Abstract
Proinflammatory cytokines induce nitric oxide-dependent DNA damage and ultimately β-cell death. Not only does nitric oxide cause β-cell damage, it also activates a functional repair process. In this study, the mechanisms activated by nitric oxide that facilitate the repair of damaged β-cell DNA are examined. JNK plays a central regulatory role because inhibition of this kinase attenuates the repair of nitric oxide-induced DNA damage. p53 is a logical target of JNK-dependent DNA repair; however, nitric oxide does not stimulate p53 activation or accumulation in β-cells. Further, knockdown of basal p53 levels does not affect DNA repair. In contrast, expression of growth arrest and DNA damage (GADD) 45α, a DNA repair gene that can be regulated by p53-dependent and p53-independent pathways, is stimulated by nitric oxide in a JNK-dependent manner, and knockdown of GADD45α expression attenuates the repair of nitric oxide-induced β-cell DNA damage. These findings show that β-cells have the ability to repair nitric oxide-damaged DNA and that JNK and GADD45α mediate the p53-independent repair of this DNA damage.
Insulin-dependent diabetes mellitus is an autoimmune disease characterized by the selective destruction of insulin-secreting pancreatic β-cells found in the islets of Langerhans (1). Cytokines, released from invading leukocytes during insulitis, are believed to participate in the initial destruction of β-cells, precipitating the autoimmune response (2, 3). Treatment of rat islets with the macrophage-derived cytokine interleukin-1 (IL-1)2 results in the inhibition of glucose-stimulated insulin secretion and oxidative metabolism and in the induction of DNA damage that ultimately results in β-cell death (4–6). Nitric oxide, produced in micromolar levels following enhanced expression of the inducible nitric-oxide synthase in β-cells, mediates the damaging actions of cytokines on β-cell function (7–9). Nitric oxide inhibits insulin secretion by attenuating the oxidation of glucose to CO2, reducing cellular levels of ATP and, thereby, attenuating ATP-inhibited K+ channel activity (10, 11). The net effect is the inhibition of β-cell depolarization, calcium entry, and calcium-dependent exocytosis. In addition to the inhibition of β-cell function, nitric oxide induces DNA damage in β-cells (4, 12, 13). Nitric oxide or the oxidation products N2O3 and ONOO− induce DNA damage through direct strand breaks and base modification (14–16) and by inhibition of DNA repair enzymes, thereby enhancing the damaging actions of nitric oxide (17, 18).
Recent studies have shown that β-cells maintain a limited ability to recover from cytokine-mediated damage (19, 20). The addition of a nitric-oxide synthase inhibitor to islets treated for 24 h with cytokine and continued culture with the nitric-oxide synthase inhibitor and cytokine results in a time-dependent restoration of insulin secretion, mitochondrial aconitase activity, and the repair of nitric oxide-damaged DNA (20, 21). Nitric oxide plays a dual role in modifying β-cell responses to cytokines. Nitric oxide induces β-cell damage and also activates a JNK-dependent recovery response that requires new gene expression (22). The ability of β-cells to recover from cytokine-mediated damage is temporally limited because cytokine-induced β-cell damage becomes irreversible following a 36-h incubation, and islets at this point are committed to degeneration (19).
The purpose of this study was to determine the mechanisms by which β-cells repair nitric oxide-damaged DNA. Previous reports have shown that DNA damage induced by oxidizing agents, such as nitric oxide, is repaired through the base excision repair pathway (23), but how this pathway is activated in response to nitric oxide is unknown. Similar to the recovery of metabolic function, we now show that the activation of JNK by nitric oxide is required for repair of cytokine-induced DNA damage in β-cells. p53 is a logical candidate to mediate this repair because it plays a central role in DNA repair, is a target of JNK, and is activated by nitric oxide (24–27). However, we show that cytokines do not stimulate p53 phosphorylation, and nitric oxide fails to stimulate p53 accumulation and phosphorylation. Growth arrest and DNA damage (GADD) 45α is a DNA damage-inducible gene that can be regulated by both p53-dependent and p53-independent mechanisms (28–31). In contrast to p53, we show that cytokines stimulate GADD45α expression in a nitric oxide- and JNK-dependent manner and that siRNA-mediated knockdown of GADD45α results in an attenuation in the repair of nitric oxide-mediated DNA damage. These findings support a role for JNK in the regulation of GADD45α-dependent and p53-independent repair of nitric oxide-damaged β-cell DNA.
EXPERIMENTAL PROCEDURES
Materials and Animals
Male Sprague-Dawley rats (250–300 g) were purchased from Harlan (Indianapolis, IN). INS 832/13 cells were obtained from Chris Newgard (Duke University, Durham NC). RPMI 1640 medium, CMRL-1066 tissue culture medium, l-glutamine, streptomycin, and penicillin were from Mediatech, Inc. (Manassas, VA). Fetal calf serum was from Sigma. Human recombinant IL-1 was purchased from PeproTech (Rocky Hill, NJ). NG-Monomethyl-l-arginine (NMMA) and (Z)-1(N,N-diethylamino)diazen-1-ium-1,2-diolate (DEANO) were purchased from Axxora (San Diego, CA). Camptothecin was from Sigma. SP600125 and TAT-TI-JIP153–163 were obtained from Calbiochem. p53, phospho-p53, phospho-ATF-2, and phospho-c-Jun antibodies were from Cell Signaling (Beverly, MA). GAPDH antibody was from Ambion (Austin, TX). Horseradish peroxidase-conjugated donkey anti-rabbit and donkey anti-mouse antibodies were from Jackson Immunoresearch Laboratories, Inc. (West Grove, PA). GADD45α and GAPDH primers were from IDT DNA Technologies (Coralville, IA).
