Abstract
The saturated fatty acids acylated on Lipid A of lipopolysaccharide (LPS) or bacterial lipoproteins play critical roles in ligand recognition and receptor activation for Toll-like Receptor 4 (TLR4) and TLR2. The results from our previous studies demonstrated that saturated and polyunsaturated fatty acids reciprocally modulate the activation of TLR4. However, the underlying mechanism has not been understood. Here, we report for the first time that the saturated fatty acid lauric acid induced dimerization and recruitment of TLR4 into lipid rafts, however, dimerization was not observed in non-lipid raft fractions. Similarly, LPS and lauric acid enhanced the association of TLR4 with MD-2 and downstream adaptor molecules, TRIF and MyD88, into lipid rafts leading to the activation of downstream signaling pathways and target gene expression. However, docosahexaenoic acid (DHA), an n-3 polyunsaturated fatty acid, inhibited LPS- or lauric acid-induced dimerization and recruitment of TLR4 into lipid raft fractions. Together, these results demonstrate that lauric acid and DHA reciprocally modulate TLR4 activation by regulation of the dimerization and recruitment of TLR4 into lipid rafts. In addition, we showed that TLR4 recruitment to lipid rafts and dimerization were coupled events mediated at least in part by NADPH oxidase-dependent reactive oxygen species generation. These results provide a new insight in understanding the mechanism by which fatty acids differentially modulate TLR4-mediated signaling pathway and consequent inflammatory responses which are implicated in the development and progression of many chronic diseases.
Toll-like receptors (TLRs)3 are one of the key pattern recognition receptor families that play a critical role in inducing innate and adaptive immune responses in mammals by recognizing conserved pathogen-associated molecular pattern of invading microbes. So far, at least thirteen TLRs have been identified in mammalian species (1, 2).
Lipopolysaccharide (LPS) from Gram-negative bacteria is the ligand for the TLR4 complex (3), whereas, TLR2 can recognize lipoproteins/lipopeptides of Gram-positive bacteria and mycoplasma (1, 2). LPS forms a complex with LPS-binding protein in serum leading to the conversion of oligomeric micelles of LPS to monomers, which are delivered to CD14. Monomeric LPS is known to bind TLR4/MD-2/CD14 complex (4). Lipid A, which possesses most of the biological activities of LPS, is acylated with hydroxy saturated fatty acids. The 3-hydroxyl groups of these saturated fatty acids are further 3-Ο-acylated by saturated fatty acids. Removal of these Ο-acylated saturated fatty acids from Lipid A not only results in complete loss of endotoxic activity, but also makes Lipid A act as an antagonist against the native Lipid A (5, 6). One or more Lipid As containing unsaturated fatty acids are known to be non-toxic and act as an antagonist against endotoxin (7, 8). In addition, deacylated lipoproteins are unable to activate TLR2 and to induce cytokine expression in monocytes (9). Together, these results suggest that saturated fatty acids acylated on Lipid A or bacterial lipoproteins play critical roles in ligand recognition and receptor activation for TLR4 and TLR2. Indeed, it is suggested that the rapid interaction of bacterial lipopeptides with plasma membrane of macrophages occurs via insertion of their acylated saturated fatty acids as determined by electron energy loss spectroscopy and freeze-fracture techniques (10, 11). TLR2 can form a heterodimer with TLR1 or TLR6, which can discriminate the molecular structure of triacyl or diacyl lipopeptides (12–14). So far there is no evidence that microbial ligands for other TLRs are acylated by saturated fatty acids.
Results from our previous studies demonstrated that saturated fatty acids activate TLR4 and polyunsaturated fatty acids (PUFA) inhibit both saturated fatty acid- and LPS-induced activation of TLR4 (15, 16). In addition, the saturated fatty acid lauric acid potentiates, but the n-3 PUFA docosahexaenoic acid (DHA) inhibits lipopeptide (TLR2 agonist)-induced TLR2 activation (17). Together, these results suggest that both TLR2 and TLR4 signaling pathways and target gene expression are reciprocally modulated by saturated and polyunsaturated fatty acids. However, the mechanism for this modulation by fatty acids is not understood.
TLR4 is recruited to lipid raft factions after cells are treated with LPS and subsequently induces tumor necrosis factor-α expression in RAW264.7 cells (18). This process occurs in an ROS-dependent manner because inhibition of NADPH oxidase suppresses TLR4 recruitment to lipid rafts (19). Methyl-β-dextrin, a lipid raft inhibitor, significantly inhibits the LPS-induced expression of cytokine (19), suggesting that lipid rafts are essential for TLR4-mediated signal transduction and target gene expression. Lipid rafts are a collection of lipid membrane microdomains characterized by insolubility in non-ionic detergents. Lipid rafts serve as a platform where receptor-mediated signal transduction is initiated (20). Lipid rafts have a special lipid composition that is rich in cholesterol, sphingomyelin, and glycolipids (21). The polar lipids in detergent-resistant membrane contain predominantly saturated fatty acyl residues with underrepresented PUFAs (22–24), suggesting that saturated fatty acyl chains favor lipid raft association. On the other hand, n-3 PUFAs displace signaling proteins from lipid rafts by altering lipid composition, and the displacement leads to the suppression of T-cell receptor-mediated signaling (25). It is now well documented that TLRs form homo- or hetero-oligomers (1, 2). TLR4 homodimerization is the initial step of the receptor activation. Results from our previous studies suggest that the molecular target by which saturated fatty acids and n-3 PUFAs reciprocally modulate TLR4 activation is the receptor complex itself or the event leading to the receptor activation instead of the downstream signaling components (15, 16). Therefore, we determined whether the reciprocal modulation of TLR4 activation is mediated by regulation of the dimerization and recruitment of TLR4 into lipid rafts, and if these processes occur in an ROS-dependent manner.
EXPERIMENTAL PROCEDURES
Reagents
Purified LPS was obtained from List Biological Lab. Inc. (Campbell, CA). Antibodies for MyD88, TRIF, and green fluorescent protein (GFP) were purchased from eBioscience Inc. (San Diego, CA), Abcam Inc. (Cambridge, MA), and Clontech Laboratories Inc. (Mountain View, CA), respectively. Antibodies for flotillin-1, gp91[phox], and p47[phox] were purchased from BD Biosciences Inc. (San Jose, CA). Antibodies for TLR4 were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). All other reagents were purchased from Sigma-Aldrich unless otherwise described.
Cell Culture
Ba/F3 cells, an interleukin-3-dependent murine pro-B cell line, expressing TLR4 (FLAG- or GFP-tagged), CD14, MD-2 (FLAG-tagged), and NF-κB luciferase reporter gene were described previously (26). Cells were cultured in RPMI1640 medium (Invitrogen) containing recombinant murine interleukin-3 (70 units/ml), 10% (v/v) heat-inactivated fetal bovine serum (FBS, Invitrogen), 100 units/ml penicillin, and 100 mg/ml streptomycin (Invitrogen). RAW264.7 cells (a murine monocytic cell line, ATCC TIB-71) and 293T cells (human embryonic kidney) were cultured in DMEM (Invitrogen) containing 10% (v/v) FBS, 100 units/ml penicillin, and 100 mg/ml streptomycin. Cells were maintained at 37 °C in a 5% CO2/air environment. Cells were then further incubated overnight in serum-poor medium prior to the treatment with fatty acids as described previously (16).
