Abstract
CD8+ T-cells specific for MART-1-(26–35), a dominant melanoma epitope restricted by human leukocyte antigen (HLA)-A*0201, are exceptionally common in the naive T-cell repertoire. Remarkably, the TRAV12-2 gene is used to encode the T-cell receptor α (TCRα) chain in >87% of these T-cells. Here, the molecular basis for this genetic bias is revealed from the structural and thermodynamic properties of an archetypal TRAV12-2-encoded TCR complexed to the clinically relevant heteroclitic peptide, ELAGIGILTV, bound to HLA-A*0201 (A2-ELA). Unusually, the TRAV12-2 germ line-encoded regions of the TCR dominate the major atomic contacts with the peptide at the TCR/A2-ELA interface. This “innate” pattern of antigen recognition probably explains the unique characteristics and extraordinary frequencies of CD8+ T-cell responses to this epitope.
Malignant melanoma is responsible for 75% of all skin cancer-related deaths worldwide, and the global incidence is rising. The MART-1 (1) protein, also known as Melan-A (2), is expressed by virtually all fresh melanoma tumor specimens and elicits natural CD8+ T-cell responses (3, 4) that can lead to spontaneous disease regression (reviewed in Ref. 5). Consequently, CD8+ T-cell responses directed against the MART-1 protein have been investigated extensively (reviewed in Refs. 2, 6, and 7), and heteroclitic forms of the dominant MART-1-(26–35) peptide epitope (8, 9), which is restricted by human leukocyte antigen (HLA)-A*0201, are currently being used in a number of clinical trials (10–12). In recent developments, adoptive T-cell therapy directed against the MART-1 protein has been used to mediate cancer regression in ∼50% of late stage melanoma patients (13). However, these approaches have not proved to be universally effective, and there remains considerable scope for improvement. In order to design more effective immune-based therapies against the MART-1 protein, it is essential to understand the precise molecular rules that govern the interaction between T-cell receptors (TCRs)6 and the HLA-A*0201·MART-1-(26–35) complex. Previous structural studies of human TCR/peptide major histocompatibility complex (pMHC) interactions (14–16) indicate that specific regions of the TCR have different roles during antigen engagement; thus, the germ line-encoded complementarity-determining region 1 and 2 (CDR1 and -2) loops contact mainly the conserved helical region of the MHC surface, and the more variable somatically rearranged CDR3 loops contact mainly the antigenic peptide. Dissecting the nature of these contacts, which have been shown to be highly variable for individual TCR/pMHC interactions (17–19), is an important step toward understanding the principles of antigen recognition and for the development of improved T-cell vaccines (20). However, the current data base of human TCR·pMHC complexes reported in the literature is limited (∼16), compared with >100 antibody-antigen structures. This has made it difficult to ascertain whether there are conserved binding modes for TCR/pMHC interactions dictated by a number of specific contacts or whether there are potentially unlimited numbers of TCR docking orientations dependent on the nature of individual recognition events. Furthermore, there are no examples to date of human TCR·pMHC class I structures in which the bound peptide is a decamer; this represents a substantial deficiency in our current knowledge, given the preponderance with which decamer peptides are processed, presented, and recognized. The low number of TCR·pMHC complex structures solved to date reflects technical difficulties inherent in the production of soluble TCR and pMHC molecules that retain stability and challenges related to the crystallization of complexes with relatively low binding affinities (KD = 0.1–500 μm) (21, 22). In general, TCRs specific for tumor-derived epitopes bind in the weaker range of TCR/pMHC affinities (21). This obstacle to the generation of high quality co-complex crystals is underscored by the fact that only one other tumor-specific human TCR·pMHCI complex structure has been documented previously (23).
In this study, we expressed a soluble TCR (MEL5) specific for ELAGIGILTV, the common MART-1-(26–35) heteroclitic peptide, complexed to HLA-A*0201 (A2-ELA). Notably, HLA-A*0201 is the most common HLA allele in the human population (24). The CDR1 and CDR2 loops of this TCR are encoded by the TRAV12-2 and TRBV30 genes (International Immunogenetics (IMGT) nomenclature). Interestingly, the TRAV12-2 gene is expressed in the vast majority of CD8+ T-cell populations specific for HLA-A*0201·MART-1-(26–35) across multiple individuals (25, 26). To resolve the enigma of the dominant TRAV12-2 gene and determine the molecular characteristics that govern CD8+ T-cell recognition of the HLA-A*0201·MART-1-(26–35) antigen, we performed a biophysical, thermodynamic, and structural analysis of MEL5 TCR binding to A2-ELA. The data provide a molecular basis for biased TCR gene product selection in the CD8+ T-cell response to HLA-A*0201·MART-1-(26–35) and indicate that pMHC antigens can be subject to “innate-like” binding modes within adaptive immune responses.
EXPERIMENTAL PROCEDURES
Generation of CD8+ T-cell Clones Specific for HLA-A* 0201·MART-1-(26–35)
CD8+ T-cell clones were generated as described previously (27). Briefly, peripheral blood mononuclear cells, isolated from an HLA-A*0201+ healthy donor, were stimulated with 1 nm ELAGIGILTV peptide and cloned via limiting dilution. These cells were then screened for A2-ELA tetramer binding. The MEL5 CD8+ T-cell clone isolated from these experiments activated typically in response to HLA-A*0201+ target cells pulsed with the ELAGIGILTV peptide, exhibiting specific degranulation (CD107a mobilization) and the production of interferon-γ, interleukin-2, and tumor necrosis factor-α (data not shown). The MEL5 TCR was derived from the MEL5 CD8+ T-cell clone. Two independent CD8+ T-cell clones grown in the same way, MEL11 and MEL13, were shown to express an identical TCR.
Generation of Expression Plasmids
The MEL5 TCR, HLA-A*0201 α chain and β2m sequences were generated by PCR mutagenesis (Stratagene) and PCR cloning. All sequences were confirmed by automated DNA sequencing (Lark Technologies). For MEL5, a disulfide-linked construct was used to produce the soluble domains (variable and constant) for both the α and β chains (28, 29). The soluble HLA-A*0201 α chain (α1, α2, and α3 chain domains), tagged with a biotinylation sequence, and β2m were also cloned and used to make the HLA-A*0201 protein. The TCR, HLA-A*0201 α chain, and β2m sequences were inserted into separate pGMT7 expression plasmids under the control of the T7 promoter (28).
