Abstract
Inducible nitric-oxide (NO) synthase (iNOS) plays a critical role in the eradication of intracellular pathogens. However, the excessive production of NO by iNOS has also been implicated in the pathogenesis of septic shock syndrome. Previously, we have demonstrated that mice deficient in mitogen-activated protein kinase phosphatase-1 (MKP-1) exhibit exaggerated inflammatory responses and rapidly succumb to lipopolysaccharide (LPS). In response to LPS, MKP-1−/− mice produce greater amounts of inflammatory cytokines and NO than do wild-type mice, and the MKP-1−/− mice exhibit severe hypotension. To understand the molecular basis for the increase in NO production, we studied the role of MKP-1 in the regulation of iNOS expression. We found that LPS challenge elicited a stronger iNOS induction in MKP-1 knock-out mice than in wild-type mice. Likewise, LPS treatment also resulted in greater iNOS expression in macrophages from MKP-1−/− mice than in macrophages from wild-type mice. Both accelerated gene transcription and enhanced mRNA stability contribute to the increases in iNOS expression in LPS-stimulated MKP-1−/− macrophages. We found that STAT-1, a transcription factor known to mediate iNOS induction by interferon-γ, was more potently activated by LPS in MKP-1−/− macrophages than in wild-type cells. MicroRNA array analysis indicated that microRNA (miR)-155 expression was increased in MKP-1-deficient macrophages compared with wild-type macrophages. Transfection of miR-155 attenuated the expression of Suppressor of Cytokine Signal (SOCS)-1 and enhanced the expression of iNOS. Our results suggest that MKP-1 may negatively regulate iNOS expression by controlling the expression of miR-155 and consequently the STAT pathway via SOCS-1.
Nitric oxide (NO) plays an important role in the immune defense against microbial pathogens (1). In response to bacterial infection, tissue resident macrophages are activated via pathways mediated by Toll-like receptors and serve as the first line of host defense by engulfment of bacterial pathogens through phagocytosis (2). Once the microbial pathogens are ingested, macrophages can produce a variety of toxic products, including NO, that kill the engulfed microorganisms. NO is produced by the high-output inducible NO synthase (iNOS)3 during microbial infection (1, 2). Certain microbial pathogens have developed strategies to circumvent this killing by macrophages (3). Because these pathogens can actually propagate in the phagosomes of macrophages, they are shielded from the effects of antibodies and complement. The adaptive immune cells, particularly Th-1 lymphocytes, can boost the antimicrobial activity of macrophages by interacting with the infected macrophages and producing cytokines such as interferon-γ (IFN-γ). Such interaction robustly enhances the production of NO and reactive oxygen species as well as proinflammatory cytokines in macrophages (4). It has been shown that priming primary resident macrophages with IFN-γ synergistically enhances the induction of iNOS by LPS, leading to a dramatic increase in NO production (2, 4, 5). IFN-γ acts on macrophages by activating the signal transduction cascade mediated by Janus tyrosine kinase and the signal transducer and activator of transcription (STAT) family of transcription factors. Previous studies have demonstrated that STAT-1 is critical for the induction of iNOS by IFN-γ in macrophages (6, 7). In addition to the microbicidal function, NO also acts as a potent vasodilator, facilitating the mobilization of antibodies and effector leukocytes into the site of infection (1). Although NO production is crucial for host defense, overzealous release of NO can disrupt the circulatory system and cause shock and ischemic injury to vital organs, leading to multi-organ failure (8). In the United States septic shock accounts for ∼250,000 deaths annually (9, 10).
Previously, we have identified mitogen-activated protein (MAP) kinase phosphatase-1 (MKP-1) as a crucial negative regulator of the inflammatory response during bacterial infection (11–15). We have shown that MKP-1 acts as a negative feedback control regulator for p38 and JNK MAP kinases. By dephosphorylating these MAP kinases, the inducible MKP-1 protein terminates the p38 and JNK pathways in a delayed manner and switches off the inflammatory cascade. MKP-1-deficient mice exhibit substantially enhanced inflammatory responses (14–16). They produce dramatically greater amounts of inflammatory cytokines than do wild-type mice when challenged with endotoxin and promptly succumb to severe hypotension and shock. The severe hypotension and vasodilatory shock in MKP-1-deficient mice were consistent with the observation that they had substantially elevated NO levels in their blood after endotoxin challenge (14). We have since studied the effects of MKP-1 on iNOS induction during the innate immune response to endotoxin. We found that iNOS expression was dramatically enhanced in MKP-1-deficient mice (16). In this report we describe studies on the mechanisms responsible for the dramatic increases in iNOS expression in MKP-1-deficient mice. Our studies suggest that MKP-1 regulates iNOS expression through post-transcriptional mRNA stabilization and transcriptional induction mediated by the STAT signaling pathway.
EXPERIMENTAL PROCEDURES
Animals
MKP-1+/+ and MKP-1−/− mice were described previously (13, 14, 17). All animal experiments were carried out in accordance with the guidelines of the National Institutes of Health. The experimental protocols were approved by the Institutional Animal Care and Use Committee of the Research Institute at Nationwide Children's Hospital.
Isolation and Treatment of Murine Peritoneal Macrophages
Resident and thioglycollate-elicited peritoneal macrophages were isolated from mice by peritoneal lavage as previously described (14). The macrophages were cultured overnight in complete medium before being stimulated with 100 ng/ml LPS (Escherichia coli O55:B55; Calbiochem).
Western Blotting and Immunoprecipitation
Western blot analyses were carried out as previously described (11). To extract protein from tissues, murine lung and liver tissues were weighed and homogenized in ice-cold tissue homogenizing buffer as previously described (15). The homogenates were placed on ice for 15 min and then centrifuged at 15,000 rpm for 20 min at 4 °C. Supernatant was collected, and protein concentrations were determined before separation by electrophoresis. iNOS protein levels in cells or tissues were determined by Western blot analysis using a rabbit polyclonal antibody purchased from BD Transduction Laboratories. Phosphorylated p38, JNK, ERK, and STAT-1 (Tyr701) were detected using rabbit polyclonal phosphor-specific antibodies from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal antibody against phospho-STAT-3 (Tyr-705) and mouse monoclonal antibody against total STAT-1 were purchased from BD Transduction Laboratories. The rabbit polyclonal antibody against total STAT-3 was purchased from Cell Signaling Technology. Total p38 or β-actin levels were determined using a mouse monoclonal antibody against total p38 (Santa Cruz Biotechnology, Santa Cruz, CA) or against β-actin (Sigma), respectively. Total ERK2 and JNK1 were detected using a mouse monoclonal antibody against total ERK2 (BD Transduction Laboratories) or a rabbit polyclonal antibody against JNK1 (Santa Cruz Biotechnology), respectively.
Immunoprecipitation was carried out essentially as described previously (18). Briefly, soluble proteins were extracted from RAW264.7 cells stably transfected with an empty vector, a vector expressing MKP-1 cDNA, or MKP-1 siRNA described previously (11, 12). To precipitate Suppressor of Cytokine Signaling (SOCS)-1 protein, 5 μg of anti-SOCS-1 antibody (MBL International Co., Woburn, MA) was incubated with 2 mg of soluble cell lysate protein for 2 h at 4 °C. The mixtures were then incubated with 20 μl of protein G-Sepharose beads at 4 °C overnight on a rotator. The-Sepharose beads were recovered by centrifugation at 2500 × g for 10 s, and the supernatant was removed. The beads were washed 5 times with ice-cold lysis buffer. The proteins precipitated by the antibody were then resolved by SDS-polyacrylamide gel electrophoresis using 12% NuPAGE gels (Invitrogen). The amount of SOCS-1 in the immunoprecipitates was assessed by Western blot analysis using a biotin-labeled anti-SOCS-1 antibody purchased from MBL International Co.
Northern Blot and MicroRNA Microarray
Total RNA was isolated with TRIzol (Invitrogen). Northern blot analysis was carried out using a mouse iNOS cDNA as a probe as previously described (11, 12). The membrane was stripped and reprobed with 18 S rRNA or GAPDH cDNA to normalize for RNA loading. Quantitative analysis of mRNA expression was performed with a Storm 860 system and ImageQuant TL software (Amersham Biosciences).
Micro-RNA expression profiling on arrays were performed as described in detail elsewhere (19). To examine the effect of MKP-1 on miR-155 expression by Northern blot analysis, total RNA was either isolated from stably transfected RAW264.7 cells that harbor an empty vector, a vector expressing MKP-1 siRNA, or a vector expressing MKP-1 cDNA (11, 12). Alternatively, total RNA was isolated from wild-type or MKP-1−/− primary peritoneal macrophages. Total RNA (20 μg) was separated by electrophoresis on a 12% denaturing urea polyacrylamide gel. RNA was transferred onto Hybond N+ membranes (Amersham Biosciences) using a Trans-blot cell (Bio-Rad). RNA was cross-linked to the membrane using UV irradiation. The antisense miR-155 and U6 probes were labeled with [α-32P]dATP using StarFire Nucleic Acid Labeling System (IDT, Coralville, IA). The membrane was hybridized overnight at 37 °C in a solution containing 7% SDS and 200 mm Na2HPO4, pH 7.2. After washing 3 times at 37 °C in a solution containing 2× saline/sodium phosphate/EDTA and 0.1% SDS, the membrane was exposed to a phosphorimaging screen at room temperature for 1 h. The same membrane was stripped and rehybridized with a probe for the small nuclear RNA U6 for normalization.
