Abstract
TDP-43 is a nuclear protein involved in exon skipping and alternative splicing. Recently, TDP-43 has been identified as the pathological signature protein in frontotemporal lobar degeneration with ubiquitin-positive inclusions and in amyotrophic lateral sclerosis. In addition, TDP-43-positive inclusions are present in Parkinson disease, dementia with Lewy bodies, and 30% of Alzheimer disease cases. Pathological TDP-43 is redistributed from the nucleus to the cytoplasm, where it accumulates. An ∼25-kDa C-terminal fragment of TDP-43 accumulates in affected brain regions, suggesting that it may be involved in the disease pathogenesis. Here, we show that overexpression of the 25-kDa C-terminal fragment is sufficient to cause the mislocalization and cytoplasmic accumulation of endogenous full-length TDP-43 in two different cell lines, thus recapitulating a key biochemical characteristic of TDP-43 proteinopathies. We also found that TDP-43 mislocalization is associated with a reduction in the low molecular mass neurofilament mRNA levels. Notably, we show that the autophagic system plays a role in TDP-43 metabolism. Specifically, we found that autophagy inhibition increases the accumulation of the C-terminal fragments of TDP-43, whereas inhibition of mTOR, a key protein kinase involved in autophagy regulation, reduces the 25-kDa C-terminal fragment accumulation and restores TDP-43 localization. Our results suggest that autophagy induction may be a valid therapeutic target for TDP-43 proteinopathies.
TDP-43 (transactive response DNA-binding protein 43) is a conserved and ubiquitously expressed nuclear protein with a theoretical molecular mass of ∼44 kDa. It is encoded by the TARDBP gene on chromosome 1, which is made of six exons that can be alternatively spliced to yield 11 different isoforms, with the mRNA encoding TDP-43 being the major species (1). Functionally, TDP-43 appears to be involved in exon skipping and alternative splicing (2, 3), and it has also been shown to link different types of nuclear bodies (4). Structural studies have confirmed the presence of two RNA recognition motifs (RRM1 and RRM2) and a glycine-rich C-terminal tail, which is thought to mediate protein-protein interaction (5).
Recently, TDP-43 has been shown to be the major pathological protein in a wide range of disorders referred to as TDP-43 proteinopathies (6–8). These include frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U),2 motor neuron disease, and amyotrophic lateral sclerosis (ALS). These last two disorders have been directly linked to mutations in TDP-43 (9, 10). In addition, TDP-43-positive inclusions are present in Parkinson disease, dementia with Lewy bodies, and 30% of Alzheimer disease cases (11–14). Sporadic and familial forms of FTLD-U and ALS are characterized by cytoplasmic accumulation of insoluble, hyperphosphorylated, ubiquitinated, and proteolytically cleaved C-terminal fragments in affected brain and spinal cord regions. The cytoplasmic accumulation of TDP-43 is associated with a depletion of nuclear TDP-43 (8, 15–21). These data suggest that some of these TDP-43 proteinopathies may share common mechanisms of pathogenesis.
FTLD-U is caused by loss-of-function mutations in the progranulin gene, which lead, by an unknown mechanism, to the accumulation of cytoplasmic TDP-43 inclusions (22, 23). Notably, the TDP-43 inclusions in the ALS and FTLD-U brains are enriched with TDP-43 C-terminal fragments (8, 19). It has been suggested that the C-terminal fragments can be obtained by caspase-dependent cleavage of the full-length protein (24). However, it remains to be established if these fragments play a role in the disease pathogenesis.
TDP-43 proteinopathies are characterized by the accumulation of abnormally modified TDP-43, suggesting that dysfunction in the intracellular quality control systems (ubiquitin-proteasome system and the autophagy-lysosome system) may be involved in the disease pathogenesis. The autophagic system is a conserved intracellular system designed for the degradation of long-lived proteins and organelles in lysosomes (25, 26). Three types of autophagy have been described: macroautophagy, microautophagy, and chaperon-mediated autophagy. Whereas macroautophagy and microautophagy involve the “in bulk” degradation of regions of the cytosol (27, 28), chaperon-mediated autophagy is a more selective pathway, and only proteins with a lysosomal targeting sequence are degraded (29). Cumulative evidence has suggested that an age-dependent decrease in the autophagy-lysosome system may account for the accumulation of abnormal proteins during aging (30, 31).
Macroautophagy is induced when an isolation membrane is formed surrounding cytosolic components, forming an autophagic vacuole, which will eventually fuse with lysosomes for protein/organelle degradation. Induction of the isolation membrane is negatively regulated by mTOR (mammalian target of rapamycin) (32). It has been shown that increasing autophagy activation by mTOR inhibitors has beneficial effects in neurodegeneration (33–35).
EXPERIMENTAL PROCEDURES
Cell Culture
N2A and SH-SY5Y cells were purchased from the American Type Culture Collection. Cell culture methods were adapted from Ref. 36. Briefly, cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum at 37 °C with a humidified environment (5% CO2 + 95% atmosphere). For Western blot analysis, cells were grown on 6-well plates to 75% confluency before transfection or drug administration. For immunohistochemistry, cells were grown on slides. Transfection was done using BioT (Bioland Scientific) following the manufacturer's instructions. As specified under “Results,” 24 h after transfections, rapamycin (0.5 μg/ml) was added to the medium for 24 h.
Western Blots
To extract proteins, cells were centrifuged at 3000 × g for 4 min and washed twice with PBS. Cells were subsequently incubated in ice for 20 min in M-PER (Pierce) with protease and phosphatase inhibitors. After incubation, the lysate was centrifuged at 4 °C for 30 min at maximum speed, and the supernatant was removed and stored for subsequent analysis. Proteins were resolved by SDS-PAGE (10% BisTris from Invitrogen) under reducing conditions and transferred to nitrocellulose membranes, which were then incubated in a 5% solution of not-fat milk for 1 h at 20 °C. After overnight incubation at 4 °C with the appropriate primary antibody, the blots were washed with Tween/Tris-buffered saline for 20 min and incubated at 20 °C with secondary antibody. Blots were subsequently washed with Tween/Tris-buffered saline for 20 min and incubated for 5 min with SuperSignal (Pierce).