Islet Isolation and Cell Culture
Islets were isolated from male Sprague-Dawley rats (250–300 g) by collagenase digestion as described previously (32). Islets were cultured overnight in CMRL-1066 (containing 2 mm l-glutamine, 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin) at 37 °C under an atmosphere of 95% air and 5% CO2 before experimentation. INS 832/13 cells were removed from growth flasks by treatment with 0.05% trypsin and 0.02% EDTA for 5 min at 37 °C, washed twice with RPMI 1640 medium, and plated at the indicated cell densities.
Comet Assay
DNA damage was assessed using the comet assay (single-cell gel electrophoresis) as described previously (4, 33). Briefly, cells were harvested and embedded in 0.6% low melting agarose on slides precoated with 1.0% agar. Samples were then incubated in lysing solution (2.5 m NaCl, 100 mm EDTA, 10 mm Trizma (Tris base), 1% Triton X-100) overnight. Following lysis, the slides were incubated in an alkaline electrophoresis buffer (0.3 m NaOH, 1 mm EDTA (pH > 13)) for 40 min followed by electrophoresis at 25 volts/300 mA for 20 min. Slides were washed three times in 0.4 m Tris (pH 7.5) and stained with ethidium bromide (2 μg/ml). Comet images were captured using a Nikon eclipse 90I, and the CASP program (34) was used to quantify the mean tail moment from 30 to 50 cells/condition.
Western Blot Analysis
Cells were lysed in Laemmli buffer, and proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane (Amersham Biosciences) under semidry transfer conditions as described previously (35). Blots were blocked with 5% milk in Tris-buffered saline/Tween 20 for 1 h and incubated overnight at 4 °C with p53 (1:1000), phospho-p53 (1:1000), phospho-ATF-2 (1:1000), phospho-c-Jun (1:1000), or GAPDH (1:50,000) antibodies according to the manufacturer's instructions. Membranes were incubated with horseradish peroxidase-conjugated donkey anti-rabbit (1:7000) or mouse (1:5000) secondary antibodies for 1 h. Luminol chemiluminescence was used to detect the protein of interest (36).
Nitrite Determination
Nitrite production was determined from culture supernatants using the Greiss assay as described previously (37). Fifty microliters of the Greiss reagents were incubated with 50 μl of the culture supernatants, and the absorbance was measured at 540 nm using a Power Wave X-340 plate reader (Biotek Instruments). Nitrite concentrations were calculated using a sodium nitrite standard curve.
siRNA Transfection
siRNA transfection was performed using NeoFX transfection reagent (Ambion) according to the manufacturer's instructions. NeoFX transfection reagent and siRNA were diluted to give a final concentration of 30 nm siRNA. The diluted NeoFX-siRNA complex was added to each well and overlaid with 200,000 cells in 450 μl. Experiments were started 48 h following transfection. The following Silencer® Select predesigned siRNAs were obtained from Ambion: p53 sense, 5′-CAAUUUCCCUCAAUAAGCUtt-3′ and GADD45α sense, 5′-CGUGCUUUCUGUUGCGAGAtt-3′.
Real Time PCR
RNA was isolated using the RNeasy kit (Qiagen). cDNA synthesis was performed using oligo(dT) and the reverse transcriptase Superscript preamplification system according to the manufacturer's instructions (Invitrogen). Real time PCR was performed using the Light Cycler 280 (Roche Applied Biosciences) to detect SYBR Green incorporation, according to the manufacturer's instructions. The fold increase of GADD45α mRNA accumulation was normalized to the housekeeping gene GAPDH. Primer sequences for GADD45α were: forward, 5′-TGGCTGCGGATGAAGATGAC-3′ and reverse, 5′-GTGGGGAGTGACTGCTTGAGTAAC-3′. Sequences for GAPDH primers were: forward, 5′-GCTGGGGCTCACCTGAAGGG-3′ and reverse, 5′-GGATGACCTTGCCCACAGCC-3′.