Protein Expression, Refolding, and Purification
Competent Rosetta DE3 Escherichia coli cells were used to produce the MEL5 α and β chains and the HLA-A*0201 α and β2m chains in the form of inclusion bodies, using 0.5 mm isopropyl 1-thio-β-d-galactopyranoside to induce expression, as described previously (28). For a 1-liter refold, 30 mg of MEL5 α chain inclusion bodies were incubated at 37 °C for 15 min with 10 mm dithiothreitol and added to cold refold buffer (50 mm Tris, pH 8.1, 2 mm EDTA, 2.5 m urea, 6 mm cysteamine hydrochloride, and 4 mm cystamine). After 15 min, 30 mg of MEL5 β chain, incubated at 37 °C for 15 min with 10 mm dithiothreitol, was added. For a 1-liter A2-ELA refold, 30 mg of α chain was mixed with 30 mg of β2m and 4 mg of the ELAGIGILTV peptide at 37 °C for 15 min. This was then added to cold refold buffer (50 mm Tris, pH 8, 2 mm EDTA, 400 mm l-arginine, 6 mm cysteamine hydrochloride, and 4 mm cystamine). Refolds were mixed at 4 °C for >1 h. Dialysis was carried out against 10 mm Tris, pH 8.1, until the conductivity of the refolds was under 2 millisiemens/cm. The refolds were then filtered in preparation for the purification steps. Refolded proteins were purified initially by ion exchange using a Poros50HQTM column and finally gel-filtered into BIAcore buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA, and 0.005% (v/v) Surfactant P20) or crystallization buffer (10 mm Tris, pH 8.1, 10 mm NaCl) using a Superdex200HRTM column. Protein quality was analyzed by Coomassie-stained SDS-PAGE.
pMHC Biotinylation
Biotinylated pMHC was prepared as described previously (30).
Surface Plasmon Resonance Analysis
Binding analysis was performed independently using a BIAcore T100TM equipped with a CM5 sensor chip, as reported previously (31). Between 200 and 400 response units of biotinylated pMHC was immobilized to streptavidin, which was chemically linked to the chip surface. The pMHC was injected at a slow flow rate (10 μl/min) to ensure uniform distribution on the chip surface. Combined with the small amount of pMHC bound to the chip surface, this reduced the likelihood of off-rate-limiting mass transfer effects. MEL5 was purified and concentrated to ∼100 μm on the same day of surface plasmon resonance analysis to reduce the likelihood of TCR aggregation affecting the results. For equilibrium and kinetic analysis, 10 serial dilutions were carefully prepared in triplicate for each sample and injected over the relevant sensor chips at 25 °C. MEL5 was injected over the chip surface using kinetic injections at a flow rate of 45 μl/min. For the thermodynamic experiments, this method was repeated at the following temperatures; 5, 13, 21, 25, 29, and 37 °C. Results were analyzed using BIAevaluation 3.1TM, Microsoft ExcelTM, and Origin 6.1TM. The equilibrium binding constant (KD) values were calculated using a nonlinear curve fit (y = (P1x)/(P2 + x)). The thermodynamic parameters were calculated using the Gibbs equation (y = dH + dCp × (x − 298) − x × dS − x × dCp × ln(x/298)).
Crystallization
The MEL5·A2-ELA complex was crystallized as reported previously (32).
Diffraction Data Collection and Model Refinement
A data set up to 3.0 Å was collected with the rotation method at Synchrotron Radiation Source Station 14.2 (Daresbury, UK) using an Area Detection Systems Corporation Quantum 4 CCD-detector system. The wavelength (λ) was set to 0.979 Å. A total of 160 frames were recorded, each covering 0.5° of rotation. The crystal was maintained at 100 K in an Oxford cryostream. Reflection intensities were estimated with the MOSFLM package (33), and the data were scaled, reduced, and analyzed with SCALA and the CCP4 package (34). The structure was solved with molecular replacement using PHASER (35). Despite close similarity to NY-ESO-1 complexes solved recently (e.g. 2P5E or 2P5W), a solution could be obtained only with a composite search model constructed with CHAINSAW as follows. the HLA-A*0201 α chain (A) was taken from Protein Data Bank entry 2F53, the β2m chain (B) was taken from Protein Data Bank entry 2BCK, the TCRα chain (D) was taken from Protein Data Bank entry 1AO7, and the TCRβ chain (E) was taken from Protein Data Bank entry 2F53. The peptide was not included at this stage, since it was only a small fraction of the diffracting material. The model sequence was adjusted with COOT (36), and the model was refined with REFMAC5, version 5.2.0019 (37). The model was split into eight domains for TLS refinement (A2α1α2, A2α3, β2m, ELA peptide, MEL5α constant, MEL5α variable, MEL5β constant and MEL5β variable). After convergence, one round of restrained thermal parameters was completed, in order to obtain the full B values for atoms. Graphical representations were prepared with PyMOL (38). Data collection and reduction statistics and refinement statistics are shown in Table 1.
TABLE 1.
Data collection and refinement statistics (molecular replacement)
One crystal was used for data collection.
| Parameters | Values |
|---|---|
| Data set statisticsa | |
| Space group | P43 |
| Unit cell parameters (Å) | a = b = 120.9, c = 82.0 |
| Radiation source | SRS 14.2 |
| Wavelength (Å) | 0.978 |
| Resolution (Å) | 3.0 (3.16-3.0) |
| Reflections observed | 80,204 (11,687) |
| Unique reflections | 23,764 (3,464) |
| Completeness (%) | 99.5 (100.0) |
| Multiplicity | 3.4 (3.4) |
| I/σ(I) | 8.0 (1.6) |
| Rmerge (%) | 15.1 (79.4) |
| Refinement statisticsa | |
| Measured resolution range (Å) | 48.7–3.0 |
| No. of reflections used | 22,525 (1,652) |
| No. of reflections in the Rfree set | 1,211 (88) |
| Rcryst (no cut-off) (%) | 22.5 |
| Rfree (%) | 30.4 |
| Root mean square deviation from ideal geometryb | |
| Bond lengths (Å) | 0.02 (0.021) |
| Bond angles (degrees) | 1.2 (1.936) |
| Mean B value (Å2) | 40.5 |
| Wilson B-factor (Å2) | 47.6 |
| Overall coordinate error (Å) | 0.4 |
a Values in parentheses are for the highest resolution shell.
b Values in parentheses are target values.
RESULTS
Structure Determination and Analysis of MEL5 in Complex with A2-ELA
To investigate the structural basis for dominant TRAV12-2 gene usage in CD8+ T-cell populations specific for HLA-A*0201·MART-1-(26–35), we solved the atomic structure of MEL5 in complex with A2-ELA (Table 1; Fig. 1A). Molecular replacement was successful only in space group P43, consistent with the presence of one molecule of the complex per asymmetric unit, and the resolution was sufficiently high to show that the interface between the two molecules was well ordered and contained well defined electron density. The final model showed ∼96% of residues in the preferred and allowed regions of the Ramachandran plot and geometry consistent with the data resolution. The crystallographic R/Rfree factors were 22.5 and 30.4%. There was enough ordered density around the protein model to allow the identification of two glycerol molecules, 2 sulfate ions, and 44 solvent (water) molecules. The C terminus of the TCRα chain was disordered beyond residue 195, with no apparent electron density beyond that point.
