Abstract
Bone morphogenetic proteins (BMPs) require proteolytic activation by members of the proprotein convertase (PC) family. Pro-BMP4 is initially cleaved at a site adjacent to the mature ligand domain (S1) and then at an upstream site (S2) within the prodomain. Cleavage at the S2 site, which appears to occur in a tissue-specific fashion, regulates the activity and signaling range of mature BMP4. To test the hypothesis that tissue-specific cleavage of pro-BMP4 is regulated by differential expression of a site-specific protease, we identified the PCs that cleave each site in vivo. In Xenopus oocytes, furin and PC6 function redundantly to cleave both the S1 and S2 sites of pro-BMP4, as evidenced by the results of antisense-mediated gene knockdown and the use of the furin- and PC6-selective inhibitor α1-PDX. By contrast, α1-PDX blocked cleavage of the S2 but not the S1 site of pro-BMP4 in embryos, suggesting the existence of a developmentally regulated S1 site-specific convertase. This protease is likely to be PC7 based on knowledge of its required substrate cleavage motif and resistance to α1-PDX. Consistent with this prediction, an α1-PDX variant engineered to target PC7, in addition to furin and PC6, completely inhibited cleavage of BMP4 in oocytes and embryos. Further studies showed that pc7 transcripts are expressed and polyadenylated, and that the PC7 precursor protein undergoes efficient autocatalytic activation in both oocytes and embryos. These results suggest that PC7, or a convertase with similar substrate specificity, functions to selectively cleave the S1 site of pro-BMP4 in a developmentally regulated fashion.
BMP4 (bone morphogenetic protein 4) is a cell to cell signaling molecule that was originally isolated for its ability to induce ectopic bone formation (1). More recent studies demonstrate diverse roles for BMP4 during development of the skeleton and other organs and in bone homeostasis after birth (2).
The bioactivity of BMP4 is regulated post-translationally, at the level of proteolytic activation. BMP4 originates as an inactive dimeric precursor that is cleaved by specific members of the proprotein convertase (PC)2 family of endoproteases (3, 4) to yield prodomain fragments along with the carboxyl-terminal mature ligand. In mammals, seven members of the PC family have been characterized. Among these only furin (also known as PACE, SPC1, or PCSK3), PACE4 (also named SPC4 or PCSK6), PC6 (also called PC5, SPC6, or PCSK5), and PC7 (also known as PC8, LPC, SPC7, or PCSK7) are broadly expressed and active within the constitutive, as opposed to the regulated, secretory pathway, making them appropriate candidates for endogenous BMP4 convertases (5, 6). Furin prefers to cleave proproteins at the carboxyl-terminal side of the optimal consensus sequence -RX(R/K)R-, but can also cleave following the minimal sequence -RXXR- (7). PACE4 and PC6 (also commonly called PC5) recognize the same optimal and minimal furin consensus motifs, whereas PC7 has a strict requirement for a basic residue in the P2 position (counting back from the cleavage site), and can thus only cleave substrates containing an optimal furin motif (8, 9).
We have shown that BMP4 is sequentially cleaved at two sites within its prodomain and that this ordered proteolysis regulates both the activity and signaling range of mature BMP4 (10). BMP4 is cleaved first following an optimal furin motif adjacent to the mature domain (-RXKR-, the S1 site), and this allows for subsequent cleavage at an upstream minimal site within the prodomain (-RXXR-, the S2 site). In Xenopus embryos, pro-BMP4 carrying a point mutation that renders the upstream site noncleavable generates a ligand that accumulates at lower levels, and thus signals over a shorter range, compared with BMP4 cleaved from wild type precursor.
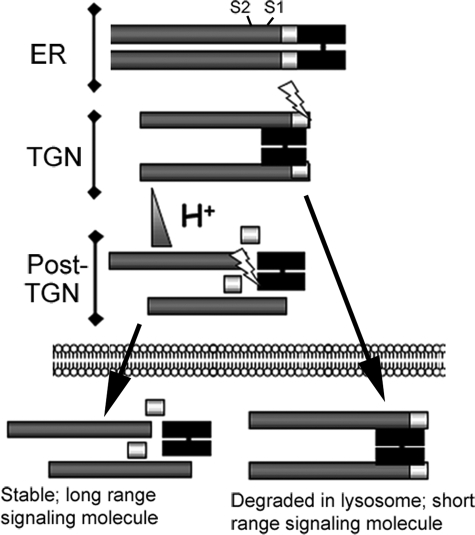
More recent studies from our laboratory suggest that differential cleavage of pro-BMP4 regulates the activity of the mature ligand by directing its intracellular trafficking to either degradatory or secretory/recycling pathways (illustrated schematically in Fig. 1) (11). Cleavage of pro-BMP4 at the S1 site, which is presumed to occur in the trans-Golgi network (TGN), generates a noncovalently associated ligand-prodomain complex that is then trafficked to a post-TGN compartment where the more acidic environment makes the S2 site accessible, possibly by triggering a conformational change. Cleavage at the S2 site liberates mature BMP4 from the prodomain, which promotes protein stability. If cleavage of the upstream S2 site does not occur, the prodomain-ligand complex is preferentially targeted to the lysosome for degradation, either directly within the biosynthetic pathway or via the endocytic pathway following secretion, receptor activation, and reuptake. As a consequence, less mature ligand is available, resulting in lower levels of BMP4 activity.
FIGURE 1.
Model for regulation of BMP4 activity by sequential cleavage. See text for details. ER, endoplasmic reticulum; TGN, trans-Golgi network.
Analysis of mice carrying a knock-in point mutation in Bmp4 (Bmp4S2G) that allows for cleavage at the S1 site (generating a wild type ligand), but prevents S2 processing, has shown that cleavage of the S2 site is essential for normal development and, more importantly, suggests that this site might be selectively cleaved in a tissue-specific fashion (12). Specifically, these mice display phenotypic defects in only a subset of tissues where full Bmp4 dosage is known to be important. For example, Bmp4S2G/S2G mice show a greater loss of primordial germ cells and more severe testicular degeneration than that observed in Bmp4 null heterozygotes (Bmp4+/−) (13). Bmp4S2G/S2G mice, however, never display polycystic kidneys, polydactyly, or other skeletal abnormalities that are observed in Bmp4+/− mice. Furthermore, levels of mature BMP4 protein are decreased in the testes but not in the kidneys of Bmp4S2G/S2G mice relative to wild type siblings.
The above studies illustrate the importance of proteolytic processing in regulating BMP4 activity, yet the PCs that cleave the S1 and/or S2 site have not been definitively identified. Recombinant furin, PACE4, and PC6 are all capable of cleaving the S1 and S2 sites of pro-BMP4 in vitro, whereas PC7 can only recognize and cleave following the optimal motif present at the S1 site (4). Ectopic expression of the selective PC inhibitor, α1-PDX, in Xenopus embryos phenocopies loss of BMP4 function. Furthermore, this same inhibitor prevents cleavage of BMP4 in an oocyte translation assay (4). α1-PDX is a genetically engineered mutant form of the naturally occurring serine protease inhibitor (serpin), α1-antitrypsin (α1-AT), that contains in its reactive site the amino acids -RIPR-, the minimal consensus motif for efficient processing by furin (14). As with all serpins, α1-PDX functions as a suicide substrate inhibitor, forming a tight, stable complex with and eventually being cleaved by the protease(s) it targets. α1-PDX is a potent and selective inhibitor of furin and PC6 in vitro, with Ki values of 0.6 and 2.3 nm, respectively (15). It can also inhibit the activity of ectopically expressed PACE4 (16), but not PC7 (15), when overexpressed in cultured cells. Collectively, these studies identify furin and PC6 as the most likely endogenous BMP4 convertases, but they do not rule out the possibility that PACE4 and/or PC7 contribute to cleavage of one or both sites.