RESULTS
JNK Activation Is Required for the Repair of Nitric Oxide-induced DNA Damage
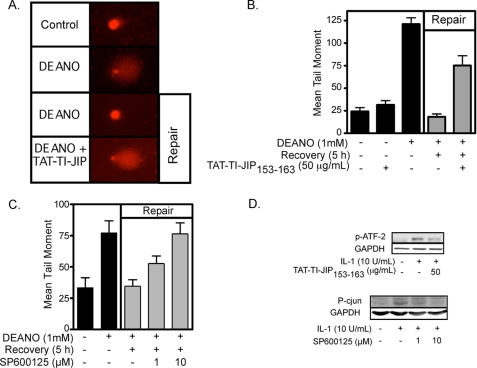
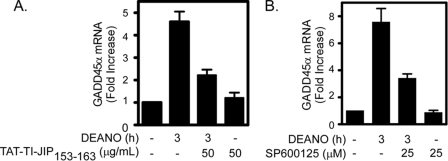
β-Cells have the ability to repair nitric oxide-mediated DNA damage (21). Using the comet assay to measure DNA damage, we show that 1-h treatment with the nitric oxide donor DEANO results in DNA comet formation (Fig. 1A) and a 5-fold increase in the mean tail moment (Fig. 1B). Removal of DEANO by washing and continued culture for 5 h results in the repair of DNA damage as evidenced by the absence of a comet tail (Fig. 1A) and the return of the mean tail moment to control levels (Fig. 1B). Because JNK plays an essential role in the recovery of metabolic function in cytokine-treated islets (22), the effects of two chemically distinct inhibitors of JNK (the peptide inhibitor TAT-TI-JIP153–163 and the pharmacological inhibitor SP600125) on DNA repair were examined. Treatment of INS 832/13 cells with TAT-TI-JIP153–163 or SP600125 during the DEANO treatment and the recovery period results in the attenuation of β-cell DNA repair (Fig. 1A for comet and Fig. 1, B and C, for quantification of comets). As a control for JNK inhibition, TAT-TI-JIP153–163 inhibits IL-1-induced ATF-2 phosphorylation (JNK substrate) at a concentration that attenuates DNA repair (Fig. 1D). In addition, SP600125 attenuates IL-1-induced c-Jun phosphorylation in a concentration-related fashion that is similar to the concentration-related inhibition of DNA repair in INS 832/13 cells (Fig. 1D). Consistent with our previous findings related to the recovery of metabolic function (22), these findings support a role for JNK in the repair of nitric oxide-damaged β-cell DNA.
FIGURE 1.
JNK activation is required for the repair of nitric oxide-induced DNA damage. INS 832/13 cells were pretreated for 30 min with TAT-TI-JIP153–163 (A and B) or SP600125 (C) at the indicated concentrations. DEANO (1 mm) was added, and the cells were cultured for 1 h, or they were cultured for 1 h, washed to remove the nitric oxide donor, and cultured for 5 additional h (repair conditions). The cells were harvested, and DNA damage was determined using the comet assay (A). This damage was quantified as the mean tail moment on 30–50 cells/condition (B and C). JNK inhibitor activity was assessed by ATF-2 or c-Jun phosphorylation in INS 832/13 cells preincubated for 30 min with TAT-TI-JIP153–163 or SP600125, respectively, followed by a 30-min treatment with IL-1 (D). Results are representative of three independent experiments.
Nitric Oxide Fails to Activate p53 in INS 832/13 Cells or Rat Islets
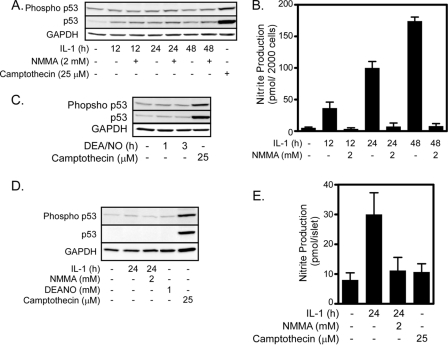
p53 is a transcription factor that participates in DNA repair through direct interaction with DNA repair enzymes, such as polymerase β (24, 38), and through the enhanced expression of genes required for DNA repair, such as GADD45α (39). p53 is regulated by phosphorylation, resulting in its stabilization and activation (40). p53 is a candidate to mediate the repair of nitric oxide-damaged DNA because nitric oxide has been shown to activate p53 and JNK directly phosphorylates p53 (26, 27, 41). However, p53 is not activated by nitric oxide in β-cells. IL-1 fails to stimulate p53 phosphorylation in both INS 832/13 cells (Fig. 2A) and rat islets (Fig. 2D). Although IL-1 fails to induce p53 phosphorylation, it does stimulate the accumulation of p53 to low levels in INS 832/13 cells (Fig. 2A), whereas p53 levels remain below the limits of detection in rat islets (Fig. 2D). The level to which p53 accumulates in response to IL-1 in INS 832/13 cells is much lower than the levels observed in response to apoptotic stimuli, such as camptothecin (Fig. 2A). Further, IL-1-induced p53 expression does not depend on nitric oxide. The nitric-oxide synthase inhibitor, NMMA, which attenuates nitric oxide production (Fig. 2, B and E), does not modulate IL-1-induced p53 accumulation (Fig. 2A), and DEANO fails to stimulate p53 phosphorylation or accumulation in INS 832/13 cells (Fig. 2C) or rat islets (Fig. 2D). Camptothecin, which has been shown to activate p53 (42), stimulates p53 phosphorylation and accumulation in INS 832/13 cells (Fig. 2, A and C). These findings indicate that p53 is not activated by nitric oxide in β-cells.
FIGURE 2.
