horseradish peroxidase-linked anti-mouse IgG antibodies (CHEMICON International) were used to detect primary antibodies.
FIGURE 6.
DPCs induce phosphorylation of CHK1 and CHK2. A, protocols of cell treatment with UV, FA, and azadC. Broken lines indicate post-repair incubation without damaging agents, and WB indicates the time point where the sample was taken for Western blotting analysis. B, Western blotting analysis of phosphorylation of CHK1 and CHK2. WI38VA13 cells were treated with FA (0.4 mm for 3 h), azadC (2 μm for 24 h), or UV (50 J/m2). After the indicated periods of treatment or repair incubation, cells were collected and lysed. Proteins (150 μg) were separated by 10% SDS-PAGE; blotted on a membrane; and probed with CHK1, CHK2, phospho-CHK1 (Ser317), phospho-CHK2 (Ser19), and actin antibodies.
RESULTS
Upper Size Limit of CLPs Amenable to Mammalian NER Is 8–10 kDa in Vitro
It has been shown by using CFEs that the mammalian NER system barely makes damage-specific 5′ incisions for DPCs containing 16–37-kDa CLPs, whereas it does so for DPCs containing short peptides of 0.57 and 1.5 kDa (5, 10, 11). However, the upper size limit of CLPs amenable to mammalian NER has not been rigorously determined. The size limit is crucial to predicting whether NER can participate in the repair of DPCs in vivo. To determine the upper size limit, we prepared a 150-mer oligonucleotide containing oxanine (150OXA), and peptides and proteins of various sizes were tethered to oxanine to prepare defined DPC substrates (150OXA-DPCs; supplemental Fig. S1). Oxanine reacts with proteins and polyamines to form stable adducts (7, 14, 19, 20). Duplex 150OXA-DPCs (the DPC-containing strand was 5′-end 32P-labeled) were incubated with CFEs from NER-proficient HeLa cells. PAGE analysis of reaction products revealed that the CFEs made clear incisions around the 21st phosphodiester bond 5′ to DPCs (Fig. 1, A and B (left)). The 5′ incision efficiency initially rose with the CLP size up to 1.6 kDa and then decreased thereafter (Fig. 1C and supplemental Fig. S2), with activity being negligible for the 11-kDa protein. The plot of the 5′ incision efficiency against the CLP size (Fig. 1C) indicates that the upper size limit of CLPs amenable to human NER is around 8–10 kDa in vitro. The HeLa CFEs also made CLP size-dependent 3′ incisions at the 6th phosphodiester bond 3′ to DPCs (Fig. 1, A and B (right)). Thus, human NER excises 26–29-mer fragments containing DPCs, with the incision sites being independent of the size of CLPs. It was confirmed that 5′ incisions for DPCs were attributable to NER activity; the 5′ incision activity for DPCs was not observed with CFEs from NER-deficient human xeroderma pigmentosum XPA or XPF cells, but it was restored when CFEs from both cells were combined (supplemental Fig. S3).
FIGURE 1.
Upper size limit of CLPs amenable to mammalian NER is 8–10 kDa in vitro. A, partial sequence of substrates (150OXA-DPCs). Proteins listed in supplemental Table S1 were tethered to oxanine (O). The arrows indicate the incision sites with HeLa CFEs. B, PAGE analysis of incision products. Left, analysis of 5′ incision. 150OXA-DPCs (5′-end 32P-labeled) were incubated with HeLa CFEs (100 μg) for 30 min. After incubation, samples were treated with proteinase K and separated by 10% denaturing PAGE. Right, analysis of 3′ incisions. 150OXA-DPCs (3′-end 32P-dC-labeled) were treated, and products were analyzed as on the left. The leftmost lanes (M) indicate 59-mer and 64-mer markers in the left and right panels, respectively. C, variations of the DPC incision efficiency with the size of CLPs. Left, incision with HeLa CFEs. The amounts of 5′-nicked products were quantified from the left panel in B and supplemental Fig. S2 and are plotted against the size of CLPs. Right, incision with UvrABC. 60OXA-DPCs were incubated with B. caldotenax UvrA and UvrB and T. maritima UvrC for 30 min at 37 or 55 °C. Products were analyzed by 12% denaturing PAGE (not shown). The amounts of 5′-nicked products are plotted against the size of CLPs.
We also compared the size limits of CLPs amenable to bacterial and human NER. UvrAB and UvrC from thermophilic bacteria (B. caldotenax UvrAB and T. maritime UvrC) were used for bacterial NER, and 5′ incision activity was assayed with 60-mer oligonucleotides containing DPCs (60OXA-DPC), as reported previously (7). The size limit of CLPs incised by UvrABC was 16 kDa at 37 °C, but it was shifted to around 20–22 kDa at 55 °C, a suboptimal temperature for thermophilic UvrABC (Fig. 1C, right). At the same time, as for human NER, a clear activity peak was observed for the 1.6-kDa CLP. Comparison of in vitro incision activity for DPCs between bacterial and human NER indicates that the upper size limit of CLPs amenable to human NER (8–10 kDa) is considerably lower than that of bacterial NER (16 kDa at 37 °C or 20–22 kDa at 55 °C), although both NER systems share the activity peak for 1.6-kDa CLPs.
