Abstract
The killer cell lectin-like receptor G1, KLRG1, is a cell surface receptor expressed on subsets of natural killer (NK) cells and T cells. KLRG1 was recently found to recognize E-cadherin and thus inhibit immune responses by regulating the effector function and the developmental processes of NK and T cells. E-cadherin is expressed on epithelial cells and exhibits Ca2+-dependent homophilic interactions that contribute to cell-cell junctions. However, the mechanism underlying the molecular recognition of KLRG1 by E-cadherin remains unclear. Here, we report structural, binding, and functional analyses of this interaction using multiple methods. Surface plasmon resonance demonstrated that KLRG1 binds the E-cadherin N-terminal domains 1 and 2 with low affinity (Kd ∼7–12 μm), typical of cell-cell recognition receptors. NMR binding studies showed that only a limited N-terminal region of E-cadherin, comprising the homodimer interface, exhibited spectrum perturbation upon KLRG1 complex formation. It was confirmed by binding studies using a series of E-cadherin mutants. Furthermore, killing assays using KLRG1+NK cells and reporter cell assays demonstrated the functional significance of the N-terminal region of E-cadherin. These results suggest that KLRG1 recognizes the N-terminal homodimeric interface of domain 1 of E-cadherin and binds only the monomeric form of E-cadherin to inhibit the immune response. This raises the possibility that KLRG1 detects monomeric E-cadherin at exposed cell surfaces to control the activation threshold of NK and T cells.
Natural killer (NK)3 cells play a critical role in the innate immune system because of their ability to kill other cells. For example, NK cells can kill virus-infected cells and tumor cells without presensitization to a specific antigen, and they produce various cytokines, including interferon-γ and tumor necrosis factor-α (1). NK cells are controlled by both inhibitory and activating receptors that are expressed on their surfaces (2). The killer cell Ig-like receptor, Ly49, CD94/NKG2, and paired Ig-like type 2 receptor families include both inhibitory and activating members and thus are designated as paired receptor families. On the other hand, some inhibitory receptors, including KLRG1 (killer cell lectin-like receptor G1), and activating receptors, such as NKG2D, also exist. The integration of the signals from these receptors determines the final functional outcome of NK cells.
These inhibitory and activating receptors can also be divided into two structurally different groups, the Ig-like receptors and the C-type lectin-like receptors, based on the structural aspects of their extracellular regions. The Ig-like receptors include killer cell Ig-like receptors and the leukocyte Ig-like receptors, and the C-type lectin-like receptors include CD94/NKG2(KLRD/KLRC), Ly49(KLRA), NKG2D(KLRK), NKR-P1(KLRB), and KLRG1. Many of these immune receptors recognize major histocompatibility complex class I molecules or their relatives (2–4), but there are still many orphan receptors expressed on NK cells. KLRG1 was one such orphan receptor; however, E-cadherin was recently found to be a ligand of KLRG1 (5, 6). Although major histocompatibility complex-receptor interactions have been extensively examined, the molecular basis of non-major histocompatibility complex ligand-receptor recognition is poorly understood.
KLRG1 is a type II membrane protein, with one C-type lectin domain in the extracellular region, one transmembrane region, and one immunoreceptor tyrosine-based inhibitory motif. KLRG1 is expressed on a subset of mature NK cells in spleen, lungs, and peripheral blood during normal development. KLRG1 expression is induced on the surface of NK cells during viral responses (7, 8). NK cells expressing KLRG1 produce low levels of interferon-γ and cytokines and have a slow in vivo turnover rate and low proliferative responsiveness to interleukin-15 (9). Furthermore, KLRG1 is recognized as a marker of some T cell subsets, as follows. KLRG1 defines a subset of T cells, short lived effector CD8 T cells (SLECs), which are mature effector cells that express high levels of KLRG1 and cannot be differentiated into long lived memory CD8 T cells. In addition, memory precursor effector cells express low levels of KLRG1 and harbor the potential to become long lived memory CD8 T cells (10). Since SLECs exhibit stronger effector function than memory precursor effector cells, it is potentially beneficial, in terms of preventing harmful excess cytotoxicity, that SLECs express KLRG1 at a higher level to inhibit the immune response. Taken together, the expression of KLRG1 during the viral response and normal development might confer the inhibition of effector function and the regulation of NK and T cell proliferation (9).
E-cadherin plays a pivotal role in Ca2+-dependent cell-cell adhesion and also contributes to tissue organization and development (11–14). E-cadherin is primarily expressed on epithelial cells, and its extracellular region consists of several domains that include cadherin motifs (15, 16). These domains mediate Ca2+-dependent homophilic interactions to facilitate cell adhesion. When E-cadherins form cis- or trans-homodimers, they utilize their N-terminal regions as an interface, which can dock with domain 1 of another E-cadherin to form strand exchange (17). Therefore, the N-terminal region plays important roles in homophilic binding and cell adhesion.