Immunofluorescence
For fluorescent labeling, cells were first washed with PBS and then fixed with 4% paraformaldehyde for 20 min at room temperature. Cells were washed three times with PBS and treated with antigen retrieval buffer (100 mm Tris and 5% urea, pH 9.5) for 10 min at 95 °C. After being washed three times with PBS, cells were incubated for 15 min at room temperature in 0.1% Triton X-100, followed by a 1-h incubation at room temperature in 10% goat serum. Cells were then placed in the appropriate primary antibody diluted in blocking solution overnight at 4 °C. Following three rinses with PBS, cells were placed in the appropriate secondary antibodies conjugated to Alexa 488, 555, or 619 fluorophores (Molecular Probes). Following a 1-h incubation, cells were rinsed three times and coverslipped using Fluoromount-G (SouthernBiotech).
Cloning
Full-length TDP-43 (FL-TDP) in pcDNA3.1 plasmid was a gift from Dr. Michael J. Strong (University of Western Ontario, London, Ontario, Canada). To clone the sequence encoding the last 199 amino acids of TDP-43 (C199-TDP) under the control of the cytomegalovirus promoter, we used a homologous recombination-based approach (BD Biosciences InFusion system). C199-TDP was amplified by PCR using a proof-checking DNA polymerase with the following primers: forward, 5′-GTTTAAACTTAAGCTTCCACCATGGATGTGATGGATGTCTTC-3′; and reverse, 5′-GCCCTCTAGACTCGAGCTAGTGATTCATTCCCCAGCCAG-3′. Both primers included 16 bp of homology to the site of insertion within the pcDNA3.1 multiple cloning site. In addition, the forward primers included a Kozak consensus site (CCACC) and the ATG start codon. To elicit recombination between the linearized pcDNA3.1 plasmid and the C199-TDP fragment, we combined 100 ng of the linearized pcDNA3.1 plasmid and 50 ng of the C199-TDP fragment with 10 μl of water; the mixture was added to the InFusion Dry-Down mixture (BD Biosciences) and incubated at room temperature for 30 min. The InFusion mixture contains a proprietary recombinase enzyme that catalyzes the specific recombination of two double-stranded homologous regions of DNA.
Metabolic Labeling and Immunoprecipitation
Pulse-chase labeling experiments with [35S]methionine were performed as described (37). Briefly, N2A cells stably transfected with FL-TDP or C199-TDP were plated on 6-well plates (100,000 cells/well), incubated for 1–2 days, and washed with methionine-free Dulbecco's modified Eagle's medium. Cells were subsequently pulse-labeled with [35S]methionine (0.1 mCi/well) at 37 °C for 2 h. Thereafter, cells were washed and subjected to 3-, 6-, and 24-h chases in complete medium with or without 3-methyladenine (3-MA) or rapamycin. TDP-43 was immunoprecipitated from cell lysates using an anti-TDP-43 polyclonal antibody (Proteintech Group).
Real-time PCR
Total RNA was extracted using an RT2 qPCR-grade RNA isolation kit (SABiosciences) according to the manufacturer's protocol and quantified using a BioSpec-1601 spectrophotometer (Shimadzu). 1 μg of total RNA was used to make the cDNA by using an RT2 First Strand kit (SABiosciences) in a total volume of 20 μl according to the manufacturer's protocol. After reverse transcription-PCR, 1 μl of cDNA was amplified and quantified in 96-well plates using an ABI Prism 7000 sequence detection system (Applied Biosystems, Foster City, CA). For PCR, the amplification conditions consisted of an initial activation step at 95 °C for 10 min and 40 cycles at 95 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s. The fluorescence of the double-stranded products accumulated was monitored in real time. The relative mRNA levels were normalized to the glyceraldehyde-3-phosphate dehydrogenase reference housekeeping gene. For the low molecular mass neurofilament (NFL) gene, the forward primer was 5′-GAGTGGCTTTCGGCTTGCT-3′, and the reverse primer was 5′-CACATTGCCGTAGATCCTGAACT-3′. Glyceraldehyde-3-phosphate dehydrogenase was amplified using primers from SABiosciences.
Statistical Analyses
Statistical analyses were conducted using multifactor analysis of variance including appropriate variables or t test when suitable. Results were considered significant when p < 0.05.