Chromosomal DPCs Are Not Removed by NER in Mammalian Cells
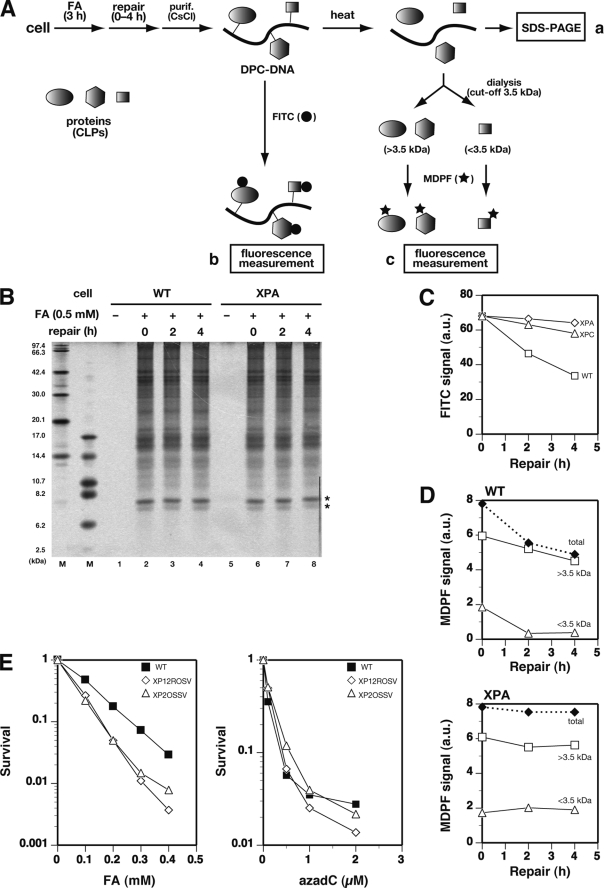
We have previously shown that in Escherichia coli cells, DPCs containing proteins of sizes less than 11 kDa are excised from chromosomal DNA in wild type (WT) but not in NER-deficient uvrA cells that had been exposed to FA, a typical DPC-inducing agent (7). To assess the role of NER in the repair of DPCs in mammalian cells, human NER-proficient (WI38VA13) and NER-deficient XPA (XP12ROSV) cells were treated with 0.5 mm FA for 3 h and then subjected to post-repair incubation in FA-free media for up to 4 h. The chromosomal DNA was isolated by CsCl density gradient centrifugation. FA-induced cross-links were reversed by heat treatment, and released proteins were analyzed by SDS-PAGE (Fig. 2A). SDS-PAGE analysis revealed various CLPs that were common to WT and XPA cells (Fig. 2B, lanes 2 and 6). Such CLPs were absent without FA treatment (lanes 1 and 5). The smallest proteins cross-linked by FA were 7.4 and 8.0 kDa (indicated by the asterisks in Fig. 2B). Importantly, in both WT and XPA cells, the amounts of individual CLPs barely changed during the course of post-repair incubation for up to 4 h (Fig. 2B, lanes 2–4 and 6–8), indicating that NER was unable to excise FA-induced DPCs containing even the smallest CLPs (7.4 and 8.0 kDa). This contrasts with our previous observation with E. coli, where FA-induced chromosomal DPCs containing CLPs smaller than 11 kDa were actively removed by NER in vivo (7).
FIGURE 2.
Chromosomal DPCs are not removed by NER in mammalian cells. A, scheme for analysis of FA-induced CLPs and CLXs using SDS-PAGE (a) and fluorescence labeling with FITC (b) and MDPF (c). B, SDS-PAGE analysis of CLPs. WT (WI38VA13) and XPA (XP12ROSV) cells were treated with 0.5 mm FA for 3 h, and chromosomal DNA was isolated after the indicated period of repair incubation. CLPs in 40 μg of DNA were released by heat and separated by 10% SDS-PAGE. Bands were visualized by silver staining. The two leftmost lanes show size markers. C, release of CLPs measured with FITC. WT, XPA, and XPC (XP4PASV) cells were treated with FA, and DNA was isolated as in B. CLPs in 15 μg of DNA were labeled by FITC, and their fluorescence was measured. Data points are means of two independent experiments. D, release of small adducts (CLXs) measured with MDPF. WT and XPA cells were treated with FA, and DNA was isolated as in B. Cross-linked adducts in 150 μg of DNA were released by heat and fractionated by dialysis (3.5 kDa cut-off). The adducts (>3.5 and <3.5 kDa) were separately labeled by MDPF, and their fluorescence was measured. Note that the fraction with >3.5 kDa contained CLPs and the large component of CLXs, whereas that with <3.5 kDa contained the small component of CLXs. Data points are means of two independent experiments. E, survival of WT and XPA (XP12ROSV and XP2OSSV) cells treated with FA (left) or azadC (right). Cells were incubated with the indicated concentrations of FA for 3 h or azadC for 24 h. Cell survival was assayed by colony formation. Data points are means of three independent experiments.
We also assessed the removal of chromosomal DPCs by fluorescence labeling (Fig. 2A). The DNA isolated from FA-treated cells (without heat treatment) was incubated with FITC that reacts with the amino group of proteins but not with DNA. After removing free FITC, the fluorescence signal was measured. Surprisingly, the FITC signal decreased significantly during the course of repair incubation for WT but not for XPA and XPC cells (Fig. 2C). These results suggest that NER removed certain FA-induced adducts (cross-link adducts (CLXs)) that were smaller than 7.4 kDa and not detectable by the SDS-PAGE analysis (Fig. 2B). The adducts were reversibly released from DNA upon heat treatment (see below), indicating that the adducts were cross-links between DNA and nonprotein chromatin components. The CLXs were further characterized. The DNA isolated from FA-treated cells was heated to release CLPs and CLXs, and the sample was dialyzed extensively using a dialysis membrane with a cut-off size of 3.5 kDa. The compounds retained in the dialysis cup (>3.5 kDa) and those released from it (<3.5 kDa) were treated separately with MDPF, which became fluorescent after reacting with primary amines. The change in the MDPF signal with repair incubation time showed that in WT cells, the adducts smaller than 3.5 kDa were rapidly removed within 2 h, whereas those greater than 3.5 kDa were removed very slowly (Fig. 2D, top). This result implies two types of CLXs that were removed by NER, one smaller than 3.5 kDa and the other between 3.5 and 7.4 kDa. Neither type of CLXs (<3.5 and >3.5 kDa) was detected in the 2.5–6.2-kDa region of SDS-PAGE (Fig. 2B). It is possible that adducts were diffused or eluted out of the gel during electrophoresis or staining (smaller CLXs) or produced in low yields (larger CLXs). The detailed nature of CLXs remains to be elucidated, but a candidate for the smaller component (<3.5 kDa) is a cross-link between the DNA base and polyamines present in chromatin. In XPA cells, neither type of CLXs (<3.5 and >3.5 kDa) was removed during repair incubation (Fig. 2D, bottom). The SDS-PAGE and MDPF data (Fig. 2, B and D) together suggest that NER is directed to the repair of CLXs but not to CLPs in FA-treated cells. The removal of CLXs (but not CLPs) by NER probably contributes to the survival of FA-treated cells, since XPA cells exhibited a moderate sensitivity to FA as compared with WT cells (Fig. 2E, left). The absence of a role of NER in the removal of CLPs was substantiated by the lack of sensitivity of XPA cells to azadC (Fig. 2E, right), which specifically induces DPCs containing 33–183-kDa DNMTs (21).