Quantitative Real-time PCR
Lungs and livers isolated from MKP-1+/+ or MKP-1−/− mice after challenge with LPS (E. coli O127:B8) at a dose of 1.5 mg/kg body weight were homogenized in TRIzol to isolate total RNA. Total RNA was then digested with RNase-free DNase I (Promega, Madison, WI) for 30 min at 37 °C. RNA was further purified with RNeasy MinElute Cleanup Kit from Qiagen (Valencia, CA). First-strand cDNA was synthesized with 2 μg of total RNA using Moloney murine leukemia virus reverse transcriptase (Promega). Quantitative real-time PCR assays were performed in triplicate using SYBR Green master mix (Superarray, Frederick, MD) on an Applied Biosystems Model 7700 instrument (Foster City, CA). All assays were performed with the comparative threshold cycle (ΔCT) method, and all data were normalized to an internal standard, GAPDH. iNOS expression levels were assayed using primers and parameters described previously (20). The primers used for assessing GAPDH expression were 5′-CCAGGTTGTCTCCTGCGACT-3′ and 5′-ATACCAGGAAATGAGCTTGACAAAGT-3′.
iNOS mRNA Stability Assay
Thioglycollate-elicited peritoneal macrophages from MKP-1+/+ and MKP-1−/− mice were stimulated with LPS (100 ng/ml) for a given time followed by treatment with the transcription blocker actinomycin D (5 μg/ml) for different periods of time. Total RNA was isolated from the cells for Northern blot analysis. To examine the effect of p38 on iNOS mRNA stability, thioglycollate-elicited peritoneal macrophages from MKP-1−/− mice were stimulated with LPS (100 ng/ml) for 23 h followed by treatment with 10 μm SB203580 for 1 h. iNOS mRNA decay was then analyzed.
Electrophoretic Mobility Shift Assay
Thioglycollate-elicited peritoneal macrophages were treated with LPS (100 ng/ml) for different periods of time. Cells were harvested and lysed in a sucrose buffer containing 0.32 m sucrose, 10 mm Tris-HCl, pH 8.0, 3 mm CaCl2, 2 mm MgOAc, 0.1 mm EDTA, 0.5% Nonidet P-40, 1 mm DTT, and 0.5 mm phenylmethylsulfonyl fluoride. The nuclei were recovered by centrifugation for 5 min at 500 × g at 4 °C, washed once with sucrose buffer, and then resuspended in a hypotonic buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 20 mm KCl, 0.2 mm EDTA, 25% glycerol (v/v), 0.5 mm DTT, and 0.5 mm phenylmethylsulfonyl fluoride). A hypertonic buffer (20 mm HEPES, pH 7.9, 1.5 mm MgCl2, 800 mm KCl, 0.2 mm EDTA, 25% glycerol (v/v), 1% Nonidet P-40, 0.5 mm DTT, 0.5 mm phenylmethylsulfonyl fluoride, and 4.0 μg/ml leupeptin, aprotinin, and pepstatin) was added slowly to extract nuclear proteins. Protein concentrations of the nuclear extracts were determined using the Bio-Rad protein assay kit. DNA binding assays were performed in a binding buffer containing 50 mm Tris-HCl, pH 8.0, 750 mm KCl, 2.5 mm EDTA, 0.5%Triton X-100, 62.5% glycerol (v/v), 1 mm DTT, 2 mg poly(dI-dC), 2 ng of 32P-labeled double-stranded oligonucleotide, and nuclear extract that contained 5 μg of protein. Binding reaction mixtures were prepared on ice, incubated in room temperature for 30 min, and then resolved on 4% polyacrylamide gels in 0.5× Tris borate-EDTA. Oligonucleotides for electrophoretic mobility shift assay were as follows: NF-κB (wild-type), 5′-AGTTGAGGGGACTTTCCCAGGC-3′; NF-κB (mutant)m 5′-AGTTGAGGCGACTTTCCCAGGC-3′.
Nuclear Run-on Assay
Thioglycollate-elicited peritoneal macrophages were stimulated with LPS (100 ng/ml) for different periods of time and harvested in phosphate-buffered saline. Cells were lysed in lysis buffer (10 mm Tris-HCl, pH 7.4, 10 mm NaCl, 3 mm MgCl2, 0.5% Nonidet P-40) and centrifuged at 500 × g for 5 min to recover the nuclei. Nuclei were resuspended in nuclei storage buffer (50 mm Tris-HCl, pH 8.5, 40% glycerol (v/v), 5 mm MgCl2, 0.1 mm EDTA) and stored at −80 °C. Transcription reactions were resumed by incubating the nuclei (purified from 5 × 106 cells) with 50 μCi of [α-32P]UTP in a buffer (25 mm HEPES, pH 7.5, 2.5 mm MgCl2, 2.5 mm DTT, 75 mm KCl, 5% glycerol, 2.8 mm ATP, 2.8 mm GTP, and 2.8 mm CTP) at room temperature for 20 min. The reaction was stopped by adding 2 units of DNase I and incubating at 37 °C for 10 min followed by a 2-h incubation at 45 °C in a digestion mixture containing 4.3 mm Tris-HCl, pH 8.0, 0.86% SDS, 3 m urea, 0.15 m NaCl, 0.43 mm EDTA, 300 μg of proteinase K, and 100 μg of tRNA (Roche Applied Science). RNA was extracted using phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated with 100% ethanol. Murine iNOS, TNF-α, IL-6, IL-1β, and β-actin cDNA fragments were excised from IMAGE clones purchased from Invitrogen and dot-blotted onto nitrocellulose filters. Radiolabeled RNA was hybridized with 5 μg of filter-bond cDNA at 42 °C for at least 72 h in hybridization buffer (50% formamide, 6× SSC (1× SSC = 0.15 m NaCl and 0.015 m sodium citrate), 10×Denhardt's solution, and 0.2% SDS) using standard molecular biology techniques (21). The hybridized signals were quantitated using a Storm system and ImageQuant TL software. The signal of a given gene was normalized to that of β-actin, and the ratio was used as a surrogate for gene transcription rate.
Luciferase Reporter Assays
Two oligonucleotides (5′-CTAGAGAAATAATAAGGCGCCCCCACTTAATGCTGCGGCACAGCGGCAGCCGG-3′ and 5′-CTAGAGAAATAATAAGGCGCCCCCACTTAATGCTGGGCACAGCGGCAGCCGG-3′) corresponding a region containing a putative miR-155 target site at the 3′-untranslated region (UTR) of murine SOCS-1 mRNA were synthesized. They were annealed and cloned into the XbaI site of the luciferase reporter pGL3-control vector (Promega). The XbaI site is located downstream of the luciferase open reading frame. The orientation of the insertion was confirmed by sequencing. When expressed, this reporter construct will produce a luciferase reporter mRNA harboring a SOCS-1-derived putative miR-155 targeting sequence at the 3′-UTR. This reporter (0.2 μg) was co-transfected with different concentrations of miR-155 together with a Renilla luciferase reporter (10 ng) into HEK293 cells using FuGENE 6 transfection reagent (Roche Applied Science). Assays were performed 24 h after transfection using the dual luciferase reporter assay system (Promega), as previously described (22). Firefly luciferase activity was normalized to Renilla luciferase activity. The experiments were performed three times in six replicates each. Statistical differences were determined using Student's t test.
