Abstract
AMP-activated protein kinase (AMPK) is a heterotrimeric complex playing a crucial role in maintaining cellular energy homeostasis. Recently, homodimerization of mammalian AMPK and yeast ortholog SNF1 was shown by us and others. In SNF1, it involved specific hydrophobic residues in the kinase domain αG-helix. Mutation of the corresponding AMPK α-subunit residues (Val-219 and Phe-223) to glutamate reduced the tendency of the kinase to form higher order homo-oligomers, as was determined by the following three independent techniques in vitro: (i) small angle x-ray scattering, (ii) surface plasmon resonance spectroscopy, and (iii) two-dimensional blue native/SDS-PAGE. Recombinant protein as well as AMPK in cell lysates of primary cells revealed distinct complexes of various sizes. In particular, the assembly of very high molecular mass complexes was dependent on both the αG-helix-mediated hydrophobic interactions and kinase activation. In vitro and when overexpressed in double knock-out (α1−/−, α2−/−) mouse embryonic fibroblast cells, activation of mutant AMPK was impaired, indicating a critical role of the αG-helix residues for AMPK activation via its upstream kinases. Also inactivation by protein phosphatase 2Cα was affected in mutant AMPK. Importantly, activation of mutant AMPK by LKB1 was restored by exchanging the corresponding and conserved hydrophobic αG-helix residues of LKB1 (Ile-260 and Phe-264) to positively charged amino acids. These mutations functionally rescued LKB1-dependent activation of mutant AMPK in vitro and in cell culture. Our data suggest a physiological role for the hydrophobic αG-helix residues in homo-oligomerization of heterotrimers and cellular interactions, in particular with upstream kinases, indicating an additional level of AMPK regulation.
The maintenance of energy homeostasis is a basic requirement of all living organisms. The AMP-activated protein kinase (AMPK)2 is crucially involved in this essential process by playing a central role in sensing and regulating energy metabolism on the cellular and whole body level (1–6). AMPK is also participating in several signaling pathways associated with cancer and metabolic diseases, like type 2 diabetes mellitus, obesity, and other metabolic disorders (7–9).
Mammalian AMPK belongs to a highly conserved family of serine/threonine protein kinases with homologs found in all eukaryotic organisms examined (1, 3, 10). Its heterotrimeric structure includes a catalytic α-subunit and regulatory β- and γ-subunits. These subunits exist in different isoforms (α1, α2, β1, β2, γ1, γ2, and γ3) and splice variants (for γ2 and γ3) and can thus assemble to a broad variety of heterotrimeric isoform combinations. The α- and β-subunits possess multiple autophosphorylation sites, which have been implicated in regulation of subcellular localization and kinase activation (11–15). The most critical step of AMPK activation, however, is phosphorylation of Thr-172 within the activation segment of the α-subunit kinase domain. At least two AMPK upstream kinases (AMPKKs) have been identified so far, namely the tumor suppressor kinase LKB1 in complex with MO25 and STRAD (16) and Ca2+/calmodulin-dependent protein kinase kinase-2 (CamKK2) (17). Furthermore, the transforming growth factor-β-activated kinase 1 was also shown to activate AMPK using a variety of in vitro approaches (18), but the physiological relevance of these findings remains unclear. Besides direct phosphorylation of Thr-172, AMPK activity is stimulated by the allosteric activator AMP, which can bind to two Bateman domains formed by two pairs of CBS domains within the γ-subunit (19–22). Hereby bound AMP not only allosterically stimulates AMPK but also protects Thr-172 from dephosphorylation by protein phosphatase 2Cα (PP2Cα) and thus hinders inactivation of the kinase (19, 22, 23). Consequently, on the cellular level, AMPK is activated upon metabolic stress increasing the AMP/ATP ratio. Furthermore, AMPK activation can also be induced by several chemical compounds, like nucleoside 5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside (24) and the anti-diabetic drug Metformin (25–28). In addition, the small molecule compound A-769662 was recently developed as a direct allosteric activator of AMPK (29, 30).
Previous work in our groups proposed a model of AMPK regulation by AMP, which incorporates the major functional features and the latest structural information (31). The latter mainly included truncated core complexes of AMPK from different species (32–35). Further valuable structural information is provided by the x-ray structures of the isolated catalytic domains, in particular of the human AMPK α2-subunit (Protein Data Bank code 2H6D) and its yeast ortholog SNF1 (36, 37). The kinase domain of SNF1 is capable of forming homodimers in the protein crystal, as well as in vitro in solution, in a unique way, which has not been observed previously in any other kinase (36). The dimer interface is predominantly formed by hydrophobic interactions of the loop-αG region, also known as subdomain X situated on the large kinase lobe (36, 38, 39), and it mainly involves Ile-257 and Phe-261. Because the T-loop activation segment was buried within the dimer interface, it was suggested that the dimeric state of the SNF1 catalytic domain represents the inactive form of the kinase. Intriguingly, it was shown in our groups by small angle x-ray scattering that AMPK self-organizes in a concentration-dependent manner to form homo-oligomers in solution (31). However, the interface responsible for oligomerization of the AMPK heterotrimer has remained elusive.
Here we further investigate the distinct oligomeric states of the AMPK heterotrimer and suggest a possible regulatory function for this process. Most importantly, we provide conclusive evidence for participation of αG-helix residues in the recognition of AMPK by its upstream kinases LKB1 and CamKK2.
EXPERIMENTAL PROCEDURES
Plasmids
Bacterial expression plasmids encoding for nontagged AMPK α1β1γ1 (40), hexahistidine-tagged LKB1-MO25α-STRADα (41), GST-tagged CamKK2 comprising residues 84–434 (42), hexahistidine-tagged protein phosphatase 2Cα (PP2Cα) (43), and the mammalian expression constructs encoding for the Myc-tagged AMPK α1 subunit (44) and FLAG-tagged LKB1 (45) have been described (31, 40, 41).
Mutagenesis
Site-directed mutagenesis (QuikChange, Stratagene) was performed to replace both Val-219 and Phe-223 of the AMPKα1 kinase domain with glutamate and both Ile-260 and Phe-264 of the LKB1 kinase domain with lysine. We refer to the corresponding double mutants as VEFE-AMPK and IKFK-LKB1 (as opposed to WT-AMPK and WT-LKB1) for the remainder of this report. Mutations were introduced into tricistronic bacterial expression plasmids encoding for nontagged AMPK α1β1γ1 and hexahistidine-tagged LKB1-MO25α-STRADα, respectively, as well as in mammalian expression plasmids encoding for Myc-tagged AMPK α1 and FLAG-tagged LKB1. Primer sequences are available from the author upon request. All plasmids were verified by sequencing (Microsynth AG, Balgach, Switzerland).
Bacterial Expression and Purification
Heterotrimeric AMPK and LKB1-MO25α-STRADα complexes were expressed in Tuner (DE3) and Rosetta2 (DE3) Escherichia coli cells, respectively (Novagen). Bacteria were grown in 4 liters of auto-induction medium (46) for 2 h at 37 °C and subsequently for 22 h at room temperature with shaking at 220 rpm. Cells were harvested by centrifugation, and proteins were extracted as described recently (31). Heterotrimeric AMPK was purified using either of two strategies both leading to high quality protein. (i) The fully automated four-dimensional purification protocol, as described previously (31, 47), was suitable for the large amounts of highly purified protein needed in SAXS measurements. (ii) The two-dimensional purification protocol (adapted from Ref. 40) was applied for purification of AMPK that was used in all other experiments. Briefly, 1 ml of nickel-Sepharose HP (GE Healthcare) was used in a batch affinity purification procedure at room temperature for 1 h before washing and elution. In both protocols the size exclusion chromatography step was carried out using a Superdex 200 16/60 column (GE Healthcare) equilibrated in gel filtration buffer containing 200 mm NaCl, 50 mm Tris-Cl, and Hepes-Cl, respectively, 4 mm MgCl2, 8 mm EDTA, pH 8.0, at 7 °C. Peak fractions were pooled and concentrated in a Vivaspin 20 centrifugal concentrator with a 10,000 molecular weight cutoff (Sartorius AG, Goettingen, Germany) at 470 × g and 4 °C. Protein stock solutions were supplemented with 2 mm tris(2-carboxyethyl)phosphine hydrochloride and stored either at 4 °C or in 50% glycerol at −20 °C. His-LKB1-MO25α-STRADα wild type and mutant complexes were purified as described recently (41). Extraction and Ni2+-affinity purification of LKB1 complexes was performed analogous to the two-dimensional purification procedure for AMPK. CamKK2 and PP2Cα were expressed and purified as published previously (22, 43).
SAXS
Data collection of the synchrotron SAXS measurements was performed at DESY (Hamburg, Germany) on the X33 camera of the EMBL analogous to the procedure reported earlier (31). WT-AMPK and VEFE-AMPK were purified by the fully automated four-dimensional purification protocol, and scattering was recorded in a concentration range of 0.5 to 12 mg/ml. To check for radiation damage, two successive 2-min exposures were compared, but no radiation effects were observed. The data were reduced and processed, and the overall parameters were computed following standard procedures of the software package PRIMUS (48). The radius of gyration (Rg) was evaluated using the Guinier approximation (49) assuming that at very small angles (s < 1.3/Rg) the intensity is represented by I(s) = I(0) exp(−1/3(sRg)2) and calculated from the entire scattering patterns using the program GNOM (50).