RESULTS
TDP-43 Is Degraded by the Autophagic System
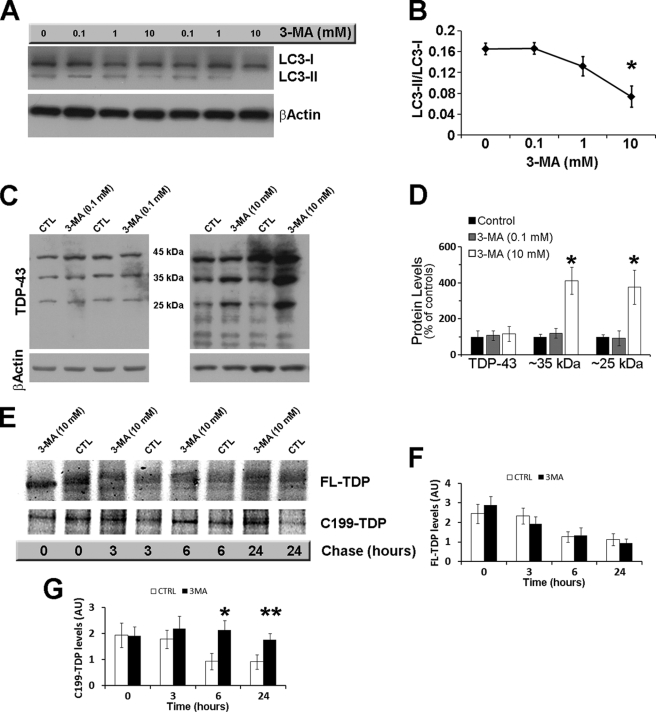
The accumulation of abnormal TDP-43 in ALS and FTLD-U patients suggests that alterations of the protein quality control mechanisms may be involved in the disease pathogenesis. Autophagy is one of the major cellular mechanisms involved in protein turnover and degradation. To determine the role of autophagy in TDP-43 turnover, autophagy induction was inhibited in N2A and SH-SY5Y cells with 3-MA. To find the optimal dose of 3-MA, we initially treated cells for 24 h with 0.1, 1, 10, and 100 mm 3-MA or vehicle only. The effects on autophagy were determined by measuring the levels of LC3-I and LC3-II: the latter is formed by a post-translational modification of LC3-I and is associated mainly with autophagosomal membranes, serving as a good indicator of autophagosome generation (38). Although 100 mm 3-MA was toxic (data not shown), we found that a stronger reduction in autophagy induction was obtained with 10 mm 3-MA (Fig. 1, A and B). In contrast, no changes in autophagy induction were detected in cells treated with 0.1 mm 3-MA (Fig. 1, A and B). These data are consistent with previous reports (39).
FIGURE 1.
The autophagic system is involved in TDP-43 metabolism. Cells transfected with FL-TDP were treated with different concentrations of the autophagy inhibitor 3-MA or vehicle. A, representative Western blot showing the effects of 3-MA on the autophagy marker LC3. β-Actin was used as a loading control. B, quantitative assessment of the LC3-II/LC3-I ratio, which is a reflection of autophagy induction, shows a dose-dependent decline with a maximum and significant decrease at 10 mm (p = 0.0096). C, representative Western blots probed with anti-TDP-43 monoclonal antibody 2E2-D3. β-Actin was used as loading control. Note the increase in the steady-state levels of low molecular mass TDP-43 fragments after the administration of 10 mm 3-MA (right panel) that was not evident after administration of 0.1 mm 3-MA (left panel). CTL, control. D, quantitative assessment of the percentage changes in FL-TDP and the ∼35- and ∼25-kDa C-terminal fragments after administration of 0.1 and 10 mm 3-MA. *, p < 0.001. E, representative Western blot after pulse-chase experiments. After being pulse-labeled for 2 h, cells were chased for the indicated time points. F and G, quantitative assessment of the FL-TDP and C199-TDP levels after the pulse-chase experiments indicates that the FL-TDP levels decreased similarly in the control and 3-MA-treated cells (p > 0.05). In contrast, the C199-TDP levels were significantly higher in 3-MA-treated cells compared with control cells at 6 and 24 h. *, p < 0.01; **, p < 0.05. Data were analyzed using one-way analysis of variance, followed by the post hoc Bonferroni test to determine individual differences in groups. AU, arbitrary units.
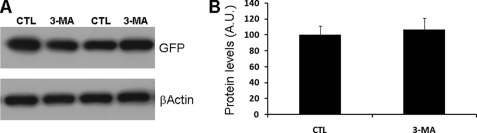
To determine the effects of autophagy inhibition on TDP-43 metabolism, N2A and SH-SY5Y cells cotransfected for 24 h with FL-TDP and green fluorescent protein (GFP) were subsequently treated with 10 mm 3-MA for an additional 24 h. As controls, cells were also treated with 0.1 mm 3-MA. (At this concentration 3-MA did not induce autophagy, as shown in Fig. 1 (A and B).) Although the steady-state levels of FL-TDP were not altered by 3-MA, we found that the ∼35- and ∼25-kDa C-terminal fragments of TDP-43 were significantly higher in cells treated with 10 mm 3-MA compared with controls (Fig. 1, C and D). Significantly, 0.1 mm 3-MA, which had no effect on autophagy, did not alter TDP-43 metabolism (Fig. 1, C and D). Similar results were obtained with two different anti-TDP-43 antibodies (supplemental Fig. S1). Notably, the ∼35- and ∼25-kDa C-terminal fragments of TDP-43 selectively accumulate in affected brain regions in FTLD-U and ALS patients (8, 19). To better understand the effects of 3-MA on TDP-43 metabolism, N2A cells stably transfected with FL-TDP were pulse-labeled with [35S]methionine for 2 h and chased in excess amounts of unlabeled methionine with or without 10 mm 3-MA. Subsequently, the cell lysate was immunoprecipitated with a polyclonal antibody raised against TDP-43. Although 3-MA had no effects on FL-TDP-43, these experiments showed that 3-MA significantly decreased the turnover of the ∼25-kDa C-terminal fragment of TDP-43 (Fig. 1, E–G). Taken together, these data suggest that, under the conditions used here, autophagy plays a primary role in TDP-43 metabolism. GFP levels were unchanged after 10 mm 3-MA administration, suggesting that blocking autophagy selectively increased TDP-43 levels (Fig. 2).
FIGURE 2.
GFP levels are not altered by 3-MA treatment. Proteins extracted from cells double-transfected with FL-TDP and GFP were treated with 3-MA or vehicle as described under “Experimental Procedures.” A, representative Western blot probed with anti-GFP antibody. β-Actin was used as a loading control. B, quantitative assessment of the steady-state levels of GFP indicated. Note that the 3-MA treatment did not significantly affect GFP levels as determined by t test analysis (p > 0.05). CTL, control; A.U., arbitrary units.