Phenotypic analysis of furin or pc6 mutant mice is consistent with the possibility that these enzymes contribute to proteolytic activation of BMP4 in vivo. Furin null mutants die by embryonic day 11.5 and show an early defect in chorioallantoic fusion (17) similar to that observed in Bmp4-deficient mice (12, 18). Loss of furin has less severe consequences than loss of Bmp4 (19), however, suggesting that additional PCs function redundantly to cleave this precursor. Mice homozygous for null alleles of pc6 die between embryonic day 15.5 and birth because of defects in multiple organ systems (20, 21). Some defects in pc6 mutants have been attributed to deficiencies in GDF11, whereas others, including defects in ventral body wall closure and cardiac abnormalities, phenocopy those observed in Bmp4 mutants (12, 18, 22, 23). By contrast, mice lacking pace4 show laterality defects that have been attributed primarily to loss of nodal processing (24). pc7 mutants develop normally (25), demonstrating that this protease does not function independently to activate pro-BMP4.
The mechanism through which tissue-specific cleavage of the S2 site of BMP4 is achieved is unknown, but one possibility involves differential expression of site-specific convertases. For example, selective cleavage of the S2 site could be mediated by tissue-specific expression of a convertase that resides in the more acidic post-TGN compartment in which the S2, but not the S1 site, is cleaved. Furin is broadly expressed and resides predominantly in the TGN (6), whereas PC6B (a membrane-bound isoform of PC6) is expressed in only a few tissues (20) and is localized to a distinct, post-TGN compartment (26). Thus, it is possible that the S1 site is constitutively cleaved by furin, whereas the S2 site is cleaved only in tissues that co-express PC6B. Alternatively, it is possible that furin, PC6, and/or PACE4 cleave both sites, whereas PC7 cleaves only the S1 site in vivo, similar to what has been demonstrated in vitro. In this scenario, BMP4 would be cleaved at both sites in all tissues except for those in which PC7 is the sole convertase. To begin to test potential mechanisms for tissue-specific regulation of S2 cleavage, we used loss-of-function approaches along with protein-based inhibitors to ask which PCs cleave the S1 and S2 sites of BMP4 in vivo.
EXPERIMENTAL PROCEDURES
cDNA Constructs and Antisense Oligonucleotides
cDNAs encoding HA- and Myc-tagged native and cleavage mutant forms of pro-BMP4 have been described previously (11, 27). cDNAs encoding other cleavage site variants were made using the splicing by overlap extension method (28) or by PCR-mediated introduction of appropriate restriction sites. A full-length Xenopus PC7 cDNA (accession number NM_00196550) was identified by searching the NCBI data base (ncbi.nlm.nih.gov) and obtained from Open BioSystems on behalf of the IMAGE consortium (29). The coding region of Xenopus PC7 or furin (30) was amplified and sequence encoding a FLAG tag inserted at the extreme carboxyl terminus using PCR-based approaches. Cleavage mutant forms of furin and PC7, in which the consensus cleavage motif (-RXKR-) was changed to -RXKG- and -GXKR-, respectively, were generated using a QuickChange mutagenesis kit (Stratagene). All cDNAs were subcloned into the vector pCS2+, and regions of cDNAs generated by PCR were sequenced. Antisense oligonucleotides specific for furin (5′-A*C*T*TGCTGCTCCAACC*A*G), all isoforms of PC6 (5′-A*T*A*CCACATGCTTGGC*C*A), and PC7 (5′-T*C*C*AGCAAGTTCATCA*G*G) (where residues with phosphorothioate bonds are indicated by an asterisk), were purchased from Invitrogen. The antisense oligonucleotide specific for PACE4 has been described previously (31).
Embryo Culture and Manipulation
Ovulation was induced by injecting female frogs with human chorionic gonadotropin (Sigma). Embryonic stages are according to Nieuwkoop and Faber (32). Capped synthetic mRNA was synthesized by in vitro transcription of linearized template cDNAs using a MegaScript kit (Ambion) and injected into embryos as described previously (33). Xenopus ventralization and ectodermal explant assays were performed as described (10).
Expression and Analysis of Radiolabeled Proteins in PC-depleted Oocytes
Xenopus oocytes (stage VI) were isolated and cultured at 18 °C as described (11). For PC depletion experiments, oocytes were injected with antisense oligonucleotides (10 ng), cultured for 3 days, and then injected with RNA encoding BMP4 (10 ng) together with [35S]Met/Cys (700 nCi). To analyze secreted BMP4 cleavage products in the presence of α1-PDX, oocytes were injected with [35S]Met/Cys (700 nCi) together with RNA encoding BMP4 (1 ng) alone or with increasing doses of RNA encoding α1-PDX (0.05–2.5 ng). In each case, oocytes were cultured overnight, and then radiolabeled BMP4 precursor proteins and cleavage products were immunoprecipitated from clarified lysates by incubation with antibodies specific for HA (12CA5 hybridoma cell supernatant, 1:10) or Myc epitope tags (9E10 hybridoma cell supernatant, 1:10) and protein A-Sepharose 4B beads as described (11). Samples immunoprecipitated with anti-HA antibodies were deglycosylated using peptide:N-glycosidase F (PNGase F) according to the manufacturer's instructions (New England Biolabs). Immunoprecipitated proteins were analyzed by SDS-PAGE (11% acrylamide) under nonreducing conditions and imaged by autoradiography. Radiolabeled bands were visualized with a GE Healthcare PhosphorImager and quantified using the Macintosh IP Lab Gel program.
Western Blot Analysis of Proteins Expressed in Oocytes and Embryos
Oocytes were injected with appropriate RNAs and cultured overnight. Ten oocytes in each group were triturated in 100 μl of lysis buffer (50 mm Tris-HCl, pH 7.5; 150 mm NaCl; 1 mm EDTA; 1% Triton X-100; 0.1% SDS; 2.5% Igepal CA-630 (Sigma), 1× complete mini protease inhibitor mixture (Roche Applied Science)). Lysates were centrifuged twice in a microcentrifuge for 10 min each. Embryos were injected with RNA and cultured to the gastrula stage (stage 11) at which time fluid was aspirated and pooled from the blastocoel of 10 embryos in each group, as described (31). Proteins were deglycosylated by incubation with PNGase F, resolved by SDS-PAGE (12% acrylamide), and transferred onto polyvinylidene difluoride membrane. Membranes were probed with anti-Myc (9E10 hybridoma cell supernatant, 1:250), anti-HA (3F10, 1:000, Roche Applied Science), anti-FLAG (M2, 1:1000, Sigma), and/or anti-α1-AT antibodies (1:1000, Calbiochem). Immunoreactive proteins were detected using enhanced chemiluminescence reagent (Pierce).