KLRG1 recognizes E-cadherins (and other class I cadherins), which are widely expressed in tissues and form tight adhesive cell-cell junctions, and Ito et al. (5) demonstrated that E-cadherin binding by KLRG1 inhibits NK cytotoxicity. Further, Gründermann et al. (6) showed that the E-cadherin-KLRG1 interaction inhibits the antigen-induced proliferation and induction of the cytolytic activity of CD8 T cells. Therefore, it is plausible that E-cadherin recognition by KLRG1, expressed on the surfaces of NK cells and T cells, may raise their activation thresholds by transducing inhibitory signals. Such an inhibition would prevent the excess injury of normal cells, which might result in inflammatory autoimmune diseases. KLRG1 may also have an important role in monitoring and removing cancer cells that lose E-cadherin expression. A recent report demonstrated that N-terminal domains 1 and 2 of E-cadherin are critical for KLRG1 recognition (18); however, despite accumulating evidence supporting the functional importance of the E-cadherin-KLRG1 interaction, the molecular basis of this interaction is poorly understood. Here, we report that the N-terminal region of E-cadherin, comprising the dimer interface, is the binding site for KLRG1. This suggests that KLRG1 does not recognize the dimeric form of E-cadherin but rather recognizes the monomeric form, which is exposed on the cell surfaces of disrupted or infected cells. This may suppress excess immune responses.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant Proteins
The plasmid pET15bEC-D1D2(His10Xa), encoding domains 1 and 2 of E-cadherin, with a His10 tag, a spacer sequence, and a Factor Xa recognition site at the N terminus (EC-D1D2(His10Xa)), and pET15bEC-D2D3(His10) encoding domains 2 and 3 (residues 109–332) with an additional His10 tag (EC-D2D3(His10)) were used to express recombinant proteins in Escherichia coli strain BL21 (DE3) pLysS. Soluble EC-D1D2(His10Xa) was subjected to Ni2+-nitrilotriacetic acid affinity chromatography (HisTrapFF, 5 ml; GE Healthcare), or the inclusion bodies of EC-D1D2(His10Xa) were dissolved in guanidine buffer (6 m guanidine HCl, 50 mm MES-NaOH, pH 6.5, 100 mm NaCl, 10 mm EDTA). To refold the recombinant protein, 10–20 mg of solubilized inclusion bodies were gradually diluted by the addition of refolding buffer (20 mm Tris-HCl, pH 7.9, 300 mm NaCl, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride) at 4 °C, into a final volume of 1 liter. The resulting solution was stirred for 2 days and was concentrated to 5–10 ml by a VIVA FLOW system and an Amicon Ultra filter (Millipore). The protein was purified by gel filtration chromatography (HiLoad26/60 Superdex 75 pg; GE Healthcare). After purification, EC-D1D2(His10Xa) was treated with Factor Xa to cleave the extra amino acids. The resulting EC-D1D2 protein was designated as EC-D1D2. EC-D2D3(His10) was prepared by the method of the refolded EC-D1D2.
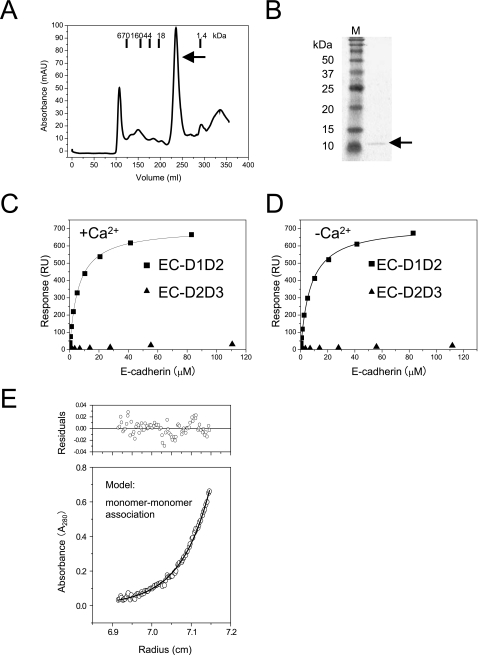
The DNA encoding the extracellular region (residues 73–188) of Mus musculus KLRG1 was ligated into pET3c, to create the plasmid pET3cKLRG1. The recombinant protein was expressed as inclusion bodies and was refolded, in a similar manner as EC-D1D2 and EC-D2D3. The refolded protein was purified by gel filtration chromatography (Fig. 1). For surface plasmon resonance (SPR) analysis, a biotinylated version of KLRG1 was prepared, as described previously (5).
FIGURE 1.
Binding analysis of KLRG1 to E-cadherin using SPR and AUC. A, gel filtration chromatogram of KLRG1 on HiLoad26/60 Superdex 75 pg (GE Healthcare). The bars indicate the elution positions of the molecular mass markers (kDa). KLRG1 was eluted at the peak indicated by the black arrow. B, this peak was analyzed by SDS-PAGE, which indicated that it contained KLRG1 (indicated by the arrow on the right of the gel). Lane M contained protein markers with standard molecular masses. C and D, binding of E-cadherin variants to KLRG1 in the presence of 10 mm calcium chloride (C) and in the absence of calcium chloride (D). Black squares and black circles indicate EC-D1D2 and EC-D2D3, respectively. KLRG1 was immobilized on research grade CM5 chips (BIAcore) at 1,000 response units (RU). E, analytical ultracentrifugation indicated that the molecular mass of the EC-D1D2-KLRG1 complex is 36 kDa and the binding affinity is 9 μm. In the lower panel, the open circles show the actual values obtained in this experiment, and the solid line indicates the fitted data based on the monomer-monomer binding model. In the upper panel, the residuals derived from the fitted data are shown.
SPR
Recombinant cadherins, including the Ala-scanning mutants, were dissolved in HBS-P buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 0.005% Surfactant P20) (BIAcore AB). SPR experiments were performed with a BIAcore2000 (Biacore AB). Biotinylated KLRG1 was immobilized on the CM5 sensor chip (BIAcore AB), onto which streptavidin had been covalently coupled. Biotinylated bovine serum albumin was used as a negative control protein. All cadherin samples were injected over the immobilized KLRG1 protein, at a flow rate of 10 μl/min, in HBS-P buffer with 10 mm CaCl2 or 3 mm EDTA (for the Ca2+-free conditions). The binding response at each concentration was calculated by subtracting the equilibrium response measured in the control flow cell from the response in each sample flow cell. The data were analyzed using the BIA evaluation version 4.1 (Biacore AB) and ORIGIN version 7 software (Microcal Inc.). Affinity constants (Kd) were derived by nonlinear curve fitting of the standard Langmuir binding isotherm.
