Abstract
Legionella pneumophila possesses several phospholipases capable of host cell manipulation and lung damage. Recently, we discovered that the major cell-associated hemolytic phospholipase A (PlaB) shares no homology to described phospholipases and is dispensable for intracellular replication in vitro. Nevertheless, here we show that PlaB is the major lipolytic activity in L. pneumophila cell infections and that PlaB utilizes a typical catalytic triad of Ser-Asp-His for effective hydrolysis of phospholipid substrates. Crucial residues were found to be located within the N-terminal half of the protein, and amino acids embedding these active sites were unique for PlaB and homologs. We further showed that catalytic activity toward phosphatidylcholine but not phosphatidylglycerol is directly linked to hemolytic potential of PlaB. Although the function of the prolonged PlaB C terminus remains to be elucidated, it is essential for lipolysis, since the removal of 15 amino acids already abolishes enzyme activity. Additionally, we determined that PlaB preferentially hydrolyzes long-chain fatty acid substrates containing 12 or more carbon atoms. Since phospholipases play an important role as bacterial virulence factors, we examined cell-associated enzymatic activities among L. pneumophila clinical isolates and non-pneumophila species. All tested clinical isolates showed comparable activities, whereas of the non-pneumophila species, only Legionella gormanii and Legionella spiritensis possessed lipolytic activities similar to those of L. pneumophila and comprised plaB-like genes. Interestingly, phosphatidylcholine-specific phospholipase A activity and hemolytic potential were more pronounced in L. pneumophila. Therefore, hydrolysis of the eukaryotic membrane constituent phosphatidylcholine triggered by PlaB could be an important virulence tool for Legionella pathogenicity.
Bacterial phospholipases are involved in many disease-promoting processes ranging from production of bioactive molecules influencing cellular signal transduction pathways to formation of membrane pores and depletion of essential components from lipid layers (1–3). They often induce massive membrane destruction, as shown for Pseudomonas aeruginosa type III secreted cytotoxin ExoU, a phospholipase A (PLA)2 /lysophospholipase A (LPLA), or Clostridium perfringens α- toxin, a phospholipase C (4–8). We recently identified PlaB, the major cell-associated PLA/LPLA of Legionella pneumophila, to be hemolytic and thereby a membrane-destroying enzyme (9). Accordingly, activity of PlaB fits into the picture of Legionnaires' disease, the severe pneumonia caused by L. pneumophila bacteria, showing loss of a functional epithelial cell layer in the lung as well as degradation of lung macrophages in infected individuals (10, 11). Now it is indeed clear that PlaB contributes to pathogenesis in a guinea pig infection model (1),3 although it is dispensable for intracellular replication in macrophage and amoeba host cells (1, 9).3 This clearly underlines the importance of PlaB as a virulence factor of L. pneumophila; however, its mechanism of action has not yet been studied in detail.
PlaB was initially detected by screening an L. pneumophila genomic library in Escherichia coli, thus yielding a clone with remarkable hemolytic activity on human red blood agar (9). Subsequently, plaB insertion mutagenesis revealed that the gene encodes the most prominent cell-associated PLA/LPLA activity, being ∼100-fold more active than secreted phospholipolytic activities of L. pneumophila. Interestingly, PlaB has no significant protein homology to any established lipase/phospholipase at all. Further, considering the putative domains responsible for catalytic activity, it does not align significantly to any defined class of lipolytic enzymes. As proposed by Arpigny and Jaeger (12), lipases can be divided into eight families characterized by conserved amino acid motifs. Members of those families in general exhibit a conserved pentapeptide of GXSXG surrounding the catalytically important serine residue except of proteins belonging to GDSL hydrolases, which harbor the catalytic active serine within a differing GDSL motif (13). Further, lipases or phospholipases usually constitute a catalytic triad of serine, aspartate, and histidine; however, combinations of Ser-Glu-His or catalytic dyads have been reported as well (12, 14, 15). The latter one is present within a recently discovered group of bacterial lipolytic enzymes, the patatin-like proteins (16, 17). Although variability in amino acid homology between phospholipases is quite elevated, we propose PlaB and homologs to represent a novel division of lipolytic proteins.
To understand the biochemical nature of L. pneumophila PlaB, in this study we (a) determined expression of PlaB during host cell infection, (b) identified the catalytically important residues and further protein domains crucial for lipid hydrolysis, (c) analyzed the contribution of the catalytic and other important sites to hemolytic activity, and (d) investigated lipid substrate specificity with respect to fatty acid chain length. We additionally addressed the questions of whether cell-associated phospholipase activity is found in various clinical isolates and whether PlaB is unique to L. pneumophila or also present within non-pneumophila species.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
L. pneumophila, environmental and clinical isolates, and different E. coli strains used in this study are listed in Table 1 and were grown as described previously (9, 18).
TABLE 1.
Bacterial strains and plasmids used in this study
Complementation of the plaB Deletion Mutant
Amplification of L. pneumophila Corby plaB (lpc_1029) was carried out by using primers plaB_SalIF and plaB_EagIr (oligonucleotides were purchased from TIB MOLBIOL (Berlin) or Eurofins MWG Operon (Ebersberg)). The resulting fragment was cloned into SalI and EagI (New England Biolabs) restricted pBCKS vector to yield pJB04. This additional complementation vector was constructed because the plaB gene within pKH192 harbors an amino acid conversion, although not influencing lipolytic activities.