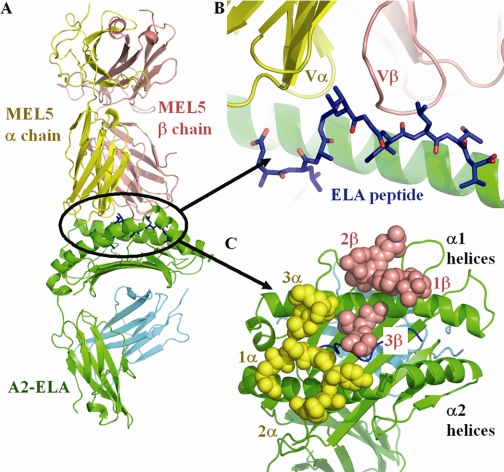
FIGURE 1.
A, the co-crystal structure of MEL5 (α chain shown as a yellow schematic diagram, β chain shown as a salmon schematic diagram) bound to the HLA-A*0201 (shown as green and blue schematic diagrams) molecule complexed with the ELAGIGILTV peptide (shown as blue sticks). B, expanded view of the interface between the MEL5 variable domain bound to the A2-ELA surface (colors as in A). The overall conformation of the ELAGIGILTV peptide (N to C terminus, left to right), including the central peptide bulge, is displayed. C, view from above of the MEL5 CDR loops bound to the A2-ELA surface (colors as in A; MEL5 CDR loops shown as spheres). The MEL5 TCR binds toward the N terminus of the peptide, making contacts with the peptide via its CDR1 and CDR3 loops and contacts with the MHC surface via its CDR1 and CDR2 loops.
The MEL5 docking angle with A2-ELA was 35° (calculated as in Ref. 14), with the TCRα chain contacting the α2 domain and the TCRβ chain contacting the α1 domain of A2-ELA (Fig. 1, A–C). The docking angle in this complex lies within the range observed for other human TCR·pMHC complexes, with the A6 TCR·A2-Tax complex representing one extreme (32°) (28) and the 1G4 TCR·A2-NY-ESO (23) complex representing the other extreme (69°). The TCR was located toward the N terminus of the MHC peptide binding groove, as observed in the A6 TCR·A2-Tax and B7 TCR·A2-Tax complexes (39, 40), and centered over the solvent-exposed bulge of the ELA peptide (Fig. 1, B and C).
The total buried surface area of the interaction was ∼1226 Å2, the lowest for any human TCR/pMHC interaction reported to date. This supports the observation that MEL5 makes significantly fewer contacts with the pMHC surface compared with other TCR·pMHCI complexes, which indicates that T-cell recognition of antigen can occur with a lower number of specific contacts than previously thought. Although the ELA peptide is a decamer, the contribution of the peptide (29% of the interface area) to TCR binding was within the normal range observed for other TCR·pMHCI complexes (18–34%) that typically contain nonamers in the peptide binding groove. The contribution of the TCRα and TCRβ chains has been shown to vary markedly for different human TCR·pMHC complexes, the largest difference being observed for the JM22 TCR·A2-Flu complex (41) (α chain, 33%; β chain, 67%). For the MEL5·A2-ELA complex, the interaction was split relatively evenly (α chain, 50.4%; β chain, 49.6%). The surface complementarity across the interface as a whole was 0.63, with a slightly lower score of 0.58 between the MHC and the TCR alone. However, the score increased to 0.76 when the TCR and the peptide interface were considered, indicating a much closer and more directed match.
MEL5/A2 Contacts
MEL5 forms 12 contacts, comprising nine electrostatic interactions and three van der Waals (vdW) close interactions, with the conserved MHC α helices of HLA-A*0201 that constitute the sides of the peptide binding groove (Table 2). These contacts are dominated by the CDR1α loop, which is located over the N terminus of the ELA peptide and makes a significant contribution to the TCR/MHC α2 helix interface, forming a dense network of four electrostatic interactions between the TCRα chain residue Arg28 and the MHC α2 residues Glu166 and Trp167. Notably, the CDR1β loop plays no part in contacting the A2-ELA antigen (Table 2). Further contacts were evident between the MHC α1 helix and the TCR CDR3α, CDR2β, and CDR3β loops, as detailed in Table 2. A number of conserved, or “gatekeeper,” interactions that are present in the majority of TCR·pMHCI complex structures solved to date (14), were also present between MEL5 and the MHC surface. These include electrostatic interactions between the TCR residue Glu59 and the MHC residue Arg65 and between the TCR residues Gly99 and Thr100 and the MHC residue Gln155. Interestingly, MEL5 does not contact MHC α1 domain residue Arg69 or Gln72, which represent the other “gatekeeper” TCR/MHC contacts (14).
TABLE 2.
MEL5/A2-ELA contacts
Contacts were calculated using a 4Å cut-off.
| CDR loop | TCR residue | Peptide residue | MHC residue | Bond type | Bond distance |
|---|---|---|---|---|---|
| Å | |||||
| CDR1α | Arg28 Nϵ | Glu166 Oϵ2 | Electrostatica | 3.4 | |
| Arg28 NH2 | Glu166 Oϵ2 | Electrostatic | 3.1 | ||
| Arg28 O | Trp167 Nϵ1 | Electrostatic | 3.7 | ||
| Arg28 NH2 | Glu166 Oϵ1 | Electrostatic | 3.5 | ||
| Gln31 Nϵ2 | Glu1 Oϵ2 | Electrostatic | 3.4 | ||
| Gln31 Nϵ2 | Leu2 O | Electrostatic | 3.4 | ||
| Gln31 Cδ | Gly4 N | vdWb | 3.3 | ||
| Gln31 Oϵ1 | Gly4 Cα | vdW | 3.0 | ||
| Gln31 Oϵ1 | Gly4 N | Electrostatic | 2.5 | ||
| Gln31 Oϵ1 | Ile5 N | Electrostatic | 3.5 | ||
| CDR2α | Tyr51 OH | His151 O | Electrostatic | 3.9 | |
| CDR3α | Asn92 Nδ2 | Gly4 O | Electrostatic | 3.2 | |
| CDR2β | Gln55 Oϵ1 | Arg75 NH1 | Electrostatic | 3.3 | |
| Glu59 Oϵ1 | Arg65 NH1 | Electrostatic | 2.8 | ||
| Glu59 Oϵ2 | Arg65 NH1 | Electrostatic | 3.5 | ||
| Glu59 Cδ | Arg65 NH1 | vdW | 3.4 | ||
| CDR3β | Gly97 Cα | Thr73 Cγ2 | vdW | 2.9 | |
| Gly99 Cα | Gln155 Oϵ1 | vdW | 3.4 | ||
| Thr100 N | Gln155 Oϵ1 | Electrostatic | 2.9 | ||
| Thr96 Oγ1 | Thr9 Oγ1 | Electrostatic | 3.8 | ||
| Leu98 Cγ2 | Ala3 O | vdW | 3.3 | ||
| Leu98 O | Gly4 O | vdW | 3.2 | ||
| Leu98 O | Ile5 Cα | vdW | 3.2 | ||
| Leu98 O | Gly6 N | Electrostatic | 3.9 | ||
| Leu98 O | Ile7 N | Electrostatic | 3.8 | ||
| Leu98 N | Ile7 O | Electrostatic | 3.2 | ||
| Gly99 N | Ile7 O | Electrostatic | 3.4 | ||
| Gly99 O | Ile5 Cα | vdW | 3.3 | ||
| Gly99 O | Ile5 Cγ2 | vdW | 3.3 |
a Electrostatic, polar contacts.
b vdW, non-polar van der Waals contacts.