Analytical Ultracentrifugation (AUC)
AUC was carried out using a Beckman Optima XL-I analytical ultracentrifuge with absorption optics, an An-50 Ti rotor, and standard double-sector centerpiece cells. Sedimentation equilibrium measurements were performed at 4 °C, and concentration profiles were recorded at 0, 20, 23, 26, and 29 h after the velocity of the rotor reached 9,000, 14,000, and 20,000 rpm. At each time and velocity, one scan was collected. Data were analyzed using the standard Optima XL-I data analysis software. All of the protein samples were in 20 mm Tris-Cl buffer, pH 7.4, with 100 mm NaCl. Monomeric EC-D1D2 and monomeric KLRG1 were mixed at a molar ratio of 1:1 without calcium.
NMR Analysis of E-cadherin Binding to KLRG1
Uniformly 15N-labeled EC-D1D2 was expressed in E. coli BL21(DE3) pLysS bearing the plasmid pET15bEC-D1D2(His10Xa), grown in M9 minimal medium containing 1 g/liter 15NH4Cl, and was prepared by the same method used for the unlabeled EC-D1D2. KLRG1 was prepared as described previously. The KLRG1 and EC-D1D2 proteins were both dissolved in the HBS-E buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 3 mm EDTA). For the direct observation of KLRG1 binding to 15N-labeled EC-D1D2, a series of 1H-15N heteronuclear sequential quantum correlation (HSQC) spectra at 1:0 (EC-D1D2/KLRG1, 70 μm:0 μm), 1:0.5 (70 μm:35 μm), 1:1 (70 μm:70 μm), and 1:2 (70 μm:140 μm) ratios were measured at 25 °C. All HSQC spectra were measured at 25 °C and collected on 600-MHz spectrometers (Bruker) equipped with cryoprobes. The collected data were analyzed by NMRView. Chemical shift changes were defined by the formula, ((δ1H2 + (δ15N/5)2)/2)½ (19).
Mutant Preparation
Alanine-scanning and D90K mutagenesis of EC-D1D2 were performed using a QuikChange (Stratagene) kit with pET15bEC-D1D2(His10Xa) as the template. All of the EC-D1D2 mutants accumulated as soluble protein and purified in the manner of EC-D1D2(His10Xa). SPR experiments for all mutants were performed in HBS-P buffer with 3 mm EDTA.
Cell Lines and Reagents
Mouse thymoma cell line BW5147 and mouse NK cell line NK 03 expressing KLRG1, KLRG1-NK03, were prepared as described previously (5). The retrovirus vector, pMXs-IRES-GFP, and retrovirus packaging cell line, Plat-E, were kindly provided by T. Kitamura (University of Tokyo).
Culture supernatant from anti-mouse E-cadherin monoclonal antibody ECCD-2 hybridoma cells was kindly provided by M. Takeichi (Institute of Physical and Chemical Research CDB, Hyogo, Japan) and was used to assess E-cadherin expression. Goat F(ab′)2 fragment of anti-mouse IgG(H+L)-phycoerythrin (PE) was purchased from Beckman Coulter.
Expression of E-cadherin on BW5147 Cells by Retroviral Transduction
DNA encoding the entire extracellular region of wild-type and mutant M. musculus E-cadherin was ligated into the pMXs-IRES-GFP vector using XhoI and NotI restriction enzyme sites to produce pMXs-WT mEC-IRES-GFP. These vectors were transfected into retrovirus packaging Plat-E cells, using Lipofectamine 2000 reagent (Invitrogen). Culture supernatants containing retrovirus were used to transduce BW5147 cells. BW5147 cells were stained with anti-mouse E-cadherin antibody (ECCD-2) and goat F(ab′)2 fragment anti-mouse IgG(H+L)-PE to confirm the expression of E-cadherin. Data were acquired with a FACSCalibur system (BD Biosciences) and analyzed with FlowJo software (TreeStar, Inc.).
KLRG1 Tetramer Binding Assay
KLRG1 tetramer was formed by incubating soluble biotinylated KLRG1 with PE-coupled streptavidin, as previously reported (20). The concentration of KLRG1 tetramer used in the binding studies was 20 μg/ml. BW5147 cells expressing E-cadherin were incubated with KLRG1 tetramer or PE-coupled streptavidin in Hanks' solution 2 (Nissui Pharmaceuticals) containing 0.1% bovine serum albumin and 0.1% NaN3 (fluorescence-activated cell sorting buffer) and were then analyzed by FACSCalibur.
KLRG1 Reporter Assay
The KLRG1 reporter cell line was established as described previously (5). The reporter cells were stimulated by target cells expressing E-cadherin for 16 h at 37 °C in 5% CO2, and then accumulated β-galactosidase activity was determined by a colorimetric assay using chlorophenol red-β-d-galactopyranoside (Wako Pure Chemicals) as substrate.
Killing Assay
The cytotoxicity of KLRG1-expressing NK03 cells against target cells expressing E-cadherin was examined using 5,6-carboxyl-succinimidyl-fluorescein ester (Dojindo) and 7-amino-actinomycin (Sigma). First, BW5147 cells expressing E-cadherin were incubated with 6 μm 6-carboxyl-succinimidyl-fluorescein ester at room temperature for 5 min in the dark. Afterward, 6-carboxyl-succinimidyl-fluorescein ester-labeled target cells were incubated with effector KLRG1-expressing NK03 cells in the presence or absence of anti-KLRG1 3D4 F(ab′)2 in a CO2 incubator for 6 h in the dark. The expression of E-cadherin protected the target cells from killing by KLRG1-expressing NK03 cells, and the protection was reversed by inhibiting the KLRG1-E-cadherin interaction with anti-KLRG1 3D4 F(ab′)2. Then 1 μg/ml 7-amino-actinomycin was added to the reaction solution, and cells were incubated on ice for 15 min. Data were collected using the FACSCalibur system and analyzed on FlowJo.