Site-directed Mutagenesis
Mutagenesis of important amino acids was carried out by utilizing the combined chain reaction method, as described elsewhere (19). Minor modifications included performance of PCR with 20 ng of each primer and 20 ng of pJB04 plasmid DNA (Table 1), a 0.25 mm concentration of each dNTP, 2.5 units of Pfu polymerase (Fermentas), and 3 units of Ampligase (Epicenter). Mutagenesis primers were phosphorylated in advance by T4 polynucleotide kinase (New England Biolabs). Plasmid pJB04, carrying wild-type plaB, served as template for single amino acid exchanges, whereas pJB06, which harbors already plaB S85A, was used for creating loss of function double mutants S85A/D203N and S85A/H251N. Amplification of the designated fragments was performed by using plaB_SalIF and plaB_EagIr primers and the appropriate mutagenesis primer (Table 2). Restriction endonucleases SalI and EagI were used for site-specific integration of the desired fragments into vector pBCKS. Mutants were verified by sequencing both DNA strands, and resulting plasmids were electroporated into L. pneumophila plaB mutant strain. Table 1 gives a summary of all generated plasmids and respective plaB mutants.
TABLE 2.
Primers used for site-directed mutagenesis, cloning, and sequence analysis
Construction of C-terminally Truncated Versions of PlaB
For construction of full-length glutathione S-transferase-PlaB fusion protein, pGEX-5X-1 vector backbone was linearized by EcoRI and EagI restriction enzymes and ligated to the appropriate digested plaB fragment, amplified by using gene-specific primers plaB_EagIr and plaB_Ecof2 (Table 2). Truncated versions were cloned into vector pGEX-6P-1. Genes for PlaB-(1–459) and PlaB-(1–388) were amplified by PlaB_truncC1_P1 and PlaB_truncC2_P2, respectively, in combination with plaB_TOPf2. Both fragments were inserted over SmaI restriction sites into vector pGEX-6P-1. The truncated version PlaB-(1–307) was amplified by means of plaB_Ecof2 and plaB_trunc3Eag primers and ligated to pGEX-6P-1 via EcoRI and EagI restriction sites. All clones were verified by sequencing before electroporation into protein expression host E. coli BL21.
Cloning of Legionella spiritensis and Legionella gormanii plaB
The plaB gene from L. spiritensis was amplified from genomic DNA using primers plaB_pBADf and plaB_c1-r. Subsequently, the product was cloned into pGEMT-EZ, resulting in vector pKR8 before primers PlaBspir_BamHI and PlaBspir_NotI were used to precisely ligate the amplicon into digested pBCKS backbone vector, yielding pJB37 (Tables 1 and 2). plaB gene sequences from strains L. spiritensis and partially for L. gormanii were determined from pKR8 and pKR10, respectively. The latter vector resulted from an amplification product generated from L. gormanii genomic DNA by using primers plaB_pBADf and plaB_g1_r and cloned into pGEMT-EZ.
Preparation of Culture Supernatants and Cell Lysates and Determination of Lipolytic Activities
Samples were obtained from cultures grown to an A660 of 2. For assessment of lipolytic activities, via release of fatty acids, bacterial cell lysates and supernatants were prepared and incubated with deduced lipids as described previously (9). Briefly, incubation of bacterial cell lysates or supernatants with different lipids (dipalmitoylphosphatidylcholine (DPPC), dipalmitoylphosphatidylglycerol (DPPG), 1-monopalmitoyllysophosphatidylcholine (MPLPC), 1-monopalmitoyllysophosphatidylglycerol (MPLPG), 1-monopalmitoylglycerol, 1-monolauroyllysophosphatidylcholine, or 1-monooctanoyllysophosphatidylcholine) was performed at 37 °C with continuous agitation at 100 rpm for overnight incubations and at 150 rpm for various shorter time points. Final concentration of each lipid was set to 6.7 mm. Free fatty acids as a marker of lipid hydrolysis by PLA/LPLA/lipase were determined by means of the NEFA-C-kit® (WAKO Chemicals, Neuss, Germany) according to the instructions of the manufacturer. Depending upon the nature of the experiment, BYE-broth, LB-broth, or 40 mm Tris-HCl, pH 7.5 (25 °C), was incubated, treated like the cultures, and subsequently used as a negative control. All lipids were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL) or Sigma.
Expression of PlaB
E. coli BL21 cells harboring the designated vectors were grown overnight prior to inoculation into fresh media. At an A660 of 0.6–0.8, protein expression was induced by using isopropyl 1-thio-β-d-galactopyranoside to a final concentration of 1 mm. Bacterial cells were harvested at an A660 of 2 and centrifuged for 5 min at 5,000 × g, and pellets were stored overnight at −20 °C. Cell lysates were obtained according to our previously published protocol and subsequently used for determination of lipolytic activity (9).
Hemolysis Assay
For detection of hemolytic activity, E. coli strains were grown for 1 day on LB agar plates. Bacteria were inoculated into PBS medium and set to a final A660 of 0.3. As described by Kirby et al. (20), human red blood cells were mixed with an equal volume of bacterial suspension, centrifuged at 16,000 × g for 2 min before incubation was carried out at 37 °C. At the desired time points, the suspension was vortexed and repelleted by centrifugation, and release of hemoglobin into the supernatant was determined at an optical density of 415 nm.
Determination of Lipolytic Activities during Intracellular Infection of Host Cells by L. pneumophila
106/ml U937 macrophages were infected with L. pneumophila Corby (9, 21, 22). For detection of enzymatic activities, samples were taken from co-incubations as well as from uninfected U937 cells and divided into culture supernatant and cell pellet. Culture supernatant was furthermore passed through a 0.2-μm filter, and cell pellet was treated for 10 min by ultrasonication, stored at −20 °C for several days, thawed, and then resuspended in 20 mm Tris-HCl, pH 7.5 (25 °C), to original culture volume of the U937 infections. Culture supernatants and cell lysates were then incubated for 3 h with different lipid substrates, and enzymatic activities were determined. Hydrolysis in the presence of 6.25 mm EDTA accounted for bacterial PLA and LPLA activities.