Western Blot Analysis of PCs in Crude Membrane Extracts from Oocytes and Embryos
Oocytes and embryos were injected with appropriate RNAs and cultured overnight. Twenty oocytes or embryos were triturated in 200 μl of homogenization buffer (0.25 m sucrose; 50 mm Tris-HCl, pH 7.5; 5 mm KOAc; 5 mm MgOAc; 1× complete mini protease inhibitor mixture (Roche Applied Science)), and lysates were centrifuged at 800 × g for 5 min at 4 °C to remove nuclei. Supernatants were then centrifuged at 70,000 rpm for 30 min at 4 °C in a TLA-100 rotor (∼180,000 × g). Pelleted proteins were deglycosylated with PNGase F, resolved by SDS-PAGE (8% acrylamide), and transferred onto polyvinylidene difluoride membrane. Membranes were probed with anti-FLAG antibodies (M2, 1:1000, Sigma). Immunoreactive proteins were detected using enhanced chemiluminescence reagent (Pierce).
Northern Blot Analysis
RNA was extracted from oocytes or embryos and Northern blots hybridized with antisense riboprobes as described previously (34).
Poly(A) Tail Assay
The linkage-mediated poly(A) tail assay was performed as described (35). Briefly, total RNA was extracted from oocytes or embryos as described (34), and 250 ng of RNA was incubated with phosphorylated oligo(dT)12–18 at 42 °C for 30 min in the presence of T4 DNA ligase. Oligo(dT)-linker primer (5′-GAGAGAACTAGTCTCGAG(T)18) was added to the reaction, which was then incubated at 14 °C for 2 h before addition of avian myeloblastosis virus-reverse transcriptase and further incubated for 1 h at 42 °C. The resulting cDNAs were subjected to PCR amplification using furin-specific primers (5′-TATTGCATTGCACAGAGACATATC) or PC7-specific primers (5′-CCTATTGGACCAACTGGGATG), which are targeted 314 and 241 nucleotides, respectively, upstream of the 3′ end of their respective mRNAs, together with a primer specific to the oligo(dT) linker (5′-GAGAGAACTAGTCTCGAGT) or together with primers complementary to the extreme 3′ end of furin (5′-CTATTCAATTGTATATCCACATCC) or PC7 transcripts (5′-CAGGTTTGCCTCAGTTTCTAGG), respectively. The following PCR conditions were used: 94 °C for 5 min, followed by 30 cycles at 94 °C for 30 s; 60 °C for 30 s; and 72 °C for 1 min, followed by incubation at 72 °C for 7 min. PCR products were separated by agarose gel electrophoresis, and Southern blots of PCR products were hybridized with radiolabeled cDNA probes specific for the 3′-untranslated region of furin or pc7 as described (36).
RESULTS
Furin and PC6 Function Redundantly to Cleave Both Sites of BMP4 in Oocytes
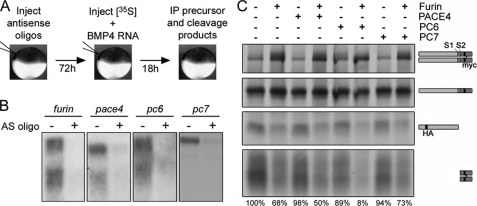
We developed an in vivo assay in oocytes to definitively identify the endogenous PCs that cleave the S1 and/or S2 site of BMP4 (illustrated in Fig. 2A). Oocytes were injected with phosphorothioate-modified antisense oligonucleotides specific for furin, pc6, pace4, and pc7, and these were shown to reduce the levels of their target mRNAs by 90–95% (Fig. 2B). Injected oocytes were cultured for 3 days to allow for turnover of endogenous PC proteins and then injected with RNA encoding epitope-tagged BMP4 together with [35S]Met/Cys. The following day, precursor protein and cleavage products were immunoprecipitated from oocyte lysates using antibodies specific for the HA tag in the prodomain or the Myc tag in the mature domain. Immunoprecipitated proteins were then analyzed by SDS-PAGE under nonreducing conditions and imaged by autoradiography (Fig. 2C, position of precursor dimer, precursor monomer, and S1 + S2 cleaved prodomain and mature ligand are illustrated to the right of the gel). Because dimerization and folding of pro-BMP4 is relatively slow and is rate-limiting for cleavage, levels of precursor monomer were not affected by the depletion of endogenous convertases over the short time course of this analysis. Thus, the precursor monomer band (Fig. 2C, 2nd panel from the top) served as a loading control. Levels of dimerized BMP4 precursor increased significantly in oocytes depleted of furin, and this effect was accompanied by a corresponding decrease in the levels of both the fully cleaved prodomain and mature BMP4 (Fig. 2C). Levels of cleaved prodomain and mature BMP4 were slightly decreased in oocytes depleted of PC6 alone and were strongly decreased in oocytes depleted of both furin and PC6. By contrast, there was no significant loss of BMP4 cleavage in oocytes depleted of PACE4 or PC7. These data show that furin and PC6 function redundantly to cleave both the S1 and the S2 sites of Pro-BMP4 in oocytes and rule out a model in which PC6B selectively cleaves the S2 site.
FIGURE 2.
Furin and PC6 function redundantly to cleave both sites of BMP4 in oocytes. A, schematic illustration of PC knockdown assay. B, Northern analysis of pc expression in oocytes 24 h after injection of antisense oligonucleotides (AS oligo) specific for each PC as indicated. C, radiolabeled pro-BMP4 precursor dimers and monomers, prodomain, and mature domain fragments (indicated to right of gel) were immunoprecipitated (IP) from oocytes injected with antisense oligonucleotides specific for furin, pace4, pc6, and/or pc7 as indicated and separated by SDS-PAGE under nonreducing conditions. Mature BMP4 dimers appear as diffuse bands because of variable glycosylation (bottom panel). Below each lane, levels of cleaved mature BMP4 normalized to precursor monomer are expressed as percent of that observed in uninjected oocytes. Results were reproduced in at least three experiments.