RESULTS
Preparation of Recombinant Proteins
N-terminal domains 1 and 2 of M. musculus E-cadherin (residues 1–221) with a His10 tag, a spacer sequence, and a Factor Xa recognition site at the N terminus were expressed in E. coli either as a soluble protein or inclusion bodies. The soluble protein was purified using Ni2+-nitrilotriacetic acid chromatography, and the inclusion bodies were refolded by the standard dilution method. For both proteins, the extra N-terminal amino acids were removed with Factor Xa, resulting in the authentic N-terminal sequence (Asp1-Trp2-Val3-) (designated as EC-D1D2). EC-D1D2 was further purified by ion exchange chromatography. A final yield of 2–4 mg of purified EC-D1D2 was obtained from 1 liter of culture. N-terminal domains 2 and 3 of M. musculus E-cadherin (designated as EC-D2D3; residues 109–332) was also produced as inclusion bodies in E. coli and refolded by the standard dilution method. The refolded protein was purified by gel filtration chromatography.
Next, the extracellular region of M. musculus KLRG1 (residues 73–188) was expressed in E. coli as inclusion bodies. KLRG1 was refolded by the dilution method, in a similar manner as E-cadherin. The refolded protein was purified by gel filtration chromatography (Fig. 1, A and B), and the final yield was 1 mg of KLRG1 from 1 liter of culture.
Surface Plasmon Resonance Analysis of KLRG1-E-cadherin Binding
To clarify the molecular interaction between KLRG1 and E-cadherin, we performed SPR analysis, using recombinant E-cadherins EC-D1D2 and EC-D2D3. Recombinant E-cadherin variants EC-D1D2 and EC-D2D3 were injected over the flow cells, in which biotinylated KLRG1 had been immobilized at a level of 1,000 response units. As a negative control, biotinylated bovine serum albumin was immobilized on one of the flow cells, at a level similar to that of KLRG1. The response derived from the negative control was subtracted from each response derived from the recombinant E-cadherins. Fig. 1, C and D, shows the conventional plots of these binding data. In accordance with recent results showing that the deletion of either domain 1 or 2 abolished reactivity in KLRG1 reporter cell assays (18), Fig. 1C indicates that KLRG1 can bind to domains 1 and 2 of E-cadherin (EC-D1D2) but not to domains 2 and 3 (EC-D2D3). The KLRG1-EC-D1D2 interaction conforms to a simple 1:1 (Langmuir) binding model, and its dissociation constant (Kd) was 7 μm at 25 °C in the presence of Ca2+. This affinity is within the range of Kd values measured for other cell-cell recognition molecules (supplemental Table S1). Furthermore, the KLRG1-E-cadherin interaction in the absence of Ca2+ exhibited a Kd of 12 μm, indicating that binding was independent of Ca2. Thus, Ca2+-induced reduction of interdomain flexibility does not have a significant effect on KLRG1 binding (Fig. 1, C and D; summarized in supplemental Table S1). These results imply that KLRG1 binding is dependent on the N-terminal domain of E-cadherin.
Characterization of the EC-D1D2-KLRG1 Complex
The SPR data indicated that the EC-D1D2-KLRG1 interaction occurs at 1:1 stoichiometry. To confirm this, we performed AUC. Solutions of EC-D1D2 and KLRG1 were mixed at a molar ratio of 1:1. The experimental data fit well to the monomer-monomer association model (Fig. 1E). The estimated molecular mass of the EC-D1D2-KLRG1 complex was 36 kDa, which corresponds to the molecular mass of a complex between monomeric KLRG1 (13 kDa) and monomeric EC-D1D2 (24 kDa) (Fig. 1E). Furthermore, gel filtration analysis demonstrated that, when EC-D1D2 and KLRG1 were mixed under the same conditions as for AUC, the complex eluted at the position of the 1:1 complex (data not shown). These results provide clear evidence for the 1:1 binding stoichiometry of the KLRG1-E-cadherin interaction.
NMR Analysis of EC-D1D2 Binding to KLRG1
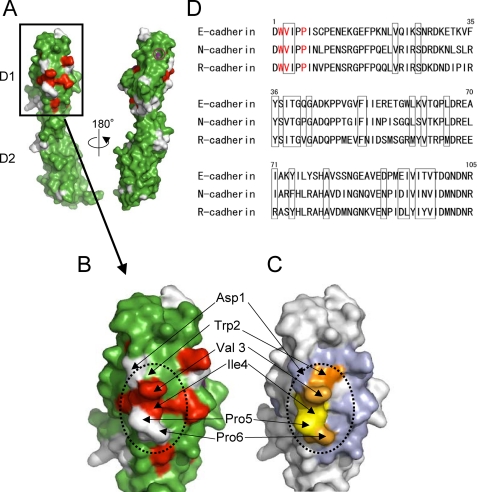
Next, to identify the binding site of KLRG1 on EC-D1D2, we performed an NMR chemical shift perturbation study. 70 μm 15N-labeled EC-D1D2 was mixed with nonlabeled KLRG1 at concentrations of 0, 35, 70, and 140 μm. 1H-15N HSQC spectra of the mixed samples were measured (supplemental Fig. S1). Chemical shift changes were observed in the HSQC spectrum when unlabeled KLRG1 bound to 15N-labeled EC-D1D2, as compared with the HSQC spectrum of free 15N-labeled EC-D1D2. These chemical shift changes were saturated at 70 μm KLRG1, corresponding to a 1:1 binding stoichiometry. The amino acid residues whose HSQC peaks were largely shifted or disappeared upon the complex formation are localized to the N-terminal region of EC-D1D2, as depicted in Fig. 2, A and B. This result indicates that KLRG1 can recognize the N-terminal region of EC-D1D2. The opposite face of the N-terminal region did not have any residues that showed large chemical shift changes (Fig. 2A).
