Abstract
Syndecan-2, a transmembrane heparan sulfate proteoglycan, is a critical mediator in the tumorigenesis of colon carcinoma cells. We explored the function of syndecan-2 in melanoma, one of the most invasive types of cancers, and found that the expression of this protein was elevated in tissue samples from both nevus and malignant human melanomas but not in melanocytes of the normal human skin tissues. Similarly, elevated syndecan-2 expression was observed in various melanoma cell lines. Overexpression of syndecan-2 enhanced migration and invasion of melanoma cells, whereas the opposite was observed when syndecan-2 levels were knocked down using small inhibitory RNAs. Syndecan-2 expression was enhanced by fibroblast growth factor-2, which is known to stimulate melanoma cell migration; however, α-melanocyte-stimulating hormone decreased syndecan-2 expression and melanoma cell migration and invasion in a melanin synthesis-independent manner. Furthermore, syndecan-2 overexpression rescued the migration defects induced by α-melanocyte-stimulating hormone treatment. Together, these data strongly suggest that syndecan-2 plays a crucial role in the migratory potential of melanoma cells.
The syndecans, a family of four transmembrane cell surface heparan sulfate proteoglycans, mainly serving as a co-receptor, regulate the adhesion-dependent signal transduction of a variety of cell types, including cancer cells (1, 2). Cell adhesion receptors or co-receptors play a critical role in the neoplastic transformation of normal cells by regulating the induction of cancer-specific cellular behavior and morphology. Thus, cancer cells probably express and utilize a distinct set of syndecans in the regulation of cancer cell growth.
Several reports have linked altered syndecan expression to various elements of cancer cell growth. Loss of syndecan-1 correlates with shorter survival times in patients with squamous cell carcinoma of the head, neck, and lung (3) as well as multiple myeloma (4); loss of syndecan-1 is also associated with an elevated potential for metastasis in patients with hepatocellular and colorectal carcinomas (5, 6). Previous studies have shown that syndecan-1 regulates tumor activity in pancreatic (7), gastric (8), and breast carcinomas (9). Syndecan-1 may thus play multiple roles in tumorigenic activity and perform various tissue- and/or tumor stage-specific functions (10). Syndecan-4 expression is reduced in colon carcinoma cells (11, 12) and appears to correlate with increased tumorigenic activity (e.g. cell migration and invasion (13)), implying that syndecan-4 functions as a tumor suppressor.
Syndecan-2 is also known to play a crucial role in the regulation of cancer activity. Increased levels of syndecan-2 confer an invasive phenotype in lung (14) and colon cancer cells (15). Reduction in syndecan-2 expression induces cells to switch from the transformed phenotype to flattened monolayers (8) and reduces tumorigenic activity in colon adenocarcinoma and fibrosarcoma cells (8, 16). In addition, syndecan-2 is highly expressed in the microvasculature of mouse gliomas and has been shown to regulate angiogenesis in microvascular endothelial cells (17). On the other hand, an inverse correlation between syndecan-2 expression and metastatic potential has been found in Lewis lung carcinoma cell lines (6). Therefore, changes in syndecan-2 expression may directly or indirectly regulate cancer growth.
Melanoma is the most aggressive malignant tumor of melanocytes. Although found predominantly in the skin, primary melanomas are also known to occur in the bowel and eye (18). Malignant melanoma is notoriously one of the most difficult cancers to treat (19). Therefore, identifying and understanding molecules that regulate the aggressive melanoma phenotype is essential for predicting the likelihood of metastasis. Interestingly, previous studies have shown that melanoma cells acquire the ability to recognize components of the extracellular matrix (ECM)2 via the ectopic expression of different ECM receptors during invasion of the basement membrane (20). Indeed, invadopodia, the dynamic organelle-like structures that form actin-rich protrusions with ECM proteolytic activity, adhere to and digest collagens, laminins, and fibronectin (21). The adhesive properties of invadopodia are primarily attributed to integrins, a large family of heterodimeric transmembrane receptors composed of α and β subunits (22). For example, β1 integrins localize within the invadopodia of melanoma cells (23), and the α5β1 integrins are enriched peripherally in invadopodia, where they stabilize invadopodia protrusion (24). Ectopic stimulation of α6β1 integrin with laminin peptides or with β1 or α6 integrin stimulatory antibodies increases invadopodia activity and melanoma invasiveness (23). The invasive behavior of melanoma cells can be attributed to increased cell motility caused by changes in cytoskeletal organization and altered contacts with the ECM. Thus, cell adhesion receptors may play a crucial role in the acquisition of highly migratory behavior.
Syndecan-2 acts as a key regulator of cancer cells, suggesting that syndecan-2 may contribute to the aggressive phenotype and metastatic potential of melanoma. Here, we report that syndecan-2 plays a pivotal role in the migratory activity of melanoma cells.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Monoclonal antibodies (mAbs) to vinculin, paxillin, and phosphotyrosine (4G10) were purchased from Upstate Biotechnology, Inc. (Lake Placid, NY). mAb to focal adhesion kinase (FAK) and polyclonal antibodies to phosphorylation site-specific FAK(Tyr(P)397) and FAK(Tyr(P)861) were purchased from BioSource Quality Controlled Biochemicals, Inc. (Morgan Hill, CA). Monoclonal antibody to human melanosome (clone HMB-45) was purchased from Dako (Denmark). Effectene was from Qiagen (Hilden, Germany), bovine pituitary extract (BPE) was from Lonza (Visp, Switzerland), α-melanocyte-stimulating hormone (MSH) was from Sigma, and the anti-Ig horseradish peroxidase detection kit was from BD Biosciences.