RESULTS
Upon LPS Challenge, MKP-1 Knock-out Mice Exhibit Profoundly Greater MAP Kinase Activities and iNOS Expression Than Do Wild-type Mice
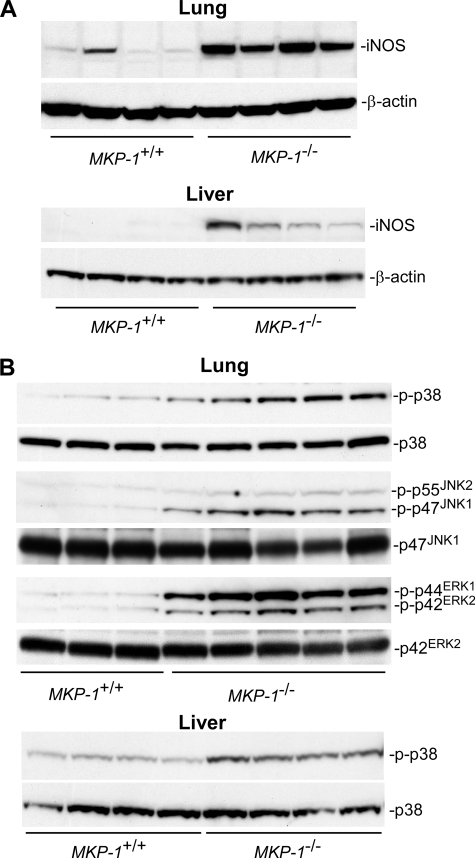
NO is an important mediator in host defense during microbial infection, serving as both a potent microbicide and a powerful vasodilator. iNOS is the primary producer of NO during sepsis. We have previously shown that plasma nitrate levels in MKP-1 knock-out mice were significantly higher than those in the wild-type mice when challenged with LPS (14). To examine the effect of MKP-1 on iNOS induction in mice after LPS challenge, wild-type and MKP-1 knock-out mice were challenged intraperitoneal with either vehicle (phosphate-buffered saline) or LPS (1.5 mg/kg body weight). Organs were excised 3 or 24 h post-LPS administration. Protein levels of iNOS in lung and liver homogenates were examined via Western blot analysis. iNOS protein was not detectable in either lung or liver homogenates from vehicle-treated wild-type or knock-out mice (data not shown). Even 3 h post-LPS administration, iNOS protein levels were still below the limit of detection in both lung and liver tissues (data not shown). At 24 h post-LPS challenge, markedly elevated levels of iNOS protein were observed in the lungs from both wild-type and MKP-1 knock-out mice (Fig. 1A). Importantly, the lung iNOS protein levels were substantially higher in the MKP-1 knock-out mice than in the wild-type mice. Overall, the iNOS protein levels in the liver tissues were substantially lower than those in the lung tissues. Nevertheless, significant amounts of iNOS protein were detected in the livers of MKP-1 knock-out mice 24 h after LPS challenge, whereas iNOS protein was only barely detectable in the livers of similarly treated wild-type mice.
FIGURE 1.
LPS challenge results in enhanced iNOS induction and greater MAP kinase activity in MKP-1−/− mice than in wild-type mice. MKP-1+/+ and MKP-1−/− mice were injected intraperitoneal with LPS (1.5 mg/kg body weight) and sacrificed 3 or 24 h later. Lung and liver tissues were collected, and tissue homogenates were analyzed for the levels of iNOS and phospho-MAP kinases by Western blotting. The membranes were stripped and reblotted with an antibody for β-actin or total MAP kinase as loading controls. A, iNOS protein levels in the lungs and livers of MKP-1+/+ and MKP-1−/− mice at 24 h post-LPS challenge. B, MAP kinase activities in lung and liver homogenates from MKP-1+/+ and MKP-1−/− mice 3 h post-LPS challenge.
The activities of MAP kinases in the mouse tissue homogenates were examined by Western blot analysis using phospho-specific MAP kinase antibodies. MAP kinase activity was similar in both the lungs and livers of wild-type and MKP-1 knock-out mice injected with vehicle (data not shown). The lungs from MKP-1 knock-out mice 3 h post-LPS challenge exhibited markedly higher p38, JNK, and ERK activities than did the lungs from similarly treated wild-type mice (Fig. 1B). The activity of p38 in the livers of MKP-1−/− mice 3 h post-LPS challenge was also higher than that in similarly treated wild-type mice, whereas no difference in either liver ERK or JNK activity was detected between wild-type and MKP-1−/− mice (data not shown). By 24 h post-LPS challenge, the two groups of mice had similar MAP kinase activities in both lung and liver tissues (data not shown). These results indicate that loss of MKP-1 results in enhanced iNOS induction associated with more robust and sustained MAP kinase activation after LPS challenge.
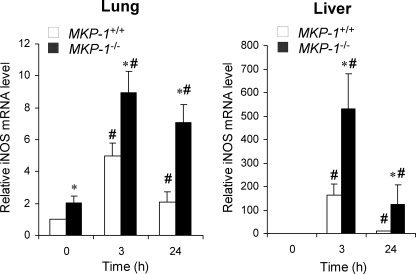
To examine the effect of MKP-1 deficiency on iNOS gene expression, total RNA was isolated from the lungs and livers. The expression of the iNOS gene was measured by quantitative real-time PCR. In the lungs iNOS mRNA levels were significantly increased after LPS treatment in both wild-type and MKP-1 knock-out mice (Fig. 2). Although iNOS protein levels were not detectable at 3 h post-LPS challenge, iNOS mRNA levels were already increased by 3 h. Importantly, iNOS mRNA levels were higher in the livers and lungs of MKP-1 knock-out mice than those in the wild-type mice. Moreover, compared with wild-type animals, basal iNOS mRNA expression in the lungs of MKP-1 knock-out mice was significantly higher (p = 0.02). In livers, LPS treatment elicited a substantial increase in iNOS expression. Compared with wild-type animals, livers from MKP-1 knock-out mice had a greater expression of iNOS at both 3 and 24 h. Taken together, these results indicate that MKP-1 negatively regulates iNOS expression at both the mRNA and the protein level during the response to LPS.
FIGURE 2.
MKP-1 knock-out mice exhibit greater iNOS mRNA expression in both lung and liver in response to LPS challenge than do wild-type mice. MKP-1+/+ and MKP-1−/− mice were challenged intraperitoneal with phosphate-buffered saline (0 h) or with LPS (1.5 mg/kg body weight) and sacrificed 3 or 24 h post-challenge. Total RNA was isolated from the lung and liver of these animals. Quantitative real-time PCR was carried out to determine the iNOS mRNA levels in tissues. GAPDH mRNA was used as an internal control for normalization between samples. Values are expressed as -fold increase relative to the iNOS mRNA levels in the lung or liver tissues isolated from vehicle-treated wild-type mice. Data are presented as the mean ± S.E. (n = 3). *, MKP-1−/− different from MKP-1+/+ at same time point, p < 0.05; #, different from vehicle-treated mice of the same genotype, p < 0.05. Left, iNOS mRNA levels in the lungs from MKP-1+/+ and MKP-1−/− mice. Right, iNOS mRNA levels in livers from MKP-1+/+ and MKP-1−/− mice.
LPS Elicits a Stronger iNOS Induction in MKP-1-deficient Macrophages
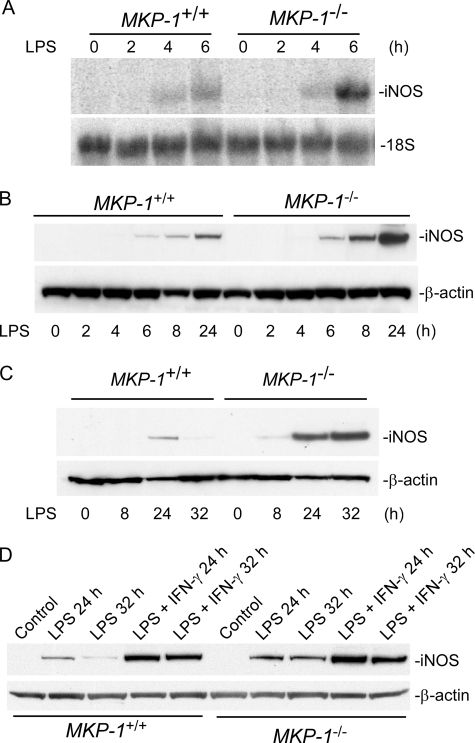
To understand the mechanism involved in the regulation of iNOS by MKP-1, we studied iNOS induction using primary peritoneal macrophages from wild-type and MKP-1 knock-out mice. Thioglycollate-elicited macrophages were treated with 100 ng/ml LPS for different periods of time. The time-course of iNOS mRNA expression was assessed by Northern blot analysis (Fig. 3A). As expected, the levels of iNOS mRNA in unstimulated macrophages were essentially undetectable. Upon LPS stimulation, iNOS mRNA was increased in both wild-type and MKP-1−/− cells; iNOS mRNA became detectable within 4 h, and the levels were substantially elevated by 6 h in both wild-type and MKP-1-deficient cells. Furthermore, 6 h post-LPS the iNOS mRNA levels in MKP-1−/− cells were substantially higher than those in the wild-type cells (Fig. 3A). Consistent with the increases in iNOS mRNA levels in the LPS-stimulated MKP-1−/− macrophages, iNOS protein levels were also markedly higher in MKP-1-deficient cells than in wild-type cells (Fig. 3B).
FIGURE 3.
LPS stimulation results in greater iNOS induction in MKP-1-deficient peritoneal macrophages than in wild-type cells. A, augmented iNOS mRNA induction by LPS in thioglycollate-elicited MKP-1-deficient peritoneal macrophages. Thioglycollate-elicited peritoneal macrophages were isolated from MKP-1+/+ and MKP-1−/− mice. Cells were treated with 100 ng/ml LPS for the indicated periods. Northern blot analysis was carried out to access the expression of iNOS mRNA. post- ribosomal RNA was used as a loading control. B, more robust iNOS protein increases in MKP-1-deficient macrophages upon LPS stimulation. Thioglycollate-elicited peritoneal macrophages were treated as in A, and Western blotting was carried out to determine levels of iNOS protein. C, the effect of MKP-1 knock-out on iNOS protein induction in resident peritoneal macrophages stimulated with LPS. Resident peritoneal macrophages were stimulated with 100 ng/ml LPS. D, the effect of IFN-γ on iNOS expression in LPS-stimulated wild-type and MKP-1-deficient resident peritoneal macrophages. Cells were treated with 100 ng/ml LPS and 50 IU/ml IFN-γ. iNOS and β-actin levels were assessed by Western blotting. Results from representative experiments are shown.