RESULTS
PlaB Is Expressed during Host Cell Infections
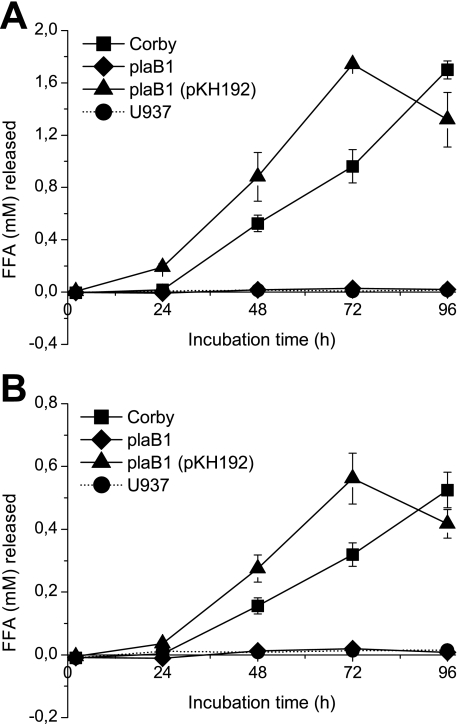
Although PlaB was shown to be dispensable for intracellular replication (9), we were interested whether PlaB-dependent lipolytic activities are expressed during the infection process. To address this question, U937 macrophages were infected with wild-type L. pneumophila Corby, plaB mutant plaB1, and the complementing strain plaB1 (pKH192). Both replication of bacteria and PLA (via hydrolysis of DPPG)/LPLA (via hydrolysis of monopalmitoyllysophosphatidylcholine (MPLPC)) activities associated with cells and present in the culture supernatant were monitored. Although bacterial replication was comparable for all three strains (data not shown), wild-type infection comprised high PLA/LPLA activities, whereas the plaB mutant infection or uninfected U937 cells did not exhibit these activities (Fig. 1, A and B). A similar pattern was observed for cell culture supernatants (data not shown). The introduction of a functional copy of plaB into the plaB1 mutant complemented demonstrated activity defects (Fig. 1, A and B). Expression of plaB was additionally confirmed by reverse transcription-PCR analysis (not shown). Comparable data were obtained both for infections of U937 macrophages and Acanthamoeba castellanii amoebae with L. pneumophila strain 130b, plaB mutant, and the complementing strain (data not shown). These data demonstrate that L. pneumophila PlaB is indeed expressed and presumably the most prominent PLA/LPLA of the pathogen during intracellular infection of eukaryotic host cells.
FIGURE 1.
Phospholipase A and lysophospholipase A activities upon intracellular infection of macrophages by L. pneumophila wild-type, plaB mutant and complementing strain. Wild-type Corby, mutant plaB1, and complementing strain plaB1 (pKH192) were used to infect monolayers of U937 cells, and PLA activity toward DPPG (A) and LPLA activity toward MPLPC (B) were determined for cell lysates at various time points postinoculation and after an additional 3 h of lipid hydrolysis. As a control, PLA and LPLA activities were monitored from uninfected U937 cells (U937).
PlaB Utilizes a Typical Catalytic Triad for Effective Hydrolysis
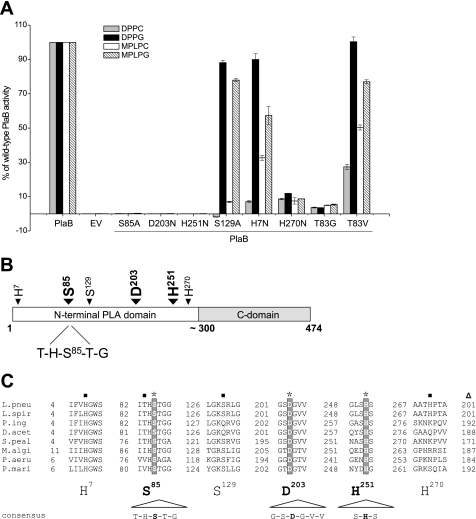
Lipase or phospholipase families typically contain a catalytic triad of Ser, Asp, and His, mostly arranged within conserved amino acid blocks. Assuming that PlaB exhibits a catalytic triad as well, we determined conserved and reasonable residues of PlaB and homologs to be used for site-directed mutagenesis. Interestingly, none of the potential catalytically active site serines is located within the typical pentapeptide GXSXG of true lipases or patatin-like proteins (12, 16); however, five serines were found to be part of similar amino acid motifs, such as Ser-85 (THSTG), Ser-129 (GKSRL), Ser-200 (GESGS), Ser-229 (GESLV), and Ser-250 (GLSHS). On the basis of this reasoning, serines were replaced by alanine, whereas reasonable aspartates and histidines were replaced by asparagine residues (Table 1). Resulting genes were expressed in a L. pneumophila plaB-deficient background, and cell lysates were subsequently tested for their ability to release fatty acids from various lipid substrates. In particular, dipalmitoylphosphatidylcholine (DPPC) and DPPG were used to check for PLA activity, and MPLPC and MPLPG were used to examine LPLA activity. All of those activities are usually comprised by the PlaB enzyme. We thereby could identify Ser-85, Asp-203, and His-251 to form the catalytic triad, since corresponding mutants were reduced in >99% of activity against all tested substrates (Fig. 2A).
FIGURE 2.