CLPs Are Not Subjected to Proteasomal Degradation Prior to NER
The mammalian NER system exhibited no measurable repair activity for FA-induced chromosomal DPCs in vivo (Fig. 2B). However, it shows a robust activity for DPCs containing short peptides in vitro (Fig. 1) (10), from which an alternative repair model for DPCs in mammalian cells has been proposed by other laboratories; CLPs are initially degraded to peptides by the proteasome, and the resulting DNA-peptide cross-links are repaired by NER (3, 9–11). Additional in vivo data supporting this model are that the repair of the green fluorescent protein (GFP) gene containing a HhaI DNMT-DPC was partially inhibited (about 50% relative to control) by a 26 S proteasome inhibitor MG132, when a plasmid containing the DPC was transfected into CHO cells (11). To elucidate whether the proteosomal degradation of CLPs plays any role in the repair of DPCs in vivo, we analyzed the polyubiquitination status of CLPs. Polyubiquitination targets proteins for recognition and degradation by the 26 S proteasome (22). WI38VA13 cells were treated with FA and subjected to post-repair incubation for up to 4 h without or with parallel treatment with MG132 (Fig. 3A). The isolated chromosomal DNA (50 μg) was blotted on a membrane. The ubiquitination status of CLPs was analyzed with the FK1 monoclonal antibody that is specific for polyubiquitinated proteins (23). Polyubiquitination signals were observed for control polyubiquitinated lysozyme, and those were not affected by coexistence of DNA isolated from FA-untreated cells (Fig. 3B, lanes 3–6). However, no signals were detected for DNA isolated immediately after or at 4 h after FA treatment (lanes 7 and 8, upper slots). The parallel treatment of cells with MG132 had no effect on the results (lanes 7 and 8, lower slots). Calibrating the chemiluminescence intensity with known amounts of polyubiquitinated lysozyme indicated that the detection limit of polyubiquitinated proteins was 0.1 ng under these conditions (data not shown). Thus, even if present, the amount of polyubiquitinated CLPs per assayed DNA (50 μg) was below 0.1 ng (∼0.1% of total estimated CLPs), strongly suggesting that CLPs are not polyubiquitinated and hence are not subjected to proteasomal degradation.
FIGURE 3.
CLPs are not polyubiquitinated for proteasomal degradation. A, protocols of cell treatment. B, Western blotting analysis of polyubiquitinated CLPs. In the presence or absence of 10 μm MG132, WI38VA13 cells were treated with 0.5 mm FA for 3 h and subjected to repair incubation for 4 h as shown in A. DNA (50 μg) isolated from mock-treated cells (−FA; lanes 1, 5, and 6) or FA-treated cells (+FA; lanes 7 and 8) were slot-blotted on a membrane and probed with polyubiquitin-specific FK1 antibodies. Lysozyme (Lyso) and ubiquitinated lysozyme ((Ub)n-Lyso)) were also slot-blotted as control and analyzed similarly (lanes 2–4). C, SDS-PAGE analysis of polyubiquitinated CLPs. WI38VA13 cells were treated with 0.5 mm FA in the presence of 10 μm MG132 as described in B. DNA (50 μg) isolated from cells was heated to release CLPs and mixed with an affinity matrix for ubiquitinated proteins. The supernatant was removed (unbound fraction), and the matrix was washed twice (wash-1 and wash-2 fractions). Finally, bound proteins were eluted with SDS-loading buffer (elute fraction). Proteins were separated by 10% SDS-PAGE and visualized by silver staining. The two leftmost lanes (M) show size markers.
We also confirmed the absence of polyubiquitination of CLPs by an alternative experiment. Cells were treated with FA as shown in Fig. 3A. The CLPs were released from DNA and incubated with an affinity matrix (UbiQapture) for ubiquitinated proteins. Unbound proteins were removed by repeated washing, and bound proteins were eluted by an SDS-polyacrylamide gel loading buffer. SDS-PAGE analysis of the fractions showed that CLPs were recovered in the unbound and first wash fractions but not in the eluted fraction. Thus, no protein bands were detected in the eluted fraction. Conversely, control polyubiquitinated lysozyme was mostly recovered in the eluted fraction. Typical data from SDS-PAGE analyses of the fractions are shown in Fig. 3C when binding assays were performed with CLPs (lanes 6–10) and ubiquitinated lysozyme (lanes 1–5). These results further confirm the absence of polyubiquitination of CLPs in FA-treated cells.