Transfection and Reporter Gene Luciferase Assay
NF-κB luciferase reporter gene assays were performed as described previously (15–17). Cells were co-transfected with a luciferase plasmid and heat shock protein 70-β-galactosidase plasmid as an internal control using SuperFect transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. Luciferase enzyme activities were determined using the luciferase assay system (Promega, Madison, WI) according to the manufacturer's instructions. Luciferase activity was normalized by β-galactosidase activity.
Immunoblotting
Immunoblotting was performed as previously described (17). Cell extracts were subjected to 10% SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane for immunoblot analyses. The membrane was blocked in phosphate-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk. Immunoblotting was performed with the indicated antibodies and secondary antibodies conjugated to horseradish peroxidase (Amersham Biosciences). The reactive bands were visualized with ECL Western blot detection reagents (Amersham Biosciences).
Immunoprecipitation
Immunoprecipitation was performed as previously described (17, 26). Protein extracts from Ba/F3 cells expressing TLR4 (FLAG- or GFP-tagged), CD14, MD-2 (FLAG-tagged), and NF-κB luciferase reporter gene for immunoprecipitation were prepared as described before (26). The samples were immunoprecipitated with rabbit anti-GFP polyclonal antibodies for 3 h and protein-A-Sepharose for 1 h. The solubilized immune complex was resolved on SDS-PAGE and transferred to polyvinylidene difluoride membrane. The membrane was blocked with phosphate-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk and was probed with anti-FLAG antibodies overnight. Thereafter, the blot was exposed to horseradish peroxidase-conjugated secondary antibodies for 1 h and detected with ECL Western blot detection reagents. The blot was reprobed with mouse anti-GFP antibodies.
Isolation of Lipid Rafts Using Sucrose Gradient Centrifugation
RAW264.7 or Ba/F3 cells (1.5 × 108 cells) stimulated with LPS or lauric acid, in the presence or absence of DHA, were used to isolate lipid rafts. RAW 264.7 cells were incubated in the serum-poor DMEM (1% FBS) for 15 h prior to the treatment with fatty acids and LPS. Lipid rafts from RAW264.7 cells were isolated by lysing cells in 750 μl of HNE lysis buffer (25 mm HEPES, pH 6.5, 150 mm sodium chloride, and 5 mm EDTA, supplemented with 1.5% Triton X-100, 1 mm sodium orthovanadate, 5 mm sodium fluoride, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each of aprotinin and leupeptin) for 15 min on ice followed by 10 rounds of Dounce homogenization. Lysates were mixed with an equal volume of 80% sucrose in HNE buffer. 2 ml of 30% sucrose in HNE buffer was overlaid followed by 1 ml of 5% sucrose in HNE buffer. Samples were ultracentrifuged in an SW60 rotor for 17 h at 40,000 rpm using maximum acceleration and no brake conditions. Eight 500-μl fractions were collected from the top. For immunoblotting, fractionated samples from RAW264.7 cells were subjected to trichloroacetic acid precipitation. Ba/F3 cells were incubated in serum-free RPMI 1640 medium containing IL-3 for 15 h prior to the treatment with fatty acids and LPS. Ba/F3 cells were lysed on ice for 10 min with 1 ml of TNE lysis buffer (25 mm Tris, pH 7.5, 150 mm sodium chloride, and 5 mm EDTA, supplemented with 1% Triton X-100, 1 mm sodium orthovanadate, 5 mm sodium fluoride, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml each of aprotinin and leupeptin) followed by 10 rounds of Dounce homogenization. Lysates were mixed with an equal volume of 85% sucrose in TNE buffer to adjust the final concentration to 42.5% sucrose. Two milliliters of the lysate-sucrose mixture was transferred to the bottom of Beckman ultracentrifuge tubes and overlaid with 6 ml of 35% sucrose in TNE buffer and 4 ml of 5% sucrose in TNE buffer. Samples were ultracentrifuged for 17 h at 38,000 rpm using a Beckman SW41 rotor using maximum acceleration and no brake conditions. Twelve 1-ml fractions were collected from the top of the gradient. For the immunoprecipitation, fractions 4 and 5, which were the lipid raft fractions, were pooled.
Dimerization of TLR4
To assess the dimerization of TLR4 or the association of TLR4 with MD-2, TLR4 from cell lysates or lipid rafts fractions were immunoprecipitated with anti-GFP and immunoblotted with anti-FLAG antibodies. To assess recruitment of TLR4, TRIF, and MyD88 into lipid rafts, immunoblotting was performed with corresponding antibodies. Flotillin-1 was used as a lipid raft marker. To examine association of TLR4, MyD88, and TRIF in lipid rafts, trichloroacetic acid precipitation was performed on lipid rafts fractions in RAW264.7 cells and subjected to immunoblotting with anti-TLR4, anti-TRIF, anti-MyD88, and anti-flotillin-1 antibodies.
Confocal Microscopy Analysis
Confocal microscopy was performed according to the procedure described by Nakahira et al. (19), but with modifications. RAW264.7 cells were seeded onto coverslips for 12 h in either DMEM containing 10% FBS for LPS-treated cells or serum-poor DMEM (1% FBS) for lauric acid-treated cells. Cells were treated with LPS (100 ng/ml) for 10 min or lauric acid (100 μm) for 1 h in the presence or absence of DHA (20 μm). Cells were washed with serum-free DMEM and incubated with 8 μg/ml fluorescein isothiocyanate-conjugated cholera toxin B (Sigma-Aldrich) on ice for 10 min. Cells were fixed with 4% paraformaldehyde for 45 min followed by incubation with 50 mm ammonium hydroxide for 10 min and were permeabilized with 0.1% Triton X-100 for 15 min. Samples were washed three times with bovine serum albumin (BSA) solution (0.5% BSA, 0.15% glycine in phosphate-buffered saline). Coverslips were blocked with 5% goat serum (Zymed Laboratories Inc., South San Francisco, CA) for 45 min followed by washing with BSA solution. Samples were incubated for 1 h with 1/100 dilution of anti-TLR4 (H-80, Santa Cruz Biotechnology) in BSA solution followed by a 1-h incubation with 1/500 dilution in BSA solution of Alexa Fluor 546-conjugated F(ab′)2 fragment of goat anti-rabbit IgG (Invitrogen). Coverslips were washed three times with BSA solution and phosphate-buffered saline and mounted onto glass slides. For ROS analysis, RAW264.7 cells were incubated with 10 μm 5-(and 6-)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetylester (CM-H2DCFDA) (Invitrogen) for 30 min. Cells were preincubated with 20 μm DHA, 2 μm diphenyleneiodonium chloride (DPI) or 10 mm N-acetyl-l-cysteine (NAc) for 30 min followed by incubation with 150 μm lauric acid or 100 ng/ml LPS for 30 min. Confocal microscopy was performed with the Zeiss LSM 510 microscope with 63× objective lens and using laser excitation at 488 and 543 nm.