Enzymatic activities of PlaB and its site-directed mutant derivatives and distribution of crucial amino acids within the PlaB protein of L. pneumophila and homologs. A, mutagenesis of several amino acids severely influences PlaB activity. Values are expressed as percentages of wild-type PlaB activity. B, PlaB can be divided into an N-terminal region (amino acids 1 to ∼300) containing the catalytic triad of Ser-85/Asp-203/His-251 and further amino acids important for hydrolytic activity and a C-terminal domain with unknown function (amino acids ∼300–474). C, sequence alignment of L. pneumophila PlaB to potential phospholipases or hypothetical proteins of several bacterial species with an expect value of <0.01. Amino acids comprising the catalytic triad are marked with an asterisk and are highlighted by a gray background; further amino acids important for L. pneumophila PlaB activity are labeled with black squares (■). The last digits indicate the number of amino acids representing the residual C-terminal domain and are marked with a triangle. The consensus sequence is based on the amino acids most frequently found at this position. L.pneu, L. pneumophila; L.spir, L. spiritensis; P.ing, Psychromonas ingrahamii; D.acet, Desulfuromonas acetoxidans; S.peal, Shewanella pealeana; M.algi, Marinobacter algicola; P.aeru, P. aeruginosa PA7; P.mari, Persephonella marina. EV, empty vector pBCKS.
Further, mutant S129A was severely reduced in hydrolysis of choline-containing phospholipids, such as both DPPC and MPLPC, but not remarkably against substrates containing glycerol esterified to the phosphate residue instead (Fig. 2A). Moreover, we found that H270N mutants retained only ∼10% of their original hydrolytic properties and found His-7 to be particularly linked to substrate specificity regarding choline-containing lipids as well, since the mutant was about 10-fold less active against DPPC when compared with wild-type PlaB (Fig. 2A). Other mutants (S200A, S229A, S250A, D75N, D167N, D342N, D381N, H339N, and H433N) retained their ability to hydrolyze deduced substrates (data not shown). Reduction of enzymatic activity of described PlaB mutants was not due to second site mutations or decreased expression levels, as determined by sequencing and reverse transcription-PCR analysis (data not shown).
Together, amino acids crucial for PlaB enzymatic activity constitute a typical lipase Ser-Asp-His triad, arranged within the N-terminal part of the protein, thus leaving an extension of about 174 amino acids at the C-terminal domain, a function which still requires further investigation (Fig. 2B).
The Crucial Active Sites Are Embedded into a New Amino Acid Environment
Amino acid sequence alignment to related proteins (ClustalW2; EMBL-EBI) revealed that the designated residues composing the catalytic triad and embedding amino acid motifs are well conserved throughout PlaB homologs (Fig. 2C), underlining their necessity for enzymatic activity. Interestingly, the crucial Ser-85 in PlaB is surrounded by similar amino acids, as is the case for most lipases; however, a threonine substitutes for the usually comprised glycine residue within the GXSXG motif. Omitting Thr in the immediate vicinity of the active site Ser through Gly substitution strongly reduces PlaB activity by 95%, revealing its particular importance for PlaB enzyme activity (Fig. 2A). Additional replacement of Thr-83 by Val, a residue found in a PlaB homolog of P. aeruginosa (Fig. 2C) underlined this observation, since mutant T83V was reduced in DPPC/MPLPC hydrolysis to 27 or 50% of wild-type activity, respectively (Fig. 2A).
Ultimately, PlaB utilizes a well known catalytic triad, although conserved motifs embedding the crucial catalytic amino acids, designated THSTG, GSDGVV, and SHS (Fig. 2C), are unique among described phospholipases. PlaB thus represents the first described member of a new class of lipolytic families.
Phosphatidylcholine (PC)-specific Catalytic Activity of PlaB Is Responsible for plaB-dependent Hemolysis
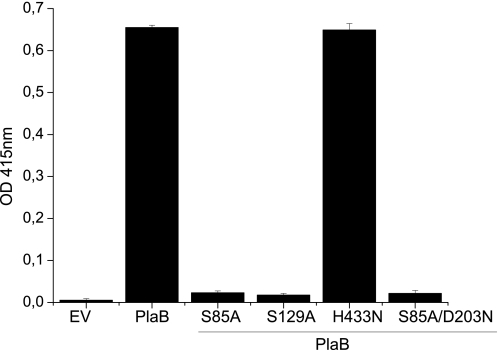
PlaB has been shown to contribute to hemolytic activity of L. pneumophila (9). Although it seems reasonable to assume that extensive phospholipase activity correlates with cytolytic activity, we aimed to clarify whether the identified catalytic domains are indeed essential for hemolysis. Corresponding PlaB site-directed mutant genes were expressed in non-hemolytic E. coli DH5α. Disrupting catalytically important amino acids, such as Ser-85 or Ser-85/Asp-203, a double knock-out without residual lipolytic activity, diminished hemolytic potential of plaB to the level of the empty vector-containing strain (Fig. 3). Mutant H433N did not show altered lipolytic quantities and profiles and therefore lysed human erythrocytes to the same extent as E. coli DH5α cells containing wild-type plaB. Interestingly, when we expressed mutant Ser-129, which was shown to hydrolyze DPPC/MPLPC less efficiently, hemolysis was comparable with vector control level (Fig. 3). Thus, PC-hydrolyzing activity of PlaB was directly responsible for its hemolytic effects.
FIGURE 3.
Contact-dependent hemolysis of human red blood cells upon incubation of E. coli strains expressing mutated variants of PlaB. Bacteria, obtained from 1-day-old agar plates, were set to an equal A660 before added to human erythrocytes. Cell lysis was determined by measuring absorbance after 18 h of incubation at 37 °C. EV, empty vector pBCKS.