A2-ELA Conformation in Uncomplexed and TCR-complexed Forms
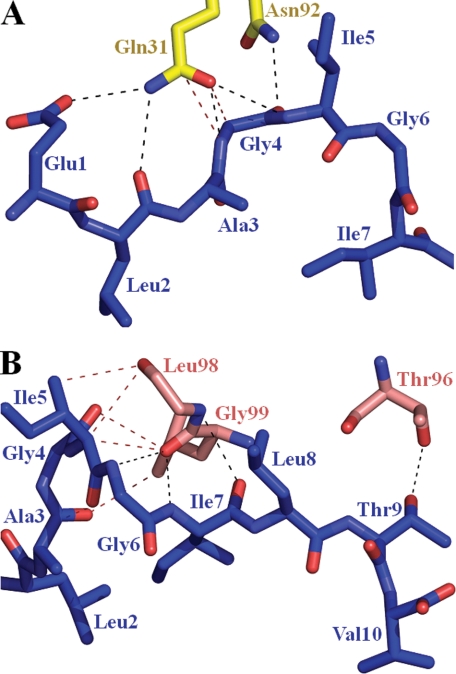
The crystal structure of uncomplexed A2-ELA has been solved (42). Although the ELA peptide is a decamer, it adopts a similar conformation to nonamer peptides with a central bulge between residues 4 and 6 (Fig. 2A). In the case of the ELA peptide, the extra residue is accommodated, not through a more prominent central bulge but by an extension of the peptide Cα chain toward the α2 domain of the HLA-A*0201 binding groove. Superposition of the uncomplexed A2-ELA and the MEL5-complexed A2-ELA structures shows that the peptide termini are located at virtually identical positions. This peptide conformation results in the availability of a number of residues for specific TCR contacts; these include GluP1, LeuP2, AlaP3, GlyP4, IleP5, GlyP6, IleP7, LeuP8, and ThrP9 (Fig. 2). Compared with the uncomplexed A2-ELA structure, the conformation of the ELA peptide upon docking with the TCR is very similar, with a root mean square deviation value of 0.369 between the peptides in uncomplexed and TCR-complexed forms. There are some conformation differences in the side chains of LeuP8, and ThrP9, which adopt visually different conformations in the uncomplexed A2-ELA structure; however, these differences are within experimental error and may not represent changes in peptide conformation due to stabilization of the peptide upon TCR docking.
FIGURE 2.
A, the interactions between the CDR loops of MEL5 α chain (shown as yellow sticks) and the ELAGIGILTV peptide (shown as blue sticks). Electrostatic interactions are depicted as black dotted lines, and vdW interactions are shown as red dotted lines. B, the interactions between the CDR loops of MEL5 β chain (shown as salmon sticks) and the ELAGIGILTV peptide (shown as blue sticks). Electrostatic interactions are depicted as black dotted lines, and vdW interactions are shown as red dotted lines.
The Dominant Role of the TRAV12-2-encoded CDR1 Loop during Peptide Binding
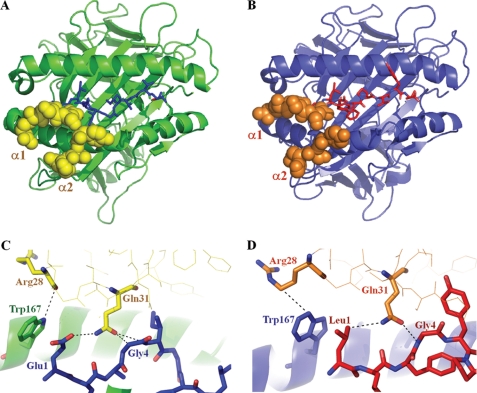
MEL5 contacts eight of the A2-ELA peptide residues (Table 2 and Fig. 2B), compared with only three for the immunodominant LC13 TCR·B8-EBNA (43) and the four for JM22 TCR·A2-Flu (41) complexes. Unusually, the CDR3α loop has a minimal role in contacting the ELA peptide, making only one electrostatic interaction between TCR residue Asn92 and the ELA peptide at GlyP4. A substantial number of contacts at the interface are formed between Gln31 in the CDR1α loop, encoded by the TRAV12-2 gene, and the antigenic peptide (Table 2). The Gln31 residue makes a dense network of contacts to GluP1, LeuP2, GlyP4, and IleP5 and is therefore likely to have a chief role in peptide recognition (Table 2 and Fig. 2B). In this unusual TCR binding mode, the CDR1α loop acts in a manner comparable with that of a classical CDR3 loop with respect to peptide contacts. The dominance of the CDR1α loop in terms of contacting both the surface of HLA-A*0201 and the bound ELA peptide could explain the prevalent expression of the TRAV12-2 gene in CD8+ T-cell responses specific for MART-1-(26–35) (26). Furthermore, the TRAV12-2 gene, which encodes the CDR1α and CDR2α loops of MEL5, is also expressed by the A6 TCR, which is specific for the Tax-(11–19) peptide (LLFGYPVYV) bound to HLA-A*0201 (A2-Tax) (28). The CDR1α and CDR2α loops of the A6 TCR utilize an antigen binding mode virtually identical to that seen in the MEL5·A2-ELA complex, with both TCRs making contacts between CDR1α loop residue Arg28 and MHC α2 residue Trp167 and between CDR1α loop residue Gln31 and peptide positions 1 and 4 (GluP1 and GlyP4 for A2-ELA; LeuP1 and GlyP4 for A2-Tax) (Fig. 3). In all other published TCR·pMHC complexes, the CDR1α and CDR2α loops, which are not encoded by the TRAV12-2 gene, form unique contacts with their respective pMHC complexes compared with the MEL5·A2-ELA complex; this observation lends credence to the idea that TCRs expressing the TRAV12-2 gene could have a selective advantage when binding to cognate antigen restricted by HLA-A*0201 due to germ line-encoded, or “innate,” recognition of residues on the MHC surface and in the bound peptide. Consequently, although the TCRα and TCRβ chains contribute relatively equally in both the A6 TCR and MEL5 complexes, the conservation in the position of the TCRα chain in both complexes indicates that the A6 TCR and MEL5 bind to their cognate pMHCs in an “α-centric” manner (i.e. the binding position of these TCRs is governed by the α chain).
FIGURE 3.