Reverse Transcription-PCR
Preparation of total RNA, cDNA synthesis, and PCR were conducted as described (26). The mouse primers used in reverse transcription-PCR are described as following: tumor necrosis factor-α (5′-CTCTTCTCATTCCTGCTTGT-3′ and 5′-TTGAAGAGAACCTGGGAGTA-3′), COX-2 (5′-CACTACATCCTGACCCACTT-3′ and 5′-GTCCTCGCTTATGATCTGTC-3′), and IRF-3 (5′-CAATAGCAAGGACCCTTATG-3′ and 5′-AAAAGAGTGTCTGCTGGAAG-3′). The primers used for hypoxanthine phosphoribosyltransferase were described previously (27).
NADPH Oxidase Activation
RAW264.7 cells were seeded into 10-cm dishes and grown to near confluency. Cells were preincubated with DPI for 1 h, NAc for 2 h, or DHA for 30 min followed by incubation with lauric acid or LPS for 30 min. Cells were lysed in TNE buffer containing 1.5% Triton X-100 for 30 min. Immunoprecipitation was performed with anti-gp91[phox] antibody, run on SDS-PAGE, and immunoblotted with anti-p47[phox] and anti-gp91[phox] antibodies.
RESULTS
Lauric Acid Enhances Recruitment of TLR4 and Its Downstream Adaptor Molecules into Lipid Rafts
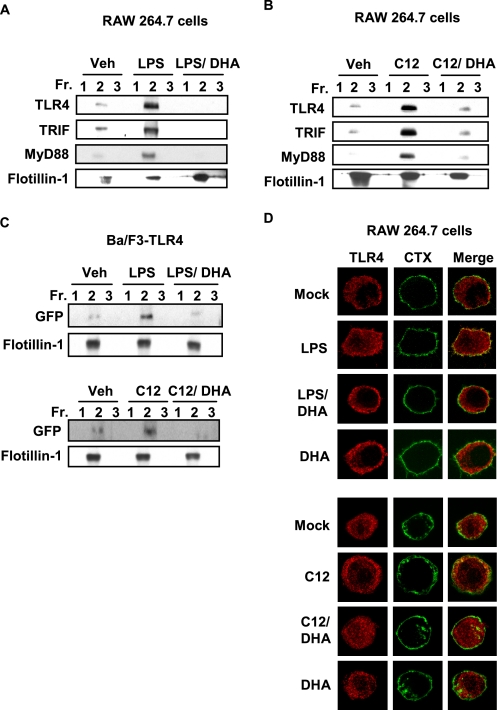
It was shown that LPS-induced TLR4 activation is initiated by recruitment of TLR4 into lipid rafts (19). We have previously reported that fatty acids differentially modulate TLR4 activation (15, 17, 28, 29). Based on these results, we investigated whether fatty acids also modulate the recruitment of TLR4 into lipid rafts. The cells treated with LPS showed increased amounts of TLR4 in the fractions 2 and 3 containing flotillin-1, a lipid raft marker, when compared with vehicle treated cells (Fig. 1). Fractions 2 and 3 were highly enriched with lipid rafts as evidenced by detection of flotillin-1. These results demonstrate that LPS enhances the recruitment of TLR4 into lipid raft fractions (Fig. 1). Similarly, lauric acid (C12) enhanced the recruitment of TLR4 into lipid raft fractions (Fig. 1). Because TRIF and MyD88 are downstream adaptor molecules for TLR4, we further investigated whether the recruitment of these adaptor molecules into the lipid rafts is also induced by LPS or lauric acid. As shown in Fig. 1, the amounts of TRIF and MyD88 detected in the fraction 2 were increased in cells treated with LPS or lauric acid compared with those from control cells. In contrast, DHA did not show such effects (Fig. 1). Together, these results suggest that lauric acid or LPS induce the recruitment of TLR4 and downstream adaptor molecules into lipid rafts.
FIGURE 1.
Lauric acid enhances the recruitment of TLR4 and its adaptor molecules, TRIF and MyD88, into the lipid rafts. RAW264.7 cells were stimulated with LPS (100 ng/ml) or lauric acid (100 μm, C12) for 7 min or DHA (20 μm) for 1 h. The cells were lysed and lipid rafts (Fractions 2 and 3) and non-lipid rafts fractions (Fractions 5–8) were separated by sucrose-gradient ultracentrifugation. Each fraction was trichloroacetic acid precipitated and subjected to immunoblotting using anti-TLR4, anti-TRIF, anti-MyD88, or anti-flotillin-1 antibodies.
DHA Inhibits LPS- or Lauric Acid-induced Recruitment of TLR4 into Lipid Rafts
We previously demonstrated that DHA inhibits LPS- or lauric acid-induced TLR4 activation (29). Thus, we investigated whether DHA inhibits LPS- or lauric acid-induced recruitment of TLR4 into lipid rafts. As shown in Fig. 2(A and B), DHA inhibited the recruitment of TLR4, TRIF, and MyD88 into the lipid rafts in RAW264.7 cells. To further confirm the results, we employed Ba/F3 cells stably transfected with GFP-TLR4, FLAG-TLR4, and FLAG-MD-2 constructs. TLR4 was recruited into the lipid rafts by the treatment of LPS or lauric acid, but the recruitment was attenuated by DHA (Fig. 2C).
FIGURE 2.
DHA inhibits LPS- or lauric acid-induced recruitment of TLR4 into the lipid rafts. RAW264.7 cells were treated with LPS (A) or lauric acid (B) for 7 min in the presence or absence of DHA (20 μm). The cells were lysed, and lysates were fractionated using sucrose-gradient ultracentrifugation as described in Fig. 1, lipid rafts collected in Fractions 1–3 and trichloroacetic acid-precipitated. Fractions were immunoblotted with anti-TLR4, TRIF, MyD88, or flotillin-1. C, Ba/F3 cells stably transfected with GFP/FLAG-tagged TLR4 were treated in identical manner as in A and B, but immunoblotted with anti-GFP or anti-flotillin-1. D, RAW264.7 cells were stimulated with LPS or lauric acid for 1 h with or without DHA (20 μm) or DHA alone for 1 h and followed by incubation with fluorescein isothiocyanate-conjugated cholera toxin B on ice for 10 min. LPS-stimulated cells were incubated in DMEM with 10% FBS, whereas, lauric acid-stimulated cells were incubated in serum-poor DMEM. Cells were analyzed for fluorescein isothiocyanate-conjugated cholera toxin B-stained glycosphingolipid 1 (GM1, green), which is enriched in lipid rafts and TLR4 (red) by confocal microscopy.