Unlike inflammatory macrophages such as those elicited by thioglycollate, resident peritoneal macrophages mount a weaker inflammatory response after stimulation with LPS (4). To examine how MKP-1 knock-out affects iNOS induction in resident macrophages, we compared the iNOS induction between wild-type and MKP-1−/− resident macrophages after LPS stimulation. The stimulation of wild-type resident macrophages with LPS resulted in a weak iNOS expression, as indicated by Western blot analysis (Fig. 3C). iNOS protein was not detectable at 8 h post-LPS stimulation in wild-type resident peritoneal macrophages. Although a low level of iNOS can be seen in these cells 24 h after LPS stimulation, iNOS proteins became undetectable by 32 h in wild-type resident peritoneal macrophages. In contrast, LPS stimulation of resident peritoneal macrophages isolated from MKP-1−/− mice resulted in a robust iNOS expression (Fig. 3C). The expression of iNOS was detectable at as early as 8 h post-LPS stimulation and reached substantially higher levels at 24 h that lasted at least to 32 h.
It has been shown that IFN-γ enhances the anti-microbial activity of macrophages (23) and enhances the induction of iNOS by LPS in resident macrophages (5). Previously, we have demonstrated that IFN-γ inhibits MKP-1 expression (14). To determine the role of MKP-1 in the enhancing effect of IFN-γ on iNOS induction by LPS, we examined the effect of MKP-1 deficiency on iNOS induction (Fig. 3D). As expected, LPS alone caused a weak and transient induction of iNOS in wild-type macrophages. Knock-out of the MKP-1 gene significantly enhanced iNOS induction by LPS in wild-type macrophages. IFN-γ not only substantially enhanced iNOS induction by LPS in both wild-type and MKP-1−/− macrophages but also prolonged LPS-induced iNOS expression. Taken together these results indicate that knock-out of the MKP-1 gene sensitizes resident macrophages to LPS stimulation. The results suggest that the signal provided by IFN-γ to enhance LPS-stimulated iNOS expression is partially activated by the knock-out of the MKP-1 gene. It is clear that iNOS induction by LPS plus IFN-γ in wild-type macrophages was stronger than in MKP-1−/− macrophages stimulated by LPS alone (Fig. 3D). This result indicates that IFN-γ enhances LPS-induced iNOS expression through mechanisms beyond simply inhibiting MKP-1 expression.
Messenger RNA of iNOS Is More Stable in MKP-1-deficient Macrophages Than in Wild-type Cells When Stimulated with LPS
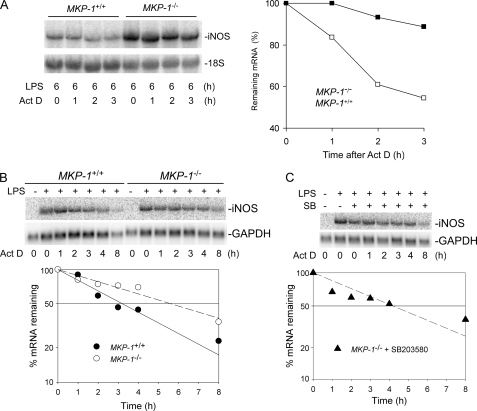
It has been shown that iNOS mRNA stability can be regulated post-transcriptionally through a JNK-mediated pathway (24, 25). We and others have recently demonstrated that MKP-1 deficiency leads to prolonged JNK and p38 activation (13, 14, 26–28). Thus, we examined whether the stabilities of iNOS mRNA were different between macrophages isolated from wild-type mice and those isolated from MKP-1 knock-out mice. Peritoneal macrophages were treated with LPS for 6 h and then exposed to actinomycin D to stop gene transcription and allow RNA to decay. Total RNA was isolated at various times, and the iNOS mRNA decay was assessed by Northern blotting. As shown in Fig. 4A, iNOS mRNA underwent a more rapid decay in wild-type macrophages than in MKP-1 knock-out cells. The half-life of iNOS mRNA in wild-type cells was ∼3 h. In contrast, the half-life of iNOS mRNA in MKP-1-deficient macrophages was estimated to be ∼17 h. The difference in half-lives between the two groups of macrophages was also detected 24 h after LPS, although the half-life of iNOS mRNA in MKP-1−/− macrophages decreased to ∼5.5 h (Fig. 4B). To answer the question of whether p38 plays a role in the regulation of iNOS mRNA stability, we first stimulated MKP-1−/− macrophages with LPS for 23 h and then treated these cells with a pharmacological inhibitor of p38, SB203580, for 60 min. The decay of iNOS mRNA in the presence of the p38 inhibitor was then examined (Fig. 4C). The presence of p38 inhibitor decreased the half-life of iNOS mRNA to ∼4 h. These results indicate that the enhanced stability of iNOS mRNA in MKP-1-deficient macrophages is at least partially because of elevated p38 activity in MKP-1-deficient cells.
FIGURE 4.
Knock-out of MKP-1 enhances the stability of iNOS mRNA, and inhibition of p38 decreases iNOS mRNA stability. Thioglycollate-elicited peritoneal macrophages were treated with LPS (100 ng/ml) for 6 h (A), 24 h (B), or 23 h (C), and actinomycin D (5 μg/ml) was added in the medium to inhibit transcription. Total RNA was isolated at the indicated time points after the addition of actinomycin D (Act D). Northern blot analysis was used to determine iNOS mRNA levels. Levels of iNOS mRNA were normalized to 18 S, and the quantified data are presented graphically. A, iNOS mRNA decay assay following 6 h of LPS stimulation. B, iNOS mRNA decay assay after 24 h of LPS stimulation. C, effects of p38 inhibitor on iNOS mRNA stability. MKP-1-deficient peritoneal macrophages were first stimulated with LPS for 23 h, then treated with SB203580 (SB, 10 μm) for 1 h. Actinomycin D was added thereafter and iNOS mRNA decay was examined. The experiments were performed at least three times with similar results. Data presented in the panels are from representative experiments.
MKP-1 Deficiency Enhances iNOS Gene Transcription in Response to LPS
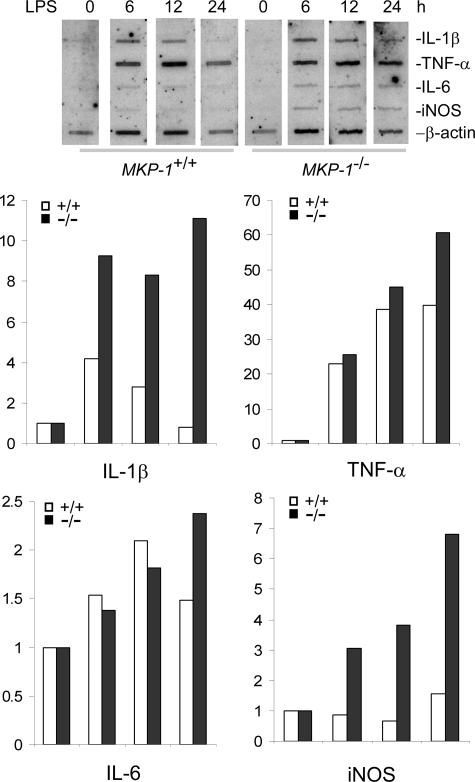
To understand the molecular mechanism underlying enhanced iNOS induction in MKP-1-deficient macrophages, nuclear run-on assays were performed to assess the rate of gene transcription in wild-type and MKP-1−/− macrophages. Primary macrophages were stimulated with LPS for different periods of time, and nuclei were harvested. These nuclei were then incubated with nucleic acids including [α-32P]UTP to resume gene transcription. The synthesized radiolabeled RNAs were then hybridized to blots containing iNOS (Fig. 5). As controls, a housekeeping gene, β-actin, as well as several pro-inflammatory cytokine genes known to be regulated by MKP-1 were also included in the experiment. In unstimulated controls, transcription of iNOS as well as the cytokine genes was barely detectable. The transcription rates of these genes were markedly enhanced upon LPS stimulation. The transcription of both iNOS and IL-1β was considerably more robust in MKP-1−/− macrophages than in wild-type cells, whereas transcription rates of TNF-α and IL-6 were similar in the two groups of cells. These studies suggest that MKP-1 regulates the expression of LPS-induced genes through distinct mechanisms.
FIGURE 5.
Knock-out of MKP-1 enhances iNOS transcription triggered by LPS in primary macrophages. Thioglycollate-elicited peritoneal macrophages isolated from MKP-1+/+ and MKP-1−/− mice were stimulated with LPS (100 ng/ml) for the indicated times, and the nuclei were isolated. Gene transcription was resumed by incubating the nuclei with nucleic acids including [α-32P]UTP to make nascent RNAs as radioactive probes. These probes were then hybridized with membrane-bound mouse cDNAs corresponding to different genes. The signals of given genes were normalized to that of the housekeeping gene β-actin. Data are expressed as -fold induction relative to unstimulated controls (0 h). The experiments were performed twice, with similar results. Data presented are from a representative experiment.