The C Terminus of PlaB Is Important for Catalytic Activity
As demonstrated in Fig. 2B, PlaB catalytically important residues are present within the N-terminal region; therefore, the protein exhibits an extended C terminus of additional 174 amino acids, a function that remains to be elucidated. To this end, we constructed three truncated versions of the protein, ranging from 15 to 167 amino acids less than wild-type PlaB. Deletion of only 15 or more amino acids completely abolished enzymatic activities of PlaB, whereas protein expression levels of the truncated enzymes were comparable with the full-length construct (data not shown). Thus, catalytic activity of PlaB depends on both the N-terminal sequence harboring the crucial amino acids and the C-terminal region.
PlaB Mainly Hydrolyzes Long-chain Fatty Acid Substrates
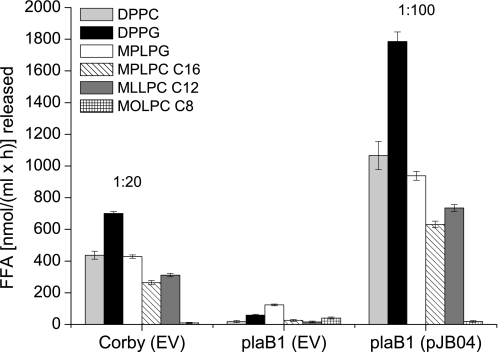
To further examine biochemical properties, we analyzed acyl chain length selectivity of wild-type PlaB enzyme. To this end, cell lysates of L. pneumophila Corby wild-type, the plaB mutant, and complementing strain were exposed to comparable concentrations of lysophophatidylcholines varying in chain length from C8 to C16, and the release of fatty acids was quantified. We demonstrate here that PlaB almost solely hydrolyzed long fatty acid chained substrates, such as MPLPC (C16) and 1-monolauroyllysophosphatidylcholine (C12) (Fig. 4). 1-Monooctanoyllysophosphatidylcholine (C8) was not affected by PlaB-dependent lipolysis under our experimental conditions (Fig. 4). This observation was verified by using p-nitrophenyl-based substrates esterified to carboxylic acids from C4, C12, and C16 in chain length (data not shown). Thus, PlaB acts mostly against long-chain fatty acid substrates with preferences for lipids containing fatty acyl residues larger than eight carbon atoms.
FIGURE 4.
Acyl chain length selectivity of wild-type L. pneumophila PlaB. Cell lysates of Legionella pneumophila strain Corby wild-type (empty vector pBCKS), plaB1 (empty vector pBCKS) mutant, or the complementing strain plaB1 (pJB04), diluted prior to incubation as indicated, were exposed to various lipids ranging from 8 (1-monooctanoyllysophosphatidylcholine) to 12 (1-monolauroyllysophosphatidylcholine) to 16 (MPLPC) carbon atoms in chain length, and the release of fatty acids was quantified. As an internal control, DPPG, DPPC, and MPLPG were carried along under equal conditions. EV, empty vector pBCKS.
Distribution of Cell-associated Lipolytic Activities among L. pneumophila Clinical Isolates
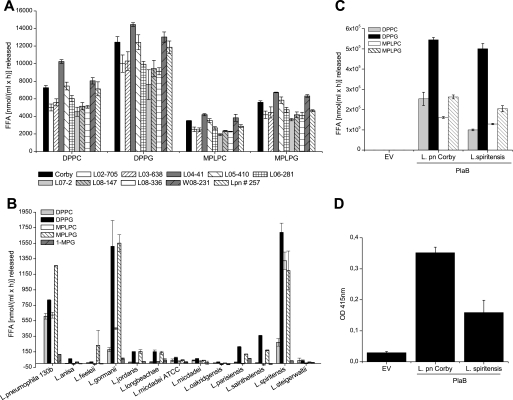
We demonstrated above that PlaB is a potent hemolytic agent and is expressed during intracellular replication, which probably contributes to extensive lung destruction during Legionnaires' disease. Thus, we investigated the distribution of cell-associated PLA/LPLA activity within nine clinical isolates. We observed comparable hydrolytic activities for various L. pneumophila strains, including different serogroups (Fig. 5A). Investigation of putative plaB expression by reverse transcription-PCR analysis revealed that these isolates indeed possess and actively transcribe plaB-like genes (not shown). This suggests that PlaB is widely spread among L. pneumophila bacteria isolated from various patients. Interestingly, an environmental L. pneumophila strain (Lpn 257), obtained from a water conduit, showed lipolytic potential comparable with that of the clinical isolates tested (Fig. 5A).
FIGURE 5.
Cell-associated PLA activity within clinical isolates of L. pneumophila and PlaB-like enzyme properties of non-pneumophila Legionellae. A, cell lysates of various clinical isolates (Table 1) were incubated with PLA (DPPG and DPPC) and LPLA (MPLPG and MPLPC) substrates. Nine clinical isolates were compared with L. pneumophila Corby and with the water conduit strain Legionella 257. B, determination of released fatty acids by L. pneumophila 130b and 12 different non-pneumophila strains. Cells were grown to an A660 of 1.5, pelleted, lysed, and added to different lipids, followed by quantification of released fatty acids. C, cell lysates of E. coli DH5α expressing either L. pneumophila or L. spiritensis plaB were incubated with different lipid substrates, and released fatty acids were quantified. DPPC hydrolysis of L. spiritensis PlaB was significantly decreased (p < 0.05) compared with L. pneumophila Corby PlaB. D, lysis of human red blood cells by E. coli strains as mentioned in C was measured at 415 nm after the bacteria-cell suspension was incubated for 4 h at 37 °C. EV, empty vector pBCKS.