Overexpression of C199-TDP Leads to Cytoplasmic TDP-43 Accumulation
A recent work by Petrucelli and co-workers (24) showed that progranulin mediates the proteolytic cleavage of TDP-43 to generate ∼35- and ∼25-kDa species, a process mediated by caspase activation. Notably, the ∼25-kDa fragment has been consistently isolated from affected brain regions of FTLD-U patients, indicating that it may be involved in the disease pathogenesis.
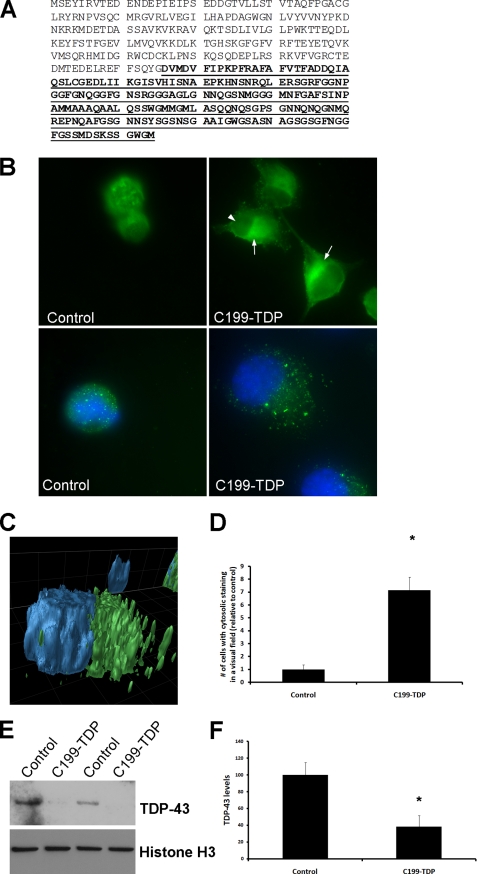
We therefore sought to determine the role of the ∼25-kDa C-terminal fragment (encoded by the last 199 amino acids of TDP-43, referred to as C199-TDP) (Fig. 3A) in TDP-43 aggregation. Using a homologous recombination-based approach, we cloned the sequence encoding the last 199 amino acids of TDP-43 under the control of the cytomegalovirus promoter. An ATG start codon and a Kozak sequence were also added at the 5′-end of the sequence (data not shown). We used this construct to transfect N2A and SH-SY5Y cells. Twenty-four hours after transfection, cells were stained with anti-TDP-43 monoclonal antibody 2E2-D3. We found that overexpression of C199-TDP led to an increase in cytoplasmic TDP-43 immunoreactivity, which was associated with a reduction in nuclear TDP-43 staining (Fig. 3, B–D). These data are consistent with the results showing that accumulation of cytoplasmic TDP-43, after deletion of its nuclear localization signals, leads to a redistribution of endogenous TDP-43 from the nucleus to the cytoplasm (40).
FIGURE 3.
C199-TDP is sufficient to induce nuclear TDP-43 depletion and cytoplasmic TDP-43 accumulation. A, amino acid sequence of TDP-43 (NCBI accession number NP_031401). The putative 199-amino acid sequence of the ∼25-kDa fragment is underlined and in boldface. Notably, this fragment has been isolated from FTLD-U and ALS brains. B, representative microphotographs of cells transfected with C199-TDP or vector alone. The arrows point to cytoplasmic TDP-43-positive accumulation. Note the reduction in nuclear staining (arrowhead), which is also evident by reduced TDP-43 localization with 4′,6-diamidino-2-phenylindole. C, three-dimensional reconstruction of a z-stack showing that the TDP-43 immunoreactivity (green) is predominately outside the nucleus (blue). D, quantitative assessment of the number of cells with cytosolic TDP-43 staining after C199-TDP expression. *, p < 0.01 as determined by t test analysis. E, representative Western blot of the nuclear fraction of cells transfected with C199-TDP, further confirming the decrease in endogenous TDP-43 levels in the nucleus. F, quantitative assessment of the Western blots shows an ∼60% decrease in nuclear steady-state levels of TDP-43 after C199-TDP transfection. *, p < 0.01 as determined by t test analysis.
Notably, we found that the accumulation of cytoplasmic TDP-43 was associated with a decrease in endogenous nuclear TDP-43 immunoreactivity (Fig. 3B, arrowhead). To better quantify these changes, we directly measured TDP-43 levels in nuclear fractions of C199-TDP-transfected and control cells. Although the cytoplasmic levels of TDP-43 did not change after expression of C199-TDP (data not shown), we found an ∼60% decrease in nuclear TDP-43 steady-state levels (Fig. 3, E and F). These data suggest that the overexpression of C199-TDP is sufficient to cause cytoplasmic TDP-43 accumulation, which leads to a redistribution of endogenous TDP-43 from the nucleus to the cytoplasm. This is directly relevant to FTLD-U and ALS, as it has been shown that, in these diseases, there is a significant reduction in the levels of nuclear TDP-43 (8).
Rapamycin Reduces TDP-43 Mislocalization
Taken together, the data presented here provide compelling evidence for a role of autophagy in the turnover of C-terminal fragments of TDP-43, which are sufficient to initiate TDP-43 aggregation. We next sought to determine the effect of increasing autophagy on TDP-43 aggregation.