A, the position of the CDR1α and CDR2α loops, encoded by the TRAV12-2 gene, of MEL5 (shown as yellow spheres) in the HLA-A*0201 (shown as a green schematic diagram) ELAGIGILTV peptide (shown as blue sticks) complex. B, the positions of the CDR1α and CDR2α loops, encoded by the TRAV12-2 gene, of the A6 TCR (shown as orange spheres) in the HLA-A*0201 (shown as a blue schematic diagram) LLFGYPVYV peptide (shown as red sticks) complex. C, the conserved interactions (shown as dotted lines) between the CDR1α and CDR2α loops, encoded by the TRAV12-2 gene, of MEL5 (shown as yellow sticks) and the HLA-A*0201 (shown as green sticks) ELAGIGILTV peptide (shown as blue sticks) complex. D, the conserved interactions (shown as dotted lines) between the CDR1α and CDR2α loops, encoded by the TRAV12-2 gene, of the A6 TCR (shown as orange sticks) and the HLA-A*0201 (shown as blue sticks) LLFGYPVYV peptide (shown as red sticks) complex.
The Role of the TCRβ Chain during Peptide Binding
The TCRβ chain utilizes a classical peptide binding mode, interacting with the ELA peptide solely through contacts made by the CDR3β loop. In total, the CDR3β loop makes five hydrogen bonds and five vdW interactions between Thr96, Leu98, and Gly99 of the TCR and AlaP3, GlyP4, IleP5, GlyP6, IleP7, and ThrP9 of the peptide. However, all of the hydrogen bonds, which constitute the majority of the binding energy, are located between the CDR3β loop and the C terminus of the peptide (GlyP6, IleP7, and ThrP9). Interestingly, the position of the CDR3β loop enables a dense network of interactions, including three hydrogen bonds, between TCR residues Leu98 and Gly99 and peptide residue IleP7. These contacts are noteworthy, because the side chain of IleP7 acts like an anchor, pointing down toward the MHC surface, thereby making it an unusual candidate for TCR binding. Thus, the contacts between the CDR3β loop and the ELA peptide contribute significantly to the binding stability of the MEL5·A2-ELA complex.
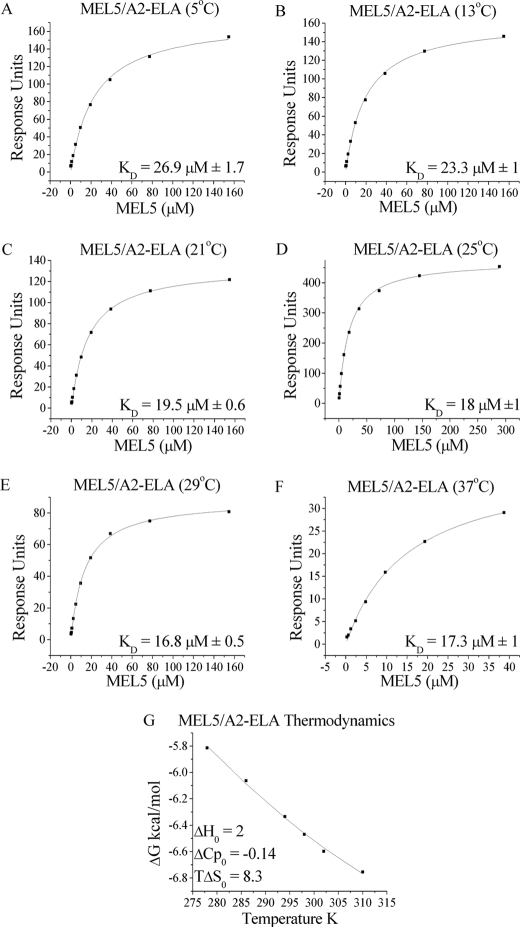
Binding Affinity and Thermodynamics of the MEL5·A2-ELA Complex
We have previously shown that MEL5 binds to A2-ELA with a comparatively weak affinity and extremely fast kinetics compared with other TCR/pMHC interactions (KD = 18 μm; kinetics too fast to measure) (21). To investigate the thermodynamic properties of this interaction, we measured the binding of MEL5 to A2-ELA at 5, 13, 21, 25, 29, and 37 °C using a BIAcore T100TM surface plasmon resonance detection system (Fig. 4 and Table 3). The affinity of the MEL5/A2-ELA interaction increased from KD = 26.9 μm at 5 °C to KD = 17.3 μm at 37 °C (Fig. 4, A–F). The affinity data were plotted as binding ΔG0 versus temperature using nonlinear regression to fit the three-parameter equation to the curve in order to calculate ΔH0, TΔS0, and ΔCp0 (Fig. 4G). At 25 °C, the MEL5/A2-ELA interaction was characterized by a ΔG0 of −6.5 kcal/mol, which is within the normal range for TCR/pMHC interactions (Table 3). The MEL5/A2-ELA interaction is strongly entropically driven, with a favorable TΔS0 of 8.3 kcal/mol. This favorable entropy is likely to be derived mainly from the expulsion of ordered water molecules upon complex formation, allowing the TCR to contact the pMHC surface directly and form the electrostatic and vdW interactions evident in the co-complex structure. Interestingly, this is the first published instance of a TCR/pMHC interaction that is enthalpically unfavorable, with a ΔH0 of 2 kcal/mol. This indicates that there is a net decrease in the number of favorable non-covalent bonds (hydrogen bonds and vdW contacts) during complex formation and reinforces the importance of the favorable entropic contribution to the binding of MEL5 to A2-ELA. Furthermore, this observation could explain the extremely fast kinetics of the MEL5/A2-ELA interaction (i.e. the net loss of bond formation could lead to the observed low stability, and hence the fast off-rate, of the complex). During complex formation, there is normally a net increase in the total buried surface area, which results in a negative ΔCp0 value. In the case of the MEL5/A2-ELA interaction, however, a relatively small ΔCp0 value (−0.14 kcal/mol·K) was observed compared with other TCR·pMHC complexes. This supports the observation that the total buried surface area of 1226 Å2 for the MEL5·A2-ELA complex is the smallest reported to date for any TCR·pMHC complex.
FIGURE 4.
Binding affinity and thermodynamics of the MEL5/A2-ELA interaction. These data were produced by surface plasmon resonance experiments using a BIAcore T100TM machine, which were then analyzed using equilibrium analysis and thermodynamic analysis using the Gibbs equation. The raw data and the fits are shown in A–G. These data illustrate the unusual thermodynamic properties of the MEL5/A2-ELA interaction (G).
TABLE 3.