An α1-PDX-insensitive Enzyme Acts Redundantly with Furin and PC6 to Cleave the Optimal Site of BMP4 in Embryos but Not Oocytes
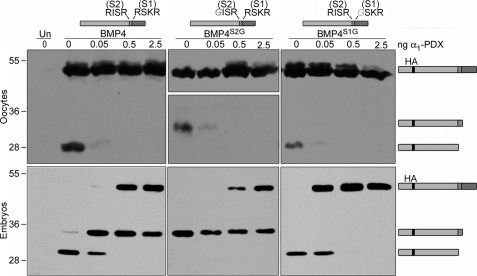
To ask whether furin and PC6 are the sole convertases responsible for cleaving pro-BMP4 throughout development, we analyzed BMP4 cleavage products in oocytes and embryos in the presence of the selective PC inhibitor α1-PDX. As described previously, α1-PDX is a potent inhibitor of furin and PC6 but is ineffective against PC7. Xenopus oocytes were injected with 1 ng of RNA encoding epitope-tagged pro-BMP4, pro-BMP4S2G (which can only be cleaved at the S1 site), or pro-BMP4S1G (which can only be cleaved at the S2 site) either alone or together with increasing amounts of RNA encoding α1-PDX. Western blots of oocyte extracts were then probed with antibodies specific for the HA tag in the prodomain of BMP4. When we expressed α1-PDX in oocytes at doses of 0.5 ng or greater, cleavage of both the S1 and the S2 site of wild type BMP4 was blocked (Fig. 3, top panels; position of precursor, S1-only cleaved prodomain, and S1 + S2 cleaved prodomain are illustrated to the right of the gel). Cleavage of pro-BMP4S2G and pro-BMP4S1G at the S1 or S2 sites, respectively, was ablated at similar doses of α1-PDX. We do not detect any increase in BMP4 precursor levels concomitant with the loss of cleavage products in oocyte lysates because pro-BMP4 is present primarily in monomeric form inside of cells, and this pool is impervious to inhibitors of cleavage. These findings are consistent with the results of our antisense depletion experiments (Fig. 2C), and with our previous studies using this inhibitor (4) in showing that furin and PC6 are fully responsible for cleavage of BMP4 in oocytes. A different result was obtained, however, when we analyzed BMP4 cleavage in embryos (Fig. 3, lower panels). In these experiments, RNA encoding epitope-tagged pro-BMP4, pro-BMP4S2G, or pro-BMP4S1G (1 ng) was injected near the animal pole of four-cell embryos either alone or together with RNA encoding increasing doses of α1-PDX. At the mid-gastrula stage, fluid was aspirated and pooled from the blastocoel of 10 embryos in each group, and Western blots of blastocoel fluid were probed with antibodies specific for the HA tag in the prodomain of BMP4. Pro-BMP4 is cleaved intracellularly, and thus precursor proteins are detected only in the intact cells of embryo homogenates, whereas the cleaved prodomain and mature ligand are secreted into the blastocoel. In the absence of α1-PDX, BMP4 precursor protein was not detected in the blastocoel fluid, but a prodomain fragment generated by cleavage at both the S1 and the S2 sites was observed (Fig. 3, lower panels). In the presence of increasing doses of α1-PDX, uncleaved precursor began to accumulate in the blastocoel, consistent with a loss of furin- and PC6-mediated cleavage of at least a fraction of pro-BMP4. This was accompanied by complete loss of the S1 + S2-cleaved prodomain band and a concomitant increase in the level of S1-only cleaved prodomain. The S1-only cleaved prodomain fragment generated from wild type BMP4 or BMP4S2G persisted even at the highest doses of α1-PDX. By contrast, cleavage of the S2 site of wild type BMP4 or of BMP4S1G was completely inhibited by α1-PDX at doses of 0.5 ng or greater. Taken together, these results suggest that furin and PC6 are the only active BMP4 convertases in oocytes and that they cleave both the S1 and the S2 sites of this precursor. By the mid-gastrula stage, however, an additional α1-PDX-insensitive convertase, which is either not present or not active in oocytes, functions redundantly with furin and PC6 to cleave the optimal (-RSKR-) motif at the S1 site, but not the minimal (-RISR-) motif at the S2 site, of BMP4. We hypothesize that this enzyme could be PC7 based on previous studies showing that PC7 is unique among PC family members in having a stringent requirement for a P2 basic residue (which is present at the S1 but not the S2 site of BMP4), and based on its known insensitivity to inhibition by α1-PDX (9, 15).
FIGURE 3.
An α1-PDX-insensitive enzyme acts redundantly with furin and PC6 to cleave the optimal site of BMP4 in embryos but not oocytes. Western blot of oocyte lysates (top panels) or blastocoel fluid from embryos (bottom panels) not injected (Un) or injected with RNA encoding wild type or cleavage mutant forms of pro-BMP4 (illustrated above each panel) either alone or together with increasing doses of α1-PDX are as indicated. Blots were probed with HA-specific antibodies to detect precursor proteins and cleaved prodomain fragments (illustrated schematically to right of gel). Monomeric forms of pro-BMP4 are present at significantly higher levels than cleavage products in oocyte lysates, and thus when Western blots are exposed for sufficient periods of time to visualize cleavage products, bands corresponding to precursor proteins are overexposed. This is particularly true in the case of BMP4S2G, because cleavage products are unstable, and thus two different exposures of the same Western blot are shown. Results were reproduced in at least three experiments.
The α1-PDX-resistant Protease Present in Embryos Requires an Optimal PC7 Motif for Cleavage
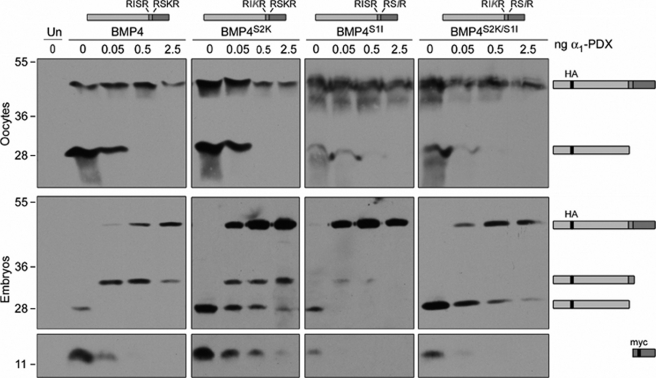
To more stringently test whether the α1-PDX-resistant activity we observe in embryos but not oocytes is only able to cleave substrates following an optimal (-RX(K/R)R-) motif, consistent with the known minimal consensus site for cleavage by PC7, we analyzed processing of BMP4 cleavage site variants in the presence and absence of α1-PDX. Specifically, we assayed cleavage of BMP4 variants (illustrated at the top of Fig. 4) that contain optimal (BMP4S2K) or minimal (BMP4S1I) furin motifs at both the S1 and the S2 sites or in which the order of the optimal and minimal motifs is reversed (BMP4S2K/S1I). Oocytes or embryos were injected with RNA encoding epitope-tagged wild type or cleavage variant pro-BMP4 (1 ng), either alone or together with RNA (0.05–2.5 ng) encoding α1-PDX. Precursor proteins and cleavage products present in oocyte homogenates or blastocoel fluid were analyzed by Western blot as described above, using antibodies directed against the HA tag to detect precursor and cleaved prodomain, or against the Myc tag to detect the cleaved mature domain. In the absence of α1-PDX, all precursor proteins were efficiently cleaved at both the S1 and the S2 site to generate prodomain and mature BMP4 (Fig. 4, precursor and cleavage products are illustrated to the right of the gel). Ligand generated from each variant was active in vivo, as assayed by the ability to ventralize Xenopus embryos and induce expression of the BMP target gene, Xbra, in Xenopus ectodermal explants (data not shown and see Ref. 10).
FIGURE 4.
The α1-PDX-resistant protease present in embryos requires an optimal PC7 motif for cleavage. Western blot of oocyte lysates (top panels) or blastocoel fluid from embryos (bottom panels) not injected (Un) or injected with RNA encoding wild type or cleavage variant forms of pro-BMP4 (illustrated above each panel) either alone or together with increasing doses of α1-PDX are shown. Blots were probed with HA-specific antibodies to detect precursor proteins and cleaved prodomain fragments or with Myc-specific antibodies to detect mature BMP4 (illustrated to right of gel). Results were reproduced in at least three experiments.