Regarding the data on the inhibition by MG132 of the host cell reactivation of the DPC-containing GFP gene in the transfecting plasmid (11), we noticed that in the reported study, a critical control experiment was missing to assess whether MG132 affected the transfection efficiency of the intact plasmid. To clarify this, we transfected an intact GFP plasmid into WI38VA13 cells in the presence and absence of MG132 and compared the transfection efficiencies after 24 h of incubation (supplemental Fig. S4A). The fraction of GFP-positive cells was 16.3% without MG132 but decreased markedly to 2.8 and 1.0% at 0.5 and 2 μm of MG132, respectively (supplemental Fig. S4, B and C), corresponding to 6- and 16-fold decreases in the transfection efficiencies. Accordingly, we attributed the apparent reduction of the host cell reactivation efficiency of the GFP gene containing the HhaI DNMT-DPC (one-half at 0.412 μm MG132) to the compromised transfection of plasmid rather than to compromised repair of the DPC per se due to the absence of proteasomal degradation. This conclusion is also consistent with the absence of proteasomal degradation of chromosomal CLPs in vivo (see above).
HR Constitutes the Major Pathway in Tolerance of DPCs
In E. coli cells, HR affords a pivotal mechanism in tolerance of DPCs containing oversized CLPs (7). The inability of NER to process chromosomal DPCs containing even the smallest CLPs (Fig. 2) in mammalian cells suggests that tolerance of all DPCs relies on HR. To elucidate the role of HR, we assessed the sensitivity to DPC-inducing agents of CHO cells deficient in HR, together with those deficient in nonhomologous end joining (NHEJ) and NER. As expected, 51D1 and irs1SF cells deficient in RAD51D and XRCC3, respectively, were 2 and 3 orders of magnitude more sensitive to FA and azadC than the parental WT AA8 cell (Table 1 and supplemental Fig. 5A). The sensitivity of 51D1 cells to FA and azadC was completely corrected by introducing hamster RAD51D cDNA, as shown for 51D1.3 cells (supplemental Fig. S5A). Conversely, V3 cells deficient in DNA-dependent protein kinase catalytic subunit exhibited no sensitivity to FA and azadC, eliminating the role of NHEJ in DPC tolerance (survival curves not shown). As for NER-deficient human cells (XPA in Fig. 2E and XPD in Table 1), NER-deficient UV5 cells (ERCC2/XPD) exhibited a mild sensitivity to FA but not azadC (Table 1 and supplemental Fig. S5B). UV41 cells (ERCC4/XPF) were more sensitive to FA than were UV5 cells, and interestingly, they exhibited a weak but measurable sensitivity to azadC (2.8-fold relative to AA8 cells). As demonstrated for human cells (Fig. 2), the sensitivity of UV5 and UV41 cells to FA is at least partly attributable to compromised repair of CLXs rather than to that of CLPs per se. It is not clear why UV41 cells are sensitive to azadC. It is known that XPF-ERCC1 is involved in the repair of DNA interstrand cross-links and has an additional repair role outside NER (24). To our knowledge, the induction of DNA interstrand cross-links by azadC has not been reported. Recent data have implicated a role of XPF-ERCC1 in HR (24). However, given that the sensitivity to azadC that induces large DPCs exclusively differs markedly between HR- and XPF-deficient CHO cells (Table 1), it is evident that HR plays a dominant role in tolerance of DPCs, although the XPF-dependent pathway might make a small contribution.
TABLE 1.
Sensitivity of mammalian cells to FA and azadC
| Cell | Mutation | Defect | Sensitivitya |
|
|---|---|---|---|---|
| FA | azadC | |||
| CHO cells | ||||
| AA8 | WT | None | Control | Control |
| 51D1 | RAD51D | HR | ++ | ++ |
| 51D1.3 | RAD51 + cDNAb | HR corrected | − | − |
| irs1SF | XRCC3 | HR | +++ | +++ |
| V3 | DNA-PKcs | NHEJ | − | − |
| UV5 | ERCC2 (XPD) | NER | + | −c |
| UV41 | ERCC4 (XPF) | NER | ++ | +d |
| Human cells | ||||
| WI38VA13 | WT | None | Control | Control |
| XP12ROSV | XPA | NER | + | − |
| XP2OSSV | XPA | NER | + | − |
| XP6BESV | XPD | NER | + | − |
a Symbols denote sensitivity relative to WT cells for 0.3 mm FA or 2 μm azadC as follows: no sensitivity (−) and sensitivity increased by 1 order (+), 2 orders (++), and 3 orders (+++) of magnitude. Data were from survival curves in Fig. 2E for human cells and in supplemental Fig. S5 for CHO cells. Survival curves for V3 and XP6BESV cells are not shown.
b Complemented with hamster RAD51D cDNA.
c Slightly resistant (2.3-fold) relative to WT cells.
d Slightly sensitive (2.8-fold) relative to WT cells.
FA and azadC Treatments Induce RAD51 and γ-H2AX Nuclear Foci
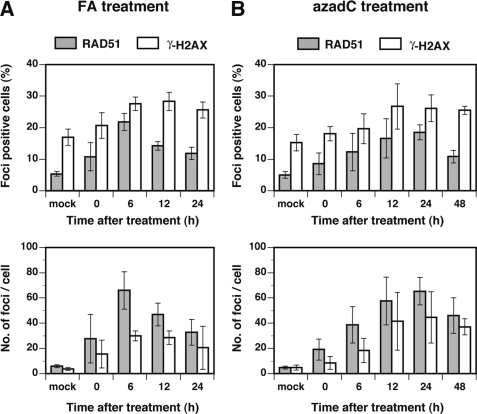
To confirm the involvement of HR in tolerance of DPCs, we analyzed the nuclear foci formation of RAD51, which is the central factor of HR, together with that of phosphorylated histone H2AX (γ-H2AX), which is generally believed to occur in conjunction with chromatin remodeling. MRC5SV cells were treated with FA (0.1 mm for 3 h) or azadC (1 μm for 24 h), and the cells were stained with RAD51 and γ-H2AX antibodies after appropriate periods of incubation. In both FA and azadC treatments, the fraction of RAD51-positive cells and the number of RAD51 nuclear foci per cell increased with incubation time, peaking at 6 h (FA) or 24 h (azadC) after treatment (Fig. 4). Similar increases were observed for γ-H2AX foci (Fig. 4), which colocalized partially with RAD51 (data not shown). The formation of RAD51 and γ-H2AX nuclear foci, together with the hypersensitivity of HR-deficient cells (Table 1), unambiguously demonstrates the indispensable role of HR in mitigating the toxic effects of DPCs.