Thermodynamic analysis of the MEL5/A2-ELA interaction
| Parameters | Values |
|---|---|
| Temperature (°C) | 25 |
| KD (μm) | 18 |
| ΔG0 (kcal/mol) | −6.5 |
| ΔH0 (kcal/mol) | 2 |
| TΔS0 (kcal/mol) | 8.3 |
| ΔCp0 (kcal/mol·K) | −0.14 |
DISCUSSION
Here, we report the structure of an archetypal TRAV12-2-encoded TCR (MEL5), derived from a CD8+ T-cell clone specific for MART-1-(26–35) in complex with A2-ELA. A2-ELA is currently the most studied heteroclitic peptide in the literature and is central to a number of clinical trials (10–12). Although the A2-ELA peptide (ELAGIGILTV) is a heteroclitic version of the MART-1-(26–35) peptide (EAAGIGILTV), structural evidence suggests that the substitution of Ala to Leu at the peptide anchor position 2 does not substantially alter the peptide conformation (42, 44). Thus, an understanding of the molecular basis for T-cell recognition of this antigen should inform the rational improvement of current melanoma therapies.
Previous investigations have shown that CD8+ T-cells engage the HLA-A*0201·MART-1-(26–35) antigen with predictable TCR gene usage patterns (26, 45). Thus, these particular TCRs show classic class II bias (shared TRBV and/or TRAV usage among individuals bearing the same MHCI allele) and class III bias (identical TCRs among individuals bearing the same MHCI allele) (46); interestingly, they can also be detected both in tumor-infiltrated lymph nodes and in the blood of healthy individuals and newborns (26, 45). T-cells specific for the HLA-A*0201 MART-1-(26–35) antigen display striking bias in TRAV gene usage. A recent study revealed 47 of 53 CD8+ T-cell clones (87%) raised on the HLA-A*0201 MART-1-(26–35) target expressed a TCR of the TRAV12-2 family (26). This bias was observed across numerous individuals. Since the TRAV12-2 gene is only expressed on the surface of ∼3% of circulating lymphocytes (47), the very high prevalence of TRAV12-2 usage in these melanoma-specific CD8+ T-cell responses indicates that this particular gene is advantageous during antigen-driven selection. The TRAV12-2-encoded regions of the MEL5 TCR play a dominant role at both the peptide and MHC interface. The CDR1α residue Arg28 helps to fix the TCR to the MHC α2 helix via four electrostatic interactions, whereas the CDR1α residue Gln31 appears integral to peptide recognition through two vdW bonds and four electrostatic interactions to GluP1, LeuP2, GlyP4, and IleP5. In general, the CDR1α loop plays a role comparable with that of the CDR3 loop in other TCR·pMHC structures, given its central position above the N terminus of the peptide and the extensive binding networks between the CDR1α loop and bound peptide. It is striking to note that residues encoded by the TRAV12-2 gene make 9 of 19 of the electrostatic interactions with A2-ELA, more than any of the other TCR regions in this structure. Furthermore, when the bonding networks are taken into account, it becomes clear why the TRAV12-2 subfamily is heavily selected in vivo. For instance, only 3 of 47 TRAV genes encode Arg at position 28, and only 3 of 47 TRAV genes encode Gln at position 31; only the TRAV12-2 gene encodes both. In fact, even the highly homologous sister genes of TRAV12-2, specifically TRAV12-1 and TRAV12-3, probably could not be substituted, given that they encode a polar, uncharged Ser at position 28. This single substitution would probably result in the loss of four electrostatic interactions at the pMHC interface. In support of this idea, no TCRs specific for the HLA-A*0201·MART-1-(26–35) complex have so far been found that use either the TRAV12-1 or the TRAV12-3 genes (26). Thus, the dominant selection of this “quasi-innate,” self-reactive TCR probably represents an advantageous role during host responses against the HLA-A*0201·MART-1-(26–35) antigen that cannot be fulfilled by TCRs of alternate specificities.
Further inspection of the MEL5·A2-ELA complex reveals additional features that could contribute to the dominant expression of the TRAV12-2-encoded CDR1α loop in CD8+ T-cell responses specific for HLA-A*0201·MART-1-(26–35). Strikingly, MEL5 contacts primarily the main chain of the ELA peptide; indeed, only 3 of 17 peptide contacts are side chain interactions. This is disparate from the more equal distribution of side chain versus main chain peptide interactions observed in most other TCR·pMHC structures and implies that MEL5 may be less sensitive to peptide sequence relative to peptide conformation. Moreover, the somatically rearranged CDR3β loop principally utilizes main chain atoms to contact the peptide, whereas the germ line-encoded CDR1α loop uses predominantly side chain atoms to contact the peptide. This interesting dichotomy of binding strategies between the CDR3β and CDR1α loops in the MEL5·A2-ELA complex may have implications for antigen specificity; thus, changes in the sequence of the CDR3β loop may be tolerated when binding to A2-ELA as long as the overall conformation of the loop is maintained, whereas changes in the sequence of the CDR1α loop may disrupt T-cell recognition of A2-ELA due to loss of side chain-specific interactions.
Interestingly, the TRAV12-2 gene is also used by the A6 TCR, which is specific for the A2-Tax complex; this is the first structural example in which an identical gene is shared between TCRs specific for a tumor antigen (MEL5·A2-ELA) and a viral antigen (A6 TCR·A2-Tax). In both of these TCR·pMHCI complexes, the TRAV12-2 gene-encoded CDR1α loops are suspended over the N terminus of their respective cognate pMHCI molecules. This feature enables the CDR1α loops of each TCR to form far more contacts with the antigenic peptide compared with the corresponding CDR3α loops. In both complexes, the CDRβ domains conform to the classical model of TCR·pMHC binding, with the CDR2β loop contributing mainly to MHC contacts and the CDR3β loop contributing mainly to peptide interactions. Furthermore, despite the fact that MEL5 and the A6 TCR are encoded by unique TCRβ chain genes (TRBV30 and TRBV6-5, respectively), the TCRs align in virtually identical positions and orientations over the respective cognate pMHCI molecules in both the MEL5·A2-ELA and the A6 TCR·A2-Tax complexes, thereby indicating that the TCRα chain encoded by the TRAV12-2 gene has a dominant role during TCR·pMHCI docking compared with the TCRβ chain. This observation lends support to the idea that MEL5 and the A6 TCR are “α-centric” (i.e. their binding orientation is governed by common contacts between the TCRα chain and the MHC surface) (18). Moreover, despite the unique sequences of the peptides in the A2-ELA (ELAGIGILTV) and A2-Tax (LLFGYPVYV) complexes, the binding mode implemented by the CDR1α loop encoded by the germ line TRAV12-2 gene in both the MEL5·A2-ELA complex and the A6 TCR·A2-Tax complex is virtually identical; thus, contacts between the CDR1α loop residue Arg28 and the MHC α2 residue Trp167 and, most notably, between the CDR1α loop residue Gln31 and peptide positions 1 and 4 (GluP1 and GlyP4 for A2-ELA; LeuP1 and GlyP4 for A2-Tax) are evident in both complex structures (Fig. 3, C and D). Thus, the specificity of MEL5 for the HLA-A*0201·MART-1-(26–35) antigen is achieved not only through the highly variable somatically rearranged CDR3 loops, as observed for other TCR·pMHC complexes, but also through the germ line-derived CDR1α loop, which contributes significantly to peptide binding. Although considerable contacts between the TCR CDR1 loops and the antigenic peptide have been observed in some other TCR·pMHC complexes (48), this is the first example of specific antigen recognition via identical germ line-encoded receptor loops shared by both anti-viral (A6 TCR·A2-Tax) and anti-self (MEL5·A2-ELA) T-cells.