Deletion of MKP-1 Augments LPS-stimulated Tyrosine Phosphorylation of STAT-1 and STAT-3 but Has No Effects on NF-κB Activity
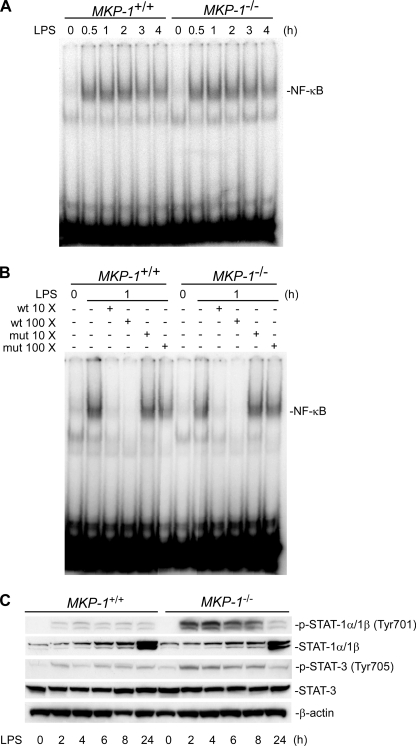
To understand the mechanism involved in the regulation of iNOS by MKP-1, we studied transcription factors known to regulate iNOS induction. NF-κB and STAT-1 have been shown to play pivotal roles in the induction of iNOS (29–32). To investigate if MKP-1 deficiency alters the activity of NF-κB, thioglycollate-elicited peritoneal macrophages from both wild-type and MKP-1 knock-out mice were treated with LPS for different period of time. Nuclear protein was extracted, and an electrophoretic gel shift assay was performed to assess the DNA binding abilities of NF-κB. Upon LPS treatment, NF-κB binding ability increased in both wild-type and MKP-1 knock-out macrophages (Fig. 6A). No differences in NF-κB DNA binding ability were observed between wild-type and MKP-1 knock-out macrophages either before or after LPS stimulation (Fig. 6A). The specificity of the DNA binding activity of NF-κB was confirmed by competition assays, as NF-κB DNA binding activity was competed by excessive cold oligonucleotides containing a wild-type but not a mutated NF-κB-binding consensus sequence (Fig. 6B).
FIGURE 6.
MKP-1 deficiency leads to more robust STAT-1 and STAT-3 tyrosine phosphorylation but has no effect on NF-κB DNA binding activity in primary macrophages stimulated with LPS. A, NF-κB DNA binding activity in wild-type and MKP-1−/− primary macrophages in response to LPS. Nuclear protein was isolated from thioglycollate-elicited peritoneal macrophages stimulated with LPS (100 ng/ml) for the indicated times. A gel shift assay using the 32P-labeled consensus NF-κB oligonucleotide probes was utilized to determine the DNA binding ability of NF-κB. B, specificity of the gel shift assays. Different concentrations of cold wild-type (wt) or mutant (mut) NF-κB oligonucleotides were utilized to compete with the radio-labeled NF-κB probe. C, enhanced STAT-1 and STAT-3 tyrosine phosphorylation upon LPS stimulation in MKP-1-deficient macrophages. Thioglycollate-elicited peritoneal macrophages isolated from MKP-1+/+ and MKP-1−/− mice were treated with 100 ng/ml LPS for the indicated times. Western blotting was carried out to determine levels of STAT-1 (Tyr-701) and STAT-3 (Tyr-705) phosphorylation. The blots were striped and blotted with STAT-1, STAT-3, or β-actin antibody. Data shown are from representative experiments.
IFN-γ is a potent cytokine that interacts synergistically with LPS to induce transcription of iNOS (33). Because MKP-1 deficiency at least partially alleviated the requirement for IFN-γ for iNOS induction by LPS in resident peritoneal macrophages (Fig. 3C), we explored the possibility of a functional interaction between MKP-1 and the well characterized IFN-γ-regulated pathway in the regulation of iNOS induction. IFN-γ stimulates iNOS expression largely through a signal transduction pathway mediated by the Janus kinase/STAT pathway (34). To understand the molecular mechanisms for the enhanced iNOS induction in MKP-1-deficient macrophages, we examined STAT tyrosine phosphorylation in thioglycollate-elicited primary peritoneal macrophages (Fig. 6C). Before LPS stimulation, STAT-1 was not phosphorylated in either wild-type or MKP-1-deficient macrophages. Upon LPS stimulation, STAT-1 was phosphorylated at tyrosine 701 within 2 h. This phosphorylation lasted for at least 24 h in both wild-type and MKP-1-deficient macrophages. Compared with wild-type macrophages, tyrosine phosphorylation of STAT-1 in MKP-1-deficient macrophages was substantially greater. Although LPS stimulation resulted in increases in the STAT-1 protein levels in both MKP-1−/− and wild-type macrophages, no differences in the STAT-1 protein levels were found between these two groups. STAT-3 also underwent phosphorylation at tyrosine 706 upon LPS stimulation in both genotypes, although STAT-3 phosphorylation appears to be less robust than that of STAT-1. Nevertheless, tyrosine phosphorylation of STAT-3 was also greater in MKP-1-deficient macrophages than in wild-type cells. The protein levels of STAT-3 were comparable in both wild-type and MKP-1−/− macrophages and did not change over time. The fact that tyrosine phosphorylation of STAT-1 and STAT-3 precede iNOS induction suggests that these transcription factors may be involved in the dramatic iNOS induction observed in MKP-1-deficient macrophages. Taken together, our results suggest that signal transduction pathways mediated by STATs rather than NF-κB are involved in the enhanced iNOS induction in MKP-1-deficient macrophages after LPS challenge.
MKP-1−/− Macrophages Express Higher Levels of miR-155 Than Do Wild-type Macrophages
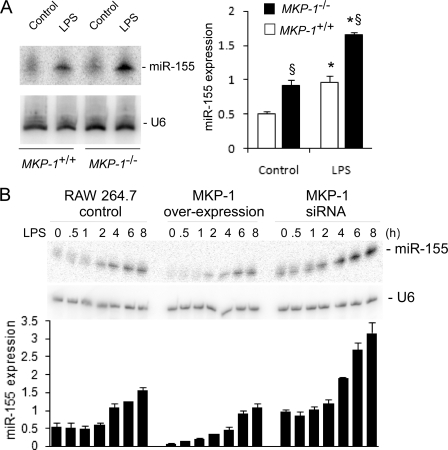
Recent studies have demonstrated that microRNA plays a significant role in inflammatory responses (35, 36). Because MKP-1-deficient macrophages exhibit dramatic changes in their response to inflammation, we analyzed the microRNA expression profile using microRNA array. miR-155 was identified as one of the microRNAs whose expression was enhanced by MKP-1 knock-out. Northern blot analysis confirmed that miR-155 expression was enhanced in response to LPS stimulation. Furthermore, miR-155 expression was higher in MKP-1−/− macrophages than in wild-type cells (Fig. 7A). The effect of MKP-1 on miR-155 expression was also clearly seen in RAW264.7 macrophages expressing MKP-1 cDNA or small interfering MKP-1 RNA (siRNA). miR-155 expression was induced by LPS in RAW264.7 harboring a control plasmid (Fig. 7B). Both the basal and LPS-induced miR-155 expression levels were attenuated by overexpression of MKP-1 cDNA. Compared with RAW264.7 cells carrying an empty vector, RAW264.7 cells expressing MKP-1 siRNA produced markedly greater levels of miR-155.
FIGURE 7.
MKP-1 negatively regulates the expression of miR-155. A, miR-155 expression in MKP-1+/+ and MKP-1−/− macrophages. Thioglycollate-elicited peritoneal macrophages isolated from MKP-1+/+ and MKP-1−/− mice were stimulated with LPS (100 ng/ml) for 4 h, and total RNA was subjected to Northern blot analysis. U6 small nuclear RNA was used as a loading control for signal normalization. miR-155 expression is presented in the graph on the right as -fold relative to miR-155 expression level in unstimulated wild-type macrophages. The graph on the right represents the means ± S.E. of the relative miR-155 expression calculated from three separate experiments. Relative expression was calculated as (miR-155 valuegiven sample/average miR-155 valueall samples)/(U6 valuegiven sample/average U6 Valueall samples). *, p < 0.05, relative to control; §, p < 0.05, relative to similarly treated wild-type cells. Data were analyzed using a Student's t test. B, effects of MKP-1 overexpression or knock-down on miR-155 induction by LPS in RAW264.7 macrophages. The establishment of RAW264.7 cells harboring an empty vector, a MKP-1 overexpression construct, or a construct expressing a MKP-1 siRNA were previously described (11, 12). These stable clones were stimulated with LPS (100 ng/ml) or left untreated. miR-155 expression was examined by Northern blot analysis. The graph underneath the images represents means ± S.E. of the relative miR-155 expression calculated from three experiments. By two-way ANOVA there was an effect of time (p < 0.05), cell line (p < 0.05) and a statistically significant interaction among these groups (p < 0.05).