Nitric oxide fails to activate p53 in INS 832/13 cells or rat islets. INS 832/13 cells (A–C) or rat islets (D and E) were treated with IL-1 (10 units/ml) ± NMMA (2 mm) or DEANO (1 mm) for the indicated times. Following the treatments, culture supernatants were harvested, and nitrite production was determined (B and E). Cells were harvested, and phopsho-p53 and total p53 were determined by Western blot analysis. Camptothecin was used as a positive control for p53 activation, and GAPDH was used as a loading control. Results are representative of at least three experiments.
p53 Is Not Required for Repair of Nitric Oxide-induced DNA Damage
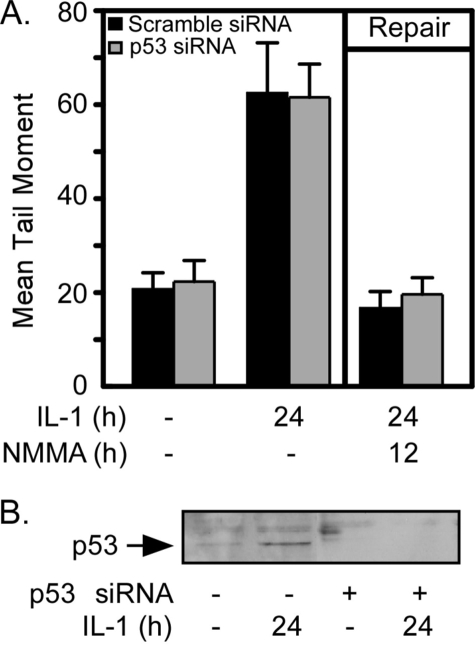
The lack of p53 expression or activation in response to nitric oxide in β-cells suggests that DNA repair in β-cells (which is activated by nitric oxide) does not require p53. To examine this issue directly, DNA repair was examined in cells in which p53 expression was attenuated using siRNA. INS 832/13 cells transfected with scramble siRNA or p53 siRNA were treated with IL-1 for 24 h or treated with IL-1 for 24 h followed by the addition of the nitric-oxide synthase inhibitor, NMMA, for an additional 12-h incubation to stimulate DNA repair. IL-1 induces DNA damage in both scramble or p53 siRNA-transfected cells as indicated by the 3-fold increase in the mean tail moment (Fig. 3A). The addition of NMMA followed by 12 additional h of incubation (in the presence of IL-1) results in the complete repair of nitric oxide-induced DNA damage in both the scramble and p53 siRNA-transfected cells. As shown by Western blot analysis, p53 accumulation in response to IL-1 treatment is attenuated in cells transfected with p53 siRNA (Fig. 3B). Scramble siRNA has no effect on IL-1-induced p53 expression (data not shown). These findings indicate that p53 is not required for the repair of nitric oxide-damaged DNA in β-cells because nitric oxide fails to stimulate p53 activation, and siRNA knockdown of p53 does not modify DNA repair in IL-1-treated β-cells.
FIGURE 3.
p53 is not required for repair of nitric oxide-induced DNA damage. INS 832/13 cells (200,000 cells/400 μl) were transfected with scrambled siRNA or siRNA targeted for p53 (30 nm). Forty-eight hours after transfection, cells were treated with IL-1 (10 units/ml) for 24 h or treated with IL-1 for 24 h followed by the addition of NMMA (2 mm) and 12-h incubation with both IL-1 + NMMA (repair conditions). A, the cells were harvested, and DNA damage was determined using the comet assay. B, alternatively, p53 levels were determined by Western blot analysis to control for gene knockdown. Results are the average ± S.E. (A) or representative of three experiments.
Nitric Oxide Induces GADD45α Expression in INS 832/13 Cells and Rat Islets
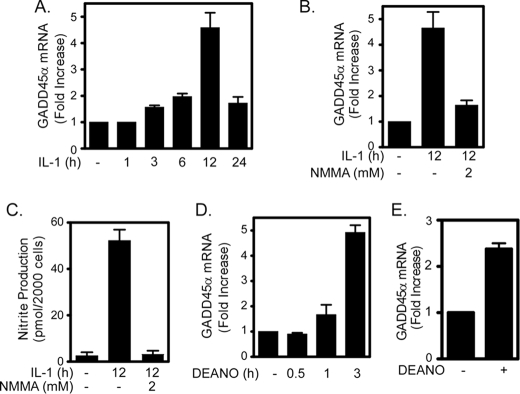
While we examined the potential role of p53 in mediating DNA repair in β-cells, we also examined the expression of GADD45α, a DNA damage-inducible gene that can be regulated by p53-dependent and p53-independent pathways (29–31). IL-1 induces GADD45α mRNA accumulation in a time-dependent fashion that is maximal following 12-h incubation (Fig. 4, A and B). The expression of GADD45α is nitric oxide-dependent because the nitric-oxide synthase inhibitor, NMMA, which attenuates nitric oxide production (Fig. 4C), inhibits IL-1-induced GADD45α mRNA accumulation (Fig. 4B). The nitric oxide donor DEANO also induces GADD45α mRNA accumulation in INS 832/13 cells (Fig. 4D) and rat islets (Fig. 4E). Similarly, treatment of human islets for 3 h with DEANO results in a >2-fold increase in GADD45α expression as measured by Affymetrix gene chip analysis (data not shown).
FIGURE 4.
Nitric oxide induces GADD45α expression in INS 832/13 cells and rat islets. A, IL-1 (10 units/ml) stimulates GADD45α mRNA accumulation in INS 832/13 cells in a time-dependent manner that is maximal following 12-h incubation. B, GADD45α mRNA accumulation in response to a 12-h incubation with IL-1 is prevented by NMMA. C, the accumulation of GADD45α mRNA correlates with nitrite production by INS 832/13 cells. D and E, exogenous addition of nitric oxide using the donor DEANO (1 mm) stimulates GADD45α mRNA accumulation in INS 832/13 cells (D) and rat islets (E). mRNA accumulation was quantified by real time PCR and normalized to GAPDH. Results are the average of three experiments ± S.E.