FIGURE 4.
DPCs induce nuclear RAD51 and γ-H2AX foci. MRC5SV cells were treated with 0.1 mm FA for 3 h or 1 μm azadC for 24 h and subjected to repair incubation for the indicated periods. Cells were fixed, probed with RAD51 and γ-H2AX antibodies, and analyzed for nuclear foci. A, FA treatment. B, azadC treatment. The upper panels show the fraction of foci-positive cells, and lower panels show the number of foci/cell. Cells containing more than 10 RAD51 or 20 γ-H2AX foci were counted as foci-positive cells. Data points are means of three or four independent experiments with S.D.
DPCs Results in Accumulation of Double Strand Breaks (DSBs) in HR-deficient but Not HR-proficient Cells
To elucidate how DPCs initiate HR, the fate of replication forks stalled at DPCs was analyzed by comet assays. The neutral comet assay detects DNA DSBs exclusively, whereas the alkaline comet assay detects the combination of DNA double and single strand breaks and alkali-labile sites (25). The tail moment is a measure of strand breaks in both assays. AA8 (WT) and irs1SF (XRCC3) cells were treated with azadC (1 μm, 24 h) or FA (0.2 mm, 3 h) and further incubated without azadC for 12 h or FA for 6 h. Tail moments were analyzed by neutral and alkaline comet assays. With azadC-treated AA8 cells, no increases in tail moments were observed in either assay (Fig. 5A). In contrast, with HR-deficient irs1SF cells, tail moments in neutral as well as alkaline comet assays increased significantly following azadC treatment and remained high at 12 h after treatment (Fig. 5A). In FA treatment, the tail moments of irs1SF but not AA8 cells increased at 6 h after treatment in both neutral and alkaline comet assays (Fig. 5B). These results strongly suggest that replication forks stalled by DPCs underwent breakage to yield DSBs during azadC treatment or after FA treatment. The resulting DSBs were efficiently repaired in HR-proficient cells but not in HR-deficient cells, leading to the accumulation of DSBs. It is noted that the background tail moments of FA-untreated cells (AA8 and irs1SF) were a few-fold higher than those of the corresponding azadC-untreated cells, probably due to the extra proteinase K treatment of lysed cells that was necessary in the comet assay of FA-treated cells (18).
FIGURE 5.
DPCs result in accumulation of DSBs in HR-deficient but not HR-proficient cells. AA8 (WT) and irs1SF (XRCC3) cells were treated with azadC (1 μm, 24 h) or FA (0.2 mm, 3 h) and incubated without azadC for 12 h or FA for 6 h. Tail moment as a measure of strand breaks was analyzed by neutral and alkaline comet assays as described under “Experimental Procedures.” Data points are means of three independent experiments with S.D. A, azadC treatment. B, FA treatment. The upper and lower panels show the results of neutral and alkaline comet assays, respectively.
HR of DPCs Requires FANCD1/BRCA2 and FANCD2, and DPCs Activate ATR and ATM Damage Response Pathways
HR plays a pivotal role in the tolerance of DPCs (Table 1) and provides support for DNA replication in the recovery of replication forks stalled at DPCs (Fig. 5). An increasing number of proteins are being identified as mediators of HR, including BRCA1, FANCD1/BRCA2, FANCD2, and NBS1 together with RAD51 and RAD51 paralogues (RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3) (26). In addition, DNA damage and replication fork stalling activate ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) kinases, respectively (27), which further activate the downstream checkpoint kinases CHK2 and CHK1. To gain insight into the role of the HR factors and damage response pathways in the tolerance of DPCs, we analyzed the sensitivity of FANCD1/BRCA2, FANCD2, and ataxia telangiectasia cells to azadC that produces DPCs exclusively processed by HR. The sensitivities of FANCD1/BRCA2, FANCD2, and ataxia telangiectasia cells were corrected by introducing the respective cDNAs (supplemental Fig. S6). The survival increases of the cDNA-complemented cells as well as the WT (MRC5SV) cell relative to mutant ones were small but significant, suggesting the role of FANCD1/BRCA2 and FANCD2 in the HR of DPCs and ATM in the damage response to DPCs.
To elucidate the damage response pathway, WI38VA13 cells were treated with FA or azadC, and the phosphorylation of CHK1 and CHK2 was analyzed by Western blotting (Fig. 6A). Treatment with FA and azadC resulted in the phosphorylation of CHK1 (Ser317) and CHK2 (Ser19) (Fig. 6B), suggesting that both ATR and ATM pathways were activated by DPCs. The phosphorylation of CHK1 occurred during the course of FA treatment for 3 h, whereas that of CHK2 occurred after 12 h of FA treatment (lanes 2 and 3), indicating a time lag between the activation of CHK1 and CHK2. Interestingly, FA and azadC treatments led to strong activation of both CHK1 and CHK2 (lanes 3 and 4), whereas UV treatment led to strong activation of CHK1 but only marginal activation of CHK2 (lane 5). It is likely that the arrest of replication machinery by genomic DPCs or UV lesions rapidly activates the ATR kinase, transducing signals to promote repair as well as to halt the S phase cell cycle progression, but the damage responses might differ significantly between UV and DPC lesions that act as polymerase and putative replicative helicase blocks, respectively (28).