Surface Plasmon Resonance (SPR)
Nontagged WT-AMPK and VEFE-AMPK were purified by the two-dimensional purification protocol. AMPK was dialyzed overnight at 4 °C against 10 mm Hepes-Cl, 100 mm NaCl, 50 μm EDTA, 2 mm dithiothreitol, pH 7.4. Samples were supplemented with 0.005% Surfactant P20 before being injected at concentrations of 0.5 and 5.6 μm. The stability of AMPK oligomers was analyzed by SPR using a BIAcore instrument (GE Healthcare) equipped with an NTA sensor chip that was loaded with Ni2+ according to the manufacturer's instructions. Because of the intrinsic affinity of AMPK to Ni-NTA (31), a kinetically stable immobilization of monomeric AMPK heterotrimers (e.g. at concentrations below 0.5 mg/ml) can be achieved.3 In addition, because AMPK oligomerization is a readily reversible process upon dilution (31), the recorded dissociation kinetics upon injection were a measure of dissociating AMPK-AMPK but not AMPK-chip interactions. WT-AMPK and VEFE-AMPK were injected at 1.2 ml/h and 25 °C directly onto the chip, and association and dissociation curves were recorded. The chip surface was recovered by injection of regeneration buffer (10 mm Hepes-Cl, 150 mm NaCl, 350 mm EDTA, 0,005% Surfactant P20, pH 8.3). Although the association kinetics were similar at a given protein concentration, dissociation showed marked differences. However, the latter could not be fitted by one or two exponential rate equations, indicating a complex interaction. Curves were instead normalized to dissociation start (just subsequent to a bulk refractive index shift, if occurring), and an end dissociation point at 100 s was defined to analyze the relative extent of dissociation. Experiments were done at least in triplicate to calculate average and standard deviation.
Two-dimensional Blue Native (BN)/SDS-PAGE
Samples containing AMPK were separated by gradient BN-PAGE in the first dimension and then denatured and separated again using SDS-PAGE in the second dimension. The AMPK α-subunit was subsequently detected by Western blot analysis. BN-PAGE was conducted similar to published protocols (51, 52), except for the cathode buffer (15 mm BisTris-Cl, 50 mm Bicine, pH 7.0, at 4 °C) and the acrylamide/bisacrylamide mixtures (AB mix). The AB mix consisted either of 33% T, 3% C or 30% T, 2.6% C (53). Gradient gels (4–13%) with a stacking layer (3%) were prepared with a gradient mixer (Bio-Rad) using 4% T and 13% T, 20% glycerol solutions and gel plates with a 1-mm spacer. Molecular mass markers (all from Sigma) were thyroglobulin (669 kDa), apoferritin (443 kDa), and albumin (monomer 66 kDa and dimer 132 kDa). Markers and samples were supplemented with 10% glycerol and Coomassie Brilliant Blue G-250 at a concentration of 0.025%. For comparison of activated with nonactivated recombinant α1β1γ1 AMPK, 10 μg were incubated in kinase buffer (10 mm Hepes-Cl, pH 7.5, 50 mm MgCl2, 1 mm ATP) in the presence or absence of 90 μg/ml recombinant His-LKB1-MO25α-STRADα for 1.5 h at 30 °C. After BN PAGE at 4 °C (200 V, 3 h), gels were stained with Coomassie Blue, and individual lanes were excised. The strips were soaked in 2% SDS and 1% β-mercaptoethanol for 1 h at room temperature and subjected to SDS-PAGE as indicated in the figure legends. In Tricine/SDS-PAGE (54, 55), the BN-PAGE stripes were embedded in a 6% native stacking layer on top of a 6% denaturing stacking layer and an 8% denaturing resolving layer. In Laemmli SDS-PAGE (56), a 5% stacking layer embedding the BN-PAGE strips were layered on top of a 12 or 15% resolving layer. Both methods gave similar results. For comparison of two different samples (e.g. WT-AMPK and VEFE-AMPK), these were separated in parallel in the same first dimension gel (mini gel) and second dimension gel (16.5 × 22-cm glass plates) before being transferred to a single membrane and analyzed by immunodetection.
SDS-PAGE and Western Blotting
Analyses by SDS-PAGE and Western blotting were performed as reported earlier (22). AMPKα (2532) and Thr(P)-172 (2535) antibodies were from Cell Signaling Inc. Signals were quantified by densitometry using the software Quantity One, version 4.6.3.088 (Bio-Rad).
Activation of AMPK by Upstream Kinases and Activity Determination
AMPK (25 μg/ml) was activated in kinase buffer by recombinant His-LKB1-MO25α-STRADα (187.5 μg/ml) or recombinant GST-CamKK2 (15 μg/ml) for 20 min at 30 °C or as indicated in the figure. AMPK activity was probed simultaneously by Western blot analysis of Thr-172 phosphorylation and by determination of kinase activity as described (22). Kinase activity of AMPK from transfected immortalized AMPKα subunit double knock-out (α1−/−, α2−/−) MEFs was determined following immunoprecipitation. Briefly, cells were lysed, and AMPK was immunoprecipitated from 0.5 mg of precleared cell lysate overnight at 4 °C using anti-AMPKα1 antibody (kindly provided by Professor D. G. Hardie, University of Dundee, Dundee, Scotland, UK). The immunoprecipitate was washed twice in lysis buffer and once with lysis buffer lacking Triton X-100. Subsequently, the immunoprecipitated AMPK was incubated with SAMS peptide at 37 °C for 30 min, and kinase activity was determined as described (22).
Cell Culture Methods
Immortalized AMPKα subunit double knock-out (α1−/−, α2−/−) MEFs were described previously (57). Cells were cultured for about 24 h in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 4.5 g/liter glucose and 10% fetal calf serum (BioConcept, Allschwil, Switzerland) to 50–70% confluence prior to transfection. After 12 h of transfection (Metafectene Pro; Biontex, Germany), cells were starved in fetal calf serum-free Dulbecco's modified Eagle's medium for 1 h, followed by AMPK activation for 5 h with 5 mm Metformin (Sigma). Cells were washed once with phosphate-buffered saline and lysed by addition of 50 μl of lysis buffer (20 mm Tris-Cl, pH 7.4, 50 mm NaCl, 5 mm EDTA, 250 mm sucrose, 1% Triton X-100, 50 mm NaF, 5 mm Na4P2O7, Complete Mini EDTA-free protease inhibitor mixture (Roche Applied Science), 1 mm dithiothreitol, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride), and lysates were immediately frozen in liquid nitrogen. After thawing, cell lysis was completed by incubation on ice for 30 min and three pulses of sonication (each 1 min) in a sonication bath. After centrifugation at 17,000 × g for 5 min at 4 °C, the supernatant was analyzed by Western blotting or subjected to immunoprecipitation for kinase activity determination.
Hepatocytes were prepared from 3-month-old male Wistar rats (58) kept in controlled conditions with ad libitum access to food and water. Lysates were prepared from 10 mg (dry weight) of primary hepatocytes dissolved in 500 μl of lysis buffer (50 mm sodium phosphate buffer, pH 8.0, 0.5 m sucrose, 30% glycerol, 250 mm imidazole). Soluble hepatocyte proteins for PAGE were obtained after three freeze/thaw cycles in liquid nitrogen and subsequent centrifugation for 30 min at 20,000 × g and 4 °C.
Human primary keratinocytes were isolated and cultured as described earlier (59). Cells were cultured to 80–90% confluence in K-SFM (Invitrogen) supplemented with epidermal growth factor and bovine pituitary extract and then incubated in K-SFM without supplements for 4 h prior to treatment with 1 mm H2O2.
Dephosphorylation of AMPK at Thr-172
Purified recombinant WT- and VEFE-AMPK were first activated and then repurified and finally subjected to dephosphorylation. GST-CamKK2 was used in excess (protein amount of 1:3 in relation to AMPK) for 80 min at 30 °C to ensure maximal phosphorylation at Thr-172. The GST fusion protein was removed by batch affinity purification using glutathione-Sepharose (GE Healthcare) for 2 h at room temperature, and AMPK was recovered in the supernatant of the resin after centrifugation. For removal of ATP remnants, AMPK was dialyzed at 4 °C against 50 mm Hepes-Cl, pH 8.0, and 200 mm NaCl. Repurified active AMPK (25 μg/ml) was supplemented with 2 mm tris(2-carboxyethyl)phosphine hydrochloride and dephosphorylated by incubation with PP2Cα (30 μg/ml) in 10 mm Hepes-Cl, pH 7.5, 50 mm MgCl2 at 37 °C. Samples were taken at the indicated time points, and the degree of AMPK dephosphorylation at Thr-172 was determined by Western blot analysis using the corresponding Thr(P)-172 antibody.
Statistical Analysis
Results are expressed as mean values ± S.D. Statistical analysis was performed using the analysis of variance data analysis toolbox (OATBRAN project). p values smaller than 0.05 were considered statistically significant. The different levels of significance such as p < 0.01 and p < 0.001 were not distinguished.