In oocytes, α1-PDX blocked proteolysis of both the S1 and the S2 site of wild type and cleavage variant pro-BMP4 at doses of 0.5 ng of RNA or greater (Fig. 4, top panel). In embryos, however, α1-PDX blocked cleavage of minimal furin motifs, whereas cleavage at optimal motifs was partially preserved even in the presence of the highest doses of α1-PDX, regardless of their position (S1 versus S2) within the protein. Specifically, prodomain fragments generated by cleavage at both of the optimal (-RXKR-) motifs in BMP4S2K and at the S2 optimal motif of BMP4S2K/S1I persisted even at the highest doses of α1-PDX. By contrast, α1-PDX completely inhibited cleavage at both of the minimal (-RXXR-) motifs present in BMP4S1I and at the S1 minimal motif in BMP4S2K/S1I. These results confirm that the α1-PDX-resistant protease present in embryos requires a P2 basic residue to cleave a substrate, consistent with our hypothesis that this protease is PC7.
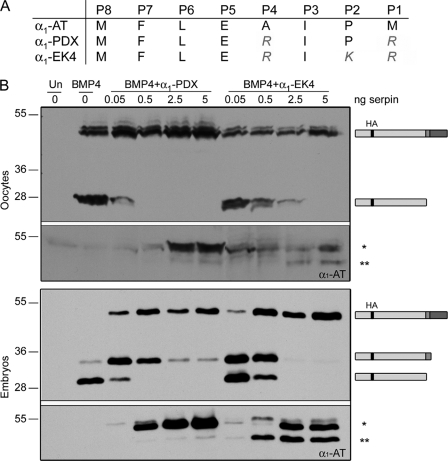
An α1-PDX Variant Predicted to Target PC7 as Well as Furin and PC6 Fully Inhibits BMP4 Cleavage in Oocytes and Embryos
To begin to test our hypothesis that PC7 functions redundantly with furin and PC6 to cleave the S1 site of BMP4, we initially attempted to knock down expression of all three proteases individually and in combination in embryos using translation-blocking morpholino antisense oligonucleotides. We were not, however, able to deplete BMP4 convertase activity in early (gastrula stage) embryos to any detectable degree using this technology,3 presumably because all three of these enzymes are expressed maternally (Fig. 2B) and have relatively long half-lives. As an independent method of testing whether PC7, or an enzyme with similar substrate selectivity, contributes to cleavage of the S1 site of BMP4 in embryos, we asked whether an α1-PDX variant containing a P2 basic residue in its reactive site loop could fully inhibit cleavage of BMP4 in embryos. As described previously, α1-PDX is modeled after the naturally occurring serpin, α1-AT, but contains in its reactive site the minimal consensus motif for efficient processing by furin (illustrated in Fig. 5A). This serpin is a potent inhibitor of furin and PC6, but it does not bind or inhibit PC7, most likely due to the lack of the P2 basic residue that is required for recognition by PC7 (14, 15). The serpin α1-EK4, which has been further modified to include a P2 basic residue (Fig. 5A), has been shown to interact with and inhibit furin (37) and is predicted to inhibit PACE4, PC6, and PC7 as well. Oocytes or embryos were injected with RNA encoding epitope-tagged pro-BMP4 (1 ng), either alone or together with increasing amounts (0.05–5 ng) of RNA encoding α1-PDX or α1-EK4. Precursor proteins and cleavage products present in oocyte homogenates or blastocoel fluid were analyzed by Western blot as described above. As shown in Fig. 5B, α1-PDX and α1-EK4 were both able to fully inhibit the endogenous convertases that cleave the S1 and S2 sites of pro-BMP4 in oocytes. In embryos, however, α1-PDX inhibited the endogenous convertases responsible for cleavage of the S2 but not the S1 site, whereas α1-EK4 inhibited the convertases responsible for cleaving both of these sites. To test the possibility that α1-EK4 is merely more effective because it is expressed at higher levels than α1-PDX, Western blots were stripped and reprobed with antibody specific for α1-AT. In both oocytes (Fig. 5B, top section, lower panel) and embryos (Fig. 5B, bottom section, lower panel), steady state levels of α1-PDX protein were reproducibly higher than those of α1-EK4 following injection of equivalent amounts of RNA. Furthermore, although the vast majority of the α1-PDX that was detected was the fully active form (Fig. 5B, asterisk), ∼50% of α1-EK4 had been converted to the inactive, cleaved form (double asterisk). This finding is consistent with previous studies showing that although α1-PDX forms a highly stable, SDS-resistant complex with its substrates (15), α1-EK4 more readily dissociates to yield the active enzyme and cleaved inhibitor (37). Thus, the ability of α1-EK4, but not α1-PDX, to block cleavage of the S1 site of pro-BMP4 in vivo likely reflects differences in substrate selectivity rather than potency. These results further support our hypothesis that PC7, or an enzyme with similar selectivity, functions redundantly with furin and PC6 to cleave the S1 site of BMP4 in embryos but not in oocytes.
FIGURE 5.
α1-EK4 fully inhibits cleavage of both the optimal and minimal sites of BMP4 in oocytes and embryos. A, sequence surrounding the reactive site loop of α1-AT and genetically engineered variants. B, Western blot of oocyte lysates (top two panels) or blastocoel fluid from embryos (bottom two panels) not injected (Un) or injected with RNA encoding pro-BMP4 alone or together with increasing doses of α1-PDX or α1-EK4. Blots were probed with HA-specific antibodies to detect precursor proteins and cleaved prodomain fragments (illustrated to right of gels). Blots were reprobed with antibodies specific for α1-AT to detect the intact (asterisk) and cleaved form (double asterisk) of each serpin. Results were reproduced in at least two experiments.
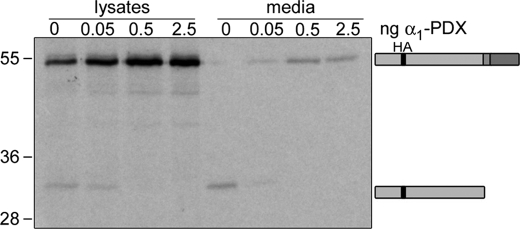
PC7-like Activity Is Not Detected in Oocytes Even When Extracellular BMP4 Cleavage Products Are Analyzed
One possible explanation for why PC7-like activity is detected in embryos but not oocytes is that this enzyme might cleave substrates at the cell surface, as they are being secreted. If this were the case, cleavage products generated by this enzyme would only be present in embryonic blastocoel fluid or in culture media from oocytes but not in oocyte lysates. We are unable to detect cleavage products present in oocyte media by Western blot because of technical limitations. Thus, we used a more sensitive assay to ask whether prodomain fragments generated by PC7-like cleavage of pro-BMP4 at the S1 site alone can be detected in culture media from oocytes made to express α1-PDX, similar to what is observed in embryos. Oocytes were injected with [35S]Met/Cys and RNA encoding epitope-tagged pro-BMP4 (1 ng), either alone or together with increasing amounts (0.05–2.5 ng) of RNA encoding α1-PDX. The following day, BMP4 precursor protein and cleavage products were immunoprecipitated from oocyte lysates or media using antibodies specific for the HA tag in the prodomain. Immunoprecipitated proteins were then analyzed by SDS-PAGE under reducing conditions and imaged by autoradiography. In the absence of α1-PDX, BMP4 precursor protein was detected only in oocyte lysates, whereas a prodomain fragment generated by cleavage at both the S1 and the S2 site was observed in oocyte lysates and media (Fig. 6). In the presence of increasing doses of α1-PDX, uncleaved precursor protein began to accumulate in the media, and this was accompanied by complete loss of all prodomain fragments in the culture media, similar to what is observed in oocyte lysates. We did not detect the appearance of an S1-only cleaved prodomain fragment in the media from oocytes made to express α1-PDX, even when the autoradiograph was overexposed. These results further confirm that the PC7-like activity we detect in embryos made to express α1-PDX is either not present or not active in oocytes.