FIGURE 2.
NMR and mutagenesis analyses of E-cadherin binding to KLRG1. A and B, a map of the amino acids with HSQC peaks that shifted or disappeared upon complex formation with KLRG1 is shown (red, disappeared; purple (Tyr36, small open circle to the right of A), >0.06ppm chemical shift change; green, <0.06 ppm chemical shift change; white, unassigned). The dotted line indicates the putative KLRG1 binding area. The surfaces of EC-D1D2 (A) and EC-D1 (B) are shown. C, mapping of the significant residues that showed no or reduced binding affinity on the structure of E-cadherin domain 1. Trp2, colored orange, did not bind to KLRG1. Light orange coloring of the residues (Val3 and Pro6) indicates that these mutations reduced the KLRG1 binding by 5–6-fold, relative to the wild type. Light blue coloring indicates that the mutations of these residues, including the D90K mutation, did not affect KLRG1 binding. Yellow coloring of the residues (Ile4 and Pro5) indicates that these mutations abolished KLRG1 tetramer binding. All mutations of orange, light orange, and yellow residues abrogated KLRG1-mediated inhibition of NK cell cytotoxic activity. D, amino acid sequence alignment of E-cadherin and other type I classical cadherins. The amino acid numbers above the alignment are derived from E-cadherin. The red characters show the positions of the Ala mutations that reduced KLRG1 binding. Residues that showed large chemical shift changes or disappeared upon complex formation in the HSQC spectra are boxed.
Mutagenesis Mapping of the KLRG1 Binding Site on E-cadherin by Tetramer Staining and Reporter Cell Assays
To confirm the results of the NMR binding study, alanine-scanning mutagenesis was performed. The mutants of EC-D1D2, shown in Table 1, were prepared as described under “Experimental Procedures.” SPR analysis using these mutants showed that the W2A mutant cannot bind to KLRG1. In addition, both the V3A and P6A mutants showed significantly reduced KLRG1 binding. All of these amino acids are in the N-terminal region of E-cadherin (Fig. 2, C and D), consistent with the results of the NMR analysis. The other mutants, including the D90K mutant, bound to KLRG1 at levels comparable that of the wild-type protein. The mutagenesis data are summarized in Table 1.
TABLE 1.
SPR study of KLRG1 binding to wild-type and mutant E-cadherin
The ligand was KLRG1, and the analytes were EC-D1D2 mutants, including D90K. The values are means ± range derived from two experiments. NB, no binding.
| Analytes | Kd (25 °C) |
|---|---|
| μm | |
| Wild type | 12 ± 3.0 |
| D1A | 19 ± 0.0 |
| W2A | NB |
| V3A | 66 ± 2.1 |
| P6A | 86 ± 9.2 |
| N20A | 14 ± 0.7 |
| F35A | 7.8 ± 0.1 |
| S37A | 6.2 ± 0.3 |
| T39A | 8.8 ± 0.3 |
| Q64A | 13 ± 0.7 |
| K73A | 5.5 ± 0.7 |
| I75A | 8.0 ± 0.0 |
| Y77A | 22 ± 0.0 |
| E89A | 30 ± 0.7 |
| D90K | 20 ± 2.1 |
| P91A | 8.6 ± 0.2 |
| E93A | 24 ± 2.8 |
| V95A | 22 ± 0.0 |
| T97A | 19 ± 0.7 |
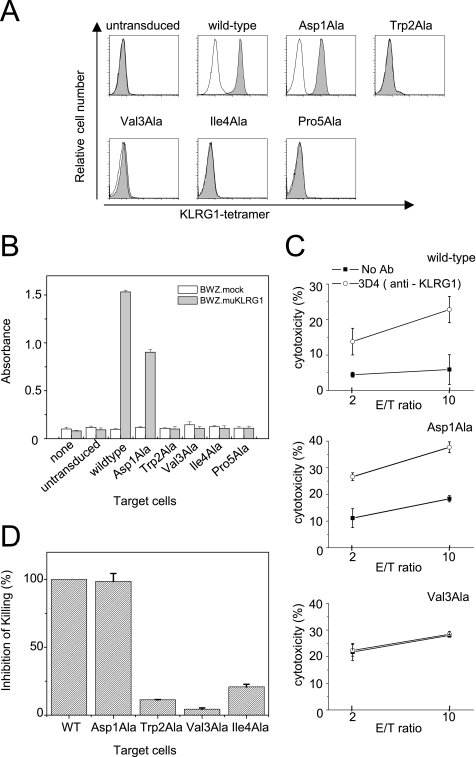
To further examine the KLRG1 binding site on the N-terminal region of E-cadherin, KLRG1 tetramer binding to wild type or a series of mutant E-cadherins (D1A, W2A, V3A, I4A, and P5A) expressed on the cell surface was examined by flow cytometry (Fig. 3A). BW5147 cells expressing wild-type or mutant E-cadherin were generated by retroviral transduction, and E-cadherin expression was confirmed by anti-E-cadherin staining with monoclonal antibody, ECCD2, as shown in supplemental Fig. S2. PE-conjugated KLRG1 tetramer was prepared as described under “Experimental Procedures.” The W2A, V3A, I4A, and P5A mutants failed to bind the KLRG1 tetramer. These data indicate that KLRG1 recognizes a relatively large area of the N-terminal region of E-cadherin that is responsible for homophilic association, supporting the idea that KLRG1 can only bind to monomeric E-cadherin. Interestingly, the D1A mutant E-cadherin bound to the KLRG1 tetramer at levels comparable with that of wild type (Fig. 3A).