To visualize the co-localization of TLR4 with lipid rafts, confocal microscopy images of fixed RAW264.7 cells were analyzed. Lipid rafts were stained with the fluorescein isothiocyanate-conjugated cholera toxin B conjugate, which binds to monosialoganglioside (GM1), a protein enriched in lipid rafts (30). In these confocal microscopy experiments, lipid rafts were stained green, TLR4 was stained red, and merging these two colors resulted in a yellow staining when the lipid rafts and TLR4 were co-localized. Staining of lipid rafts and TLR4 in resting RAW264.7 cells did not show co-localization of TLR4 with lipid rafts. TLR4 was localized in condensed formations that were distributed throughout the cytoplasm (Fig. 2D). When cells were stimulated with LPS, TLR4 became co-localized with lipid rafts. This was observed as a diffuse yellow staining around the plasma membrane when images were merged. When RAW264.7 cells were pretreated with DHA before LPS treatment, the co-localization of TLR4 with lipid rafts was diminished. Lauric acid produced similar effects as LPS- and lauric acid-induced co-localization of TLR4 with lipid rafts was also diminished by the pretreatment of DHA.
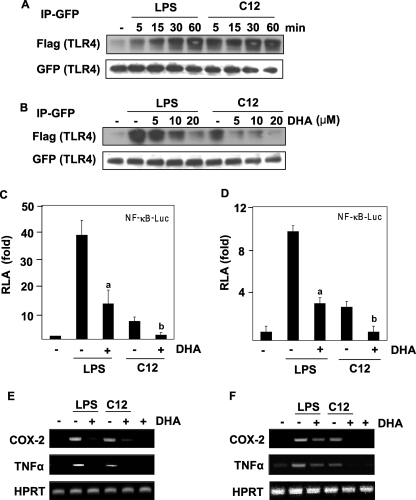
Lauric Acid Induces, but DHA Inhibits the Homodimerization of TLR4
It is known that some TLRs function as homo- or heterodimers (31). Homodimerization of TLR4 is an initial step for receptor activation (32–34). Therefore, we determined whether lauric acid promotes the dimerization of TLR4. The dimerization assay was performed as previously described with Ba/F3 cells stably transfected with GFP-TLR4, FLAG-TLR4, and FLAG-MD-2 constructs (26). Briefly, GFP-TLR4 was immunoprecipitated, and dimerization was determined by the co-immunoprecipitation of FLAG-TLR4. As shown in Fig. 3A, the amount of dimerized TLR4 was low in the Ba/F3-TLR4 cells in resting state, but gradually increased with LPS treatment, reaching peak dimerization at 60 min. Similarly, the amount of dimerized TLR4 increased with the treatment of lauric acid, although the time course differed from that of LPS (Fig. 3A). However, the amount of dimerized TLR4 diminished in the presence of DHA in a dose-dependent manner (Fig. 3B). These results suggest that LPS or lauric acid induced TLR4 dimerization, but DHA inhibited LPS- or lauric acid-induced dimerization. The enhanced dimerization of TLR4 correlated with NF-κB activation and target gene expression induced by LPS or lauric acid (Fig. 3, C–F). DHA inhibited LPS- or lauric acid-induced NF-κB activation and target gene expression in both RAW264.7 cells with endogenously expressed TLR4 (Fig. 3, C and E), and Ba/F3 cells stably transfected with TLR4 (Fig. 3, D and F). Taken together, these results demonstrate the reciprocal modulation of TLR4 dimerization by lauric acid and DHA.
FIGURE 3.
LPS or lauric acid induces, but DHA inhibits the homodimerization of TLR4. A, Ba/F3 whole cell lysates were immunoprecipitated with an anti-GFP antibody and then immunoblotted with an anti-FLAG antibody. The membrane was stripped and reprobed with anti-GFP. B, dose-dependent response of Ba/F3 cells toward pretreatment with DHA. Cells were stimulated with LPS or C12, and cell lysates were processed as described in Fig. 2A. For the analysis of NF-κB promoter activity, Ba/F3-TLR4 cells transfected with NF-κB- luciferase construct (C) or RAW264.7 cells transfected with NF-κB- luciferase construct (D) were used. Cells were treated with LPS or lauric acid for 8 h in the presence or absence of DHA. Cells were lysed to determine the luciferase activity. The results were expressed as relative luciferase activity (RLA) against the value of the vehicle treatment. Values are means ± S.E. of the mean of at least three independent experiments. a and b were significantly different from the values for the control group without DHA treatment (p < 0.05). For the analysis of mRNA levels of indicated genes, Ba/F3-TLR4 cells (E) or RAW264.7 cells (F) were treated with LPS or lauric acid in the presence or absence of DHA for 6 h. RNAs were prepared, and reverse transcription-PCR was performed as described under “Experimental Procedures.” Hypoxanthine phosphoribosyltransferase (HPRT) was used as a control.
Lauric Acid and DHA Reciprocally Modulate Recruitment of TLR4 into Lipid Rafts and Association of TLR4 with MD-2
To determine the effects of lauric acid and DHA on the homodimerization of TLR4 and association of TLR4 with MD-2 in lipid rafts, lipid rafts were isolated in Ba/F3 cells. Cell lysis in 1% Triton X-100 for 10 min allowed for the isolation of the lipid raft marker flotillin-1 in fractions 4 and 5. Markers for non-lipid raft membrane proteins (transferrin receptor), internal membranes (Bcl-2), and cytoplasmic proteins (p65) were found at the bottom of the gradient (fractions 10–12, data not shown). GFP-TLR4 from lipid raft fractions was immunoprecipitated with anti-GFP antibodies. Subsequent immunoblotting with anti-FLAG antibodies should reflect the extent to which FLAG-TLR4 is dimerized with GFP-TLR4. LPS or lauric acid enhanced the amount of dimerized TLR4 in the lipid raft fraction, but the effect was attenuated by DHA (Fig. 4A). The total amount of TLR4 in the lipid raft fraction significantly increased after the treatment of the cells with LPS or lauric acid, but DHA diminished the effects of LPS or lauric acid as shown in Figs. 1 and 2. Together, these results suggest that the recruitment of TLR4 into lipid rafts was increased by LPS or lauric acid. Because TLR4 is known to associate with MD-2 to initiate signaling, we determined whether LPS or lauric acid enhances the association of TLR4 with MD-2 in lipid rafts fractions. The association of TLR4 with MD-2 was enhanced by treatment of LPS or lauric acid, but DHA inhibited the association of TLR4 with MD-2 in lipid rafts (Fig. 4A). However, flotillin-1 levels in the lipid raft fraction were not changed by the treatment either with LPS or lauric acid (Fig. 4A). In contrast, FLAG-TLR4 was not co-immunoprecipitated with GFP-TLR4 in non-lipid raft fractions (Fig. 4B). Together, these results suggest that the dimerization of TLR4 is coupled with the recruitment of TLR4 into lipid rafts and that the dimerization and recruitment of TLR4 into lipid rafts are enhanced by LPS or lauric acid. The effects of LPS or lauric acid were diminished by DHA treatment.
FIGURE 4.