Production of Monoclonal Antibody against the Extracellular Domain of Syndecan-2
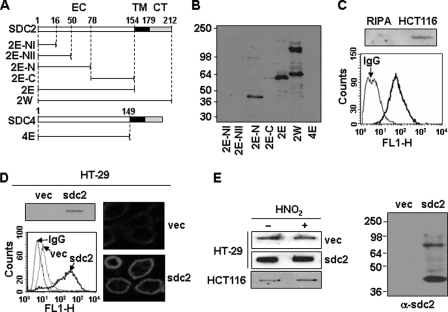
mAb to syndecan-2 was produced by AdipoGen Inc. A portion of rat syndecan-2 cDNA representing the extracellular domain (S2E) was amplified by PCR and cloned upstream of human immunoglobulin heavy chain constant region (Fc) constructed in both pAGCF and pAGPCF2; the former utilized the natural leader sequence, whereas the latter used the plasminogen activator inhibitor-1 leader peptide. The Fc fusion constructs were transfected into human embryonic kidney 293 (HEK293) cells. DMEM serum-free medium was used to collect rat syndecan-2-Fc fusion protein from confluent cells for 2 days. The medium was harvested and spun down to remove cells, and the pH was adjusted to 8.0 with 1 m Tris. The medium was then filtered with a 0.4-μm filter and loaded onto a Protein A column, eluted with 100 mm glycine, pH 3.0, and was immediately neutralized with one-tenth volume of 1 m Tris, pH 8.0. Both constructs yielded similar amounts of rat syndecan-2-Fc. BALB/c mice were repeatedly immunized with S2E-Fc fusion protein until the polyclonal sera from the immunized mice exhibited strong immune responses. Splenocytes were isolated from the mice and fused to mouse myeloma Sp2/0. Screening and single cell cloning were conformed to the established standard protocol. Ascites were prepared from the immunocompromised BALB/c mice. Immunoglobulin fractions were prepared through Protein G. One stable hybridoma clone (clone 197) secreting mAbs against syndecan-2 were obtained (Fig. 1). Specificity and cross-reactivity of the mAbs were determined by Western blot analysis, slot blot analysis, fluorescence-activated cell sorting analysis, and immunohistochemistry. Note that antibody 197 recognizes the extracellular domain of both human and rat syndecan-2.
FIGURE 1.
Characterization of monoclonal antibody against syndecan-2. A, schematic representation of GST-syndecan-2 and -4 core proteins. The extracellular domain is represented by the white box (EC), the transmembrane domain by the black box (TM), and the cytoplasmic domain by the gray box (CT). Syndecan-2 and -4 are labeled with the amino acid number to show the location of deletion mutants. Shown are the whole extracellular domain of syndecan-2 (2E) and each deletion mutant (2E-NI (amino acids 1–16), 2E-NII (amino acids 1–50), 2E-N (amino acids 1–78), and 2E-C (amino acids 78–154)). B, GST-syndecan-2 core proteins were expressed in E. coli. Purified recombinant proteins were separated on 10% SDS-PAGE and subjected to immunoblotting with anti-syndecan-2 antibodies. C, total cell lysates from HCT116 colon carcinoma cells were analyzed by slot blotting with anti-syndecan-2 antibody (top panel). HCT116 cells were incubated with anti-syndecan-2 antibodies, and the cell surface expression levels of syndecan-2 were analyzed by flow cytometry. IgG was used as a control (bottom). D, total cell lysates from HT-29 colon adenocarcinoma cells and HT-29 stably expressing syndecan-2 (HT-29-sdc2) were analyzed by slot blotting with anti-syndecan-2 antibody (left, top). Cells were incubated with anti-syndecan-2 antibody, and the cell surface expression levels of syndecan-2 were analyzed by flow cytometry. IgG was used as a control (left, bottom). Cells were incubated with anti-syndecan-2 antibody, stained with a Texas Red-conjugated secondary antibody, and photographed under confocal microscopy (right). E, partially purified proteoglycans from HT-29, HT-29-sdc2, and HCT116 cells were treated with nitrous acid (HNO2) to degrade glycosaminoglycan chains. Syndecan-2 was analyzed by either slot blotting (left) or Western blotting (right) with anti-syndecan-2 antibody. Note that the proteins were recognized by the syndecan-2 antibody even after degradation of glycosaminoglycan chains with nitrous acid.
Partial Purification of Proteoglycan
Cells were lysed with cold matrix buffer (25 mm Tris, pH 7.5, 4 m urea, 20 mm EDTA, pH 8.0, 4 mm benz-HCl, 2 mm phenylmethylsulfonyl fluoride, 1% (v/v) Tween 20). After centrifugation (14,000 rpm, 4 °C, 10 min), the supernatant was applied onto DEAE-Sepharose columns (Amersham Biosciences) in 50 mm Tris, pH 8.0, containing 4 m urea, 200 mm NaCl, 0.1% Tween 20, 5 mm EDTA, 2 mm phenylmethylsulfonyl fluoride, washed with 30 mm sodium acetate, pH 4.0, containing 4 m urea, 200 mm NaCl, 0.1% Tween 20, 25 mm EDTA, 2 mm phenylmethylsulfonyl fluoride, pH 4.0, and then eluted with 15 mm sodium acetate buffer, pH 4.0, containing 4 m guanidine HCl, 0.1% Tween 20, pH 4.0. Fractions were collected, and absorbance at 280 nm was measured. Protein-containing fractions were pooled and dialyzed against PBS. To remove glycosaminoglycan chains, samples were incubated with nitrous acid (HNO2) solution at pH 1.5, as previously described (25).
Synthesis of Small Interfering RNA Constructs
Oligonucleotides were designed targeting either human syndecan-2 or mouse syndecan-2, containing a 9-bp hairpin loop. Oligonucleotides were annealed and cloned into a pSUPER vector. Sequences of the primers are as follows: human syndecan-2 sense primer, 5′-GATCCCCTGACGATGACTACGCTTCTTTCAAGAGAACTGCTACTGATGCGAAGATTTTTGGAAA-3′; human syndecan-2 antisense primer, 5′-AGCTTTTCCAAAAATGACGATGACTACGCTTCTTCTCTTGAAACTGCTACTGATGCGAAGAGGG-3′; mouse syndecan-2 sense sequence, 5′-GGAGAAACAUUCAGACAAU-3′; mouse syndecan-2 antisense sequence, 5′-AUUGUCUGAAUGUUUCUCC-3′. Boldface characters indicate syndecan-2 sequences, and italics indicate the hairpin loop. Scrambled small interfering RNA (siGENOME non-targeting siRNA 2) was purchased from Dharmacon, Inc. (Chicago, IL).
RNA Extraction and Reverse Transcription-PCR
Total RNA extracted from cultured cells was used as template for reverse transcriptase reaction. Aliquots of cDNA were amplified using the following primers: rat syndecan-2 (forward), 5′-ATGCGGGTACGAGCCACGTC-3′; rat syndecan-2 (reverse) 5′-CGGGAGCAGCACTAGTGAGG-3′; rat β-actin (forward), 5′-TGGAATCCTGTGGCATCCATGAAA-3′; rat β-actin (reverse), 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′. After the initial denaturation at 94 °C for 5 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 60 s were carried out. The reaction products were analyzed in 1.5% agarose gels. The amplified DNA fragments were cloned and sequenced to confirm the PCR products.