L. gormanii and L. spiritensis Possess Reduced Potential for Phosphatidylcholine Hydrolysis
L. pneumophila has been most often associated with Legionella infections, and therefore researchers seek to identify virulence factors that are present only in pathogenic but not in less pathogenic species (23, 24). Although we previously determined plaB genetically to be restricted to L. pneumophila (9), we now wanted to verify whether non-pneumophila strains possess cell-associated PLA/LPLA activities. To this end, we tested L. pneumophila 130b and 12 non-pneumophila (Table 1) cell lysates for fatty acid release from phospholipids, lysophospholipids, and a lipase substrate, 1-MPG. Only L. gormanii and L. spiritensis possessed a spectrum and quantity of PLA/LPLA/lipase activities comparable with that of L. pneumophila 130b (Fig. 5B). However, both L. gormanii and L. spiritensis showed a lower PLA activity toward DPPC than L. pneumophila strains (Fig. 5, A and B). Additionally, by increasing the incubation time, Legionella anisa, Legionella jordanis, Legionella parisiensis, and Legionella sainthelensis very predominantly hydrolyzed phospho- and lysophospholipids with an attached glycerol (DPPG and MPLPG) instead of choline (DPPC and MPLPC) (data not shown).
L. gormanii and L. spiritensis Possess Genes Encoding PlaB Homologs
To address the question of plaB-like gene presence in L. gormanii and in L. spiritensis, a pool of primers were used for PCR with L. gormanii and L. spiritensis genomic DNA. For L. spiritensis amplification, products of a size comparable with that of L. pneumophila DNA were obtained and subsequently sequenced. The resulting L. spiritensis plaB gene (GenBankTM accession number EF408871) showed 86% identity, and its protein product showed 88% protein identity and 94% similarity to L. pneumophila Corby plaB or PlaB, respectively. Some of the primers yielded PCR products from L. gormanii DNA. The resulting translated partial sequence also showed 86% of amino acid identity and 95% similarity to the L. pneumophila Corby PlaB sequence (amino acids 16–256; data not shown). As demonstrated for L. spiritensis (Fig. 2C), L. gormanii as well comprises the crucial amino acids representing the catalytic active triad of serine, aspartate, and histidine, as well as important Ser-129 responsible for substrate specificity.
Furthermore, we cloned L. spiritensis plaB into vector pBCKS and compared lipolytic and hemolytic potential of transformed E. coli DH5α to L. pneumophila Corby plaB. Although no major differences in DPPG hydrolysis were observed, L. spiritensis was about 2.5-fold less able to hydrolyze DPPC than L. pneumophila Corby PlaB protein (Fig. 5C) (p < 0.05). Furthermore, lysis of human red blood cells agreed with this observation, since L. spiritensis plaB expressing DH5α cells were 2.2-fold reduced in hemolytic potential against human erythrocytes (Fig. 5D). These results emphasize again that only L. pneumophila-derived PlaB is most efficiently able to hydrolyze PC, the most abundant lipid in the outer leaflet of eukaryotic membranes.
DISCUSSION
L. pneumophila possesses a variety of phospholipid-hydrolyzing enzyme activities both of a secreted, such as PlaA and PlaC, and cell-associated nature, such as PlaB (9, 17, 25–28). Legionella phospholipases A may contribute to bacterial virulence in several ways. Those activities destroy lung surfactant phospholipids of mainly PC and phosphatidylglycerol thereby endangering lung function by increasing breathing effort. On the other hand, as shown for PlaB, they contribute to cell destruction by means of cytolytic/hemolytic properties and additionally by releasing toxic lysophospholipids, thus perturbing membrane integrity (9, 26, 29, 30).
As demonstrated recently, the cell-associated PLA activity of L. pneumophila is about 100-fold more active than the total of secreted PLA activity present in the culture supernatant (9). Since an L. pneumophila plaB knock-out mutant basically loses all of its cell-associated lipolytic activity, PlaB is a very potent and most importantly the major phospholipase A of the lung pathogen. PlaB is not essential for intracellular replication under laboratory conditions; nevertheless it represents the most prominently expressed PLA/LPLA activity throughout bacterial host cell infections. Although macrophage or amoeba cell infection rates were not influenced by the presence or absence of PlaB, guinea pig infection experiments show its importance for Legionella replication and dissemination in vivo (1).3
PlaB does not comprise hydrophobic stretches to form β-barrel structures; nevertheless, it could resemble E. coli or Helicobacter pylori outer membrane phospholipase A function, to allow bacterial adaptation through membrane alteration during the infection process (31, 32). However, PlaB does not show protein homology to outer membrane phospholipase A enzymes or to any other phospholipase described so far. We therefore analyzed the role of several putative active sites and clarified that PlaB and homologs exhibit a “classical” catalytic triad of Ser, Asp, and His (Ser-85, Asp-203, His-251). Lipases mostly share a common α/β-hydrolase folding pattern, at which conserved motifs surround the nucleophilic residue (Ser, Asp, or Cys), the catalytic acid (Asp or Glu) and histidine (33). However, PlaB and homologs differ from previously published enzymes, since they do not exhibit a GXSXG or a GDSL motif, characteristically displayed by eukaryotic and prokaryotic lipases and esterases as well (12, 34). Instead, PlaB constitutes the designated motif THSTG, surrounding the catalytically active serine. Reconstitution of the lipase-based amino acid sequence by changing Thr-83 to Gly severely affected hydrolysis. Interestingly, replacing Thr-83 by Val, as found in PlaB homologs of several P. aeruginosa strains, only affected substrate specificity regarding choline-containing phospholipids. Thus, to our knowledge, this is the first description of an amino acid other than glycine to be of major importance in the immediate vicinity of the active site Ser residue, which probably favors special steric conditions for effective hydrolysis of deduced substrates. This distinguishes PlaB from other lipases, such as Vst of Vibrio harveyi, since the usually invariant glycine was shown to be indispensable for catalytic activity (35).