RESULTS
Analysis of the AMPK αG-helix
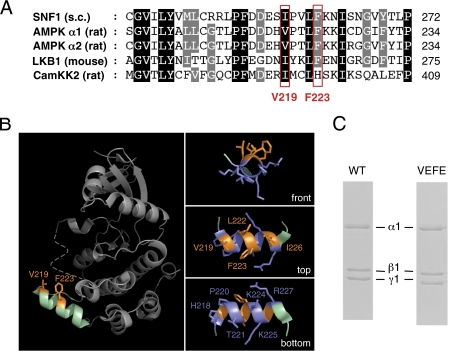
It has been shown previously that the kinase domain of the Saccharomyces cerevisiae SNF1 protein is capable of dimerizing via hydrophobic interactions involving Ile-257 and Phe-261 of the αG-helix region (36). The corresponding phenylalanine residue is conserved in the mammalian AMPK α1- and α2-subunit isoforms (Phe-223), as well as in LKB1 (Phe-264), whereas the isoleucine residue is either conserved or conservatively replaced by valine in α1-/α2-subunits (Val-219), LKB1 (Ile-260), and also CamKK2 (Ile-394) (Fig. 1A). Focused analyses of the αG-helix region, using the structure of the human α2 kinase domain (Protein Data Bank code 2H6D), revealed that both hydrophobic residues are located in the amphipathic αG-helix within a hydrophobic patch and are well surface-exposed and thus accessible for intermolecular interactions (Fig. 1B) (60). To investigate the role of the αG-helix for oligomerization of the heterotrimeric AMPK complex, mutations were introduced in AMPK altering residues analogous to the SNF1 dimerization-deficient mutants (36). Thus, the substitution of Val-219 and Phe-223 by glutamate residues was expected to disrupt homodimerization of AMPK by electrostatic charge repellence. Heterotrimeric, nontagged α1β1γ1 wild type (WT-AMPK) and α1(V219E,F223E)β1γ1 (VEFE-AMPK) mutant AMPK protein complexes were bacterially expressed and purified and both yielding high purity and the expected 1:1:1 stoichiometry (Fig. 1C). In addition, similar to the analysis of WT-AMPK (31), the formation of VEFE-AMPK heterotrimers was confirmed by mass spectrometry using specific cross-linkers.4 Furthermore, both complexes showed similar specific activities when maximally activated (see below), indicating functional integrity of the mutant enzyme. Such recombinant AMPK complexes were used for the in vitro experiments throughout this study.
FIGURE 1.
Analysis of the amphipathic αG-helix of the AMPK kinase domain. A, alignment of the αG-helix region sequences of Snf1 from S. cerevisiae, AMPK α1 and α2 subunits and CamKK2 from Rattus norvegicus, and LKB1 from Mus musculus (GenBankTM accession numbers P06782, AAC52355, CAA82620, NP_112628, and NP_035622). Residues highlighted in black show conservation in all sequences. Residues highlighted in gray show partial conservation. Conserved hydrophobic residues corresponding to AMPK α1 and α2 residues Val-219 and Phe-223 are boxed in red. B, focused analysis of the AMPK α2-subunit kinase domain (Protein Data Bank code 2H6D). Left panel, surface-exposed location of αG-helix (green) Right panels, front, top, and bottom view of the αG-helix. Hydrophobic residues are highlighted in orange (hydropathy index >2.5), and rather hydrophilic residues are highlighted in blue (hydropathy index < −0.5). C, Coomassie Blue-stained gels after SDS-PAGE showing similar purity of WT- and VEFE-AMPK.
Reduced Concentration-dependent Oligomerization of VEFE-AMPK Versus WT-AMPK
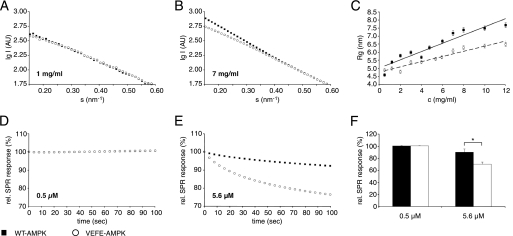
The radius of gyration (Rg), a parameter characterizing particle size, was determined by SAXS to compare concentration-dependent oligomerization of WT- and VEFE-AMPK in solution. For monodisperse systems, the radius of gyration computed from the SAXS data correspond to a single molecule species. For mixtures containing multiple species, the experimental scattering curve is a linear combination of the scattering from individual components weighted by their relative concentrations (i.e. their volume fractions in the solution). In this case the effective value of Rg is also average over the ensemble weighted by the volume fractions of the species. Earlier work has shown that at low concentrations, i.e. 0.5 mg/ml, AMPK solutions behave as ideal, monodisperse solutions in SAXS, thus representing a homogeneous population of AMPK heterotrimers (31). WT- and VEFE-AMPK both exhibited a concentration-dependent increase of the Rg (Fig. 2, A–C), corresponding to the formation of homo-oligomers of the AMPK heterotrimers. When compared with WT-AMPK, the concentration-dependent tendency to form oligomers, especially large ones, was significantly smaller for VEFE-AMPK (Fig. 2C). Although these differences are apparent at smaller scattering angles, the overlapping scattering curves of WT- and VEFE-AMPK at larger angles are a measure for the high similarity of protein concentrations and monomer structures of both AMPK species (Fig. 2, A and B). Upon dilution of such concentrated samples, the Rg decreased to values similar to those observed for low concentrated protein solutions before concentration (data not shown). In agreement with previously published data (31), our results confirm that oligomerization of AMPK is a reversible, concentration-dependent process.
FIGURE 2.
Analysis of concentration-dependent oligomerization of WT- and VEFE-AMPK. A and B, representative SAXS scattering curves of AMPK in solution at 1 mg/ml (A) and 7 mg/ml (B). The log of radially symmetric scattered x-ray intensity in arbitrary units (AU) is plotted as a function of the momentum transfers (0.15 nm−1 < s < 0.59 nm−1) to visualize significant differences between wild type and mutant at small angles. The momentum transfer is s = 4πsinθ/λ, where 2θ is the scattering angle and λ is the radiation wavelength. At larger angles the scattering curves of wild type and mutant were equal. C, Rg in nanometers as a function of AMPK protein concentration. Rg was calculated using the Guinier approximation at small angles. D and E, representative surface plasmon resonance (BIAcore) kinetics showing the dissociation of AMPK complexes from Ni-NTA sensor chips at injected protein concentrations of 0.5 μm (D) and 5.6 μm (E). The data were normalized and the dissociation is given as percentage relative to dissociation start (for details see “Experimental Procedures”; relative SPR response (%)). F, relative SPR signal after 100 s of dissociation as a function of protein concentration (n = 3; S.D.; *, p < 0.05). Filled squares and bars, nontagged WT-AMPK; open circles and bars, nontagged VEFE-AMPK. Note: for clarity of the presentation not every data point is shown.
SPR is capable of directly monitoring protein complex formation or dissociation. Because nontagged AMPK complexes exhibit an intrinsic high affinity for Ni-NTA (31), AMPK can be stably immobilized from solution onto NTA sensor chips,3 and the subsequent dissociation signal is mainly a direct measure for the disintegration of higher order AMPK complexes. Injection at different protein concentrations revealed a concentration-dependent difference between WT- and VEFE-AMPK (Fig. 2, D–F). Because the dissociation kinetics were highly complex and escaped reasonable kinetic analysis, a simple dissociation end point at 100 s after injection was determined. At a concentration of 0.5 μm AMPK, corresponding to monodisperse heterotrimers (31), almost no dissociation occurred, and this was identical for WT- and VEFE-AMPK (Fig. 2D). Thus, both proteins show a high affinity interaction with the Ni-NTA chip surface. At protein concentrations favoring the formation of higher order AMPK complexes, e.g. 5.6 μm, dissociation behavior of both proteins differed significantly (Fig. 2E). Although WT-AMPK showed only a low signal decrease of about 8% during 100 s, the dissociation of VEFE-AMPK was increased to 25% (Fig. 2F). Given the equal affinity of both proteins to Ni-NTA, the difference reveals a strongly decreased stability of the larger complexes because of the VEFE mutation, entirely corroborating results obtained with SAXS.
Oligomerization Pattern of AMPK
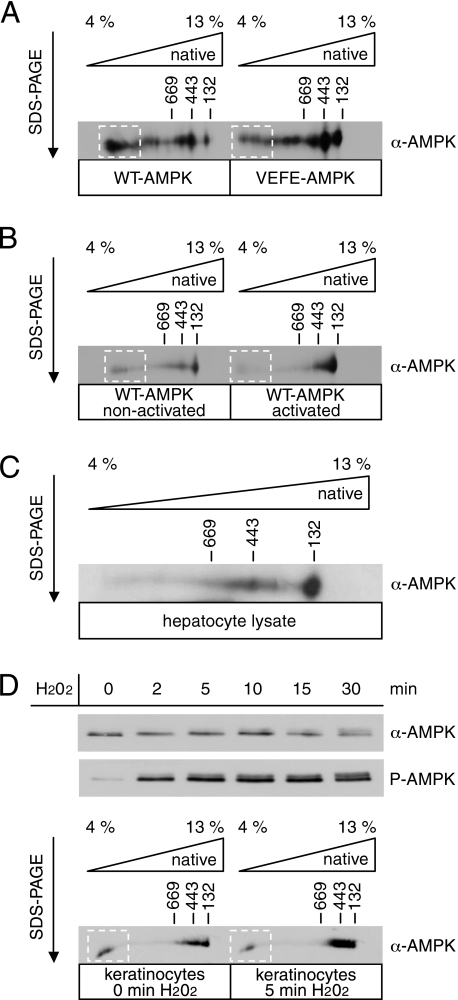
To further investigate higher order AMPK oligomers, BN/SDS-PAGE was applied. In the first dimension, BN-PAGE, the organization of AMPK subunits into heterotrimers and homo-oligomers thereof are expected to remain undisturbed. In contrast, separation in the second dimension, SDS-PAGE, should disrupt all protein-protein interactions and thus visualize the stoichiometric composition of the subunits. After BN/SDS-PAGE of recombinant AMPK, the silver-stained gel indeed confirmed the separation into distinct complexes of various sizes each truly consisting of heterotrimers with the catalytic α-subunit and the regulatory β- and the γ-subunits (supplemental Fig. S1). To allow for direct comparison of two different AMPK samples, both were subjected to first and second dimension by applying the samples to the same gel in parallel, which was then further analyzed by Western blotting using antibodies recognizing the AMPK α-subunit (Fig. 3, A–D). Based on comparison with molecular weight markers, the size of recombinant AMPK heterotrimers reached from about 180 kDa (in the high percentage area of the gel) up to very high order oligomers in the MDa range (in the low percentage area of the gel) (Fig. 3A, left panel). The lowest apparent molecular mass (MM) likely represents the monomer of the heterotrimeric complex, albeit migrating at a somewhat higher MM than calculated from the primary sequence (∼130 kDa). This is in agreement with previous data and likely relates to the elongated, nonglobular shape of native heterotrimeric AMPK particles (31). Several distinct signals are detectable at higher MM, indicating the existence of defined homo-oligomers each consisting of a certain number of AMPK heterotrimers. Using BN/SDS-PAGE, we furthermore investigated the involvement of αG-helix residues in the emergence of oligomers. As expected from the SAXS and SPR data, mutation of Val-219 and Phe-223 to glutamate reduced the occurrence of very high order oligomers and increased the abundance of complexes with MM ∼370 and ∼180 kDa, respectively (Fig. 3A, right panel). These results substantiate the critical role for Phe-219 and Val-223 in homo-oligomerization of AMPK. One can assume that the species at MM ∼370 kDa corresponds to a dimer of heterotrimers, as suggested by the approximate doubling of MM. Thus, the data would favor a model where the αG-helix residues are important for the formation of very large homo-oligomeric complexes but would not mediate the dimerization of AMPK heterotrimers, because the mutant VEFE-AMPK more abundantly forms dimers than WT-AMPK. This appears to be contrary to the observed αG-helix-mediated dimerization of SNF1 (36). But here it is important to note that our study is focusing on the full-length heterotrimer of mammalian AMPK and not the isolated kinase domain.