Cell Culture and Transfection
Human colon adenocarcinoma HT-29 and HCT116, human melanoma A375 and SK-MEL-28, mouse melanoma cells B16 and B16F10, and human fibrosarcoma HT1080 cells were purchased from the Korean Cell Line Bank. HT-29 cells were maintained in McCoy's 5A complete medium (Invitrogen); A375, B16, B16F10, and HT1080 cells were maintained in DMEM (Invitrogen); and SK-MEL-28 cells were maintained in RPMI (Invitrogen), supplemented with 10% (v/v) fetal bovine serum (FBS) with gentamicin (50 μg/ml; Sigma) at 37 °C in 5% CO2 in a humidified atmosphere. Transient transfections were carried out using either Effectene reagent (Qiagen) or Lipofectamine 2000 (Invitrogen) according to the provided manufacturer's instructions.
Stable Transfection of Syndecan-2 cDNA
HT29 (3 × 106) cells were trypsinized, harvested, washed twice with cold PBS, and then incubated with syndecan-2 cDNA on ice for 20 min. Transfections were carried out by electroporation using 0.2 kV, one pulse time, and 15 s of length). The G148-resistant clones were selected in 3 weeks. Syndecan-2 expression was assessed by reverse transcription-PCR using syndecan-2-specific primer.
Immunoprecipitation and Immunoblotting
The cultures were washed twice with PBS, and the cells were lysed in radioimmune precipitation buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 1% Nonidet P-40, 10 mm NaF, 2 mm Na3VO4) containing a protease inhibitor mixture (1 μg/ml aprotinin, 1 μg/ml antipain, 5 μg/ml leupeptin, 1 μg/ml pepstatin A, 20 μg/ml phenylmethylsulfonyl fluoride). The lysates were clarified by centrifugation at 13,000 rpm for 15 min at 4 °C, denatured with SDS sample buffer, boiled, and analyzed by SDS-PAGE. For immunoprecipitations, each sample containing 200–1,000 μg of total protein was incubated with the relevant antibody for 2 h at 4 °C, followed by incubation with protein G-Sepharose beads for 1 h. Immune complexes were collected by centrifugation. The proteins were transferred onto polyvinylidene difluoride membranes (Amersham Biosciences) and probed with appropriate antibodies. The signals were detected by ECL (Amersham Biosciences).
Flow Cytometry
Cells cultured in 60-mm diameter tissue culture dishes were washed with PBS and released by 1 mm EDTA, followed by the addition of 5% FBS in PBS. After pelleting, cells were resuspended in PBS and counted. Cells (1 × 106/ml) were incubated with anti-syndecan-2 antibody in 10% FBS in PBS for 1 h on 4 °C. After washing three times with PBS containing 0.05% Tween 20, cells were incubated with FITC-conjugated anti-mouse antibody in 10% FBS in PBS for 30 min. Syndecan-2 expression was analyzed by flow cytometry.
Migration Assay
Gelatin (10 μg/ml) was added to each well of a Transwell plate (Costar; 8-μm pore size), and then the membranes were allowed to dry at 25 °C for 1 h. The Transwell plates were assembled in a 24-well plate, and the lower chambers were filled with FGF-2 (100 ng/ml) in fresh media. Cells (5 × 104) were added to each upper chamber, and the plates were incubated at 37 °C in 5% CO2 for 12 h. The cells that had migrated to the lower surface of the filters were stained with 0.6% hematoxylin and 0.5% eosin and counted. For in vitro invasion assays, 24-well Transwell plates (Costar; 8-μm pore size) were coated with gelatin (10 μg/ml) on the lower side of the membrane and with Matrigel (30 μg/μl) on the upper side.
Tumor Metastasis
For in vivo experimental pulmonary metastasis assays, B16F10 melanoma cells (1 × 105 cells/mouse) were injected into a tail vein of C57BL/6 mice (n = 10) on day 0. On day 13, lungs were excised, and metastatic nodules were photographed.
Biopsy
Two millimeter punch biopsies were obtained from patients who gave their informed consent in accordance with the policy of the Catholic Research Medical Institutional Review Board. The punch biopsies were taken from the malignant lesions and normal dermis of patients who visited the out-patient clinic. Biopsies were immediately frozen in the liquid nitrogen and kept at −70 °C until used for further analysis.
Immunohistochemistry
Five sets of biopsies from patients were fixed in 10% neutralized buffered formalin for 24 h, embedded in paraffin, sectioned, and deparaffinized for staining. The sections were stained with hematoxylin and eosin. For immunostaining, the sections were rinsed, and endogenous peroxidase activity was blocked with 3% hydrogen peroxide and 10% methanol in PBS, followed by preincubation in PBS containing 0.2% Triton X-100 (PBS-T) and 3% bovine serum albumin for 2 h. The sections were then incubated for 16 h in PBS-T containing either anti-SDC2 antibody (1:1000) or anti-melanosome HMB-45 antibody (1:50). Following a wash with PBS, they were then rinsed and incubated with biotinylated secondary antibody (1:1,000; Dako, Denmark) in PBS-T containing 3% bovine serum albumin for 60 min at room temperature. The sections were subsequently rinsed and incubated with peroxidase-labeled streptavidin (Dako) for 60 min, followed by reaction with diaminobenzidine (0.5 mg/ml) and hydrogen peroxide. After rinsing, the sections were mounted, dehydrated, and covered with coverslips. All images were acquired with a Carl Zeiss Axioskop2.
Wound-healing Assay
Melanoma cells were plated 60-mm tissue culture plates and incubated overnight. After one streak was made in the middle of the monolayer using a sterile pipette tip, the cells were washed twice with PBS and cultured in fresh media. Pictures of the wound distance were taken at various time points.
Measurement of Melanin Content
Melanin contents were measured as reported by Kazuomi et al. (26). Briefly, cells were treated with either 1.0 μm α-MSH or 200 μm Kojic acid in DMEM containing 10% heat-inactivated FBS for 48 h and then detached by incubation with trypsin/EDTA. After pelleting, cells were resuspended in DMEM for cell counting or solubilized into boiling 2 m NaOH for 20 min. After cooling to room temperature, the absorbance of the solution was measured at 405 nm for melanin content determination.
Cell Proliferation Assay
Cell proliferation was measured by a colorimetric assay using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide as described (3).