DISCUSSION
The present study has shown that mammalian cells employ HR as a primary mechanism to mitigate the genotoxic effects conferred by DPCs (Fig. 7). Conversely, analysis of the removal of chromosomal DPCs revealed that NER provides no alternative or additional pathway for this task in mammalian cells (Fig. 7), although it provides a crucial repair pathway for secondary lesions (CLXs) generated by FA (Fig. 2). Thus, the present and previous (7) studies show the contrasting involvement of NER in the repair of DPCs in bacterial and mammalian cells, which is attributable to the difference in the intrinsic repair capacities of the NER systems regarding the size limit of CLPs amenable to NER (Fig. 1C). The FA sensitivity of a panel of repair-deficient chicken DT40 mutants was recently reported (29). The effects of targeted mutations of repair genes were in the following order: HR > translesion synthesis > base excision repair and NER > NHEJ. Among the HR-related genes, the mutation of FANCD2 and FANCD1/BRCA2 conferred an extreme FA sensitivity on DT40 cells. Although we observed a qualitatively similar phenotype with human FANCD2 and FANCD1/BRCA2 cells upon treatment with azadC, there were only small differences in sensitivities between mutant and cDNA-complemented (and WT) cells (supplemental Fig. S6). Moreover, the previous study (29) ruled out both ATR and ATM pathways in the DNA damage response to FA-induced DPCs. In contrast, with human cells, we observed the phosphorylation of CHK1 and CHK2 upon induction of DPCs (Fig. 6), strongly suggesting that DPCs activate both ATR and ATM pathways (Fig. 7). Accordingly, the data from the mammalian and DT40 cells combined together point to the pivotal role of HR in tolerance of DPCs in higher organisms, but the precise phenotype and DNA damage response pathways to DPCs appear to differ significantly between mammalian and chicken DT40 cells.
FIGURE 7.
Responses of mammalian cells to DPCs. DPCs induce fork stalling and rapidly activate the ATR damage response pathway. Subsequently, forks stalled at DPCs undergo breakage, initiating HR that reestablishes replication forks. It is not clear whether the ATM damage response pathway is activated by fork breakage (DSBs) or ATR. Neither NER nor NER coupled with the proteasomal degradation of CLPs participates in the repair of DPCs. DSBs resulting from fork breakage are not repaired by NHEJ.
It has been proposed that mammalian cells use proteosomal degradation of CLPs to target DPC for NER (3, 9–11). The DNA-peptide cross-links are nearly as good substrates as (6-4) photoproducts for mammalian NER (10). Also, the loss of FA-induced DPCs from human chromosomal DNA is attenuated by a proteasome inhibitor, although NER does not appear to be involved in the removal of DPCs (30). Similarly, proteasomal degradation of topoisomerase I covalently trapped to the 3′-end of DNA is suggested, since tyrosine-DNA phosphodiesterase involved in repair excises intact topoisomerase I very poorly (31). Accordingly, degradation of covalently trapped proteins to peptide adducts would relieve the steric hindrance of CLPs and thereby permit repair factors to access the damage site. Although this is an attractive model for the repair of unusually bulky DNA lesions, the present data show that CLPs on uninterrupted DNA are not polyubiquitinated and hence are not targeted for degradation of the proteasome (Fig. 3). This conclusion is consistent with our previous result that cytosolic ATP-dependent proteases, which are considered counterparts of eukaryotic proteasomes, played no in vivo role in the NER of DPCs in E. coli (7).
Previous studies showed that cell treatments with replication inhibitors, such as hydroxyurea (nucleotide depletion) and aphidicolin (chain termination), resulted in the accumulation of DSBs at stalled forks in HR-proficient WT cells and that both HR and NHEJ contributed to cell survival following treatments with replication inhibitors (32, 33). However, in the present study, we obtained different results for replication inhibition by DPCs. First, accumulation of DSBs attributable to fork breakage was observed only in HR-deficient cells and not in HR-proficient cells (Fig. 5). Thus, accumulation of DSBs was HR-dependent for fork stalling induced by DPCs but not replication inhibitors. It remains to be elucidated why HR can rapidly repair fork breakage induced by DPCs but not that by replication inhibitors, although HR plays a crucial role in cell survival following treatments with both DPC-inducing and replication inhibiting agents. Second, NHEJ did not contribute to cell survival following treatment with DPC-inducing agents (Table 1 and Fig. 7), indicating the differential involvement of NHEJ in the repair of fork breakage induced by DPCs and replication inhibitors. It is tempting to speculate that structurally simple DSBs induced by replication inhibitors are readily processed by NHEJ, whereas DPCs proximal to fork breakage hamper the binding or assembly of NHEJ factors by steric hindrance. Further studies are necessary to answer this question. It was also shown by other studies that prolonged treatment (48–72 h) of A549, HeLa, and HCT116 cells with azadC resulted in DSB formation (34, 35) and that cells became apoptotic after 24–48 h of treatment with azadC (36, 37). We infer that DSBs observed in these studies are related to apoptosis occurring in a later stage of cell response rather than to breakage of the stalled fork for the following reasons. First, we found that accumulation of DSBs occurred only in HR-deficient cells. Accordingly, HR-proficient A549, HeLa, and HCT116 cells will not accumulate DSBs resulting from fork breakage. Second, we also found that accumulation of DSBs due to fork breakage occurred in a relatively early stage after the induction of chromosomal DPCs. DSBs were observed at 6 h after a 3-h FA treatment or during 24-h azadC treatment. In E. coli, the HR-mediated recovery of stalled forks at DPCs proceeds through the RecBCD pathway and requires RuvABC and RecG (7). Interestingly, breakage of the chromosome was not observed in FA-treated E. coli cells, suggesting that the stalled fork at DPCs does not undergo fork breakage. Thus, HR plays a pivotal role in the recovery of stalled forks at DPCs and hence contributes to cell survival both in bacterial and mammalian cells. However, the precise mechanism of HR-mediated fork recovery at the sites of DPCs seems to be different in bacterial and mammalian cells.