Lauric acid induces but DHA inhibits TLR4 homodimerization and association of TLR4 with MD-2 in lipid rafts. A, Ba/F3 cells stably transfected with GFP/FLAG-tagged TLR4 and FLAG-tagged MD-2 were treated with LPS or lauric acid in the presence or absence of DHA (20 μm). For the immunoprecipitation, lipid raft Fractions 4 and 5 were pooled from the sucrose gradient. One half of the lipid raft fraction was immunoprecipitated with anti-GFP antibodies and then immunoblotted with anti-FLAG antibodies. The membranes were reprobed with anti-GFP antibodies. The other half of the samples was immunoblotted with anti-flotillin-1 antibodies to show the presence of the lipid raft marker. B, samples from pooled non-lipid raft Fractions 10–12 were immunoprecipitated and immunoblotted as described above in A.
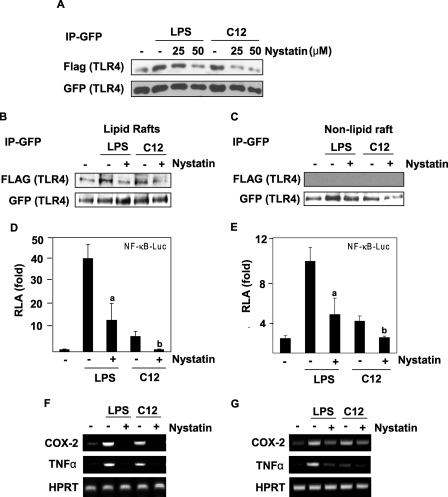
Lipid Rafts Are Essential for TLR4-mediated Signal Transduction
To further understand the role of lipid rafts for TLR4 activation, we employed the lipid raft inhibitor nystatin, which is known to inhibit the formation of lipid rafts by sequestering cholesterol. TLR4 dimerization induced by LPS or lauric acid was inhibited by nystatin (Fig. 5A). The dimerization of TLR4 in the lipid raft fraction was also inhibited by nystatin (Fig. 5B). Dimerized TLR4 was only observed in lipid rafts but not in the non-lipid raft fractions (Fig. 5C). Methyl-β-cyclodextrin, another lipid raft inhibitor, had the same effect as nystatin (data not shown). Nystatin also inhibited LPS- or lauric acid-induced NF-κB activation and target gene expression (Fig. 5, D–G). Taken together, these results suggest that the formation of intact lipid rafts, recruitment of TLR4, and dimerization of TLR4 into lipid rafts are critical for initiating LPS or lauric acid-induced TLR4 signal transduction.
FIGURE 5.
Lipid raft inhibitor nystatin inhibits LPS- or lauric acid-induced homodimerization of TLR4, and the activation of NF-κB and TLR4 target gene expression. A, Ba/F3 cells were treated with nystatin prior to treatment with LPS or lauric acid for 30 min. Cell lysates were immunoprecipitated with anti-GFP antibodies and then immunoblotted with anti-FLAG antibodies. B, Ba/F3 cells were treated as in A but were lysed and fractionated by the sucrose gradient. GFP-TLR4 was immunoprecipitated with anti-GFP antibodies from lipid raft fractions and immunoblotted with anti-FLAG antibodies. C, samples from the non-lipid raft fractions were immunoprecipitated and immunoblotted as in B. For the analysis of NF-κB-luciferase activity, Ba/F3-TLR4 cells (D) or RAW264.7 cells (E) transfected with NF-κB-luciferase construct were treated with nystatin for 8 h before the treatment with LPS or lauric acid. Cells were lysed to determine the luciferase activity. a and b were significantly different from the values for the control group without nystatin treatment. For the analysis of mRNA of indicated genes, Ba/F3-TLR4 cells (F) or RAW264.7 cells (G) were treated with nystatin for 6 h before treatment with LPS or lauric acid. RNAs were prepared and reverse transcription-PCR was performed as described under “Experimental Procedures.”
Lauric Acid Activates but DHA Inhibits NADPH Oxidase
A previous report demonstrated that LPS stimulates recruitment of TLR4 to lipid rafts and is dependent on NADPH oxidase-mediated by reactive oxygen species (ROS) (19). To determine if lauric acid activates TLR4 in a similar mechanism as LPS, ROS levels and NADPH oxidase were analyzed. RAW264.7 cells were stimulated with lauric acid, and ROS were analyzed by confocal microscopy after staining with a CM-H2DCFDA, a ROS-specific fluorescent probe. Lauric acid, like LPS, increased ROS in cells. This increase in ROS by lauric acid and LPS was inhibited by DHA. The NADPH oxidase inhibitor, DPI, and the antioxidant NAc inhibited lauric acid-induced ROS increase in RAW264.7 cells (Fig. 6A).
FIGURE 6.
Lauric acid activates, but DHA inhibits NADPH oxidase and ROS generation. A, RAW264.7 cells were preincubated with 10 μm CM-H2DCFDA for 30 min followed by incubation with DHA (20 μm), DPI (2 μm), NAc (10 mm), or vehicle for 30 min. LPS (100 ng/ml) or lauric acid (150 μm) was incubated with the cells for an additional 30 min. Cells were fixed and imaged by confocal microscopy. B, RAW264.7 cells were incubated with 50 μm lauric acid for the indicated time periods, cells were lysed, and gp91[phox] was immunoprecipitated and immunoblotted with anti-gp91[phox] and anti-p47[phox] antibodies. C, similar to B, cells were incubated with indicated amounts of lauric acid for 30 min. D, RAW264.7 cells were pretreated with DHA for 30 min, followed by incubation with 50 μm lauric acid or 100 ng/ml LPS for 30 min (E). F, RAW264.7 cells were pretreated with NAc (10 mm) for 2 h or DPI (2 μm) for 1 h followed by incubation with 50 μm lauric acid.
To determine if lauric acids activates NADPH oxidase similar to LPS (19), formation of the NADPH complex was analyzed. When active, NADPH oxidase component gp91[phox], a membrane protein, binds to p47[phox], a cytosolic protein (19). Lauric acid added to RAW264.7 cells increased the interaction between p47[phox] and gp91[phox] in a time- and dose-dependent manner (Fig. 6, B and C). When RAW264.7 cells were pre-treated with DHA, NADPH oxidase activation by lauric acid was inhibited in a dose-dependent manner (Fig. 6D). LPS-induced NADPH oxidase activation was also inhibited by DHA (Fig. 6E). In addition, lauric acid-induced activation of NADPH oxidase was inhibited by DPI and NAc (Fig. 6F).