FIGURE 3.
Effects of E-cadherin mutagenesis on KLRG1 tetramer binding, reporter cell assays, and KLRG1-expressing NK cell killing assays. A, untransduced BW5147 cells or BW5147 cells transduced with wild-type or mutant E-cadherin (D1A, W2A, V3A, I4A, and P5A) were stained with KLRG1 tetramer (shaded histograms) or PE-streptavidin (thin lines). B, KLRG1 reporter cells were stimulated with the indicated target cells for 16 h and assayed for β-galactosidase activity. The bars represent the relative activities. C, KLRG1-expressing NK03 cell killing assays against BW5147 cells expressing wild-type or mutant E-cadherins. The assays were performed in the presence or absence of the anti-KLRG1 3D4 F(ab′)2. Representative data for wild-type, D1A, and V3A E-cadherins are shown. D, bar (mean ± S.D.) indicates the differences of KLRG1-expressing NK03 cell cytotoxicity against target cells expressing the indicated E-cadherins in the presence and absence of the anti-KLRG1 3D4 F(ab′)2. The difference of target cells expressing wild-type E-cadherin was set as 100%.
We next performed KLRG1 reporter cell assays using the KLRG1-expressing cell line BWZ.muKLRG1 and BW5147 cells expressing either wild-type or mutant E-cadherins. The BWZ.muKLRG1 cells express a chimeric receptor that has the extracellular and transmembrane regions of KLRG1 and the cytoplasmic region of mouse T cell receptor ζ chain, which can mediate the activating signal to induce interleukin-2 gene expression. This cell line also harbors a β-galactosidase reporter gene under the control of the interleukin-2 promoter. Thus, the E-cadherin binding to the chimera KLRG1 can stimulate the β-galactosidase expression. The W2A, V3A, I4A, and P5A E-cadherin mutants failed to induce β-galactosidase expression (Fig. 3B). In contrast, the wild-type and D1A E-cadherins induced the expression of β-galactosidase (Fig. 3B). These results are consistent with the SPR and tetramer binding experiments and indicate that the N-terminal region of E-cadherin plays an essential role in KLRG1-mediated cellular signaling.
Killing Assay Using KLRG1-expressing NK Cells
To investigate how much the N-terminal mutations of E-cadherin affect NK cell function, we performed NK cell cytotoxicity assays using the KLRG1-expressing mouse NK cell line, NK03. BW5147 cells expressing wild-type or mutant E-cadherin were used as target cells in the killing assays, which were performed as described under “Experimental Procedures.” The expression of wild-type E-cadherin protected target cells from killing by KLRG1-expressing NK03 cells. This protection was reversed in the presence of the F(ab′)2 fragment of anti-KLRG1 antibody 3D4, which inhibits the KLRG1-E-cadherin interaction (Fig. 3, C and D). Target cells expressing wild-type E-cadherin did not show any difference of killing by KLRG1-deficient NK03 cells in the presence or absence of the anti-KLRG1 3D4 F(ab′)2 (data not shown). Similar data were obtained using cells expressing the D1A mutant E-cadherin, consistent with the KLRG1-E-cadherin binding and reporter cell assays. On the other hand, target cells expressing the W2A, V3A, and I4A mutant E-cadherins were killed by KLRG1-expressing NK03 cells at similar levels both in the presence and absence of the anti-KLRG1 3D4 F(ab′)2. These results show that E-cadherin mutants that abrogate binding also abolish KLRG1-mediated inhibition of killing by KLRG1-expressing NK03 cells (Fig. 3, C and D). These results indicate that the N-terminal amino acids of E-cadherin are critical for the KLRG1-mediated inhibition of NK cell cytotoxicity.
DISCUSSION
E-cadherin is expressed mainly by epithelial cells and mediates Ca2+-dependent cell-cell adhesion. The N-terminal regions of cadherins are known to play an important role in homotypic adhesion (14, 17). The N-terminal region of E-cadherin can adopt two conformations. In the intramolecular conformation, Trp2 interacts with Glu89 and Met92. In the intermolecular conformation, the N-terminal region can dock with domain 1 of another cadherin to form an adhesive dimer, in which strand exchange occurs (Fig. 4, left) (21). This study shows that KLRG1 recognizes a relatively broad area of the N-terminal region of E-cadherin (Trp2–Pro6), which largely overlaps with the strand-exchanged homodimer interface. In addition, cells expressing E-cadherin mutants (W2A, V3A, I4A, and P5A) could not interact with KLRG1 tetramer or induce intracellular signaling of KLRG1 reporter cell lines. Further, the expression of the N-terminal mutant E-cadherins (W2A, V3A, I4A) on target cells did not inhibit the cytotoxic activity of KLRG1-expressing NK cells. These results demonstrate that KLRG1 specifically recognizes the strand-exchanged homodimeric interface of E-cadherin, indicating that KLRG1 only binds to the monomeric form of E-cadherin, with an exposed N-terminal region, to regulate immune function, such as NK cell cytotoxicity.