DNMT-DPCs induced by azacytidine block plasmid replication in E. coli cells (6). Likewise, DPCs generated by azadC and FA used in this study would also block the progression of the replication fork in mammalian cells. This notion is supported by the facts that murine embryonic stem cells deficient in DNMTs are resistant to azadC compared with WT cells (38, 39) and that azadC induces cell cycle arrest (34, 40). It is known that fork stalling results in the activation of ATR kinase for downstream signaling (28). We found that induction of DPCs by azadC and FA resulted in the activation not only of CHK1 (downstream kinase of ATR) but also of CHK2 (downstream kinase of ATM). Interestingly, CHK1 was phosphorylated as early as at the end of FA treatment for 3 h, whereas the phosphorylation of CHK2 occurred in a much later stage during the post-repair incubation for 12 h (Fig. 6B). With FA, breakage of the replication fork (Fig. 5B) and the formation of RAD51 and γ-H2AX nuclear foci (Fig. 4A) occurred during the post-repair incubation for 6 h, intervening between the two events of phosphorylation. Although these results were obtained with different cell lines and hence should be interpreted carefully, they suggest the following sequence of cell responses to FA (Fig. 7). First, FA produces chromosomal DPCs and induces replication fork stalling at DPCs. Second, fork stalling results in the prompt activation of ATR and the downstream kinase CHK1, as was seen in the rapid phosphorylation of CHK1 during FA treatment. The third stage involves the initiation of HR coupled with fork breakage, as indicated by the concomitant formation of DSBs and RAD51 and γ-H2AX nuclear foci during the post-repair incubation for 6 h. Finally, ATM and the downstream kinase CHK2 are activated. There are two possible mechanisms of the activation of ATM in response to DPCs. ATM can be activated directly by DSBs (27) that are generated by fork breakage. Alternatively, ATR may function upstream of ATM and activates it in the absence of DSB formation (41). It is not clear which is true for FA-induced DPCs. With azadC, incorporation of azadC into the genome and subsequent DNMT-DPC induction occur with cell cycle progression. Thus, all cell responses mentioned above are already initiated at the end of azadC treatment for 24 h (Figs. 4B, 5A, and 6B).
Supplementary Material
Acknowledgments
We thank Kiyoji Tanaka (Osaka University), Takashi Yagi (Osaka Prefecture University), Akira Yasui (Tohoku University), Kenshi Komatsu (Kyoto University), Toshiyasu Taniguchi (Fred Hutchinson Cancer Research Center), Larry H. Thompson (Lawrence Livermore National Laboratory), John Thacker (Medical Research Council), and Yosef Shiloh (Tel Aviv University) for generously providing mammalian cells and expression vectors. We also thank Haruna Nishimura and Kazumasa Fujita (both from Hiroshima University) for assisting with the cell survival assay and fluorescence measurement of GFP-positive cells and Naoaki Sakamoto (Hiroshima University) for the pcDNA-EGFP plasmid.
This work was supported in part by Grants-in-aid for Scientific Research from the Japan Society for the Promotion of Science (to T. N., H. T., and H. I.) and by a Grant-in-aid for the Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology (to H. I.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S6.
- DPC
- DNA-protein cross-link
- DSB
- double strand break
- azadC
- 5-aza-2′-deoxycytidine
- FA
- formaldehyde
- CLP
- cross-linked protein
- CLX
- small cross-link adduct induced by FA
- NER
- nucleotide excision repair
- HR
- homologous recombination
- NHEJ
- nonhomologous end joining
- FITC
- fluorescein isothiocyanate
- MDPF
- 2-methoxy-2,4-diphenyl-3(2H)-furanone
- DNMT
- DNA cytosine methyltransferase
- CFEs
- cell-free extracts
- CHO
- Chinese hamster ovary
- ATM
- ataxia telangiectasia mutated
- ATR
- ataxia telangiectasia and Rad3-related
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- MEM
- minimal essential medium
- WT
- wild type
- GFP
- green fluorescent protein.
REFERENCES
- 1.Barker S., Weinfeld M., Murray D. (2005) Mutat. Res. 589, 111–135 [DOI] [PubMed] [Google Scholar]
- 2.Gowher H., Jeltsch A. (2004) Cancer Biol. Ther. 3, 1062–1068 [DOI] [PubMed] [Google Scholar]
- 3.Reardon J. T., Cheng Y., Sancar A. (2006) Cell Cycle 5, 1366–1370 [DOI] [PubMed] [Google Scholar]
- 4.Chválová K., Brabec V., Kaspárková J. (2007) Nucleic Acids Res. 35, 1812–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novakova O., Kasparkova J., Malina J., Natile G., Brabec V. (2003) Nucleic Acids Res. 31, 6450–6460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo H. K., Griffith J. D., Kreuzer K. N. (2007) Cancer Res. 67, 8248–8254 [DOI] [PubMed] [Google Scholar]
- 7.Nakano T., Morishita S., Katafuchi A., Matsubara M., Horikawa Y., Terato H., Salem A. M., Izumi S., Pack S. P., Makino K., Ide H. (2007) Mol. Cell 28, 147–158 [DOI] [PubMed] [Google Scholar]