JNK Is Required for Nitric Oxide-induced GADD45α Expression
Because inhibitors of JNK attenuate the repair of nitric oxide-induced DNA damage, the effect of JNK inhibition on nitric oxide-induced GADD45α expression was evaluated. Treatment of INS 832/13 cells with DEANO for 3 h results in the accumulation of GADD45α mRNA. At concentrations that prevent DNA repair, the JNK inhibitors TAT-TI-JIP153–163 (Fig. 5A) and SP600125 (Fig. 5B) attenuate DEANO-induced GADD45α mRNA accumulation. These results indicate that nitric oxide-induced GADD45α expression requires the activation of JNK.
FIGURE 5.
JNK is required for nitric oxide induced GADD45α expression. Inhibitors of JNK, TAT-TI-JIP153–163 (A), and SP600125 (B) prevent 1 mm DEANO-induced GADD45α mRNA accumulation by INS 832/13 cells. Cells were pretreated for 30 min prior to the addition of the nitric oxide donor. mRNA accumulation was quantified by real time PCR and was normalized to GAPDH. Results are the average of three experiments ± S.E.
GADD45α Is Essential for Repair of Nitric Oxide-induced DNA Damage
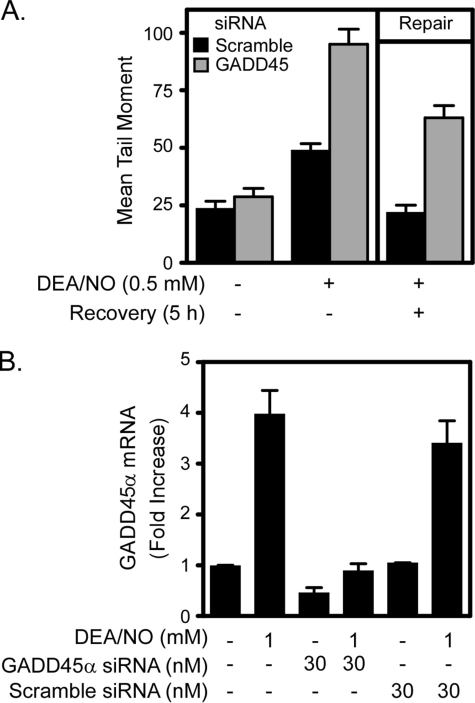
To determine whether GADD45α participates in the repair of nitric oxide-damaged DNA in β-cells, the effects of GADD45α siRNA gene knockdown on DNA repair were examined. Treatment of INS 832/13 cells for 1 h with DEANO results in a 2-fold increase in DNA damage. This DNA damage is repaired completely following removal of nitric oxide by washing and continued culture for 5 h in the absence of the nitric oxide donor. Importantly, DEANO-induced DNA damage following a 1-h incubation is enhanced by ∼2-fold in INS 832/13 cells transfected with GADD45α siRNA, and the ability of these cells to repair damaged DNA during the 5-h recovery period is reduced by >50% compared with scramble siRNA-transfected control cells (Fig. 6A). The induction of GADD45α mRNA expression by DEANO is attenuated with GADD45α siRNA and not affected by scramble siRNA (Fig. 6B). These results indicate that the induction of GADD45α by nitric oxide participates in the repair of nitric oxide-induced DNA damage.
FIGURE 6.
GADD45α is essential for repair of nitric oxide-induced DNA damage. INS 832/13 cells were transfected with 30 nm GADD45α or scramble (negative control) siRNA as indicated. Forty-eight hours after transfection, INS 832/13 cells were treated with DEANO for 1 h or with DEANO for 1 h followed by washing to remove the nitric oxide donor, and continued culture for 5 additional h (repair conditions). A, the cells were harvested, and DNA damage was quantified by the comet assay. B, cells were also harvested, and real time PCR was used to confirm knockdown of GADD45α mRNA using siRNA. Results are the average ± S.E. of three independent experiments.
DISCUSSION
It has been shown that nitric oxide, produced in micromolar levels by the β-cell in response to cytokines, mediates the damaging effects of cytokines through the inhibition of mitochondrial enzymes, inhibition of glucose-stimulated insulin secretion, and the induction of DNA strand breaks and base modifications (8, 9). We have established that β-cells possess an active defense or repair process that affords these cells a limited capacity to recover from these damaging effects of nitric oxide (20). Although nitric oxide activates multiple MAP kinases in β-cells, JNK activation is required for the recovery of oxidative metabolism following nitric oxide-mediated damage (22). In this report, the mechanism underlying the induction of DNA repair in response to nitric oxide-mediated damage in β-cells was evaluated. A number of mechanisms have been proposed to explain the repair of oxidative DNA damage induced by free radicals (23). Damage induced by the oxidized products of nitric oxide, N2O3 or ONOO−, results primarily in the deamination and oxidation of bases (14, 16, 43). These altered bases are repaired by base excision repair in which specific glycosylases first recognize and remove the damaged base. An apurinic/apyridimic endonuclease then recognizes the abasic site and cleaves the phosphate-sugar backbone. The resulting gap is filled by polymerase β and annealed by DNA ligase III (23). Although these mechanisms explain the repair of oxidized DNA damage, the mechanisms controlling how cells respond to nitric oxide to activate or enhance DNA repair pathways are not well understood. Most studies thus far have focused on the damaging actions of nitric oxide on DNA and fail to examine the protective mechanisms activated in cells that are designed to repair the damage.