RESULTS
Syndecan-2 Is Up-regulated in Melanoma Cells
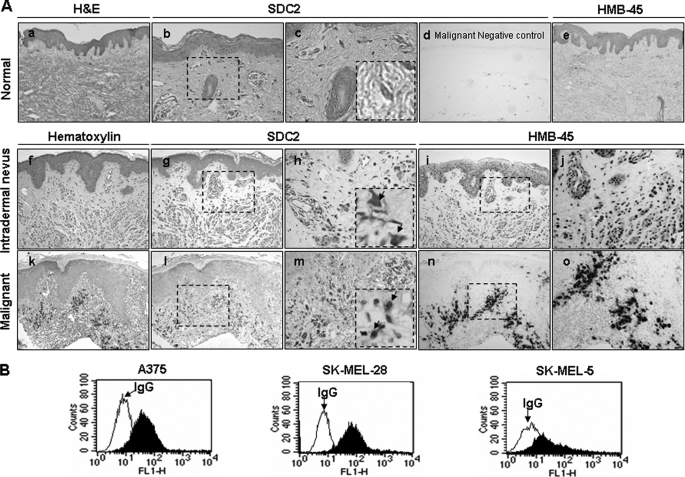
To investigate the role of syndecan-2 in the development of human skin melanoma, the expression of syndecan-2 was compared in tissues obtained from cutaneous malignancies and normal skin. Immunohistochemical studies revealed the absence of syndecan-2 expression in normal skin (Fig. 2A, b and c). On the other hand, we found that there was increased syndecan-2 expression in intradermal nevus (Fig. 2A, g and h). Interestingly, syndecan-2 expression was significantly increased in malignant melanomas (Fig. 2A, l and m) stained positively with melanoma marker HMB-45 in the adjacent slide (Fig. 2A, e, i, j, n, and o), whereas preimmune serum showed no staining except for the melanin granules in the malignant melanoma (Fig. 2A, d). Consistent with this observation, elevated cell surface expression of syndecan-2 was observed in human melanoma cell lines (Fig. 2B). These data suggest that syndecan-2 expression is increased in human melanoma cells.
FIGURE 2.
Syndecan-2 is highly expressed in human skin melanoma cells. A, paraffin-embedded tissue sections were either stained with hematoxylin and eosin (H&E) (a) or hematoxylin (f and k) or immunostained with an anti-syndecan-2 antibody (SDC2) (b, g, and l). d, negative control for immunostaining with the preimmune serum. The immunoreactivities were visualized by 3,3′-diaminobenzidine (DAB) intensified with nickel. c, h, and m, higher magnifications of b, g, and l, respectively. Immunostaining with an anti-HMB-45 antibody was detected using the same methods (e, i, and n). j and o are higher magnifications of i and n. The arrowheads indicate the melanocyte with immunoreactivities of syndecan-2 and HMB-45, respectively. Representative images from one of five experiments are shown. B, cells from human melanoma cell lines were incubated with anti-syndecan-2 antibody, and protein expression level was analyzed by flow cytometry. IgG was used as the control.
Syndecan-2 Regulates Melanoma Cell Migration
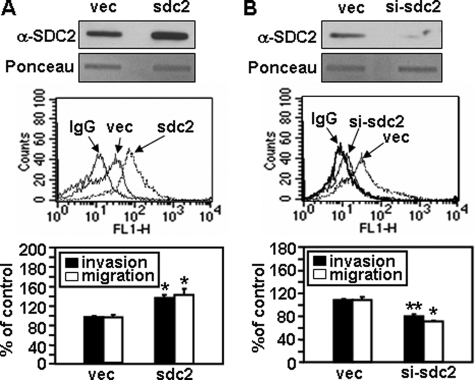
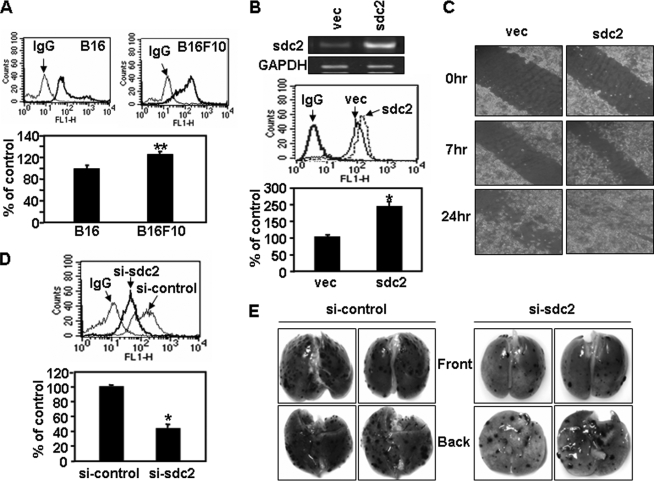
Syndecan-2 is known to play a critical role as an adhesion receptor during cell migration. To more directly assess the role of syndecan-2 in the regulation of tumorigenic activity, we examined the effects of syndecan-2 expression on melanoma cell migration. Human melanoma A375 cells were transfected with sdc2 (syndecan-2) cDNA, and a transwell migration assay was performed (Fig. 3A). After transfection with sdc2, both cell migration and invasion were significantly higher in A375 cells than in vector-transfected cells (vec in Fig. 3). In contrast, cell migration/invasion was decreased when syndecan-2 expression was suppressed by small interfering RNAs (siRNAs) (Fig. 3B). Consistent with the human melanoma cells, elevated cell surface expression of syndecan-2 was also observed in mouse melanoma cell lines (Fig. 4). Interestingly, B16F10 cells expressing higher levels of syndecan-2 migrated faster than B16 cells (Fig. 4A). Syndecan-2 overexpression enhanced the migration of mouse melanoma B16 cells, based on transwell migration (Fig. 4B) and wound healing assays (Fig. 4C), whereas cell migration decreased when syndecan-2 expression was suppressed by siRNAs (Fig. 4D). Similar results were observed using B16F10 cells (data not shown). Consistently, knockdown of syndecan-2 expression by siRNA reduced metastatic potential of B16F10 melanoma cells in mice (Fig. 4E). These findings strongly suggest that syndecan-2 regulates cell migration in melanoma cells.
FIGURE 3.