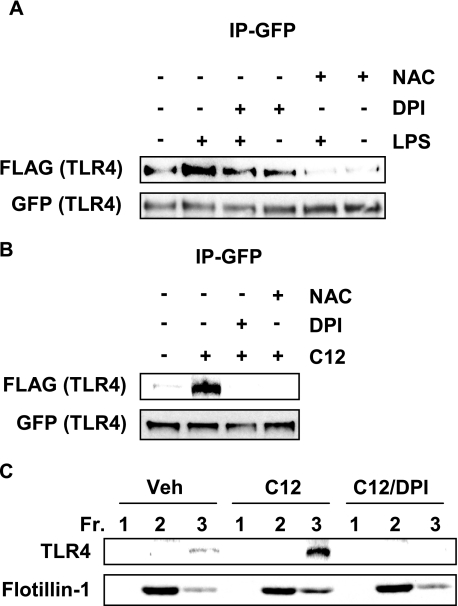
Inhibition of NADPH Oxidase Suppresses LPS- or Lauric Acid-induced TLR4 Dimerization and Recruitment to Lipid Rafts
NADPH oxidase was differentially modulated by lauric acid and DHA similar to TLR4, suggesting that both are involved in the same signaling pathway. We wanted to determine if inhibiting NADPH oxidase would affect TLR4 dimerization. DPI and NAc inhibited TLR4 dimerization when stimulated by LPS or lauric acid (Fig. 7, A and B). RAW264.7 cells pretreated with DPI also inhibited lauric acid-induced recruitment of TLR4 to lipid rafts (Fig. 7C). Together, these results suggest that lauric acid induces dimerization and recruitment to lipid rafts in a ROS-dependent manner as does LPS.
FIGURE 7.
Inhibition of NADPH oxidase suppresses LPS or lauric acid-induced TLR4 dimerization. A, Ba/F3 cells were pretreated with DPI (2 μm) for 1 h or NAc (10 mm) for 2 h and then stimulated with LPS (100 ng/ml) or (B) 100 μm lauric acid for 30 min. Cells were lysed, and GFP-TLR4 was immunoprecipitated and immunoblotted with anti-FLAG and anti-GFP antibodies. C, RAW264.7 cells were pretreated with 2 μm DPI for 1 h and treated with 100 μm lauric acid for 30 min. Cells were lysed, and lysates were fractionated by sucrose gradient fractionation. Lipid raft fractions (1–3) were collected, trichloroacetic acid-precipitated, and immunoblotted with anti-TLR4 and anti-flotillin-1 antibodies.
DISCUSSION
It is now recognized that chronic inflammation is one of the key etiological conditions for the development and progression of many chronic diseases, including cancer, atherosclerosis, and insulin resistance, suggesting that the inhibition of inflammation can reduce the risk of development of such diseases. Inflammation can be induced by microbial infection. Toll-like receptors mediate infection-induced inflammation by recognizing invading pathogens and activating downstream signaling pathways that lead to the expression of diverse arrays of pro-inflammatory marker gene products (1, 2). However, it is not well understood what causes and mediates “sterile inflammation”: inflammation from a non-infectious origin. Recent evidence now suggests that certain TLRs can be activated by endogenous molecules derived from tissue injury and saturated fatty acids (17, 35). This suggests that TLRs mediate not only infection-induced inflammation but also sterile inflammation induced by endogenous molecules. It further suggests that TLRs and their downstream signaling pathways can provide important targets for preventive and therapeutic strategies against the chronic diseases. Results from epidemiological and genetic studies linked TLRs to the development and progression of many chronic diseases, including insulin resistance, atherosclerosis, and cancer (17, 36–38). Our recent studies demonstrated for the first time that dietary saturated fatty acids and n-3 polyunsaturated fatty acids reciprocally modulate TLR4 activation (14–17). The animal model of diet-induced obesity also revealed that saturated fatty acid-induced insulin resistance and vascular or adipocyte inflammation are mediated by the activation of TLR4 (36, 38–41). However, the molecular mechanism by which these fatty acids modulate the activation of TLRs is not known.
Results from our previous studies further suggest that the molecular target of the n-3 polyunsaturated fatty acid DHA in inhibiting TLR4 activation is the receptor itself or the upstream events (instead of downstream components) leading to the activation of TLR4 (14–17). It was demonstrated that TLR4 is recruited to lipid raft fractions after cells are treated with the LPS (18). The activation of TLR leads to recruitment of immediate downstream adaptor protein MyD88 (42, 43). Therefore, if TLRs are activated after being recruited to lipid rafts, this activation can be assessed by enhanced recruitment of MyD88 into lipid raft, which can be determined by co-immunoprecipitation of MyD88 and TLRs. TLRs form homo- or hetero-oligomers (32, 34, 44–47). TLR4 homodimerizes, whereas TLR2 heterodimerizes with TLR6 or TLR1. The TLR2/6 dimer recognizes peptidoglycan and diacylated lipopeptide (MALP-2), whereas TLR2/1 dimer recognizes triacylated lipopeptides (12–14). Thus, receptor dimerization and recruitment of TLR4 into lipid rafts appear to be two of the initial events leading to the activation of TLR4 signaling pathways.
Our results presented here demonstrate that saturated fatty acid lauric acid stimulates, but the n-3 polyunsaturated fatty acid DHA inhibits the dimerization and recruitment of TLR4 into lipid rafts. In addition, lauric acid stimulates but DHA inhibits the association of TLR4 with MD-2 and downstream adaptor molecules, and the activation of NF-κB and target gene expression. The treatment of cells with the lipid raft disrupting agent nystatin caused significant reduction of the amount of TLR4 homodimer in whole cell lysates and lipid rafts. Nystatin also inhibited LPS- or lauric acid-induced activation of NF-κB and the expression of TLR4 target genes. Lauric acid, like LPS, activated NADPH oxidase and increased ROS generation while DHA inhibited NADPH oxidase. Inhibiting NADPH oxidase with DPI and NAc suppressed TLR4 dimerization induced by LPS or lauric acid. Together, these results suggest that the recruitment and dimerization of TLR4 into lipid rafts are the initial steps required for the activation of the downstream signaling pathways and that lauric acid and DHA reciprocally modulate these processes. Dimerized TLR4 was not detected in non-lipid raft fractions even when stimulated with LPS or lauric acid, suggesting that agonist-induced dimerization of TLR4 occurs in the lipid rafts rather than the non-lipid raft compartment. These results further suggest that the recruitment of TLR4 into lipid rafts is coupled with the dimerization of TLR4. The results from our previous studies also showed that saturated fatty acids and n-3 PUFAs reciprocally modulate the dimerization of cytoplasmic pattern recognition receptor family Nod2 that also recognizes bacterial components (48).
Previous studies showed that LPS induces recruitment of TLR4 to lipid rafts through NADPH oxidase-dependent ROS production (19). It was also shown that oxidative stress generated by hemorrhagic shock recruits TLR4 to lipid rafts in macrophages (50). In our studies, lauric acid, like LPS, activated, but DHA inhibited NADPH oxidase. The NADPH oxidase inhibitor DPI or the antioxidant NAc abrogated TLR4 dimerization induced by LPS or lauric acid. These results suggest that lauric acid, like LPS, induces, but DHA inhibits dimerization and recruitment of TLR4 to lipid rafts in a ROS-dependent manner. It was shown that NADPH oxidase 4 isozyme directly interacts with TLR4 in human embryonic kidney cells transfected with TLR4, MD-2, and CD14 (51). However, it was shown that MyD88, the downstream component of TLR4, controls NADPH oxidase function in primary macrophages (52). Thus, it is not clear where NADPH oxidase is located in the hierarchical order of TLR4 signaling pathways. Because both DPI and NAc inhibit dimerization and recruitment of TLR4 to lipid rafts induced by LPS or lauric acid, it is conceivable that these one or more enzymes may be a part of the signaling complex directly interacting with TLR4 in lipid rafts.