In this report we identify a role for JNK in the repair of nitric oxide-induced DNA damage in β-cells. Using chemical and peptide inhibitors, we demonstrate that JNK is required for the repair of nitric oxide-induced DNA damage. This result may seem counterintuitive because JNK is generally considered a proapoptotic MAP kinase; however, recent studies by Andreka et al. (44) have shown that JNK activation is protective against nitric oxide-induced cardiac myocyte death. Additionally, Mercola and co-workers (45–48) have described a role for JNK in DNA repair from genotoxic stresses. Although this report presents evidence of a protective role for JNK activation in nitric oxide-induced β-cell damage, others have suggested that JNK activation participates in cytokine-induced β-cell apoptosis (49). We do not observe apoptosis in response to DEANO or IL-1 under the culture conditions used in this study (13; and data not shown). Ammendrup et al. (49) have shown that prolonged incubations with IL-1 result in β-cell apoptosis and that the inhibition of JNK protects against cytokine-mediated apoptosis. These seemingly opposing results could be explained by the differences in cytokine exposures. In support of this hypothesis, we have recently shown that short exposures to cytokine stimulate β-cell necrosis, but prolonged incubations with IL-1 for >36 h result in a shift in the mechanism of death from necrosis to apoptosis.3 This shift in the type of cell death from necrosis to apoptosis is associated with irreversible DNA damage. Similar to the temporal effects of cytokines on β-cell viability, recent studies suggest that the temporal activation of JNK regulates the cellular responses to cytokines such as tumor necrosis factor α-induced damage (50). Tumor necrosis factor α-induced JNK activation has been shown to be biphasic, where early transient activation of JNK is protective, while prolonged activation induces apoptosis (50). This mechanism of JNK regulation may apply to nitric oxide-induced JNK activation where transient early activation stimulates protective responses, whereas sustained JNK activation contributes to cytokine-induced apoptosis.
To investigate further how JNK regulates DNA repair, we examined the activation of p53 by nitric oxide in β-cells. p53 is a transcription factor shown to be involved directly in base excision repair through the interaction with polymerase β and indirectly through the transcriptional regulation of factors involved in DNA repair, such as GADD45α (28, 38). We originally hypothesized that JNK would induce DNA repair through the activation of p53 because JNK has been shown to activate p53 directly and nitric oxide activates p53 in other cell types (25–27, 41). However, nitric oxide, produced endogenously in response to IL-1 or exogenous using DEANO, fails to induce p53 phosphorylation or accumulation in β-cells. IL-1 does enhance p53 accumulation to low levels in INS 832/13 cells, but this induction does not regulate DNA repair because INS 832/13 cells with attenuated p53 levels maintain the ability to repair DNA damage.
Although p53 is not induced by nitric oxide in β-cells, the expression of GADD45α, a target of p53 that can be regulated by p53-independent mechanisms, is enhanced by nitric oxide. GADD45α is an inducible gene activated by various DNA-damaging agents that participates in base excision repair by interacting directly with DNA repair enzymes, including proliferating cell nuclear antigen, an auxiliary factor for polymerase β, and apurinic/apyridimic endonuclease (28, 31, 39). GADD45α maintains nuclear localization of these enzymes, thereby enhancing DNA repair (28). In addition to p53, GADD45α expression is regulated by various transcription factors, including FoxO1, Oct-1, and NF-YA (29, 30, 39, 51). The accumulation of GADD45α mRNA in response to nitric oxide depends on JNK activation. These findings correlate JNK- and nitric oxide-dependent GADD45α expression with JNK-dependent DNA repair. Using siRNA gene knockdown, the role of GADD45α in the repair of nitric oxide-damaged DNA in β-cells was examined. Importantly, the siRNA knockdown of nitric oxide-induced GADD45α expression results not only in an attenuation in the repair of damaged DNA, it also makes β-cells more susceptible to nitric oxide-mediated DNA damage (Fig. 6A). Knockdown of GADD45α does not completely prevent the repair of damaged DNA. This result may be due to an incomplete knockdown of GADD45α, or it may be a consequence of the activation of additional pathways that participate in the repair the damaged DNA. Although the inhibition of DNA repair is not complete, our findings support a role for GADD45α in maintaining and restoring DNA integrity under conditions of stress induced by nitric oxide in β-cells.
These findings describe a repair mechanism activated by β-cells in response to nitric oxide-induced DNA damage. We show that JNK activation by nitric oxide plays a central role in the repair process through the p53-independent regulation of GADD45α. Currently, the mechanism by which JNK stimulates GADD45α expression remains unknown; however, we speculate that ATF-2 activation may play a central role. ATF-2, a JNK substrate that we show to be phosphorylated in response to cytokines (Fig. 1D), has been shown to contribute to the transcriptional regulation of GADD45α expression and to participate in JNK-dependent DNA repair (46, 52).
Acknowledgment
We thank Kimberly Jaimes for expert technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK52194 and AI44458 (to J. A. C.).