FIGURE 3.
Detection of WT- and VEFE-AMPK oligomers by two-dimensional BN/SDS-PAGE. A, purified, recombinant WT- and VEFE-AMPK were simultaneously analyzed by 4–13% (% T) gradient BN-PAGE in the first dimension and SDS-PAGE according to Laemmli in the second dimension. AMPK was detected by Western blotting using an anti-AMPKα antibody. B, nonactivated and activated WT-AMPK were simultaneously analyzed as in A. To ensure full activation of the kinase, AMPK heterotrimers were incubated with an excess of recombinant His-LKB1-MO25α-STRADα. C, primary hepatocyte lysate was analyzed as in A using Tricine/SDS-PAGE for the second dimension. D, human primary keratinocytes were treated with 1 mm H2O2, and cells were lysed at different time points as indicated. Lysates were analyzed by SDS-PAGE and Western blotting with anti-AMPKα and phospho-specific anti-AMPKα Thr-172 (P-AMPK) antibodies thus revealing rapid and sustained activation of AMPK in these cells. Lysates of nontreated (0 min) and treated human primary keratinocytes (5 min) were simultaneously analyzed by two-dimensional BN/SDS-PAGE as in A. Western blots shown are representatives of multiple experiments.
To analyze the effect of AMPK activation on the oligomerization process, recombinant WT-AMPK was incubated with or without CamKK2 prior to analysis by BN/SDS-PAGE. Upon activation, WT-AMPK showed a significantly reduced formation of very high order oligomers and a clear shift to lower oligomeric states, comparable with the pattern seen with nonactivated VEFE-AMPK (Fig. 3B). Activation of AMPK is known to induce autophosphorylation at several sites and is also altering conformation of the kinase, notably by rearrangement of the activation segment. Thus, both changes could influence accessibility of the amphipathic αG-helix and therefore could account for the observed differences (see under “Discussion” and supplemental Fig. S2).
After electrophoretic separation of cell lysates of cultured primary hepatocytes, AMPK shows a size distribution ranging from very high to low MM complexes (Fig. 3C), thus suggesting the existence of higher oligomeric states of AMPK heterotrimers also on the cellular level. Furthermore, we compared cell lysates of primary keratinocytes before and after treatment with H2O2. H2O2 markedly activates AMPK in these cells as shown by Western blotting using an antibody recognizing phosphorylated Thr-172 (Fig. 3D, upper panel). The analysis of AMPK size distribution shows a shift from very high to low MM complexes upon activation that is reminiscent of the observed differences between active and inactive recombinant AMPK (compare Fig. 3, B and D, lower panel).
In conclusion, oligomer formation of AMPK heterotrimers (i) occurs with recombinant enzymes in vitro as well as in cellular extracts in situ, (ii) yields multiple and distinct oligomeric states, (iii) depends on hydrophobic interactions involving Val-219 and Phe-223 in the αG-helix, and (iv) shows an inverse relation to AMPK activation.
Participation of αG-helix Residues in Recognition of AMPK by Upstream Kinases
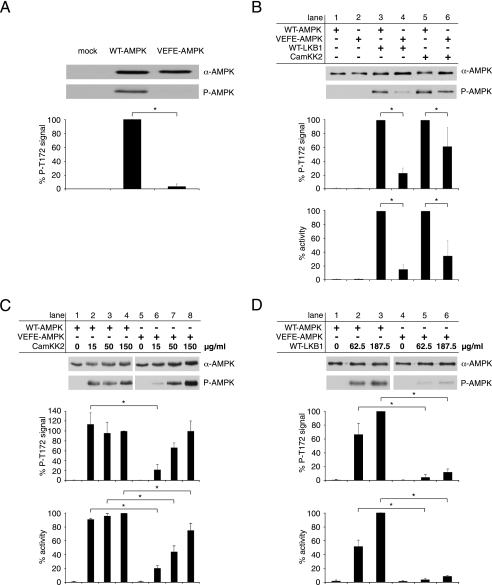
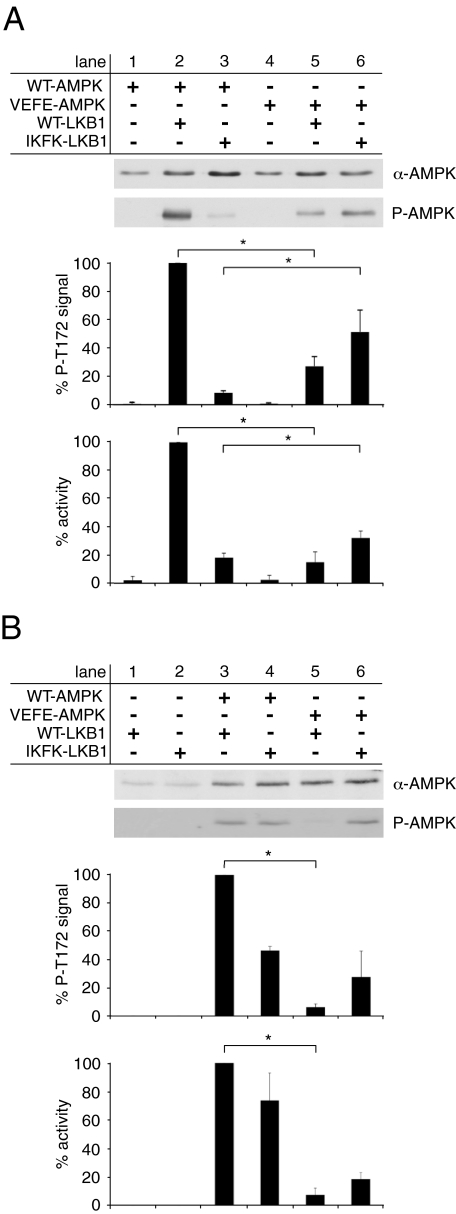
To further evaluate the physiological relevance of AMPK oligomerization, AMPKα subunit double knock-out (α1−/− and α2−/−) mouse embryonic fibroblasts (MEFs) were transfected with FLAG-tagged WT-AMPKα1 and VEFE-AMPKα1 expression plasmids. AMPK activation was induced with Metformin and evaluated by Western blot analysis of Thr-172 phosphorylation. Both AMPK subunits were equally expressed, but mutation of the hydrophobic residues Val-219 and Phe-223 to glutamate resulted in an ∼15-fold reduction of AMPK activation (Fig. 4A). This result is unexpected, because a lower degree of oligomerization was observed with VEFE-AMPK (Figs. 2 and 3) and thus would be predicted to rather favor activation, if presuming that the Thr-172 residue is buried in higher oligomeric states. As illustrated in Fig. 1A, the hydrophobic couple homologous to Val-219 and Phe-223 of AMPK is also conserved within the activating upstream kinases LKB1 and, partially, also in CamKK2. Therefore, we considered a possible involvement of these hydrophobic residues in transient heterodimerization between AMPK and LKB1 or CamKK2. As being part of the recognition process, such interactions would directly affect the process of AMPK activation. To test this hypothesis, we took advantage of the defined in vitro system. Recombinant WT- and VEFE-AMPK were incubated with either recombinant LKB1-MO25α-STRADα or recombinant CamKK2. The degree of activation was evaluated by activity determination, as well as Western blot analysis of Thr-172 phosphorylation. The concentration of upstream kinases was adjusted to yield just about saturation of WT-AMPK activation. Under these conditions, activation of the VEFE mutant was significantly lower compared with wild type (Fig. 4B). As compared with WT-AMPK, the enzymatic activity of VEFE-AMPK was reduced to ∼15 and ∼40% after activation with LKB1 complex and CamKK2, respectively. However, activation levels of VEFE-AMPK similar to those of WT-AMPK could be achieved by simply increasing the concentration of CamKK2 (Fig. 4C). However, when using higher amounts of LKB1 complex, activation of VEFE-AMPK was only partially recovered (Fig. 4D). This difference between both AMPKKs correlates with the degree of conservation of the hydrophobic Val-219/Phe-223 couple in CamKK2 and LKB1. Both residues are highly conserved in the αG-helix of LKB1, but only Val-219 and not Phe-223 is conserved in CamKK2. The achievable activity of VEFE-AMPK, which is almost reaching the level of WT-AMPK (Fig. 4C), is also confirming the functional integrity of VEFE-AMPK. The functional differences of WT- and VEFE-AMPK, as observed in various experiments, are therefore attributable to the αG-helix region, as we can exclude a general impairment of the catalytic properties of VEFE-AMPK.