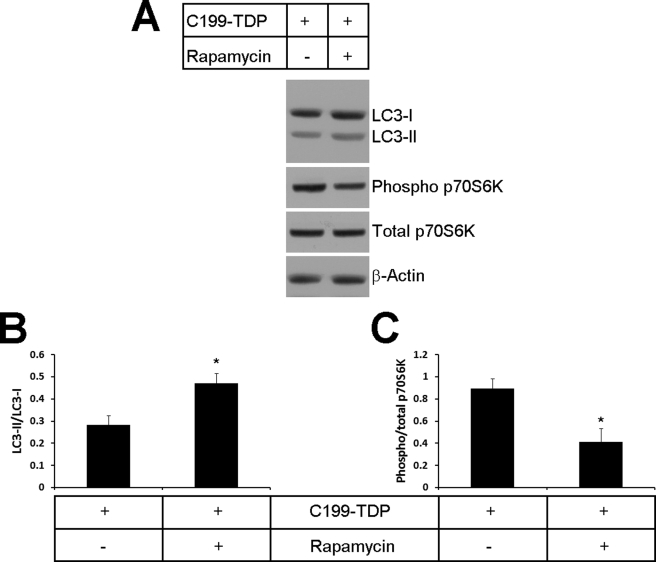
Twenty-four hours after being transfected with C199-TDP, cells were treated for 24 h with rapamycin (0.5 μg/ml). Rapamycin is a selective inhibitor of mTOR, a negative regulator of autophagy. We found that, after 24 h of rapamycin treatment, the LC3-II/LC3-I ratio was significantly increased in the rapamycin-treated cells compared with control cells, indicating an increase in autophagosomal formation (Fig. 4, A and B). Phospho-p70S6K is a mitogen-activated Ser/Thr protein kinase whose activity is directly controlled by mTOR (41). Indeed, mTOR-mediated phosphorylation of p70S6K at Thr389 closely correlates with p70S6K activity in vivo (42), and it is normally used to infer mTOR activity (43). To determine whether the increase in autophagy after rapamycin treatment was mediated by mTOR, we measured the levels of phospho-p70S6K and total p70S6K and found that the ratio of phospho-p70S6K to total p70S6K was significantly reduced in cells treated with rapamycin, indicating a decrease in mTOR activity (Fig. 4, A and C). Taken together, these data show that rapamycin administration increases autophagy via an mTOR-mediated mechanism.
FIGURE 4.
Rapamycin administration increases autophagy by decreasing mTOR activity. After transfection with C199-TDP, cells were treated with rapamycin or vehicle as described under “Experimental Procedures.” A, representative Western blots probed with LC3 (to measure autophagy induction) and phospho-p70S6K and total p70S6K (to measure mTOR activity). β-Actin was used as a loading control. B, quantitative assessment of the Western blots shows that, after rapamycin administration, there was a significant increase in the LC3-II/LC3-I ratio, which indicates an increase in autophagy induction. p = 0.0250 as determined by t test analysis. C, quantitative assessment of the Western blots shows that, after rapamycin administration, there was a significant decrease in p70S6K, which indicates a decrease in mTOR activity. p = 0.0343 as determined by t test analysis.
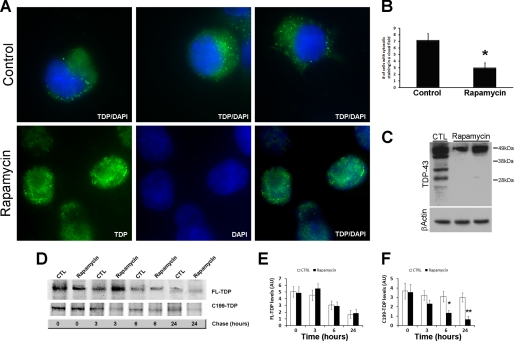
We next determined the effect of the rapamycin-mediated increase in autophagy on TDP-43 mislocalization. Cells transfected with C199-TDP and treated with rapamycin or vehicle were stained with a TDP-43-specific antibody. We found a significant decrease in the number of cells with prominent cytoplasmic TDP immunoreactivity (Fig. 5, A and B). The reduction in cytosolic TDP-43 immunoreactivity was associated with a reduction in C199-TDP steady-state levels as measured by Western blotting (Fig. 5C). Notably, after rapamycin administration, we could not detect TDP-43 low molecular mass species (Fig. 5C).
FIGURE 5.
Rapamycin reduces cytosolic TDP-43 immunoreactivity. Cells transfected with C199-TDP were treated with rapamycin or vehicle as described under “Experimental Procedures.” A, representative microphotographs of cells stained with 2E2-D3. Remarkably, rapamycin administration significantly reduced TDP-43 mislocalization as denoted by the increased TDP-43 immunoreactivity (green) in the nucleus (blue). B, quantitative assessment of the number of cells showing cytosolic TDP-43 immunoreactivity after C199-TDP expression. Rapamycin reduced the number of cells showing cytosolic TDP-43 immunoreactivity by ∼60%. p = 0.0016 as determined by t test analysis. C, representative Western blot probed with anti-TDP-43 monoclonal antibody 2E2-D3 showing a significant decrease in the steady-state levels of the C-terminal fragments of TDP-43. D, representative Western blot after pulse-chase experiments. After being pulse-labeled for 2 h, cells were chased for the indicated time points. E and F, quantitative assessment of the FL-TDP and C199-TDP levels after the pulse-chase experiments indicates that the FL-TDP levels decreased similarly in the control (CTL) and rapamycin-treated cells (p > 0.05). In contrast, C199-TDP levels were significantly lower in rapamycin-treated cells compared with control cells at 6 and 24 h, indicating that rapamycin facilitates the turnover of C199-TDP. *, p < 0.01; **, p < 0.001 as determined using one-way analysis of variance, followed by the post hoc Bonferroni test to determine individual differences in groups. DAPI, 4′,6-diamidino-2-phenylindole; AU, arbitrary units.