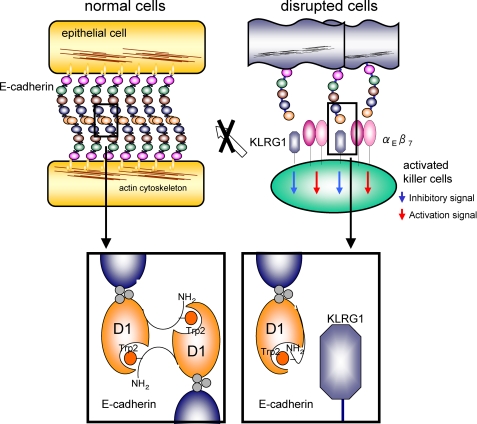
FIGURE 4.
Predicted mechanism of E-cadherin recognition by KLRG1. Top, schematic representation of a normal adherens junction (left), comprising the homodimeric interactions of E-cadherins (colored circles indicate cadherin domains) and disrupted or infected tissues (right), which are recognized by NK or T cells (large green circle). Bottom, the strand-exchanged homodimeric interactions of EC-D1 (black line, N-terminal strand; orange ball, Trp2; orange circle, EC-D1) are shown at the bottom left. KLRG1 (deep blue angular shapes), expressed on NK or T cells, recognizes monomeric E-cadherin on disrupted cells. The αEβ7 integrin, shown in pink circles, may be expressed on KLRG1-expressing lymphocytes to compete with E-cadherin binding and signaling by KLRG1.
In normal epithelial tissues, two E-cadherin molecules interact with each other to form strand-exchanged homodimeric complexes constituting adherent junctions, as described above, and thus they are not easily accessible to NK cells or T cells (Fig. 4, left). However, E-cadherin may be exposed on the surfaces of disrupted or infected epithelial cells. In turn, NK cells and T cells can directly detect E-cadherin by utilizing KLRG1, but this recognition occurs in a more sophisticated manner. Here, we show that KLRG1 recognizes the N-terminal strand-exchanged homodimer interface of E-cadherin and thus can only bind to the monomeric form to inhibit KLRG1-mediated cytotoxicity. This monomeric form is thought to be expressed on abnormal epithelial tissues. Therefore, NK cells and T cells can mediate suitable inhibitory responses via KLRG1 binding to only monomeric E-cadherin. The KLRG1-E-cadherin interaction is presumably safe and efficient for inhibiting the excess functional responses of NK cells as well as those of the SLECs (Fig. 4, right). This function is similar to that of inhibitory costimulatory molecules, such as CTLA4 (cytotoxic T-lymphocyte antigen 4).
The αEβ7 integrin (CD103) receptor also recognizes E-cadherin (22). The αEβ7 integrin is found on intraepithelial lymphocytes, where it mediates their adhesion to epithelial cells and enhances their function (23). The αEβ7 integrin recognizes the conserved residue Glu31 and its surrounding area at the top of E-cadherin domain 1 (supplemental Fig. S3) (23), adjacent to the KLRG1 binding site. Thus, KLRG1 may physically interfere with αEβ7 integrin binding. Although the expression of KLRG1 in activated intraepithelial lymphocytes has not been determined, if KLRG1 is expressed simultaneously with αEβ7 integrin, it may inhibit excess immune responses both by transducing E-cadherin-mediated inhibitory signals and by competing for αEβ7 integrin binding. Interestingly, αEβ7 integrin can bind to both monomeric and homodimeric E-cadherin, whereas KLRG1 binds only monomeric E-cadherin. This functional difference may have a sophisticated potential to finely modulate mucosal immunity (24) as well as NK and T cell development/differentiation.
Several types of cancer cells express mutant E-cadherin. One alteration of the E-cadherin gene is induced by the in-frame skipping of either exon 8 or exon 9. Interestingly, these alterations result in the deletion of either domain 2 or domain 3 of E-cadherin and are reported to abolish KLRG1 binding (25). However, our data suggest that the KLRG1 binding site is located in the N-terminal region, far from domains 2 and 3. Thus, these mutations may indirectly disrupt the conformation of the N-terminal region that is required for KLRG1 recognition. A definitive conclusion will require future investigations of the structural characteristics of the E-cadherin mutants and KLRG1 binding interaction.
The mutation or lack of E-cadherin in some cancer cells causes the loss of cell adhesion and the subsequent acquisition of cellular motility, facilitating tumor metastasis. Such abnormal cells may be susceptible to NK cells and T cells, because of the lack of KLRG1-mediated inhibitory signals. On the other hand, some tumors may exploit the KLRG1-E-cadherin interaction to their advantage. Such tumor cells might initially down-regulate E-cadherin to acquire motility and metastatic potential but then re-express E-cadherin to establish adherent metastatic foci and avoid immune attack by NK cells and T cells (24–27). Therefore, the development of inhibitors of KLRG1-E-cadherin recognition is important for eliminating these cancer cells. Our results demonstrate that KLRG1 recognizes the N-terminal amino acids of monomeric E-cadherin domain 1, which are well conserved in the type I classical cadherins (Fig. 2D). In fact, KLRG1 can also bind to the other type I classical cadherins, neuronal and retinal (N- and R-cadherins) (5), strongly suggesting that the N-terminal amino acids are a major determinant for the KLRG1 binding. Therefore, the N-terminal region may be a useful template for designing inhibitors of the KLRG1-E-cadherin interaction.
CONCLUSIONS
This study demonstrates that KLRG1 recognizes the N-terminal region of E-cadherin, known to be critical for homophilic adhesion. Therefore, we propose that KLRG1 cannot bind to the homodimeric form of E-cadherin but rather binds to the monomeric form. The monomeric form of E-cadherin is exposed on the adherens junction in special conditions, such as damaged epithelial tissues. Under these circumstances, it may serve as a target for KLRG1 recognition to mediate inhibitory signals, raising the activation threshold of NK cells and T cells and preventing excessive immune response.