FIGURE 6.
PC7-like activity is not detected in oocytes, even when extracellular BMP4 cleavage products are analyzed. Radiolabeled pro-BMP4 precursor and prodomain fragments (indicated to right of gel) were immunoprecipitated from lysates and media of uninjected oocytes or oocytes injected with increasing doses of α1-PDX, and separated by SDS-PAGE under reducing conditions.
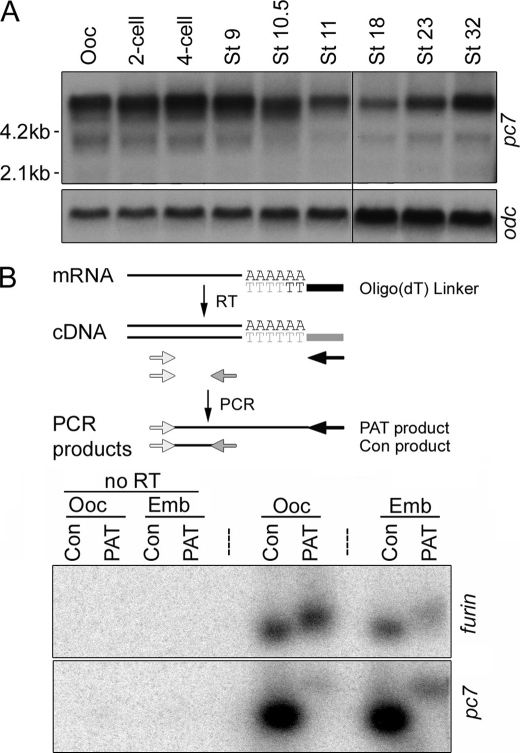
PC7 Transcripts Are Present and Polyadenylated in Both Oocytes and Early Embryos
To begin to ask whether pc7 expression is developmentally regulated at the level of transcription, which might account for our results showing that a PC7-like activity is absent in oocytes but present by mid-gastrula stages, we analyzed the temporal and spatial patterns of expression of this enzyme. pc7 transcripts of ∼5.8, ∼3.8, and ∼2 kb were present in oocytes and persisted at fairly equivalent levels at least through early gastrula stages (St. 10.5) as assayed by Northern blot analysis (Fig. 7A). The largest and most abundant transcript corresponds in size to full-length ESTs published in the NCBI data base. The abundance of pc7 transcripts decreased by the mid-gastrula stage (St. 11) and remained at these levels until the tailbud stage (St. 32). Transcripts were ubiquitously distributed in embryos as analyzed by in situ hybridization of PC7 probes to developmentally staged embryos and by Northern blot analysis of dissected embryos (data not shown). Thus, the acquisition of PC7-like activity during gastrula stages cannot be accounted for by an increase in pc7 transcription or localized accumulation of transcripts in embryos relative to oocytes.
FIGURE 7.
pc7 transcripts are expressed and polyadenylated to a similar extent in oocytes and embryos. A, Northern blot analysis of pc7 and ornithine decarboxylase (odc, as a loading control) expression in Xenopus oocytes (Ooc) and developmentally staged embryos. Black vertical line between St. 11 and St. 18 indicates position where a lane was removed because of degraded RNA. B, analysis of poly(A) tail length of furin and pc7 transcripts in oocytes and embryos (Emb) using the PCR-based assay illustrated schematically above the gel (see text for details). cDNA was synthesized in the presence or absence (no RT) of reverse transcriptase and amplified via PCR using gene-specific forward and reverse primers (light and dark shaded arrows) to generate a control band (Con) or the same gene-specific forward primer with a common reverse primer that anneals to the linker (black arrow) to analyze poly(A) tail (PAT) length. Southern blots of PCR products were hybridized with radiolabeled cDNA probes specific for the 3′-UTR of furin or pc7. For control lanes, 1/10th reaction was loaded. Results were reproduced in at least two experiments.
An alternative explanation for the appearance of PC7-like activity in early embryos might be that maternally inherited pc7 transcripts are not translated in oocytes but are translated in embryos. Early animal development is controlled in part by maternal RNAs that are initially stored in a dormant state, but then undergo elongation of their poly(A) tail and mobilization onto polysomes upon oocyte maturation or after fertilization (38). To test whether PC7 is post-transcriptionally regulated via this mechanism, we used a PCR-based poly(A) tail (PAT) assay (35) (illustrated in Fig. 7B) to compare the polyadenylation status of endogenous pc7 transcripts in Xenopus oocytes and gastrula stage embryos. Polyadenylation of endogenous furin was analyzed in parallel as a positive control because our data show that this enzyme is active in oocytes. RNA was isolated from Xenopus oocytes and gastrula stage embryos and reverse-transcribed using an oligo(dT) linker primer. The resultant cDNA was then amplified using primers that anneal to sequences located ∼300 nucleotides from the end of the 3′-untranslated region (UTR) of the PC of interest (Fig. 7B, lightly shaded arrows) in combination with primers that anneal to either the 3′ linker sequence (black arrow), to generate PAT PCR products, or to the extreme 3′ end of the UTR (darkly shaded arrow), to generate control PCR products. Specific products were then detected by hybridizing Southern blots with a radiolabeled probe generated from the 3′-UTR of each PC. As shown in Fig. 7B, the length of the poly(A) tail on pc7 transcripts was identical in oocytes relative to gastrula stage embryos, and the same was true for furin transcripts. Antibodies that recognize endogenous PC7 are not available, and thus we are unable to definitively rule out the possibility that maternally inherited pc7 transcripts are not translated until after fertilization. The above results, however, demonstrate that maternal pc7 is not translationally regulated at the level of poly(A) tail elongation.
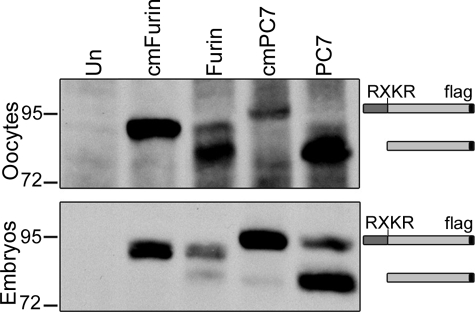
Cleaved Form of PC7 Is Present in Both Embryos and Oocytes
An alternate mechanism by which PC7 might be differentially regulated in oocytes versus embryos is at the level of proteolytic activation. All members of the PC family are generated as inactive zymogens that must be autocatalytically cleaved within the biosynthetic pathway to generate an enzyme that is capable of cleaving substrates in trans (6). To analyze the ability of furin and PC7 to undergo autocatalytic cleavage, we injected RNA encoding FLAG-tagged forms of Xenopus furin and PC7 into oocytes and embryos and cultured them overnight. We then looked at whether the mature, cleaved form of each enzyme was generated in vivo by probing Western blots of membrane-enriched extracts with antibodies directed against the FLAG tag. As a size control for the uncleaved zymogen form, we analyzed cleavage mutant forms of furin and PC7 (in which the consensus motif cleavage forms) for cleavage had been disrupted) in parallel. As shown in Fig. 8, all of the PC7 and most of the furin detected in oocytes was the mature form, demonstrating that autocatalytic cleavage is rapid and nearly complete. In embryos, a small fraction of both furin and PC7 persisted in the uncleaved zymogen form, but the mature, cleaved form of PC7 was the predominant species in membrane extracts from embryos (Fig. 8), although mature cleaved furin was detected primarily as the shed form in blastocoel fluid (data not shown), rather than membrane extracts. Thus, the activity of PC7 is not developmentally regulated at the level of autocatalytic activation. Notably, the presence of cleaved PC7 does not definitively prove that the enzyme is active in either oocytes or embryos.