K. J. Hughes and J. A. Corbett, unpublished data.
- IL
- interleukin
- ATF-2
- activating transcription factor-2
- DEANO
- (Z)-1(N,N-diethylamino)diazen-1-ium-1,2-diolate
- GADD
- growth arrest and DNA damage
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- JNK
- c-Jun N-terminal kinase
- MAP
- mitogen-activated protein
- NMMA
- NG-monomethyl-l-arginine
- siRNA
- small interfering RNA.
REFERENCES
- 1.Gepts W. (1965) Diabetes 14, 619–633 [DOI] [PubMed] [Google Scholar]
- 2.Mandrup-Poulsen T. (1996) Diabetologia 39, 1005–1029 [DOI] [PubMed] [Google Scholar]
- 3.Rabinovitch A., Suarez-Pinzon W. L. (1998) Biochem. Pharmacol. 55, 1139–1149 [DOI] [PubMed] [Google Scholar]
- 4.Delaney C. A., Green M. H., Lowe J. E., Green I. C. (1993) FEBS Lett. 333, 291–295 [DOI] [PubMed] [Google Scholar]
- 5.Corbett J. A., Lancaster J. R., Jr., Sweetland M. A., McDaniel M. L. (1991) J. Biol. Chem. 266, 21351–21354 [PubMed] [Google Scholar]
- 6.Welsh N., Eizirik D. L., Bendtzen K., Sandler S. (1991) Endocrinology 129, 3167–3173 [DOI] [PubMed] [Google Scholar]
- 7.Corbett J. A., Wang J. L., Sweetland M. A., Lancaster J. R., Jr., McDaniel M. L. (1992) J. Clin. Invest. 90, 2384–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbett J. A., Wang J. L., Hughes J. H., Wolf B. A., Sweetland M. A., Lancaster J. R., Jr., McDaniel M. L. (1992) Biochem. J. 287, 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Southern C., Schulster D., Green I. C. (1990) FEBS Lett. 276, 42–44 [DOI] [PubMed] [Google Scholar]
- 10.Misler S., Barnett D. W., Gillis K. D., Pressel D. M. (1992) Diabetes 41, 1221–1228 [DOI] [PubMed] [Google Scholar]
- 11.Hughes J. H., Colca J. R., Easom R. A., Turk J., McDaniel M. L. (1990) J. Clin. Invest. 86, 856–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eizirik D. L., Delaney C. A., Green M. H., Cunningham J. M., Thorpe J. R., Pipeleers D. G., Hellerström C., Green I. C. (1996) Mol. Cell. Endocrinol. 118, 71–83 [DOI] [PubMed] [Google Scholar]
- 13.Steer S. A., Scarim A. L., Chambers K. T., Corbett J. A. (2006) PLoS Med. 3, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burney S., Caulfield J. L., Niles J. C., Wishnok J. S., Tannenbaum S. R. (1999) Mutat. Res. 424, 37–49 [DOI] [PubMed] [Google Scholar]
- 15.Tamir S., Burney S., Tannenbaum S. R. (1996) Chem. Res. Toxicol. 9, 821–827 [DOI] [PubMed] [Google Scholar]
- 16.Niles J. C., Wishnok J. S., Tannenbaum S. R. (2006) Nitric Oxide 14, 109–121 [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal M., LaRusso N. F., Nishioka N., Nakabeppu Y., Gores G. J. (2001) Cancer Res. 61, 6388–6393 [PubMed] [Google Scholar]
- 18.Graziewicz M., Wink D. A., Laval F. (1996) Carcinogenesis 17, 2501–2505 [DOI] [PubMed] [Google Scholar]
- 19.Scarim A. L., Heitmeier M. R., Corbett J. A. (1997) Endocrinology 138, 5301–5307 [DOI] [PubMed] [Google Scholar]
- 20.Corbett J. A., McDaniel M. L. (1994) Biochem. J. 299, 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosales A. L., Cunningham J. M., Bone A. J., Green I. C., Green M. H. (2004) Free Radic. Res. 38, 665–674 [DOI] [PubMed] [Google Scholar]
- 22.Scarim A. L., Nishimoto S. Y., Weber S. M., Corbett J. A. (2003) Endocrinology 144, 3415–3422 [DOI] [PubMed] [Google Scholar]
- 23.Seeberg E., Eide L., Bjørås M. (1995) Trends Biochem. Sci. 20, 391–397 [DOI] [PubMed] [Google Scholar]
- 24.Smith M. L., Seo Y. R. (2002) Mutagenesis 17, 149–156 [DOI] [PubMed] [Google Scholar]
- 25.Messmer U. K., Ankarcrona M., Nicotera P., Brüne B. (1994) FEBS Lett. 355, 23–26 [DOI] [PubMed] [Google Scholar]
- 26.Hofseth L. J., Saito S., Hussain S. P., Espey M. G., Miranda K. M., Araki Y., Jhappan C., Higashimoto Y., He P., Linke S. P., Quezado M. M., Zurer I., Rotter V., Wink D. A., Appella E., Harris C. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 143–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., Michael D., de Murcia G., Oren M. (2002) J. Biol. Chem. 277, 15697–15702 [DOI] [PubMed] [Google Scholar]