FIGURE 4.
Activation of WT- and VEFE-AMPK by upstream kinases. A, Western blot analysis of mock- and single-transfected (α1−/− and α2−/−) MEF cell lysates with anti-AMPKα and phospho-specific anti-AMPKα Thr-172 antibodies (P-AMPK). The background signal of mock-transfected cells was subtracted, and data were normalized relative to the WT-AMPK single-transfected cells (n = 3; S.D.; *, p < 0.05). B, in vitro activation of recombinant nontagged WT- and VEFE-AMPK by recombinant WT-LKB1 complex and CamKK2. Activity was determined by a HPLC- and SAMS-peptide-based activity assay. Data were normalized to signal of WT-AMPK activation (n = 5; S.D.; *, p < 0.05). C, in vitro activation of recombinant WT- and VEFE-AMPK with increasing concentrations of recombinant CamKK2 as indicated. Data were normalized relative to WT-AMPK activation with 150 μg/ml CamKK2 (n = 3; S.D.; *, p < 0.05). D, as in C with increasing concentrations of recombinant WT-LKB1 complex. Data were normalized relative to WT-AMPK activation with 187.5 μg/ml WT-LKB1 (n = 3; S.D.; *, p < 0.05).
Taken together, the in vitro data and the experiments in cultured cells provide evidence for an involvement of Val-219 and Phe-223 within the αG-helix of AMPK in the recognition process by LKB1 and, to a lesser extent, by CamKK2.
Functional Recovery of VEFE-AMPK by Mutant LKB1
To further verify and corroborate the above findings, we aimed at designing an LKB1 mutant capable of rescuing the low rate of activation of VEFE-AMPK. The following ideas provided a rationale for the construction of a suitable LKB1 mutant. (i) AMPK recognition by LKB1 is mediated via hydrophobic interactions between Val-219/Phe-223 within the αG-helix of the AMPK kinase domain and Ile-260/Phe-264 within the αG-helix of the LKB1 kinase domain. (ii) VEFE-AMPK is refractory to recognition by LKB1 because the negatively charged glutamate residues abolish the hydrophobic interplay between the two kinases. (iii) Substitution of Ile-260/Phe-264 in the LKB1 kinase domain with complementary, positively charged lysine residues should therefore restore this interaction by electrostatic attraction between the two domains of AMPK and LKB1 (see under “Discussion” and Fig. 7D).
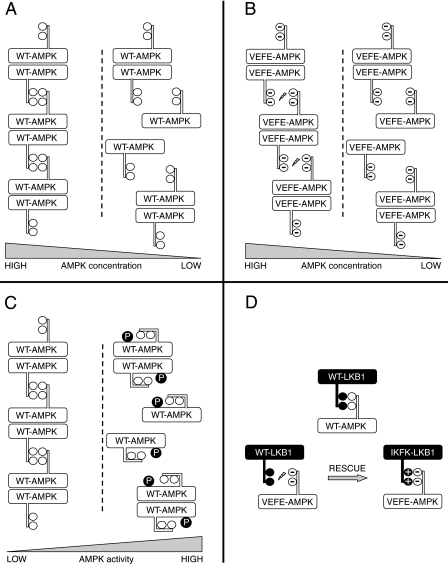
FIGURE 7.
Model of AMPK oligomerization and role of αG-helix interaction. A, WT-AMPK heterotrimers form higher oligomers via at least two independent interaction sites, including the hydrophobic αG-helix residues Val-219/Phe-223. At low protein concentration, higher oligomers of WT-AMPK dissociate to monomers and dimers. B, VEFE-AMPK is unable to interact via the hydrophobic αG-helix residues, because of electrostatic repulsion. Thus, formation of higher oligomers is reduced at high concentration, whereas the formation of monomeric and dimeric AMPK species at low concentrations remains unaffected. C, upon activation of WT-AMPK a conformational change of the αG-helix decreases the accessibility of Val-219/Phe-223 and thus prevents higher order oligomerization. D, hydrophobic interaction between Val-219/Phe-223 of the AMPKα1 subunit and Ile-260/Phe-264 of LKB1 is required for recognition and subsequent activation of AMPK by LKB1. Top, WT-AMPK and WT-LKB1 interact via hydrophobic residues of their solvent-exposed αG-helices. Bottom left, negatively charged hydrophilic residues prevent the hydrophobic interaction between WT-LKB1 and VEFE-AMPK. Bottom right, recognition of VEFE-AMPK is restored when using IKFK-LKB1 complex, which creates electrostatic attraction of the respective αG-helices.
The presumed capability of LKB1(I260K,F264K)-MO25α-STRADα (IKFK-LKB1) to rescue impaired activation of VEFE-AMPK was first investigated in vitro using the purified enzymes and second in transfected cell culture. Equal amounts of recombinant wild type (WT-) or IKFK-LKB1 were allowed to phosphorylate both WT- and VEFE-AMPK. Subsequently, the activity of AMPK was assessed by Thr(P)-172 immunoblotting and phosphorylation of the SAMS peptide. Again, WT-LKB1 readily activated WT-AMPK but only poorly recognized VEFE-AMPK (Fig. 5A, lanes 2 and 5). On the contrary, IKFK-LKB1 scarcely phosphorylated WT-AMPK and considerably activated VEFE-AMPK (Fig. 5A, lanes 3 and 6). Thus, IKFK-LKB1, by acting as a complementing mutant, indeed recovered activation of VEFE-AMPK. These results also emphasize the decisive role of the αG-helices, both of AMPK and LKB1, for their mutual recognition. To study the recognition of AMPK by LKB1 via αG-helix residues in a more physiological setting, AMPKα subunit double knock-out (α1−/− and α2−/−) MEFs were transfected or co-transfected with mammalian expression plasmids encoding for WT-AMPKα1, VEFE-AMPKα1, WT-LKB1, and IKFK-LKB1. AMPK activation was induced with 5 mm Metformin and evaluated by Western blot analysis of Thr-172 phosphorylation and activity determination (Fig. 5B). Consistently, activation of WT-AMPK was more efficient in cells expressing WT-LKB1 compared with IKFK-LKB1 (Fig. 5B, lanes 3 and 4). On the other hand, activation of VEFE-AMPK by WT-LKB1 was barely detectable, whereas IKFK-LKB1 more effectively activated VEFE-AMPK (Fig. 5B, lanes 5 and 6).
FIGURE 5.
Functional recovery of VEFE-AMPK by LKB1 rescue mutant. A, in vitro activation of recombinant heterotrimeric WT-AMPK and VEFE-AMPK by recombinant WT-LKB1 and IKFK-LKB1, respectively. Activity was determined by a HPLC- and SAMS-peptide-based activity assay. Data were normalized to WT-AMPK activation by WT-LKB1 (n = 3; S.D.; *, p < 0.05). B, Western blot analysis of transfected (α1−/− and α2−/−) MEF cell lysates using anti-AMPKα and phospho-specific anti-AMPKα Thr-172 antibodies (P-AMPK). Transfected plasmids encoded for myc-AMPKα1 or FLAG-LKB1 wild type and mutant proteins. AMPK activity was determined after immunoprecipitation using an anti-AMPKα1 antibody. The background signal of LKB1 single-transfected cells was subtracted, and data were normalized to the WT-AMPKα1/WT-LKB1 double-transfected cells (n = 3; S.D.; *, p < 0.05).
The in vitro and cell culture data both provide solid evidence for the activation of AMPK by LKB1 being strongly dependent on interaction of residues Val-219/Phe-223 in AMPK with residues Ile-260/Phe-264 in LKB1. Accordingly, WT-AMPK and WT-LKB1 interact via hydrophobic residues of their exposed αG-helices, whereas VEFE-AMPK does not harmonize with WT-LKB1 thus repelling one another. On the contrary, VEFE-AMPK interacts with IKFK-LKB1 by electrostatic attraction via opposite charges (see also Fig. 7D).
Impact of the αG-helix on Dephosphorylation of AMPK at Thr-172
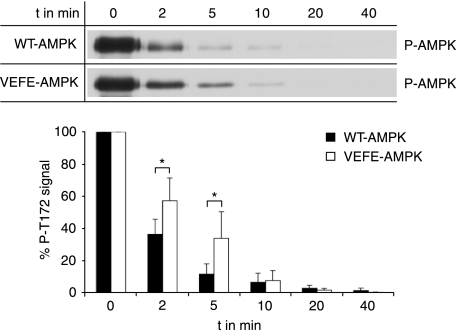
Because the amphipathic αG-helix of AMPK was found to mediate interaction with activating upstream kinases (LKB1 and CamKKβ), we further asked whether it also represents a key structural element for recognition by the inactivating protein phosphatase PP2Cα. Phosphorylation of WT- and VEFE-AMPK at Thr-172 was saturated by incubation with excessive amount of recombinant GST-tagged CamKK2. AMPK was repurified, and dephosphorylation by PP2Cα was monitored by Western blot analysis of Thr(P)-172. Before addition of phosphatase, WT- and VEFE-AMPK were equally phosphorylated (Fig. 6, t = 0 min), which again confirms that high amounts of upstream kinase can compensate for the reduced activation rate of VEFE-AMPK. Within the first few minutes of phosphatase action, dephosphorylation of VEFE-AMPK was slower compared with WT-AMPK (p < 0.05). After 10 min, the difference between WT- and VEFE-AMPK diminished, and Thr-172 was almost completely dephosphorylated in both proteins (Fig. 6).