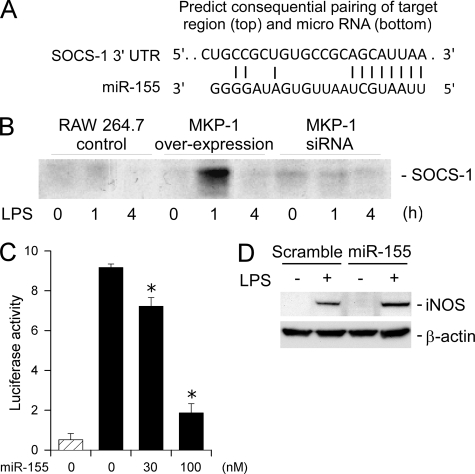
To understand the functional significance of miR-155 elevation in MKP-1 knock-out cells, we searched the TargetScan data base for putative miR-155 targets that potentially could regulate STAT activation and iNOS induction. SOCS-1 mRNA was found to contain a putative miR-155 targeting sequence at the 3′-UTR (Fig. 8A). To address if MKP-1 was capable of affecting SOCS-1 protein levels, RAW264.7 cells expressing MKP-1 cDNA or siRNA were stimulated with 100 ng/ml LPS. SOCS-1 protein levels in these cells were assessed by immunoprecipitation followed by Western blot analysis. RAW264.7 cells were chosen because a large quantity of cells was readily available to overcome the difficulty in SOCS-1 detection, largely because of the relatively low titer of the SOCS-1 antibody. SOCS-1 protein level was significantly increased at 1-h post-LPS stimulation in RAW264.7 cells expressing MKP-1 cDNA (Fig. 8B). However, the expression of SOCS-1 was barely detectable during the experimental period in RAW264.7 cells transfected with an empty vector or expressing MKP-1 siRNA. These data indicate that overexpression of MKP-1 enhances SOCS-1 induction by LPS.
FIGURE 8.
miR-155 attenuates the expression of SOCS-1 and enhances the expression of iNOS. A, the miR-155 targeting sequence in the 3′-UTR of murine SOCS-1 mRNA. Paring with miR-155 was also illustrated. B, the effect of MKP-1 overexpression or knockdown on SOCS-1 expression in LPS-treated RAW264.7 macrophages. Stable clones of RAW264.7 macrophages expressing a MKP-1 cDNA, a MKP-1 siRNA, or carrying an empty vector were stimulated with LPS (100 ng/ml) for the indicated periods. SOCS-1 expression was assessed by immunoprecipitation followed by Western blotting. C, effects of miR-155 on a luciferase reporter containing a SOCS-1-derived putative miR-155 targeting sequence. Luciferase reporter containing a SOCS-1-derived putative miR-155 targeting sequence together with an internal control reporter (Renilla luciferase) and different concentrations of miR-155 was transfected into HEK 293 cells. Luciferase activity was measured 48 h post-transfection, and values were normalized to Renilla luciferase activity. Data represent the ratio between the firefly and Renilla luciferase activities. The striped bar represents a background reading from a promoterless firefly luciferase reporter. Data were presented as means ± S.E. of three independent experiments. D, effect of miR-155 transfection on iNOS induction by LPS in RAW264.7 macrophages. RAW264.7 cells were transiently transfected with miR-155 or scramble RNA and then stimulated with LPS (100 ng/ml) 24 h later. iNOS protein levels in these cells were assessed by Western blotting. Data shown are from a representative experiment.
We then examined the mechanism via which miR-155 regulates SOCS-1 expression. Northern blot analysis of SOCS-1 mRNA revealed no significant differences between wild-type and MKP-1−/− macrophages (data not shown), excluding the possibility of regulation at the level of transcription or mRNA stability. Because siRNA often inhibits protein translation through its potential targeting site without affecting mRNA levels, we cloned the putative miR-155 targeting sequence into a luciferase reporter and performed reporter assays. Transfection of a promoter-less luciferase reporter into HEK293 cells yielded no luciferase activity (Fig. 8C). In contrast, transient transfection of a functional reporter construct harboring the miR-155 targeting sequence yielded a substantial level of luciferase activity. Co-transfection of miR-155 resulted in a gradual decrease in luciferase activity in a dose-dependent manner, suggesting that the SOCS-1 mRNA-derived miR-155 targeting sequence is functional, at least in this in vitro assay system. To assess whether the expression of miR-155 affects iNOS induction, RAW264.7 macrophages were transfected with either miR-155 or a scramble control RNA. Twenty-four hours later these RAW264.7 cells were stimulated with 100 ng/ml LPS. After 24 h iNOS protein levels in these cells were assessed. iNOS induction by LPS was augmented in RAW264.7 cells transfected with miR-155 compared with RAW264.7 cells transfected with the scramble RNA (Fig. 8D), indicating that miR-155 is capable of regulating iNOS induction by LPS. Taken together, our results provided a plausible explanation for the enhanced STAT activation and the more robust iNOS induction in response to LPS in MKP-1-deficient macrophages.
DISCUSSION
We have previously shown that in response to LPS challenge, mice deficient in MKP-1 more readily develop severe hypotension and shock than do MKP-1-sufficient mice (14). To understand the molecular mechanism underlying the increased sensitivity of MKP-1 knock-out mice to endotoxin, we studied the role of MKP-1 in the induction of iNOS. The induction of iNOS plays a critical role in the production of NO underlying vasodilatory shock. We found that iNOS was more robustly induced by LPS in MKP-1 knock-out mice than in wild-type mice and enhanced iNOS induction was associated with elevated activities of the MAP kinases (Fig. 1). The elevated iNOS expression after LPS stimulation was detected at the mRNA levels in both tissues and primary macrophages derived from MKP-1 knock-out mice (Figs. 2 and 3). Both increased gene transcription (Fig. 5) and enhanced iNOS mRNA stability (Fig. 4) appear to contribute to the elevated iNOS expression in MKP-1-deficient macrophages. To delineate the mechanism responsible for the enhanced iNOS induction, we analyzed the molecular events known to regulate iNOS expression. We found that STAT1 and, to a lesser extent, STAT3 underwent more robust activation in response to LPS stimulation in MKP-1−/− macrophages than in wild-type cells (Fig. 6C), whereas NF-κB activation did not differ between the two groups of cells (Fig. 6A). Because STAT proteins have been shown to play an important role in the induction of iNOS in macrophages in response to IFN-γ (6, 7), the increases in STAT-1 and STAT-3 activity provide a logical explanation for the dramatic augmentation in iNOS induction in MKP-1-deficient mice and macrophages.
Enhanced STAT Activation in MKP-1-deficient Macrophages
STAT transcription factors have been shown to play a critical role in the regulation of a variety of genes in response to cytokines (37). STAT-1, in particular, is crucial for the induction of iNOS in macrophages by IFN-γ (6, 7). Previously, we have shown that priming of resident peritoneal macrophages with IFN-γ attenuates the induction of MKP-1 by LPS and enhances the activity of MAP kinases (14). It has been shown previously that without priming with interferons, resident peritoneal macrophages isolated from naïve healthy animals produce very small quantities of cytokines and express very low levels of iNOS after LPS stimulation (4, 23). The resident peritoneal macrophages isolated from the MKP-1-deficient mice displayed more robust inflammatory responses to LPS, expressing substantial iNOS protein (Fig. 3) and producing considerable amounts of cytokines (14). These observations suggest that one important function of IFN-γ priming is to attenuate MKP-1 expression, thus prolonging the window of MAP kinase activation and allowing epigenetic changes to occur at the loci of inflammatory genes, such as iNOS. It is plausible that MKP-1 deletion in the resident macrophages partially alleviates the requirement for IFN-γ, allowing for modest induction of iNOS in the absence of IFN-γ priming. Given that IFN-γ enhances iNOS expression through a complex mechanism (38), including the activation of STAT-mediated iNOS transcription, it is not surprising that attenuation of MKP-1 expression accounts for only a part of the mechanism. This notion is supported by the observation that iNOS expression was greater in wild-type macrophages stimulated with LPS plus INF-γ than in MKP-1−/− macrophages stimulated with LPS alone (Fig. 3D).