To better understand the effects of rapamycin on TDP-43 metabolism, N2A cells stably transfected with C199-TDP were pulse-labeled with [35S]methionine for 2 h and chased in excess amounts of unlabeled methionine with or without rapamycin. Subsequently, the cell lysate was immunoprecipitated with a polyclonal antibody raised against TDP-43. Although rapamycin had no effects on FL-TDP, these experiments showed that rapamycin significantly facilitated the turnover of the ∼25-kDa C-terminal fragment of TDP-43 (Fig. 5, D and F). Taken together, these results show the involvement of the autophagic system in TDP-43 metabolism and suggest a potential beneficial effect for rapamycin.
Rapamycin Administration Increases NFL mRNA Levels
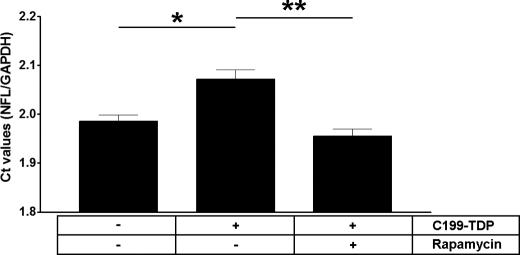
Although the physiological functions of TDP-43 are not completely understood, its structure strongly suggests that it may participate in mRNA processing. Toward this end, it has been shown that TDP-43 interacts with and stabilizes the NFL mRNA (44). To determine the functional consequences of TDP-43 mislocalization induced by C199-TDP, we measured the levels of NFL mRNA in cells stably transfected with C199-TDP using real-time PCR. We found that C199-TDP overexpression led to a significant decrease in the levels of NFL mRNA as reflected by the increase in Ct values (Fig. 6). Notably, rapamycin administration rescued the C199-TDP-induced deficit in NFL mRNA (Fig. 6). Although we could not determine whether the reduction in NFL mRNA levels was due to nuclear depletion of TDP-43 or to its cytoplasmic accumulation, our data clearly indicate that C199-TDP is sufficient to impair this specific function of TDP-43.
FIGURE 6.
Rapamycin administration increases NFL mRNA levels. The mRNA levels of NFL were measured using real-time PCR in cells stably transfected with C199-TDP and treated with rapamycin or vehicle only. The NFL mRNA levels were compared with control cells. We found that expression of C199-TDP was sufficient to reduce the levels of NFL mRNA as indicated by higher Ct values. Notably, rapamycin administration rescued the C199-TDP-induced changes in mRNA levels. *, p < 0.01; **, p < 0.001 as determined using one-way analysis of variance, followed by the post hoc Bonferroni test to determine individual differences in groups. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
The presence of C-terminal TDP-43 fragments in selected brain regions of FTLD-U and ALS patients (8, 19) suggests the possibility that these fragments may be involved in TDP-43 pathogenesis. It has been suggested that a caspase-dependent process may be responsible for the cleavage of TDP-43 and the generation of the 42-, 35-, and 25-kDa C-terminal fragments (24). Here, we have shown that overexpression of the 25-kDa fragment (C199-TDP) is sufficient to induce the cytoplasmic accumulation of TDP-43 and its depletion from the nucleus, which is consistent with a previous report (45). Although it remains to be determined whether the C-terminal fragments are necessary for TDP-43 accumulation, the results presented here highlight the importance of these low molecular mass fragments of TDP-43 in the disease pathogenesis.
The accumulation of abnormally modified TDP-43 in FTLD-U and ALS patients suggests that dysfunction in the intracellular quality control systems may be involved in the disease pathogenesis. Indeed, recent data suggest that ubiquilin, a ubiquitin-like protein involved in other proteinopathies (46–50), binds to TDP-43, forming ubiquilin-TDP-43 aggregates that resemble autophagosomes and colocalize with the autophagosome marker LC3 (51). Consistent with these data, we have shown here for the first time that the autophagic system is involved in TDP-43 metabolism. Both FTLD-U and ALS are age-dependent disorders, and even the familial forms manifest in an age-dependent manner. This indicates that one or more age-related changes in brain function may act as a co-trigger and induce TDP-43 accumulation. It is tempting to speculate that the age-dependent decline in autophagy function may contribute to TDP-43 accumulation, as it may alter the balance between production and degradation of TDP-43 and its C-terminal fragments.
Another important finding reported here, which may have profound clinical implication, is that increasing autophagy with rapamycin reduces the cytoplasmic TDP-43 accumulation. Rapamycin has already been shown to be effective in reducing toxicity of polyglutamine expansions in fly and mouse models of Huntington disease (34) and to increase life span in a variety of animal models, including Saccharomyces cerevisiae, Caenorhabditis elegans, and Drosophila melanogaster (52–54) and mice.3 Moreover, rapamycin is already clinically used in transplant medicine (55, 56), and a recent clinical trial showed that it may be useful for the treatment of certain types of cancer (57). If the effects of rapamycin on TDP-43 accumulation are confirmed in vivo (when an animal model of TDP-43 proteinopathies will be available), there will be strong supporting data for the use of rapamycin in a clinical trial.
Supplementary Material
Acknowledgment
We thank Dr. Michael J. Strong for the TDP-43 plasmid.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
R. Strong, unpublished data.
- FTLD-U
- frontotemporal lobar degeneration with ubiquitin-positive inclusions
- ALS
- amyotrophic lateral sclerosis
- PBS
- phosphate-buffered saline
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- FL-TDP
- full-length TDP-43
- 3-MA
- 3-methyladenine
- NFL
- low molecular mass neurofilament
- GFP
- green fluorescent protein.