Supplementary Material
Acknowledgments
We thank T. Saito, Y. Ishino, D. Kohda, T. Sugi, P. Bowness, and K. Morikawa for helpful discussions. We thank S. Grzesick for providing the NMR assignment data for E-cadherin. We thank T. Matsukura for providing anti-KLRG1 monoclonal antibody.
This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology and the Japan Bio-oriented Technology Research Advancement Institute (BRAIN).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- NK
- natural killer
- SLEC
- short lived effector CD8 T cell
- MES
- 4-morpholineethanesulfonic acid
- SPR
- surface plasmon resonance
- AUC
- analytical ultracentrifugation
- HSQC
- heteronuclear sequential quantum correlation
- PE
- phycoerythrin.
REFERENCES
- 1.Biron C. A., Nguyen K. B., Pien G. C., Cousens L. P., Salazar-Mather T. P. (1999) Annu. Rev. Immunol. 17, 189–220 [DOI] [PubMed] [Google Scholar]
- 2.Lanier L. L. (2005) Annu. Rev. Immunol. 23, 225–274 [DOI] [PubMed] [Google Scholar]
- 3.Moretta L., Moretta A. (2004) Curr. Opin. Immunol. 16, 626–633 [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama W. M. (1998) Curr. Opin. Immunol. 10, 298–305 [DOI] [PubMed] [Google Scholar]
- 5.Ito M., Maruyama T., Saito N., Koganei S., Yamamoto K., Matsumoto N. (2006) J. Exp. Med. 203, 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gründemann C., Bauer M., Schweier O., von Oppen N., Lässing U., Saudan P., Becker K. F., Karp K., Hanke T., Bachmann M. F., Pircher H. (2006) J. Immunol. 176, 1311–1315 [DOI] [PubMed] [Google Scholar]
- 7.Robbins S. H., Tessmer M. S., Mikayama T., Brossay L. (2004) J. Immunol. 173, 259–266 [DOI] [PubMed] [Google Scholar]
- 8.Robbins S. H., Nguyen K. B., Takahashi N., Mikayama T., Biron C. A., Brossay L. (2002) J. Immunol. 168, 2585–2589 [DOI] [PubMed] [Google Scholar]
- 9.Huntington N. D., Tabarias H., Fairfax K., Brady J., Hayakawa Y., Degli-Esposti M. A., Smyth M. J., Tarlinton D. M., Nutt S. L. (2007) J. Immunol. 178, 4764–4770 [DOI] [PubMed] [Google Scholar]
- 10.Joshi N. S., Cui W., Chandele A., Lee H. K., Urso D. R., Hagman J., Gapin L., Kaech S. M. (2007) Immunity 27, 281–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gooding J. M., Yap K. L., Ikura M. (2004) BioEssays 26, 497–511 [DOI] [PubMed] [Google Scholar]
- 12.Takeichi M. (1991) Science 251, 1451–1455 [DOI] [PubMed] [Google Scholar]
- 13.Gumbiner B. M. (2000) J. Cell Biol. 148, 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pokutta S., Weis W. I. (2007) Annu. Rev. Cell Dev. Biol. 23, 237–261 [DOI] [PubMed] [Google Scholar]
- 15.Overduin M., Harvey T. S., Bagby S., Tong K. I., Yau P., Takeichi M., Ikura M. (1995) Science 267, 386–389 [DOI] [PubMed] [Google Scholar]
- 16.Shapiro L., Fannon A. M., Kwong P. D., Thompson A., Lehmann M. S., Grübel G., Legrand J. F., Als-Nielsen J., Colman D. R., Hendrickson W. A. (1995) Nature 374, 327–337 [DOI] [PubMed] [Google Scholar]
- 17.Nose A., Tsuji K., Takeichi M. (1990) Cell 61, 147–155 [DOI] [PubMed] [Google Scholar]
- 18.Rosshart S., Hofmann M., Schweier O., Pfaff A. K., Yoshimoto K., Takeuchi T., Molnar E., Schamel W. W., Pircher H. (2008) Eur. J. Immunol. 38, 3354–3364 [DOI] [PubMed] [Google Scholar]
- 19.Ohki I., Shimotake N., Fujita N., Nakao M., Shirakawa M. (1999) EMBO J. 18, 6653–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumoto N., Mitsuki M., Tajima K., Yokoyama W. M., Yamamoto K. (2001) J. Exp. Med. 193, 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parisini E., Higgins J. M., Liu J. H., Brenner M. B., Wang J. H. (2007) J. Mol. Biol. 373, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cepek K. L., Shaw S. K., Parker C. M., Russell G. J., Morrow J. S., Rimm D. L., Brenner M. B. (1994) Nature 372, 190–193 [DOI] [PubMed] [Google Scholar]
- 23.Agace W. W., Higgins J. M., Sadasivan B., Brenner M. B., Parker C. M. (2000) Curr. Opin. Cell Biol. 12, 563–568 [DOI] [PubMed] [Google Scholar]
- 24.Colonna M. (2006) J. Exp. Med. 203, 261–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartzkopff S., Gründemann C., Schweier O., Rosshart S., Karjalainen K. E., Becker K. F., Pircher H. (2007) J. Immunol. 179, 1022–1029 [DOI] [PubMed] [Google Scholar]
- 26.Takeichi M. (1993) Curr. Opin. Cell Biol. 5, 806–811 [DOI] [PubMed] [Google Scholar]
- 27.Cavallaro U., Christofori G. (2004) Nat. Rev. Cancer 4, 118–132 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.