FIGURE 8.
Cleaved form of PC7 is present in both embryos and oocytes. Western blot of membrane proteins from oocytes or embryos that were injected with wild type or cleavage mutant (cm) forms of FLAG-tagged Xenopus furin or PC7. Blots were probed with FLAG-specific antibodies. Approximate position of precursor and cleaved mature proteins is indicated to the right. Results were reproduced in at least three experiments. Left two lanes of oocyte panel are from the same blot washed 2 extra days to remove background signal and redeveloped. Un, untreated.
DISCUSSION
In this study, we provide the first definitive evidence that furin and PC6 are the primary endogenous BMP4 convertases and that they function redundantly to cleave both the S1 and the S2 site of this precursor protein. These two enzymes are necessary and sufficient to activate BMP4 in Xenopus oocytes. By the early gastrula stage, however, an additional convertase activity that selectively cleaves only the optimal motif at the S1 site of BMP4 becomes apparent. This site-specific protease is likely to be PC7, or a closely related enzyme, based on its substrate selectivity and inhibitor sensitivity. This study provides insight into a potential mechanism by which the S2 site of BMP4 might be cleaved in a tissue-specific fashion. In addition, this study highlights the fact that substrate selectivity of PCs is highly regulated at levels other than cleavage motif recognition or tissue-specific transcription and provides essential information that sets the stage for further studies into how this occurs.
This study does not definitively rule out the possibility that the S1-selective cleavage activity detected in embryos but not oocytes is PACE4 rather than PC7. Because PACE4 is capable of cleaving both the S1 and the S2 sites of BMP4 in vitro, however (4), it is improbable that it cleaves only the S1 site in vivo. Furthermore, the phenotypes of mouse or Xenopus embryos depleted of pace4 are not consistent with a major role for this enzyme in proteolytic activation of BMP4 or related family members (24, 31). Finally, pro-BMP4 is still cleaved in early gastrula stage mice lacking both pace4 and furin, thereby eliminating PACE4 as an essential BMP4 convertase in the early embryo (39).
pc7 is maternally expressed, and yet our data suggest that this convertase is first active in embryos. This implies that PC7 expression or activity is regulated at a post-transcriptional level, although initial analyses revealed that this does not occur at the level of polyadenylation or autocatalytic activation. At least three alternative mechanisms by which PC7 expression or activity might be developmentally regulated can be envisioned. First, it is possible that PC7 expression is regulated at the level of translation, but that this occurs independent of poly(A) tail lengthening. Definitive evidence for or against translational control will require the development of antibodies that can detect endogenous Xenopus PC7. Second, PC7 might require a binding protein that is expressed in embryos but not oocytes to generate an active enzyme following autocatalytic cleavage. Precedent for the necessity of accessory proteins for PC activation is seen in the case of PC2, which requires its binding partner 7B2 for production of an active enzyme species (40). In the absence of 7B2, pro-PC2 can fold, transit the secretory system, and undergo cleavage, but the mature enzyme that is generated remains incapable of cleaving substrates in trans. Recent studies suggest that 7B2 functions as a chaperone that prevents partially unfolded pro-PC2 from aggregating (41). It is likely that other PC family members utilize similar chaperones, and our data are consistent with the possibility that a PC7-specific chaperone is expressed in embryos but not oocytes. A third potential mechanism by which PC7 expression or activity could be developmentally regulated is at the level of post-translational modifications. PC7 is unique among members of the PC family in that it is reversibly modified by palmitoylation of its cytoplasmic tail (9). In the context of transmembrane proteins such as PC7, palmitoylation serves a variety of functions, including binding to specific lipid or protein domains in the membrane compartment in which the palmitoylated protein resides, aiding in folding, preventing aggregation, and/or stabilizing the protein (42). The functional consequences of palmitoylation of PC7 are unknown, but it appears to promote protein stability because a form of PC7 that cannot be palmitoylated is more rapidly degraded (9). Alternatively, or in addition, palmitoylation may be required to anchor PC7 to the subcellular compartments or plasma membrane domains where it encounters its substrates. If PC7 is palmitoylated in embryos, but not oocytes, this might explain why PC7 activity can be detected in the former but not the latter. Further studies, which are beyond the scope of the current work, will be required to determine whether PC7 requires specific binding partners or post-translational modifications to become active.
This study suggesting that furin and PC6 function redundantly to cleave both the S1 and S2 sites of BMP4, whereas PC7 contributes to cleavage of the S1 site alone, is consistent with the possibility that tissue-specific cleavage of the S2 site of BMP4 is regulated by differential expression of active furin, PC6, and PC7. Specifically, it is possible that furin and PC6 function redundantly to cleave both the S1 and the S2 site of BMP4 in all tissues where one or both of these enzymes are expressed, whereas BMP4 is cleaved at the S1 site alone in tissues that express PC7 exclusively. Analysis of Bmp4S2G/S2G mice, which carry a knock-in point mutation that allows for cleavage at the S1 site but prevents cleavage at the upstream S2 site, suggests that cleavage at both sites is essential for normal BMP4 function in many tissues (12, 18). This is consistent with the observation that both furin and pc6 are broadly expressed, and their expression domains often overlap with those of Bmp4 (43). The two tissues in which cleavage at the S2 site appears to be dispensable for vertebrate BMP4 function are the skeleton and the kidneys (12). If BMP4 is cleaved at the S1 site alone in these tissues, and if, as hypothesized, this occurs because active PC7 (but not furin or PC6) is co-expressed with BMP4 in these tissues, then pc7 mutants would be expected to show phenotypic abnormalities in the skeleton and kidneys similar to those observed in Bmp4 mutants. The skeletal and kidney phenotypes in mice with reduced Bmp4 dosage are incompletely penetrant and fairly subtle, however (12, 13). Thus, although pc7 null mutants are reported to have no overt abnormalities, a detailed analysis has not been published (25), and similar defects may exist in pc7 mutants. Closer examination of pc7 null mice, or of compound mutants that lack potentially compensatory PCs, may define more clearly the role of PC7 in development in general and in BMP4 cleavage in particular.
Acknowledgments
We thank Richard Leduc for the ±1-EK4 cDNA and for helpful discussions and T. O'Hare, M. Wong, and D. Goldman for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1HD037976 (to J. L. C.). This work was also supported by the Shriners Hospitals for Children Grant 8530 and by a predoctoral fellowship from the American Heart Association (to S. M. N.).