REFERENCES
- 1.Wang H. Y., Wang I. F., Bose J., Shen C. K. (2004) Genomics 83, 130–139 [DOI] [PubMed] [Google Scholar]
- 2.Buratti E., Baralle F. E. (2001) J. Biol. Chem. 276, 36337–36343 [DOI] [PubMed] [Google Scholar]
- 3.Buratti E., Dörk T., Zuccato E., Pagani F., Romano M., Baralle F. E. (2001) EMBO J. 20, 1774–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang I. F., Reddy N. M., Shen C. K. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13583–13588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buratti E., Baralle F. E. (2008) Front. Biosci. 13, 867–878 [DOI] [PubMed] [Google Scholar]
- 6.Kwong L. K., Neumann M., Sampathu D. M., Lee V. M., Trojanowski J. Q. (2007) Acta Neuropathol. 114, 63–70 [DOI] [PubMed] [Google Scholar]
- 7.Neumann M., Kwong L. K., Sampathu D. M., Trojanowski J. Q., Lee V. M. (2007) Arch. Neurol. 64, 1388–1394 [DOI] [PubMed] [Google Scholar]
- 8.Neumann M., Sampathu D. M., Kwong L. K., Truax A. C., Micsenyi M. C., Chou T. T., Bruce J., Schuck T., Grossman M., Clark C. M., McCluskey L. F., Miller B. L., Masliah E., Mackenzie I. R., Feldman H., Feiden W., Kretzschmar H. A., Trojanowski J. Q., Lee V. M. (2006) Science 314, 130–133 [DOI] [PubMed] [Google Scholar]
- 9.Gitcho M. A., Baloh R. H., Chakraverty S., Mayo K., Norton J. B., Levitch D., Hatanpaa K. J., White C. L., 3rd, Bigio E. H., Caselli R., Baker M., Al-Lozi M. T., Morris J. C., Pestronk A., Rademakers R., Goate A. M., Cairns N. J. (2008) Ann. Neurol. 63, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutherford N. J., Zhang Y. J., Baker M., Gass J. M., Finch N. A., Xu Y. F., Stewart H., Kelley B. J., Kuntz K., Crook R. J., Sreedharan J., Vance C., Sorenson E., Lippa C., Bigio E. H., Geschwind D. H., Knopman D. S., Mitsumoto H., Petersen R. C., Cashman N. R., Hutton M., Shaw C. E., Boylan K. B., Boeve B., Graff-Radford N. R., Wszolek Z. K., Caselli R. J., Dickson D. W., Mackenzie I. R., Petrucelli L., Rademakers R. (2008) PLoS Genet. 4, e1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amador-Ortiz C., Lin W. L., Ahmed Z., Personett D., Davies P., Duara R., Graff-Radford N. R., Hutton M. L., Dickson D. W. (2007) Ann. Neurol. 61, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bigio E. H. (2008) Acta Neuropathol. 116, 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higashi S., Iseki E., Yamamoto R., Minegishi M., Hino H., Fujisawa K., Togo T., Katsuse O., Uchikado H., Furukawa Y., Kosaka K., Arai H. (2007) Brain Res. 1184, 284–294 [DOI] [PubMed] [Google Scholar]
- 14.Uryu K., Nakashima-Yasuda H., Forman M. S., Kwong L. K., Clark C. M., Grossman M., Miller B. L., Kretzschmar H. A., Lee V. M., Trojanowski J. Q., Neumann M. (2008) J. Neuropathol. Exp. Neurol. 67, 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seelaar H., Schelhaas H. J., Azmani A., Küsters B., Rosso S., Majoor-Krakauer D., de Rijik M. C., Rizzu P., ten Brummelhuis M., van Doorn P. A., Kamphorst W., Willemsen R., van Swieten J. C. (2007) Brain 130, 1375–1385 [DOI] [PubMed] [Google Scholar]
- 16.Neumann M., Mackenzie I. R., Cairns N. J., Boyer P. J., Markesbery W. R., Smith C. D., Taylor J. P., Kretzschmar H. A., Kimonis V. E., Forman M. S. (2007) J. Neuropathol. Exp. Neurol. 66, 152–157 [DOI] [PubMed] [Google Scholar]
- 17.Davidson Y., Kelley T., Mackenzie I. R., Pickering-Brown S., Du Plessis D., Neary D., Snowden J. S., Mann D. M. (2007) Acta Neuropathol. 113, 521–533 [DOI] [PubMed] [Google Scholar]
- 18.Cairns N. J., Bigio E. H., Mackenzie I. R., Neumann M., Lee V. M., Hatanpaa K. J., White C. L., 3rd, Schneider J. A., Grinberg L. T., Halliday G., Duyckaerts C., Lowe J. S., Holm I. E., Tolnay M., Okamoto K., Yokoo H., Murayama S., Woulfe J., Munoz D. G., Dickson D. W., Ince P. G., Trojanowski J. Q., Mann D. M. (2007) Acta Neuropathol. 114, 5–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igaz L. M., Kwong L. K., Xu Y., Truax A. C., Uryu K., Neumann M., Clark C. M., Elman L. B., Miller B. L., Grossman M., McCluskey L. F., Trojanowski J. Q., Lee V. M. (2008) Am. J. Pathol. 173, 182–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F., Morita M., Nakano I., Oda T., Tsuchiya K., Akiyama H. (2008) Ann. Neurol. 64, 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inukai Y., Nonaka T., Arai T., Yoshida M., Hashizume Y., Beach T. G., Buratti E., Baralle F. E., Akiyama H., Hisanaga S., Hasegawa M. (2008) FEBS Lett. 582, 2899–2904 [DOI] [PubMed] [Google Scholar]
- 22.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J. J., van Duijn C., Peeters K., Sciot R., Santens P., De Pooter T., Mattheijssens M., Van den Broeck M., Cuijt I., Vennekens K., De Deyn P. P., Kumar-Singh S., Van Broeckhoven C. (2006) Nature 442, 920–924 [DOI] [PubMed] [Google Scholar]