How the saturated fatty acid lauric acid induces, but DHA inhibits dimerization and recruitment of TLR4 to lipid rafts, and how lauric acid and DHA reciprocally modulate NADPH oxidase are intriguing questions. The facts that Lipid A is acylated with saturated fatty acids, that removal of saturated fatty acids from Lipid A results in complete loss of endotoxic activity (15, 16), and that Lipid A(s) containing unsaturated fatty acids are nontoxic or act as an antagonist against endotoxins (17, 18) suggest that saturated fatty acids, but not unsaturated fatty acids, play the key role in LPS signaling. Unlike phospholipids in plasma membrane, polar lipids in lipid rafts are predominantly acylated with saturated fatty acids that facilitate the formation of liquid-ordered lipid rafts. Our results showing that both LPS and lauric acid induce dimerization and recruitment of TLR4 to lipid rafts suggest the possibility that the saturated fatty acids acylated in Lipid A may help initiate the formation of lipid rafts, and thereby, recruit TLR4 to lipid rafts where TLR4 interacts with other signaling molecules to activate the downstream signaling pathways.
It was demonstrated that treatment of cells with n-3 PUFA eicosapentaenoic acid causes incorporation of this fatty acid into lipids in lipid rafts, which are highly enriched with saturated fatty acids (25). Such an alteration of the fatty acid composition of lipids in lipid rafts by n-3 PUFAs leads to displacement of signaling molecules from lipid rafts, and thus inhibits downstream signaling pathways (25, 49). Our results demonstrate that DHA inhibits LPS- or saturated fatty acid-induced dimerization and recruitment of TLR4 to lipid rafts, which is the initial step of TLR4 signaling pathways, suggesting a possibility that n-3 PUFAs inhibit the formation of lipid rafts by altering the fatty acid composition of polar lipids that are required to be acylated with saturated fatty acid for the formation of liquid-ordered lipid rafts. Therefore, the inhibition of the formation of lipid rafts by DHA may lead to reduction of signaling molecules recruited to lipid rafts.
Our results provide a new paradigm in understanding the mechanism by which saturated fatty acids and n-3 polyunsaturated fatty acids differentially modulate TLR4-induced inflammatory responses through the regulation of ROS-dependent dimerization and recruitment of TLR4 to lipid rafts.
Acknowledgment
We thank Dr. Roger Adamson in the Dept. of Physiology & Membrane Biology at the University of California at Davis for help with confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grants DK064007, DK41868, and CA75613. This work was also supported by Grant 2001-35200-10721 from the United States Dept. of Agriculture (USDA), Grant 01A095Rev from the American Institutes for Cancer Research, and program funds from the Western Human Nutrition Research Center/ARS/USDA (to D. H. H.) and by NIH Grants HL079904, HL55330, and HL6234 (to A. M. K. C.). This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grants C06 RR17348-01 and C06 RR12088-01 from the National Center for Research Resources, NIH.
- TLR
- Toll-like receptor
- LPS
- lipopolysaccharide
- PUFA
- polyunsaturated fatty acid
- DHA
- docosahexaenoic acid
- ROS
- reactive oxygen species
- GFP
- green fluorescent protein
- FBS
- fetal bovine serum
- DMEM
- Dulbecco's modified Eagle's medium
- BSA
- bovine serum albumin
- DPI
- diphenyleneiodonium chloride
- NAc
- N-acetyl-l-cysteine
- MyD88
- myeloid differentiation factor 88
- TIR
- Toll/IL-1 receptor
- TRIF
- TIR domain-containing adaptor inducing interferon-β
- CM-H2DCFDA
- 5-(and 6-)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetylester.
REFERENCES
- 1.Uematsu S., Akira S. (2006) J. Mol. Med. 84, 712–725 [DOI] [PubMed] [Google Scholar]
- 2.Kawai T., Akira S. (2007) Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 3.Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 4.Saitoh S., Miyake K. (2006) Chem. Rec. 6, 311–319 [DOI] [PubMed] [Google Scholar]
- 5.Munford R. S., Hall C. L. (1986) Science 234, 203–205 [DOI] [PubMed] [Google Scholar]
- 6.Kitchens R. L., Ulevitch R. J., Munford R. S. (1992) J. Exp. Med. 176, 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krauss J. H., Seydel U., Weckesser J., Mayer H. (1989) Eur. J. Biochem. 180, 519–526 [DOI] [PubMed] [Google Scholar]
- 8.Qureshi N., Takayama K., Kurtz R. (1991) Infect. Immun. 59, 441–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brightbill H. D., Libraty D. H., Krutzik S. R., Yang R. B., Belisle J. T., Bleharski J. R., Maitland M., Norgard M. V., Plevy S. E., Smale S. T., Brennan P. J., Bloom B. R., Godowski P. J., Modlin R. L. (1999) Science 285, 732–736 [DOI] [PubMed] [Google Scholar]
- 10.Wolf B., Hauschildt S., Uhl B., Metzger J., Jung G., Bessler W. G. (1989) Immunol. Lett. 20, 121–126 [DOI] [PubMed] [Google Scholar]
- 11.Uhl B., Speth V., Wolf B., Jung G., Bessler W. G., Hauschildt S. (1992) Eur. J. Cell Biol. 58, 90–98 [PubMed] [Google Scholar]
- 12.Takeuchi O., Kawai T., Mühlradt P. F., Morr M., Radolf J. D., Zychlinsky A., Takeda K., Akira S. (2001) Int. Immunol. 13, 933–940 [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., Modlin R. L., Akira S. (2002) J. Immunol. 169, 10–14 [DOI] [PubMed] [Google Scholar]
- 14.Alexopoulou L., Thomas V., Schnare M., Lobet Y., Anguita J., Schoen R. T., Medzhitov R., Fikrig E., Flavell R. A. (2002) Nat. Med. 8, 878–884 [DOI] [PubMed] [Google Scholar]