Although it is clear that the STAT pathway is augmented in MKP-1-deficient macrophages, the underlying mechanism is still unclear. Because STAT-1 is not a substrate for MKP-1 in vitro (39), the up-regulation of STAT pathways in MKP-1-deficient macrophages is most likely not because of defects in STAT dephosphorylation. At least two mechanisms could lead to augmented STAT signaling, and both of these mechanisms may contribute to the augmented STAT activation. First, it is well established that MKP-1-deficient macrophages produce greater quantities of cytokines, including TNF-α, IL-1, IL-6, and IL-10, after pro-inflammatory stimulation (14, 27, 28, 40). These cytokines may activate their cognate receptors on the cell surface in an autocrine manner resulting in activation of the STAT pathways, either directly or indirectly. It is our opinion that this mechanism certainly contributes to the augmented iNOS induction in the MKP-1 knock-out cells and mice. Second, the regulatory circuits controlling the STAT pathways could be altered as a result of MKP-1 deletion. Consistent with such an intrinsic alteration in the STAT pathway, we found that miR-155 expression was enhanced in MKP-1-deficient or MKP-1-attenuated macrophages (Fig. 7). One potential target of miR-155 is SOCS-1 mRNA, as it contains a putative miR-155-targeting sequence in its 3′-UTR (Fig. 8A). Because miR-155 is predicted to down-regulate the expression of SOCS-1, we examined whether MKP-1 deficiency affects SOCS-1 expression. We found that SOCS-1 mRNA levels were similar in wild-type and MKP-1-deficient macrophages (data not shown), suggesting that miR-155 does not inhibit SOCS-1 expression through the promotion of mRNA decay. To investigate the role of MKP-1 on SOCS-1 protein expression, we took advantage of the RAW264.7 macrophage cell lines modified to either overexpress or knock down MKP-1 (11, 12). We demonstrated that RAW264.7 cells overexpressing MKP-1 exhibited attenuated miR-155 expression and increased SOCS-1 protein levels, indicating that MKP-1 has the capacity to attenuate SOCS-1 expression. However, we were unable to detect a reliable SOCS-1 signal in either RAW264.7 cells expressing MKP-1 siRNA or in primary peritoneal macrophages. This is most likely because of the relatively low titer of the SOCS-1 antibody and/or relatively low abundance of SOCS-1 protein in macrophages. Because both knockdown and knock-out of MKP-1 are predicted to result in a further decrease in SOCS-1 protein levels, it is not surprising that SOCS-1 is more difficult to detect in these cells. However, using a luciferase reporter carrying a putative miR-155 targeting sequence derived from the 3′-UTR of SOCS-1 mRNA, we demonstrated that the putative miR-155 found on the SOCS-1 mRNA was functional. We showed that miR-155 inhibited the luciferase reporter function through the SOCS-1 mRNA-derived miR-155 targeting sequence, at least in this in vitro system (Fig. 7). Furthermore, an increase in iNOS protein was observed in macrophages transiently transfected with miR-155 (Fig. 8C). These results suggest that miR-155 may inhibit the translation of SOCS-1 mRNA, thus weakening the negative regulatory mechanism that normally restrains the STAT pathways. In other words, miR-155-mediated down-regulation of SOCS-1 could be partially responsible for the augmented STAT-1 and STAT-3 activity and for the enhanced iNOS induction in MKP-1-deficient macrophages. While this manuscript was in revision, Rudensky and co-workers (41) demonstrated that the SOCS-1 protein levels were increased in miR-155-deficient Treg cells compared with wild-type Treg cells, suggesting that miR-155 targets SOCS-1 in Treg cells. Their findings are consistent with the model proposed herein. Interestingly, a very recent study has reported that miR-19 also down-regulates SOCS-1 expression in human cells (42).
Previously, O'Connell et al. (43) have shown that miR-155 is highly induced by Toll-like receptor ligands through a mechanism mediated by TNF-α and JNK. We have previously shown augmentation of JNK activity in MKP-1-deficient macrophages relative to wild-type macrophages after LPS stimulation (14, 15). We and others have also shown that LPS-induced TNF-α production is substantially more robust in MKP-1-deficient macrophages than in wild-type macrophages (14, 27, 28, 40). After the initial LPS stimulation, MKP-1−/− macrophages will have greater JNK activity because of the absence of the MKP-1-mediated negative feedback control mechanism. Moreover, because these macrophages produce larger quantities of cytokines, such as TNF-α, these cells are also subject to a stronger secondary stimulation by autocrine cytokines than wild-type macrophages. Therefore, it is not surprising that miR-155 expression is significantly enhanced in MKP-1-deficient macrophages. Consistent with this model, we found that the RAW264.7 macrophages that overexpress MKP-1 exhibited a substantial decrease in miR-155 (Fig. 7B).
Stabilization of iNOS mRNA in MKP-1-deficient Cells
Numerous reports have previously shown that stabilization of iNOS mRNA contributes to its induction in macrophages (25, 44–47). JNK in particular has been shown to be involved in the stabilization of iNOS mRNA in macrophages (25). We found that iNOS mRNA stability was significantly increased in MKP-1-deficient macrophages (Fig. 4). Furthermore, we found that pharmacological inhibition of p38 leads to a decrease in iNOS mRNA half-life in MKP-1-deficient macrophages, suggesting that the increased iNOS mRNA stability is at least partially because of the elevated p38 activity in these cells (Fig. 4C). The mouse iNOS mRNAs contain a number of AUUUA elements in their 3′-UTR. Although JNK and HuR have been implicated in the stabilization of iNOS (25, 45), our study is the first to implicate MKP-1 and p38 in the regulation of iNOS mRNA stability. We observed that iNOS mRNA stability changes over time in MKP-1-deficient macrophages. In these cells iNOS mRNA was more stable at earlier time points than at later time points; e.g. half-life >16 h at 6 h of post-LPS stimulation versus only 5.5 h at 24 h of post-LPS stimulation. In contrast, the iNOS mRNA half-life stayed roughly the same (∼3 h) in MKP-1+/+ macrophages at both 6- and 24-h post-LPS stimulation (Fig. 4, A and B). Perhaps this can be explained by the less effective inactivation of p38 and JNK in the MKP-1−/− cells. Whereas p38 and JNK activities in LPS-stimulated wild-type macrophages return quickly to basal levels, the activities of p38 and JNK in MKP-1-deficient macrophages last significantly longer. As a result, iNOS is more stable in MKP-1-deficient cells than in wild-type cells shortly (e.g. 6 h) after LPS stimulation. We speculate that by 24 h post-LPS addition, the differences in p38 and JNK activities between the MKP-1+/+ and MKP-1−/− groups are considerably smaller than at 6 h. This explains the relatively smaller difference in iNOS mRNA half-life between the MKP-1-sufficient and the MKP-1-deficient macrophages at the later time point (Fig. 4).
The Substrate Specificity of MKP-1 in Vivo
We and others have shown that in macrophages the absence of the MKP-1 gene leads to prolonged p38 and JNK activities but has little effect on the activation kinetics of ERK (14, 27, 28, 40). However, in this study we found that at least in the lung, the activities of all three MAP kinases in the LPS-challenged MKP-1−/− mice were greater than those in similarly treated wild-type mice (Fig. 1B). This observation raises the question of whether, like p38 and JNK, ERK is also a physiological substrate for MKP-1. There are several possibilities that could account for this observation. First, it is possible that MKP-1 could play an important role in the inactivation of ERK in the lung. In transfection assays, MKP-1 is effective in inactivating ERK (48, 49), likely because of the relatively high expression levels in such assays. In titration assays, it has been clearly shown that p38 and JNK are the preferred substrates for MKP-1 (50). However, when MKP-1 expression is high, ERK can also be effectively inactivated (50). It is likely that the expression level of MKP-1 in primary macrophages is modest compared with transfected cells, thus enabling us to observe the relative selectivity of MKP-1 toward p38 and JNK (14). Compared with other tissues, the expression level of MKP-1 in the lung is considerably higher (51). In fact, Charles et al. (51) have shown that in adult mice MKP-1 is expressed predominantly in the lung. The high level of MKP-1 expression could compensate for the modest enzymatic activity toward ERK, making its contribution to ERK inactivation substantial. The second possibility is that the elevated ERK activity in the lung of MKP-1−/− mice is not because of the defective inactivation of ERK but rather because of potent ERK activation secondary to the more robust inflammatory responses in these knock-out mice. For example, we and several other laboratories have found that compared with wild-type mice, MKP-1−/− mice produce greater quantities of a large number of cytokines, including TNF-α, IL-6, and IL-10 as well as many chemokines (14, 27, 28, 40). These MKP-1 knock-out mice also develop more severe pulmonary edema (14). These cytokines or physical changes in the lung as the result of pulmonary edema could, in principle, trigger activation of the MAP kinase cascades. Finally, we have previously shown that the MKP-1−/− mice exhibit more robust leukocyte infiltration than do wild-type mice (14), and this type of change in cellularity in the lung may also lead to signaling changes. Immunohistochemical analysis of lung tissues for the phosphorylated MAP kinases may provide useful information on the molecular mechanisms underlying the changes in the ERK MAP kinases in the MKP-1 knock-out mice.
Acknowledgments
We thank Bristol-Myers Squibb Co. for providing MKP-1 knock-out mice. We thank the veterinary staff led by Dr. James Cooper for enthusiastic support. We thank Joshua Kuhlman for technical support. We are grateful to George A. Calin for advice in microRNA analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants AI 57798 and AI 68956 (to Y. L.) and HL 75261 (to L. D. N.).
- iNOS
- inducible nitric-oxide synthase
- LPS
- lipopolysaccharide
- STAT
- signal transducers and activators of transcription
- TNF
- tumor necrosis factor
- IL
- interleukin
- IFN
- interferon
- UTR
- untranslated region
- MAP
- mitogen-activated protein
- MKP
- MAP kinase phosphatase
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- SOCS
- suppressor of cytokine signaling
- miR
- microRNA
- siRNA
- small interfering RNA
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- DTT
- dithiothreitol.