We further determined His-7 and Ser-129 to be involved in substrate specificity of the protein. Alignment to PlaB homologs displays His-7 to be as conserved as residues constituting the catalytic triad. Adjacent to a central glycine, which most likely represents a part of the oxyanion hole, His-7 probably facilitates hydrolysis of deduced substrates through stabilization of the tetrahedral intermediate (34). Regarding conservation of Ser-129, PlaB-like proteins of Psychromonas or Desulfuromonas exhibit a Gln residue at the designated position. Both amino acids, His-7 and Ser-129, selectively contribute to hydrolysis of phospholipids harboring choline side chains. Similar results were obtained by replacing Thr-83 by Val. Apparently, PC is not the main substrate used by the PlaB enzyme, because minor modifications beyond the catalytic triad severely decrease corresponding hydrolysis. Instead, Legionella might additionally benefit from PlaB action by using its force to destruct this most abundant lipid within the outer leaflet of eukaryotic membranes (36). Further, lung surfactant is mainly composed of PC, therefore representing a perfect target for PlaB to deplete this important monolayer in the lung and contribute to the loss of lung function as observed in Legionnaires' disease (29). This assumption was underlined by the observation that hemolytic properties of PlaB almost solely depend on its ability to hydrolyze PC.
Another potent phospholipase and virulence factor, ExoU (exotoxin U) of P. aeruginosa, was shown to be dispensable for bacterial infection, since some clinical isolates lack the exoU encoding gene (37). Nevertheless, increased virulence was directly associated with secretion of the enzyme (38). As demonstrated within this study, different clinical isolates of L. penumophila serogroup 1–3 were tested positive for cell-associated PLA activity. Taking into account host cell and guinea pig infection studies (Fig. 1) (1),3 the presence of PlaB-like activity within all clinical and environmental isolates tested encourages our assumption of PlaB being valuable for pathogenic L. pneumophila.
Regarding distribution of PlaB throughout Legionellae other than L. pneumophila, we determined by means of biochemical activity assays that two of the 12 tested non-pneumophila strains, L. gormanii and L. spiritensis, showed cell-associated PLA and LPLA activities in quantities comparable with L. pneumophila. We additionally found that the plaB genes and deduced proteins of L. gormanii or L. spiritensis share a high degree of similarity to L. pneumophila plaB or PlaB, respectively. This shows that plaB paralogs are present in certain other Legionella species and may contribute to bacterial virulence as described for L. pneumophila PlaB. Indeed L. gormanii is a known but rare agent of Legionnaires' disease; however, this has not yet been demonstrated for L. spiritensis (23, 24). As already mentioned, L. pneumophila cell-associated PLA is strongly directed against phosphatidylglycerol, which is also the case for L. gormanii, L. spiritensis, and other non-pneumophila species showing cell-associated lipolytic activities. Of all tested Legionella species, L. pneumophila interestingly hydrolyzed the highest percentage of PC. This was verified when the plaB genes of L. pneumophila and L. spiritensis were expressed in E. coli DH5α cells. Since L. pneumophila is responsible for more than 90% of Legionnaires' disease cases (24), specificity for cleavage of typical eukaryotic phospholipids, like PC, could be a potent tool for bacterial pathogenicity.
To summarize, we have shown here that cell-associated L. pneumophila PlaB is the major PLA and LPLA of the lung pathogen expressed during host cell infection. The highly active enzyme exhibits a “classical” catalytic triad of serine hydrolases, although special motifs surrounding the active sites distinguish PlaB and homologs from previously discovered phospholipases. Thus, PlaB represents the first described member of a new subfamily of lipolytic enzymes. Although it is not restricted to the most prominent pathogenic species, L. pneumophila, it probably confers additional colonization advantages through hydrolysis of certain phospholipids, such as PC, the most abundant lipid within the outer leaflet of eukaryotic membranes.
Acknowledgments
We are grateful to Dave Matthews for critically reading the manuscript and to Ferda Ersöz for excellent technical laboratory support. We further thank Birgid Neumeister (Tübingen University, Germany), Christian Lück (German Reference Laboratory for Legionella, Dresden, Germany), and Barry Fields (Centers for Disease Control and Prevention, Atlanta, GA) for providing non-pneumophila strains and clinical isolates, respectively.
This work was supported by Deutsche Forschungsgemeinschaft (Bonn, Germany) Grants HE 2845/4-1 and FL 359/4-1 (to K. H. and A. F., respectively).
E. Schunder, P. Adam, F. Higa, K. A. Remer, U. Lorenz, J. Bender, A. Flieger, M. Steinert, J. Hacker, and K. Heuner, submitted for publication.
- PLA
- phospholipase
- LPLA
- lysophospholipase
- DPPG
- dipalmitoylphosphatidylglycerol
- DPPC
- dipalmitoylphosphatidylcholine
- MPLPG
- 1-monopalmitoylphosphatidylglycerol
- MPLPC
- 1-monopalmitoyphosphatidylcholine
- PC
- phosphatidylcholine
- 1-MPG
- 1-monopalmitoylglycerol.