- 23.Baker M., Mackenzie I. R., Pickering-Brown S. M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A. D., Rollinson S., Cannon A., Dwosh E., Neary D., Melquist S., Richardson A., Dickson D., Berger Z., Eriksen J., Robinson T., Zehr C., Dickey C. A., Crook R., McGowan E., Mann D., Boeve B., Feldman H., Hutton M. (2006) Nature 442, 916–919 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y. J., Xu Y. F., Dickey C. A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L. (2007) J. Neurosci. 27, 10530–10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Vicente M., Cuervo A. M. (2007) Lancet Neurol. 6, 352–361 [DOI] [PubMed] [Google Scholar]
- 26.Cuervo A. M., Bergamini E., Brunk U. T., Dröge W., Ffrench M., Terman A. (2005) Autophagy 1, 131–140 [DOI] [PubMed] [Google Scholar]
- 27.Yorimitsu T., Klionsky D. J. (2005) Cell Death Differ. 12, Suppl. 2, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 29.Dice J. F. (2007) Autophagy 3, 295–299 [DOI] [PubMed] [Google Scholar]
- 30.Ward W. F. (2002) Prog. Mol. Subcell. Biol. 29, 35–42 [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Vicente M., Sovak G., Cuervo A. M. (2005) Exp. Gerontol. 40, 622–633 [DOI] [PubMed] [Google Scholar]
- 32.Díaz-Troya S., Pérez-Pérez M. E., Florencio F. J., Crespo J. L. (2008) Autophagy 4, 851–865 [DOI] [PubMed] [Google Scholar]
- 33.Sarkar S., Ravikumar B., Floto R. A., Rubinsztein D. C. (2009) Cell Death Differ. 16, 46–56 [DOI] [PubMed] [Google Scholar]
- 34.Ravikumar B., Vacher C., Berger Z., Davies J. E., Luo S., Oroz L. G., Scaravilli F., Easton D. F., Duden R., O'Kane C. J., Rubinsztein D. C. (2004) Nat. Genet. 36, 585–595 [DOI] [PubMed] [Google Scholar]
- 35.Ehninger D., Han S., Shilyansky C., Zhou Y., Li W., Kwiatkowski D. J., Ramesh V., Silva A. J. (2008) Nat. Med. 14, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oddo S., Caccamo A., Smith I. F., Green K. N., LaFerla F. M. (2006) Am. J. Pathol. 168, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chomyn A. (1996) Methods Enzymol. 264, 197–211 [DOI] [PubMed] [Google Scholar]
- 38.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cui Q., Tashiro S., Onodera S., Minami M., Ikejima T. (2007) J. Pharmacol. Sci. 105, 317–325 [DOI] [PubMed] [Google Scholar]
- 40.Winton M. J., Igaz L. M., Wong M. M., Kwong L. K., Trojanowski J. Q., Lee V. M. (2008) J. Biol. Chem. 283, 13302–13309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pullen N., Thomas G. (1997) FEBS Lett. 410, 78–82 [DOI] [PubMed] [Google Scholar]
- 42.Weng Q. P., Kozlowski M., Belham C., Zhang A., Comb M. J., Avruch J. (1998) J. Biol. Chem. 273, 16621–16629 [DOI] [PubMed] [Google Scholar]
- 43.Schmelzle T., Hall M. N. (2000) Cell 103, 253–262 [DOI] [PubMed] [Google Scholar]
- 44.Strong M. J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. (2007) Mol. Cell. Neurosci. 35, 320–327 [DOI] [PubMed] [Google Scholar]
- 45.Igaz L. M., Kwong L. K., Chen-Plotkin A., Winton M. J., Unger T. L., Xu Y., Neumann M., Trojanowski J. Q., Lee V. M. (2009) J. Biol. Chem. 284, 8516–8524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arias-Vásquez A., de Lau L., Pardo L., Liu F., Feng B. J., Bertoli-Avella A., Isaacs A., Aulchenko Y., Hofman A., Oostra B., Breteler M., van Duijn C. (2007) Neurosci. Lett. 424, 1–5 [DOI] [PubMed] [Google Scholar]
- 47.Doi H., Mitsui K., Kurosawa M., Machida Y., Kuroiwa Y., Nukina N. (2004) FEBS Lett. 571, 171–176 [DOI] [PubMed] [Google Scholar]
- 48.Lu A., Hiltunen M., Romano D. M., Soininen H., Hyman B. T., Bertram L., Tanzi R. E. (2009) J. Mol. Neurosci. 38, 19–30 [DOI] [PubMed] [Google Scholar]
- 49.Mah A. L., Perry G., Smith M. A., Monteiro M. J. (2000) J. Cell Biol. 151, 847–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H., Lim P. J., Yin C., Rieckher M., Vogel B. E., Monteiro M. J. (2006) Hum. Mol. Genet. 15, 1025–1041 [DOI] [PubMed] [Google Scholar]
- 51.Kim S. H., Shi Y., Hanson K. A., Williams L. M., Sakasai R., Bowler M. J., Tibbetts R. S. (2009) J. Biol. Chem. 284, 8083–8092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia K., Chen D., Riddle D. L. (2004) Development 131, 3897–3906 [DOI] [PubMed] [Google Scholar]
- 53.Kaeberlein M., Powers R. W., 3rd, Steffen K. K., Westman E. A., Hu D., Dang N., Kerr E. O., Kirkland K. T., Fields S., Kennedy B. K. (2005) Science 310, 1193–1196 [DOI] [PubMed] [Google Scholar]
- 54.Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., Benzer S. (2004) Curr. Biol. 14, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Monaco A. P. (2009) Transplantation 87, 157–163 [DOI] [PubMed] [Google Scholar]
- 56.Wali R. K., Weir M. R. (2008) Curr. Opin. Organ Transplant. 13, 614–621 [DOI] [PubMed] [Google Scholar]
- 57.Easton J. B., Houghton P. J. (2006) Oncogene 25, 6436–6446 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.