FIGURE 6.
Dephosphorylation of WT- and VEFE-AMPK by PP2Cα. Activated and repurified recombinant WT- and VEFE-AMPK were incubated with protein phosphatase PP2Cα, and samples were taken at indicated time points. The degree of dephosphorylation was determined by Western blotting using a phospho-specific anti-AMPKα Thr-172 antibody (upper panel, P-AMPK). In addition, P-AMPK signals were densitometrically quantified and shown by percentage relative to Thr-172 phosphorylation at time point 0. Black bars, WT-AMPK; white bars, VEFE-AMPK (n = 5; S.D.; *, p < 0.05).
Thus, the exchange of hydrophobic residues of the αG-helix to glutamate shows a reduced dephosphorylation rate of AMPK at Thr(P)-172 by PP2Cα. In comparison to the marked suppression of activation by LKB1, the phosphatase action is much less affected by these mutations.
DISCUSSION
This study assigns several new features to the αG-helix located within subdomain X of the kinase domain of mammalian AMPK. In living cells and in vitro, we provide evidence for the control of AMPK function by the kinase domain αG-helix at different levels as follows: (i) oligomerization of the inactive kinase, (ii) activation by upstream kinases, and (iii) inactivation by PP2Cα.
The molecular structures of SNF1/AMPK reveal a solvent-accessible hydrophobic face of the amphipathic αG-helix in subdomain X within the large kinase lobe. This hydrophobic patch is involved in dimerization of the yeast ortholog SNF1 (36) and is dominated in AMPKs by the conserved hydrophobic couple Val-219/Phe-223 (Fig. 1). Exchanging these residues in AMPK by glutamate did not alter AMPK structure nor kinase activity, as demonstrated by intact heterotrimerization and full enzymatic activity of the AMPK mutant, respectively. However, as shown by three independent methods, namely small angle x-ray scattering, surface plasmon resonance, and two-dimensional BN/SDS-PAGE, the V219E,F223E double mutation clearly reduced the concentration-dependent oligomerization of AMPK. Consequently, the hydrophobic surface of the αG-helix represents a crucial element for the assembly of large homo-oligomers of AMPK via its kinase domain. A certain degree of oligomerization still persisting in VEFE-AMPK indicates the existence of additional unknown elements apart from the αG-helix that promote interaction between heterotrimers. Similarly, the homologous I257E,F261E mutation in the SNF1 kinase domain disrupts the kinase domain interaction in vitro but fails to abolish self-association of SNF1 proteins in vivo (36). Unfortunately, no further information can be obtained from the published x-ray structures of AMPK heterotrimers, because these core structures all lack the kinase domain containing the αG-helix. Thus, mammalian AMPK was hitherto crystallized as a monomer, whereas the Saccharomyces orthologs form crystallographic dimers of trimers, albeit at different interfaces with questionable physiological significance, involving either γ-subunits (SNF4) or additional residues of the α- and β-subunits (32, 34).
Kinase domain dimerization is a common regulatory mechanism in kinase autoactivation and autoinhibition (61, 62). The latter is more likely to apply to AMPK. First, AMPK is not known to perform autoactivation. Second, in the structure of the orthologous SNF1 kinase domain, the activation loop is buried within the homodimerization interface, suggesting an inactive state of this kinase in the crystal (36). Another structurally characterized kinase, the PAK1 (p21-activated kinase 1), also forms a homodimeric and autoinhibited complex, whereas the mode of dimerization is different from the one seen in SNF1 (63). Interestingly, Leu-473 in human PAK1, a hydrophobic residue homologous to Phe-223 of AMPK, is in direct contact between the kinase domain and a so-called inhibitory switch domain (64). Furthermore, the solvent-exposed αG-helix is involved in the interaction between the catalytic and the regulatory subunits of cAMP-dependent protein kinase and thus was suggested to serve as a potential docking motif for most protein kinases (65, 66). In conclusion, our results further support the idea that hydrophobic residues of the αG-helix are important mediators of protein-protein interactions.
AMPK oligomerization in solution was studied by SAXS and occurs above 1 mg/ml, as shown here and in our earlier study (31). SPR and BN/SDS-PAGE both reveal a similar oligomerization behavior of AMPK heterotrimers. However, the applied protein concentrations that induced oligomerization are difficult to compare with these used in SAXS measurements because the effective concentration here is higher than deployed. This is because of concentration effects on the sensor chip surface in SPR and also within the stacking layer of the native gel in BN/SDS-PAGE. Similarly, in living cells with cytoplasmic protein concentrations up to 200–400 mg/ml (67–69), macromolecular crowding effects can promote microcompartmentation and association or self-assembly of macromolecules (69, 70). For example, AMPK can be targeted to a glycogen microcompartment via its glycogen-binding domain in the β-subunit (71, 72), possibly leading to higher local concentrations, and thus could in fact allow for in vivo oligomerization. Such multiple, distinct AMPK heterotrimers were indeed directly visualized here for the first time with the recombinant protein and, most importantly, also in lysates from primary hepatocytes and keratinocytes using BN/SDS-PAGE. The formation of large homo-oligomers is another frequent intracellular mechanism, occurring among others within the kinase family, e.g. calmodulin-dependent protein kinase 2 (CaMK2) forms dodecameric assemblies of ∼700,000 kDa, which is believed to strongly support its functions (73, 74). In AMPK, the V219E,F223E double mutation mainly affected large oligomeric complexes that were shifted to lower MM, thus indicating that additional, unknown interaction sites give rise to smaller assemblies such as dimers. Intriguingly, a similar change of the oligomeric state showing the decreased emergence of higher MM species was also observed upon activation of recombinant AMPK and of endogenous AMPK in primary keratinocytes. To further analyze this effect, we compared published active and inactive conformations of kinase domains, including Aurora A (75, 76), protein kinase B (77, 78), p38γ (79, 80), and insulin receptor tyrosine kinase (81–83) (supplemental Fig. S2). All inactive conformations show the αG-helix surface exposed, whereas in the active kinase the same helix is moved toward the core of the large kinase lobe thus reducing accessibility. We presume this could explain the decreased formation of very high order oligomers of AMPK upon activation, because a lowered solvent exposure of the αG-helix would impair self-interaction and thus reduce formation of higher oligomers.
The amphipathic αG-helix of AMPK is even more directly linked to AMPK activation by providing a docking site for its upstream kinases. Sequence alignment of AMPK with the upstream kinases LKB1 and CamKK2 disclosed a significant degree of homology of hydrophobic residues in both of their αG-helices. The hydrophobic couple Val-219/Phe-223 was either entirely (LKB1) or at least partially (CamKK2) conserved. Accordingly, activation of VEFE-AMPK was diminished with WT-LKB1 and somewhat less impaired with CamKK2. Thus, we assumed the Val-219/Phe-223 residues of AMPK could potentially represent a recognition site for upstream kinases distinct from the linear sequence surrounding Thr-172. The most striking piece of evidence for such distal interaction comes from the experiments using IKFK-LKB1. Although IKFK-LKB1 was barely able to activate WT-AMPK, we observed a clearly increased activity of IKFK-LKB1 only in conjunction with VEFE-AMPK. Thus, IKFK-LKB1 represents a rescue mutant for the functionally impaired VEFE-AMPK, as demonstrated with recombinant proteins in vitro and in vivo using transfected cell culture (Fig. 5). The specific interaction of αG-helix residues therefore is required for recognition of AMPK by its upstream kinases, especially LKB1, and vice versa.
The critical participation of subdomain X in signaling of MAPK has been described recently (84, 85). Selected alanine mutations within subdomain X of the MAPK kinase kinase (MAP3K) MEKK1, especially of two hydrophobic residues (F1443A and I1445A), diminished binding and activation of its downstream target MAPK kinase (MAP2K) MKK4 and activation of other downstream kinases (MKK7, IKK, and MEK1) (85). Furthermore, hydrophobic residues of subdomain X were also required for proper MEKK2-dependent activation of c-Jun NH2-terminal kinase (JNK) and ERK5 pathways (84). Together with our results, these findings argue for a more general role of residues located in subdomain X of protein kinases that would facilitate target recognition.
Our data further suggest an involvement of the αG-helix region in AMPK inactivation by protein phosphatase PP2Cα. Dephosphorylation of Thr-172 by PP2Cα was decelerated in VEFE-AMPK relative to WT. However, complete inactivation of the mutant could still be achieved, indicating that alternative or adjacent recognition sites between AMPK and protein phosphatase PP2Cα are likely to exist. In general, little is known about the molecular basis of substrate specificity of protein phosphatases. Moreover, the metal-dependent PP2C phosphatases reportedly share a broad and overlapping substrate specificity (86–88) and probably recognize their respective targets by molecular determinants involving regions different from that surrounding the phosphorylated residue (88).
Although the specificity of target protein recognition by their respective kinase is often described by a consensus sequence, the interaction also frequently requires distal interactions between the kinase and its substrate (89). Earlier, residues of the loop connecting helices G and F in the kinase domain were suggested to mediate interaction between AMPKα1 and an acetyl-CoA carboxylase-1/α model substrate (90). Preliminary results indicate that the VEFE-AMPK mutant more often shows reduced ability to phosphorylate downstream target proteins if compared with WT-AMPK, but the observed differences were varying between targets.5 Further studies are required to clarify this issue.