In summary, this first structure of a human TCR in complex with a decamer peptide bound to MHCI considerably extends our knowledge of TCR/pMHC interactions. First, the MEL5/A2-ELA interaction is the only enthalpically unfavorable TCR interaction ever described (49). Second, the MEL5·A2-ELA complex has the lowest buried surface area (1,226 Å2) of any published TCR·pMHC complex, an observation that is supported by the relatively small ΔCp0 value (−0.14 kcal/mol·K). Third, this structure shows that the specificity of MEL5 for A2-ELA is achieved in large part through interactions with the CDR1α loop, which contributes significantly to peptide binding and whose function is akin to that of classical CDR3 loops during pMHC binding. The dominance of the germ line-encoded regions in this α-centric TCR during pMHC binding indicates an important role for the TRAV12-2 gene product in antigen recognition. Thus, TCRs that use the TRAV12-2 gene may enable CD8+ T-cells to recognize and respond to the HLA-A*0201·MART-1-(26–35) antigen through a “quasi-innate” recognition system. These observations probably explain the biased TCR usage that has been observed in CD8+ T-cell populations specific for HLA-A*0201·MART-1-(26–35) and further reported in a variety of other human diseases (reviewed in Refs. 46 and 50).
Acknowledgments
We thank the staff at the Synchrotron Radiation Source for use of facilities, the UK Research Councils for supplying beamtime, and Professor Eleanor Dodson for assistance with PHASER for structure solution with molecular replacement.
This work was supported in part by the Cardiff University Link Chair Scheme and by Biotechnology and Biological Sciences Research Council (BBSRC) UK Grant BB/H001085/1, which enabled the establishment of protein crystallography at Cardiff University School of Medicine.
The atomic coordinates and structure factors (code 3HG1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TCR
- T-cell receptor
- MHC
- major histocompatibility complex
- pMHC
- peptide major histocompatibility complex
- CDR
- complementarity-determining region
- HLA
- human leukocyte antigen
- vdW
- van der Waals
- A2-ELA
- ELAGIGILTV bound to HLA-A*0201
- pMHCI
- pMHC class I
- β2m
- β2-microglobulin.
REFERENCES
- 1.Kawakami Y., Eliyahu S., Sakaguchi K., Robbins P. F., Rivoltini L., Yannelli J. R., Appella E., Rosenberg S. A. (1994) J. Exp. Med. 180, 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Romero P., Valmori D., Pittet M. J., Zippelius A., Rimoldi D., Lévy F., Dutoit V., Ayyoub M., Rubio-Godoy V., Michielin O., Guillaume P., Batard P., Luescher I. F., Lejeune F., Liénard D., Rufer N., Dietrich P. Y., Speiser D. E., Cerottini J. C. (2002) Immunol. Rev. 188, 81–96 [DOI] [PubMed] [Google Scholar]
- 3.Voelter V., Rufer N., Reynard S., Greub G., Brookes R., Guillaume P., Grosjean F., Fagerberg T., Michelin O., Rowland-Jones S., Pinilla C., Leyvraz S., Romero P., Appay V. (2008) Int. Immunol. 20, 1087–1096 [DOI] [PubMed] [Google Scholar]
- 4.Pittet M. J., Valmori D., Dunbar P. R., Speiser D. E., Liénard D., Lejeune F., Fleischhauer K., Cerundolo V., Cerottini J. C., Romero P. (1999) J. Exp. Med. 190, 705–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chodorowski Z., Anand J. S., Wiœniewski M., Madaliñski M., Wierzba K., Wiœniewski J. (2007) Przegl. Lek. 64, 380–382 [PubMed] [Google Scholar]
- 6.Pittet M. J., Zippelius A., Valmori D., Speiser D. E., Cerottini J. C., Romero P. (2002) Trends Immunol. 23, 325–328 [DOI] [PubMed] [Google Scholar]
- 7.Kawakami Y., Rosenberg S. A. (1997) Int. Rev. Immunol. 14, 173–192 [DOI] [PubMed] [Google Scholar]
- 8.Speiser D. E., Liénard D., Pittet M. J., Batard P., Rimoldi D., Guillaume P., Cerottini J. C., Romero P. (2002) Eur. J. Immunol. 32, 731–741 [DOI] [PubMed] [Google Scholar]
- 9.Valmori D., Fonteneau J. F., Lizana C. M., Gervois N., Liénard D., Rimoldi D., Jongeneel V., Jotereau F., Cerottini J. C., Romero P. (1998) J. Immunol. 160, 1750–1758 [PubMed] [Google Scholar]
- 10.Bins A., Mallo H., Sein J., van den Bogaard C., Nooijen W., Vyth-Dreese F., Nuijen B., de Gast G. C., Haanen J. B. (2007) J. Immunother. 30, 234–239 [DOI] [PubMed] [Google Scholar]
- 11.Chen Q., Jackson H., Shackleton M., Parente P., Hopkins W., Sturrock S., MacGregor D., Maraskovsky E., Tai T. Y., Dimopoulos N., Masterman K. A., Luke T., Davis I. D., Chen W., Cebon J. (2005) Cancer Immun. 5, 5. [PubMed] [Google Scholar]
- 12.Meidenbauer N., Marienhagen J., Laumer M., Vogl S., Heymann J., Andreesen R., Mackensen A. (2003) J. Immunol. 170, 2161–2169 [DOI] [PubMed] [Google Scholar]
- 13.Morgan R. A., Dudley M. E., Wunderlich J. R., Hughes M. S., Yang J. C., Sherry R. M., Royal R. E., Topalian S. L., Kammula U. S., Restifo N. P., Zheng Z., Nahvi A., de Vries C. R., Rogers-Freezer L. J., Mavroukakis S. A., Rosenberg S. A. (2006) Science 314, 126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 15.Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., Burrows S. R., McCluskey J., Rossjohn J. (2007) Nat. Immunol. 8, 268–276 [DOI] [PubMed] [Google Scholar]
- 16.Armstrong K. M., Piepenbrink K. H., Baker B. M. (2008) Biochem. J. 415, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borg N. A., Ely L. K., Beddoe T., Macdonald W. A., Reid H. H., Clements C. S., Purcell A. W., Kjer-Nielsen L., Miles J. J., Burrows S. R., McCluskey J., Rossjohn J. (2005) Nat. Immunol. 6, 171–180 [DOI] [PubMed] [Google Scholar]