Syndecan-2 regulates human melanoma cell migration. A375 cells were transfected with 2 μg of either empty vector (vec) or vectors encoding sdc2 (A) and either pSUPER vector (vec) or si-syndecan-2 RNA (si-sdc2) (B). Total cell lysates were analyzed by slot blotting with an anti-syndecan-2 antibody (top). Cells were incubated with an anti-syndecan-2 antibody, and the cell surface expression levels of syndecan-2 were analyzed by flow cytometry. IgG was used as a control (middle). Transwell migration assays were done with the indicated A375 cells using FGF-2 as a chemoattractant in the lower chamber. Cells (5 × 104) were allowed to migrate on 10 μg/ml gelatin-coated Transwell plates for 12 h. After fixing and staining with 0.6% hematoxylin and 0.5% eosin, the number of migrated cells was counted. For an invasion assay, cells were loaded onto the upper compartments of Matrigel (30 μg/ml)-coated plates and incubated for 12 h (bottom). Columns, average of at least three independent experiments. *, p < 0.01; **, p < 0.05 versus control.
FIGURE 4.
Syndecan-2 regulates mouse melanoma cell migration. A, either B16 or B16F10 cells were incubated with an anti-syndecan-2 antibody, and the cell surface expression levels of syndecan-2 were analyzed by flow cytometry. IgG was used as a control (top). Cells (5 × 104) were loaded to migrate on Transwell plates, as described in the legend to Fig. 3 (bottom). B, B16 cells were transfected with sdc2 cDNA. Total RNA was extracted, and expression of syndecan-2 was analyzed by reverse transcription-PCR. Glyceraldehyde-3-phosphate dehydrogenase mRNA was used as the loading control (top). Cells were incubated with an anti-syndecan-2 antibody, and the cell surface expression levels of syndecan-2 were analyzed by flow cytometry. IgG was used as a control (middle). Transwell migration assays were done with B16 cells transfected with 2 μg of the indicated cDNA, as described in the legend to Fig. 3 (bottom). C, B16 cells were transfected with sdc2 cDNA, and 24 h later, wound-healing assays were performed. One streak was made in the middle of the monolayer using a sterile pipette tip and pictures of the wound distance were taken at each time point. D, B16 cells were transfected with syndecan-2 siRNA. Cell surface expression levels of syndecan-2 were analyzed by flow cytometry. IgG was used as a control (top). Cells (5 × 104) were allowed to migrate on Transwell plates as described in the legend to Fig. 3 (bottom). E, B16F10 cells were transfected with syndecan-2 siRNA. A metastasis assay was performed as detailed under “Experimental Procedures.” Two independent experiments were performed, and each experiment was conducted using three mice per group. Two representative photographs of both the front and back side of each lung are shown. Columns, average of three independent experiments. *, p < 0.01; **, p < 0.05 versus control.
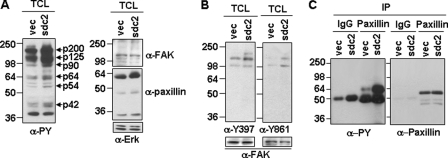
FAK is known to play a pivotal role in cancer cell migration (27, 28). Accordingly, we found that total cellular tyrosine phosphorylation was enhanced in sdc2-overexpressing B16 melanoma cells. In particular, tyrosine phosphorylation of seven different polypeptides (i.e. p200, p125, p90, p64, p54, p44, and p42) was higher in cells transfected with sdc2 than in cells transfected with the control vector (Fig. 5A, left). Since enhanced tyrosyl phosphorylation mediated by FAK often happens in response to stimulation of cell migration (29), the same membrane was stripped and reprobed with antibodies against FAK or paxillin, a well known substrate of FAK (30). FAK was detected at the position of p125, and paxillin was detected at the position of p64 (Fig. 5A, right), suggesting involvement of FAK tyrosine phosphorylation. Indeed, sdc2 expression increased autophosphorylation at tyrosine 397 (i.e. Tyr397), phosphorylation of FAK at tyrosine 861 (known to regulate cell transformation) (Fig. 5B), and tyrosyl phosphorylation of paxillin (Fig. 5C). These data indicate that syndecan-2-induced migration activity is intimately associated with the activation of FAK-mediated signaling.
FIGURE 5.
Focal adhesion kinase is involved in the regulation of syndecan-2-mediated melanoma cell migration. A, B16 cells transfected with either the empty vector (vec) or sdc2 cDNA were lysed with radioimmune precipitation buffer and analyzed by Western blotting with anti-phosphotyrosine (α-PY) antibody (left). The same membrane was stripped and reprobed with antibodies against FAK (α-FAK), paxillin (α-paxillin), or Erk (α-Erk). B, B16 cells were lysed, and site-specific FAK phosphorylation was analyzed with anti-FAK(Tyr(P)397) and anti-FAK(Tyr(P)861) antibodies, followed by stripping and reprobing with anti-FAK antibody. C, B16 cell extracts were immunoprecipitated with an antibody against paxillin, and the phosphorylation state of proteins was determined by immunoblotting anti-phosphotyrosine (PY) antibody. The levels of protein in each immunoprecipitate were determined by immunoblotting with anti-paxillin antibody.
Syndecan-2 Expression Correlates with Melanoma Cell Migration
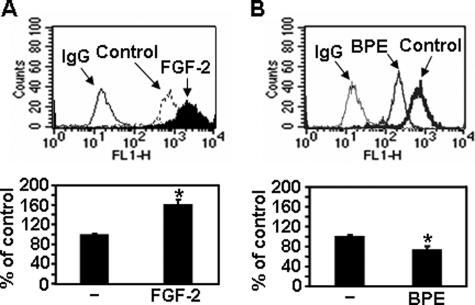
The previous study demonstrated the stimulatory effects of FGF-2 on melanoma cell migration (31). In parallel with increased cell migration, FGF-2 caused increased cell surface expression of syndecan-2 (Fig. 6A), suggesting that syndecan-2 expression correlates with FGF-2-enhanced cell migration. BPE is routinely used as a mitogenic supplement in growth media for human melanocytes (32). In contrast to FGF-2 treatment, BPE treatment reduced cell surface expression of syndecan-2 and melanoma cell migration (Fig. 6B), supporting the idea that syndecan-2 expression correlates with melanoma cell migration.
FIGURE 6.
Syndecan-2 expression correlates with melanoma cell migration. B16 cells were treated with either FGF-2 (300 ng/ml) (A) or BPE (0.4% v/v) (B) for 24 h. Cells were incubated with anti-syndecan-2 antibody, and the protein expression level was analyzed by flow cytometry. IgG was used as the control (top). B16 cells (5 × 104) treated either with BPE or FGF-2 were allowed to migrate on Transwell plates as described in the legend to Fig. 3 (bottom). Columns, average of three independent experiments. *, p < 0.01 versus control.