J. L. Christian, unpublished data.
- PC
- proprotein convertase
- α1-AT
- α1-antitrypsin
- TGN
- trans-Golgi network
- PNGase F
- peptide:N-glycosidase F
- PAT
- poly(A) tail
- UTR
- untranslated region
- HA
- hemagglutinin
- St.
- stage
- BMP
- bone morphogenetic protein.
REFERENCES
- 1.Wozney J. M. (1992) Mol. Reprod. Dev. 32, 160–167 [DOI] [PubMed] [Google Scholar]
- 2.Hogan B. L. (1996) Genes Dev. 10, 1580–1594 [DOI] [PubMed] [Google Scholar]
- 3.Constam D. B., Robertson E. J. (1999) J. Cell Biol. 144, 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui Y., Jean F., Thomas G., Christian J. L. (1998) EMBO J. 17, 4735–4743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steiner D. F. (1998) Curr. Opin. Chem. Biol. 2, 31–39 [DOI] [PubMed] [Google Scholar]
- 6.Thomas G. (2002) Nat. Rev. Mol. Cell Biol. 3, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molloy S. S., Bresnahan P. A., Leppla S. H., Klimpel K. R., Thomas G. (1992) J. Biol. Chem. 267, 16396–16402 [PubMed] [Google Scholar]
- 8.Rockwell N. C., Krysan D. J., Komiyama T., Fuller R. S. (2002) Chem. Rev. 102, 4525–4548 [DOI] [PubMed] [Google Scholar]
- 9.van de Loo J. W., Creemers J. W., Bright N. A., Young B. D., Roebroek A. J., Van de Ven W. J. (1997) J. Biol. Chem. 272, 27116–27123 [DOI] [PubMed] [Google Scholar]
- 10.Cui Y., Hackenmiller R., Berg L., Jean F., Nakayama T., Thomas G., Christian J. L. (2001) Genes Dev. 15, 2797–2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degnin C., Jean F., Thomas G., Christian J. L. (2004) Mol. Biol. Cell 15, 5012–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldman D. C., Hackenmiller R., Nakayama T., Sopory S., Wong C., Kulessa H., Christian J. L. (2006) Development 133, 1933–1942 [DOI] [PubMed] [Google Scholar]
- 13.Dunn N. R., Winnier G. E., Hargett L. K., Schrick J. J., Fogo A. B., Hogan B. L. (1997) Dev. Biol. 188, 235–247 [DOI] [PubMed] [Google Scholar]
- 14.Anderson E. D., Thomas L., Hayflick J. S., Thomas G. (1993) J. Biol. Chem. 268, 24887–24891 [PubMed] [Google Scholar]
- 15.Jean F., Stella K., Thomas L., Liu G., Xiang Y., Reason A. J., Thomas G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsuji A., Hashimoto E., Ikoma T., Taniguchi T., Mori K., Nagahama M., Matsuda Y. (1999) J. Biochem. 126, 591–603 [DOI] [PubMed] [Google Scholar]
- 17.Roebroek A. J., Umans L., Pauli I. G., Robertson E. J., van Leuven F., Van de Ven W. J., Constam D. B. (1998) Development 125, 4863–4876 [DOI] [PubMed] [Google Scholar]
- 18.Goldman D. C., Donley N., Christian J. L. (2009) Mech. Dev. 126, 117–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winnier G., Blessing M., Labosky P. A., Hogan B. L. (1995) Genes Dev. 9, 2105–2116 [DOI] [PubMed] [Google Scholar]
- 20.Essalmani R., Zaid A., Marcinkiewicz J., Chamberland A., Pasquato A., Seidah N. G., Prat A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5750–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szumska D., Pieles G., Essalmani R., Bilski M., Mesnard D., Kaur K., Franklyn A., El Omari K., Jefferis J., Bentham J., Taylor J. M., Schneider J. E., Arnold S. J., Johnson P., Tymowska-Lalanne Z., Stammers D., Clarke K., Neubauer S., Morris A., Brown S. D., Shaw-Smith C., Cama A., Capra V., Ragoussis J., Constam D., Seidah N. G., Prat A., Bhattacharya S. (2008) Genes Dev. 22, 1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiao K., Kulessa H., Tompkins K., Zhou Y., Batts L., Baldwin H. S., Hogan B. L. (2003) Genes Dev. 17, 2362–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W., Selever J., Wang D., Lu M. F., Moses K. A., Schwartz R. J., Martin J. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 4489–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Constam D. B., Robertson E. J. (2000) Genes Dev. 14, 1146–1155 [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor N. A., Van De Ven W. J., Creemers J. W. (2003) FASEB J. 17, 1215–1227 [DOI] [PubMed] [Google Scholar]
- 26.Xiang Y., Molloy S. S., Thomas L., Thomas G. (2000) Mol. Biol. Cell 11, 1257–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sopory S., Nelsen S. M., Degnin C., Wong C., Christian J. L. (2006) J. Biol. Chem. 281, 34021–34031 [DOI] [PubMed] [Google Scholar]
- 28.Horton R. M., Cai Z. L., Ho S. N., Pease L. R. (1990) BioTechniques 8, 528–5352357375 [Google Scholar]
- 29.Lennon G., Auffray C., Polymeropoulos M., Soares M. B. (1996) Genomics 33, 151–152 [DOI] [PubMed] [Google Scholar]
- 30.Nelsen S., Berg L., Wong C., Christian J. L. (2005) Dev. Dyn. 233, 1038–1044 [DOI] [PubMed] [Google Scholar]
- 31.Birsoy B., Berg L., Williams P. H., Smith J. C., Wylie C. C., Christian J. L., Heasman J. (2005) Development 132, 591–602 [DOI] [PubMed] [Google Scholar]
- 32.Nieuwkoop P. D., Faber J. (1967) Normal Table of Xenopus laevis, North Holland Publishing Co., Amsterdam [Google Scholar]
- 33.Moon R. T., Christian J. L. (1989) Technique 1, 76–89 [Google Scholar]
- 34.Christian J. L., Moon R. T. (1993) Genes Dev. 7, 13–28 [DOI] [PubMed] [Google Scholar]
- 35.Sallés F. J., Strickland S. (1999) Methods Mol. Biol. 118, 441–448 [DOI] [PubMed] [Google Scholar]
- 36.Tian Q., Nakayama T., Dixon M. P., Christian J. L. (1999) Development 126, 3371–3380 [DOI] [PubMed] [Google Scholar]
- 37.Dufour E. K., Denault J. B., Bissonnette L., Hopkins P. C., Lavigne P., Leduc R. (2001) J. Biol. Chem. 276, 38971–38979 [DOI] [PubMed] [Google Scholar]
- 38.Radford H. E., Meijer H. A., de Moor C. H. (2008) Biochim. Biophys. Acta 1779, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beck S., Le Good J. A., Guzman M., Ben Haim N., Roy K., Beermann F., Constam D. B. (2002) Nat. Cell Biol. 4, 981–985 [DOI] [PubMed] [Google Scholar]
- 40.Mbikay M., Seidah N. G., Chrétien M. (2001) Biochem. J. 357, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S. N., Lindberg I. (2008) Endocrinology 149, 4116–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 43.Constam D. B., Calfon M., Robertson E. J. (1996) J. Cell Biol. 134, 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]