- 18.Feng D., Bond C. J., Ely L. K., Maynard J., Garcia K. C. (2007) Nat. Immunol. 8, 975–983 [DOI] [PubMed] [Google Scholar]
- 19.Tynan F. E., Borg N. A., Miles J. J., Beddoe T., El-Hassen D., Silins S. L., van Zuylen W. J., Purcell A. W., Kjer-Nielsen L., McCluskey J., Burrows S. R., Rossjohn J. (2005) J. Biol. Chem. 280, 23900–23909 [DOI] [PubMed] [Google Scholar]
- 20.Borbulevych O. Y., Baxter T. K., Yu Z., Restifo N. P., Baker B. M. (2005) J. Immunol. 174, 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole D. K., Pumphrey N. J., Boulter J. M., Sami M., Bell J. I., Gostick E., Price D. A., Gao G. F., Sewell A. K., Jakobsen B. K. (2007) J. Immunol. 178, 5727–5734 [DOI] [PubMed] [Google Scholar]
- 22.Varela-Rohena A., Molloy P. E., Dunn S. M., Li Y., Suhoski M. M., Carroll R. G., Milicic A., Mahon T., Sutton D. H., Laugel B. E., Moysey R., Cameron B. J., Vuidepot A., Purbhoo M. E., Cole D. K., Phillips R. E., June C. H., Jakobsen B. K., Sewell A. K., Riley J. L. (2008) Nat. Med. 14, 1390–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen J. L., Stewart-Jones G., Bossi G., Lissin N. M., Wooldridge L., Choi E. M., Held G., Dunbar P. R., Esnouf R. M., Sami M., Boulter J. M., Rizkallah P., Renner C., Sewell A., van der Merwe P. A., Jakobsen B. K., Griffiths G., Jones E. Y., Cerundolo V. (2005) J. Exp. Med. 201, 1243–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krausa P., Brywka M., 3rd, Savage D., Hui K. M., Bunce M., Ngai J. L., Teo D. L., Ong Y. W., Barouch D., Allsop C. E. (1995) Tissue Antigens 45, 223–231 [DOI] [PubMed] [Google Scholar]
- 25.Trautmann L., Labarrière N., Jotereau F., Karanikas V., Gervois N., Connerotte T., Coulie P., Bonneville M. (2002) Eur. J. Immunol. 32, 3181–3190 [DOI] [PubMed] [Google Scholar]
- 26.Dietrich P. Y., Le Gal F. A., Dutoit V., Pittet M. J., Trautman L., Zippelius A., Cognet I., Widmer V., Walker P. R., Michielin O., Guillaume P., Connerotte T., Jotereau F., Coulie P. G., Romero P., Cerottini J. C., Bonneville M., Valmori D. (2003) J. Immunol. 170, 5103–5109 [DOI] [PubMed] [Google Scholar]
- 27.Laugel B., van den Berg H. A., Gostick E., Cole D. K., Wooldridge L., Boulter J., Milicic A., Price D. A., Sewell A. K. (2007) J. Biol. Chem. 282, 23799–23810 [DOI] [PubMed] [Google Scholar]
- 28.Garboczi D. N., Ghosh P., Utz U., Fan Q. R., Biddison W. E., Wiley D. C. (1996) Nature 384, 134–141 [DOI] [PubMed] [Google Scholar]
- 29.Boulter J. M., Glick M., Todorov P. T., Baston E., Sami M., Rizkallah P., Jakobsen B. K. (2003) Protein Eng. 16, 707–711 [DOI] [PubMed] [Google Scholar]
- 30.Wyer J. R., Willcox B. E., Gao G. F., Gerth U. C., Davis S. J., Bell J. I., van der Merwe P. A., Jakobsen B. K. (1999) Immunity 10, 219–225 [DOI] [PubMed] [Google Scholar]
- 31.Cole D. K., Dunn S. M., Sami M., Boulter J. M., Jakobsen B. K., Sewell A. K. (2008) Mol. Immunol. 45, 2700–2709 [DOI] [PubMed] [Google Scholar]
- 32.Yuan F., Georgiou T., Hillon T., Gostick E., Price D. A., Sewell A. K., Moysey R., Gavarret J., Vuidepot A., Sami M., Bell J. I., Gao G. F., Rizkallah P. J., Jakobsen B. K. (2007) Acta. Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 63, 758–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leslie A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography, Vol. 26, Daresbury Laboratory, Warrington, UK [Google Scholar]
- 34.Collaborative Computational Project 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 35.McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 36.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 39.Ding Y. H., Smith K. J., Garboczi D. N., Utz U., Biddison W. E., Wiley D. C. (1998) Immunity 8, 403–411 [DOI] [PubMed] [Google Scholar]
- 40.Ding Y. H., Baker B. M., Garboczi D. N., Biddison W. E., Wiley D. C. (1999) Immunity 11, 45–56 [DOI] [PubMed] [Google Scholar]
- 41.Stewart-Jones G. B., McMichael A. J., Bell J. I., Stuart D. I., Jones E. Y. (2003) Nat. Immunol. 4, 657–663 [DOI] [PubMed] [Google Scholar]
- 42.Sliz P., Michielin O., Cerottini J. C., Luescher I., Romero P., Karplus M., Wiley D. C. (2001) J. Immunol. 167, 3276–3284 [DOI] [PubMed] [Google Scholar]
- 43.Kjer-Nielsen L., Clements C. S., Purcell A. W., Brooks A. G., Whisstock J. C., Burrows S. R., McCluskey J., Rossjohn J. (2003) Immunity 18, 53–64 [DOI] [PubMed] [Google Scholar]
- 44.Borbulevych O. Y., Insaidoo F. K., Baxter T. K., Powell D. J., Jr., Johnson L. A., Restifo N. P., Baker B. M. (2007) J. Mol. Biol. 372, 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valmori D., Dutoit V., Liénard D., Lejeune F., Speiser D., Rimoldi D., Cerundolo V., Dietrich P. Y., Cerottini J. C., Romero P. (2000) J. Immunol. 165, 533–538 [DOI] [PubMed] [Google Scholar]
- 46.Turner S. J., Doherty P. C., McCluskey J., Rossjohn J. (2006) Nat. Rev. Immunol. 6, 883–894 [DOI] [PubMed] [Google Scholar]
- 47.Yoshioka T., Matsutani T., Iwagami S., Tsuruta Y., Kaneshige T., Toyosaki T., Suzuki R. (1997) J. Immunol. Methods 201, 145–155 [DOI] [PubMed] [Google Scholar]
- 48.Tynan F. E., Burrows S. R., Buckle A. M., Clements C. S., Borg N. A., Miles J. J., Beddoe T., Whisstock J. C., Wilce M. C., Silins S. L., Burrows J. M., Kjer-Nielsen L., Kostenko L., Purcell A. W., McCluskey J., Rossjohn J. (2005) Nat. Immunol. 6, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 49.Armstrong K. M., Insaidoo F. K., Baker B. M. (2008) J. Mol. Recognit. 21, 275–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miles J. J., Silins S. L., Burrows S. R. (2006) Curr. Med. Chem. 13, 2725–2736 [DOI] [PubMed] [Google Scholar]