REFERENCES
- 1.Bogdan C. (2001) Nat. Immunol. 2, 907–916 [DOI] [PubMed] [Google Scholar]
- 2.Janeway C. A. J., Travers P., Walport M., Shlomchik M. J. (2005) Immunobiology: The Immune System in Health and Disease, 6th Ed., pp. 37–100, Garland Publishing, New York [Google Scholar]
- 3.Bogdan C., Röllinghoff M., Diefenbach A. (2000) Immunol. Rev. 173, 17–26 [DOI] [PubMed] [Google Scholar]
- 4.Gordon S. (1995) BioEssays 17, 977–986 [DOI] [PubMed] [Google Scholar]
- 5.Malu S., Srinivasan S., Kumar Maiti P., Rajagopal D., John B., Nandi D. (2003) J. Immunol. Methods 272, 55–65 [DOI] [PubMed] [Google Scholar]
- 6.Ohmori Y., Hamilton T. A. (2001) J. Leukocyte Biol. 69, 598–604 [PubMed] [Google Scholar]
- 7.Heitmeier M. R., Scarim A. L., Corbett J. A. (1999) J. Biol. Chem. 274, 29266–29273 [DOI] [PubMed] [Google Scholar]
- 8.Parrillo J. E. (1993) N. Engl. J. Med. 328, 1471–1477 [DOI] [PubMed] [Google Scholar]
- 9.Martin G. S., Mannino D. M., Eaton S., Moss M. (2003) N. Engl. J. Med. 348, 1546–1554 [DOI] [PubMed] [Google Scholar]
- 10.Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., Pinsky M. R. (2001) Crit. Care Med. 29, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 11.Chen P., Li J., Barnes J., Kokkonen G. C., Lee J. C., Liu Y. (2002) J. Immunol. 169, 6408–6416 [DOI] [PubMed] [Google Scholar]
- 12.Shepherd E. G., Zhao Q., Welty S. E., Hansen T. N., Smith C. V., Liu Y. (2004) J. Biol. Chem. 279, 54023–54031 [DOI] [PubMed] [Google Scholar]
- 13.Zhao Q., Shepherd E. G., Manson M. E., Nelin L. D., Sorokin A., Liu Y. (2005) J. Biol. Chem. 280, 8101–8108 [DOI] [PubMed] [Google Scholar]
- 14.Zhao Q., Wang X., Nelin L. D., Yao Y., Matta R., Manson M. E., Baliga R. S., Meng X., Smith C. V., Bauer J. A., Chang C. H., Liu Y. (2006) J. Exp. Med. 203, 131–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Meng X., Kuhlman J. R., Nelin L. D., Nicol K. K., English B. K., Liu Y. (2007) J. Immunol. 178, 5312–5320 [DOI] [PubMed] [Google Scholar]
- 16.Calvert T. J., Chicoine L. G., Liu Y., Nelin L. D. (2008) Am. J. Physiol. Heart Circ. Physiol 294, H1621–H1629 [DOI] [PubMed] [Google Scholar]
- 17.Dorfman K., Carrasco D., Gruda M., Ryan C., Lira S. A., Bravo R. (1996) Oncogene 13, 925–931 [PubMed] [Google Scholar]
- 18.Chen W., Martindale J. L., Holbrook N. J., Liu Y. (1998) Mol. Cell. Biol. 18, 5178–5188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu C. G., Calin G. A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C. D., Shimizu M., Zupo S., Dono M., Alder H., Bullrich F., Negrini M., Croce C. M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9740–9744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prabhu K. S., Zamamiri-Davis F., Stewart J. B., Thompson J. T., Sordillo L. M., Reddy C. C. (2002) Biochem. J. 366, 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., pp. 9.47–9.58, Cold Spring Harbor Press, New York [Google Scholar]
- 22.Tili E., Michaille J. J., Cimino A., Costinean S., Dumitru C. D., Adair B., Fabbri M., Alder H., Liu C. G., Calin G. A., Croce C. M. (2007) J. Immunol. 179, 5082–5089 [DOI] [PubMed] [Google Scholar]
- 23.Collart M. A., Belin D., Vassalli J. D., de Kossodo S., Vassalli P. (1986) J. Exp. Med. 164, 2113–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fechir M., Linker K., Pautz A., Hubrich T., Förstermann U., Rodriguez-Pascual F., Kleinert H. (2005) Mol. Pharmacol. 67, 2148–2161 [DOI] [PubMed] [Google Scholar]
- 25.Lahti A., Jalonen U., Kankaanranta H., Moilanen E. (2003) Mol. Pharmacol. 64, 308–315 [DOI] [PubMed] [Google Scholar]
- 26.Hammer M., Mages J., Dietrich H., Schmitz F., Striebel F., Murray P. J., Wagner H., Lang R. (2005) Eur. J. Immunol. 35, 2991–3001 [DOI] [PubMed] [Google Scholar]
- 27.Chi H., Barry S. P., Roth R. J., Wu J. J., Jones E. A., Bennett A. M., Flavell R. A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2274–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salojin K. V., Owusu I. B., Millerchip K. A., Potter M., Platt K. A., Oravecz T. (2006) J. Immunol. 176, 1899–1907 [DOI] [PubMed] [Google Scholar]
- 29.Kamijo R., Harada H., Matsuyama T., Bosland M., Gerecitano J., Shapiro D., Le J., Koh S. I., Kimura T., Green S. J. (1994) Science 263, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 30.Martin E., Nathan C., Xie Q. W. (1994) J. Exp. Med. 180, 977–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hecker M., Preiss C., Klemm P., Busse R. (1996) Br. J. Pharmacol. 118, 2178–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salkowski C. A., Detore G., McNally R., van Rooijen N., Vogel S. N. (1997) J. Immunol. 158, 905–912 [PubMed] [Google Scholar]
- 33.Lorsbach R. B., Murphy W. J., Lowenstein C. J., Snyder S. H., Russell S. W. (1993) J. Biol. Chem. 268, 1908–1913 [PubMed] [Google Scholar]
- 34.Gavrilescu L. C., Butcher B. A., Del Rio L., Taylor G. A., Denkers E. Y. (2004) Infect. Immun. 72, 1257–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindsay M. A. (2008) Trends Immunol. 29, 343–351 [DOI] [PubMed] [Google Scholar]
- 36.Hoefig K. P., Heissmeyer V. (2008) Curr. Opin. Immunol. 20, 281–287 [DOI] [PubMed] [Google Scholar]
- 37.O'Shea J. J., Murray P. J. (2008) Immunity 28, 477–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu X., Chakravarty S. D., Ivashkiv L. B. (2008) Immunol. Rev. 226, 41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slack D. N., Seternes O. M., Gabrielsen M., Keyse S. M. (2001) J. Biol. Chem. 276, 16491–16500 [DOI] [PubMed] [Google Scholar]
- 40.Hammer M., Mages J., Dietrich H., Servatius A., Howells N., Cato A. C., Lang R. (2006) J. Exp. Med. 203, 15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu L. F., Thai T. H., Calado D. P., Chaudhry A., Kubo M., Tanaka K., Loeb G. B., Lee H., Yoshimura A., Rajewsky K., Rudensky A. Y. (2009) Immunity 30, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pichiorri F., Suh S. S., Ladetto M., Kuehl M., Palumbo T., Drandi D., Taccioli C., Zanesi N., Alder H., Hagan J. P., Munker R., Volinia S., Boccadoro M., Garzon R., Palumbo A., Aqeilan R. I., Croce C. M. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connell R. M., Taganov K. D., Boldin M. P., Cheng G., Baltimore D. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1604–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. H., Murphy W. J., Russell S. W., Morrison D. C., Koide N., Yoshida T., Yokochi T. (2005) Cell. Immunol. 234, 16–22 [DOI] [PubMed] [Google Scholar]
- 45.Di Marco S., Mazroui R., Dallaire P., Chittur S., Tenenbaum S. A., Radzioch D., Marette A., Gallouzi I. E. (2005) Mol. Cell. Biol. 25, 6533–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor L. S., Cox G. W., Melillo G., Bosco M. C., Espinoza-Delgado I. (1997) Cancer Res. 57, 2468–2473 [PubMed] [Google Scholar]
- 47.Jun C. D., Choi B. M., Hoon-Ryu Um J. Y., Kwak H. J., Lee B. S., Paik S. G., Kim H. M., Chung H. T. (1994) J. Immunol. 153, 3684–3690 [PubMed] [Google Scholar]
- 48.Charles C. H., Sun H., Lau L. F., Tonks N. K. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5292–5296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun H., Charles C. H., Lau L. F., Tonks N. K. (1993) Cell 75, 487–493 [DOI] [PubMed] [Google Scholar]
- 50.Franklin C. C., Kraft A. S. (1997) J. Biol. Chem. 272, 16917–16923 [DOI] [PubMed] [Google Scholar]
- 51.Charles C. H., Abler A. S., Lau L. F. (1992) Oncogene 7, 187–190 [PubMed] [Google Scholar]