- 8.Minko I. G., Zou Y., Lloyd R. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1905–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minko I. G., Kurtz A. J., Croteau D. L., Van Houten B., Harris T. M., Lloyd R. S. (2005) Biochemistry 44, 3000–3009 [DOI] [PubMed] [Google Scholar]
- 10.Reardon J. T., Sancar A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4056–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker D. J., Wuenschell G., Xia L., Termini J., Bates S. E., Riggs A. D., O'Connor T. R. (2007) J. Biol. Chem. 282, 22592–22604 [DOI] [PubMed] [Google Scholar]
- 12.Skorvaga M., Theis K., Mandavilli B. S., Kisker C., Van Houten B. (2002) J. Biol. Chem. 277, 1553–1559 [DOI] [PubMed] [Google Scholar]
- 13.Truglio J. J., Croteau D. L., Skorvaga M., DellaVecchia M. J., Theis K., Mandavilli B. S., Van Houten B., Kisker C. (2004) EMBO J. 23, 2498–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano T., Katafuchi A., Shimizu R., Terato H., Suzuki T., Tauchi H., Makino K., Skorvaga M., Van Houten B., Ide H. (2005) Nucleic Acids Res. 33, 2181–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi J., Tauchi H., Sakamoto S., Nakamura A., Morishima K., Matsuura S., Kobayashi T., Tamai K., Tanimoto K., Komatsu K. (2002) Curr. Biol. 12, 1846–1851 [DOI] [PubMed] [Google Scholar]
- 16.Morishima K., Sakamoto S., Kobayashi J., Izumi H., Suda T., Matsumoto Y., Tauchi H., Ide H., Komatsu K., Matsuura S. (2007) Biochem. Biophys. Res. Commun. 362, 872–879 [DOI] [PubMed] [Google Scholar]
- 17.Speit G., Hartmann A. (2006) Methods Mol. Biol. 314, 275–286 [DOI] [PubMed] [Google Scholar]
- 18.Speit G., Schütz P., Högel J., Schmid O. (2007) Mutagenesis 22, 387–394 [DOI] [PubMed] [Google Scholar]
- 19.Nakano T., Terato H., Asagoshi K., Masaoka A., Mukuta M., Ohyama Y., Suzuki T., Makino K., Ide H. (2003) J. Biol. Chem. 278, 25264–25272 [DOI] [PubMed] [Google Scholar]
- 20.Nakano T., Asagoshi K., Terato H., Suzuki T., Ide H. (2005) Mutagenesis 20, 209–216 [DOI] [PubMed] [Google Scholar]
- 21.Liu K., Wang Y. F., Cantemir C., Muller M. T. (2003) Mol. Cell. Biol. 23, 2709–2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mani A., Gelmann E. P. (2005) J. Clin. Oncol. 23, 4776–4789 [DOI] [PubMed] [Google Scholar]
- 23.Fujimuro M., Sawada H., Yokosawa H. (1994) FEBS Lett. 349, 173–180 [DOI] [PubMed] [Google Scholar]
- 24.Bergstralh D. T., Sekelsky J. (2008) Trends Genet. 24, 70–76 [DOI] [PubMed] [Google Scholar]
- 25.Olive P. L., Banáth J. P. (2006) Nat. Protoc. 1, 23–29 [DOI] [PubMed] [Google Scholar]
- 26.Li X., Heyer W. D. (2008) Cell Res. 18, 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niida H., Nakanishi M. (2006) Mutagenesis 21, 3–9 [DOI] [PubMed] [Google Scholar]
- 28.Lambert S., Carr A. M. (2005) Biochimie 87, 591–602 [DOI] [PubMed] [Google Scholar]
- 29.Ridpath J. R., Nakamura A., Tano K., Luke A. M., Sonoda E., Arakawa H., Buerstedde J. M., Gillespie D. A., Sale J. E., Yamazoe M., Bishop D. K., Takata M., Takeda S., Watanabe M., Swenberg J. A., Nakamura J. (2007) Cancer Res. 67, 11117–11122 [DOI] [PubMed] [Google Scholar]
- 30.Quievryn G., Zhitkovich A. (2000) Carcinogenesis 21, 1573–1580 [PubMed] [Google Scholar]
- 31.Debéthune L., Kohlhagen G., Grandas A., Pommier Y. (2002) Nucleic Acids Res. 30, 1198–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saintigny Y., Delacôte F., Varès G., Petitot F., Lambert S., Averbeck D., Lopez B. S. (2001) EMBO J. 20, 3861–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundin C., Erixon K., Arnaudeau C., Schultz N., Jenssen D., Meuth M., Helleday T. (2002) Mol. Cell. Biol. 22, 5869–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W. G., Hileman T., Ke Y., Wang P., Lu S., Duan W., Dai Z., Tong T., Villalona-Calero M. A., Plass C., Otterson G. A. (2004) J. Biol. Chem. 279, 15161–15166 [DOI] [PubMed] [Google Scholar]
- 35.Palii S. S., Van Emburgh B. O., Sankpal U. T., Brown K. D., Robertson K. D. (2008) Mol. Cell. Biol. 28, 752–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider-Stock R., Diab-Assef M., Rohrbeck A., Foltzer-Jourdainne C., Boltze C., Hartig R., Schönfeld P., Roessner A., Gali-Muhtasib H. (2005) J. Pharmacol. Exp. Ther. 312, 525–536 [DOI] [PubMed] [Google Scholar]
- 37.Kiziltepe T., Hideshima T., Catley L., Raje N., Yasui H., Shiraishi N., Okawa Y., Ikeda H., Vallet S., Pozzi S., Ishitsuka K., Ocio E. M., Chauhan D., Anderson K. C. (2007) Mol. Cancer Ther. 6, 1718–1727 [DOI] [PubMed] [Google Scholar]
- 38.Jüttermann R., Li E., Jaenisch R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 11797–11801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oka M., Meacham A. M., Hamazaki T., Rodić N., Chang L. J., Terada N. (2005) Oncogene 24, 3091–3099 [DOI] [PubMed] [Google Scholar]
- 40.Karpf A. R., Moore B. C., Ririe T. O., Jones D. A. (2001) Mol. Pharmacol. 59, 751–757 [PubMed] [Google Scholar]
- 41.Stiff T., Walker S. A., Cerosaletti K., Goodarzi A. A., Petermann E., Concannon P., O'Driscoll M., Jeggo P. A. (2006) EMBO J. 25, 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.