α-MSH Inhibits Melanoma Cell Migration through Reduced Expression of Syndecan-2
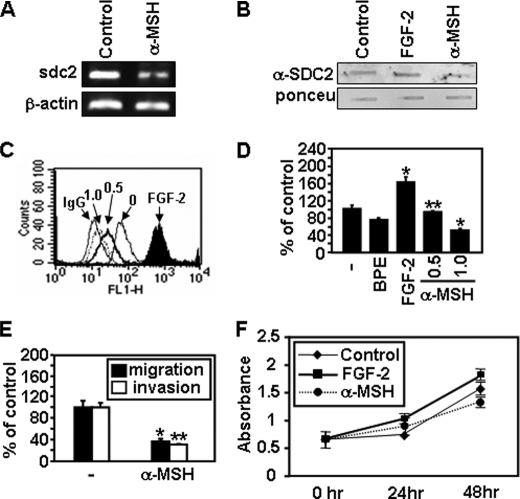
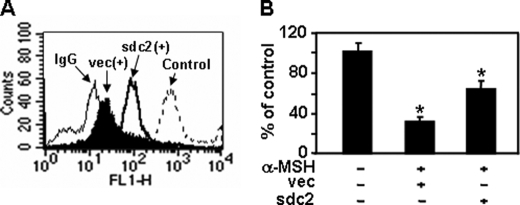
MSH are a class of peptides produced by cells in the intermediate lobe of the pituitary gland (33). These peptides are known to inhibit murine melanoma cell migration (26). Since BPE contains α-MSH, we investigated whether α-MSH inhibits syndecan-2 expression in melanoma cells. As expected, α-MSH decreased mRNA and cell surface protein expression of syndecan-2 (Fig. 7, A–C). Consistent with reduced syndecan-2 expression, α-MSH caused reduced migration and invasion of melanoma cells (Fig. 7, D and E), whereas cell proliferation was not significantly affected by α-MSH treatment during 24 h (Fig. 7F). Furthermore, syndecan-2 overexpression rescued the migration defects induced by α-MSH treatment (Fig. 8). Collectively, these results suggest that α-MSH inhibits melanoma cell migration through reduced expression of syndecan-2 and support the idea that syndecan-2 expression plays a crucial role in melanoma cell migration.
FIGURE 7.
α-MSH inhibits expression of syndecan-2 paralleled with reduced cell migration. A, B16 cells were treated with α-MSH (1.0 μm) for 24 h. Total RNA was extracted, and expression of syndecan-2 was analyzed by reverse transcription-PCR. β-Actin mRNA was used as the loading control. B, B16 cells were treated with either FGF-2 (300 ng/ml) or α-MSH (1.0 μm). After 24 h, total cell lysates were analyzed by slot blotting. C, B16 cells treated with the indicated amount of α-MSH (μm) for 24 h were incubated with anti-syndecan-2 antibody, and the protein expression level was analyzed by flow cytometry. IgG was used as the negative control. D, B16 cells (5 × 104) pretreated with either FGF-2 (300 ng/ml) or α-MSH (0.5 μm or 1.0 μm) for 24 h were allowed to migrate on Transwell plates as described in the legend to Fig. 3. Columns, average of three independent experiments. E, B16 cells were pretreated with α-MSH (1.0 μm) for 24 h. Transwell cell migration or invasion assay was performed as described in Fig. 3. F, B16 cells were treated with either FGF-2 (300 ng/ml) or α-MSH (1.0 μm), and then 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays were performed. Columns, average of three independent experiments. *, p < 0.01; **, p < 0.05 versus control.
FIGURE 8.
α-MSH inhibits melanoma cell migration through reduced expression of syndecan-2. B16 cells transfected with either the empty vector or sdc2 cDNA were treated with α-MSH (1.0 μm) for 24 h. Syndecan-2 expression levels (A) and cell migration (B) were analyzed as described in the legend to Fig. 3. Columns, average of three independent experiments. *, p < 0.01 versus control.
Syndecan-2-mediated Cell Migration Occurs Independently of Melanin Synthesis
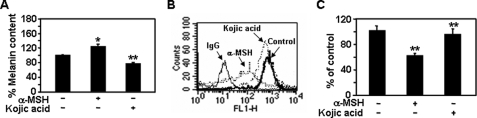
The production of melanin pigments is stimulated by α-MSH in the skin, hair, and eye (34). α-MSH stimulates up-regulation of intracellular cAMP and sequential activation of various pigmentation enzymes (e.g. tyrosinase, which catalyzes a rate-limiting step in melanin synthesis) (35). Therefore, we investigated whether melanin synthesis plays a role in the syndecan-2-mediated cell migration of melanoma cells. Consistent with previous findings (36), α-MSH enhanced the melanin content of melanoma cells (i.e. by ∼121%), whereas melanin production was reduced by treatment with the tyrosinase inhibitor, kojic acid (i.e. by ∼76%) (Fig. 9A) (26). In contrast, syndecan-2 expression decreased in response to treatment with α-MSH but did not change in response to kojic acid (Fig. 9B). Accordingly, α-MSH-mediated reductions in cell migration corresponded with reduced syndecan-2 expression, whereas inhibition of tyrosinase activity did not influence melanoma cells migration (Fig. 9C). Therefore, it appears that α-MSH regulates syndecan-2-mediated cell migration independent of the melanin biosynthesis pathway.
FIGURE 9.
Syndecan-2-mediated cell migration is independent of melanin synthesis. A, after B16F10 cells were treated with either α-MSH (1.0 μm) or kojic acid (200 μM) for 48 h, the melanin content of the cells was measured as described under “Experimental Procedures.” Cell surface expression of syndecan-2 (B) and cell migration activity (C) were analyzed as described in the legend to Fig. 3. Columns, average of three independent experiments. *, p < 0.05; **, p < 0.01 versus control.
DISCUSSION
Cell migration is a critical step in tumor invasion and metastasis, two major causes of death among cancer patients. We previously reported that elevated syndecan-2 expression is crucial for tumorigenic activity in colon carcinoma and fibrosarcoma cells (3, 29). Here, we provide further evidence for the tumorigenic activity of syndecan-2 using melanoma, one of the most highly invasive types of cancers. Consistent with our previous findings in colon carcinoma and fibrosarcoma cells, we identified syndecan-2 expression in tissue samples from both nevus and malignant human melanomas but not in normal melanocytes (Fig. 2A). Similarly, syndecan-2 expression was elevated in all melanoma cell lines tested (Fig. 2B). Increased syndecan-2 expression correlates with increased melanoma cell migration, whereas the opposite effects were observed when syndecan-2 expression was suppressed using siRNAs (Figs. 3 and 4). Therefore, syndecan-2 appears to act as a general protumorigenic receptor in cancer cells.