In conclusion, our study provides evidence that the small amphipathic αG-helix of the kinase domain plays a crucial role in targeting regulatory proteins to AMPK. In particular, its major involvement in reversible AMPK self-assembly and activation by AMPK upstream kinases discloses a new regulatory layer of this crucial cellular fuel gauge. The molecular functioning of the AMPK αG-helix is summarized in a simplified model (Fig. 7). Accordingly, in an inactive (dephosphorylated) state, AMPK can associate into inert, large oligomers of heterotrimers, which promote autoinhibition by burying the active sites in the interaction interfaces. Whereas the self-assembly of very high order oligomers is mediated by the αG-helix, the formation of dimers clearly depends on other so far unknown interaction sites. Upon activation (phosphorylation) by its upstream kinases via the αG-helix, the large oligomers disaggregate, leading to dimers and monomers of the AMPK heterotrimer. These may be more diffusible and expose their active sites allowing for further regulatory control of the kinase. Our findings might open new perspectives for the design of small chemical compounds that could trigger AMPK activity by affecting the oligomeric state or upstream kinase recognition and thus would be expected to prove beneficial for AMPK-related human disorders.
Supplementary Material
Acknowledgments
We thank Uwe Riek and Corinne Sidler (Cell Biology, ETH Zurich, Zurich, Switzerland) and Sacnicte Ramirez and Guillaume Vial (Inserm U884, Université Joseph Fournier, Grenoble, France) for their contribution to different experiments. We also thank Hans-Dietmar Beer and Gabriel Sollberger (Cell Biology, ETH Zurich, Zurich, Switzerland) for providing human primary keratinocytes and D. G. Hardie (University of Dundee, Dundee, Scotland, United Kingdom) for the provision of anti-AMPKα1 antibody. We greatly appreciate the gift of plasmids from H. Tokumitsu (Kagawa Medical University, Kagawa, Japan), A. Balendran (AstraZeneca, Sweden), D. Carling (Imperial College London, London, United Kingdom), and D. R. Alessi (University of Dundee, Dundee, Scotland, United Kingdom).
This work was supported by Swiss National Science Foundation Grant 3100A0-114137, ETH graduate student grants (to R. F. T. and R. S.), the Krebsliga Zentralschweiz (to D. N.), FP-6 European Union Framework Programme Project LSHM-CT-2004-005272 EXGENESIS, INSERM, and the French Agence Nationale de Recherche (“Chaire d'Excellence” given to U. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
C. Polge and U. Schlattner, unpublished data.
A. Nazabal and R. Zenobi, personal communication.
R. Scholz and D. Neumann, unpublished data.
- AMPK
- AMP-activated protein kinase
- BN-PAGE
- gradient blue native PAGE
- BN/SDS-PAGE
- two-dimensional gradient blue native/SDS-PAGE
- CamKK2
- Ca2+/calmodulin-dependent kinase kinase 2
- IKFK-LKB1
- LKB1(I260K,F264K)-MO25α-STRADα
- LKB1
- serine/threonine kinase 11 (STK11)
- MAPK
- mitogen-activated protein kinase
- MEF
- mouse embryonic fibroblast
- MM
- molecular mass
- MO25
- mouse protein 25
- PP2Cα
- protein phosphatases 2Cα
- SAMS
- synthetic peptide HMRSAMSGLHLVKRR
- SAXS
- small angle x-ray scattering
- SNF1
- sucrose nonfermenting kinase 1
- SPR
- surface plasmon resonance
- STRAD
- STE20-related adaptor protein
- VEFE-AMPK
- AMPK α1(V219E,F223E)β1γ1
- HPLC
- high performance liquid chromatography
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- Bicine
- N,N-bis(2-hydroxyethyl)glycine
- Ni-NTA
- nickel-nitrilotriacetic acid
- WT
- wild type
- GST
- glutathione S-transferase.
REFERENCES
- 1.Carling D. (2005) Biochimie 87, 87–91 [DOI] [PubMed] [Google Scholar]
- 2.Hardie D. G., Hawley S. A., Scott J. W. (2006) J. Physiol. 574, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn B. B., Alquier T., Carling D., Hardie D. G. (2005) Cell Metab. 1, 15–25 [DOI] [PubMed] [Google Scholar]
- 4.Sanz P. (2008) Curr. Protein Pept. Sci. 9, 478–492 [DOI] [PubMed] [Google Scholar]
- 5.Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 6.Neumann D., Schlattner U., Wallimann T. (2003) Biochem. Soc. Trans. 31, 169–174 [DOI] [PubMed] [Google Scholar]
- 7.Hardie D. G. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 185–210 [DOI] [PubMed] [Google Scholar]
- 8.Hardie D. G. (2008) FEBS Lett. 582, 81–89 [DOI] [PubMed] [Google Scholar]
- 9.Luo Z., Saha A. K., Xiang X., Ruderman N. B. (2005) Trends Pharmacol. Sci. 26, 69–76 [DOI] [PubMed] [Google Scholar]
- 10.Hardie D. G. (1999) Biochem. Soc. Symp. 64, 13–27 [PubMed] [Google Scholar]
- 11.Tuerk R. D., Auchli Y., Thali R. F., Scholz R., Wallimann T., Brunisholz R. A., Neumann D. (2009) Anal. Biochem. 390, 141–148 [DOI] [PubMed] [Google Scholar]
- 12.Chen Z., Heierhorst J., Mann R. J., Mitchelhill K. I., Michell B. J., Witters L. A., Lynch G. S., Kemp B. E., Stapleton D. (1999) FEBS Lett. 460, 343–348 [DOI] [PubMed] [Google Scholar]
- 13.Horman S., Vertommen D., Heath R., Neumann D., Mouton V., Woods A., Schlattner U., Wallimann T., Carling D., Hue L., Rider M. H. (2006) J. Biol. Chem. 281, 5335–5340 [DOI] [PubMed] [Google Scholar]
- 14.Mitchelhill K. I., Michell B. J., House C. M., Stapleton D., Dyck J., Gamble J., Ullrich C., Witters L. A., Kemp B. E. (1997) J. Biol. Chem. 272, 24475–24479 [DOI] [PubMed] [Google Scholar]
- 15.Woods A., Vertommen D., Neumann D., Turk R., Bayliss J., Schlattner U., Wallimann T., Carling D., Rider M. H. (2003) J. Biol. Chem. 278, 28434–28442 [DOI] [PubMed] [Google Scholar]
- 16.Hawley S. A., Boudeau J., Reid J. L., Mustard K. J., Udd L., Mäkelä T. P., Alessi D. R., Hardie D. G. (2003) J. Biol. 2, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woods A., Dickerson K., Heath R., Hong S. P., Momcilovic M., Johnstone S. R., Carlson M., Carling D. (2005) Cell Metab. 2, 21–33 [DOI] [PubMed] [Google Scholar]
- 18.Momcilovic M., Hong S. P., Carlson M. (2006) J. Biol. Chem. 281, 25336–25343 [DOI] [PubMed] [Google Scholar]
- 19.Davies S. P., Helps N. R., Cohen P. T., Hardie D. G. (1995) FEBS Lett. 377, 421–425 [DOI] [PubMed] [Google Scholar]
- 20.Hardie D. G., Carling D. (1997) Eur. J. Biochem. 246, 259–273 [DOI] [PubMed] [Google Scholar]
- 21.Scott J. W., Hawley S. A., Green K. A., Anis M., Stewart G., Scullion G. A., Norman D. G., Hardie D. G. (2004) J. Clin. Invest. 113, 274–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suter M., Riek U., Tuerk R., Schlattner U., Wallimann T., Neumann D. (2006) J. Biol. Chem. 281, 32207–32216 [DOI] [PubMed] [Google Scholar]
- 23.Sanders M. J., Grondin P. O., Hegarty B. D., Snowden M. A., Carling D. (2007) Biochem. J. 403, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corton J. M., Gillespie J. G., Hawley S. A., Hardie D. G. (1995) Eur. J. Biochem. 229, 558–565 [DOI] [PubMed] [Google Scholar]
- 25.Fryer L. G., Parbu-Patel A., Carling D. (2002) J. Biol. Chem. 277, 25226–25232 [DOI] [PubMed] [Google Scholar]
- 26.Hawley S. A., Gadalla A. E., Olsen G. S., Hardie D. G. (2002) Diabetes 51, 2420–2425 [DOI] [PubMed] [Google Scholar]
- 27.Shaw R. J., Lamia K. A., Vasquez D., Koo S. H., Bardeesy N., Depinho R. A., Montminy M., Cantley L. C. (2005) Science 310, 1642–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou G., Myers R., Li Y., Chen Y., Shen X., Fenyk-Melody J., Wu M., Ventre J., Doebber T., Fujii N., Musi N., Hirshman M. F., Goodyear L. J., Moller D. E. (2001) J. Clin. Invest. 108, 1167–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cool B., Zinker B., Chiou W., Kifle L., Cao N., Perham M., Dickinson R., Adler A., Gagne G., Iyengar R., Zhao G., Marsh K., Kym P., Jung P., Camp H. S., Frevert E. (2006) Cell Metab. 3, 403–416 [DOI] [PubMed] [Google Scholar]