- 28.Jung H. J., Kim E. H., Mun J. Y., Park S., Smith M. L., Han S. S., Seo Y. R. (2007) Oncogene 26, 7517–7525 [DOI] [PubMed] [Google Scholar]
- 29.Tran H., Brunet A., Grenier J. M., Datta S. R., Fornace A. J., Jr., DiStefano P. S., Chiang L. W., Greenberg M. E. (2002) Science 296, 530–534 [DOI] [PubMed] [Google Scholar]
- 30.Tong T., Fan W., Zhao H., Jin S., Fan F., Blanck P., Alomo I., Rajasekaran B., Liu Y., Holbrook N. J., Zhan Q. (2001) Exp. Cell Res. 269, 64–72 [DOI] [PubMed] [Google Scholar]
- 31.Smith M. L., Chen I. T., Zhan Q., Bae I., Chen C. Y., Gilmer T. M., Kastan M. B., O'Connor P. M., Fornace A. J., Jr. (1994) Science 266, 1376–1380 [DOI] [PubMed] [Google Scholar]
- 32.Kelly C. B., Blair L. A., Corbett J. A., Scarim A. L. (2003) Methods Mol. Med. 83, 3–14 [DOI] [PubMed] [Google Scholar]
- 33.Singh N. P., McCoy M. T., Tice R. R., Schneider E. L. (1988) Exp. Cell Res. 175, 184–191 [DOI] [PubMed] [Google Scholar]
- 34.Koñca K., Lankoff A., Banasik A., Lisowska H., Kuszewski T., GóŸdŸ S., Koza Z., Wojcik A. (2003) Mutat. Res. 534, 15–20 [DOI] [PubMed] [Google Scholar]
- 35.Heitmeier M. R., Scarim A. L., Corbett J. A. (1997) J. Biol. Chem. 272, 13697–13704 [DOI] [PubMed] [Google Scholar]
- 36.White B. H., Kaczmarek L. K. (1997) J. Neurosci. 17, 1582–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Green L. C., Wagner D. A., Glogowski J., Skipper P. L., Wishnok J. S., Tannenbaum S. R. (1982) Anal. Biochem. 126, 131–138 [DOI] [PubMed] [Google Scholar]
- 38.Zhou J., Ahn J., Wilson S. H., Prives C. (2001) EMBO J. 20, 914–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrier F., Smith M. L., Bae I., Kilpatrick K. E., Lansing T. J., Chen C. Y., Engelstein M., Friend S. H., Henner W. D., Gilmer T. M., Kastani M. B., Kournace A. J., Jr. (1994) J. Biol. Chem. 269, 32672–32677 [PubMed] [Google Scholar]
- 40.Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 [DOI] [PubMed] [Google Scholar]
- 41.Buschmann T., Potapova O., Bar-Shira A., Ivanov V. N., Fuchs S. Y., Henderson S., Fried V. A., Minamoto T., Alarcon-Vargas D., Pincus M. R., Gaarde W. A., Holbrook N. J., Shiloh Y., Ronai Z. (2001) Mol. Cell. Biol. 21, 2743–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watcharasit P., Bijur G. N., Zmijewski J. W., Song L., Zmijewska A., Chen X., Johnson G. V., Jope R. S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 7951–7955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kow Y. W. (2002) Free Radic. Biol. Med. 33, 886–893 [DOI] [PubMed] [Google Scholar]
- 44.Andreka P., Zang J., Dougherty C., Slepak T. I., Webster K. A., Bishopric N. H. (2001) Circ. Res. 88, 305–312 [DOI] [PubMed] [Google Scholar]
- 45.Gjerset R. A., Lebedeva S., Haghighi A., Turla S. T., Mercola D. (1999) Cell Growth Differ. 10, 545–554 [PubMed] [Google Scholar]
- 46.Hayakawa J., Depatie C., Ohmichi M., Mercola D. (2003) J. Biol. Chem. 278, 20582–20592 [DOI] [PubMed] [Google Scholar]
- 47.Potapova O., Basu S., Mercola D., Holbrook N. J. (2001) J. Biol. Chem. 276, 28546–28553 [DOI] [PubMed] [Google Scholar]
- 48.Potapova O., Haghighi A., Bost F., Liu C., Birrer M. J., Gjerset R., Mercola D. (1997) J. Biol. Chem. 272, 14041–14044 [DOI] [PubMed] [Google Scholar]
- 49.Ammendrup A., Maillard A., Nielsen K., Aabenhus Andersen N., Serup P., Dragsbaek Madsen O., Mandrup-Poulsen T., Bonny C. (2000) Diabetes 49, 1468–1476 [DOI] [PubMed] [Google Scholar]
- 50.Ventura J. J., Hübner A., Zhang C., Flavell R. A., Shokat K. M., Davis R. J. (2006) Mol. Cell 21, 701–710 [DOI] [PubMed] [Google Scholar]
- 51.Jin S., Fan F., Fan W., Zhao H., Tong T., Blanck P., Alomo I., Rajasekaran B., Zhan Q. (2001) Oncogene 20, 2683–2690 [DOI] [PubMed] [Google Scholar]
- 52.Maekawa T., Sano Y., Shinagawa T., Rahman Z., Sakuma T., Nomura S., Licht J. D., Ishii S. (2008) Oncogene 27, 1045–1054 [DOI] [PubMed] [Google Scholar]