Although melanoma cell migration is associated with syndecan-2 expression, little is known about the mechanism of syndecan-2-mediated regulation of melanoma cell migration. Our results indicate that enhanced FAK signaling plays a role in the regulation of syndecan-2-mediated cell migration. Overexpression of syndecan-2 increased phosphorylation of FAK and its substrate, paxillin (Fig. 5). Boukerche et al. (37) recently reported that the invasion and metastatic spread of melanoma cells is regulated by elevated syntenin expression and enhanced via FAK- and p38-induced activation of NF-κB. Syntenin is an adaptor molecule that binds to the cytoplasmic domain of syndecans; thus, it also may play an important role in the syndecan-2-mediated migration of melanoma cells.
Melanocytes and melanoma cells may differ with regard to the cell surface expression of syndecan-2 that appears to play a critical role in the migration of melanoma cells. Syndecan-1 is required for the induction of mammary tumor formation by the proto-oncogene, Wnt-1, which mediates expression of oncogenes, such as Tcf-1, LEF1, Axin-2, and matrix metalloproteinase 7 (38). However, we found that Wnt3a did not influence syndecan-2 expression (data not shown), implying that the Wnt/β-catenin pathway is not involved in the regulation of syndecan-2-mediated cell migration. Interestingly, syndecan-2 expression was regulated by α-MSH, a pituitary-derived peptide that stimulates melanin production. Previous studies have found that increased α-MSH levels are associated with melanoma progression (34, 39). However, α-MSH is also known to reduce the abilities of human cutaneous and ocular melanoma cells to invade the ECM (40) and is a potent inhibitor of invasion by highly invasive B16-BL6 murine melanoma cells (39, 41). Our findings suggest that α-MSH negatively regulate melanoma cell migration via reduced syndecan-2 expression (Fig. 7). Indeed, FGF-2 up-regulated syndecan-2 expression and increased melanoma cell migration (Fig. 6), whereas BPE or α-MSH down-regulated syndecan-2 expression and reduced melanoma cell migration (Figs. 6–8). Although the exact role for α-MSH in melanoma cell migration remains unclear, our findings suggest that regulation syndecan-2 expression by α-MSH plays a crucial role in the migration of melanoma cells. Recent clinical studies have shown that a potent synthetic analogue of α-MSH, NDP-MSH, significantly increases the eumelanin content of human skin (42). Interestingly, α-MSH-mediated syndecan-2 expression occurs independently of melanin synthesis (Fig. 9). Therefore, α-MSH may play independent roles in the regulation of syndecan-2 expression, in cell migration, and in the melanin biosynthesis pathway. In addition, it has been known that α-MSH suppresses melanoma cell migration induced by the inflammatory cytokine tumor necrosis factor α (43). It has been also known that α-MSH inhibits the production of MMP-2 and MMP-9 and invasion of Matrigel by B16 melanoma cells (36). Therefore, we cannot exclude the possibility that some other factors, unrelated to reduced syndecan-2 expression, could contribute to some extent to reduced cell migration via α-MSH.
In summary, syndecan-2, a cell surface heparan sulfate proteoglycan, plays a critical role in regulating the migratory potential of melanoma cells. In addition, α-MSH negatively regulates the expression of syndecan-2 independently of the melanin biosynthesis pathway and reduces concomitant cell migration. Since the treatment of melanoma remains extremely challenging, our evidence regarding the role of syndecan-2 in melanoma cell migration merits further investigation. These findings may provide clues as to the metastatic potential of the cancer and may pave the way for the development of novel therapies to reduce the metastasis of melanoma cells.
This study was supported by a grant of the Korea Healthcare Technology R&D Projects, Ministry for Health, Welfare, and Family Affairs, Republic of Korea (A084292).
- ECM
- extracellular matrix
- MSH
- melanocyte-stimulating hormone(s)
- FGF
- fibroblast growth factor
- DMEM
- Dulbecco's modified Eagle's medium
- FAK
- focal adhesion kinase
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- mAb
- monoclonal antibody
- BPE
- bovine pituitary extract
- siRNA
- small interfering RNA.
REFERENCES
- 1.Bass M. D., Humphries M. J. (2002) Biochem. J. 368, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sebestyén A., Tótth A., Mihalik R., Szakács O., Paku S., Kopper L. (2000) Tumour Biol. 21, 349–357 [DOI] [PubMed] [Google Scholar]
- 3.Park H., Kim Y., Lim Y., Han I., Oh E. S. (2002) J. Biol. Chem. 277, 29730–29736 [DOI] [PubMed] [Google Scholar]
- 4.Purushothaman A., Chen L., Yang Y., Sanderson R. D. (2008) J. Biol. Chem. 283, 32628–32636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras H. R., Fabre M., Granés F., Casaroli-Marano R., Rocamora N., Herreros A. G., Reina M., Vilaró S. (2001) Biochem. Biophys. Res. Commun. 286, 742–751 [DOI] [PubMed] [Google Scholar]
- 6.Munesue S., Kusano Y., Oguri K., Itano N., Yoshitomi Y., Nakanishi H., Yamashina I., Okayama M. (2002) Biochem. J. 363, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beauvais D. M., Rapraeger A. C. (2004) Reprod. Biol. Endocrinol. 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim Y., Park H., Lim Y., Han I., Kwon H. J., Woods A., Oh E. S. (2003) Oncogene 22, 826–830 [DOI] [PubMed] [Google Scholar]