REFERENCES
- 1.Bender J., Flieger A. (2009) Microbiology of Hydrocarbons, Oils, Lipids, and Derived Compounds, Springer-Verlag GmBH, Heidelberg, Germany, in press [Google Scholar]
- 2.Istivan T. S., Coloe P. J. (2006) Microbiology 152, 1263–1274 [DOI] [PubMed] [Google Scholar]
- 3.Schmiel D. H., Miller V. L. (1999) Microbes Infect. 1, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 4.Jepson M., Titball R. (2000) Microbes Infect. 2, 1277–1284 [DOI] [PubMed] [Google Scholar]
- 5.Phillips R. M., Six D. A., Dennis E. A., Ghosh P. (2003) J. Biol. Chem. 278, 41326–41332 [DOI] [PubMed] [Google Scholar]
- 6.Sato H., Frank D. W., Hillard C. J., Feix J. B., Pankhaniya R. R., Moriyama K., Finck-Barbançon V., Buchaklian A., Lei M., Long R. M., Wiener-Kronish J., Sawa T. (2003) EMBO J. 22, 2959–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato H., Frank D. W. (2004) Mol. Microbiol. 53, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 8.Titball R. W., Naylor C. E., Basak A. K. (1999) Anaerobe 5, 51–64 [DOI] [PubMed] [Google Scholar]
- 9.Flieger A., Rydzewski K., Banerji S., Broich M., Heuner K. (2004) Infect. Immun. 72, 2648–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winn W. C., Jr., Myerowitz R. L. (1981) Hum. Pathol. 12, 401–422 [DOI] [PubMed] [Google Scholar]
- 11.Blackmon J. A., Chandler F. W., Cherry W. B., England A. C., 3rd, Feeley J. C., Hicklin M. D., McKinney R. M., Wilkinson H. W. (1981) Am. J. Pathol. 103, 429–465 [PMC free article] [PubMed] [Google Scholar]
- 12.Arpigny J. L., Jaeger K. E. (1999) Biochem. J. 343, 177–183 [PMC free article] [PubMed] [Google Scholar]
- 13.Upton C., Buckley J. T. (1995) Trends Biochem. Sci. 20, 178–179 [DOI] [PubMed] [Google Scholar]
- 14.Rydel T. J., Williams J. M., Krieger E., Moshiri F., Stallings W. C., Brown S. M., Pershing J. C., Purcell J. P., Alibhai M. F. (2003) Biochemistry 42, 6696–6708 [DOI] [PubMed] [Google Scholar]
- 15.Vernet T., Ziomek E., Recktenwald A., Schrag J. D., de Montigny C., Tessier D. C., Thomas D. Y., Cygler M. (1993) J. Biol. Chem. 268, 26212–26219 [PubMed] [Google Scholar]
- 16.Banerji S., Flieger A. (2004) Microbiology 150, 522–525 [DOI] [PubMed] [Google Scholar]
- 17.Banerji S., Aurass P., Flieger A. (2008) Int. J. Med. Microbiol. 298, 169–181 [DOI] [PubMed] [Google Scholar]
- 18.Edelstein P. H. (1981) J. Clin. Microbiol. 14, 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bi W., Stambrook P. J. (1998) Anal. Biochem. 256, 137–140 [DOI] [PubMed] [Google Scholar]
- 20.Kirby J. E., Vogel J. P., Andrews H. L., Isberg R. R. (1998) Mol. Microbiol. 27, 323–336 [DOI] [PubMed] [Google Scholar]
- 21.Cianciotto N. P., Fields B. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 5188–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liles M. R., Edelstein P. H., Cianciotto N. P. (1999) Mol. Microbiol. 31, 959–970 [DOI] [PubMed] [Google Scholar]
- 23.Fang G. D., Yu V. L., Vickers R. M. (1989) Medicine 68, 116–132 [DOI] [PubMed] [Google Scholar]
- 24.Muder R. R., Yu V. L. (2002) Clin. Infect. Dis. 35, 990–998 [DOI] [PubMed] [Google Scholar]
- 25.Banerji S., Bewersdorff M., Hermes B., Cianciotto N. P., Flieger A. (2005) Infect. Immun. 73, 2899–2909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flieger A., Gong S., Faigle M., Deeg M., Bartmann P., Neumeister B. (2000) J. Bacteriol. 182, 1321–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flieger A., Gong S., Faigle M., Stevanovic S., Cianciotto N. P., Neumeister B. (2001) J. Bacteriol. 183, 2121–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flieger A., Neumeister B., Cianciotto N. P. (2002) Infect. Immun. 70, 6094–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flieger A., Gongab S., Faigle M., Mayer H. A., Kehrer U., Mussotter J., Bartmann P., Neumeister B. (2000) FEMS Microbiol. Lett. 188, 129–133 [DOI] [PubMed] [Google Scholar]
- 30.Weltzien H. U. (1979) Biochim. Biophys. Acta 559, 259–287 [DOI] [PubMed] [Google Scholar]
- 31.Snijder H. J., Dijkstra B. W. (2000) Biochim. Biophys. Acta 1488, 91–101 [DOI] [PubMed] [Google Scholar]
- 32.Tannaes T., Dekker N., Bukholm G., Bijlsma J. J., Appelmelk B. J. (2001) Infect. Immun. 69, 7334–7340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J. (1992) Protein Eng. 5, 197–211 [DOI] [PubMed] [Google Scholar]
- 34.Jaeger K. E., Ransac S., Koch H. B., Ferrato F., Dijkstra B. W. (1993) FEBS Lett. 332, 143–149 [DOI] [PubMed] [Google Scholar]
- 35.Teo J. W., Zhang L. H., Poh C. L. (2003) Gene 312, 181–188 [DOI] [PubMed] [Google Scholar]
- 36.Devaux P. F., Morris R. (2004) Traffic 5, 241–246 [DOI] [PubMed] [Google Scholar]
- 37.Hirakata Y., Finlay B. B., Simpson D. A., Kohno S., Kamihira S., Speert D. P. (2000) J. Infect. Dis. 181, 765–769 [DOI] [PubMed] [Google Scholar]
- 38.Schulert G. S., Feltman H., Rabin S. D., Martin C. G., Battle S. E., Rello J., Hauser A. R. (2003) J. Infect. Dis. 188, 1695–1706 [DOI] [PubMed] [Google Scholar]
- 39.Jepras R. I., Fitzgeorge R. B., Baskerville A. (1985) J. Hyg. (Lond.) 95, 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]