- 30.Sanders M. J., Ali Z. S., Hegarty B. D., Heath R., Snowden M. A., Carling D. (2007) J. Biol. Chem. 282, 32539–32548 [DOI] [PubMed] [Google Scholar]
- 31.Riek U., Scholz R., Konarev P., Rufer A., Suter M., Nazabal A., Ringler P., Chami M., Müller S. A., Neumann D., Forstner M., Hennig M., Zenobi R., Engel A., Svergun D., Schlattner U., Wallimann T. (2008) J. Biol. Chem. 283, 18331–18343 [DOI] [PubMed] [Google Scholar]
- 32.Amodeo G. A., Rudolph M. J., Tong L. (2007) Nature 449, 492–495 [DOI] [PubMed] [Google Scholar]
- 33.Jin X., Townley R., Shapiro L. (2007) Structure 15, 1285–1295 [DOI] [PubMed] [Google Scholar]
- 34.Townley R., Shapiro L. (2007) Science 315, 1726–1729 [DOI] [PubMed] [Google Scholar]
- 35.Xiao B., Heath R., Saiu P., Leiper F. C., Leone P., Jing C., Walker P. A., Haire L., Eccleston J. F., Davis C. T., Martin S. R., Carling D., Gamblin S. J. (2007) Nature 449, 496–500 [DOI] [PubMed] [Google Scholar]
- 36.Nayak V., Zhao K., Wyce A., Schwartz M. F., Lo W. S., Berger S. L., Marmorstein R. (2006) Structure 14, 477–485 [DOI] [PubMed] [Google Scholar]
- 37.Rudolph M. J., Amodeo G. A., Bai Y., Tong L. (2005) Biochem. Biophys. Res. Commun. 337, 1224–1228 [DOI] [PubMed] [Google Scholar]
- 38.Hanks S. K., Quinn A. M. (1991) Methods Enzymol. 200, 38–62 [DOI] [PubMed] [Google Scholar]
- 39.Knighton D. R., Zheng J. H., Ten Eyck L. F., Ashford V. A., Xuong N. H., Taylor S. S., Sowadski J. M. (1991) Science 253, 407–414 [DOI] [PubMed] [Google Scholar]
- 40.Neumann D., Woods A., Carling D., Wallimann T., Schlattner U. (2003) Protein Expr. Purif. 30, 230–237 [DOI] [PubMed] [Google Scholar]
- 41.Neumann D., Suter M., Tuerk R., Riek U., Wallimann T. (2007) Mol. Biotechnol. 36, 220–231 [DOI] [PubMed] [Google Scholar]
- 42.Tokumitsu H., Muramatsu M., Ikura M., Kobayashi R. (2000) J. Biol. Chem. 275, 20090–20095 [DOI] [PubMed] [Google Scholar]
- 43.Marley A. E., Sullivan J. E., Carling D., Abbott W. M., Smith G. J., Taylor I. W., Carey F., Beri R. K. (1996) Biochem. J. 320, 801–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woods A., Salt I., Scott J., Hardie D. G., Carling D. (1996) FEBS Lett. 397, 347–351 [DOI] [PubMed] [Google Scholar]
- 45.Sapkota G. P., Kieloch A., Lizcano J. M., Lain S., Arthur J. S., Williams M. R., Morrice N., Deak M., Alessi D. R. (2001) J. Biol. Chem. 276, 19469–19482 [DOI] [PubMed] [Google Scholar]
- 46.Studier F. W. (2005) Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 47.Riek U., Ramirez S., Wallimann T., Schlattner U. (2009) BioTechniques 46, ix–xii [DOI] [PubMed] [Google Scholar]
- 48.Konarev P. V., Petoukhov M. V., Volkov V. V., Svergun D. I. (2006) J. Appl. Crystallogr. 39, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guinier A. (1939) Ann. Phys. 12, 161–237 [Google Scholar]
- 50.Svergun D. I. (1992) J. Appl. Crystallogr. 25, 495–503 [Google Scholar]
- 51.Schägger H., von Jagow G. (1991) Anal. Biochem. 199, 223–231 [DOI] [PubMed] [Google Scholar]
- 52.Wittig I., Braun H. P., Schägger H. (2006) Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 53.Hjerten S. (1962) Arch. Biochem. Biophys. 99, 466–475 [DOI] [PubMed] [Google Scholar]
- 54.Schägger H. (2006) Nat. Protoc. 1, 16–22 [DOI] [PubMed] [Google Scholar]
- 55.Schägger H., von Jagow G. (1987) Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 56.Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 57.Laderoute K. R., Amin K., Calaoagan J. M., Knapp M., Le T., Orduna J., Foretz M., Viollet B. (2006) Mol. Cell. Biol. 26, 5336–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Groen A. K., Sips H. J., Vervoorn R. C., Tager J. M. (1982) Eur. J. Biochem. 122, 87–93 [DOI] [PubMed] [Google Scholar]
- 59.Feldmeyer L., Keller M., Niklaus G., Hohl D., Werner S., Beer H. D. (2007) Curr. Biol. 17, 1140–1145 [DOI] [PubMed] [Google Scholar]
- 60.Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 61.Pellicena P., Kuriyan J. (2006) Curr. Opin. Struct. Biol. 16, 702–709 [DOI] [PubMed] [Google Scholar]
- 62.Pike A. C., Rellos P., Niesen F. H., Turnbull A., Oliver A. W., Parker S. A., Turk B. E., Pearl L. H., Knapp S. (2008) EMBO J. 27, 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parrini M. C., Lei M., Harrison S. C., Mayer B. J. (2002) Mol. Cell 9, 73–83 [DOI] [PubMed] [Google Scholar]
- 64.Lei M., Lu W., Meng W., Parrini M. C., Eck M. J., Mayer B. J., Harrison S. C. (2000) Cell 102, 387–397 [DOI] [PubMed] [Google Scholar]
- 65.Taylor S. S., Haste N. M., Ghosh G. (2005) Cell 122, 823–825 [DOI] [PubMed] [Google Scholar]
- 66.Taylor S. S., Kim C., Cheng C. Y., Brown S. H., Wu J., Kannan N. (2008) Biochim. Biophys. Acta 1784, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellis R. J., Minton A. P. (2003) Nature 425, 27–28 [DOI] [PubMed] [Google Scholar]
- 68.Fulton A. B. (1982) Cell 30, 345–347 [DOI] [PubMed] [Google Scholar]
- 69.Saks V., Beraud N., Wallimann T. (2008) Int. J. Mol. Sci. 9, 751–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chebotareva N. A., Kurganov B. I., Livanova N. B. (2004) Biochemistry 69, 1239–1251 [DOI] [PubMed] [Google Scholar]
- 71.Hudson E. R., Pan D. A., James J., Lucocq J. M., Hawley S. A., Green K. A., Baba O., Terashima T., Hardie D. G. (2003) Curr. Biol. 13, 861–866 [DOI] [PubMed] [Google Scholar]
- 72.Polekhina G., Gupta A., Michell B. J., van Denderen B., Murthy S., Feil S. C., Jennings I. G., Campbell D. J., Witters L. A., Parker M. W., Kemp B. E., Stapleton D. (2003) Curr. Biol. 13, 867–871 [DOI] [PubMed] [Google Scholar]
- 73.Rosenberg O. S., Deindl S., Comolli L. R., Hoelz A., Downing K. H., Nairn A. C., Kuriyan J. (2006) FEBS J. 273, 682–694 [DOI] [PubMed] [Google Scholar]
- 74.Rosenberg O. S., Deindl S., Sung R. J., Nairn A. C., Kuriyan J. (2005) Cell 123, 849–860 [DOI] [PubMed] [Google Scholar]
- 75.Bayliss R., Sardon T., Vernos I., Conti E. (2003) Mol. Cell 12, 851–862 [DOI] [PubMed] [Google Scholar]
- 76.Cheetham G. M., Knegtel R. M., Coll J. T., Renwick S. B., Swenson L., Weber P., Lippke J. A., Austen D. A. (2002) J. Biol. Chem. 277, 42419–42422 [DOI] [PubMed] [Google Scholar]
- 77.Huang X., Begley M., Morgenstern K. A., Gu Y., Rose P., Zhao H., Zhu X. (2003) Structure 11, 21–30 [DOI] [PubMed] [Google Scholar]
- 78.Yang J., Cron P., Good V. M., Thompson V., Hemmings B. A., Barford D. (2002) Nat. Struct. Biol. 9, 940–944 [DOI] [PubMed] [Google Scholar]
- 79.Bellon S., Fitzgibbon M. J., Fox T., Hsiao H. M., Wilson K. P. (1999) Structure 7, 1057–1065 [DOI] [PubMed] [Google Scholar]
- 80.Wang Z., Harkins P. C., Ulevitch R. J., Han J., Cobb M. H., Goldsmith E. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2327–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hubbard S. R. (1997) EMBO J. 16, 5572–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hubbard S. R., Wei L., Ellis L., Hendrickson W. A. (1994) Nature 372, 746–754 [DOI] [PubMed] [Google Scholar]
- 83.Nolen B., Taylor S., Ghosh G. (2004) Mol. Cell 15, 661–675 [DOI] [PubMed] [Google Scholar]
- 84.Huang J., Tu Z., Lee F. S. (2003) Biochem. Biophys. Res. Commun. 303, 532–540 [DOI] [PubMed] [Google Scholar]
- 85.Tu Z., Mooney S. M., Lee F. S. (2003) Cell. Signal. 15, 65–77 [DOI] [PubMed] [Google Scholar]
- 86.Klumpp S., Thissen M. C., Krieglstein J. (2006) Biochem. Soc. Trans. 34, 1370–1375 [DOI] [PubMed] [Google Scholar]
- 87.Lammers T., Lavi S. (2007) Crit. Rev. Biochem. Mol. Biol. 42, 437–461 [DOI] [PubMed] [Google Scholar]
- 88.Mumby M. C., Walter G. (1993) Physiol. Rev. 73, 673–699 [DOI] [PubMed] [Google Scholar]
- 89.Ubersax J. A., Ferrell J. E., Jr. (2007) Nat. Rev. Mol. Cell Biol. 8, 530–541 [DOI] [PubMed] [Google Scholar]
- 90.Scott J. W., Norman D. G., Hawley S. A., Kontogiannis L., Hardie D. G. (2002) J. Mol. Biol. 317, 309–323 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.