- 9.Modrowski D., Baslé M., Lomri A., Marie P. J. (2000) J. Biol. Chem. 275, 9178–9185 [DOI] [PubMed] [Google Scholar]
- 10.Kusano Y., Oguri K., Nagayasu Y., Munesue S., Ishihara M., Saiki I., Yonekura H., Yamamoto H., Okayama M. (2000) Exp. Cell Res. 256, 434–444 [DOI] [PubMed] [Google Scholar]
- 11.Wang H. B., Dembo M., Hanks S. K., Wang Y. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11295–11300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan J. L. (1997) Matrix Biol. 16, 195–200 [DOI] [PubMed] [Google Scholar]
- 13.Hauck C. R., Sieg D. J., Hsia D. A., Loftus J. C., Gaarde W. A., Monia B. P., Schlaepfer D. D. (2001) Cancer Res. 61, 7079–7090 [PubMed] [Google Scholar]
- 14.Nackaerts K., Verbeken E., Deneffe G., Vanderschueren B., Demedts M., David G. (1997) Int. J. Cancer 74, 335–345 [DOI] [PubMed] [Google Scholar]
- 15.Agochiya M., Brunton V. G., Owens D. W., Parkinson E. K., Paraskeva C., Keith W. N., Frame M. C. (1999) Oncogene 18, 5646–5653 [DOI] [PubMed] [Google Scholar]
- 16.Jayson G. C., Vives C., Paraskeva C., Schofield K., Coutts J., Fleetwood A., Gallagher J. T. (1999) Int. J. Cancer 82, 298–304 [DOI] [PubMed] [Google Scholar]
- 17.Fears C. Y., Gladson C. L., Woods A. (2006) J. Biol. Chem. 281, 14533–14536 [DOI] [PubMed] [Google Scholar]
- 18.Markovic S. N., Erickson L. A., Rao R. D., Weenig R. H., Pockaj B. A., Bardia A., Vachon C. M., Schild S. E., McWilliams R. R., Hand J. L., Laman S. D., Kottschade L. A., Maples W. J., Pittelkow M. R., Pulido J. S., Cameron J. D., Creagan E. T. (2007) Mayo Clin. Proc. 82, 364–380 [DOI] [PubMed] [Google Scholar]
- 19.Hocker T. L., Singh M. K., Tsao H. (2008) J. Invest. Dermatol. 128, 2575–2595 [DOI] [PubMed] [Google Scholar]
- 20.Hess A. R., Postovit L. M., Margaryan N. V., Seftor E. A., Schneider G. B., Seftor R. E., Nickoloff B. J., Hendrix M. J. (2005) Cancer Res. 65, 9851–9860 [DOI] [PubMed] [Google Scholar]
- 21.Stylli S. S., Kaye A. H., Lock P. (2008) J. Clin. Neurosci. 15, 725–737 [DOI] [PubMed] [Google Scholar]
- 22.Linder S., Aepfelbacher M. (2003) Trends Cell Biol. 13, 376–385 [DOI] [PubMed] [Google Scholar]
- 23.Nakahara H., Nomizu M., Akiyama S. K., Yamada Y., Yeh Y., Chen W. T. (1996) J. Biol. Chem. 271, 27221–27224 [DOI] [PubMed] [Google Scholar]
- 24.Mueller S. C., Ghersi G., Akiyama S. K., Sang Q. X., Howard L., Pineiro-Sanchez M., Nakahara H., Yeh Y., Chen W. T. (1999) J. Biol. Chem. 274, 24947–24952 [DOI] [PubMed] [Google Scholar]
- 25.Shively J. E., Conrad H. E. (1976) Biochemistry 15, 3932–3942 [DOI] [PubMed] [Google Scholar]
- 26.Sato K., Takahashi H., Iraha R., Toriyama M. (2008) Biol. Pharm. Bull. 31, 33–37 [DOI] [PubMed] [Google Scholar]
- 27.Owens L. V., Xu L., Dent G. A., Yang X., Sturge G. C., Craven R. J., Cance W. G. (1996) Ann. Surg. Oncol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 28.Owens L. V., Xu L., Craven R. J., Dent G. A., Weiner T. M., Kornberg L., Liu E. T., Cance W. G. (1995) Cancer Res. 55, 2752–2755 [PubMed] [Google Scholar]
- 29.Park H., Han I., Kwon H. J., Oh E. S. (2005) Cancer Res. 65, 9899–9905 [DOI] [PubMed] [Google Scholar]
- 30.Schaller M. D. (2001) Oncogene 20, 6459–6472 [DOI] [PubMed] [Google Scholar]
- 31.Meier F., Caroli U., Satyamoorthy K., Schittek B., Bauer J., Berking C., Möller H., Maczey E., Rassner G., Herlyn M., Garbe C. (2003) Exp. Dermatol. 12, 296–306 [DOI] [PubMed] [Google Scholar]
- 32.Donatien P., Surlève-Bazeille J. E., Thody A. J., Taïeb A. (1993) Arch. Dermatol. Res. 285, 385–392 [DOI] [PubMed] [Google Scholar]
- 33.Böhm M., Luger T. A. (2004) Hautarzt 55, 436–445 [DOI] [PubMed] [Google Scholar]
- 34.Price E. R., Horstmann M. A., Wells A. G., Weilbaecher K. N., Takemoto C. M., Landis M. W., Fisher D. E. (1998) J. Biol. Chem. 273, 33042–33047 [DOI] [PubMed] [Google Scholar]
- 35.Passeron T., Valencia J. C., Bertolotto C., Hoashi T., Le Pape E., Takahashi K., Ballotti R., Hearing V. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13984–13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata J., Ayukawa K., Ogasawara M., Fujii H., Saiki I. (1997) Invasion Metastasis 17, 82–93 [PubMed] [Google Scholar]
- 37.Boukerche H., Su Z. Z., Emdad L., Sarkar D., Fisher P. B. (2007) Cancer Res. 67, 1812–1822 [DOI] [PubMed] [Google Scholar]
- 38.Alexander C. M., Reichsman F., Hinkes M. T., Lincecum J., Becker K. A., Cumberledge S., Bernfield M. (2000) Nat. Genet. 25, 329–332 [DOI] [PubMed] [Google Scholar]
- 39.Nagahama M., Funasaka Y., Fernandez-Frez M. L., Ohashi A., Chakraborty A. K., Ueda M., Ichihashi M. (1998) Br. J. Dermatol. 138, 981–985 [DOI] [PubMed] [Google Scholar]
- 40.Liu P. Y., Johansson O. (1995) J. Dermatol. Sci. 10, 203–212 [DOI] [PubMed] [Google Scholar]
- 41.Kameyama K., Vieira W. D., Tsukamoto K., Law L. W., Hearing V. J. (1990) Int. J. Cancer 45, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 42.Dorr R. T., Ertl G., Levine N., Brooks C., Bangert J. L., Powell M. B., Humphrey S., Alberts D. S. (2004) Arch. Dermatol. 140, 827–835 [DOI] [PubMed] [Google Scholar]
- 43.Zhu N., Lalla R., Eves P., Brown T. L., King A., Kemp E. H., Haycock J. W., MacNeil S. (2004) Br. J. Cancer 90, 1457–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]