- 15.Lee J. Y., Ye J., Gao Z., Youn H. S., Lee W. H., Zhao L., Sizemore N., Hwang D. H. (2003) J. Biol. Chem. 278, 37041–37051 [DOI] [PubMed] [Google Scholar]
- 16.Lee J. Y., Plakidas A., Lee W. H., Heikkinen A., Chanmugam P., Bray G., Hwang D. H. (2003) J. Lipid Res. 44, 479–486 [DOI] [PubMed] [Google Scholar]
- 17.Lee J. Y., Zhao L., Youn H. S., Weatherill A. R., Tapping R., Feng L., Lee W. H., Fitzgerald K. A., Hwang D. H. (2004) J. Biol. Chem. 279, 16971–16979 [DOI] [PubMed] [Google Scholar]
- 18.Triantafilou M., Miyake K., Golenbock D. T., Triantafilou K. (2002) J. Cell Sci. 115, 2603–2611 [DOI] [PubMed] [Google Scholar]
- 19.Nakahira K., Kim H. P., Geng X. H., Nakao A., Wang X., Murase N., Drain P. F., Wang X., Sasidhar M., Nabel E. G., Takahashi T., Lukacs N. W., Ryter S. W., Morita K., Choi A. M. (2006) J. Exp. Med. 203, 2377–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons K., Toomre D. (2000) Nat. Rev. Mol. Cell Biol. 1, 31–39 [DOI] [PubMed] [Google Scholar]
- 21.Anderson R. G., Jacobson K. (2002) Science 296, 1821–1825 [DOI] [PubMed] [Google Scholar]
- 22.Hanada K., Nishijima M., Akamatsu Y., Pagano R. E. (1995) J. Biol. Chem. 270, 6254–6260 [DOI] [PubMed] [Google Scholar]
- 23.Schroeder R., London E., Brown D. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 12130–12134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown D. A., London E. (1998) Annu. Rev. Cell Dev. Biol. 14, 111–136 [DOI] [PubMed] [Google Scholar]
- 25.Stulnig T. M., Huber J., Leitinger N., Imre E. M., Angelisova P., Nowotny P., Waldhausl W. (2001) J. Biol. Chem. 276, 37335–37340 [DOI] [PubMed] [Google Scholar]
- 26.Saitoh S., Akashi S., Yamada T., Tanimura N., Kobayashi M., Konno K., Matsumoto F., Fukase K., Kusumoto S., Nagai Y., Kusumoto Y., Kosugi A., Miyake K. (2004) Int. Immunol. 16, 961–969 [DOI] [PubMed] [Google Scholar]
- 27.Kwon M. J., Soh J. W., Chang C. H. (2006) J. Immunol. 177, 950–956 [DOI] [PubMed] [Google Scholar]
- 28.Hwang D. (2001) FASEB J. 15, 2556–2564 [DOI] [PubMed] [Google Scholar]
- 29.Lee J. Y., Sohn K. H., Rhee S. H., Hwang D. (2001) J. Biol. Chem. 276, 16683–16689 [DOI] [PubMed] [Google Scholar]
- 30.Middlebrook J. L., Dorland R. B. (1984) Microbiol. Rev. 48, 199–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gay N. J., Gangloff M., Weber A. N. (2006) Nat. Rev. Immunol. 6, 693–698 [DOI] [PubMed] [Google Scholar]
- 32.Ozinsky A., Underhill D. M., Fontenot J. D., Hajjar A. M., Smith K. D., Wilson C. B., Schroeder L., Aderem A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13766–13771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J. Y., Lowell C. A., Lemay D. G., Youn H. S., Rhee S. H., Sohn K. H., Jang B., Ye J., Chung J. H., Hwang D. H. (2005) Biochem. Pharmacol. 70, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 34.Zhang H., Tay P. N., Cao W., Li W., Lu J. (2002) FEBS Lett. 532, 171–176 [DOI] [PubMed] [Google Scholar]
- 35.Brunn G. J., Bungum M. K., Johnson G. B., Platt J. L. (2005) FASEB J. 19, 872–874 [DOI] [PubMed] [Google Scholar]
- 36.Kim F., Pham M., Luttrell I., Bannerman D. D., Tupper J., Thaler J., Hawn T. R., Raines E. W., Schwartz M. W. (2007) Circ. Res. 100, 1589–1596 [DOI] [PubMed] [Google Scholar]
- 37.Michelsen K. S., Wong M. H., Shah P. K., Zhang W., Yano J., Doherty T. M., Akira S., Rajavashisth T. B., Arditi M. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10679–10684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi H., Kokoeva M. V., Inouye K., Tzameli I., Yin H., Flier J. S. (2006) J. Clin. Invest. 116, 3015–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis J. E., Gabler N. K., Walker-Daniels J., Spurlock M. E. (2008) Obesity 16, 1248–1255 [DOI] [PubMed] [Google Scholar]
- 40.Nguyen M. T., Favelyukis S., Nguyen A. K., Reichart D., Scott P. A., Jenn A., Liu-Bryan R., Glass C. K., Neels J. G., Olefsky J. M. (2007) J. Biol. Chem. 282, 35279–35292 [DOI] [PubMed] [Google Scholar]
- 41.Suganami T., Tanimoto-Koyama K., Nishida J., Itoh M., Yuan X., Mizuarai S., Kotani H., Yamaoka S., Miyake K., Aoe S., Kamei Y., Ogawa Y. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 84–91 [DOI] [PubMed] [Google Scholar]
- 42.Medzhitov R., Janeway C. A., Jr. (1997) Cell 91, 295–298 [DOI] [PubMed] [Google Scholar]
- 43.O'Neill L. A., Greene C. (1998) J. Leukoc. Biol. 63, 650–657 [PubMed] [Google Scholar]
- 44.Tao X., Xu Y., Zheng Y., Beg A. A., Tong L. (2002) Biochem. Biophys. Res. Commun. 299, 216–221 [DOI] [PubMed] [Google Scholar]
- 45.Xu Y., Tao X., Shen B., Horng T., Medzhitov R., Manley J. L., Tong L. (2000) Nature 408, 111–115 [DOI] [PubMed] [Google Scholar]
- 46.Hajjar A. M., O'Mahony D. S., Ozinsky A., Underhill D. M., Aderem A., Klebanoff S. J., Wilson C. B. (2001) J. Immunol. 166, 15–19 [DOI] [PubMed] [Google Scholar]
- 47.Alexopoulou L., Holt A. C., Medzhitov R., Flavell R. A. (2001) Nature 413, 732–738 [DOI] [PubMed] [Google Scholar]
- 48.Zhao L., Kwon M. J., Huang S., Lee J. Y., Fukase K., Inohara N., Hwang D. H. (2007) J. Biol. Chem. 282, 11618–11628 [DOI] [PubMed] [Google Scholar]
- 49.Fan Y. Y., Ly L. H., Barhoumi R., McMurray D. N., Chapkin R. S. (2004) J. Immunol. 173, 6151–6160 [DOI] [PubMed] [Google Scholar]
- 50.Powers K. A., Szászi K., Khadaroo R. G., Tawadros P. S., Marshall J. C., Kapus A., Rotstein O. D. (2006) J. Exp. Med. 203, 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park H. S., Jung H. Y., Park E. Y., Kim J., Lee W. J., Bae Y. S. (2004) J. Immunol. 173, 3589–3593 [DOI] [PubMed] [Google Scholar]
- 52.Laroux F. S., Romero X., Wetzler L., Engel P., Terhorst C. (2005) J. Immunol. 175, 5596–5600 [DOI] [PubMed] [Google Scholar]